SUMMARY
C9orf72 repeat expansions are the most common genetic cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). Poly(GR) proteins are toxic to neurons by forming cytoplasmic inclusions that sequester RNA-binding proteins including stress granule (SG) proteins. However, little is known of the factors governing poly(GR) inclusion formation. Here, we show that poly(GR) infiltrates a finely tuned network of protein-RNA interactions underpinning SG formation. It interacts with G3BP1, the key driver of SG assembly and a protein we found is critical for poly(GR) inclusion formation. Moreover, we discovered that N6-methyladenosine (m6A)-modified mRNAs and m6A-binding YTHDF proteins not only co-localize with poly(GR) inclusions in brains of c9FTD/ALS mouse models and patients with c9FTD, they promote poly(GR) inclusion formation via the incorporation of RNA into the inclusions. Our findings thus suggest that interrupting interactions between poly(GR) and G3BP1 or YTHDF1 proteins or decreasing poly(GR) altogether represent promising therapeutic strategies to combat c9FTD/ALS pathogenesis.
In brief
Park et al. show that poly(GR) and G3BP1 interactions initiate toxic poly(GR) inclusion formation. The recruitment of N6-methyladenosine (m6A)-modified mRNAs and m6A-binding YTHDF proteins potentiates inclusion formation. Interrupting these protein-RNA interactions may represent a therapeutic strategy to combat the most common genetic forms of frontotemporal dementia and amyotrophic lateral sclerosis.
Graphical abstract
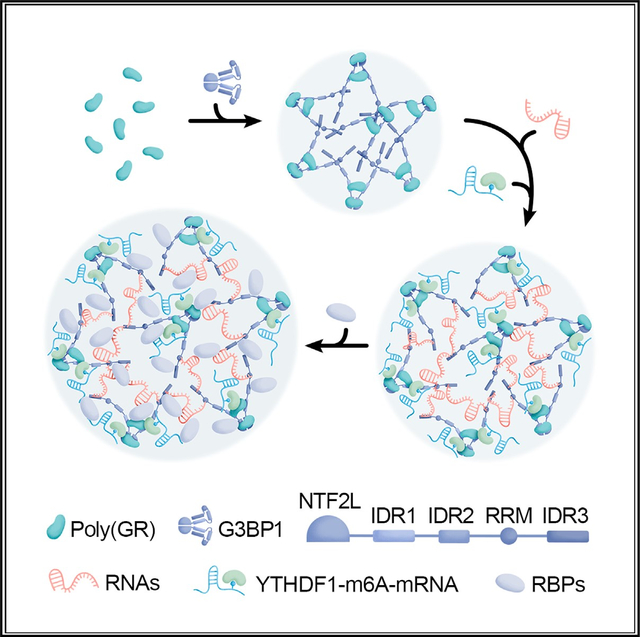
INTRODUCTION
A G4C2 repeat expansion in chromosome 9 open reading frame 72 (C9orf72) is the most common genetic cause of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS), two fatal neurodegenerative diseases. While it remains unclear how C9orf72 repeat expansions cause FTD and ALS (c9FTD/ALS), mounting evidence indicates that dipeptide repeat (DPR) proteins atypically translated from the expanded repeat play a role. Inclusions composed of DPR proteins are considered a pathognomonic feature of c9FTD/ALS, and preclinical studies spanning multiple models have shown that the positively charged arginine-rich DPR proteins, poly(GR) and poly(PR), are especially toxic.1–5 They trigger nucleolar toxicity, inhibit protein synthesis, impair ribosomal RNA processing and ribosome biogenesis, interact with RNA-binding proteins, and alter the liquid-liquid phase separation of various membraneless organelles.1–8 For example, we recently reported that cytoplasmic poly(GR) inclusions sequester RNA-binding proteins like TDP-43 and stress granule (SG) proteins.9 Not only may this promote the formation of TDP-43 inclusions, a pathological hallmark of c9FTD/ALS, it may also impair SG biology. SGs are dynamic, phase-separated, cytoplasmic membraneless assemblies composed of mRNA and protein that form in response to cellular stress to promote cell survival. Because a cell’s ability to mount an effective stress response is paramount to its health, the sequestration of SG proteins by poly(GR) inclusions is expected to have dire consequences. As such, we investigated key factors driving the formation of cytoplasmic poly(GR) inclusions with the purpose of elucidating the molecular mechanism underpinning this detrimental phenomenon.
On the basis of recent reports that G3BP1 and G3BP2 proteins are essential for arsenite-induced SG assembly,10–12 and based on our findings that cytoplasmic poly(GR) inclusions contain SG proteins, we examined whether G3BP1/2 participate in poly(GR) inclusion formation. In addition, since N6-methyladenosine (m6A)-modified mRNAs are enriched in SGs, and since m6A-binding YTHDF proteins are critical for SG formation,13,14 we investigated whether they too regulate the formation of cytoplasmic poly(GR) inclusions. m6A, the most common post-transcriptional RNA modification, participates in multiple RNA metabolism pathways such as RNA splicing, localization, translation, and stability. Of note, mounting evidence suggests that m6A-modified RNAs and m6A-binding proteins are involved in neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease.15–18 For instance, it was recently shown that interactions of tau with HNRNPA2B1 and m6A-modified RNA mediates tauopathy progression.19
Here, we demonstrate that cytoplasmic poly(GR) inclusions are not formed haphazardly. Rather, poly(GR) infiltrates itself into a well-described network of protein-RNA interactions having as a central node the G3BP1 protein.10–12 Indeed, we found that poly(GR) interacts with G3BP1 and triggers RNA recruitment. Furthermore, we observed that m6A-modified RNA and YTHDF proteins co-localize with cytoplasmic poly(GR) inclusions in mammalian cells, mice, and human patients with c9FTD, and we show that m6A-modified RNA and YTHDF proteins potentiate the formation of cytoplasmic poly(GR) inclusions via the incorporation of mRNA into the inclusions. Overall, our findings suggest that the formation of cytoplasmic poly(GR) inclusions is achieved by usurping otherwise finely tuned mechanisms underpinning conventional SG formation.
RESULTS
G3BP1 and G3BP2 are critical for cytoplasmic poly(GR) inclusion formation
Given that cytoplasmic poly(GR) inclusions contain SG proteins,4,9,20 and that G3BP1 and G3BP2 (G3BP1/2) trigger arsenite-induced SG assembly,10–12 we investigated whether G3BP1/2 similarly play a critical role in cytoplasmic poly(GR) inclusion formation. In wild-type U2OS cells expressing green fluorescent protein (GFP)-tagged (GR)100, large, cytoplasmic poly(GR) inclusions immunopositive for G3BP1 or other SG proteins (ataxin 2, eIF3h, or TIA-1) were seen in approximately 22% of GFP-(GR)100-positive cells (Figures 1A, 1B, and S1A). In contrast, cytoplasmic poly(GR) inclusions were virtually absent in G3BP1/2 knockout U2OS cells expressing GFP-(GR)100 (Figures 1A, 1B, and S1A). Notably, unlike cytoplasmic poly(GR) inclusions, nucleolar poly(GR) accumulation was not affected by G3BP1/2 knockout (Figures 1A and S1A). In addition, G3BP1/2 knockout did not affect the formation of poly(GA) inclusions in U2OS cells expressing GFP-(GA)100 (Figures S1B and S1C). Since G3BP1/2 proteins are essential for poly(GR) inclusion formation, we examined how they mediate this process. The G3BP1 protein harbors a nuclear transport factor 2-like (NTF2L) domain suggested to play a role in protein-protein interactions including G3BP1 dimerization, an RNA recognition motif (RRM), and three distinct intrinsically disordered regions (IDRs) that regulate G3BP1 liquid-liquid phase separation (LLPS) propensity (Figure S1D). Under physiological conditions, G3BP1 reportedly adopts a closed conformation through electrostatic interactions of the negatively charged IDR1 and the positively charged IDR3.11,12 However, in response to ARS treatment, polysome disassembly causes an increase in free, negatively charged cytosolic RNA that competes with the IDR1 for binding to the IDR3.11,12 These G3BP1-RNA complexes are believed to promote LLPS and SG assembly.11,12 Given that poly(GR) is highly positively charged, we hypothesized that poly(GR) binds to the negatively charged IDR1 domain of G3BP1, thus promoting cytoplasmic poly(GR) inclusion formation. To test this possibility, we generated cell lines stably expressing different mCherry-tagged G3BP1 species (i.e., full-length G3BP1 [FL] or G3BP1 with a deleted [D] NTF2L, IDR1, IDR3, or RRM) under a G3BP1/2 knockout background. Immunoblot analysis confirmed that these exogenous G3BP1 species were expressed comparably in cells (Figure S1E). Like endogenous G3BP1,10,11 immunofluorescence analysis showed that they were diffusely distributed throughout the cytoplasm under basal conditions (Figure S1F). Using these cell lines, we conducted a proximity ligation assay (PLA) in which a fluorescent PLA signal is indicative of GFP-(GR)100 binding to a given G3BP1 species. Compared to the mCherry control, a significant increase in PLA fluorescence was detected within the cytosol and in poly(GR) inclusions in cells expressing mCherry-tagged FL G3BP1 (Figures 1C and 1D). Of note, deletion of IDR1, the domain we hypothesized to bind poly(GR), significantly decreased the PLA signal compared to that in cells expressing FL G3BP1 (Figures 1C and 1D), indicating that ΔIDR1 interacts poorly with poly(GR). The poly(GR)-G3BP1 interaction was also decreased when deleting the NTF2L domain of G3BP1 but not the IDR3 or RRM domain (Figures 1C and 1D). These results indicate that NTF2L and IDR1 domains mediate poly(GR)-G3BP1 interaction.
Figure 1. Poly(GR) inclusion formation requires its interaction with G3BP1 and RNA recruitment.
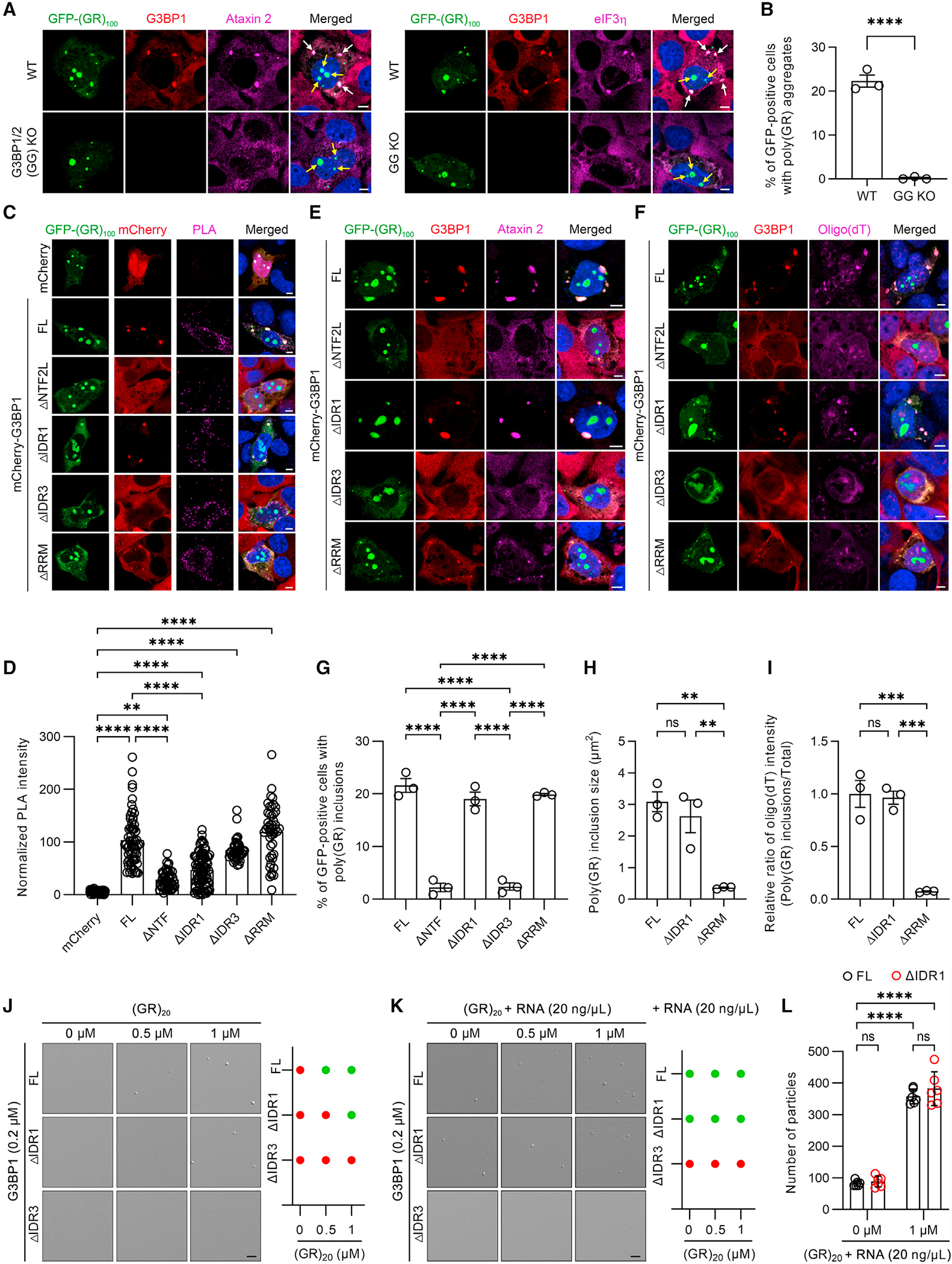
(A) Double-immunofluorescence staining for G3BP1 and either ataxin 2 or eIF3h in wild-type (WT) or G3BP1/2 knockout (GG KO) U2OS cells expressing GFP-(GR)100 48 h post transfection. White arrows indicate cytoplasmic poly(GR) inclusions, and yellow arrows indicate nucleolar poly(GR) accumulation. Scale bars, 5 μm.
