Abstract
Neuraminidase inhibitors (NAIs) are recommended for influenza treatment and prevention worldwide. The most widely prescribed NAI is oral oseltamivir, while inhaled zanamivir is less commonly used. Using phenotypic neuraminidase (NA) enzymatic assays and molecular modeling approaches, we examined the ability of the investigational orally-dosed NAI AV5080 to inhibit viruses of the influenza A(H1N1)pdm09, A(H3N2), A(H5N1), and A(H7N9) subtypes and the influenza B/Victoria- and B/Yamagata-lineages containing NA substitutions conferring oseltamivir or zanamivir resistance including: NA-R292K, NA-E119G/V, NA-H274Y, NA-I122L/N, and NA-R150K. Broadly, AV5080 showed enhanced in vitro efficacy when compared with oseltamivir and/or zanamivir. Reduced AV5080 inhibition was determined for influenza A viruses with NA-E119G and NA-R292K, and for B/Victoria-lineage viruses with NA-I122N/L and B/Yamagata-lineage virus with NA-R150K. Molecular modeling suggested loss of the short hydrogen bond to the carboxyl group of AV5080 affected inhibition of NA-R292K viruses, whereas loss of the salt bridge with the guanidine group of AV5080 affected inhibition of NA-E119G. The resistance profiles and predicted binding modes of AV5080 and zanamivir are most similar, but dissimilar to those of oseltamivir, in part because of a guanidine moiety compensatory binding effect. Overall, our data suggests that AV5080 is a promising oral-dosed NAI that exhibited similar or superior in vitro efficacy against viruses with reduced or highly reduced inhibition phenotypes with respect to currently approved NAIs.
Keywords: AV5080, oseltamivir, zanamivir, influenza virus, neuraminidase, reduced susceptibility, resistance, NAI
Influenza viruses are important human pathogens, causing annual epidemics and significant morbidity worldwide (Adlhoch et al., 2021; Iuliano et al., 2018; Olsen et al., 2021). Vaccination is the most effective influenza control strategy, but efficacy varies from 10% to 60% because of antigenic mismatch between vaccine composition and novel antigenic variants (Centers for Disease Control and Prevention; Tenforde et al., 2021; U.S. Federal Food and Drug Administration). Influenza antivirals provide alternative avenues for disease management and comprise two classes of direct-acting drugs: the neuraminidase (NA) protein inhibitors (NAIs) that target enzymatic activity of surface NA to inhibit virus budding, and a polymerase acidic (PA) cap-dependent endonuclease (CEN) inhibitor baloxavir marboxil. Baloxavir is a new and potent option, but it is not widely used outside Japan (Ison et al., 2021), while the NAIs are widely prescribed worldwide. Currently, there are four NAIs available that are approved in various countries. Oral oseltamivir and inhaled zanamivir have been available worldwide since 1999–2000, intravenous peramivir is approved in China, Japan, South Korea, and the United States, whereas inhaled laninamivir octanoate is approved only in Japan (Beigel and Hayden, 2021). With only one oral NAI available, the development of novel NAIs continues.
AV5080 is an investigational oral NAI (Fig. 1) for which phase 2 clinical study was completed, and phase 3 clinical study has been approved in Russia (ClinicalTrials.gov, 2021). It was designed to have improved activity against the NAs of influenza A and B viruses (IAVs and IBVs), demonstrated 50% inhibitory concentrations (IC50s) at low nM range, and molecular docking showed that AV5080 binding modes within A/Duck/Minnesota/1525/1981 (H5N1) NA active site are similar to oseltamivir and zanamivir (Ivachtchenko et al., 2014).
Fig. 1. Chemical structures of anti-influenza NAI drugs:
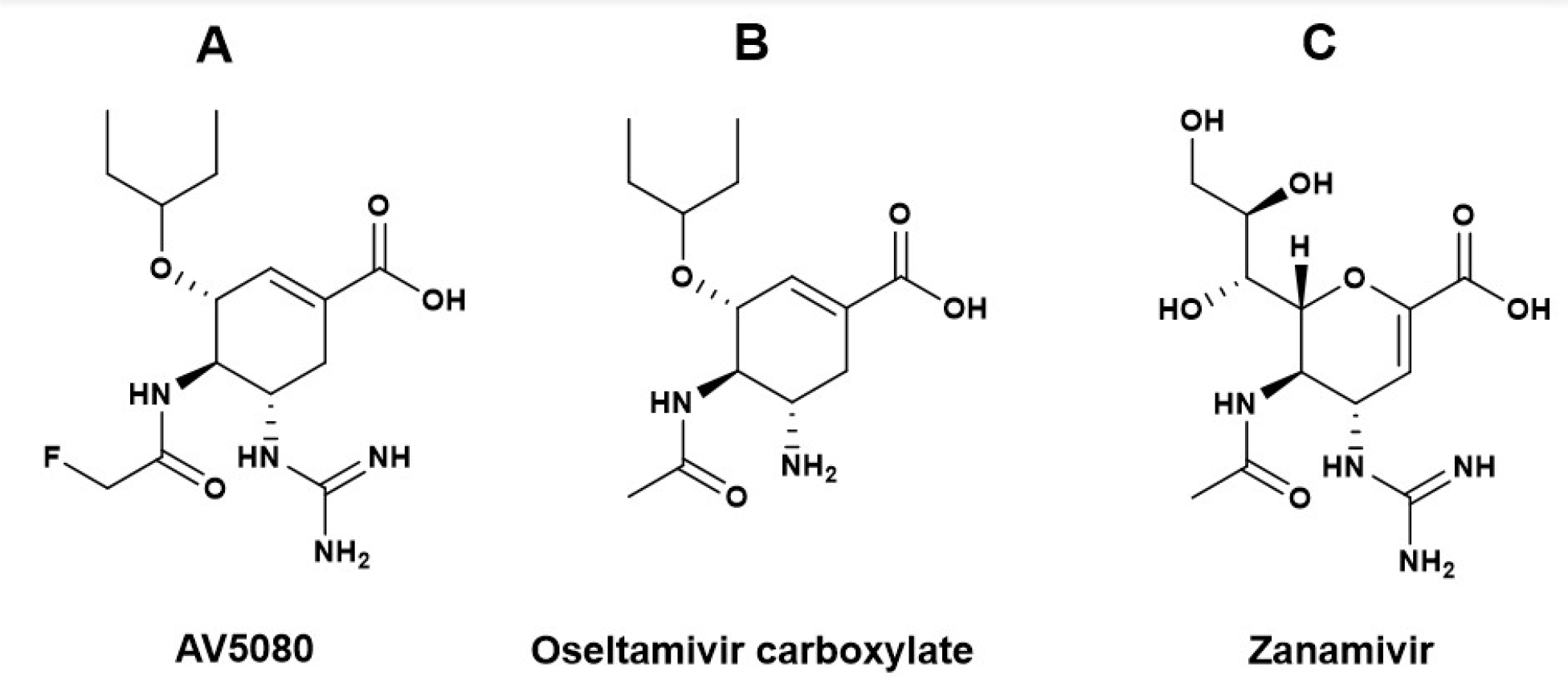
(A) Investigational drug AV5080, (B) oseltamivir carboxylate (the active metabolite of prodrug oseltamivir phosphate), (C) zanamivir.
