SUMMARY
Centromeres direct genetic inheritance but are not themselves genetically encoded. Instead, centromeres are defined epigenetically by the presence of a histone H3 variant, CENP-A1. In cultured somatic cells, an established paradigm of cell cycle-coupled propagation maintains centromere identity: CENP-A is partitioned between sisters during replication and replenished by new assembly, which is restricted to G1. The mammalian female germline challenges this model because of the cell cycle arrest between pre-meiotic S-phase and the subsequent G1, which can last for the entire reproductive lifespan (months to decades). New CENP-A chromatin assembly maintains centromeres during prophase I in worm and starfish oocytes2,3, suggesting that a similar process may be required for centromere inheritance in mammals. To test this hypothesis, we developed an oocyte specific conditional knockout mouse for Mis18α, an essential component of the assembly machinery. We find that embryos derived from Mis18α knockout oocytes fail to assemble CENP-A nucleosomes prior to zygotic genome activation, validating the knockout model. We show that deletion of Mis18α in the female germline at the time of birth has no impact on centromeric CENP-A nucleosome abundance even after 6–8 months of aging. In addition, there is no detectable detriment to fertility. Thus, centromere chromatin is maintained long-term independent of new assembly during the extended prophase I arrest in mouse oocytes.
Keywords: centromeres, meiosis, reproductive aging, oocytes
eTOC Blurb
In brief
To test if centromeres are maintained by new assembly in aging mouse oocytes, Das et al. conditionally delete Mis18α, a centromere assembly component. They find that centromere-specifying nucleosomes containing the histone H3 variant, CENP-A, are stably maintained even after 6–8 months of aging, with no detriment to fertility.
RESULTS AND DISCUSSION
In contrast to genetic information encoded in our genome, minimal epigenetic information is inherited because most parental epigenetic marks are removed and reprogrammed in germ cells and the early embryo4,5. A key exception is the centromeric histone H3 variant, CENP-A6–8. Although centromeres direct the process of genetic inheritance by connecting chromosomes to spindle microtubules, they are not encoded in DNA, but rather epigenetically specified by nucleosomes containing CENP-A9–12. Thus, CENP-A nucleosomes are inherited to preserve centromere identity.
Studies in cycling somatic cells have established a general paradigm for propagation of epigenetic information between cell cycles. DNA or histone modifications are partitioned between sister chromatids during DNA replication, and then replenished by “reader” proteins that recognize the modification and “writers” that extend it to adjacent nucleosomes13. CENP-A, follows this paradigm, with new assembly restricted to G1 by CDK1/2 activity14–16. CENP-A and its bound partner histone H4 molecules are remarkably stable in tissue culture cells17,18, with measured CENP-A turnover rates explained entirely through the dilution when existing CENP-A is partitioned to replicated centromeric DNA during S-phase each cell cycle19–21. This paradigm poses a challenge in the mammalian female germline because of the extended prophase I cell cycle arrest, after replication but before an opportunity for new G1 assembly, which can last for the entire reproductive lifespan of the animal22 (Figure 1A). Centromeres are preserved throughout the arrest (>1 year in mouse) in the absence of new Cenpa transcription, as shown by conditional knockout of the Cenpa gene23. CENP-A nucleosomes are therefore either replenished by new assembly in contrast to the somatic cell paradigm, drawing on a stable pool of CENP-A protein, or stable for the entire duration of the arrest (Figure 1B).
Figure 1: Testing models for stable retention of CENP-A chromatin in oocytes.
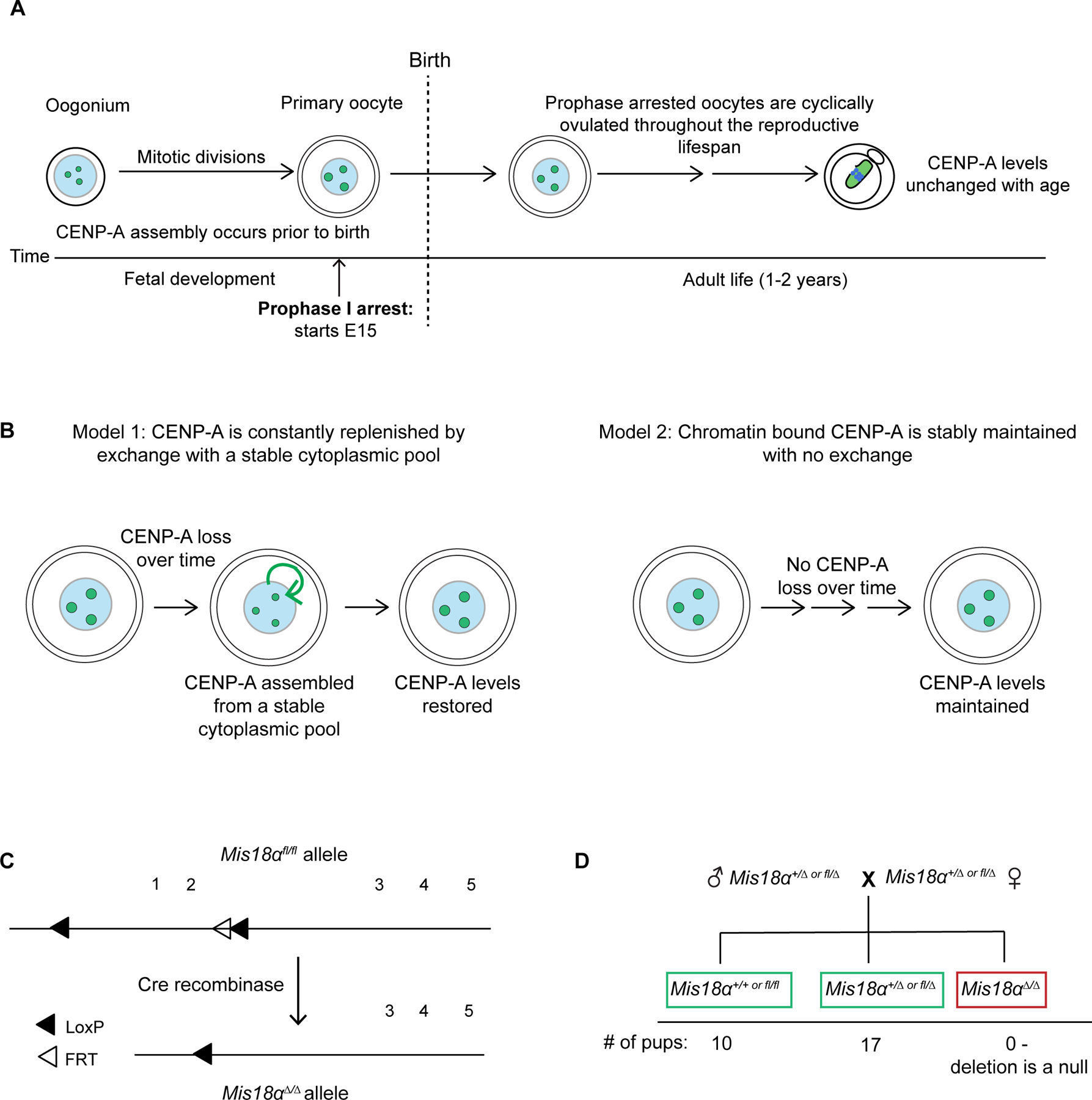
(A) Schematic showing maintenance of CENP-A chromatin across the oogenesis timeline. Oocytes arrested in prophase I are cyclically recruited to begin growth before ovulation and maturation. Prophase arrest prior to recruitment can last up to 2 years in mice. Centromeric CENP-A levels are stable over time even if the Cenpa gene is eliminated shortly after birth23.
