Abstract
Actin plays many well-known roles in cells, and understanding any specific role is often confounded by the overlap of multiple actin-based structures in space and time. Here, we review our rapidly expanding understanding of actin in mitochondrial biology, where actin plays multiple distinct roles, exemplifying the versatility of actin and its functions in cell biology. One well-studied role of actin in mitochondrial biology is its role in mitochondrial fission, where actin polymerization from the endoplasmic reticulum through the formin INF2 has been shown to stimulate two distinct steps. However, roles for actin during other types of mitochondrial fission, dependent on the Arp2/3 complex, have also been described. In addition, actin performs functions independent of mitochondrial fission. During mitochondrial dysfunction, two distinct phases of Arp2/3 complex-mediated actin polymerization can be triggered. First, within 5 min of dysfunction, rapid actin assembly around mitochondria serves to suppress mitochondrial shape changes and to stimulate glycolysis. At a later time point, at more than 1 h post-dysfunction, a second round of actin polymerization prepares mitochondria for mitophagy. Finally, actin can both stimulate and inhibit mitochondrial motility depending on the context. These motility effects can either be through the polymerization of actin itself or through myosin-based processes, with myosin 19 being an important mitochondrially attached myosin. Overall, distinct actin structures assemble in response to diverse stimuli to affect specific changes to mitochondria.
Introduction
In cell biology, the interface between two fields can be a challenging place. Every field is complicated in itself, loaded with details at once esoteric and vital, without which one can have a general idea of how the system works but not a full understanding of the implications and possibilities. Crossing from one field to another requires learning a whole new set of terms, rules and mindset. To the cell, however, these boundaries do not exist, with one field blending seamlessly into the other.
This Review addresses the developing links between mitochondria and the actin cytoskeleton. Since the first evidence for functional interactions in mammals almost 20 years ago1, this association has expanded to include multiple distinct contexts and effects (Table 1), with the full implications of these interactions still unclear. Both fields are well developed, with detailed understanding of biophysics, biochemistry and cell biology associated with both mitochondria and actin cytoskeleton. For actin, form often dictates function. In other words, organization of the actin filaments into specific higher-order structures is necessary to provide motile or resistive force for other cellular structures. Actin polymerization is not an end in itself but a means to an end by assisting other processes. By contrast, the mitochondrion is a fundamental hub in eukaryotic biology, and its roles in both energy generation and homeotic signalling require extensive communication with the rest of the cell2.
Table 1 ∣.
Roles for actin in mitochondrial biology
| Mitochondrial fission | Mitochondrial damage response | Mitochondrial motility | ||||||
|---|---|---|---|---|---|---|---|---|
| Process | CIA | Actin waves | ADA | PDA | Actin-based tethering | Myosin-based tethering | Actin-based motility (comet tail) | Myosin-based motility |
| Stimulus | Cytoplasmic calcium increase | Unknown | Mitochondrial ATP decrease | Parkin recruitment or induction of mitophagy | Increased glucose | Mitosis, mitocytosis, neuronal mitochondrial transport | Mitosis, mtDNA damage | Cell growth (budding yeast), glucose starvation (mammals) |
| Actin nucleators | INF2 formin (activator – calmodulin) | Arp2/3 complex (activator unknown) | Arp2/3 complex (activator – WAVE), FMNL formins (activator – CDC42) | Arp2/3 complex (activators – WASP, N-WASP) | Unknown | Unknown | Arp2/3 complex (activator unknown) | Formins (budding yeast), unknown (mammals) |
| Other actin-binding proteins | SPIRE1C, myosin II, fascin, myosin 19 | Unknown | Unknown | Myosin VI | FHL2 | Myosin 19, myosin V, myosin VI | Unknown | Myosin V (budding yeast), myosin 19 (mammals) |
| Effects | Increased mitochondrial calcium, DRP1 recruitment, mitochondrial fission | DRP1 recruitment, mitochondrial fission | Increased glycolysis, inhibits mitochondrial dynamics | Inhibits fusion, disperses mitochondrial aggregates, links to autophagy factors | Inhibits microtubule-based motility | Inhibits motility, supports balanced mitotic inheritance during cell division | Randomizes mitochondrial inheritance | Movement to bud (myosin V), movement in filopodia (myosin 19) |
Additional roles for actin are discussed in this Review, but we focus here on the best-understood examples. ADA, acute damage-induced actin; CIA, calcium-induced actin; FHL2, four and a half LIM domains protein 2; mtDNA, mitochondrial DNA; PDA, prolonged damage-induced actin.
We first provide background on both actin and mitochondria to emphasize key elements pertaining to their interactions. For more details on any of these elements, we reference relevant reviews and primary literature.
The actin cytoskeleton
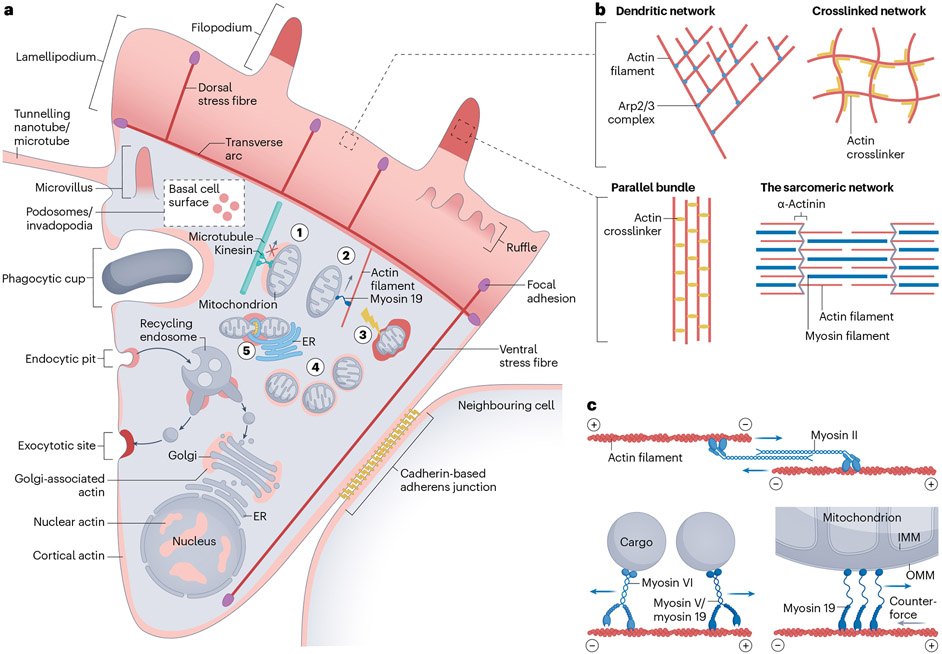
A key feature of actin is its near-ubiquitous cellular presence. Actin plays roles on most cellular membranes as well as in the bulk cytoplasm and in the nucleus (Fig. 1a). The specific function of actin varies, from providing force through its polymerization, to acting as a dynamic scaffold to maintain cellular structure, to serving as a surface for myosin motors3. To carry out these multiple functions, actin filaments can be organized in multiple ways, including single filaments, crosslinked filament arrays, filament bundles, dendritic networks and sarcomeric contractile networks (Fig. 1b). Actin monomers can also play roles without polymerizing, such as in transcriptional regulation4 and as components of chromatin remodelling complexes5. An important question when investigating a new role for actin is, which function is actin providing?
Fig. 1 ∣. Actin-based structures.
a, A hypothetical mammalian cell, migrating upwards and remaining in contact with another cell on the right. Cellular structures or processes known to use actin are labelled, with actin indicated by red shading. Actin-based functions discussed in this Review are: (1) inhibition of microtubule-mediated mitochondrial translocation, caused either by actin polymerization or by myosin-based tethering (not shown here); (2) myosin 19-based mitochondrial motility along an actin filament; (3) actin polymerization around a damaged mitochondrion, inducing several downstream responses (damage indicated by a lightning bolt); (4) cycling actin polymerization around several mitochondria; and (5) actin polymerization during endoplasmic reticulum (ER)-associated mitochondrial fission. b, Varying architecture of actin networks. Dendritic networks are mediated by the Arp2/3 complex (blue) and parallel-bundled actin filaments can be mediated by fascin, plastins or high concentrations of α-actinin or filamin (yellow). Crosslinked networks can be mediated by α-actinin or filamin (yellow). Non-muscle sarcomeric networks are antiparallel actin structures bridged by myosin II filaments (blue) with actin barbed ends at α-actinin-enriched regions (grey). Sarcomeric structures in non-muscle cells (for example, ventral stress fibres, cytokinetic ring, components of cortical actin) consist of smaller oligomers of myosin II and are generally less organized than muscle sarcomeres. c, Roles of myosins. Three types of myosin-based force are discussed in this Review: contractile force on antiparallel actin filaments (myosin II), translocation of cargo towards the actin filament barbed (“+”) end (myosin V, 19) or pointed (“−”) end (myosin VI), and static force, which can serve to resist a counter-force or to maintain the mitochondrion in a certain location (myosin 19). IMM, inner mitochondrial membrane; OMM, outer mitochondrial membrane.
Another feature of actin is its dynamics. Extensive networks of actin filaments can assemble in seconds, and disassemble equally quickly. Examples of this transience include actin at endocytic pits6 and in leading-edge lamellipodia7. To a large extent, all cellular actin filaments are built from a common actin monomer pool, and the rapid polymerization dynamics means that an actin monomer in one filament can find itself in a completely different filament, at the other end of the cell for a completely different purpose, 30 s later.
Finally, cellular actin-based structures can coexist in close proximity, an example being the overlapping localization of lamellipodia, filopodia, and focal adhesions or stress fibres in many cells3. These co-existing structures can cooperate for a common purpose or function for quasi-independent purposes. It is easy to confuse one structure for another.
To carry out its multiple functions, actin is aided by a myriad of binding proteins8. We focus on the proteins involved in mitochondrially associated actin and discussed in this Review. Box 1 provides more detail on actin-binding protein classes in general.
Box 1 Quick overview of actin dynamics and organization.
Actin and its polymerization
Actin is a 43-kDa monomeric protein that polymerizes into two-stranded helical filaments with two distinct ends: the barbed end and the pointed end (Supplementary Fig. 1). The barbed end is almost always the growing end in cells8,10. The high cellular concentration of actin (50–200 μM)73,101 makes it readily available for polymerization, although monomer-binding proteins like profilin and thymosin act as buffers to prevent spurious polymerization.
Actin is an ATPase, but actin monomers display essentially no ATP hydrolysis. Polymerization activates ATP hydrolysis, but this rate is still slow compared with elongation so a cap of ATP-actin exists towards the barbed end. Release of the phosphate product of hydrolysis is even slower so the central region of filaments is often rich in ADP–phosphate-bound actin. Phosphate release from actin triggers two changes that favour depolymerization: increased actin off-rate from filament ends and increased affinity for cofilin. Cofilin-mediated severing increases the number of ends, allowing increased depolymerization.
A general function of actin is to generate force, which it does in two ways: (1) by barbed end growth to push a cargo forward or (2) as the substrate for myosins, which provide a pulling force. Actin-binding proteins control the biological functions of actin and we divide these proteins into three classes: actin dynamics proteins, actin-organizing proteins and myosins. Our discussion focuses on mammals. An important point is that actin-binding proteins rarely act alone and generally work together to create or disassemble a particular actin-based structure.
Proteins controlling actin dynamics
A handful of proteins are used repeatedly to control actin polymerization and depolymerization so that specific structures assemble precisely when and where needed3,8-10 (Supplementary Figs. 1 and 2). Nucleation factors initiate new filaments from the monomer pool and include five classes: the Arp2/3 complex, formins (15 in mammals), tandem WH2 motif-containing proteins, leiomodins, and adenomatous polyposis coli (APC) protein. Several proteins can activate Arp2/3 complex, including WASP, N-WASP, WAVE, WASH, WHAMM, JMY and Spin90–DIP–WISH9. Capping proteins block barbed end growth, terminating polymerization189. Elongation factors counteract capping proteins, allowing continued barbed end growth, and include formins (subsequent to nucleation) and Ena–VASP proteins. The interaction between formins and capping proteins is more intricate than simple competitive binding of the barbed end190,191, but the net result is that barbed end elongation can continue. Tropomodulins cap pointed ends. Thymosin is a monomer-binding protein that prevents all dynamics (nucleation, barbed end and pointed end elongation). Profilin is a monomer-binding protein that allows barbed end (but not pointed end) elongation and can work with formins in this function8. Profilin also accelerates nucleotide exchange on actin, allowing rapid re-charging of depolymerized monomers with ATP. Nucleotide exchange can also be accelerated by cyclase-associated proteins. Cofilin severs aged actin filaments (ADP-bound regions generally near the pointed end), which is often important for filament depolymerization or turnover but also can be used for new polymerization in some circumstances192. A number of other proteins, including cyclase-associated proteins193,194, AIP1 (ref. 195) and coronins195,196, work with cofilin to accelerate actin turnover. In addition, cofilin and a related protein called GMF can accelerate de-branching of Arp2/3 complex-mediated branches9. Some coronin proteins can also de-branch197. Twinfilin accelerates depolymerization in a cofilin-independent manner198,199.
