SUMMARY:
Type VIIb secretion systems (T7SSb) in Gram-positive bacteria facilitate physiology, interbacterial competition, and/or virulence via EssC ATPase-driven secretion of small ɑ-helical proteins and toxins. Recently, we characterized T7SSb in group B Streptococcus (GBS), a leading cause of infection in newborns and immunocompromised adults. GBS T7SS comprises four subtypes based on variation in the C-terminus of EssC and the repertoire of downstream effectors; however, the intra-species diversity of GBS T7SS and impact on GBS-host interactions remains unknown. Bioinformatic analysis indicates that GBS T7SS loci encode subtype-specific putative effectors, which have low inter-species and inter-subtype homology but contain similar domains/motifs and therefore may serve similar functions. We further identify orphaned GBS WXG100 proteins. Functionally, we show that GBS T7SS subtype I and III strains secrete EsxA in vitro and that in subtype I strain CJB111, esxA1 appears to be differentially transcribed from the T7SS operon. Further, we observe subtype-specific effects of GBS T7SS on host colonization, as CJB111 subtype I but not CNCTC 10/84 subtype III T7SS promotes GBS vaginal colonization. Finally, we observe that T7SS subtypes I and II are the predominant subtypes in clinical GBS isolates. This study highlights the potential impact of T7SS heterogeneity on host-GBS interactions.
Keywords: Streptococcus agalactiae, group B Streptococcus, GBS, Type VII Secretion System, genetic diversity, colonization, operon, effectors
Graphical Abstract
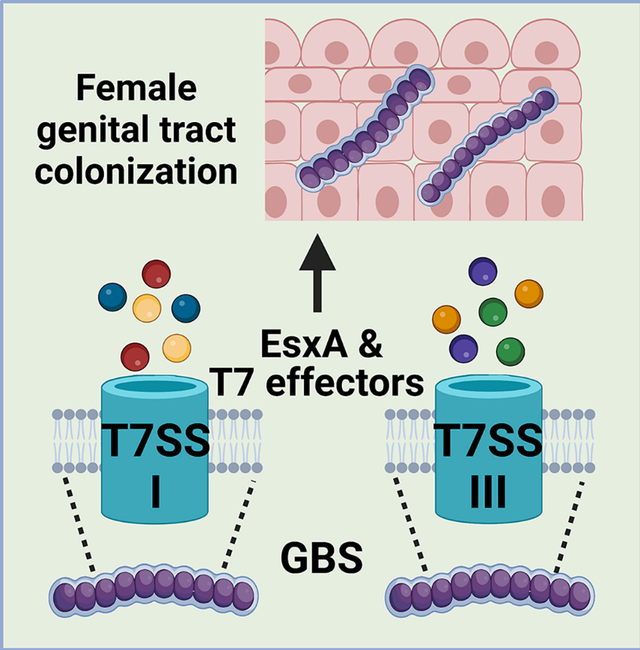
The group B streptococcal type VII secretion system (T7SS) comprises four subtypes, which encode similar machinery but unique effectors, including putative LXG toxins. We show that subtypes I and II are prevalent amongst GBS clinical isolates and hypothesize that T7SS subtype-specific effectors may impact GBS female genital tract colonization.
1 |. INTRODUCTION
Type VII secretion systems (T7SS) are encoded by both Actinobacteria and Gram-positive organisms (designated T7SSa and T7SSb, respectively). T7SSb in Gram-positive bacteria contribute to interbacterial competition as well as virulence by damaging host cells and modulating immune responses (Unnikrishnan et al., 2017, Tran et al., 2021, Bowman & Palmer, 2021). While T7SSb machinery varies in sequence and genomic arrangement across species, common components are cytoplasmic protein EsaB and membrane proteins EsaA, EssA, EssB, and EssC, an ATPase that powers secretion of substrates across the bacterial membrane (Bowman & Palmer, 2021). Another distinguished feature of T7SSb is the presence of canonical T7SS substrate WXG100 protein EsxA (named for its 100 amino acid sequence and central Trp-X-Gly [WXG] motif), other small ɑ-helical proteins, and T7SS-associated toxins (Warne et al., 2016, Bowran & Palmer, 2021). These toxins, which sometimes contain an N-terminal LXG motif, encode unique C-terminal toxin domains, and therefore have biochemically diverse functions. Some T7SS toxins function in interbacterial competition and are frequently co-transcribed with chaperones that facilitate their secretion and immunity factors that prevent self-toxicity. These functions have been described in Staphylococcus aureus, Streptococcus intermedius, Bacillus subtilis, Enterococcus faecalis, and recently Streptococcus gallolyticus (Cao et al., 2016, Whitney et al., 2017, Klein et al., 2018, Ulhuq et al., 2020, Kobayashi, 2021, Chatterjee et al., 2021, Teh et al., 2022, Klein et al., 2022). Interestingly, in some cases, these toxins also promote virulence and modulate immune responses within the host (Dai et al., 2017, Ohr et al., 2017, Ulhuq et al., 2020).
Despite these common traits, the T7SSb encodes largely unique effectors across species. Extensive intra-species T7SSb diversity has also been characterized within S. aureus, Listeria monocytogenes, and Staphylococcus lugdunensis based on EssC C-terminus variants and downstream effectors (Warne et al., 2016, Lebeurre et al., 2019, Bowran & Palmer, 2021), further dividing T7SSb into species-specific “subtypes”. These EssC variants are thought to determine specificity for substrate recognition and secretion and define species-specific subtypes. In S. aureus, although all four EssC variants are capable of secreting common T7SS substrate EsxA, the EssC2, EssC3 and EssC4 variants are incapable of secreting non-cognate substrate EsxC (which is encoded for downstream of essC1 only) (Jager et al., 2018). We recently showed intra-species diversity in T7SS downstream effectors in group B Streptococcus (GBS) (Spencer et al., 2021b), and hypothesized that variant-specific T7SSb effectors may promote differing bacterial interactions with the host or other microbes (Spencer & Doran, 2022).
As an opportunistic pathogen, GBS (or Streptococcus agalactiae) asymptomatically resides in the gastrointestinal and/or female genital tract of 25–30% of healthy adults (Wilkinson, 1978, Regan et al., 1991) but can cause severe infections in some individuals, such as pregnant people and newborns, the elderly, and patients living with cancer or diabetes (Nandyal, 2008, Pimentel et al., 2016, Russell et al., 2017, Patras & Nizet, 2018, van Kassel et al., 2019, Navarro-Torne et al., 2021). Within the female genital tract, GBS coexists and/or competes with the vaginal microbiota and, therefore, has evolved mechanisms to survive these encounters while also avoiding immune clearance (Okumura & Nizet, 2014, Vrbanac et al., 2018, Coleman et al., 2021). We showed previously that GBS T7SS subtype I plays a role in virulence, cytotoxicity, and pore formation via the secreted effector EsxA (Spencer et al., 2021b), and that four different GBS T7SS subtypes can be delineated based on differing number of copies of esxA, a unique EssC ATPase C-terminus, and a unique repertoire of downstream genes/putative effectors. However, the full heterogeneity of the GBS T7SS operon and the functions of the putative T7SS effectors in GBS-host and GBS-microbe interactions has not yet been investigated.
We hypothesized that GBS T7SS subtypes encode unique downstream effectors that differ from previously studied T7SS substrates in other species and may modulate GBS fitness within the host. Using bioinformatic analyses of the T7SS locus across the four subtypes, we found that the GBS T7SS encodes putative effector proteins with high genetic variability but with similar domains and motifs, suggesting some conserved functions. Functionally, we confirmed that GBS T7SS subtype I and III isolates secrete EsxA in vitro and we observed variable impacts of essC deficiency across T7SS subtypes in vivo using a murine model of female genital tract colonization. This study highlights T7SSb diversity across GBS strains and indicates that the T7SS subtype-specific effector repertoire may differentially modulate host phenotypes.
2 |. RESULTS
2.1 |. Bioinformatic analysis of GBS T7SS subtypes I-IV
In our previous analysis of GBS whole-genome sequences, we identified four GBS T7SS subtypes based on variation in the EssC C-terminus, T7SS substrate EsxA, and several genes encoded downstream of essC (Spencer et al., 2021b). However, in other species, T7SS loci can encode 10–20 or more genes following essC (Bowran & Palmer, 2021, Lebeurre et al., 2019, Warne et al., 2016). To investigate the full range of GBS T7SS diversity, we extended bioinformatic analysis to the entire T7SS region encoded by 80 T7SSb+ GBS genomes deposited in GenBank. Using CJB111, 2603V/R, CNCTC 10/84, and COH1 as example strains for GBS T7SS subtypes I, II, III, and IV respectively, we analyzed each gene within the putative T7SS locus for conserved domains [via InterPro (Blum et al., 2021) and Conserved Domain Architecture Retrieval Tool (CDART) analysis (Geer et al., 2002)] and for predicted protein topology [Protter analysis (Omasits et al., 2014)] (Supp. Table 1). As discussed in detail below, we found high conservation of GBS T7SS genomic location and machinery between subtypes, but variability in the copy number of locus-associated and orphaned esxA genes and heterogeneity in the EssC ATPase and associated putative effector repertoire (Fig. 1). We further assessed homology of the T7SS locus to eight other Gram-positive T7SSb-containing species (Supp. Fig. 1 and Supp. Table 2).
Fig. 1. Intra-species diversity of the GBS T7SS.
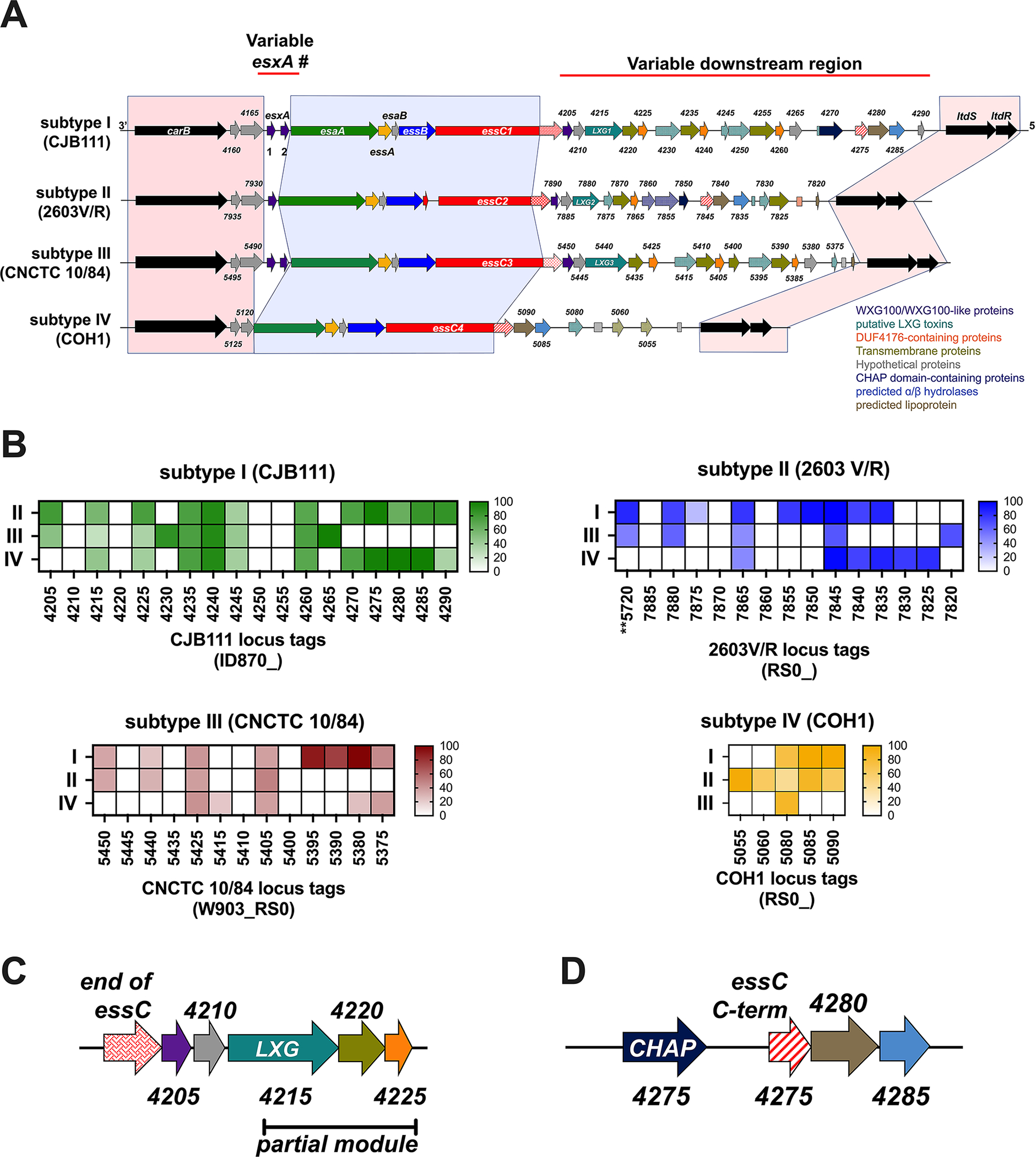
A) Example GBS T7SS loci by subtype (roughly to scale). GBS T7SS loci are flanked by carbamoyl phosphate synthesis genes and two-component system genes ltdS/ltdR (light red shading). GBS T7SS encode conserved machinery genes (light blue shading) but subtypes vary in copies of esxA (purple) and in putative downstream effectors, including WXG100-like proteins (purple), putative LXG toxins (teal), transmembrane proteins (olive green), hypothetical proteins (gray), DUF4176 proteins (orange), CHAP domain-containing proteins (navy blue), ɑβ hydrolases (light blue), and lipoproteins (light brown). Arrows with patterns indicate fragmented genes. Incompletely filled arrows indicate genes encoding prematurely truncated products (e.g., CNCTC 10/84 esxA2). B) Heatmaps indicating homology of T7SS-associated genes downstream of essC across T7SS subtype against whole genomes of other representative GBS subtypes via BLAST analysis. Color intensity based on Geneious alignment grade, which considers query coverage and percent identity. Many putative downstream GBS T7SS effectors have little homology to genes encoded by other GBS representative subtype strains or are entirely T7SS subtype specific. C) Commonly occurring “LXG modules” are encoded for downstream of essC in subtypes I-III, which include putative chaperones (WXG100-like proteins, purple), a putative LXG toxin (teal), a transmembrane protein (olive green), and a DUF4176 protein (orange). Partial LXG modules were also identified, which contain only a fragmented LXG gene (patterned teal) and the subsequent transmembrane and DUF4176 protein encoding genes. D) Another commonly found module within T7SS loci consists of a gene fragment encoding the C-terminal end of the COH1 essC (red/white stripe), followed by ɑβ hydrolase (light blue) and lipoprotein (light brown) encoding genes. This module is often preceded by an amidase encoding gene (navy blue).
