Summary
Exposure to ultraviolet radiation (UVR) is harmful to living cells, leading organisms to evolve protective mechanisms against UVR-induced cellular damage and stress1,2. UVR, particularly UVB (280–320nm), can damage proteins and DNA, leading to errors during DNA repair and replication. Excessive UVR can induce cellular death. Aquatic organisms face risk of UV exposure as biologically harmful levels of UVB can penetrate >10 meters in clear water3. While melanin is the only known sunscreen in vertebrates, it often emerges late in embryonic development, rendering embryos of many species vulnerable during the earlier stages. Algae and microbes produce a class of sunscreening compounds known as mycosporine-like amino acids (MAAs)4. Fish eggs contain a similar compound called gadusol, whose role as a sunscreen has yet to be tested despite its discovery over 40 years ago5. The recent finding that many vertebrate genomes contain a biosynthetic pathway for gadusol suggests that fish may produce and use this molecule as a sunscreen6. We generated a gadusol-deficient mutant zebrafish to investigate the role of gadusol in protecting fish embryos and larvae from UVR. Our results demonstrate that maternally provided gadusol is the primary sunscreen in embryonic and larval development, while melanin provides modest secondary protection. The gadusol biosynthetic pathway is retained in the vast majority of teleost genomes but is repeatedly lost in species whose young are no longer exposed to UVR. Our data demonstrate that gadusol is a maternally provided sunscreen that is critical for early-life survival in the most species-rich branch of the vertebrate phylogeny.
eTOC
Sunscreens have evolved to mitigate ultraviolet radiation (UVR) induced stress. Here Rice et al. show that a maternally provided transparent compound called gadusol is a powerful sunscreen that protects fish embryos. They find that gadusol synthesis genes have been repeatedly lost in fish species whose young are not exposed to UVR.
Graphical Abstract
Results
Gadusol is maternally provided and protects embryos and larvae from UVR
To test if gadusol is a sunscreen in vertebrate embryos, we used CRISPR-Cas9 to delete most of exon 2 of zebrafish eevs, which encodes the enzyme essential for the first step in gadusol biosynthesis (Figures 1A, S1A, S1C, Data S1). We chose zebrafish for these experiments because they live and spawn in shallow sunlit waters, they are known to produce gadusol6, and they are genetically tractable. Grown in our animal facility, where they are protected from UVR, homozygous eevs mutant females and males survived to fertile adulthood like their wild-type peers. Using reciprocal crosses between homozygous mutant adults (eevs−/−) and wild-type adults (eevs+/+), we generated heterozygous mutant embryos that lack maternal contribution of gadusol (hereafter referred to as Meevs) and heterozygous mutant embryos that retain this maternal contribution (referred to as eevs+/−) (Figure 1B). Notably, Meevs and eevs+/− embryos have identical genotypes but either lack or possess maternally provided gadusol, as judged by mass spectrometry (Figure 1B) and UV-spectrophotometry (Figure S1C). We generated maternal-zygotic homozygous mutant embryos (referred to as MZeevs) from in-crosses of homozygous mutant parents. Immediately after fertilization, gadusol was nearly absent in MZeevs embryos and indistinguishable from Meevs (Figure 1C, Figure S1C). We next asked how long maternally provided gadusol persisted in embryos and larvae. We compared gadusol abundances from whole embryos and larvae with the following genotypes: eevs+/+ (wild-type), Meevs, and MZeevs. We found only a modest increase in gadusol abundance in Meevs relative to MZeevs at 5 days post-fertilization (dpf) (Figure 1C). Together with transcriptomic data that shows eevs mRNA is only present during early stages of oogenesis7 (Figure S1B) and absent from embryos6,8, our data suggests that maternally synthesized and deposited gadusol is the source of nearly all gadusol in the developing zebrafish. This is an example of a maternal effect, where disruption of the eevs gene in mothers eliminates deposition of gadusol presence in their embryos, regardless of embryo genotype.
Figure 1. Gadusol is maternally provided and protects zebrafish embryos and larvae from UVB.
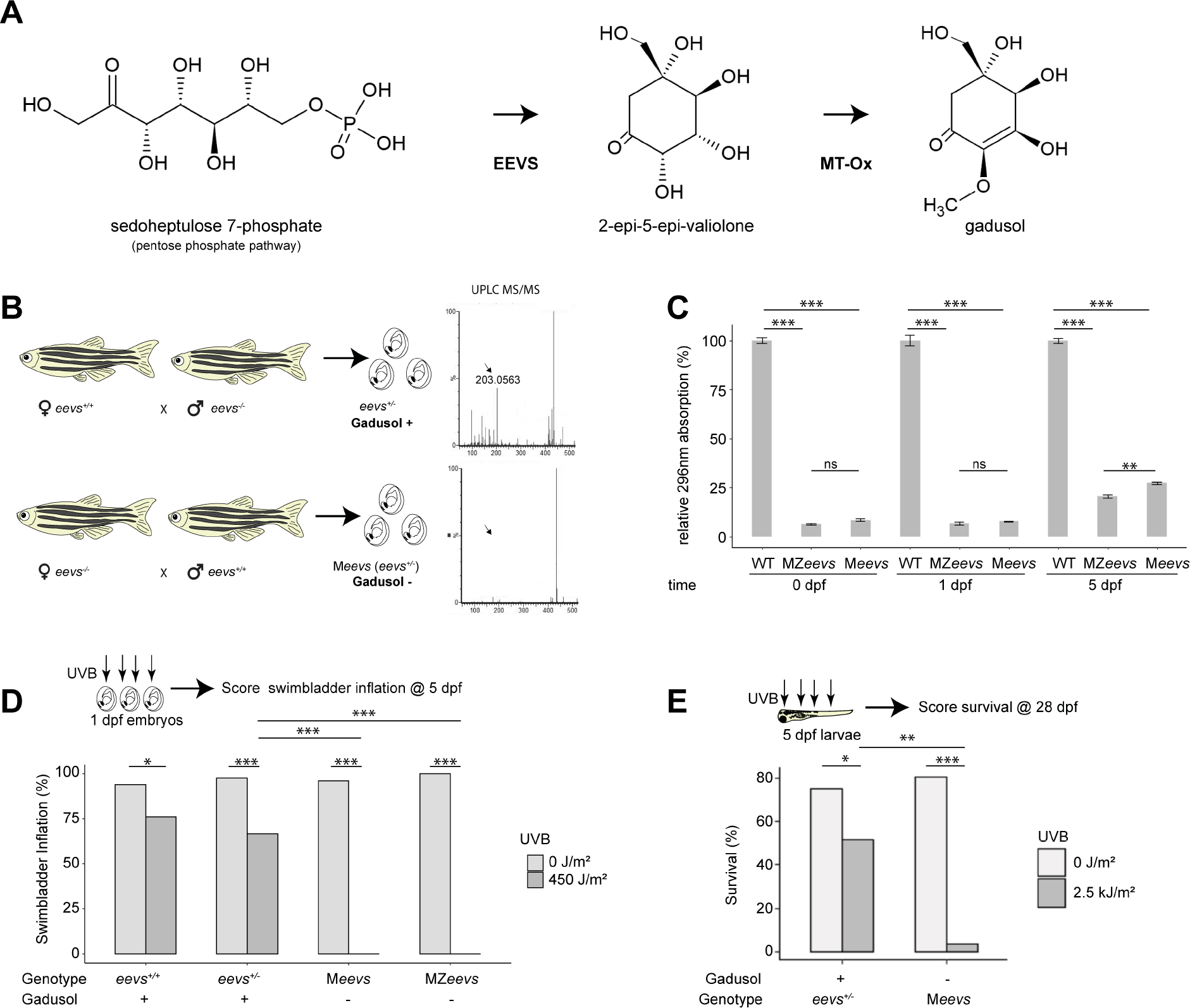
A, The biosynthetic pathway for gadusol production.
B, Experimental diagram for generating heterozygous mutant eevs+/− embryos and larvae with identical genotypes but containing maternal contribution of gadusol (top) or depleted of maternally provided gadusol (bottom). On the right, UPLC mass spectra of 0 hpf egg extracts from each genetic cross; arrow indicates gadusol mass.
