Abstract
Prenatal screening using sequencing of circulating cell-free DNA has transformed obstetric care over the past decade and significantly reduced the number of invasive diagnostic procedures like amniocentesis for genetic disorders. Nonetheless, emergency care remains the only option for complications like preeclampsia and preterm birth, two of the most prevalent obstetrical syndromes. Advances in noninvasive prenatal testing expand the scope of precision medicine in obstetric care. In this review, we discuss advances, challenges, and possibilities toward the goal of providing proactive, personalized prenatal care. The highlighted advances focus mainly on cell-free nucleic acids; however, we also review research that uses signals from metabolomics, proteomics, intact cells, and the microbiome. We discuss ethical challenges in providing care. Finally, we look to future possibilities, including redefining disease taxonomy and moving from biomarker correlation to biological causation.
Keywords: liquid biopsy, prenatal care, cfDNA, cfRNA, genomics, noninvasive prenatal testing, NIPT
INTRODUCTION
Over the past decade, noninvasive prenatal testing (NIPT) employing circulating cell-free DNA (cfDNA) to screen for fetal aneuploidy has transformed prenatal care by dramatically reducing the use of and risks associated with invasive approaches such as amniocentesis and chorionic villus sampling (1–3). NIPT is covered by both private and public insurance and is typically administered before 12 weeks of gestation (4). The success of noninvasive screening for common fetal aneuploidies has led to expanded fetal genetic screening that includes rare autosomal trisomies and microdeletions. However, major obstetrical syndromes, like preterm birth and preeclampsia, which are leading causes of fetal and maternal complications and death, respectively, remain underserved and understudied (5, 6). They also disproportionately affect Black and American Indian/Alaska Native pregnant individuals in the United States who not only experience these complications at higher rates but also use a healthcare system that fails them in important respects (7, 8). For complications like preeclampsia or spontaneous preterm birth, emergency care can often be a clinician’s only option, which stands in stark contrast with the promise of proactive clinical care. One step toward bridging this gap is to identify at-risk pregnancies early and deliver on the promise of precision medicine.
Precision medicine requires that we have sufficient tools to predict an individual’s risk of a given condition, monitor its progression, and provide treatment. Liquid biopsies that measure macromolecules or whole cells present in blood, including DNA, RNA, white blood cells, proteins, lipids, and metabolites, are one possible solution to predict risk and monitor progression (Figure 1a; also see the section titled Advances) (9–13). Indeed, more recently, liquid biopsy tests that measure other macromolecules or quantify cfDNA differently (e.g., for single-gene disorders) have been demonstrated to be clinically useful in proof-of-principle studies and hold promise for the future (14, 15). Such tests provide a unique lens into the relationship between the health of the pregnant individual and the health of the fetus; however, this insight can be a double-edged sword, as an abnormal finding can screen for but not definitively diagnose a health complication, leading to ethical concerns (see the section titled Challenges). The distinction between screening to detect potential health issues prior to symptom presentation versus diagnosis to definitively identify illness based on symptoms has been part of the promise and risk of NIPT since its inception.
Figure 1.
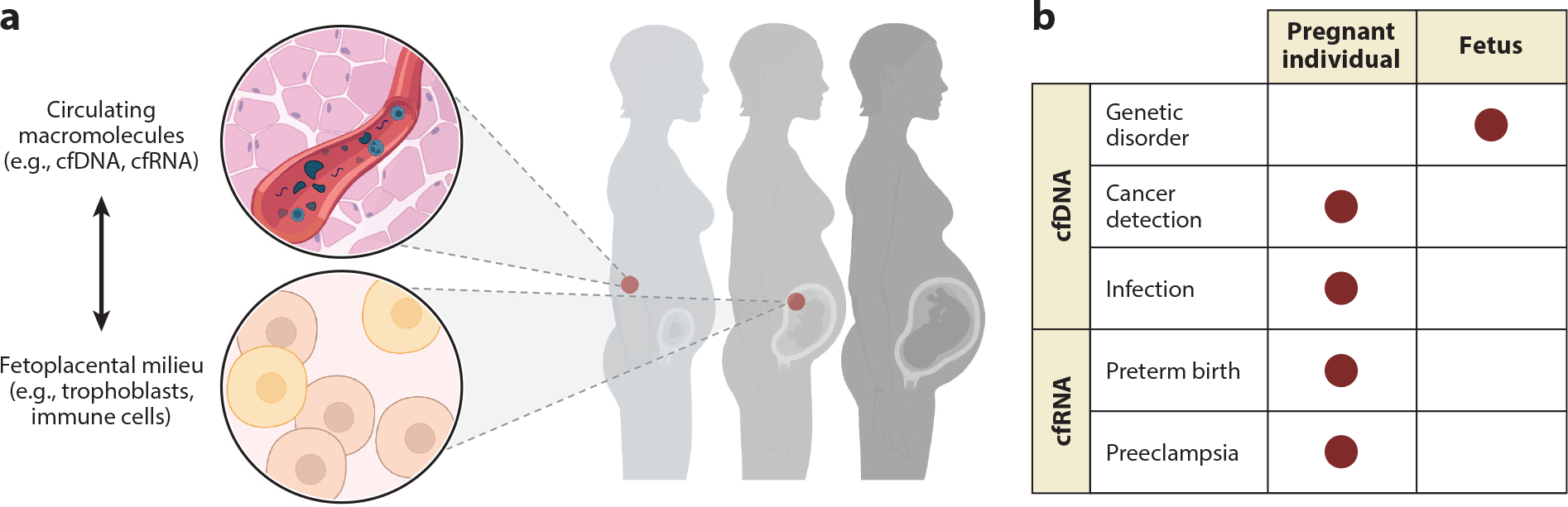
Noninvasive prenatal testing provides a lens into the health of the pregnant individual, fetus, and placenta. (a) Across pregnancy, liquid biopsy tests can measure macromolecules or whole cells present in blood, including cell-free DNA and RNA (cfDNA and cfRNA), white blood cells, proteins, lipids, and metabolites. (b) This approach is useful for the screening and diagnosis of numerous pregnancy-related conditions including genetic disorders, parental cancers and infections, and obstetric syndromes like preeclampsia and preterm birth, thereby helping to guide clinical practice. Figure adapted from images created with BioRender.com.
Information derived from both screening and diagnostic testing depends on the selected data modality. For example, the fetal fraction of cfDNA is derived mainly from syncytiotrophoblast cell turnover and is therefore placental in origin. Because the placenta consists of contributions from both the fetus and the pregnant individual, one can quantify the fetal fraction from cfDNA. Fetal contributions to cfDNA can range between 10 and 20%; however, this estimate is sensitive to the pregnant individual’s age, body mass index (BMI), and gestational age at sampling (16). In contrast, cell-free RNA (cfRNA) is derived from diverse sources, including from the fetus (up to 14% of total cfRNA), placenta, and various other tissues of the pregnant individual. It is also not affected by BMI (13). Owing to its multifold origin in pregnancy, cfRNA can provide clues about the interplay among the pregnant individual, fetus, and placenta. Gene expression levels measured using cfRNA change during gestation in predictable ways that map closely with their placental and fetal gene expression and represent a physiological snapshot in time (17–23). Measuring immune cells present in circulation can also provide functional information about immunophenotypes and related protein expression, such as insight into feto-maternal tolerance and immune contributions to the onset of labor (24). Measuring fetal cell karyotypes may point to a more direct means of screening for copy number variation and single-gene disorders (25–28). Finally, protein and metabolite measurements can point to a role for proteomic or hormonal signaling over pregnancy.
In addition to screening applications, liquid biopsy studies to date have also been investigative in nature. The identification of new molecular measurements (biomarkers) has therefore been a consequence of discovery-focused research. As a result, risk-associated macromolecules may provide clues to disease pathogenesis. Syndromes like preeclampsia, which to date have been defined by a loose categorization of symptoms that have changed over the past two decades, would benefit from a molecular definition of disease and more broadly an improved understanding of disease pathogenesis (29, 30). Concepts like “full-term birth” versus “preterm birth” (delivery before 37 weeks) will also be well served by more precise molecular definitions of biological gestational age. Ultrasound examination, which has become a standard of prenatal care to detect multiple gestations and congenital abnormalities, can at best predict gestational age within two weeks on either side of the stated due date. This has potential medical consequences. Infants born after 37 weeks are considered full term, but they may exhibit preterm-like symptoms (e.g., under-developed lungs) and consequently require ventilatory support. There are no current means of identifying neonates who will require more intensive care. It is also clear that defining “full-term pregnancy” as a binary with a cutoff of 37 weeks is inconsistent with clinical observation. Instead, full-term pregnancy may exist on a continuum and require a new definition. Thus, studies that examine the relationship of molecular measurements to obstetric dogma may help to redefine or refine disease taxonomy and add to the clinician’s toolbox (see the section titled Possibilities).
In this review, we discuss advances, challenges, and possibilities in NIPT, focusing mainly on cell-free nucleic acids, but also noting exciting advances using signals from metabolomics, proteomics, intact cells, and the microbiome. We focus on advances by obstetric complication as opposed to data modality (e.g., cfDNA or cfRNA), as the appropriate data modality will depend on the biological question asked (Figure 1b). We also highlight ethical, technical, and biological challenges. Finally, we look to future possibilities, including redefining disease taxonomy and moving from biomarker correlation to biological causation.
ADVANCES
Genetic Disorders: Aneuploidies and Mendelian Disease
Genetic disorders can arise in many ways. Aneuploidies such as trisomy 21 and Klinefelter syndrome (also known as 47,XXY) are caused by abnormal total chromosome numbers based on missing or extra chromosomes. They are primarily but not exclusively due to errors in meiosis. While many aneuploidies, such as monosomy X, underlie fetal loss (miscarriage), others are observed in fetuses surviving later in gestation (31). In contrast, Mendelian disorders such as sickle cell anemia, Duchenne muscular dystrophy, and cystic fibrosis are the result of single-gene germline or de novo mutations and do not generally result in fetal loss.
Advances in noninvasive testing and clinical translation.
