ABSTRACT
Objective:
Feeding restriction in rats alters the oscillators in suprachiasmatic, paraventricular, and arcuate nuclei, hypothalamic areas involved in food intake. In the present study, using the same animals and experimental protocol, we aimed to analyze if food restriction could reset clock genes ( Clock, Bmal1 ) and genes involved in lipid metabolism ( Pgc1a, Pparg, Ucp2 ) through nutrient-sensing pathways ( Sirt1, Ampk, Nampt ) in peripheral tissues.
Materials and methods:
Rats were grouped according to food access: Control group (CG, food ad libitum ), Restricted night-fed (RF-n, food access during 2 h at night), Restricted day-fed (RF-d, food access during 2 h in the daytime), and Day-fed (DF, food access during 12 h in the daytime). After 21 days, rats were decapitated at ZT3 (0900-1000 h), ZT11 (1700-1800 h), or ZT17 (2300-2400 h). Blood, liver, brown (BAT) and peri-epididymal (PAT) adipose tissues were collected. Plasma corticosterone and gene expression were evaluated by radioimmunoassay and qPCR, respectively.
Results:
In the liver, the expression pattern of Clock and Bmal1 shifted when food access was dissociated from rat nocturnal activity; this phenomenon was attenuated in adipose tissues. Daytime feeding also inverted the profile of energy-sensing and lipid metabolism-related genes in the liver, whereas calorie restriction induced a pre-feeding increased expression of these genes. In adipose tissues, Sirt1 expression was modified by daytime feeding and calorie restriction, with concomitant expression of Pgc1a , Pparg , and Ucp2 but not Ampk and Nampt .
Conclusion:
Feeding restriction reset clock genes and genes involved in lipid metabolism through nutrient-sensing-related genes in rat liver, brown, and peri-epididymal adipose tissues.
Keywords: Caloric restriction, feeding behavior, biological clocks, circadian rhythm, sirtuin 1
INTRODUCTION
Biological circadian rhythms allow organisms to prepare for recurrent daily changes in the light-dark cycle and food availability. In mammals, the circadian rhythms are controlled by the circadian clock system, comprising a central pacemaker localized in the suprachiasmatic nucleus (SCN) that synchronizes peripheral pacemakers in different tissues. The circadian clock system consists of a complex of transcriptional-translational feedback loops regulated by the “clock genes.” In the main loop, the heterodimeric transcription factors CLOCK and BMAL1 promote the transcription of their inhibitors, the Cryptochromes ( Cry1 , Cry2 ) and Periods ( Per1 , Per2 and Per3) genes. Bmal1 and Clock genes also increase the mRNA levels of Rev-erba and Rora , which compete for binding to the retinoic acid-related orphan receptor response elements (ROREs) repressing or activating the expression of Bmal1 ( 1 ). The circadian clocks are self-sustained, but they are synchronized by external zeitgebers. Whereas the SCN is synchronized mainly by photic stimuli, the peripheral oscillators also respond to other cues, such as feeding pattern and temperature ( 1 , 2 ).
The molecular circadian clock is an important regulator of metabolism and feeding behavior. Moreover, feeding time affects the circadian clock. Clock and Rev-erba mutant mice develop obesity ( 3 , 4 ). However, obesity is avoided in Clock mutant mice submitted to time-restricted feeding, even without altering levels of food consumption or activity ( 5 ).
In mice, daytime feeding is capable of uncoupling the peripheral oscillators from the SCN ( 6 ). In humans, night shift workers present increased risk of type 2 diabetes mellitus (T2DM) and metabolic syndrome (MetS) compared with day shift workers ( 7 ). Currently, animal models have been used to mimic shiftwork conditions. Studies using these animals have enabled researchers to evaluate the consequences of circadian disruption ( 8 ) and possible mechanisms responsible for metabolic disease development. Moreover, they allow researchers to evaluate the arising potential therapies for MetS and T2DM targeting the circadian molecular clock, such as REV-ERB agonists, ROR agonists, and CRY stabilizers ( 7 ).
To date, the mechanisms responsible for the resetting of peripheral clocks in response to changes in feeding time are not completely understood. Glucocorticoids are plausible candidates because they are secreted in daily cycles and are able to modulate the expression of clock genes causing phase shifts exclusively in peripheral cells ( 9 ). The importance of the glucocorticoid receptor for the regulation of clock genes by feeding entrainment was recently demonstrated in zebrafish, a non-mammalian species ( 10 ). In adrenalectomized mice submitted to daytime feeding, larger phase shifts were observed compared with controls, suggesting that glucocorticoids in fact counteract the re-phasing of peripheral clocks ( 11 ).
Nutrient-sensing molecules, such as SIRT1 (sirtuin-1) and AMPK (AMP-activated protein kinase), act as clock resetting signals ( 12 ). SIRT1 is a deacetylase, which requires the coenzyme NAD+. The NAD+/NADH status serves as a surrogate marker for the cellular redox state. According to the redox status, SIRT1 targets key metabolic proteins and can deacetylase histones, possibly counteracting the histone acetyltransferase activity of CLOCK ( 13 ). Furthermore, SIRT1 promotes deacetylation of important clock machinery proteins: the circadian repressor PER2, leading to its degradation, and BMAL1, which is recruited by the CLOCK/BMAL1 complex, consequently regulating gene expression ( 12 ).
AMPK is a heterotrimeric enzyme with a glycogen-binding domain and sites for the binding of AMP, ADP, and ATP. In fact, its activity is increased by ATP depletion, and some of the clock system components contain potential AMPK target phosphorylation sites ( 14 ). SIRT1 and AMPK interact with each other and might also regulate different metabolic pathways, including lipid storage, synthesis, and usage. AMPK also influences SIRT1 by increasing cellular nicotinamide phosphoribosyl-transferase (NAMPT) expression, thereby increasing NAD+ levels and consequently SIRT1 activity ( 15 ). Therefore, SIRT1 and AMPK may be capable of transducing signals related to feeding inputs to the clock machinery.
In a previous study from our lab, we demonstrated that the master oscillator in the SCN – as well as the oscillator in paraventricular and arcuate hypothalamic nuclei, brain areas involved in food intake – responds in a tissue-specific manner to feeding restriction in rats ( 16 ). In the present study, we took advantage of the same animals submitted to that experimental protocol and hypothesized that different feeding restriction patterns could reset clock genes and genes involved in lipid metabolism ( Pgc1a , Pparg , Ucp2 ) through nutrient-sensing-related genes ( Sirt1 , Ampk , Nampt ) in rat peripheral tissues, such as liver, brown, and peri-epididymal adipose tissues.