Although viruses with reduced NAI susceptibility (by phenotypic or genotypic evaluation) have been only rarely detected since 2010 (Govorkova et al., 2022; Takashita et al., 2020), emergence of drug-resistant viruses can affect the clinical management of influenza. Continued preclinical and clinical evaluation of AV5080 in the presence of defined NAI resistance markers is important. Previously, AV5080 showed activity against oseltamivir-resistant influenza A(H1N1)pdm09 with NA-H274Y (N2 numbering) and IBVs with NA-D197E (B numbering) (Ivachtchenko et al., 2014). Here, we comprehensively defined the activity of AV5080 against both naturally occurring and reverse genetics (rg)-derived IAVs and IBVs carrying NA substitutions mediating reduced inhibition (RI) (10- to 100-fold over wildtype [WT] for IAVs; 5- to 50-fold for IBVs) and highly reduced inhibition (HRI) (>100-fold for IAVs; >50-fold for IBVs) by oseltamivir and zanamivir (World Health Organization, 2012). Our virus panel included clinically relevant and naturally occurring A(H1N1)pdm09 and/or A(H3N2) viruses with NA-H274Y(Hurt et al., 2012), NA-E119X (Gaymard et al., 2016), NA-R294K (Gaymard et al., 2016), as well as recombinant viruses based upon naturally occurring A(H5N1) NA-H274Y and NA-H294S (Le et al., 2005; McKimm-Breschkin et al., 2018), and A(H7N9) NA-R292K (Watanabe et al., 2013). Additionally, rg-generation of clinically relevant NA substitutions in novel subtypes like A(H5N1) NA-E119G or in IBV genotypes including NA-I221X, NA-N294X, and NA-D197E (Abed and Boivin, 2017; Burnham et al., 2014; Farrukee et al., 2015) were also rescued to enhance the panel and provide access to viruses not otherwise readily available for us to test.
Phenotypic NAI susceptibilities were determined using a MUNANA fluorescence-based NA inhibition assay (Govorkova et al., 2013; Potier et al., 1979) to generate IC50 values. Virus isolates with naturally occurring NA substitutions were used for A(H1N1)pdm09 and A(H3N2) subtypes. An 8-plasmid reverse-genetics system (Hoffmann et al., 2002a; Hoffmann et al., 2002b) was used to introduce NA substitutions into zoonotic A(H5N1) and A(H7N9) subtypes and Yamagata-lineage and Victoria-lineage IBVs (Pascua et al., 2020; Pascua et al., 2017) (Supplemental Methods).
We first determined oseltamivir, zanamivir, and AV5080 IC50 values and fold-changes over WT for NAI-resistant IAVs (Table 1). NA-H274Y in A(H1N1)pdm09 and A(H5N1) caused HRI for oseltamivir but normal inhibition (NI) for zanamivir and AV5080. NA-E119V in A(H3N2) virus (L’Huillier et al., 2015) yielded similar results, causing HRI for oseltamivir but NI for zanamivir and AV5080.However, NA-E119G in A(H5N1) virus caused HRI for zanamivir, while only RI for oseltamivir and AV5080. NA-N294S caused only RI for oseltamivir in A(H5N1), but no negative effects for zanamivir or AV5080. Finally, NA-R292K caused HRI for oseltamivir in A(H3N2) but only RI for zanamivir and AV5080. In all cases where oseltamivir/zanamivir showed HRI/RI, AV5080 showed RI or NI. The exception was NA-R292K in A(H7N9), which caused HRI for oseltamivir and AV5080, but RI for zanamivir. However, IC50 fold-changes for oseltamivir vs. AV5080 were dramatically different, with AV5080 nearly 80-fold more effective (Table 1).
Table 1.