WXG100 proteins and putative GBS T7SS machinery encoding genes
Similar to several other Gram-positive bacteria, most GBS T7SS loci contain putative core component genes (esxA, esaA, essA, esaB, essB, and essC) followed by putative effector and chaperone-encoding genes (Warne et al., 2016, Cao et al., 2016, Lai et al., 2017, Taylor et al., 2021, Chatterjee et al., 2021). In GBS subtypes I-III, the T7SS locus begins with gene(s) encoding the canonical substrate, WXG100 protein EsxA, which has been hypothesized to also function as a core machinery component of the T7SSb (Sundaramoorthy et al., 2008, Kneuper et al., 2014). GBS subtype I and III isolates encode two copies of esxA (Spencer et al., 2021b), while subtype II encodes one copy, and subtype IV strains do not encode a locus-associated esxA (Fig. 1). The EsxA sequence is highly conserved across GBS T7SS subtypes (≥94% identity) and both EsxA1 and EsxA2 contain the canonical central WXG motif (Spencer et al., 2021b).
Downstream of esxA (or an WxcM-domain protein encoding gene in subtype IV), putative GBS T7SS machinery genes include esaA, essA, esaB, essB, and ATPase-encoding essC. These first four core components are highly conserved across GBS T7SS subtypes I-IV, with 97–100% identity at the protein level (Supp. Table 2) and appear in the same genomic arrangement as S. aureus and L. monocytogenes T7SS loci (Kneuper et al., 2014, Bowran & Palmer, 2021). Despite this similarity in arrangement, T7SS core machinery sequences exhibit extremely low homology across a panel of Gram-positive species. For example, GBS EsaA is only ×15–20% identical to EsaA homologs in Bacillus, Enterococcus, Staphylococcus, and Listeria spp., and only 30–40% identical to streptococcal EsaA homologs in S. gallolyticus, S. intermedius, and S. suis. Similarly, GBS EssA, EsaB, and EssB proteins exhibited only 11–34% identity to homologs in other genera and 21–54% identity to homologs in other streptococci. The EssC ATPase exhibited the highest inter-species identity, but the lowest intra-species protein sequence identity of the T7SS core proteins. For example, GBS EssC variants were 34–48% identical to non-streptococcal EssC proteins, and 56–80% identical to other streptococcal EssC proteins (Supp. Table 2). Within GBS, EssC variants shared 89–98% identity, with sequence variation primarily restricted to the EssC C-terminal 225 amino acids (Spencer et al., 2021b). As these variants are associated with unique effector repertoires, it is likely that a given GBS EssC may only export substrates encoded by their cognate subtype, as has been demonstrated in S. aureus (Jager et al., 2018).
Putative T7SS effectors, chaperones, and immunity factors:
As many coding sequences within the hypervariable T7SS effector region are annotated as hypothetical, we compared these proteins across GBS subtypes using InterPRO, NCBI CDART, and Protter transmembrane analysis (Supp. Table 1). Subtypes I-III encode SACOL2603 T7SS effector proteins (DUF3130-containing or TIGR04197 family proteins), putative LXG toxins, DUF4176-containing proteins, and transmembrane proteins. In addition, amidase domain-containing proteins, C-terminal fragments of EssC, putative toxin fragments, and lipoproteins are also found in some, but not all, GBS T7SS loci (Fig. 1, Supp. Table 1). Although similar domains and motifs are detected in different GBS T7SS loci, many proteins differ significantly in sequence homology across subtypes I-III and some lack any identifiable homolog in GBS genomes representative of other T7SS subtypes (Fig. 1B), suggesting they are distinct proteins with possibly biochemically diverse activities. Interestingly, subtype IV strains do not encode locus associated esxA or many common T7SS effectors (SACOL2603, DUF4176, etc.) and encode the shortest GBS T7SS locus, with just 5 hypothetical genes downstream of essC that share the highest homology with the subtype II locus (Fig. 1B).
Within GBS subtypes I-III variable regions, a commonly occurring module consists of two adjacent WXG100-like proteins (the first containing DUF3130 and the second predicted to contain coil domains), a putative LXG toxin, and one or two hypothetical protein(s), followed by a DUF4176-domain containing protein – hereafter termed an “LXG module” (Fig. 1C). A similar gene cluster was recently described in S. intermedius where the DUF3130 gene and the adjacent gene encode accessory proteins/chaperones required for secretion of the LXG toxins TelC and TelD (Klein et al., 2022). Consequently, these chaperones were named LXG-associated α-helical proteins (Lap). Genes encoding DUF4176 proteins were also observed near this LXG module in S. intermedius, although a role for these proteins in T7SS has yet to be shown in any species. A second commonly occurring module in GBS loci consists of a fragment of the subtype IV essC, followed by lipoprotein and ɑβ hydrolase encoding genes (Fig. 1D).
The first two GBS WXG100-like proteins encoded within the LXG module, hereafter termed putative Lap1 and Lap2, are small and ɑ-helical but lack the canonical central WXG motif; however, Lap1 does display a central FXG motif and a FxxxD/E motif in many GBS strains (Supp. Fig. 2). GBS Lap1 proteins exhibit some homology across subtypes I - III (33–75% identity; Supp. Fig. 2A) but generally low homology to their Gram-positive counterparts, with ×40% identity to S. gallolyticus, S. intermedius, and S. suis (Supp. Fig. 1). While neither CDART nor InterPro analyses indicate any putative function for Lap2, these GBS proteins are predicted to have coil domains and to be ɑ-helical in nature (Fig. 2A, Supp. Table 1), similar to those described in S. intermedius (Klein et al., 2022). GBS Lap2 proteins are not well conserved across subtypes I-III, with ≤41% identity between any two (Supp. Fig. 2D–F). This suggests that specific Lap/LXG protein pairs may also be required for GBS LXG protein secretion. Although it is currently unknown whether GBS Laps associate with the putative downstream LXG toxin as chaperones, using AlphaFold modeling and predictions, putative Lap1 and Lap2 across GBS T7SS subtypes seem likely to interact with their cognate LXG protein (Fig. 2A) as their counterparts do in S. intermedius.
Fig. 2. Subtype-specific LXG toxins are encoded by GBS T7SS.
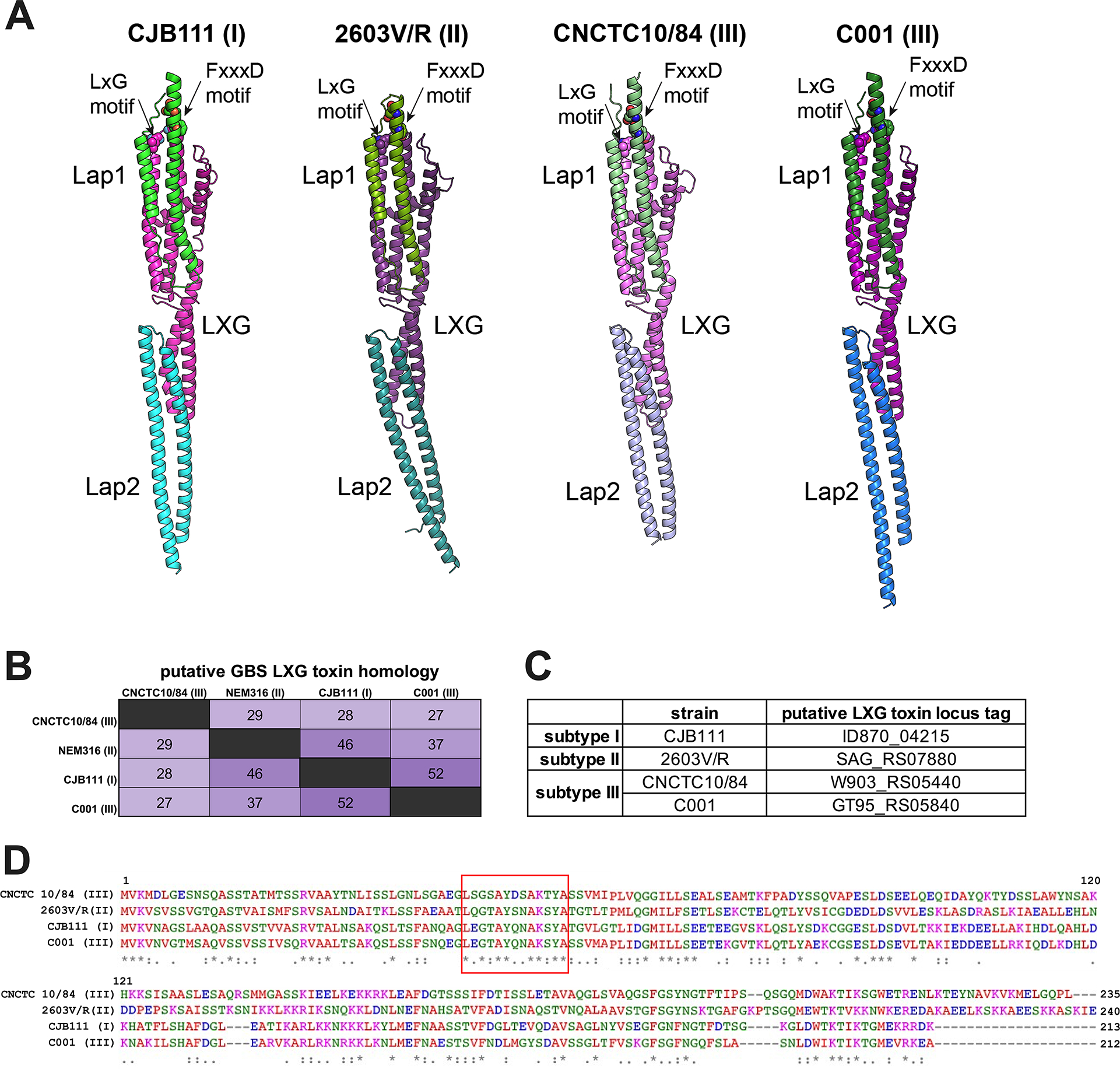
A) Predicted models of complexes between specific Lap1 (green) and Lap2 (blue/purple) pairs with the LXG domain (pink) of their cognate LXG protein. The putative effector domains of LXG proteins and the predicted disordered regions of Lap proteins are omitted for clarity. The conserved residues of LxG and FxxxD motifs are shown as spheres. The modelling confidence metrics are presented in Supp. Fig. 3. B–D) Percent identity matrix of full-length LXG protein sequences and Clustal Omega alignment of the LXG domain from CJB111 (subtype I), 2603V/R (subtype II), CNCTC 10/84 (subtype III), and C001 (subtype III). In the above matrix, the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity. The longer motif found in all GBS LXG proteins is denoted by a red box in D.
Similar to the Lap-encoding genes, GBS putative LXG toxins exhibit minimal identity across T7SS subtypes, with homology concentrating towards the N-terminal LXG domain and sequences diverging in the C-terminal domains (Fig. 2B–D) indicating potential differing biochemical properties. We identified four putative full-length LXG toxins encoded within GBS T7SS loci: one in subtype I (CJB111, ID870_4215), one in subtype II (2603V/R, SAG_RS07880), and two within subtype III strains (CNCTC 10/84, W903_RS05440 and C001, GT95_RS05840) (Fig. 2B–D; Supp. Fig. 4A). Upon intra-subtype comparisons, subtype I and II LXG proteins were highly conserved across strains. In 44 of the 46 subtype I strains, the LXG protein exhibited 99.8–100% identity, with just one amino acid substitution (A157E) in the N-terminal LXG domain in some strains. Similarly, the subtype II LXG protein demonstrated 99–100% identity across isolates, with just two of the 15 strains encoding a nonsense mutation at amino acid 218 (G218*) resulting in premature truncation after the LXG domain. As subtype III contains only five completely sequenced strains, further assessment of the two full-length LXG proteins was not possible.
GBS T7SS LXG toxins are unique from those encoded by other Gram-positive organisms and any limited homology is restricted to the N-terminal LXG domain within the first ×200 amino acids of the protein. An exception to this was ×40% homology observed between the CJB111 LXG C-terminus (ID870_4215) and that of S. intermedius TelD (Supp. Fig. 1). As CDART/InterPro failed to identify domains within the C-terminus of these proteins (Supp. Table 1), the functions of GBS LXG proteins remain unknown and must be determined experimentally. Finally, we observed that the four full-length GBS LXG toxins commonly encode a longer LXG motif: LxGxAYxxAKxYA (Fig. 2D). This full motif was conserved across other streptococcal LXG proteins from S. gallolyticus (TX20005 JGX27_RS02965) and S. suis (WUSS351 E8M06_RS09920). Other Gram-positive LXG proteins contain a similar but slightly abbreviated/modified version of this motif (LxGxAYxxA[K/R]), including those encoded by S. aureus (TspA), S. intermedius (TelC and TelD), S. lugdunensis (HKU09–01 SLGD_RS02660), S. suis (WUSS351 E8M06_RS09940 and RS09970), and S. gallolyticus (JGX27_RS04665, RS08265, RS03950, and RS11360). This longer and more specific conserved motif may facilitate easier identification of streptococcal LXG toxins in the future.
In several species, T7SS toxins are produced in tandem with an immunity factor to prevent self-toxicity. Downstream of the putative LXG toxins, GBS subtypes I and III encode one hypothetical gene followed by a DUF4176 gene and GBS subtype II encodes two hypothetical genes (a LXG protein fragment [7875] and a predicted transmembrane protein encoding gene [7870]) followed by a DUF4176 gene (Fig. 1). We hypothesize that one of these downstream genes may function as an immunity factor for each subtype’s unique LXG protein. In support of this, these hypothetical proteins are highly subtype specific and have low homology to their counterparts across GBS T7SS subtypes (Supp. Fig. 4B) or to any proteins found in other Gram-positive T7SSb (CJB111 ID870_4220, Supp. Fig. 1). Further, no functional domains were identified in these proteins by cDART or InterPro but, across subtypes I-III, all were predicted to contain two to four transmembrane domains (Supp. Table 1, see olive green arrows following teal LXG genes in Fig. 1).