C, Absorption values at 296nm from the indicated genotypes at the indicated timepoints. All absorption values normalized to wild type. Error bars indicate standard deviation from biological replicates.
D, Distribution of swimbladder inflation scored in 5 dpf larvae, with genotypes and gadusol presence indicated, after mock exposure (grey) or UVB exposure (dark grey) at 24 hpf stage. All embryos resulted from crosses between TU and AB strain parents, except the TU in-cross that generated MZeevs embryos. From left to right, n = 50, 50, 75, 75, 100, 97, 50, 50; N = 2, 2, 3, 3, 4, 4, 2, 2.
E, Survival distribution scored at 28 dpf, with genotypes and gadusol presence indicated, after mock exposure (grey) or UVB exposure (dark grey) at 5 dpf. From L-R n = 100, 95, 100, 97, N = 4 for all groups.
n = embryos/larvae. N = clutches. statistics: student t test C, Fisher’s Exact t-test D, E, *p<0.05; **p<0.01; ***p<0.0001. See also Figures S1 and S2, Data S1 and S3.
To determine if gadusol protects zebrafish embryos against UVB, we developed an assay to deliver precise doses of UVB to embryos and measure the effect on swim bladder inflation at 5 dpf (a hallmark of healthy development essential for survival, Figures S2A–S2D). We found that 450 joules (J)/m2 of UVB (fluence rate: 2.5 W/m2, see Methods) delivered at 24 hours post-fertilization (hpf) resulted in ~75% swim bladder inflation in wild-type and eevs+/− embryos, respectively, but did not result in gross developmental defects (Figures 1D, S2C). In stark contrast, MZeevs and Meevs embryos were extremely vulnerable to the same dose of UVB; all embryos failed to inflate their swim bladders (Figure 1D).
Since zygotic production of gadusol was still minimal at 5 dpf (Figure 1C), we hypothesized that larvae lacking maternal gadusol should be highly sensitive to UVB at this later stage. We repeated UVB dosage curves on 5 dpf larvae and identified 2.5 kJ/m2 for a significant impact on wild-type larvae survival (Figure S2E). We grew UV-exposed and control larvae in our fish facility nursery to 28 dpf, which requires developing animals to forage for food to survive. We found that only 2% of exposed Meevs larvae survived, compared to ~50% of controls exposed to the same dose of UVB (Figure 1E). Together, these data demonstrate that maternally provided gadusol provides powerful UVB protection to early embryos and older larvae.
Gadusol prevents DNA damage and apoptosis
Next, we sought to understand the mechanism by which gadusol protects embryos from UVB. In other species, gadusol and related molecules were hypothesized to function as antioxidants as well as sunscreens5,6,9. To test if gadusol serves as an antioxidant in zebrafish embryos, we exposed 24 hpf embryos to hydrogen peroxide to induce oxidative stress. At 5 dpf, gadusol-depleted Meevs and control eevs+/− embryos had similar responses to oxidative stress, suggesting that gadusol does not function as an antioxidant in vivo (Figures S3A–S3B).
To test if gadusol serves as a sunscreen by absorbing UVB, we measured the production of cyclobutane pyrimidine dimers (CPDs), a signature of UVB-induced DNA damage10. If gadusol acts as a sunscreen, then it would absorb UVB photons and shield the underlying DNA from CPD formation. We exposed 24 hpf embryos to UVB and used immunohistochemistry to detect CPDs and quantify fluorescence intensity. Embryos that lacked gadusol had significantly higher levels of CPD formation after UVB exposure compared to controls containing gadusol (Figures 2A–2B). CPDs are cytotoxic and induce apoptosis at high abundance. We used immunohistochemistry to detect a fast-acting apoptotic marker (activated caspase-3) in embryos exposed to UVB11 (Figure 2C, Figure S3C). We found that embryos lacking gadusol had increased levels of apoptotic nuclei, relative to controls (Figure 2C), supporting a role for gadusol in absorbing UVB and preventing DNA damage.
Figure 2. Gadusol functions as a sunscreen preventing DNA damage and apoptosis.
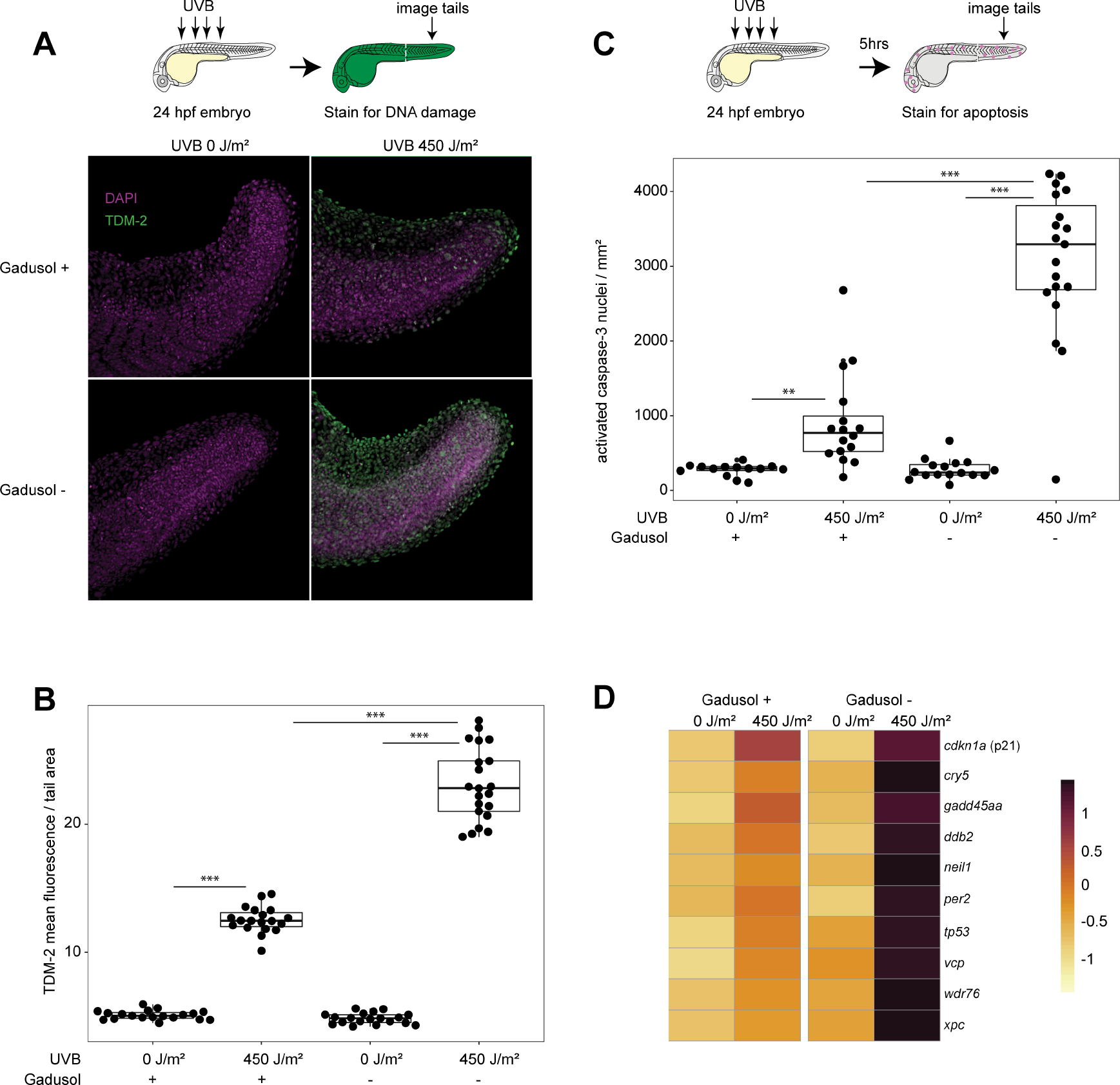
A, Immunohistochemistry, using an antibody that recognizes CPDs (TDM-2), on 24 hpf embryos immediately after mock or UVB exposure. Representative images shown.
B, Quantification of CPD labeling normalized to tail area (mm2). From left to right, n = 19, 19, 19, 21; N = 2 for all groups.
