Abstract
Optical microscopes are an important imaging tool that have effectively advanced the development of modern biomedicine. In recent years, super-resolution microscopy (SRM) has become one of the most popular techniques in the life sciences, especially in the field of living cell imaging. SRM has been used to solve many problems in basic biological research and has great potential in clinical application. In particular, the use of SRM to study drug delivery and kinetics at the subcellular level enables researchers to better study drugs’ mechanisms of action and to assess the efficacy of their targets in vivo. The purpose of this paper is to review the recent advances in SRM and to highlight some of its applications in assessing subcellular drug dynamics.
Keywords: super-resolution microscopy, absorption and transport, distribution, metabolism and excretion, drug design, nanoparticles
Graphical Abstract
1. Introduction
Many drugs need to enter specific sub-organelles in order to reach their targets. Furthermore, the uptake and movement of these micro- and macromolecules into cells significantly control some of the pharmacokinetic and pharmacodynamic parameters thus regulating cell responses to exogenous and endogenous stimuli[1–3]. Classical pharmacokinetic theory is based on plasma drug concentration measurement; however, with this approach it is often difficult to predict drug efficacy in vivo, as many drugs must penetrate multiple biological barriers and bind to targets within cells to be effective[4–6]. Therefore, since the subcellular operation and metabolism of drugs are the basis of drug function[7], it is urgently needed for pharmacokinetic studies to go from the “macroscopic” plasma drug concentration to the “microscopic” cellular/subcellular level. Furthermore, this ability of pharmacologically active molecules to selectively discover their targets is closely related to their potential as successful therapeutic agents[8, 9]. In addition to potentially achieving a higher therapeutic index than targeting tissues and cells, subcellular targeting also considers the limitations of systemic drug delivery and significantly improves drug dynamics[10, 11]. Thus, the study of drug dynamics at the subcellular level has become a strong basis of the development of clinical pharmacokinetics.
In recent years, optical microscopy has been widely used to study pharmacokinetics at the subcellular level[12–14]. However, conventional optical microscopes are limited by Abbe diffraction and lack sufficient spatial resolution to observe smaller biomolecules and structures[15–17]. With the advent of scanning electron microscopy, scanning tunneling microscopy, and atomic force microscopy, nanoscale resolution has become possible[18–20]. However, these techniques are not suitable for the observation of biological samples, especially living samples, because they are destructive and can only observe the sample surface. In recent decades, a series of super-resolution microscopy (SRM) techniques that break the optical diffraction limit have become a powerful tool in cell biology to study the fine structure of organelles, the interactions between organelles, and the function of proteins[21–24].
There are three main types of SRM techniques: stimulated emission depletion microscopy (STED), which is imaged by point scanning with two beams of light[25], single-molecule localization microscopy (SMLM)[26], represented by photoactivated localization microscopy (PALM) and random optical reconstruction microscopy (STORM)[27, 28], and structured illumination microscopy (SIM), which is based on structured light illumination imaging[29]. Using these advanced technologies, researchers not only locate and monitor the behavior of drugs at the subcellular level but also track drugs’ changes within different organelles and their effects on organelle interactions[30–32]. This includes (1) observing drugs’ route and ease of transmembrane entry[33, 34], (2) evaluating the accumulation and metabolism of drugs in different organelles[30], and (3) determining the key proteins of regulation and organelle specific targeting[35]. However, in pharmacokinetic studies at the subcellular level, the systematic presentation of imaging strategies for each organelle is rare. Here, we give a brief review of these techniques and their applications in pharmacokinetics. We expect that this review will help researchers identify the technical advantages and relevant applications of various SRM techniques, thereby helping researchers successfully utilize these novel technologies in their future work.
2. SRM techniques
2.1. The advent of SRM
Light microscopes are an important tool that help people see the microscopic world[36]. In order to advance this exploration, the resolution of microscopes continuously advances so that people can see smaller and smaller things[37]. However, the diffraction of light limits the resolution of the optical microscope, making it unable to distinguish the microstructure within 200 nm[38]. To overcome the diffraction limit, starting in the 1990s, the unremitting efforts of scientists such as Stefan Hell, William E. Moerner, Eric Betzig, Xiaowei Zhuang, and Mats G. L. Gustafsson led to the maturity of SRM technology—including STED, SMLM, and SIM—around 2008[39–41]. In the past 20 years, SRM technology has been successfully applied to major research projects in the life and material sciences, and it has shown prospects of broad application[42–47].
2.2. Development of SRMs
2.2.1. STED
The STED technique was the first far-field super-resolution imaging method to be proposed theoretically and realized experimentally[48]. Drawing on Einstein’s theory of stimulated emission, German scientist Stefan W. Hell created STED in 1999 by suppressing spontaneous fluorescence radiation through stimulated radiation[49]. The basic principle of STED is that two laser beams are used to irradiate the sample at the same time. One laser beam spurs the transition of the fluorescence molecules within the Airy disc in the focus of the objective lens from the ground state to the excited state. Another circular quenching light (also known as STED light) makes the excited fluorescent molecules in the edge region of the Airy disc in the focus of the objective lens return to the ground state through the process of radiation loss; the excited molecules in the Airy disc in the focus are not affected and continue to return to the ground state by way of spontaneous fluorescence. In this way, the excited fluorescent molecules in the center can be distinguished from the ground state fluorescent molecules in the surrounding edge. The fluorescence spots beyond the diffraction limit are obtained by reducing the diffraction spots of the fluorescence point, thereby improving the resolution of the microscope[50]. This principle can actually be illustrated by a more graphic example. As shown in Fig. 1A, the green circles represent excited light, while the red circles with a white center symbolize quenching light. The red doughnut-shaped ring with a white center works as an eraser, blocking the emitting photons from the edge of the green circles. As a result, only the central points of the green circle remain illuminated and observable, hence, the resolution of the microscope has been greatly increased.
Figure 1.
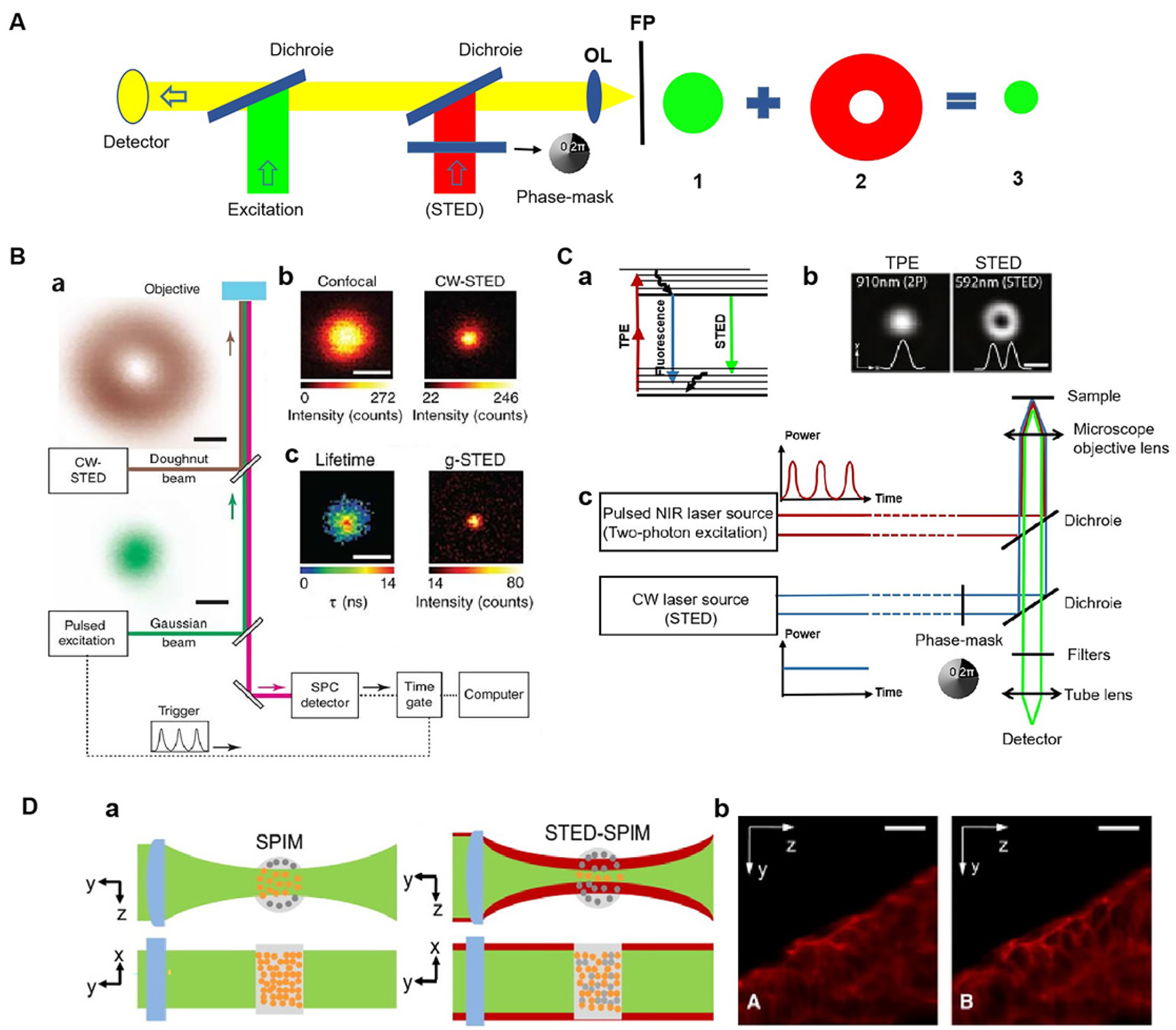
Principle and application of SRM based on STED microscope. (A) Principle of STED. (B) Principle of g-STED. (a) Microscope setup with pulsed excitation and CW-STED lasers. (b) Fluorescence images of a single isolated NV center for confocal (left) and CW-STED (right, PSTED=47 mW). (c) g-STED (right, Tg=15 ns) and a fluorescence lifetime image for the CW-STED recording (left). Fig. 1B reproduced from Ref [53]. (C) Principles of the reported implementation of TPE-STED microscopy. (a) Energy diagram of the 3 processes involved. (b) Light intensity distribution of the two-photon excitation spot (left) and of the STED doughnut (right) in the focal plan. (c) Setup of TPE-STED. Fig. 1C (b) reproduced from Ref [58]. (D) (a) Principles of SPIM and STED-SPIM. (b) Comparison of SPIM and STED-SPIM imaging of actin in zebrafish embryos stained with ATTO647 phalloidin. Fig. 1D reproduced from Ref [61]. Scale bars: (B) 200 nm, (C) 500 nm, (D) 20 μm.
Some advantages of STED technology include that samples can be imaged by scanning without a need for later image reconstruction and real-time changes in living cells can be observed quickly[25]. In addition, STED data processing only arranges corresponding points in specific locations of the images[51], so it has no false positives and is more reliable. At present, the main drawback of STED imaging is that the laser power depletion is too high, which makes it easily susceptible to photobleaching[52]. In response to this problem, in 2011, Vicidomini et al. proposed time-gate-based STED (g-STED) microscopy technology (Fig 1A)[53]. g-STED removes short-lived spontaneous fluorescence at the edge of the excitation light through real-time filtering of fluorescence signals. This greatly reduces the required fluorescence intensity (attenuation by a factor of 3) and improves the system resolution[53]. Then, in 2014, Hao et al. proposed a method to control the spot size in the middle of the ring depletion light: they used a pair of prosthesis prisms to realize image inversion and phase modulation to control the size of the middle spot of the ring depletion light, so that the size of the middle spot can vary between 0.555 and 0.830[54, 55]. If the resolution does not need to be improved, the intensity of the depletion light can be reduced by 33% compared to the common depletion light.
Moreover, an additional drawback of STED microscopy is that it uses single-photon excitation (linear excitation) of fluorescent probes, which has poor optical penetration. This results in the production of large aberrations and overall reduction in the system’s resolution when imaging thick biological samples. Consequently, STED microscopy can only be used for tissue surface imaging (<100 μm)[56]. To address the imaging needs of thicker biological samples, in 2009, Hell et al. developed TPE-STED microscopy by combining two-photon excited fluorescence microscopy with STED, thereby replacing the single-photon excitation in conventional STED microscopy with two-photon excitation [57]. The spatial resolution was greatly improved to less than 50 nm for all sides in the focal plane, and the imaging depth was extended (Fig 1B)[58]. Without an emphasis on super resolution, the resolution of general STED can be obtained by reducing the intensity of light depletion. In addition, two-photon excitation and stimulated emission depletion can be achieved by using a laser of the same wavelength instead of single-molecule excitation. Thus, only one laser is needed to realize high-resolution imaging, which not only reduces the system cost but also simplifies the imaging system[59]. Then, in 2011, Harms et al. were the first to combine selective planar microscopy (SPIM) with STED[60]; this maximized the advantages of fast STED imaging speed, deep SPIM imaging depth, and low light intensity, and it improved the axial resolution by 60%. In this way, rapid and high-resolution imaging of living organisms with thickness of >100 μm can be achieved (Fig 1C)[61]. In 2012, Hell’s group achieved a lateral resolution of 2.4 nm using solid-state immersion technology (NA = 2.2), which is the highest resolution ever reported for STED microscopy[62].
2.2.2. STORM/PALM
SMLM is an imaging technique that uses fluorescence to obtain super-resolution images of biological samples. SMLM includes PALM, STORM and imaging technologies derived from these two technologies. In 1994, Eric Betzig proposed PALM and achieved PALM SRM imaging in 2006[63]. In the same year, Zhuang’s group developed STORM, a super-resolution imaging technique based on single-molecule fluorescence detection and applied it to achieve 3D super-resolution imaging[63]. The basis of SMLM is total internal reflection illuminated light path. First, a specific wavelength of laser is used to activate the fluorescent molecules, and then another wavelength of laser is used to excite them. Because the intensity of the laser is controlled, only a few random, discrete fluorescent molecules are activated in the imaging region at a time. A single molecule positioning algorithm (such as two-dimensional Gaussian fitting) is used to accurately locate the central position of a single molecule, and super-resolution images can be obtained by superimposing and reconstructing the collected image information (Fig 2A)[63]. In simple terms, high-resolution imaging is achieved by using the scintillation signal emitted by a single fluorescent molecule. This signal helps pinpoint the location of a single molecule within a sample. The main difference between PALM and STORM is the labeling method with PALM technology utilizing the photoactivation effect of fluorescent proteins, while STORM technology uses fluorescent probes and immunofluorescence [64].
Figure 2.