DUF4176 proteins are also commonly encoded in the vicinity of T7SSb LXG proteins but their roles in T7SSb are unclear. GBS T7SS subtype I, II, and III strains encode two to three loci associated DUF4176 genes, and most encode an orphaned DUF4176 elsewhere in the genome (occasionally fragmented or annotated as pseudogenes) (Supp. Fig. 4C). Of the genes downstream of GBS essC, DUF4176 exhibited the most overall homology across subtypes (Fig. 1B) as well as to proteins in other T7SSb+ Gram-positive bacteria (30–60% identity across several species; Supp. Fig. 1). Further, all GBS DUF4176 proteins encode a central “FXG” motif (Supp. Fig. 4D). Interestingly, the orphaned DUF4176 proteins encoded by subtype II, III, and IV strains were almost identical (94–100% identity; subtype I’s orphaned DUF4176 is a pseudogene). The locus-associated DUF4176 protein encoded by ID870_4240 in subtype I strain CJB111 also exhibited more homology to the orphan DUF4176 proteins (81–85% identity) compared to its locus-associated DUF4176 proteins (35–48% identity) (Supp. Fig. 4E). Using AlphaFold, we sought to predict if these putative transmembrane proteins or DUF4176 proteins might interact with their cognate GBS LXG proteins. In CJB111, the transmembrane protein encoded for downstream of LXG (by ID870_4220) yielded the highest confidence score (0.8) indicating there may be a stable interaction between this protein pair, but this would need to be confirmed experimentally.
Some T7SS loci encode additional similarly arranged partial LXG modules further downstream, but these additional modules typically encode a fragmented LXG protein (usually retaining only the central linker region or the C-terminal toxic domain (Zhang et al., 2012, Chatterjee et al., 2021)). We observed fragmented LXG proteins in all subtypes, including subtype I strain CJB111 (ID870_4230, 4245, and 4250) and in subtype III CNCTC 10/84 (W903_RS05415, RS05395), the majority of which were similarly followed by genes encoding for hypothetical transmembrane and DUF4176 proteins (Fig. 1A,C). Following these partial LXG modules, the furthest downstream regions of T7SS loci are more variable but sometimes contain blocks of T7SS genes that can be found across subtypes (Fig. 1), indicating that this region of the putative T7SS locus may be prone to recombination, as recently shown in S. aureus (Garrett et al., 2022). This region includes genes encoding hypothetical and transmembrane proteins, CHAP domain containing proteins, FtsK domain containing proteins (C-terminal essC fragments), lipoproteins, and ɑβ hydrolases (Supp. Table 1, Fig. 1A,D).
Genomic location and genes flanking GBS T7SSb
In all GBS isolates examined, T7SS genes are located between carbamoyl phosphate synthase genes (carB; CJB111 ID870_4155) and the LtdRS two component system (Deng et al., 2018, Faralla et al., 2014) (Fig. 1). Two highly conserved genes downstream of carB encode for a 107 amino acid hypothetical protein with no predicted function or domains (CJB111 ID870_4160) and a WxcM domain-containing protein predicted to contain ɑβ hydrolase and/or lipase-like domains (CJB111 ID870_4165). While this WxcM protein is prematurely truncated in subtype IV strains, it maintains >97% identity to the subtype I - III homologs within the first 60% of the amino acid sequence. ID870_4160 homologs were not found within other T7SSb containing Gram-positive species, and the GBS WxcM domain containing protein exhibited minimal identity to homologs in E. faecalis, S. intermedius, and Streptococcus suis, indicating that these genes are fairly specific to GBS.
To confirm the boundaries of the T7SS operon in subtype I strain CJB111, we assessed which genes are co-transcribed within this genomic region by performing RT-PCR using primers spanning each T7SS gene junction. Primers were confirmed using gDNA and non-specific amplification was assessed using no-RT controls. We observed bands for almost every cDNA RT-PCR reaction, indicating that most genes are capable of being co-transcribed; however, esxA1 consistently appears to be transcribed separately from the rest of the T7SS operon, indicated by a lack of amplicon from cDNA using primers spanning the esxA1 - esxA2 junction (Supp. Fig. 5). Bands were observed between esxA2 and machinery genes of the T7SS indicating they are transcribed together, as has been shown in S. aureus and S. gallolyticus (Kneuper et al., 2014, Taylor et al., 2021). Further, we observed bands for all gene junctions until the junction spanning the ltdS operon for reactions including cDNA as template (but no bands were detected in the no-RT controls), indicating that the genes within the T7SS locus from esxA2 through the gene upstream of ltdS are capable of being co-transcribed. Although, as RT-PCR can detect low levels of transcript, more sensitive methods should be used in the future to assess potential differential regulation within the T7SS locus.
2.2 |. Intra-subtype diversity of the GBS T7SS
While the above bioinformatic analysis assessed diversity of the GBS T7SS between subtypes and between Gram-positive species, we also observed extensive intra-subtype diversity. Within subtype I strains, the region from carB through the 5 genes downstream of essC (CJB111 ID870_4225) was highly conserved (×99% nucleotide identity). However, heterogeneity in the form of insertion or loss of gene blocks occurred further downstream (Fig. 3A). For example, 12 out of 46 subtype I strains (representative strain CJB111) encode 18 genes downstream of essC. In other subtype I strains (12/46, representative strain A909), the T7SS locus is truncated to just 11 genes downstream of essC (ID870_4245 through ID870_4285 have been lost). Additional subtype I T7SS loci (10/46 strains) are almost identical to A909 but have a 73 bp deletion after the first DUF4176 encoding gene (which deletes a putative terminator) and a 69 bp deletion in the intergenic region before A909 gene SAK_RS05570. Finally, compared to the A909 T7SS arrangement, a few strains (representative strain Sag153) have undergone further reductive evolution, losing additional genes in the T7SS locus.
Fig. 3. Intra-subtype diversity of the GBS T7SS.
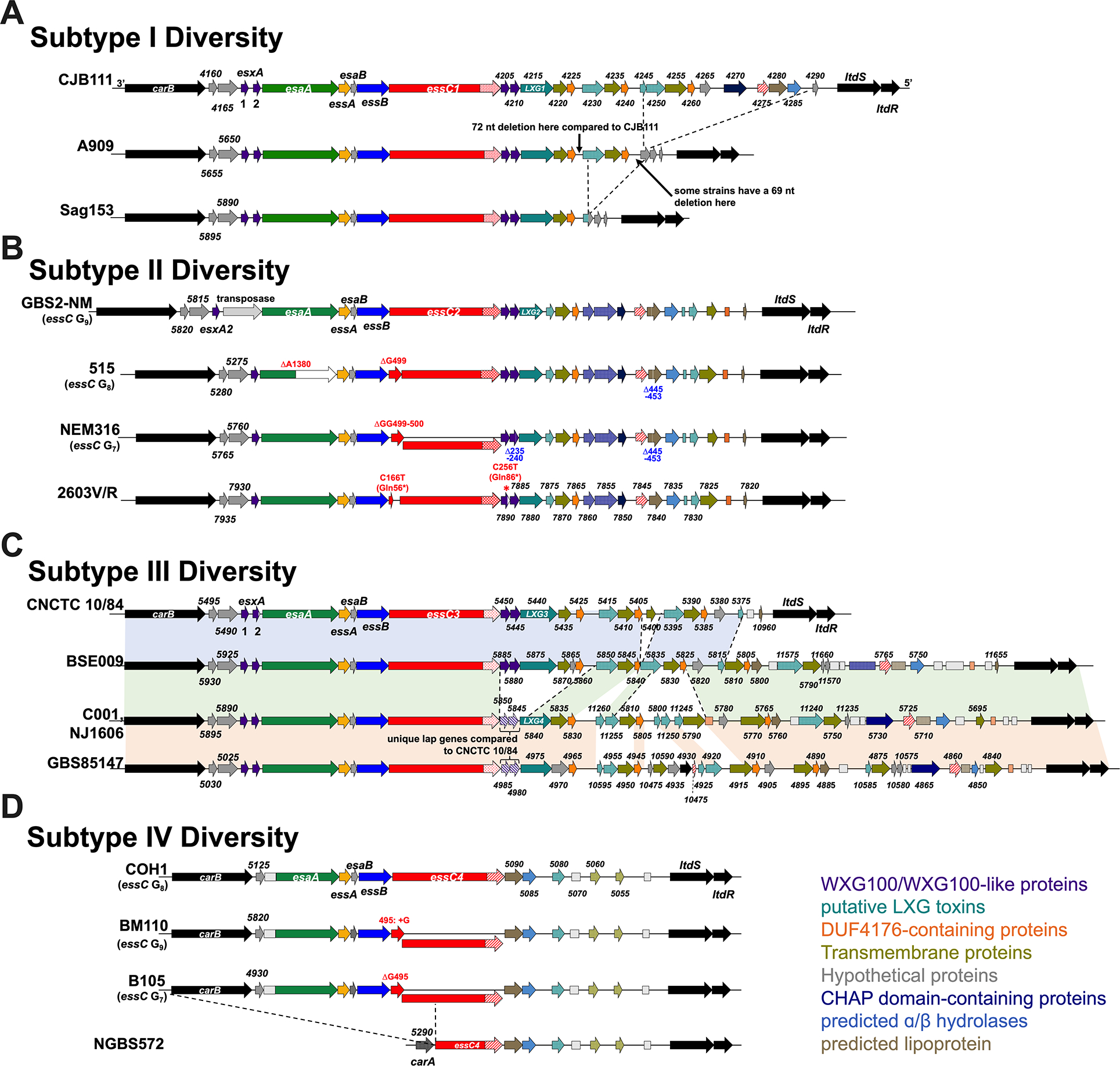
GBS T7SS loci representing genomic diversity within A) subtype I, B) subtype II, C) subtype III, and D) subtype IV. GBS T7SS encode putative downstream effectors including WXG100-like proteins (purple), putative LXG toxins (teal), transmembrane proteins (olive green), hypothetical proteins (gray), DUF4176 proteins (orange), CHAP domain-containing proteins (navy blue), ɑβ hydrolases (light blue), and lipoproteins (light brown). Arrows with patterns indicate fragmented genes. Incompletely filled arrows indicate genes encoding prematurely truncated products (e.g., CNCTC 10/84 esxA2). Genes upstream of T7SS loci are labeled with each strains’ unique “RS0_X” locus tag numbers for reference (see Supp. Table 3 for strains’ GenBank accession numbers). Subtype I and III strains differ is presence/absence of downstream genes (A, C). Subtype II and IV strains differ in mutations within essC (B, D) and the length of the homopolymeric tract within essC is annotated as Gn.
While most GBS T7SS subtype II strains encode the same set of genes downstream of essC, diversity within subtype II is most commonly based on nonsense and frameshift mutations in essC and esaA (Fig. 3B). Of the 15 closed GBS genomes that encode GBS T7SS subtype II, 13 encode a putatively truncated/multiple-CDS EssC. The majority of these are due to a slip-strand mutation within a homopolymeric Gn tract in essC, resulting in the loss of either one or two nucleotides, and frameshift and truncation of EssC to 166 or 173 amino acids, respectively. In some subtype II strains, an additional nonsense mutation occurs before this homopolymeric tract resulting in an EssC truncation to 56 amino acids (C166T Gln56*). However, NCBI ORF Finder predicts that the remainder of this EssC may still be encoded in these subtype II strains as a second ORF, resulting in a 1291 amino acid protein (of the usual 1469 amino acid protein), which may still allow T7SSb activity. A second common area for mutation within subtype II strains is esaA (in 5 of 15 subtype II strains, including strain 515), also potentially due to slippage within a homopolymeric An tract, resulting in deletion of one nucleotide, frameshift, and truncation of EsaA at 465 amino acids (full length EsaA is 1005 amino acids). Similar to EssC truncation, NCBI ORF Finder predicts that the remainder of EsaA may be encoded as a second ORF. It remains to be seen how these mutations impact the T7SS. Despite the presence of these essC and esaA homopolymeric tracts across GBS T7SS subtypes, we have not observed these putative slippage-induced frameshift mutations within T7SS machinery genes in our GBS subtype I and III strains, and only rarely observe these essC mutations in our subtype IV strains (only 2 of 14 isolates).
Subtype III is the least prevalent subtype of GBS T7SS and encompasses immense diversity, with four unique versions of the downstream T7SS region existing in just five isolates (Fig. 3C). Similar to subtype I, diversity of subtype III is due to the presence/absence of downstream genes between essC and ltdS, with some subtype III strains encoding different LXG modules (e.g., strains CNCTC 10/84 and C001 encode different full length LXG proteins). Because these strains are unique in their downstream T7SS gene arrangement, each strain’s downstream repertoire has been independently analyzed in Supp. Table 1. Subtype IV strains are also rarer, which impairs intra-subtype comparisons; however, two of 14 subtype IV strains encode similar slip-strand mutations in essC as seen in subtype II strains (Fig. 3D).
2.3 |. GBS T7SS orphaned WXG100 modules
In addition to locus associated T7SS genes, many GBS strains encode for “orphaned” WXG100 proteins elsewhere in the genome. Comparative genomic analysis of example strains from each T7SS subtype revealed two orphaned WXG100 groups (here termed Orphaned Modules 1 and 2) based on genomic location/neighboring genes and WXG100 protein sequence alignment (Fig. 4A and Supp. Fig. 6). Module 1 WXG100 orphan proteins are encoded in 64 of the 80 T7SS+ GenBank isolate cohort (Supp. Table 4), often upstream of a MutR family transcriptional regulator (Fig. 4A) and are well conserved, exhibiting 97–100% identity across strains (Supp. Fig. 6A–B). Module I WXG100 genes are often encoded either in proximity to a RelE/ParE family toxin protein and plasmid recombinase genes in some strains (as in NEM316) or downstream of a DEAD/DEAH box helicase gene in others (CJB111 and 515). Furthermore, Module 1 WXG100 orphan genes are commonly followed by a cluster of conserved hypothetical genes. In a subset of subtype IV strains, the orphan WXG100 gene is encoded downstream of rpsI (sometimes with restriction-modification and abortive infection system genes immediately preceding the WXG100 gene) and this “Module 1b” contains a ×15kb insertion (encoding for tyrosine recombinases, sigma factors, and tetM) following the second hypothetical gene of the cluster downstream of the WXG100 gene.
Fig. 4. GBS T7SS orphaned modules.