MATERIALS AND METHODS
Animals and housing
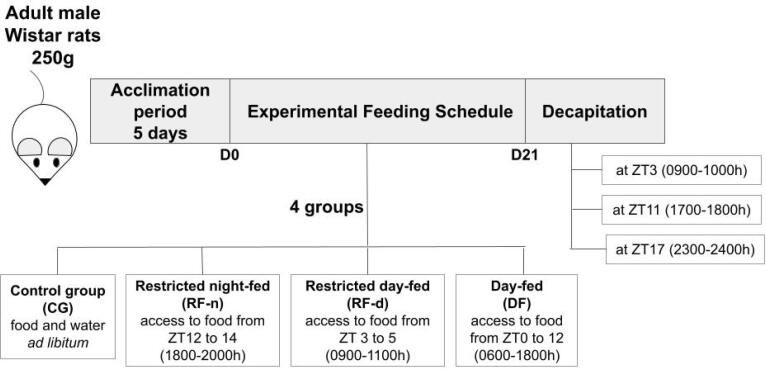
This study was approved by the Animal Ethics Committee of the Ribeirao Preto Medical School of the University of Sao Paulo, Brazil (Protocol n° 077/2011). The animals used in the present study were submitted to the experimental procedure performed in our lab and previously published ( 16 ). Briefly, young adult male Wistar rats aged approximately 8 weeks, weighing between 200 g and 300 g (average weight = 250 g), obtained from the Animal Facility of the Ribeirao Preto Medical School, were housed for 5 days in individual hanging wire cages with food and water ad libitum , under controlled conditions of temperature (23 °C with a maximum variability range of 4 °C) and humidity (range between 40 and 60%). The light was regulated on a 12/12 h light/dark cycle, with lights on at 6:00 A.M. (zeitgeber time [ZT] 0). All animals were fed with standard chow. As shown in Figure 1 , after five days of acclimation, the rats were divided into four groups with different dietary patterns for 20 days – Control group (CG, total of 16 rats): food and water ad libitum ; Restricted night-fed (RF-n, total of 19 rats): access to food from ZT12 to 14 (1800-2000 h); Restricted day-fed (RF-d, total of 22 rats): access to food from ZT3 to 5 (0900-1100 h); and Day-fed (DF, total number of 21 rats): access to food from ZT0 to ZT12 (0600-1800 h) – as previously published by our group ( 16 ). To avoid unspecific or stress-related elevations of corticosterone secretion, animals were handled by the same investigator during the experiment, and, on the last day, animals were decapitated within 60 seconds. Animals of the control group decapitated at ZT3, which showed plasma concentrations of corticosterone above 3 ug/dL, were excluded because of undesirable stress conditions, as previously published by our laboratory ( 17 ).
Figure 1. Schematic design of the experiment.
D: Day of experiment; ZT: zeitgeber time.
Experimental design
Body weight and food intake were measured daily throughout the experiment. On the 21st day, rats were decapitated per time point at ZT3 (0900-1000 h), ZT11 (1700-1800 h), or ZT17 (2300-2400 h).
Trunk blood was collected for plasma corticosterone measurement. Tissues (liver, brown adipose tissue [BAT], and peri-epididymal adipose tissue [PAT]) were collected, flash-frozen in dry ice, and stored at -80 °C until RNA isolation.
Corticosterone assay
Plasma corticosterone (B) was measured by radioimmunoassay as previously described ( 16 ). The assay sensitivity was 0.4 μg/dL, and the inter- and intra-assay variations were 4.8% and 6.7%, respectively.
RNA isolation, cDNA synthesis and amplification, and real-time PCR
RNA from the liver, BAT, and PAT obtained from animals submitted to the previous experimental protocol ( 16 ) was isolated using TRIzol® reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA). Sample integrity was evaluated by spectrophotometry at an absorbance of 260/280 nm using NanoDropTM 2000/2000c (Thermo Fisher Scientific, Wilmington, Delaware, USA) and by agarose gel electrophoresis.
cDNA was obtained using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Life Technologies, Foster City, CA, USA). qPCR was performed by the 7500 RT-PCR System (Applied Biosystems, Life Technologies, Foster City, CA, USA) using TaqMan® Gene Expression Assays: Clock ( Rn00573120_m1 ), Bmal1 ( Rn00577590_m1 ), Sirt1 ( Rn01428096_m1 ), Ampk ( Rn00576935_m1 ), Nampt ( Rn00822046_m1 ), Pparg ( Rn00440945_m1 ), Pgc1a ( Rn00580241_m1 ), Ucp2 ( Rn01754856_m1 ), and reference genes Gapdh ( 4352338E ) and Actb ( 4352340E ). Reactions were incubated in a 96-well optical plate at 95 °C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60 °C for 1 min. The cycle threshold (Ct) was defined as the fractional cycle number at which the fluorescence surpasses the fixed threshold. Data were presented as the Ct-mean of each sample of each target gene normalized by the reference genes Ct-mean expression and calibrated by the ΔCt-median value obtained from animals decapitated at ZT3 (0900 h) of the control group. Relative expression was calculated using the 2−ΔΔCt method, as previously detailed ( 16 ).
Statistical analysis
Continuous variables were expressed as mean and standard deviation. Kruskal-Wallis with Dunn’s post-test was used for continuous variables. Data were analyzed by GraphPadPrism 5 software (GraphPad, San Diego, CA), and differences were considered significant at p < 0.05.
RESULTS
In a previous study from our lab, rats were submitted to different restricted feeding schedules during light or dark phases to address clock gene expression in the SCN and other hypothalamic nuclei ( 16 ). Material collected from rats of that protocol were used for a gene expression study in the liver, BAT, and PAT.
Body weight, food intake, and corticosterone measurement
Data on body weight, food intake, and corticosterone levels were published in a previous work from our lab ( 16 ). Briefly, all groups presented similar body weight at the beginning of the experiment. After 21 days, we observed a decreased in body weight (g) in the RF-n (228.1 ± 29.24) and RF-d (223.9 ± 44.43) groups compared with the CG (370.4 ± 47.95) and DF (369.6 ± 36.55) groups (p < 0.0001). Similarly, lower daily food intake (g) was observed in the RF-n (13.01 ± 1.14) and RF-d (13.52 ± 1.75) groups compared with the CG (31.05 ± 3.12) and DF (24.72 ± 2.82) groups (p < 0.0001). Consequently, lower caloric intake (kcal) was observed in the RF-n (51.74 ± 4.54) and RF-d (53.74 ± 6.96) compared with the CG (123.4 ± 12.42) and DF (98.28 ± 11.23) groups (p < 0.0001). In addition, the CG and RF-n groups presented higher corticosterone levels at ZT11 and ZT17 compared with ZT3, with no difference between ZT3 and ZT17. On the other hand, the RF-d group showed an inverted daily pattern of corticosterone secretion compared with the CG and RF-n groups, with higher levels at ZT3, whereas the DF group showed higher corticosterone levels at ZT11.