Aneuploidies have been identified in fetuses during gestation since the 1960s using amniocentesis (32). Over time, the technologies for prenatal screening for fetal aneuploidies have improved dramatically to include far less invasive approaches such as ultrasound examination and the measurement of serum analytes and cfDNA. In the past decade, NIPT screening for fetal aneuploidy has become routine and adopted by clinicians across the world (33, 34). Tens of millions of pregnant individuals have now received NIPT for fetal aneuploidy since the first proofs of concept were published in 2008 (35, 36). The clinical impact of the implementation of NIPT can also be seen on the provider side: A generation of obstetricians has not been trained to perform invasive procedures. As the number of requested invasive procedures continues to fall, the next generation will be trained to perform such procedures via simulation (37). Academic cytogenetic laboratories have also closed owing to fewer prenatal diagnostic procedures. From providers to patients, the impact of NIPT is clear, with more clinical tests coming online as the technology continues to develop.
Today, most clinical NIPT screens for aneuploidy by quantifying the fetal contribution to cfDNA, which is primarily derived from placental trophoblast turnover. Testing based on prenatal cfDNA uses coverage-based methods like copy number analysis to determine the relative amounts of various chromosome fragments in the blood. An abnormal ratio of various DNA fragments mapping to a particular chromosome indicates aneuploidy in the presence of a predominant cfDNA background from the pregnant individual (Figure 2). To avoid a false negative result, one often uses a threshold of at least 2% of total cfDNA, termed the fetal fraction (38–40). Computationally, these techniques rely on binning reads based on where they map to the genome. For aneuploidies, it is sufficient to use shallow-depth whole-genome sequencing (0.2–1× coverage, 10 million reads per sample) and bin reads per chromosome (41, 42). The counting principle is then used to estimate overrepresentation of potentially aneuploid chromosomes in the fetus. This approach assumes that the sequencing measurement is unbiased; however, various biochemical steps involved in sequencing introduce systematic errors, including dependence on fragment length and guanine–cytosine (GC) content. It has been crucial in making practical diagnostics to correct for the GC bias in sequencers, and computational approaches that do so have led to the conclusion that it is possible to create algorithms that are limited only by counting statistics and not by systematic errors (43).
Figure 2.
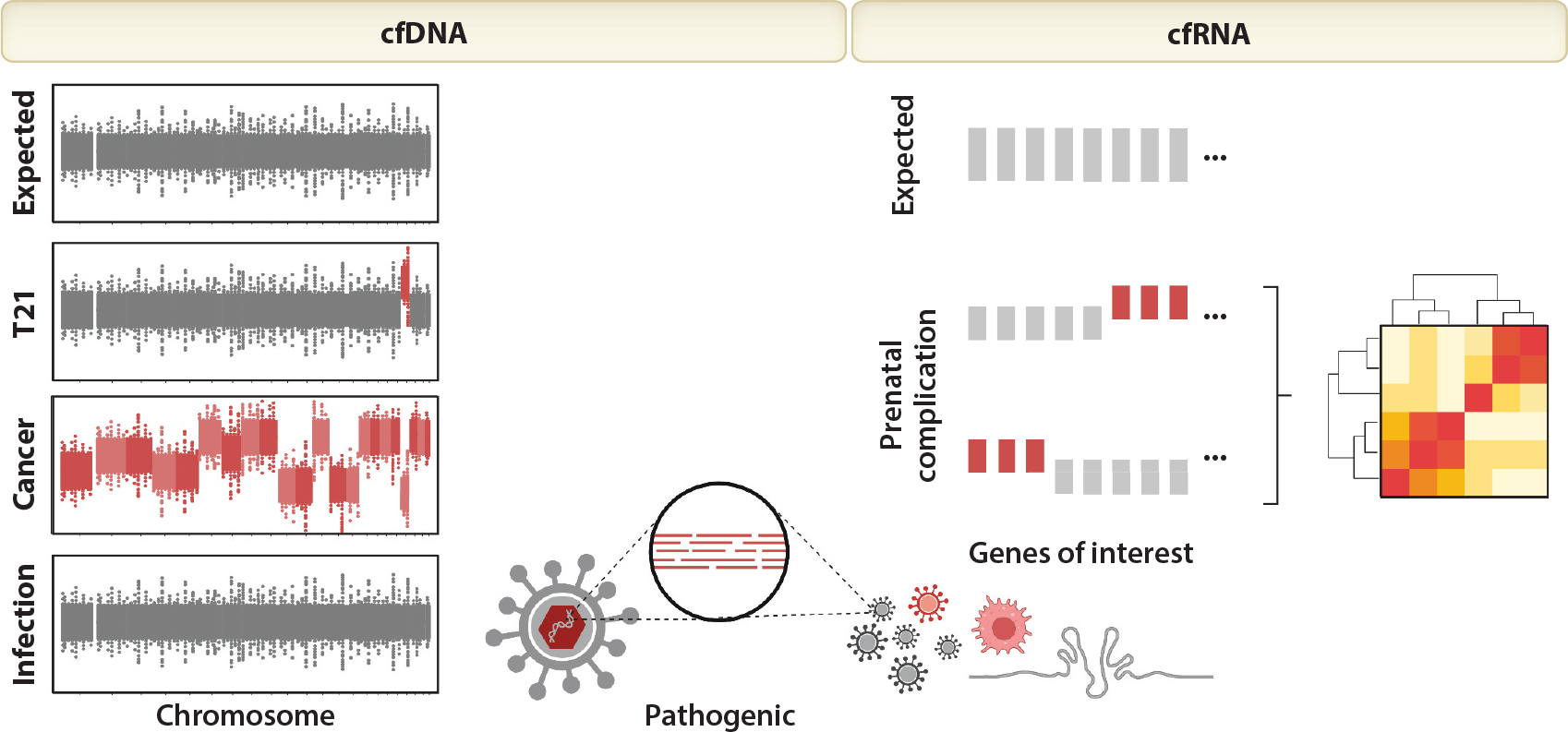
Representative results for each noninvasive prenatal testing (NIPT) test. NIPT tests that measure cell-free DNA (cfDNA) expect equal representation from all chromosomes. Individual chromosomal imbalances suggest fetal aneuploidy [e.g., trisomy 21 (T21)], whereas genome-wide imbalances involving more than one chromosome suggest maternal malignancy (e.g., cancer). The detection of pathogenic cfDNA or cell-free RNA (cfRNA) sequences suggests infection; the detection of elevated host immune genes or pathogenic RNA suggests active infection. NIPT for prenatal complications typically assess the under- or overrepresentation of specific genes of interest; in combination using predictive models like machine learning, these unusual gene levels can suggest preeclampsia or preterm birth. Figure adapted from images created with BioRender.com.
Today, these screening tests have seen broad success and worldwide adoption with reported sensitivities and specificities of 99%. It is also useful to define cfDNA-based clinical utility as compared to other noninvasive methods using positive and negative predictive values (PPV and NPV, respectively) and the risk that a given procedure poses to a pregnancy. PPV and NPV implicitly consider the prevalence of a condition and better reflect a test result’s real-world interpretation. PPV drops with decreasing prevalence and decreasing parental age at pregnancy while NPV remains constant at 99.99% (44, 45).
Using cfDNA, the PPVs for more prevalent conditions such as trisomy 21, which has a PPV of 96% (46, 47), are quite remarkable, especially when compared to other standard noninvasive prenatal screening methods such as measurements of nuchal translucency and biochemical analytes, which have PPVs on the order of 3–4% for trisomy 21 (4, 48). Low PPVs translate to high false positive results and subsequent unnecessary, invasive prenatal diagnostic procedures such as amniocentesis and chorionic villus sampling. These invasive procedures have an increased risk of pregnancy loss. The emotional cost of pregnancy loss is deeply personal and related to a specific person’s experience around conception (e.g., infertility or difficulty conceiving due to age or other personal health issues). Consequently, individuals may opt out of invasive procedures to avoid any additional risk of pregnancy loss. Indeed, it has been observed that older pregnant individuals opt out of invasive procedures more frequently (49). Providers should also consider the risk of fetal loss for an invasive procedure (~1 in 200) (see the subsection on Ethics in the section titled Challenges). Alternatively, a negative result may also influence a person’s choice to proceed with an invasive follow-up. In practice, the very high NPV associated with NIPT has influenced many pregnant individuals to decline invasive diagnostic testing. This is reflected by the 60% decrease globally in invasive diagnostic procedures.
Given the successful implementation of cfDNA sequencing into prenatal care, it is not surprising that expanded noninvasive testing menus have been developed. Initially these included testing for fetal sex and sex chromosome aneuploidies, but later they have grown to include detection of chromosome microdeletions (50, 51) and rare autosomal trisomies (RATs) (31) and genome-wide detection of copy number variants (52). Microdeletion syndromes, although clinically significant, have lower prevalences, and consequently tests for these conditions have lower PPVs. These findings have raised ethical questions around how actionable a result can be for conditions that have very low prevalence (see the subsection on Ethics in the section titled Challenges). By contrast, RATs are collectively not so rare and associated with late miscarriages, intrauterine growth restriction, true fetal mosaicism, and uniparental disomy; their collective association with adverse pregnancy outcomes may help triage high-risk pregnancies (31).
Noninvasive molecular techniques have also advanced in the past decade to facilitate noninvasive prenatal diagnosis (NIPD) of single-gene disorders. These methods typically rely on the absolute quantification of cfDNA using digital PCR (polymerase chain reaction) methods, sequencing with unique molecular identifiers, or exclusion of paternal mutations (14, 15, 53–57). The line between NIPD and NIPT can be defined broadly based on Mendelian disorder inheritance patterns and consequently whether results can be considered definitive and therefore sufficient for decisive diagnosis (58–60). Paternally inherited or de novo autosomal dominant conditions like skeletal dysplasia (e.g., achondroplasia and thanatophoric dysplasia) are the most straightforward to diagnose, as fetal mutations can be discerned from the absence of a maternal background signal and correlated with sonographic findings (61–63). Identifying single-gene disorders becomes progressively harder as background signal from the pregnant individual increases. The most technically challenging cases occur when both parents are carriers for the same autosomal recessive or X-linked mutation. In these cases, several approaches have been developed to quantify the fetal fraction although only one meets the criteria for diagnosis. The diagnostic approach relies on relative haplotype dosage analysis, which requires blood samples from both parents and uses single-nucleotide polymorphisms to develop phasing for alleles associated with high or low risk for the genetic condition (64). The screening approach requires only a blood sample from the pregnant individual and estimates the fetal fraction and a corresponding confidence interval using a panel of single-nucleotide polymorphisms quantified with digital PCR (55, 65). NIPD is routinely used in the United Kingdom to diagnose single-gene disorders such as cystic fibrosis, Rhesus D blood group, and several of the muscular dystrophies, but it is less widely adopted in the United States (54, 66). As gene therapy and gene editing becomes more routine, these tests will become more actionable (60).