Susceptibility to oseltamivir, zanamivir, and investigational compound AV5080 of IAV carrying NAI resistance–associated substitutions.
| Influenza A virus | NA amino acid substitutiona | Susceptibility to NAIs, mean IC50 ± SD (nM)b | NAI susceptibility phenotype (fold-change)c | ||||
|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | AV5080 | Oseltamivir | Zanamivir | AV5080 | ||
| A(H1N1)pdm09 | |||||||
| A/Perth/265/2009 | WT | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.1 ± 0.0 | NI | NI | NI |
| A/Perth/261/2009 | NA-H274Y | 118.8 ± 8.6 | 0.3 ± 0.0 | 0.7 ± 0.8 | HRI (396) | NI (0.6) | NI (7) |
| A/Denmark/524/2009 | WT | 0.2 ± 0.0 | 0.3 ± 0.0 | 0.1 ± 0.0 | NI | NI | NI |
| A/Denmark/528/2009 | NA-H274Y | 118.5 ± 11.5 | 0.3 ± 0.0 | 0.9 ± 0.0 | HRI (592.5) | NI (1.0) | NI (9.0) |
| A(H3N2) | |||||||
| A/Washington/01/2007 | WT | 0.3 ± 0.0 | 1.1 ± 0.1 | 0.5 ± 0.0 | NI | NI | NI |
| A/Texas/12/2007 | NA-E119V | 96.4 ± 7.0 | 1.3 ± 0.0 | 0.7 ± 0.1 | HRI (321.3) | NI (1.3) | NI (0.9) |
| A/Bethesda/956/2006 | NA-R292K | 593.0 ± 16.8 | 10.6 ± 1.4 | 6.6 ± 0.3 | HRI (1976.7) | ~RI (9.6) | RI (13.2) |
| A/Fukui/20/2004 | WT | 0.4 ± 0.0 | 1.5 ± 0.1 | 1.1 ± 0.0 | NI | NI | NI |
| A/Fukui/45/2004 | NA-E119V | 68.0 ± 1.1 | 1.2 ± 0.7 | 0.6 ± 0.0 | HRI (170.0) | NI (0.8) | NI (0.6) |
| A(H5N1) | |||||||
| rg-A/Vietnam/1203/2004 d | WT | 0.1 ± 0.0 | 0.4 ± 0.0 | 0.1 ± 0.0 | NI | NI | NI |
| rg-A/Vietnam/1203/2004-H274Yd | NA-H274Y | 162.3 ± 6.1 | 0.6 ± 0.0 | 1.6 ± 0.6 | HRI (1552.5) | NI (1.7) | NI (9) |
| rg-A/Vietnam/1203/2004-N294Sd | NA-N294S | 5.3 ± 0.3 | 1.4 ± 0.2 | 0.3 ± 0.0 | RI (50.0) | NI (4.0) | NI (2.6) |
| rg-A/Vietnam/1203/2004 e | WT | 0.3 ± 0.1 | 0.9 ± 0.2 | 0.2 ± 0.0 | NI | NI | NI |
| rg-A/Vietnam/1203/2004-E119Ge | NA-E119G | 3.2 ± 0.4 | 582.2 ± 308.7 | 3.3 ± 1.3 | RI (13.0) | HRI (643.0) | RI (17.1) |
| A(H7N9) | |||||||
| rg-A/Anhui/1/2013f | WT | 0.7 ± 0.0 | 0.7 ± 0.2 | 0.4 ± 0.0 | NI | NI | NI |
| rg-A/Anhui/1/2013 × A/Shanghai/1/2013 g | NA-R292K | 10,820.0 ± 70.7 | 51.9 ± 9.6 | 77.4 ± 7.6 | HRI (15457.1) | RI (74.1) | HRI (193.5) |
Amino acid residue substitution in NA indicated by N2 NA numbering.
Concentration of NAI that reduced viral NA activity by 50% (IC50) relative to viral NA activity without inhibitor in a fluorescence-based assay with MUNANA substrate (100 μM final concentration). Values represent three independent experiments ± SD.
Fold-change relative to the mean IC50 of the WT. The criteria recommended by the World Health Organization Antiviral Working Group for interpreting data for the reduced inhibition of IAVs by NAIs are based on the fold-change in IC50 compared with that for the susceptible virus: NI, NAI-susceptible IAV < 10-fold; RI, 10- to 100-fold; HRI, >100-fold (World Health Organization, 2012). RI and HRI fold-changes are indicated in boldface.
A recombinant A/Vietnam/1203/2004 (H5N1) influenza viruses carrying either WT NA or NA-H274Y or NA-N294S.
A recombinant A(H5N1) influenza viruses carrying either WT NA or NA-E119G, HA genes from A/Vietnam/1203/2004 (H5N1) virus and the six internal genes from A/Puerto Rico/8/34 (H1N1) virus.
A recombinant A/Anhui/1/2013 (H7N9) influenza virus carrying WT NA.
A recombinant A(H7N9) influenza virus carrying HA gene from A/Anhui/1/2013 (H7N9) virus, NA gene from A/Shanghai/1/2013 (H7N9) virus and the six internal genes from A/Puerto Rico/8/34 (H1N1) virus.
Abbreviations: IC50, 50% inhibitory concentration; NA, neuraminidase; NAI, NA inhibitor; NI, normal inhibition; RI, reduced inhibition; HRI, highly reduced inhibition; WT, wild-type; rg, recombinant.
