Abstract
The β-barrel assembly machinery (Bam) complex facilitates the assembly of outer membrane proteins (OMPs) in gram-negative bacteria. The Bam complex is conserved and essential for bacterial viability and consists of five subunits, BamA-E. BamA is the transmembrane component, and its β-barrel domain opens laterally to allow folding and insertion of incoming OMPs. The remaining components are regulatory, among which only BamD is essential. Previous studies suggested that BamB regulates BamA directly, while BamE and BamC serve as BamD regulators. However, specific molecular details of their functions remain unknown.
Our previous research demonstrated that BamE plays a specialized role in assembling the complex between the lipoprotein RcsF and its OMP partners, required for the Regulator of Capsule Synthesis (Rcs) stress response. Here, we used RcsF/OmpA as a model substrate to investigate BamE function. Our results challenge the current view that BamE only serves as a BamD regulator. We show that BamE also directly interacts with BamA. BamE interaction with both BamA and BamD are important for function. Our genetic and biochemical analysis show that BamE stabilizes the Bam complex and promotes bidirectional signaling interaction between BamA and BamD. This BamE function becomes essential when direct BamA/BamD communication is impeded.
Keywords: outer membrane biogenesis, bacterial cell envelope, membrane proteins
Graphical Abstract
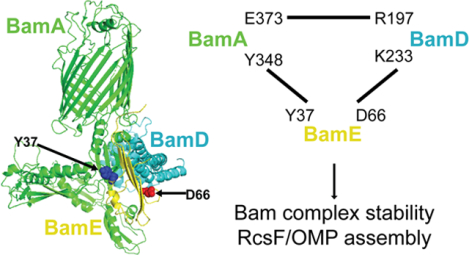
The Bam complex is essential for folding and inserting outer membrane proteins in gram-negative bacteria and is a key target for antibiotic development. However, the specific functions of its components, especially BamE, are poorly understood. Our study demonstrates that BamE directly interacts with essential components BamA and BamD, and plays a critical role in coordinating their activities, promoting the assembly of challenging substrates, such as the stress-sensing RcsF/OMP complex.
INTRODUCTION
Cells of gram-negative bacteria are surrounded by two membranes, a cytoplasmic and an outer membrane (OM) (Saha et al., 2021). The outer membrane has a distinct structure, being an asymmetrical bilayer and containing a large number of β-barrel OM proteins (OMPs) (Sun et al., 2022). The assembly of OMPs is facilitated by the β-barrel assembly machinery (Bam) complex (Tomasek & Kahne, 2021). Bam complex is highly conserved and essential for bacterial viability, making it an attractive target for antibiotic development (Overly Cottom et al., 2023).
In the best-studied Escherichia coli model system, the Bam complex is a stable heteropentamer formed by BamA-E subunits (Wu et al., 2005, Sklar et al., 2007). Most of the Bam complex research thus far has been focused on BamA. BamA is the only transmembrane component of the Bam complex as it contains a β-barrel domain. Its β -barrel domain is quite unusual because it can open up laterally, providing a site for folding and insertion of the incoming OMP substrates (Shen et al., 2023, Doyle et al., 2022, Lee et al., 2019, Tomasek et al., 2020, Wu et al., 2021). Additionally, BamA is the most conserved component of the complex and is found in all gram-negative bacteria, and also two-membrane organelles of bacterial origin, such as mitochondria and chloroplasts (Misra, 2012, Diederichs et al., 2021, Webb et al., 2012). BamA also contains a periplasmic region made up by five POTRA (polypeptide transport–associated) domains. The POTRA domains scaffold four lipoprotein components BamB, C, D, and E, tethered to OM periphery by their N-terminal lipid modifications. In E. coli, BamD is the only essential component (Malinverni et al., 2006); BamB,C,E are individually dispensable but collectively essential for viability (Tellez & Misra, 2012, Sklar et al., 2007, Rigel et al., 2012, Hart & Silhavy, 2020). Mutant variants of BamA have been reported that allow it to function in the absence of the Bam lipoproteins (Hart & Silhavy, 2020, Tellez & Misra, 2012, Hart et al., 2020), indicating that BamA alone is sufficient for the OMP assembly, while Bam lipoproteins likely play a regulatory role, although their specific mechanistic functions remain unknown. As the lipoprotein components are also less conserved in other species (Webb et al., 2012), they likely have specialized functions.
Previous studies have suggested that OMP assembly involves a cycle of coordinated conformational changes in BamA and BamD (Lee et al., 2018, Hagan et al., 2015, Hagan et al., 2013, Pavlova et al., 2013, McCabe et al., 2017). During this cycle, the transition between active conformation states of both proteins is facilitated by a signaling interaction that relies on a dynamic electrostatic network between BamA’s POTRA5 domain and BamD (McCabe et al., 2017, Sinnige et al., 2015). Introducing mutations at the BamA/BamD interface deregulates the two essential components, preventing them from adopting functionally compatible conformations (Ricci et al., 2012, McCabe et al., 2017).
Mutations at the BamA/BamD interface also separate the Bam complex into two subcomplexes, BamAB and BamDCE (Malinverni et al., 2006, Ricci et al., 2012, McCabe et al., 2017). While the stability of the heteropentameric Bam complex is not required for its function (Ricci et al., 2012, McCabe et al., 2017), this observation laid a foundation for the widely accepted model, in which BamB regulates BamA directly, while BamE and BamC serve as BamD regulators (Hart & Silhavy, 2020). BamB increases the efficiency of the OMP assembly in vivo and in vitro (Hagan et al., 2010, Hagan & Kahne, 2011, Roman-Hernandez et al., 2014, Mahoney et al., 2016). BamE and BamC remain the most mysterious components of the Bam complex, as their deletion does not affect OMP assembly.
Our previous research provided the first direct evidence of the specialized functions of Bam lipoproteins by demonstrating that BamE is uniquely required for the assembly of the OM complex between the lipoprotein RcsF and its OMP partners (Tata & Konovalova, 2019, Konovalova et al., 2016). The RcsF/OMP complexes function in the Regulator of Capsule Synthesis (Rcs) stress response, allowing RcsF to monitor integrity of the OM (Lach et al., 2023, Tata et al., 2021). Identification of the RcsF/OMP complexes as the first substrate of the Bam complex that requires BamE activity, presented an opportunity to study a molecular function of BamE. Our work suggested that BamE coordinates BamA and BamD to prevent a formation of an aberrant off-pathway dead-end complex between RcsF and BamA, that can sequester BamA from its function in the OMP assembly (Tata et al., 2021, Tata & Konovalova, 2019). This BamA sequestration by RcsF is a molecular reason for synthetic lethal interactions of ΔbamE with mutations that lower Bam complex levels or efficiency, for example, those caused by bamB null mutations(Hart et al., 2019, Tata & Konovalova, 2019).
The purpose of this study is to investigate how BamE coordinates the activities of BamA and BamD. Our results challenge the current view that BamE only serves as a BamD regulator. We demonstrate that BamE can directly interact with BamA, independently of other Bam components. Using genetic and biochemical analysis, we show that BamE stabilizes BamA/BamD complex and facilitates bidirectional signaling interaction among them. BamE function becomes essential when mutations at the BamA/BamD interface disrupt their direct communication, and this essentiality is dependent on RcsF. As BamE and its BamA and BamD interaction interface are more broadly conserved than RcsF, we propose that BamE is particularly important for assembling challenging substrates that impede the ability of BamA and BamD to communicate directly during their assembly pathway.
RESULTS
BamE stabilizes BamA-BamD interaction in vivo and in vitro.