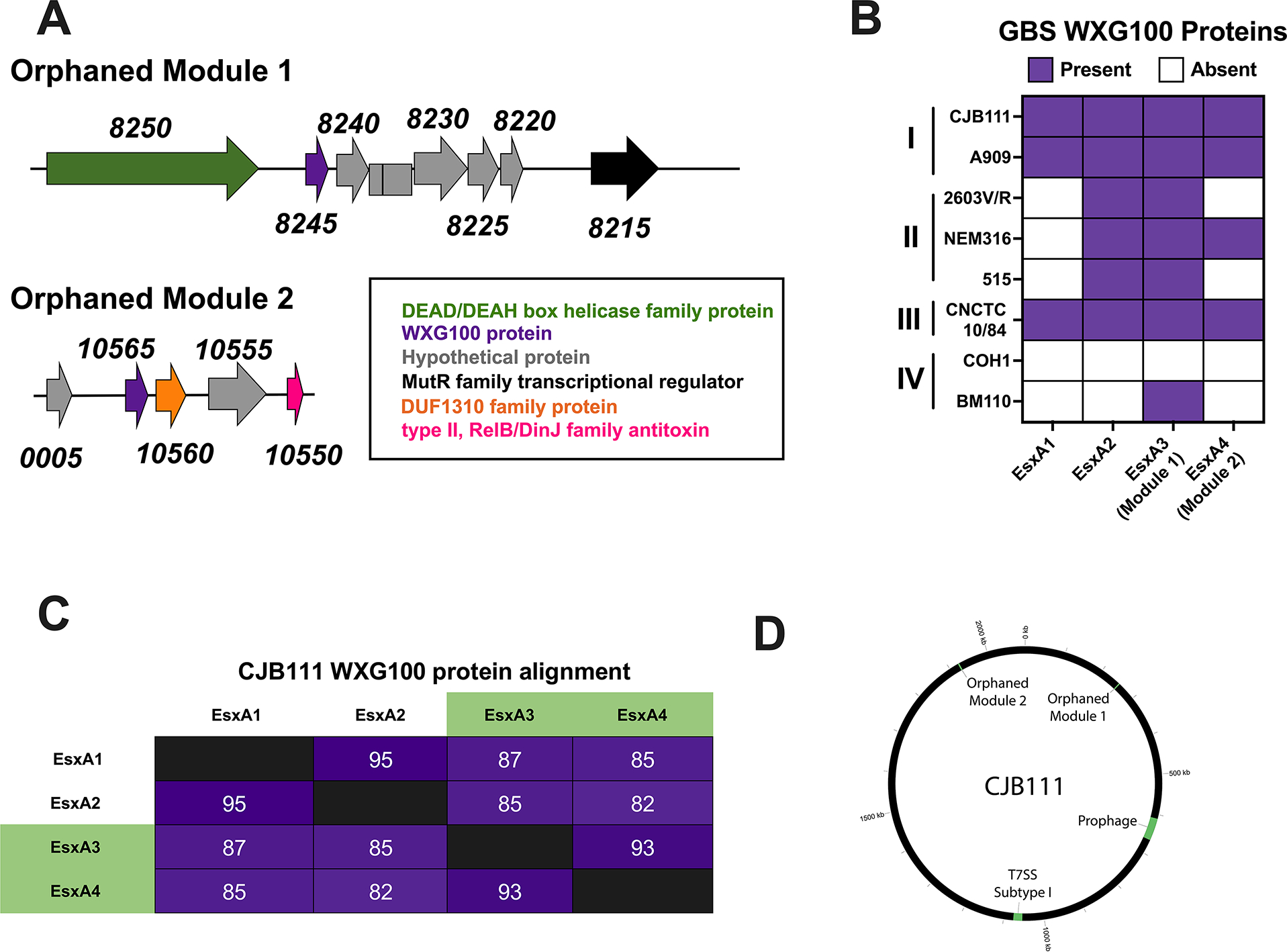
A) Representative GBS T7SS orphaned modules 1 and 2. The subtype I strain CJB111 Module 1 region includes genes encoding a DEAH/DEAD box helicase (green), WXG100 protein (purple), hypothetical proteins (gray), and a MutR family transcriptional regulator (black). The Module 2 region includes genes encoding hypothetical proteins (gray), a WXG100 protein (purple), DUF1310 protein(s) (orange), and a type II RelB/DinJ family antitoxin (pink). B) Heatmap indicating presence (purple) or absence (white) of locus associated EsxA1 and EsxA2 and of GBS T7SS orphaned modules 1 and 2 (containing EsxA3 and EsxA4, respectively) in various GBS clinical isolates. C) Percent identity matrix of EsxA1–4 protein sequences from CJB111 (subtype I). In the above matrix, the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity. D) Circos plot showing distinct genomic locations of the T7SS locus, orphaned modules, and prophage in subtype I strain CJB111.
Module 2 WXG100 orphans are less common (encoded in just 45 of the 80 T7SS+ GenBank isolates) and are only observed in strains encoding the Module 1 WXG100 orphan (Fig 4B, Supp. Table 4). While the genomic region surrounding Module 2 orphans are more variable, this WXG100 gene (ID870_10565 in subtype I strain CJB111) is consistently encoded upstream of at least one DUF1310 gene and a few genes upstream of a Type II toxin-antitoxin system RelB/DinJ family antitoxin fragment (Fig. 4A). Module 2 WXG100 orphans are well-conserved at the protein sequence level (Supp. Fig. 6C) and orphaned WXG100 proteins are more similar to each other compared to locus associated EsxA in subtype I strain CJB111 (Fig. 4C). However, because orphaned WXG100 proteins are distinct from WXG100 proteins found in other species, we have continued the esxA nomenclature and have annotated these genes as esxA3 (Module 1 orphan) and esxA4 (Module 2 orphan). Integrase/recombinase genes and toxin-antitoxin system genes are often associated with phage (Harms et al., 2018), which are known to mediate horizontal gene transfer and to modulate bacterial fitness and virulence (Borodovich et al., 2022, Basler et al., 2012, Hobbs & Mattick, 1993); therefore, we investigated whether GBS T7SS genes may be encoded within prophage. Interestingly, neither T7SS loci nor orphaned WXG100 proteins appear within prophage regions (Fig. 4D).
2.4 |. EsxA secretion across T7SS subtypes
We next sought to determine GBS T7SS activity across subtypes by measuring EsxA secretion in vitro as previously described (Spencer et al., 2021b). We observed EssC-dependent secretion in subtype I strains CJB111 and A909 and subtype III strain CNCTC 10/84 but could not detect EsxA in the supernatant or cell pellet of subtype II strain 2603V/R (Fig. 5, Supp. Fig 7A). This lack of detection was not due to inability of the anti-EsxA antibody (raised against CJB111 EsxA1) to bind the 95% identical subtype II EsxA2, since it was cross-reactive against subtype II strain NEM316 EsxA in both the supernatant and cell pellet. These results indicate that T7SS may be repressed or that EsxA may be degraded or unstable in the 2603V/R background. Alternatively, some T7SS subtype II strains may require different inducing conditions to express and secrete EsxA in vitro. Subtype IV strain COH1 served as a negative control in these studies as it does not encode any EsxA homolog.
Fig. 5. EsxA secretion across GBS T7SS subtypes.

Western blot showing EssC-dependent secretion of EsxA from subtype I strain CJB111 and subtype III strain CNCTC 10/84, but not from subtype II strain 2603V/R, in vitro. EsxA was also shown to be secreted in subtype I strain A909 and subtype II strain NEM316. Subtype IV strain COH1 does not encode EsxA and served as a negative control. Blot pictured is representative of 3 independent experiments.
2.5 |. Impact of GBS T7SS subtype on vaginal colonization
Secretion systems are often induced by stress, competing bacteria, and/or host pressures. We have previously shown that GBS T7SS promotes systemic infection but hypothesized that T7SS may also play a role during vaginal colonization and ascending infection, niches in which GBS must encounter host immune responses and compete with the native vaginal microbiota. As each GBS T7SS subtype expresses unique putative effectors, we further hypothesized that subtypes may have varying impacts on GBS interaction with the host. As subtypes I and III T7SS are active in vitro (as seen by EsxA secretion in Fig. 5 and Supp. Fig. 7A), we utilized our essC deletion mutants in subtype I (CJB111 and A909) and III (CNCTC 10/84) to assess the role of T7SS in vaginal persistence and ascending female genital tract infection in a murine model of colonization. CD1 mice were vaginally inoculated with parental or ΔessC mutant strains and GBS persistence over time was assessed by vaginal lavage or swab and tissue burdens were evaluated at the experimental endpoint. Loss of essC did not impact subtype I GBS persistence in the vaginal lumen as indicated by percentages of mice vaginally colonized between parental and ΔessC colonized groups in CJB111 (Fig. 6A) over time. However, we recovered increased parental CJB111 from vaginal, cervical, and uterine tissues at the experimental endpoint compared to the CJB111ΔessC mutant (Fig. 6B–D), indicating that the subtype I CJB111 T7SS promotes tissue invasion and ascending infection in the female genital tract. The loss of essC in subtype I strain A909 did not significantly impact vaginal persistence or tissue burden (Supp. Fig. 7B–E). Interestingly, for GBS T7SS subtype III, the CNCTC 10/84ΔessC mutant persisted better than the parental CNCTC 10/84 strain in the murine vaginal lumen (Fig. 6E). At the experimental endpoint, the CNCTC 10/84ΔessC mutant exhibited higher tissue burdens in the vagina, cervix, and uterus compared to parental CNCTC 10/84-colonized mice (Fig. 6F–H). Together these data indicate that GBS T7SS in subtype I strain CJB111 may provide an advantage in colonization of the female genital tract while T7SS in subtype III strain CNCTC 10/84 may be disadvantageous in this environment.
Fig. 6. Varying impact of EssC deficiency on GBS vaginal colonization by T7SS subtype I and III strains.
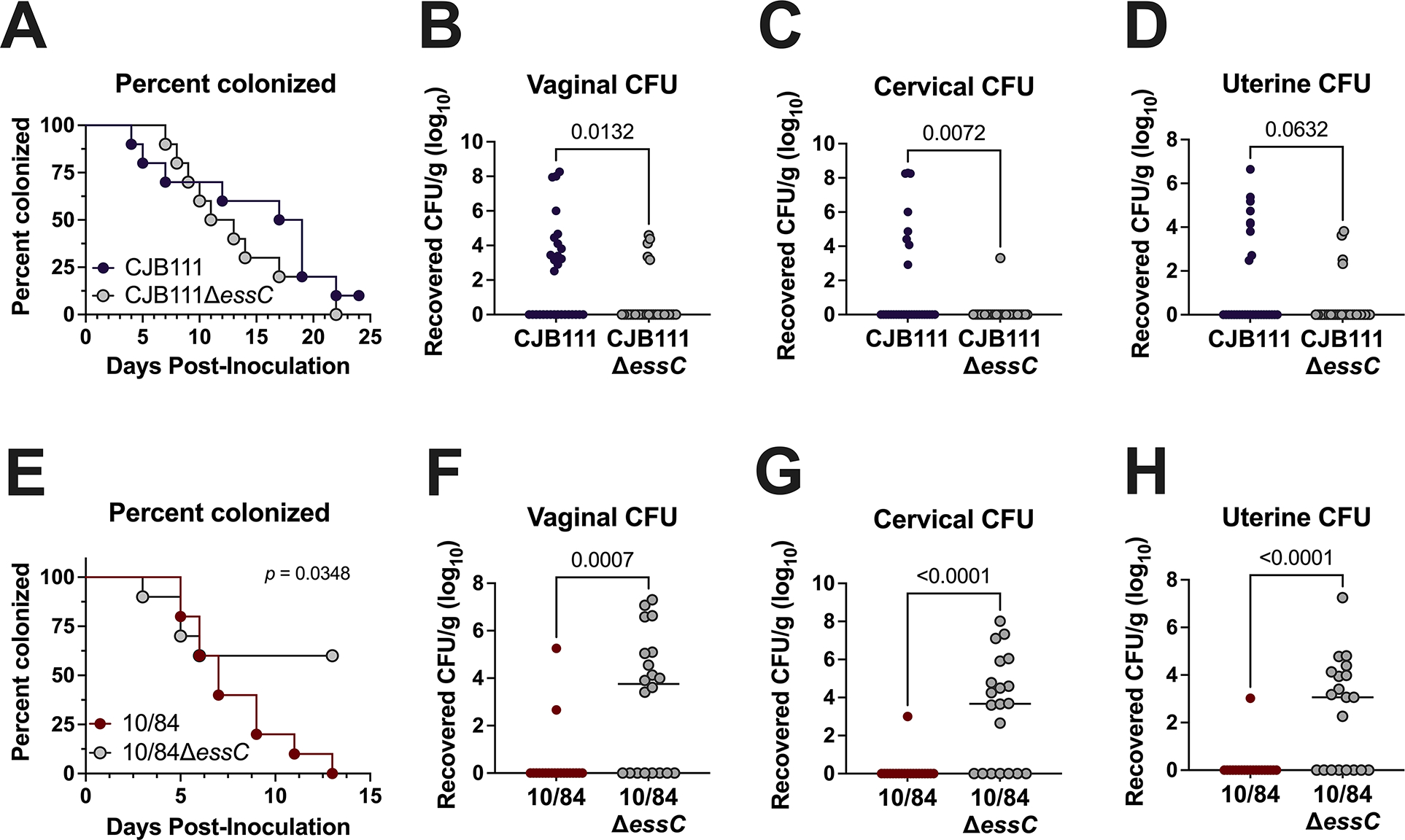
A, E) Percent colonization curves of 8-week-old CD1 female mice vaginally inoculated with A) subtype I strain CJB111 or CJB111ΔessC or E) subtype III strain CNCTC 10/84 or CNCTC 10/84ΔessC. Graphs are representative of three or two independent experiments, respectively (n = 10/group). Statistics reflect the Log rank (Mantel-Cox) test. Recovered CFU counts from the B, F) vaginal C, G) cervical, and D, H) uterine tissue of colonized mice. In panels B-D and F-H, each dot represents one mouse and all independent experiments’ data are combined in these figures (n = 30/group for CJB111 experiments and n = 20/group for CNCTC 10/84 experiments). The bars in these plots show the median and statistics represent the Mann Whitney U test.