Through molecular modeling, we examined structural explanations for these NAI reduced-susceptibility phenotypes (Fig. 2 and detailed explanations in Supplementary Table 1). For oseltamivir, NA-H274Y shifts Glu277 ≤3Å up towards a bulky pentane moiety, causing a tangible clash (distance between the O- of Glu277 and the H of the terminal methyl group of oseltamivir is <1.5 Å), moving it out of the sub-pocket. This is accompanied by the partial loss of xH-π contacts with lone pairs of Asn295 and Arg293, significantly reducing potential binding affinity and allowing no compensatory effect via additional contacts within the binding site. In contrast, the guanidine moiety of zanamivir and AV5080 forms a tight interaction ensemble with the carbonyl groups of Trp179 and Asp151 and with carboxylic fragments of Asp151 and Glu228 by H-bonds and electrostatic interactions (salt bridges). This results in compensatory effects sufficient to retain binding, and likely reflects the efficacy phenotypes. NA-E119G resulted in complete loss of salt bridge contact with AV5080 and zanamivir’s guanidine moiety (Fig. 2A). In addition, this space was left unfilled by the amino acid side chain, allowing the guanidine more mobility, potentially deviating from the most efficient binding mode. NA-E119G also decreases oseltamivir affinity, as loss of hydrogen bonding occurs with the drug’s amino group. Our models suggest NA-E119V should decrease affinity for all three NAIs tested, as there are salt bridge losses similarly to those in NA-E119G (Supplementary Table 1). However, we only observed oseltamivir efficacy loss in vitro. AV5080 and zanamivir inhibited normally, possibly because of pose stabilizations by H-π interaction of the valine methyl group protons and their guanidine group (Supplementary Table 1).
Fig. 2. Binding modes of AV5080 (the carbon backbone is shown in purple) and oseltamivir (the carbon backbone is shown in orange) in the active site of NA of influenza A and B viruses with different NA substitutions (in green).
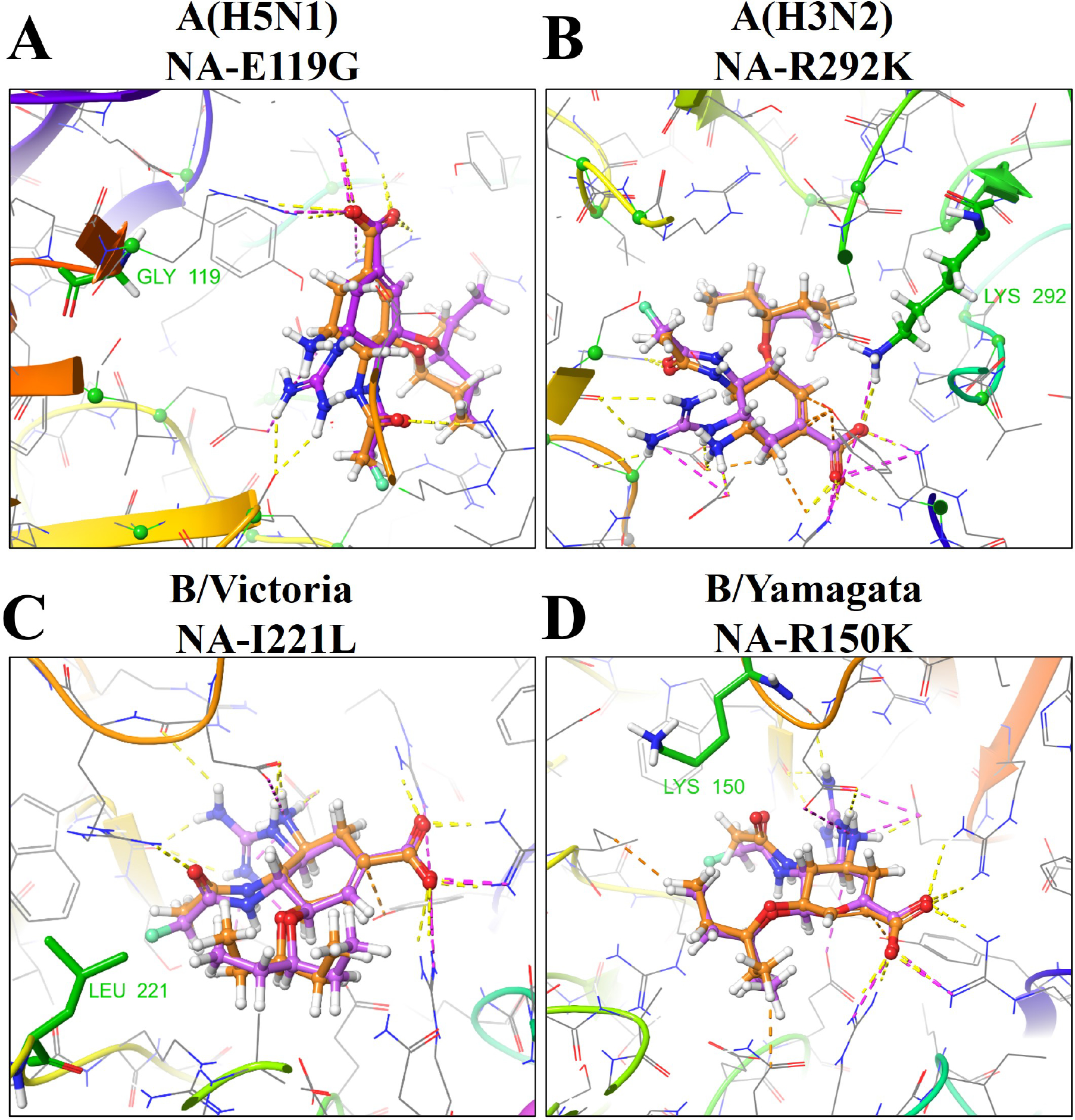
(A) The model of A/Vietnam/1203/2004 (H5N1) (PDB code: 2HU0) NA-E119G substitution leads to the loss of salt bridge contact between the NA amino acid residue and the guanidine moiety of zanamivir or AV5080. (B) The model of A/Tanzania/205/2010 (H3N2) (PDB code: 4GZP) NA-R292K substitution leads to the loss of hydrogen bonding stability and xH-π interactions with AV5080. (C) The model of Victoria Lineage B/Lyon/CHU/15.216/2011 (PDB code: 4CPZ) NA-I221L substitution can make the Ile221–Arg151–Asp197 cluster more labile within the NA active site. (D) The model of Yamagata lineage B/Brisbane/60/2008 (PDB code: 4CPN) NA-R150K substitution likely causes loss of contact with the carbonyl oxygen of AV5080.