Proteins controlling actin organization
The Arp2/3 complex, in addition to its nucleation activity, automatically assembles dendritic networks through its ability to bind both pointed ends and filament sides. Arp2/3 complex-assembled networks can be extensive, such as lamellipodia200, or more limited such as around endosomes160. Cortactin stabilizes Arp2/3 complex-mediated branches. Crosslinking and bundling proteins bind two actin filaments to create either networks or bundles. α-Actinin and filamin favour networks (but can bundle at higher concentrations), while fascin creates parallel bundles. Some formins (for example, FMNL formins) and capping proteins (for example, villin) also display actin-bundling activity. ERM proteins and several I-BAR proteins mediate interactions between filaments and the plasma membrane201. Tropomyosins bind along the filament and modulate filament stability and/or interaction with myosins or other proteins202. Tropomodulins bind tightly to tropomyosin-bound filaments, cap pointed ends, and protect filaments from cofilin binding or severing but can also have additional functions203. While tropomyosin and tropomodulin function is well studied in skeletal muscle, the cellular functions of the multiple tropomyosins and tropomodulins in non-muscle cells are less clear202,203.
Myosins
The wide variety of myosin motors serves three general purposes: contraction, translocation, and anchoring or tethering. Myosin I, myosin II, myosin V, myosin VI and myosin 19 are barbed end-directed motors, while myosin VI is a pointed end-directed motor. Myosin II assembles into bipolar filaments that pull actin filaments in opposite directions, creating contractile force114. However, studies in fission yeast and mammalian cells suggest that myosin II can also assume organizations that are not bipolar109-111. Myosins can translocate cargo along actin filaments, with an example from this Review being myosin 19. Nevertheless, in some situations, these myosins can also be used to restrict motility, with myosin 19 again being an example in this Review.
A key step in actin-based structure assembly is the initial nucleation of new filaments, which is controlled through nucleation factors. Major actin nucleation factors are the Arp2/3 complex, formin proteins and tandem WH2-containing proteins9. The Arp2/3 complex nucleates filaments in a distinct branched pattern, which often elaborates into a dendritic network (Fig. 1b). In this Review, we will discuss four distinct Arp2/3 complex-mediated processes influencing mitochondria. Formins nucleate actin through a fundamentally different mechanism. After nucleation, formins remain on the elongating filament end and allow prolonged filament growth8 (Box 1). There are multiple formins (15 genes) in mammals. Two formin classes important to this Review are INF2 and FMNL formins. Tandem WH2-containing proteins nucleate actin filaments through a third mechanism10 (Box 1). The member associated with mitochondria and relevant to this Review, SPIRE1, can directly interact with several formins and works with these to mediate actin polymerization11 (Box 1). A splice variant of SPIRE1, called SPIRE1c, is tightly bound to mitochondria.
A myriad of other actin-binding proteins mediate the assembly of higher-order actin structures suitable for a specific function (fascin and filamin in this Review) or sever filaments either for depolymerization or increased elongation (cofilin in this Review) (Box 1). Finally, myosins are important actin-binding proteins; the specific myosin motors mentioned herein serve three distinct purposes (Fig. 1b): to generate contractile force (myosin II), to move mitochondria (myosin V and myosin 19) and to restrict mitochondrial motility (myosin VI and myosin 19).
Mitochondrial structure, dynamics and heterogeneity
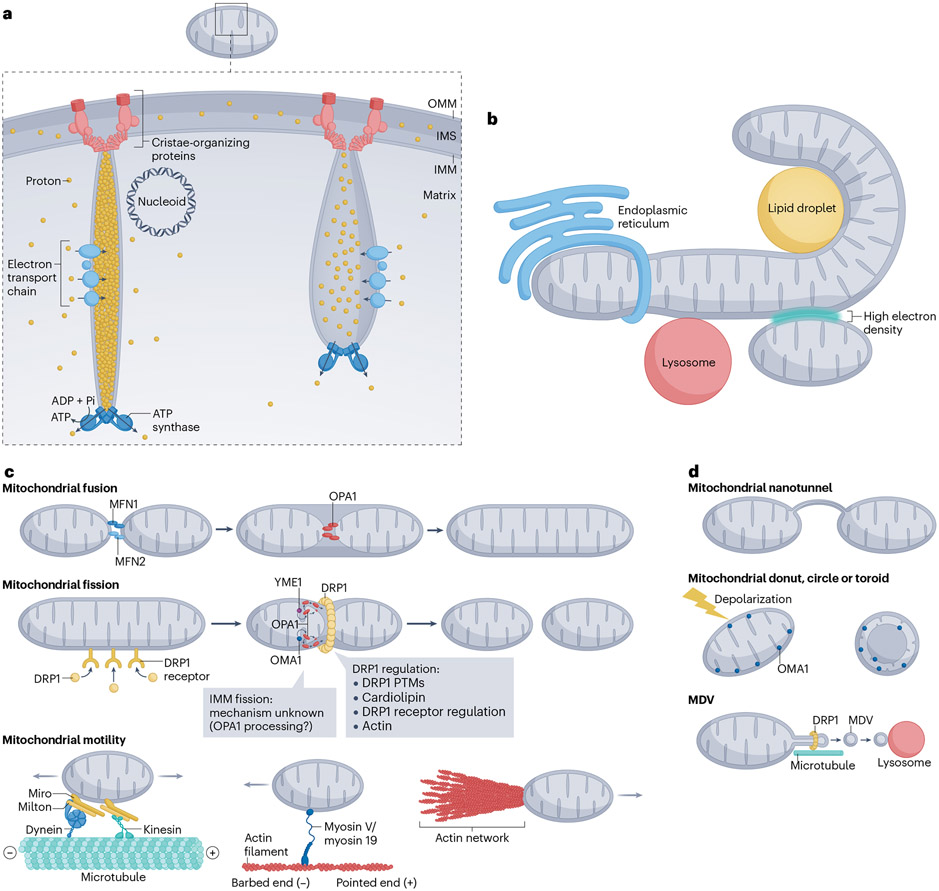
Mitochondria are double-membrane organelles, with an outer mitochondrial membrane (OMM), which is quite porous to small molecules, and an inner mitochondrial membrane (IMM) that is a much tighter barrier (Fig. 2a). These membranes separate two aqueous compartments: the matrix within the IMM and the inter-membrane space between the IMM and OMM. A key mitochondrial feature is its circular genome in the matrix, which is a remnant of its bacterial origins but has been highly adapted in the ensuing billion years12. In humans, the 16.6-kilobase circular genome is compacted into a nearly spherical structure, ~100 nm in diameter, called a nucleoid13. Most mitochondria contain multiple nucleoids, and a cultured cell typically contains several hundred nucleoids14,15.
Fig. 2 ∣. Mitochondrial structure and dynamics.
a, Mitochondrial structure. The double-membrane structure (outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM)) results in two aqueous compartments: the inter-membrane space (IMS) and the matrix. The IMM is segregated into the cristae membrane and inner-boundary membrane. Cristae junction proteins (light red) maintain a tight opening at the cristae base and interact with proteins mediating OMM-IMM tethering (dark red). The four complexes of the electron transport chain (light blue) enrich on the side of the cristae, whereas ATP synthase (dark blue) enriches at the cristae tip. A nucleoid, containing compacted mitochondrial DNA, might attach to the cristae side. Shown here are two cristae, one with a tight cristae junction and high proton gradient (left), and the other with a more open cristae junction and lower proton gradient (right). b, Mitochondrial contacts with the endoplasmic reticulum, lipid droplets, lysosomes and other mitochondria. c, Core components of mitochondrial fusion (MFN1, MFN2, OPA1), fission (DRP1, DRP1 receptors) and motility (microtubule based by kinesins or dynein, myosin based by myosin V or myosin 19, actin network based). Hypothetical mechanism of IMM fission, mediated through cleaved OPA1 (red) generated by either the YME1 (purple) or OMA1 (blue) protease are also shown. Dynactin complex for dynein motility is not shown for simplicity. Actin network for polymerization-based motility is not drawn with a specific architecture, which is undefined at this point. d, Other forms of mitochondrial dynamics. Top: nanotunnels between two mitochondria, with both OMM and IMM in a nanotunnel. Middle: mitochondrial IMM reorganization into toroids (also called donuts or circles), without OMM fission. This rearrangement occurs upon acute mitochondrial depolarization and is dependent on OMA1. Bottom: mitochondrial-derived vesicle (MDV) formation through membrane tube extrusion (microtubule-dependent)and DRP1-mediated constriction. PTMs, post-translational modifications.
A major mitochondrial function is ATP production through oxidative phosphorylation, in which fuels are oxidized in the matrix and the resulting electrons are used by the electron transport chain (ETC) to move protons across the IMM, creating a proton gradient that drives ATP synthase. The ETC and ATP synthase are enriched in tube-like cristae of the IMM, which are segregated from the non-cristae IMM by cristae junctions (Fig. 2a). In mitochondria with an active ETC, the matrix pH is >8 while cristae lumen pH can be <7 (ref. 16). The proton gradients of neighbouring cristae can be semi-autonomous17, suggesting the importance of the limited cristae volume (<10 nm cristae diameter) and cristae junctions in maintaining this difference.
Mitochondria serve many purposes besides ATP production such as biomolecule synthesis, redox balance and calcium homeostasis2. In addition, mitochondria are important regulators of cellular homeostasis, with dissipation of the mitochondrial proton gradient (also known as mitochondrial depolarization) being an easily monitored alarm for cellular dysfunction. Mitochondrial depolarization triggers disposal of dysfunctional mitochondria through mitophagy, and defects in mitophagy lead to a wide variety of diseases18. In addition, the release of factors like cytochrome c, SMAC and OMI from the inter-membrane space can trigger apoptosis19.
Mitochondria communicate extensively with other cellular compartments. An important communication mechanism is the assembly of close contacts with other organelles (Fig. 2b). One important contact is between mitochondria and the endoplasmic reticulum (ER), referred to as ER-mitochondria contact (ERMC) sites20. Many proteins have been identified or implied as mediating ERMC, which might reflect the variety of ERMC functions. One ERMC function is the facilitation of calcium transfer from ER to mitochondria21. Mitochondria also associate with lipid droplets, lysosomes and Golgi-derived vesicles for a variety of purposes22-24. Finally, mitochondria can make close connections with each other, called inter-mitochondrial junctions, which are especially prominent in cardiomyocytes and in oxidative phosphorylation-rich skeletal muscle25,26.
Mitochondrial fusion, fission and motility
Contrary to their textbook depictions as ‘pill-like’, mitochondria vary significantly in length12, which is dynamically controlled by mitochondrial fission and fusion in response to changing cell state or metabolic needs27. In addition, mitochondrial motility enables proper cellular distribution. These three processes (fission, fusion, motility) are commonly referred to as mitochondrial dynamics, and defects in dynamics link to several pathologies28.
Mitochondrial fusion is mediated by two sets of dynamin GTPases: mitofusins (MFN1 and MFN2) on the OMM and OPA11 on the IMM (Fig. 2c). Fusion requires a proton gradient across the IMM (in other words, healthy mitochondria)29,30, although fusion in which only one of the mitochondria is polarized can occur31. While many mechanistic issues concerning mitochondrial fusion are unresolved, the process is not known to be associated with actin and is not a focus of this Review. We refer readers to excellent recent publications27,28,32.
Mitochondrial fission is driven by the dynamin GTPase DRP1 (ref. 33), a cytoplasmic protein that is recruited to the OMM, where it oligomerizes into a constricting ring (Fig. 2c). Multiple OMM proteins act as DRP1 receptors, including MFF, MID49 and MID51. Another protein, FIS1, is a DRP1 receptor in budding yeast but serves other roles in mammals23. Two events often precede DRP1 recruitment: replication of the mitochondrial genome near the fission site34, and ERMC assembly at the fission site35. ERMC assembly is associated with pre-constriction of the mitochondrion prior to DRP1-mediated constriction35,36. Intriguingly, ER contact with endosomes influences the ability of ER to stimulate mitochondrial fission37. Contacts with lysosomes can also stimulate fission38. Finally, additional steps are likely required after DRP1. Another dynamin, dynamin 2, has been proposed to play such a role39 but this function is not universally accepted40. An alternative is that contact with Golgi-derived vesicles stimulates a final fission step downstream of DRP1 (ref. 24).