(B) Models for retention of CENP-A in meiotic prophase I arrested eggs.
(C) Schematic of the Mis18α conditional knockout gene locus36. The 1st and 2nd Mis18α protein coding exons are flanked by LoxP sites (floxed, fl). The FRT site is a remnant from FLP mediated excision of the Neomycin cassette in the original construct used to generate the KO animals (see Figure S1 for genotyping).
(D) Genotype frequencies of the progeny from a cross of Mis18α+/∆ or fl/∆ heterozygotes (See Figure S2 for generation of heterozygotes). Number of litters = 6, number of pups = 27.
Studies in worm and starfish oocytes show nascent CENP-A chromatin assembly in prophase I (akin to a G2 biochemical cell cycle state in a somatic cell)2,3. In mouse, continual deposition of nucleosomes containing another H3 variant, H3.3, during oocyte development is required for oocyte genome integrity and, ultimately, for fertility24. These studies suggest that new assembly may maintain CENP-A chromatin through the prophase I arrest. To test this prediction, we created an oocyte specific conditional knockout (cKO) of an essential component of the CENP-A deposition machinery, Mis18α (Mis18a in mouse but referred to as Mis18α for simplicity)25–29,31, to prevent nascent CENP-A chromatin assembly. Mis18α is part of the Mis18 complex, which recruits the CENP-A chaperone, HJURP, bound to nascent CENP-A/histone H4 dimers30,32–34, to centromeres. The Mis18 complex is required for both nascent CENP-A chromatin assembly in G1 and replication-coupled CENP-A chromatin assembly in S phase in cycling somatic cells21,35. Specifically relevant to our test of ongoing chromatin assembly in the mouse oocyte, prophase I assembly in both worms and starfish oocytes requires the Mis18 complex2,3. Thus, if CENP-A chromatin is replenished by new assembly, CENP-A nucleosome levels would decay in Mis18α knockout oocytes.
For conditional knockout, we used a floxed Mis18α allele in which the first two exons, encoding the YIPPEE domain necessary for CENP-A deposition, are flanked by LoxP sites36,37 (Figure 1C, S1). To confirm that Cre-mediated excision generates a null allele, we crossed Mis18α heterozygous parents and did not recover any progeny homozygous for the deletion, as expected because Mis18α is an essential gene36 (Figures 1D, S2). Combining the floxed allele with Cre recombinase driven by oocyte specific promoters, we generated cKOs of Mis18α either early or late in prophase I (Figure S2). The early Cre driver (Gdf9-Cre) deletes Mis18α in arrested oocytes 2 days post birth, preventing assembly of new CENP-A nucleosomes for nearly the entire lifespan of the animal38. The late Cre driver (Zp3-Cre) deletes the gene during oocyte growth, 2–3 weeks prior to ovulation38 (Figure 2A).
Figure 2: Maternally deposited Mis18α protein is eliminated in late KO oocytes.
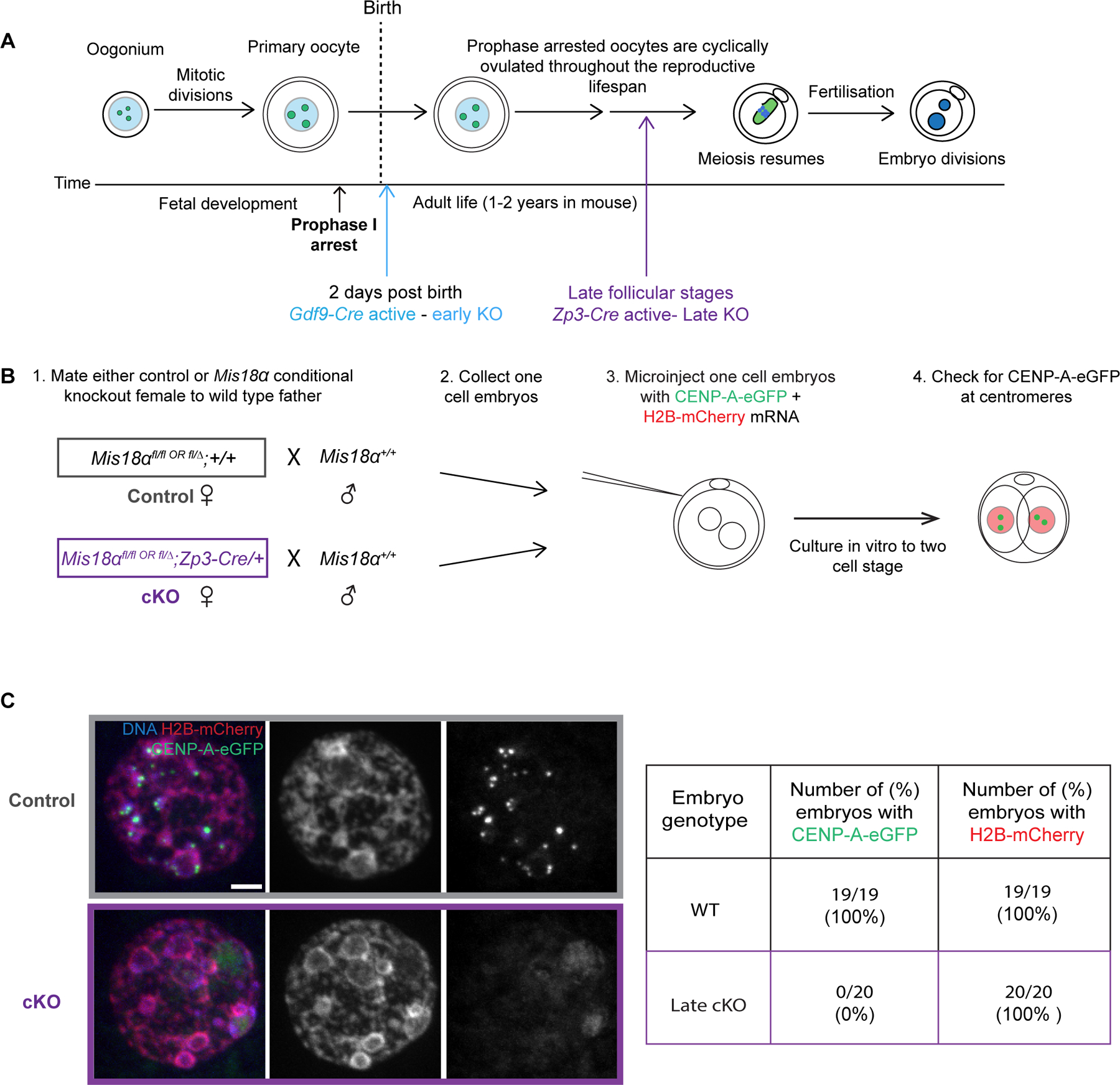
(A) Schematic showing early and late Mis18α knockout depending on the timing of Cre recombinase expression driven by either Gdf9 or Zp3 promoters (colored arrows).