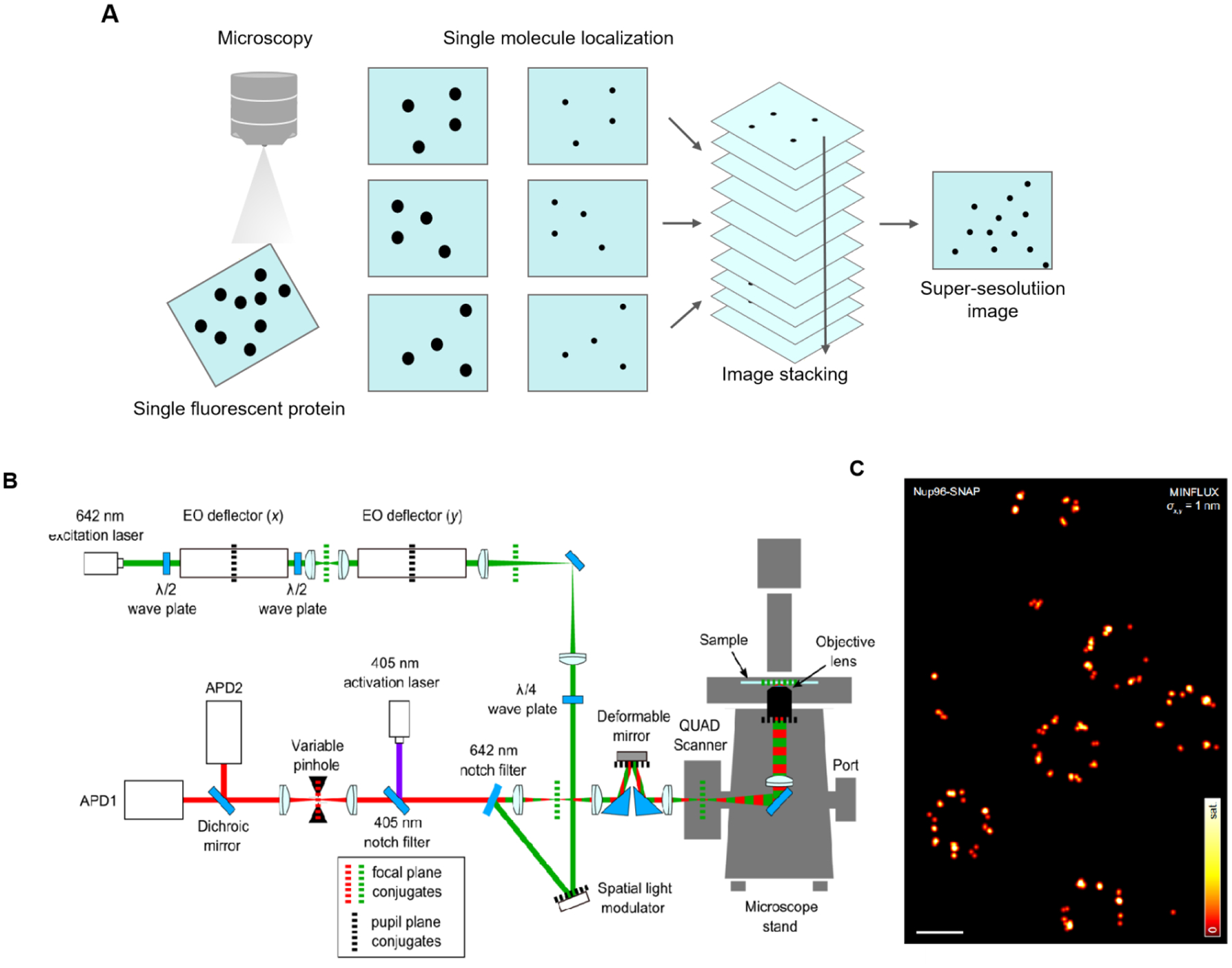
Principle and application of SRM based on SMLM. (A) Principle of SMLM. (B) MINFLUX optical arrangement. An excitation beam (shown in green) is electro-optically deflected in x,y plane, spatially phase-modulated for conversion into a donut-shape, overlapped with an activation beam (purple), and after passing a deformable mirror and a galvanometer scanner in a 3D scanning assembly, it is focused into the sample on top of an all-purpose inverted microscope stand. Fluorescence from the sample (red) is descanned by the scanning assembly and passed to a variable confocal pinhole for detection using two avalanche photodiodes (APD). A stabilization unit based on both near-infrared scattering from fiducial markers and active-feedback correction provides sub-nanometer stability. (C) Principle of MINFLUX nanoscopy. MINFLUX fluorescence imaging of labeled cellular ultrastructure down to 1 nm (standard deviation) in fluorophore precision. σ: the precision obtained from a combined analysis of the statistical localization spread (standard deviation) in x and y. Fig. 2 B, C reproduced from Ref [70]. Scale bars: 200 nm.
SMLM technology has the highest spatial resolution of the current super-resolution imaging methods[65]. Its spatial resolution primarily depends on the positioning accuracy of monomolecular imaging and in theory can reach 1 nm. Yet, limited by many factors, the current transverse and longitudinal resolutions of SMLM can only reach 20 nm and 50 nm, respectively. Additionally, the cyclic imaging process requires repeated activation and quenching of fluorescent molecules, which leads to a longer imaging time and a lower temporal resolution[65].
In recent years, researchers have worked hard to improve its resolution. In terms of spatial resolution, in 2008, Zhuang et al. realized 3D-STORM imaging by using the cylindrical mirror diffusion method to make the transverse and longitudinal ratio of point spread function (PSF) change with the axial depth for the first time. At present, this is the most commonly used 3D-SMLM imaging method, with an imaging depth of approximately 1μm and an axial resolution of about 50 nm. In the same year, Huang et al. used optical astigmatism to achieve a resolution of 20 ~ 30 nm on the 3D-STORM xy-plane[66]. In 2010, Matsuda et al. used PALM technology to pre-process the acquired images through denoising and deconvolution, and they observed filament-like substances at 30 nm in the mitotic chromosomes of Drosophila melanogaster, greatly improving the spatial resolution of super-resolution imaging[67]. In 2017, Liu et al. developed a method that can effectively reduce autofluorescence and photobleaching of cyanobacteria and plant cells, and they obtained a transverse resolution of ~10 nm in Prochlorococcus spheroides and the flowering plant Arabidopsis using STORM[68]. In 2019, Ji et al. proposed a novel single-molecule interferometric localization super-resolution imaging method, which determines the precise position of fluorescent molecules through the phase relationship between fluorescence molecular intensity and interference fringe. DNA origami arrays with 5-nm spacing were successfully resolved at a resolution of approximately 3 nm[69]. Combining STED and SMLM, Stefan W Hell’s group proposed a super-resolution imaging technique with Minimal Photon Fluxes (MINFLUX) (Fig 2B), which could achieve a location accuracy of ~1 nm (Fig 2C) and an imaging accuracy of ~6 nm. This is currently the highest resolution single molecule localization imaging technology[70].
Meanwhile, the time resolution can be improved by increasing the laser intensity, but too high of a laser intensity will cause photobleaching and even damage living cell samples. In 2011, Zhuang et al. achieved 3D imaging of living cells by tagging cell transferrins with SNAP tags using a photoconversion dye; they achieved a lateral resolution of 30 nm, an axial resolution of 50 nm, and a temporal resolution of 1~2 s[71]. In 2017, Xu et al. developed Quick-3B algorithm, which increased the computing speed by a factor of 17 and greatly improved the time resolution[72]. Additionally, this algorithm can be applied to various microscopy techniques, such as total internal reflection fluorescence microscopy, PALM, STORM, and laminar microscopy.
2.2.3. SIM
In 2000, Gustafsson et al. experimentally obtained transverse spatial resolution of approximately 115 nm, which increased the resolution of super-resolution imaging to twice the diffraction limit resolution[73]. In 2005, Gustafsson’s group developed a SRM imaging technique called SIM[74]. The basic principle of SIM imaging is the use of a spatially structured light beam to excite fluorescence and modulate the high frequency signal. Fluorescence results are determined by changing the direction and phase of the pattern. Since the high frequency signal cannot be detected directly by the imaging objective to the detectable low pass band of its optical transfer function (OTF), the target high frequency signal is extracted from the original data and reconstructed by the algorithm to obtain the final super-resolution image. In short, two high frequency signals are superimposed to obtain a low frequency signal, and then the method is to use the low frequency signal and a known high frequency signal to restore the unknown high frequency signal to achieve high precision detection of a fine structure. The complete SIM imaging process includes high-quality raw data acquisition, PSF/OTF calibration of the system to match the imaging conditions, and SIM algorithm reconstruction processing. SIM’s resolution in the focal plane is about twice that of conventional fluorescence microscopy, i.e., about 100 nm[75].
Compared with STED and SMLM, SIM has the advantages of fast imaging speed, strong dye compatibility, and simple sample preparation. It is a super-resolution imaging technology that is especially suitable for dynamic imaging of living cells[76, 77]. However, its resolution is far behind that of STED and SMLM[21].
Recent years have seen some important improvements in SIM resolution. In 2005, Gustafsson et al. proposed saturated structured illumination microscopy (SSIM), which is based on the nonlinear effects of fluorophorescence under laser light[78]. The horizontal resolution of traditional SIM is improved to about 40 nm; however, due to the long excitation time and the high excitation power, the use of this technique is limited in live cell imaging applications. In 2008, Gustafsson et al. developed 3D-SIM technology, which increased the horizontal resolution to about 100 nm and the vertical resolution to about 300 nm (Fig 3)[79]. Then in 2012, Gustafsson et al. developed a nonlinear structured illumination microtechnique (NLSIM) by capitalizing on the fluorescent protein Dronpa’s ability to reversibly activate and deactivate its signal. In comparison to fluorescence saturation, this simultaneously reduces the excitation light intensity and realizes nonlinear structural light illumination. They used this reversible fluorescent protein to observe purified microtubules, nuclear pore complexes, and actin fibers in stationary cells. This technique simultaneously reduced the required excitation light intensity by six orders of magnitude and achieved a spatial resolution of approximately 50 nm thus revealing the possibility of imaging living cells of < 50 nm[80]. In 2015, Li et al. proposed a novel nonlinear SIM technique combined with pattern activation, which achieved a spatial resolution of 62 nm in imaging of living cells[81].
Figure 3.
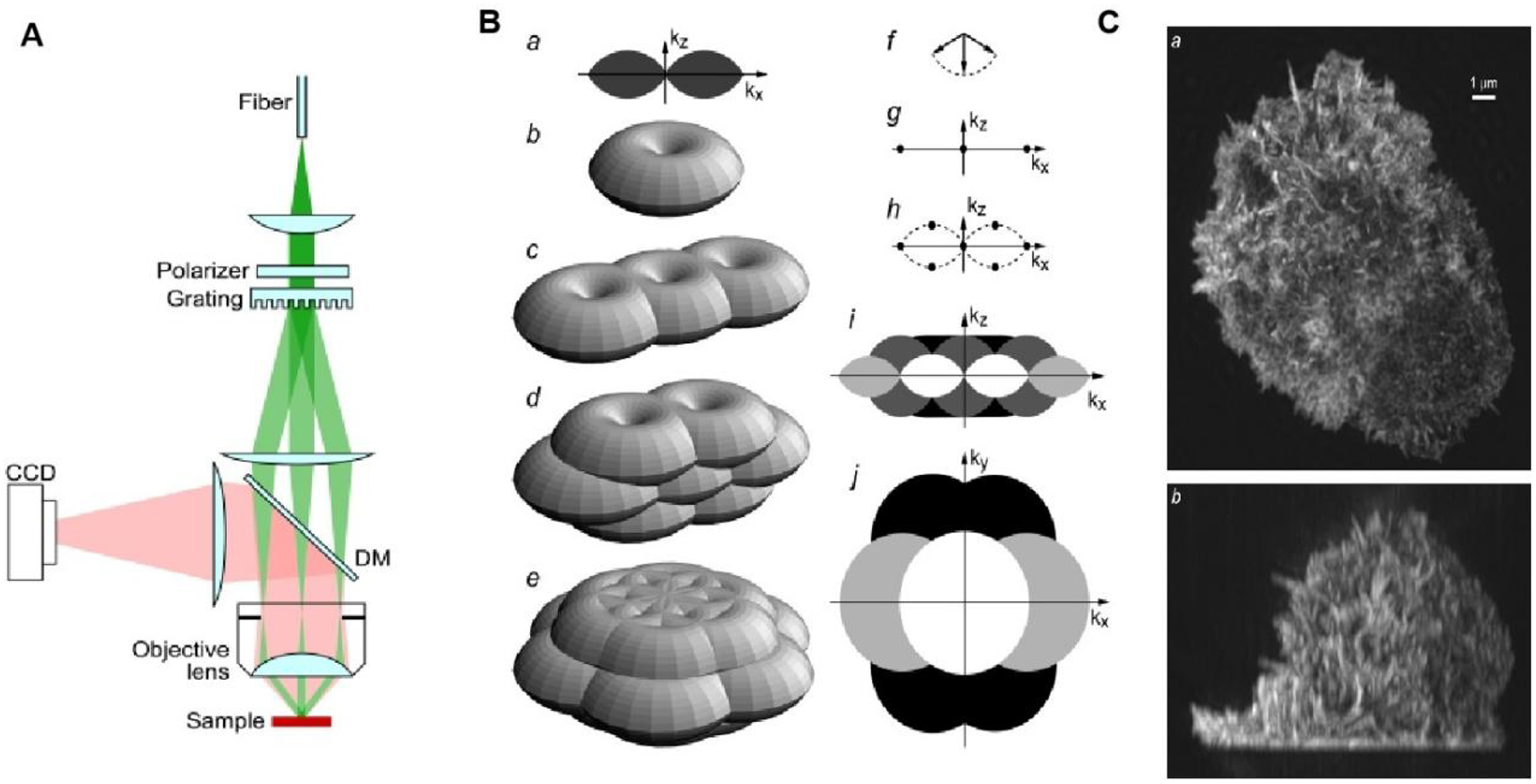
Principle and application of SRM based on SIM. (A) Simplified diagram of the structured illumination apparatus. Scrambled laser light from a multimode fiber is collimated onto a linear phase grating. Diffraction orders −1, 0 and +1 are refocused into the back focal plane of an objective lens. The beams, recollimated by the objective lens, intersect at the focal plane in the sample, where they interfere and generate an intensity pattern with both lateral and axial structure. (B) Principle of three-dimensional SIM. (a–e) Observable regions for (a and b) the conventional microscope, and for structured illumination microscopy using two illumination beams (c), and three illumination beams in one (d) or three (e) sequential orientations. (f) The three amplitude wave vectors corresponding to the three illumination beam directions. All three wave vectors have the same magnitude 1/l. (g–h) The resulting spatial frequency components of the illumination intensity for the two-beam (g) and three-beam (h) case. The dotted outline in panel h indicates the set of spatial frequencies that are possible to generate by illumination through the objective lens; compare with the observable region in panel a. An intensity component occurs at each pairwise difference frequency between two of the amplitude wave vectors. (i, j): xz (i) and xy (j) sections through the OTF supports in panel b (shown in white), panel c (light shaded), panel d (dark shaded), and panel e (solid). The darker regions fully contain the lighter ones. (C) Maximum intensity projections through a three-dimensional structured illumination reconstruction of the actin cytoskeleton in an HL-60 cell, shown in top view (a) and side view (b). Fig. 3 reproduced from Ref [79].
However, as a widefield-based SRM technology, SIM still contains some challenges. To start, the energy density of the excitation mode based on widefield illumination is low, which makes it easily affected by sample scattering during SIM imaging. In addition, its low tissue penetration depth mostly confines its application to the single-cell level. To address some of these limitations, in 2016, Legant et al. developed LS-Paint by combining lattice layer optical microscopy with point accumulation of nanoscale morphology microscopy imaging. This technique achieved high-precision localization of nerve mound in 20 μm-thick dense samples (including cell division and zebrafish embryos), greatly improving SIM penetration in tissue imaging[82].
Another flaw that conventional SIM exhibits is the difference observed between experimental data acquisition and post-acquisition data processing[38]. In particular, SIM image reconstruction is a pathological inverse process, which easily produces artifacts. To address this limitation and reduce the artifacts of reconstruction, almost all SIM algorithms have stringent requirements on the quality of the original image, the matching degree of the PSF used, and the parameters set by users. In 2023, Li et al. developed a high-fidelity SIM reconstruction algorithm based on PSF engineering[83]. By processing raw data of different quality, this method effectively overcomes the algorithm problems of conventional SIM that result in reconstruction artifacts and poor optical layer cutting ability, thereby producing high-quality super-resolution images and improving the fidelity of SIM imaging.
Yet despite these drawbacks, SIM imaging has become the most popular technology in the life sciences particularly for the multiple advantages it presents in live cell imaging.