(B) Quantification of the percentage of cells containing poly(GR) inclusions in WT or GG KO U2OS cells expressing GFP-(GR)100 48 h post transfection (n = 3 independent experiments).
(C) Representative proximity ligation assay (PLA) images for GFP-(GR)100 and mCherry in GG KO U2OS cells co-expressing GFP-(GR)100 and mCherry or for GFP-(GR)100 and mCherry-G3BP1 species in GG KO U2OS cells stably expressing mCherry-G3BP1 species. The PLA signal is indicative of these protein interactions. Scale bars, 5 μm.
(D) Quantification of the intensity for PLA signal in GG KO U2OS cells co-expressing GFP-(GR)100 and mCherry or for GFP-(GR)100 and mCherry-G3BP1 species in GG KO U2OS cells stably expressing mCherry-G3BP1 species (n = 38–87 cells).
(E) Double-immunofluorescence staining for G3BP1 and ataxin 2 in GG KO U2OS cells expressing GFP-(GR)100 and stably expressing mCherry-G3BP1 species. Scale bars, 5 μm.
(F) Immunofluorescence staining for G3BP1 followed by RNA-FISH for oligo(dT) in GG KO U2OS cells expressing GFP-(GR)100 and stably expressing mCherry-G3BP1 species. Scale bars, 5 μm.
(G) Quantification of the percentage of poly(GR)-positive cells containing poly(GR) inclusions in GG KO U2OS cells stably expressing mCherry-G3BP1 species (n = 3 independent experiments).
(H) Quantification of the size of poly(GR) inclusions in GG KO U2OS cells stably expressing mCherry-G3BP1 species (n = 3 independent experiments).
(I) Quantification of the relative ratio of oligo(dT) intensity (poly(GR) inclusions/total) in GG KO U2OS cells stably expressing mCherry-G3BP1 species (n = 3 independent experiments).
(J) Representative images (left) and phase diagram (right) of 0.2 μM of the indicated recombinant G3BP1 protein species mixed with (GR)20 peptides (0–1 μM). Scale bars, 5 μm.
(K) Representative images (left) and phase diagram (right) of 0.2 μM recombinant G3BP1 protein species mixed with (GR)20 peptides (0–1 μM) and total RNA (20 ng/μL). Scale bars, 5 μm.
(L) Quantification of the numbers of droplets of the indicated recombinant G3BP1 protein species mixed with (GR)20 peptides (0, 1 μM) and total RNA (20 ng/μL) (n = 6 regions).
Data represent the mean ± SEM. In (B), ****p < 0.0001, unpaired two-tailed t test. In (D), **p = 0.0032 and ****p < 0.0001, one-way ANOVA, Tukey’s post hoc analysis. In (G), ****p < 0.0001, one-way ANOVA, Tukey’s post hoc analysis. In (H), ns (not significant) p = 0.6381, ** (left to right) p = 0.0036 and p = 0.0089, one-way ANOVA, Tukey’s post hoc analysis. In (I), ns = 0.9485, *** (left to right) p = 0.0005 and p = 0.0006, one-way ANOVA, Tukey’s post hoc analysis. In (L), (left to right) ns = 0.9884 and ns = 0.5207, ****p < 0.0001, two-way ANOVA, Tukey’s post hoc analysis.
To study the G3BP1 domains important for poly(GR) inclusion formation, we expressed GFP-(GR)100 in cell lines stably expressing different mCherry-tagged G3BP1 species under a G3BP1/2 knockout background. We found that ΔNTF2L and ΔIDR3 failed to rescue cytoplasmic GFP-(GR)100 inclusion formation, whereas FL, ΔIDR1, or ΔRRM G3BP1 did rescue the formation of GFP-(GR)100 inclusions (Figures 1C, 1E–1G, S1G, and S1H). Regarding the latter, it must nonetheless be noted that the size of cytoplasmic poly(GR) inclusions was significantly decreased in ΔRRM-expressing G3BP1/2 knockout cells compared to G3BP1/2 knockout cells expressing FL or ΔIDR1 (Figures 1C, 1E, 1F, 1H, S1G, and S1H). Given that the RRM domain of G3BP1 modulates the size of cytoplasmic poly(GR) inclusions, we investigated whether RNA recruitment is critical for maintaining poly(GR) inclusion size by probing for oligo(dT) in poly(GR) inclusions using fluorescence in situ hybridization. In G3BP1/2 knockout cell lines stably expressing FL, ΔIDR1, or ΔRRM, oligo(dT)-positive mRNA co-localized with cytoplasmic GFP-(GR)100 inclusions (Figure 1F); however, compared to the FL and ΔIDR1 cell lines, the oligo(dT) fluorescence intensity within the inclusions vs. within the whole cell was significantly decreased in ΔRRM-expressing cells (Figure 1I). We also noted that, while ΔIDR1 interacted less strongly with poly(GR) in comparison to FL G3BP1, it nonetheless rescued poly(GR) inclusion formation to a similar degree as FL G3BP1. In contrast, while ΔIDR3 interacted with poly(GR) at a similar level as FL G3BP1, it failed to rescue cytoplasmic GFP-(GR)100 inclusion formation. To elucidate the potential mechanism for this phenomenon, we tested whether poly(GR) peptides influence ΔIDR1 and ΔIDR3 LLPS given recent in vitro studies showing that poly(GR) peptides induce recombinant FL G3BP1 protein LLPS.21 We found that synthetic (GR)20 peptides, at either 0.5 or 1 μM, caused FL G3BP1 to phase separate into condensates (Figure 1J). However, ΔIDR1 G3BP1 condensate formation was only observed in the presence of 1 μM (GR)20 peptides (Figure 1J), indicating that deleting IDR1 attenuates poly(GR)-mediated LLPS of G3BP1, which is likely due to the decreased interaction of ΔIDR1 with (GR)20 peptides. We also observed that ΔIDR3 G3BP1 failed to form condensates at either (GR)20 peptide concentration, indicating that IDR3 is required for poly(GR)-mediated LLPS of G3BP1 (Figure 1J). Given that deleting IDR1 from G3BP1 facilitates RNA recruitment and thereby induces LLPS of G3BP1,11 we tested whether the addition of RNA would rescue poly(GR)-induced ΔIDR1 condensate formation. Consistent with prior findings,11 we found that adding RNA induced LLPS of FL and ΔIDR1 in the absence of (GR)20 peptides (Figure 1K). Moreover, adding RNA rescued ΔIDR1 condensate formation in the presence of 0.5 μM (GR)20 peptide (Figure 1K). Compared to RNA alone, adding (GR)20 peptides along with RNA significantly enhanced the number of FL and ΔIDR1 condensates (Figure 1L). As the number of ΔIDR1 and FL G3BP1 condensates were comparable, ΔIDR1 and FL G3BP1 exhibit similar LLPS activity in the presence of both RNA and (GR)20 peptides. In contrast, the addition of RNA failed to rescue poly(GR)-induced LLPS of ΔIDR3 G3BP1 because the IDR3 domain mediates G3BP1 binding to RNA (Figure 1K).11 Collectively, our results suggest that poly(GR) interacts with the NTF2L and IDR1 domains of G3BP1, and the resulting complex recruits RNA and initiates cytoplasmic poly(GR) inclusion formation. In addition, NTF2L and/or IDR3 domains are required for poly(GR) inclusion formation, and RNA recruitment is also critical for maintaining proper poly(GR) inclusion size.
m6A-modified RNA and the m6A-binding YTHDF1 protein co-localize with poly(GR) inclusions in vitro and in vivo
On the basis of our finding that RNA is recruited to cytoplasmic poly(GR) inclusions via the SG protein G3BP1, we investigated the role of m6A-modified RNA and m6A-binding YTHDF proteins in poly(GR) inclusion formation. Both m6A-modified RNA—the most abundant post-transcriptional mRNA modification—and YTHDF proteins are enriched in SGs.22,23 Indeed, m6A-modified RNA potentiates the condensation of YTHDF proteins and guides them to SGs, thus contributing to mRNA partitioning in SGs. We thus examined whether these SG components are enriched in cytoplasmic poly(GR) inclusions in vitro and in vivo. In HEK293T cells expressing GFP-(GR)100, we observed cytoplasmic poly(GR) inclusions immunopositive for G3BP1 both 24 and 48 h post transfection with the number and size of poly(GR) inclusions being greater at the 48-h time point (Figures S2A–S2C). Of note, YTHDF1 co-localized with cytoplasmic poly(GR) inclusions but not with nucleolar poly(GR) (Figure S2A), and m6A-modified RNA was also enriched in cytoplasmic poly(GR) inclusions (Figures S2D and S2E).
We subsequently examined whether mice expressing 149 G4C2 repeats, which develop DPR protein pathology in the brain, show YTHDF1 or m6A-modified RNA aberrations using anti-YTHDF1 and anti-m6A antibodies, respectively. Immunohistochemical analysis revealed diffuse, cytoplasmic YTHDF1 in control mice expressing only two G4C2 repeats, whereas cytoplasmic inclusions immunopositive for YTHDF1 were seen in the cortex of (G4C2)149 mice (Figure 2A). Immunofluorescence studies confirmed that these YTHDF1-containing inclusions were immunopositive for poly(GR) (Figure 2B). Moreover, cytoplasmic poly(GR) inclusions in (G4C2)149 mice additionally contained m6A-modified RNA (Figure 2C). To determine whether YTHDF1 and m6A specifically co-localize to poly(GR) inclusions, we performed co-immunofluorescence staining for YTHDF1 or m6A using mice transduced to express GFP-(GR)200 or (GA)100-V5 in the brain. As observed in (G4C2)149 mice, YTHDF1 and m6A-modified RNA co-localized with cytoplasmic poly(GR) inclusions in the cortex of GFP-(GR)200 mice. However, no YTHDF1- or m6A-positive inclusions were seen in cells with only diffuse poly(GR) (Figures 2D–2G). In contrast to our findings in GFP-(GR)200 mice, YTHDF1 and m6A did not co-localize with cytoplasmic poly(GA) inclusions in (GA)100-V5 mice (Figures 2H and 2I). Finally, we confirmed that m6A-modified RNA and YTHDF1 were enriched in cytoplasmic poly(GR) inclusions in postmortem brain tissues from patients with c9FTD (Figures 2J and 2K). Collectively, our data demonstrate that YTHDF1 and m6A-modified RNA co-localize with cytoplasmic poly(GR) inclusions.
Figure 2. YTHDF1 and m6A-modified RNAs co-localize with cytoplasmic poly(GR) inclusions.