Gene expression
Data on the gene expression of Clock , Bmal1 , Sirt1 , Ampk , Nampt , Pgc1a , Pparg , and Ucp2 obtained from the liver, BAT, and PAT are presented as mean ± SD, median, and interquartile interval in Supplementary Tables 1 , 2 , and 3 , respectively.
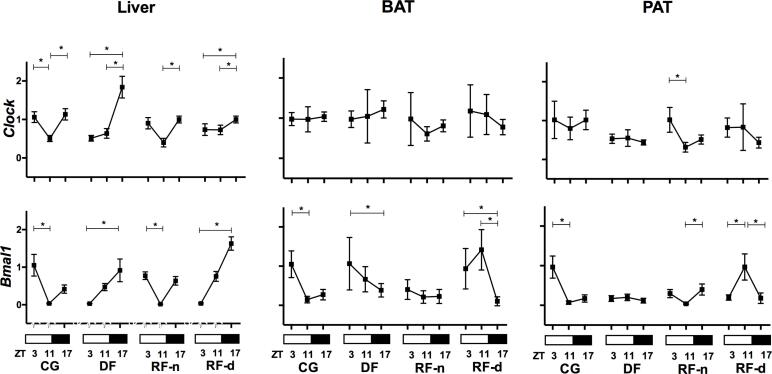
Figure 2 shows the expression of Clock and Bmal1 in the different tissues of the studied groups. In the CG, lower expressions of Clock (p = 0.0003) and Bmal1 (p < 0.0001) were observed in the afternoon (ZT11) in the liver. This pattern was also seen for Bmal1 in the PAT (p < 0.0001) and BAT (p < 0.0001), whereas Clock presented no difference among morning, afternoon, and night in BAT and PAT. In the RF-n group, lower expressions of Clock (p = 0.0006 and p < 0.0001) and Bmal1 (p < 0.0001 and p = 0.0003) were observed in the afternoon (ZT11) in the liver and PAT, but not in the BAT. In the liver, the RF-d group showed an altered profile compared with the CG, with a lower expression of Clock (p = 0.002) and Bmal1 (p < 0.0001) in the morning (ZT3) and a higher expression at night (ZT17), whereas higher expression of Bmal1 was observed in the afternoon (ZT11) in the PAT and BAT (p = 0.0003). In contrast, in the PAT and BAT, no difference among morning, afternoon, and night was observed for Clock expression. Similar to the RF-d, the DF group also presented a lower expression of Clock (p = 0.0003) and Bmal1 (p < 0.0001) in the liver in the morning (ZT3) and a higher expression at night (ZT17). In the BAT, this profile was inverted, with lower expression of Bmal1 (p = 0.03) at night, whereas Clock presented no differential expression among ZT3, ZT11, and ZT17 in the PAT and BAT. Therefore, in the liver, food restriction during the night (RF-n) preserved the pattern of Clock and Bmal1 expression observed in controls (CG), but when the food access occurred in dissociation with the rat’s nocturnal activity (RF-d and DF), we observed a shift of this profile. In the adipose tissues, we observed the same phenomenon at a lower magnitude. In rats submitted to daytime feeding (RF-d in BAT, RF-d and DF in PAT), the Bmal1 expression pattern observed in the controls was lost or inverted. In addition, calorie restriction per se (RF-n) modified Bmal1 expression in the BAT, but not in the PAT.
Figure 2. Clock and Bmal1 gene expression (2-ΔΔCt) in the liver, BAT (brown adipose tissue), and PAT (peri-epididymal adipose tissue) of Control group (CG), Day-fed (DF), Restricted night-fed (RF-n), and Restricted day-fed (RF-d) rats at ZT3 (0900 h), ZT11 (1700 h), and ZT17 (2300 h).
* p < 0.05
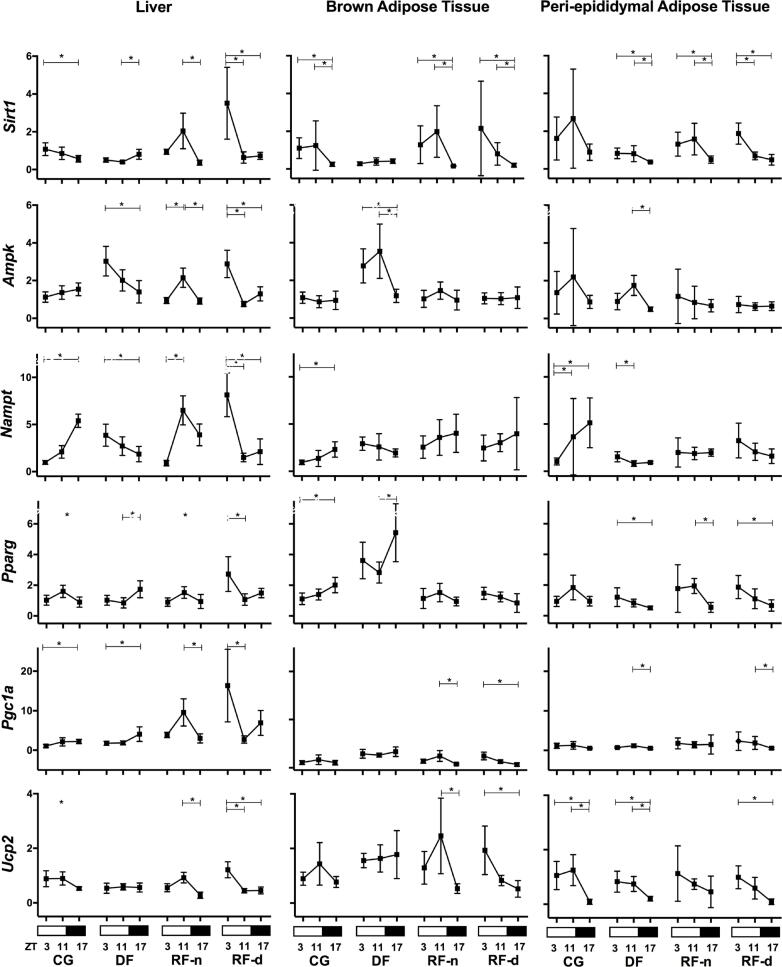
Figure 3 shows the expression of energy-sensing and lipid metabolism-related genes in the liver, BAT, and PAT of all studied groups. In the liver of the CG, the expression of Sirt1 (p = 0.03) and Ucp2 (p = 0.02) was higher in the morning (ZT3) and lower at night (ZT17), in contrast to the higher expression of Nampt (p < 0.0001) and Pgc1a (p = 0.02) that occurred at night. A higher expression of Pparg was observed in the afternoon (p = 0.03), whereas no differential expression of Ampk was observed among any time points. In the RF-n group, differently from the CG, higher expressions of Sirt1 (p < 0.0001), Ampk (p = 0.001), Pparg (p = 0.04), Pgc1a (p = 0.0007), Ucp2 (p < 0.0001), and Nampt (p < 0.0001) were observed in the afternoon (ZT11). Compared with RF-n, the RF-d group exhibited an inverse pattern, with lower expression at ZT11 for Sirt1 (p < 0.0001), Ampk (p < 0.0001), Pparg (p < 0.0001), Pgc1a (p < 0.0001), Ucp2 (p < 0.0001), and Nampt (p = 0.0007). The DF group showed higher expression of Sirt1 (p = 0.0001), Pparg (p = 0.004), and Pgc1a (p = 0.01) at ZT17 and Ampk (p = 0.0005) and Nampt (p = 0.01) at ZT3, whereas Ucp2 exhibited no differential expression among any time points. Thus, in the liver, the food availability during the 12 hours of the light phase (DF) inverted the profile of the energy-sensing and lipid metabolism-related genes observed in controls, except for Pgc1a . In addition, the calorie restriction (RF-d and RF-n) induced an increased expression of these genes, mainly in anticipation of food availability.