2.3. Comparison of SRM techniques
By retaining the advantages of fluorescent microscopy while simultaneously surpassing the diffraction limit, SRM has revolutionized biological imaging and theoretically should have unrestricted spatial resolution[84]. The various techniques are essentially the same: each is photostressed and manipulates fluorescent radiation by switching fluorescent dye molecules on and off nonlinearly, and then records the details of the entire observation area in sequence over time[30]. Each of these technologies can achieve lateral resolution of more than 100 nm, and when combined with 3D technology, they can greatly improve axial resolution[85].
However, these technologies have obvious differences in their parameters and performance, and each has its own advantages and disadvantages[21]. First, laser scanning confocal microscopy technology is the basis of STED super-resolution imaging, so STED technology inherits the former’s characteristics of fast imaging speed and no need for later image reconstruction[25]. However, STED technology requires high-energy laser power, which not only increases the complexity of the optical system but also limits the types of dyes that can be used for imaging as well as its application in live-cell imaging[86].
Meanwhile, PALM/STORM technology can achieve much higher resolution than STED and SIM; however, its wide-field imaging requires the reconstruction of multiple original images to obtain a super-resolution image, resulting in poor temporal resolution, thus affecting its ability to observe dynamic processes in living cells[26, 65]. Moreover, the effectiveness of PALM/STORM technology varies based on application with the image acquisition of particles and fine fibers exhibiting higher quality than that of color slices.
Regarding SIM technology, it has been shown to be useful for 3D real-time imaging in multicellular organisms, with the advantages of fast speed, low phototoxicity, and the need for special fluorescent labeling[87–89]. However, compared to the other two technologies, its low horizontal and vertical resolutions still need to be further optimized[90].
At present, each of the three technologies has its own advantages and disadvantages (Fig 4), and none of them shows a clear lead: the choice of technology depends on the biological phenomenon under study and the characteristics of the sample. Yet, across the board, the highly specific and clear cellular and subcellular resolution offered by these techniques has realized high-quality imaging of intracellular ions, organelles, and living cells, leading to continuous leaps in biology. Of particular note here, the continuous improvement of these technologies has led more and more researchers to apply imaging towards the study of subcellular pharmacokinetics, making SRM a powerful tool for drug development and evaluation.
Figure 4.
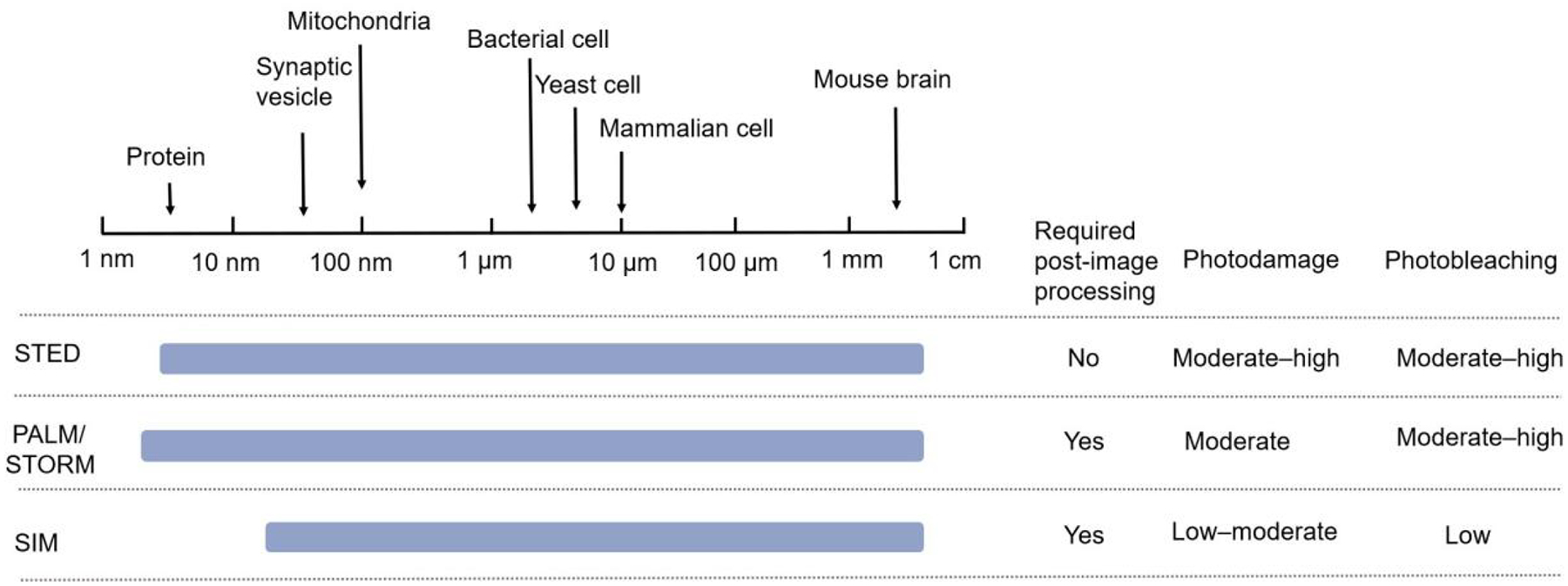
Comparison of SRM technologies.
3. SRM techniques for tracking drug dynamics at the subcellular level
3.1. Application of SRM techniques for tracking drug dynamics at the subcellular level
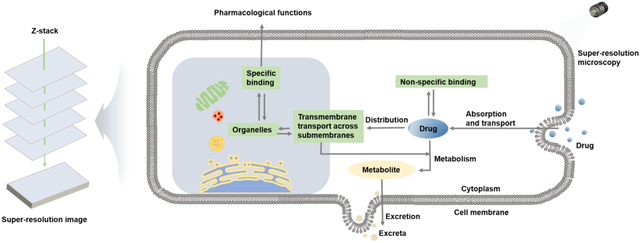
The effect of a drug is not only related to its concentration in the target tissue but also to its release at the specific target site, as a drug can only be effective if it is released at a specific site and binds to its target[3, 91]. Therefore, SRM technology is used to visualize the drug’s route of entry into the cell and its content in intracellular and subcellular targets as well as to analyze the kinetic processes of drug absorption, transport, distribution, metabolism, and excretion in the cell (Table 1). The ability to illuminate these features through SRM technology is of great significance, as elucidating drugs’ mechanisms of action in cells can permit the prediction and evaluation of drug efficacy thereby increasing the success rate of drug research and development.
Table 1.
Current studies of drug dynamics using SRM techniques.
| Drug Dynamic mode | Types of drugs and drug carriers | SRMs | Mechanisms | Refs. |
|---|---|---|---|---|
| Absorption and transport | Protracted release drug, LL-37-TAMRA | STED | The uptake of LL-37 may be an active process rather than passive diffusion. STED microscopy also showed distinct clusters of LL-37 in the ring structure, demonstrating the advantage of STED microscopy in identifying different subcellular structures. | [144] |
| Protracted release drugs, NU-1000 and NU-901 | SIM | The uptake of drug (NU-1000 and NU-901) carrier MOFs in cells can occur through different active transport mechanisms depending on the surface chemistry of the MOF and the charge and size of the MOF complex. | [105] | |
| Drug carrier, silica particles | STED | The size of particles affects the uptake of fluorescent drug carrier silica particles (diameters 32 and 83 nm) into cells (Caco-2) and particles with a size of 32 nm exhibited faster migration into cells compared to particles sized 83 nm. | [106] | |
| Polymer capsules, PMASH | SIM | The uptake of PMASH in HeLa, RAW, and dTHP-1 cells under SIM demonstrated that various cell types produce different mechanical forces during the process of capsule ingestion. | [107] | |
| Distribution | Drug carrier, SiO2-Atto647N | STED | After a long incubation, SiO2-Atto647N is only distributed in the cytoplasm due to the influence of particle size. | [114] |
| natural fluorescent drug molecule, MF | SIM | MF is distributed in mitochondria and binds to hypochlorite (ClO) targets during iron poisoning, suggesting that the specificity of the drug can affect its distribution in cells. | [35] | |
| Therapeutic polymers, HPMA polymer | dSTORM | Affected by the protein CD20, the internalized polymer conjugate was localized in clusters for 4 hours. But after 24 hours, the polymer was released into a fluorophore in the cytoplasm via enzymatic degradation of the peptide. | [117] | |
| Photoactivated prodrug | SIM | Pt2L is distributed in autophagolysosomes before photostimulation. Then it escapes from the autophagolysosomes to the nucleus after photostimulation due to the influence of pH. | [119] | |
| Mitochondrial localization inhibitor of apoptosis, (vMIA) proteins | Confocal microscopy, gSTED, MSIM, PALM | It is not possible to determine the distribution of vMIA in the submitochondrial chamber via confocal microscopy. However, gSTED, MSIM and PALM imaging showed that vMIA exists in clusters of ~100 ~ 150nm, indicating that SRMs have as higher resolution for studying pharmacokinetics. | [120] | |
| Metabolism and excretion | Immunofluores cently labeled exosomes, SKBR3 exosomes-Alexa Fluor 647 | PALM/STORM | The clear presence of exosomes in lysosomes provides compelling evidence supporting the conclusion that exosomes are transported to lysosomes for further degradation after their binding to recipient cells. | [130] |
| Drug carrier, silica particles | STED | The silica particles accumulated in the nucleus and formed 300 nm adhesion after 72 hours, indicating that the fluorescent silica particles of this size had a long half-life and were suitable for use as long-acting slow-release nanoparticle drug carriers. | [106] |
3.1.1. SRM techniques for tracking absorption and transport of drug at the subcellular level
The dynamic process of a drug in the body includes absorption (except in intravascular administration), distribution, metabolism, and excretion[8, 92]. These processes involve the absorption and transport of drugs by biofilms, such as cell membrane and intracellular membrane[93]. High drug absorption and transport efficiency can improve the efficacy of drugs or reduce their side effects[94, 95]. With conventional drug absorption and transport detection methods, it is difficult to visually see drugs’ dynamic process at the subcellular level. In recent years, more and more researchers have used SRM imaging technology to visualize the route and efficiency of drug implantation.
Modifying drug molecules with specific and sensitive ligands targeting biomolecules is the key to achieving super-resolution visualization of the drug’s dynamics. To this end, the most commonly used method involves combining fluorescent molecules with effective drug molecules or drug carriers to form a drug-carrying fluorescence system[96].
Dhruva et al. incubated macrophages with LL-37-TAMRA, an antimicrobial peptide drug with fluorescent molecules, and observed the uptake and intracellular localization of macrophages 30 min later via confocal microscopy and STED microscopy. Both imaging techniques clearly show effective uptake of LL-37 by macrophages. The uptake of LL-37 may be an active process rather than passive diffusion, as a significant reduction in intracellular peptides was observed on flow cytometry when the culture was incubated at 4 °C. The confocal imaging shows spot-like but uniform markings within the macrophages, indicating that LL-37 is internalized. STED microscopy not only showed the uptake of LL-37, but also found distinct clusters of LL-37 in the ring structure, demonstrating the advantage of STED microscopy in identifying different subcellular structures[97].
In terms of drug delivery, the ideal system would have high membrane penetration and be able to efficiently reach specific subcellular targets[8, 98, 99]. Yet, since most drugs are organic compound molecules, their water solubility and biocompatibility are poor[100]. To address this challenge, the nanodrug-carrying system makes drug delivery more compatible with biological tissues by enclosing the drug within a lipophilic capsule followed by a hydrophilic exterior[101]. This increases the biocompatibility of the system while protecting the drug from hydrolysis or enzymatic hydrolysis, thus increasing the biological stability of the drug. Therefore, the use of nanoparticles as drug carriers that enter cells through endocytosis and other mechanisms can improve drug penetration through membrane tissues, promote drug transdermal absorption, and enhance drug efficacy in cells.
More generally, relevant considerations for the drug carrier system include the carrier properties, particle size, shape, etc.[102, 103]. For example, Metal-organic frameworks (MOFs)—which have the advantages of clear structure, high specific surface area and porosity, adjustable pore size, and easy chemical functionalization[104]—are a very advantageous nano-drug carriers. Teplensky et al. developed a MOF to slow the release rate of model drugs (calcein) from pores through temperature treatment of Zrbased organometer-metal frames NU-1000 and NU-901 and used SIM to study the route of MOF cellular entry (Fig 5)[105]. The results suggest that uptake of MOFs by HeLa cells can occur through different active transport mechanisms depending on the surface chemistry of the MOF and the charge and size of the MOF complex. For both MOFs tested in their study, the fosse-mediated pathway plays an important role in cellular uptake. This information can be used in the future to design MOFs as nano-carriers as to maximize the amount of MOF absorbed by cells and to ensure the most effective targeted drug delivery.
Figure 5.
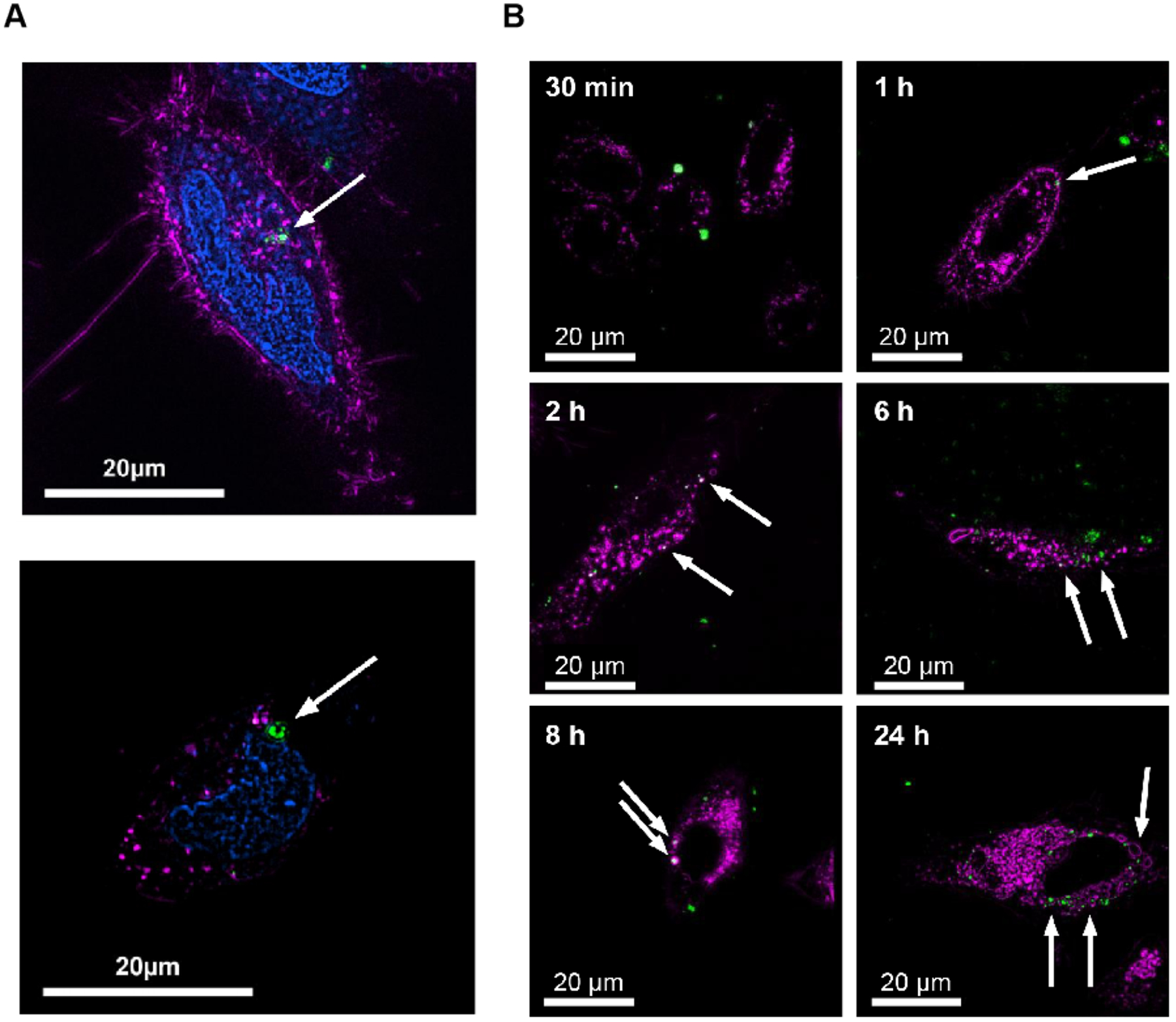
Super-resolution microscopy techniques for tracking absorption and transport of drug at the subcellular level. (A) SIM 3-color image of HeLa cells demonstrating NU-1000 (150 nm) uptake into cellular boundary. Nucleus colored in blue, early endosomes colored in magenta, NU-1000 colored in green. (B) SIM images of HeLa cells incubated with calcein-loaded temperature-treated NU-901 (cal@t.t.NU-901) at 30 min, 1 h, 2 h, 6 h, 8 h and 24 h. Lysosomes colored in magenta, cal@t.t.NU-901 colored in green. Fig. 5 reproduced from Ref [105].