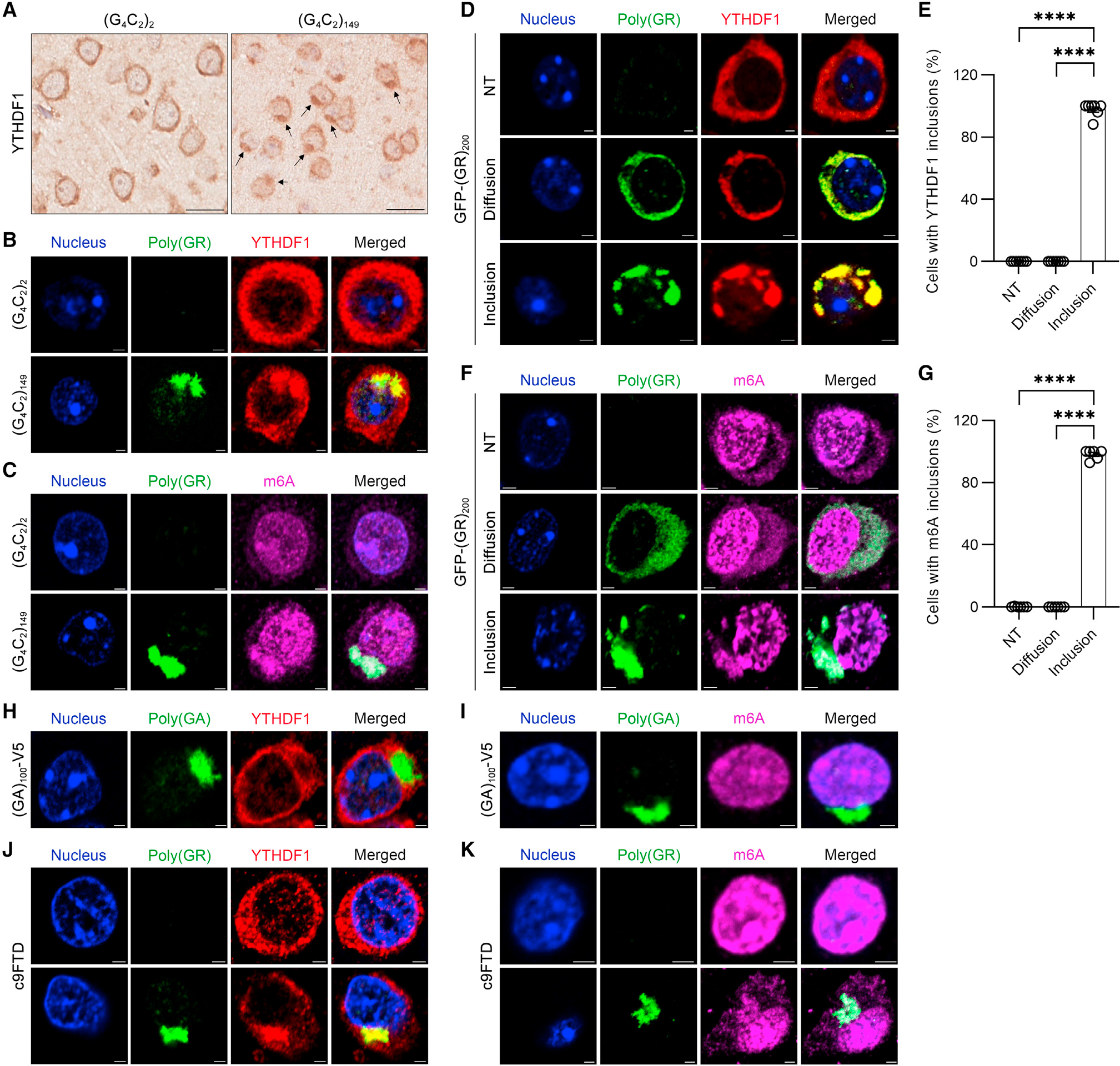
(A) Representative images of immunohistochemical analysis of YTHDF1 in the cortex of 12-month-old (G4C2)2 or (G4C2)149 mice (n = 6 per group). Black arrows indicate inclusions. Scale bars, 20 μm.
(B) Double-immunofluorescence staining for poly(GR) and YTHDF1 in the cortex of 12-month-old (G4C2)2 or (G4C2)149 mice (n = 6 per group). Scale bars, 2 μm.
(C) Double-immunofluorescence staining for poly(GR) and m6A-modified RNAs in the cortex of 12-month-old (G4C2)2 or (G4C2)149 mice. Scale bars, 2 μm.
(D) Double-immunofluorescence staining for poly(GR) and YTHDF1 in the cortex of 2-week-old GFP-(GR)200 mice. Scale bars, 2 μm. NT, non-transduced cells; Diffusion, cells with diffuse poly(GR); Inclusion, cells with poly(GR) inclusions.
(E) Quantification of the percentage of NT cells and transduced cells with diffuse poly(GR) or poly(GR) inclusions with YTHDF1 inclusions (n = 6).
(F) Double-immunofluorescence staining for poly(GR) and m6A-modified RNAs in the cortex of 2-week-old GFP-(GR)200 mice. Scale bars, 2 μm.
(G) Quantification of the percentage of NT cells or transduced cells with diffuse poly(GR) or poly(GR) inclusion with m6A-modified RNA-containing inclusions (n = 6).
(H) Double-immunofluorescence staining for poly(GA) and YTHDF1 in the cortex of 3.5-month-old (GA)100-V5 mice (n = 3). Scale bars, 2 μm.
(I) Double-immunofluorescence staining for poly(GA) and m6A-modified RNAs in the cortex of 3.5-month-old (GA)100-V5 mice (n = 3). Scale bars, 2 μm.
(J) Double-immunofluorescence staining for poly(GR) and YTHDF1 in the mid-frontal cortex of patients with c9FTD. The intracellular localization of YTHDF1 is shown for cells without or with poly(GR) inclusions (n = 6). Scale bars, 2 μm.
(K) Double-immunofluorescence staining for poly(GR) and m6A-modified RNAs in the mid-frontal cortex of patients with c9FTD. The intracellular localization of m6A-modified RNA is shown for cells without and with poly(GR) inclusions (n = 6). Scale bars, 2 μm.
Data are shown as the mean ± SEM. In (E), ****p < 0.0001, one-way ANOVA, Tukey’s post hoc analysis. In (G), ****p < 0.0001, one-way ANOVA, Tukey’s post hoc analysis.
m6A-binding YTHDF proteins enhance poly(GR) inclusion formation
Given that m6A-binding YTHDF proteins and m6A-modified RNA co-localize with cytoplasmic poly(GR) inclusions, we evaluated whether YTHDF proteins and/or m6A-modified RNA influence poly(GR) inclusion formation. To do so, we first depleted YTHDF1 and YTHDF3 in HEK293T cells expressing GFP-(GR)100 (Figure S3A), and we assessed poly(GR) expression and inclusion formation 48 h after transfection. Even though YTHDF proteins are involved in protein synthesis,24,25 knocking down both YTHDF1 and YTHDF3 did not alter poly(GR) expression as determined by immunoassay (Figure S3B). However, we did observe significant decreases in the number of GFP-positive cells containing cytoplasmic poly(GR) inclusions and in inclusion size when YTHDF1 and YTHDF3 were knocked down (Figures 3A–3C). In contrast, neither the number of GFP-positive cells containing cytoplasmic poly(GA) inclusions nor inclusion size was affected by knockdown of YTHDF1 and YTHDF3 (Figures S3C–S3E). We next overexpressed V5-tagged YTHDF1 in HEK293T cells co-expressing either GFP or GFP-(GR)100 for 24 h (Figure S3F). Consistent with our data above, modulating YTHDF1 expression did not change GFP-(GR)100 abundance (Figure S3G). We did nonetheless see significant increases in the number of GFP-positive cells containing cytoplasmic poly(GR) inclusions and an increase in inclusion size in V5- and GFP-positive cells when YTHDF1 was overexpressed (Figures 3D–3F). These data indicate that YTHDF proteins promote poly(GR) inclusion formation without altering poly(GR) protein expression.
Figure 3. YTHDF proteins and m6A-modified RNAs promote cytoplasmic poly(GR) inclusion formation.
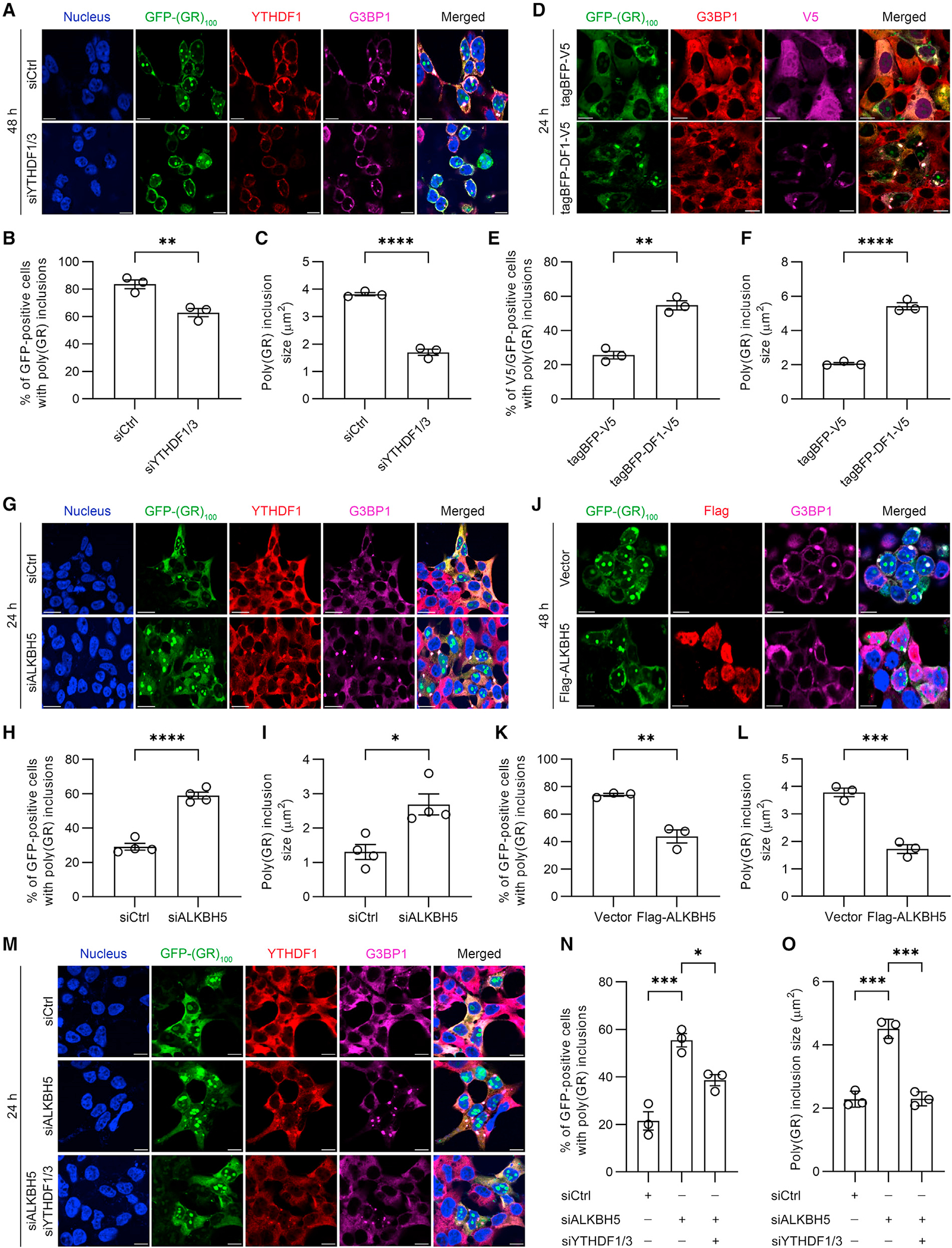
(A) Double-immunofluorescence staining for YTHDF1 and G3BP1 in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 48 h post transfection. Scale bars, 10 μm.
(B) Quantification of the percentage of GFP-positive cells containing poly(GR) inclusions in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 (n = 3 independent experiments).
(C) Quantification of the size of poly(GR) inclusions in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 (n = 3 independent experiments).
(D) Double-immunofluorescence staining for V5 and G3BP1 in HEK293T cells co-expressing GFP-(GR)100 and either tagBFP-V5 or tagBFP/V5 tagged wild-type YTHDF1 (tagBFP-DF1-V5). Scale bars, 10 μm.
(E) Quantification of the percentage of GFP-positive cells containing poly(GR) inclusions in HEK293T cells co-expressing GFP-(GR)100 and either tagBFP-V5 or tagBFP-DF1-V5 (n = 3 independent experiments).
(F) Quantification of the size of poly(GR) inclusions in GFP-(GR)100 overexpressing HEK293T cells co-expressing either tagBFP-V5 or tagBFP-DF1-V5 (n = 3 independent experiments).
(G) Double-immunofluorescence staining for YTHDF1 and G3BP1 in ALKBH5-depleted HEK293T cells expressing GFP-(GR)100. Staining was performed 24 h after GFP-(GR)100 transfection. Scale bars, 20 μm.
