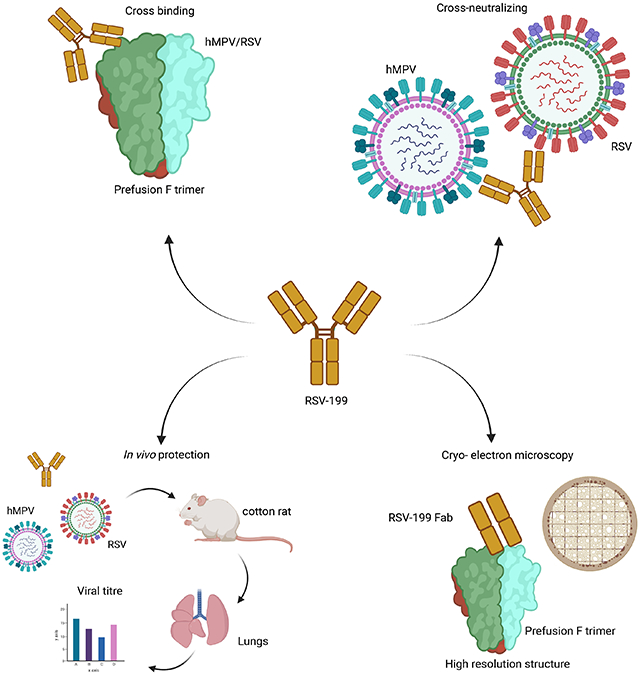
SUMMARY
Respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) infections pose a significant health burden. Using pre-fusion conformation fusion (F) proteins, we isolated a panel of anti-F antibodies from a human donor. One antibody (RSV-199) potently cross-neutralized 8 RSV and hMPV strains by recognizing antigenic site III, which is partially conserved in RSV and hMPV F. Next, we determined the cryo-EM structures of RSV-199 bound to RSV F trimers, hMPV F monomers and an unexpected dimeric form of hMPV F. These structures revealed how RSV-199 engages both RSV and hMPV F proteins through conserved interactions of the antibody heavy chain variable region and how variability within heavy-chain complementarity-determining region 3 (HCDR3) can be accommodated at the F protein interface in site-III-directed antibodies. Furthermore, RSV-199 offered enhanced protection against RSV A and B strains and hMPV in cotton rats. These findings highlight the mechanisms of broad neutralization and therapeutic potential of RSV-199.
Keywords: Respiratory Syncytial Virus, Human, Antibodies, Neutralizing, Antibodies, Monoclonal, Human, Adaptive Immunity
Graphical Abstract
eTOC blurb
Respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) cause widespread and significant infections, particularly in children and the elderly. Wen et al. describe a naturally occurring human monoclonal antibody RSV-199 that recognizes, neutralizes and protects against both RSV and hMPV.
INTRODUCTION
Respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) are members of the Pneumoviridae family that cause widespread and repeated infections worldwide 1. Both viruses infect a larger percentage of individuals, with primary morbidity in children and the elderly. RSV infections are responsible for a greater health burden in young children, causing up to 30-60% of hospitalizations for respiratory infections, while hMPV infections result in ~5% 1-3. While there are no approved vaccines for either RSV or hMPV, several clinical trials for RSV vaccines are in progress and have shown promising results 4-8. In addition, some antibodies targeting the RSV fusion (F) glycoprotein protect high-risk infants against severe disease resulting in hospitalization. Palivizumab was approved in 1998 to prevent severe respiratory disease in children 9 and more recently MEDI8897 (nirsevimab) and MK-1654 are progressing in clinical trials for the treatment of RSV 10,11. Antibody and vaccine development for hMPV also has progressed but remains predominantly in preclinical stages.
The RSV and hMPV F proteins fold to a trimeric, metastable conformation, and stabilization of the pre-fusion structure of F has propelled both vaccine and therapeutic antibody development 12. Stabilized pre-fusion forms of the RSV F protein induce more potently neutralizing antibody responses, as compared to the post post-fusion form, and have helped clarify how antigenic sites are targeted by neutralizing antibodies 4,12,13 Stabilization of the prefusion hMPV F protein has shown more mixed improvements as a vaccine antigen, relative to its post post-fusion form, which may in part be due to increased dynamics and opening of the F trimer 14,15. Antibodies that recognize the pre-fusion hMPV F protein have been isolated from the circulating B cells of human donors and mapped to six primary antigenic sites (sites Ø to V). Some of these antibodies target the interior interfaces of the trimer, consistent with exposure of these buried surfaces during a natural infection 16-19. The antigenic landscapes of RSV and hMPV F proteins have been compared and shown to share some neutralizing regions, although the important neutralization site Ø in RSV F is blocked by a glycosylation site in hMPV F 12,20. RSV and hMPV F strain variation also impacts different major antigenic sites differentially. At least one candidate anti-RSV antibody therapeutic mAb (suptavamab) failed in a phase III clinical trial due to the circulation of naturally occurring viruses with polymorphisms in the RSV F antigenic site V that resisted neutralization by suptavamab 10,21.
Broadly neutralizing antibodies that recognize both RSV and hMPV F proteins have been identified 22-25, despite the relatively low amino acid identity of the RSV and hMPV F proteins of ~35%. The first such antibody identified, designated MPE8, is representative of a class of cross-reactive antibodies encoded by the IGHV1-23 heavy chain gene and exhibiting restricted usage of light chain variable genes 22-24 This class of site III antibodies recognize RSV/ hMPV F at the interface of two adjacent F protomers and can accommodate antibody heavy chain CDR3 regions of diverse sequence and length 23,24. Such cross-reactive antibodies have the potential to provide protection against both RSV and hMPV infection and to resist escape by virus strain variants.
Here we isolated and characterized a panel of antibodies from a single human donor that recognize the hMPV or RSV F protein and identify a potent cross-neutralizing antibody (RSV-199) that recognizes antigenic site III. RSV-199 neutralizes 9 different RSV A, RSV B, hMPV A2, or hMPV B2 strains with overall potencies comparable to RSV-specific antibodies. RSV-199 is resistant to escape by natural sequence variants that disrupt antibodies recognizing F protein antigenic site V. We mapped the interactions of these antibodies on the surface of the F protein using competition-binding assays and electron microscopy and determined the high-resolution structures of RSV-199 bound to RSV and hMPV F. The structures reveal how RSV-199 engages RSV and hMPV F proteins through conserved interactions of the antibody heavy chain variable region, how HCDR3 regions are accommodated at the F protein interface, and how the RSV-199 binding pose differs from previously studied antibodies recognizing site III. In addition, we unexpectedly observed both monomer and dimer forms of hMPV F + Fab RSV-199 complexes, consistent with an inherent flexibility in the oligomeric states that hMPV F protein can adopt. Finally, we show that RSV-199 protects in vivo against representative RSV subgroup A and B strains in cotton rats.
RESULTS
Isolation and characterization of anti-F antibodies from a human donor.
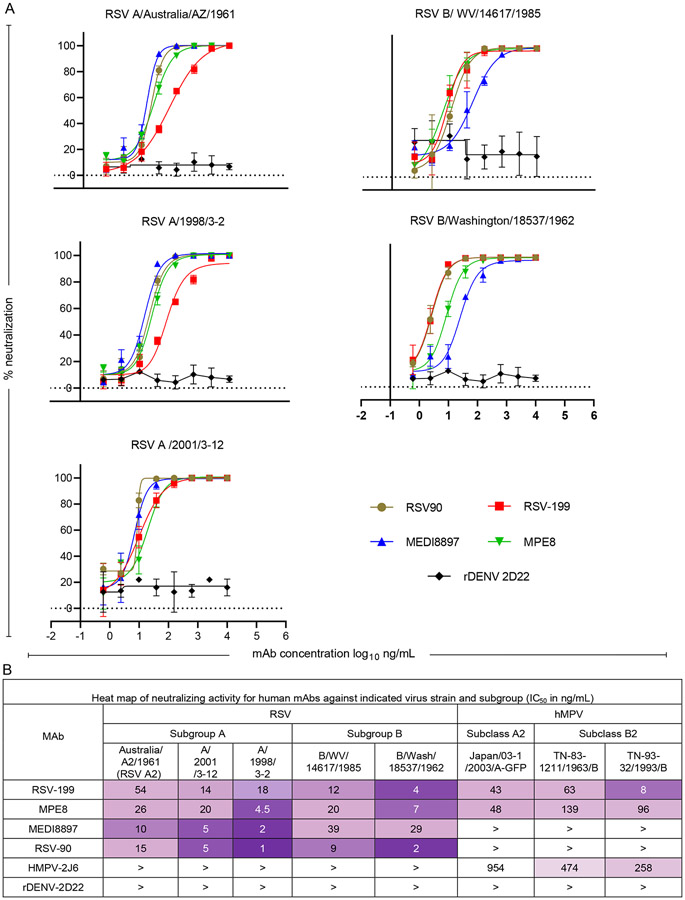
To identify RSV- and hMPV-cross-neutralizing mAbs, we screened a panel of recently isolated hMPV F mAbs (HMPV-2J6, HMPV-4I3, HMPV-4G14, and RSV-199) in neutralization assays. We used three RSV A, three RSV B strains and two hMPV A2 or B2 strains to compare antibody neutralization breadth and included other previously described anti-RSV antibodies, RSV90 26, MEDI8897 27 and MPE8 22,23 (, Fig. 1A&B). As demonstrated previously MEDI8897 was able to neutralize both RSV A and B strains 28 but failed to neutralize hMPV virus strains in our workflow. Similarly, the MPE8 antibody, which previously demonstrated pneumovirus cross neutralizing activity, was able to neutralize both RSV and hMPV viral strains that we tested 29. RSV90, similar to MEDI8897, neutralized all the RSV subgroup A or B strains tested but failed to neutralize any of the hMPV strains (, Fig. 1A&B).
Figure 1.
A. Neutralization curves for RSV-reactive or control antibodies against diverse strains of RSV. B. Neutralizing activity of RSV-, hMPV-, or RSV- and hMPV-reactive mAbs against diverse strains of RSV and hMPV.
In contrast, RSV-199 exhibited the most interesting profile and neutralized diverse hMPV and RSV strains, with half maximal inhibitory concentration (IC50) values in the 10 to 40 ng/mL range. Both MPE8 and RSV-199 neutralized each of the eight RSV and hMPV strains tested, but RSV-199 exhibited greater overall potency and breadth (, Fig. 1A&B).
