Abstract
Inflammasomes are cytoplasmic organelles that stimulate inflammation upon cellular detection of infectious or non-infectious stress. While much foundational work has focused on the infection-associated aspects of inflammasome activities, recent studies have highlighted the role of inflammasomes in non-infectious cellular and organismal functions. Herein, we discuss the evolution of inflammasome components and highlight characteristics that permit inflammasome regulation of physiologic processes. We focus on emerging data that highlight the importance of inflammasome proteins in the regulation of reproduction, development, and malignancy. A framework is proposed to contextualize these findings.
Keywords: inflammasome, innate immunity, NLRs, interleukin-1, caspase, reproduction, development, malignancy, cancer, inflammation, pyroptosis
1. Introduction
Inflammation is a component of an organism’s response to noxious or potentially noxious stimuli. Much of our knowledge in this area derives from studies of the immune response to infection, which results in a rapid, coordinated inflammatory assault designed to eradicate the invading microorganism. Inflammation is also associated with cellular responses to metabolic and endoplasmic reticulum stress, development, and malignancy[1–4]. A notable distinction between infection-induced and other inflammatory responses is the sensitivity of the inflammatory network to its trigger. Non-infectious stresses can be detected and resolved without inducing inflammation. For example, endoplasmic reticulum (ER) stress response pathways are activated in the absence of an inflammatory response within pancreatic acinar cells to promote digestive enzyme secretion[5]. If the ER stress response is overwhelmed, acinar cells induce inflammation via the secretion of chemokines that recruit macrophages to remove cells experiencing unresolved stress[6]. Thus, inflammatory activities in response to non-infectious stress are only initiated if existing homeostatic control systems are insufficient to restore homeostasis[7]. In contrast, the presence of a single microbe initiates an inflammatory response[8]. Within individual macrophages, single cell analysis revealed that innate immune pathways activated by bacteria operate in an all-or-nothing fashion (i.e., macrophages either respond to a bacterial encounter or they do not)[8–10]. While detection of infection operates in a binary fashion, the output of macrophage activation varies based upon the number of microorganisms or concentration of pathogen associated molecular patterns, indicating that the inflammatory response is calibrated to the severity of the threat[11,12]. Regardless of the threshold associated with an inflammatory network, inflammation ultimately contributes to a restoration of homeostasis by altering the function of tissues throughout the organism. A key feature of this response is that it comes at the expense of normal tissue function and leads to a transient deviation from homeostasis[13]. For example, local inflammation increases vascular permeability to promote extravasation of leukocytes and serum proteins into the stressed, infected, or damaged tissue[14]. This process comes at the cost of impaired local circulation and hemostasis. The cost of this trade-off between host defense and homeostasis disruption may be proportional to the severity of the insult. Thus, the ability of an organism to survive a noxious stimulus depends on the severity of the stimulus and the organism’s tolerance to inflammation-induced deviation from normal function[15,16]. The reduction in tolerance to homeostasis disruption that occurs with age may explain why older individuals are more likely to succumb to infections that are cleared by younger individuals[13,17].
Central to discussions of inflammation are the inflammasomes, which are key mediators of the inflammatory response to infection and injury[18]. Inflammasomes are cytoplasmic protein complexes that serves as the principal site of inflammatory caspase activities that promote the release of several interleukin-1 (IL-1) family cytokines[18]. Inflammasome activities are stimulated by pattern recognition receptors (PRRs) and guard proteins of the innate immune system (Figure 1). These proteins bind to microbial products directly (PRRs) or detect homeostatic disruptions (guards)[19,20]. Upon detection of either of these types of threats, an activated PRR or guard seeds the assembly of inflammasomes that can promote IL-1 release from either living or dead (i.e. pyroptotic) cells[21]. Diverse PRRs and guards can seed inflammasome assembly and activities, with well-described examples being members of the nucleotide-binding domain leucine rich repeat containing (NLR) family, the AIM2-like receptor family, the neuronal apoptosis inhibitor protein (NAIP) family, and the protein Pyrin. PRRs include AIM2 and NAIP5, which respectively detect viral double stranded DNA or bacterial flagellin subunits in the host cytosol, and the guard protein Pyrin, which detects dysregulation of Rho family GTPase activities[22–26]. NLRP3 is another guard protein which can be activated by diverse ion imbalances in the cell that lead to alterations in endosomal membrane dynamics[27]. PRRs and guards seed inflammasome assembly through the process of protein oligomerization, which results in the assembly of a micron-sized supramolecular organizing center (SMOC) that recruits and activates caspase-1 through auto-catalytic cleavage[19,28]. Activated caspase-1 cleaves the cytoplasmic protein gasdermin D (GSDMD), liberating the N-terminal domain which then oligomerizes within the plasma membrane to form a pore[21,29]. These pores serve as membrane channels that facilitate ion exchange and the secretion of bioactive IL-1 family cytokines, which have been cleaved by caspase-1[30]. If GSDMD pores are not repaired by the cell, pyroptosis can ensue, resulting in membrane rupture of the cellular corpse via the membrane protein Ninjurin1[31,32].
Figure 1. Pattern recognition receptors and guard proteins activate the inflammasome.
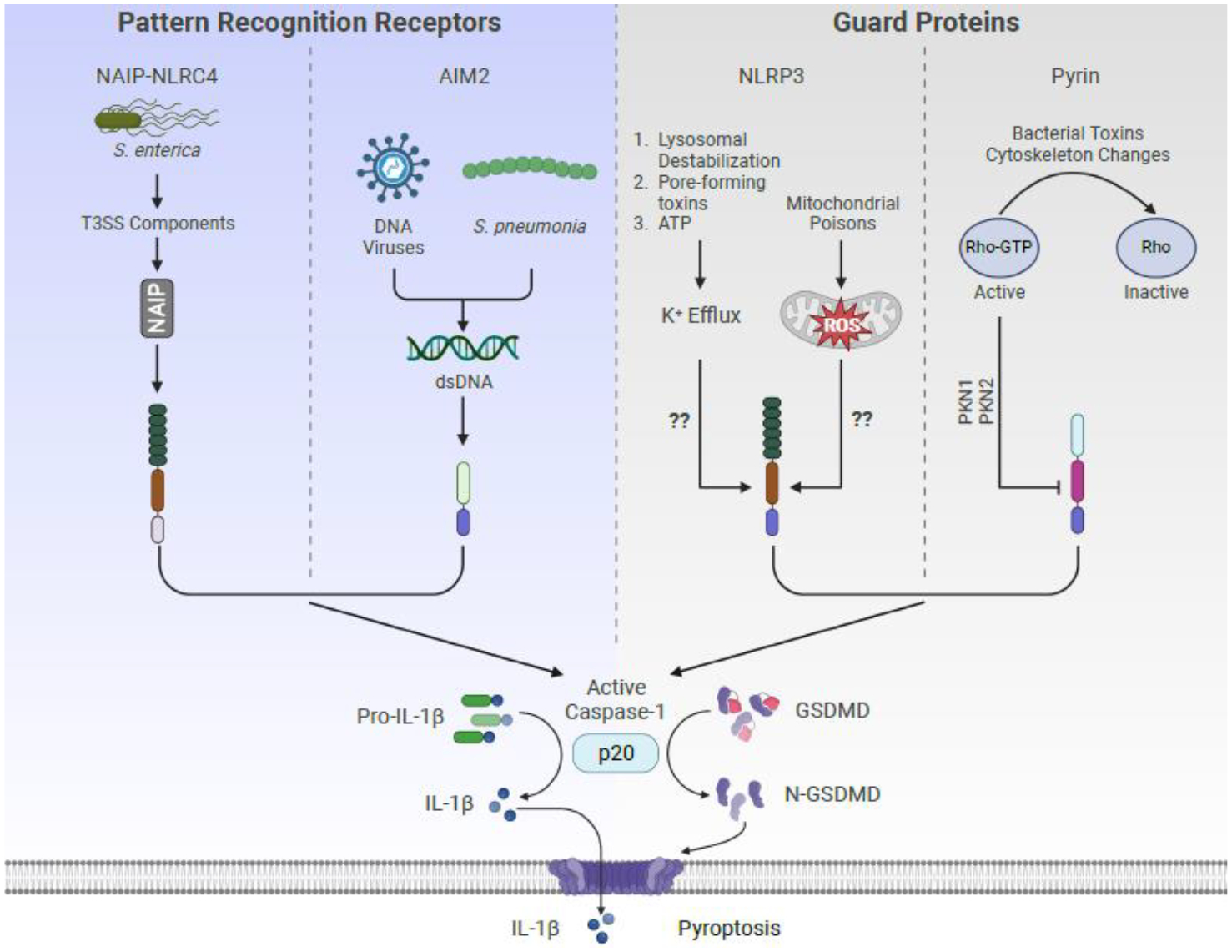
Two classes of proteins can activate the inflammasome. Pattern recognition receptors (e.g., NAIP-NLRC4 or AIM2) directly detect microbial products while guard proteins (e.g., NLRP3 or Pyrin) detect perturbation in specific cellular processes.
The molecular details of how distinct microbial products, virulence factors, and disruptions of homeostasis promote inflammasome and anti-infective activities have been reviewed elsewhere and will not be described in detail here[18–20,25,27,33,34]. In this review, we will discuss the contribution of inflammasomes to the regulation of physiologic processes beyond infection and across the species. We begin by discussing the evolution of inflammasome regulatory proteins (NLRs and caspases) and highlight how features conserved over evolutionary time prime these proteins to regulate homeostasis outside of their role in immunity. We will then discuss examples of the inflammasomes in the non-infectious contexts that span the stages of life, from development to reproduction to malignancy.
2. Diversity of NLRs and Caspases Across the Species
Proteins of the NLR family are present in several domains of life[19,35–38]. NLRs in plants and metazoans appear to have arisen separately during evolutionary time from domains that originated in bacteria[37]. Metazoan and plant NLRs contain C-terminal leucine-rich repeat (LRR) domains and an N-terminal effector domain but differ in the central regions between these domains[39]. Several metazoan NLRs contain a central NACHT domain (NAIP, major histocompatibility complex [MHC] transactivator [CIITA], HET-E, and TP1) whereas plant NLRs contain a central NB-ARC (Nucleotide-Binding, Apaf1, Resistance, CED4) domain[39]. In plants the N-terminal effector domain is comprised of either the Toll/IL-1 receptor/resistance (TIR) or coiled-coiled (CC) domain, while in metazoans it is comprised of either a caspase activation and recruitment domain (CARD) or Pyrin domain (PYD). The functional consequences of using different nucleotide-binding domains in plants and metazoans is unclear but may reflect evolution from distinct lineages[39].