Mitochondrial fusion and fission can occur in quick succession, in a process called ‘kiss-and-run’, resulting in rapid exchange of mitochondrial components41,42. Kiss-and-run may be a result of machineries for both processes accumulating at ERMCs31.
Mitochondrial motility can be based either on microtubules or actin (Fig. 2c). In budding yeast, actin-based motility through type V myosins mediates many aspects of mitochondrial translocation, although details are still debated43-45. In mammals, microtubule-based kinesin and dynein motors mediate most long-range mitochondrial motility46. However, actin-based mitochondrial motility is emerging in several contexts, both through a mitochondrially bound myosin (myosin 19) and through actin polymerization47-49. Actin and myosins are also used to oppose motility in specific circumstances.
Mitochondrial dynamics are integral to cellular function in several ways. During cell division, mitochondrial fission and motility are important for proper distribution to daughter cells49,50. Mitochondrial motility is also required for appropriate mitochondrial distribution in interphase cells, particularly in highly polarized and energy-demanding cells such as neurons51. Mitochondrial fission and fusion are also intimately linked to mitochondrial homeostasis27. Depolarized regions of mitochondria can be separated from polarized regions by fission, with the depolarized daughter mitochondrion having a low probability of re-fusion and a high probability of mitophagy41. By contrast, nutrient depletion can induce increased mitochondrial length, inhibiting mitophagy52. Mutations in key mitochondrial dynamics genes, such as DRP1, MFN2 and OPA1, lead to several human pathologies, particularly neurodegenerative diseases27. Mutations to INF2 also have disease links, as discussed later.
Mitochondrial dynamics are not confined to fission, fusion and motility, with other dynamic processes including protrusion of nanotunnels between mitochondria53,54, mitochondrial branching52 and cristae remodelling (Fig. 2d). A key player in cristae remodelling is OPA1 which, in addition to its role in mitochondrial fusion, maintains cristae junctions55,56. Finally, small vesicles can bud from mitochondria, termed mitochondrial-derived vesicles (MDVs)57 or mitochondrial-derived compartments58. MDV assembly is a DRP1-dependent process, thus related to mitochondrial fission58,59.
Mitochondrial heterogeneity
One important mitochondrial feature is their heterogeneity at multiple levels: within an individual mitochondrion in changing cellular conditions, between mitochondrial populations within a single cell, and between mitochondria in different cell types. Mitochondria can vary greatly in fuel preference, such as the varying ability of muscle mitochondria to catabolize fatty acids60. However, mitochondrial function spreads far beyond ATP production. For example, mitochondria are key centres of steroid hormone biosynthesis in specific cells61, brown adipocytes use uncoupled mitochondrial metabolism for heat generation62 and hepatocyte mitochondria perform ketogenesis63. In addition, mitochondria are important biosynthetic centres and, in cancer cells, which predominantly use glycolysis for ATP production, the biosynthetic function of mitochondria appears to be dominant64. Finally, mitochondria can vary functionally with age in primary human immune cells65. Given these functional differences, it is not surprising that mitochondria vary significantly in protein composition across multiple mouse tissues66, in neighbouring cells in the brain67 or sub-cellularly68.
Morphologically, mitochondria vary in length from fragmented to highly elongated. While there is some evidence that mitochondrial length might correspond to metabolic state, it might be prudent not to take this correlation as a general rule27,69-71. A second morphological variation is mitochondrial ‘networking’, through branches between adjacent mitochondria52,72. Finally, mitochondria can vary significantly in diameter. Our laboratory has observed location-dependent mitochondrial diameter variation in U2OS human osteosarcoma cells73. A more dramatic example is within axons of cultured neurons, in which mitochondria narrow to diameters of 20 nm in places74, which is barely enough space for the OMM and IMM bilayers alone.
One experimental issue that may lead to varying mitochondrial characteristics is media composition in cell culture. Differences in fuel source can lead to dramatic differences in mitochondrial morphology75. Important parameters in this respect are glucose, pyruvate and glutamine. Two commonly used media are hyperglycaemic DMEM (25 mM) and RPMI1640 (11 mM), allowing significant ATP production through glycolysis and potentially lessening the ATP production role of mitochondria. By contrast, Leibovitz L-15 medium contains galactose instead of glucose, making mitochondria essential for ATP production.
The features above are clear examples of mitochondrial heterogeneity that likely manifests in more subtle ways in many cellular situations. Given these examples, intracellular mitochondrial heterogeneity should be considered the rule, not the exception, and this heterogeneity may come with differing roles for actin.
Actin and mitochondrial fission
A role for actin in mitochondrial fission was first suggested almost 20 years ago1, with molecular players starting to emerge about 10 years after. Actin is now associated with mitochondrial fission in several contexts as will be discussed in this section. An important point is that actin might not play the same mechanistic role in all cases. Before addressing these processes, we raise several questions concerning fission in general, the answers for which relate to actin.
Outstanding questions in mitochondrial fission
In terms of basic fission mechanisms, there are numerous knowledge gaps. Of relevance here, what is the mechanism by which the IMM undergoes fission? As opposed to fusion, there is no IMM dynamin linked to fission. IMM fission might accompany the pre-constriction that occurs prior to DRP1 recruitment and corresponds with ER-mitochondria interaction35,36. There is evidence for actin involvement in pre-constriction76.
Another question is: how many varieties of fission exist? DRP1 recruitment is regulated by many factors, including multiple DRP1 post-translational modifications, MFF phosphorylation, OMM lipid composition, mechanical forces and actin33,77. A recent study shows that two distinct DRP1-dependent mitochondrial fission pathways exist in the same cell: one for healthy mitochondria and another for damaged mitochondria78. DRP1-independent mitochondrial fission can also occur79,80. We postulate that there are multiple routes to mitochondrial fission and that actin participates in a subset of these routes.
On a broader scale, are all the events currently referred to as fission really fission events? For example, cells treated with uncouplers, such as carbonyl cyanide-m-chlorophenylhydrazone (CCCP) and carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), are commonly referred to as undergoing mitochondrial fragmentation, in which the entire mitochondrial population abruptly converts from elongated to punctate structures. While most studies observe this fragmentation after prolonged treatment29,81, some studies report fragmentation within minutes in an actin-dependent manner82,83. However, work from multiple groups suggests that the rapid mitochondrial shape changes occurring after depolarization are not due to fission but to a reorganization of the IMM, with the resulting mitochondria referred to as donut-shaped, toroidal or circular1,84-88. The OMM remains intact during toroid formation, while IMM reorganizes extensively in a process requiring the IMM protease OMA1 (refs. 86-88) (Fig. 2d). Importantly, toroid formation does not require actin1 and is, in fact, inhibited by it87,88. To be clear, damaged mitochondria can clearly undergo fission, which facilitates their disposal by mitophagy41,78; however, the fission event does not necessarily occur early after depolarization. There is also evidence that mitochondrial fission occurs later, concurrent with autophagosome assembly and independent of DRP1, in specific types of mitophagy79,89. We discuss damage-induced actin responses later in the Review.
CIA and mitochondrial fission
In the most well-described mechanism of actin-assisted mitochondrial fission, an ER-bound splice variant of the formin INF2 nucleates a network of actin filaments, some of which interact with mitochondria76,90-92. We refer to this actin polymerization as calcium-induced actin (CIA) because increased cytoplasmic calcium activates INF2 (refs. 76,91,93,94). A second actin nucleation factor, a mitochondrially bound splice variant of SPIRE1 (SPIRE1C), might also be involved in CIA95. Two myosins, myosin II73,96,97 and myosin 19 (ref. 98), as well as the actin-bundling protein fascin99, have been linked to CIA-mediated mitochondrial fission. Notably, the Arp2/3 complex is not required for CIA87, as opposed to its central importance in actin polymerization associated with mitochondrial damage (discussed later).
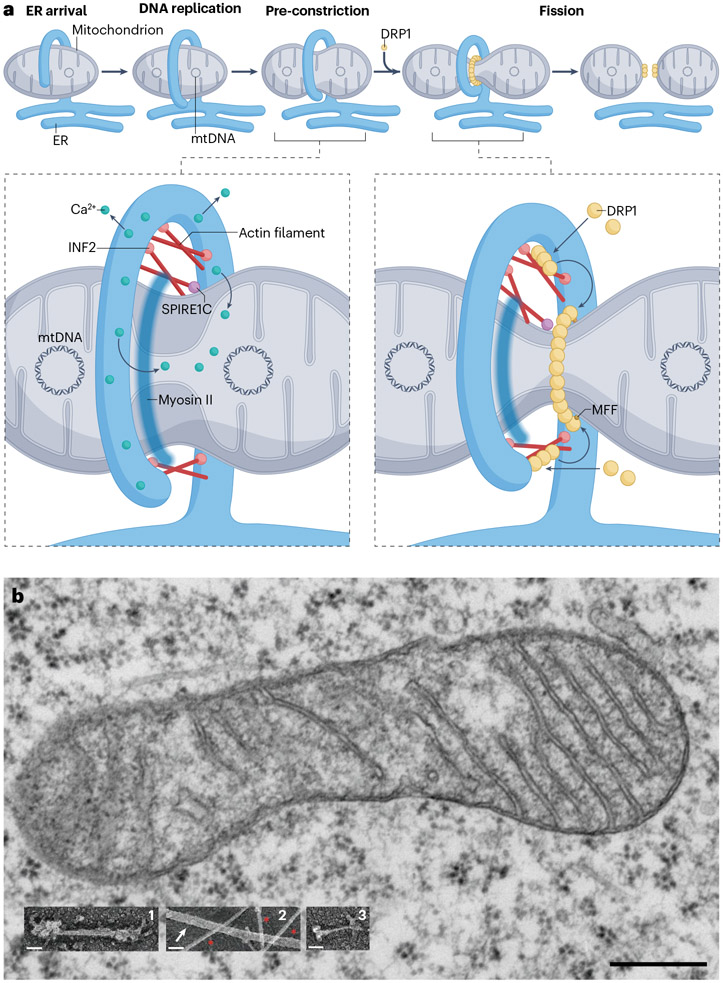
CIA stimulates two steps in mitochondrial fission (Fig. 3a). First, CIA increases ER–mitochondria calcium transfer by enhancing ERMC76. The increased mitochondrial calcium causes IMM constriction in a DRP1-independent manner76,100, perhaps explaining why pre-constriction precedes recruitment of DRP1. Second, CIA stimulates mitochondrial DRP1 recruitment90. DRP1 binds actin filaments directly91,101 and actin synergizes with MFF in DRP1 activation102. In addition, some CIA-stimulated DRP1 oligomerization may initiate on the ER103. A current model is that CIA initiates DRP1 oligomerization, with DRP1 oligomers being passed to MFF for assembly of a productive contractile ring102, corresponding with the preference of MFF for larger DRP1 oligomers104.
Fig. 3 ∣. Actin and mitochondrial fission.
a, Calcium-induced actin, endoplasmic reticulum (ER) and mitochondrial fission. The top row shows a progression of mitochondrial fission at ER–mitochondria contacts. The mitochondrial genome replicates at the contact site, followed by the emergence of DRP1-independent pre-constriction. DRP1 is then recruited, followed by fission. The bottom row shows models for actin involvement at two fission steps. Left: actin polymerized by ER-bound INF2 leads to enhanced ER-mitochondria contact in a myosin II-dependent manner. Enhanced ER–mitochondria contact allows more efficient calcium transfer from ER to mitochondria, leading to pre-constriction of both the inner mitochondrial membrane (IMM) and outer mitochondrial membrane (OMM). Pre-constriction may be driven by forces on the IMM, with the OMM being pulled behind it due to proteins mediating IMM–OMM tethering. SPIRE1C might be involved at this stage but this has not been tested directly. Right: actin filaments recruit DRP1, allowing transfer of these small oligomers to OMM receptors for further oligomerization, leading to full ring assembly and OMM constriction. Although myosin II participates in both pre-constriction and DRP1 recruitment, its localization is unclear, and its large size puts limits on its possible mechanistic roles. We depict myosin II as a haze for this reason. b, Size relationship between mitochondria, myosin II and actin. The non-muscle myosin II filament (~300 nm) is of similar length to the mitochondrial diameter. Electron micrographs at the same scale for all images. Main figure: mitochondrion from U2OS cell. Inset 1: myosin II filament from REF52 cell. Inset 2: microtubule (white arrow) and actin filaments (red asterisks) from B35 neuroblastoma cell. Inset 3: myosin monomer from REF52 cell, decorated with gold-conjugated antibody to the tail (motor heads to right). Main figure: thin-section electron micrograph taken by RC and Radu Stan (Dartmouth). Insets 1 and 3: platinum replica electron micrographs taken by Maria Shutova & Tatyana Svitkina (University of Pennsylvania). Inset 2: platinum replica electron micrograph taken by Tatyana Svitkina. Scale bars: 300 nm (main figure), 50 nm (insets).