For NA-N294S, Asn294 is implicated in stabilizing the enzymes’ Ca2+-dependent cluster forming H-bonds with proximal Trp296 and Tyr316. Considering that Arg292 is the next amino acid to Asn294 and that Arg292 plays a significant role in binding (Fig. 2B), NA-N294S may lead to spatial rearrangement of this substitution ensemble. However, the substitution of Asp for Ser at residue 294 presumably maintains cluster stability because of a similar H-bonding interface and may not cause a significant reduction in binding. Nevertheless, our phenotypic data (Table 1) suggest oseltamivir is more adversely affected by such minor shifts than AV5080 or zanamivir. Finally, NA-R292K is a very impactful substitution within the NA catalytic residues, causing cross-resistance to at least three NAIs (oseltamivir, zanamivir, and peramivir) in group 2 IAVs. NA-R292K is a part of a highly conserved arginine triad (Arg118, Arg292, and ArgR371) shared by all sialidases (Colman et al., 1983). Modeling demonstrated reduced affinity of oseltamivir, zanamivir, and AV5080 for NA-R292K, although the salt bridge with acid functionality (a free carboxylic acid- moiety) of the drugs is mainly retained. Moreover, xH-π interactions between Arg lone pairs and protons of a bulky aliphatic substituent in position 3 of the drug core are lost (Fig. 2B). Of the three NAIs tested, oseltamivir was the most negatively affected by NA-R292K because of the lack of a guanidine-related compensatory effect on the NA binding affinity. In contrast, the zanamivir and AV5080 guanidine moieties form tight interaction ensembles with the carbonyl groups of Trp179 and Asp151 and with the carboxylic fragments of Asp151 and Glu228 by H-bonds and electrostatic interactions (salt bridges). The resulting compensatory effect is likely sufficient to retain interactions and mediating their lower IC50 values compared to oseltamivir.
We next determined oseltamivir, zanamivir, and AV5080 IC50 values and fold-changes over WT and NAI-resistant IBVs (Table 2). RI/HRI for oseltamivir and zanamivir are reported at low incidence in B/Victoria-lineage and B/Yamagata-lineage viruses across influenza seasons (Govorkova et al., 2022). Several NA substitutions in clinical specimens are included in our panel, but changes at the Asp197 and Ile221 residues were most common (Burnham et al., 2014; Govorkova et al., 2022; Takashita et al., 2020). Using a previously generated IBVs with NA substitutions, (Pascua et al., 2020), we assessed the susceptibilities to oseltamivir, zanamivir, and AV5080 of rg-B/Brisbane/60/2008 (B/Victoria-lineage) and rg-B/Phuket/3073/2013 (B/Yamagata-lineage) viruses carrying NA-E105K, NA-G145E, NA-R150K, NA-D197N, NA-I221L/N/T/V, NA-H273Y, NA-N294S, or NA-G407S.
Table 2.
Susceptibility to oseltamivir, zanamivir, and investigational AV5080 of recombinant IBV carrying NAI resistance–associated substitutions.
| NA amino acid substitutiona | Susceptibility to NAIs, mean IC50 ± SD (nM)b | NAI susceptibility phenotype (fold-change)c | ||||
|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | AV5080 | Oseltamivir | Zanamivir | AV5080 | |
| rg-B/Brisbane/60/2008 (B/Victoria-lineage) | ||||||
| WT | 8.4 ± 0.8 | 3.0 ± 0.3 | 3.1± 0.0 | NI | NI | NI |
| NA-G145E | 5.1 ± 0.4 | 1.8 ± 0.2 | 1.2 ± 0.1 | NI (0.6) | NI (0.6) | NI (0.4) |
| NA-D197N | 52.1 ± 2.1 | 12.4 ± 0.1 | 8.3 ± 0.7 | RI (6.2) | NI (4.2) | NI (2.7) |
| NA-I221L | 1247.0 ± 49.5 | 52.2 ± 11.2 | 27.1 ± 0.7 | HRI (149) | RI (17.7) | RI (8.7) |
| NA-I221N | 468.1 ± 10.3 | 18.7 ± 1.5 | 18.0 ± 2.1 | HRI (55.9) | RI (6.3) | RI (5.8) |
| NA-I221T | 47.8 ± 9.6 | 5.5 ± 0.0 | 6.4 ± 0.7 | RI (5.7) | NI (1.9) | NI (2.1) |
| NA-I221V | 12.6 ± 3.8 | 2.90 ± 0.8 | 6.8 ± 0.4 | NI (1.5) | NI (1) | NI (2.2) |
| NA-H273Y | 37.0 ± 1.7 | 1.5 ± 0.1 | 1.7 ± 0.0 | NI (4.4) | NI (0.5) | NI (0.5) |
| NA-N294S | 115.2 ± 23.6 | 8.4 ± 1.9 | 5.3 ± 0.5 | RI (13.8) | NI (2.8) | NI (1.7) |
| NA-G407S | 27.7 ± 4.3 | 5.6 ± 0.4 | 3.7 ± 0.3 | NI (3.3) | NI (1.9) | NI (1.2) |
| rg-B/Phuket/3073/2013 (B/Yamagata-lineage) | ||||||
| WT | 5.7 ± 0.9 | 1.3 ± 0.2 | 1.4 ± 0.0 | NI | NI | NI |
| NA-G145E | 7.5 ± 2.1 | 0.7 ± 0.1 | 0.9 ± 0.0 | NI (1.3) | NI (0.5) | NI (0.6) |
| NA-R150K | 910.2 ± 47.5 | 30.1 ± 0.6 | 29.0 ± 1.7 | HRI (161.1) | RI (23.5) | RI (20.7) |
| NA-D197N | 57.2 ± 6.4 | 4.1 ± 0.7 | 3.4 ± 0.2 | RI (10.0) | NI (3.2) | NI (2.4) |
| NA-I221L | 199.4 ± 41.9 | 8.4 ± 4.1 | 2.3 ± 0.2 | RI (35.3) | RI (6.6) | NI (1.6) |
| NA-I221N | 377.0 ± 60.9 | 17.2 ± 0.9 | 3.6 ± 0.0 | HRI (66.7) | RI (13.2) | NI (2.6) |
| NA-I221T | 57.0 ± 14.2 | 2.1 ± 0.2 | 2.6 ± 0.1 | RI (10.1) | NI (1.6) | NI (1.9) |
| NA-I221V | 2.7 ± 0.3 | 3.1 ± 0.2 | 2.0 ± 0.2 | NI (0.5) | NI (2.4) | NI (1.4) |
| NA-H273Y | 34.5 ± 2.9 | 0.8 ± 0.1 | 1.0 ± 0.1 | RI (6.1) | NI (0.6) | NI (0.7) |
| NA-N294S | 62.0 ± 14.5 | 2.4 ± 0.2 | 1.9 ± 0.2 | RI (11.0) | NI (1.9) | NI (1.4) |
| NA-G407S | 13.4 ± 2.5 | 1.8 ± 0.2 | 1.2 ± 0.0 | NI (2.4) | NI (1.4) | NI (0.9) |
Amino acid residue substitution in NA indicated by IBV numbering.