Caspases are the effector proteins downstream of metazoan NLRs. Caspases are a class of cysteine-dependent proteases that cleave target proteins at conserved sequences containing aspartic acid and are required for the regulation of programmed cell death (PCD) and induction of inflammation[40]. Caspases are categorized based upon their ability to initiate or execute apoptotic cell death and their involvement in inflammation[40]. A related group of proteins, termed metacaspases, exist in bacteria, plants, fungi, and other eukaryotes, and are postulated to fill the same regulatory function as metazoan caspases, however this idea remains debated[41,42]. Metacaspases share with metazoan caspases a conserved cystine-histidine catalytic site but differ in that they do not require dimerization for activation and cleave after arginine or lysine residues[41]. Despite these functional differences, metacaspases are involved in PCD in plants and protists, although it remains unknown if the mechanisms are shared with metazoan caspases[43].
Apoptotic caspases are also required for development independent of their ability to induce PCD. The most convincing evidence exists for caspases being required for differentiation and maintenance of stem cell state[43]. Importantly, this developmental role shares a key feature with the role in PCD, namely that caspases regulate irreversible cell fate decisions. For example, caspase-3 is critical for differentiation of embryonic stem cells and hematopoietic stem cells in mice[44,45]. Similarly, drICE (the Drosophila caspase-3 homolog) is necessary for spermatid maturation and reproduction, the metacaspase Yca1 regulates proteostasis in yeast, and CED-3 (a C. elegans caspase-3 homolog) regulates asymmetric division of seam cells (neuroectodermal stem cells)[46–49]. Furthermore, transient activation of caspase-3 and caspase-9 is required for the differentiation of myotubes and formation of murine skeletal muscle[50–52]. Notably, the kinetics and intensity of caspase activation during differentiation is different than what is observed during PCD. Whereas caspase activation in PCD is rapid and maximal, the same caspase is only transiently and mildly activated during development[43]. The mechanisms of caspase regulation are just being uncovered but so far appear to center on posttranslational modification (PTM) of caspases and their target proteins alongside spatial control of caspase activation[43]. Thus, caspase activation occurs along a spectrum, from most intense which is responsible for PCD to least intense which is important for regulating the functions of living cells.
Inflammatory caspases are the primary effector enzymes within inflammasomes, which cleave select IL-1 family members to bioactive components and promote GSDMD pore formation[21]. While activation of inflammatory caspases was initially described to result in pyroptosis, recent work has indicated that inflammasomes can process inflammatory cytokines in the absence of cell death[53]. Cells that use inflammasomes to secrete IL-1β while remaining viable are potent inducers of adaptive immunity[54]. These cells are referred to as hyperactive, as compared to inflammatory cells that release cytokines other than IL-1ß (which are commonly referred to as active)[53,55–60]. The discovery of inflammasome activities in living cells indicates that while activation of the inflammasome may be digital, the output is not. Whether inflammasome activities lead to pyroptosis or cell hyperactivation remains an active area of research, and there are at least four possible underlying mechanisms. First, posttranslational modifications of inflammasome seed proteins may allow diverse cellular processes to regulate inflammasome output [27]. Alternatively, a secondary process may regulate the threshold of inflammasome signaling, allowing the output to be tuned to the specific activating signal. Third, differential regulation of membrane repair could determine the balance between pyroptosis and hyperactivation[32,61]. Finally, it is possible that, like apoptotic caspases that operate in living cells during development, the kinetics, localization, and activities of inflammasome-associated caspase activities dictates pyroptosis verses cell hyperactivation. Overall, despite notable gaps in our mechanistic understanding of these processes, caspases have the capability to control cellular function in cell deathdependent and -independent manners. In the next section, we will discuss three physiologic processes whereby inflammasomes and their effector caspases regulate restoration of homeostasis after non-infectious stress.
3. Physiologic Roles of Inflammasomes
3.1. NLRs in Reproduction
There are several NLR family members for which the upstream activators and downstream effectors remain unknown. A group of these orphan NLRs (NLRP2, −5, −7, −9, −14) regulate reproduction in mammals (Figure 2A)[62]. These reproductive NLRs are the result of a tandem duplication event that phylogenetically separates them from other NLR proteins[63]. Each reproductive NLR can be classified by which gamete expresses it: oocytes express NLRP2, −5, −7, and −9 while developing spermatocytes express NLRP14[62]. Deficiency of either NLRP2 or NLRP5 is associated with embryonic arrest at the 2-cell stage in mice and humans[64–69]. Embryonic arrest in NLRP5 deficient ova has been linked to increased mitochondrial activity and generation of reactive oxygen species (ROS)[70]. NLRP9 underwent gene duplication during rodent evolution such that there are three isoforms in rodents and one isoform in humans[71]. In humans, NLRP9 is expressed in oocytes, and knock down leads to embryonic arrest at the 2 or 4-cell stage[72]. Oocytes isolated from mice deficient for all three isoforms of NLRP9 similarly undergo embryonic arrest at the 2- or 4-cell stage[73]. Intriguingly, the gene duplication event in rodent evolution permitted the diversification of NLRP9 function, and mice deficient in NLRP9b are more susceptible to rotavirus infection[74]. Rotaviral RNA binds to DHX9 in the cytosol of infected cells, which then triggers NLRP9b oligomerization and activation[74]. However, it is unclear if this mechanism is conserved in humans, as crystal structures of the human NLRP9 pyrin domain suggests that it is unlikely to oligomerize[75,76]. Despite this uncertainty, the ability of viral RNA to activate NLRP9b in mice suggests that specific RNA species could be sensed by the NLRP9 inflammasome to control development.
Figure 2. Inflammasomes regulate physiologic processes.
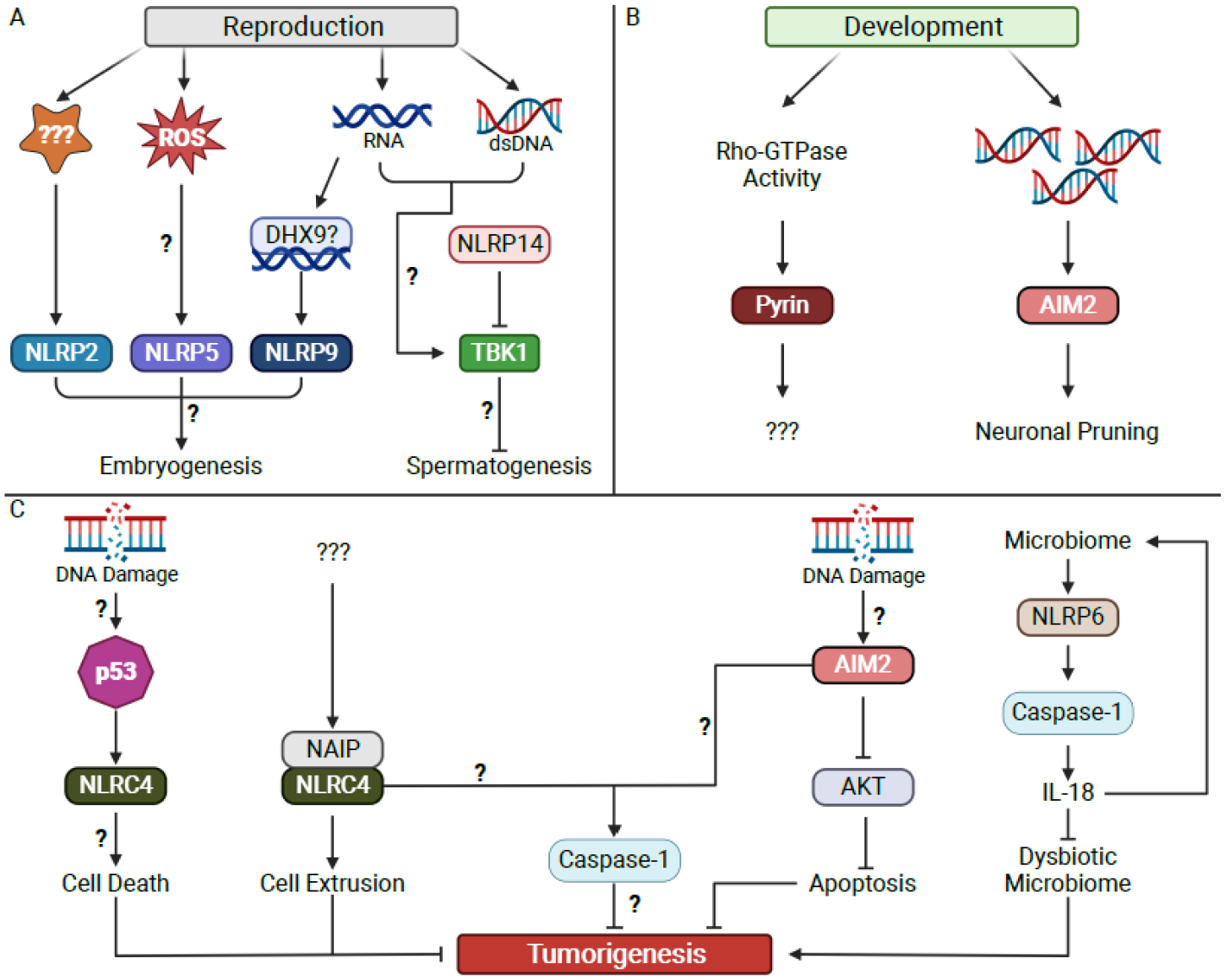
(A) Embryogenesis is regulated by NLRP2, NLRP5, and NLRP9, while spermatogenesis is regulated by NLRP14. Sensing of cytosolic nucleic acids may serve as a central regulator of both reproductive processes.
(B) Development generates cytosolic dsDNA which signals through AIM2 to control neuronal pruning. It is proposed that Pyrin senses changing Rho-GTPase activity to regulate unknown features of development.
(C) Multiple inflammasomes are proposed to regulate tumorigenesis through diverse pathways, some of which may be independent of inflammatory caspase activity.