Many questions remain unanswered for CIA. The fundamental mechanism by which CIA stimulates IMM constriction is not understood. Myosin II clearly plays a role in both establishing pre-constriction and DRP1 recruitment73,76,92,97. An early model was that an actomyosin structure akin to the cytokinetic ring might assemble between the ER and mitochondria, constricting the OMM36. More recent findings suggest a less direct effect, with actin/myosin-stimulated calcium transfer into the mitochondrial matrix stimulating constriction mechanisms within the ER73,76. One possibility is that increased mitochondrial calcium could stimulate IMM constriction by activating oxidative phosphorylation105,106, with the resulting ATP increase stimulating the IMM protease YME1L. YME1L releases OPA1 from the IMM, and this short form of OPA1 mediates IMM constriction. This mechanism is speculative but based on the correlation between increased levels of short OPA1 and increased fission107.
How might actin and myosin II be organized to mediate this ERMC? One issue is that the functional oligomers of non-muscle myosin II are ~300-nm long108, which is close to the width of the unconstricted mitochondrion (Fig. 3b). It is possible that non-polymerized myosin II is the active motor in this process as has been suggested for other myosin II-based processes109-111. Even in its non-polymerized form, myosin II is a long molecule (150 nm), so this size must be considered when developing mechanistic models. Another alternative is that myosin II might not be acting directly at the fission site but at a distance to this site, putting the fission site filaments under tension as suggested by ultrastructural studies92. This ultrastructural work also raises the possibility that the two proposed functions for myosin II (promoting ERMC or directly driving mitochondrial constriction) are not mutually exclusive.
Similar mechanistic questions arise for SPIRE1C, myosin 19 and fascin. Functions for non-mitochondrially bound forms of SPIRE1 include cooperation with the FMN family of formin proteins in oocyte development in both Drosophila and mammals11,112. Mitochondrially bound SPIRE1C might cooperate with ER-bound INF2 in a similar manner95. How these filaments would act with myosin II to enhance ERMC or DRP1 recruitment are unknown. Interestingly, in mammalian oogenesis, which depends on FMN formins and SPIRE, the FMN formin is ER bound and polymerizes the actin that generates a pushing force around the spindle, apparently by pushing on mitochondria113, which could suggest functional similarities.
A recent publication suggests that myosin 19 acts as a tether to mediate ERMC during INF2–SPIRE1C-mediated mitochondrial fission98. Fascin is an actin-bundling protein also shown to be recruited to mitochondria in an INF2-dependent manner99. It is not clear whether myosin II, myosin 19 and fascin act together or at different stages of the process. In addition, fascin causes parallel-bundled actin filaments, and it is unclear how these would function with myosin II, which acts on antiparallel filaments114.
It is also unclear why DRP1 binds preferentially to actin near the fission site over other actin-based structures90,103. The architecture of CIA networks could have a high affinity for DRP1, perhaps dictated by myosin II, myosin 19, fascin, SPIRE1 or other as-yet unidentified actin-associated proteins.
Other types of mitochondrial fission involving actin
Several publications have found evidence for actin-dependent mitochondrial fission in an apparently INF2-independent manner. For example, some cultured cells display mitochondria that are surrounded by a ‘cloud’ of actin filaments. These clouds travel around the cell in a wave with a periodicity of ~15 min. As a result, at any given time, only a subset of mitochondria is surrounded by the actin clouds, and the presence of the wave correlates with increased DRP1-dependent mitochondrial fission (Fig. 5). Mitochondria often re-fuse after the actin wave passes, which might suggest kiss-and-run mechanisms of fusion–fission dynamics41,42. These interphase waves are dependent on the Arp2/3 complex, and thus distinct from CIA.
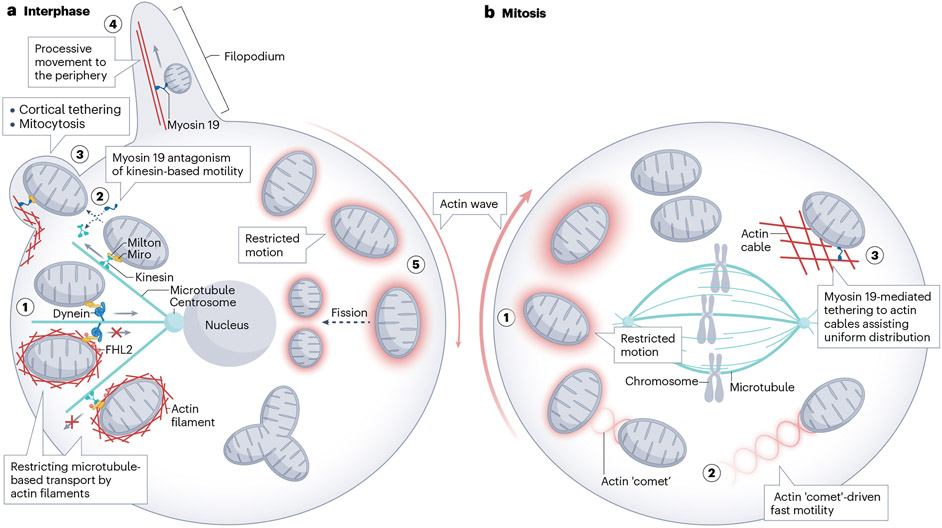
Fig. 5 ∣. Actin and mitochondrial motility.
a, Interphase cell. Mitochondrial motility arrest by actin (1). Increased O-GlcNAcylation of the protein Milton (which together with Miro attaches mitochondria to microtubule-based motors) leads to actin polymerization around the translocating mitochondrion, resulting in motility arrest. In a separate mechanism, myosin 19 can arrest plus-end microtubule-mediated mitochondrial transport by competing with kinesin for binding to Miro (2) and attaching mitochondria to cortical actin (3). In some circumstances, cortically attached mitochondria can be expelled from cells via mitocytosis (3). In other situations, myosin 19 can also mediate mitochondrial motility towards filopodial tips (4). Given the narrow filopodial diameter (100–200 nm), it is unclear how mitochondria adapt for this transit. Actin clouds assemble around sub-sets of mitochondria (5), with the cloud moving between mitochondria in a uniform direction over time (here shown clockwise, although both directions are possible). The actin cloud does not enhance mitochondrial motility but does correspond to an increase in mitochondrial fission. b, Mitotic cell. The actin cloud shown in the interphase cell can transition to a mitotic actin cloud, which increases in speed of rotation around the cell. As with interphase clouds, the associated mitochondria are less motile (1). However, some clouds (13%) can transform into actin ‘comets’, which cause rapid (250 nm/s) translocation of the associated mitochondrion to randomize mitochondrial inheritance (2). As a separate event, a network of actin filaments assembles throughout the mitotic cytoplasm, outside of the spindle zone (3). This network allows myosin 19-mediated mitochondrial tethering to the network, facilitating appropriate mitochondrial distribution to daughter cells. FHL2, Four and a half LIM domains protein 2.
A recent study115 revealed that cells cultured on soft substrates display small mitochondria with increased peri-mitochondrial actin filaments, increased mitochondrial DRP1 accumulation and increased mitochondrial reactive oxygen species (mtROS). These effects are reversed by DRP1 inhibition, suggesting that mitochondrial fission is important in this phenotype. Arp2/3 complex inhibition decreases mitochondrially associated actin and DRP1. Dominant-negative SPIRE1C constructs inhibit the increase in mtROS but INF2 suppression has no effect. These results suggest that actin might participate in mitochondrial fission under conditions of low cellular tension in an INF2-independent but SPIRE1C-dependent manner. Myosin II inhibition causes some of the same effects elicited by soft substrates, suggesting that inhibiting myosin II-based contraction in stress fibres is sufficient to induce this type of fission but that myosin II is not actually involved in this fission mechanism.
Another study shows that an alternately expressed short version of the gap junction protein connexin 43, called GJA1-20K, is enriched on mitochondria and, in turn, causes peri-mitochondrial actin enrichment80. GJA1-20K expression also results in a short mitochondria phenotype in a DRP1-independent manner. Curiously, the actin polymerization inhibitor latrunculin A does not reverse these effects. The nucleation factor involved in this process is unknown.
Finally, the actin-crosslinking protein filamin A has been shown to play a role in mitochondrial fission through direct interaction with DRP1 (ref. 116). The actin-binding property of filamin might also be required, but direct evidence for actin filaments at these fission sites is lacking. It is unclear whether this filamin effect is associated with one of the above actin-associated processes. The context of this fission is hypoxia–reoxygenation of cardiomyocytes, a process that induces both mitochondrial damage (the subject of the next section of this Review) and increased cytoplasmic calcium117,118, so either the Arp2/3 complex or INF2 could be involved.
Conclusions and outstanding questions for actin and mitochondrial fission
While it is now clear that actin can participate in mitochondrial fission, many questions loom. Perhaps the largest of which is, what percentage of mitochondrial fission events are stimulated by some form of actin? Considering that there could be multiple mechanisms of actin-stimulated mitochondrial fission (for example, INF2-dependent CIA versus INF2-independent mechanisms), this question becomes more complicated, and raises the general question of how many mechanisms of mitochondrial fission exist (both dependent on and independent of actin), as discussed above. Quantification from the seminal study on mitochondrial fission at ERMC sites showed that 64% of mitochondrial constriction events are accompanied by ERMCs35, suggesting that a significant proportion of fission occurs in the absence of ER contact.
A related question is, in what physiological processes is CIA-induced mitochondrial fission important? Most work on CIA-induced fission has been conducted using either ionomycin or histamine as stimuli (both increase cytoplasmic calcium)76,91. Would CIA play a role in the absence of a stimulated increase in cytoplasmic calcium? It is possible that localized calcium oscillations routinely occur in the absence of stimulation as has been shown in several cell types119,120. A recent study suggests that INF2 is required for fission of healthy mitochondria78 in dividing cells, possibly to enhance mitochondrial expansion during cell growth. This study was conducted without stimulation of cytoplasmic calcium increases, suggesting either that perhaps local calcium release at ERMCs is occurring or that there are alternate mechanisms of INF2 activation.
Other suggestions for the physiological importance of CIA come from links between INF2 and several processes. Dominant INF2 mutations link to two diseases: focal segmental glomerulosclerosis, which is a kidney disease, and Charcot–Marie–Tooth disease, a peripheral neuropathy121. Charcot–Marie–Tooth disease has other associations with mitochondrial dynamics through mutations to MFN2, the gene encoding the mitochondrial fusion protein MFN2 (ref. 122). In addition, INF2 regulation of mitochondrial size is important for placental development123. INF2 also has a role in oxidative stress responses to ischaemia–reperfusion124,125. Finally, calcium-dependent plasma membrane repair requires mitochondrial fission near the wound site126, which is consistent with a role for CIA in this context.
Finally, do the non-CIA types of actin-associated mitochondrial fission discussed above use fundamentally different mechanisms to stimulate mitochondrial fission? Considering that actin can serve several mechanical functions (generation of pushing force, track for myosins, scaffold), the mechanisms need not be identical. In addition, it is certainly possible that these non-CIA types of actin differ from each other in their mechanisms of stimulating fission.
Actin and mitochondrial damage
Oxidative phosphorylation can result in the production of mtROS that damage mitochondrial DNA, lipids and proteins, often manifesting as decreased mitochondrial ATP production or membrane polarization127. Mutations to nuclear genes encoding mitochondrial proteins can have similar effects, leading to a range of mitochondrially associated diseases128. More acute mitochondrial damage can be induced by ETC inhibitors (antimycin, rotenone) or ATP synthase inhibitors (oligomycin). While these treatments are generally used as research tools, the widely prescribed anti-diabetes drug metformin has been shown to act as an ETC inhibitor129. Hypoxia–ischaemia also induces a form of mitochondrial damage, cutting off a key ETC substrate (oxygen)130. Finally, the drugs CCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone and DNP induce rapid mitochondrial uncoupling (depolarization)131.
Damaged mitochondria are removed by mitophagy, and defective mitophagy is associated with several pathological conditions132. A key step in mitophagy is recognizing the damaged mitochondrion and targeting it to the autophagic pathway, which mammals conduct through several pathways. The PINK–PARKIN pathway is a particularly elegant recognition mechanism, in which loss of the proton gradient stabilizes the protein kinase PINK on the OMM, leading to recruitment of the E3 ubiquitin ligase PARKIN to initiate mitophagy132. Other mitophagy receptors include BNIP3–NIX, FUNDC1, BCL2L13 and FKBP8 (ref. 133). It is worth considering that quality control through MDVs might also occur in response to mitochondrial damage59.