Another critical factor is particle size. Schubbe et al. used STED to determine the polymer size of fluorescent drug carrier silica particles (diameters 32 and 83 nm) in human colon cancer cells (Caco-2) and compared the differences in particle absorption into the cells across different particle sizes[106]. The results showed that the content of particles with a size of 32 nm was significantly higher than that of particles sized 83 nm, and particles with a size of 32 nm migrated into cells more quickly. This indicates that the particle size is a key factor affecting the cell entry velocity.
The characteristics of the drug delivery system are also related to cell type. Chen et al. prepared thiolated poly (methacrylic acid) (PMASH) polymer capsules with fluorescent labeling, then tracked their uptake in HeLa, RAW, and dTHP-1 cells using SIM[107]. The results showed that PMASH capsules could be absorbed by all three types of cells and could also bind to cell membranes. The polymer capsules were also compared during ingestion and found to undergo different degrees of deformation as a result of the different mechanical forces generated by the different cell types. This proved that the intracellular forces exerted during internalization of particles is one of the key parameters for successful delivery.
According to the results of cellular drug and drug delivery system dynamics, researchers can try to change the drug absorption and transport pathway by changing the properties of the preparation, such as carrier type and particle size when designing carrier drugs so as to improve the drug’s absorption in the body.
3.1.2. SRM techniques for tracking distribution of drug at the subcellular level
Biodistribution and pharmacodynamic biodistribution of a drug is the phenomenon wherein a drug is absorbed into the blood and transported to the tissues. In order to be effective, a drug needs to pass the blood-tissue barrier as well as successfully locate its target organ[108–110]. However, the in vivo study of drug distribution ignores the differences between individual cells and cannot accurately describe the characteristics of drug distribution within a single cell. In recent years, researchers have studied the distribution characteristics of different drugs and drug carriers in cells by means of SRM imaging.
Factors that affect the intracellular distribution of a drug include the properties of the drug carrier (such as the size of the drug carrier), the properties of the drug itself (such as the specificity) and the regulatory role of related proteins[111–113]. For example, Kraegeloh et al. combined fluorescent molecule Atto647N with drug carrier SiO2 using the covalent method and assembled SiO2 drug carrier nanoparticles with a particle size of 128 ± 7 nm. They used STED to monitor the distribution of the SiO2 drug carrier nanoparticles after entering the cells[114]. Although particle aggregation was visible in the cytoplasm after a long incubation, no agglomeration or particles were detected in the nucleus. Previous reports have shown that it is only possible to transport nanoparticles to the nucleus when they are smaller than 40 nm[115]. Therefore, the inability of the SiO2 drug carrier nanoparticles to enter the nucleus may be caused by the size of the particles used. This indicates that the size of the drug carrier is one of the factors affecting its entry into the cell.
Chen et al. applied SIM to capture the distribution of Cy5-Dextran at different incubation periods in living cells. They examined the distribution of Cy5-Dextran in cells at 0.5, 1, 3, 6, and 12 h. The SIM image of the cell taken 0.5 h after Cy5-Dextran treatment demonstrated a weak fluorescence that was evenly distributed throughout the cytoplasm. During the subsequent 3 h incubation period, there was no significant change observed in the distribution of fluorescence. Three hours after Cy5-Dextran treatment, part of the subcellular localization of Cy5-Dextran could be attributed to localization within the lysosomes of the cell. However, 6 h following treatment, the Cy5-Dextran was localized within the mitochondria[116].
Additionally, Wei et al. observed that MF was present in HeLa cells via using fluorescence confocal microscopy. However, due to the limited resolution of traditional microscopes, the visualization of MF in living cells at the nano-level remains unsolved. Later, SIM was used to trace the distribution of the natural fluorescent drug molecule magnoflorine (MF) in cells. Recent research found that MF was distributed in mitochondria and it could bind to hypochlorite (ClO) targets during the process of ferroptosis (Fig 6)[35]. This finding shows that the specificity of a drug can affect its distribution in cells. In addition, to observe the effect of MF on ferroptosis and the morphological distribution of mitochondria under SIM, researchers used a commercial inducer called erastin to treat HeLa cells, aiming to initiate ferroptosis within HeLa cells. Compared to the mitochondrial morphology in untreated HeLa cells, mitochondria during ferroptosis appeared to be a thinner and more fibrous morphology, accompanied by the disappearance of the mitochondrial cristae structure. These distinct subcellular nanoscale changes cannot be observed in conventional confocal microscopy.
Figure 6.
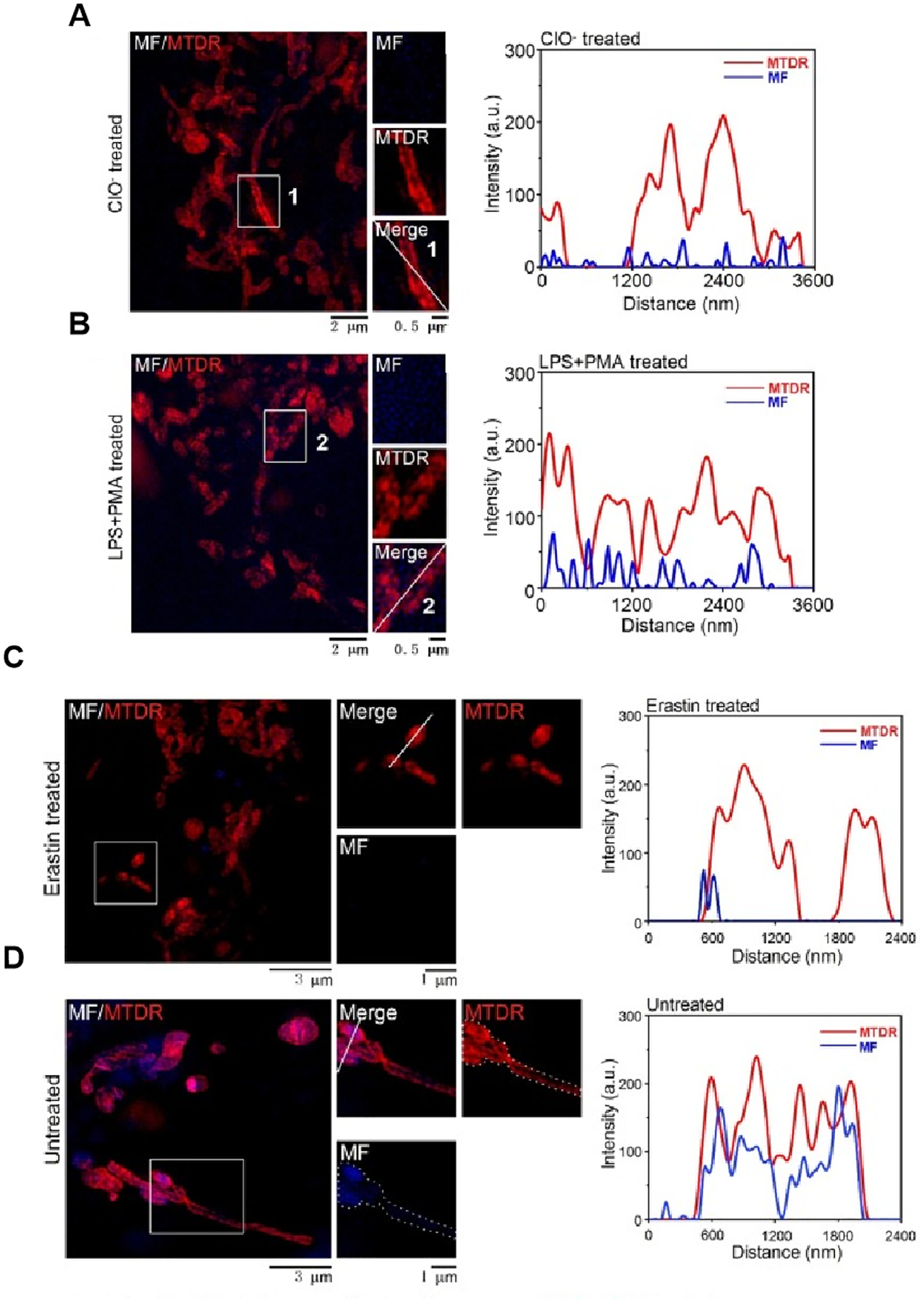
Super-resolution microscopy techniques for tracking distribution of drug at the subcellular level. (A-B) MF targets the mitochondrial ClO- altering its subcellular distribution. (A) Fluorescence images of mitochondria in HeLa cells incubated with MTDR and MF (10 μM) for 30 min, followed by incubation with exogenous ClO- (50 μM) for another 30 min. (B) Fluorescence images of mitochondria in HeLa cells incubated with lipopolysaccharide (LPS, 1 μg/mL) for 12 h and phorbol myristate acetate (PMA, 1 μg/mL) for 30 min. (C-D) MF show a definite response and bind to its target in ferroptosis. (C) Fluorescence images of mitochondria in HeLa cells incubated with erastin (20 μM) for 12 h and MTDR (λex=640 nm) and MF (10 μM) (λex=405 nm) for 30 min at 37 °C. (D) Fluorescence images of mitochondria in HeLa cells incubated with MTDR and MF (10 μM) for 30 min at 37 °C. Fig. 6 reproduced from Ref [35].
Michael Hartley et al. labeled therapeutic polymers (HPMA polymer) with four different synthetic fluorophores and used dSTORM to track the location of therapeutic components at different time points. Affected by the protein CD20, the internalized polymer conjugate was localized in clusters at 4 hours. After 24 hours, the polymer was released into a fluorophore in the cytoplasm by enzymatic degradation of the peptide. The pair correlation function of the dye attached to the polymer and the released dye indicated a change in attenuation length between 4 and 24 h, and the pair correlation function of the released dye showed a random distribution after 24 h [117].
Another important consideration is that different targets and different drug properties may cause changes in intracellular transport paths, which cause drugs to have different effects. Let us take DOX and paclitaxel as examples: their physical and chemical properties are relatively stable, and they can be effectively delivered into the cell to have an effect[118]. However, biomolecular drugs (peptides, proteins, siRNA) are unstable in a low-pH microenvironment with abundant enzymes in lysosomes. Liu et al. designed and synthesized a platinum-based complex Pt2L with photoactivated prodrug potential and tracked the distribution of Pt2L in cells using SIM[119]. The results showed that Pt2L was distributed in autolysosomes before light stimulation and escaped from the autolysosomes to the nucleus after light stimulation. This indicates that the properties of Pt2L are affected by light exposure which in turn indicates that the photoselectivity of Pt2L has the potential to improve the treatment’s specificity. Therefore, stability (such as lysosome escape) should be considered in the design of drug molecules in accordance with the therapeutic purpose of the drug.
It is worth noting that different microscopes have different visualizations of intracellular pharmacokinetics. Shivaprasad et al. visualized the distribution of cytomegalovirus (HCMV) viral mitochondrial localization inhibitor of apoptosis (vMIA) proteins in cells via using a combination of confocal microscopy and SRMs. Deconvolution of confocal microscopy images revealed that vMIA was located at the mitochondria-endoplasmic reticulum interface, away from the mitochondrial matrix. It is not possible to determine vMIA distribution within sub-mitochondrial compartments using conventional confocal microscopy. To do this, the researchers turned to SRMs, which enable imaging beyond the diffraction limitations, providing improved insights into vMIA’s distribution in sub-mitochondrial compartments. By using gSTED imaging, they found that vMIA distributed in clusters along the interface. Using a multi-color and multi-focal structure illumination microscope (MSIM), researchers found that the vMIA cluster was positioned away from MitoTracker Red, indicating its OMM localization. GSTED and MSIM imaging reveal that vMIA is present in clusters of ~100 to 150 nm, which is consistent with cluster sizes determined by PALM. With these different super-resolution methods, the researchers visualized the cluster distribution of vMIA at OMM near the ER. The obtained results directly compare the relative advantages of each super-resolution imaging modality of MAM and submitochondrial compartmentimaging components. These findings provide valuable references for the study of the pharmacokinetics at the subcellular level[120].
Given that not all drugs work equally well at the same plasma concentration and that a drug’s effect is closely related to its mechanism of action and intracellular distribution, SRM imaging has demonstrated many advantages for uncovering these important features. Moreover, this knowledge about the intracellular environment can be applied to guide drug design for controlled subcellular distribution.
3.1.3. SRM techniques for tracking metabolism and excretion of drug at the subcellular level
After a drug enters the body as a foreign matter, the body must mobilize various tissues and organs to create a reaction. For most drugs, this process involves being catalyzed by specific enzymes before they can have their relevant effect[121]. At the subcellular level, these enzymes are mainly located on lysosomes, mitochondria, the endoplasmic reticulum, the nuclear membrane, and the plasma membrane[122, 123]. After breakdown, excess drug material should be quickly excreted out of the body [124]. This elimination process involves both metabolism and excretion[125, 126]. Through the study of the metabolism and excretion of intracellular drugs with SRM imaging technology, the utilization and accumulation of drugs in cells can be observed more directly.
Some drugs modified or labeled with fluorescent groups may exhibit changes to their transport pathway after entering cells as well as changes to their elimination process and efficacy. Wang et al. designed a pH-responsive prodox micelle and quantified its distribution in both the endocytosomes and lysosomes[127]. The micelles showed different intracellular transport pathways from DOX. Free DOX first diffused through the cell membrane into the cytoplasm. While most drugs accumulate in the nucleus at early time points and are slower to accumulate in the lysosome, drug-carrying micelles first formed endocytic vesicles through endocytosis and then fused with lysosomes. DOX was released in response to an acid trigger in the lysosome, diffused through the lysosome membrane into the cytoplasm, and then reached the nucleus to exert its efficacy.
Nevertheless, the metabolic path of many drugs will not change after being modified or labeled with fluorescent groups. Previous studies have shown that cellular uptake of exosomes is dependent on endocytosis and phagocytosis[128]; once endocytosis occurs, large amounts of exosomes are then transported to lysosomes for degradation[129]. Chen et al. labeled exosomes of SKBR 3 using Alexa Fluor 647 IF and used LysoTracker Red to label lysosomes of MRC-5 cells. PALM/STORM imaging was used to monitor the metabolism of SKBR 3 exosomes in MRC-5 cells. They found that most exosomes were localized in lysosomes. It provides more powerful evidence for the conclusion that after binding to recipient cells, exosomes will be transported into lysosomes for further degradation. This indicates that the metabolic process of fluorescently labeled exosomes does not change, a factor that is very important for the maintenance of the original drug effect[130].