C, Quantification of immunohistochemistry, using an antibody that recognizes activated caspase-3. n = 14, 16, 16, 20. N = 2 for all groups.
D, Significant upregulation of select UVR response and DNA damage GO term-associated genes measured from the indicated conditions and genotypes using RNAseq on 24 hpf embryos after mock exposure or UVB exposure. RNA was collected 5 hours post mock or UVB exposure. Gene expression is scaled by rows. Significance determined via Fishenricher.26.
Student’s T-test P*<0.05; P**<0.01; P***<0.0001. n = number of embryos. N = number of clutches. See also Figure S3, Data S3, and Table S1.
To characterize transcriptional responses to UVR in the absence of gadusol, we performed RNAseq comparing gadusol-depleted Meevs and wild-type embryos. Five hours after exposure to UVB, embryos lacking gadusol had significantly higher expression of many key stress response genes (tp53, gadd45aa, ddb2, and cdkn1a) relative to UVB-treated controls (Figure 2D). GO terms enriched in UV-exposed gadusol-depleted embryos included response to UV, response to DNA damage, response to light, and other stress response terms (Figure S3D, Table S1). Several of these genes were also modestly induced by visible light (Figure S3E), consistent with previous reports12,13, but their induction was similar between Meevs and wild type embryos. Together, our imaging and gene expression data confirm that gadusol in zebrafish embryos acts as a true sunscreen to provide efficient protection against UV-induced DNA damage, cellular stress, and cell death.
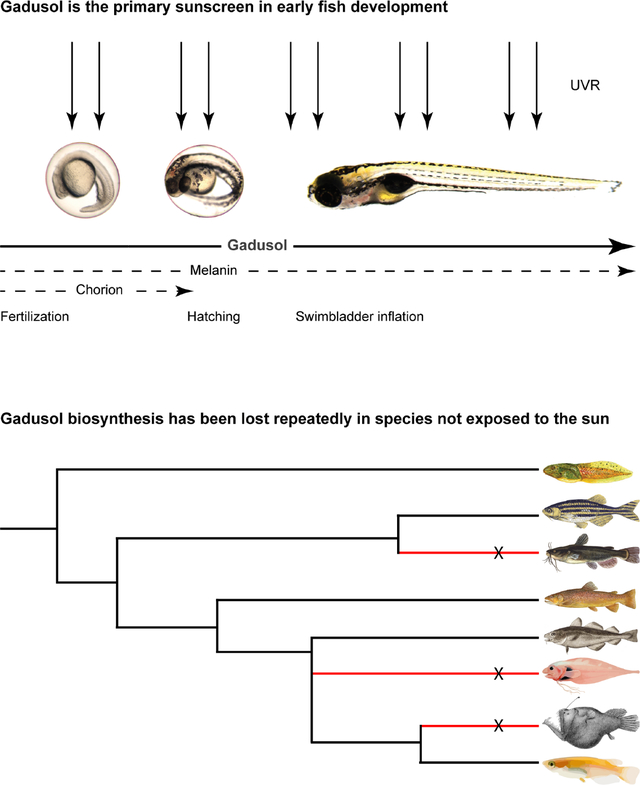
Gadusol is the primary sunscreen in early fish development
In light of our finding that gadusol acts as a sunscreen, we compared the relative sunscreening potency of gadusol with that of other potential UV-blocking/absorbing mechanisms in larval zebrafish. Melanin is a well-known sunscreen in many organisms including humans. In zebrafish, melanophores become pigmented around 36 hpf, ultimately forming stripes that partially cover the larval brain and body, a pattern that is stable until ~14 dpf14,15. Melanophores protect the hematopoietic niche in larval zebrafish16, but their role as a whole-body sunscreen remains untested. The nacre/mitfa mutant disrupts a key melanophore master regulator and lacks melanophores. We generated two groups of larvae, each with pigmented and unpigmented siblings. One group contained no maternal gadusol, while the other group contained gadusol (Figure 3A). We treated all 5 dpf larvae with 2.5 kJ/m2 of UVB and assessed survival in the nursery at 28 dpf. Larvae with gadusol were highly resistant to UVB stress, regardless of pigmentation status (Figure 3B). All larvae that lacked gadusol were highly sensitive to UVB, and larvae that lacked both gadusol and melanin were slightly more sensitive to UVB than their pigmented siblings. At a lower UVB dose (1.5 kJ/m2), we also found a modest but significant effect of melanophores in protecting against UVB (Figure S3F). We conclude that while melanin plays a minor role in UVR protection, gadusol is the primary sunscreen in early fish development.
Figure 3. Melanin and the chorion serve as secondary UV-shielding mechanisms in embryonic and larval fish.
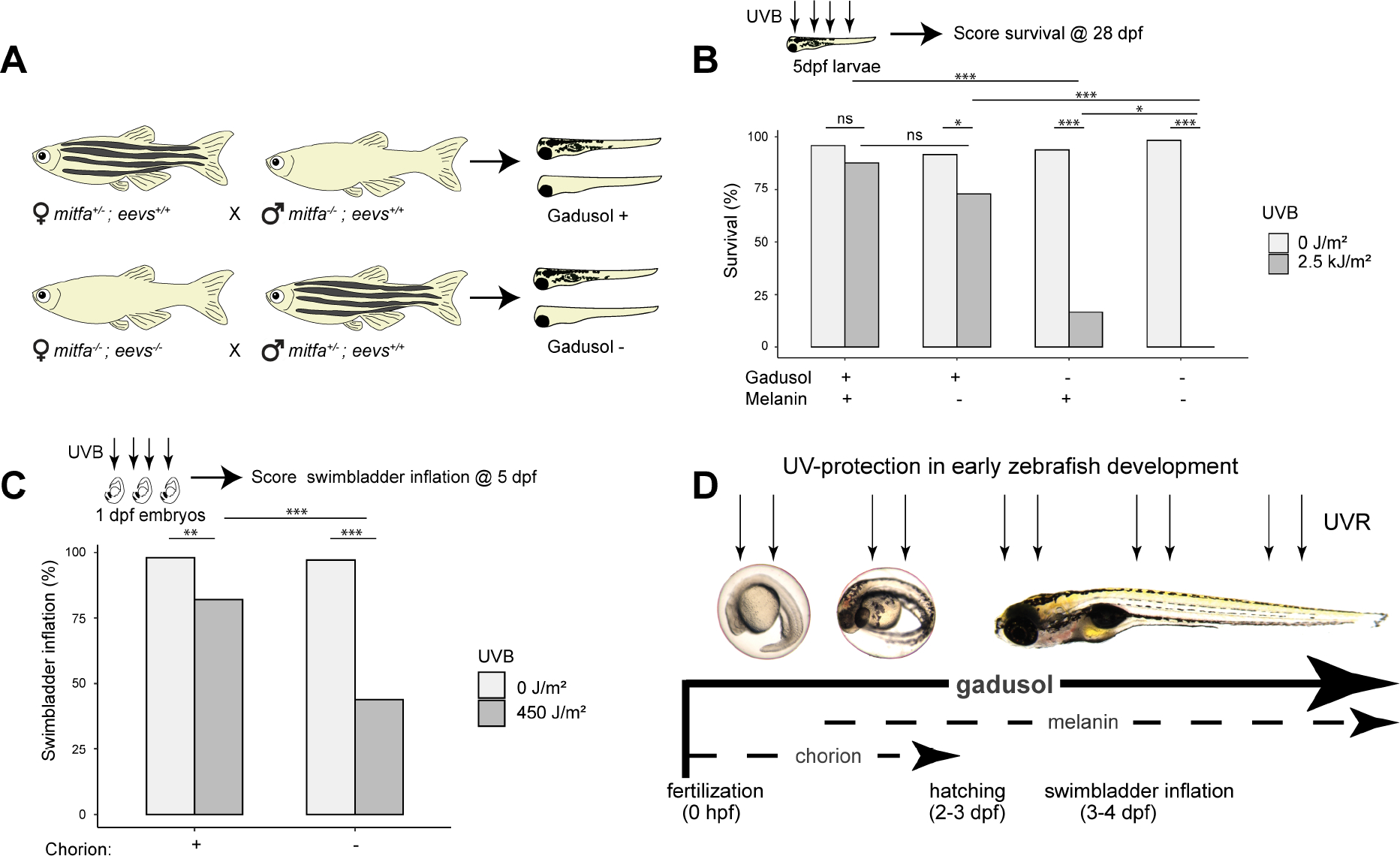
A, Experimental diagram for generating embryos that lack either melanin, maternally provided gadusol, or both.