Actin polymerization at two distinct stages after damage contributes in somewhat opposing ways to the mitochondrial damage response (Fig. 4). In addition, actin appears to play multiple roles in downstream steps shared with other forms of autophagy. We focus on the mitochondria-specific actin processes here, with autophagy-associated actin being nicely discussed elsewhere134,135.
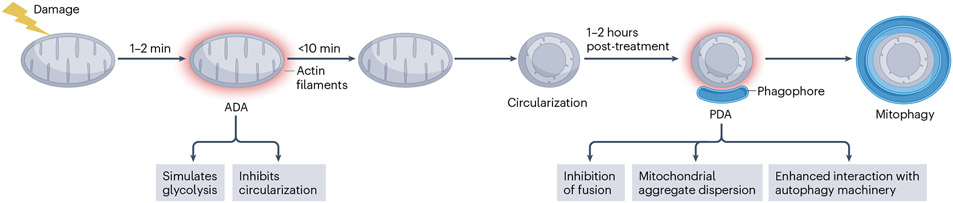
Fig. 4 ∣. Two types of actin induced by mitochondrial damage.
Treatment with a number of mitochondrial inhibitors (CCCP, antimycin, rotenone, oligomycin, hypoxia, metformin) leads to rapid actin polymerization around the mitochondrion, which we refer to as acute damaged-induced actin (ADA). Appearance of ADA generally starts within 1–2 min of treatment and depolymerizes within 10 min of assembly. ADA has two effects: glycolytic stimulation and inhibition of inner mitochondrial membrane rearrangements (circularization). Mitochondrial circulation ensues after actin depolymerization. After approximately 1 h, a second round of actin polymerization occurs, which we refer to as prolonged damage-induced actin (PDA). PDA has three demonstrated effects that favour mitophagy: inhibition of fusion, even if the damaged mitochondrion repolarizes; dispersal of mitochondrial aggregates; and recruitment of core autophagy components.
Acute damage-induced actin
Actin filaments assemble around damaged mitochondria within minutes, as first reported in mouse embryonic fibroblasts82 and subsequently in multiple cell types87,88,136,137. We call this rapidly assembling actin ‘acute damage-induced actin’ (ADA). ADA is caused by a range of treatments, including CCCP, antimycin, rotenone, oligomycin, metformin and hypoxia mimetics88,137.
ADA requires two initial signals: decreased ATP and increased cytoplasmic calcium88. The calcium increase is small, compared to that elicited by stimuli such as histamine or ionomycin88, and insufficient to activate CIA. The mitochondrial sodium–calcium antiporter NCLX is required for ADA, suggesting that initial calcium release is from the mitochondrion itself, while subsequent calcium release from the ER is also required. Increased calcium activates the Arp2/3 complex, through a pathway including conventional protein kinase C (cPKC), the RAC-GEF TRIO, the Rho GTPase RAC and, finally, the Arp2/3 complex activator WAVE. Decreased ATP levels activate the FMNL family of formin proteins through a pathway including AMPK (through LKB1), the CDC42 GEF FGD1 and the Rho GTPase CDC42.
What might be the function of ADA? One recently identified ADA function is to rapidly stimulate glycolysis137. This glycolytic increase is not observed in hyperglycaemic media (such as DMEM and RPMI1640) but becomes apparent in normoglycaemic medium and accentuated in hypoglycaemic medium. It is unclear which glycolytic step is enhanced by ADA. Links have been made previously between actin and several glycolytic proteins, including aldolase138, phosphofructokinase139 and glucose channels140. The rapidity of ADA-activated glycolysis might rule out some of these mechanisms, which take place over hours.
A second consequence of ADA is to suppress mitochondrial dynamics. The ADA-derived actin filaments surround mitochondria but do not promote mitochondrial fission. In fact, as discussed earlier, the mitochondrial dynamics that occur rapidly after mitochondrial depolarization do not appear to be fission events but are associated with reorganization of the IMM, resulting in toroid or circular mitochondria. The purpose of circularization is unclear but may prime the organelle for downstream mitophagy. ADA inhibits these mitochondrial shape changes87,88,137.
Overall, the two effects of ADA may serve as acute responses to loss of mitochondrial function. By upregulating glycolysis, ADA helps to maintain ATP levels. By inhibiting circularization, ADA might delay mitophagic clearance, allowing for mitochondrial recovery. The mitophagic delay is hypothetical at this point.
Prolonged damage-induced actin
A second round of peri-mitochondrial actin polymerization, which we term ‘prolonged damage-induced actin’ (PDA) occurs 1–2 h after damage136,141 (Fig. 4) and has very different functions to ADA. Like ADA, PDA is Arp2/3 complex dependent but its activation is regulated by a different set of factors, including Parkin, myosin 6 and CDC42 (ref. 136). The direct Arp2/3 complex activator in PDA is WASP in haematopoietic cells142 and its homologue N-WASP in non-haematopoietic cells136. Macrophages from patients carrying WASP mutants display aberrant mitochondria with decreased oxidative phosphorylation142.
Three non-mutually exclusive functions have been ascribed to PDA. One function is to inhibit re-fusion of damaged mitochondria with healthy mitochondria even if the damaged mitochondrion recovers membrane potential136, sealing the fate of this mitochondrion for destruction. In this sense, actin appears to serve a ‘cage’ function. A second function is to disperse tightly packed clumps of mitochondria, which also aids mitophagy. Interestingly, myosin II might be involved in this dispersal141.
A third PDA function may be to link mitochondria with general autophagy factors like Atg14L141. Actin is clearly involved in macroautophagy in general, with the Arp2/3 complex being a key polymerization factor, possibly during multiple steps134,135,143,144. It is unclear whether all actin-based functions identified in general macroautophagy are required for mitophagy specifically.
Actin around chronically damaged mitochondria
Peri-mitochondrial actin resembling ADA or PDA occurs in more chronic forms of mitochondrial dysfunction caused by mitochondrial DNA depletion, knockout of the ETC complex I subunit NDUFS4 and in cells from patients with Leigh syndrome (a rare inherited neurometabolic disorder)137. Although this mitochondrially associated actin is constitutive, Arp2/3 complex inhibition causes its removal within 5 min, suggesting constant actin turnover. In addition, Arp2/3 complex inhibition causes a rapid drop in glycolysis137, suggesting a similar function to ADA. It is not clear, however, whether the signalling pathways used to establish these mitochondria-associated actin networks resemble those for ADA or for PDA.
Conclusions and outstanding questions for actin and mitochondrial damage
Elucidating the roles of actin polymerization during mitochondrial damage has been confounded by several issues. First, there has been a tendency to immediately associate this actin with mitochondrial fission, whereas there is no direct evidence for this function. Second, there are at least two independent mitochondria-associated actin networks responding to acute or prolonged mitochondrial damage, respectively, making it challenging to deconvolve their specific effects. Understanding the distinct pathways that trigger ADA and PDA should aid in assessing their specific functions.
A number of immediate questions can be raised concerning ADA and PDA. How do the two assembly factors (Arp2/3 complex and FMNL formins) work together in ADA? Possibilities include formin-mediated supply of mother filaments for Arp2/3 complex activation or formin-mediated elongation of Arp2/3 complex-nucleated filaments. It is possible that both roles occur, considering that depletion of all three FMNL formins is necessary to inhibit ADA88. How different are ADA and PDA in terms of polymerization mechanism and architecture of the resulting actin filaments? To address this question, higher-resolution imaging of the networks is necessary.
Another question concerns the factors controlling mitochondrial localization of actin during ADA or PDA. The Arp2/3 complex, a WAVE complex subunit and the Arp2/3 complex-interacting protein cortactin are rapidly recruited to mitochondria during CCCP-induced ADA88. The initial localization signal, however, is unclear. Additionally, it is unclear whether components of the second branch of the ADA pathway (FMNL formins, CDC42 or the CDC42 GEF FDG1) are also mitochondrially recruited. For PDA, it is unclear whether the Arp2/3 complex or WASP–N-WASP are recruited directly to mitochondria, and whether PARKIN mediates this direct recruitment.
An additional question is how ADA or PDA might relate to other major cellular actin-dependent processes, many of which are also regulated by Rho GTPases. The work on ADA suggests that two signals stemming from mitochondrial dysfunction (mitochondrial calcium release and a drop in cytoplasmic ATP) provide the initial stimuli directing actin polymerization specifically around mitochondria88, while PARKIN recruitment to mitochondria might be a key event in PDA activation136. It is unclear at present whether other signals more commonly used in actin cytoskeletal regulation, such as specific phosphoinositide generation, also play roles in either process.
Finally, are there cell type variabilities in ADA and PDA? This question might relate to the issue of mitochondrial heterogeneity, discussed earlier. The work on both ADA and PDA has been conducted in a limited number of cell types, and specifics in both the signalling pathways and responses may vary. For example, ADA requires two Rho GEFs in U2OS cells (Trio and FGD1)88, but these are members of the large GEF family for which function is known to vary between cell types145. For the ADA–glycolysis connection, it is unclear whether cancer cells respond in the same manner as the mouse embryonic fibroblasts and T effector cells that have previously been studied137 considering the constitutive dependence on glycolysis displayed by many cancers64.
Actin and mitochondrial motility
Actin both promotes and inhibits mitochondrial motility, depending on the cellular context (cell type, stimulus) through mechanisms involving either myosins or the polymerization of actin itself to exert force (Fig. 5).
Actin inhibition of microtubule-based mitochondrial motility
An important mitochondrial motility mechanism is kinesin-based or dynein-based movement along microtubules, in which these motors interface with mitochondria through two intermediaries: Miro and Milton (also called TRAK)46. Regulation of these interactions is important in directional motility control, especially in highly polarized cells such as neurons. A recently identified regulatory mechanism is that increased cytoplasmic glucose causes post-translational modification of Milton by N-acetylglucosamine (O-GlcNAcylation)146, enabling Milton binding to four and a half LIM domains protein 2 (FHL2). The Milton–FHL2 interaction results in mitochondrial binding to actin filaments, arresting microtubule-based movement147. Overexpression of the enzyme mediating O-GlcNAcylation also causes an impressive increase in peri-mitochondrial actin filaments, which are likely the filaments causing motility restriction147. The assembly mechanisms for this actin are not known.
Myosin effects on mitochondrial motility
In contrast to the widespread use of microtubule motors for mitochondrial motility, myosins are less frequently used, with notable exceptions. In budding yeast, type V myosins translocate mitochondria and other organelles from mother to bud43-45. In mammals, the mitochondrially associated myosin 19 can cause processive mitochondrial movement towards the cell periphery in some situations, notably upon cell stress47,48,148,149.
Intriguingly, myosin 19 is also used to restrict motility, a function that has been described in four distinct contexts. We have already discussed involvement of myosin 19 in mitochondrial fission98, where it may act as a tether between mitochondria and ER. In a second context, myosin 19 might tether mitochondria to the plasma membrane during a novel quality control process that expels damaged mitochondria, termed mitocytosis150. Myosin 19 mutants that bind actin but cannot move along filaments are capable of participating in mitocytosis, suggesting that the role of myosin 19 is to restrict motility. A third such role for myosin 19 is during mitosis when a network of actin cables assembles outside of the spindle area49. The nucleator of mitotic cables is unknown, but nucleating activity is not provided by the Arp2/3 complex. Mitochondria orient along these cables in a myosin 19-dependent manner, suggesting that myosin 19 attaches mitochondria to cables49. This association with actin cables appears important for appropriate mitochondrial distribution to daughter cells49,151,152, possibly by providing a uniform distribution of mitochondria. Depletion of myosin 19 results in asymmetric mitochondrial inheritance. Finally, a recent study shows that myosin 19 motor activity is required to stabilize cristae architecture, through interactions with SAM50 (part of the sorting and assembly machinery (SAM) necessary for the assembly of β-barrel proteins) on the OMM153. While the mechanism by which myosin 19 stabilizes cristae architecture is unclear, myosin 19 restricts motility during the process and does not move mitochondria. In addition to these motility-restricting functions, myosin 19 might be directly antagonistic to microtubule motor-based mitochondrial translocation since it also interacts with Miro proteins in a competitive manner to Milton154-156.
Two other myosins, myosin V and myosin VI, can oppose microtubule-based mitochondrial transport, thus restricting motility157,158. We have previously discussed myosin II function in mitochondrial fission and myosin VI function upon mitochondrial damage. In both cases, the myosin is not acting to translocate mitochondria. In summary, while myosins have the capacity to affect mitochondrial motility in some cases, often myosins serve other functions in mitochondrial biology, with one function being to restrict motility. An excellent recent review covers mitochondrial functions for myosins, kinesins and dynein in detail159.