Next, we compared the heavy and light chain gene usages of these antibodies and observed that RSV-199 shares VH gene usage with MPE8 (Supplementary Table 1). RSV-199 (IGHV3-21*01/ IGHJ5*02 and MPE8 (IGHV3-21*01/ IGHJ4*02) use same IGHV genes but their IGHJ genes are different. Furthermore, their light chain IGLJ genes and CDRH3 sequences are also different (see Supplementary Table 1). RSV-199 is clonally related to 25P13, which we previously showed binds to both RSV and hMPV F proteins at antigenic site III and neutralizes both viruses 23, although we did not characterize its activity using a panel of RSV and hMPV strains. HMPV-2J6 and the previously published neutralizing mAb MEDI8897 both use the commonly found IGHV1-69*01 gene that was identified in many other anti-viral infections 23,30-32.
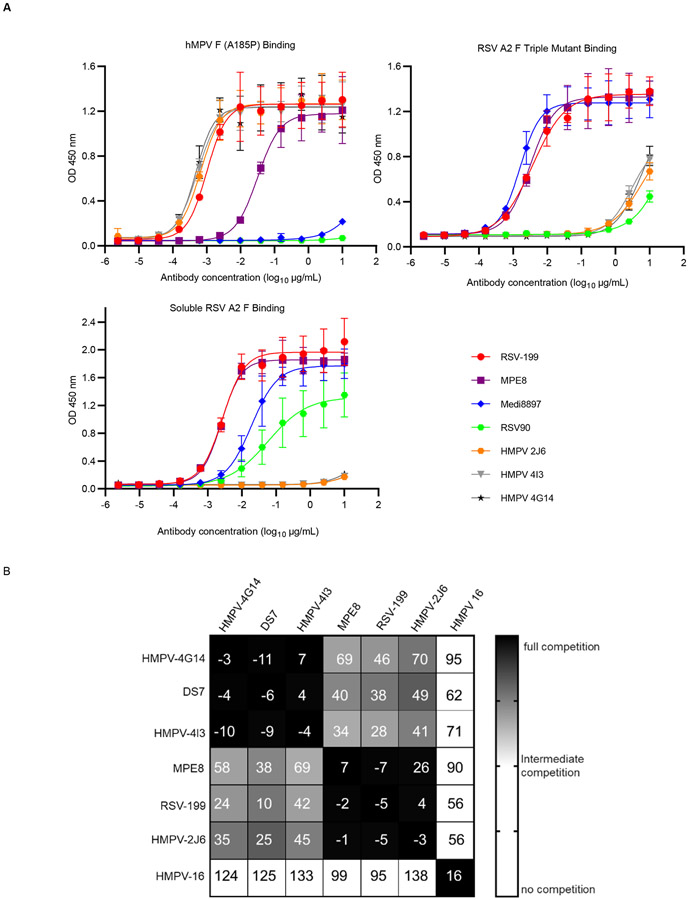
To biochemically characterize the F protein binding specificities of the antibodies, we carried out ELISA assays using RSV strain A2 pre-fusion F protein, an RSV strain A2 F protein antigenic site V triple mutant (pre-fusion) and hMPV A185P (pre-fusion) F proteins. The results confirmed that RSV-199 binds to the pre-fusion conformations of both RSV and hMPV F proteins with EC50 values of 2 ng/mL to both (Fig. 2A). When binding to RSV F protein, RSV-199 has a very similar half maximal effective concentration (EC50) value for binding as did MPE8 (EC50 = 2 ng/mL) and somewhat higher affinity than MEDI8897 (EC50 = 19 ng/mL). For binding to hMPV F, RSV-199 (EC50 =2 ng/mL) has almost the same EC50 value as hMPV-specific antibodies such as HMPV-2J6, HMPV-4I3, and HMPV-4G14. In contrast, MPE8 (EC50 = 30 ng/mL) exhibits lower affinity for binding to the hMPV A2 pre-fusion F protein than RSV-199. We examined the binding of these antibodies to a triple mutant of RSV F that represents strain variants that reduce the binding of site targeted antibodies such as suptavamab and RSV-90. RSV-199 retains its high affinity binding to the triple mutant, similar to MPE8 and MEDI8897 (Fig. 2A). We, next tested RSV-199 and MPE8 Fab binding to RSV-F protein using biolayer interferometry. We noted that the RSV-199 Fab had a higher Kon value compared to that of the MPE8 Fab on RSV F (Supplementary Fig. 1A) in agreement with our ELISA results. Taken together, these data suggest RSV-199 exhibits comparable binding and neutralization capacity with other known best-in-class RSV mAbs.
Figure 2. Two human mAbs strongly bind to both RSV and hMPV F proteins and compete for binding.
(A) ELISA binding of RSV-199, MEDI8897, MPE8, RSV-90 or rDENV-2D22 to hMPV prefusion F protein, RSV strain A2 pre-fusion F protein (SC-TM) or RSV strain A2 post-fusion F protein. Data are mean ± standard deviations (S.D.) of technical triplicates from a representative experiment repeated twice. (B) Competition-binding of monoclonal antibodies reactive with hMPV prefusion F protein. Values in squares are the percent binding of the monoclonal antibody in the presence of the competing monoclonal antibody relative to a mock-competition control. Black squares indicate full competition (<33% relative binding); grey squares indicate intermediate competition; white squares indicate no competition (>67% relative binding).
To investigate the antigenic binding sites for our mAbs, we performed competition-binding assays using hMPV pre-fusion F protein (Fig. 2B). Our previous studies of RSV 25P13 and MPE8 indicated that both of those antibodies recognize F protein antigenic site III, overlapping antigenic sites II and IV 23. Here, we tested competition-binding for RSV-199 in ELISA with a panel of hMPV-specific antibodies. RSV-199 competed fully for binding with HMPV-2J6 and MPE8, while it partially competed with HMPV-4G14 and HMPV-4I3. RSV-199 also partially competed with an hMPV-specific mAb we previously described called DS7 33 (Fig. 2B). The similar competition-binding patterns for MPE8, RSV 25P13, and HMPV-2J6, indicate that these antibodies compete for binding to antigenic site III, while HMPV-2J6, HMPV-4G14, and DS7 bind to a unique molecular footprint in the bottom half of the hMPV F protein.
Overall, the results indicate that RSV-199 mediates broad neutralizing activity for diverse RSV and hMPV strains by binding to antigenic site III. RSV-199 is potently neutralizing for RSV B isolates than the MEDI8897 antibody that is being tested in clinical trials, and more potently neutralizing than MPE8 for hMPV B2 isolates (Fig. 1B).
Structures of hMPV F bound to RSV-199, HMPV-2J6 and HMPV-4I3 antibodies.
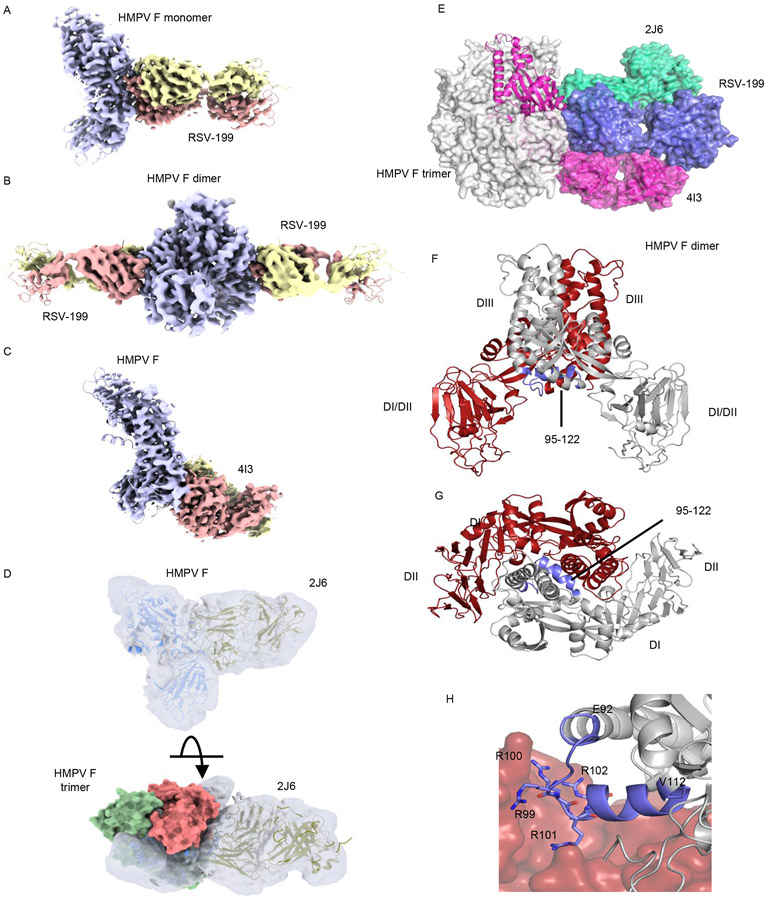
The structures of hMPV F protein bound to RSV-199, HMPV-4I3 and HMPV-2J6 were determined using cryo-EM (Fig. 3A-D). Although hMPV formed stable trimeric complexes with each of the antibodies as observed by gel filtration chromatography (Supplementary Fig. 1B), hMPV F protein trimers were not observed in the cryo-EM grids. Instead, each of the antibodies formed 1:1 complex with individual hMPV F protomers (Fig. 3E and Supplementary Fig. 2). In addition to these 1:1 complex, a subset of hMPV F + Fab RSV-199 complexes also were observed to form a population of dimers, with two RSV-199 Fabs arranged symmetrically around two F protomers (Supplementary Fig. 2, Fig. 3B). The structures of the hMPV F complexes with RSV-199 were determined to resolutions of 3.48Å or 3.38Å, for monomer or dimer structures, respectively, using the same set of micrographs (Fig. 3A, B; Supplementary Figs. 2-4; Supplementary Tables 2, ). The final monomer reconstruction was generated from a set of 165,267 particles and the RSV-199 dimer complexes with a set of 563,257 particles, suggesting that RSV-199 dimers were more prevalent in the micrographs (Supplementary Figs. 2-4). However, processing steps during the data analysis may also impact this relative estimate. The HMPV-4I3 or HMPV-2J6 structures were determined to resolutions of ~3.3Å or ~7Å, respectively (Figs. 3C, D; Supplementary Figs. 2, 5, 6; Supplementary Tables 2, 3).
Figure 3. Structural insights into anti-F antibody epitopes and hMPV F dimerization.