NLRP7 is unique to primates and has been linked to the development of molar pregnancies[77]. Multiple non-synonymous mutations in NLRP7 have been linked to the development of hydatiform mole, potentially due to disruption of oligomerization[78–80]. How NLRP7 regulates pregnancy remains uncertain, however it appears important for both oocyte development and placental development[77]. One possibility is that mutations in NLRP7 impair oligomerization in response to an endogenous or exogenous stimulus. However, the ability of NLRP7 to activate the inflammasome remains unclear, with disparate studies reporting distinct mechanisms of regulation[81–84].
NLRP14 is uniquely expressed in the testes and mutations in it are linked to failure of spermatogenesis[85]. NLRP14 appears to inhibit cytosolic nucleic acid sensing by interfering with the activity of a central kinase that regulates interferon expression, TBK1[86]. It is unclear how nucleic acid production and the consequent TBK1 signaling are important in spermatogenesis and subsequent fertilization, however this is reminiscent of the ability of the Drosophila caspase-3 homolog drICE to control spermatogenesis[46]. We do not know if NLRP14 can oligomerize and form a functional inflammasome.
How and why are NLR proteins essential for normal reproduction? It is unlikely NLRs control reproduction through production of IL-1 or induction of pyroptosis, as mice deficient in MyD88 and GSDMD are viable. The existing data suggest the possibility that cytosolic nucleic acid sensing may be the upstream activator of these diverse NLRs (Figure 2A). Whether there is specificity beyond the type of nucleic acid or if this is a purposefully controlled process remains unknown. Additionally, there is no evidence supporting the role of inflammatory caspases as essential downstream effectors of NLR activation during reproduction, nor have ASC or IL-1ß been implicated. It is known that mice deficient in caspase-1 and caspase-11 have normal fecundity, however subtler developmental differences may not manifest in mice raised in a vivarium.
3.2. Inflammasomes in Development
Development is characterized by exponential cell division. This process results in the generation of multiple byproducts including ATP, dsDNA, and mitochondrial ROS, all of which regulate inflammasomes[18]. Production of these inflammasome regulatory factors serves as a readout of a specific cellular function, with ATP and ROS indicating the health of mitochondrial function and dsDNA indicating nuclear integrity. Substantial data indicate the evolutionarily conserved role of apoptotic caspases in normal development[43–46,50–52]. However, it is unknown if byproducts of development activate the inflammasome to regulate developmental programs.
Neurodevelopment is characterized by high rates of replicative stress resulting in DNA damage and the generation of cytosolic dsDNA[87]. Several neurologic diseases are caused by mutations in genes involved in DNA damage repair[88,89]. AIM2 is predominantly expressed myeloid cells and is activated by cytosolic dsDNA larger than 80 base pairs without sequence specificity[22–24,90]. AIM2 deficient mice develop increased anxious behaviors and decreased associative memory[91]. These behavioral differences are associated with increased dendrite length and branching, suggestive of inadequate neuronal pruning during development[91]. Subsequent work demonstrated that formation of AIM2 inflammasomes in neurons accompanies normal development[92]. Neuronal AIM2 may detect DNA damage during neurodevelopment to control cell pruning, a process that requires caspase-1 activation and GSDMD pore formation (Figure 2B)[92]. Neither microglial AIM2 nor total body IL-1 signaling are required for control of anxious behaviors and associative memory, suggesting that inflammation is not responsible for the observed impairment in neurodevelopment[92]. The inflammation-independent role of AIM2 in control of anxious behaviors and memory is distinct from the described importance of TLR signaling, cytokines, and microglia in the control of other neurodevelopmental processes (reviewed in [93–95]). In addition to its role in neurodevelopment, AIM2-deficiency protects against the development of Parkinson’s Disease in mice and reduces the severity of experimental autoimmune encephalomyelitis independent of inflammasome activation[96,97]. However, AIM2 may contribute to the pathology of Alzheimer’s Disease and to worsened outcomes after ischemic and hemorrhage stroke[98]. Thus, AIM2 may support beneficial developmental processes but be accompanied by a subsequent vulnerability to certain acute or chronic injuries that manifest later in life. A prediction would be that AIM2 serves other important roles in development outside of the nervous system.
Pyrin is another inflammasome seed protein that regulates development. Pyrin is encoded by the MEFV gene, mutations in which cause Familial Mediterranean Fever[99]. Under homeostatic conditions, Rho GTPases activate protein kinases N1 and N2 which phosphorylate and inactivate Pyrin (Figure 1)[100]. Upon disruption of Rho GTPase function, Pyrin becomes dephosphorylated, reducing its interaction with 14-3-3 proteins and permitting oligomerization and formation of an active inflammasome[26,101–105]. These findings indicate Pyrin is a sensor of Rho GTPase function while other studies have indicated Pyrin also senses cytoskeletal function[106]. Rho GTPases regulate numerous cellular processes including the actin cytoskeleton, cell polarity, and gene transcription, and thus their activity is controlled by over 130 proteins[107]. Rho GTPase activity can thus serve as a proxy for a variety of cellular functions important for development. Furthermore, migration is a key feature of the developmental program for neural crest cells and depends on a functional cytoskeleton[108]. These observations raise the question of whether the Pyrin inflammasome contributes to development. No studies have specifically evaluated this question, and Pyrin deficient mice appear phenotypically normal. However, caspase-1 deficient mice also initially appeared phenotypically normal until subtle neurologic developmental defects were identified in these animals[92]. Thus, we suggest that Pyrin may serve an important developmental role due to its ability to detect changes in Rho GTPase function (Figure 2B).
3.3. Inflammasomes in Malignancy
There is an intimate relationship between the immune system and the development of malignancy. While the immune system can exert anti-tumorigenic effects, local and systemic inflammation can promote tumorigenesis[1,109–111]. For example, in the azoxymethane (AOM) and dextran sodium sulfate (DSS) model of colitis-associated colorectal cancer (CAC) in mice, AOM triggers DNA damage, resulting in cellular transformation[112]. Recurrent exposure to DSS induces chronic inflammation and accelerates tumorigenesis[112]. This AOM-DSS model recapitulates the phenotypic appearance of human colorectal cancer (CRC)[113].
The inflammasome is a central mediator of the inflammatory component driving tumorigenesis[109,111]. Inflammatory activities are a likely link between inflammasomes and cancer, and chemical induction of pyroptosis directly in tumor cells can trigger a robust anti-tumor immune response[114,115]. However, inflammasome signaling independent of inflammation also contributes to controlling tumorigenesis. Below, we will discuss the roles of inflammasomes in the regulation of tumorigenesis and integrate that into our understanding of inflammasome biology.
NLRC4 is expressed in intestinal epithelial cells (IECs)[116]. Upon activation by NAIPs during Salmonella enterica serovar Typhimurium infection, the NAIP-NLRC4 inflammasome induces rapid extrusion of infected IECs into the intestinal lumen[116,117]. NLRC4 activation triggers polymerization of actin around the infected cell which is required for cell extrusion from the epithelial monolayer[117]. IEC extrusion does not require cell death[117]. The control of IEC survival and residence within the epithelial layer suggests a role for the NAIP-NLRC4 inflammasome as an epithelial cell quality control sensor. In the AOM-DSS CAC model, NLRC4 deficiency leads to increased tumor burden without changes in inflammation[118]. The precise activator of NLRC4 in the AOM-DSS model is unclear, however NAIPs are required for the anti-tumorigenic process of NLRC4. Mice deficient in all 6 NAIPs develop increased tumor burden in the AOM-DSS CAC model[119]. NAIPs may play a protective role early in tumorigenesis, as NAIP deficient mice are protected from AOM induced tumorigenesis in the absence of DSS[119]. NAIP deficiency within the IEC compartment leads to increased tumorigenesis in the AOM-DSS CAC model, suggesting a cell-intrinsic effect of the NAIP-NLRC4 inflammasome in CRC tumorigenesis[119]. Additionally, NLRC4 has been shown to reduce growth in a cutaneous melanoma model independent of caspase-1, however recent studies have shown conflicting results[120–122]. As in the studies on AOM-DSS CAC, the agonist of NLRC4 in melanoma remains unknown. Taken together, these data suggest that NAIPs respond to changes in cellular homeostasis that occur along the path to tumor formation and activate NLRC4 to eliminate the potentially tumorigenic focus (Figure 2C). It remains unknown whether the tumor suppressive role of NLRC4 always requires caspase-1 or if the cell extrusion seen during Salmonella infection is the underlying mechanism. One clue may reside in a study demonstrating that NLRC4 has antitumorigenic effects independent of caspase-1[123]. This study found that NLRC4 was required for p53-mediated cell death, suggesting that p53 senses genotoxic stress and activates the NLRC4 inflammasome to initiate cell extrusion[123]. One prediction of these observations is that NAIPs and NLRC4 are important for tumor surveillance in addition to their role protecting against enteric pathogens. A few studies demonstrate that lower expression of NLRC4 correlates with CRC development and mortality; however further mechanistic studies are needed to confirm these findings[124,125].
AIM2 was identified originally as a tumor suppressor in melanoma and has additionally been found to suppress tumor growth in hepatocellular carcinoma, renal cell carcinoma, breast cancer, prostate cancer, and colon cancer[126,127]. Detailed examination of 394 human CRC samples revealed that expression of AIM2 within colorectal tumors is correlated with survival[128]. In this study, over 75% of patients had reduction in AIM2 expression in CRC cells compared to adjacent normal tissue[128]. Additionally, complete loss of AIM2 expression within the malignant cell was associated with a 3-fold increase in mortality[128]. Two studies have suggested a mechanism by which AIM2 can regulate CRC tumorigenesis. In the murine AOM-DSS model of CAC, mice lacking AIM2 develop more numerous, larger, and higher-grade tumors[129]. This process occurs independently of inflammasomes, as caspase-1 cleavage and secretion of IL-1ß and IL-18 are unchanged in the absence of AIM2[129]. The authors suggest that AIM2 suppresses enterocyte proliferation through suppression of AKT phosphorylation on serine 473 and consequent promotion of cell-intrinsic apoptosis (Figure 2C)[129]. Intriguingly, AIM2 expression in bone marrow-derived and stromal cells appears to be important[129]. A second study demonstrated an identical phenotype in AIM2 deficient animals using the AOM-DSS model and confirmed their findings in the ApcMin/+ model of CRC[130]. Mechanistic insights from this study suggest that AIM2 suppresses DNA-dependent protein kinase phosphorylation of AKT on serine 473 to regulate cellular growth[130]. Inhibition of AKT in AIM2 deficient animals reduced tumor burdens in the AOM-DSS model without affecting tumorigenesis in wild type mice[130]. In both studies, the assumption is that AIM2 detects cytosolic dsDNA produced during the transformation of a cell from benign to malignant. It is unknown whether AIM2 activation of caspase-1 within the malignant cell is required for its tumor suppressive function or if there are caspase-independent functions for AIM2 in CRC.