Figure 3. Relative expression (2-ΔΔCt) of energy-sensing genes ( Sirt1 , Ampk , and Nampt ) and lipid metabolism-related genes ( Pparg , Pgc1a , and Ucp2 ) in the liver, BAT (brown adipose tissue), and PAT (peri-epididymal adipose tissue) of Control group (CG), Day-fed (DF), Restricted night-fed (RF-n), and Restricted day-fed (RF-d) rats at ZT3 (0900 h), ZT11 (1700 h), and ZT17 (2300 h).
* p < 0.05
In the BAT, the CG exhibited higher expression of Sirt1 at ZT11 (p = 0.002) and higher expression of Nampt (p = 0.02) and Pparg (p = 0.003) at ZT17, whereas Ampk , Pgc1a, and Ucp2 exhibited no differential expression among any time points. The RF-n group also exhibited higher expression of Sirt1 (p = 0.0001) at ZT11; the same pattern was observed for Pgc1a (p = 0.01) and Ucp2 (p = 0.008), whereas no differential expression among morning, afternoon, and night was observed for Ampk , Nampt , and Pparg. In the RF-d group, higher expression of Sirt1 (p = 0.0002), Pgc1a (p < 0.0001), and Ucp2 (p = 0.0001) occurred at ZT3, and no differential expression among time points was observed for Ampk , Nampt , and Pparg . The DF group showed higher expression of Ampk (p = 0.0001) at ZT11 and of Pparg (p = 0.02) at ZT17.
In the PAT, the CG exhibited no difference in gene expression of Sirt1 , Ampk , Pparg , and Pgc1a among the time points. Lower expressions of Nampt (p = 0.0001) and Ucp2 (p = 0.002) occurred at ZT3 and ZT17, respectively. The RF-n group presented no difference in the expression of Ampk , Pgc1a, Nampt, and Ucp2 between morning, afternoon, and night. A higher expression of Sirt1 (p = 0.0002) and Pparg (p = 0.003) were observed at ZT11. The RF-d group showed lower expression of Sirt1 (p = 0.0002), Pparg (p = 0.004), Pgc1a (p = 0.02), and Ucp2 (p < 0.0001) at ZT17, whereas no differential expression of Ampk and Nampt was observed among all time points. The DF group exhibited higher expression of Sirt1 (p = 0.003), Pparg (p = 0.02), Nampt (p = 0.03), and Ucp2 (p = 0.0008) at ZT3. Ampk (p < 0.0001) and Pgc1a (p = 0.01) exhibited higher expression at ZT11. In summary, in the adipose tissues, daytime feeding modified the Sirt1 expression pattern, and calorie restriction increased its gene expression, preceding mealtime. Differently from our findings observed regarding calorie restriction in the liver, in adipose tissues, there was a concurrent expression pattern of Sirt1 , Pparg , Pgc1a , and Ucp2 but not of Sirt1 , Ampk, and Nampt .
DISCUSSION
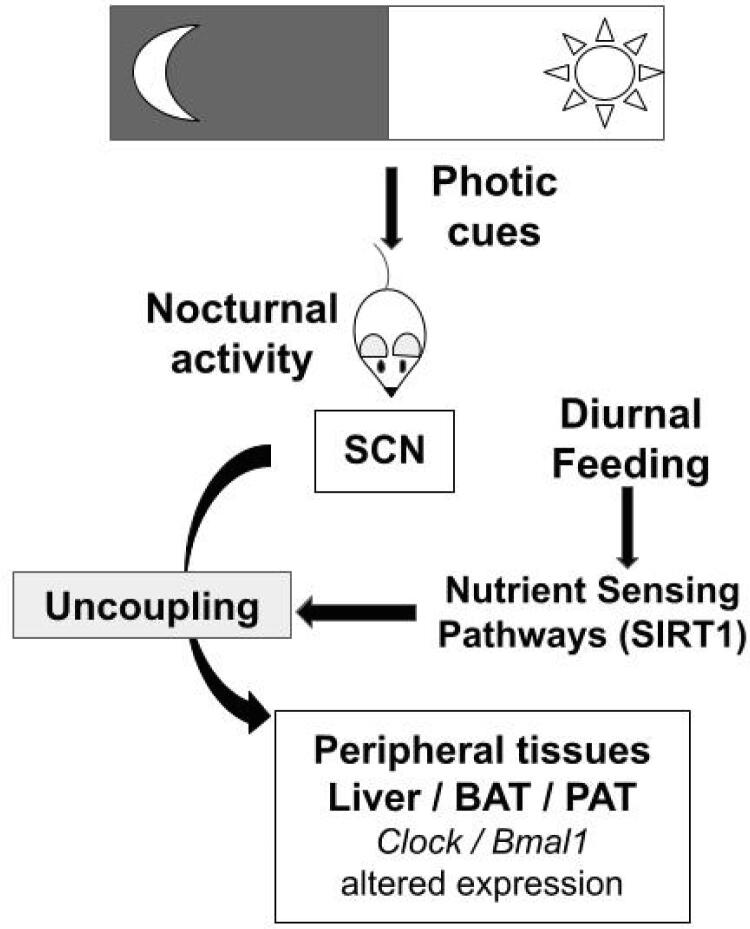
In the present study, we hypothesized that different feeding restriction patterns could reset clock genes ( Clock and Bmal1 ) and lipid metabolism-involved genes ( Pgc1a , Pparg , Ucp2 ) through nutrient-sensing-related genes ( Sirt1 , Ampk , Nampt ). In our study, food restriction during the night preserved the expression pattern of clock genes in the liver, but there was a shift of this profile when the food access occurred in dissociation with the rat’s nocturnal activity, as illustrated in Figure 4 . In the adipose tissues, the shift phenomenon occurred at a lower magnitude. In addition, in the liver as well as in adipose tissues, food availability during the light phase inverted the profile of energy-sensing and lipid metabolism-related genes, whereas the calorie restriction induced an increased expression of these genes, mainly in anticipation of food availability.