BamA/D interaction was reported to be destabilized in vivo in the ΔbamE mutant strain (Sklar et al., 2007). We confirmed this finding using pull-down experiments with Strep-BamA as the bait; BamD co-purified with BamA in a BamE-dependent manner (Fig. 1A). BamE is required for BamA/BamD coordination only in the presence of RcsF, suggesting that RcsF binding to BamA may prevent direct BamA/BamD communication. Therefore, we sought to clarify the underlying relationship between BamE, RcsF, and the stability of the BamAD complex. For this, we tested whether BamA/D destabilization results from the absence of BamE or stalling of RcsF on BamA by comparing the stability of the BamA/D complex with or without RcsF. We observed that BamE was required for BamA/BamD co-purification regardless of the presence of RcsF (Fig. 1A).
Figure1. BamE stabilizes BamA-BamD interaction in vivo and in vitro.
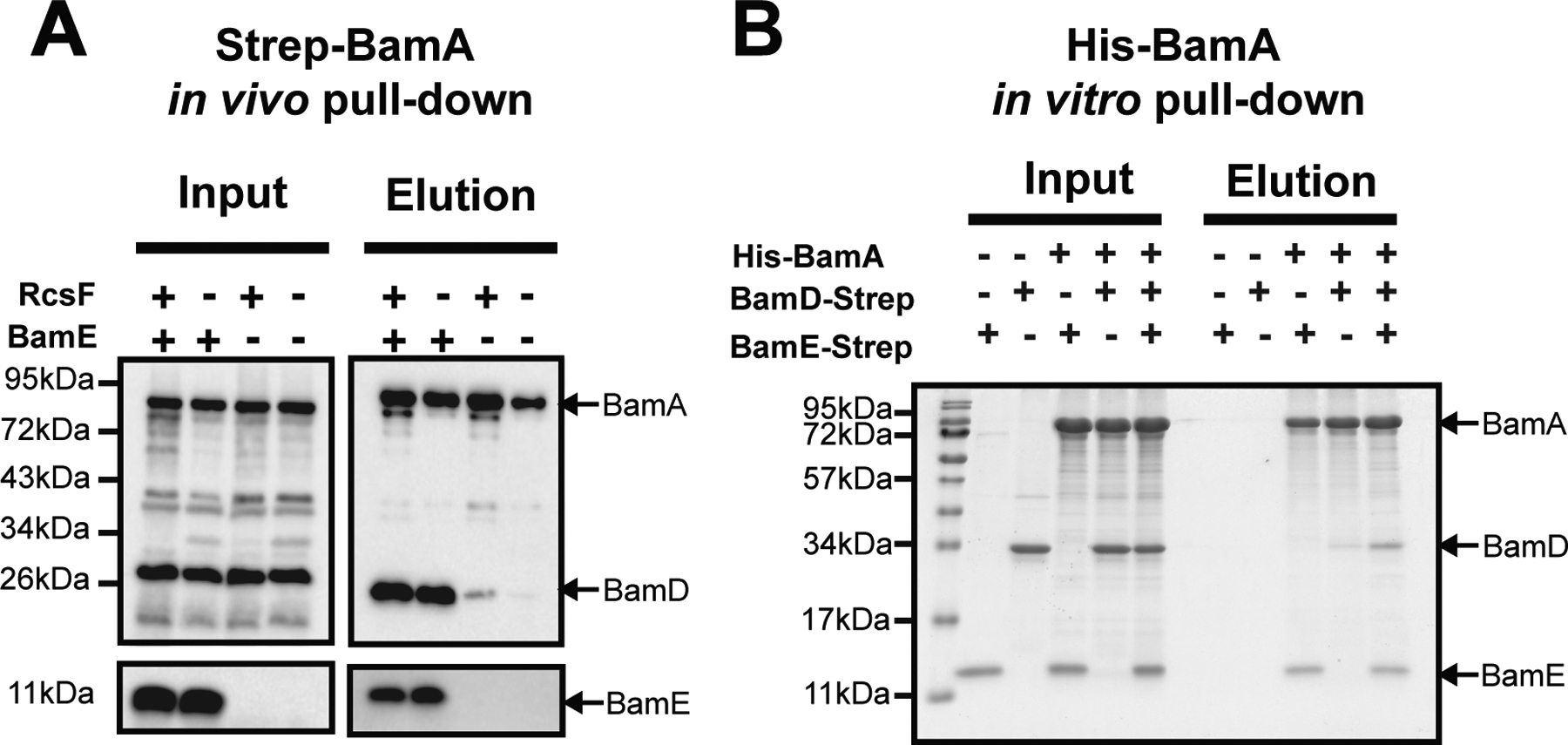
(A) Effect of BamE and RcsF on the stability of BamA and BamD interaction using in vivo pull-down assay with Strep-BamA expressed from the low copy pZS21 plasmid (Tata et al., 2021). Total cell lysates containing solubilized membranes were subjected to Streptacin purification. The figure represents the immunoblot analysis of input and elution fractions with α-BamA, BamD, and BamE antibodies. For immunoblot quantifications of independent biological replicates, see Fig. S1A. (B) In vitro interaction assay using purified components. Individual proteins were purified separately, mixed at the equimolar ratio, and subjected to a Ni-NTA purification. The figure represents the SDS-PAGE analysis of input and elution fractions visualized by the Imperial Protein Stain. For immunoblot quantifications of independent biological replicates, see Fig S1B.
Previous studies suggested that BamE only interacts with BamD based on in vivo pull-down experiments (Malinverni et al., 2006, Ricci et al., 2012). However, when the first Bam complex structures became available, they revealed direct BamA/BamE contact sites (Bakelar et al., 2016, Iadanza et al., 2016). Nonetheless, whether BamE can interact with BamA directly in the absence of the other Bam components, namely BamD, has not been tested. Therefore, we decided to examine the interactions between the BamA/BamE/BamD subunits in vitro (Fig. 1B). We expressed and purified soluble (without lipid modification) BamE-Strep and BamD-Strep proteins and the detergent-refolded His-BamA, mixed the components at the equimolar ratio, and subjected them to the Ni-NTA purification. To our surprise, we observed that BamE co-purified with BamA (Fig. 1B), providing the first experimental evidence for direct BamE/BamA interaction. In contrast, BamD did not efficiently co-purified with BamA (Fig. 1B) despite a large interaction interface between the two proteins observed in the various Bam complex structures. However, the efficiency of BamD co-purification with BamA improved significantly in the presence of BamE (Fig. 1B), demonstrating that BamE stabilizes BamA/BamD interaction in vitro.
Examination of BamE/BamA and BamE/BamD interface.
To facilitate further functional studies, we investigated BamA/BamE and BamD/BamE interaction interface more closely to identify mutations that disrupt corresponding interactions. In various E. coli Bam complex structures, these interfaces are nearly identical: BamE interaction with the Potra 5 domain of BamA is facilitated by several hydrogen bonds (Fig 2A, and S2, S3 Table S1), while BamE interaction with BamD involves both hydrogen bonds and a salt bridge interaction (Fig. 2A and S2, S3, and Table S2). Although the interaction residues are conserved in many but not all species (Fig. S3), Collab Fold Multimer modeling (Mirdita et al., 2022) revealed a high degree of conservation of interaction interfaces between the corresponding protein pairs, suggesting that the proteins likely co-evolved (Fig. S2, Tables S1,S2).
Figure 2. BamE/BamA and BamE/BamD interaction interface.