Due to the actions of various efflux proteins, drugs can easily enter normal cells but have difficulty aggregating in cells, resulting in a short half-life[93]. This can be improved through drug modification. Mu et al. compared the drug accumulations of Taxotere and docetaxel micelles in sensitive human oral epidermal carcinoma (KB) cells and resistant oral epidermal carcinoma (KBv) cells[131]. The results showed that in KB cells, there was no significant difference in the intracellular content of docetaxel between the two preparations at 4 h. However, in KBv cells, the intracellular content of docetaxel micelles at 4 h was approximately 1.5 times higher than that of its free drug. The researchers believed that this result could be explained by the carrier material (Pluronic) of the docetaxel micelles inhibiting P-pg protein and affecting drug efflux, thus increasing the intracellular content of docetaxel micelles.
Additionally, using STED, Schubbe et al. observed that 32-nm fluorescent drug carrier silica particles entered the nucleus and formed 200 nm adherents 48 hours later, and then accumulated in the nucleus and formed 300 nm adherents 72 hours later (Fig 7)[106]. The results indicate that the fluorescent silica particles of this size had a long half-life and were suitable to be used as long-acting sustained-release nanoparticle drug carriers.
Figure 7.
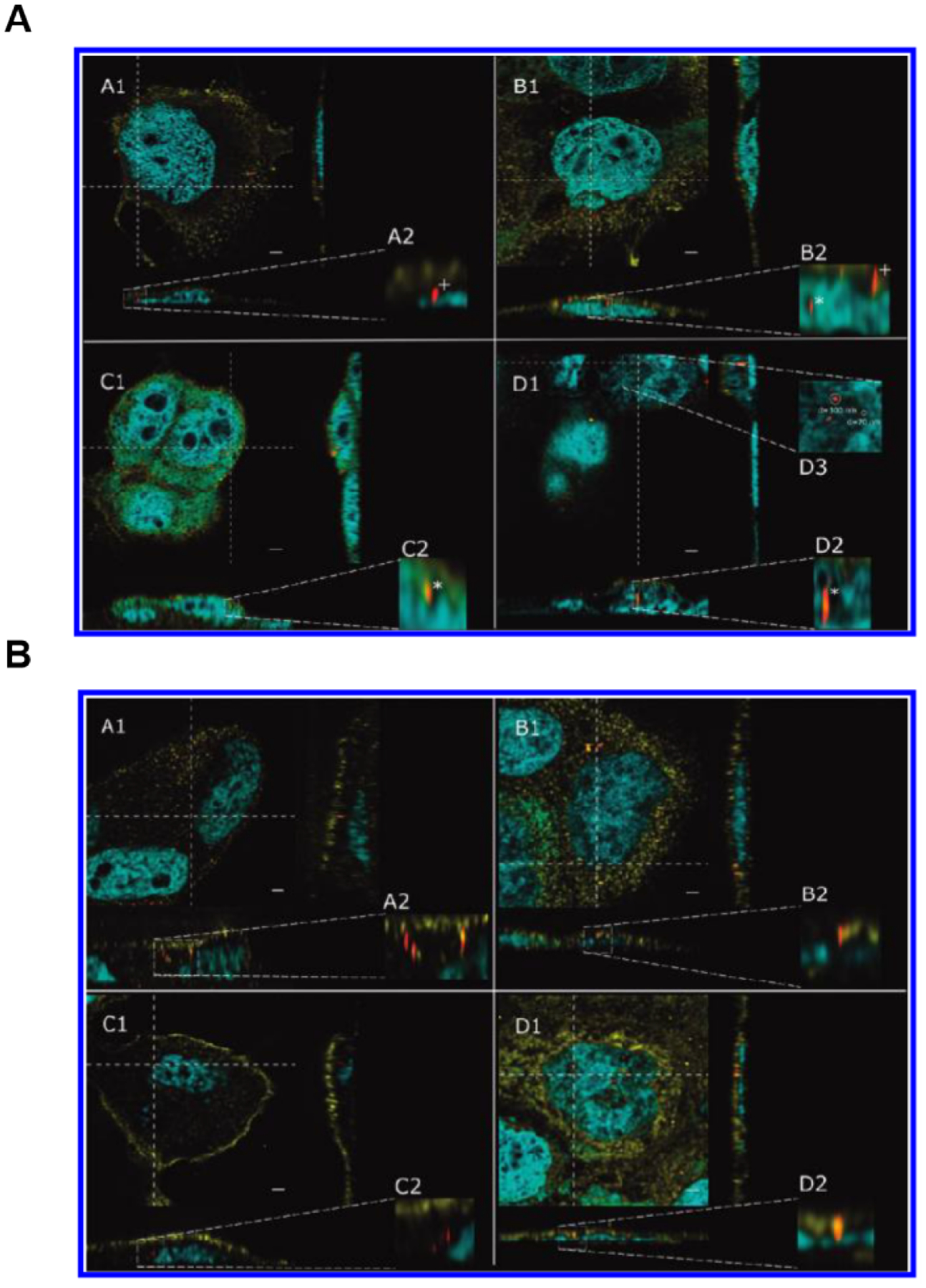
Super-resolution microscopy techniques for tracking metabolism and excretion of drug at the sub-cellular level. (A-B) High resolution STED confocal fluorescence images of Caco-2 cells incubated with 1 μg mL−1 32 nm silica particles for 3 days. Samples were taken after 5 h (A1), 24 h (B1), 48 h (C1), and 72 h (D1). (A) A2–D2 display enlargements of the orthogonal view to visualize nanoparticles inside (*) and outside (+) the nucleus. D3 depicts an enlargement of the aggregated particles shown in D1. (B) A2–D2 depict enlargements of the orthogonal view to demonstrate that nanoparticles are outside the nucleus. Particles that appear to be localized inside the nucleus in the xy section actually represent signals from an image plane above the nucleus, as can be seen in the enlargements of the orthogonal view (B2 and D2). The nucleus is shown in blue, the cell membrane in yellow, and silica nanoparticles in red. Images are deconvolved and oversaturated for better visualization. The scale bar represents 2 μm. Fig. 7 reproduced from Ref [106].
Therefore, it is of great importance to study the elimination process of drugs at the organelle level in order to improve drug efficacy and design new preparations. Although SRM imaging technology can determine the elimination of drugs, due to the limitation of strong excitation light intensity, it still faces great challenges in tracking drugs’ and drug carriers’ dynamic changes in living cells over a long time period.
3.2. How to select appropriate SRM techniques for tracking drug dynamics at the subcellular level
The imaging of drug dynamics at the subcellular level using SRM is of great significance to understanding the structural design of drug molecules as well as the interactions between drug molecules and cells.
SIM uses a lower intensity of excitation light than other SRM technologies; therefore, SIM is the best choice when imaging drug molecules in living cells in 3D or over long periods of time[21, 43, 132]. However, its resolution is relatively low, which requires stable calibration of the system. In addition, postprocessing requires a quality check to avoid artifacts. Drugs that require high resolution and high imaging system stability are not suitable for SIM imaging.
STED does not have these drawbacks because it does not need image reconstruction and can avoid errors caused by later image processing[25, 43, 133]. However, STED still uses a point-scan method, and its high laser intensity can cause light damage. Thus, its fast dynamic imaging is limited to small frame sizes[134]. In addition, STED’s high-intensity laser irradiation may cause drug molecules containing nanomaterials (such as metal nanoparticles) to generate heat. For drug molecules with poor stability, this may affect the dynamic process of the drug and the drug’s effect. Therefore, STED can be used for imaging when the fluorophore is relatively stable and high-quality resolution is required.
Currently, SMLM provides the highest resolution for observing nanoscale cellular interactions at the single-molecule level[135]. The resolution of SMLM is primarily determined by two factors: localization accuracy and molecular density. The localization, or positioning, accuracy is the position of the target molecule on the transverse dimension, and the molecular density is the density of the target molecule in the sample; both are equally important in determining resolution. On the basis of higher molecular localization, drugs with higher molecular density are suitable for SMLM imaging. For applications with live cells, PALM using fluorescent proteins is the preferred method[136]. Ideally, each protein of interest would be quantified with a fluorescent protein[137]; however, fluorescent proteins exhibit lower light stability and photon counting than organic dyes, which reduces positioning accuracy and generally requires longer acquisition times. With the use of SMLM, the selection of fluorescent molecules, excitation wavelength, and the drug label density are crucial and must be extensively optimized. Nevertheless, SMLM can image living cells in three dimensions with high spatial and temporal resolution.
All in all, each of these techniques has its own advantages and disadvantages; therefore, it is very important to select a suitable SRM technique for observing intracellular drug dynamics according to the imaging requirements and the characteristics of the drug molecules.
3.3. The scope of application of SRM techniques for tracking drug dynamics at the subcellular level
An extension of traditional drug dynamics is the study of cellular drug dynamics using SRM. However, this research method has its own scope of application, as using SRM technology for studying cell drug dynamics is not suitable for all drugs. A drug for which SRM can be used for cellular pharmacokinetic study must have a number of characteristics. First, the drug should have groups that emit fluorescence (the drug itself can emit fluorescence, or the drug must be labeled with fluorescent molecules)[138]. Second, the target of a drug’s action should be inside the cell, not outside the cell or on the membrane, as only when a drug enters the cell through the biofilm to complete the kinetic process of intracellular absorption, transport, distribution, metabolism, and efflux can it have a basis for the study of its kinetic parameters[139]. Finally, the dynamics of the drug in the cell and the combination of the drug with the target should be the determinants of drug efficacy[18, 91, 140]. Therefore, the intracellular targets of the drugs should be specific rather than widespread—that is, the drug has a specific target when it enters the cell and binds with organelles, nuclear receptors, various kinases, DNA, metabolic enzymes, etc. before exerts its effect.
3.4. Significance of tracking drug dynamics at the subcellular level using SRM techniques
Optimize pharmacokinetic properties of drugs.
Pharmacokinetic studies have proven that changing drug dosage forms can prolong drug retention time in vivo, enhance a drug’s passive targeting, and improve its pharmacokinetic behavior in vivo[141–143]. Teplensky et al. delayed the release of the model compound caleflavin and the anticancer therapy alpha-cyano-4-hydroxycinnamic acid (alpha-Chc) by causing partial collapse of the pores of NU-1000 and NU-901 through mild temperature treatment. To better understand the release of MOF payloads in vitro, they used SIM to visualize the interaction of MOF with living cells[105]. This study provides a solid theoretical and technical basis for the modification of dosage form and the optimization of kinetic parameters.
Accelerate drug screening and research and development.
In the process of new drug development, approximately 40% of drugs are eliminated due to poor pharmacokinetic parameters[144]. Screening and optimization of drug dynamics at the subcellular level using SRM imaging can not only provide analysis indexes that cannot be detected by traditional pharmacokinetics but can also reduce analysis errors caused by individual cell differences as well as provides more comprehensive parameters to screen out ineffective drug molecules that are difficult to detect with traditional pharmacokinetics. The PALM/STORM technique has been used to achieve super-resolution imaging of exosomes of cancer origin. Compared to traditional field microscopes, PALM/STORM provides better spatial resolution in the observation of exosomes at scales up to nanometers. Researchers can thus more precisely visualize the intracellular location of the added exosomes (Fig 8)[130]. This provides a firm foundation for the study of drugs’ action in cells and drug screening which greatly reduces the cost and risk of enterprise research and development. Drug screening based on cell models has been widely used, and the study of cellular pharmacokinetics using SRM imaging technology can accelerate the process of drug screening and drug development.
Figure 8.
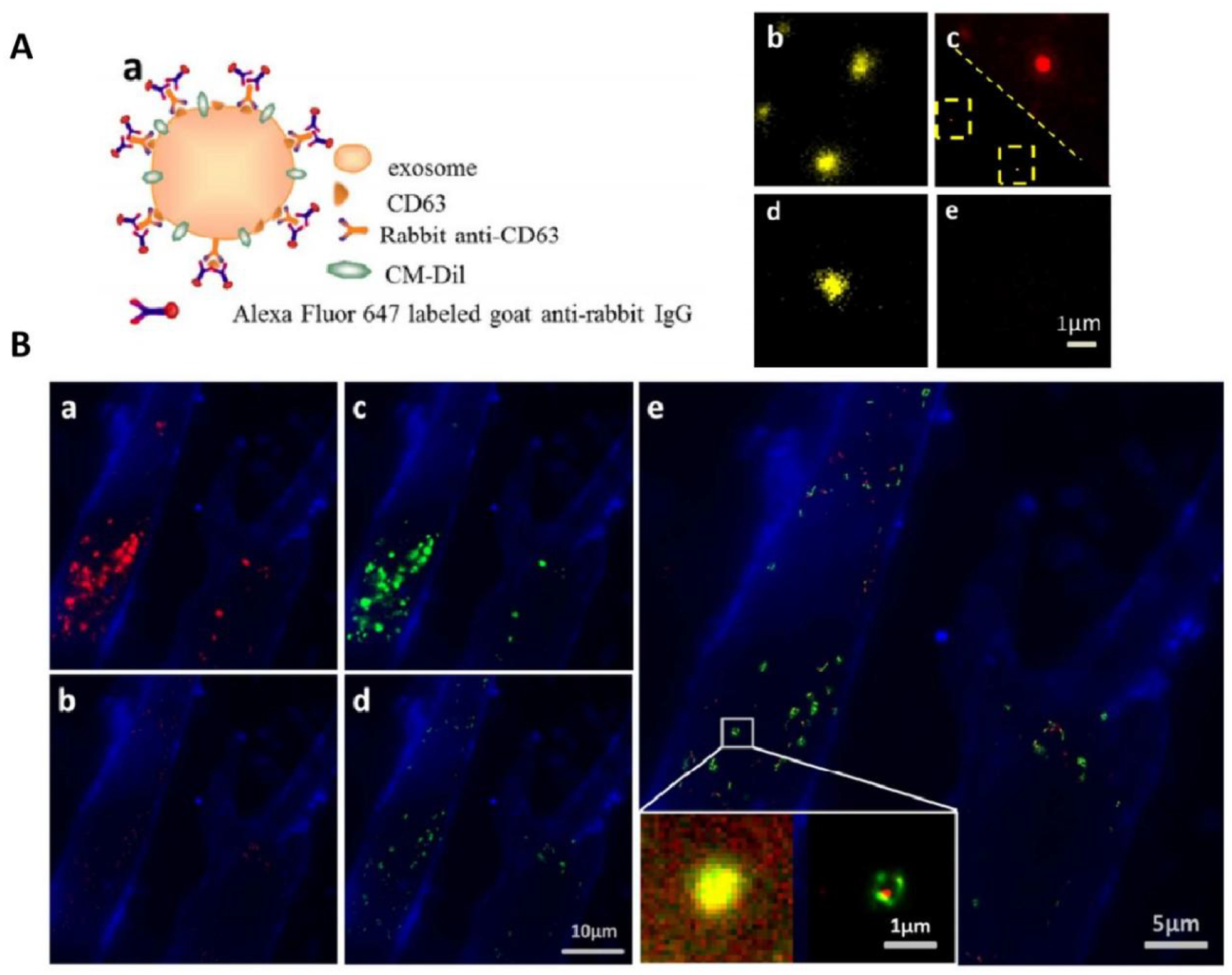
TIRF and PALM/STORM imaging of HeLa exosomes. (A) (a) Schematic illustration of indirect IF labeling of CD63 and exosome membrane stained with CM-Dil. (b) TIRF image of HeLa-derived exosomes stained with CM-Dil. (c) IF labeling of CD63 with Alexa Fluor 647, conventional diffractionlimited image (upper right) and PALM/STORM image (low left). (d) Immunofluorescence control sample without primary antibody for specificity detection, CM-Dil channel. (e) Immunofluorescence control sample without primary antibody for specificity detection, Alexa Fluor 647 channel. (B) Colocalization of SKBR3 exosomes and MRC-5 lysosomes. (a) TIRF image of internalized SKBR3 exosomes (red) and MRC-5 membrane (blue). (b) PALM/STORM image of the same region as shown in (a). (c) TIRF image of MRC-5 lysosomes (green) and MRC-5 membrane (blue). (d) PALM/STORM image of the same region as shown in (c). (e) Colocalization of MRC-5 lysosomes (green) and internalized SKBR3 exosomes (red). Fig. 8 reproduced from Refs [130].