Concentration of NAI that reduced viral NA activity by 50% (IC50) relative to viral NA activity without inhibitor in a fluorescence-based assay with MUNANA substrate (100 μM final concentration). Values represent three independent experiments ± SD.
Fold-change relative to the mean IC50 of the rg-WT. The criteria recommended by the World Health Organization Antiviral Working Group for interpreting data for the reduced inhibition of IBVs by NAIs are based on the fold-change in IC50 compared with that of the susceptible virus: NI, NAI-susceptible IBV < 5-fold; RI, 5- to 50-fold; HRI, >50-fold (World Health Organization, 2012). RI and HRI fold-changes are indicated in boldface.
Abbreviations: IBV, influenza B virus; HRI, highly reduced inhibition; IC50, 50% inhibitory concentration; NA, neuraminidase; NAI, NA inhibitor; ND, not determined; NI, normal inhibition; RI, reduced inhibition; HRI, highly reduced inhibition; WT, wild-type; rg, recombinant.
For B/Victoria-lineage viruses, the RI oseltamivir phenotype was determined for viruses with NA-D197N, NA-I221T, and NA-N294S, and the HRI phenotype for viruses with NA-I221L/N, recapitulating previous results (Pascua et al., 2020). NA-R150K detrimentally affected NA activity of rg-B/Brisbane/60/2008 virus such that the virus was successfully rescued, but the IC50 values could not be determined from the NA activity of the propagated stocks. For zanamivir, the RI phenotype was exhibited by the presence of NA-I221L/N in rg-B/Brisbane/60/2008 viruses. Notably, AV5080 lost efficacy only against B/Victoria-lineage viruses carrying NA-I221L or NA-I221N (RI: 8.7-fold and 5.8-fold changes in IC50, respectively). Modeling poses (in detail in Supplementary Table 2) containing NA-I221N or NA-I221L substitutions led to a reduction in Ile221–Arg151–Asp197 cluster stability and resulted in significant clashes with the -O(CH2CH3)2 moiety of AV5080 and oseltamivir (Fig. 2C). We propose that the H-bond between Arg151 and the carbonyl oxygen is weakened by these rearrangements.
For B/Yamagata-lineage viruses, a slightly different pattern of oseltamivir susceptibility was observed than for B/Victoria-lineage viruses: the RI phenotype was determined for viruses with NA-D197N, NA-I221L/T, NA-H273Y, and NA-N294S, and the HRI phenotype for viruses with NA-R150K and NA-I221N (Table 2). For zanamivir, the RI phenotype was exhibited by the presence of NA-I221L/N. Additionally, RI by this NAI was observed in the B/Yamagata-lineage virus possessing the NA-R150K substitution. AV5080 lost efficacy only against B/Yamagata-lineage virus carrying NA-R150K, indicating that B/Yamagata-lineage viruses that acquire this marker are potentially resistant to AV5080. This substitution may lead to a loss of contact with the carbonyl oxygen of AV5080, oseltamivir, and zanamivir (Fig. 2D). Moreover, it can influence the stability of the Ile221 and Asp197 cluster; consequently, the interaction between Ile221 and the methyl group of the substituent in position 3 of the drug scaffold may be negatively altered.