2.6 |. Multiplex PCR to T7SS type GBS clinical isolates
Given this striking strain-specific phenotype between subtype I CJB111 and subtype III CNCTC 10/84, we sought to validate whether the T7SS subtype distribution observed in our GenBank GBS isolates is representative of recent clinical GBS isolates using collections of vaginal isolates from pregnant women and from diabetic foot ulcers that our laboratory has characterized previously (Burcham et al., 2019, Keogh et al., 2022). To determine GBS T7SS subtypes, we developed a multiplex PCR assay using primers against subtype specific transmembrane encoding genes, the amplicons of which could be distinguished by size (Fig. 7A; Supp. Table 5). Within the clinical isolate cohorts, most encode T7SS subtype I and subtype II (24/64, 37.5% and 33/64, 51.6%, respectively) (Fig. 7B–C). A small percentage of isolates encode subtype IV (7/64, 10.9%); however, we did not identify any subtype III in our clinical isolate cohorts, corroborating its low prevalence in whole genome sequences available on GenBank. We compiled relevant information on these strains as well as those from GenBank including sequence type, serotype, T7SS locus subtype, prophage cluster, and orphan modules encoded to determine if any associations exist between these metrics using Fisher’s exact tests (Supp. Tables 3 and 5). Similar to a previous report on GBS T7SS genomic arrangement (Zhou et al., 2022) we found that GBS T7SS subtype is associated with sequence-type (p < 2.2e-16) and is also associated with GBS serotype (p < 2.158E-13). Within our clinical isolate cohorts, trends were particularly observed between T7SS subtype I and serotype Ib, T7SS subtype II and serotypes Ia and II, and T7SS subtype IV with serotype III sequence type 17 (ST-17) (Supp. Tables 3 and 5). While we also observed significant associations between prophage cluster, orphaned modules, and T7SS subtype in our vaginal isolate cohort, it is possible these associations may be artifacts of T7SS association with sequence type.
Fig. 7. Multiplex PCR to identify T7SS subtype amongst GBS isolates.
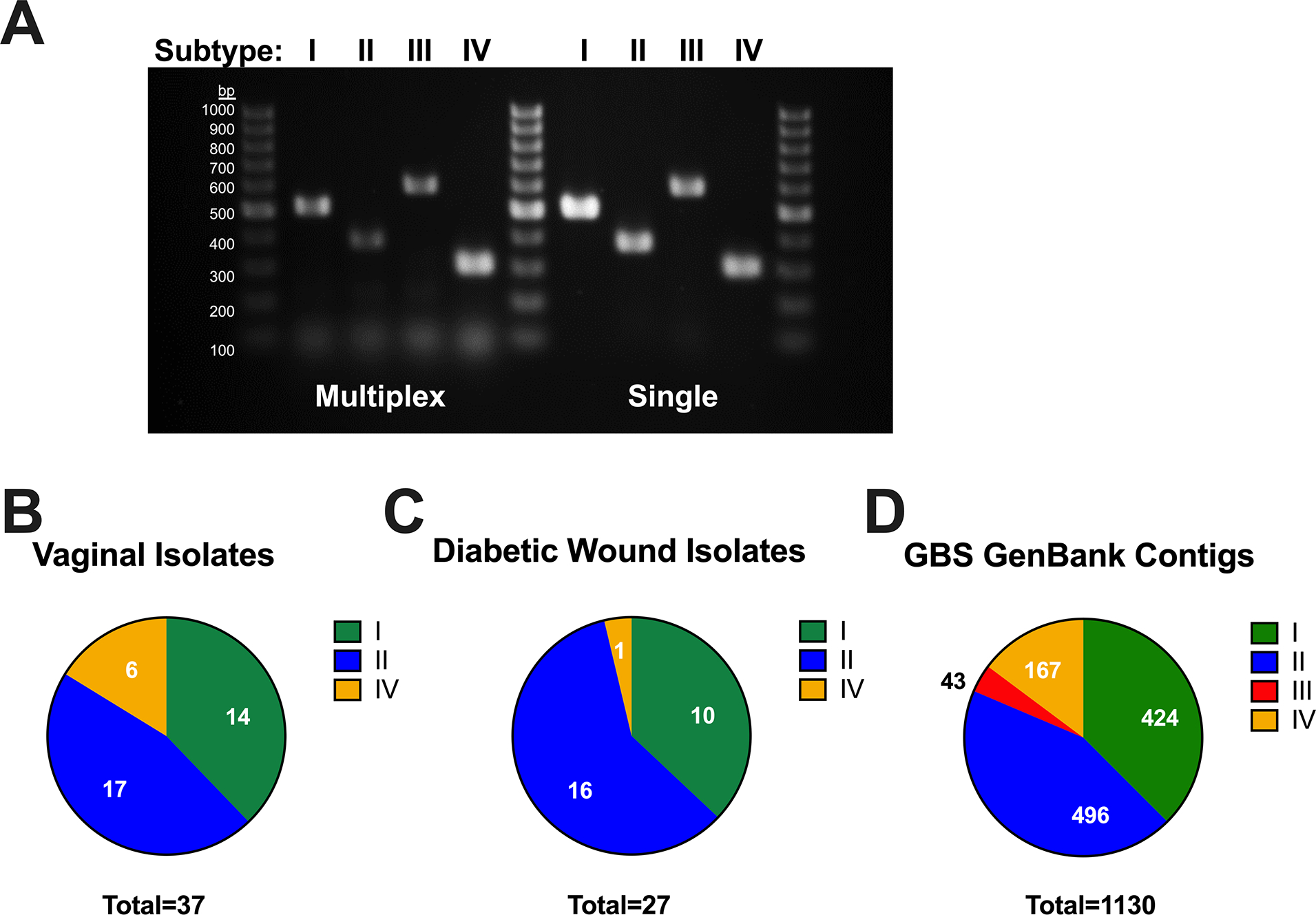
A multiplex PCR was developed for GBS T7SS typing using primers that amplify subtype specific genes, designed to yield distinct amplicon sizes across T7SS subtypes (see Supp. Table 5). A) Agarose gel showing subtype specific amplicon products from single-plex and multiplex PCR (of GBS genomic DNA), which can be differentiated by size using a 100 bp DNA ladder. This multiplexed PCR system was utilized in the laboratory or in silico using Geneious to T7SS type GBS B) vaginal (n =37) and C) diabetic wound isolates (n = 27) or D) GBS contig sequences available from GenBank (n =1130), respectively.
To achieve a larger dataset and to ensure that our isolate banks are not biased due to collection, we performed in silico T7SS subtyping of WGS GBS sequences available in contigs. Of 1342 sequences, 1130 were able to be typed using BLAST of the 225 C-terminal amino acids of EssC and the subtype specific gene used for physical typing of isolates in Supp. Table 5 and Fig. 7. Strains were only considered typable if positive for both a specific subtype’s EssC and downstream subtype-specific gene. Upon screening of these WGS sequence contigs of GBS isolates with typable T7SS, 424 were subtype I (37.5%), 496 were subtype II (43.9%), 43 were subtype III (3.8%), and 167 were subtype IV (14.8%) (Fig. 7D). As these sequences are in contigs, T7SS typing was not possible for all isolates (sequences may have gaps within the genomic region containing the T7SS locus or, occasionally, some strains do not encode a T7SS). Sequence analysis of these additional strains further confirmed that subtypes I and II are the most prevalent and that extensive diversity exists within GBS T7SS.
3 |. DISCUSSION
This study highlights the heterogeneity of GBS T7SS subtypes, which could affect GBS interactions in the host. One purpose of this work was to compare GBS T7SS to T7SS loci encoded by other Gram-positive species. Despite having a similar arrangement of T7SS genes, GBS core proteins and putative effectors were largely unique compared to T7SS proteins described in eight other Gram-positive organisms. The secreted substrate/putative core protein EsxA and ATPase EssC were the most conserved T7SS components, consistent with previous observations that these two proteins are general features of T7SS (Pallen, 2002). Despite low sequence homology, GBS T7SS components are commonly found within T7SSb in other species, including WXG-like DUF3130 containing proteins, putative LXG toxins, putative immunity factors and chaperones, CHAP domain containing putative lysins, transmembrane proteins, lipoproteins, fragments of essC, and DUF4176 proteins.
Within GBS, T7SS loci reside in a common genomic location, contain homologous machinery, subtype-specific putative effectors/immunity factors/chaperones downstream of essC, and, with the exception of subtype IV, encode at least one copy of esxA upstream of T7SS machinery. However, we also observed extensive intra-subtype GBS T7SS heterogeneity. Within subtype I specifically, strains encoding the A909 type locus lack approximately eight genes compared to the subtype I CJB111 locus; yet A909 maintains functional EsxA secretion (Fig. 3A, Fig. 5, Supp. Fig. 7A). In vaginal colonization experiments, parental CJB111 exhibited higher tissue invasion compared to an isogenic ΔessC mutant but tissue burden differences were not observed between parental A909 and A909ΔessC-colonized groups (Fig. 6B–D, Supp. Fig. 7C–E). It is unclear whether this eight gene cluster encoded by the CJB111 locus contributes to T7SS activity and future work will investigate these strain differences in different mouse lines that have longer GBS persistence and varied microbiota. GBS T7SS subtype II loci differ from each other based on truncations of T7SS machinery, particularly within essC due to either nonsense or slip strand mutations (Fig 3B). Although subtype II strain 2603V/R encodes a truncated essC, we also could not detect EsxA in the cell pellet, suggesting that EsxA is not well expressed in this strain under the conditions used. Further, it is possible that these putative truncated EssC proteins may actually retain function in some cases as subtype II strain NEM316 is capable of EsxA secretion (Fig. 5). It is possible that a second EssC ORF is transcribed and translated, and whether this second CDS works with EssC CDS1 or is functional on its own would be interesting to investigate.
Across species and strains, T7SSb proteins also commonly contain T7-associated motifs, including the [W/F]XG motif (found within EsxA and other WXG100-like proteins), the LXG motif (found within toxins that promote interbacterial competition), and the YxxxD/E motif. The YxxxD/E motif is thought to form one part of a bipartite signal required for homing of a substrate to the machinery for export in mycobacterial T7SSa and Firmicute T7SSb (Champion et al., 2006, Daleke et al., 2012, Anderson et al., 2013, Sysoeva et al., 2014). Permutations of these motifs also exist, as a C-terminal FxxxD/E motif was found within GBS Lap1 and “FXG” motifs were identified in GBS Lap1 and DUF4176 proteins. While the presence of a given motif does not necessitate a role in T7SS, we observed that many machinery and putative effector proteins encode YxxxD/E motifs, which may be important for substrate recognition. Interestingly, unlike mycobacterial and staphylococcal EsxA homologs, GBS EsxA1 and EsxA2 do not encode a C-terminal YxxxD/E motif. Previously, Poulsen et al. showed that some WXG100 proteins may encode a less specific C-terminal motif, which might direct these substrates to the T7SS machinery: HxxxD/ExxhxxxH (in which the H and h, stand for high and low conservation of hydrophobic resides, respectively (Poulsen et al., 2014)). Both GBS EsxA1 and EsxA2 C-terminal sequences include an HxxxD/ExxhxxxH motif, as does the S. gallolyticus EsxA (Taylor et al., 2021); however, further studies are needed to determine whether this C-terminal motif is required for GBS EsxA secretion.
LXG modules (as depicted in Fig. 1C) are widespread in T7SSb+ Gram-positive species and often encode unique upstream genes (putative chaperones) and unique downstream genes (putative immunity factors). In S. aureus, WXG-like proteins EsxC and EsxD heterodimerize with WXG100 proteins and are required for their export (Anderson et al., 2013). Further, in S. aureus it was recently shown that WXG100 EsxB and WXG-like EsxC and EsxD chaperone export of T7SS nuclease toxin EsaD (Yang et al., 2023). Similarly, S. intermedius Lap1 and Lap2 WXG-like proteins also facilitate secretion of a downstream LXG toxin (Klein et al., 2022). We have also observed WXG-like genes downstream of the GBS essC that are predicted to interact with the LXG N-terminus by AlphaFold 2 (Fig. 2A). While deletion of these WXG-like genes in subtype I strain CJB111 did not affect EsxA secretion (data not shown), we hypothesize that GBS Lap proteins may function in LXG protein secretion, similar to S. intermedius. Transmembrane protein and DUF4176 protein encoding genes, commonly observed in the vicinity of T7SS loci and toxins, appeared downstream of the LXG genes in GBS subtypes I-III. In S. aureus, cognate immunity factor esaG (DUF600) is encoded downstream of nuclease EsaD (Cao et al., 2016) and in S. intermedius, Tel immunity proteins are encoded either adjacent to (in the case of TelB, TelC, TelD) or one gene separated (in the case of TelA) from the LXG toxin (Whitney et al., 2017, Klein et al., 2022). We therefore expect that one of these downstream GBS genes encodes a subtype specific immunity factor and determining these LXG toxin-immunity factor pairs is the subject of our future work.
Similar to other T7SSb, GBS LXG proteins have highly conserved ɑ-helical N-termini, which are structurally similar to the WXG100 proteins (as originally described by Aravind et al., 2011), but have unique, globular C-terminal (and putatively toxic) domains (Supp. Fig. 4A) with unique LXG toxins encoded across GBS subtypes I-III. Our bioinformatic analysis failed to identify domains/predicted functions for these C-terminal putatively toxic regions; thus, our future work will determine the putatively toxic activities of these proteins experimentally. Many T7SS+ Gram positive species also encode orphaned LXG proteins elsewhere in the genome (e.g., S. aureus TspA (Ulhuq et al., 2020)). L. monocytogenes, B. subtilis, S. gallolyticus, S. intermedius and S. suis also encode multiple full-length LXG toxins, not all of which are associated with the T7SS locus (Bowran & Palmer, 2021, Whitney et al., 2017, Kobayashi, 2021, Teh et al., 2022, Liang et al., 2022). While we did not identify any orphaned full-length GBS LXG proteins in our representative strains based on presence of an LXG motif with the first 100 amino acids of the protein, N-terminal homology to other proteins, or by searching specifically in genomic regions that encode orphaned DUF4176 or WXG100 proteins, it is possible that orphaned C-terminal toxin fragments may exist in GBS strains.