(B) Experimental design to test for a stable pool of Mis18α protein in the maternal cytoplasm. CENP-A-eGFP and H2B-mCherry mRNA are injected into one cell embryos, and CENP-A foci are assessed after the first embryonic mitosis when assembly is expected to occur but before zygotic genome activation.
(C) Images show H2B-mCherry, CENP-A-eGFP, and DNA (DAPI) in interphase two cell embryos from control or cKO mothers. Table shows frequencies of detectable assembly for CENP-A-eGFP and H2B-mCherry, obtained from 2 independent matings. Scale bars = 5 μm. Also see Figure S2 and S3.
As a functional assay for Mis18α deletion, we tested for CENP-A chromatin assembly in embryos from late KO mothers (driven by Zp3-Cre) and a wild type father (Figure 2B). Prior to zygotic genome activation (ZGA) at the two-cell stage embryos, CENP-A deposition during the early embryonic mitotic cycles depends solely on maternal Mis18α protein. To assess new chromatin assembly, we injected mRNAs encoding CENP-A-eGFP and H2B-mCherry into one-cell embryos derived from either KO mothers or wild type mothers as controls (Figure 2B). H2B-mCherry serves as a positive control for chromatin assembly because it utilizes a deposition pathway distinct from that of CENP-A and therefore does not require the Mis18 complex. Based on the paradigm established in cycling somatic cells, we expected new CENP-A chromatin assembly in G1 after the first embryonic mitosis. Any Mis18α protein present in the embryo prior to ZGA would be solely contributed maternally from the oocyte. We expect embryos derived from control oocytes would be assembly proficient whereas embryos from KO oocytes lacking Mis18α would fail to assemble. Indeed, CENP-A-eGFP localized to centromeres in all control embryos (100%, N = 19) from wild type mothers. In contrast, none of the embryos from late Mis18α KO mothers (0%, N = 20) contained detectable centromeric CENP-A-eGFP (Figure 2C). H2B-mCherry was present in chromatin in 100% of both control and KO injected embryos as expected. The all-or-none effect of Mis18α KO is due to the high efficiency of Cre excision and the relatively fast turnover of Mis18α mRNA in oocytes following excision (Figure S3). This finding establishes that even after a relatively short duration KO, Mis18α deletion in oocytes abrogates nascent CENP-A chromatin assembly in early embryos.
Next, we measured fertility of Mis18α KO mothers as the ultimate test of centromere function, because centromeres are required for the meiotic divisions and for maternal centromere inheritance, and even partial reduction of maternal CENP-A chromatin lowers fertility8. All Mis18α early and late KO mothers were fertile when crossed to wild type males (Figure 3A). We also confirmed that both Cre recombinases have a 100% deletion efficiency as we did not recover a floxed allele of Mis18α inherited from a cKO mother (Figure 3B, C). Combined with our embryo experiments, this result confirms that every oocyte with a combination of floxed alleles and Cre lacks Mis18α protein and that fertility is not a consequence of inefficient Cre activity in some oocytes. Thus, centromere identity is maintained in aging oocytes without Mis18α protein or nascent CENP-A assembly.
Figure 3: Mis18α knockout mothers can support fertility.