(H) Quantification of the percentage of GFP-positive cells containing poly(GR) inclusions in ALKBH5-depleted HEK293T cells expressing GFP-(GR)100 (n = 4 independent experiments).
(I) Quantification of the size of poly(GR) inclusions in ALKBH5-depleted HEK293T cells expressing GFP-(GR)100 (n = 4 independent experiments).
(J) Double-immunofluorescence staining for Flag and G3BP1 in Flag-ALKBH5 overexpressing HEK293T cells co-expressing GFP-(GR)100. Scale bars, 20 μm.
(K) Quantification of the percentage of GFP-positive cells with poly(GR) inclusions in Flag-ALKBH5 overexpressing HEK293T cells co-expressing GFP-(GR)100 (n = 3 independent experiments).
(L) Quantification of the size of poly(GR) inclusions in Flag-ALKBH5 and GFP-(GR)100 co-expressing HEK293T cells (n = 3 independent experiments).
(M) Double-immunofluorescence staining for YTHDF1 and G3BP1 in HEK293T cells expressing GFP-(GR)100 in which only ALKBH5 was depleted or in which ALKBH5 and YTHDF1/3 were depleted. Scale bars, 20 μm.
(N) Quantification of the percentage of GFP-positive cells containing poly(GR) inclusions in HEK293T cells expressing GFP-(GR)100 in which only ALKBH5 was depleted or in which ALKBH5 and YTHDF1/3 were depleted (n = 3 independent experiments).
(O) Quantification of the size of poly(GR) inclusions in ALKBH5-depleted or ALKBH5 and YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 (n = 3 independent experiments).
Data are shown as mean ± SEM. In (B), **p = 0.0095, unpaired two-tailed t test. In (C), ****p < 0.0001, unpaired two-tailed t test. In (E), **p = 0.0011, unpaired two-tailed t test. In (F), ****p < 0.0001, unpaired two-tailed t test. In (H), ****p < 0.0001, unpaired two-tailed t test. In (I), *p = 0.0101, unpaired two-tailed t test. In (K), **p = 0.0033, unpaired two-tailed t test. In (L), ***p = 0.0007, unpaired two-tailed t test. In (N), ***p = 0.0006 and *p = 0.0193, one-way ANOVA, Tukey’s post hoc analysis. In (O), *** (left to right) p = 0.0001 and p = 0.0001, one-way ANOVA, Tukey’s post hoc analysis.
m6A-modified RNA promotes poly(GR) inclusion formation
To study the effect of m6A-modified RNA on poly(GR) inclusion formation, we knocked down the m6A demethylase, ALKBH5, in HEK293T cells expressing GFP-(GR)100 for 24 h (Figure S3H). While ALKBH5 depletion and the resulting increase in m6A-modified RNA (Figures S3I–S3K) did not alter GFP-(GR)100 expression (Figure S3L) nor the expression of YTHDF1 or YTHDF3 proteins (Figure S3H), it did increase both the number of GFP-positive cells containing cytoplasmic poly(GR) inclusions and inclusion size (Figures 3G–3I). In contrast, neither the number of GFP-positive cells containing cytoplasmic poly(GA) inclusions nor inclusion size was affected by ALKBH5 knockdown (Figures S3M–S3O). We next overexpressed Flag-tagged ALKBH5 in cells co-expressing GFP or GFP-(GR)100 for 48 h and found that m6A-modified RNA levels were decreased as were the number of cells with cytoplasmic poly(GR) inclusions and inclusion size despite no change in GFP-(GR)100, YTHDF1, or YTHDF3 expression (Figures 3J–3L and S3P–S3T). These results demonstrate that m6A-modified RNA enhances poly(GR) inclusion formation without altering GFP-(GR)100 or YTHDF protein abundance.
YTHDF proteins are required for m6A-modified RNA to promote poly(GR) inclusion formation
Given that m6A-modified RNA induces YTHDF protein condensation and partitioning to SGs,22,23 and that both m6A-modified RNA and YTHDF proteins enhance cytoplasmic poly(GR) inclusion formation, we investigated whether m6A-modified RNA and YTHDF proteins work in concert to promote cytoplasmic poly(GR) inclusion formation. Toward this end, we expressed GFP or GFP-(GR)100 in HEK293T cells either depleted only of ALKBH5 or depleted of ALKBH5, YTHDF1, and YTHDF3 (Figure S3U). As above, m6A-modified RNA was increased upon ALKBH5 depletion; however, additionally depleting YTHDF1 and YTHDF3 did not alter m6A-modified RNA abundance (Figure S3V). Although GFP-(GR)100 expression did not change among any of the tested conditions (Figure S3W), the number of GFP-positive cells containing cytoplasmic poly(GR) inclusions as well as inclusion size were significantly greater in cells depleted of ALKBH5 alone compared to control cells or cells depleted of ALKBH5, YTHDF1, and YTHDF3 (Figures 3M–3O). These results indicate that YTHDF proteins are required for m6A-modified RNA to promote cytoplasmic poly(GR) inclusion formation.
YTHDF proteins and m6A-modified RNA carry mRNA to poly(GR) inclusions
We show above that m6A-modified RNA is enriched in cytoplasmic poly(GR) inclusions as has been observed in SGs formed in response to oxidative stress (Figures 2C, 2F, 2G, S2D, and S2E).13,14 Since we demonstrated that RNA partitioning affects poly(GR) inclusions (Figures 1E–1I), we examined whether complexes of m6A-modified RNA and YTHDF proteins incorporate RNA into poly(GR) inclusions. To test this, we fist examined whether the binding of m6A-modified RNA to YTHDF proteins is required for YTHDF proteins to interact with poly(GR). To do so, we performed PLA studies in HEK293T cells expressing GFP-(GR)100 and either wild-type YTHDF1 (YTHDF1-WT) or YTHDF1 with a mutation in the YTH domain that disrupts m6A-modified RNA binding (YTHDF1-mut) (Figure 4A).26 Compared to the tagBFP-V5 control, a significant increase of the fluorescent PLA signal was detected in the cytoplasm and in cytoplasmic poly(GR) inclusions in cells expressing tagBFP-V5 tagged YTHDF1-WT and YTHDF1-mut (Figures 4B and 4C), indicating that both YTHDF1-WT and YTHDF1-mut interact with GFP-(GR)100 independently of m6A-modified RNA binding. When overexpressing YTHDF1-WT or YTHDF1-mut in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100, we observed that YTHDF1-WT, but not YTHDF1-mut, significantly increased the number and size of cytoplasmic poly(GR) inclusions compared to controls (Figures 4D–4F). These findings demonstrate that the ability of YTHDF1 to bind m6A-modified RNA does not influence its binding to poly(GR) but is crucial for it to promote poly(GR) inclusion formation.
Figure 4. YTHDF proteins incorporate mRNA into poly(GR) inclusions via m6A-modified RNA binding.
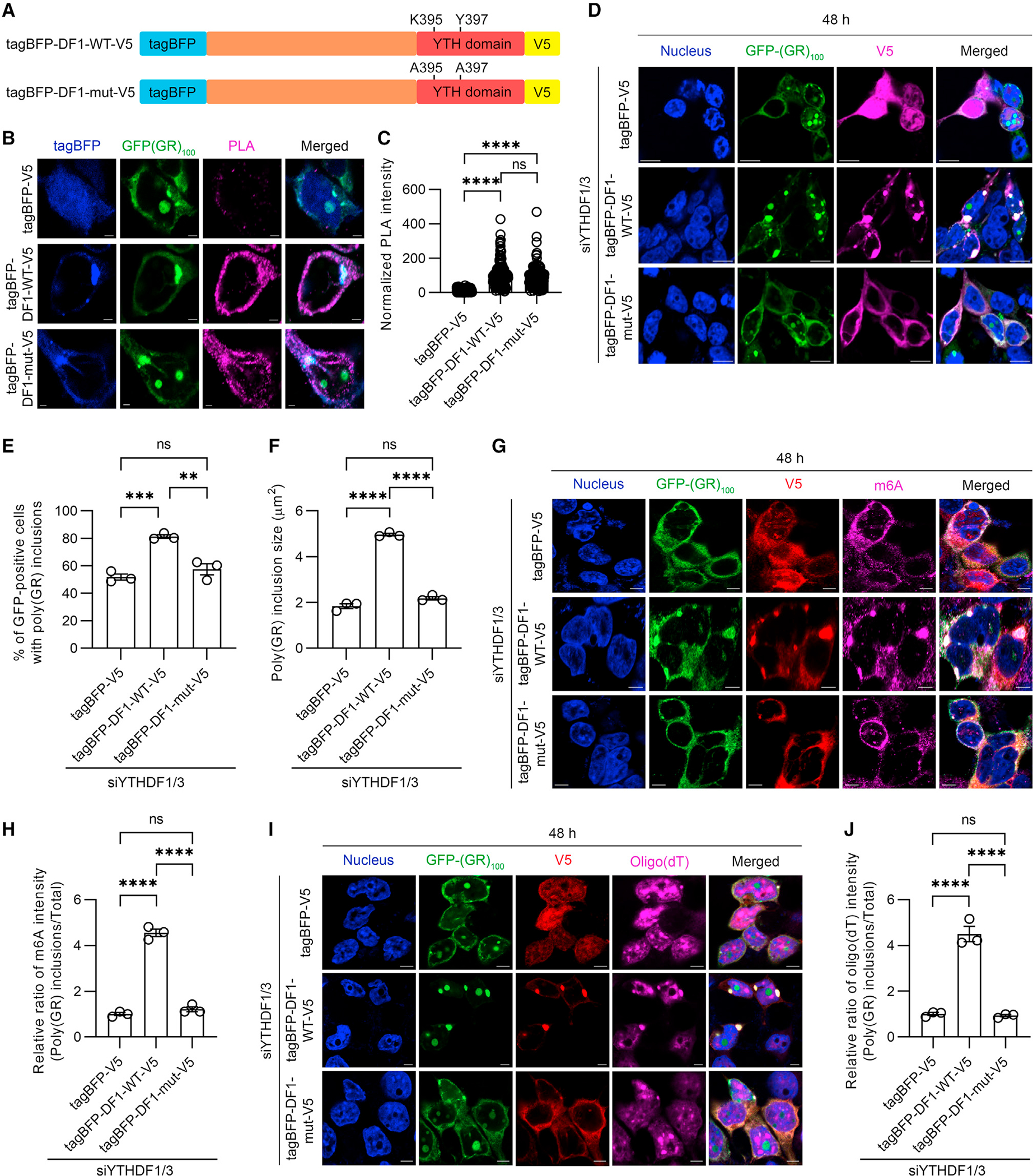
(A) Schematic of the tagBFP/V5 tagged YTHDF1 wild-type (tagBFP-DF1-WT-V5) and mutant (tagBFP-DF1-mut-V5) constructs, the latter having mutations in the YTH domain that impair the ability of YTHDF1 to bind m6A-modified RNA (top).
(B) Representative images of proximity ligation assay (PLA) for GFP-(GR)100 and tagBFP or tagBFP-YTHDF1 species in HEK293T cells co-expressing GFP-(GR)100 and tagBFP or tagBFP-YTHDF1 species. Scale bars, 2 μm.
(C) Quantification of the intensity for PLA signal in HEK293T cells co-expressing GFP-(GR)100 and tagBFP or tagBFP-YTHDF1 species (n = 152–176 cells).
(D) Immunofluorescence staining for V5 in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 and either tagBFP-V5, tagBFP-DF1-WT-V5 or tagBFP-DF1-mut-V5. Scale bars, 10 μm.
(E) Quantification of the percentage of cells with poly(GR) inclusions in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 and either tagBFP-V5, tagBFP-DF1-WT-V5, or tagBFP-DF1-mut-V5 (n = 3 independent experiments).
(F) Quantification of the size of poly(GR) inclusions in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 and either tagBFP-V5, tagBFP-DF1-WT-V5, or tagBFP-DF1-mut-V5 (n = 3 independent experiments).
(G) Triple-immunofluorescence staining for GFP, V5, and m6A in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 and either tagBFP-V5, tagBFP-DF1-WT-V5, or tagBFP-DF1-mut-V5. Scale bars, 5 μm.
(H) Quantification of the relative ratio of m6A intensity (poly(GR) inclusions/total) from YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 and either tagBFP-V5, tagBFP-DF1-WT-V5, or tagBFP-DF1-mut-V5 (n = 3 independent experiments).