In conclusion, the candidate drug AV5080 showed favorable resistance profiles in IAVs and IBVs, including activity similar or superior to oseltamivir against A(H5N1) and A(H7N9) subtypes, which are associated with human zoonotic infections and severe outcomes. CDC interim analyses suggest that patients (outpatient or hospitalized) presenting with infections by these subtypes be administered oseltamivir, although clinical guidance based on randomized clinical trials is lacking. That AV5080 worked as well as or better than oseltamivir against viruses with NAI-resistant substitutions in our study is a powerful preclinical indicator of its efficacy against these and possibly other novel viruses. However, future clinical implementation of AV5080 must be accompanied by active monitoring of treatment-emergent NA substitutions in seasonal and zoonotic subtypes. Overall, the pattern of RI/HRI by AV5080 most resembled that of zanamivir, and in some cases, AV5080 performed better than zanamivir [with NA-E119G in A(H5N1) and NA-I221L/N in B/Yamagata-lineage]. Our docking data demonstrate the importance of guanidine moieties for the binding capacity of both zanamivir and AV5080, which partially explains the similar phenotypes. Zanamivir and AV5080 are analogs of DANA, with a positively charged guanidino group replacing a hydroxyl group linked to C-4, better approximating the influenza NA natural substrate. We propose a guanidine moiety is the most important fragment of AV5080 responsible for sustainability against NA substitutions causing the RI/HRI phenotype. With NA-R292K, which caused the most significant reduction in AV5080 efficacy, switching from Arg to Lys may reduce binding between NA’s positively charged amino groups and AV5080. The distance between the two interacting groups increases, leading to H-bond loss and/or an ineffective binding mode (Supplementary Tables 1 and 2). Although phenotypic and structural properties of AV5080 and zanamivir are similar, their routes of administration differ. Zanamivir is administered via aerosol nebulizer rather than orally (as with oseltamivir), potentially limiting its clinical implementation. In contrast, oral administration of AV5080 in mice lethally challenged with A(H1N1) or A(H3N2) viruses yielded survival outcomes similar to those with oseltamivir (Ivachtchenko et al., 2014). The preclinical efficacy of oral AV5080, combined with its zanamivir-like efficacy in vitro against NAI-resistant viruses, suggests that AV5080 has potential for further development as an NAI.
Supplementary Material
HIGHLIGHTS.
AV5080 is a novel neuraminidase inhibitor (NAI) for influenza A and B viruses
AV5080 exhibits a different resistance profile compared to NAI oseltamivir
This profile is partially due to AV5080’s guanidine moiety compensatory effect(s)
Acknowledgments and funding
We thank Keith A. Laycock for excellent editing of the manuscript. This project was supported by the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, and the Department of Health and Human Services [Contract no. HHSN272201400006C]. The content of this manuscript is solely the responsibility of the authors. It does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of competing interest A.A.I. and A.V.I. are the founders of ChemDiv, managing members of ASAVI LCC. A.V.I. is the author of patent application US 8895613. All other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
Data Availability Statement:
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.
REFERENCES
- Abed Y, Boivin G, 2017. A Review of Clinical Influenza A and B Infections With Reduced Susceptibility to Both Oseltamivir and Zanamivir. Open Forum Infect Dis 4, ofx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlhoch C, Mook P, Lamb F, Ferland L, Melidou A, Amato-Gauci AJ, Pebody R, European Influenza Surveillance, N., 2021. Very little influenza in the WHO European Region during the 2020/21 season, weeks 40 2020 to 8 2021. Euro Surveill 26. [Google Scholar]
- Beigel JH, Hayden FG, 2021. Influenza Therapeutics in Clinical Practice-Challenges and Recent Advances. Cold Spring Harb Perspect Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham AJ, Baranovich T, Marathe BM, Armstrong J, Webster RG, Govorkova EA, 2014. Fitness costs for Influenza B viruses carrying neuraminidase inhibitor-resistant substitutions: underscoring the importance of E119A and H274Y. Antimicrob Agents Chemother 58, 2718–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, N.C.f.I.a.R.D.N., Effectiveness of seasonal Flu vaccines from the 2004–2021 Flu seasons., CDC Seasonal Flu Vaccine Effectiveness Studies. [Google Scholar]
- ClinicalTrials.gov, 2021. Identifier NCT05093998. Study evaluating the efficacy and safety of AV5080 in patients with uncomplicated influenza. Phase 3 National Library of Medicine; (US), Bethesda (MD). [Google Scholar]
- Colman PM, Varghese JN, Laver WG, 1983. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature 303, 41–44. [DOI] [PubMed] [Google Scholar]
- Farrukee R, Leang SK, Butler J, Lee RT, Maurer-Stroh S, Tilmanis D, Sullivan S, Mosse J, Barr IG, Hurt AC, 2015. Influenza viruses with B/Yamagata- and B/Victoria-like neuraminidases are differentially affected by mutations that alter antiviral susceptibility. J Antimicrob Chemother 70, 2004–2012. [DOI] [PubMed] [Google Scholar]
- Gaymard A, Charles-Dufant A, Sabatier M, Cortay JC, Frobert E, Picard C, Casalegno JS, Rosa-Calatrava M, Ferraris O, Valette M, Ottmann M, Lina B, Escuret V, 2016. Impact on antiviral resistance of E119V, I222L and R292K substitutions in influenza A viruses bearing a group 2 neuraminidase (N2, N3, N6, N7 and N9). J Antimicrob Chemother 71, 3036–3045. [DOI] [PubMed] [Google Scholar]