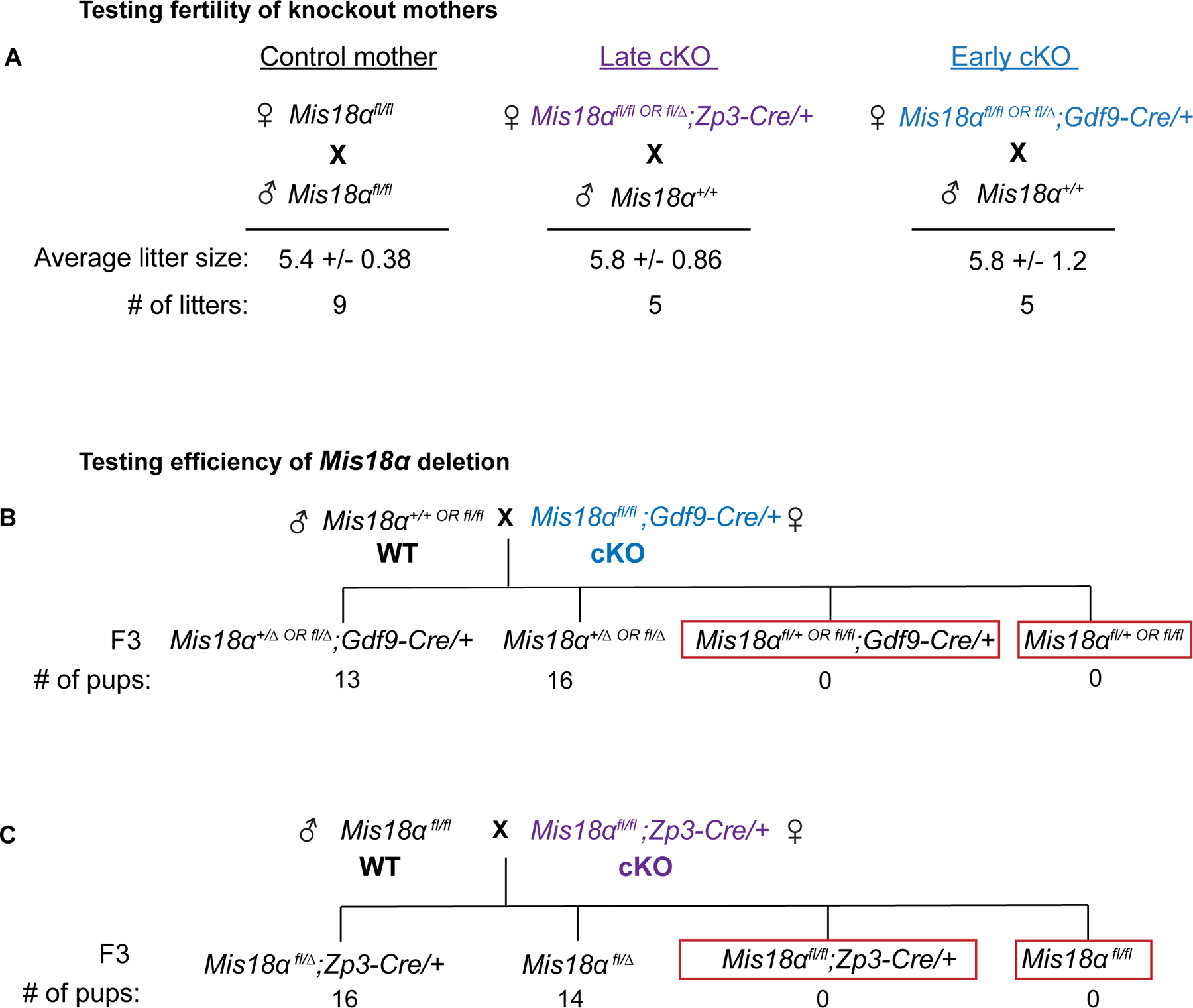
(A) Litter sizes for control, early cKO, and late cKO mothers (age 2–4 months) crossed to wild type males. See Figure S2 for generating cKO mothers.
(B, C) Genotype frequencies of the progeny from a cross between a WT or Mis18αfl/fl father and an early or late cKO mother respectively (N= 29 pups, 5 litters; N = 30 pups, 5 litters).
Even though the cKO mice are fertile, there could still be a reduction in CENP-A nucleosomes over time. Therefore, we tested if CENP-A levels decay in the late KO oocytes. We predict that if CENP-A nucleosomes are continually replenished by new assembly from a stable pool of CENP-A protein, centromeric CENP-A levels would decline in the KO oocytes compared to control oocytes. We did not see any reduction in centromeric CENP-A levels in late KO oocytes compared to controls (Figures 4A, B). However, Mis18α is deleted for only 2–3 weeks before ovulation in the late KO, leaving open the possibility of a more dramatic reduction in CENP-A nucleosomes on longer timescales.
Figure 4: Pre-meiotically assembled CENP-A chromatin is maintained in prophase arrested oocytes without new assembly.
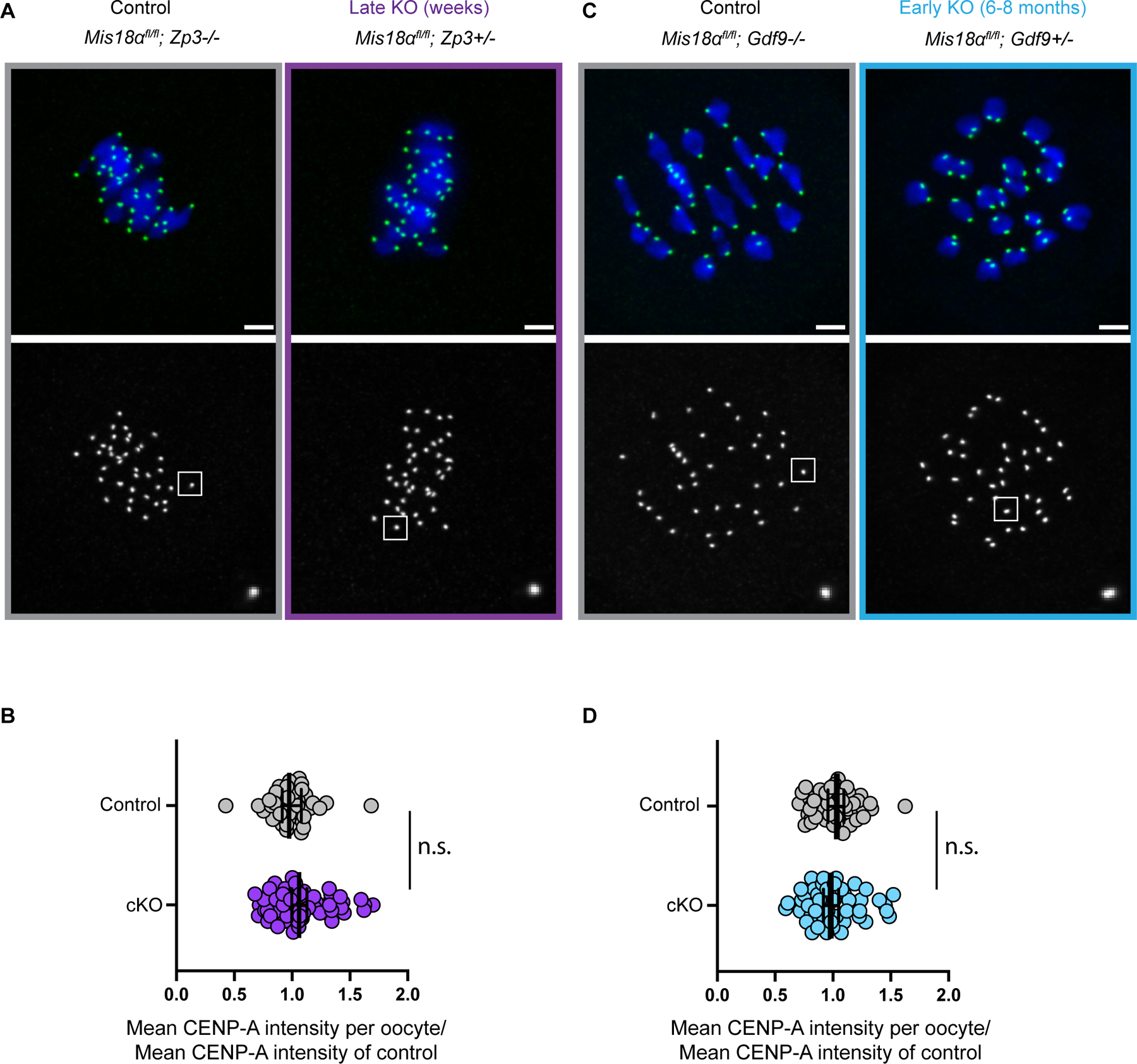
(A, C) Images show metaphase I oocytes stained for CENP-A (green) and DNA (DAPI, blue) from late cKO (A) or early cKO (C) mothers. Scale bars = 5 µm.