Challenges remaining.
Performing noninvasive whole-genome analysis of the fetus via sequencing of circulating plasma DNA during pregnancy has been proven to be technically feasible (56, 57, 67), but whether it has clinical utility and whether it is affordable remain to be proven. As long-read, portable sequencing technologies become more widely available, this landscape may change. These low-cost, easy-to-use technologies (e.g., Oxford Nanopore MinION) have improved in accuracy over the past five years and allow for the rapid identification of structural DNA variants and the characterization of DNA methylation and fragment length. It has proven challenging to define differences in length for fetal and maternal DNA using short-read sequencing (68). One recent application of long-read sequencing identified ultralong fetal DNA fragments in circulation (69). Applications of long-read sequencing to NIPT have yet to be fully explored, but we expect that they may change the landscape of what is possible and at what price point, just as short-read sequencing technologies did before.
Sequencing fetal (nucleated red blood cells or trophoblasts) cells presents an alternative, earlier-stage approach to cfDNA-based NIPT that may be diagnostic for aneuploidy and single-gene disorders. Although it is not a new concept (70–73), this initiative has gained new traction owing to technical advances that may overcome key hurdles—namely, a high false positive rate for aneuploidy and sex determination. Fetal cells in circulation or in the cervix are rare and potentially mosaic in the case of trophoblasts. Like cfDNA, their prevalence in circulation or cervical tissue correlates with gestational age. Newer methods like microfluidics, capillary-based methods, and laser microdissection have paved the way for enrichment of very rare cell populations. In parallel, sequencing methods have advanced and opened the door to whole-genome amplification from low-input (74) and even single-cell whole-genome analysis (75, 76). These methodological advances have led to several proof-of-concept studies that identify genetic abnormalities ranging from aneuploidy to Mendelian single-gene mutations using fetal cells from the cervix (25, 28, 77, 78) or circulation (26, 27, 79, 80) collected as early as five weeks of gestation. The clinical utility (PPV and NPV) of fetal-cell-based technologies remains to be seen. It also remains challenging to define and isolate rare fetal cells in the background of whole blood or cervical tissue during pregnancy. In practice, to ensure the isolation of at least two fetal cells, these tests may require relatively large blood draws or cervical tissue samples that will have to fit within the paradigm of obstetric care.
Cancer Detection
In late 2011, when cfDNA screening for fetal aneuploidy was first incorporated into prenatal care, it challenged screening paradigms that had been in place for several decades. At that time, the sensitivity of massively parallel sequencing of circulating DNA to additionally detect conditions in the pregnant individual was not fully appreciated. It is now known that cfDNA NIPT platforms can detect uterine fibroids (81), sex chromosome and autosomal aneuploidies, autoimmune conditions, and clinically silent malignancies (82). These incidental findings can confound fetal risk assessment for aneuploidy (83).
Advances in noninvasive testing and clinical translation.
At least six large-scale studies using data from commercial or national laboratories have demonstrated the correlation between multiple aneuploidies detected by cfDNA screening and cancer in pregnant individuals (84–89). These studies have primarily been retrospective and reanalyze NIPT results from pregnant individuals with known cancer diagnoses. The types of cancers detected are those that would be expected in people of reproductive age: lymphoma (Hodgkin and B cell), colorectal cancer, leukemia, ovarian cancer, and breast carcinoma.
In pregnant individuals that also harbor an unknown cancer diagnosis, the tumor sheds cfDNA, which contributes to the sequenced background, potentially distorting NIPT results and their interpretation. Tumor-derived cfDNA, along with placental and other background DNA from the pregnant individual, is sequenced and mapped to the human genome. In European laboratories that use whole-genome sequencing and open source bioinformatics algorithms such as WISECONDOR (41), genome-wide imbalance involving more than one chromosome is immediately obvious and suggests parental malignancy (Figure 2). In the United States, however, NIPT results often rely on proprietary algorithms that use reference chromosomes and mask nontarget chromosomes. Instead of an obvious pattern that suggests malignancy, the analysis generates a nonreportable result. There are currently no professional guidelines for clinical management following a nonreportable result or for multiple aneuploidies other than to confirm the fetal diagnosis with amniocentesis or chorionic villus sampling. Indeed, it is a matter of debate whether cfDNA results that suggest parental malignancy should even be reported back to the pregnant individual (90).
Challenges remaining.
Challenges to the identification and management of nonreportable or multiple aneuploidy NIPT results are numerous and largely stem from a lack of consensus on how to report these types of results. There is also a lack of evidence-based guidelines on whether the results should be disclosed to the pregnant individual and what is the appropriate posttest workup. In addition, insurance companies refuse to pay for subsequent management of a person who is otherwise asymptomatic (83). Furthermore, in the United States, there is a need to educate both primary obstetrical providers and oncologists regarding the significance of these results and the need for timely follow-up. To address the need for evidence-informed clinical guidelines, researchers at the National Institutes of Health’s Clinical Center in Bethesda, Maryland, are currently conducting a prospective longitudinal research study (83). Known as IDENTIFY (Incidental DEtection of maternal Neoplasia Through non-Invasive cell-Free DNA analysis) (https://clinicaltrials.gov identifier: NCT04049604), the study brings participants free of charge to the NIH for a comprehensive workup that includes laboratory studies and whole-body imaging. Results of the research studies are disclosed to the participants and are actionable. A parallel qualitative study is evaluating the benefits and limitations of disclosing the unusual NIPT results in persons who do and do not have cancer. Lastly, although we hypothesize the early detection and treatment of cancer will ultimately result in improved outcomes, it will be a challenge to prove this.
Infection During Pregnancy
Pregnant individuals and their offspring are susceptible to many infectious agents that can cross the placenta and be directly transmitted to the fetus or indirectly affect placental function through inflammation and sepsis. Infection can result in preterm rupture of membranes, preterm birth, preeclampsia, and fetal abnormalities including growth restriction, developmental delay, hearing loss, and fetal demise. Evidence is accumulating that clinically silent but active herpes infections are the underlying etiology behind many of these complications (91).
Advances in noninvasive testing and clinical translation.
The detection of viral, bacterial, and fungal DNA sequences from cfDNA shotgun sequencing was first appreciated in the context of noninvasive monitoring of organ transplantation (92–94). Tests that leverage this approach to noninvasively detect infection have since been shown to be clinically useful in several disease contexts (95, 96). More recently, this approach was extended to pregnancy using retrospective NIPT samples from national biobanks (91, 97–100). In a complementary approach, it has also been demonstrated that prenatal cfRNA sequencing can detect infection. Pathogen-related RNA, as opposed to DNA, can point to active infection (19). For both RNA-and DNA-based methods, it is straightforward to identify pathogen-related nucleic acids by querying reads that do not align to the human genome against the National Center for Biotechnology Information’s bacterial, viral, and fungal databases (Figure 2). To avoid false positive results, it is important to consider sources of contamination during sample processing (e.g., sequence a negative control) and consider the human relevance of identified pathogens (101). Importantly for cfDNA-based NIPT, detection of viral DNA sequences does not affect the accuracy of screening for fetal aneuploidy (97, 98). For cfRNA-based NIPT, the detection of pathogenic RNA can be paired with measurements of human immune-specific RNA that reflects the pregnant individual’s immune response (19).
The most frequently reported DNA viruses during pregnancy include human herpes viruses, parvovirus B19, hepatitis B, papillomavirus, adenovirus, polyomavirus, and human torque teno virus (91). Some of these, like human herpesvirus, are associated with known risks during pregnancy. For example, inherited chromosomally integrated (ici) human herpesvirus 6 (HHV6) can be integrated into the telomeres of each cell and transmitted to offspring in a dominant manner. The presence of maternal iciHHV6 sequence carries a threefold increased risk of preeclampsia and an increased chance of miscarriage (102, 103). Interestingly, the studies performed to date show different prevalence of the same viruses in different geographic and ethnic populations. For example, in one Chinese study, the prevalence of HHV7 sequences was only 0.3% (99). By contrast, in that same Chinese population, hepatitis B viral sequences were found in 2.5% of 141,431 plasma samples from pregnant individuals, whereas in a Dutch population, the prevalence of hepatitis B was only 0.12% (97). Focusing on healthy pregnancy, Tong et al. (100) reanalyzed cfDNA whole-genome sequencing data from a large cohort of 107,763 samples to determine the microbial baseline. All except two had positive reads for at least one microorganism. Of the nonhuman DNA sequences detected, 95% were from bacteria, 3% from eukaryotes, and 0.4% from viruses. There were 1,524 different bacterial species identified. Those that were commonly detected included Acinetobacter johnsonii (found in 47.4% of samples), Enhydrobacter aerosaccus (found in 35.9%), and Delftia acidovorans (34.3%). The clinical significance of finding these organisms in the blood of pregnant individuals is currently unknown.
Challenges remaining.
The clinical utility of tests that leverage this approach to noninvasively detect infection has been demonstrated outside of pregnancy (95, 96). For prenatal care, several proof-of-concept studies have now suggested that pathogenic DNA can be detected using standard NIPT cfDNA sequencing; however, the clinical utility of such a platform remains to be seen. To identify potentially harmful infections during pregnancy, clinicians currently use the panel test TORCH (Toxoplasmosis gondii; Other, including syphilis, hepatitis B virus, and parvovirus; Rubella; Cytomegalovirus; and Herpes simplex virus/HIV). The test relies on the measurement of serum antibodies to identify acute and past infections. All the organisms in the TORCH panel, except for rubella, an RNA virus, could in theory be detected using NIPT cfDNA sequencing for fetal chromosome abnormalities. Rubella could be detected by a parallel cfRNA sequencing approach.