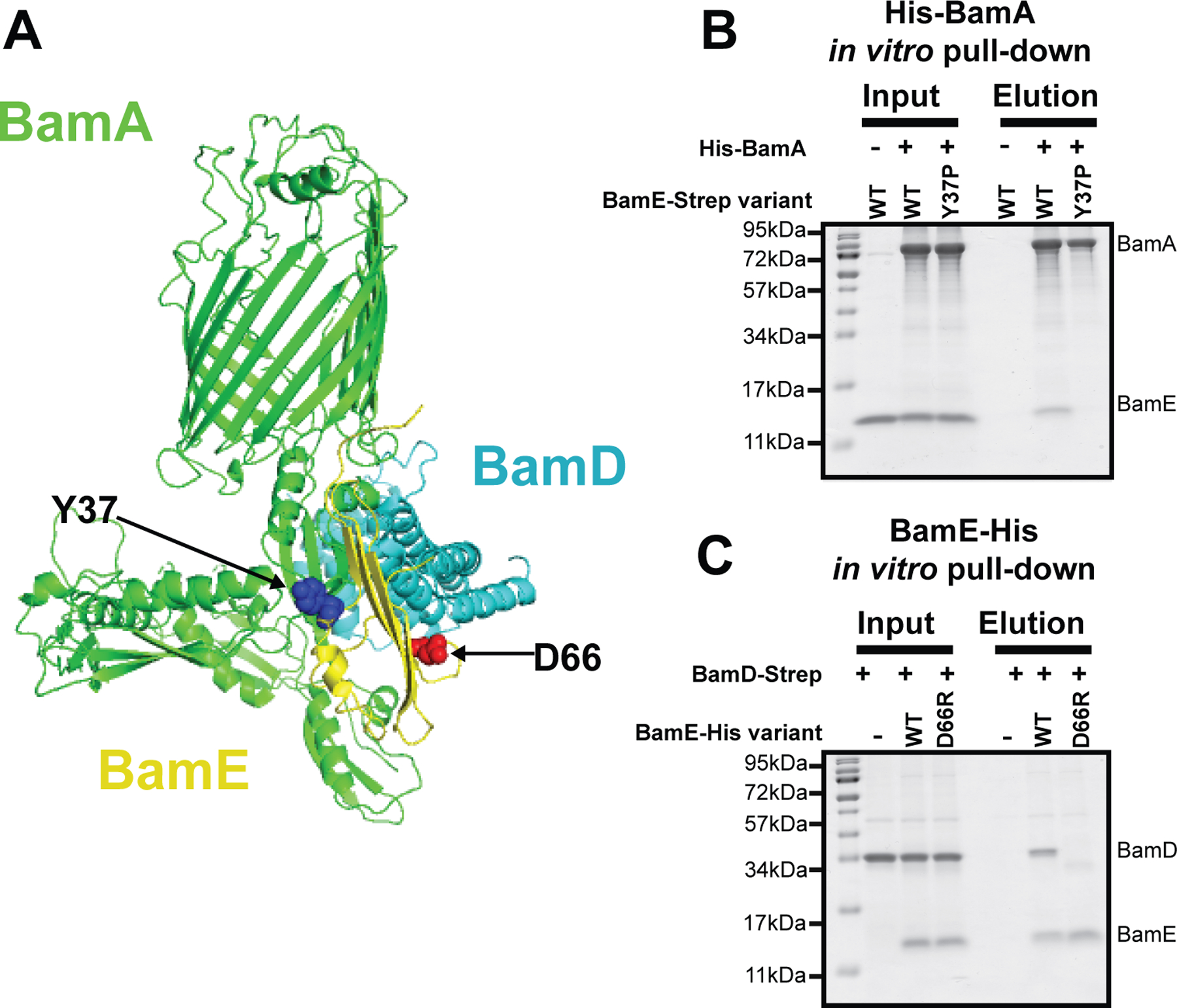
(A) The structure of Bam Complex (PDB: 5ekq (Bakelar et al., 2016)), BamA (green), BamE (yellow), BamD (blue) are shown with the remaining components omitted for clarity. Y37 and D66 residues of BamE chosen for this study are visualized by blue and red spheres (B, C). In vitro interaction assay using purified components. Individual proteins and mutant derivatives were purified separately, mixed at the equimolar ratio, and subjected to Ni-NTA purification. The figure represents the SDS-PAGE analysis of input and elution fractions visualized by the Imperial Protein Stain. (B) In vitro interaction between His-BamA and BamE-Strep variants. (C) In vitro interaction between BamD-Strep and BamE-His variants.
We then selected several possible residues on BamE that facilitate BamA or BamD interaction for further analysis using the following criteria: the residues established polar contacts in several independent Bam complex structures; structure-based in silico mutagenesis of the residues disfavored the corresponding interaction; the residues were well separated from each other on the BamE structure and away from the BamA/BamD interaction interface to facilitate interpretation of the downstream genetic analysis. Here, we report on two of such BamE residues, Y37 and D66 (Fig. 2A).
BamE Y37 residue falls on the BamA interface and interacts with several hydrogen bonds with Y348 residue of BamA (Fig. 2A, Fig. S4). Y37 residue is within an unstructured loop of BamE, and its mutation is not expected to disrupt BamE folding (Fig. S4). We generated bamE(Y37P) mutation because, based on the in silico mutagenesis, proline was the only substitution that resulted in a decrease in hydrogen bonding at the interface and therefore is most likely a disruptor of BamE/BamA interaction (Fig. S4). Indeed, when we purified the BamE(Y37P) variant, it was unable to bind to His-BamA in vitro (Fig. 2B).
BamE D66 falls within the BamD interface (Fig. 2A, Fig. S5)) and forms the salt bridge with BamD K233 residue. D66 is also located in an unstructured loop on the opposite surface of the BamE molecule, and its mutation was also not expected to disrupt BamE folding but weakened BamE/BamD interface (Fig. S5). Introducing charge-substitution mutation, D66R, resulted in a loss of the BamE/BamD interaction in vitro (Fig. 2C).
We concluded that the BamE interactions with both BamA and BamD we observed in vitro are facilitated by the same interface that has been observed in the Bam complex structures. Furthermore, these interfaces are conserved in many other bacterial species, which suggests that they are functionally significant.
Both BamA/BamE and BamE/BamD interactions is required for function.
Our initial observation revealed that BamE directly interacts with BamA and BamD, promoting the stability and function of the Bam complex. To further investigate the mechanism by which BamE achieves this, we aimed to distinguish between BamE’s role as a structural bridge between these essential components and its ability to regulate their conformations. If BamE functions solely as a structural bridge, disrupting either the BamE/A or BamE/D interfaces should result in a loss-of-function phenotype, mimicking the phenotype observed in the ΔbamE mutant. However, if BamE plays a regulatory role for BamA or BamD, mutations in the BamE/A or BamE/D interfaces may produce distinct phenotypes, suggesting that one interface is more crucial for complex stability and/or RcsF/OMP assembly than the other.
To address the BamE function, we conducted in vivo phenotypic characterization of bamE(Y37P), bamE(D66R), and the double mutants. For this, we cloned bamE with or without corresponding mutations into a pTrc99a vector (Amann et al., 1988). We employed a leaky expression from the trc promoter (without an inducer) to ensure physiological protein levels (Fig. S6), and tested phenotypes in the complementation experiments in the ΔbamE background.
First, we examined the impact of the mutations on the Bam complex stability in vivo using pull-down experiments (Fig. 3). Surprisingly, we found that not only the BamE(D66R) variant failed to co-purify with BamD or BamA (Fig. 3A), but also disrupted the interaction between BamA and BamD evident from the reciprocal Strep-BamA pull-down (Fig. 3B). On the other hand, bamE(Y37P) mutation had no impact the complex stability, and the bamE(D66R Y37P) mutant phenocopied bamE(D66R) (Fig. 3A,B). Because the Y37P and D66R mutations resulted in different outcomes, we concluded that rather than serving as a bridge BamE, BamE acts as an allosteric regulator, stimulating BamD to interact with BamA, thereby stabilizing the BamADE complex.
Figure 3. Characterization of bamE mutants.