(B, D) Quantifications of CENP-A intensities in metaphase I oocytes from late cKO (B) or early cKO (D) mothers. Each data point represents average CENP-A levels at centromeres in each oocyte normalized to the control mean. Late cKO (Zp3-Cre, purple): mean ± S.E.M. = 1.1 ± 0.034 compared to control, number of oocytes = 58, number of centromeres = 1512, N = 7 females) and Cre negative control oocytes (number of oocytes = 40, number of centromeres = 962, N = 5 females). Early cKO (Gdf9-Cre, blue): mean ± S.E.M. = 0.97 ± 0.068 (number of oocytes = 47, number of centromeres = 1746, N = 7 females) relative to control (number of oocytes = 50, number of centromeres = 2019, N = 8 females); n.s.: Mann-Whitney U Test. Error bars: geometric mean ± 95% confidence interval. The early cKO mothers are aged 6–8 months, and their oocytes lack Mis18α for the entire lifespan. Also see Figure S2 for generation of cKO animals.
Thus, we leveraged the early KO (Gdf9-Cre) oocytes that delete Mis18α shortly after birth, to test if centromeric CENP-A levels decline after months without new assembly (Figures 4C, D). We aged control and early KO females for 6–8 months, representing most of the fertile lifespan, and found that CENP-A levels were not significantly reduced compared to the controls. Although not statistically significant, we measured a ~3% decrease in CENP-A levels in the KO oocytes relative to control (Figure 4D) that equates to loss at a rate of ~0.02% per day over 180 days. Such loss in signal would be in the range of one nucleosome per month, assuming ~200 CENP-A nucleosomes per centromere as estimated in human cells19. In comparison, new assembly in starfish oocytes is estimated at a rate of 2% per day, based on centromere localization of GFP-tagged exogenous CENP-A in cells cultured in vitro for 10 days3. Our result is consistent with our previous study, where we conditionally deleted the Cenpa gene using Gdf9-Cre and followed CENP-A nucleosome stability further and measured no substantial decrease23.
CONCLUSION
In conclusion, these results provide clear evidence supporting long term retention of CENP-A nucleosomes assembled prior to birth as the dominant pathway for maintaining centromere identity in mammalian oocytes. This is in stark contrast to H3.3, whose ongoing deposition into bulk chromatin during prophase I is essential for normal oocyte chromatin structure and fertility24. In addition, our findings have implications for human female meiosis, which is inherently error prone and especially vulnerable to aging. With advancing maternal age at childbirth, mechanisms that preserve centromeres in aging oocytes gain increasing significance. Although maternal knockout of Mis18α in mice preserves fertility due to early zygotic genome activation, maternal depletion of Mis18α is expected to have more severe consequences in human embryos, where activation occurs later. Our findings can now direct future research into the mechanisms that underlie CENP-A retention in mammalian oocytes. Previous studies of centromere chromatin suggest multiple potential molecular mechanisms that could contribute to its stability: the relatively low level of transcription of centromeric DNA39–42 relative to genic regions harboring histone H3.3 nucleosomes43; structural features that differ from canonical nucleosomes, including internal structural rigidity at the CENP-A/histone H4 interface44,45; and non-histone CCAN components that bind and stabilize CENP-A nucleosomes in tissue culture cells17,18,46. In sum, it remains to be seen if the mechanisms that function to retain CENP-A chromatin over short periods of time in cycling cells also contribute to extreme stability during oogenesis.
STAR METHODS:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ben E. Black (blackbe@pennmedicine.upenn.edu).
Materials availability
Mouse lines generated in this study are available upon request.
Data and code availability
All imaging data reported in this paper will be shared by the lead contact upon request. This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
All animal experiments and protocols were approved by the Institutional Animal Use and Care Committee of the University of Pennsylvania and were consistent with National Institutes of Health guidelines (protocol #803994). Experimental animals were compared to age, gender and genetic background matched controls. The Mis18afl/fl strain is a previously published and validated knockout strain36. This strain is in a mixed genetic background of C57BL6/J/129Sv/CBA/J (generated by Sung Hee Baek, Seoul National University and obtained from Kikue Tachibana, Max Planck Institute). Oocyte specific conditional knockout strains Mis18afl/fl;Zp3-Cre (Mis18α late KO) and Mis18afl/fl;Gdf9-iCre (Mis18α early KO) were generated by crossing the original Mis18afl/fl strain to either C57BL/6-Tg(Zp3-cre)93Knw/J (RRID:IMSR_JAX:003651, Jackson Laboratory) or Tg(Gdf9-icre)5092Coo/J (RRID:IMSR_JAX:011062, Jackson Laboratory). The following primers were used to genotype the animals: 1) Mis18afl/fl: SDL-Forward: TGC CTA TTG GTG TAC CTT CCA GTG, LOXP-Reverse: CCT AAG TCG TTG ACC TGA CCG AGG, 2) Mis18afl/Δ : Mis18-DeltaAT2 Reverse: GGA CAG GAA TAG GAC ACT TTC AAC combined with SDL-Forward (Figure S1). For testing fertility, age matched single conditional knockout (Mis18α early or late KO) or control mothers (Mis18afl/fl) were mated in cages to single males (either Mis18afl/fl or Mis18afl/fl ). Litter sizes were determined for multiple mating pairs per cross. Oocytes and embryos were collected from multiple mothers for all experiments.