Cryo-EM structures of complexes of (A) monomeric hMPV F+ Fab RSV-199, (B) dimeric hMPV F + Fab RSV-199, (C) hMPV F + Fab hMPV-4I3 and (D) hMPV F + Fab hMPV-2J6. The proximity of 2J6 to adjacent F subunits in a modeled trimer is shown below. (E) docking of hMPV-4I3, hMPV-2J6 and RSV-199 Fabs onto a model of the hMPV F trimer. (F, G) hMPV F dimer observed in complexes with Fab RSV-199; one protomer is colored grey the other is colored red. Residues surrounding the furin cleavage site at the dimer interface are colored in blue. (H) Asymmetric packing of residues 92 to 112 comprising the furin cleavage site and N-terminal residues of the fusion peptide into the neighboring F protomer.
The three antibodies cluster around the midsection of the F head (Fig. 3E). RSV-199 binds to the conserved site III epitope, analogous to the interaction formed by MPE8 12,22-24. HMPV-4I3 and the clonally related HMPV-4G14 bind to an epitope in the DI beta sheet domain that overlaps the previously identified DS7 epitope 33. The HMPV-4I3 and RSV-199 epitopes and the position of those bound Fab domains do not overlap with each other, consistent with the lack of competition in the epitope binning studies (Fig. 2B). HMPV-4I3 engages a surface region that is partially conserved between hMPV and RSV F, accounting for its weak cross-reactivity to RSV F (Fig. 2B). While RSV-199 potently neutralizes RSV and hMPV, HMPV-4I3 and HMPV-4G14 both neutralize hMPV poorly. HMPV-2J6, which also neutralizes hMPV weakly, engages an epitope that is adjacent to that of RSV-199 and hMPV-4I3 (Fig. 3E). The lower resolution of this structure precludes establishing the orientation of the heavy and light chains, but the Fab shows significant structural overlap with RSV-199 and less overlap with HMPV-4I3, consistent with the strong competition with RSV-199 and weaker competition with HMPV-4I3 or HMPV-4G14 (Fig. 2). HMPV-2J6 binds to the α2 helix of the helix-turn-helix motif of site II, forming interactions that are C-terminal to those formed by RSV-199. The HMPV-2J6 epitope lies at the edge of the hMPV F subunit and has the potential to form interactions across subunits in the trimer (Fig. 2D). However, 2J6 was observed only in complexes with single F subunits, suggesting that its binding interactions do not significantly stabilize the hMPV trimer as part of its neutralization mechanism.
Our observation that recombinant hMPV F protein formed both monomers and dimers in the RSV-199 cryo-EM dataset was surprising, as the dimer represents an unexpected oligomeric state for F. The monomer and dimeric complexes with hMPV F are highly similar, with overall RMSDs of 0.64Å. The structures of the RSV-199 complementarity determining region (CDR) loops and the hMPV F epitope did not differ appreciably between the dimer and monomer structures (Supplementary Fig. 8A&B). Comparison of these hMPV F protein structures with the crystal structure of the hMPV F trimer indicates that RSV-199 could be accommodated readily in the hMPV F trimer and is not likely to cause the opening or dimerization of the hMPV F subunits.
The hMPV F dimer (Fig. 3F, G) forms through subunit contacts that overlap contacts seen the hMPV F trimer (Supplementary Fig. 8C-F). At the core of the dimer interface, the subunits exhibit an asymmetric interaction in which residues 95 to 122 from an uncleaved F subunit pack against the adjacent protomer (Fig. 3F, G) in which these residues are disordered and not visible in the cryo-EM maps. Residues 95 to 122 are central to the inter -subunit interaction (Fig. 3H) and this area includes residues that were mutated to form a consensus furin cleavage site (residues 99 to 102), as well as the N-terminal end of the fusion peptide (residues 123 to 122). The dimer interaction appears to require at least one of the protomers to be uncleaved at the furin site and could potentially preferentially form between one cleaved and one uncleaved chain. In addition to these unique packing interactions in the dimer, both the monomer and dimeric F structures exhibit an extension of the third (or “C”) heptad repeat region (HRC) helix through residues 84 to 92 forming a helical structure that is not present in the hMPV F trimer. The helical extension of these residues may promote the opening of the hMPV F subunits from the compact trimeric state, as observed here and in other studies 15-17,19. It is unclear whether the HMPV F dimer observed here could form during F biosynthesis in cells or be incorporated into virions during natural infection.
Structure of RSV-199 bound to RSV F provides insights into its cross-neutralizing activity.
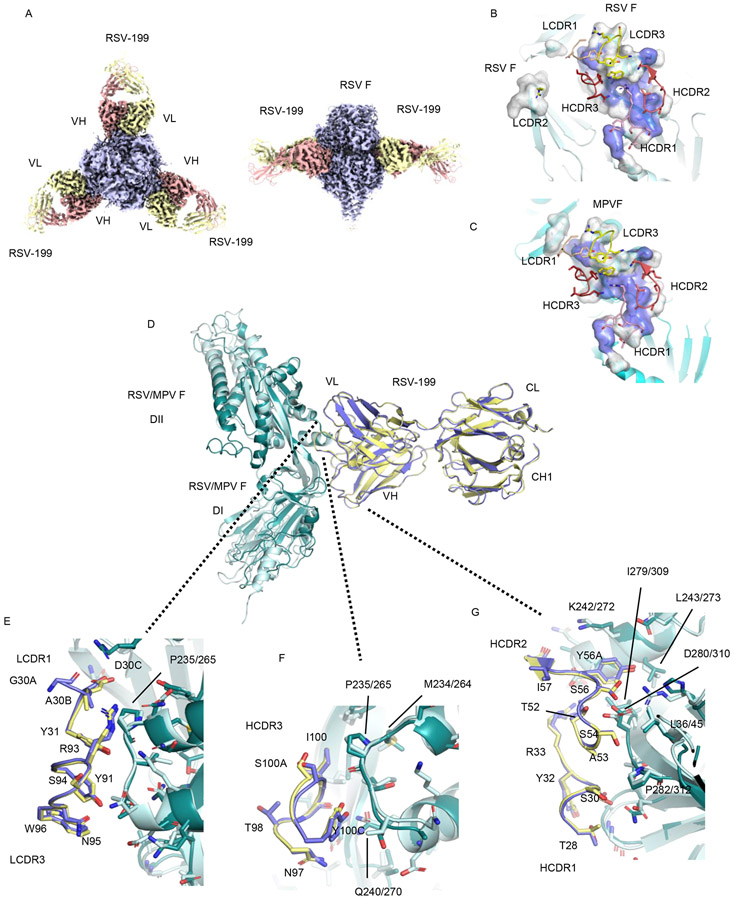
To examine the structural basis for the cross-neutralization of RSV and hMPV by RSV-199, we determined the cryo-EM structure of RSV-199 in complex with recombinant RSV F protein to a resolution of 2.46 Å (Fig. 4A, B; Supplementary Figs. 7G, 8; Supplementary Table 2). In contrast to hMPV F, RSV F retained its trimeric structure, with three RSV-199 Fabs binding to the three site III epitopes in the trimer (Fig. 4A, B). Comparisons of the footprints of the RSV-199 CDRs on RSV and hMPV F (Fig. 4B, C), show common contacts with conserved residues across the two F proteins. The RSV-199 HCDR1 and HCDR2 loop engage the largest patch of conserved residues, with HCDR3 making additional peripheral contacts, anchoring the antibodies to F (Fig. 4B, C). While LCDR2 makes contacts to a neighboring RSV F subunit, this interaction is not formed in either the hMPV F monomer or dimer structures.
Figure 4. Structural basis for cross-neutralization of hMPV and RSV by RSV-199.
(A) Cryo-EM structure of RSV F protein + mAb RSV-199 trimers. (B) RSV-199 epitope on the RSV F protein surface. Conserved residue surfaces are shown in blue. (C) RSV-199 epitope on the hMPV F protein surface. Conserved residue surfaces are shown in blue. (D) Superposition of single protomer complexes of hMPV F and RSV F RSV-199 complexes. (E) Interactions of LCDR1 and LCDR3 at the RSV or hMPV F interface. (F) Interactions of the antibody HCDR3 at the RSV or hMPV F interface. (G) Interactions of the antibody HCDR1 and HCDR2 at the RSV or hMPV F interface. (VH, VL indicates heavy chain variable region and light chain variable region; DI and DII indicate domain-I and domain-II)
Superposition of the RSV and hMPV F complexes shows that the detailed interactions of residues across the two interfaces are remarkably well conserved despite local sequence differences in F (Fig. 4D-G). The RSV-199 CDR loops retain essentially identical conformations, with only minor adjustments observed in the HCDR3 loop (Fig 4F). Similarly, despite sequence differences in the F proteins, the overall conformation of the epitope is well conserved with most sequence changes leading predominantly to changes in side-chain chemistry and orientation. Of the conserved F residues, amino acid hMPV/RSV P235/P265 makes van der Waals contacts with LCDR1 (Fig 4E), while two residues (hMPV/RSV P235/P265 and Q240/Q270) contact HCDR3 (Fig 4F). Most of the conserved interactions involve the HCDR1 and HCDR2 interface (Fig. 4G), containing 6 conserved residues in F (hMPV/RSV L36/L45, K242/K272, L243/L273, I279/I309, D280/D310 and P282/P312). Studies of the infant B cell repertoire response to RSV demonstrated that site-III-directed antibodies preferentially use predominantly IGHV3-21:IGLV1-40 or IGHV3-11:IGLV1-40 pairings 23,24 In the light chain variable region, residues Y31 (LCDR1) and Y91 (LCDR3) are important for binding RSV F and appear similarly positioned to recognize hMPV F. Germline gene-encoded residues, including serines S54, S56 and Y56A, in the HCDR2 specified by IGHV3-21 also are important contributors to F binding and make similar contacts in both RSV and hMPV F complexes with RSV-199.
Accommodation of HCDR3 sequence and length diversity in site III anti-F antibodies.