GBS subtype I and II were the most commonly identified T7SS subtypes based on multiplex PCR typing of cohorts of clinical isolates or by in silico typing of whole genome sequences and contigs. Subtype IV strains, the next most common subtype, do not encode many common T7SS components, such as locus associated WXG100, DUF3130/SACOL2603, or full-length LXG or DUF4176 proteins. While the genes encoded between the COH1 subtype IV essC and ltdS are all annotated as hypothetical, interestingly, some of these COH1 genes (the essC-lipoprotein-hydrolase module depicted in Fig. 1D) also appear in subtype I, II, and III loci; see Fig 1 and Fig. 3), indicating that homologous recombination may occur in T7SS loci as has been reported in other species. While the biological purpose of the GBS T7SS subtype IV is currently unclear, we have observed in subtype IV strain COH1 background that T7SS genes are modulated in certain conditions, such as in a cas9 deletion mutant (Spencer et al., 2019), or during incubation with mucins (Burcham et al., 2022b), thus indicating that the COH1 T7SS may play a role during stress. As most subtype IV strains identified are from the hypervirulent ST-17, serotype III lineage, which is associated with neonatal meningitis, they may have lost some T7SS components due to acquisition of additional virulence factors. Lastly, while T7SS promoted subtype I strain CJB111 colonization of genital tract tissues, the T7SS encoded by subtype III strain CNCTC 10/84 appeared to be detrimental for vaginal colonization. We propose that these phenotypes may be due to subtype specific effectors encoded by each strain. Our future work will investigate whether this detrimental role for the CNCTC 10/84 T7SS is due to modulation of the vaginal microbiota or due to modulation of immune responses, resulting in parental CNCTC 10/84 clearance. This disadvantage of the CNCTC 10/84 T7SS in the vaginal tract may account for subtype III T7SS’s rarity across GBS isolates. Indeed, subtype III constitutes just ×4% of T7SSb+ GBS contig sequences (n =1130).
Bacterial secretion systems are often heavily regulated and may be minimally expressed in vitro. We have similarly observed that the GBS T7SS is lowly expressed in vitro; yet, we observe striking GBS T7SS-dependent phenotypes in cell and animal models of infection. Notably, staphylococcal T7SS genes are upregulated in the female genital tract (Deng et al., 2019) and the E. faecalis T7SS was shown to be required for colonization of the vaginal tract (Alhajjar et al., 2020), further indicating that this system may be important in host environments. Interestingly, transcriptomic studies comparing GBS isolated from the murine vaginal lumen to in vitro broth culture indicated that T7SS machinery genes were not significantly dysregulated or were even down regulated (Cook et al., 2018) (Burcham et al., 2022a), which may explain why we do not observe significant T7SS-dependent differences in vaginal lumen persistence (Fig. 6A).
Regulation of bacterial virulence factors commonly occurs via phase variation, which can involve transcriptional slippage resulting in expression of a given factor in some environments and under certain conditions (Phillips et al., 2019). For example, transcriptional slippage within genes encoding pneumococcal capsule modifying enzymes, facilitates pneumococcal evasion of vaccine elicited antibodies (van Selm et al., 2003, Rajam et al., 2007, Spencer et al., 2017). We and others have observed many homopolymeric tracts within the GBS T7SS locus (Janulczyk et al., 2010), with the most common T7SS slip-strand mutation occurring within essC in subtypes II and IV, due to a homopolymeric Gn tract. This tract occurs within all subtypes; therefore, it is unclear why these essC mutations are not observed in subtype I strains. It is possible that subtype specific effectors may induce host pressure, therefore necessitating stringent regulation of the subtype II locus specifically. Although more work is needed to understand EssC regulation and expression, we observed numerous homopolymeric tracts within GBS T7SS loci and hypothesize that these may regulate T7SS gene expression. Finally, numerous single and two-component regulators control T7SSb loci, and this has been most extensively studied in S. aureus (Bowman & Palmer, 2021). While GBS T7SS gene expression is induced upon deletion of cas9 (Spencer et al., 2019), in the presence of mucins (Burcham et al., 2022b), in amniotic fluid (Sitkiewicz et al., 2009), and other stress conditions including removal of nutrients, exposure to serum, and oxygen deprivation (Avican et al., 2021), our investigation of a specific GBS T7SS regulator is ongoing.
Secretion systems are commonly associated with phage, with a well-established connection between T6SS and bacteriophage machinery in particular. A link between T7SS and bacteriophage infection may also exist as T7SS WXG100 and putative toxins have been identified on Mycobacterium abscessus prophage (Dedrick et al., 2021) and phage infection induces E. faecalis T7SS (Chatterjee et al., 2021). Although we do not observe T7SS proteins encoded within GBS prophage, we did observe that some GBS T7SS proteins exhibit homology to phage proteins and that many phage proteins contain T7SS-associated motifs. It is known that encoding of prophage can modulate bacterial resistance to bacteriophage infection and antibiotics (Wendling et al., 2021). Recently, integration of a temperate phage into S. aureus was shown to increase virulence, not by encoding virulence factors itself, but instead due to upregulation of various bacterial virulence factors including EsxA (Yang et al., 2022). This was also recently shown in GBS, in which loss of prophage from CNCTC 10/84 modulated gene expression (Wiafe-Kwakye and Neely, unpublished observation). Therefore, future investigation of a link between GBS T7SS and prophage is warranted.
In summary, this study bioinformatically characterizes the diversity encoded within the GBS T7SS and indicates that GBS T7SS contains similar, but unique, individual effectors compared to T7SSb in other Gram-positive species. GBS can be classified into four subtypes, which encode unique effectors and appear to modulate GBS T7SS-dependent interactions within the host, such as during vaginal colonization. This study also identifies orphaned modules containing potential T7SS-associated WXG100 proteins. Taken together, this study suggests a “one size does not fit all” approach to studying T7SS as phenotypes and implications for host and interbacterial phenotypes likely depend on the specific effectors encoded across and even within species. Future studies are warranted to further characterize GBS T7SS effectors and their impact on colonization and disease.
4 |. MATERIALS AND METHODS
4.1 |. Bacterial strains
Example strains from each of the GBS T7SS subtypes were used in this study (subtype I-CJB111; GenBank accession CP063198.2 (01-APR-2021 version) (Spencer et al., 2021a), subtype II- 2603V/R; GenBank accession NC_004116.1 (Tettelin et al., 2002), subtype III- CNCTC 10/84; GenBank accession NZ_CP006910.1 (Nizet et al., 1996, Hooven et al., 2014, Wilkinson, 1977), subtype IV- COH1; GenBank accession NZ_HG939456.1 (Nizet et al., 1996, Da Cunha et al., 2014)). Further, previously described cohorts of clinical GBS isolates were utilized for molecular T7SS subtyping and analysis of T7SS activity in vitro. Vaginal isolates were obtained from Melody Neely from the Detroit Medical Center as described previously (Burcham et al., 2019). Diabetic wound isolates were obtained from Elizabeth Grice (University of Pennsylvania) as well as the CU-Anschutz Medical center (Keogh et al., 2022). GBS was grown statically in Todd Hewitt Broth (THB; Research Products International, RPI) at 37°C. All strains used in this study can be found in Supp. Table 5.
4.2 |. Bioinformatic analysis of GBS T7SS
All comparative genomics were performed in Geneious Prime 2022.0.2 using genomes of Streptococcus agalactiae downloaded from NIH GenBank (as described in Supplementary Table 1 of (Spencer et al., 2021b)) as well as using 1342 WGS contigs of Streptococcus agalactiae downloaded from NIH GenBank (as of December 2020). Protein BLAST was utilized in Geneious to determine presence/absence or conservation of T7SS-associated proteins across GBS subtypes and across other Gram-positive species. Protein sequences yielding a grade of 30% or less were considered not homologous. Protein alignments were performed using the EMBL-EBI (European Molecular Biology Laboratory-European Bioinformatics Institute) Clustal Omega (v.1.2.4). CDART (Geer et al., 2002), and InterPro (Blum et al., 2021) were used to identify domains within T7SS locus-associated proteins. Protter was used to characterize protein topology (Omasits et al., 2014) and amino acid sequences were scanned manually for T7SS-associated motifs.
4.2 |. Modelling of GBS T7SS proteins
The modelling of LXG proteins and Lap chaperones was performed using original or multimer AlphaFold2 weights (Jumper et al., 2021, Evans et al., 2022) as implemented in ColabFold (Mirdita et al., 2022) or using Robetta (Kim et al., 2004). Structural illustrations were generated using PyMol (The PyMOL Molecular Graphics System, Version 2.3.5 Schrödinger, LLC. (Schrodinger, 2015)).
4.3 |. Cloning
Clean essC deletion mutants were created via allelic exchange in CJB111, A909, 2603V/R, CNCTC 10/84, and COH1 backgrounds using the temperature sensitive plasmid pHY304 (Spencer et al., 2019) and using a gene encoding spectinomycin resistance (aad9) in the knockout construct. Second crossover mutants were screened for erythromycin sensitivity and spectinomycin resistance. Strains created in this study, essC locus tags for each strain, and primers used in this study can be found in Supp. Table 5.
4.4 |. T7SS operon mapping
Mapping of the T7SS operon in the CJB111 strain background was performed similar to that described previously (Kneuper et al., 2014, Taylor et al., 2021). Briefly, primers were designed to span gene junctions within the putative T7SS locus and surrounding genomic area. These primers sets were used in PCR reactions with cDNA template (50 ng/ reaction) to determine which adjacent genes were co-transcribed. Genomic DNA (50 ng/ reaction) was used as a positive control for the primer sets and no-reverse transcriptase cDNA was generated and diluted equivalently to control for possible genomic DNA contamination of the cDNA. cDNA and no-RT cDNA were generated as previously described (Spencer et al., 2021b). Briefly, GBS strains were grown to mid-log (OD600 = 0.4–0.6) and RNA was purified using the MACHEREY-NAGEL NucleoSpin kit (catalog# 740955.250) according to manufacturer instructions with the addition of three bead beating steps (30 sec × 3, with one minute rest on ice between each) following the resuspension of bacterial pellets in RA1 buffer + β-mercaptoethanol. Purified RNA was treated with the Turbo DNase kit (Invitrogen, catalog# AM1907) according to manufacturer instructions. cDNA was synthesized using the SuperScript cDNA synthesis kit (QuantaBio, catalog# 95047–500), per manufacturer instructions. All RT-PCR reactions were performed using Q5 polymerase (New England Biolabs) under the following cycling conditions on a Bio-Rad T100 thermal cycler: 98°C, 2-min hot start; 34 cycles (98°C, 10 seconds; 60 °C, 20 seconds; 72°C, 30-second extension/kb); and 72°C, 10-min extension. Primers used for these experiments can be found in Supp. Table 5. PCR amplicons were visualized via gel electrophoresis using 1% agarose gels and GeneRuler 1 kb Plus DNA ladder (Thermo Scientific, SM1331). Three independent replicates were performed.
4.5 |. EsxA secretion assay
Secretion of EsxA during growth in THB was assessed for GBS clinical isolates as described previously (Spencer et al., 2021b). Briefly, overnight cultures of GBS isolates were sub-cultured into 5 mL of THB and grown statically for 24 hours at 37°C. Bacteria were pelleted at 3214 × g for 10 minutes at 4°C. Supernatants were removed from pellets, filtered (Millex Low Protein Binding Durapore PVDF Membrane 0.22μm filters, catalog #SLGVR33RS), and supplemented with an EDTA-free protease inhibitor cocktail (Millipore-Sigma set III, catalog # 539134; 1:250 dilution). Supernatants were then precipitated overnight with trichloroacetic acid (TCA) at 4°C. Precipitated proteins were centrifuged for 15 minutes at 13K × g and resulting pellets were gently washed with acetone. Pellets were then centrifuged again at the same settings. Acetone was removed and pellets were allowed to dry before being resuspended in Tris buffer (50 mM Tris HCl, 10% glycerol, 500 mM NaCl, pH 7). Bacterial pellets were washed once with PBS, frozen overnight, and resuspended in Tris buffer + protease inhibitor. Pellets were then bead beaten (2 × one minute) using 0.1mm zirconia/silica beads (BioSpec). Triton-X-100 was then added to lysates at a final concentration of 1% to solubilize membrane proteins and vortexed to mix.
Supernatant and pellet samples were mixed 1:1 with Laemmli buffer + beta-mercaptoethanol, boiled 10 minutes, and run on SDS-PAGE for Western blotting. Proteins were transferred to membranes via the BioRad Trans-Blot Turbo Transfer System (high molecular weight settings). Membranes were washed three times in TBST and blocked in LI-COR’s Intercept Blocking Buffer (catalog# 927–60001) for one hour at room temperature. Membranes were probed with an anti-EsxA1 rabbit polyclonal antibody (0.5 μg/ml; GenScript) in the above LI-COR blocking buffer overnight at 4°C. Following washes in TBST, membranes were incubated with IRDye 680RD goat anti-rabbit IgG (H + L) secondary antibodies from LI-COR (1:10,000 dilution; 1 hour, room temperature; catalog# 926–68071). Following washes in TBST and water, western blots were imaged using the LI-COR Odyssey.
4.6 |. Murine Model of GBS Vaginal Colonization
GBS vaginal colonization was assessed using a previously described murine model of vaginal persistence and ascending infection (Patras & Doran, 2016). Briefly, 8–10-week-old female CD1 (Charles River) mice were synced with beta-estradiol at day −1 and inoculated with mid-log grown GBS (approximately 1 × 107 CFU) in PBS on day 0. Post-inoculation, mice were lavaged with PBS daily, and the samples were serially diluted and plated for CFU counts to determine bacterial persistence on differential and selective GBS CHROMagar [catalog# SB282(B)]. At experimental end points, mice were euthanized, and female genital tract tissues (vagina, cervix, and uterus) were collected. Tissues were homogenized and samples were serially diluted and plated on CHROMagar for CFU enumeration. Bacterial counts were normalized to the tissue weight. Vaginal colonization experiments with CJB111 and CNCTC 10/84 were ended upon vaginal clearance of one strain by 80–90% of the mice. Experiments with A909 were ended upon vaginal clearance by 50% of mice. These experiments were approved by the committee on the use and care of animals at the University of Colorado-Anschutz Medical Campus (protocol #00316).