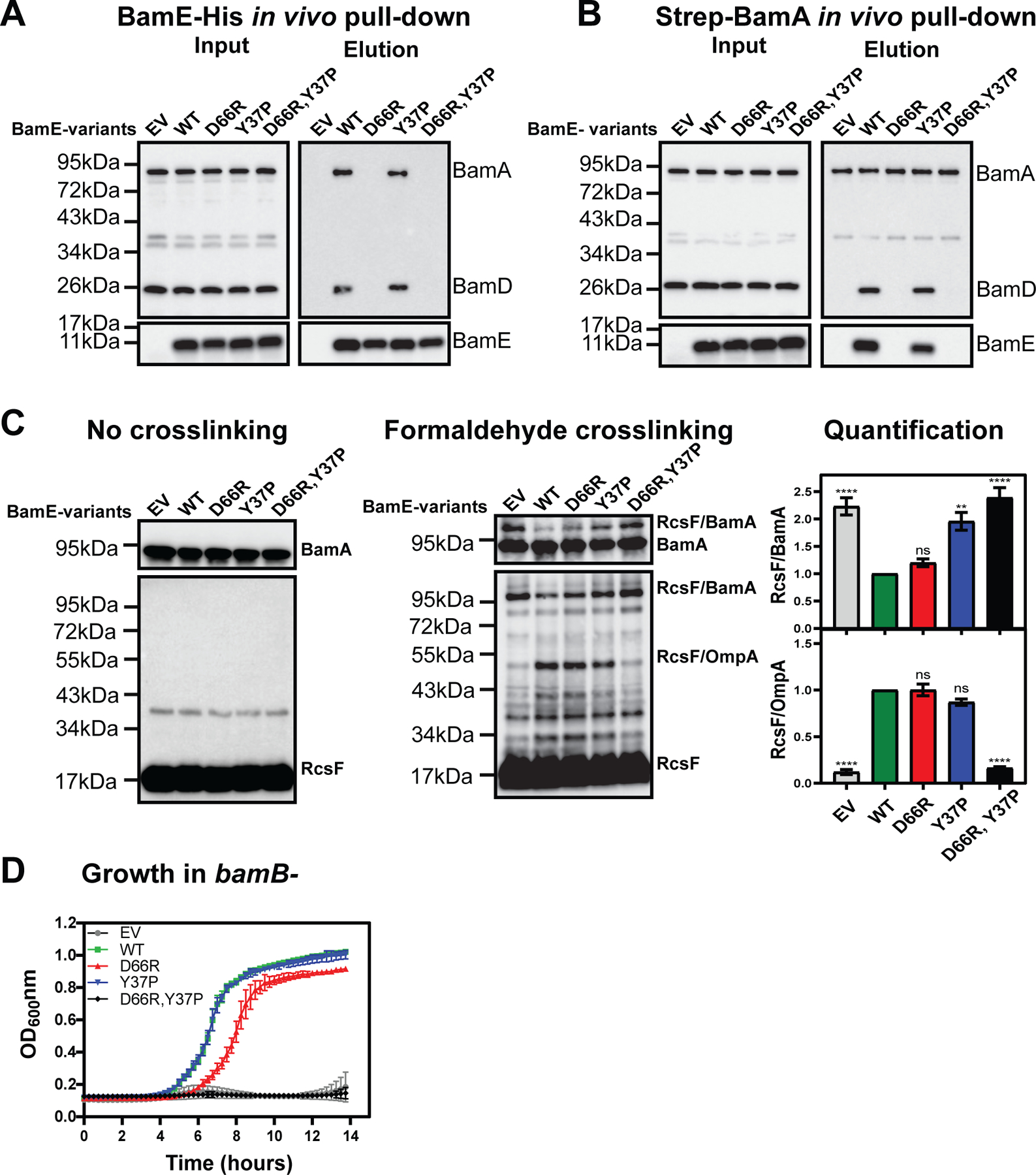
(A, B) Effect of bamE mutations on the stability of BamA/BamD/BamE interaction in vivo using Ni-NTA pull-down of BamE-His variants (A) or Streptactin pull-down of Strep-BamA in the presence of BamE-His variants (B). Empty vector (EV) pTrc99a, and its derivatives with indicated bamE-His alleles were transformed in the MT-567 [ΔbamE ΔbamA//pZS21::Strep-bamA] background, and resulting strains were subjected to the in vivo pull-down followed by immunoblot analysis as described in Fig. 1. For immunoblot quantifications of independent biological replicates, see Fig S7. (C) Immunoblot analysis of in vivo formaldehyde crosslinked samples probed with α-BamA (Top) and α-RcsF antibodies (bottom). For band validation, see (Tata & Konovalova, 2019) Quantification of RcsF/BamA and RcsF/OmpA crosslinking bands based on three independent biological replicates relative to the WT control, mean+/− standard error of the mean (SEM). Statistical analysis was performed by using one-way ANOVA in comparison with the WT control. n.s. = P ≥ 0.05, **P <0.01, ****P < 0.0001. (D) Growth curve analysis. EV and plasmids with indicated bamE-His8 alleles were transformed in the AK-1255 [ΔbamE bamB8 PBAD-bamE] background and propagated in the presence of arabinose to allow BamE expression from the chromosomal locus. Overnight cultures were used to inoculate media without arabinose at the approximate cell density of 105 cell/ml to allow depletion of BamE expressed from the chromosomal locus. Growth was monitored continuously at 37°C in the Bioteck plate reader.
Because the Bam complex stability does not always correlate with function (McCabe et al., 2017), we used RcsF/OMP assembly as a direct functional readout. In the absence of BamE cells fail to assemble RcsF complex with partner OMPs (Lach et al., 2023), but we use OmpA as a convenient readout in the formaldehyde crosslinking experiments (Tata & Konovalova, 2019, Tata et al., 2021). RcsF/OmpA complex is significantly reduced in the absence of BamE, and instead, RcsF accumulated on BamA (Fig. 3C). Even though bamE(D66R) mutation led to the destabilization of BamA/D interaction, it had no impact on RcsF crosslinking (Fig. 3C). In contrast, bamE(Y37P) displayed increased RcsF/BamA crosslinking, but still assembled RcsF/OmpA complexes (Fig. 3C). We interpret this intermediate phenotype as an indication that in the bamE(Y37P) strain, a significant fraction of BamA exists in the conformation prone to RcsF binding in the off-pathway conformation, while the remaining fraction of still capable of assembling RcsF/OmpA complex. Importantly, in the bamE(D66R Y37P) mutant RcsF/OmpA assembly was significantly reduced, and RcsF/BamA crosslinking was increased, phenocopying ΔbamE strain (Fig. 3C).
The formation of off-pathway RcsF/BamA complex leads to the BamA sequestration from its function in the OMP assembly, underlying the RcsF-dependent synthetic lethal interaction of ΔbamE with bamB null and several other bam mutant alleles (Tata & Konovalova, 2019, Hart et al., 2019, Tata et al., 2021). Therefore, we also tested the bamE mutants for their ability to complement bamE bamB synthetic lethality (Fig. 3D). We introduced the plasmids into the AK-1255 [ΔbamE bamB8 PBAD-bamE] background that contains a loss-of-function allele of bamB and bamE under control of arabinose inducible promoter (Tata et al., 2021). Growth of this strain in the absence of arabinose requires the presence of a functional bamE allele on a plasmid (Fig. 3D). Consistent with our crosslinking experiments, both bamE(Y37P) and bamE(D66R) mutants were able to grow, while the double mutant behaved like the EV control (Fig. 3D).
We also tested bamE(Y37G) and bamE(D66G) mutants to establish whether the observed phenotypes were due to specific amino acid substitution. bamE(Y37G) phenocopied bamE(Y37P) in every assay (Fig. S8). bamE(D66G) mutant had a somewhat different phenotype, as, unlike bamE(D66R), it did not destabilize the BamADE complex (Fig. S9). However, when combined with Y37P mutation, it resulted in the same outcome in complex stability, RcsF assembly, and growth assays (Fig. S9). It suggests that D66G is a milder version of D66R mutation and uncovers that BamA co-purification with BamE(Y37P) was likely facilitated by BamD.