B, Survival distribution scored at 28 dpf, with presence of melanin and gadusol indicated, after mock exposure (grey) or UVB exposure (dark grey) at 5 dpf. From left to right, n = 48, 48, 48, 48, 48, 36, 60, 36; N = 2 for each group.
C, Distribution of swimbladder inflation scored in 5 dpf larvae after mock exposure (grey) or UVB exposure (dark grey) at 24 hpf stage, with or without chorions. n = 100 for each group. N = 3 for each group.
D, Model illustrating the relative importance and timing of multiple UV-shielding mechanisms used in early zebrafish development.
Fisher’s Exact T-test *p<0.05 ** p<0.01 ***p<0.0001. n = total number of individual embryos/larvae. N = total number of clutches. See also Figure S1 and Data S3.
Another potential UV-protective mechanism is the chorion, the nearly transparent eggshell that contains perivitelline fluid and the embryo from fertilization until 2–3 dpf. We tested the sunscreening role of the chorion by mechanically removing it with forceps and exposing these embryos, and sibling controls that retained the chorion, to 450 J/m2 of UVB at 24 hpf. We found that the chorion does provides significant protection from UVB as ~60% of dechorionated embryos failed to inflate their swim bladders, significantly less than sibling controls (Figure 3C). We examined if gadusol was present in the chorion or in the perivitelline fluid within the chorion but found little to none (Figure S1D). These results suggest that the chorion structure itself can shield some incoming UVB. However, we conclude that the chorion provides less UV protection than gadusol, as gadusol-depleted embryos – even with intact chorions - all failed to inflate their swim bladders when challenged with the same dose of UVB (Figure 1D).
Together, our findings support a model where embryonic and larval fish are protected by multiple layers of UVB protection that span early development (Figure 3D). The egg is maternally loaded with gadusol, which provides the primary and most important layer of UV protection from fertilization until at least 5 dpf. The chorion and melanophores are secondary, and less effective, means of UVR protection. The chorion protects the developing embryo between fertilization and hatching (2–3 dpf), when pigmented melanophores emerge and modestly protect the growing larval fish.
Gadusol has been repeatedly lost in fish species whose embryos are no longer exposed to sunlight
The two-enzyme biosynthetic pathway necessary for gadusol production (Eevs and MT-Ox) is encoded in numerous vertebrate genomes, including fish, birds, reptiles, and amphibians6. Osborn et al. identified the loss of the gadusol biosynthetic pathway in the coelacanth genome, and suggested the loss might be attributable to lack of UV penetration in the deep-sea habitat of this species6. To test for broader patterns of conservation and loss among fish, we surveyed additional genomes, including many species that live in habitats not exposed to UVR. We hypothesized that gadusol synthesis genes would not be required in species that live in deep waters, caves, are live bearers, or use electroreception to navigate habitats with poor light penetrance17. To test this hypothesis, we searched 136 teleost genomes for inactivation or loss of either eevs or MT-Ox. In all species, we identified a syntenic genomic region demarcated by highly conserved flanking genes and assessed the presence or absence of intact ORFs encoding functional copies of eevs and MT-Ox. Our approach largely confirmed that the vast majority of teleosts have functional copies of eevs and MT-Ox6. However, our survey identified 16 independent losses of either the eevs or MT-Ox genes across the teleost phylogeny (Figure 4A, red species). Most of these genomes had lost orthologs of both eevs and MT-Ox, while others had lost only one gene or had pseudogene remnants (Figure 4B). The loss of genes involved in gadusol production was significantly correlated with lifestyle traits that identified species that live or spawn in habitats protected from the sun (p = 0.012) (Figures 4, S4, and Data S2). To corroborate the link between loss of eevs or MT-Ox and loss of gadusol, we measured gadusol levels in medaka embryos, which have intact eevs and MT-Ox genes, and ovaries of channel catfish, which have lost eevs and MT-Ox. We found a strict correlation between the presence of intact genes and maternally provided gadusol (Figure S1E). We conclude that gadusol production has been repeatedly lost during evolution in teleost species whose lifestyles protect them from UVR.
Figure 4. Gadusol production has been lost in several species no longer exposed to UVR.
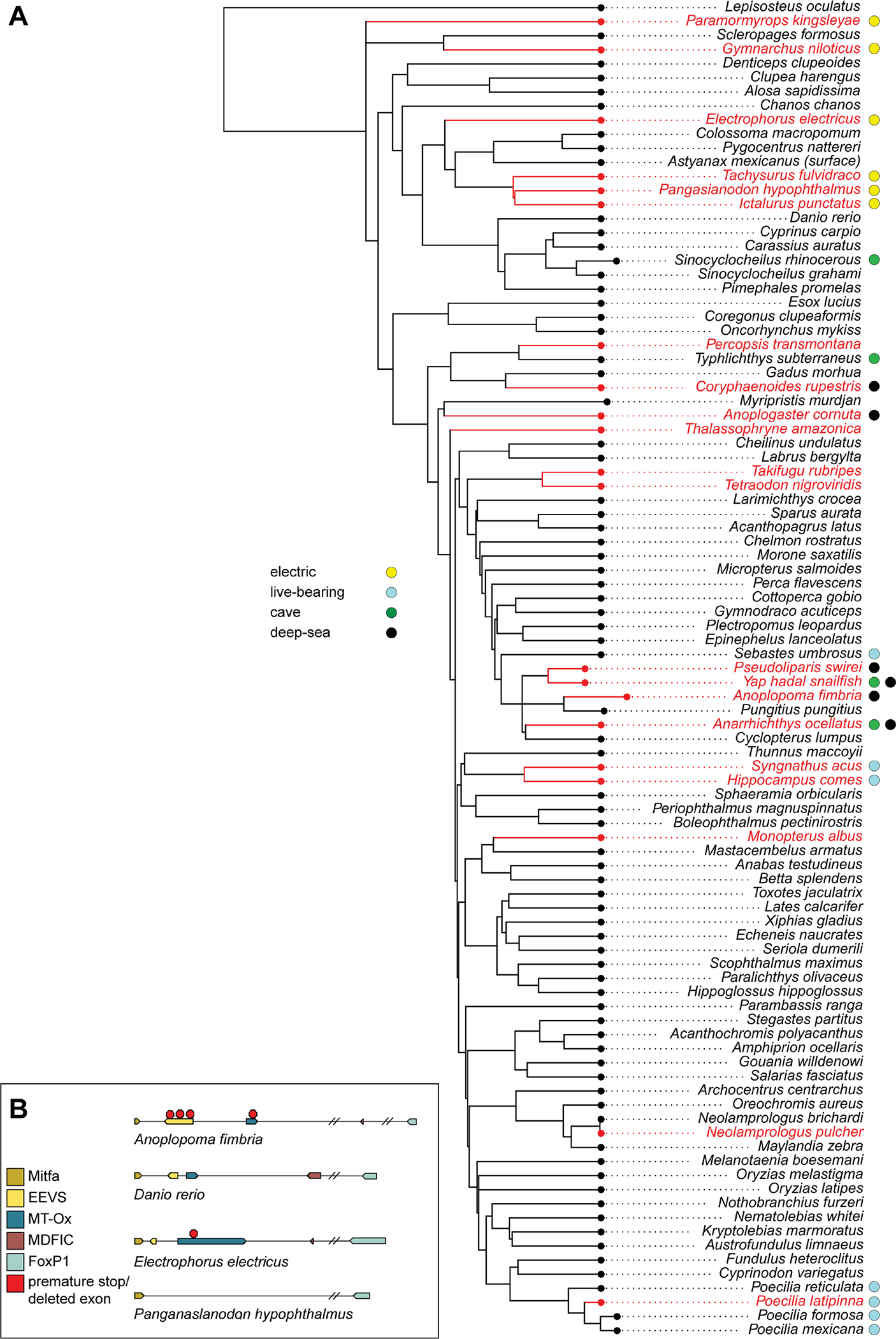
A, For each of 136 teleost species (full tree in Figure S4), we assessed various life history traits that identify habitats that may not require embryonic protection from UVR, including electroreception, live-bearing, cave dwelling, and deep-sea dwelling, indicated with colors in the legend to the left of the phylogeny. For each species, we identified the presence of intact open reading frames for eevs and/or MT-Ox. Species that have lost the genes required for gadusol production are indicated in red. We found 16 independent losses across this phylogeny. We found that fish with these traits are more likely than by chance to lose gadusol (p=0.012).