(I) Immunofluorescence staining for V5 followed by RNA-FISH for oligo(dT) in YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 and tagBFP-V5, tagBFP-DF1-WT-V5, or tagBFP-DF1-mut-V5. Scale bars, 5 μm.
(J) Quantification of the relative ratio of oligo(dT) intensity (poly(GR) inclusions/total) from YTHDF1/3-depleted HEK293T cells expressing GFP-(GR)100 and either tagBFP-V5, tagBFP-DF1-WT-V5, or tagBFP-DF1-mut-V5 (n = 3 independent experiments).
Data are shown as mean ± SEM. In (C), ****p < 0.0001 and ns p = 0.0547, one-way ANOVA, Tukey’s post hoc analysis. In (E), ***p = 0.0007, **p = 0.0022, and ns (not significant) p = 0.3670, one-way ANOVA, Tukey’s post hoc analysis. In (F), ****p < 0.0001 and ns p = 0.0576, one-way ANOVA, Tukey’s post hoc analysis. In (H), ****p < 0.0001 and ns p = 0.4376, one-way ANOVA, Tukey’s post hoc analysis. In (J), ****p < 0.0001 and ns p = 0.9690, one-way ANOVA, Tukey’s post hoc analysis.
To investigate whether mRNA incorporation into cytoplasmic poly(GR) inclusions is affected by m6A-modified RNA and YTHDF proteins, we analyzed m6A and oligo(dT) intensity in cytoplasmic GFP-(GR)100 inclusions in YTHDF-depleted HEK293T cells overexpressing YTHDF1-WT or YTHDF1-mut. Consistent with the fact that the YTHDF1-mut has an impaired ability to bind m6A-modified RNA, the abundance of m6A-modified RNA and oligo(dT) in poly(GR) inclusions was significantly lower in cells expressing YTHDF1-mut compared to cells expressing YTHDF1-WT (Figures 4G–4J). These data show that the ability of YTHDF proteins to bind m6A-modified RNA is important for mRNA incorporation into cytoplasmic poly(GR) inclusions.
To examine the role of m6A-modified RNA in poly(GR) inclusion formation further, we assessed mRNA incorporation into cytoplasmic poly(GR) inclusions in ALKBH5-depleted or ALKBH5-overexpressing cells. As knocking down ALKBH5 increased m6A-modified RNA, and overexpression of ALKBH5 decreased it (Figures S3I–S3K and S3Q–S3S), Oligo(dT) intensity in poly(GR) inclusions was significantly increased in ALKBH5-depleted cells and significantly decreased in cells overexpressing ALKBH5 (Figures S4A–S4D). Our data thus demonstrate that m6A-modified RNA and YTHDF proteins function to carry mRNA to cytoplasmic poly(GR) inclusions.
DISCUSSION
In this study, we uncovered a key mechanism driving cytoplasmic poly(GR) inclusion formation: poly(GR) binding to G3BP1, a molecular switch known to trigger SG assembly.10–12 Of importance, we also discovered that m6A-modified RNAs and m6A-binding YTHDF proteins not only co-localize with cytoplasmic poly(GR) inclusions in vitro and in vivo but also promote poly(GR) inclusion formation via the incorporation of RNA to the inclusions.
Recently, G3BP1/2 proteins were identified as key molecules regulating arsenite-induced SG assembly.10–12 Notably, even though poly(GR) spontaneously forms cytoplasmic inclusions containing SG proteins in the absence of conventional stressors, G3BP1/2 was nonetheless required for cytoplasmic poly(GR) inclusion formation. Indeed, in the absence of G3BP1/2, cytoplasmic poly(GR) inclusions were not formed in poly(GR)-expressing cells. The IDR domains of G3BP1 are reported to influence its conformation and propensity to initiate SG formation.11,12 More specifically, under non-stress conditions, intramolecular interactions between the acidic IDR1 and the basic IDR3 of G3BP1 are believed to create a compact or closed conformation. However, stress-induced increases in free mRNA result in the binding of RNA to IDR3; this displaces IDR1 from IDR3 and putatively permits the G3BP1-RNA complex to adopt an open conformation capable of initiating SG assembly.11,12 In the present study, we found that the highly positively charged poly(GR) interacts with G3BP1, and this may promote poly(GR) inclusion formation, at least in part, by its binding to the IDR1 of G3BP1. In this manner, poly(GR) likely prevents IDR1 from interacting with IDR3, allowing G3BP1 to initiate LLPS. Our PLA studies also revealed that, in addition to the IDR1 domain, poly(GR) binds the NTF2L domain of G3BP1. We investigated the importance of these interactions on cytoplasmic poly(GR) inclusion formation using G3BP1/2 knockout cells stably expressing G3BP1 protein species lacking specific domains. Whereas exogenous FL G3BP1 restored poly(GR) inclusion formation in G3BP1/2 knockout cells, G3BP1 lacking the NTF2L domain failed to rescue poly(GR) inclusion formation. These findings highlight a crucial role for the G3BP1 NTF2L domain in cytoplasmic poly(GR) inclusion formation, as has been observed for SG formation.10,11 Also of interest, although G3BP1 lacking the IDR3 domain has a similar activity as FL G3BP1 to bind poly(GR), it failed to rescue poly(GR) inclusion formation. This is likely because G3BP1 lacking the IDR3 domain changes the conformation of the poly(GR)-G3BP1 complex and/or reduces RNA recruitment. Moreover, G3BP1 lacking the IDR1 domain interacted less strongly with poly(GR) in comparison to FL G3BP1 but nonetheless rescued poly(GR) inclusion formation to a similar degree as FL G3BP1. In G3BP1/2 knockout cells expressing G3BP1 without the IDR1 domain, cytoplasmic poly(GR) inclusions were comparable in size to those in cells expressing FL G3BP1. The deletion of IDR1 from G3BP1 likely facilitated RNA recruitment to poly(GR) inclusions by exposing IDR3 and RRM domains and enabling RNA binding.11,12 Indeed, our in vitro LLPS studies showed that addition of RNA rescues (GR)20 peptide-induced ΔIDR1 condensate formation to a similar extent as FL G3BP1, suggesting that interaction of RNA and ΔIDR1 G3BP1 enhances poly(GR) inclusion formation when the interactions between poly(GR) and G3BP1 are compromised. Furthermore, the cytoplasmic poly(GR) inclusions in G3BP1/2 knockout cells expressing ΔRRM contained less RNA and were smaller, further supporting the critical roles of RNA in mediating poly(GR) inclusion formation. Collectively, these results suggest that both poly(GR)-G3BP1 interactions and the recruitment of RNA into this protein complex are required for cytoplasmic poly(GR) inclusion formation and to regulate inclusion size.
Further bolstering the notion that RNA plays an important role in cytoplasmic poly(GR) inclusion formation, we demonstrate that m6A-binding YTHDF proteins and m6A-modified RNAs co-localize with poly(GR) in cultured cell and mouse models and in tissues from patients with c9FTD/ALS, as well as enhancing poly(GR) inclusion formation. Regarding the latter, we found that depleting YTHDF proteins in cells expressing poly(GR) significantly compromised poly(GR) inclusion formation and resulted in smaller cytoplasmic inclusions. Additionally, increasing m6A-modified RNAs by knocking down ALKBH5, an m6A-eraser,27–29 also increased both the percentage of cells with poly(GR) inclusions and inclusion size. We also observed that YTHDF proteins and m6A-modified RNAs work in concert to promote cytoplasmic poly(GR) inclusion formation. Indeed, increases in m6A-modified RNAs failed to appreciably enhance poly(GR) inclusion formation when YTHDF proteins were depleted, and a m6A-recognition deficient YTHDF1 mutant failed to promote poly(GR) inclusion formation despite its ability to interact with poly(GR). Based on these findings, we propose the following mechanism through which YTHDF proteins and m6A-modified RNAs promote cytoplasmic poly(GR) inclusion formation: the binding of m6A-modified RNAs to YTHDF proteins causes the resulting RNA-YTHDF complex to be incorporated into cytoplasmic poly(GR) inclusions through YTFDF1 and poly(GR) interactions, thereby enhancing poly(GR) inclusion formation.
Studies examining the partitioning of m6A-modified RNAs to SGs have yielded conflicting results with some studies,22,23 but not all,30 showing that m6A-modified RNAs and YTHDF proteins are present in SGs formed in response to oxidative stress. Despite these differences in findings, that cytoplasmic poly(GR) inclusions contain m6A-modified RNAs and YTHDF proteins along with SG proteins points once again to shared mechanisms between SG assembly and poly(GR) inclusion formation. It must nonetheless be noted that poly(GR) inclusions are more stable than conventional SGs induced by acute stressors such as sodium arsenite and oxidate stress.4,7 The persistent sequestration of RNA-binding proteins and mRNAs in poly(GR) inclusions is expected to impair gene expression and RNA quality control and thus have dire consequences on cellular health. For example, we have shown that, in the brain of GFP-(GR)200 mice, TDP-43 is sequestered into cytoplasmic poly(GR) inclusions, and we also observed a greater age-dependent loss of cells containing poly(GR) inclusions versus cells with diffuse poly(GR).9 These findings highlight the detrimental impact caused by the abnormal sequestration and loss of function of RNA-binding proteins like TDP-43, a major pathological protein in c9FTD/ALS, by poly(GR) inclusions.
The present study sheds valuable light on the mechanisms driving cytoplasmic poly(GR) inclusion formation, information important in our quest to combat c9FTD/ALS. Our data suggest that cytoplasmic poly(GR) inclusions are formed by poly(GR) interacting with G3BP1, which triggers RNA binding and recruitment to the cytoplasmic poly(GR)-G3BP1-RNA complexes. We further show that m6A-modified RNAs and YTHDF1 are key regulators of cytoplasmic poly(GR) inclusion formation. Our findings thus indicate that interrupting interactions between poly(GR) and G3BP1 or YTHDF1 proteins or decreasing poly(GR) altogether represent promising therapeutic strategies to combat c9FTD/ALS pathogenesis.
Limitations of the study
Along with the knowledge gained from our findings above, limitations with the present study must be recognized. Although we observed that G3BP1, m6A-modified mRNAs, and YTHDF proteins are crucial for cytoplasmic poly(GR) inclusion formation in cultured cells, validation studies utilizing a mouse model to confirm these findings in vivo are warranted. Moreover, future investigations aimed at deciphering which m6A-modified mRNAs accumulate in cytoplasmic poly(GR) inclusions are needed as they may provide insight on the harmful influence of poly(GR) on RNA metabolism.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Leonard Petrucelli (Petrucelli.Leonard@mayo.edu).