METHOD DETAILS
Oocyte collection, meiotic maturation, and culture
Female mice were hormonally primed with 5 U of pregnant mare serum gonadotropin (PMSG, Peptides International) 44–48 h before oocyte collection. Germinal vesicle intact oocytes were collected in bicarbonate-free minimal essential medium (M2, Sigma), denuded from cumulus cells, and cultured in Chatot–Ziomek–Bavister47 (CZB, Fisher Scientific) medium covered with mineral oil (Sigma, BioXTRA) in a humidified atmosphere of 5% CO2 in air at 37 °C. During collection, meiotic resumption was inhibited by addition of 2.5 mM milrinone (Sigma). Milrinone was subsequently washed out to allow meiotic resumption and oocytes were fixed 6–7 h later at metaphase I.
Oocyte immunocytochemistry
Oocytes were fixed in 2% paraformaldehyde (Sigma) in phosphate buffered saline (PBS) with 0.1% Triton X-100 (Sigma), pH 7.4, for 20 min at room temperature (r.t.), permeabilized in PBS with 0.2% Triton X-100 for 15 min at r.t., placed in blocking solution (PBS containing 0.3% bovine serum albumin (BSA) and 0.01% Tween-20) for 20 minutes at r.t., treated with λ-phosphatase (1,600 U, NEB) for 1 h at 30 °C for CENP-A staining, incubated for 1 h with primary antibody in blocking solution, washed three times for 10 min each, incubated for 1 h with secondary antibody, washed three times for 10 min each, and mounted in Vectashield with 4′,6-diamidino-2-phenylindole (DAPI; Vector) to visualize the chromosomes. The primary antibody was rabbit anti-mouse CENP-A (1:200, Cell Signaling, C51A7). The secondary antibody was donkey anti-rabbit Alexa Fluor 488 (1:500, Invitrogen).
Microscopy
Confocal images were collected as z-stacks with 0.5-μm intervals, using a microscope (DMI4000 B; Leica) equipped with a 63× 1.3-NA glycerol-immersion objective lens, an x–y piezo Z stage (Applied Scientific Instrumentation), a spinning disk confocal scanner (Yokogawa Corporation of America), an electron-multiplying charge-coupled device camera (ImageEM C9100–13; Hamamatsu Photonics) and either an LMM5 (Spectral Applied Research) or Versalase (Vortran Laser Technology) laser merge module, controlled by MetaMorph software (Molecular Devices, v7.10.3.294). Images were acquired using the same laser settings and all images in a panel were scaled the same. Single channels are shown wherever quantifications were performed.
Embryo collection, microinjection and culture
Mis18afl/fl or Mis18afl/fl;Zp3-Cre (Mis18α late KO) females were hormonally primed with 5 U of PMSG (Peptides International) and the oocytes were matured in vivo with 5 U of human chorionic gonadotropin (hCG; Sigma) before mating with B6D2F1/J males (F1 hybrid of a cross between C57BL6/J and DBA2/J; RRID:IMSR_JAX:100006, Jackson Laboratory). Males were fed a special low soymeal diet (5LG4 irradiated diet, Labdiet) and housed singly. Embryos were collected 14–16 h post hCG in M2 containing hyaluronidase (0.3 mg ml−1) to remove cumulus cells and subsequently washed in M2 (Sigma). and cultured in EmbryoMax Advanced KSOM (AKSOM, Millipore Sigma) with humidified air and 5% CO2.
Embryos were then subjected to inter-cytoplasmic microinjection in M2 medium covered with mineral oil (Sigma, BioXtra) at r.t. with a micromanipulator (Narishige) and a picoinjector (Medical Systems Corp). Each embryo was injected with 2 pl of cRNA, then cultured in EmbryoMax Advanced KSOM (AKSOM, Millipore Sigma) with humidified air and 5% CO2 until 2 cell stage and fixed in 2% paraformaldehyde. The following cRNAs were used for microinjection: H2B-mCherry (human histone H2B with mCherry at the C-terminus) at 25 ng/ul, CENP-A-EGFP (mouse CENP-A with EGFP at the C-terminus) at 20 ng/ul. The cRNAs were synthesized using the T7 mScript Standard mRNA kit (Thermo Fisher Scientific) and purified by phenol-chloroform extraction.
mRNA quantification in oocytes
Total RNA was extracted from at least 20 full-grown oocytes from two females each for control and late conditional knockout (Zp3-Cre mediated) using Arcturus Picopure RNA isolation kit (Thermofisher Scientific), and cDNA was prepared by reverse transcription of total RNA with Superscript III First Strand Synthesis system (Thermofisher) using oligo dT primers. Mis18α was amplified for standard PCR using Kapa polymerase (Roche) from 1 µg of cDNA. Real time PCR was performed using Mis18α Taqman probes and H2A serving as the endogenous control. Each sample was run twice in triplicate. Quantification was performed using the comparative Ct method (Livak method) on an Applied Biosystems ViiA 7 machine.
Quantification and statistical analysis
To quantify centromere signal ratios, a sum intensity Z-projection was made using Fiji/ImageJ software. Circles of constant diameter were drawn around individual centromeres and the average intensity was calculated for each centromere after subtracting background, obtained from nearby regions. Raw centromere intensities were obtained from several controlled independent experiments and multiple cells were analyzed from each animal. Normalization of centromere intensities was performed using age- and gender-matched controls for each independent experiment. Statistical tests (Mann-Whitney U test) were performed using the Graphpad Prism software. Details of the p-values and error bars are provided in figure legends.