Figure 4. Schematic summary of study hypothesis and main results. Food restriction, in dissociation with rat nocturnal activity, alters the expression pattern of clock genes in peripheral tissues, such as liver and adipose tissues. These interactions may occur through Sirt1 , a nutrient-sensing gene. BAT: brown adipose tissue; PAT: peri-epididymal adipose tissue; SCN: suprachiasmatic nucleus.
Daytime feeding in murine is an important zeitgeber for peripheral clock genes and is capable of uncoupling the peripheral oscillators from the SCN ( 2 , 6 ). Previously, using the same protocol, we observed that restricted food availability led to decreased body weight, mostly due to the amount of food eaten and not the time of feeding. We also observed a pre-feeding corticosterone peak when animals were submitted to restricted food availability independently of the feeding time ( 16 ).
In the present study, our results demonstrate that in rats submitted to daytime feeding (RF-d or DF), independently of the amount of food eaten, the expression pattern of Clock and Bmal1 was modified in the liver compared with control and RF-n groups, in which food intake was in accordance with rat nocturnal activities. These data are in accordance with a previous study evaluating daytime feeding in mice, in which an inverted phase of Per1 , Per2 , and Dbp clock genes was observed in the liver ( 18 ). In rats submitted to daytime feeding, higher expression of Bmal1 at the end of the day (or the timing of feeding) has also been described, independently of feeding or fasting conditions ( 19 ). One recent study involving transcriptomic analysis indicated that, besides Bmal1 and Clock , nearly all rhythmic transcripts exhibited a 12-hour phase shift in the liver of mice under diurnal feeding, called reverse-phase feeding, suggesting a dominant role of feeding on rhythmic phase regulation ( 20 ).
Similarly to the observed expression in the liver, higher Bmal1 expression in the PAT was observed in the control and RF-n groups in the morning. Bmal1 expression was modified exclusively by daytime feeding (DF and RF-d groups). However, in the BAT, both daytime feeding and calorie restriction at night (RF-n) modified Bmal1 expression. Our data confirm previously reported higher Bmal1 expression in the morning in brown and epididymal white adipose tissues from control animals and its phase shifting in daytime-fed animals ( 21 ). Furthermore, our data expand information about Bmal1 phase shifting in adipose tissues from animals submitted to restricted feeding during a few hours of the light phase. Interestingly, Bmal1 has been associated with regulation of brown and white adipose tissue differentiation ( 22 , 23 ).
In fact, the molecular mechanisms by which feeding affects the circadian clock remain not completely defined. Recently, Liu and cols. demonstrated that mechanistic target of rapamycin (mTOR) may modulate the hepatic circadian clock via tight junction protein 1 (TJP1). Upon feeding, mTOR phosphorylates TJP1, disrupting the TJP1/PER1 association and promoting PER1 nuclear translocation to inhibit the expression of CLOCK/BMAL1 target genes ( 24 ). In food-restricted rats, the food anticipatory activity was also associated with a significant reduction of liver glycogen levels, an increase of circulating free fatty acids and ketone bodies, and the occurrence of an oxidized cytoplasmic and mitochondrial redox state, which could cooperate to induce a new status of handling and distribution of nutrients by the liver upon food restriction ( 25 ).
Few studies have addressed the role of Sirt1 and Ampk as possible candidates to entrain the peripheral clocks in response to feeding time ( 12 ). Our data support that Sirt1 expression in the liver and in adipose tissues is modified by dietary restriction, with higher levels preceding mealtime. We also observed that Ampk and Nampt expressions in the liver were modified by both daytime feeding and dietary restriction. Ampk and Nampt expression patterns were concurrent with Sirt1 only in the liver of animals submitted to intense calorie restriction. These findings reinforce the importance of SIRT1 as a nutrient-sensing molecule in regulating the circadian clock, specifically in the liver. It has been shown that mice treated with metformin, a commonly used drug whose mechanism of action has been linked to the activation of AMPK, exhibited a 3-h phase advance in liver clock genes ( Per1 , Clock , Bmal1, and Rora and muscle Bmal1 and Rev-erba ) ( 26 ). Furthermore, mice submitted to a high-fat diet exhibited decreased hepatic AMPK expression and activity, accompanied by disturbance of hepatic clock genes’ circadian rhythm ( 27 ). In accordance with our data, Chaix and cols. observed, in Clock mutant mice submitted to feeding time restriction, altered liver nicotinate and nicotinamide, two precursors of NAD+. Both are upstream of Nampt, the expression of which was increased in animals submitted to food restriction ( 5 ).
Ampk and Nampt presented no differential expression in restricted feeding groups in the BAT and PAT, suggesting that the metabolic changes evoked by food restriction in these tissues might not be mediated by the interaction between Sirt1 and Ampk or Nampt , as suggested in the liver. In the BAT, other sirtuins, such as SIRT2 and SIRT3, may be relevant ( 28 ). Moreover, SIRT1 deacetylase activity can also vary in a circadian manner, regardless of its expression ( 13 ), which is a limitation in studies evaluating gene or protein expression. In addition, fasting conditions are associated with higher expression of Sirt1 mRNA ( 19 , 29 ). Thus, the influence of fasting cannot be totally excluded in our study. Additionally, experimental animals might adapt to restricted levels of energy intake, mainly by a reduction in basal metabolic rate and in total heat production, with either reduction or no difference in spontaneous activity compared with ad libitum-fed rats. However, in the present study, spontaneous activity was not evaluated. Interestingly, a study on calorie restriction and weight loss in humans demonstrated an increase in SIRT1 expression in subcutaneous adipose tissue, concomitant with an increased glucose uptake in the BAT, which could indicate that the increase in SIRT1 could indirectly improve BAT metabolic efficiency upon weight loss ( 30 ).