Optimize the parameters of SRM techniques.
The design and synthesis of fluorescent drug molecules with excellent optical properties can reduce the impact of SRM technology drawbacks, such as photobleaching and fluorescent background as well as improve the imaging resolution[90, 138]. Stefen W. Hell et al. designed and synthesized fluorescent tubulin probes consisting of fluorescent dyes (510R, 580CP, GeR, and SiR) and chemotherapy drugs (taxane, docetaxel, carpacel, and larotaxel). Optimization of the probe enabled the authors to achieve a resolution of 29 ± 11 nm using STED microscopy images of microtubule networks in living fibroblasts[145]. This is the highest resolution of STED imaging of living cells to date.
4. Prospects and Conclusion
The study of drug dynamics infuses the whole process of new drug discovery, preclinical research, and clinical research, and it plays an important role in the development of innovative drugs[146–148]. It has been reported that more than one-third of drug targets are located within cells[35]. Drugs with such targets must penetrate multiple barriers, such as tissues and cell membranes, to effectively deliver the drugs to the in-cell targets in order to have an effect[149–151]. Therefore, it is of great importance to improve the efficiency and success rate of new drug research and development by more accurately studying the dynamic behavior of drugs in cells/sub-organelles so as to reflect the process and mechanism of drug action.
Recent years have seen the development of a variety of SRM techniques and specialized fluorescent probes that enable direct observation of subcellular structures and protein arrangements at a resolution that exceeds that of conventional light microscopy[42, 152–155]. They have revealed the absorption, distribution, metabolism, and transport processes of drugs at the cellular/subcellular level in an intuitive and visual way, which has great significance for drug research and development, screening, and clinical application[30, 154, 156]. Here, we introduce several classical SRMs, systematically elaborate their principles and development, and compare their advantages and disadvantages. We also describe the application of these techniques in the study of drug dynamics at the subcellular level. By summarizing previous studies that detailed the absorption, distribution, metabolism, and excretion of drugs in cells using SRM, this article provides the basis for studying drug metabolism at the subcellular level, the design of drug molecules, and the selection of SRM techniques.
Nevertheless, there are still many problems to be overcome in the research of pharmacokinetics at the subcellular level using SRM. In terms of drug molecules, the properties of the drug itself—such as targeting, chemical modification, size, and polarity—will affect its metabolism in the cell[8, 10, 94, 157]. Drug metabolism may also significantly differ between cell lines (such as intestinal cells and liver cells), exert different pathways in tumor cells, and be affected by a host of other factors such as differing metabolic enzymes and pH environment[158, 159]. For the drugs whose targets contain cellular localization sites, it is necessary to avoid the interference of these factors with the pharmacokinetics as much as possible in the process of SRM imaging: only in this way can we more effectively and comprehensively evaluate and predict the efficacy and possible side effects of drugs as well as provide important references for the preliminary design of drug molecules.
In terms of super-resolution imaging, the study of pharmacokinetics at the subcellular level using SRM requires long-term dynamic imaging in living cells. However, SRM technology still has the disadvantage of requiring high laser emissions for imaging, a factor that greatly limits its use in research[160]. In addition, live-cell imaging also has very stringent requirements for the imaging environment, which demands strict control of experimental temperature, humidity, and other conditions[161]. Cells can only maintain cell activity during a very short exposure time with long exposure resulting in cell death. In experiments, the selection of light intensity must avoid excessive irradiation and light damage in order to maintain cell activity for a sufficient time period. In addition, the sampling frequency during imaging must be high enough to observe cell dynamics without losing information. Therefore, many challenges remain in the use of SRM for subcellular drug dynamic imaging.
Although some concerns remain, SRM, like other fluorescence microscopy techniques, relies on the expression of fluorescent molecules within cells[162, 163]. Imaging is usually performed by introducing external groups that directly affect cell physiology, and therefore the process may not reflect “true” drug molecular action. Nevertheless, the development of super-resolution technology represents a major advance and a powerful tool for understanding how drug molecules behave on their journey through biological substances[33, 154, 164, 165]. Comprehensive study of the intracellular disposal process of drugs can effectively screen and evaluate new intracellular-targeting drugs, improve the accuracy and efficiency of drug evaluation, and reduce the risks involved in drug development. It also guides the design of targeted prodrugs or preparations to improve targeting, reduce toxic side effects, increase the success rate of new drug research and development, clarify the interactions and mechanisms in cells, and provide a basis for clinical rational drug use.
Supplementary Material
Highlights:
Super-resolution microscopy (SRM) techniques can track drug dynamics at the subcellular level
Significance of tracking drug dynamics at the subcellular level using SRM techniques
Selection of appropriate SRM techniques for various tracking systems.
Acknowledgements
J.D. was supported by the National Institutes of Health (NIH R35GM128837).
Abbreviations
- SRM
super resolution microscopy
- STED
stimulated emission depletion microscopy
- SMLM
stochastic optical reconstruction microscopy
- PALM
photoactivated localization microscopy
- STORM
random optical reconstruction microscopy
- SIM
structured illumination microscopy
- g-STED
time-gate based STED
- MINFLUX
Minimal Photon Fluxes
- OTF
optical transfer function
- NL-SIM
nonlinear structure illumination obvious microtechnique
- PSF
point spread function
- PSF engineering
point spread function engineering
- MF
magnoflorine
- ClO
hypochlorite
- alpha-Chc
alpha-cyano-4-hydroxycinnamic acid
- MOF
Metal-organic framework
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Babikova RKD, Momekova D, Ugrinova I, Momekov G, Dimitrov I, Multifunctional polymer nanocarrier for efficient targeted cellular and subcellular anticancer drug delivery, ACS Biomater Sci Eng, 5 (2019) 2271–2283. [DOI] [PubMed] [Google Scholar]
- [2].Bae Y, Preparation and biological characterization of polymeric micelle drug carriers with intracellular pH-triggered drug Release property tumor permeability, Bioconjugate Chem, 16 (2005) 122–130. [DOI] [PubMed] [Google Scholar]
- [3].Bucko CKLPJ, Rathbun L, Garcia I, Bhat A, Wordeman L, Smith FD, Maly DJ, Hehnly H, Scott JD, Subcellular drug targeting illuminates local action of polo-like kinase 1 and aurora a during mitosis, FASEB J, 34 (2020) 1–1. [Google Scholar]
- [4].Elgart V, Lin JR, Loscalzo J, Determinants of drug-target interactions at the single cell level, PLoS Comput Biol, 14 (2018) e1006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gorman BL, Brunet MA, Pham SN, Kraft ML, Measurement of Absolute Concentration at the Subcellular Scale, ACS Nano, 14 (2020) 6414–6419. [DOI] [PubMed] [Google Scholar]
- [6].Pelkonen ABO, Reichel Editors A, Pharmacokinetic Challenges in Drug Discovery, Ernst Schering Research Foundation Workshop, 37 (2002) 213–234. [Google Scholar]
- [7].Kumar A, Ahmad A, Vyawahare A, Khan R, Membrane Trafficking and Subcellular Drug Targeting Pathways, Front Pharmacol, 11 (2020) 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].D’Souza GG, Weissig V, Subcellular targeting: a new frontier for drug-loaded pharmaceutical nanocarriers and the concept of the magic bullet, Expert Opin Drug Deliv, 6 (2009) 1135–1148. [DOI] [PubMed] [Google Scholar]
- [9].Dubach JM, Vinegoni C, Mazitschek R, Fumene Feruglio P, Cameron LA, Weissleder R, In vivo imaging of specific drug-target binding at subcellular resolution, Nat Commun, 5 (2014) 3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gao J, Dutta K, Zhuang J, Thayumanavan S, Cellular- and Subcellular-Targeted Delivery Using a Simple All-in-One Polymeric Nanoassembly, Angew Chem Int Ed Engl, 59 (2020) 23466–23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guo X, Wei X, Chen Z, Zhang X, Yang G, Zhou S, Multifunctional nanoplatforms for subcellular delivery of drugs in cancer therapy, Progress in Materials Science, 107 (2020) 100599. [Google Scholar]
- [12].Ward ES, Velmurugan R, Ober RJ, Targeting FcRn for therapy: from live cell imaging to in vivo studies in mice, Immunol Lett, 160 (2014) 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Rebeccah JAT Marsh E, Sawyer Michael B., Vos Kenneth J.E., Emergence of power laws in the pharmacokinetics of paclitaxel due to competing saturable processes, J Pharm Pharmaceut Sci, 11 (2008) 77–96. [DOI] [PubMed] [Google Scholar]
- [14].Joshi S, Reif R, Wang M, Zhang J, Ergin A, Bruce JN, Fine RL, Bigio IJ, Intra-arterial mitoxantrone delivery in rabbits: an optical pharmacokinetic study, Neurosurgery, 69 (2011) 706–712; discussion 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yu X, Wen X, Araki Y, Iwami K, Umeda N, Measurement of nano-particles size by evanescent interference field with conventional optical microscope, International Society for Optics and Photonics, 7133 (2008) 71333C. [Google Scholar]
- [16].Hao X, Liu X, Kuang C, Li Y, Ku Y, Zhang H, Li H, Tong L, Far-field super-resolution imaging using near-field illumination by micro-fiber, Applied Physics Letters, 102 (2013) 013104. [Google Scholar]
- [17].Iwanaga M, Hyperlens-array-implemented optical microscopy, Applied Physics Letters, 105 (2014) 053112. [Google Scholar]
- [18].Legin AA, Schintlmeister A, Jakupec MA, Galanski MS, Lichtscheidl I, Wagner M, Keppler BK, NanoSIMS combined with fluorescence microscopy as a tool for subcellular imaging of isotopically labeled platinum-based anticancer drugs, Chemical Science, 5 (2014) 3135–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu H, Chen S, Analysis of Sonoporation on Cell Membrane by Scanning Electron Microscopy, (2012) 968–971.
- [20].Legleiter J, Kowalewski T, Tapping, pulling, probing: atomic force microscopy in drug discovery, Drug Discov Today Technol, 1 (2004) 163–169. [DOI] [PubMed] [Google Scholar]
- [21].Godin AG, Lounis B, Cognet L, Super-resolution microscopy approaches for live cell imaging, Biophys J, 107 (2014) 1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang R, He X, Niu G, Meng F, Lu Q, Liu Z, Yu X, A Single Fluorescent pH Probe for Simultaneous Two-Color Visualization of Nuclei and Mitochondria and Monitoring Cell Apoptosis, ACS Sens., 6 (2021) 1552–1559. [DOI] [PubMed] [Google Scholar]
- [23].Wang KN, Shao X, Tian Z, Liu LY, Zhang C, Tan CP, Zhang J, Ling P, Liu F, Chen Q, Diao J, Mao ZW, A Continuous Add-On Probe Reveals the Nonlinear Enlargement of Mitochondria in Light-Activated Oncosis, Adv Sci (Weinh), 8 (2021) e2004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang L, Chen R, Han G, Liu X, Huang T, Diao J, Sun Y, Super-resolution analyzing spatial organization of lysosomes with an organic fluorescent probe, Exploration (Beijing), 2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vicidomini G, Bianchini P, Diaspro A, STED super-resolved microscopy, Nat Methods, 15 (2018) 173–182. [DOI] [PubMed] [Google Scholar]
- [26].Henriques R, Griffiths C, Hesper Rego E, Mhlanga MM, PALM and STORM: unlocking live-cell super-resolution, Biopolymers, 95 (2011) 322–331. [DOI] [PubMed] [Google Scholar]
- [27].Dudok B, Barna L, Ledri M, Szabo SI, Szabadits E, Pinter B, Woodhams SG, Henstridge CM, Balla GY, Nyilas R, Varga C, Lee SH, Matolcsi M, Cervenak J, Kacskovics I, Watanabe M, Sagheddu C, Melis M, Pistis M, Soltesz I, Katona I, Cell-specific STORM super-resolution imaging reveals nanoscale organization of cannabinoid signaling, Nat Neurosci, 18 (2015) 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Enderlein J, Ngo VA, Gryczynski ZK, Law YN, Hariharan S, Erdmann R, Koberling F, Lee HK, Ahmed S, Gregor I, Accurate single-molecule localization of super-resolution microscopy images using multiscale products, 8228 (2012) 822813. [Google Scholar]
- [29].Richter V, Piper M, Wagner M, Schneckenburger H, Increasing Resolution in Live Cell Microscopy by Structured Illumination (SIM), Applied Sciences, 9 (2019) 1188. [Google Scholar]
- [30].Cardellini J, Balestri A, Montis C, Berti D, Advanced Static and Dynamic Fluorescence Microscopy Techniques to Investigate Drug Delivery Systems, Pharmaceutics, 13 (2021) 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen Q, Fang H, Shao X, Tian Z, Geng S, Zhang Y, Fan H, Xiang P, Zhang J, Tian X, Zhang K, He W, Guo Z, Diao J, A dual-labeling probe to track functional mitochondria-lysosome interactions in live cells, Nat Commun, 11 (2020) 6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen Q, Jin C, Shao X, Guan R, Tian Z, Wang C, Liu F, Ling P, Guan JL, Ji L, Wang F, Chao H, Diao J, Super-resolution tracking of mitochondrial dynamics with an Iridium(III) luminophore, Small, 14 (2018) e1802166. [DOI] [PubMed] [Google Scholar]
- [33].van der Zwaag D, Vanparijs N, Wijnands S, De Rycke R, De Geest BG, Albertazzi L, Super Resolution Imaging of Nanoparticles Cellular Uptake and Trafficking, ACS Appl Mater Interfaces, 8 (2016) 6391–6399. [DOI] [PubMed] [Google Scholar]
- [34].Lefman J, Cheuk T, Metaferia B, Scott K, Khan J, Stranick SJ, Super-Resolution Imaging of Drug Delivery to Live Cells using Real-Time Structured Illumination Microscopy, Microscopy and Microanalysis, 16 (2010) 666–667. [Google Scholar]
- [35].Wei Y, Kong L, Chen H, Liu Y, Xu Y, Wang H, Fang G, Shao X, Liu F, Wang Y, Chen Q, Super-resolution image-based tracking of drug distribution in mitochondria of a label-free naturally derived drug molecules, Chemical Engineering Journal, 429 (2022) 132134. [Google Scholar]
- [36].Greulich KO, Weber G, The light microscope on its way from an analytical to a preparative tool, Journal of Microscopy, 167 (1992) 127–151. [Google Scholar]
- [37].Conchello J-A, Chao J, Cogswell CJ, Ram S, Abraham AV, Wilson T, Brown TG, Ward ES, Ober RJ, Resolution in 3D in multifocal plane microscopy, 6861 (2008) 68610Q. [Google Scholar]
- [38].Hell SW, Sahl SJ, Bates M, Zhuang X, Heintzmann R, Booth MJ, Bewersdorf J, Shtengel G, Hess H, Tinnefeld P, Honigmann A, Jakobs S, Testa I, Cognet L, Lounis B, Ewers H, Davis SJ, Eggeling C, Klenerman D, Willig KI, Vicidomini G, Castello M, Diaspro A, Cordes T, The 2015 super-resolution microscopy roadmap, Journal of Physics D: Applied Physics, 48 (2015) 443001. [Google Scholar]
- [39].Orrit M, Celebrating optical nanoscopy, Nature Photonics, 8 (2014) 887–888. [Google Scholar]
- [40].Leonhard Mçckl DCL, and Bruchle Christoph, Super-resolved Fluorescence Microscopy Nobel Prize in., Angew. Chem. Int. Ed, 53 (2014). [DOI] [PubMed] [Google Scholar]
- [41].Escorihuela J, Gonzalez-Martinez MA, Lopez-Paz JL, Puchades R, Maquieira A, Gimenez-Romero D, Dualpolarization interferometry: a novel technique to light up the nanomolecular world, Chem Rev, 115 (2015) 265–294. [DOI] [PubMed] [Google Scholar]
- [42].Chen Q, Shao X, Tian Z, Chen Y, Mondal P, Liu F, Wang F, Ling P, He W, Zhang K, Guo Z, Diao J, Nanoscale monitoring of mitochondria and lysosome interactions for drug screening and discovery, Nano Res, 12 (2019) 1009–1015. [Google Scholar]