Together, our results demonstrated that both BamE/BamA and BamE/BamD interactions are functionally important but not equivalent. BamE/BamD interaction is more important for promoting BamA/BamD interaction and overall BamADE complex stability, as D66R mutation alone destabilized BamADE complex. However, even without the stable association, BamE can directly promote BamA function, as only when Y37P was introduced cells lost the ability to assemble RcsF.
BamE becomes essential when direct communication between BamA and BamD is compromised.
To ensure proper OMP assembly, BamA and BamD interact to undergo coordinated conformational changes (McCabe et al., 2017, Ricci et al., 2012). The stable BamA/BamD association is not required for OMP assembly, however, the signaling interaction between BamA and BamD that regulates the necessary conformational changes in both components is essential. As our results suggest that BamE plays a similar function, we hypothesized that BamE may become more important when BamA/BamD interaction is altered by the mutations at the BamA/BamD interface.
The interaction between BamA and BamD is facilitated by an electrostatic network involving the E373 and R197 residues (Hagan et al., 2015, Hart & Silhavy, 2020). The bamA(E373K) mutation is lethal because it prevents the activation of BamD, and BamD remains in the functionally incompatible conformation. The bamA(E373A) mutation is less severe, allowing for some interaction between BamA and BamD (Ricci et al., 2012).
We observed that bamA(E373A) exhibits synthetic lethal interaction with bamE null (Fig. 4A, Fig. S10, S11A), which is consistent with previous studies (Rigel et al., 2012). bamE null derivative strains could not grow under standard laboratory conditions (LB media at 37°C), but could grow under permissive conditions (glucose minimal medium at 30°C) (Fig. S10, S11) because slower growth lowers the demand for the Bam complexes. We found that bamA(E373A) bamE synthetic lethal phenotype was also rcsF-dependent (Fig. S10), suggesting that the underlying reason is the RcsF-sequestration of BamA. To further investigate the role of BamE, we introduced bamE point mutants and tested them in complementation experiments (Fig. 4A, Fig. 5, Fig. S11A). bamA(E373A) bamE(D66R) mutant was not viable under standard conditions (Fig. 4a, Fig. S1A). This result suggests that BamE and BamA play partially overlapping functions in regulating BamD conformations.
Figure 4. Characterization of bamE mutants in the bamA(E373A) and bamD(R197L) backgrounds.
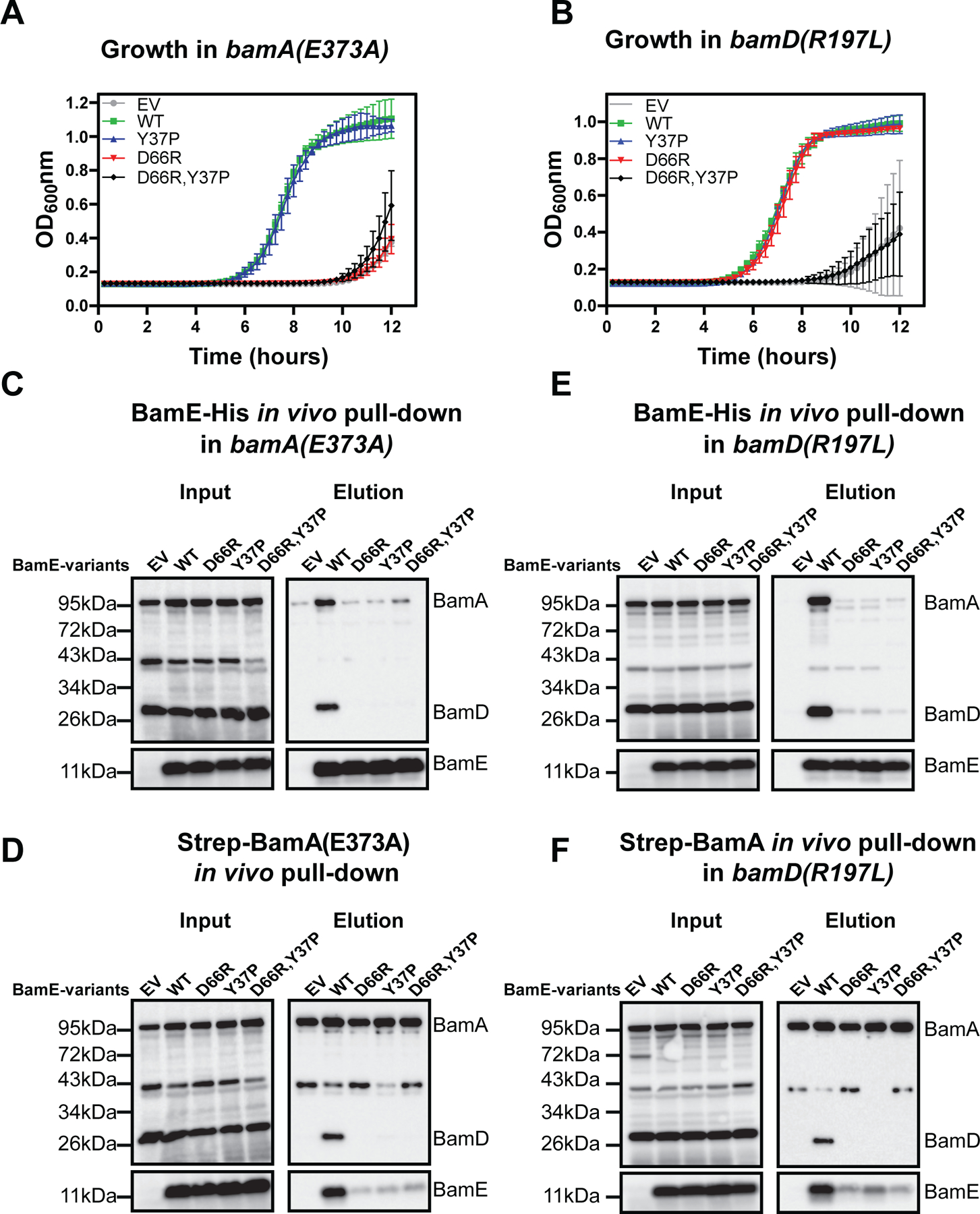
EV and vectors with indicated bamE alleles were transformed in the SK-130 [ΔbamE ΔbamA PBAD-bamA // pZS21:Strep-bamA(373A)] and MT-171 [bamE::cm bamD(R197L) nadB::Tn10] and propagated under permissive conditions (M9 glucose minimal media at 30°C). (A,B) Growth curves of bamA(E373A) (A) and bamD(R197L)(B) derivate strains in LB at 37°C. (C-F) in vivo BamA/BamD/BamE complex stability assayed using Ni-NTA pull-down of BamE-His variants (C, E) or Streptactin pull-down of Strep-BamA in the presence of BamE-His variants (D, F). Experiments were performed using culture grown under permissive conditions and processed as described in Fig. 3. For immunoblot quantifications of independent biological replicates see Fig S12.
Figure 5. BamE interacts with both BamA and BamD to promote their stable association and functional coordination.