We also observed a concurrent Sirt1 expression pattern with the expression patterns of Pparg , Pgc1a , and Ucp2 in liver and adipose tissues in different feeding schedules, reinforcing the control exerted by Sirt1 protein in lipid metabolism. Upon fasting, SIRT1 is recruited to the promoters of PPARG target genes in adipose tissue, counteracting this important adipogenesis regulator and promoting mobilization of fat stores during food deprivation ( 31 ). SIRT1 has also been identified as a PPARG deacetylase, whose effects could mimic outcomes of ligand-dependent PPARG activation ( 32 ). Indeed, resveratrol, through activation of the AMPK/SIRT1, prevents liver fat accumulation induced by high-fat and high-sucrose diets in rats by increasing fatty acid oxidation and decreasing lipogenesis ( 33 ). In addition, SIRT1 liver-specific deleted or knocked-down mice exhibited impaired fatty acid oxidation and increased hepatic free fatty acids ( 34 , 35 ). In myotubes, elevated SITR1 promotes deacetylation and activation of PGC1A, resulting in increased expression of molecules involved in mitochondrial biogenesis, such as UCP2 ( 36 ).
In conclusion, food restriction in dissociation with rat nocturnal activity alters the expression pattern of clock genes mainly in the liver. In addition, food availability during the 12 hours of the light phase inverted the profile of the energy-sensing and lipid metabolism-related genes, whereas the calorie restriction induced an increased expression of these genes in anticipation of food availability, also mainly in the liver. These interactions in liver and adipose tissues may occur through Sirt1 , but the interaction with Ampk and Nampt might also be involved in the liver. Thus, dietary restriction resets clock and lipid metabolism-related genes in liver and adipose tissues through nutrient-sensing genes.
Acknowledgements:
the authors thank Rogerio Lenotti Zuliani and Jose Roberto Silva for technical assistance.
SUPPLEMENTARY
Supplementary Table 1. Relative expression of clock genes, energy-sensing and metabolism-related genes in rat liver on restricted feeding schedules.
| Gene | Liver | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CG | DF | RF-n | RF-d | |||||||||
| ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | |
| Clock | 1.1 ± 0.1 | 0.5 ± 0.1 | 1.1 ± 0.1 | 0.5 ± 0.1 | 0.6 ± 0.1 | 1.8 ± 0.3 | 0.9 ± 0.1 | 0.4 ± 0.1 | 1.0 ± 0.1 | 0.7 ± 0.2 | 0.7 ± 0.1 | 1.0 ± 0.1 |
| 1.0 | 0.5 | 1.2 | 0.5 | 0.6 | 1.8 | 0.9 | 0.4 | 1.0 | 0.7 | 0.7 | 1.0 | |
| [1.0-1.2] | [0.4-0.6] | [1.0-1.3] | [0.4-0.6] | [0.5-0.8] | [1.6-2.1] | [0.8-1.0] | [0.3-0.5] | [0.9-1.0] | [0.6-0.9] | [0.6-0.8] | [0.9-1.0] | |
| Bmal1 | 1.0 ± 0.3 | 0.03 ± 0.01 | 0.4 ± 0.1 | 0.03 ± 0.01 | 0.5 ± 0.1 | 0.9 ± 0.3 | 0.8 ± 0.1 | 0.02 ± 0.01 | 0.6 ± 0.1 | 0.04 ± 0.02 | 0.8 ± 0.1 | 1.6 ± 0.2 |
| 1.0 | 0.03 | 0.4 | 0.03 | 0.4 | 0.9 | 0.8 | 0.01 | 0.6 | 0.04 | 0.8 | 1.6 | |
| [0.8-1.3] | [0.03-0.04] | [0.3-0.5] | [0.03-0.04] | [0.4-0.5] | [0.7-1.0] | [0.7-0.9] | [0.01-0.02] | [0.6-0.7] | [0.02-0.04] | [0.7-0.9] | [1.5-1.8] | |
| Silt1 | 1.1 ± 0.3 | 0.9 ± 0.3 | 0.6 ± 0.2 | 0.5 ± 0.1 | 0.4 ± 0.1 | 0.8 ± 0.3 | 0.9 ± 0.1 | 2.0 ± 0.9 | 0.4 ± 0.1 | 3.5 ± 1.9 | 0.6 ± 0.3 | 0.7 ± 0.2 |
| 1.0 | 0.8 | 0.6 | 0.5 | 0.4 | 0.8 | 1.0 | 1.8 | 0.3 | 3.0 | 0.6 | 0.7 | |
| [0.8-1.4] | [0.6-1.2] | [0.4-0.7] | [0.4-0.6] | [0.4-0.5] | [0.6-0.9] | [0.8-1.0] | [1.4-2.8] | [0.3-0.5] | [2.0-4.7] | [0.4-0.9] | [0.6-0.8] | |