- Govorkova EA, Baranovich T, Seiler P, Armstrong J, Burnham A, Guan Y, Peiris M, Webby RJ, Webster RG, 2013. Antiviral resistance among highly pathogenic influenza A (H5N1) viruses isolated worldwide in 2002–2012 shows need for continued monitoring. Antiviral Res 98, 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorkova EA, Takashita E, Daniels RS, Fujisaki S, Presser LD, Patel MC, Huang W, Lackenby A, Nguyen HT, Pereyaslov D, Rattigan A, Brown SK, Samaan M, Subbarao K, Wong S, Wang D, Webby RJ, Yen HL, Zhang W, Meijer A, Gubareva LV, 2022. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2018–2020. Antiviral Res 200, 105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Krauss S, Perez D, Webby R, Webster RG, 2002a. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20, 3165–3170. [DOI] [PubMed] [Google Scholar]
- Hoffmann E, Mahmood K, Yang CF, Webster RG, Greenberg HB, Kemble G, 2002b. Rescue of influenza B virus from eight plasmids. Proc Natl Acad Sci U S A 99, 11411–11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt AC, Chotpitayasunondh T, Cox NJ, Daniels R, Fry AM, Gubareva LV, Hayden FG, Hui DS, Hungnes O, Lackenby A, Lim W, Meijer A, Penn C, Tashiro M, Uyeki TM, Zambon M, Antivirals WHOCo.P.I.A.V.R.t., 2012. Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 12, 240–248. [DOI] [PubMed] [Google Scholar]
- Ison MG, Hayden FG, Hay AJ, Gubareva LV, Govorkova EA, Takashita E, McKimm-Breschkin JL, 2021. Influenza polymerase inhibitor resistance: Assessment of the current state of the art - A report of the isirv Antiviral group. Antiviral Res 194, 105158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson MA, Bresee JS, Global Seasonal Influenza-associated Mortality Collaborator, N., 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391, 1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivachtchenko AV, Ivanenkov YA, Mitkin OD, Yamanushkin PM, Bichko VV, Shevkun NA, Karapetian RN, Leneva IA, Borisova OV, Veselov MS, 2014. Novel oral anti-influenza drug candidate AV5080. J Antimicrob Chemother 69, 1892–1902. [DOI] [PubMed] [Google Scholar]
- L’Huillier AG, Abed Y, Petty TJ, Cordey S, Thomas Y, Bouhy X, Schibler M, Simon A, Chalandon Y, van Delden C, Zdobnov E, Boquete-Suter P, Boivin G, Kaiser L, 2015. E119D Neuraminidase Mutation Conferring Pan-Resistance to Neuraminidase Inhibitors in an A(H1N1)pdm09 Isolate From a Stem-Cell Transplant Recipient. J Infect Dis 212, 1726–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le QM, Kiso M, Someya K, Sakai YT, Nguyen TH, Nguyen KH, Pham ND, Ngyen HH, Yamada S, Muramoto Y, Horimoto T, Takada A, Goto H, Suzuki T, Suzuki Y, Kawaoka Y, 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437, 1108. [DOI] [PubMed] [Google Scholar]
- McKimm-Breschkin JL, Barrett S, Wong FYK, Pudjiatmoko, Azhar M., Selleck P., Davies KR., Hartaningsih N., McGrane J., 2018. Identification of Indonesian clade 2.1 highly pathogenic influenza A(H5N1) viruses with N294S and S246N neuraminidase substitutions which further reduce oseltamivir susceptibility. Antiviral Res 153, 95–100. [DOI] [PubMed] [Google Scholar]
- Olsen SJ, Winn AK, Budd AP, Prill MM, Steel J, Midgley CM, Kniss K, Burns E, Rowe T, Foust A, Jasso G, Merced-Morales A, Davis CT, Jang Y, Jones J, Daly P, Gubareva L, Barnes J, Kondor R, Sessions W, Smith C, Wentworth DE, Garg S, Havers FP, Fry AM, Hall AJ, Brammer L, Silk BJ, 2021. Changes in Influenza and Other Respiratory Virus Activity During the COVID-19 Pandemic - United States, 2020–2021. MMWR Morb Mortal Wkly Rep 70, 1013–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascua PNQ, Marathe BM, Bisen S, Webby RJ, Govorkova EA, 2020. Influenza B viruses from different genetic backgrounds are variably impaired by neuraminidase inhibitor resistance-associated substitutions. Antiviral Res 173, 104669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascua PNQ, Mostafa HH, Marathe BM, Vogel P, Russell CJ, Webby RJ, Govorkova EA, 2017. Pathogenicity and peramivir efficacy in immunocompromised murine models of influenza B virus infection. Sci Rep 7, 7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB, 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal Biochem 94, 287–296. [DOI] [PubMed] [Google Scholar]
- Takashita E, Daniels RS, Fujisaki S, Gregory V, Gubareva LV, Huang W, Hurt AC, Lackenby A, Nguyen HT, Pereyaslov D, Roe M, Samaan M, Subbarao K, Tse H, Wang D, Yen HL, Zhang W, Meijer A, 2020. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2017–2018. Antiviral Res 175, 104718. [DOI] [PubMed] [Google Scholar]
- Tenforde MW, Kondor RJG, Chung JR, Zimmerman RK, Nowalk MP, Jackson ML, Jackson LA, Monto AS, Martin ET, Belongia EA, McLean HQ, Gaglani M, Rao A, Kim SS, Stark TJ, Barnes JR, Wentworth DE, Patel MM, Flannery B, 2021. Effect of Antigenic Drift on Influenza Vaccine Effectiveness in the United States-2019–2020. Clin Infect Dis 73, e4244–e4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Federal Food and Drug Administration, Influenza Vaccine for the 2021–2022 Season.. Watanabe T., Kiso M., Fukuyama S., Nakajima N., Imai M., Yamada S., Murakami S., Yamayoshi S., Iwatsuki-Horimoto K., Sakoda Y., Takashita E., McBride R., Noda T., Hatta M., Imai H., Zhao D., Kishida N., Shirakura M., de Vries RP., Shichinohe S., Okamatsu M., Tamura T, Tomita Y, Fujimoto N, Goto K, Katsura H, Kawakami E, Ishikawa I, Watanabe S, Ito M, Sakai-Tagawa Y, Sugita Y, Uraki R, Yamaji R, Eisfeld AJ, Zhong G, Fan S, Ping J, Maher EA, Hanson A, Uchida Y, Saito T, Ozawa M, Neumann G, Kida H, Odagiri T, Paulson JC, Hasegawa H, Tashiro M, Kawaoka Y, 2013. Characterization of H7N9 influenza A viruses isolated from humans. Nature 501, 551–555. World Health Organization, 2012. Laboratory methodologies for testing the antiviral susceptibility of influenza viruses: neuraminidase inhibitor (NAI). World Health Organization, Genevea, Switzerland. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.