Materials availability
All unique plasmids and reagents used in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell culture, transfections, and treatments
HEK293T cells and U2OS cells were grown in Opti-Mem supplemented with 10% FBS and 1% penicillin–streptomycin. Cells were incubated and grown at 5% CO2 and 37°C. Cells grown in 6-well plates or on glass coverslips in 24-well plates were transfected with the indicated plasmids using Lipofectamine 2000 (Thermo Fisher Scientific) for HEK293T cells or FuGENE HD Transfection Reagent (Promega) for U2OS cells according to the manufacturers’ instructions. When HEK293T cells were transfected with RNAi and the plasmids, RNAi was transfected 24 h after plating cells using Lipofectamine RNAimax (Thermo Fisher Scientific) followed by plasmid transfection using Lipofectamine 2000 (Thermo Fisher Scientific) 24 h after that. Cells were harvested or fixed for Western blot and immunofluorescence staining, respectively, 24 or 48 h post-transfection. siRNAs used in this study are summarized in key resources table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| anti-G3BP1 | Proteintech | Cat#13057-2-AP; RRID:AB_2232034 |
| anti-GFP | Thermo Fisher Scientific | Cat#33-2600; RRID:AB_2533111 |
| anti-G3BP1 | BD Biosciences | Cat#611126; RRID:AB_398437 |
| anti-mCherry | Novus biologicals | Cat#NBP1-96752; RRID:AB_11034849 |
| anti-m6A | Synaptic Systems (SySy) | Cat#202 003; RRID:AB_2279214 |
| anti-GFP | Millipore | Cat#MAB3580; RRID:AB_94936 |
| anti-ALKBH5 | Proteintech | Cat#16837-1-AP; RRID:AB_2242665 |
| anti-YTHDF1 | Proteintech | Cat#17479-1-AP; RRID:AB_2217473 |
| anti-YTHDF3 | Proteintech | Cat#25537-1-AP; RRID:AB_2847817 |
| anti-V5 | Thermo Fisher Scientific | Cat#R960-25; RRID:AB_2556564 |
| anti-Flag | Sigma | Cat#F1804; RRID:AB_262044 |
| anti-GAPDH | Meridian Life Science | Cat#H86504M; RRID:AB_151542 |
| anti-eIF3η | Santa Cruz | Cat#sc-137214; RRID:AB_2277705 |
| anti-Ataxin 2 | BD Biosciences | Cat#611378; RRID:AB_398900 |
| anti-TIA-1 | Abcam | Cat#ab40693; RRID:AB_2201438 |
| anti-GR | Millipore | Cat#MABN778; RRID:AB_2728664 |
| anti-eIF3η | Santa Cruz | Cat#sc-16377; RRID:AB_671941 |
| Anti-GA | Millipore | Cat#MABN889; RRID:AB_272866 |
| anti-GR (Rb7810) | Gendron, T.F., et al.31 | N/A |
| BIOTIN-Anti-C9RANTV2. 9259 Purified antibody | Zhang, Y.J., et al.4 | N/A |
| Sulfo-Affinity Purified 7810 (Anti-GR) | Zhang, Y.J., et al.4 | N/A |
| Peroxidase AffiniPure F(ab’)2 Fragment Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat#715-036-150; RRID:AB_2340773 |
| Peroxidase AffiniPure F(ab’)2 Fragment Donkey Anti-Rabbit IgG (H+L) |
Jackson ImmunoResearch | Cat#711-036-152; RRID:AB_2340590 |
| AlexaFluor 488 (Donkey anti-Rabbit IgG (H+L)) | Invitrogen | Cat#A32790; RRID:AB_2762833 |
| AlexaFluor 488 (Donkey anti-Rat IgG (H+L)) | Invitrogen | Cat#A48269; RRID:AB_2893137 |
| AlexaFluor 568 (Donkey anti-Mouse IgG (H+L)) | Invitrogen | Cat#A10037; RRID:AB_2534013 |
| AlexaFluor 568 (Goat anti-Rabbit IgG (H+L)) | Invitrogen | Cat#A11034; RRID:AB_2576217 |
| AlexaFluor 647 (Donkey anti-Mouse IgG (H+L)) | Invitrogen | Cat#A31571; RRID:AB_162542 |
| AlexaFluor 647 (Donkey anti-Rabbit IgG (H+L)) | Invitrogen | Cat#A31573; RRID:AB_2536183 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| RosettaTM(DE3)pLysS competent cells | Millipore | Cat#70956 |
| rAAV9-(GA)100-V5 | Shao W., et al.32 | N/A |
| rAAV9-GFP-(GR)200 | Cook, C.N., et al.9 | N/A |
| rAAV9-(G4C2)2 | Chew, J., et al.20 | N/A |
| rAAV9-(G4C2)149 | Chew, J., et al.20 | N/A |
|
| ||
| Biological samples | ||
|
| ||
| Postmortem mid-frontal cortex tissues from c9FTD patients | Mayo clinic Florida Brain Bank | Table S1 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| DNase | Sigma-Aldrich | Cat#11284932001 |
| RNase | Sigma-Aldrich | Cat#R6748 |
| Protease inhibitor | Roche | Cat#A32965 |
| Glutathione Sepharose 4B resin | GE Health | Cat#17-0756-01 |
| PreScission Protease | Cytiva | Cat#27084301 |
| TRIzol | Thermo Fisher Scientific | Cat#15596026 |
| RNeasy Plus Mini Kit | Qiagen | Cat#74034 |
| SecureSeal Imaging Spacers | Grace Bio-laboratories | Cat#470352 |
| Opti-MEM | Thermo Fisher Scientific | Cat#51985034 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat#11668500 |
| FuGENE HD | Promega | Cat#E2311 |
| Lipofectamine RNAimax | Thermo Fisher Scientific | Cat#13778500 |
| Sodium Deoxycholate | Sigma-Aldrich | Cat#D6750-100G |
| Benzonase | Sigma-Aldrich | Cat#E8263 |
| Polyethylenimine | Polyscience, Inc. | Cat#23966 |
| Isopropyl β-d-1-thiogalactopyranoside (IPTG) | Thermo Fisher Scientific | Cat#BP1755 |
| Triton X-100 | Sigma-Aldrich | Cat#T9284-1L |
| Phosphate Buffered Saline | Thermo Fisher Scientific, | Cat#BP3994 |
| Dulbecco’s Phosphate Buffered Saline | Thermo Fisher Scientific, | Cat#14190144 |
| Dithiothreitol (DTT) | Genesee | Cat#18-203 |
| Coomassie Brilliant Blue R-250 | Bio-Rad | Cat#1610400 |
| Direct-zol RNA microprep kit | Zymo Research | Cat#R2062 |
| Dynabead mRNA purification kit | Thermo Fisher Scientific | Cat#61006 |
| G418 Sulfate | Gemini Bioproducts | Cat#400-111P |
| Protease inhibitor tablets | Roche | Cat#A32965 |
| RNase inhibitor | Invitrogen | Cat#AM2696 |
| Wash buffer A for sequential staining | Biosearch Technologies | Cat#SMF-WA1-60 |
| Hybridization buffer for sequential staining | Biosearch Technologies | Cat#SMF-HB1-10 |
| Cy5-Oligo d(T)20 | Genelink | Cat#26-4420-02 |
| Wash buffer B for sequential staining | Biosearch Technologies | Cat#SMF-WB1-20 |
| Dako Peroxidase Block | DAKO | Cat#S2001 |
| Dako Protein Block Serum-Free | DAKO | Cat#X0909 |
| Dako Envision-Plus anti-rabbit | DAKO | Cat#K4003 |
| Liquid DAB+ substrate Chromogen System | DAKO | Cat#K3468 |
| Gill’s Hematoxylin | Statlab (SLMP LLC) | Cat#HXGHE1LT |
| (GR)20 peptide | Peptide 2.0 Inc | N/A |
| G3BP1 FL | This study | N/A |
| G3BP1 ΔIDR1 | This study | N/A |
| G3BP1ΔIDR3 | This study | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| Duolink In Situ PLA® Probe Anti-Rabbit PLUS | Sigma-Aldrich | Cat#DUO92002 |
| Duolink In Situ PLA® Probe Anti-Mouse MINUS | Sigma-Aldrich | Cat#DUO92004 |
| Duolink In Situ Detection Reagents FarRed | Sigma-Aldrich | Cat#DUO92013 |
| Duolink In Situ Wash Buffers, Fluorescence | Sigma-Aldrich | Cat#DUO82049 |
| EpiQuik m6A RNA methylation quantification Kit | EpigenTek | Cat#P-9005 |
| Coomassie Plus (Bradford) Assay Kit | Thermo Fisher Scientific | Cat#23236 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T cell line | ATCC | Cat#CRL-3216 |
| G3BP1/2 KO U2OS cell line | Sanders, D.W. et al.10 | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6J | The Jackson Laboratory | Cat#000664; RRID:IMSR_JAX:000664 |
|
| ||
| Oligonucleotides | ||
|
| ||
| siALKBH5 ON-TARGETplus human ALKBH5 | Dharmacon | Cat#L-004281-01 |
| siYTHDF1 ON-TARGETplus human YTHDF1 | Dharmacon | Cat#L-018095-02 |
| siYTHDF3 ON-TARGETplus human YTHDF3 | Dharmacon | Cat#L-017080-01 |
| siYTHDF1 ON-TARGETplus human YTHDF1 3’UTR | Dharmacon | Cat#J-018095-19 |
| siYTHDF3 ON-TARGETplus human YTHDF3 3’UTR | Dharmacon | Cat#J-017080-10 |
| Primers for site-directed mutagenesis of YTHDF1 | Li, Q., et al.26 | Table S2 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pmCherry-C1 | Clontech Laboratories | Cat#632524 |
| pmCherry-C1 G3BP1 FL | This study | N/A |
| pmCherry-C1 G3BP1 ΔNTF2L | This study | N/A |
| pmCherry-C1 G3BP1 ΔIDR1 | This study | N/A |
| pmCherry-C1 G3BP1 ΔIDR3 | This study | N/A |
| pmCherry-C1 G3BP1 ΔRRM | This study | N/A |
| pGEX-6P-1 | GE Health | Cat#28-9546-48 |
| pGEX-6P-1 G3BP1 FL | This study | N/A |
| pGEX-6P-1 G3BP1 ΔNTF2L | This study | N/A |
| pGEX-6P-1 G3BP1 ΔIDR1 | This study | N/A |
| pGEX-6P-1 G3BP1 ΔIDR3 | This study | N/A |
| pGEX-6P-1 G3BP1 ΔRRM | This study | N/A |
| pGEX-4T-1 YTHDF1 | Wang, X. et al.33 | Addgene Cat#70087 |
| pcDNA6 V5-His A | Invitrogen | Cat#V22120 |
| mTagBFP2-ER-5 | Subach, O.M., et al.34 | Addgene Cat#55294 |
| pAAV-tagBFP | This study | N/A |
| pcDNA6 V5-tagBFP | This study | N/A |
| pcDNA6 tagBFP-YTHDF1-WT-V5 | This study | N/A |
| pcDNA6 tagBFP-YTHDF1-mut-V5 | This study | N/A |
| pCMV-Flag-ALKBH5 | SinoBiological | Cat#HG24078-CF |
| pEGFP-(GR)100 | Cook, C.N.,etal.9 | N/A |
| pEGFP-(GA)100 | Cook, C.N.,etal.9 | N/A |
| pFDelta6 | Chakrabarty, P., et al.35 | N/A |
| pCap9 | Chakrabarty, P., et al.35 | N/A |
| pAM/CBA-pl-WPRE-BGH (pAAV) | Chakrabarty, P., et al.35 | N/A |
| pAAV-(GA)100-V5 | Shao W., et al.32 | N/A |
| pAAV-GFP-(GR)200 | Cook, C.N.,etal.9 | N/A |
| pAAV-(G4C2)2 | Chew, J., et al.20 | N/A |
| pAAV-(G4C2)149 | Chew, J., et al.20 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Graphpad Prism v9 | GraphPad Software, LLC | https://www.graphpad.com/scientificsoftware/prism/ |
| ImageScope® software | Leica Biosystems | https://www.leicabiosystems.com/us/digital-pathology/manage/aperioimagescope/ |
| Zeiss Zen 2.3 | Carl Zeiss | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| CellProfiler | Lamprecht, M.R., et al.36 | https://cellprofiler.org/ |
|
| ||
| Others | ||
|
| ||
| Zeiss AxioObserver A1 microscope | Zeiss | https://www.zeiss.com/microscopy/en/products/light-microscopes/widefieldmicroscopes/axio-observer-for-lifescience-research.html |
| Zeiss LSM980 laser scanning confocal microscope | Zeiss | https://www.zeiss.com/microscopy/en/products/light-microscopes/confocalmicroscopes/lsm-980-withairyscan-2.html |
| MESO QUICKPLEX SQ 120 | Meso Scale Diagnostics | https://www.mesoscale.com/en/products_and_services/instrumentation/quickplex_sq_120mm |
| Amersham ImageQuant 800 | Cytiva | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/molecular-imagingfor-proteins/imaging-systems/amershamimagequant-800-systems-p-11546 |
Animal studies
All procedures in this study using mice were performed in accordance with the National Institutes of Health Guide for Care and Use of Experimental Animals and approved by the Mayo Clinic Institutional Animal Care and Use Committee (IACUC) (Protocol number A00005246–20). Male and female C57BL/6J mice (Jackson Laboratories) were maintained in the animal facilities at Mayo Clinic Florida on a 12-hour light/dark cycle in standard housing. When in their home cage, animals had access to standard mouse chow and water ad libitum. Mouse pups on postnatal day 0 were used for neonatal viral injection of rAAV9-(GA)100-V5, rAAV9-GFP-(GR)200, rAAV9-(G4C2)2 or rAAV9-(G4C2)149. The 2-week-old GFP-(GR)200 mice (n = 6, 4 males and 2 females), 3.5-month-old (GA)100-V5 mice (n = 3, 2 males and 1 female), 12-month-old (G4C2)2 (n = 6, 3 males and 3 females) or (G4C2)149 (n = 6, 3 males and 3 females) mice were used in this study.
Human tissues
Postmortem mid-frontal cortical tissues from patients with frontotemporal lobar degeneration, the neuropathological diagnosis of frontotemporal dementia (FTD) with a C9orf72 repeat expansion were obtained from the Mayo Clinic Florida Brain Bank. Information on human patients is provided in Table S1. Written informed consent was obtained before study entry from all subjects or their legal next of kin if they were unable to give written consent, and biological samples were obtained with Mayo Clinic Institutional Review Board (IRB) approval.