- [43].Qiu K, Du Y, Liu J, Guan JL, Chao H, Diao J, Super-resolution observation of lysosomal dynamics with fluorescent gold nanoparticles, Theranostics, 10 (2020) 6072–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Doksani Y, Wu JY, de Lange T, Zhuang X, Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation, Cell, 155 (2013) 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Qiao C, Li D, Guo Y, Liu C, Jiang T, Dai Q, Li D, Evaluation and development of deep neural networks for image super-resolution in optical microscopy, Nat Methods, 18 (2021) 194–202. [DOI] [PubMed] [Google Scholar]
- [46].Chen Q, Shao X, Ling P, Diao J, Quantitative Analysis of Interactive Behavior of Mitochondria and Lysosomes using Structured Illumination Microscopy, Biophysical Journal, 116 (2019) 267a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fang H, Geng S, Hao M, Chen Q, Liu M, Liu C, Tian Z, Wang C, Takebe T, Guan JL, Chen Y, Guo Z, He W, Diao J, Simultaneous Zn(2+) tracking in multiple organelles using super-resolution morphology-correlated organelle identification in living cells, Nat Commun, 12 (2021) 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Muller T, Schumann C, Kraegeloh A, STED microscopy and its applications: new insights into cellular processes on the nanoscale, Chemphyschem, 13 (2012) 1986–2000. [DOI] [PubMed] [Google Scholar]
- [49].Dominik Wildanger ER, Kastrup Lars and Hell Stefan W., STED microscopy with a supercontinuum laser source., Optics express, 16 (2008) 9614–9621. [DOI] [PubMed] [Google Scholar]
- [50].Jahr W, Velicky P, Danzl JG, Strategies to maximize performance in STimulated Emission Depletion (STED) nanoscopy of biological specimens, Methods, 174 (2020) 27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang P, Goodwin PM, Werner JH, Fast, super resolution imaging via Bessel-beam stimulated emission depletion microscopy, Opt Express, 22 (2014) 12398–12409. [DOI] [PubMed] [Google Scholar]
- [52].Sharma R, Singh M, Sharma R, Recent advances in STED and RESOLFT super-resolution imaging techniques, Spectrochim Acta A Mol Biomol Spectrosc, 231 (2020) 117715. [DOI] [PubMed] [Google Scholar]
- [53].Vicidomini G, Moneron G, Han KY, Westphal V, Ta H, Reuss M, Engelhardt J, Eggeling C, Hell SW, Sharper low-power STED nanoscopy by time gating, Nat Methods, 8 (2011) 571–573. [DOI] [PubMed] [Google Scholar]
- [54].Hao X, Kuang C, Wang T, Liu X, Effects of polarization on the de-excitation dark focal spot in STED microscopy, Journal of Optics, 12 (2010) 115707. [Google Scholar]
- [55].Vicidomini G, Hernandez IC, d’Amora M, Zanacchi FC, Bianchini P, Diaspro A, Gated CW-STED microscopy: a versatile tool for biological nanometer scale investigation, Methods, 66 (2014) 124–130. [DOI] [PubMed] [Google Scholar]
- [56].Helmchen F, Denk W, Deep tissue two-photon microscopy, Nat Methods, 2 (2005) 932–940. [DOI] [PubMed] [Google Scholar]
- [57].Gael Moneron SWH, Two-photon excitation STED microscopy., Opt Express, 17 (2009) 14567–14573. [DOI] [PubMed] [Google Scholar]
- [58].Bethge P, Chereau R, Avignone E, Marsicano G, Nagerl UV, Two-photon excitation STED microscopy in two colors in acute brain slices, Biophys J, 104 (2013) 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Teodora Scheul CDA, Wang Irène, and Vial Jean-Claude, Two-photon excitation and stimulated emission depletion by a single wavelength., Opt Express, 19 (2011) 18036–18048. [DOI] [PubMed] [Google Scholar]
- [60].Friedrich M, Gan Q, Ermolayev V, Harms GS, STED-SPIM: Stimulated emission depletion improves sheet illumination microscopy resolution, Biophys J, 100 (2011) L43–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Friedrich M, Harms GS, Axial resolution beyond the diffraction limit of a sheet illumination microscope with stimulated emission depletion, J Biomed Opt, 20 (2015) 106006. [DOI] [PubMed] [Google Scholar]
- [62].Arroyo-Camejo S, Lazariev A, Hell SW, Balasubramanian G, Room temperature high-fidelity holonomic singlequbit gate on a solid-state spin, Nat Commun, 5 (2014) 4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF, Imaging intracellular fluorescent proteins at nanometer resolution, Science, 313 (2006) 1642–1645. [DOI] [PubMed] [Google Scholar]
- [64].Rust MJ, Bates M, Zhuang X, Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM), Nat Methods, 3 (2006) 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Solomon O, Mutzafi M, Segev M, Eldar YC, Sparsity-based super-resolution microscopy from correlation information, Opt Express, 26 (2018) 18238–18269. [DOI] [PubMed] [Google Scholar]
- [66].Bo Huang WW, Bates Mark, Zhuang Xiaowei, Reconstruction of super-resolution STORM images using compressed sensing based on low-resolution raw images and interpolation., Science, 319 (2008) 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Matsuda A, Shao L, Boulanger J, Kervrann C, Carlton PM, Kner P, Agard D, Sedat JW, Condensed mitotic chromosome structure at nanometer resolution using PALM and EGFP-histones, PLoS One, 5 (2010) e12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liu R, Liu Y, Liu S, Wang Y, Li K, Li N, Xu D, Zeng Q, Three-Dimensional Superresolution Imaging of the FtsZ Ring during Cell Division of the Cyanobacterium Prochlorococcus, mBio, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gu L, Li Y, Zhang S, Xue Y, Li W, Li D, Xu T, Ji W, Molecular resolution imaging by repetitive optical selective exposure, Nat Methods, 16 (2019) 1114–1118. [DOI] [PubMed] [Google Scholar]
- [70].Schmidt R, Weihs T, Wurm CA, Jansen I, Rehman J, Sahl SJ, Hell SW, MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope, Nat Commun, 12 (2021) 1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jones SA, Shim SH, He J, Zhuang X, Fast, three-dimensional super-resolution imaging of live cells, Nat Methods, 8 (2011) 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Fan Xu MZ, He Wenting, Han Renmin, Xue Fudong, Liu Zhiyong, Zhang Fa, Lippincott-Schwartz Jennifer, Xu Pingyong, Live cell single molecule-guided Bayesian localization., Nat Methods (2013) 96–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mats D.A.A.a.J.W.S. Gustafsson GL, Doubling the lateral resolution of wide-field fluorescence microscopy using structured illumination., 3919 (2000) 141–150. [Google Scholar]
- [74].Gustafsson MGL, Extended-Resolution Reconstruction of Structured Illumination Microscopy Data., Optical Society of America, (2005). [Google Scholar]
- [75].Heintzmann R, Huser T, Super-Resolution Structured Illumination Microscopy, Chem Rev, 117 (2017) 13890–13908. [DOI] [PubMed] [Google Scholar]
- [76].Fang H, Hu L, Chen Q, Geng S, Qiu K, Wang C, Hao M, Tian Z, Chen H, Liu L, Guan JL, Chen Y, Dong L, Guo Z, He W, Diao J, An ER-targeted “reserve-release” fluorogen for topological quantification of reticulophagy, Biomaterials, 292 (2023) 121929. [DOI] [PubMed] [Google Scholar]
- [77].K Qiu AY, Tian Z, Guo Z, Shi D, Nandi CK, Diao Jiajie, Ultralong-term super-resolution tracking of lysosomes in brain organoids by nearInfrared noble metal nanoclusters, ACS Materials Lett, 4 (2022) 1565–1573. [Google Scholar]
- [78].Gustafsson MGL, Nonlinear structured-illumination microscopywide-field fluorescence imaging with theoretically unlimited resolution., PNAS, 102 (2005) 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW, Threedimensional resolution doubling in wide-field fluorescence microscopy by structured illumination, Biophys J, 94 (2008) 4957–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Rego EH, Shao L, Macklin JJ, Winoto L, Johansson GA, Kamps-Hughes N, Davidson MW, Gustafsson MG, Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution, Proc Natl Acad Sci U S A, 109 (2012) E135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Betzig DLRE, Non-linear structured illumination microscopy, 2019.
- [82].Tingwei Quan HZ, Liu Xiaomao, Liu Yongfeng,Ding Jiuping, Zeng Shaoqun, and Huang Zhen-Li, High-density localization of active molecules using Structured Sparse Model and Bayesian Information Criterion., Optics express, 19 (2011) 16963–16974. [DOI] [PubMed] [Google Scholar]
- [83].Wen G, Li S, Wang L, Chen X, Sun Z, Liang Y, Jin X, Xing Y, Jiu Y, Tang Y, Li H, High-fidelity structured illumination microscopy by point-spread-function engineering, Light Sci Appl, 10 (2021) 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Chi KR, Super-resolution microscopy: breaking the limits, Nature Methods, 6 (2008) 15–18. [Google Scholar]
- [85].Huang B, Babcock H, Zhuang X, Breaking the diffraction barrier: super-resolution imaging of cells, Cell, 143 (2010) 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yong Wu X.W.a.E.S., Reducing Photobleaching in STED Microscopy with Higher Scanning Speed., Biophysical Journal, 108 (2015) 477a. [Google Scholar]
- [87].Fiolka R, Shao L, Rego EH, Davidson MW, Gustafsson MG, Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination, Proc Natl Acad Sci U S A, 109 (2012) 5311–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fang H, Yao S, Chen Q, Liu C, Cai Y, Geng S, Bai Y, Tian Z, Zacharias AL, Takebe T, Chen Y, Guo Z, He W, Diao J, De Novo-Designed Near-Infrared Nanoaggregates for Super-Resolution Monitoring of Lysosomes in Cells, in Whole Organoids, and in Vivo, ACS Nano, 13 (2019) 14426–14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Qiu RSK, Han G, Ishiyama M, Ueno Y, Tian Z, Sun Y, Diao J, De Novo Design of A Membrane-Anchored Probe for Multidimensional Quantification of Endocytic Dynamics, Adv Healthc Mater, 11 (2022) e2102185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Yang Z, Sharma A, Qi J, Peng X, Lee DY, Hu R, Lin D, Qu J, Kim JS, Super-resolution fluorescent materials: an insight into design and bioimaging applications, Chem Soc Rev, 45 (2016) 4651–4667. [DOI] [PubMed] [Google Scholar]
- [91].Li Q, Zhou T, Wu F, Li N, Wang R, Zhao Q, Ma YM, Zhang JQ, Ma BL, Subcellular drug distribution: mechanisms and roles in drug efficacy, toxicity, resistance, and targeted delivery, Drug Metab Rev, 50 (2018) 430–447. [DOI] [PubMed] [Google Scholar]
- [92].Varma MV, El-Kattan AF, Transporter-Enzyme Interplay: Deconvoluting Effects of Hepatic Transporters and Enzymes on Drug Disposition Using Static and Dynamic Mechanistic Models, J Clin Pharmacol, 56 Suppl 7 (2016) S99–S109. [DOI] [PubMed] [Google Scholar]
- [93].Balaz S, Subcellular pharmacokinetics and its potential for library focusing, Journal of Molecular Graphics and Modelling (2002). [DOI] [PubMed] [Google Scholar]
- [94].Fretz M, Jin J, Conibere R, Penning NA, Al-Taei S, Storm G, Futaki S, Takeuchi T, Nakase I, Jones AT, Effects of Na+/H+ exchanger inhibitors on subcellular localisation of endocytic organelles and intracellular dynamics of protein transduction domains HIV-TAT peptide and octaarginine, J Control Release, 116 (2006) 247–254. [DOI] [PubMed] [Google Scholar]
- [95].Guan M, Zhu QL, Liu Y, Bei YY, Gu ZL, Zhang XN, Zhang Q, Uptake and transport of a novel anticancer drug-delivery system: lactosyl-norcantharidin-associated N-trimethyl chitosan nanoparticles across intestinal Caco-2 cell monolayers, Int J Nanomedicine, 7 (2012) 1921–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rajendran L, Knolker HJ, Simons K, Subcellular targeting strategies for drug design and delivery, Nat Rev Drug Discov, 9 (2010) 29–42. [DOI] [PubMed] [Google Scholar]
- [97].Deshpande D, Grieshober M, Wondany F, Gerbl F, Noschka R, Michaelis J, Stenger S, Super-Resolution Microscopy Reveals a Direct Interaction of Intracellular Mycobacterium tuberculosis with the Antimicrobial Peptide LL-37, Int J Mol Sci, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Davison HR, Taylor S, Drake C, Phuan PW, Derichs N, Yao C, Jones EF, Sutcliffe J, Verkman AS, Kurth MJ, Functional fluorescently labeled bithiazole DeltaF508-CFTR corrector imaged in whole body slices in mice, Bioconjug Chem, 22 (2011) 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Marquardt K, Eicher AC, Dobler D, Mader U, Schmidts T, Renz H, Runkel F, Development of a protective dermal drug delivery system for therapeutic DNAzymes, Int J Pharm, 479 (2015) 150–158. [DOI] [PubMed] [Google Scholar]
- [100].Phuong Ha-Lien Tran TT-DT, Lee Kyoung-Ho,, Dong-Jin Kim B-JL, Dissolution-modulating mechanism of pH modifiers in solid dispersion containing weakly acidic or basic drugs with poor water solubility., Expert Opinion on Drug Delivery, 7 (2010) 647–661. [DOI] [PubMed] [Google Scholar]
- [101].Fuchigami T, Kitamoto Y, Namiki Y, Size-tunable drug-delivery capsules composed of a magnetic nanoshell, Biomatter, 2 (2012) 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Podczeck F, The relationship between physical properties of lactose monohydrate and the aerodynamic behaviour of adhered drug particles, International Journal of Pharmaceutics, 160 (1998) 119–130. [Google Scholar]
- [103].Podczeck F, The relationship between particulate properties of carrier materials and the adhesion force of drug particles in interactive powder mixtures, Journal of Adhesion Science and Technology, 11 (1997) 1089–1104. [Google Scholar]
- [104].Lv M, Zhou W, Tavakoli H, Bautista C, Xia J, Wang Z, Li X, Aptamer-functionalized metal-organic frameworks (MOFs) for biosensing, Biosens Bioelectron, 176 (2021) 112947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].ichelle H Teplensky MF, Li Peng, Wang Timothy C., Mehta Joshua P., Young Laurence J., Moghadam Peyman Z., Hupp Joseph T., Farha Omar K., Kaminski Clemens F., and Fairen-Jimene David, Temperature Treatment of Highly Porous Zirconium-Containing Metal-Organic Frameworks Extends Drug Delivery Release, J. Am. Chem. Soc, (2017). [DOI] [PubMed] [Google Scholar]
- [106].Schübbe S, Schumann C, Cavelius C, Koch M, Müller T, Kraegeloh A, Size-Dependent Localization and Quantitative Evaluation of the Intracellular Migration of Silica Nanoparticles in Caco-2 Cells, Chemistry of Materials, 24 (2011) 914–923. [Google Scholar]
- [107].Chen X, Cui J, Sun H, Mullner M, Yan Y, Noi KF, Ping Y, Caruso F, Analysing intracellular deformation of polymer capsules using structured illumination microscopy, Nanoscale, 8 (2016) 11924–11931. [DOI] [PubMed] [Google Scholar]
- [108].Sobolev AS, Modular transporters for subcellular cell-specific targeting of anti-tumor drugs, Bioessays, 30 (2008) 278–287. [DOI] [PubMed] [Google Scholar]
- [109].Zhang P, Yao J, Wang B, Qin L, Microfluidics-Based Single-Cell Protrusion Analysis for Screening Drugs Targeting Subcellular Mitochondrial Trafficking in Cancer Progression, Anal Chem, 92 (2020) 3095–3102. [DOI] [PubMed] [Google Scholar]
- [110].Cohen-Erez I, Rapaport H, Coassemblies of the Anionic Polypeptide gamma-PGA and Cationic beta-Sheet Peptides for Drug Delivery to Mitochondria, Biomacromolecules, 16 (2015) 3827–3835. [DOI] [PubMed] [Google Scholar]
- [111].Madni A, Rehman S, Sultan H, Khan MM, Ahmad F, Raza MR, Rai N, Parveen F, Mechanistic Approaches of Internalization, Subcellular Trafficking, and Cytotoxicity of Nanoparticles for Targeting the Small Intestine, AAPS PharmSciTech, 22 (2020) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].C.O.i. Anaesthesiology, Slow-release formulations of local anaesthetics and opioids, Current Opinion in Anaesthesiology, 9 (1996) 399–403. [Google Scholar]
- [113].Michael BMM Roberts S, Burczynski Frank J. and Weiss Michael, Enterohepatic circulation: Physiological, pharmacokinetic and clinical implications, Clinical Pharmacokinetics, 41 (2002) 751–790. [DOI] [PubMed] [Google Scholar]
- [114].Schübbe S, Cavelius C, Schumann C, Koch M, Kraegeloh A, STED Microscopy to Monitor Agglomeration of Silica Particles Inside A549 Cells, Advanced Engineering Materials, 12 (2010) 417–422. [Google Scholar]
- [115].Xin C, Cheng C, Hou K, Bao M, Zhang H, Wang Z, A novel melanin complex displayed the affinity to HepG2 cell membrane and nucleus, Mater Sci Eng C Mater Biol Appl, 122 (2021) 111923. [DOI] [PubMed] [Google Scholar]
- [116].Chen H, Wang H, Wei Y, Hu M, Dong B, Fang H, Chen Q, Super-resolution imaging reveals the subcellular distribution of dextran at the nanoscale in living cells, Chinese Chemical Letters, 33 (2022) 1865–1869. [Google Scholar]
- [117].Super-resolution imaging of drug-free macromolecular therapeutics, (2016).