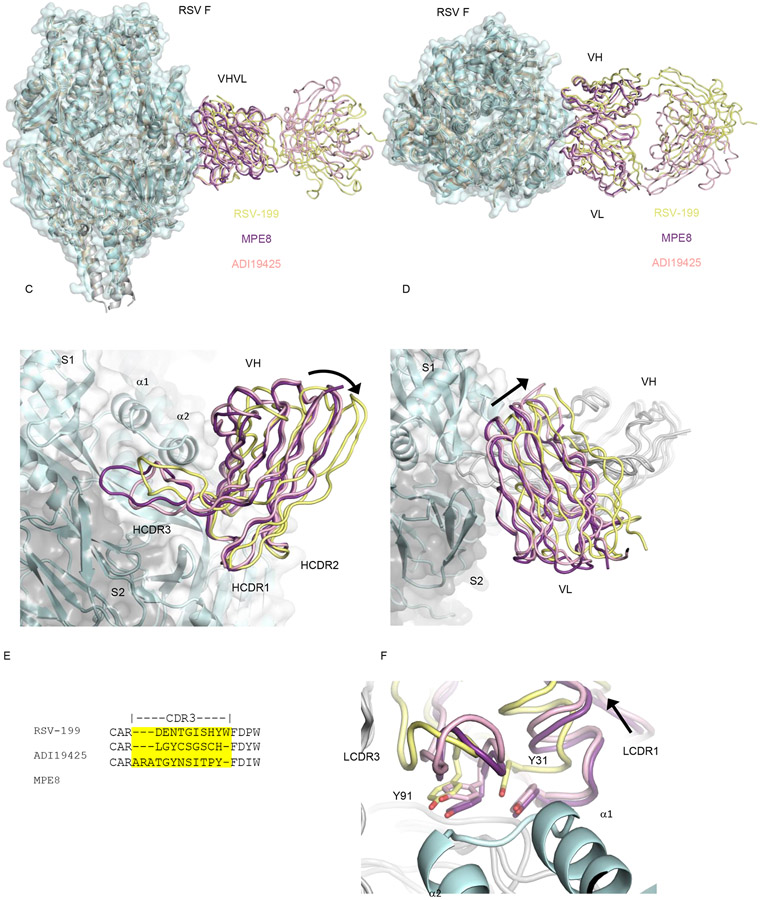
Site III antibodies are unusual in their ability to engage RSV and hMPV F with extensive HCDR3 sequence and length diversity 23,24. To better understand how different antibodies engage site III, we compared the structure of the RSV-199 complex with previously determined structures of RSV F with MPE8 23 23and RSV F with ADI19425 24. Superpositions of the antibodies show an overall conserved angle of approach to the F epitope (Fig. 5A, B), consistent with the restricted variable gene usage common to all three antibodies. The heavy and light chains of MPE8 and ADI19425 superimpose more closely to each other as compared to RSV-199. The RSV-199 pose pivots around an anchor point in the HCDR1/2 interactions (Fig. 5C), causing both the heavy chain and light chain domains to shift significantly relative to the F trimer. In the heavy chain, this difference is evident in a rotation of the core beta strands of the heavy chain and a relocation of the HCDR3 region (Fig. 5C). In the light chain, this pivoting results in an even more significant translocation of the variable domains away from the F trimer and inter -subunit interface (Fig. 5D). This pivoting occurs around the conserved HCDR1/2 contact interface, highlighting its importance in nucleating antigenic site III recognition. In addition, this reorientation may lessen interactions across the F trimer subunits, indicating that neutralization at this site does not require substantial stabilization of the inter -subunit pre-fusion interfaces. RSV-199, MPE8 and ADI19425 HCDR3 sequences differ in both amino acid sequence and length (Fig. 5E). MPE8 has the longest HCDR3 with 12 residues, RSV-199 is intermediate with 10 HCDR3 residues, and ADI19425 shortest with 9 residues. Each of these CDR3 regions adopts a unique conformation and interactions with RSV F (Fig. 5C) that likely bolster the intrinsic interactions of the germline contacts made by other residues in the heavy and light chains variable domains. Even with the substantial pivoting of RSV-199 around the conserved HCDR1/2 interactions, we observe that the positioning of the key light chain tyrosines important for F binding (Y31 and Y91) remain similarly positioned at the F interfaces with in three antibodies (Fig. 5F).
Figure 5. Accommodation of the antibody HCDR3 sequence for structural diversity in F protein antigenic site III.
(A, B) Superposition of RSV-199, MPE8 and ADI19425 Fabs on an RSV F trimer. (C) The heavy chain variable region of RSV-199 adopts a rotated pose compared to that of MPE8 and ADI19425, pivoting about conserved interactions mediated by the HCDR1 and HCDR2 loops. HCDR3 loops adopt distinct structures and interactions to accommodate their different lengths and sequences. (D) The light chain variable region of RSV-199 shifts away from the RSV F trimer surface relative to MPE8 and ADI19425 Fabs. (E) HCDR3 sequence and length diversity of RSV-199, MPE8 and ADI19425 Fabs. (F) Y91/93 and Y31/33, which are important for affinity of binding to F protein, adopt similar positions in RSV-199 despite shifts in the Fab pose on F.
RSV-199 reduces RSV and HMPV replication in cotton rats.
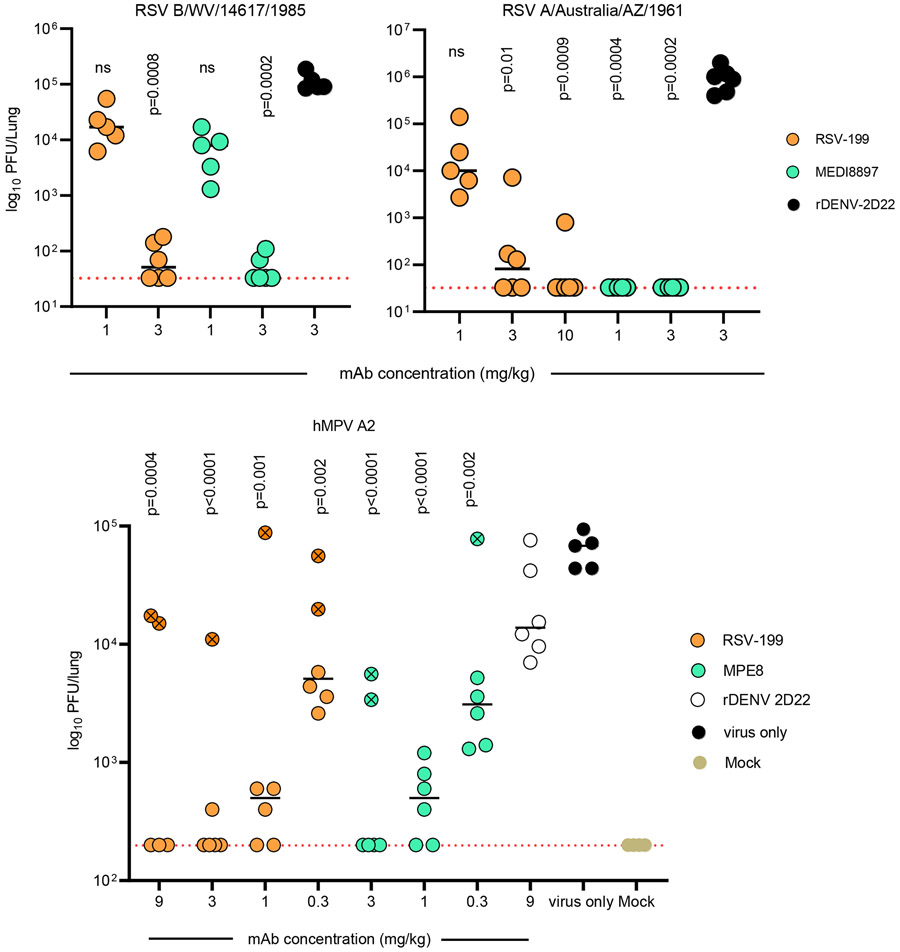
Next, we evaluated the prophylactic efficacy of RSV-199 in the cotton rat animal model (Fig. 6). Cotton rats were administered 1 or 3 mg/kg of RSV-199, MEDI8897 or 3 mg/kg of rDENV-2D22 as isotype control by the i.p. route and challenged by RSV B/WV/14617/1985 virus intranasally. Lungs were harvested on day 4 after challenge to assess viral titers. Cotton rat groups treated with 3 mg/kg of either RSV-199 or MEDI8897 had significantly reduced viral titers, while cotton rat groups treated with 1 mg/kg provided no protection against challenge (Fig. 6).
Figure 6. In vivo efficacy of RSV-199 in cotton rats against challenge with RSV subgroup A or B and hMPV A2 virus strains.
Cotton rats were treated with the indicated concentrations (mg/kg) of mAb, and then inoculated intranasally with the RSV strain indicated. Viral burden at 4 dpi for RSV and 5 dpi for hMPV in the lungs, measured by plaque assay; comparisons were made using a one-way ANOVA with Dunn’s post hoc test (n = 5 cotton rats per group). Animals with an “X” in the circle indicates below detectable levels of serum human IgG.
Similarly, we also tested the in vivo efficacy of RSV-199 and MEDI8897 against RSV A/Australia/A2/1961 strain except that we increased the dose of RSV-199 to 10 mg/kg. RSV-199 provided complete protection in 5/6 animals (Fig. 6). RSV-199 antibody given at a dose of 10 mg/kg greatly reduced RSV strain A2 replication in cotton rats, while the protective capacity was reduced gradually with a stepwise reduction of antibody dosage. RSV-199 prophylaxis with 1 mg/kg did not protect against virus challenge. MEDI8897 protected cotton rats from detectable RSV A2 replication at either dosage, consistent with its more potent in vitro neutralization of RSV strain A2 (Fig. 1). Analogously, we also tested the in vivo efficacy of RSV-199 and MPE8 against hMPV. 6–8-week-old female cotton rats were inoculated with different doses of either RSV-199 or MPE8 via intraperitoneal injection (i.p.) a day before viral challenge. At a timepoint 24 hrs later, 105 pfu of hMPV/A2 was administered intranasally (i.n.). Doses of 9, 3 or 1 mg/kg of RSV-199 mediated complete protection in the lungs from hMPV A2 challenge (Fig. 6). Similarly, 3 or 1 mg/kg doses of MPE8 also offered protection (Fig. 6). Although there were outliers for lung viral titers in each group, low serum human IgG quantification in the outlier animals reflected the reason for those animals to be outliers (Supplementary Fig. 9.) Thus, RSV-199 not only broadly neutralizes strains of RSV and hMPV but also provides in vivo protection in cotton rats.