B, Examples of gadusol synthesis gene loss and pseudogenization in select species. Note Danio rerio has intact eevs and MT-Ox genes and is capable of gadusol production. See also Figures S1, S4 and Data S2).
Discussion
Plants and microorganisms use numerous UV-absorbing compounds as sunscreens2,4. However, other than melanin, the repertoire of vertebrate sunscreens – especially compounds that protect the most vulnerable early stages of development – remain essentially unknown. Here, we provide experimental and phylogenomic evidence that gadusol is an ancient sunscreen essential for protecting fish embryos from UVR. First, we use a CRISPR mutant that disrupts gadusol biosynthesis to show that gadusol is produced during oogenesis and persists in the embryo until at least 5 dpf. Second, we demonstrate that maternally deposited gadusol safeguards embryonic and larval development by preventing UV-induced developmental defects and improving survival. Third, we find that gadusol acts as a true sunscreen preventing the formation of CPDs, a signature of UVB-induced DNA damage, and consequently reducing levels of cell and organismal death. Gadusol does not have any obvious functions beyond protecting against UVR, as mutants survive to adulthood and are fertile. Together, these data demonstrate that gadusol is a maternally provided sunscreen employed during early fish development.
Our work explores two alternative mechanisms of UV protection during early development. We find that the chorion, a transparent eggshell that shields the developing embryo, also provides modest UV protection during embryogenesis. This protection is short lived (zebrafish hatch by 2–3 dpf) but may provide secondary protection during the most vulnerable stages of development. Melanin pigmentation emerges around embryo hatching and serves a relatively modest role as a whole-body sunscreen in 5 dpf larvae. Together, our results show that gadusol is the primary sunscreen across embryonic and larval development, while melanin and the chorion play secondary roles during distinct phases of development.
Finally, our phylogenetic analysis of gadusol biosynthetic genes, building on a previous study6, suggest that gadusol is an ancient sunscreen conserved broadly to protect teleost embryos. However, gadusol production has been repeatedly lost during teleost evolution. Intriguingly, these genes are absent in many fish species whose embryos are not exposed to UVR, including deep sea-dwelling and electroreceptive fish. We suggest that similar to our protected fish facility environment, gadusol is also dispensable for embryonic development in natural environments that lack UVR. In microorganisms, the production of sunscreening compounds have been estimated to require >10% of all metabolic activity4. Perhaps the loss of gadusol production in nutrient-poor dark habitats provides some evolutionary advantage, analogous to the energy conservation hypothesis invoked to explain the repeated loss of eyes in Mexican cavefish18,19. Similar to loss of UV-responsive gene expression in a cavefish12,13, gadusol appears dispensable in species not exposed to sunlight. Once these genes have been lost, descendent species may enter an evolutionary fitness trap where they are confined to breeding environments lacking UVR.
It remains unclear what role gadusol might play in other tetrapods. Functional copies of eevs and MT-Ox have been found in numerous vertebrate genomes6, but to our knowledge the presence of gadusol has never been reported in vertebrates other than fish. Gadusol has been detected in the eggs or embryos of several aquatic invertebrates, including sponge20, starfish20, sea urchin21, and brine shrimp22. We hypothesize that gadusol may also protect early development in these diverse aquatic organisms.
Oxybenzone and octinoxate, two common ingredients in commercial sunscreens, have been recently banned in Hawaii due to concerns of toxicity to coral reefs23. Others have suggested gadusol might be a safe natural sunscreen replacement6,24. Our data supports a role for gadusol as an effective natural sunscreen that warrants further investigation as a preventative agent.
Here, we show that aquatic vertebrates produce and employ an additional sunscreen to melanin. Melanin and gadusol both absorb well in the UVB spectrum. However, melanin also absorbs most wavelengths in the visible light spectrum, making it opaque and conspicuous while gadusol is transparent and invisible. Transparency as camouflage is a common trait in aquatic animals, especially in the open ocean where there is nothing to hide behind25. To date, gadusol has only been detected in aquatic organisms. We speculate that gadusol has been particularly advantageous to these animals as it offers protection from UVR, enabling an organism to stay in nutrient-rich sunlit areas, while remaining optically inconspicuous. We propose that aquatic ecosystems exhibit unique ecological challenges that have selected for the use of a transparent sunscreen.
STAR Methods
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, James A. Gagnon (james.gagnon@utah.edu)
Materials availability
The fish eevs knockout mutant lines generated in this paper (zj2 and zj5) are maintained in the laboratory of James A. Gagnon and are available upon request.
Data and code availability
RNA-seq data have been deposited at GEO under accession number GSE229587 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal Anti-cyclobutane pyrimidine dimers | Cosmo Bio | Cat# NM-DND-001 |
| Rabbit anti-active Caspase-3 | BD Biosciences | Cat# 559565 |
| Alexa Fluor 546 goat anti-mouse IgG | ThermoFisher | Cat# A-11030 |
| Alexa Fluor 594 goat anti-rabbit IgG | ThermoFisher | Cat# A-11012 |
| Biological samples | ||
| Danio rerio embryos | This paper | N/A |
| Danio rerio larvae | This paper | N/A |
| Danio rerio ovaries | This Paper | N/A |
| O. latipes embryos | This Paper | N/A |
| I. punctatus ovaries | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Methanol | Sigma-Aldrich | Cat# 34860 |
| Critical Commercial Assays | ||
| QuanTiTect Reverse Transcription Kit | Qiagen | Cat# 205311 |
| PowerUp SYBR Green Master Mix | ThermoFisher | Cat# A25742 |
| Deposited data | ||
| Code for bioinformatic exploration of loss of gadusol | This paper | https://github.com/nclark-lab/gadusol |
| RNAseq data (raw and analyzed) | GEO | GSE229587 |
| Experimental models: Organisms/strains | ||
| Zebrafish D. rerio (Tübingen strain) | ZIRC | ZL57 |
| Zebrafish D. rerio (AB strain) | ZIRC | ZL1 |
| Zebrafish D. rerio (mitfaw2/w2) | ZIRC | ZL2104 |
| Zebrafish D. rerio (eevs zj2/zj2) | This paper | N/A |
| Zebrafish D. rerio (eevs zj5/zj5) | This paper | N/A |
| Oligonucleotides | ||
| Primers for sequences cloning, see Table S1 | This paper | N/A |
| Primers for qRT-PCR, see Table S1 | This paper | N/A |
| Software and algorithms | ||
| GraphPad | GraphPad Software | https://graphpad.com |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Microsoft Excel Data Analysis Student t test | Microsoft | N/A |
| Other | ||
| Benchtop UV Transilluminator | UVP | M-15V P/N 95–0456-01 |
| UVB broad band bulb (306nm) | Ushio | G8T5E |
| Digital UV Radiometer UVB | Solarmeter | 6.0 UVB |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
in vivo animal studies
zebrafish D. rerio (Tübingen and AB strains) embryos and larvae
All zebrafish work was performed at University of Utah’s CBRZ zebrafish facility. This study was conducted under the approval of the Office of Institutional Animal Care and Use Committee (IACUC no. 18–2008) of the University of Utah’s animal care and use program. Zebrafish were maintained in a water circulation system at 28° C with a 14hr light and 10hr dark cycle. Fish were fed twice daily. Embryos were either exposed to UVB at 1 dpf or 5 dpf.