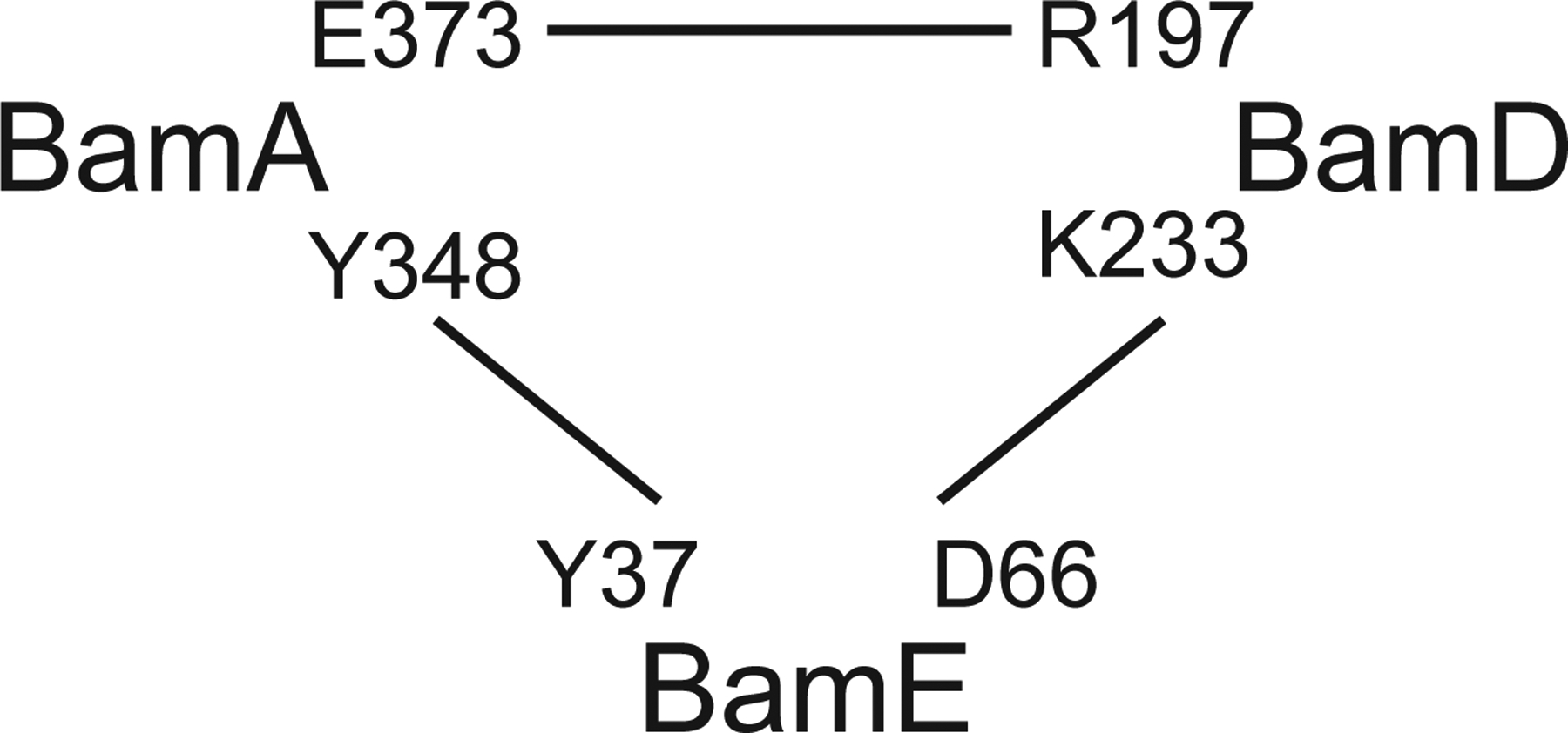
The residues utilized for genetic analysis to explore the functionality of the protein interfaces are indicated.
bamD(R197L) is a gain-of-function mutation that enables BamD to adopt a conformation that is compatible with BamA(E373K), circumventing BamA regulation (Ricci et al., 2012). bamD(R197L) restores the viablity of bamA(E373K) mutant without restoring Bam complex stability (Hagan et al., 2015). bamD(R197L) is also compatible with bamA(WT) allele and does not appear to cause OMP assembly defects (McCabe et al., 2017, Ricci et al., 2012). However, bamD(R197L) strain cannot assemble RcsF, similar to the ΔbamE strain (Tata & Konovalova, 2019), suggesting that it is at least somewhat defective in coordinating with BamA.
Despite its gain of function nature, bamD(R197L) mutation is also synthetic lethal with ΔbamE (Rigel et al., 2012), and this phenotype is also rcsF-dependent (Tata & Konovalova, 2019)(Fig. 4B, Fig. S11B). We then introduced bamE point mutants in this background, we observed that unlike bamA(E373A) bamE(D66R), the bamD(R197L) bamE(D66R) mutant was viable (Fig. 4B, S11B), indicating that BamD(R197L) bypasses not only regulation by BamA but also by BamE. However, bamD(R197L) bamE(D66R Y37P) strain remained not viable (Fig. 4B, S11B), suggesting that the regulation of BamA by BamE was still required. This result suggests that BamE and BamD play a partially overlapping function in regulating BamA conformations.
When we assayed the Bam complex stability in both backgrounds under the permissive conditions, we observed BamADE complex destabilization in all mutants, including bamE(Y37P) (Fig. 4C–F), demonstrating that BamE/BamA interaction becomes more important for complex stability when the BamA/BamD interface is weakened by either bamA(E373A) or bamD(R197L) mutations. Additionally, these result explains the discrepancy between the in vitro and in vivo pull-down assays with BamE(Y37P) (Fig. 2 and Fig. 3), as the co-elution of BamA with BamE(Y37P) in vivo is mediated by BamD (Fig. 4C–F).
Taken together, our analysis summarized in Table 1 and Fig. 5 supports the model that BamE does not simply bridge BamA and BamD for complex stability but rather acts as a bifunctional regulator. BamE enables bidirectional signaling between BamA and BamD, and this function becomes essential when the direct communication between BamA and BamD is compromised.
Table 1.
Summary phenotypes of strains used in this study.
| bamE/rcsF allele | RcsF crosslinking | Growth in LB at 37°C | BamAD complex stability | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OmpA | BamA | bam+1 | bamB- |
bamA
E373A |
bamD
R197L |
bam+1 |
bamA
E373A |
bamD
R197L |
|
| bamE(WT) | +++ | + | + | + | + | + | + | + | + |
| ΔbamE | + | +++ | + | − | − | − | − | − | − |
| ΔbamE ΔrcsF | NA | NA | + | + | + | + | − | ND3 | ND3 |
| bamE(D66R) | +++ | + | + | + | − | + | − | − | − |
| bamE(Y37P) | +++ | ++ | + | + | + | + | + | − | − |
| bamE(D66R Y37P) | + | +++ | + | − | − | − | − | − | − |
bam+ indicated WT alleles for all BamA-D components.
NA, not applicable.
ND, not determined.
DISCUSSION
BamE was the last component of the Bam complex to be identified because of its small size, which made it difficult to detect during the initial Bam complex purification (Wu et al., 2005, Sklar et al., 2007). However, the lack of significant phenotypes in the ΔbamE strain hindered its functional studies. Nonetheless, BamE quickly emerged as a BamD regulator because of reported mutations in bamD or bamA that separated the stable Bam complex into two separate BamAB and BamCDE subcomplexes (Malinverni et al., 2006, Ricci et al., 2012). This suggested that BamE associated with the Bam complex exclusively through BamD. All subsequent studies interpreted the role of BamE in regulating BamA conformations indirectly through BamD, despite the fact that pairwise subunit interactions were never tested. When the structures of the Bam complex emerged, they uncovered a direct BamA/BamE interaction interface. However, it remained unclear whether this interaction was simply driven by adjacent BamD or had any functional significance. Here, we provide the first direct evidence that BamE interacts directly with BamA and BamD, and both interactions are important for function. We show that despite a large interface predicted by the Bam complex structures, BamA and BamD cannot stably associate with each other in the absence of BamE. BamE promotes the stability of the Bam complex, but our study presents compelling evidence, discussed below, that BamE achieves this by regulating the conformations of BamA and BamD, rather than simply bridging the two components.