| Ampk | 1.1 ± 0.3 | 1.4 ± 0.4 | 1.5 ± 0.3 | 3.0 ± 0.8 | 2.0 ± 0.6 | 1.4 ± 0.6 | 0.9 ± 0.2 | 2.2 ± 0.5 | 0.9 ± 0.2 | 2.9 ± 0.7 | 0.8 ± 0.1 | 1.3 ± 0.4 |
| 1.0 | 1.3 | 1.6 | 3.0 | 1.9 | 1.4 | 1.0 | 2.1 | 0.9 | 3.1 | 0.7 | 1.3 | |
| [0.9-1.4] | [1.1-1.6] | [1.2-1.8] | [2.3-3.5] | [1.6-2.6] | [0.8-1.9] | [0.8-1.1] | [1.7-2.6] | [0.8-1.0] | [2.2-3.4] | [0.6-0.9] | [1.0-1.6] | |
| Nampt | 1.0 ± 0.2 | 2.1 ± 0.7 | 5.4 ± 0.7 | 3.9 ± 1.2 | 2.7 ± 1.0 | 1.9 ± 0.8 | 0.9 ± 0.3 | 6.5 ± 1.5 | 3.9 ± 1.1 | 8.2 ± 2.3 | 1.5 ± 0.5 | 2.1 ± 1.4 |
| 1.0 | 2.1 | 5.6 | 3.8 | 2.4 | 1.6 | 0.8 | 7.0 | 3.7 | 8.3 | 1.4 | 1.7 | |
| [0.8-1.2] | [1.6-2.5] | [4.7-6.0] | [3.5-4.5] | [1.7-3.6] | [1.1-2.8] | [0.7-1.1] | [5.1-7.7] | [3.0-4.8] | [6.0-10.2] | [1.2-1.7] | [1.1-3.3] | |
| Pparg | 1.0 ± 0.3 | 1.6 ± 0.4 | 0.9 ± 0.4 | 1.0 ± 0.3 | 0.9 ± 0.3 | 1.7 ± 0.5 | 0.9 ± 0.3 | 1.5 ± 0.4 | 0.9 ± 0.5 | 2.7 ± 1.1 | 1.1 ± 0.4 | 1.5 ± 0.3 |
| 1.0 | 1.7 | 1.0 | 1.0 | 0.7 | 1.6 | 0.9 | 1.5 | 0.7 | 2.6 | 1.2 | 1.4 | |
| [0.7-1.3] | [1.4-1.8] | [0.6-1.2] | [0.8-1.4] | [0.7-1.0] | [1.2-2.2] | [0.7-1.1] | [1.2-1.8] | [0.6-1.3] | [2.0-3.3] | [0.6-1.2] | [1.2-1.8] | |
| Pgc1a | 1.1 ± 0.4 | 2.1 ± 1.1 | 2.2 ± 0.6 | 1.8 ± 0.5 | 1.9 ± 0.5 | 4.0 ± 1.8 | 3.9 ± 0.7 | 9.6 ± 3.4 | 3.0 ± 1.2 | 16.4 ± 9.2 | 2.7 ± 0.9 | 7.0 ± 3.1 |
| 1.0 | 1.6 | 2.0 | 1.7 | 1.7 | 4.3 | 4.1 | 10.1 | 2.7 | 15.5 | 2.5 | 7.3 | |
| [0.7-1.5] | [1.4-3.0] | [1.8-2.7] | [1.4-2.4] | [1.6-2.5] | [2.1-5.6] | [3.1-4.5] | [6.6-12.2] | [2.0-4.1] | [7.5-24.1] | [2.2-2.9] | [5.1-8.0] | |
| Ucp2 | 0.9 ± 0.3 | 0.9 ± 0.2 | 0.5 ± 0.06 | 0.5 ± 0.2 | 0.6 ± 0.1 | 0.6 ± 0.2 | 0.6 ± 0.2 | 0.9 ± 0.2 | 0.3 ± 0.1 | 1.2 ± 0.3 | 0.4 ± 0.1 | 0.4 ± 0.1 |
| 1.0 | 0.9 | 0.5 | 0.5 | 0.6 | 0.5 | 0.6 | 1.0 | 0.3 | 1.1 | 0.4 | 0.4 | |
| [0.6-1.1] | [0.7-1.1] | [0.5-0.6] | [0.4-0.6] | [0.5-0.7] | [0.4-0.7] | [0.4-0.7] | [0.8-1.1] | [0.2-0.4] | [0.9-1.6] | [0.4-0.5] | [0.3-0.6] | |
Data (2-ΔΔCt) are shown as mean ±; standard deviation, median and [interquartile interval]. CG: control group, DF: day-fed, RF-n: restricted night-fed, and RF-d: restricted day-fed. ZT3, ZT11 and ZT17: Zeitgeber times 3, 11 and 17, corresponding to 0900, 1700, and 2300h, respectively.
Supplementary Table 2. Relative expression of clock genes, energy-sensing and metabolism-related genes in rat brown adipose tissue on restricted feeding schedules.
| Gene | Brown adipose tissue | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CG | DF | RF-n | RF-d | |||||||||
| ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | |
| Clock | 1.0 ± 0.2 | 1.0 ± 0.3 | 1.0 ± 0.1 | 1.0 ± 0.2 | 1.0 ± 0.7 | 1.2 ± 0.2 | 1.0 ± 0.7 | 0.6 ± 0.2 | 0.8 ± 0.1 | 1.2 ± 0.6 | 1.0 ± 0.5 | 0.8 ± 0.2 |
| 1.0 | 0.9 | 1.0 | 1.0 | 0.9 | 1.3 | 0.6 | 0.5 | 0.8 | 0.9 | 0.8 | 0.7 | |
| [0.8-1.2] | [0.8-1.2] | [0.9-1.1] | [0.7-1.1] | [0.7-1.0] | [1.0-1.4] | [0.5-1.6] | [0.5-0.8] | [0.7-1.0] | [0.7-1.8] | [0.7-1.6] | [0.6-0.9] | |
| Bmal1 | 1.0 ± 0.3 | 0.1 ± 0.08 | 0.3 ± 0.1 | 1.0 ± 0.7 | 0.7 ± 0.3 | 0.4 ± 0.2 | 0.4 ± 0.2 | 0.2 ± 0.2 | 0.2 ± 0.2 | 0.9 ± 0.5 | 1.4 ± 0.5 | 0.1 ± 0.1 |
| 1.0 | 0.1 | 0.3 | 1.0 | 0.6 | 0.3 | 0.5 | 0.2 | 0.2 | 0.8 | 1.2 | 0.07 | |
| [0.8-1.3] | [0.06-0.2] | [0.1-0.4] | [0.4-1.6] | [0.4-0.8] | [0.2-0.5] | [0.1-0.6] | [0.07-0.4] | [0.08-0.4] | [0.6-1.0] | [0.9-1.9] | [0.03-0.1] | |
| Silt1 | 1.1 ± 0.5 | 1.2 ± 1.3 | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.4 ± 0.2 | 0.4 ± 0.1 | 1.3 ± 1.0 | 2.0 ± 1.4 | 0.2 ± 0.02 | 2.2 ± 2.5 | 0.8 ± 0.6 | 0.2 ± 0.1 |
| 1.0 | 0.6 | 0.2 | 0.3 | 0.3 | 0.4 | 1.1 | 2.1 | 0.2 | 1.0 | 0.5 | 0.2 | |