METHOD DETAILS
Generation of eevs mutant lines
To generate a stable gadusol-depleted mutant line, eevs was targeted using CRISPR-Cas9 mutagenesis. Four gRNAs (Data S1) were designed using ChopChop27, targeting exon 2 (Figure S1A) due to the lack of suitable target sites within the small exon 1. Guide RNAs were synthesized from DNA oligos using standard protocols28. Freshly laid wild-type TU-strain embryos were injected with SpCas9 protein (NEB) mixed with gRNAs (~300 ng/ul), KCl, and phenol red. 1–2 nanoliters were injected into each embryo. Mosaic mutant embryos were raised to adulthood and outcrossed to wild-type Tübingen strain. Primers designed from ChopchopV227 were used to amplify the region targeted for CRISPR editing and to select for edited alleles with large deletions. A compound deletion allele was identified by Sanger sequencing that removes 393 bp from the eevs open reading frame (Figure S1A, sequences in Data S1) (Genewiz). This eevs mutant allele was given the designation zj2 and can be genotyped using PCR with allele specific primers (Data S1). Sibling fish with the zj2 allele were crossed to produce homozygous eevszj2/zj2 fish, labeled as eevs−/− in Figures 1B–1E and Figures 2A–2D. Because mitfa and eevs are adjacent genes in the zebrafish genome, an additional eevs mutant line was generated in the mitfaw2/w2; mpv17−/− mutant background using the CRISPR protocol described above. A compound deletion allele was identified by Sanger sequencing that removes 161 bp from the eevs open reading frame (Figure S1A, sequences in Data S1). This mitfa; eevs double mutant allele was given the designation zj5, and was used in Figures 3A–3B. Embryos resulting from crosses of eevs−/− mothers had little to no gadusol compared to wild-type embryos, confirming the successful generation of gadusol-depleted lines.
Gadusol extraction and UPLC MS/MS detection
Gadusol was extracted twice from embryos (7.5mg of vacuum dried egg material, crushed with a microfuge pestle) using 150 ul of a (80:20, v/v) methanol:water solution. The extraction supernatant was analyzed using ultraperformance liquid chromatography (Waters Acquity I-Class, 2.1 × 100 mm BEH Amide column) and mass spectrometry (Waters Xevo G2 QToF) (UPLC-MS) in negative ionization mode (detector range of 50–2000 Da). We used a regular phase chromatography method starting with 95% acetonitrile (+0.1% formic acid) and 5 % water (+0.1% formic acid) following a linear gradient over 12 minutes ending with 30% acetonitrile (+0.1% formic acid). Analytical standards of pure gadusol were run during the same acquisition run to match the retention time and observed mass between embryo samples and the pure standard.
Gadusol detection via Nanodrop
To monitor gadusol production the UV-vis spectrometry on a Nanodrop was employed to determine relative gadusol concentrations. Briefly, 25 embryos/larvae were placed in a microfuge tube. All excess water was removed with a Pasteur pipette. 100 ul of 80:20 (v:v) methanol:water was added to embryos. Embryos were mashed with a microfuge pestle for 15 seconds. Samples were left to extract for at least 15 minutes, and then centrifuged at 12,000 g. Clear supernatant, containing polar compounds such as gadusol, was separated and analyzed on the nanodrop.
UV exposure, swim bladder inflation, and survival assays
24 hpf embryos were exposed to 450 J of UVB as measured on a radiometer (Solarmeter UVB) at a fluence rate of 2.5 W/m2 in 30ml of clear E3 media. This is a conservative estimate of a physiologically relevant UVB dose that fish embryos would routinely experience in the wild16. A raised and inverted UVP transilluminator with 306 nm broadband UVB bulbs was used (Ushio G8T5E) on the “low” setting (see Figures S2A–S2B). Embryos were returned to the incubator and kept in the dark after mock or UV exposure. Swim bladder inflation was scored at 5 dpf by adding ice to the petri dish to stun the larvae, followed by manual counting on a dissection scope. A standard dose curve was conducted to determine that 450 J/m2 was an appropriate dose (Figure S2C). 5 dpf larvae were exposed to a dose curve to determine that 2.5 kJ/m2 was an appropriate dose (Figure S2E). After mock or UV exposure, larvae were placed in an incubator for 1 day (dark) and then placed in the nursery at 6 dpf. Survival was scored at 28 days post-fertilization to ensure that all living juveniles could feed on their own and were not being sustained on maternal yolk. See also Data S3.
Determination of CPDs in 24 hpf embryos
24 hpf embryos were dechorionated to obtain more consistent UV exposure. Embryos were exposed to 450 J/m2 of UVB and then immediately fixed after exposure in 4% PFA for 1 hour at 25°C. After exposure to UVR, embryos were kept in the dark and covered in tin foil while being fixed in PFA to avoid photoreactivation. Fixed embryos were then washed in PBST. Embryos were exposed to 2 M HCl for 1 hour to break apart dsDNA and expose CPD epitopes. Samples were blocked in 5% NGS + PBST. Mouse anti-CPD primary antibody (TDM-2, Cosmo Bio) was used to stain for CPDs. Goat anti-mouse AF546 secondary antibody (Invitrogen) was used to visualize CPDs. Embryos were also stained with DAPI to visualize nuclei. Prior to imaging on a confocal microscope, tails were removed from embryos and placed on a flat glass slide with a small drop of PBST. A cover slip was mounted over the tails and sealed with nail polish. Tails were then imaged on an inverted confocal microscope with a 20x objective (Zeiss 880). Images were analyzed using ImageJ29 to determine mean fluorescence intensity / tail area using the DAPI channel to create a mask for the tail. See also Data S3.
Apoptosis assay
24 hpf embryos within chorions were exposed to 450 J of UVB and then placed in the incubator for 5 hours. Chorions were removed and embryos were fixed for 1 hour in 4% PFA. Embryos were stained with an activated caspase-3 antibody (BD Biosciences, anti:Rabbit) to mark apoptotic cells. Goat anti-rabbit AF594 secondary antibody (Invitrogen) was used to visualize apoptotic cells. Embryo tails were removed, processed, and imaged as above. ImageJ was used to process images and count the number of activated caspase-3 positive nuclei/mm2. See also Data S3.
RNAseq sample prep, library prep, sequencing, and analysis
After 5 or 24hrs post UV exposure embryos were smashed with a microfuge pestle (MTC Bio) and RNA extracted using TRI Reagent (Zymo) and purified via Direct-zol RNA Miniprep Plus (Zymo). Library prepared using NEBNext Ultra II Directional RNA Library Prep with poly(A) mRNA Isolation. Samples then sequenced with Total RNA (eukaryote) NovaSeq SP Reagent Kit v1.5_50×50 bp. Each sample sequenced to a depth of 25 million reads. Reads aligned using STAR30 and zebrafish reference genome (GRCz11). Optical duplicates removed and adapters trimmed. Differential expression analysis conducted with DESeq231 and specifically the Bioconductor package32. See also Table S1.
qRT-PCR
24 hpf embryos were exposed to 5hrs of cool white LED light while control embryos were kept in constant darkness. RNA was extracted as described above. cDNA was synthesized using QuanTiTect Reverse Transcription Kit (Qiagen). PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) was used for qPCR reactions in a QuantStudio 3 (Thermo Fisher Scientific). Primers for cry5, ddb2, per2, and xpc were obtained from Weger et al.33. Neil1 was obtained from Zhao et al.12. Housekeeping control gene elfa was obtained from McCurly and Callard34. Fold expression was calculated using the 2(-Delta Delta C(T)) method35. Primers listed in Data S1C.
Generating embryos that lack melanin and gadusol
To generate embryos that lacked melanin, mitfaw2/w2 fish were crossed with mitfa+/w2 fish to produce clutches of 1:1 pigmented:unpigmented siblings, all with maternally provided gadusol (Figure 3A). To generate embryos that lack both melanin and gadusol, mitfa+/w2; mpv17−/−; eevszj5/zj5 females were crossed to mitfaw2/w2; eevs+/+ males to produce 1:1 pigmented:unpigmented siblings that all lacked maternal gadusol. Lack of gadusol was confirmed using Nanodrop.
Chorion UV protection assay
24 hpf wild-type TU-strain embryos were manually dechorionated with forceps in a dish with a thin film of 0.5% agar on the base of the dish. Embryos were moved with a fire-smoothened Pasteur pipette. Embryos were exposed to 450 J of UVB as described above and then placed in incubator and swim bladder inflation was scored at 5 dpf. See also Data S3.