The first line of evidence comes from the analysis of Bam complex stability. The mutations that disrupt pairwise BamE/BamA and BamE/BamD interactions in vitro do not have the same phenotypic outcomes in vivo. BamE/BamD interaction is clearly more important for overall complex stability, suggesting that BamE allosterically regulates BamD’s ability to associate with BamA. BamE(Y37P) does not interact with BamA in vitro, but in vivo BamA still co-purifies with BamE(Y37P) because of the indirect interaction facilitated by BamD. Only when BamA/BamD or BamE/BamD interfaces are weakened by corresponding bamA(E373A), bamD(R197L), or bamE(D66G) mutations, BamA no longer co-purified with BamE(Y37P).
The second line of evidence comes from the functional assays using RcsF/OMP assembly as a readout. Our previous work suggested that the underlying defect in the ΔbamE strain is the inability of BamD and BamA to functionally coordinate with each other, as depletion of bamD phenocopied bamE null (Tata & Konovalova, 2019). Here we show that destabilization of BamA/BamD interaction is not sufficient to cause a phenotype as the bamE(D66R) mutation does not affect RcsF/OMP assembly. Only when BamE/BamA regulation is additionally perturbed cells lose the ability to assemble RcsF/OMP.
The third line of evidence comes from the genetic interaction of bamE mutations with mutations affecting the BamA/BamD interface (Fig. 5). bamA(E373A) bamE(D66R) synthetic lethality suggests that BamE and BamA play a partially overlapping function in regulating BamD. On the other hand, bamD(R197L) bypasses bamE(D66R) but not bamE(D66R Y37P), suggesting that BamE and BamD play a partially overlapping function in regulating BamA. The phenotypic similarity between bamD(R197L) and the ΔbamE strain, including the accumulation of RcsF on BamA (Tata & Konovalova, 2019), indicates that BamA adopts a similar defective conformation in both strains, and these findings are consistent with in vivo biochemical evidence (Rigel et al., 2012, Rigel et al., 2013). Together these results provide evidence that BamE can promote bidirectional signaling interaction from BamA to BamD and from BamD to BamA, thereby providing an alternative route for BamA/BamD coordination. This is why BamE becomes essential when direct BamA/BamD coordination is impaired.
Several independent studies have isolated suppressors of bamE synthetic lethal interactions (Hart et al., 2020, Hart & Silhavy, 2020, Tata & Konovalova, 2019, Tellez & Misra, 2012, Tata et al., 2021). Despite the prevalent model of BamE being a BamD regulator, these suppressor mutations were always found in bamA, never in bamD. The bifunctional regulatory model of BamE proposed in this study provides an explanation for this paradox. Our model suggests that BamE regulates BamA both directly and indirectly via BamD. Therefore, bamE suppressors need to bypass both arms of regulation. Indeed, several bamA suppressors that we found to alleviate RcsF-sequestration of BamA were independently reported to be partial bypass suppressors of bamD (Tellez & Misra, 2012, Misra et al., 2015, Hart & Silhavy, 2020).
It is important to emphasize that BamE is only essential for BamA/BamD coordination in the presence of RcsF and is otherwise completely dispensable for viability or general OMP assembly in a ΔrcsF background (Hart et al., 2019, Tata & Konovalova, 2019). So, why is BamE specifically required for RcsF/OMP assembly? The RcsF/OMP complex is clearly a challenging substrate for the Bam complex, as it requires coordination of OMP folding and RcsF surface exposure. At some point during RcsF/OMP complex assembly, BamA and BamD may lose the ability to communicate directly, relying on BamE as an alternative route for functional coordination. While RcsF sequestration of BamA can explain the phenotypes of the bamE mutant in E. coli (Hart et al., 2019, Tata & Konovalova, 2019), it is interesting that BamE, including its BamA and BamD interfacing residues, is more widely conserved than RcsF, suggesting the presence of additional substrates.
The lack of viability observed in bam mutants is often attributed to severely impaired OMP assembly consistent with the loss of function of the Bam complex. However, the example of RcsF illustrates how a defect in the assembly of a single substrate can affect overall OMP assembly indirectly by sequestering BamA. To better understand the function(s) of the Bam complex, it is crucial to identify additional model substrates rather than relying solely on growth phenotypes. The Bam complex is responsible for assembling a structurally diverse set of OMPs. Some OMPs exist as homo- or hetero-oligomers and may have periplasmic, plug, or large extracellular domains that are translocated during the folding of a β-barrel (Konovalova et al., 2017). Assembly of these challenging substrates may require specialized activities of the Bam complex. Identifying such substrates will be crucial for studying the functional roles of individual Bam components.
MATERIALS AND METHODS.
All the bacterial strains used in this study are listed in Table S3. For media and growth conditions, refer to the Supplemental Materials and Methods.
In vitro pull-down experiments.
Equimolar amount of each purified protein (5μM) was incubated in 200 μl total reaction volume in buffer C (25mM Tris-HCl pH8.0, 150mM NaCl, 0.05% n-Dodecyl-beta-Maltoside (DDM) for 1 hour at room temperature, and then mixed with 100 μL of Ni-NTA His Bind® Resin (EMD Millipore), pre-equilibrated with buffer C. Beads were loaded into Pierce Centrifuge Columns (Thermo Fisher Scientific), washed four times with 500 μL of buffer C, and eluted three times with 50 μL buffer C with 250 mM imidazole. Each fraction (20 μL each) was analyzed on a 15% SDS-PAGE (SDS–polyacrylamide gel electrophoresis) and visualized with Imperial Protein Stain (Thermo Fisher Scientific). Bio-Rad Image Lab was used to quantify gels from independent replicates. For quantification, the intensity of corresponding bands was normalized to the level of the bait protein His-BamA. Graphs represent mean +/− SD. Statistical analysis was performed by using one-way ANOVA using GraphPad Prism.
In vivo pull-down experiments.
Cells from mid-log cultures grown in LB at 37°C or under permissive conditions as indicated were harvested by centrifugation, washed twice with phosphate buffer saline (PBS) and resuspended in 400μL of membrane solubilizing buffer (25 mM Tris–HCl pH 8.0, 150 mM NaCl, 20mM MgCl2, 1% DDM, 0.1mM PMSF, and 1X Protease Inhibitor Cocktail, 2mg/mL lysozyme, and 8U/mL benzonase). Samples were incubated at room temperature for 60 min with mild rotation and cell debris were removed by high-speed centrifugation 15000g for 15min.
For BamE-His pull-down, the clarified lysate (200 μl) was mixed with 100μl of 50% suspension of Ni-NTA His Bind® Resin (EMD Millipore), pre-equilibrated with buffer C and incubated for 1 h at 4°C with mild rotation. Beads were loaded into Pierce Centrifuge Columns (Thermo Fisher Scientific), columns were washed with 10 times of 400 μL of buffer C and eluted with buffer C containing 250mM of imidazole.
For Strep-BamA pull-down, the clarified lysate (200 μl) was mixed with 100μl of 50% suspension of Strep-Tactin beads (IBA Life sciences), pre-equilibrated with buffer C (25 mM Tris–HCl pH 8.0, 150 mM NaCl and 0.05% DDM), and treated as described above. The proteins were eluted with buffer C containing 5mM deshthiobiotin.
All the input and elution fractions were analyzed by immunoblotting.
In vivo crosslinking and immunoblot analysis.