DISCUSSION
RSV-199 exhibited a broad and potent neutralization profile when tested against 8 different RSV and hMPV strains, with potencies that rival RSV or hMPV mono-specific antibodies, suggesting its potential for development as a therapeutic to prevent diverse pneumovirus infections. The RSV-199 antibody was identified in a screen of B cell clones initially tested for binding to hMPV F and it was found to bind with high affinity to wild-type or antigenic site V mutant RSV F proteins. RSV-199 is a clonal relative of the previously described RSV 25P13 antibody, and it belongs to a class of antibodies encoded by the IGVH1-23 gene segment. These antibodies bind the F antigenic site III through a relatively conserved set of interactions 22-24 Structural studies of RSV-199 bound to both RSV and hMPV F proteins reveals details of its interaction and how it differs from previously determined structures of other site-III-directed antibodies. RSV-199 pivots about the conserved heavy chain + F interaction surface and swings away from the adjacent F protomer in the trimer, eliminating cross-subunit interactions observed with other antibodies. Specific contacts made within the single F protomer may enhance RSV-199 binding affinity, compensating for inter -subunit contacts that may form with other antibodies in this class. This subunit-focused interaction also precludes a fusion inhibition mechanism whereby the antibody stabilizes the pre-fusion F conformation by binding across two subunits. The unusual packing of the RSV-199 HCDR3 loop along the surface of one F protomer allows flexible accommodation of both the length and the sequence of this loop in related antibodies that engage antigenic site III. Specific features of HCDR3 may impact the overall antibody pose and requirement for cross-trimer contacts mediated through the light chain variable domain. The ability of RSV-199 to bind to a single F subunit and neutralize virus may enhance its activity for hMPV F, which appears to form less stable trimers than RSV F and adopts open conformations that allow antibody access to the trimer interior 14-16,33. While RSV F forms trimeric complexes with RSV-199, we observe that hMPV F forms both monomeric and unexpected dimeric complexes with the antibody. The significance of the hMPV F dimers is unclear, but the dimer does not involve direct cross-dimer contacts between RSV-199 antibodies and therefore does not seem to be directly stabilized by antibody binding. These dimers may arise spontaneously in the recombinantly expressed F protein, which contains a C-terminal trimerization domain, or they could potentially form dynamically in vivo in virions or on cells. Given that the F dimer interface involves interactions mediated by an uncleaved subunit, F dimers could form transiently during biosynthesis along an assembly path to trimers. Future studies to address the biological significance of the hMPV F dimer could include cryo-electron tomography of virions and the development of reagents and approaches to detecting HMPV F dimers in cells. Finally, we demonstrated that RSV-199 protects cotton rats against challenge with both RSV A and B strains, consistent with its activity in in vitro neutralization assays.
We also isolated 4 other hMPV F specific antibodies that do not neutralize or only weakly neutralize, HMPV-16, HMPV-2J6, HMPV-4I3 and HMPV-4G14. The HMPV-4I3 and HMPV-4G14 antibodies bind to antigenic site DI-domain and compete for binding to F protein with the previously identified DS7 antibody. The HMPV-16 antibody does not compete for binding with any of the other antibodies tested and may engage alternative antigenic sites that have recently been identified 16-19. The HMPF-2J6 antibody competes with site III antibodies MPE8 and RSV-199, but it does not bind to RSV F and is ~20-fold less potently neutralizing for hMPV than RSV-199. EM mapping of the HMPVF-2J6 complex shows that it engages an epitope adjacent to site III, placing the antibody in a position to sterically compete with site III antibodies. This shift in binding site from RSV-199 to HMPVF-2J6 corresponds with a loss in both neutralization activity and breadth of binding to diverse F proteins.
RSV and hMPV infections place significant health burdens on young and old populations with particularly severe impact on those in lower income settings and with underlying risk factors, such as pre-existing heart and lung diseases. While vaccines for RSV show promise, antibody therapeutics offer an alternative approach for those who may not be eligible for vaccination, and hMPV vaccine development lags that of RSV. Recent clinical studies with the extended half-life antibody MEDI8897 demonstrate that a single antibody dose can protect preterm infants from severe RSV for up to 5 months, spanning a typical RSV season 11. MEDI8897 targets the RSV F protein antigenic site Ø, and its effectiveness has so far not been compromised by natural RSV strain variations, although this remains a possibility 10,34 MEDI8897 is not active against hMPV infection. RSV-199 and other site-III-targeted antibodies with broad neutralization profiles offer the opportunity to inhibit both RSV and hMPV infections and may better prevent resistance of variant strains by targeting this highly conserved antigenic site on the F protein.
STAR METHODS
RESOURCE AVAILABILITY
LEAD CONTACT.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, James E. Crowe, Jr. (james.crowe@vumc.org).
MATERIALS AVAILABILITY.
Materials described in this paper are available for distribution for nonprofit use using templated documents from Association of University Technology Managers “Toolkit MTAs”, available at: https://autm.net/surveys-and-tools/agreements/material-transfer-agreements/mta-toolkit.
DATA AND CODE AVAILABILITY.
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplemental Information. Protein structures and EM maps have been deposited in the Protein Data Bank with the following deposition codes: hMPVF + HPMV-4I3 −8EAY; hMPV F protein monomer + Fab RSV-199 −8E2U; hMPV protein dimer + Fab RSV-199 −8EBP; RSV F protein trimer + Fab RSV-199 −8DZW.
This work dis not use or generate any new code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Research participants.
Participation of the research participant from whom HMPV antibodies were isolated was an otherwise healthy human adult subject male individual. Peripheral blood was obtained for isolation of peripheral blood mononuclear cells by phlebotomy after written informed consent was obtained. The study was approved by the Vanderbilt University Institutional Review Board, and blood samples were obtained only after informed written consent of Vanderbilt University Medical Center, Nashville, TN, USA. RSV-199 was isolated from the peripheral blood mononuclear cells (PBMCs) in the leukofiltration filter of a fully de-identified individual who was a Nashville Red Cross blood donor. Use of de-identified cells from Red Cross leukofiltration filters was approved by the Vanderbilt and Nashville Red Cross Institutional Review Boards.
Cell lines.
HEp-2 host cells (ATCC CCL-23, Cell Lines Services) or LLC-MK2 cell monolayer cultures were cultured in DMEM supplemented with 10% (v/v) FBS, 1x MEM non-essential amino acids solution, 1x MEM vitamin solution and 50 units/mL penicillin; 50 μg/mL streptomycin; and 2 mM Glutamax.
ExpiCHO cells (Thermo Fisher Scientific, A29127) were maintained at 37 °C in 8% CO2 in ExpiCHO Expression Medium (Thermo Fisher Scientific, A2910002). Mycoplasma testing of Expi293F and ExpiCHO cultures was performed monthly using a PCR-based mycoplasma detection kit (ATCC, 30-1012K)
Animal models.
8 weeks old female Sigmodon hispidus cotton rats was ordered from inotivco. Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the VUMC. Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
METHOD DETAILS
Antigen purification.
The gene encoding the stabilized prefusion DS-CAV1 RSV F trimer 35 was synthesized (GeneWiz) and cloned into the PTT5 expression vector (National Research Council (NRC), Canada) with a T4-fibritin trimerization domain in-frame with the heptad repeat of the C-terminal HRB, and with Thrombin cleavage site. The wt MPE8 single chain Fv (scFv) gene was synthesized by Life Technologies as a GeneArt® Strings™ DNA Fragment, was cloned into the pCEP4 expression vector (Invitrogen) and contains a C-terminal TEV cleavage site and His6 tag. The RSV F and scFv expression plasmids were prepared using the Plasmid Mega Kit (Qiagen) and transfected into suspension 293-6E cells (NRC) at a density of 1.8 to 2.0 million cells/mL using 25-kDa linear polyethylenimine (Polysciences), (PEI) following 293-6E cell protocols. Supernatants were harvested five days post-transfection by centrifugation (20 min at 8,000 × g at room temperature), filtered through 0.45 μm filters and dialyzed against 200 mM NaCl, 50 mM Na2HPO4 pH 7.4. The RSV F protein was purified by Co2+ affinity chromatography (TALON Resin, BD Biosciences) and size exclusion chromatography using a Superdex-200 column equilibrated in 25 mM sodium phosphate, pH 7.4, and 100 mM NaCl, 100 mM imidazole. The purified protein was concentrated with Amicon Ultra centrifugal filters with a 10 kD molecular weight cut-off (Millipore).
Hybridoma.
Peripheral blood mononuclear cells were isolated from a single eight-year-old human donor by Ficoll-gradient centrifugation and the cells were frozen for later use. For B cell screening, thawed cells were transformed with Epstein–Barr virus as previously described36 and plated in 384-well plates to generate immortalized lymphoblastoid cell lines. Supernatants from the transformed cells were screened for antibodies binding to a highly stable pre-fusion conformation of RSV strain A2 F protein, using the single-chain 6 triple mutant (SC-TM) construct. B cells from cultures producing antibodies reactive with the pre-fusion F protein were electro fused with the HMMA2.5 myeloma cell line to generate stable hybridoma cell lines. To obtain homogeneous antibody secretions, hybridoma cells were cloned biologically by single cell flow cytometric sorting. Cells expanded after clonal selection were submitted for antibody variable gene sequence analysis of cDNA from total cell RNA processed with reverse transcription and 5′RACE.
MAb production and purification.
cDNAs encoding mAbs of interest were synthesized (Twist Bioscience) and cloned into an IgG1 monocistronic expression vector (designated as pTwist-mCis_G1) or Fab expression vector (designated as pTwist-mCis_FAB) and used for production in mammalian cell culture. This vector contains an enhanced 2A sequence and GSG linker that allows for the simultaneous expression of mAb heavy and light chain genes from a single construct upon transfection 6. For antibody production, we performed transfection of ExpiCHO cell cultures using the Gibco ExpiCHO Expression System as described by the vendor. IgG molecules were purified from culture supernatants using HiTrap MabSelect SuRe (Cytiva) on a 24-column parallel protein chromatography system (Protein BioSolutions).
Fab proteins were purified using CaptureSelect column (Thermo Fisher Scientific). Purified antibodies were buffer-exchanged into PBS, concentrated using Amicon Ultra-4 50-kDa (IgG) or 30 kDa (Fab) centrifugal filter units (Millipore Sigma) and stored at 4°C until use. F(ab′)2 fragments were generated after cleavage of IgG with IdeS protease (Promega) and then purified using TALON metal affinity resin (Takara) to remove the enzyme and protein A agarose (Pierce) to remove the Fc fragment. Purified mAbs were tested routinely for endotoxin levels and found to be less than 30 EU per mg IgG. Endotoxin testing was performed using the PTS201F cartridge (Charles River), with a sensitivity range from 10 to 0.1 EU per mL, and an Endosafe Nexgen-MCS instrument (Charles River).
ELISA binding assays.
Wells of 96-well microtiter plates were coated with purified recombinant F protein at 4°C overnight. Plates were blocked with 2% non-fat dry milk and 2% normal goat serum in Dulbecco’s phosphate-buffered saline (DPBS) containing 0.05% Tween-20 (DPBS-T) for 1 h. The bound antibodies were detected using goat anti-human IgG conjugated with horseradish peroxidase (HRP) (Southern Biotech, cat. 2040-05, lot B3919-XD29, 1:5,000 dilution) and a 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific). Color development was monitored, 1 M HCl was added to stop the reaction, and the absorbance was measured at 450 nm using a spectrophotometer (Biotek). For dose–response assays, serial dilutions of purified mAbs were applied to the wells in triplicate, and antibody binding was detected as detailed above. Half maximal effective concentration (EC50) values for binding were determined using Prism v.8.0 software (GraphPad) after log transformation of the mAb concentration using sigmoidal dose–response nonlinear regression analysis.