Mitochondrial motility through actin polymerization
Actin polymerization by the Arp2/3 complex can drive translocation of sub-cellular structures such as endosomes160. Intracellular pathogenic microbes, such as Listeria, Shigella and Rickettsia, are particularly adept at harnessing this actin-based motility161, often (but not always) via Arp2/3 complex-mediated networks that form ‘comets’ or ‘tails’ behind the moving microorganism.
Evidence for Arp2/3 complex-mediated mitochondrial motility was first suggested in budding yeast162, but the purpose of this motility is not known. Recently, Arp2/3 complex-dependent motility was shown for mammalian mitochondria during mitosis49. Similar to the previously discussed waves of actin clouds around interphase mitochondria163, actin clouds can develop around mitotic mitochondria, often transitioning from the interphase actin structures but circling the cell at greater speed (full revolution in 6 min). There is no evidence for mitotic actin clouds around other organelles.
Similar to interphase clouds163, mitotic clouds restrict mitochondrial motility49. However, in a minority of cases (13%), the cloud converts into an actin tail behind the mitochondrion, with the mitochondrion rapidly moving directionally away from the tail (~250 nm/s). Interestingly, the tail contains two main strands that, at times, are helically entwined, similar to the distinctive tails behind Rickettsia at a specific stage in its infection cycle161.
While this actin-based motility is not important for the overall symmetrical distribution of mitotic mitochondria, it does increase mixing of mitochondrial populations, which randomizes the inheritance of damaged mitochondria to ensure both daughters receive similarly functional mitochondrial populations. Overall, two actin-based processes govern mitotic mitochondrial distribution: myosin 19-mediated tethering to actin cables, assisting uniform distribution; and Arp2/3 complex-mediated motility, mixing mitochondrial populations49.
Actin and other mitochondria-associated processes
We briefly discuss possible roles for actin in two other mitochondrial contexts, both of which present uncertainties: roles in cell death and functions of actin inside the mitochondrion.
Actin, mitochondria and cell death
The literature regarding the role of actin in apoptotic or necrotic cell death pathways is mixed. The Arp2/3 complex activator WAVE1 has been linked with mitochondria in both pro-apoptotic164,165 and anti-apoptotic events166, but actin is not implicated directly. A recent study provides evidence for two other Arp2/3 complex activators, JMY and WHAMM, in promoting mitochondrial cytochrome c release during DNA damage-induced apoptosis167. The role of JMY depends on the Arp2/3 complex, but the resulting actin is not nucleated on mitochondria. The localization of the WHAMM-nucleated actin is unclear. In addition, many papers have documented the translocation of cofilin (factor regulating depolymerization and dynamics) to mitochondria upon treatments that stimulate apoptotic or necrotic cell death168-174. Most of these papers show a stimulatory effect of this cofilin on cell death, while two papers find no effect. Cofilin oxidation might be important for its mitochondrial translocation, possibly linking ROS to this process173-175. The papers vary in their identification of actin as being implicated together with cofilin in these events. Clearly, more investigation is needed on potential roles for actin in mitochondrial-based apoptosis.
Actin within the mitochondrion
A suitable way to end this Review is with the most speculative potential mitochondrial role for actin. There is no compelling a priori reason to imagine a role for actin inside the mitochondrion since actin does not possess a clear N-terminal mitochondrial translocation pre-sequence. However, such a signal can be variable176-178 and is not present on all mitochondrial proteins179. In addition, mitochondrial populations of proteins formerly considered to reside elsewhere have been identified180. In other words, the absence of signal sequence does not necessarily equate with the absence of mitochondrial actin.
Three studies suggest roles for actin within mitochondria181-183. One study found biochemical evidence for mitochondrial actin and myosin IIA and further showed their association with mitochondrial DNA in HEK cells181. Another study used super-resolution microscopy images of fixed cells as evidence for intra-mitochondrial actin182. A third study, using isolated brain mitochondria, suggests a role for actin in ETC complex IV function183.
Clearly, there is a long way to go in determining the validity of intra-mitochondrial actin. The potential for cytoplasmic actin contamination (or actin associated with the cytoplasmic face of mitochondria) is a concern in studies involving isolated mitochondria183. However, it took a long time to accept the now well-established presence and function of actin in the nucleus184, so time will tell on mitochondrial actin.
Conclusions and perspectives
Elucidating the function of actin in any cellular context is complicated by its abundance, participation in multiple processes, and overlapping functions in space and time. In our opinion, there are certainly more functions for actin in mitochondrial biology than those reported here. Here are a few things to keep in mind in further investigation. First, actin-binding proteins are unlikely to act alone, with any actin-based structure requiring multiple proteins to mediate its assembly, organization and disassembly. If one actin-binding protein is identified in a process, others should be expected. Second, if a role for actin is suspected based on long-term treatment (1 h or longer), it is possible that more than one actin-mediated event might have occurred in that period. An example from this Review is ADA and PDA after mitochondrial damage. Third, it is often difficult to observe significant actin filament accumulation by microscopy for several reasons: the actin is transient (for example, CIA or ADA in this Review), the actin is operating at a distance from the actual structure (for example, myosin II-based constriction might be occurring over a broader region), the actin is obscured by other actin-based structures or the actin is not well resolved by the technique used. For this last point, the initial inability to observe clear actin in the nucleus is an excellent example and novel probes were necessary for nuclear actin imaging184. Additional challenges arise when using electron microscopy due to destruction of all but the most stable actin filaments by traditional thin-section electron microscopy processing185,186. Actin can even be difficult to detect by cryo-electron microscopy techniques if the filaments are short187,188. Other electron microscopy techniques have been more successful in observing actin around mitochondria83,92.
Finally, as actin biologists, we have learned that any understanding of mitochondrial roles for actin requires an understanding of mitochondria themselves, an understanding we are still working to acquire. Similarly, those coming from the mitochondrial field must acquire an understanding of the actin cytoskeleton. Without an appreciation of both fields, findings are likely to be superficial, making overly simple conclusions about complex and heterogeneous processes.
Supplementary Material
Acknowledgements
The authors thank the following individuals for their valuable input: A. Akhtar, H. Aydin, L Blanchoin, T. Bretscher, P. Chinnery, D. Colon-Ramos, J. Cooper, R. Dominguez, S. Eustermann, V. Fowler, J. Gautier, E. Gingmer, B. Goode, R. Grosse, A. Henn, R. Kay, L. Kiss, J. Kollman, M. Mietalobs, D. Mullins, M. Ostap, K. Pfanner, M. Picard, T. Pollard, L. Pon, M. Quinlan, K. Rottner, Y. Sancak, M. Schuldiner, B. Schulman, T. Schwarz, O. Shirihai, T. Svitkina, C. Thompson, C. Toseland, M. Vartiainen & B. Webb. During the revision process, the material related to many of these contributions was removed, but the correspondence is still valued. This work was supported by NIH grant R35 GM122545.
Footnotes
Competing interests
The authors declare no competing financial or non-financial interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41580-023-00613-y.
References
- 1. De Vos KJ, Allan VJ, Grierson AJ & Sheetz MP Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr. Biol 15, 678–683 (2005). This paper is the first to clearly show an effect of actin on mitochondrial division, and also shows that mitochondrial depolarization does not cause rapid mitochondrial fission.
- 2. Picard M & Shirihai OS Mitochondrial signal transduction. Cell Metab. 34, 1620–1653 (2022). An excellent recent review on the diverse functions of mammalian mitochondria.
- 3.Blanchoin L, Boujemaa-Paterski R, Sykes C & Plastino J Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev 94, 235–263 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Posern G & Treisman R Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16, 588–596 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Jungblut A, Hopfner KP & Eustermann S Megadalton chromatin remodelers: common principles for versatile functions. Curr. Opin. Struct. Biol 64, 134–144 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Kaksonen M & Roux A Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol 19, 313–326 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Krause M & Gautreau A Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol 15, 577–590 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Pollard TD Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol 10.1101/cshperspect.a018226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautreau AM, Fregoso FE, Simanov G & Dominguez R Nucleation, stabilization, and disassembly of branched actin networks. Trends Cell Biol. 32, 421–432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez R. The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem. Sci 41, 478–490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinlan ME Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development 140, 4417–4425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vafai SB & Mootha VK Mitochondrial disorders as windows into an ancient organelle. Nature 491, 374–383 (2012). We regard this review as a ‘classic’, providing clear insights into many aspects of mitochondrial biology that are still relevant 11 years later.
- 13.Kukat C. et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl Acad. Sci. USA 112, 11288–11293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogenhagen D & Clayton DA The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem 249, 7991–7995 (1974). [PubMed] [Google Scholar]
- 15.Satoh M & Kuroiwa T Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res 196, 137–140 (1991). [DOI] [PubMed] [Google Scholar]
- 16.Rieger B, Arroum T, Borowski MT, Villalta J & Busch KB Mitochondrial F(1) F(O) ATP synthase determines the local proton motive force at cristae rims. EMBO Rep. 22, e52727 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolf DM et al. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J. 38, e101056 (2019). An elegant study showing the variation in polarization between neighbouring cristae, suggesting that a single mitochondrion can have considerable functional diversity along its length.
- 18.Tan JX & Finkel T Mitochondria as intracellular signaling platforms in health and disease. J. Cell Biol 10.1083/jcb.202002179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R, Letai A & Sarosiek K Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol 20, 175–193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H, Carvalho P & Voeltz GK Here, there, and everywhere: the importance of ER membrane contact sites. Science 10.1126/science.aan5835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csordás G, Weaver D & Hajnóczky G Endoplasmic reticulum-mitochondrial contactology: structure and signaling functions. Trends Cell Biol. 28, 523–540 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veliova M, Petcherski A, Liesa M & Shirihai OS The biology of lipid droplet-bound mitochondria. Semin. Cell Dev. Biol 108, 55–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong YC, Kim S, Peng W & Krainc D Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 29, 500–513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagashima S. et al. Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science 367, 1366–1371 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Picard M. et al. Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat. Commun 6, 6259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glancy B. et al. Power grid protection of the muscle mitochondrial reticulum. Cell Rep. 19, 487–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan DC Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. Mech. Dis 15, 235–259 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Yapa NMB, Lisnyak V, Reljic B & Ryan MT Mitochondrial dynamics in health and disease. FEBS Lett. 595, 1184–1204 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Legros F, Lombès A, Frachon P & Rojo M Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol. Biol. Cell 13, 4343–4354 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Z, Chen H, Fiket M, Alexander C & Chan DC OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol 178, 749–755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abrisch RG, Gumbin SC, Wisniewski BT, Lackner LL & Voeltz GK Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J. Cell Biol 10.1083/jcb.201911122 (2020). This paper shows that the ERMC sites might mark sites of both MFN1 and DRP1 to these sites, and that rescue of mitochondrial membrane potential can happen at these sites.
- 32.Gao S & Hu J Mitochondrial fusion: the machineries in and out. Trends Cell Biol. 31, 62–74 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Kraus F, Roy K, Pucadyil TJ & Ryan MT Function and regulation of the divisome for mitochondrial fission. Nature 590, 57–66 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Lewis SC, Uchiyama LF & Nunnari J ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman JR et al. ER tubules mark sites of mitochondrial division. Science 334, 358–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatch AL, Gurel PS & Higgs HN Novel roles for actin in mitochondrial fission. J. Cell Sci 127, 4549–4560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang W. et al. Endosomal lipid signaling reshapes the endoplasmic reticulum to control mitochondrial function. Science 378, eabq5209 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Wong YC, Ysselstein D & Krainc D Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JE, Westrate LM, Wu H, Page C & Voeltz GK Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540, 139–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fonseca TB, Sánchez-Guerrero Á, Milosevic I & Raimundo N Mitochondrial fission requires DRP1 but not dynamins. Nature 570, E34–E42 (2019). [DOI] [PubMed] [Google Scholar]
- 41. Twig G. et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 (2008). This paper shows evidence for rapid sequential fusion then fission (termed ‘kiss-and-run’), and daughter mitochondria after fission can have different membrane potentials, with the lower potential daughter undergoing mitophagy.