Preeclampsia
Preeclampsia affects 3–5% of pregnancies in the United States and up to 8% of all pregnancies globally (104). It accounts for 10–15% of maternal deaths (105) and 15–20% of preterm births (106). The pathophysiology of preeclampsia remains mysterious, although abnormal placentation early in pregnancy and an inflammatory response are very likely important contributing causes (29, 30, 107). Formally diagnosed after 20 weeks of gestation, the multiorgan syndrome may, in fact, be more than one disease, imperfectly divided into early or late onset disease—or, alternatively, into disease with or without severe symptoms. However, a final common pathway probably involves endothelial dysfunction and end-organ damage, and in some cases it leads to stroke and even death. Although delivery is the only present treatment, prediction of risk early in gestation (prior to 16 weeks) can guide the prophylactic use of risk-reducing agents like low-dose aspirin (108).
Advances in noninvasive testing and clinical translation.
While the specific biological processes involved in the development of preeclampsia are not yet completely understood, early prediction would be desirable to prevent emergency medical care. Different noninvasive modalities have proven useful here, and recent studies have also integrated high-throughput omics analyses (e.g., genome, transcriptome, proteome, and metabolome) on the same biological sample (109). In a single-site study, urine metabolite and plasma proteomic measurements predicted preeclampsia before 16 weeks of gestation using a stacked logistic regression model; if further validated, these results would pave the way for a urine-based screen for preeclampsia risk (109). Separately in several multicenter studies, cfRNA has also shown considerable promise as a standalone measure that can predict risk of preeclampsia early in gestation, including prior to 16 weeks of gestation and long before there are any clinical signs of preeclampsia, allowing for prevention (9, 10, 22, 23) (Figure 2). Each of these predictive models first identifies changes associated with preeclampsia (e.g., machine learning with regularization penalty, differential expression) to build a test. To address interpersonal differences, these models often integrate information from multiple sources (e.g., different data modalities, multiple genes) to yield a summary statistic (e.g., probability) that represents a pregnant individual’s risk of developing preeclampsia. If broader sample sets are available, these studies next check that the test generalizes beyond a single laboratory, hospital, or cohort. Importantly, the predictive capacity of (machine learning–based) preeclampsia risk models that employed cfRNA measurements were not affected by parental race and worked just as well for pregnant individuals of races and ethnicities not included during model training (22, 23).
Interrogating the biological pathways involved in these predictions confirms a role for placental and endothelial-linked pathways (22, 23, 109). In fact, the most striking changes in preeclampsia risk–related cfRNA profiles occur as early as five weeks of gestation, consistent with the idea that preeclampsia has its origins in the first trimester. These findings also suggest that the identified molecules may point to new clues about disease pathogenesis (30, 110, 111). These studies also provide a molecular means by which preeclampsia can be grouped into subsets. For example, future diagnostic criteria may rely on measuring pathway signatures that are common to preeclampsia, like abnormal innate and adaptive immune signaling (23, 112). The organ health of pregnant individuals may also be monitored in a noninvasive manner using cell-type-specific molecular signatures (20, 22, 23, 113).
Challenges remaining.
These proof-of-concept studies suggest that preeclampsia risk might be predicted using a liquid biopsy test but must be clinically validated in large, multicenter trials to better understand their clinical utility (PPV and NPV). There also remains more to be done to definitively connect these biomarkers to disease pathogenesis as opposed to correlation alone.
Preterm Birth
Preterm birth is now the primary worldwide cause of death under five years of age, and in about half of cases, it is spontaneous and enigmatic. Understanding how immunologic tolerance is initiated and maintained, and then disrupted, sometimes too soon, might provide clues to understand not only normal parturition and preterm birth but potentially also cancer, autoimmunity, and a variety of other inflammatory conditions such as cardiovascular disease, or even aging itself.
Advances in noninvasive testing and clinical translation.
Is preterm birth a calendar event or a broad biologic anomaly? Indeed, it is the latter. Using a single blood draw that measures cfRNA, Ngo et al. (20) demonstrated that it is possible to predict the onset of preterm labor months in advance of the event. To do so, candidate differentially expressed genes were identified and combined to form a heuristic test that predicted an individual’s risk of developing preterm birth up to two months in advance of delivery (Figure 2). The predictive power of cfRNA was also recently validated in a larger, multicenter trial (114). As the transcriptomic clock suggested, many of the genes changing over pregnancy are immune genes in the innate immune system. The immune clock of pregnancy has been further described by using time-of-flight mass cytometry (CyTOF) (115), allowing apparent disruptions in the immune clock to be identified in pregnancies destined for preterm birth. The immunology of normal gestation has also been characterized, as well as an immunologic switch that signals the impending onset of normal labor and delivery (24).
Work from several groups has also highlighted an association between the vaginal microbiome and metabolome and preterm birth (100, 116–120). Pregnant individuals presenting with low-lactobacilli vaginal microenvironment are at an increased risk (odds ratio = 1.69) of preterm delivery, as highlighted in a recent systematic review and meta-analysis (116). Some studies have also described a connection between the microbiome, metabolome, and host immune system. Microbiota harbor biosynthetic pathways that yield metabolites that can shape the organism’s microenvironment and elicit a host immune response (117, 118). The correlation between the vaginal microbiome and signals from cfRNA, metabolomics, and proteomics in peripheral blood plasma has also been explored (11, 121).
Challenges remaining.
New liquid biopsy tests to predict risk of preterm birth have not yet entered the clinic. Although multicenter studies like that run by Camunas-Soler and colleagues are encouraging (114), the clinical utility of these tests remains to be proven. Such trials are currently underway and should publish results in the next year or two. There is also the question of adoption: Cultural norms across the globe may lead to different test type preferences. For example, although it could be argued that vaginal swabbing can easily be done at home, the cultural taboo that persists around female anatomy makes such a test socially unacceptable in some parts of the world. In contrast, a blood test would require a trained phlebotomist to draw blood and either ship it to a central facility or process the sample on site. Recent evidence suggests that it may be possible to ship blood samples overnight or even five days postcollection at room temperature without any effect on test results (21, 122); however, this again will require further testing.
CHALLENGES
Ethics
NIPT is a field that conceptually could include a variety of minimally invasive tests, including imaging studies, to determine risk for certain developmental abnormalities. Conventional cfDNA testing provides fundamentally genetic information. It is also accurate enough to be clinically useful for many prevalent chromosomal abnormalities and avoids the miscarriage risk associated with more invasive methods. Its accuracy varies for the different chromosomal anomalies according to their prevalence, with prediction for trisomy 21 being better than that for trisomy 18 or 13. NIPT has also been used to identify single-gene defects (see the section titled Advances), but generally confirmatory diagnosis is necessary for both chromosomal and any other suspected genetic disorders. Genetic testing also comes with a variety of challenges, not all of which are technical in nature.
First, as used today, cfDNA-based NIPT is most often a screening method and not a definitive diagnostic test for the individual fetus. The line between screening and diagnosis can be defined broadly by the question: Can a test’s results be considered definitive and therefore sufficient for decisive diagnosis? For example, certain single-gene and X-linked conditions have no background prevalence. Consequently, any cfDNA signal can be definitively assigned to the fetus. On the other hand, assessing the number of chromosomes to identify aneuploidy can be obscured by the ratio of DNA from the pregnant individual relative to that from the fetus. Consequently, cfDNA-based aneuploidy tests are considered screens and not diagnostic. Nonetheless, the fetus either has the genetic condition or it does not. Still, the phenotype associated with the genetic diagnosis may vary in terms of physical and other biologic features and severity because of the expression and penetrance of the genetic variant as well as the environment. The importance of the latter is apparent in the case of metabolic disorders, often presenting in the newborn period. Some of the major ethical concerns regarding the implementation of cfDNA screening for chromosomal abnormalities include the fact that it is now technically possible to screen for conditions with lower prevalences. These abnormalities include sex chromosome aneuploidies such as Turner syndrome (also known as 45,X) and Klinefelter syndrome (47,XXY), rare autosomal trisomies such as trisomy 16 and trisomy 7, and microdeletion syndromes such as DiGeorge syndrome (22q11.2 deletion) (51, 123–126). Tests for these rare conditions have lower PPVs, which consequently raises an ethical question around how actionable this information really is. What should a clinician and patient do when they receive a positive test result for a rare condition? Such a result could very likely be a false positive. Consequently, the clinical utility of having this additional screening information may be neutral or worse detrimental. As gene therapy and gene editing becomes more routine, NIPT will become more actionable and questions around decision-making will become more pressing. How such action is regulated is an open question.
Second, decisions based on genetic diagnoses are often fraught and assume personal, social, or societal risks beyond the medical ones for the pregnant individual and fetus. Nonetheless, the primary ethical maxim should be foremost and consistent with the physician’s oath: Primum non nocere (do no harm). This principle of nonmaleficence is one of several principles that need to be applied to the use of NIPT and invasive diagnostic tests. Noninvasive tests with low PPVs translate to high false positive results and lead to invasive prenatal diagnostic procedures that have an increased risk of pregnancy loss. The emotional cost associated with potential miscarriage risk is deeply personal and should not be undervalued when providing care. Additional principles vary slightly with respect to content, but most ethicists would agree on beneficence, autonomy, and justice as well. From autonomy, we derive the ethical principles of truth-telling and informed consent. Another common principle is respect for persons. None of these principles make decision-making any easier. In fact, at least two of them are especially challenging—respect for persons and autonomy. Respect for persons requires that there is agreement about when personhood can be said to exist during pregnancy. There can be personal, cultural, political, legal, and philosophical differences of opinion in this regard, making how to use the information obtained from NIPT difficult in any case. Autonomy is also a challenging principle to apply to decision-making based on NIPT because the fetus is dependent upon others to decide what is in the best interest of the fetus.
Third, the measurement of cfRNA has now been added to NIPT. Here, the ethical challenges become even more nuanced. Whereas knowing one’s genetic makeup is like knowing the structure of a piano, knowing what genes are being expressed over time in a pregnancy is like knowing what music is playing or perhaps what will be played in the future. The sources of cfRNA include the pregnant individual, fetus, and placenta and can provide insights into normal development, for example, gestational age dating, but also acquired conditions, such as preterm birth and preeclampsia (20, 22, 23, 114). Once the changing biologic disposition of the fetus is understood, interventions might be proposed to alter the developmental trajectory of the fetus or avoid an acquired pathologic condition. On the face of it, such information would seem inherently useful. However, many conditions, like preterm birth, have social determinants that, of course, contribute to known health disparities in this regard (12). Thus, the solutions may not be solely medical but social, political, and legal in nature. Moreover, a biomarker, even when the biology underpinning the social determinant is understood, is useless without feasible tactics to ensure equal access to NIPT and the resources required to resolve the social inequalities that exist.