Competition-binding analysis.
Wells of 384-well microtiter plates were coated with 1 μg/mL purified hMPV pre-fusion F protein at 4°C overnight. Plates were blocked with 2% BSA in DPBS-T for 1 h. Purified unlabeled monoclonal antibodies were diluted tenfold in blocking buffer, added to the wells (20 μL per well) in quadruplicates and incubated for 1 h at ambient temperature. A biotinylated preparation of a recombinant monoclonal antibodies was added to each of four wells with the respective monoclonal antibody at 2.5 μg/mL in a volume of 5 μL per well (final concentration of biotinylated monoclonal antibody 0.5 μg/mL) without washing of unlabeled antibody, and then incubated for 1 h at ambient temperature. Plates were washed and bound antibodies were detected using HRP-conjugated avidin (Sigma) and a TMB substrate. The signal obtained for binding of the biotin-labelled reference antibody in the presence of the unlabeled tested antibody was expressed as a percentage of the binding of the reference antibody alone after subtracting the background signal. Tested mAbs were considered competing if their presence reduced the reference antibody binding to less than 41% of its maximal binding and non-competing if the signal was greater than 71%. A level of 40 to 70% was considered intermediate competition.
Affinity kinetics analysis using biolayer interferometry.
FAB2G biosensor tips (Forte Biosciences) on an Octet Red96 or HTX biolayer interferometry instrument (Forte Biosciences) were soaked for 10 minutes in 1× kinetics buffer (Forte Biosciences), followed by a baseline signal measurement for 60 s. Recombinant RSV-199 Fab or MPE8 (10 μg/mL; IBT BioServices) was immobilized onto the biosensor tips for 30 s. After a wash step in 1× kinetics buffer for 30 to 60 s, the recombinant RSV prefusion F protein titrated down 2-fold up to 8 dilutions starting at 100 μg/mL and was incubated with the Fab-containing biosensor for 300 s. Disassociation of antigen was done by wash step in 1× kinetics buffer for 300 s. Kinetics analysis was performed using Octet® Analysis Studio Software version 10.
Plaque reduction neutralization test (PRNT).
Serial dilutions of mAb were incubated with 4.8 x 104/mL suspension of infectious RSV, hMPV for 1 h at 37°C. The antibody-virus complexes were added to HEp-2 cell-culture monolayers in 24-well plates for 1 h at 37°C. Cells then were overlaid with 1% (w/v) methylcellulose in minimum essential medium (MEM) supplemented to contain 2% heat-inactivated FBS. Plates were fixed 30 h later by removing overlays and fixed with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature. The plates were incubated sequentially with 1 μg/mL of anti- antibody or and then HRP-conjugated goat anti-human IgG (Sigma-Aldrich, A6029) in PBS supplemented with 0.1% (w/v) saponin (Sigma) and 0.1% BSA. RSV-infected plaques were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot 5.0.37 Macro Analyzer (Cellular Technologies). Half maximal inhibitory concentration (IC50) values were determined by nonlinear regression analysis (with a variable slope) using Prism software.
Animals, infection, and measurement of viral burden.
Eight- to ten-week-old cotton rats were intranasally inoculated under isoflurane anesthesia with 105 plaque forming units in 100 μL with indicated RSV strain. Viral burden at 4 dpi in the lungs, measured by plaque assay. Serial dilutions of lung homogenates were inoculated onto HEp-2 or LLC-MK2 cell monolayer cultures, and plaque assays were performed as described above. The viral titer was determined by multiplying the number of plaques by the reciprocal sample dilution and expressed as PFU per lung.
RSV and hMPV F protein expression and purification.
The construction, expression, and purification of the stabilized pre-fusion DS-CAV1 RSV F trimer were described previously 23. Briefly, the F construct was synthesized (GENEWIZ) and cloned into the PTT5 expression vector (National Research Council (NRC), Canada) with a T4-fibritin trimerization domain after the F heptad repeat B, along with a thrombin cleavage site and His8 tag for purification. The hMPV F expression construct from the A2 strain was synthesized (Twist) in the PTT5 expression vector (National Research Council (NRC), Canada) and included C-terminal T4 fibritin and His8 tags. This construct included an RRRR furin cleavage site at the fusion peptide instead of original RQSR site, an A185P substitution and A113C/A339C substitutions as described previously (Patent WO 2016/103238A1). The RSV F and hMPV F expression plasmids were prepared using the Plasmid Maxiprep Kit (ZymoPURETM II Plasmid Maxiprep Kit) and transfected into suspension 293-6E cells (NRC) at a density of 1.8 to 2.0 million cells/mL using 25-kDa linear polyethylenimine (PEI, Polysciences) following 293-6E cell protocols. Supernatants were harvested five days post-transfection by centrifugation (20 min at 8,000 × g at 4°C) and filtered through 0.45 μm filters. The F proteins were purified by Co2+ affinity chromatography (TALON Resin, BD Biosciences) and size exclusion chromatography using a Superdex-200 column equilibrated in 25 mM sodium phosphate, pH 7.4, and 100 mM NaCl, 100 mM imidazole. The purified protein was concentrated with Amicon Ultra centrifugal filters with a 10 kD molecular weight cut-off (Millipore).
Complexes of HMPV-4I3, HMPV-2J6 or RSV-199 Fab with either the RSV or hMPV F proteins were prepared by mixing the two proteins in a 1:1.2 ratio on ice for 2 hours. The purified complexes of RSV-199 with F protein were obtained using size exclusion chromatography with a Superdex-200 column and verified by SDS–PAGE.
Cryo-EM data collection.
For cryo-EM data collection, 3 μL of complex of Fab RSV-199 with the RSV F protein at ~1.75 mg/mL with 0.075% octyl-β-d-glucopyranoside was applied to glow-discharged Quantifoil gold R2/1 200 mesh grids. The grids were blotted with Whatman filter paper for 2 s at 96% humidity using a Leica GP automatic plunge freezer and frozen in liquid ethane. A total of 7,263 movie stacks were collected on an FEI Titan Krios electron microscope operated at 300 kV with an energy filter (20-keV slit width) and a Gatan K3 Summit direct detector data collection session. Movie stacks were recorded at ×130,000 magnification, corresponding to 1.11 Å/pixel, with a total accumulated dose of 49.58 eÅ−2, 0.2 s/frame, and a total exposure time of 10 s. The cryo-EM data was processed primarily in cryoSPARC 37 The image stacks were motion corrected by patch motion, and the contrast transfer function (CTF) was estimated using patch CTF estimation in cryoSPARC. After blob picking of 4,178,389 particles, we imported these 2D templates into cryoSPARC to allow template picking of ~5,584,963 particles. After a few rounds of 2D classification to remove junk particles, the remaining ~532,893 good particles were extracted again with a box size of 320 pixels. After ab initio, heterogenous refinement to remove more junk particles, the final remaining ~476,914 very good particles. Once the map quality and resolution could no longer be improved by further reducing the particle numbers, we performed the best particles and ran the homogenous and nonuniform refinement steps.
For cryo-EM data collection, 3 μL of complex of Fab RSV-199 with the hMPV F protein at ~1.43 mg/mL with 0.075% octyl-β-d-glucopyranoside was applied to glow-discharged Quantifoil gold R2/1 200 mesh grids. The grids were blotted with Whatman filter paper for 2 s at 96% humidity using a Leica GP automatic plunge freezer and frozen in liquid ethane. A total of 5,697 movie stacks were collected on an FEI Titan Krios electron microscope operated at 300 kV with an energy filter (20-keV slit width) and a Gatan K3 Summit direct detector data collection session. Movie stacks were recorded at ×130,000 magnification, corresponding to 1.11 Å/pixel, with a total accumulated dose of 49.58 eÅ−2, 0.2 s/frame, and a total exposure time of 10 s. The cryo-EM data was processed in cryoSPARC 37
For the monomer complex structure of Fab RSV-199 with hMPV F protein, the image stacks were motion corrected by patch motion, and the contrast transfer function (CTF) was estimated using patch CTF estimation in cryoSPARC. After manual picking of ~7,521 particles. We imported these 2D templates into cryoSPARC to allow template picking, after a few rounds of template picking of ~5,603,348 particles. The picked particles were inspected, extracted with a box size of 320 pixels, and then Fourier cropped to a box size of 128 pixels. After a few rounds of 2D classification to remove junk particles, the remaining −391,164 particles were extracted again with a box size of 320 pixels. After rounds of ab initio, heterogenous refinement and homogeneous refinement were used to further clean and classify the particles, yielding ~165,267 good particles. Homogenous refinement and nonuniform refinement were used to further clean and classify the particles, Once the map quality and resolution could no longer be improved by further reducing the particle numbers, we performed local and global CTF refinement on the subset of the best particles and reran the nonuniform refinement.
For the dimer complex structure of Fab RSV-199 with the hMPV F protein, the image stacks were motion corrected by patch motion, and the contrast transfer function (CTF) was estimated using patch CTF estimation in cryoSPARC. After manual picking of ~7,521 particles. We imported these 2D templates into cryoSPARC to allow template picking of ~3,968,774 particles. The picked particles were inspected, extracted with a box size of 320 pixels, and then Fourier cropped to a box size of 128 pixels. After a few rounds of 2D classification to remove junk particles, the remaining ~1,344,628 particles were extracted again with a box size of 256 pixels. A few rounds of ab initio, heterogenous refinement and homogeneous refinement were used to further clean and classify the particles, yielding ~563,257 good particles. This subset of the best particles was further processed with nonuniform refinement step and cryoSPARC volume and sharpening tools.