- 42.Liu X, Weaver D, Shirihai O & Hajnóczky G Mitochondrial ‘kiss-and-run’: interplay between mitochondrial motility and fusion-fission dynamics. EMBO J. 28, 3074–3089 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh T, Toh EA & Matsui Y Mmr1p is a mitochondrial factor for Myo2p-dependent inheritance of mitochondria in the budding yeast. EMBO J. 23, 2520–2530 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swayne TC et al. Role for cER and Mmr1p in anchorage of mitochondria at sites of polarized surface growth in budding yeast. Curr. Biol 21, 1994–1999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chernyakov I, Santiago-Tirado F & Bretscher A Active segregation of yeast mitochondria by Myo2 is essential and mediated by Mmr1 and Ypt11. Curr. Biol 23, 1818–1824 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwarz TL Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Biol 10.1101/cshperspect.a011304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato O. et al. Mitochondria-associated myosin 19 processively transports mitochondria on actin tracks in living cells. J. Biol. Chem 298, 101883 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ušaj M & Henn A Kinetic adaptation of human Myo19 for active mitochondrial transport to growing filopodia tips. Sci. Rep 7, 11596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moore AS et al. Actin cables and comet tails organize mitochondrial networks in mitosis. Nature 591, 659–664 (2021). Two types of novel mitochondrially associated actin during mitosis are revealed here: cables that tether mitochondria and clouds that can result in rapid mitochondrial motility.
- 50.Mishra P & Chan DC Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol 15, 634–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misgeld T & Schwarz TL Mitostasis in neurons: maintaining mitochondria in an extended cellular architecture. Neuron 96, 651–666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rambold AS, Kostelecky B, Elia N & Lippincott-Schwartz J Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl Acad. Sci. USA 108, 10190–10195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang X. et al. Kissing and nanotunneling mediate intermitochondrial communication in the heart. Proc. Natl Acad. Sci. USA 110, 2846–2851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lavorato M. et al. Increased mitochondrial nanotunneling activity, induced by calcium imbalance, affects intermitochondrial matrix exchanges. Proc. Natl Acad. Sci 114, E849–E858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cogliati S, Enriquez JA & Scorrano L Mitochondrial cristae: where beauty meets functionality. Trends Biochem. Sci 41, 261–273 (2016). [DOI] [PubMed] [Google Scholar]
- 56.MacVicar T & Langer T OPA1 processing in cell death and disease — the long and short of it. J. Cell Sci 129, 2297–2306 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Soubannier V. et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol 22, 135–141 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Hughes AL, Hughes CE, Henderson KA, Yazvenko N & Gottschling DE Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. eLife 10.7554/eLife.13943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. König T. et al. MIROs and DRP1 drive mitochondrial-derived vesicle biogenesis and promote quality control. Nat. Cell Biol 23, 1271–1286 (2021). A technical tour-de-force, defining many aspects of mitochondrially derived vesicle formation in mammalian cells.
- 60.Picard M, Hepple RT & Burelle Y Mitochondrial functional specialization in glycolytic and oxidative muscle fibers: tailoring the organelle for optimal function. Am. J. Physiol. Cell Physiol 302, C629–C641 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Selvaraj V, Stocco DM & Clark BJ Current knowledge on the acute regulation of steroidogenesis. Biol. Reprod 99, 13–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosen ED & Spiegelman BM What we talk about when we talk about fat. Cell 156, 20–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morio B, Panthu B, Bassot A & Rieusset J Role of mitochondria in liver metabolic health and diseases. Cell Calcium 94, 102336 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Pavlova NN, Zhu J & Thompson CB The hallmarks of cancer metabolism: still emerging. Cell Metab. 34, 355–377 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rausser S. et al. Mitochondrial phenotypes in purified human immune cell subtypes and cell mixtures. eLife 10.7554/eLife.70899 (2021). An elegant study showing mitochondrial variation in humans on a week-to-week basis.
- 66.Pagliarini DJ et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fecher C. et al. Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nat. Neurosci 22, 1731–1742 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Stauch KL, Purnell PR & Fox HS Quantitative proteomics of synaptic and nonsynaptic mitochondria: insights for synaptic mitochondrial vulnerability. J. Proteome Res 13, 2620–2636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.El Bacha T, Luz M & Da Poian A Dynamic adaptation of nutrient utilization in humans. Nat. Educ 3, 11 (2010). [Google Scholar]
- 70.Alan L & Scorrano L Shaping fuel utilization by mitochondria. Curr. Biol 32, R618–R623 (2022). [DOI] [PubMed] [Google Scholar]
- 71.Picard M, Shirihai OS, Gentil BJ & Burelle Y Mitochondrial morphology transitions and functions: implications for retrograde signaling. Am. J. Physiol. Regul. Integr. Comp. Physiol 304, R393–R406 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steiner P, Luckner M, Kerschbaum H, Wanner G & Lütz-Meindl U Ionic stress induces fusion of mitochondria to 3-D networks: An electron tomography study. J. Struct. Biol 204, 52–63 (2018). [DOI] [PubMed] [Google Scholar]
- 73. Kage F, Vicente-Manzanares M, McEwan BC, Kettenbach AN & Higgs HN Myosin II proteins are required for organization of calcium-induced actin networks upstream of mitochondrial division. Mol. Biol. Cell 33, ar63 (2022). Figure 8 of this paper shows clear evidence for morphological heterogeneity between perinuclear and peripheral mitochondria in the same cell and differential influence of myosin II on these two populations.
- 74. Fischer TD, Dash PK, Liu J & Waxham MN Morphology of mitochondria in spatially restricted axons revealed by cryo-electron tomography. PLoS Biol. 16, e2006169 (2018). This paper shows the ability of mitochondria to narrow to 20nm diameter in certain situations.
- 75.Mishra P, Carelli V, Manfredi G & Chan DC Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19, 630–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chakrabarti R. et al. INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol 217, 251–268 (2017). Comprehensive study on CIA-induced mitochondrial calcium increase.
- 77.Helle SCJ et al. Mechanical force induces mitochondrial fission. eLife 10.7554/eLife.30292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kleele T. et al. Distinct fission signatures predict mitochondrial degradation or biogenesis. Nature 593, 435–439 (2021). A seminal study showing that two mechanistically distinct mitochondrial fission processes exist in mammalian cells.
- 79.Yamashita SI et al. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol 215, 649–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shimura D. et al. Protective mitochondrial fission induced by stress-responsive protein GJA1–20k. eLife 10.7554/eLife.69207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fu D & Lippincott-Schwartz J Monitoring the effects of pharmacological reagents on mitochondrial morphology. Curr. Protoc. Cell Biol 79, e45 (2018). [DOI] [PubMed] [Google Scholar]
- 82. Li S. et al. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol 208, 109–123 (2015). First report of rapid actin polymerization around dysfunctional mitochondria (ADA).
- 83.Mageswaran SK et al. Nanoscale details of mitochondrial fission revealed by cryo-electron tomography. bioRxiv 10.1101/2021.12.13.472487 (2021). [DOI] [Google Scholar]
- 84.Minamikawa T, Williams DA, Bowser DN & Nagley P Mitochondrial permeability transition and swelling can occur reversibly without inducing cell death in intact human cells. Exp. Cell Res 246, 26–37 (1999). [DOI] [PubMed] [Google Scholar]
- 85.Liu X & Hajnóczky G Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ. 18, 1561–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Miyazono Y. et al. Uncoupled mitochondria quickly shorten along their long axis to form indented spheroids, instead of rings, in a fission-independent manner. Sci. Rep 8, 350 (2018). A correlative fluorescence microscopy/electron microscopy study showing that mitochondrial depolarization does not result in rapid fission but in IMM rearrangement leading to circular mitochondria with an intact OMM.
- 87.Fung TS, Ji W-K, Higgs HN & Chakrabarti R Two distinct actin filament populations have effects on mitochondria, with differences in stimuli and assembly factors. J. Cell Sci 132, jcs234435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fung TS et al. Parallel kinase pathways stimulate actin polymerization at depolarized mitochondria. Curr. Biol 32, 1577–1592.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guha S. et al. Selective disruption of Drp1-independent mitophagy and mitolysosome trafficking by an Alzheimer’s disease relevant tau modification in a novel Caenorhabditis elegans model. Genetics 10.1093/genetics/iyac104 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Korobova F, Ramabhadran V & Higgs HN An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ji WK, Hatch AL, Merrill RA, Strack S & Higgs HN Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife 4, e11553 (2015). Study showing CIA-mediated recruitment of DRP1 to mitochondrial fission sites.
- 92.Yang C & Svitkina TM Ultrastructure and dynamics of the actin-myosin II cytoskeleton during mitochondrial fission. Nat. Cell Biol 21, 603–613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shao X, Li Q, Mogilner A, Bershadsky AD & Shivashankar GV Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc. Natl Acad. Sci. USA 112, E2595–E2601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wales P. et al. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. eLife 10.7554/eLife.19850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Manor U. et al. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife 10.7554/eLife.08828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.DuBoff B, Götz J & Feany MB Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron 75, 618–632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korobova F, Gauvin TJ & Higgs HN A role for myosin II in mammalian mitochondrial fission. Curr. Biol 24, 409–414 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coscia SM et al. Myo19 tethers mitochondria to endoplasmic reticulum-associated actin to promote mitochondrial fission. J. Cell Sci 10.1242/jcs.260612 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lin S. et al. Fascin controls metastatic colonization and mitochondrial oxidative phosphorylation by remodeling mitochondrial actin filaments. Cell Rep. 28, 2824–2836. e28 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cho B. et al. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun 8, 15754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hatch AL, Ji WK, Merrill RA, Strack S & Higgs HN Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol. Biol. Cell 27, 3109–3121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu A, Kage F & Higgs HN Mff oligomerization is required for Drp1 activation and synergy with actin filaments during mitochondrial division. Mol. Biol. Cell 10.1091/mbc.E21-04-0224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ji WK et al. Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J. Cell Biol. 216, 4123–4139 (2017). Study showing evidence for ER-bound pools of both MFF and FIS1 as well as actin-stimulated DRP1 recruitment to ER.
- 104.Liu R & Chan DC The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol. Biol. Cell 26, 4466–4477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Denton RM Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta 1787, 1309–1316 (2009). [DOI] [PubMed] [Google Scholar]
- 106.Ashrafi G, de Juan-Sanz J, Farrell RJ & Ryan TA Molecular tuning of the axonal mitochondrial Ca(2+) uniporter ensures metabolic flexibility of neurotransmission. Neuron 105, 678–687.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anand R. et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol 204, 919–929 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Billington N, Wang A, Mao J, Adelstein RS & Sellers JR Characterization of three full-length human nonmuscle myosin II paralogs. J. Biol. Chem 288, 33398–33410 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Laporte D, Coffman VC, Lee IJ & Wu JQ Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol 192, 1005–1021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pollard LW et al. Fission yeast myosin Myo2 is down-regulated in actin affinity by light chain phosphorylation. Proc. Natl Acad. Sci. USA 114, E7236–E7244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shutova MS, Spessott WA, Giraudo CG & Svitkina T Endogenous species of mammalian nonmuscle myosin IIA and IIB include activated monomers and heteropolymers. Curr. Biol 24, 1958–1968 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E & Schuh M Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol 21, 955–960 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Duan X. et al. Dynamic organelle distribution initiates actin-based spindle migration in mouse oocytes. Nat. Commun 11, 277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sellers JR & Heissler SM Nonmuscle myosin-2 isoforms. Curr. Biol 29, R275–R278 (2019). [DOI] [PubMed] [Google Scholar]
- 115.Romani P. et al. Mitochondrial fission links ECM mechanotransduction to metabolic redox homeostasis and metastatic chemotherapy resistance. Nat. Cell Biol 24, 168–180 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nishimura A. et al. Hypoxia-induced interaction of filamin with Drp1 causes mitochondriaL hyperfission-associated myocardial senescence. Sci. Signal 10.1126/scisignal.aat5185 (2018). [DOI] [PubMed] [Google Scholar]
- 117.Bai Y. et al. Mitochondrial quality control in cardiac ischemia/reperfusion injury: new insights into mechanisms and implications. Cell. Biol. Toxicol 10.1007/s10565-022-09716-2 (2022). [DOI] [PubMed] [Google Scholar]
- 118.Pedriali G. et al. Perspectives on mitochondrial relevance in cardiac ischemia/reperfusion injury. Front. Cell Dev. Biol 10, 1082095 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang TF, Zhou C, Tang AH, Wang SQ & Chai Z Cellular mechanism for spontaneous calcium oscillations in astrocytes. Acta Pharmacol. Sin 27, 861–868 (2006). [DOI] [PubMed] [Google Scholar]
- 120.Zhou Y. et al. Spontaneous calcium signaling of cartilage cells: from spatiotemporal features to biophysical modeling. FASEB J. 33, 4675–4687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Labat-de-Hoz L & Alonso MA The formin INF2 in disease: progress from 10 years of research. Cell Mol. Life Sci 77, 4581–4600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zaman M & Shutt TE The role of impaired mitochondrial dynamics in MFN2-mediated pathology. Front. Cell Dev. Biol 10, 858286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lamm KYB et al. Inverted formin 2 regulates intracellular trafficking, placentation, and pregnancy outcome. eLife 10.7554/eLife.31150 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang Z & Yu J Nurr1 exacerbates cerebral ischemia-reperfusion injury via modulating YAP-INF2-mitochondrial fission pathways. Int. J. Biochem. Cell Biol 104, 149–160 (2018). [DOI] [PubMed] [Google Scholar]
- 125.Chen Z. et al. INF2 regulates oxidative stress-induced apoptosis in epidermal HaCaT cells by modulating the HIF1 signaling pathway. Biomed. Pharmacother 111, 151–161 (2019). [DOI] [PubMed] [Google Scholar]
- 126.Horn A, Raavicharla S, Shah S, Cox D & Jaiswal JK Mitochondrial fragmentation enables localized signaling required for cell repair. J. Cell Biol 10.1083/jcb.201909154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murphy MP Understanding and preventing mitochondrial oxidative damage. Biochem. Soc. Trans 44, 1219–1226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Suomalainen A & Battersby BJ Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol 19, 77–92 (2018). [DOI] [PubMed] [Google Scholar]
- 129.Owen MR, Doran E & Halestrap AP Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J 348, 607–614 (2000). [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou M. et al. Myocardial ischemia-reperfusion injury: therapeutics from a mitochondria-centric perspective. Cardiology 146, 781–792 (2021). [DOI] [PubMed] [Google Scholar]
- 131.Bertholet AM et al. Mitochondrial uncouplers induce proton leak by activating AAC and UCP1. Nature 606, 180–187 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pickles S, Vigié P & Youle RJ Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol 28, R170–R185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Killackey SA, Philpott DJ & Girardin SE Mitophagy pathways in health and disease. J. Cell Biol 10.1083/jcb.202004029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kast DJ & Dominguez R The cytoskeleton-autophagy connection. Curr. Biol 27, R318–R326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu X & Mullins RD LC3 and STRAP regulate actin filament assembly by JMY during autophagosome formation. J. Cell Biol 218, 251–266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kruppa AJ et al. Myosin VI-dependent actin cages encapsulate parkin-positive damaged mitochondria. Dev. Cell 44, 484–499.e6 (2018). First report of the process we call PDA.