- [118].Wang J, Bhattacharyya J, Mastria E, Chilkoti A, A quantitative study of the intracellular fate of pH-responsive doxorubicin-polypeptide nanoparticles, J Control Release, 260 (2017) 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Liu LY, Fang H, Chen Q, Chan MH, Ng M, Wang KN, Liu W, Tian Z, Diao J, Mao ZW, Yam VW, Multiple-Color Platinum Complex with Super-Large Stokes Shift for Super-Resolution Imaging of Autolysosome Escape, Angew. Chem. Int. Ed. Engl, 59 (2020) 19229–19236. [DOI] [PubMed] [Google Scholar]
- [120].Bhuvanendran S, Salka K, Rainey K, Sreetama SC, Williams E, Leeker M, Prasad V, Boyd J, Patterson GH, Jaiswal JK, Colberg-Poley AM, Superresolution imaging of human cytomegalovirus vMIA localization in sub-mitochondrial compartments, Viruses, 6 (2014) 1612–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Sara PW Fitzsimmons A, Grever Michael, Paull Kenneth, Camalier Richard, Lewis Alexander D., Reductase enzyme expression across the National Cancer Institute Tumor cell line panel correlation with sensitivity to mitomycin C and EO9., Journal of the National Cancer Institute, 88 (1996) 259–269. [DOI] [PubMed] [Google Scholar]
- [122].Sparkes I, Lessons from optical tweezers: quantifying organelle interactions, dynamics and modelling subcellular events, Curr Opin Plant Biol, 46 (2018) 55–61. [DOI] [PubMed] [Google Scholar]
- [123].Yang Z, Zhang Z, Zhao Y, Ye Q, Li X, Meng L, Long J, Zhang S, Zhang L, Organelle Interaction and Drug Discovery: Towards Correlative Nanoscopy and Molecular Dynamics Simulation, Front Pharmacol, 13 (2022) 935898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Nose MYN, Kinetic studies on disappearance pattern of sulfaquinoxaline from the organs and eggs of chickens., Japanese Poultry Science, 22 (1985) 245–255. [Google Scholar]
- [125].Sezaki MYYKTKH, Drug elimination function of rat small intestine: Metabolism and intraluminal excretion, Biochemical Pharmacology, 33 (1984) 3131–3136. [DOI] [PubMed] [Google Scholar]
- [126].Penner N, Xu L, Prakash C, Correction to Radiolabeled Absorption, Distribution, Metabolism, and Excretion Studies in Drug Development: Why, When, and How?, Chemical Research in Toxicology, 26 (2013) 1023–1023. [DOI] [PubMed] [Google Scholar]
- [127].Liao J, Peng H, Liu C, Li D, Yin Y, Lu B, Zheng H, Wang Q, Dual pH-responsive-charge-reversal micelle platform for enhanced anticancer therapy, Mater Sci Eng C Mater Biol Appl, 118 (2021) 111527. [DOI] [PubMed] [Google Scholar]
- [128].Khanam A, Yu J, Zempleni J, Class A Scavenger Receptor-Mediated Phagocytosis of Bovine Milk Exosomes in Murine Bone Marrow-Derived Macrophages, Current Developments in Nutrition, 5 (2021) 332. [Google Scholar]
- [129].Liping Wang GL, Zuo Zhongyuan, Li Yarong, Byeon Seul Kee, Pandey Akhilesh, Bellen Hugo J., Neuronal activity induces glucosylceramide that is secreted via exosomes for lysosomal degradation in glia., Sci. Adv, 8 (2022) eabn3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chen C, Zong S, Wang Z, Lu J, Zhu D, Zhang Y, Cui Y, Imaging and Intracellular Tracking of Cancer-Derived Exosomes Using Single-Molecule Localization-Based Super-Resolution Microscope, ACS Appl Mater Interfaces, 8 (2016) 25825–25833. [DOI] [PubMed] [Google Scholar]
- [131].Mu CF, Balakrishnan P, Cui FD, Yin YM, Lee YB, Choi HG, Yong CS, Chung SJ, Shim CK, Kim DD, The effects of mixed MPEG-PLA/Pluronic copolymer micelles on the bioavailability and multidrug resistance of docetaxel, Biomaterials, 31 (2010) 2371–2379. [DOI] [PubMed] [Google Scholar]
- [132].Zhang C, Shao H, Zhang J, Guo X, Liu Y, Song Z, Liu F, Ling P, Tang L, Wang KN, Chen Q, Long-term live-cell lipid droplet-targeted biosensor development for nanoscopic tracking of lipid droplet-mitochondria contact sites, Theranostics, 11 (2021) 7767–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].gerl UVN, Live-cell imaging of dendritic spines by STED microscopy.pdf, (2008). [DOI] [PMC free article] [PubMed]
- [134].Jin D, Xi P, Wang B, Zhang L, Enderlein J, van Oijen AM, Nanoparticles for super-resolution microscopy and single-molecule tracking, Nat Methods, 15 (2018) 415–423. [DOI] [PubMed] [Google Scholar]
- [135].Kamiyama D, Huang B, Development in the STORM, Dev Cell, 23 (2012) 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Dahms TES, Super-Resolution Microscopy SIM, STED and Localization Microscopy.pdf, (2015).
- [137].Shivanandan A, Analytical Methods, Correlative Microscopy and Software Tools for Quantitative Single Molecule Localization Microscopy, ioc manuals & guides, (2015). [Google Scholar]
- [138].Fernandez-Suarez M, Ting AY, Fluorescent probes for super-resolution imaging in living cells, Nature reviews. Molecular cell biology, 9 (2008) 929–943. [DOI] [PubMed] [Google Scholar]
- [139].Liu CG, Han YH, Kankala RK, Wang SB, Chen AZ, Subcellular Performance of Nanoparticles in Cancer Therapy, Int J Nanomedicine, 15 (2020) 675–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Rovira-Clave X, Jiang S, Bai Y, Zhu B, Barlow G, Bhate S, Coskun AF, Han G, Ho CK, Hitzman C, Chen SY, Bava FA, Nolan GP, Subcellular localization of biomolecules and drug distribution by high-definition ion beam imaging, Nat Commun, 12 (2021) 4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Laulicht B, Cheifetz P, Tripathi A, Mathiowitz E, Are in vivo gastric bioadhesive forces accurately reflected by in vitro experiments?, J Control Release, 134 (2009) 103–110. [DOI] [PubMed] [Google Scholar]
- [142].Shaimaa A.A.A.R.a.A.-W.R.h. Abd Al Hammid N, Formulation and Evaluation of Floating Mefenamic Acid Drug Delivery Tablet., International Journal of Pharmacy and Medical Sciences 2(2012) 1–6. [Google Scholar]
- [143].A Singh DYK, Neelima K, Poornima B, Oral matrix formulation for floating drug delivery system to increase gastric retention time, L, 1 (2013) 32–39., JPBMA, 1 (2013) 32–39. [Google Scholar]
- [144].Ram MBA-D, Gupta P, Cytochrome P450 enzymes in chickens: characteristics and induction by xenobiotics, Comparative Biochemistry and Physiology Part C 121 (1998) 73–83. [DOI] [PubMed] [Google Scholar]
- [145].Lukinavicius G, Mitronova GY, Schnorrenberg S, Butkevich AN, Barthel H, Belov VN, Hell SW, Fluorescent dyes and probes for super-resolution microscopy of microtubules and tracheoles in living cells and tissues, Chem Sci, 9 (2018) 3324–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Morikawa Y, Kitazato M, Morita T, Mizunaga S, Mitsuyama J, Clinical significance of cerebrospinal fluid inhibitory titers of antibiotics, based on pharmacokinetic/pharmacodynamic parameters, in the treatment of bacterial meningitis, J Infect Chemother, 15 (2009) 233–238. [DOI] [PubMed] [Google Scholar]
- [147].Ogata K, Mendell-Harary J, Tachibana M, Masumoto H, Oguma T, Kojima M, Kunitada S, Clinical safety, tolerability, pharmacokinetics, and pharmacodynamics of the novel factor Xa inhibitor edoxaban in healthy volunteers, J Clin Pharmacol, 50 (2010) 743–753. [DOI] [PubMed] [Google Scholar]
- [148].Velickovic-Radovanovic R, Mikov M, Paunovic G, Djordjevic V, Stojanovic M, Cvetkovic T, Djordjevic AC, Gender differences in pharmacokinetics of tacrolimus and their clinical significance in kidney transplant recipients, Gend Med, 8 (2011) 23–31. [DOI] [PubMed] [Google Scholar]
- [149].Thimiri Govinda Raj DB, Khan NA, Surface functionalization dependent subcellular localization of Superparamagnetic nanoparticle in plasma membrane and endosome, Nano Converg, 5 (2018) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Rajendran L, Udayar V, Goodger ZV, Lipid-anchored drugs for delivery into subcellular compartments, Trends Pharmacol Sci, 33 (2012) 215–222. [DOI] [PubMed] [Google Scholar]
- [151].El-Kadi AA-MA, P-glycoprotein effects on drugs pharmacokinetics and drug-drug-interactions and their clinical implications., Libyan J Pharm & Clin Pharm, 1 (2012) 48154. [Google Scholar]
- [152].Lidke DS, Lidke KA, Advances in high-resolution imaging--techniques for three-dimensional imaging of cellular structures, J Cell Sci, 125 (2012) 2571–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Sigrist SJ, Sabatini BL, Optical super-resolution microscopy in neurobiology, Curr Opin Neurobiol, 22 (2012) 86–93. [DOI] [PubMed] [Google Scholar]
- [154].Bullen A, Microscopic imaging techniques for drug discovery, Nature Reviews Drug Discovery, 7 (2008) 54–67. [DOI] [PubMed] [Google Scholar]
- [155].Fang G, Chen H, Shao X, Wang H, Zhan D, Wang R, Meng P, Fang H, Liu F, Ling P, Wu Z, Diao J, Yao Q, Chen Q, Single Image Capture of Bioactive Ion Crosstalk within Inter-Organelle Membrane Contacts at Nanometer Resolution, Small Methods, 6 (2022) e2200321. [DOI] [PubMed] [Google Scholar]
- [156].Paillard A, Hindre F, Vignes-Colombeix C, Benoit JP, Garcion E, The importance of endo-lysosomal escape with lipid nanocapsules for drug subcellular bioavailability, Biomaterials, 31 (2010) 7542–7554. [DOI] [PubMed] [Google Scholar]
- [157].Lee SD, Lim SJ, Kim CK, Effect of chemical modification on the pharmacokinetics and biodistribution of carboxymethyl dextran as a drug carrier, Drug Deliv, 9 (2002) 187–193. [DOI] [PubMed] [Google Scholar]
- [158].Zhou F, Zhang J, Li P, Niu F, Wu X, Wang G, Roberts MS, Toward a new age of cellular pharmacokinetics in drug discovery, Drug Metab Rev, 43 (2011) 335–345. [DOI] [PubMed] [Google Scholar]
- [159].Annette AEE, Larsen K, Skladanowski Andrzej, Resistance mechanisms associated with altered intracellular distribution of anticancer agents, Pharmacology & Therapeutics, 85 (2000) 217–229. [DOI] [PubMed] [Google Scholar]
- [160].Hauser M, Wojcik M, Kim D, Mahmoudi M, Li W, Xu K, Correlative Super-Resolution Microscopy: New Dimensions and New Opportunities, Chem Rev, 117 (2017) 7428–7456. [DOI] [PubMed] [Google Scholar]
- [161].Liu W, Du G, Guo J, Wu R, Wei J, Chen H, Li Y, Zhao J, Li X, Influence of the environment and phototoxicity of the live cell imaging system at IMP microbeam facility, Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms, 404 (2017) 125–130. [Google Scholar]
- [162].AINSWORTH OH-GTD, LEGGAT W,, Imaging the fluorescence ofmarine invertebrates and their associated flora, Journal ofMicroscopy, 232 (2008) 197–199. [DOI] [PubMed] [Google Scholar]
- [163].Qiu Kangqiang TZ, Jiajie Diao, Monitoring lysosomal acidity, Nat Chem Biol, 19 (2023). [DOI] [PubMed] [Google Scholar]
- [164].Thurber GM, Yang KS, Reiner T, Kohler RH, Sorger P, Mitchison T, Weissleder R, Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo, Nat Commun, 4 (2013) 1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kim M, Chen C, Yaari Z, Frederiksen R, Randall E, Wollowitz J, Cupo C, Wu X, Shah J, Worroll D, Lagenbacher RE, Goerzen D, Li Y-M, An H, Wang Y, Heller DA, Nanosensor-based monitoring of autophagy-associated lysosomal acidification in vivo, Nat. Chem. Biol, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