Phylogenetic analysis of eevs and MT-Ox presence
123 genomes were gathered from the UCSC genome ark (GenArk) and additional 11 genomes for deep sea and electro-receptive fish were gathered NCBI genomes for all except the Yap Hadal snailfish36 and pseudoliparis swirei37. A BLAST database for each species was created by using the zebrafish sequence spanning from FRMD4B to FOXP1 to find the same region in all curated genomes. If there was no BLAST hit for FOXP1 or MITF then the genome was dropped for low quality. We then performed a tBLASTn search on the created databases for the remaining genomes, using the zebrafish EEVS and MtOX translated nucleotide sequence as the query. If there was no hit for EEVS or Mt-OX in the tblastn search, we expanded the search from the FRMD4B-FOXP1 region to the entire genome. If there were still no hits at an e-value <10e-50 that species was labeled as not having gadusol. If a species did have a tblastn hit and an e-value < 10e-50 the hits were analyzed for pseudogenization by aligning back to the zebrafish mRNA sequence. The aligned regions were checked for missing exons, frameshifts causing premature stop codons, as well as checked for potential genome masking. If the alignment showed either a deletion of an exon or a premature stop codon and had no evidence of masked regions, the gene was called a pseudogene and marked as “absent” for that species.
To correlate the presence/absence of gadusol with life history traits we first collected life history data for all species (Data S2) from information available on fishbase38. The life history traits that we annotated were: deep-sea, nocturnality, live-bearing, electro-reception, and cave dwelling. We then built a species tree using fishtree39 and added the Yap hadal snailfish36 and Pseuolapris swirei37 using the phylogenetic relationship determined in Mu et. al36. Due to gene loss in sister species not being independent, we used Bayestraits40 to perform the correlation test. We used discrete model testing and a likelihood ratios test comparing each of the five life-history traits to loss of gadusol (Data S2).
When running Bayestraits the loss of gadusol (parameter beta1 in the independent model and q31 and q42 in dependent model) was set as trait one and the various life history traits were set as trait two. The rate at which gadusol can be regained after loss was constrained to zero because we were scoring for loss of the gene, and assumed it is nearly impossible to regain the gene, especially in the short time span we are investigating. The parameters that estimate the rate of life history traits changing from absent to present (q12,q34,q21,q43) were constrained to equal to each other, under the assumption that it is unreasonable that a fish would change its lifestyle after loss of gadusol. When comparing the cave life history to gadusol loss, the parameter that estimates the rate of moving from cave to surface (q21 and q43) was constrained to zero under the assumption that species do not re-emerge from a cave after adapting to that life-style.
The significance of the correlation between life-history trait and loss of gadusol was determined using a likelihood ratio test which is calculated by 2*((dependent model likelihood)-(independent model likelihood)). The significance is then determined using a chi-sq distribution with 2 degrees of freedom.
All parameters and code to re-run these models can be found in https://github.com/nclark-lab/gadusol
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed using GraphPad (graphpad.com/quickcalcs/contingency1) when Fisher’s exact t-test was being performed. Student’s t-test was performed using Microsoft Excel. Two-tailed p values were calculated assuming equal variance in samples. P values <0.05 were considered statistically significant.
Supplementary Material
Data S1. Oligos and DNA sequences for eevs mutant. Related to STAR methods and Figures 1, S1, and S3.
A) gRNAs and Primers for eevs mutants. B) eevs wildtype and mutant sequences. C) Primers for qRT-PCR.
Data S2. Life history trait scoring and statistical analysis for gadusol loss across fish species. Related to STAR methods and Figures 4 and S4.
A) Life history traits. B) Absence/Presence of Gadusol Enzymes. C) Bayes traits statistics.
Data S3. Complete data for main figures. Related to STAR methods and Figures 1–3.
A) Figure 1C. B) Figure 1D. C) Figure 1E. D) Figure 2A. E) Figure 2C. F) Figure 3B. G) Figure 3C.
Table S1. GO term enrichment from RNAseq. Related to STAR methods and Figures 2 and S3.
GO term enrichment from RNAseq. GO terms found using fishenricher (Kuleshov et al. 2016); only GO terms with Padj. <0.05 are shown.
Highlights.
Gadusol is a powerful sunscreen that protects early stages of fish development
Gadusol is produced by the mother and deposited into the egg
Gadusol is a more efficient sunscreen during larval stages than melanin
Gadusol synthesis has been lost in many species whose young are not exposed to UVR
Acknowledgements:
We thank all members of the Gagnon lab for discussions and comments. We thank Phyliss Coley, Julie Hollien, Taifo Mahmud, and Angie Serrano for help with protocols, reagents, and equipment, and Nels Elde, Nitin Phadnis, Alex Schier, and Michael Shapiro for comments on the manuscript. We thank Tyler Jumper for sharing channel catfish ovary tissue. We thank CZAR and CBRZ staff, especially Nathan Baker, for zebrafish care. We thank the Cell Imaging and the HCI Bioinformatics core facilities. This research was conducted on the traditional and ancestral homeland of the Shoshone, Paiute, Goshute, and Ute Tribes. We affirm and support the University of Utah’s partnership with Native Nations and Urban Indian communities. This project was supported by National Institutes of Health grant R35GM142950 (JAG), by the GSRM summer program supported by National Institutes of Health grant R25HG009886 (JM), and by startup funds from the Henry Eyring Center for Cell & Genome Science and the University of Utah (JAG).
Inclusion and Diversity:
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science.
Footnotes
Declaration of interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AND SOFTWARE AVAILABILITY
RNAseq data has been deposited at GEO accession number GSE229587.
REFERENCES
- 1.Dahms H-U and Lee J-S (2010). UV radiation in marine ectotherms: Molecular effects and responses. Aquatic Toxicology 97, 3–14. [DOI] [PubMed] [Google Scholar]
- 2.Cockell CS and Knowland J (1999). Ultraviolet radiation screening compounds. Biological Reviews 74, 311–345. [DOI] [PubMed] [Google Scholar]
- 3.Williamson CE (1995). What role does UV-B radiation play in freshwater ecosystems? Limnology and Oceanography 40, 386–392. [Google Scholar]
- 4.Gao Q and Garcia-Pichel F (2011). Microbial ultraviolet sunscreens. Nature Reviews Microbiology 9, 791–802. [DOI] [PubMed] [Google Scholar]
- 5.Plack PA, Fraser NW, Grant PT, Middleton C, Mitchell AI, and Thomson RH (1981). Gadusol, an enolic derivative of cyclohexane-1,3-dione present in the roes of cod and other marine fish. Isolation, properties and occurrence compared with ascorbic acid. Biochem J 199, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborn AR, Almabruk KH, Holzwarth G, Asamizu S, LaDu J, Kean KM, Karplus PA, Tanguay RL, Bakalinsky AT and Mahmud T (2015). De novo synthesis of a sunscreen compound in vertebrates. Elife 4, e05919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Kossack ME, McFaul ME, Christensen LN, Siebert S, Wyatt SR, Kamei CN, Horst S, Arroyo N, Drummond IA and Juliano CE (2022). Single-cell transcriptome reveals insights into the development and function of the zebrafish ovary. Elife 11, e76014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sur A, Wang Y, Capar P, Margolin G and Farrell JA (2023). Single-cell analysis of shared signatures and transcriptional diversity during zebrafish development. bioRxiv 2023–03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arbeloa EM, Uez MJ, Bertolotti SG and Churio MS (2010). Antioxidant activity of gadusol and occurrence in fish roes from Argentine Sea. Food Chemistry 119, 586–591. [Google Scholar]