In vivo formaldehyde crosslinking on cells from mid-log cultures grown in LB at 37 °C, and the subsequent immunoblot analysis were performed as described (31). Immunoblots were visualized using the ChemiDoc MP Imaging System (Bio-Rad) and quantified using Image Lab (Bio-Rad). All figures are representative images from at least three independent biological replicates, graphs represent quantification of independendent replicates mean +/− SD. Statistical analysis was performed by using one-way ANOVA using GraphPad Prism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hannah Wilson for generating the pTrc99a::bamE(Y37G) plasmid. We thank all members of the Konovalova lab for helpful discussions. The research in the A.K. Lab is supported by NIGMS 5R01GM133904-04 and the Welch Foundation Research Grant AU-1998-20220331.
REFERENCES
- Amann E, Ochs B, and Abel KJ (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69: 301–315. [DOI] [PubMed] [Google Scholar]
- Bakelar J, Buchanan SK, and Noinaj N (2016) The structure of the beta-barrel assembly machinery complex. Science 351: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs KA, Buchanan SK, and Botos I (2021) Building Better Barrels - beta-barrel Biogenesis and Insertion in Bacteria and Mitochondria. J Mol Biol 433: 166894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MT, Jimah JR, Dowdy T, Ohlemacher SI, Larion M, Hinshaw JE, and Bernstein HD (2022) Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding. Cell 185: 1143–1156 e1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CL, and Kahne D (2011) The reconstituted Escherichia coli Bam complex catalyzes multiple rounds of beta-barrel assembly. Biochemistry 50: 7444–7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CL, Kim S, and Kahne D (2010) Reconstitution of outer membrane protein assembly from purified components. Science 328: 890–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CL, Westwood DB, and Kahne D (2013) bam Lipoproteins Assemble BamA in vitro. Biochemistry 52: 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan CL, Wzorek JS, and Kahne D (2015) Inhibition of the beta-barrel assembly machine by a peptide that binds BamD. Proc Natl Acad Sci U S A 112: 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart EM, Gupta M, Wuhr M, and Silhavy TJ (2019) The Synthetic Phenotype of DeltabamB DeltabamE Double Mutants Results from a Lethal Jamming of the Bam Complex by the Lipoprotein RcsF. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart EM, Gupta M, Wuhr M, and Silhavy TJ (2020) The gain-of-function allele bamA E470K bypasses the essential requirement for BamD in beta-barrel outer membrane protein assembly. Proc Natl Acad Sci U S A 117: 18737–18743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart EM, and Silhavy TJ (2020) Functions of the BamBCDE Lipoproteins Revealed by Bypass Mutations in BamA. J Bacteriol 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadanza MG, Higgins AJ, Schiffrin B, Calabrese AN, Brockwell DJ, Ashcroft AE, Radford SE, and Ranson NA (2016) Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat Commun 7: 12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Kahne DE, and Silhavy TJ (2017) Outer Membrane Biogenesis. Annual Review of Microbiology 71: 539--556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Mitchell AM, and Silhavy TJ (2016) A lipoprotein/beta-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lach SR, Kumar S, Kim S, Im W, and Konovalova A (2023) Conformational rearrangements in the sensory RcsF/OMP complex mediate signal transduction across the bacterial cell envelope. PLoS Genet 19: e1010601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Sutterlin HA, Wzorek JS, Mandler MD, Hagan CL, Grabowicz M, Tomasek D, May MD, Hart EM, Silhavy TJ, and Kahne D (2018) Substrate binding to BamD triggers a conformational change in BamA to control membrane insertion. Proc Natl Acad Sci U S A 115: 2359–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Tomasek D, Santos TM, May MD, Meuskens I, and Kahne D (2019) Formation of a beta-barrel membrane protein is catalyzed by the interior surface of the assembly machine protein BamA. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney TF, Ricci DP, and Silhavy TJ (2016) Classifying beta-Barrel Assembly Substrates by Manipulating Essential Bam Complex Members. J Bacteriol 198: 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, and Silhavy TJ (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61: 151–164. [DOI] [PubMed] [Google Scholar]
- McCabe AL, Ricci D, Adetunji M, and Silhavy TJ (2017) Conformational Changes That Coordinate the Activity of BamA and BamD Allowing beta-Barrel Assembly. J Bacteriol 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, and Steinegger M (2022) ColabFold: making protein folding accessible to all. Nat Methods 19: 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R (2012) Assembly of the beta-Barrel Outer Membrane Proteins in Gram-Negative Bacteria, Mitochondria, and Chloroplasts. ISRN Mol Biol 2012: 708203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R, Stikeleather R, and Gabriele R (2015) In vivo roles of BamA, BamB and BamD in the biogenesis of BamA, a core protein of the beta-barrel assembly machine of Escherichia coli. J Mol Biol 427: 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overly Cottom C, Stephenson R, Wilson L, and Noinaj N (2023) Targeting BAM for Novel Therapeutics against Pathogenic Gram-Negative Bacteria. Antibiotics (Basel) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova O, Peterson JH, Ieva R, and Bernstein HD (2013) Mechanistic link between beta barrel assembly and the initiation of autotransporter secretion. Proc Natl Acad Sci U S A 110: E938–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci DP, Hagan CL, Kahne D, and Silhavy TJ (2012) Activation of the Escherichia coli beta-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A 109: 3487–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigel NW, Ricci DP, and Silhavy TJ (2013) Conformation-specific labeling of BamA and suppressor analysis suggest a cyclic mechanism for beta-barrel assembly in Escherichia coli. Proc Natl Acad Sci U S A 110: 5151–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigel NW, Schwalm J, Ricci DP, and Silhavy TJ (2012) BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol 194: 1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Hernandez G, Peterson JH, and Bernstein HD (2014) Reconstitution of bacterial autotransporter assembly using purified components. Elife 3: e04234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Lach SR, and Konovalova A (2021) Homeostasis of the Gram-negative cell envelope. Curr Opin Microbiol 61: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Chang S, Luo Q, Chan KC, Zhang Z, Luo B, Xie T, Lu G, Zhu X, Wei X, Dong C, Zhou R, Zhang X, Tang X, and Dong H (2023) Structural basis of BAM-mediated outer membrane beta-barrel protein assembly. Nature 617: 185–193. [DOI] [PubMed] [Google Scholar]
- Sinnige T, Weingarth M, Daniels M, Boelens R, Bonvin AM, Houben K, and Baldus M (2015) Conformational Plasticity of the POTRA 5 Domain in the Outer Membrane Protein Assembly Factor BamA. Structure 23: 1317–1324. [DOI] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, and Silhavy TJ (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A 104: 6400–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Rutherford ST, Silhavy TJ, and Huang KC (2022) Physical properties of the bacterial outer membrane. Nat Rev Microbiol 20: 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata M, and Konovalova A (2019) Improper Coordination of BamA and BamD Results in Bam Complex Jamming by a Lipoprotein Substrate. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata M, Kumar S, Lach SR, Saha S, Hart EM, and Konovalova A (2021) High-throughput suppressor screen demonstrates that RcsF monitors outer membrane integrity and not Bam complex function. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez R Jr., and Misra R (2012) Substitutions in the BamA beta-barrel domain overcome the conditional lethal phenotype of a DeltabamB DeltabamE strain of Escherichia coli. J Bacteriol 194: 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek D, and Kahne D (2021) The assembly of beta-barrel outer membrane proteins. Curr Opin Microbiol 60: 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek D, Rawson S, Lee J, Wzorek JS, Harrison SC, Li Z, and Kahne D (2020) Structure of a nascent membrane protein as it folds on the BAM complex. Nature 583: 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Heinz E, and Lithgow T (2012) Evolution of the beta-barrel assembly machinery. Trends Microbiol 20: 612–620. [DOI] [PubMed] [Google Scholar]
- Wu R, Bakelar JW, Lundquist K, Zhang Z, Kuo KM, Ryoo D, Pang YT, Sun C, White T, Klose T, Jiang W, Gumbart JC, and Noinaj N (2021) Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM. Nat Commun 12: 7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, and Kahne D (2005) Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121: 235–245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