Computational
At their core, predictive models for prenatal complications like preeclampsia and preterm birth must first identify changes associated with the condition (e.g., machine learning with regularization penalty, differential expression) and then confirm that these identified changes generalize beyond a single laboratory, hospital, or cohort. To address interpersonal differences, these models often integrate information from multiple sources (e.g., stacked models that use different data modalities, multiple genes in a single model) to yield a summary statistic (e.g., probability) that represents a pregnant individual’s risk of developing a prenatal complication. When used with high-content omics data in which the number of features (103–105) quickly outpaces the number of samples available for training (102, 103), sparsely regularized algorithms (e.g., logistic regression with a lasso penalty) do not guarantee the selection of a consistent subset of features. So long as the predictive power of the identified features generalizes to large, diverse cohorts, sparsely regularized algorithms have proven adequate to predict the risk of prenatal complications. However, as we look to understand a given complication’s pathogenesis and ultimately develop new treatments, it will be necessary to identify a core set of biological hypotheses to test in the laboratory. Presently, this is often accomplished by interrogating the biological pathways involved using orthogonal analyses such as gene ontology. As we look ahead, a single statistical workflow that can identify predictive and consistent biomarkers would be useful.
POSSIBILITIES
Redefining Taxonomy
Historically, the maturation of the fetus and the maturity of the newborn have been used to define the risk profiles of the fetus and newborn in the respective fields of maternal–fetal medicine and neonatal–perinatal medicine. Moreover, pathologic conditions or diseases of the fetus and newborn have been defined categorically. With respect to biologic function, concentrations of electrolytes, metabolites, and various other molecules in blood, urine, or other body fluids have defined the existence of normal physiology or pathophysiology. Analyte-based medicine dominates medical textbooks and reinforces a practice of medicine that treats disease states instead of predicting or preventing them.
Precision health is an attempt to redirect medicine toward predicting and preventing disease (12). Knowing where a system is headed biologically puts the healthcare provider in a position to direct the system toward a healthy trajectory rather than waiting to make a diagnosis when the system is plagued by dysfunction and injury related to a pathologic course. Liquid biopsies provide the opportunity to know the genetic capacity of the fetus and placenta. The evolving patterns of the transcriptome can redefine maturational events in terms of gene expression, for example, gestational age dating, but also identify deviations from typical development, predicting preterm birth or preeclampsia early in pregnancy, thereby providing a window of opportunity for preventive and therapeutic interventions.
Another beneficial consequence of this kind of monitoring during pregnancy is the ability to redefine the taxonomy of prematurity, which currently is based primarily on gestational age. It follows from the latter situation that the risks for the various complications of prematurity are understood only in terms of the time of maturation. While the latter is important, it is also appreciated that not all babies of the same gestational age experience the same complications, and some babies of similar gestational ages experience no complications. Liquid biopsies show promise for providing insights into which babies are on biologic trajectories that put them at risk for the various disorders of the newborn, such as bronchopulmonary dysplasia, necrotizing enterocolitis, intraventricular hemorrhage, and retinopathy of prematurity, besides their maturational state at the time of birth. Preterm birth, preeclampsia, and many conditions of the newborn may be anticipated far in advance of their typical clinical presentation, and a better understanding of their pathophysiology may follow with new ways to intervene and prevent their occurrence altogether in most pregnancies prone to such outcomes. Liquid biopsies may provide the data for a whole new taxonomy of preterm birth, preeclampsia, and innumerable other pathologies presenting later in childhood or even later in life.
Placenta as a Window into Fetal Health
One of the advantages of measuring cfRNA in circulation during pregnancy is that cfRNA comes from the pregnant individual, fetus, and the placenta. The placenta is a hybrid of parental and fetal cells, and it clearly contributes to the normal physiologic changes of pregnancy in the individual and their fetus. Moreover, the successful functioning of the placenta is essential to fetal survival and well-being. The fetus is dependent on the placenta for gas exchange and the fluxes of various fuels for growth and development and the waste products of fetal metabolism, which change over time in preparation for extrauterine existence. Liquid biopsies can provide insights into placental function and maturation, and placental gene expression can contribute to estimates of gestational age or prediction of preterm birth or preeclampsia. Other applications may lead to a better understanding of fetal organ development (20, 22) or how drugs given to the pregnant individual may affect placental or fetal metabolism through altered gene expression of placental and fetal genes. Perhaps monitoring the gene expression patterns of the fetus and placenta would reassure providers as they consider the use of drugs in the pregnant individual who may have needs of their own during pregnancy. In any case, a window to placental health provided by liquid biopsies is also a window to fetal health.
From Correlation to Causation
Predictions are never perfect, but they should be actionable. The clinical utility of cfDNA to screen for aneuploidy has been proven. Looking toward other conditions, NIPT seems potentially useful, as is clear from preliminary studies. What is possible to learn from cfRNA, microbiome, and metabolite studies seems less clear, and not just because of some of the technical challenges with each of these modalities’ measurement and interpretation. Focusing on cfRNA as a case example, it has been established that cfRNA can serve as a biomarker for ensuing biologic events, but that is not the same as proving that a particular gene or gene pathway is causative of the predicted condition. Nonetheless, the promise of such measurements is that they are more than biomarkers; they suggest genes and gene pathways that indeed may be causative of the condition of concern. Therefore, they also suggest targets for molecular or immunologic therapies that might be preventive or curative. Thus, the source of cfRNA becomes relevant, and the relationship of cfRNA to tissue and cell type gene expression becomes important for understanding the pathogenesis of various disease states (see the sidebar titled The Immunomodulatory Effect of Circulating Macromolecules). Such studies are only beginning and will require advances in computation as well as the application of other technologies, such as single-cell CyTOF in tissues, so that the proximity of cells and their signaling over time can be understood in relation to what is observed in the circulation. This approach is most easily taken with immune and blood cells in circulation and in situ, and there are examples of this approach in cancer, neurodegeneration, and metabolic disease studies (127–129). Encouragingly, many changes in gene expression over time in pregnancy are related to, in fact, immune cells. The measurement of epigenomic and epitranscriptomic signatures may further connect cfDNA and cfRNA to its tissue and cell type of origin in pregnancy, as has been done for cfDNA in cancer (130, 131). To move from biomarker status and association to causation, we will need experiments to test the hypotheses generated by descriptive data. Some of this work can be conducted in human cells in vitro, especially when evaluating repurposed or novel drugs, but some well-designed animal experiments will also guide eventual clinical trials.
THE IMMUNOMODULATORY EFFECT OF CIRCULATING MACROMOLECULES.
It is well established that nucleic acids can interact with immune-related cell surface receptors and elicit an immune response (132). During pregnancy, the definition of self versus nonself shifts as the pregnant individual grows an entirely new organ, the placenta, to support the fetus. The interplay of fetal and parental immune cells at the placental boundary has been a subject of research interest for years, as reviewed thoroughly elsewhere (107). What remains unclear is whether and how circulating nucleic acids and macromolecules may directly interact with the immune system and how the placenta may modulate those interactions. For example, two recent studies highlight that antibodies from pregnant individuals contain unique and functionally distinct glycosylation patterns (133, 134). Rizzuto et al. (133) provided evidence that trophoblast-derived sialoglycoproteins suppress B cells to establish feto-maternal tolerance. Erickson et al. (134) separately described how antibodies are selectively modified during pregnancy and enhance protection in neonates against Listeria monocytogenes infection, an intracellular pathogen. These findings point to an underappreciated role for modifications like glycosylation in the interaction between circulating macromolecules and the immune systems during pregnancy. The extension of such findings to placentally derived circulating nucleic acids remains to be explored. We hypothesize that such modifications may also be present on cell-free DNA and RNA given recent findings that glycosylated RNA is present in cells and on their surface (135). These interactions may inform NIPT and extend its utility.
SUMMARY POINTS.
Noninvasive prenatal testing (NIPT) has transformed prenatal care over the past decade and significantly reduced the use of invasive diagnostic procedures like amniocentesis for genetic disorders.
Advances in NIPT provide new opportunities for precision medicine in prenatal care. Recent exciting advances include proof-of-principle approaches to screen for preterm birth and preeclampsia, both of which are prevalent obstetrical syndromes with major adverse outcomes.
Studies that examine the relationship of molecular measurements to obstetric dogma may help to redefine or refine disease taxonomy and add to the clinician’s toolbox.
NIPT provides a unique lens into the relationship between the pregnant individual and fetus and their health; however, this insight can be a double-edged sword, as an abnormal finding can often screen for but not definitively diagnose a health complication, leading to ethical concerns.
FUTURE ISSUES.
It remains a matter of debate whether cell-free DNA (cfDNA) results that suggest malignancy should even be reported back to the pregnant individual.
As gene therapy and gene editing becomes more routine, noninvasive prenatal diagnosis tests for Mendelian disorders will become more actionable; however, how such action is regulated is an open question.
Questions around cost and accessibility of NIPT advances such as whole-genome fetal analysis using cfDNA may shift as inexpensive, portable, long-read sequencing technologies become more prevalent and accurate.
ACKNOWLEDGMENTS
We apologize to all the investigators whose research could not be cited owing to space constraints. D.K.S. would like to acknowledge the Hess Research Fund and the Roberts Research Fund. M.N.M. would like to acknowledge the Stanford Bio-X Bowes Graduate Student Fellowship.