- 10.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, and Nikaido O (1991). Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4) photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochemistry and photobiology 54, 225–232. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita M (2003). Apoptosis in zebrafish development. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 136, 731–742. [DOI] [PubMed] [Google Scholar]
- 12.Zhao H, Di Mauro G, Lungu-Mitea S, Negrini P, Guarino AM, Frigato E, Braunbeck T, Ma H, Lamparter T, Vallone D et al. (2018). Modulation of DNA Repair Systems in Blind Cavefish during Evolution in Constant Darkness. Current Biology 28, 3229–3243.e4. [DOI] [PubMed] [Google Scholar]
- 13.Zhao H, Li H, Du J, Di Mauro G, Lungu-Mitea S, Geyer N, Vallone D, Bertolucci C and Foulkes NS (2021). Regulation of ddb2 expression in blind cavefish and zebrafish reveals plasticity in the control of sunlight-induced DNA damage repair. PLoS Genetics 17, e1009356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelsh RN (2004). Genetics and Evolution of Pigment Patterns in Fish. Pigment Cell Research 17, 326–336. [DOI] [PubMed] [Google Scholar]
- 15.Kelsh RN, Harris ML, Colanesi S and Erickson CA (2009). Stripes and belly-spots - - a review of pigment cell morphogenesis in vertebrates. Semin. Cell Dev. Biol. 20, 90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapp FG, Perlin JR, Hagedorn EJ, Gansner JM, Schwarz DE, O’Connell LA, Johnson NS, Amemiya C, Fisher DE, Wölfle U et al. (2018). Protection from UV light is an evolutionarily conserved feature of the haematopoietic niche. Nature 558, 445–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwassmann HO (1978). Ecological aspects of electroreception. in ecology Sensory, Ali MA, ed. (Boston: Springer; ), pp. 521–533. [Google Scholar]
- 18.Krishnan J and Rohner N (2017). Cavefish and the basis for eye loss. Philosophical Transactions of the Royal Society B: Biological Sciences 372, 20150487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran D, Softley R and Warrant EJ (2015). The energetic cost of vision and the evolution of eyeless Mexican cavefish. Science Advances 1, e1500363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandaranayake WM, Bourne DJ and Sim RG (1997). Chemical Composition during Maturing and Spawning of the Sponge Dysidea herbacea (Porifera: Demospongiae). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 118, 851–859. [Google Scholar]
- 21.Chioccara F, Zeuli L and Novellino E (1986). Occurrence of mycosporine related compounds in sea urchin eggs. Comp. Biochem. Physiol. 85, 459–461. [Google Scholar]
- 22.Grant PT, Middleton C, Plack PA and Thomson RH (1985). The isolation of 4 aminocyclohexenimines (mycosporines) and a structurally related derivative of cyclohexane-1–3-dione (gadusol) from the brine shrimp, Artemia. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology 80, 755–759. [Google Scholar]
- 23.Raffa RB, Pergolizzi JV Jr., Taylor R Jr., Kitzen JM and Group, for the N. R. (2019). Sunscreen bans: Coral reefs and skin cancer. Journal of Clinical Pharmacy and Therapeutics 44, 134–139. [DOI] [PubMed] [Google Scholar]
- 24.Pandika M (2018). Looking to Nature for New Sunscreens. ACS Cent. Sci. 4, 788–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnsen S (2001). Hidden in plain sight: the ecology and physiology of organismal transparency. The Biological Bulletin 201, 301–318. [DOI] [PubMed] [Google Scholar]
- 26.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labun K, Montague TG, Gagnon JA, Thyme SB and Valen E (2016). CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic acids research 44, W272–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takasugi PR, Wang S, Truong KT, Drage EP, Kanishka SN, Higbee MA, Bamidele N, Ojelabi O, Sontheimer EJ and Gagnon JA, (2022). Orthogonal CRISPR-Cas tools for genome editing, inhibition, and CRISPR recording in zebrafish embryos. Genetics 220, iyab196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abràmoff MD, Magalhães PJ and Ram SJ (2004). Image processing with ImageJ. Biophotonics international 11, 36–42. [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome biology 5, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weger BD, Sahinbas M, Otto GW, Mracek P, Armant O, Dolle D, Lahiri K, Vallone D, Ettwiller L, Geisler R et al. (2011). The Light Responsive Transcriptome of the Zebrafish: Function and Regulation. PLOS ONE 6, e17080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCurley AT and Callard GV (2008). Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Molecular Biology 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 36.Mu Y, Bian C, Liu R, Wang Y, Shao G, Li J, Qiu Y, He T, Li W, Ao J et al. (2021). Whole genome sequencing of a snailfish from the Yap Trench (~ 7,000 m) clarifies the molecular mechanisms underlying adaptation to the deep sea. PLoS genetics 17, e1009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Shen Y, Yang Y, Gan X, Liu G, Hu K, Li Y, Gao Z, Zhu L, Yan G et al. (2019). Morphology and genome of a snailfish from the Mariana Trench provide insights into deep-sea adaptation. Nature ecology & evolution 3, 823–833. [DOI] [PubMed] [Google Scholar]
- 38.Froese R and Pauly D (2010) FishBase. [Google Scholar]
- 39.Chang J, Rabosky DL, Smith SA and Alfaro ME (2019). An R package and online resource for macroevolutionary studies using the ray-finned fish tree of life. Methods in Ecology and Evolution 10, 1118–1124. [Google Scholar]
- 40.Pagel M, Meade A and Barker D (2004). Bayesian estimation of ancestral character states on phylogenies. Systematic biology 53, 673–684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Oligos and DNA sequences for eevs mutant. Related to STAR methods and Figures 1, S1, and S3.
A) gRNAs and Primers for eevs mutants. B) eevs wildtype and mutant sequences. C) Primers for qRT-PCR.
Data S2. Life history trait scoring and statistical analysis for gadusol loss across fish species. Related to STAR methods and Figures 4 and S4.
A) Life history traits. B) Absence/Presence of Gadusol Enzymes. C) Bayes traits statistics.
Data S3. Complete data for main figures. Related to STAR methods and Figures 1–3.
A) Figure 1C. B) Figure 1D. C) Figure 1E. D) Figure 2A. E) Figure 2C. F) Figure 3B. G) Figure 3C.
Table S1. GO term enrichment from RNAseq. Related to STAR methods and Figures 2 and S3.
GO term enrichment from RNAseq. GO terms found using fishenricher (Kuleshov et al. 2016); only GO terms with Padj. <0.05 are shown.
Data Availability Statement
RNA-seq data have been deposited at GEO under accession number GSE229587 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal Anti-cyclobutane pyrimidine dimers | Cosmo Bio | Cat# NM-DND-001 |
| Rabbit anti-active Caspase-3 | BD Biosciences | Cat# 559565 |
| Alexa Fluor 546 goat anti-mouse IgG | ThermoFisher | Cat# A-11030 |
| Alexa Fluor 594 goat anti-rabbit IgG | ThermoFisher | Cat# A-11012 |
| Biological samples | ||
| Danio rerio embryos | This paper | N/A |
| Danio rerio larvae | This paper | N/A |
| Danio rerio ovaries | This Paper | N/A |
| O. latipes embryos | This Paper | N/A |
| I. punctatus ovaries | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Methanol | Sigma-Aldrich | Cat# 34860 |
| Critical Commercial Assays | ||
| QuanTiTect Reverse Transcription Kit | Qiagen | Cat# 205311 |
| PowerUp SYBR Green Master Mix | ThermoFisher | Cat# A25742 |
| Deposited data | ||
| Code for bioinformatic exploration of loss of gadusol | This paper | https://github.com/nclark-lab/gadusol |
| RNAseq data (raw and analyzed) | GEO | GSE229587 |
| Experimental models: Organisms/strains | ||
| Zebrafish D. rerio (Tübingen strain) | ZIRC | ZL57 |
| Zebrafish D. rerio (AB strain) | ZIRC | ZL1 |
| Zebrafish D. rerio (mitfaw2/w2) | ZIRC | ZL2104 |
| Zebrafish D. rerio (eevs zj2/zj2) | This paper | N/A |
| Zebrafish D. rerio (eevs zj5/zj5) | This paper | N/A |
| Oligonucleotides | ||
| Primers for sequences cloning, see Table S1 | This paper | N/A |
| Primers for qRT-PCR, see Table S1 | This paper | N/A |
| Software and algorithms | ||
| GraphPad | GraphPad Software | https://graphpad.com |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Microsoft Excel Data Analysis Student t test | Microsoft | N/A |
| Other | ||
| Benchtop UV Transilluminator | UVP | M-15V P/N 95–0456-01 |
| UVB broad band bulb (306nm) | Ushio | G8T5E |
| Digital UV Radiometer UVB | Solarmeter | 6.0 UVB |
RNAseq data has been deposited at GEO accession number GSE229587.