4.7 |. Molecular T7SS typing of GBS clinical isolates
Molecular typing was performed by multiplex PCR amplification using primers within subtype-specific genes: subtype I, CJB111 ID870_4220; subtype II, 2603V/R SAG_RS07870; subtype III, CNCTC 10/84 W903_RS05410; and subtype IV, COH1 GBSCOH1_RS05060. Primers used for these experiments can be found in Supp. Table 5. Multiplex PCR reactions were performed using Q5 polymerase (New England Biolabs) under the following cycling conditions on a Bio-Rad T100 thermal cycler: 98°C, 30 second hot start; 35 cycles (98°C, 10 seconds; 59 °C, 30 seconds; 72°C, 30 second extension/kb); and 72°C, 2-minute extension. PCR amplicons were visualized via gel electrophoresis using 1.4% agarose gels and Gene Ruler 100bp DNA ladder (Thermo Fisher Scientific, SM0243).
4.8 |. Data analysis and statistics
Fisher’s exact test were performed to assess associations between T7SS subtype, T7SS orphaned modules, sequence type, serotype, and prophage cluster. Tables and R script used for Fisher’s exact tests can be found in Supp. Table 6. For vaginal colonization experiments, statistical analysis was performed using Prism version 9.4.1 (458) for macOS (GraphPad Software, La Jolla, CA, United States). Significance was defined as p < α, with α = 0.05.
Supplementary Material
Supp. Fig. 1. Comparison of GBS T7SS effectors to other Gram-positive organisms. Heat map comparing CJB111 T7SS protein homology to a panel of eight Gram-positive organisms in which the T7SSb has been studied previously: S. aureus (variants 1–4), S. lugdunensis (variants 1, 2), Listeria monocytogenes (variants 1–7), E. faecalis (strain OG1RF), Bacillus subtilis (PY79), S. gallolyticus (strain TX20005), S. intermedius (strains B196 and GC1825), and S. suis (strains GZ5065 and WUSS351). See Supp. Table 2 for individual strain information. Heat map color intensity is based on Geneious alignment grade, which considers query coverage and percent identity. Shading of the locus tag numbers on the x-axis corresponds to the key for gene color in Fig 1. Most downstream CJB111 T7SS effectors have little to no homology across species.
Supp. Fig. 2. Subtype-specific LXG-associated proteins are encoded by GBS T7SS.
Percent identity matrices and Clustal Omega alignments of A-C) LXG-associated protein 1 (Lap1) and D-F) LXG-associated protein 2 (Lap2) sequences from CJB111 (subtype I), 2603V/R (subtype II), CNCTC 10/84 (subtype III), and C001 (subtype III). In the above matrices, the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity. Conserved putative T7SS-associated FXG and FxxxD/E motifs are highlighted by red boxes in C.
Supp. Fig. 3. Confidence scores and predicted aligned error for LXG-Lap AlphaFold models in Fig. 2A. A) Predicted LXG-Lap complex models shown in Fig. 2A but with color corresponding to per-residue confidence level (predicted local distance difference test [pLDDT] score 1–100; pLDDT > 90 are expected to be modelled to high accuracy). B) Predicted aligned error for each LXG-Lap complex, with colors indicating the confidence of domain positions (higher predicted error in red, lower predicted error in blue).
Supp. Fig. 4. Modeling of full-length LXG proteins and homology of downstream putative immunity factor and DUF4176 genes
A) Robetta predicted structures for full-length putative GBS LXG toxins across subtypes I, II, and III. B) Percent identity matrix of putative immunity factors (transmembrane domain containing proteins) encoded for downstream of LXG genes in CJB111 (subtype I), 2603V/R (subtype II), CNCTC 10/84 (subtype III), and C001 (subtype III). C-E) Percent identity matrices and Clustal Omega alignments of DUF4176 protein sequences from CJB111 (subtype I), 2603V/R (subtype II), CNCTC 10/84 (subtype III), and COH1 (subtype IV). Green highlighting in C-E indicate orphaned DUF4176 proteins. A conserved central FXG motif within GBS DUF4176 proteins is highlighted by a red box in D. In all of the above percent identity matrices, the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity.
Supp. Fig. 5. Transcriptional landscape of the CJB111 T7SS locus
RT-PCR was performed to evaluate transcriptional organization of the CJB111 T7SS locus. Using cDNA as template (and no RT-cDNA and genomic DNA as negative and positive controls, respectively), PCRs were performed using primer pairs spanning every gene junction in the putative T7SS locus. Primers used for these experiments can be found in Supp. Table 5. Agarose gels shown are representative of three independent experiments.
Supp. Fig. 6. Homology of orphaned GBS WXG100 proteins.
A) Locus tag tables and percent identity matrices of B) Orphaned Module 1 WXG100 protein EsxA3 and C) Orphaned Module 2 WXG100 protein EsxA4 sequences across a panel of GBS isolates representing T7SS subtypes I-IV. Green highlighting in A indicates orphaned WXG100 proteins. In the above matrices (B-C), the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity.
Supp. Fig 7. Impact of EssC deficiency on GBS vaginal colonization by T7SS subtype I strain A909. A) Western blot showing EssC-dependent secretion of EsxA from subtype I strain A909, in vitro. B) Percent colonization curve of 8-week-old CD1 female mice vaginally inoculated with subtype I strain A909 or A909ΔessC. Statistics reflect the Log rank (Mantel-Cox) test. Recovered CFU counts from the C) vaginal D) cervical, and E) uterine tissue of colonized mice. In panels C-E, each dot represents one mouse and two independent experiments’ data are combined in these figures (n = 20/group total). Plots show the median and statistics represent the Mann Whitney U test.
Acknowledgements:
We acknowledge the National Summer Undergraduate Research Program for funding Morgan Apolonio’s virtual research project, which contributed to this work. We also thank Haider Manzer for contributing the Circos plot in Fig. 4D and Uday Tak for contributing to the Robetta models in Supp. Fig. 4A. Funding for this work was provided by NIH/NIAID F32 AI143203 to BLS, NIH/NIAID R01 AI153332 and NIH/NINDS R01 NS116716 to KSD, NIH/NIAID R21 AI169231 to KVK and KAP, and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001 to JCM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interests: The authors have declared that no competing interests exist.
Ethics statement: Animal experiments were performed using accepted veterinary standards as approved by the Institutional Animal Care and Use Committee at University of Colorado-Anschutz protocol #00316. The University of Colorado-Anschutz is AAALAC accredited, and their facilities meet and adhere to the standards in the “Guide for the Care and Use of Laboratory Animals”.
Data availability:
All relevant data are within the manuscript and its Supporting Information files.
REFERENCES
- Alhajjar N, Chatterjee A, Spencer BL, Burcham LR, Willett JLE, Dunny GM, Duerkop BA, and Doran KS (2020) Genome-Wide Mutagenesis Identifies Factors Involved in Enterococcus faecalis Vaginal Adherence and Persistence. Infect Immun 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M, Aly KA, Chen YH, and Missiakas D (2013) Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol Microbiol 90: 734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avican K, Aldahdooh J, Togninalli M, Mahmud A, Tang J, Borgwardt KM, Rhen M, and Fallman M (2021) RNA atlas of human bacterial pathogens uncovers stress dynamics linked to infection. Nat Commun 12: 3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, and Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, and Finn RD (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49: D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovich T, Shkoporov AN, Ross RP, and Hill C (2022) Phage-mediated horizontal gene transfer and its implications for the human gut microbiome. Gastroenterol Rep (Oxf) 10: goac012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman L, and Palmer T (2021) The Type VII Secretion System of Staphylococcus. Annu Rev Microbiol 75: 471–494. [DOI] [PubMed] [Google Scholar]
- Bowran K, and Palmer T (2021) Extreme genetic diversity in the type VII secretion system of Listeria monocytogenes suggests a role in bacterial antagonism. Microbiology (Reading) 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham LR, Akbari MS, Alhajjar N, Keogh RA, Radin JN, Kehl-Fie TE, Belew AT, El-Sayed NM, McIver KS, and Doran KS (2022a) Genomic Analyses Identify Manganese Homeostasis as a Driver of Group B Streptococcal Vaginal Colonization. mBio 13: e0098522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham LR, Bath JR, Werlang CA, Lyon LM, Liu N, Evans C, Ribbeck K, and Doran KS (2022b) Role of MUC5B during Group B Streptococcal Vaginal Colonization. mBio 13: e0003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burcham LR, Spencer BL, Keeler LR, Runft DL, Patras KA, Neely MN, and Doran KS (2019) Determinants of Group B streptococcal virulence potential amongst vaginal clinical isolates from pregnant women. PLoS One 14: e0226699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Casabona MG, Kneuper H, Chalmers JD, and Palmer T (2016) The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2: 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion PA, Stanley SA, Champion MM, Brown EJ, and Cox JS (2006) C-terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis. Science 313: 1632–1636. [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Willett JLE, Dunny GM, and Duerkop BA (2021) Phage infection and sub-lethal antibiotic exposure mediate Enterococcus faecalis type VII secretion system dependent inhibition of bystander bacteria. PLoS Genet 17: e1009204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M, Armistead B, Orvis A, Quach P, Brokaw A, Gendrin C, Sharma K, Ogle J, Merillat S, Dacanay M, Wu TY, Munson J, Baldessari A, Vornhagen J, Furuta A, Nguyen S, Adams Waldorf KM, and Rajagopal L (2021) Hyaluronidase Impairs Neutrophil Function and Promotes Group B Streptococcus Invasion and Preterm Labor in Nonhuman Primates. mBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook LCC, Hu H, Maienschein-Cline M, and Federle MJ (2018) A Vaginal Tract Signal Detected by the Group B Streptococcus SaeRS System Elicits Transcriptomic Changes and Enhances Murine Colonization. Infect Immun 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez MJ, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, Consortium D, Holden MTG, Poyart C, and Glaser P (2014) Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 5: 4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Wang Y, Liu Q, Gao Q, Lu H, Meng H, Qin J, Hu M, and Li M (2017) A Novel ESAT-6 Secretion System-Secreted Protein EsxX of Community-Associated Staphylococcus aureus Lineage ST398 Contributes to Immune Evasion and Virulence. Front Microbiol 8: 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CM, Luirink J, and Bitter W (2012) General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci U S A 109: 11342–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick RM, Aull HG, Jacobs-Sera D, Garlena RA, Russell DA, Smith BE, Mahalingam V, Abad L, Gauthier CH, and Hatfull GF (2021) The Prophage and Plasmid Mobilome as a Likely Driver of Mycobacterium abscessus Diversity. mBio 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Mu R, Weston TA, Spencer BL, Liles RP, and Doran KS (2018) Characterization of a Two-Component System Transcriptional Regulator, LtdR, That Impacts Group B Streptococcal Colonization and Disease. Infect Immun 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Schilcher K, Burcham LR, Kwiecinski JM, Johnson PM, Head SR, Heinrichs DE, Horswill AR, and Doran KS (2019) Identification of Key Determinants of Staphylococcus aureus Vaginal Colonization. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, Ronneberger O, Bodenstein S, Zielinski M, Bridgland A, Potapenko A, Cowie A, Tunyasuvunakool K, Jain R, Clancy E, Kohli P, Jumper J, and Hassabis D (2022) Protein complex prediction with AlphaFold-Multimer. bioRxiv. [Google Scholar]
- Faralla C, Metruccio MM, De Chiara M, Mu R, Patras KA, Muzzi A, Grandi G, Margarit I, Doran KS, and Janulczyk R (2014) Analysis of two-component systems in group B Streptococcus shows that RgfAC and the novel FspSR modulate virulence and bacterial fitness. MBio 5: e00870–00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett SR, Mariano G, Dicks J, and Palmer T (2022) Homologous recombination between tandem paralogues drives evolution of a subset of type VII secretion system immunity genes in firmicute bacteria. Microb Genom 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, and Bryant SH (2002) CDART: protein homology by domain architecture. Genome Res 12: 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Brodersen DE, Mitarai N, and Gerdes K (2018) Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol Cell 70: 768–784. [DOI] [PubMed] [Google Scholar]
- Hobbs M, and Mattick JS (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol 10: 233–243. [DOI] [PubMed] [Google Scholar]
- Hooven TA, Randis TM, Daugherty SC, Narechania A, Planet PJ, Tettelin H, and Ratner AJ (2014) Complete Genome Sequence of Streptococcus agalactiae CNCTC 10/84, a Hypervirulent Sequence Type 26 Strain. Genome Announc 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager F, Kneuper H, and Palmer T (2018) EssC is a specificity determinant for Staphylococcus aureus type VII secretion. Microbiology (Reading) 164: 816–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulczyk R, Masignani V, Maione D, Tettelin H, Grandi G, and Telford JL (2010) Simple sequence repeats and genome plasticity in Streptococcus agalactiae. J Bacteriol 192: 3990–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, and Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh RA, Haeberle AL, Langouët-Astrié CJ, Kavanaugh JS, Schmidt EP, Moore GD, Horswill AR, and Doran KS (2022) Group B Streptococcus Adaptation Promotes Survival in a Hyperinflammatory Diabetic Wound Environment. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DE, Chivian D, and Baker D (2004) Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res 32: W526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TA, Grebenc DW, Shah PY, McArthur OD, Dickson BH, Surette MG, Kim Y, and Whitney JC (2022) Dual Targeting Factors Are Required for LXG Toxin Export by the Bacterial Type VIIb Secretion System. mBio: e0213722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TA, Pazos M, Surette MG, Vollmer W, and Whitney JC (2018) Molecular Basis for Immunity Protein Recognition of a Type VII Secretion System Exported Antibacterial Toxin. J Mol Biol 430: 4344–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jager F, Cargill JS, Chalmers J, van der Kooi-Pol MM, van Dijl JM, Ryan RP, Hunter WN, and Palmer T (2014) Heterogeneity in ess transcriptional organization and variable contribution of the Ess/Type VII protein secretion system to virulence across closely related Staphylocccus aureus strains. Mol Microbiol 93: 928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K (2021) Diverse LXG toxin and antitoxin systems specifically mediate intraspecies competition in Bacillus subtilis biofilms. PLoS Genet 17: e1009682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Dai J, Tang H, Zhang S, Wu C, Qiu W, Lu C, Yao H, Fan H, and Wu Z (2017) Streptococcus suis serotype 9 strain GZ0565 contains a type VII secretion system putative substrate EsxA that contributes to bacterial virulence and a vanZ-like gene that confers resistance to teicoplanin and dalbavancin in Streptococcus agalactiae. Vet Microbiol 205: 26–33. [DOI] [PubMed] [Google Scholar]
- Lebeurre J, Dahyot S, Diene S, Paulay A, Aubourg M, Argemi X, Giard JC, Tournier I, Francois P, and Pestel-Caron M (2019) Comparative Genome Analysis of Staphylococcus lugdunensis Shows Clonal Complex-Dependent Diversity of the Putative Virulence Factor, ess/Type VII Locus. Front Microbiol 10: 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Wu H, Bian C, Chen H, Shen Y, Gao X, Ma J, Yao H, Wang L, and Wu Z (2022) The antimicrobial systems of Streptococcus suis promote niche competition in pig tonsils. Virulence 13: 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, and Steinegger M (2022) ColabFold: making protein folding accessible to all. Nat Methods 19: 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandyal RR (2008) Update on group B streptococcal infections: perinatal and neonatal periods. J Perinat Neonatal Nurs 22: 230–237. [DOI] [PubMed] [Google Scholar]
- Navarro-Torne A, Curcio D, Moisi JC, and Jodar L (2021) Burden of invasive group B Streptococcus disease in non-pregnant adults: A systematic review and meta-analysis. PLoS One 16: e0258030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Gibson RL, Chi EY, Framson PE, Hulse M, and Rubens CE (1996) Group B streptococcal beta-hemolysin expression is associated with injury of lung epithelial cells. Infect Immun 64: 3818–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohr RJ, Anderson M, Shi M, Schneewind O, and Missiakas D (2017) EssD, a Nuclease Effector of the Staphylococcus aureus ESS Pathway. J Bacteriol 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura CY, and Nizet V (2014) Subterfuge and sabotage: evasion of host innate defenses by invasive gram-positive bacterial pathogens. Annu Rev Microbiol 68: 439–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Muller S, and Wollscheid B (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30: 884–886. [DOI] [PubMed] [Google Scholar]
- Pallen MJ (2002) The ESAT-6/WXG100 superfamily -- and a new Gram-positive secretion system? Trends Microbiol 10: 209–212. [DOI] [PubMed] [Google Scholar]
- Patras KA, and Doran KS (2016) A Murine Model of Group B Streptococcus Vaginal Colonization. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patras KA, and Nizet V (2018) Group B Streptococcal Maternal Colonization and Neonatal Disease: Molecular Mechanisms and Preventative Approaches. Front Pediatr 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ZN, Tram G, Seib KL, and Atack JM (2019) Phase-variable bacterial loci: how bacteria gamble to maximise fitness in changing environments. Biochem Soc Trans 47: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Pimentel BA, Martins CA, Mendonca JC, Miranda PS, Sanches GF, Mattos-Guaraldi AL, and Nagao PE (2016) Streptococcus agalactiae infection in cancer patients: a five-year study. Eur J Clin Microbiol Infect Dis 35: 927–933. [DOI] [PubMed] [Google Scholar]
- Poulsen C, Panjikar S, Holton SJ, Wilmanns M, and Song YH (2014) WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern. PLoS One 9: e89313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajam G, Carlone GM, and Romero-Steiner S (2007) Functional antibodies to the O-acetylated pneumococcal serotype 15B capsular polysaccharide have low cross-reactivities with serotype 15C. Clin Vaccine Immunol 14: 1223–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan JA, Klebanoff MA, and Nugent RP (1991) The epidemiology of group B streptococcal colonization in pregnancy. Vaginal Infections and Prematurity Study Group. Obstet Gynecol 77: 604–610. [PubMed] [Google Scholar]
- Russell NJ, Seale AC, O’Sullivan C, Le Doare K, Heath PT, Lawn JE, Bartlett L, Cutland C, Gravett M, Ip M, Madhi SA, Rubens CE, Saha SK, Schrag S, Sobanjo-Ter Meulen A, Vekemans J, and Baker CJ (2017) Risk of Early-Onset Neonatal Group B Streptococcal Disease With Maternal Colonization Worldwide: Systematic Review and Meta-analyses. Clin Infect Dis 65: S152–S159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, LLC, (2015) The AxPyMOL Molecular Graphics Plugin for Microsoft PowerPoint, Version 1.8. In., pp.