Footnotes
DISCLOSURE STATEMENT
S.R.Q. is a founder, consultant, and shareholder of Mirvie and receives royalties from Stanford University from patents on NIPT. M.N.M. is also a shareholder of Mirvie. M.N.M., G.M.S., D.K.S., and S.R.Q. are inventors on a patent application submitted by the Chan Zuckerberg Biohub and Stanford University that covers noninvasive early prediction of preeclampsia and monitoring maternal organ health over pregnancy (US Patent and Trademark Office application numbers 63/159,400, filed on March 10, 2021, and 63/276,467, filed on November 5, 2021). M.N.M. and S.R.Q. are inventors on a patent application (number 62/578,360) submitted by the Chan Zuckerberg Biohub that covers noninvasive estimates of gestational age, delivery, and preterm birth. D.W.B. has no disclosures.
LITERATURE CITED
- 1.Chetty S, Garabedian MJ, Norton ME. 2013. Uptake of noninvasive prenatal testing (NIPT) in women following positive aneuploidy screening. Prenat. Diagn. 33(6):542–46 [DOI] [PubMed] [Google Scholar]
- 2.Larion S, Warsof SL, Romary L, Mlynarczyk M, Peleg D, Abuhamad AZ. 2014. Uptake of noninvasive prenatal testing at a large academic referral center. Am. J. Obstet. Gynecol. 211(6):651.e1–651.e7 [DOI] [PubMed] [Google Scholar]
- 3.Chan YM, Leung WC, Chan WP, Leung TY, Cheng YKY, Sahota DS. 2015. Women’s uptake of non-invasive DNA testing following a high-risk screening test for trisomy 21 within a publicly funded healthcare system: findings from a retrospective review. Prenat. Diagn. 35(4):342–47 [DOI] [PubMed] [Google Scholar]
- 4.Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, et al. 2014. DNA sequencing versus standard prenatal aneuploidy screening. N. Engl. J. Med. 370(9):799–808 [DOI] [PubMed] [Google Scholar]
- 5.Petersen EE, Davis NL, Goodman D, Cox S, Mayes N, et al. 2019. Vital signs: pregnancy-related deaths, United States, 2011–2015, and strategies for prevention, 13 states, 2013–2017. Morb. Mortal. Wkly. Rep. 68(18):423–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L, Johnson HL, Cousens S, Perin J, Scott S, et al. 2012. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379(9832):2151–61 [DOI] [PubMed] [Google Scholar]
- 7.Inst. Med. 2007. Preterm Birth: Causes, Consequences, and Prevention. Washington, DC: Natl. Acad. Press [Google Scholar]
- 8.Lisonkova S, Joseph KS. 2013. Incidence of preeclampsia: risk factors and outcomes associated with early-versus late-onset disease. Am. J. Obstet. Gynecol. 209(6):544.e1–544.e12 [DOI] [PubMed] [Google Scholar]
- 9.Bianchi DW. 2021. Function follows form: gene expression and prenatal screening. Trends Mol. Med. 27(8):725–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray KJ, Hemberg M, Karumanchi SA. 2022. Cell-free RNA transcriptome and prediction of adverse pregnancy outcomes. Clin. Chem. 68(11):1358–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson LS, Stelzer IA, Tsai AS, Ghaemi MS, Han X, et al. 2020. Multiomic immune clockworks of pregnancy. Semin. Immunopathol. 42(4):397–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevenson DK, Wong RJ, Aghaeepour N, Maric I, Angst MS, et al. 2021. Towards personalized medicine in maternal and child health: integrating biologic and social determinants. Pediatr. Res. 89(2):252–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moufarrej MN, Wong RJ, Shaw GM, Stevenson DK, Quake SR. 2020. Investigating pregnancy and its complications using circulating cell-free RNA in women’s blood during gestation. Front. Pediatr. 8:605219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsao DS, Silas S, Landry BP, Itzep NP, Nguyen AB, et al. 2019. A novel high-throughput molecular counting method with single base-pair resolution enables accurate single-gene NIPT. Sci. Rep. 9:14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Li J, Saucier JB, Feng Y, Jiang Y, et al. 2019. Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat. Med. 25(3):439–47 [DOI] [PubMed] [Google Scholar]
- 16.Taglauer ES, Wilkins-Haug L, Bianchi DW. 2014. Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 35(Suppl.):S64–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maron JL, Johnson KL, Slonim D, Lai C-Q, Ramoni M, et al. 2007. Gene expression analysis in pregnant women and their infants identifies unique fetal biomarkers that circulate in maternal blood. J. Clin. Investig. 117(10):3007–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh W, Pan W, Gawad C, Fan HC, Kerchner GA, et al. 2014. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. PNAS 111(20):7361–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan W, Ngo TTM, Camunas-Soler J, Song C-X, Kowarsky M, et al. 2017. Simultaneously monitoring immune response and microbial infections during pregnancy through plasma cfRNA sequencing. Clin. Chem. 63(11):1695–704 [DOI] [PubMed] [Google Scholar]
- 20.Ngo TTM, Moufarrej MN, Rasmussen M-LH, Camunas-Soler J, Pan W, et al. 2018. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science 360(6393):1133–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munchel S, Rohrback S, Randise-Hinchliff C, Kinnings S, Deshmukh S, et al. 2020. Circulating transcripts in maternal blood reflect a molecular signature of early-onset preeclampsia. Sci. Transl. Med. 12(550):eaaz0131. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen M, Reddy M, Nolan R, Camunas-Soler J, Khodursky A, et al. 2022. RNA profiles reveal signatures of future health and disease in pregnancy. Nature 601(7893):422–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moufarrej MN, Vorperian SK, Wong RJ, Campos AA, Quaintance CC, et al. 2022. Early prediction of preeclampsia in pregnancy with cell-free RNA. Nature 602(7898):689–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelzer IA, Ghaemi MS, Han X, Ando K, Hédou JJ, et al. 2021. Integrated trajectories of the maternal metabolome, proteome, and immunome predict labor onset. Sci. Transl. Med. 13(592):eabd9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain CV, Kadam L, van Dijk M, Kohan-Ghadr H-R, Kilburn BA, et al. 2016. Fetal genome profiling at 5 weeks of gestation after noninvasive isolation of trophoblast cells from the endocervical canal. Sci. Transl. Med. 8(363):363re4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kølvraa S, Singh R, Normand EA, Qdaisat S, van den Veyver IB, et al. 2016. Genome-wide copy number analysis on DNA from fetal cells isolated from the blood of pregnant women. Prenat. Diagn. 36(12):1127–34 [DOI] [PubMed] [Google Scholar]
- 27.Breman AM, Chow JC, U’Ren L, Normand EA, Qdaisat S, et al. 2016. Evidence for feasibility of fetal trophoblastic cell-based noninvasive prenatal testing. Prenat. Diagn. 36(11):1009–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, Ruan L, Cao L, Lu G, Hong Q, et al. 2022. Noninvasive isolation of transcervical trophoblast cells for fetal identification. J. Obstet. Gynaecol. Res. 48(7):1613–20 [DOI] [PubMed] [Google Scholar]
- 29.Phipps EA, Thadhani R, Benzing T, Karumanchi SA. 2019. Pre-eclampsia: pathogenesis, novel diagnostics and therapies. Nat. Rev. Nephrol. 15(5):275–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton GJ, Redman CW, Roberts JM, Moffett A. 2019. Pre-eclampsia: pathophysiology and clinical implications. BMJ 366:l2381. [DOI] [PubMed] [Google Scholar]
- 31.Pertile MD, Halks-Miller M, Flowers N, Barbacioru C, Kinnings SL, et al. 2017. Rare autosomal trisomies, revealed by maternal plasma DNA sequencing, suggest increased risk of feto-placental disease. Sci. Transl. Med. 9(405):eaan1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steele MW, Breg WR. 1966. Chromosome analysis of human amniotic-fluid cells. Lancet 1(7434):383–85 [DOI] [PubMed] [Google Scholar]
- 33.Bianchi DW, Chiu RWK. 2018. Sequencing of circulating cell-free DNA during pregnancy. N. Engl. J. Med. 379(5):464–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gadsbøll K, Petersen OB, Gatinois V, Strange H, Jacobsson B, et al. 2020. Current use of noninvasive prenatal testing in Europe, Australia and the USA: a graphical presentation. Acta Obstet. Gynecol. Scand. 99(6):722–30 [DOI] [PubMed] [Google Scholar]
- 35.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. 2008. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. PNAS 105(42):16266–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiu RWK, Chan KCA, Gao Y, Lau VYM, Zheng W, et al. 2008. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. PNAS 105(51):20458–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordier A-G, Fuchs F, Tassin M, Saada J, Letourneau A, et al. 2016. Teaching invasive prenatal procedures: effectiveness of two simple simulators in training. Prenat. Diagn. 36(10):905–10 [DOI] [PubMed] [Google Scholar]
- 38.Yaron Y 2016. The implications of non-invasive prenatal testing failures: a review of an under-discussed phenomenon. Prenat. Diagn. 36(5):391–96 [DOI] [PubMed] [Google Scholar]
- 39.Artieri CG, Haverty C, Evans EA, Goldberg JD, Haque IS, et al. 2017. Noninvasive prenatal screening at low fetal fraction: comparing whole-genome sequencing and single-nucleotide polymorphism methods. [DOI] [PubMed] [Google Scholar]
- 40.Hui L, Bianchi DW. 2020. Fetal fraction and noninvasive prenatal testing: what clinicians need to know. Prenat. Diagn. 40(2):155–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Straver R, Sistermans EA, Reinders MJT. 2014. Introducing WISECONDOR for noninvasive prenatal diagnostics. Expert Rev. Mol. Diagn. 14(5):513–15 [DOI] [PubMed] [Google Scholar]
- 42.Raman L, Dheedene A, De Smet M, Van Dorpe J, Menten B. 2019. WisecondorX: improved copy number detection for routine shallow whole-genome sequencing. Nucleic Acids Res. 47(4):1605–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan HC, Quake SR. 2010. Sensitivity of noninvasive prenatal detection of fetal aneuploidy from maternal plasma using shotgun sequencing is limited only by counting statistics. PLOS ONE 5(5):e10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose NC, Kaimal AJ, Dugoff L, Norton ME, Am. Coll. Obstet. Gynecol. Comm. Pract. Bull. 2020. Screening for fetal chromosomal abnormalities: ACOG Practice Bulletin summary, number 226. Obstet. Gynecol. 136(4):859–67 [DOI] [PubMed] [Google Scholar]