NLRP6 is predominantly expressed in non-hematopoietic tissues and is reportedly activated by myriad stimuli[131,132]. NLRP6 is most highly expressed at mucosal barrier tissues and appears to control secretion of IL-18 in vivo, as animals deficient in NLRP6 have lower circulating IL-18 levels at baseline and in response to DSS colitis[131,133]. Notably, IL-1ß levels are unchanged to slightly increased in NLRP6 deficient animals, possibly due to compensatory upregulation of NLRP3 activity[132]. A key function of NLRP6 is to control of the composition of the gut microbiota. Studies from three laboratories indicated that NLRP6 deficient animals have abnormal gut microbial communities and develop more severe colitis after DSS treatment due to impaired caspase-1 activation and IL-18 secretion[131,133,134]. NLRP6 may detect microbe-derived small molecules to regulate IL-18 secretion[135]. NLRP6 triggers secretion of IL-18 independent of cell death, suggesting that hyperactivation is not restricted to myeloid cells[135]. However, our understanding of the role of NLRP6 in regulation of the gut microbiota remains incomplete, as subsequent work from two laboratories found no difference in microbiota composition or DSS colitis severity in NLRP6 deficient mice[136,137]. The reasons for these differential observations are unclear. More work is needed, especially to ensure that any phenotypes observed are not linked to passenger mutations associated with distinct lines of the same murine colonies[138]. NLRP6 also has functions independent of caspase-1. NLRP6 deficient animals are more susceptible to Citrobacter rodentium infection due to impaired goblet cell mucus secretion[139]. In this context, NLRP6 regulates autophagy to control mucus secretion from goblet cells through an unknown mechanism[139]. Alternatively, NLRP6 deficient mice are resistant to infections with Listeria, Salmonella, and E. coli[140]. During infection with these organisms, NLRP6 negatively regulates ERK1/2 and NF-κB signaling, impairing the host response to infection[140]. These observations point to important caspase-dependent and caspase-independent functions of NLRP6 in the regulation of colonic microbial communities. Thus, through two distinct pathways, NLRP6 curates the gut microbial community and controls responses to infection.
NLRP6 deficient mice develop more numerous and larger colorectal tumors in the AOM-DSS model of CAC[133,134]. This process depends on caspase-mediated secretion of IL-18 (Figure 2C)[133,141]. Intriguingly, the importance of NLRP6 regulating CRC may partially explain the sexual dimorphism seen in colon cancer in humans. Females have reduced incidence of CRC compared to males[142]. Additionally, mortality is lower in women aged 18–44 compared to men of the same age or women older than 50, suggesting that estrogen may play a protective role in the development and progression of CRC[143]. Reduction in circulating estrogens due to oophorectomy increases CRC risk while taking exogenous estrogens in the form of hormone replacement therapy reduces CRC risk and mortality[143]. Furthermore, loss of estrogen receptor isoform ß (ERß) in CRC is correlated with lower survival[143]. Thus, systemic estrogen levels and local estrogen signaling are important for regulation of CRC. A mechanistic explanation of this association was unclear until a recent paper showed that ERß induces NLRP6 expression, reducing the severity of DSS colitis in mice[144]. These data suggest that NLRP6 plays a central role in curation of a pro-health microbiota, the disturbance of which contributes to multiple diseases.
The NLRP3 inflammasome predominantly controls tumorigenesis through its pro-inflammatory actions. Multiple reviews have detailed the role of NLRP3-derived inflammation in tumorigenesis[109,111]. Emerging research suggests that the NLRP3 inflammasome can regulate tumorigenesis independent of inflammation. Non-hematopoietic NLRP3 has been proposed to suppress colitis in response to DSS[145]. Additionally, non-hematopoietic NLRP3 may contribute to an intermediate suppression of tumorigenesis in the AOM-DSS CAC model, however another study did not find an effect of NLRP3 on AOM-DSS induced CAC[118,146]. Interestingly, in a CRC metastases model, both hematopoietic and non-hematopoietic NLRP3 were shown to inhibit metastases[147]. Furthermore, NLRP3 appears to directly regulate growth of hepatocellular carcinoma independent of inflammation [148]. We are just beginning to appreciate the role of NLRP3 in the regulation of tumorigenesis independent of its ability to induce inflammation.
Taken together, multiple inflammasome regulatory proteins impact tumorigenesis independent of their role in triggering inflammation. These data suggest that inflammasome biology extends beyond the ability to induce secretion of IL-1 family members. Of note, most studies have been performed in mice with total body deficiency of a gene of interest. Future work examining the tissue specific function of inflammasome regulators will be important to identify the relative roles of the inflammasome in the immune and stromal cells. While NLRP6 regulates tumorigenesis through secretion of IL-18, the precise activating signals and downstream effectors for the remaining proteins remain to be elucidated. One intriguing possibility is that inflammasomes in tumorigenesis are acting analogously to apoptotic caspases during differentiation. Tumorigenesis is an irreversible, stepwise change in cellular function that is akin to the stepwise cellular changes that occur during differentiation, and thus inflammatory caspases could regulate tumorigenesis by sensing inappropriate cell fate decisions.
4. Conclusions and Perspectives
The field is just beginning to understand how inflammasomes regulate physiologic processes in addition to infection and injury. In this review, we discussed examples where inflammasomes contribute to the control of reproduction, development, and tumorigenesis. Below, we propose a few key principles that will help guide our understanding and future research.
First, inflammasome seed proteins evolved to surveil the cytoplasmic environment. The cytoplasmic location of inflammasome seed proteins created evolutionary constraints on which specific signals they can detect. As such, two strategies to detect perturbation of cellular homeostasis evolved in inflammasomes: direct detection of microbial molecules or guarding specific cellular processes[39]. In the first strategy, inflammasome seed proteins detect the presence of microbial molecules directly (e.g., LPS) or cellular molecules associated with tissue injury (e.g., cytoplasmic DNA)[21]. These inflammasome seeds operate as PRRs. Many of the molecules detected by PRRs are also produced by physiologic cellular processes. For example, dsDNA is generated as part of the DNA damage response or cell death, creating the opportunity for inappropriate activation of the immune response[40,87]. Other PRRs that detect DNA, such as Toll-like receptor (TLR) 9, are compartmentalized into endosomes or phagosomes[149]. Compartmentalization is a strategy employed by the immune system to control the inflammatory response, yet the cytoplasmic location of inflammasomes evades this locus of control. In the second strategy, inflammasome seeds function as guard proteins and detect perturbation of specific cellular functions (e.g., ion concentration or the cytoskeleton)[20]. These cellular functions are altered by an array of stimuli besides infections. For example, neurons and myocytes see cyclical change in ion concentration while migrating cells see dramatic reorganization of the cytoskeleton. Thus, the presence of inflammasomes in the cytosol permits detection of molecules and activities that serve as a readout of cellular processes.
Second, caspases can be activated without resulting in cell death. Apoptotic caspases can regulate cell differentiation while inflammatory caspases can permit secretion of IL-1 cytokines from living (hyperactive) cells. The ability to dissociate caspase-1 activity from cell death allows for more robust T cell activation and anti-tumor immunity[54]. What leads one cell with active caspase-1 to die while another to live is unknown, but a clue from research on the control of apoptotic caspases suggests that PTMs may play a role[43]. Inflammasome seed proteins are the target of multiple kinases, phosphatases, E3 ligases, and deubiquitinating enzymes[27,150–152]. The accumulation of PTMs could thus serve as an integration of cellular function, allowing physiologic processes to fine tune inflammasome activity. Furthermore, it remains unclear if the sole effect of inflammatory caspase activation in living cells is secretion of IL-1 cytokines or if another output is contributing.
Third, each cell type appears to have a distinct mechanism of inflammasome activation. Macrophages and dendritic cells (DCs) undergo inflammasome activation with pyroptosis[19]. Hyperactivation appears to be most common in DCs, although reports of this process in macrophages and neutrophils exist[53,55,57,58,153,154]. Additionally, human monocytes secrete IL-1β in response to LPS alone while murine monocytes do not[59]. Furthermore, living non-immune cells such as adipocytes secrete IL-1 to control differentiation[155].
Taken together, these three principles suggest a model by which inflammasomes can be activated by physiologic processes. The ability of inflammasomes to detect endogenously generated signals and activate caspase-1 without inducing cell death in a cell specific manner likely underpins this capability. Many questions remain unanswered: what other processes or activities are sensed by inflammasomes, what are the other outputs of caspase-1 activation besides IL-1, and what controls whether a cell lives or dies when the inflammasome is activated? The answers to these questions promise to improve our understanding of inflammasome biology and provide the basis for design of novel therapies for diseases of dysregulated homeostasis.
Acknowledgements
We thank members of the Kagan lab for helpful discussions. We apologize to members of the community whose studies were not cited due to space constraints. All figures were created with BioRender.com.
Funding:
This work was supported by National Institutes of Health grants AI67993, AI116550 and P30DK34854 (to J.C.K) and T32HL116275 (to D.O).