Supplementary Material
Key resources table
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-CENP-A (C51A7) | Cell Signaling | 2048; RRID: AB_1147629 |
| Alexa Fluor 488 conjugate donkey anti-rabbit IgG (H+L) | Molecular Probes (Invitrogen) | A-21206 RRID:AB_2535792 |
| Chemicals, peptides, and recombinant proteins | ||
| Lambda phosphatase | New England Biolabs | P0753L |
| Milrinone | Sigma | M4659 |
| Mineral Oil (BioXtra) | Sigma | M8410 |
| Mineral Oil (Fuji) | Fuji Film Irvine | 9305-500 ml |
| Pregnant Mare Serum Gonadotropin (PMSG) | Peptides International | HOR_272-5000 IU |
| Human Chorionic Gonadotropin | Sigma | CG10-1VL |
| Vectashield Antifade (DAPI) | Vector Laboratories | H-1200 |
| CZB | Fisher Scientific | MR-019-D |
| EmbryoMax M2 medium, with Phenol Red | Millipore Sigma | MR-015 |
| Embryomax Advanced KSOM | Millipore Sigma | MR-101-D |
| Phenol (UltraPure buffer saturated) | Thermofisher Scientific | 15513039 |
| Dulbecco’s PBS (without Ca and Mg) | Corning | MT21-031-CV |
| Triton X 100 | Sigma | X100-500 ml |
| RedExtract N-Amp PCR ready Mix | Millipore Sigma | R4775 |
| Kapa Polymerase Hifi Hotstart Ready Mix | Roche | KK2602 |
| Taqman Fast Advanced Master Mix | Thermofisher Scientific | 4444557 |
| Extraction Solution | Millipore Sigma | E7526 |
| Tissue Preparation Solution | Millipore Sigma | T3073 |
| Neutralization Solution B | Millipore Sigma | N3910 |
| 1 Kb Plus ladder | Thermofisher | SM1333 |
| Stellar Competent cells | Takara Bio | 636766 |
| Nucleospin Plasmid kit | Takara Bio | 740588.250 |
| Critical commercial assays | ||
| T7 mScript Standard mRNA | Cell Script | C-MSC100625 |
| Superscript III First Strand Synthesis | Thermofisher Scientific | 18080051 |
| Arcturus Picopure RNA Isolation Kit | Thermofisher Scientific | KIT0204 |
| Taqman Assay (Mis18α) | Thermofisher Scientific | Mm01209645_m1 |
| Taqman Assay (Hist2h2) | Thermofisher Scientific | Mm0051974_s1 |
| Experimental models: Organisms/strains | ||
| Mouse: Mis18αfl/fl; B6-129-CBA-Mis18afl/fl | Kim et al, 201236 | N/A |
| Mouse: Mis18α early KO; B6-129-CBA- Mis18afl/fl;Gdf9-Cre | This paper | N/A |
| Mouse: Mis18α late KO; B6–129-CBA- Mis18afl/fl;Zp3-Cre | This paper | N/A |
| Mouse: Zp3-Cre; B6-Tg(Zp3-cre)93Knw/J | Jackson laboratory | RRID:IMSR_JAX:003651 |
| Mouse: Gdf9-Cre; B6-Tg(Gdf9-icre)5092Coo/J | Jackson laboratory | RRID:IMSR_JAX:011062 |
| Oligonucleotides | ||
| SDL-Forward: 5’-TGC CTA TTG GTG TAC CTT CCA GTG-3’ | Kim et al, 201236 | N/A |
| LOX-Reverse: 5’-CCT AAG TCG TTG ACC TGA CCG AGG-3’ | Kim et al, 201236 | N/A |
| Mis18-DeltaAT2: 5’-GGA CAG GAA TAG GAC ACT TTC AAC-3’ | Kim et al, 201236 | N/A |
| Mis18F (exons 1–4): 5’-AGA AGT GGG CAA ACA TGT CG-3’ | This paper | N/A |
| Mis18R (exons 1–4): 5’-GAG GAC CCT AAG GTG TAA CTT TCA A-3’ | This paper | N/A |
| Mis18F (exons 2–4): 5’- AGC GTC TCC TGT AAC GTC TC-3’ | This paper | N/A |
| Mis18R (exons 2–4): 5’-GTC AGG ACT TCT TCC ATC TGC TT-3’ | This paper | N/A |
| Recombinant DNA | ||
| CENPA-eGFP | Smoak et al, 201623 | N/A |
| H2B-mCherry | Akera et al, 201748 | N/A |
| Software and algorithms | ||
| GraphPad Prism 9.3.1 (350) | GraphPad | http://www.graphpad.com/ |
| FIJI/ImageJ | Schindelin et al, 201249 | https://fiji.sc/ |
Highlights.
Genetic removal of Mis18α tests current models for centromere inheritance in oocytes
Mis18a removal has no impact on CENP-A nucleosome levels in aging oocytes
Mis18a removal disrupts CENP-A nucleosome assembly in the early embryo
CENP-A assembled prior to birth is sufficient for fertility in aging mice
ACKNOWLEDGEMENTS:
We thank M.T. Levine and R. M. Schultz for feedback on the manuscript. This work was supported by the National Institute of Child Health and Development grant HD058730 (M.A.L. and B.E.B.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS:
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
References:
- 1.Kixmoeller K, Allu PK, and Black BE (2020). The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol 10, 200051. 10.1098/rsob.200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prosée RF, Wenda JM, Özdemir I, Gabus C, Delaney K, Schwager F, Gotta M, and Steiner FA (2021). Transgenerational inheritance of centromere identity requires the CENP-A N-terminal tail in the C. elegans maternal germ line. PLoS Biol 19, e3000968. 10.1371/journal.pbio.3000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartz SZ, McKay LS, Su K-C, Bury L, Padeganeh A, Maddox PS, Knouse KA, and Cheeseman IM (2019). Quiescent Cells Actively Replenish CENP-A Nucleosomes to Maintain Centromere Identity and Proliferative Potential. Dev. Cell 51, 35–48.e7. 10.1016/j.devcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ficz G, Farthing CR, and Reik W (2009). The epigenomic landscape of reprogramming in mammals. In Epigenomics 10.1007/978-1-4020-9187-2_15. [DOI] [Google Scholar]
- 5.Xu Q, and Xie W (2018). Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol 28, 237–253. 10.1016/j.tcb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Palmer DK, O’Day K, Wener MH, Andrews BS, and Margolis RL (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol 104, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer DK, O’Day K, and Margolis RL (1990). The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma 100, 32–36. [DOI] [PubMed] [Google Scholar]
- 8.Das A, Iwata-Otsubo A, Destouni A, Dawicki-McKenna JM, Boese KG, Black BE, and Lampson MA (2022). Epigenetic, genetic and maternal effects enable stable centromere inheritance. Nat. Cell Biol 24, 748–756. 10.1038/s41556-022-00897-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kixmoeller K, Allu PK, and Black BE (2020). The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol 10, 200051. 10.1098/rsob.200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black BE, and Cleveland DW (2011). Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 144, 471–479. 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernad R, Sánchez P, and Losada A (2009). Epigenetic specification of centromeres by CENP-A. Exp. Cell Res 315, 3233–3241. 10.1016/j.yexcr.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Mendiburo MJ, Padeken J, Fulop S, Schepers A, and Heun P (2011). Drosophila CENH3 Is Sufficient for Centromere Formation. Science (80-. ) 334, 686–690. 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- 13.Fitz-James MH, and Cavalli G (2022). Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet 23, 325–341. 10.1038/s41576-021-00438-5. [DOI] [PubMed] [Google Scholar]