For cryo-EM data collection, 3 μL of complex of Fab HMPV-4I3 with the hMPV F protein at ~1.56 mg/mL with 0.05% octyl-β-d-glucopyranoside was applied to glow-discharged Quantifoil gold R2/1 200 mesh grids. The grids were blotted with Whatman filter paper for 2 s at 96% humidity using a Leica GP automatic plunge freezer and frozen in liquid ethane. A total of 6,895 movie stacks were collected on an FEI Titan Krios electron microscope operated at 300 kV with an energy filter (20-keV slit width) and a Gatan K3 Summit direct detector data collection session. Movie stacks were recorded at ×130,000 magnification, corresponding to 1.11 Å/pixel, with a total accumulated dose of 49.58 eÅ−2, 0.2 s/frame, and a total exposure time of 10 s. The cryo-EM data was processed primarily in cryoSPARC 37. After manually curating the imported movies, 944 movie stacks were accepted for further analysis. The image stacks were motion corrected by patch motion, and the contrast transfer function (CTF) was estimated using patch CTF estimation in cryoSPARC. After Blob picking of ~878,284 particles, the picked particles were inspected, extracted with a box size of 256 pixels, and then Fourier cropped to a box size of 128 pixels. After a few rounds of 2D classification to remove junk particles, we imported these 2D templates into cryoSPARC to allow template picking. After rounds of 2D classification to remove junk particles, the remaining ~403,956 particles were extracted again with a box size of 256 pixels. A few rounds of ab initio, heterogenous refinement and homogeneous refinement were used to further clean and classify the particles, providing ~226,482 good particles. Once the map quality and resolution could no longer be improved by further reducing the particle numbers, a final nonuniform refinement step was used.
For cryo-EM data collection, 3 μL of complex of Fab HMPV-2J6 with the hMPV F protein at ~1.17 mg/mL with 0.05% octyl-β-d-glucopyranoside was applied to glow-discharged Quantifoil gold R2/1 200 mesh grids. The grids were blotted with Whatman filter paper for 2 s at 96% humidity using a Leica GP automatic plunge freezer and frozen in liquid ethane. A total of 1,872 movie stacks was collected on a Glacios electron microscope operated at 200 kV with a Gatan K2 Summit direct detector data. Movie stacks were recorded at 36,000× magnification, corresponding to 1.17 Å/pixel, with a total accumulated dose of 8.215 eÅ−2, 0.2 s/frame, and a total exposure time of 10 s. The cryo-EM data set was processed primarily in cryoSPARC 37. We imported a total of 1,872 movies into cryoSPARC to allow manual curation and the selection of ~851 movies stacks for further analysis. The image stacks were motion corrected by patch motion, and the contrast transfer function (CTF) was estimated using patch CTF estimation in cryoSPARC. After manual picking of ~812 particles and a few rounds of 2D classification, we imported these 2D templates into cryoSPARC to allow template picking of ~539,268 particles. After rounds of 2D classification to remove junk particles, the remaining ~502,257 particles were extracted again with a box size of 256 pixels. A few rounds of ab initio and heterogenous refinement were used to further clean and classify the particles, yielding the remaining ~100,310 good particles. Once the map quality and resolution could no longer be improved by further reducing the particle numbers, we ran homogeneous refinement and nonuniform refinement steps.
Cryo-EM model building and refinement
RSV F trimer + RSV-199 Fab complex model building and refinement.
The previously determined RSV F + scFv MPE8 structure (5U68) was used to generate the RSV F trimer model. The Fab 1AQK model was generated RSV-199 Fab model. The two models were docked into the EM density map using UCSF Chimera 38,39. RSV F protein and RSV-199 Fab were docked into the density map separately, saved as one model, and then reconnected in Coot 40. A combination of Phenix refinement41-43 and Coot was used to further refine the model.
hMPV F monomer + Fab RSV-199 complex model building and refinement.
The previously determined hMPV F + Fab DS7 structure (4DAG) was used to generate the hMPV F monomer model. The Fab 6W16 model was generated using the RSV-199 Fab model. The two models were docked into the EM density map using UCSF Chimera 38,39. HMPV F protein and RSV-199 Fab were docked into the density map separately, saved as one model, and then reconnected in Coot 40. A combination of Phenix refinement and Coot was used to further refine the model.
MPV F dimer + Fab RSV-199 complex model building and refinement.
The previously determined hMPV F protein + Fab DS7 structure (4DAG) was used to generate the hMPV F dimer model. The Fab 6W16 model was generated using the RSV-199 Fab model. The two models were docked into the EM density map using UCSF Chimera. HMPV F protein and RSV-199 Fab were docked into the density map separately, saved as one model, and then reconnected in Coot. A combination of Phenix refinement and Coot was used to further refine the model.
hMPV F protein monomer + Fab HMPV-4I3 complex model building and refinement.
The previously determined hMPV F protein + Fab DS7 structure (4DAG) was used to generate the hMPV F monomer model. The Fab 4QHK model was generated HMPV-4I3 Fab model. The two models were docked into the EM density map using UCSF Chimera. hMPV F protein and HMPV-4I3 Fab were docked into the density map separately, saved as one model, and then reconnected in Coot. A combination of Phenix refinement and Coot was used to further refine the model.
hMPV F protein + HMPV-2J6 Fab complex model building and refinement.
The previously determined hMPV F protein + Fab DS7 structure (4DAG) was used to generate the hMPV F monomer model. The Fab 4QHK model was generated HMPV-4I3 Fab model. The two models were docked into the EM density map using UCSF Chimera. hMPV F protein and HMPV-4I3 Fab were docked into the density map separately, saved as one model, and then reconnected in Coot. A combination of Phenix refinement and Coot was used to further refine the model.
Quantification and statistical analysis.
Mean ± S.E.M. or mean ± S.D. were determined for continuous variables as noted. Technical and biological replicates are described in the figure legends. For analysis of mouse studies, the comparison of weight-change curves was performed using a one-way ANOVA with Dunnett’s post hoc test of the area under the curve for days 3 to 6 post-infection, using Prism v.9.0 (GraphPad). Infectious viral loads were compared by a one-way ANOVA with Dunnett’s multiple comparisons test using Prism v.9.0 (GraphPad).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human lambda, mouse ads-UNLB | Southern Biotech | RRID: AB_2795760 |
| Goat anti-human kappa, mouse ads-UNLB | Southern Biotech | RRID: AB_2795728 |
| Goat anti-human IgG Fc, Multispecies ads-HRP | Southern Biotech | RRID: AB_2795580 |
| Goat anti-human IgG-HRP | Southern Biotech | RRID: AB_2795644 |
| Bacterial and virus strains | ||
| RSV Australia/A2/1961 (RSV A2) | BEI | NR-28529 |
| RSV B/WV/14617/1985 | BEI | NR-48831 |
| HMPV Japan/03-1/2003/A-GFP | Zhou M et.al., 2013 | N/A |
| HMPV TN-83-1211/1963/B | BEI | NR-22227 |
| RSV A/1998/3-2 | BEI | NR-44233 |
| Biological samples | ||
| Human PBMCs from phlebotomy sample | Vanderbilt Vaccine Center | N/A |
| Human PBMCs from leukofiltration filter | Nashville Red Cross | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Avidin–Peroxidase | Sigma | Cat# A3151 |
| 1-step Ultra TMB-ELISA substrate solution | Thermo Fisher | Cat# 34029 |
| ExpiCHO Expression Medium | Thermo Fisher | CAT# A2910001 |
| Freestyle 293 expression medium | Thermo Fisher | CAT# 1238002 |
| Fetal Bovine Serum, ultra-low IgG | Thermo Fisher | CAT# 16250078 |
| PEI, 25-kDa linear polyethylenimine | Polysciences Inc. | Cat# 23996 |
| TALON Resin | BD Biosciences | Cat# 635504 |
| EZ-Link™ NHS-PEG4-Biotin, No-Weigh™ Format | Thermo Fisher | CAT# A39259 |
| FabALACTICA® Fab kit | Genovis | Cat# A2-AFK-025 |
| BioLock biotin blocking | IBA Lifescience | Cat# 2-0205-050 |
| Deposited data | ||
| RSVF_RSV-199-trimer | wwPDB Deposition | 8DZW |
| MPVF_RSV-199-monomer | wwPDB Deposition | 8E2U |
| MPVF_4I3-monomer | wwPDB Deposition | 8EAY |
| MPVF_RSV-199-dimer | wwPDB Deposition | 8EBP |
| Experimental models: Cell lines | ||
| Homo sapiens, human HEp-2 | ATCC | CCL23 |
| LLC-MK2 | ATCC | CCL-7 |
| Homo sapiens, human HEK 293-6E | ATCC | CRL-1573 |
| Experimental models: Organisms/strains | ||
| Cotton rats | Inotiv | Sigmodon hispidus |
| Recombinant DNA | ||
| pTwist_DENV-2D22_mCisL_hG1 | This paper | N/A |
| pML_RSV-90_hG1 | This paper | N/A |
| pML_MEDI8897_hG1 | This paper | N/A |
| pTwist_RSV-199_mCisL_FAB | This paper | N/A |
| pVVC_RSV-199_mCisL_hG1 | This paper | N/A |
| pTwist_MPE8_mCisL_hG1 | This paper | N/A |
| pTwist_HMPV-2J6_mCisK_hG1 | This paper | N/A |
| pTwist_MPV-2J6_mCisK_FAB | This paper | N/A |
| pML_RSV-90_FAB | This paper | N/A |
| DS-CAV1 RSV F | construct was synthesized (GeneWiz) | N/A |
| hMPV F (A185P and A113C/A339C substitutions) | construct was synthesized (Twist) | N/A |
| Software and algorithms | ||
| Prism | GraphPad | v 9.0.0 |
| cryoSPARC | This paper | Live |
| Other | ||
| xCELLigence RTCA MP analyzer | Acea Biosciences, Inc | N/A |
| xCELLigence E-Plate 96 PET cell culture plates | Acea Biosciences, Inc | Cat# 300601010 |
| Equipment | ||
| Electron microscope | FEI Titan Krios | N/A |
| Electron microscope | FEI Titan Glacios | N/A |
Highlights.
RSV-199 potently cross-neutralized 8 different RSV and hMPV strains.
Cryo-EM reveals RSV-199 targeting antigenic site III of prefusion F.
RSV-199 mediates enhanced protection in vivo against RSV A and B strains and hMPV.
RSV-199 exhibited potencies comparable to RSV or hMPV mono-specific antibodies.
ACKNOWLEDGEMENTS.
We thank members of the Jardetzky and Crowe laboratories and Donghua Chen (Stanford) for his support in cryo-EM data collection and analysis. The work was supported by NIAID/NIH grant R01 AI13752. We thank the NIAID Division of Microbiology and Infectious Diseases (DMID) Preclinical Services Program for support (contract HHSN272201700028I) through a cotton rat model contract under Task Order 75N93020F00001, Task A45, awarded to Sigmovir Biosystems, Inc, which conducted the hMPV cotton rats studies in collaboration with the Vanderbilt team.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS.