- 45.Lutgendorf MA, Stoll KA. 2015. Why 99% may not be as good as you think it is: limitations of screening for rare diseases. J. Matern. Fetal. Neonatal. Med. 29(7):1187–89 [DOI] [PubMed] [Google Scholar]
- 46.Gil MM, Galeva S, Jani J, Konstantinidou L, Akolekar R, et al. 2019. Screening for trisomies by cfDNA testing of maternal blood in twin pregnancy: update of the fetal medicine foundation results and meta-analysis. Ultrasound Obstet. Gynecol. 53(6):734–42 [DOI] [PubMed] [Google Scholar]
- 47.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. 2017. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet. Gynecol. 50(3):302–14 [DOI] [PubMed] [Google Scholar]
- 48.Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, et al. 2015. Cell-free DNA analysis for noninvasive examination of trisomy. N. Engl. J. Med. 372(17):1589–97 [DOI] [PubMed] [Google Scholar]
- 49.Fajnzylber E, Hotz VJ, Sanders SG. 2010. An economic model of amniocentesis choice. Adv. Life Course Res. 15(1):11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin K, Iyengar S, Kalyan A, Lan C, Simon AL, et al. 2018. Clinical experience with a single-nucleotide polymorphism-based non-invasive prenatal test for five clinically significant microdeletions. Clin. Genet. 93(2):293–300 [DOI] [PubMed] [Google Scholar]
- 51.Dar P, Jacobsson B, Clifton R, Egbert M, Malone F, et al. 2022. Cell-free DNA screening for prenatal detection of 22q11.2 deletion syndrome. Am. J. Obstet. Gynecol. 227(1):79.e1–79.e11 [DOI] [PubMed] [Google Scholar]
- 52.Lefkowitz RB, Tynan JA, Liu T, Wu Y, Mazloom AR, et al. 2016. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. Am. J. Obstet. Gynecol. 215(2):227.e1–227.e16 [DOI] [PubMed] [Google Scholar]
- 53.Jacky L, Yurk D, Alvarado J, Leatham B, Schwartz J, et al. 2021. Virtual-partition digital PCR for high-precision chromosomal counting applications. Anal. Chem. 93(51):17020–29 [DOI] [PubMed] [Google Scholar]
- 54.Chandler NJ, Ahlfors H, Drury S, Mellis R, Hill M, et al. 2020. Noninvasive prenatal diagnosis for cystic fibrosis: implementation, uptake, outcome, and implications. Clin. Chem. 66(1):207–16 [DOI] [PubMed] [Google Scholar]
- 55.Camunas-Soler J, Lee H, Hudgins L, Hintz SR, Blumenfeld YJ, et al. 2018. Noninvasive prenatal diagnosis of single-gene disorders by use of droplet digital PCR. Clin. Chem. 64(2):336–45 [DOI] [PubMed] [Google Scholar]
- 56.Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR. 2012. Non-invasive prenatal measurement of the fetal genome. Nature 487(7407):320–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitzman JO, Snyder MW, Ventura M, Lewis AP, Qiu R, et al. 2012. Noninvasive whole-genome sequencing of a human fetus. Sci. Transl. Med. 4(137):137ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verhoef TI, Hill M, Drury S, Mason S, Jenkins L, et al. 2016. Non-invasive prenatal diagnosis (NIPD) for single gene disorders: cost analysis of NIPD and invasive testing pathways. Prenat. Diagn. 36(7):636–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jenkins LA, Deans ZC, Lewis C, Allen S. 2018. Delivering an accredited non-invasive prenatal diagnosis service for monogenic disorders and recommendations for best practice. Prenat. Diagn. 38(1):44–51 [DOI] [PubMed] [Google Scholar]
- 60.Wapner RJ, Norton ME. 2021. An introduction: prenatal screening, diagnosis, and treatment of single gene disorders. Clin. Obstet. Gynecol. 64(4):852–60 [DOI] [PubMed] [Google Scholar]
- 61.Chitty LS, Griffin DR, Meaney C, Barrett A, Khalil A, et al. 2011. New aids for the non-invasive prenatal diagnosis of achondroplasia: dysmorphic features, charts of fetal size and molecular confirmation using cell-free fetal DNA in maternal plasma. Ultrasound Obstet. Gynecol. 37(3):283–89 [DOI] [PubMed] [Google Scholar]
- 62.Chitty LS, Khalil A, Barrett AN, Pajkrt E, Griffin DR, Cole TJ. 2013. Safe, accurate, prenatal diagnosis of thanatophoric dysplasia using ultrasound and free fetal DNA. Prenat. Diagn. 33(5):416–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chitty LS, Mason S, Barrett AN, McKay F, Lench N, et al. 2015. Non-invasive prenatal diagnosis of achondroplasia and thanatophoric dysplasia: next-generation sequencing allows for a safer, more accurate, and comprehensive approach. Prenat. Diagn. 35(7):656–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanson B, Scotchman E, Chitty LS, Chandler NJ. 2022. Non-invasive prenatal diagnosis (NIPD): how analysis of cell-free DNA in maternal plasma has changed prenatal diagnosis for monogenic disorders. Clin. Sci. 136(22):1615–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu W, Koh W, Blumenfeld YJ, El-Sayed YY, Hudgins L, et al. 2014. Noninvasive prenatal diagnosis in a fetus at risk for methylmalonic acidemia. Genet. Med. 16(7):564–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parks M, Court S, Bowns B, Cleary S, Clokie S, et al. 2017. Non-invasive prenatal diagnosis of spinal muscular atrophy by relative haplotype dosage. Eur. J. Hum. Genet. 25(4):416–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lo YMD, Chan KCA, Sun H, Chen EZ, Jiang P, et al. 2010. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2(61):61ra91. [DOI] [PubMed] [Google Scholar]
- 68.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. 2010. Analysis of the size distributions of fetal and maternal cell-free DNA by paired-end sequencing. Clin. Chem. 56(8):1279–86 [DOI] [PubMed] [Google Scholar]
- 69.Yu SCY, Jiang P, Peng W, Cheng SH, Cheung YTT, et al. 2021. Single-molecule sequencing reveals a large population of long cell-free DNA molecules in maternal plasma. PNAS 118(50):e2114937118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zipursky A, Hull A, White FD, Israels LG. 1959. Fœtal erythrocytes in the maternal circulation. Lancet 273(7070):451–52 [DOI] [PubMed] [Google Scholar]
- 71.Walknowska J, Conte FA, Grumbach MM. 1969. Practical and theoretical implications of fetal-maternal lymphocyte transfer. Lancet 1(7606):1119–22 [DOI] [PubMed] [Google Scholar]
- 72.Herzenberg LA, Bianchi DW, Schröder J, Cann HM, Iverson GM. 1979. Fetal cells in the blood of pregnant women: detection and enrichment by fluorescence-activated cell sorting. PNAS 76(3):1453–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bianchi DW, Simpson JL, Jackson LG, Elias S, Holzgreve W, et al. 2002. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. Prenat. Diagn. 22(7):609–15 [DOI] [PubMed] [Google Scholar]
- 74.Huang L, Ma F, Chapman A, Lu S, Xie XS. 2015. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu. Rev. Genom. Hum. Genet. 16:79–102 [DOI] [PubMed] [Google Scholar]
- 75.Fu Y, Zhang F, Zhang X, Yin J, Du M, et al. 2019. High-throughput single-cell whole-genome amplification through centrifugal emulsification and EMDA. Commun. Biol. 2:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Biezuner T, Raz O, Amir S, Milo L, Adar R, et al. 2021. Comparison of seven single cell whole genome amplification commercial kits using targeted sequencing. Sci. Rep. 11:17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moser G, Drewlo S, Huppertz B, Armant DR. 2018. Trophoblast retrieval and isolation from the cervix: origins of cervical trophoblasts and their potential value for risk assessment of ongoing pregnancies. Hum. Reprod. Update 24(4):484–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bailey-Hytholt CM, Sayeed S, Kraus M, Joseph R, Shukla A, Tripathi A. 2019. A rapid method for label-free enrichment of rare trophoblast cells from cervical samples. Sci. Rep. 9:12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang C-E, Ma G-C, Jou H-J, Lin W-H, Lee D-J, et al. 2017. Noninvasive prenatal diagnosis of fetal aneuploidy by circulating fetal nucleated red blood cells and extravillous trophoblasts using silicon-based nanostructured microfluidics. Mol. Cytogenet. 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vossaert L, Wang Q, Salman R, McCombs AK, Patel V, et al. 2019. Validation studies for single circulating trophoblast genetic testing as a form of noninvasive prenatal diagnosis. Am. J. Hum. Genet. 105(6):1262–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scott F, Menezes M, Smet ME, Carey K, Hardy T, et al. 2022. Influence of fibroids on cell-free DNA screening accuracy. Ultrasound Obstet. Gynecol. 59(1):114–19 [DOI] [PubMed] [Google Scholar]
- 82.Bianchi DW. 2018. Cherchez la femme: Maternal incidental findings can explain discordant prenatal cell-free DNA sequencing results. Genet. Med. 20(9):910–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turriff AE, Annunziata CM, Bianchi DW. 2022. Prenatal DNA sequencing for fetal aneuploidy also detects maternal cancer: importance of timely workup and management in pregnant women. J. Clin. Oncol. 40(22):2398–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, et al. 2015. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 314(2):162–69 [DOI] [PubMed] [Google Scholar]
- 85.Dharajiya NG, Grosu DS, Farkas DH, McCullough RM, Almasri E, et al. 2018. Incidental detection of maternal neoplasia in noninvasive prenatal testing. Clin. Chem. 64(2):329–35 [DOI] [PubMed] [Google Scholar]
- 86.van der Meij KRM, Sistermans EA, Macville MVE, Stevens SJC, Bax CJ, et al. 2019. TRIDENT-2: national implementation of genome-wide non-invasive prenatal testing as a first-tier screening test in the Netherlands. Am. J. Hum. Genet. 105(6):1091–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ji X, Li J, Huang Y, Sung P-L, Yuan Y, et al. 2019. Identifying occult maternal malignancies from 1.93 million pregnant women undergoing noninvasive prenatal screening tests. Genet. Med. 21(10):2293–302 [DOI] [PubMed] [Google Scholar]