Abbreviations
- AIM2
Absent in melanoma 2
- AOM
Azoxymethane
- CAC
Colitis-associated colorectal cancer
- CARD
Caspase activation and recruitment domain
- CC
Coiled-coiled
- CRC
Colorectal cancer
- DC
Dendritic cell
- DHX9
DExH-box helicase 9
- drICE
Death related ICE-like caspase
- DSS
Dextran sodium sulfate
- ER
Endoplasmic reticulum
- ERß
Estrogen receptor ß
- ERK
Extracellular signal-related kinases
- GSDMD
Gasdermin D
- HET-E
Heterokaryon incompatibility E
- IEC
Intestinal epithelial cell
- IL
Interleukin
- LPS
Lipopolysaccharide
- MHC
Major histocompatibility complex
- NAIP
Neuronal apoptosis inhibitor protein
- NB-ARC
Nucleotide-binding Apaf1 Resistance CED4
- NF-κB
Nuclear factor Kappa B
- NLR
Nucleotide-binding domain leucine rich repeat containing
- NLRC
NLR family CARD domain containing
- NLRP
NLR family Pyrin domain containing
- PCD
Programmed cell death
- PRR
Pattern recognition receptor
- PTM
Post-translational modification
- PYD
Pyrin domain
- ROS
Reactive oxygen species
- TBK1
TANK-binding kinase 1
- TIR
Toll/Interleukin-1 receptor
- TLR
Toll-like receptor
- TP1
Telomerase associated protein 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: J.C.K consults and holds equity in Corner Therapeutics, Larkspur Biosciences and Neumora Therapeutics. None of these relationships impacted this manuscript.
References
- [1].Greten FR, Grivennikov SI, Inflammation and Cancer: Triggers, Mechanisms, and Consequences, Immunity. 51 (2019) 27–41. 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhang K, Kaufman RJ, From endoplasmic-reticulum stress to the inflammatory response, Nature. 454 (2008) 455–462. 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hotamisligil GS, Inflammation, metaflammation and immunometabolic disorders, Nature. 542 (2017) 177–185. 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
- [4].Jiang NM, Cowan M, Moonah SN, Petri WA, The Impact of Systemic Inflammation on Neurodevelopment, Trends Mol Med. 24 (2018) 794–804. 10.1016/j.molmed.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kubisch CH, Logsdon CD, Endoplasmic reticulum stress and the pancreatic acinar cell, Expert Rev Gastroent. 2 (2008) 249–260. 10.1586/17474124.2.2.249. [DOI] [PubMed] [Google Scholar]
- [6].Antonucci L, Fagman JB, Kim JY, Todoric J, Gukovsky I, Mackey M, Ellisman MH, Karin M, Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress, Proc National Acad Sci. 112 (2015) E6166–E6174. 10.1073/pnas.1519384112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kotas ME, Medzhitov R, Homeostasis, Inflammation, and Disease Susceptibility, Cell. 160 (2015) 816–827. 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Avraham R, Haseley N, Brown D, Penaranda C, Jijon HB, Trombetta JJ, Satija R, Shalek AK, Xavier RJ, Regev A, Hung DT, Pathogen Cell-to-Cell Variability Drives Heterogeneity in Host Immune Responses, Cell. 162 (2015) 1309–1321. 10.1016/j.cell.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW, Single-cell NF-κB dynamics reveal digital activation and analogue information processing, Nature. 466 (2010) 267–271. 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, Schwartz S, Fowler B, Weaver S, Wang J, Wang X, Ding R, Raychowdhury R, Friedman N, Hacohen N, Park H, May AP, Regev A, Single cell RNA Seq reveals dynamic paracrine control of cellular variation, Nature. 510 (2014) 363–369. 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Allen NC, Philip NH, Hui L, Zhou X, Franklin RA, Kong Y, Medzhitov R, Desynchronization of the molecular clock contributes to the heterogeneity of the inflammatory response, Sci Signal. 12 (2019) eaau1851. 10.1126/scisignal.aau1851. [DOI] [PubMed] [Google Scholar]
- [12].Adelaja A, Taylor B, Sheu KM, Liu Y, Luecke S, Hoffmann A, Six distinct NFκB signaling codons convey discrete information to distinguish stimuli and enable appropriate macrophage responses, Immunity. 54 (2021) 916–930.e7. 10.1016/j.immuni.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Okin D, Medzhitov R, Evolution of Inflammatory Diseases, Curr Biol. 22 (2012) R733–40. 10.1016/j.cub.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nathan C, Points of control in inflammation, Nature. 420 (2002) 846–852. 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- [15].Soares MP, Teixeira L, Moita LF, Disease tolerance and immunity in host protection against infection, Nat Rev Immunol. 17 (2017) 83–96. 10.1038/nri.2016.136. [DOI] [PubMed] [Google Scholar]
- [16].Ganeshan K, Nikkanen J, Man K, Leong YA, Sogawa Y, Maschek JA, Ry TV, Chagwedera DN, Cox JE, Chawla A, Energetic Trade-Offs and Hypometabolic States Promote Disease Tolerance, Cell. (2019). 10.1016/j.cell.2019.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Taborsky B, English S, Fawcett TW, Kuijper B, Leimar O, McNamara JM, Ruuskanen S, Sandi C, Towards an Evolutionary Theory of Stress Responses, Trends Ecol Evol. (2020). 10.1016/j.tree.2020.09.003. [DOI] [PubMed] [Google Scholar]
- [18].Schroder K, Tschopp J, The Inflammasomes, Cell. 140 (2010) 821–832. 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- [19].Nozaki K, Li L, Miao EA, Innate Sensors Trigger Regulated Cell Death to Combat Intracellular Infection, Annu Rev Immunol. 40 (2022) 469–498. 10.1146/annurev-immunol-101320-011235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Remick BC, Gaidt MM, Vance RE, Effector-Triggered Immunity, Annu Rev Immunol. 41 (2023) 453–481. 10.1146/annurev-immunol-101721-031732. [DOI] [PubMed] [Google Scholar]
- [21].Evavold CL, Kagan JC, Inflammasomes: Threat-Assessment Organelles of the Innate Immune System, Immunity. 51 (2019) 609–624. 10.1016/j.immuni.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fernandes-Alnemri T, Yu J-W, Datta P, Wu J, Alnemri ES, AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA, Nature. 458 (2009) 509–513. 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey Daniel.R., Latz E, Fitzgerald KA, AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC, Nature. 458 (2009) 514–518. 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ, HIN-200 Proteins Regulate Caspase Activation in Response to Foreign Cytoplasmic DNA, Science. 323 (2009) 1057–1060. 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- [25].Duncan JA, Canna SW, The NLRC4 Inflammasome, Immunol Rev. 281 (2018) 115–123. 10.1111/imr.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong Y-N, Peng X, Xi JJ, Chen S, Wang F, Shao F, Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome, Nature. 513 (2014) 237–241. 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- [27].Swanson KV, Deng M, Ting JP-Y, The NLRP3 inflammasome: molecular activation and regulation to therapeutics, Nat Rev Immunol. 19 (2019) 477–489. 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kagan JC, Magupalli VG, Wu H, SMOCs: supramolecular organizing centres that control innate immunity, Nature Reviews Immunology. 14 (2014). 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J, Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores, Nature. 535 (2016) 153–158. 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xia S, Zhang Z, Magupalli VG, Pablo JL, Dong Y, Vora SM, Wang L, Fu T-M, Jacobson MP, Greka A, Lieberman J, Ruan J, Wu H, Gasdermin D pore structure reveals preferential release of mature interleukin-1, Nature. 593 (2021) 607–611. 10.1038/s41586-021-03478-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kayagaki N, Kornfeld OS, Lee BL, Stowe IB, O’Rourke K, Li Q, Sandoval W, Yan D, Kang J, Xu M, Zhang J, Lee WP, McKenzie BS, Ulas G, Payandeh J, Roose-Girma M, Modrusan Z, Reja R, Sagolla M, Webster JD, Cho V, Andrews TD, Morris LX, Miosge LA, Goodnow CC, Bertram EM, Dixit VM, NINJ1 mediates plasma membrane rupture during lytic cell death, Nature. (2021) 1–6. 10.1038/s41586-021-03218-7. [DOI] [PubMed] [Google Scholar]
- [32].Rühl S, Shkarina K, Demarco B, Heilig R, Santos JC, Broz P, ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation, Science. 362 (2018) 956–960. 10.1126/science.aar7607. [DOI] [PubMed] [Google Scholar]
- [33].Broz P, Dixit VM, Inflammasomes: mechanism of assembly, regulation and signalling, Nat Rev Immunol. 16 (2016) 407–420. 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- [34].Newton K, Dixit VM, Kayagaki N, Dying cells fan the flames of inflammation, Science. 374 (2021) 1076–1080. 10.1126/science.abi5934. [DOI] [PubMed] [Google Scholar]
- [35].Mariano G, Blower TR, Conserved domains can be found across distinct phage defence systems, Mol Microbiol. (2023). 10.1111/mmi.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lange C, Hemmrich G, Klostermeier UC, López-Quintero JA, Miller DJ, Rahn T, Weiss Y, Bosch TCG, Rosenstiel P, Defining the Origins of the NOD-Like Receptor System at the Base of Animal Evolution, Mol Biol Evol. 28 (2011) 1687–1702. 10.1093/molbev/msq349. [DOI] [PubMed] [Google Scholar]
- [37].Jacob F, Vernaldi S, Maekawa T, Evolution and Conservation of Plant NLR Functions, Front Immunol. 4 (2013) 297. 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tamborski J, Krasileva KV, Evolution of Plant NLRs: From Natural History to Precise Modifications, Annu Rev Plant Biol. 71 (2020) 1–24. 10.1146/annurev-arplant-081519-035901. [DOI] [PubMed] [Google Scholar]