J.E.C. has served as a consultant for Luna Labs USA, Merck Sharp & Dohme Corporation, Emergent Biosolutions, GlaxoSmithKline and BTG International Inc, is a member of the Scientific Advisory Board of Meissa Vaccines, a former member of the Scientific Advisory Board of Gigagen (Grifols) and is founder of IDBiologics. The laboratory of J.E.C. received unrelated sponsored research agreements from AstraZeneca, Takeda, and IDBiologics during the conduct of the study. T.S.J. has served as a consultant for Pfizer. All other authors report no conflicts. Vanderbilt University has applied for patents for some of the antibodies in this paper.
REFERENCES
- 1.Collins PL, and Karron RA (2013). Respiratory syncytial virus and metapneumovirus. In Fields Virology, Knipe DM, and Howley PM, eds. (Wolters Kluwer Health Adis (ESP)). [Google Scholar]
- 2.Boivin G, De Serres G, Cote S, Gilca R, Abed Y, Rochette L, Bergeron MG, and Dery P (2003). Human metapneumovirus infections in hospitalized children. Emerg Infect Dis 9, 634–640. 10.3201/eid0906.030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freymouth F, Vabret A, Legrand L, Eterradossi N, Lafay-Delaire F, Brouard J, and Guillois B (2003). Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr Infect Dis J 22, 92–94. 10.1097/00006454-200301000-00024. [DOI] [PubMed] [Google Scholar]
- 4.Mazur NI, Terstappen J, Baral R, Bardaji A, Beutels P, Buchholz UJ, Cohen C, Crowe JE Jr., Cutland CL, Eckert L, et al. (2022). Respiratory syncytial virus prevention within reach: the vaccine and monoclonal antibody landscape. Lancet Infect Dis. 10.1016/S1473-3099(22)00291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crank MC, Ruckwardt TJ, Chen M, Morabito KM, Phung E, Costner PJ, Holman LA, Hickman SP, Berkowitz NM, Gordon IJ, et al. (2019). A proof of concept for structure-based vaccine design targeting RSV in humans. Science 365, 505–509. 10.1126/science.aav9033. [DOI] [PubMed] [Google Scholar]
- 6.Chng J, Wang T, Nian R, Lau A, Hoi KM, Ho SC, Gagnon P, Bi X, and Yang Y (2015). Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. MAbs 7, 403–412. 10.1080/19420862.2015.1008351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson KA, Rainho-Tomko JN, Williams ZP, Lanza L, Peredelchuk M, Kishko M, Pavot V, Alamares-Sapuay J, Adhikarla H, Gupta S, et al. (2020). A respiratory syncytial virus (RSV) F protein nanoparticle vaccine focuses antibody responses to a conserved neutralization domain. Sci Immunol 5. 10.1126/sciimmunol.aba6466. [DOI] [PubMed] [Google Scholar]
- 8.Williams K, Bastian AR, Feldman RA, Omoruyi E, de Paepe E, Hendriks J, van Zeeburg H, Godeaux O, Langedijk JPM, Schuitemaker H, et al. (2020). Phase 1 Safety and Immunogenicity Study of a Respiratory Syncytial Virus Vaccine With an Adenovirus 26 Vector Encoding Prefusion F (Ad26.RSV.preF) in Adults Aged >/=60 Years. J Infect Dis 222, 979–988. 10.1093/infdis/jiaa193. [DOI] [PubMed] [Google Scholar]
- 9.Scott LJ, and Lamb HM (1999). Palivizumab. Drugs 58, 305–311; discussion 312-303. [DOI] [PubMed] [Google Scholar]
- 10.Pantaleo G, Correia B, Fenwick C, Joo VS, and Perez L (2022). Antibodies to combat viral infections: development strategies and progress. Nat Rev Drug Discov 21, 676–696. 10.1038/s41573-022-00495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergeron HC, and Tripp RA (2022). Breakthrough therapy designation of nirsevimab for the prevention of lower respiratory tract illness caused by respiratory syncytial virus infections (RSV). Expert Opin Investig Drugs 31, 23–29. 10.1080/13543784.2022.2020248. [DOI] [PubMed] [Google Scholar]
- 12.Battles MB, and McLellan JS (2019). Respiratory syncytial virus entry and how to block it. Nat Rev Microbiol 17, 233–245. 10.1038/s41579-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham BS, Gilman MSA, and McLellan JS (2019). Structure-Based Vaccine Antigen Design. Annu Rev Med 70, 91–104. 10.1146/annurev-med-121217-094234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart-Jones GBE, Gorman J, Ou L, Zhang B, Joyce MG, Yang L, Cheng C, Chuang GY, Foulds KE, Kong WP, et al. (2021). Interprotomer disulfide-stabilized variants of the human metapneumovirus fusion glycoprotein induce high titer-neutralizing responses. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2106196118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh CL, Rush SA, Palomo C, Chou CW, Pickens W, Mas V, and McLellan JS (2022). Structure-based design of prefusion-stabilized human metapneumovirus fusion proteins. Nat Commun 13, 1299. 10.1038/s41467-022-28931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang J, Diaz D, and Mousa JJ (2020). Antibody recognition of the Pneumovirus fusion protein trimer interface. PLoS Pathog 16, e1008942. 10.1371/journal.ppat.1008942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee A, Huang J, Rush SA, Murray J, Gingerich AD, Royer F, Hsieh CL, Tripp RA, McLellan JS, and Mousa JJ (2022). Structural basis for ultrapotent antibody-mediated neutralization of human metapneumovirus. Proc Natl Acad Sci U S A 119, e2203326119. 10.1073/pnas.2203326119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rappazzo CG, Hsieh CL, Rush SA, Esterman ES, Delgado T, Geoghegan JC, Wec AZ, Sakharkar M, Mas V, McLellan JS, and Walker LM (2022). Potently neutralizing and protective anti-human metapneumovirus antibodies target diverse sites on the fusion glycoprotein. Immunity 55, 1710–1724 e1718. 10.1016/j.immuni.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Rush SA, Brar G, Hsieh CL, Chautard E, Rainho-Tomko JN, Slade CD, Bricault CA, Kume A, Kearns J, Groppo R, et al. (2022). Characterization of prefusion-F-specific antibodies elicited by natural infection with human metapneumovirus. Cell Rep 40, 111399. 10.1016/j.celrep.2022.111399. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, Diaz D, and Mousa JJ (2019). Antibody Epitopes of Pneumovirus Fusion Proteins. Front Immunol 10, 2778. 10.3389/fimmu.2019.02778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simoes EAF, Forleo-Neto E, Geba GP, Kamal M, Yang F, Cicirello H, Houghton MR, Rideman R, Zhao Q, Benvin SL, et al. (2021). Suptavumab for the Prevention of Medically Attended Respiratory Syncytial Virus Infection in Preterm Infants. Clin Infect Dis 73, e4400–e4408. 10.1093/cid/ciaa951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. (2013). Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501, 439–443. 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 23.Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE Jr., and Jardetzky TS (2017). Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol 2, 16272. 10.1038/nmicrobiol.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodwin E, Gilman MSA, Wrapp D, Chen M, Ngwuta JO, Moin SM, Bai P, Sivasubramanian A, Connor RI, Wright PF, et al. (2018). Infants Infected with Respiratory Syncytial Virus Generate Potent Neutralizing Antibodies that Lack Somatic Hypermutation. Immunity 48, 339–349 e335. 10.1016/j.immuni.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao X, Fridman A, Zhang L, Pristatsky P, Durr E, Minnier M, Tang A, Cox KS, Wen Z, Moore R, et al. (2022). Profiling of hMPV F-specific antibodies isolated from human memory B cells. Nat Commun 13, 2546. 10.1038/s41467-022-30205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mousa JJ, Kose N, Matta P, Gilchuk P, and Crowe JE Jr. (2017). A novel prefusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat Microbiol 2, 16271. 10.1038/nmicrobiol.2016.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Q, McLellan JS, Kallewaard NL, Ulbrandt ND, Palaszynski S, Zhang J, Moldt B, Khan A, Svabek C, McAuliffe JM, et al. (2017). A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci Transl Med 9. 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Q, McLellan JS, Kallewaard NL, Ulbrandt ND, Palaszynski S, Zhang J, Moldt B, Khan A, Svabek C, McAuliffe JM, et al. (2017). A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Science Translational Medicine 9. ARTN eaaj1928 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- 29.Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. (2013). Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501, 439-+. 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 30.Chan CH, Hadlock KG, Foung SKH, and Levy S (2001). V(H)1-69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood 97, 1023–1026. DOI 10.1182/blood.V97.4.1023. [DOI] [PubMed] [Google Scholar]
- 31.Lang SS, Xie J, Zhu XY, Wu NC, Lerner RA, and Wilson IA (2017). Antibody 27F3 Broadly Targets Influenza A Group 1 and 2 Hemagglutinins through a Further Variation in V(H)1-69 Antibody Orientation on the HA. Cell Reports 20, 2935–2943. 10.1016/j.celrep.2017.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzarum N, Giang E, Kong L, He LL, Prentoe J, Augestad E, Hua YZ, Castillo S, Lauer GM, Bukh J, et al. (2019). Genetic and structural insights into broad neutralization of hepatitis C virus by human V(H)1-69 antibodies. Sci Adv 5. ARTN eaav1882. 10.1126/sciadv.aav1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen X, Krause JC, Leser GP, Cox RG, Lamb RA, Williams JV, Crowe JE Jr., and Jardetzky TS (2012). Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nature structural & molecular biology 19, 461–463. 10.1038/nsmb.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammitt LL, Dagan R, Yuan Y, Baca Cots M, Bosheva M, Madhi SA, Muller WJ, Zar HJ, Brooks D, Grenham A, et al. (2022). Nirsevimab for Prevention of RSV in Healthy Late-Preterm and Term Infants. N Engl J Med 386, 837–846. 10.1056/NEJMoa2110275. [DOI] [PubMed] [Google Scholar]
- 35.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, et al. (2013). Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342, 592–598. 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, and Crowe JE Jr. (2012). Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol 86, 2665–2675. 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. S0907444904019158 [pii] 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. (2008). Automated structure solution with the PHENIX suite. Methods Mol Biol 426, 419–435. 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 42.Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, and Urzhumtsev A (2018). New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840. 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klaholz BP (2019). Deriving and refining atomic models in crystallography and cryo-EM: the latest Phenix tools to facilitate structure analysis. Acta Crystallogr D Struct Biol 75, 878–881. 10.1107/S2059798319013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplemental Information. Protein structures and EM maps have been deposited in the Protein Data Bank with the following deposition codes: hMPVF + HPMV-4I3 −8EAY; hMPV F protein monomer + Fab RSV-199 −8E2U; hMPV protein dimer + Fab RSV-199 −8EBP; RSV F protein trimer + Fab RSV-199 −8DZW.
This work dis not use or generate any new code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.