- 88.Lenaerts L, Brison N, Maggen C, Vancoillie L, Che H, et al. 2021. Comprehensive genome-wide analysis of routine non-invasive test data allows cancer prediction: a single-center retrospective analysis of over 85,000 pregnancies. eClinicalMedicine 35:100856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heesterbeek CJ, Aukema SM, Galjaard R-JH, Boon EMJ, Srebniak MI, et al. 2022. Noninvasive prenatal test results indicative of maternal malignancies: a nationwide genetic and clinical follow-up study. J. Clin. Oncol. 40(22):2426–35 [DOI] [PubMed] [Google Scholar]
- 90.Benn P, Plon SE, Bianchi DW. 2019. Current controversies in prenatal diagnosis 2: NIPT results suggesting maternal cancer should always be disclosed. Prenat. Diagn. 39(5):339–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Linthorst J, Welkers MRA, Sistermans EA. 2022. Clinically relevant DNA viruses in pregnancy. Prenat. Diagn. In press [DOI] [PubMed] [Google Scholar]
- 92.De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, et al. 2013. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155(5):1178–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, et al. 2015. Noninvasive monitoring of infection and rejection after lung transplantation. PNAS 112(43):13336–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kowarsky M, Camunas-Soler J, Kertesz M, De Vlaminck I, Koh W, et al. 2017. Numerous uncharacterized and highly divergent microbes which colonize humans are revealed by circulating cell-free DNA. PNAS 114(36):9623–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu J, Diaz JD, Goldstein SC, Patel RD, Varela JC, et al. 2021. Impact of next-generation sequencing cell-free pathogen DNA test on antimicrobial management in adults with hematological malignancies and transplant recipients with suspected infections. Transplant. Cell. Ther. 27(6):500.e1–500.e6 [DOI] [PubMed] [Google Scholar]
- 96.Shishido AA, Noe M, Saharia K, Luethy P. 2022. Clinical impact of a metagenomic microbial plasma cell-free DNA next-generation sequencing assay on treatment decisions: a single-center retrospective study. BMC Infect. Dis. 22:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Linthorst J, Baksi MMM, Welkers MRA, Sistermans EA. 2022. The cell-free DNA virome of 108,349 Dutch pregnant women. Prenat. Diagn. In press [DOI] [PubMed] [Google Scholar]
- 98.Chesnais V, Ott A, Chaplais E, Gabillard S, Pallares D, et al. 2018. Using massively parallel shotgun sequencing of maternal plasmatic cell-free DNA for cytomegalovirus DNA detection during pregnancy: a proof of concept study. Sci. Rep. 8:4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu S, Huang S, Chen F, Zhao L, Yuan Y, et al. 2018. Genomic analyses from non-invasive prenatal testing reveal genetic associations, patterns of viral infections, and Chinese population history. Cell 175(2):347–59.e14 [DOI] [PubMed] [Google Scholar]
- 100.Tong X, Yu X, Du Y, Su F, Liu Y, et al. 2022. Peripheral blood microbiome analysis via noninvasive prenatal testing reveals the complexity of circulating microbial cell-free DNA. Microbiol. Spectr. 10(3):e0041422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Burnham P, Gomez-Lopez N, Heyang M, Cheng AP, Lenz JS, et al. 2020. Separating the signal from the noise in metagenomic cell-free DNA sequencing. Microbiome 8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gaccioli F, Lager S, de Goffau MC, Sovio U, Dopierala J, et al. 2020. Fetal inheritance of chromosomally integrated human herpesvirus 6 predisposes the mother to pre-eclampsia. Nat. Microbiol. 5(7):901–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miura H, Kawamura Y, Ohye T, Hattori F, Kozawa K, et al. 2021. Inherited chromosomally integrated human herpesvirus 6 is a risk factor for spontaneous abortion. J. Infect. Dis. 223(10):1717–23 [DOI] [PubMed] [Google Scholar]
- 104.UNFPA (U.N. Popul. Fund), WHO (World Health Organ.), UNICEF (U.N. Child. Fund), World Bank Group, U.N. Popul. Div. 2019. Trends in Maternal Mortality 2000 to 2017: Estimates by WHO,UNIVEF, UNFPA, World Bank Group and the United Nations Population Division: Executive Summary. Geneva: WHO [Google Scholar]
- 105.Duley L 2009. The global impact of pre-eclampsia and eclampsia. Semin. Perinatol. 33(3):130–37 [DOI] [PubMed] [Google Scholar]
- 106.Jeyabalan A 2013. Epidemiology of preeclampsia: impact of obesity. Nutr. Rev. 71(Suppl. 1):S18–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moffett A, Shreeve N. 2022. Local immune recognition of trophoblast in early human pregnancy: controversies and questions. Nat. Rev. Immunol. 23:222–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberge S, Nicolaides K, Demers S, Hyett J, Chaillet N, Bujold E. 2017. The role of aspirin dose on the prevention of preeclampsia and fetal growth restriction: systematic review and meta-analysis. Am. J. Obstet. Gynecol. 216(2):110–20.e6 [DOI] [PubMed] [Google Scholar]
- 109.Marić I, Contrepois K, Moufarrej MN, Stelzer IA, Feyaerts D, et al. 2022. Early prediction and longitudinal modeling of preeclampsia from multiomics. Patterns 3(12):100655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith GCS. 2010. First-trimester determination of complications of late pregnancy. JAMA 303(6):561–62 [DOI] [PubMed] [Google Scholar]
- 111.Chappell LC, Cluver CA, Kingdom J, Tong S. 2021. Pre-eclampsia. Lancet 398(10297):341–54 [DOI] [PubMed] [Google Scholar]
- 112.Han X, Ghaemi MS, Ando K, Peterson LS, Ganio EA, et al. 2019. Differential dynamics of the maternal immune system in healthy pregnancy and preeclampsia. Front. Immunol. 10:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vorperian SK, Moufarrej MN, Tabula Sapiens Consort, Quake SR. 2022. Cell types of origin of the cell-free transcriptome. Nat. Biotechnol. 40(6):855–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Camunas-Soler J, Gee EPS, Reddy M, Mi JD, Thao M, et al. 2022. Predictive RNA profiles for early and very early spontaneous preterm birth. Am. J. Obstet. Gynecol. 227(1):72.e1–72.e16 [DOI] [PubMed] [Google Scholar]
- 115.Aghaeepour N, Ganio EA, Mcilwain D, Tsai AS, Tingle M, et al. 2017. An immune clock of human pregnancy. Sci. Immunol. 2(15):aan2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gudnadottir U, Debelius JW, Du J, Hugerth LW, Danielsson H, et al. 2022. The vaginal microbiome and the risk of preterm birth: a systematic review and network meta-analysis. Sci. Rep. 12:7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bayar E, Bennett PR, Chan D, Sykes L, MacIntyre DA. 2020. The pregnancy microbiome and preterm birth. Semin. Immunopathol. 42(4):487–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, et al. 2019. The vaginal microbiome and preterm birth. Nat. Med. 25(6):1012–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wylie KM, Wylie TN, Cahill AG, Macones GA, Tuuli MG, Stout MJ. 2018. The vaginal eukaryotic DNA virome and preterm birth. Am. J. Obstet. Gynecol. 219(2):189.e1–189.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Flaviani F, Hezelgrave NL, Kanno T, Prosdocimi EM, Chin-Smith E, et al. 2021. Cervicovaginal microbiota and metabolome predict preterm birth risk in an ethnically diverse cohort. JCI Insight 6(16):e149257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ghaemi MS, DiGiulio DB, Contrepois K, Callahan B, Ngo TTM, et al. 2019. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 35:95–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Larson MH, Pan W, Kim HJ, Mauntz RE, Stuart SM, et al. 2021. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat. Commun. 12:2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Johnston M, Warton C, Pertile MD, Taylor-Sands M, Delatycki MB, et al. 2022. Ethical issues associated with prenatal screening using non-invasive prenatal testing for sex chromosome aneuploidy. Prenat. Diagn. 43(2):226–34 [DOI] [PubMed] [Google Scholar]
- 124.Ravitsky V, Roy M-C, Haidar H, Henneman L, Marshall J, et al. 2021. The emergence and global spread of noninvasive prenatal testing. Annu. Rev. Genom. Hum. Genet. 22:309–38 [DOI] [PubMed] [Google Scholar]
- 125.Dondorp WJ, Page-Christiaens GCML, de Wert GMWR. 2016. Genomic futures of prenatal screening: ethical reflection. Clin. Genet. 89(5):531–38 [DOI] [PubMed] [Google Scholar]
- 126.Minear MA, Alessi S, Allyse M, Michie M, Chandrasekharan S. 2015. Noninvasive prenatal genetic testing: current and emerging ethical, legal, and social issues. Annu. Rev. Genom. Hum. Genet. 16:369–98 [DOI] [PubMed] [Google Scholar]
- 127.Ibarra A, Zhuang J, Zhao Y, Salathia NS, Huang V, et al. 2020. Non-invasive characterization of human bone marrow stimulation and reconstitution by cell-free messenger RNA sequencing. Nat. Commun. 11:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Toden S, Zhuang J, Acosta AD, Karns AP, Salathia NS, et al. 2020. Noninvasive characterization of Alzheimer’s disease by circulating, cell-free messenger RNA next-generation sequencing. Sci. Adv. 6(50):eabb1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chalasani N, Toden S, Sninsky JJ, Rava RP, Braun JV, et al. 2021. Noninvasive stratification of non-alcoholic fatty liver disease by whole transcriptome cell-free mRNA characterization. Am. J. Physiol. Gastrointest. Liver Physiol. 320(4):G439–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, et al. 2019. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570(7761):385–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu Y 2022. At the dawn: cell-free DNA fragmentomics and gene regulation. Br. J. Cancer 126(3):379–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schlee M, Hartmann G. 2016. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16(9):566–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rizzuto G, Brooks JF, Tuomivaara ST, McIntyre TI, Ma S, et al. 2022. Establishment of fetomaternal tolerance through glycan-mediated B cell suppression. Nature 603(7901):497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Erickson JJ, Archer-Hartmann S, Yarawsky AE, Miller JLC, Seveau S, et al. 2022. Pregnancy enables antibody protection against intracellular infection. Nature 606(7915):769–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, et al. 2021. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184(12):3109–24.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]