- 14.Silva MCC, Bodor DL, Stellfox ME, Martins NMC, Hochegger H, Foltz DR, and Jansen LET (2012). Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev. Cell 22, 52–63. 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 15.Jansen LET, Black BE, Foltz DR, and Cleveland DW (2007). Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol 176, 795–805. 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fachinetti D, Diego Folco H, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LET, et al. (2013). A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol 15, 1056–1066. 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodor DL, Valente LP, Mata JF, Black BE, and Jansen LET (2013). Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell 24, 923–932. 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. (2015). CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science (80-. ) 348, 699–703. 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, and Jansen LE (2014). The quantitative architecture of centromeric chromatin. Elife 3, e02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunleavy EM, Almouzni G, and Karpen GH (2011). H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G 1 phase. Nucleus 2, 146–157. 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zasadzińska E, and Foltz DR (2017). Orchestrating the Specific Assembly of Centromeric Nucleosomes. Prog. Mol. Subcell. Biol 56, 165–192. 10.1007/978-3-319-58592-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Von Stetina JR, and Orr-Weaver TL (2011). Developmental control of oocyte maturation and egg activation in metazoan models. Cold Spring Harb. Perspect. Biol 3, a005553. 10.1101/cshperspect.a005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smoak EM, Stein P, Schultz RM, Lampson MA, and Black BE (2016). Long-Term Retention of CENP-A Nucleosomes in Mammalian Oocytes Underpins Transgenerational Inheritance of Centromere Identity. Curr. Biol 26, 1110–1116. 10.1016/j.cub.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nashun B, Hill PWS, Smallwood SA, Dharmalingam G, Amouroux R, Clark SJ, Sharma V, Ndjetehe E, Pelczar P, Festenstein RJ, et al. (2015). Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol. Cell 60, 611–625. 10.1016/j.molcel.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, and Yanagida M (2007). Priming of Centromere for CENP-A Recruitment by Human hMis18α, hMis18β, and M18BP1. Dev. Cell 12, 17–30. 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, and Yanagida M (2004). Mis16 and Mis18 Are Required for CENP-A Loading and Histone Deacetylation at Centromeres. Cell 118, 715–729. 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Nardi IK, Zasadzińska E, Stellfox ME, Knippler CM, and Foltz DR (2016). Licensing of Centromeric Chromatin Assembly through the Mis18α-Mis18β Heterotetramer. Mol. Cell 61, 774–787. 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stellfox ME, Bailey AO, and Foltz DR (2013). Putting CENP-A in its place. Cell. Mol. Life Sci 70, 387–406. 10.1007/s00018-012-1048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moree B, Meyer CB, Fuller CJ, and Straight AF (2011). CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol 194, 855–871. 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, and Black BE (2012). HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev. Cell 22, 749–762. 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan D, Klare K, Petrovic A, Take A, Walstein K, Singh P, Rondelet A, Bird AW, and Musacchio A (2017). CDK-regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP-A loading. Elife 10.7554/eLife.23352.001. [DOI] [PMC free article] [PubMed]
- 32.Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, and Foltz DR (2011). HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol 194, 229–243. 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, and Almouzni-Pettinotti G (2009). HJURP Is a Cell-Cycle-Dependent Maintenance and Deposition Factor of CENP-A at Centromeres. Cell 137, 485–497. 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 34.Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, and Cleveland DW (2009). Centromere-Specific Assembly of CENP-A Nucleosomes Is Mediated by HJURP. Cell 137, 472–484. 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKinley KL, and Cheeseman IM (2014). Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell 158, 397–411. 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, et al. (2012). Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol. Cell 46, 260–273. 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 37.Stellfox ME, Nardi IK, Knippler CM, and Foltz DR (2016). Differential Binding Partners of the Mis18α/β YIPPEE Domains Regulate Mis18 Complex Recruitment to Centromeres. Cell Rep 15, 2127–2135. 10.1016/j.celrep.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan Z-J, Xu X, and Cooney AJ (2004). Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol. Reprod 71, 1469–1474. 10.1095/biolreprod.104.031757. [DOI] [PubMed] [Google Scholar]
- 39.Catania S, Pidoux AL, and Allshire RC (2015). Sequence features and transcriptional stalling within centromere DNA promote establishment of CENP-A chromatin. PLoS Genet 11, e1004986. 10.1371/journal.pgen.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNulty SM, Sullivan LL, and Sullivan BA (2017). Human Centromeres Produce Chromosome-Specific and Array-Specific Alpha Satellite Transcripts that Are Complexed with CENP-A and CENP-C. Dev. Cell 42, 226–240.e6. 10.1016/j.devcel.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bobkov GOM, Huang A, van den Berg SJW, Mitra S, Anselm E, Lazou V, Schunter S, Feederle R, Imhof A, Lusser A, et al. (2020). Spt6 is a maintenance factor for centromeric CENP-A. Nat. Commun 11, 2919. 10.1038/s41467-020-16695-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blower MD (2016). Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell Rep 15, 1624–1633. 10.1016/j.celrep.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banaszynski LA, Allis CD, and Lewis PW (2010). Histone variants in metazoan development. Dev. Cell 19, 662–674. 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, and Cleveland DW (2004). Structural determinants for generating centromeric chromatin. Nature 430, 578–582. 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 45.Sekulic N, Bassett EA, Rogers DJ, and Black BE (2010). The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature 467, 347–351. 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo LY, Allu PK, Zandarashvili L, McKinley KL, Sekulic N, Dawicki-McKenna JM, Fachinetti D, Logsdon GA, Jamiolkowski RM, Cleveland DW, et al. (2017). Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat. Commun 8, 15775. 10.1038/ncomms15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, and Torres I (1989). An improved culture medium supports development of random-bred 1-cel mouse embryos in vitro. J. Reprod. Fertil 86, 679–688. 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- 48.Akera T, Chmátal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, and Lampson MA (2017). Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358, 668–672. 10.1126/science.aan0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All imaging data reported in this paper will be shared by the lead contact upon request. This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