- Sitkiewicz I, Green NM, Guo N, Bongiovanni AM, Witkin SS, and Musser JM (2009) Transcriptome adaptation of group B Streptococcus to growth in human amniotic fluid. PLoS One 4: e6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer BL, Chatterjee A, Duerkop BA, Baker CJ, and Doran KS (2021a) Complete Genome Sequence of Neonatal Clinical Group B Streptococcal Isolate CJB111. Microbiol Resour Announc 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer BL, Deng L, Patras KA, Burcham ZM, Sanches GF, Nagao PE, and Doran KS (2019) Cas9 Contributes to Group B Streptococcal Colonization and Disease. Front Microbiol 10: 1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer BL, and Doran KS (2022) Evolving understanding of the type VII secretion system in Gram-positive bacteria. PLoS Pathog 18: e1010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer BL, Shenoy AT, Orihuela CJ, and Nahm MH (2017) The Pneumococcal Serotype 15C Capsule Is Partially O-Acetylated and Allows for Limited Evasion of 23-Valent Pneumococcal Polysaccharide Vaccine-Elicited Anti-Serotype 15B Antibodies. Clin Vaccine Immunol 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer BL, Tak U, Mendonca JC, Nagao PE, Niederweis M, and Doran KS (2021b) A type VII secretion system in Group B Streptococcus mediates cytotoxicity and virulence. PLoS Pathog 17: e1010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy R, Fyfe PK, and Hunter WN (2008) Structure of Staphylococcus aureus EsxA suggests a contribution to virulence by action as a transport chaperone and/or adaptor protein. J Mol Biol 383: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sysoeva TA, Zepeda-Rivera MA, Huppert LA, and Burton BM (2014) Dimer recognition and secretion by the ESX secretion system in Bacillus subtilis. Proc Natl Acad Sci U S A 111: 7653–7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JC, Gao X, Xu J, Holder M, Petrosino J, Kumar R, Liu W, Hook M, Mackenzie C, Hillhouse A, Brashear W, Nunez MP, and Xu Y (2021) A type VII secretion system of Streptococcus gallolyticus subsp. gallolyticus contributes to gut colonization and the development of colon tumors. PLoS Pathog 17: e1009182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh WK, Ding Y, Gubellini F, Filloux A, Poyart C, Dramsi S, and Givskov M (2022) Characterization of TelE, an LXG effector of Streptococcus gallolyticus, antagonized by a non-canonical immunity protein. bioRxiv. [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, Paulsen IT, Nelson KE, Margarit I, Read TD, Madoff LC, Wolf AM, Beanan MJ, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Lewis MR, Radune D, Fedorova NB, Scanlan D, Khouri H, Mulligan S, Carty HA, Cline RT, Van Aken SE, Gill J, Scarselli M, Mora M, Iacobini ET, Brettoni C, Galli G, Mariani M, Vegni F, Maione D, Rinaudo D, Rappuoli R, Telford JL, Kasper DL, Grandi G, and Fraser CM (2002) Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc Natl Acad Sci U S A 99: 12391–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HR, Grebenc DW, Klein TA, and Whitney JC (2021) Bacterial type VII secretion: An important player in host-microbe and microbe-microbe interactions. Mol Microbiol 115: 478–489. [DOI] [PubMed] [Google Scholar]
- Ulhuq FR, Gomes MC, Duggan GM, Guo M, Mendonca C, Buchanan G, Chalmers JD, Cao Z, Kneuper H, Murdoch S, Thomson S, Strahl H, Trost M, Mostowy S, and Palmer T (2020) A membrane-depolarizing toxin substrate of the Staphylococcus aureus type VII secretion system mediates intraspecies competition. Proc Natl Acad Sci U S A 117: 20836–20847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan M, Constantinidou C, Palmer T, and Pallen MJ (2017) The Enigmatic Esx Proteins: Looking Beyond Mycobacteria. Trends Microbiol 25: 192–204. [DOI] [PubMed] [Google Scholar]
- van Kassel MN, Bijlsma MW, Brouwer MC, van der Ende A, and van de Beek D (2019) Community-acquired group B streptococcal meningitis in adults: 33 cases from prospective cohort studies. J Infect 78: 54–57. [DOI] [PubMed] [Google Scholar]
- van Selm S, van Cann LM, Kolkman MA, van der Zeijst BA, and van Putten JP (2003) Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect Immun 71: 6192–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrbanac A, Riestra AM, Coady A, Knight R, Nizet V, and Patras KA (2018) The murine vaginal microbiota and its perturbation by the human pathogen group B Streptococcus. BMC Microbiol 18: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warne B, Harkins CP, Harris SR, Vatsiou A, Stanley-Wall N, Parkhill J, Peacock SJ, Palmer T, and Holden MT (2016) The Ess/Type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics 17: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling CC, Refardt D, and Hall AR (2021) Fitness benefits to bacteria of carrying prophages and prophage-encoded antibiotic-resistance genes peak in different environments. Evolution 75: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JC, Peterson SB, Kim J, Pazos M, Verster AJ, Radey MC, Kulasekara HD, Ching MQ, Bullen NP, Bryant D, Goo YA, Surette MG, Borenstein E, Vollmer W, and Mougous JD (2017) A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson HW (1977) Nontypable group B streptococci isolated from human sources. J Clin Microbiol 6: 183–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson HW (1978) Group B streptococcal infection in humans. Annu Rev Microbiol 32: 41–57. [DOI] [PubMed] [Google Scholar]
- Yang D, Wang S, Sun E, Chen Y, Hua L, Wang X, Zhou R, Chen H, Peng Z, and Wu B (2022) A temperate Siphoviridae bacteriophage isolate from Siberian tiger enhances the virulence of methicillin-resistant Staphylococcus aureus through distinct mechanisms. Virulence 13: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Boardman E, Deme J, Alcock F, Lea S, and Palmer T (2023) EsxB, EsxC and EsxD facilitate export of the antibacterial nuclease EsaD by the Staphylococcus aureus Type VII secretion system. bioRxiv: 2023.2004.2001.535202. [Google Scholar]
- Zhang D, de Souza RF, Anantharaman V, Iyer LM, and Aravind L (2012) Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Xie L, Xu X, and Sun J (2022) Comparative Genomic Analysis of Type VII Secretion System in Streptococcus agalactiae Indicates Its Possible Sequence Type-Dependent Diversity. Front Cell Infect Microbiol 12: 880943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Fig. 1. Comparison of GBS T7SS effectors to other Gram-positive organisms. Heat map comparing CJB111 T7SS protein homology to a panel of eight Gram-positive organisms in which the T7SSb has been studied previously: S. aureus (variants 1–4), S. lugdunensis (variants 1, 2), Listeria monocytogenes (variants 1–7), E. faecalis (strain OG1RF), Bacillus subtilis (PY79), S. gallolyticus (strain TX20005), S. intermedius (strains B196 and GC1825), and S. suis (strains GZ5065 and WUSS351). See Supp. Table 2 for individual strain information. Heat map color intensity is based on Geneious alignment grade, which considers query coverage and percent identity. Shading of the locus tag numbers on the x-axis corresponds to the key for gene color in Fig 1. Most downstream CJB111 T7SS effectors have little to no homology across species.
Supp. Fig. 2. Subtype-specific LXG-associated proteins are encoded by GBS T7SS.
Percent identity matrices and Clustal Omega alignments of A-C) LXG-associated protein 1 (Lap1) and D-F) LXG-associated protein 2 (Lap2) sequences from CJB111 (subtype I), 2603V/R (subtype II), CNCTC 10/84 (subtype III), and C001 (subtype III). In the above matrices, the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity. Conserved putative T7SS-associated FXG and FxxxD/E motifs are highlighted by red boxes in C.
Supp. Fig. 3. Confidence scores and predicted aligned error for LXG-Lap AlphaFold models in Fig. 2A. A) Predicted LXG-Lap complex models shown in Fig. 2A but with color corresponding to per-residue confidence level (predicted local distance difference test [pLDDT] score 1–100; pLDDT > 90 are expected to be modelled to high accuracy). B) Predicted aligned error for each LXG-Lap complex, with colors indicating the confidence of domain positions (higher predicted error in red, lower predicted error in blue).
Supp. Fig. 4. Modeling of full-length LXG proteins and homology of downstream putative immunity factor and DUF4176 genes
A) Robetta predicted structures for full-length putative GBS LXG toxins across subtypes I, II, and III. B) Percent identity matrix of putative immunity factors (transmembrane domain containing proteins) encoded for downstream of LXG genes in CJB111 (subtype I), 2603V/R (subtype II), CNCTC 10/84 (subtype III), and C001 (subtype III). C-E) Percent identity matrices and Clustal Omega alignments of DUF4176 protein sequences from CJB111 (subtype I), 2603V/R (subtype II), CNCTC 10/84 (subtype III), and COH1 (subtype IV). Green highlighting in C-E indicate orphaned DUF4176 proteins. A conserved central FXG motif within GBS DUF4176 proteins is highlighted by a red box in D. In all of the above percent identity matrices, the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity.
Supp. Fig. 5. Transcriptional landscape of the CJB111 T7SS locus
RT-PCR was performed to evaluate transcriptional organization of the CJB111 T7SS locus. Using cDNA as template (and no RT-cDNA and genomic DNA as negative and positive controls, respectively), PCRs were performed using primer pairs spanning every gene junction in the putative T7SS locus. Primers used for these experiments can be found in Supp. Table 5. Agarose gels shown are representative of three independent experiments.
Supp. Fig. 6. Homology of orphaned GBS WXG100 proteins.
A) Locus tag tables and percent identity matrices of B) Orphaned Module 1 WXG100 protein EsxA3 and C) Orphaned Module 2 WXG100 protein EsxA4 sequences across a panel of GBS isolates representing T7SS subtypes I-IV. Green highlighting in A indicates orphaned WXG100 proteins. In the above matrices (B-C), the purple shading corresponds to the level of identity between two strains (on a spectrum of 0 to 100% identity), with darker shading indicative of higher percent identity.
Supp. Fig 7. Impact of EssC deficiency on GBS vaginal colonization by T7SS subtype I strain A909. A) Western blot showing EssC-dependent secretion of EsxA from subtype I strain A909, in vitro. B) Percent colonization curve of 8-week-old CD1 female mice vaginally inoculated with subtype I strain A909 or A909ΔessC. Statistics reflect the Log rank (Mantel-Cox) test. Recovered CFU counts from the C) vaginal D) cervical, and E) uterine tissue of colonized mice. In panels C-E, each dot represents one mouse and two independent experiments’ data are combined in these figures (n = 20/group total). Plots show the median and statistics represent the Mann Whitney U test.
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.