- 137. Chakrabarti R. et al. Mitochondrial dysfunction triggers actin polymerization necessary for rapid glycolytic activation. J. Cell Biol 10.1083/jcb.202201160 (2022). This paper links peri-mitochondrial actin polymerization to glycolytic regulation upon both actute mitochondrial damage (ADA) and chronic mitochondrial dysfunction.
- 138.Hu H. et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164, 433–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Park JS et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 578, 621–626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kondo H. et al. Single-cell resolved imaging reveals intra-tumor heterogeneity in glycolysis, transitions between metabolic states, and their regulatory mechanisms. Cell Rep. 34, 108750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hsieh C-W & Yang WY Omegasome-proximal PtdIns(4,5)P2 couples F-actin mediated mitoaggregate disassembly with autophagosome formation during mitophagy. Nat. Commun 10, 969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rivers E. et al. Wiskott Aldrich syndrome protein regulates non-selective autophagy and mitochondrial homeostasis in human myeloid cells. eLife 10.7554/eLife.55547 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mathiowetz AJ et al. An Amish founder mutation disrupts a PI(3)P-WHAMM-Arp2/3 complex-driven autophagosomal remodeling pathway. Mol. Biol. Cell 28, 2492–2507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dai A, Yu L & Wang HW WHAMM initiates autolysosome tubulation by promoting actin polymerization on autolysosomes. Nat. Commun 10, 3699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Müller PM et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat. Cell Biol 22, 498–511 (2020). [DOI] [PubMed] [Google Scholar]
- 146.Pekkurnaz G, Trinidad JC, Wang X, Kong D & Schwarz TL Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 158, 54–68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Basu H. et al. FHL2 anchors mitochondria to actin and adapts mitochondrial dynamics to glucose supply. J. Cell Biol 10.1083/jcb.201912077 (2021). Actin-mediated mitochondrial motility arrest in neurons, triggered by increased cytoplasmic glucose.
- 148.Shneyer BI, Ušaj M, Wiesel-Motiuk N, Regev R & Henn A ROS induced distribution of mitochondria to filopodia by Myo19 depends on a class specific tryptophan in the motor domain. Sci. Rep 7, 11577 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shneyer BI, Ušaj M & Henn A Myo19 is an outer mitochondrial membrane motor and effector of starvation-induced filopodia. J. Cell Sci 129, 543–556 (2016). [DOI] [PubMed] [Google Scholar]
- 150.Jiao H. et al. Mitocytosis, a migrasome-mediated mitochondrial quality-control process. Cell 184, 2896–2910.e13 (2021). [DOI] [PubMed] [Google Scholar]
- 151.Rohn JL et al. Myo19 ensures symmetric partitioning of mitochondria and coupling of mitochondrial segregation to cell division. Curr. Biol 24, 2598–2605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Majstrowicz K. et al. Coordination of mitochondrial and cellular dynamics by the actin-based motor Myo19. J. Cell Sci 10.1242/jcs.255844 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shi P. et al. Mechanical instability generated by Myosin 19 contributes to mitochondria cristae architecture and OXPHOS. Nat. Commun 13, 2673 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bocanegra JL et al. The MyMOMA domain of MYO19 encodes for distinct Miro-dependent and Miro-independent mechanisms of interaction with mitochondrial membranes. Cytoskeleton 77, 149–166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lopez-Domenech G. et al. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 37, 321–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Oeding SJ et al. Identification of Miro1 and Miro2 as mitochondrial receptors for myosin XIX. J. Cell Sci 10.1242/jcs.219469 (2018). [DOI] [PubMed] [Google Scholar]
- 157.Pathak D, Sepp KJ & Hollenbeck PJ Evidence that myosin activity opposes microtubule-based axonal transport of mitochondria. J. Neurosci 30, 8984–8992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li S, Xiong GJ, Huang N & Sheng ZH The cross-talk of energy sensing and mitochondrial anchoring sustains synaptic efficacy by maintaining presynaptic metabolism. Nat. Metab 2, 1077–1095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kruppa AJ & Buss F Motor proteins at the mitochondria-cytoskeleton interface. J. Cell Sci 10.1242/jcs.226084 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chakrabarti R, Lee M & Higgs HN Multiple roles for actin in secretory and endocytic pathways. Curr. Biol 31, R603–R618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lamason RL & Welch MD Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr. Opin. Microbiol 35, 48–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boldogh IR et al. Arp2/3 complex and actin dynamics are required for actin-based mitochondrial motility in yeast. Proc. Natl Acad. Sci. USA 98, 3162–3167 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Moore AS, Wong YC, Simpson CL & Holzbaur EL Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun 7, 12886 (2016). Shows first evidence of cycling waves of actin clouds around populations of mitochondria.
- 164.Danial NN et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424, 952–956 (2003). [DOI] [PubMed] [Google Scholar]
- 165.Cheng A. et al. Pancortin-2 interacts with WAVE1 and Bcl-xL in a mitochondria-associated protein complex that mediates ischemic neuronal death. J. Neurosci 27, 1519–1528 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kang R. et al. WAVE1 regulates Bcl-2 localization and phosphorylation in leukemia cells. Leukemia 24, 177–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.King VL, Leclair NK, Coulter AM & Campellone KG The actin nucleation factors JMY and WHAMM enable a rapid Arp2/3 complex-mediated intrinsic pathway of apoptosis. PLoS Genet. 17, e1009512 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chua BT et al. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol 5, 1083–1089 (2003). [DOI] [PubMed] [Google Scholar]
- 169.Rehklau K. et al. ADF/cofilin proteins translocate to mitochondria during apoptosis but are not generally required for cell death signaling. Cell Death Differ. 19, 958–967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rehklau K. et al. Cofilin1-dependent actin dynamics control DRP1-mediated mitochondrial fission. Cell Death Dis. 8, e3063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li GB et al. Mitochondrial fission and mitophagy depend on cofilin-mediated actin depolymerization activity at the mitochondrial fission site. Oncogene 37, 1485–1502 (2018). [DOI] [PubMed] [Google Scholar]
- 172.Li GB et al. Mitochondrial translocation of cofilin is required for allyl isothiocyanate-mediated cell death via ROCK1/PTEN/PI3K signaling pathway. Cell Commun. Signal 11, 50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Klamt F. et al. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat. Cell Biol 11, 1241–1246 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wabnitz GH et al. Mitochondrial translocation of oxidized cofilin induces caspase-independent necrotic-like programmed cell death of T cells. Cell Death Dis. 1, e58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hoffmann L. et al. Cofilin1 oxidation links oxidative distress to mitochondrial demise and neuronal cell death. Cell Death Dis. 12, 953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vögtle FN et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 (2009). [DOI] [PubMed] [Google Scholar]
- 177.Calvo SE et al. Comparative analysis of mitochondrial N-termini from mouse, human, and yeast. Mol. Cell. Proteom 16, 512–523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fukasawa Y. et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom 14, 1113–1126 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bykov YS et al. Widespread use of unconventional targeting signals in mitochondrial ribosome proteins. EMBO J. 41, e109519 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chatterjee A. et al. MOF Acetyl transferase regulates transcription and respiration in mitochondria. Cell 167, 722–738.e3 (2016). [DOI] [PubMed] [Google Scholar]
- 181.Reyes A. et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 39, 5098–5108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xie X, Venit T, Drou N & Percipalle P In Mitochondria? — Actin regulates mtDNA transcription and is required for mitochondrial quality control. iScience 3, 226–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Takahashi K, Miura Y, Ohsawa I, Shirasawa T & Takahashi M In vitro rejuvenation of brain mitochondria by the inhibition of actin polymerization. Sci. Rep 8, 15585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ulferts S, Prajapati B, Grosse R & Vartiainen MK Emerging properties and functions of actin and actin filaments inside the nucleus. Cold Spring Harb. Perspect. Biol 10.1101/cshperspect.a040121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lehrer SS Damage to actin filaments by glutaraldehyde: protection by tropomyosin. J. Cell Biol 90, 459–466 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Maupin P & Pollard TD Improved preservation and staining of HeLa cell actin filaments, clathrin-coated membranes, and other cytoplasmic structures by tannic acid-glutaraldehyde-saponin fixation. J. Cell Biol 96, 51–62 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kudryashev M, Lepper S, Baumeister W, Cyrklaff M & Frischknecht F Geometric constrains for detecting short actin filaments by cryogenic electron tomography. PMC Biophys. 3, 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Svitkina T. Imaging cytoskeleton components by electron microscopy. Methods Mol. Biol 2364, 25–52 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Edwards M. et al. Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol 15, 677–689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bombardier JP et al. Single-molecule visualization of a formin-capping protein ‘decision complex’ at the actin filament barbed end. Nat. Commun 6, 8707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shekhar S. et al. Formin and capping protein together embrace the actin filament in a ménage à trois. Nat. Commun 6, 8730 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L & Condeelis J Functions of cofilin in cell locomotion and invasion. Nat. Rev. Mol. Cell Biol 14, 405–415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kotila T. et al. Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat. Commun 10, 5320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shekhar S, Chung J, Kondev J, Gelles J & Goode BL Synergy between cyclase-associated protein and Cofilin accelerates actin filament depolymerization by two orders of magnitude. Nat. Commun 10, 5319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tang VW, Nadkarni AV & Brieher WM Catastrophic actin filament bursting by cofilin, Aip1, and coronin. J. Biol. Chem 295, 13299–13313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.King ZT et al. Coro1B and Coro1C regulate lamellipodia dynamics and cell motility by tuning branched actin turnover. J. Cell Biol 10.1083/jcb.202111126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cai L, Makhov AM, Schafer DA & Bear JE Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell 134, 828–842 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hakala M. et al. Twinfilin uncaps filament barbed ends to promote turnover of lamellipodial actin networks. Nat. Cell Biol 23, 147–159 (2021). [DOI] [PubMed] [Google Scholar]
- 199.Shekhar S, Hoeprich GJ, Gelles J & Goode BL Twinfilin bypasses assembly conditions and actin filament aging to drive barbed end depolymerization. J. Cell Biol 10.1083/jcb.202006022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Svitkina TM & Borisy GG Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol 145, 1009–1026 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fehon RG, McClatchey AI & Bretscher A Organizing the cell cortex: the role of ERM proteins. Nat. Rev. Mol. Cell Biol 11, 276–287 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gunning PW, Hardeman EC, Lappalainen P & Mulvihill DP Tropomyosin — master regulator of actin filament function in the cytoskeleton. J. Cell Sci 128, 2965–2974 (2015). [DOI] [PubMed] [Google Scholar]
- 203.Ghosh A & Fowler VM Tropomodulins. Curr. Biol 31, R501–R503 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.