- [39].Jones JDG, Vance RE, Dangl JL, Intracellular innate immune surveillance devices in plants and animals, Science. 354 (2016). 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- [40].Tummers B, Green DR, The evolution of regulated cell death pathways in animals and their evasion by pathogens, Physiol Rev. 102 (2022) 411–454. 10.1152/physrev.00002.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tsiatsiani L, Breusegem FV, Gallois P, Zavialov A, Lam E, Bozhkov PV, Metacaspases, Cell Death Differ. 18 (2011) 1279–1288. 10.1038/cdd.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Minina EA, Coll NS, Tuominen H, Bozhkov PV, Metacaspases versus caspases in development and cell fate regulation, Cell Death Differ. 24 (2017) 1314–1325. 10.1038/cdd.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bell RAV, Megeney LA, Evolution of caspase-mediated cell death and differentiation: twins separated at birth, Cell Death Differ. 24 (2017) 1359–1368. 10.1038/cdd.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fujita J, Crane AM, Souza MK, Dejosez M, Kyba M, Flavell RA, Thomson JA, Zwaka TP, Caspase Activity Mediates the Differentiation of Embryonic Stem Cells, Cell Stem Cell. 2 (2008) 595–601. 10.1016/j.stem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Janzen V, Fleming HE, Riedt T, Karlsson G, Riese MJ, Celso CL, Reynolds G, Milne CD, Paige CJ, Karlsson S, Woo M, Scadden DT, Hematopoietic Stem Cell Responsiveness to Exogenous Signals Is Limited by Caspase-3, Cell Stem Cell. 2 (2008) 584–594. 10.1016/j.stem.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Arama E, Agapite J, Steller H, Caspase Activity and a Specific Cytochrome C Are Required for Sperm Differentiation in Drosophila, Dev Cell. 4 (2003) 687–697. 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- [47].Hill SM, Hao X, Liu B, Nyström T, Life-span extension by a metacaspase in the yeast Saccharomyces cerevisiae, Science. 344 (2014) 1389–1392. 10.1126/science.1252634. [DOI] [PubMed] [Google Scholar]
- [48].Lee REC, Brunette S, Puente LG, Megeney LA, Metacaspase Yca1 is required for clearance of insoluble protein aggregates, Proc National Acad Sci. 107 (2010) 13348–13353. 10.1073/pnas.1006610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Weaver BP, Zabinsky R, Weaver YM, Lee ES, Xue D, Han M, CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans, Elife. 3 (2014) e04265. 10.7554/elife.04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].van den Eijnde SM, van den Hoff MJB, Reutelingsperger CPM, van Heerde WL, Henfling MER, Vermeij-Keers C, Schutte B, Borgers M, Ramaekers FCS, Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation, J Cell Sci. 114 (2001) 3631–3642. 10.1242/jcs.114.20.3631. [DOI] [PubMed] [Google Scholar]
- [51].Fernando P, Kelly JF, Balazsi K, Slack RS, Megeney LA, Caspase 3 activity is required for skeletal muscle differentiation, Proc National Acad Sci. 99 (2002) 11025–11030. 10.1073/pnas.162172899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Murray TVA, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A, Zwacka R, Fearnhead HO, A non-apoptotic role for caspase-9 in muscle differentiation, J Cell Sci. 121 (2008) 3786–3793. 10.1242/jcs.024547. [DOI] [PubMed] [Google Scholar]
- [53].Zanoni I, Tan Y, Gioia MD, Broggi A, Ruan J, Shi J, Donado CA, Shao F, Wu H, Springstead JR, Kagan JC, An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells, Science. 352 (2016) 1232–1236. 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhivaki D, Kagan JC, NLRP3 inflammasomes that induce antitumor immunity, Trends Immunol. (2021). 10.1016/j.it.2021.05.001. [DOI] [PubMed] [Google Scholar]
- [55].Zanoni I, Tan Y, Gioia MD, Springstead JR, Kagan JC, By Capturing Inflammatory Lipids Released from Dying Cells, the Receptor CD14 Induces Inflammasome-Dependent Phagocyte Hyperactivation, Immunity. 47 (2017) 697–709.e3. 10.1016/j.immuni.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhivaki D, Borriello F, Chow OA, Doran B, Fleming I, Theisen DJ, Pallis P, Shalek AK, Sokol CL, Zanoni I, Kagan JC, Inflammasomes within Hyperactive Murine Dendritic Cells Stimulate Long-Lived T Cell-Mediated Anti-tumor Immunity, Cell Reports. 33 (2020) 108381. 10.1016/j.celrep.2020.108381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen KW, Groß CJ, Sotomayor FV, Stacey KJ, Tschopp J, Sweet MJ, Schroder K, The Neutrophil NLRC4 Inflammasome Selectively Promotes IL-1β Maturation without Pyroptosis during Acute Salmonella Challenge, Cell Reports. 8 (2014) 570–582. 10.1016/j.celrep.2014.06.028. [DOI] [PubMed] [Google Scholar]
- [58].Hatscher L, Lehmann CHK, Purbojo A, Onderka C, Liang C, Hartmann A, Cesnjevar R, Bruns H, Gross O, Nimmerjahn F, Ivanović-Burmazović I, Kunz M, Heger L, Dudziak D, Select hyperactivating NLRP3 ligands enhance the TH1- and TH17-inducing potential of human type 2 conventional dendritic cells, Sci Signal. 14 (2021) eabe1757. 10.1126/scisignal.abe1757. [DOI] [PubMed] [Google Scholar]
- [59].Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AAB, Cooper MA, Graf T, Hornung V, Human Monocytes Engage an Alternative Inflammasome Pathway, Immunity. 44 (2016). 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- [60].Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, Underhill DM, Hexokinase Is an Innate Immune Receptor for the Detection of Bacterial Peptidoglycan, Cell. 166 (2016) 624–636. 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nozaki K, Maltez VI, Rayamajhi M, Tubbs AL, Mitchell JE, Lacey CA, Harvest CK, Li L, Nash WT, Larson HN, McGlaughon BD, Moorman NJ, Brown MG, Whitmire JK, Miao EA, Caspase-7 activates ASM to repair gasdermin and perforin pores, Nature. (2022) 1–8. 10.1038/s41586-022-04825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gorp HV, Kuchmiy A, Hauwermeiren FV, Lamkanfi M, NOD-like receptors interfacing the immune and reproductive systems, Febs J. 281 (2014) 4568–4582. 10.1111/febs.13014. [DOI] [PubMed] [Google Scholar]
- [63].Tian X, Pascal G, Monget P, Evolution and functional divergence of NLRPgenes in mammalian reproductive systems, Bmc Evol Biol. 9 (2009) 202. 10.1186/1471-2148-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Peng H, Chang B, Lu C, Su J, Wu Y, Lv P, Wang Y, Liu J, Zhang B, Quan F, Guo Z, Zhang Y, Nlrp2, a Maternal Effect Gene Required for Early Embryonic Development in the Mouse, Plos One. 7 (2012) e30344. 10.1371/journal.pone.0030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kuchmiy AA, D’Hont J, Hochepied T, Lamkanfi M, NLRP2 controls age-associated maternal fertility, J Exp Med. 213 (2016) 2851–2860. 10.1084/jem.20160900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tong Z-B, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM, Mater, a maternal effect gene required for early embryonic development in mice, Nat Genet. 26 (2000) 267–268. 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- [67].Mu J, Wang W, Chen B, Wu L, Li B, Mao X, Zhang Z, Fu J, Kuang Y, Sun X, Li Q, Jin L, He L, Sang Q, Wang L, Mutations in NLRP2 and NLRP5 cause female infertility characterised by early embryonic arrest, J Med Genet. 56 (2019) 471. 10.1136/jmedgenet-2018-105936. [DOI] [PubMed] [Google Scholar]
- [68].Huang L, Wang Y, Lu F, Jin Q, Song G, Ji J, Luo L, Jin R, Tong X, Novel mutations in NLRP5 and PATL2 cause female infertility characterized by primarily oocyte maturation abnormality and consequent early embryonic arrest, J Assist Reprod Gen. 39 (2022) 711–718. 10.1007/s10815-022-02412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tong X, Jin J, Hu Z, Zhang Y, Fan H, Zhang Y, Zhang S, Mutations in OOEP and NLRP5 identified in infertile patients with early embryonic arrest, Hum. Mutat 43 (2022) 1909–1920. 10.1002/humu.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Fernandes R, Tsuda C, Perumalsamy AL, Naranian T, Chong J, Acton BM, Tong Z-B, Nelson LM, Jurisicova A, NLRP5 Mediates Mitochondrial Function in Mouse Oocytes and Embryos, Biol Reprod. 86 (2012) 138, 1–10. 10.1095/biolreprod.111.093583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mullins B, Chen J, NLRP9 in innate immunity and inflammation, Immunology. 162 (2021) 262–267. 10.1111/imm.13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Amoushahi M, Steffensen LL, Galieva A, Agger J, Heuck A, Siupka P, Ernst E, Nielsen MS, Sunde L, Lykke-Hartmann K, Maternally contributed Nlrp9b expressed in human and mouse ovarian follicles contributes to early murine preimplantation development, J Assist Reprod Gen. 37 (2020) 1355–1365. 10.1007/s10815-020-01767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kanzaki S, Tamura S, Ito T, Wakabayashi M, Saito K, Kato S, Ohta Y, Sekita Y, Kimura T, Involvement of Nlrp9a/b/c in mouse preimplantation development, Reproduction. 160 (2020) 181–191. 10.1530/rep-19-0516. [DOI] [PubMed] [Google Scholar]
- [74].Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, Yang Y, Yu H, Li H-B, Wang G, Lei X, de Zoete MR, Zhao J, Zheng Y, Chen H, Zhao Y, Jurado KA, Feng N, Shan L, Kluger Y, Lu J, Abraham C, Fikrig E, Greenberg HB, Flavell RA, Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells, Nature. 546 (2017) 667–670. 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ha HJ, Park HH, Crystal structure of the human NLRP9 pyrin domain reveals a bent N-terminal loop that may regulate inflammasome assembly, Febs Lett. 594 (2020) 2396–2405. 10.1002/1873-3468.13866. [DOI] [PubMed] [Google Scholar]
- [76].Marleaux M, Anand K, Latz E, Geyer M, Crystal structure of the human NLRP9 pyrin domain suggests a distinct mode of inflammasome assembly, Febs Lett. 594 (2020) 2383–2395. 10.1002/1873-3468.13865. [DOI] [PubMed] [Google Scholar]
- [77].Carriere J, Dorfleutner A, Stehlik C, NLRP7: From inflammasome regulation to human disease, Immunology. 163 (2021) 363–376. 10.1111/imm.13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, Hanash S, Rouleau GA, Slim R, Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans, Nat Genet. 38 (2006) 300–302. 10.1038/ng1740. [DOI] [PubMed] [Google Scholar]
- [79].Wang CM, Dixon PH, Decordova S, Hodges MD, Sebire NJ, Ozalp S, Fallahian M, Sensi A, Ashrafi F, Repiska V, Zhao J, Xiang Y, Savage PM, Seckl MJ, Fisher RA, Identification of 13 novel NLRP7 mutations in 20 families with recurrent hydatidiform mole; missense mutations cluster in the leucine-rich region, J Med Genet. 46 (2009) 569. 10.1136/jmg.2008.064196. [DOI] [PubMed] [Google Scholar]