METHOD DETAILS
Generation of plasmids
To generate plasmids of mCherry-G3BP1 and GST-G3BP1 species, the plasmids containing FL G3BP1 or ΔNTF2L, ΔIDR1, ΔIDR3 or ΔRRM G3BP1 species, which have been described previously,10 were used as templates to amplify DNA fragments. Then the fragments of G3BP1 species were cloned into pmCherry-C1 (Clontech Laboratories) using Eag1/BamH1 restriction sites or cloned into pGEX-6P-1 (GE Health) using BamH1/SalI restriction sites. To generate pAAV-tagBFP plasmid, the plasmid containing tagBFP (Addgene) were used as templates to amplify the DNA fragments. tagBFP was cloned into an AAV packaging vector [pAM/CBA-pl-WPRE-BGH (“pAAV”)] containing the CMV-enhanced chicken β-actin promoter using HindIII and Xho restriction enzymes. To generate the tagBFP-YTHDF1-V5 plasmid, the plasmids containing YTHDF1 (Addgene) was used to amplify DNA fragments. YTHDF1 was cloned into pcDNA6 V5-His A (Invitrogen) using EcoRI and XhoI restriction sites. tagBFP was amplified from pAAV-tagBFP and cloned into pcDNA6 V5-His A YTHDF1 using BamHI and EcoRI restriction sites. To generate the V5-BFP-YTHDF1 mutant plasmid (a YTH domain deficient mutant), site-directed mutagenesis was performed using V5-BFP-YTHDF1 as a template, which mutated Lysine 395 and Y397 to Alanine.26 See Table S2 for the sequence of mutagenesis primers.
Virus production
rAAV9 virus was produced as previously described.9,20,37 In brief, AAV vectors expressing (GA)100-V5, GFP-(GR)200, (G4C2)2 or (G4C2)149 were co-transfected with helper plasmids in HEK293T cells using polyethylenimine (Polysciences, Inc). Cells were harvested 48 h following transfection and lysed in the presence of 0.5% sodium deoxycholate and 50 units/mL Benzonase (Sigma-Aldrich) by freeze-thawing. The virus was isolated using a discontinuous iodixanol gradient. The genomic titer of each virus was determined by qRT-PCR, and AAV solutions were diluted in Dulbecco’s phosphate-buffered saline (DPBS; Thermo Fisher Scientific).
Purification of different recombinant G3BP1 proteins
GST-tagged G3BP1 FL, ΔIDR1 or ΔIDR3 plasmids were used for transformation in Rosetta™(DE3)pLysS competent cells (Millipore). To induce expression of recombinant proteins, bacteria were cultured overnight at 16°C in the presence of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, Thermo Fisher Scientific). After centrifugation, the bacteria pellet was washed with phosphate-buffered saline (PBS), and then lysed on ice for 30 min with PBS containing 1% Triton X-100, 1 mM DTT, DNase (1 μg/mL, Sigma-Aldrich), Rnase (10 μg/mL, Sigma-Aldrich) and a protease inhibitor tablets (Roche). After sonication, the lysates were centrifuged at 18,000g for 30 min. The resulting supernatant was applied to a Glutathione Sepharose 4B resin (GE Health). After washing resin with PBST and 50 mM Tris-HCl (pH 8.0), incubate the resin by PreScission Protease (80 unit/mL, Cytiva) in 50 mM Tris-HCl (pH 8.0) on ice for 4 hours. Afterward, the recombinant proteins were collected from the resin by centrifugation at 500 g for 5min. The recombinant proteins were concentrated using Amicon Ultra-15 centrifugal filter units (10kDa, Millipore, Cat#UFC9010). The protein concentration determined by Coomassie staining and BCA assay.
Liquid-liquid phase separation of recombinant G3BP1 proteins and (GR)20 peptides
All liquid-liquid phase separation reactions were done in phosphate buffered saline (PBS; Thermo Fisher Scientific). The recombinant G3BP1 protein species (FL, ΔIDR1or ΔIDR3) were diluted in PBS at the final concentrations of 0.2 μM. (GR)20 peptides were mixed with the recombinant G3BP1 protein species under evaluation at the final concentration of 0.5 or 1 μM. For experiments in which RNA was used, RNA was extracted from U2OS cells using TRIzol (Thermo Fisher Scientific), followed by RNA purification using the Rneasy Plus Mini kit (Qiagen). RNA was added to the G3BP1-(GR)20 mixture to a concentration of 20 ng/μL. After mixture in Eppendorf tubes, 3 μL of each reaction were spotted on a glass slide containing wells created by SecureSeal Imaging Spacers (Grace Bio-Laboratories, Cat#470352). The wells were then covered with a coverslip and inverted. After a 20 min incubation at room temperature, settled droplets on the coverslips were imaged using Zeiss AxioObserver A1 microscope with DIC at 20×. For quantification analysis, images of six regions of interest per sample were collected. The number of droplets was quantified using Cellprofiler.36 Minimum cross-entropy thresholding was used for DIC image segmentation.
Generation of U2OS G3BP1/2 knockout cell lines stably expressing mCherry-G3BP1 species
To generate stable cell lines expressing mCherry-G3BP1 species under a G3BP1/2 knockout background, previously described U2OS G3BP1/2 knockout (GG KO) cells were seeded in a 6-well plate.10 Once 90% confluency was reached, cells were transfected with 0.75 μg mCherry-G3BP1 species using FuGENE HD Transfection Reagent (Promega) according to the manufacturer’s instructions. Forty-eight hours after transfection, the cells were subcultured at 2.0 × 105 cells in a 10 cm dish. Twenty-four hours later, 600 μg/mL G418 (Gemini Bioproducts) was added to the medium to screen for clones resistant to G418. After incubating cells for 10 days, single clones were picked and seeded into a 24-well plate. The cells were grown in Opti-Mem supplemented with 10% FBS, 1% penicillin/streptomycin and 400 μg/mL G418. When the cells reached ~80% confluency, each clone was split into duplicate wells of a 6-well plate. Cells in one of the duplicate wells was subjected to Western blot analysis to examine the expression of mCherry-G3BP1 species. The other was used to make frozen cell stocks. The clones with comparable expression levels of mCherry-G3BP1 species were chosen and used throughout the study.
Preparation of cell lysates
Cell pellets were lysed in Co-IP buffer (50 mM Tris–HCl, pH 7.4, 300 mM NaCl, 1% Triton X-100, 5 mM EDTA) plus 2% SDS and both protease and phosphatase inhibitors, sonicated on ice, and then centrifuged at 16,000 × g for 20 min. Supernatants were saved as cell lysates. The protein concentration of lysates was determined by BCA assay (Thermo Fisher Scientific), and samples were then subjected to Western blot analysis.
Western blot analysis
Western blot analysis was performed as described previously.4,5,37,38 In brief, lysates were diluted with 2 × SDS-loading buffer at a 1:1 ratio (v/v), and then heated at 95°C for 5 min. Afterwards, equal amounts of protein were loaded into 10-well 10% Tris-glycine gels or 4–20% Tris-glycine gels (Novex). After transferring proteins to membranes, membranes were blocked with 5% nonfat dry milk in Tris-buffer saline (TBS) plus 0.1% Tween 20 (TBST) for 1 h, then incubated with primary antibody (key resources table) overnight at 4°C. Membranes were washed in TBST and incubated with donkey anti-rabbit or anti-mouse IgG antibodies conjugated to horse-radish peroxidase (1:5000; key resources table) for 1 h at room temperature. Protein expression was visualized by enhanced chemiluminescence treatment and exposure to film or Amersham ImageQuant 800.
MSD immunoassay
Preparation of cell lysates for MSD immunoassays to detect poly(GR) expression in HEK293T cells was done using Urea-containing buffer (7 M urea, 2 M thiourea, 4% CHAPS, 30 mM Tris, pH 8.5). After sonication and centrifugation at 16,000 × g for 20 min, the protein concentration of lysates was determined by Bradford assay (Thermo Fisher Scientific), and an equal amount of protein lysate was used for a poly(GR) sandwich immunoassay. Lysates were diluted in Tris-buffered saline (TBS). Antibodies used in this assay are summarized in key resources table. MesoScale Discovery (MSD) electrochemiluminescence detection technology was used to measure poly(GR) levels in each sample. Response values according to the intensity of emitted light upon electrochemical stimulation of the assay plate were acquired with the MSD QUICKPLEX SQ120.
N6-methyladenosine (m6A) quantification by ELISA
Quantification of m6A-modified RNA in GFP-(GR)100-expressing HEK293T cells overexpressing or depleted of YTHDF or ALKBH5 was performed using mRNA purified from cells. In brief, total RNA was prepped from the cells using Direct-zol RNA microprep kit (Zymo Research) according to manufacturer’s instructions. Ten micrograms of total RNA were used for mRNA purification using the Dynabead mRNA purification kit (Thermo Fisher), then m6A modification in 200 ng of purified mRNA was quantified using the EpiQuik m6A RNA methylation quantification Kit (EpigenTek).
Immunofluorescence staining
HEK293T or U2OS cells were fixed with 4% paraformaldehyde for 10 min at room temperature, followed by permeabilization using 0.5% Triton X-100 for 10 min, blocked with 5% nonfat dry milk in DPBS for 1 h, then incubated with primary antibody (key resources table) overnight at 4°C. After washing, cells were incubated with corresponding Alexa Fluor 488-, 568- or 647-conjugated donkey anti-species antibodies (1:500 or 1:1000, key resources table) for 2 h. Hoechst 33258 (1 μg/mL, Thermo Fisher Scientific) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 980 laser scanning confocal microscope.
For m6A immunofluorescence using the antibody against m6A, HEK293T cells were fixed and permeabilized with Methanol:AcOH (3:1) solution for 15 min at −20°C, washed 3 times with cold Wash buffer A (Biosearch Technologies) for 30 min in 37°C, then blocked with blocking solution (2% BSA, 1X DPBS, 0.05% Triton X-100, 100 U RNase inhibitor (Invitrogen) for 1 h at room temperature. After blocking, cells were incubated with m6A (SySy) and GFP antibodies (Millipore) overnight at 4°C. Secondary antibody incubations and Hoechst 33258 staining were performed as described above.
Sequential staining (immunofluorescence + fluorescence in situ hybridization)
Fixed cells were washed twice with DPBS and permeabilized with DPBS plus 0.1% Triton X-100 for 5 min at room temperature, followed by washing with DPBS. Then appropriately diluted primary antibodies (key resources table) in DPBS were added into each well and incubated at room temperature for 1 h. Cells were washed twice with DPBS for 10 min and incubated with appropriately diluted secondary antibodies at room temperature for 1 h, followed by two washes with DPBS for 10 min. Cells were then treated with fixation buffer for 10 min and washed twice with DPBS. The wash buffer A (Biosearch Technologies) was added into each well and incubated for 5 min at room temperature. During the incubation, a humidified chamber was prepared by placing a single layer of Parafilm on top of a flat water-saturated paper towel in a 150 mm tissue culture plate. Within the humidified chamber, 50 μl of the hybridization buffer (Biosearch Technologies) containing Cy5-Oligo d(T)20 (1:40, Genelink) and 10% formamide was dispensed onto the Parafilm. Coverslips where gently placed, cell side down, onto the 50 μl drop of hybridization buffer and incubated in the dark at 37°C for 16 h. After incubation, the coverslips were gently transferred to a fresh 24-well plate containing 500 μl of Wash Buffer A and incubated at 37°C for 30 min. Wash Buffer A containing Hoechst 33258 was added on the coverslips and incubated for another 30 min at 37°C. The coverslips were incubated with Wash Buffer B (Biosearch Technologies) at room temperature for 5 min. After labeling, coverslips were mounted cell side down onto slides. Images were obtained on a Zeiss LSM980 laser scanning confocal microscope.
Proximity ligation assay (PLA)
Duolink In Situ kit (Sigma-Aldrich) was used for PLA assays to measure the interaction between GFP-(GR)100 and G3BP1 species, or GFP-(GR)100 and YTHDF1 species. HEK293T cells or U2OS cells in an 8-well chamber slide (Ibidi, Cat#80826) were fixed with 4% paraformaldehyde for 10 min at room temperature, followed by permeabilization with 0.1% Triton X-100 for 10 min, blocked with Duolink® blocking solution in a heated humidity chamber for 60 min at 37°C, then incubated with primary antibody (key resources table) overnight at 4°C. After washing with PBS plus 0.05% Tween 20, cells were incubated with the PLA probes MINUS and PLUS (1:5 dilution) for 1 hour at 37°C. After washing in 1× wash buffer A, cells were incubated with the ligase (1:40 dilution) in ligation buffer for 30 min at 37°C. Cells were washed with 1× wash buffer A, and then inoculated with polymerase (1:80 dilution) in amplification buffer for 90 min at 37°C. Cells were washed in 1× wash buffer B, followed by 0.01× wash buffer B. Hoechst 33258 (1 μg/mL, Thermo Fisher Scientific) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 980 laser scanning confocal microscope.