- [80].Singer H, Biswas A, Zimmer N, Messaed C, Oldenburg J, Slim R, El-Maarri O, NLRP7 interdomain interactions: the NACHT-associated domain is the physical mediator for oligomeric assembly, Mhr Basic Sci Reproductive Medicine. 20 (2014) 990–1001. 10.1093/molehr/gau060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Khare S, Dorfleutner A, Bryan NB, Yun C, Radian AD, de Almeida L, Rojanasakul Y, Stehlik C, An NLRP7-Containing Inflammasome Mediates Recognition of Microbial Lipopeptides in Human Macrophages, Immunity. 36 (2012) 464–476. 10.1016/j.immuni.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Radian AD, Khare S, Chu LH, Dorfleutner A, Stehlik C, ATP binding by NLRP7 is required for inflammasome activation in response to bacterial lipopeptides, Mol Immunol. 67 (2015) 294–302. 10.1016/j.molimm.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T, PYPAF3, a PYRIN-containing APAF-1-like Protein, Is a Feedback Regulator of Caspase-1-dependent Interleukin-1β Secretion*, J Biol Chem. 280 (2005) 21720–21725. 10.1074/jbc.m410057200. [DOI] [PubMed] [Google Scholar]
- [84].Messaed C, Akoury E, Djuric U, Zeng J, Saleh M, Gilbert L, Seoud M, Qureshi S, Slim R, NLRP7, a Nucleotide Oligomerization Domain-like Receptor Protein, Is Required for Normal Cytokine Secretion and Co-localizes with Golgi and the Microtubule-organizing Center*, J Biol Chem. 286 (2011) 43313–43323. 10.1074/jbc.m111.306191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Westerveld GH, Korver CM, van Pelt AMM, Leschot NJ, van der Veen F, Repping S, Lombardi MP, Mutations in the testis-specific NALP14 gene in men suffering from spermatogenic failure, Hum Reprod. 21 (2006) 3178–3184. 10.1093/humrep/del293. [DOI] [PubMed] [Google Scholar]
- [86].Abe T, Lee A, Sitharam R, Kesner J, Rabadan R, Shapira SD, Germ-Cell-Specific Inflammasome Component NLRP14 Negatively Regulates Cytosolic Nucleic Acid Sensing to Promote Fertilization, Immunity. 46 (2017) 621–634. 10.1016/j.immuni.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Welch G, Tsai L, Mechanisms of DNA damage mediated neurotoxicity in neurodegenerative disease, Embo Rep. 23 (2022) e54217. 10.15252/embr.202154217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Takashima H, Boerkoel CF, John J, Saifi GM, Salih MAM, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR, Mutation of TDP1, encoding a topoisomerase I–dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy, Nat Genet. 32 (2002) 267–272. 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- [89].Rao KS, Mechanisms of Disease: DNA repair defects and neurological disease, Nat Clin Pract Neuro. 3 (2007) 162–172. 10.1038/ncpneuro0448. [DOI] [PubMed] [Google Scholar]
- [90].Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS, Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor, Immunity. 36 (2012) 561–571. 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wu P-J, Liu H-Y, Huang T-N, Hsueh Y-P, AIM 2 inflammasomes regulate neuronal morphology and influence anxiety and memory in mice, Sci Rep-Uk. 6 (2016) 32405. 10.1038/srep32405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lammert CR, Frost EL, Bellinger CE, Bolte AC, McKee CA, Hurt ME, Paysour MJ, Ennerfelt HE, Lukens JR, AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment, Nature. 580 (2020) 647–652. 10.1038/s41586-020-2174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zengeler KE, Lukens JR, Innate immunity at the crossroads of healthy brain maturation and neurodevelopmental disorders, Nat. Rev. Immunol 21 (2021) 454–468. 10.1038/s41577-020-00487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Li Q, Barres BA, Microglia and macrophages in brain homeostasis and disease, Nat. Rev. Immunol 18 (2018) 225–242. 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- [95].Lukens JR, Eyo UB, Microglia and Neurodevelopmental Disorders, Annu. Rev. Neurosci 45 (2022) 425–445. 10.1146/annurev-neuro-110920-023056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rui W, Li S, Yang L, Liu Y, Fan Y, Hu Y, Ma C, Wang B, Shi J, Microglial AIM2 alleviates antiviral-related neuro-inflammation in mouse models of Parkinson’s disease, Glia. 70 (2022) 2409–2425. 10.1002/glia.24260. [DOI] [PubMed] [Google Scholar]
- [97].Ma C, Li S, Hu Y, Ma Y, Wu Y, Wu C, Liu X, Wang B, Hu G, Zhou J, Yang S, AIM2 controls microglial inflammation to prevent experimental autoimmune encephalomyelitis, J Exp Med. 218 (2021) e20201796. 10.1084/jem.20201796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Li Y-K, Chen J-G, Wang F, The emerging roles of absent in melanoma 2 (AIM2) inflammasome in central nervous system disorders, Neurochem Int. 149 (2021) 105122. 10.1016/j.neuint.2021.105122. [DOI] [PubMed] [Google Scholar]
- [99].Consortium TFF, Bernot A, Clepet C, Dasilva C, Devaud C, Petit J-L, Caloustian C, Cruaud C, Samson D, Pulcini F, Weissenbach J, Heilig R, Notanicola C, Domingo C, Rozenbaum M, Benchetrit E, Topaloglu R, Dewalle M, Dross C, Hadjari P, Dupont M, Demaille J, Touitou I, Smaoui N, Nedelec B, Méry J-P, Chaabouni H, Delpech M, Grateau G, A candidate gene for familial Mediterranean fever, Nat Genet. 17 (1997) 25–31. 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- [100].Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I, The Pyrin Inflammasome in Health and Disease, Front Immunol. 10 (2019) 1745. 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chung LK, Park YH, Zheng Y, Brodsky IE, Hearing P, Kastner DL, Chae JJ, Bliska JB, The Yersinia Virulence Factor YopM Hijacks Host Kinases to Inhibit Type III Effector-Triggered Activation of the Pyrin Inflammasome, Cell Host Microbe. 20 (2016) 296–306. 10.1016/j.chom.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Gao W, Yang J, Liu W, Wang Y, Shao F, Site-specific phosphorylation and microtubule dynamics control Pyrin inflammasome activation, Proc National Acad Sci. 113 (2016) E4857–E4866. 10.1073/pnas.1601700113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Aubert DF, Xu H, Yang J, Shi X, Gao W, Li L, Bisaro F, Chen S, Valvano MA, Shao F, A Burkholderia Type VI Effector Deamidates Rho GTPases to Activate the Pyrin Inflammasome and Trigger Inflammation, Cell Host Microbe. 19 (2016) 664–674. 10.1016/j.chom.2016.04.004. [DOI] [PubMed] [Google Scholar]
- [104].Park YH, Wood G, Kastner DL, Chae JJ, Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS, Nat Immunol. 17 (2016) 914–921. 10.1038/ni.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kim ML, Chae JJ, Park YH, Nardo DD, Stirzaker RA, Ko H-J, Tye H, Cengia L, DiRago L, Metcalf D, Roberts AW, Kastner DL, Lew AM, Lyras D, Kile BT, Croker BA, Masters SL, Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β, J Exp Medicine. 212 (2015) 927–938. 10.1084/jem.20142384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Heilig R, Broz P, Function and mechanism of the pyrin inflammasome, Eur. J. Immunol 48 (2018) 230–238. 10.1002/eji.201746947. [DOI] [PubMed] [Google Scholar]
- [107].Etienne-Manneville S, Hall A, Rho GTPases in cell biology., Nature. 420 (2002) 629–635. 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- [108].Szabó A, Mayor R, Mechanisms of Neural Crest Migration, Annu Rev Genet. 52 (2018) 43–63. 10.1146/annurev-genet-120417-031559. [DOI] [PubMed] [Google Scholar]
- [109].Karki R, Kanneganti T-D, Diverging inflammasome signals in tumorigenesis and potential targeting, Nat Rev Cancer. 19 (2019) 197–214. 10.1038/s41568-019-0123-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kim R, Emi M, Tanabe K, Cancer immunoediting from immune surveillance to immune escape, Immunology. 121 (2007) 1–14. 10.1111/j.1365-2567.2007.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Sharma BR, Kanneganti T-D, NLRP3 inflammasome in cancer and metabolic diseases, Nat Immunol. 22 (2021) 550–559. 10.1038/s41590-021-00886-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Neufert C, Becker C, Neurath MF, An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression, Nat Protoc. 2 (2007) 1998–2004. 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- [113].Robertis MD, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, Signori E, Fazio VM, The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies, J Carcinog. 10 (2011) 9. 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wang Q, Wang Y, Ding J, Wang C, Zhou X, Gao W, Huang H, Shao F, Liu Z, A bioorthogonal system reveals antitumour immune function of pyroptosis, Nature. 579 (2020) 421–426. 10.1038/s41586-020-2079-1. [DOI] [PubMed] [Google Scholar]
- [115].Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X, Pyroptosis: mechanisms and diseases, Signal Transduct Target Ther. 6 (2021) 128. 10.1038/s41392-021-00507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Sellin ME, Müller AA, Felmy B, Dolowschiak T, Diard M, Tardivel A, Maslowski KM, Hardt W-D, Epithelium-Intrinsic NAIP/NLRC4 Inflammasome Drives Infected Enterocyte Expulsion to Restrict Salmonella Replication in the Intestinal Mucosa, Cell Host Microbe. 16 (2014) 237–248. 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- [117].Rauch I, Deets KA, Ji DX, von Moltke J, Tenthorey JL, Lee AY, Philip NH, Ayres JS, Brodsky IE, Gronert K, Vance RE, NAIP-NLRC4 Inflammasomes Coordinate Intestinal Epithelial Cell Expulsion with Eicosanoid and IL-18 Release via Activation of Caspase-1 and −8, Immunity. 46 (2017) 649–659. 10.1016/j.immuni.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C, Eisenbarth SC, Flavell RA, Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4, Proc National Acad Sci. 107 (2010) 21635–21640. 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Allam R, Maillard MH, Tardivel A, Chennupati V, Bega H, Yu CW, Velin D, Schneider P, Maslowski KM, Epithelial NAIPs protect against colonic tumorigenesis, J Exp Med. 212 (2015) 369–383. 10.1084/jem.20140474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Janowski AM, Colegio OR, Hornick EE, McNiff JM, Martin MD, Badovinac VP, Norian LA, Zhang W, Cassel SL, Sutterwala FS, NLRC4 suppresses melanoma tumor progression independently of inflammasome activation, J Clin Invest. 126 (2016) 3917–3928. 10.1172/jci86953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Tenthorey JL, Chavez RA, Thompson TW, Deets KA, Vance RE, Rauch I, NLRC4 inflammasome activation is NLRP3- and phosphorylation-independent during infection and does not protect from melanoma, J Exp Med. 217 (2020) e20191736. 10.1084/jem.20191736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Yu X, Liu W, Chen S, Cheng X, Paez PA, Sun T, Yuan F, Wei C, Landry JW, Poklepovic AS, Bear HD, Subjeck JR, Repasky E, Guo C, Wang X-Y, Immunologically programming the tumor microenvironment induces the pattern recognition receptor NLRC4-dependent antitumor immunity, J Immunother Cancer. 9 (2021) e001595. 10.1136/jitc-2020-001595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Sadasivam S, Gupta S, Radha V, Batta K, Kundu TK, Swarup G, Caspase-1 activator Ipaf is a p53-inducible gene involved in apoptosis, Oncogene. 24 (2005) 627–636. 10.1038/sj.onc.1208201. [DOI] [PubMed] [Google Scholar]
- [124].Liu R, Truax AD, Chen L, Hu P, Li Z, Chen J, Song C, Chen L, Ting JP-Y, Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: correlation with cancer stages and inflammasome components, Oncotarget. 6 (2015) 33456–33469. 10.18632/oncotarget.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Peng L, Zhu N, Wang D, Zhou Y, Liu Y, Comprehensive Analysis of Prognostic Value and Immune Infiltration of NLRC4 and CASP1 in Colorectal Cancer, Int J Gen Medicine. 15 (2022) 5425–5440. 10.2147/ijgm.s353380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, Meltzer PS, Trent JM, Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma, Oncogene. 15 (1997) 453–457. 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- [127].Zhu H, Zhao M, Chang C, Chan V, Lu Q, Wu H, The complex role of AIM2 in autoimmune diseases and cancers, Immun Inflamm Dis. 9 (2021) 649–665. 10.1002/iid3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Dihlmann S, Tao S, Echterdiek F, Herpel E, Jansen L, Chang‐Claude J, Brenner H, Hoffmeister M, Kloor M, Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients, Int. J. Cancer 135 (2014) 2387–2396. 10.1002/ijc.28891. [DOI] [PubMed] [Google Scholar]
- [129].Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RKS, Gurung P, Neale G, Olsen SR, Carter RA, McGoldrick DJ, Wu G, Finkelstein D, Vogel P, Gilbertson RJ, Kanneganti T-D, Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer, Cell. 162 (2015) 45–58. 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, Rogers AB, Brickey WJ, Wang Y, Schneider M, Mühlbauer M, Chou W-C, Barker BR, Jobin C, Allbritton NL, Ramsden DA, Davis BK, Ting JPY, Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt, Nat Med. 21 (2015) 906–913. 10.1038/nm.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA, NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis, Cell. 145 (2011) 745–757. 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Zheng D, Kern L, Elinav E, The NLRP6 inflammasome, Immunology. 162 (2021) 281–289. 10.1111/imm.13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Chen GY, Liu M, Wang F, Bertin J, Núñez G, A Functional Role for Nlrp6 in Intestinal Inflammation and Tumorigenesis, J Immunol. 186 (2011) 7187–7194. 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, Lemoine Y, Hot D, Chamaillard M, Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury, Proc National Acad Sci. 108 (2011) 9601–9606. 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, Pevsner-Fischer M, Shapiro H, Christ A, Harmelin A, Halpern Z, Latz E, Flavell RA, Amit I, Segal E, Elinav E, Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling, Cell. 163 (2015) 1428–1443. 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Mamantopoulos M, Ronchi F, Hauwermeiren FV, Vieira-Silva S, Yilmaz B, Martens L, Saeys Y, Drexler SK, Yazdi AS, Raes J, Lamkanfi M, McCoy KD, Wullaert A, Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition, Immunity. 47 (2017) 339–348.e4. 10.1016/j.immuni.2017.07.011. [DOI] [PubMed] [Google Scholar]
- [137].Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM, Muruve DA, Philpott DJ, Girardin SE, The NLR Protein NLRP6 Does Not Impact Gut Microbiota Composition, Cell Reports. 21 (2017) 3653–3661. 10.1016/j.celrep.2017.12.026. [DOI] [PubMed] [Google Scholar]
- [138].Vanden Berghe T, Hulpiau P, Martens L, Vandenbroucke RE, Van Wonterghem E, Perry SW, Bruggeman I, Divert T, Choi SM, Vuylsteke M, Shestopalov VI, Libert C, Vandenabeele P, Passenger Mutations Confound Interpretation of All Genetically Modified Congenic Mice, Immunity. 43 (2015) 200–209. 10.1016/j.immuni.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang J-P, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, Elinav E, Finlay BB, Flavell RA, NLRP6 Inflammasome Orchestrates the Colonic Host-Microbial Interface by Regulating Goblet Cell Mucus Secretion, Cell. 156 (2014) 1045–1059. 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Anand PK, Malireddi RKS, Lukens JR, Vogel P, Bertin J, Lamkanfi M, Kanneganti T-D, NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens, Nature. 488 (2012) 389–393. 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Karki R, Man SM, Kanneganti T-D, Inflammasomes and Cancer, Cancer Immunol Res. 5 (2017) 94–99. 10.1158/2326-6066.cir-16-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].White A, Ironmonger L, Steele RJC, Ormiston-Smith N, Crawford C, Seims A, A review of sex-related differences in colorectal cancer incidence, screening uptake, routes to diagnosis, cancer stage and survival in the UK, Bmc Cancer. 18 (2018) 906. 10.1186/s12885-018-4786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Abancens M, Bustos V, Harvey H, McBryan J, Harvey BJ, Sexual Dimorphism in Colon Cancer, Frontiers Oncol. 10 (2020) 607909. 10.3389/fonc.2020.607909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Fan W, Ding C, Liu S, Gao X, Shen X, Boevre MD, Gao Z, Li M, Zhang S, Miao Y, Guan W, Liu G, Yan L, Saeger SD, Song S, Estrogen receptor β activation inhibits colitis by promoting NLRP6-mediated autophagy, Cell Reports. 41 (2022) 111454. 10.1016/j.celrep.2022.111454. [DOI] [PubMed] [Google Scholar]
- [145].Zaki Md.H., Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti T-D, The NLRP3 Inflammasome Protects against Loss of Epithelial Integrity and Mortality during Experimental Colitis, Immunity. 32 (2010) 379–391. 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Allen IC, TeKippe EM, Woodford R-MT, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP-Y, The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer, J Exp Med. 207 (2010) 1045–1056. 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Dupaul-Chicoine J, Arabzadeh A, Dagenais M, Douglas T, Champagne C, Morizot A, Rodrigue-Gervais IG, Breton V, Colpitts SL, Beauchemin N, Saleh M, The Nlrp3 Inflammasome Suppresses Colorectal Cancer Metastatic Growth in the Liver by Promoting Natural Killer Cell Tumoricidal Activity, Immunity. 43 (2015) 751–763. 10.1016/j.immuni.2015.08.013. [DOI] [PubMed] [Google Scholar]
- [148].Wei Q, Guo P, Mu K, Zhang Y, Zhao W, Huai W, Qiu Y, Li T, Ma X, Liu Y, Chen X, Han L, Estrogen suppresses hepatocellular carcinoma cells through ERβ-mediated upregulation of the NLRP3 inflammasome, Lab Invest. 95 (2015) 804–816. 10.1038/labinvest.2015.63. [DOI] [PubMed] [Google Scholar]
- [149].Fitzgerald KA, Kagan JC, Toll-like Receptors and the Control of Immunity, Cell. 180 (2020) 1044–1066. 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Mukherjee S, Kumar R, Lenou ET, Basrur V, Kontoyiannis DL, Ioakeimidis F, Mosialos G, Theiss AL, Flavell RA, Venuprasad K, Deubiquitination of NLRP6 inflammasome by Cyld critically regulates intestinal inflammation, Nat Immunol. 21 (2020) 626–635. 10.1038/s41590-020-0681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Qu Y, Misaghi S, Izrael-Tomasevic A, Newton K, Gilmour LL, Lamkanfi M, Louie S, Kayagaki N, Liu J, Kömüves L, Cupp JE, Arnott D, Monack D, Dixit VM, Phosphorylation of NLRC4 is critical for inflammasome activation, Nature. 490 (2012) 539–542. 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- [152].Liu W, Liu X, Li Y, Zhao J, Liu Z, Hu Z, Wang Y, Yao Y, Miller AW, Su B, Cookson MR, Li X, Kang Z, LRRK2 promotes the activation of NLRC4 inflammasome during Salmonella Typhimurium infection, J Exp Med. 214 (2017) 3051–3066. 10.1084/jem.20170014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Evavold CL, Ruan J, Tan Y, Xia S, Wu H, Kagan JC, The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages, Immunity. 48 (2018) 35–44.e6. 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].He Y, Franchi L, Núñez G, TLR Agonists Stimulate Nlrp3-Dependent IL-1β Production Independently of the Purinergic P2X7 Receptor in Dendritic Cells and In Vivo, J Immunol. 190 (2013) 334–339. 10.4049/jimmunol.1202737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Stienstra R, Joosten LAB, Koenen T, van Tits B, van Diepen JA, van den Berg SAA, Rensen PCN, Voshol PJ, Fantuzzi G, Hijmans A, Kersten S, Müller M, van den Berg WB, van Rooijen N, Wabitsch M, Kullberg B-J, van der Meer JWM, Kanneganti T, Tack CJ, Netea MG, The Inflammasome-Mediated Caspase-1 Activation Controls Adipocyte Differentiation and Insulin Sensitivity, Cell Metab. 12 (2010) 593–605. 10.1016/j.cmet.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]