Neonatal viral injections
Intracerebroventricular injections of virus were performed as previously described.4,5,20,37,39 In brief, 2 μl (0.5×1010 or 1×1010 genomes/μl) of rAAV9-(GA)100-V5, rAAV9-GFP-(GR)200, rAAV9-(G4C2)2 or rAAV9-(G4C2)149 solution was manually injected into each lateral ventricle of cryo-anesthetized C57BL/6J mouse pups on postnatal day 0. Pups were allowed to recover from cryoanesthesia on a heating pad and were then returned to the home cage with the mother.
Tissue processing
The mice were euthanized by CO2. Brains were then harvested and cut sagittally across the midline. The brain was rapidly removed and hemisected. Sagittal half brains were immersion fixed in 4% paraformaldehyde, embedded in paraffin, sectioned (5 μm thick), and then mounted on glass slides for immunofluorescence and immunohistochemistry.
Immunohistochemistry staining
The paraffin sections were deparaffinized in xylene, and rehydrated through a series of ethanol solutions, followed by washing in dH2O. Antigen retrieval was performed by steaming slides in sodium citrate buffer (pH 6.0) for 30 min followed by a 5 min incubation in Dako Peroxidase Block (DAKO) to block endogenous peroxidase activity. The slides were blocked with Dako Protein Block Serum-Free (DAKO) for 1 h, and incubated with primary antibody (key resources table) for 45 min. After washing, sections were incubated for 30 min in Dako Envision-Plus anti-rabbit (DAKO) labeled HRP polymer. Peroxidase labeling was visualized with the Liquid DAB + Substrate Chromogen System (DAKO). Following labeling, all sections were counterstained with hematoxylin (Statlab), dehydrated through ethanol and xylene washes, and coverslipped with Cytoseal mounting medium (Thermo Fisher Scientific). Slides were scanned with a ScanScope® AT2 (Leica Biosystems), and representative images taken with ImageScope® software (v12.4.2.7000; Leica Biosystems).
Immunofluorescence in mouse and human brains
For immunofluorescence, paraffin sections (5 μm) of mouse and human brain tissues were deparaffinized, rehydrated, steamed for 30 min in sodium citrate buffer (pH 6.0), blocked with Dako Protein Block Serum-Free (DAKO) for 1 h, and incubated with primary antibody (key resources table). After washing, sections were incubated with corresponding Alexa Fluor 488-, 568- or 647-conjugated donkey anti-species antibodies (1:500, key resources table) for 2 h. Hoechst 33258 (1 μg/mL, Thermo Fisher Scientific) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 980 laser scanning confocal microscope.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of the percentage of cells with inclusions, inclusion size and the ratio of RNA fluorescence, as well as m6A and PLA fluorescence intensity
To quantify the percentage of GFP-(GR)100-transfected cells with cytoplasmic poly(GR) inclusions, we counted ~80–270 GFP-positive cells with and without poly(GR) inclusions within each group from 3 or 4 independent experiments. The same method was used when quantify the percentage of GFP-(GA)100-transfected cells with poly(GA) inclusions. To quantify the percentage cells containing YTHDF1 or m6A-modified RNA inclusions, ~140–600 cells non-transduced (NT) cells and cells with diffuse poly(GR) or poly(GR) inclusions per mouse were counted in the GFP-(GR)200 mice. To quantify the size of cytoplasmic poly(GR) or poly(GA) inclusions, Zeiss Zen 2.3 was used to measure the area of all inclusions within a cell; ~50–200 cells per group were tested across 3 or 4 independent experiments. To quantify the ratio of oligo(dT) RNA or m6A-modified RNA fluorescence intensity in cells expressing poly(GR) for 24 or 48 h, Zeiss Zen 2.3 was used to measure the sum fluorescence intensity of oligo(dT) or m6A in poly(GR) inclusions within a cell, and total cellular m6A fluorescence intensity; ~50–150 cells per group were analyzed across 3 independent experiments. The sum fluorescence intensity in poly(GR) inclusions was divided by that within cells, and then normalized to the corresponding controls. To quantify the total m6A intensity of ALKBH5-modulated HEK293T cells, Zeiss Zen 2.3 was used to measure the mean fluorescence intensity of m6A with in a cell; ~50–100 cells per group were analyzed across 3 independent experiments. To quantify PLA intensity, Zeiss Zen 2.3 was used to measure the sum fluorescence intensity of PLA signal and area of each cell: 38‒87 cells were analyzed for U2OS cells, and 152‒176 cells were analyzed for HEK293T cells. The sum fluorescence intensity of PLA within a cell was divided by the area of the cell, and then normalized to the corresponding controls (U2OS: G3BP1 FL as the control; HEK293T: YTHDF1 WT as the control). All quantification analyses were done in an unblinded fashion.
Statistics
Data are presented as mean ± standard error of the mean (SEM), and analyzed with unpaired two-tailed t test or one-way ANOVA followed by Tukey’s post-hoc analysis (Prism statistical software). P < 0.05 is considered statistically significant. All of the statistical details of experiments can be found in the figure legends. No methods were used to determine whether the data met assumptions of the statistical approach.
Supplementary Material
Highlights.
The interaction of poly(GR) with G3BP1 is critical for poly(GR) inclusion formation
RNA recruitment to poly(GR)-G3BP1 complexes regulates poly(GR) inclusion size
YTHDF1 and m6A-modified mRNAs co-localize with poly(GR) inclusions
YTHDF1 and m6A-modified mRNAs promote poly(GR) inclusion formation
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R35NS097273 [L.P.], P01NS084974 [D.W.D., T.F.G., B.O., L.P., and Y.-J.Z.], P01NS099114 [T.F.G. and L.P.], U54NS123743 [L.P.], RF1AG062077 [L.P.], RF1AG062171 [L.P.], R01NS117461 [T.F.G. and Y.-J.Z], 1R21NS127331 [Y.-J.Z], RF1AG 061706 [B.W. and A.E.], and R01AG080810 [B.W.]), Mayo Clinic Foundation (L.P.), Robert Packard Center for ALS Research at Johns Hopkins (L.P.), and Target ALS Foundation (L.P. and Y.-J.Z.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112822.
REFERENCES
- 1.Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, and McKnight SL (2014). Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345, 1139–1145. 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, et al. (2014). C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194. 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. (2014). Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84, 1213–1225. 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YJ, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN, et al. (2018). Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142. 10.1038/s41591-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang YJ, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen-West K, O’Raw AD, Pickles SR, Prudencio M, Carlomagno Y, et al. (2019). Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363, eaav2606. 10.1126/science.aav2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, et al. (2017). Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol. Cell 65, 1044–1055.e5. 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. (2016). C9orf72 Dipeptide Repeats Impair the Assembly, Dynamics, and Function of Membrane-Less Organelles. Cell 167, 774–788.e17. 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White MR, Mitrea DM, Zhang P, Stanley CB, Cassidy DE, Nourse A, Phillips AH, Tolbert M, Taylor JP, and Kriwacki RW (2019). C9orf72 Poly(PR) Dipeptide Repeats Disturb Biomolecular Phase Separation and Disrupt Nucleolar Function. Mol. Cell 74, 713–728.e6. 10.1016/j.molcel.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook CN, Wu Y, Odeh HM, Gendron TF, Jansen-West K, Del Rosso G, Yue M, Jiang P, Gomes E, Tong J, et al. (2020). C9orf72 poly(GR) aggregation induces TDP-43 proteinopathy. Sci. Transl. Med. 12, eabb3774. 10.1126/scitranslmed.abb3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324.e28. 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. (2020). G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345.e28. 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, et al. (2020). RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361.e317. 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian SB, and Ignatova Z (2018). Dynamic m(6)A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1, e201800113. 10.26508/lsa.201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, and Zhuang X (2020). m(6)A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 16, 955–963. 10.1038/s41589-020-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Yu C, Guo M, Zheng X, Ali S, Huang H, Zhang L, Wang S, Huang Y, Qie S, and Wang J (2019). Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 10, 2355–2363. 10.1021/acschemneuro.8b00657. [DOI] [PubMed] [Google Scholar]
- 16.Deng Y, Zhu H, Xiao L, Liu C, Liu YL, and Gao W (2021). Identification of the function and mechanism of m6A reader IGF2BP2 in Alzheimer’s disease. Aging (Albany NY) 13, 24086–24100. 10.18632/aging.203652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Ren Y, Mao K, Hua F, Yang Y, Wei N, Yue C, Li D, and Zhang H (2018). FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem. Biophys. Res. Commun. 498, 234–239. 10.1016/j.bbrc.2018.02.201. [DOI] [PubMed] [Google Scholar]
- 18.Qin L, Min S, Shu L, Pan H, Zhong J, Guo J, Sun Q, Yan X, Chen C, Tang B, and Xu Q (2020). Genetic analysis of N6-methyladenosine modification genes in Parkinson’s disease. Neurobiol. Aging 93, 143.e9–143.e13. 10.1016/j.neurobiolaging.2020.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Jiang L, Lin W, Zhang C, Ash PEA, Verma M, Kwan J, van Vliet E, Yang Z, Cruz AL, Boudeau S, et al. (2021). Interaction of tau with HNRNPA2B1 and N(6)-methyladenosine RNA mediates the progression of tauopathy. Mol. Cell 81, 4209–4227.e12. 10.1016/j.molcel.2021.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chew J, Cook C, Gendron TF, Jansen-West K, Del Rosso G, Daughrity LM, Castanedes-Casey M, Kurti A, Stankowski JN, Disney MD, et al. (2019). Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener. 14, 9. 10.1186/s13024-019-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Mewborne QT, Girdhar A, Sheth U, Coyne AN, Punathil R, Kang BG, Dasovich M, Veire A, DeJesus Hernandez M, et al. (2022). Poly(ADP-ribose) promotes toxicity of C9ORF72 arginine-rich dipeptide repeat proteins. Sci. Transl. Med. 14, eabq3215. 10.1126/scitranslmed.abq3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, and Jaffrey SR (2019). m(6)A enhances the phase separation potential of mRNA. Nature 571, 424–428. 10.1038/s41586-019-1374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan J Ries BFP, Poh HX, Sim N, and Jaffrey SR (2022). m6A governs length-dependent enrichment of mRNAs in stress granules. Preprint at bioRxiv. 10.1101/2022.02.18.480977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, and He C (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, Hu Y, Qiu J, Pu L, Tang J, and Wang X (2021). HIF-1alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target. Ther. 6, 76. 10.1038/s41392-020-00453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauer J, and Jaffrey SR (2018). FTO, m(6) Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 592, 2012–2022. 10.1002/1873-3468.13092. [DOI] [PubMed] [Google Scholar]
- 28.Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. (2018). Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 71, 973–985.e5. 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khong A, Matheny T, Huynh TN, Babl V, and Parker R (2022). Limited effects of m(6)A modification on mRNA partitioning into stress granules. Nat. Commun. 13, 3735. 10.1038/s41467-022-31358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PEA, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. (2013). Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844. 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao W, Todd TW, Wu Y, Jones CY, Tong J, Jansen-West K, Daughrity LM, Park J, Koike Y, Kurti A, et al. (2022). Two FTD-ALS genes converge on the endosomal pathway to induce TDP-43 pathology and degeneration. Science 378, 94–99. 10.1126/science.abq7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subach OM, Cranfill PJ, Davidson MW, and Verkhusha VV (2011). An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One 6, e28674. 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakrabarty P, Rosario A, Cruz P, Siemienski Z, Ceballos-Diaz C, Crosby K, Jansen K, Borchelt DR, Kim JY, Jankowsky JL, et al. (2013). Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One 8, e67680. 10.1371/journal.pone.0067680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamprecht MR, Sabatini DM, and Carpenter AE (2007). CellProfiler: free, versatile software for automated biological image analysis. Biotechniques 42, 71–75. 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 37.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. (2016). C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 19, 668–677. 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, et al. (2014). Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 128, 505–524. 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, et al. (2015). Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348, 1151–1154. 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.