| [0.7-1.7] | [0.4-2.4] | [0.2-0.3] | [0.2-0.3] | [0.2-0.5] | [0.4-0.5] | [0.3-2.3] | [0.6-3.3] | [0.1-0.2] | [0.5-4.2] | [0.3-1.1] | [0.1-0.3] | |
| Ampk | 1.1 ± 0.3 | 0.9 ± 0.3 | 0.9 ± 0.5 | 2.8 ± 0.9 | 3.5 ± 1.4 | 1.2 ± 0.3 | 1.0 ± 0.5 | 1.5 ± 0.5 | 1.0 ± 0.5 | 1.0 ± 0.3 | 1.0 ± 0.3 | 1.1 ± 0.6 |
| 1.0 | 0.9 | 0.9 | 2.9 | 3.7 | 1.1 | 1.0 | 1.6 | 1.0 | 1.1 | 1.0 | 1.1 | |
| [0.9-1.2] | [0.6-1.2] | [0.5-1.4] | [1.9-3.4] | [1.8-5.1] | [1.0-1.4] | [0.6-1.4] | [1.0-1.8] | [0.3-1.4] | [0.8-1.3] | [0.8-1.3] | [0.5-1.5] | |
| Nampt | 1.0 ± 0.2 | 1.4 ± 0.8 | 2.3 ± 0.8 | 2.9 ± 0.7 | 2.6 ± 1.4 | 1.9 ± 0.4 | 2.6 ± 1.2 | 3.6 ± 1.9 | 4.0 ± 2.0 | 2.5 ± 1.4 | 3.0 ± 1.0 | 4.0 ± 3.8 |
| 1.0 | 1.3 | 2.1 | 3.1 | 2.5 | 2.0 | 2.4 | 4.6 | 3.6 | 1.9 | 3.1 | 2.3 | |
| [0.8-1.1] | [0.5-2.2] | [1.8-3.0] | [2.2-3.4] | [1.4-3.8] | [1.5-2.2] | [1.6-3.7] | [1.5-5.1] | [2.3-67] | [1.5-4.1] | [2.3-4.0] | [1.7-5.2] | |
| Pparg | 1.1 ± 0.4 | 1.4 ± 0.4 | 2.0 ± 0.5 | 3.6 ± 1.2 | 2.8 ± 0.7 | 5.4 ± 1.9 | 1.1 ± 0.7 | 1.5 ± 0.6 | 0.9 ± 0.3 | 1.5 ± 0.4 | 1.2 ± 0.3 | 0.8 ± 0.6 |
| 1.0 | 1.6 | 1.9 | 3.2 | 2.6 | 5.0 | 1.0 | 1.6 | 0.8 | 1.4 | 1.3 | 0.7 | |
| [0.8-1.4] | [1.0-1.7] | [1.7-2.3] | [3.0-4.8] | [2.1-3.6] | [5.4-6.9] | [0.7-1.7] | [1.0-2.0] | [0.7-1.3] | [1.1-1.8] | [0.9-1.4] | [0.3-1.2] | |
| Pgc1a | 1.1 ± 0.4 | 1.7 ± 1.0 | 1.1 ± 0.5 | 2.9 ± 0.9 | 2.6 ± 0.4 | 3.3 ± 1.0 | 1.4 ± 0.4 | 2.4 ± 1.1 | 0.8 ± 0.2 | 2.4 ± 0.8 | 1.3 ± 0.3 | 0.7 ± 0.3 |
| 1.0 | 1.4 | 0.9 | 3.1 | 2.5 | 3.4 | 1.5 | 3.0 | 0.7 | 2.1 | 1.3 | 0.6 | |
| [0.8-1.4] | [1.1-2.4] | [0.8-1.3] | [2.0-3.7] | [2.3-3.0] | [2.5-4.0] | [0.9-1.7] | [1.3-3.3] | [0.6-0.9] | [1.7-3.4] | [1.0-1.5] | [0.4-1.0] | |
| Ucp2 | 0.9 ± 0.2 | 1.4 ± 0.8 | 0.8 ± 0.2 | 1.6 ± 0.3 | 1.6 ± 0.5 | 1.8 ± 0.9 | 1.3 ± 0.6 | 2.5 ± 1.4 | 0.6 ± 0.2 | 1.9 ± 0.9 | 0.8 ± 0.2 | 0.5 ± 0.3 |
| 1.0 | 1.5 | 0.8 | 1.5 | 1.6 | 1.6 | 1.1 | 2.4 | 0.6 | 1.7 | 0.8 | 0.4 | |
| [0.6-1.1] | [0.7-2.1] | [0.6-1.0] | [1.4-1.8] | [1.1-2.2] | [1.0-2.2] | [0.8-1.9] | [1.2-3.7] | [0.3-0.7] | [1.5-2.3] | [0.7-1.0] | [0.3-0.7] | |
Data (2-ΔΔCt) are shown as mean ±; standard deviation, median and [interquartile interval]. CG: control group, DF: day-fed, RF-n: restricted night-fed, and RF-d: restricted day-fed. ZT3, ZT11 and ZT17: Zeitgeber times 3, 11 and 17, corresponding to 0900, 1700, and 2300h, respectively.
Supplementary Table 3. Relative expression of energy-sensing and metabolism-related genes in rat peri-epididymal adipose tissue on restricted feeding schedules.
| Gene | Peri-epididymal adipose tissue | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CG | DF | RF-n | RF-d | |||||||||
| ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | ZT3 | ZT11 | ZT17 | |
| Clock | 1.0 ± 0.5 | 0.8 ± 0.3 | 1.0 ± 0.2 | 0.5 ± 0.1 | 0.5 ± 0.2 | 0.4 ± 0.1 | 1.0 ± 0.3 | 0.3 ± 0.1 | 0.5 ± 0.1 | 0.8 ± 0.2 | 0.8 ± 0.6 | 0.4 ± 0.1 |
| 1.0 | 0.8 | 0.9 | 0.5 | 0.5 | 0.4 | 0.9 | 0.3 | 0.5 | 0.8 | 0.6 | 0.4 | |
| [0.5-1.4] | [0.6-0.9] | [0.8 ± 1.2] | [0.4-0.7] | [0.4-0.7] | [0.4-0.5] | [0.8-1.3] | [0.2-0.4] | [0.4-0.6] | [0.6-1.0] | [0.4-1.6] | [0.3-0.6] | |
| Bmal1 | 1.0 ± 0.3 | 0.06 ± 0.04 | 0.2 ± 0.1 | 0.2 ± 0.07 | 0.2 ± 0.08 | 0.1 ± 0.06 | 0.3 ± 0.1 | 0.04 ± 0.02 | 0.4 ± 0.1 | 0.2 ± 0.07 | 1.0 ± 0.3 | 0.2 ± 0.1 |
| 1.0 | 0.07 | 0.1 | 0.2 | 0.2 | 0.1 | 0.3 | 0.04 | 0.4 | 0.2 | 1.0 | 0.1 | |
| [0.7-1.2] | [0.03-0.1] | [0.1-0.3] | [0.1-0.2] | [0.1-0.3] | [0.07-0.2] | [0.2-0.4] | [0.02-0.05] | [0.3-0.5] | [0.1-0.2] | [0.8-1.1] | [0.07-0.3] | |
| Silt1 | 1.6 ± 1.1 | 2.7 ± 2.6 | 0.9 ± 0.4 | 0.8 ± 0.3 | 0.8 ± 0.4 | 0.4 ± 0.04 | 1.3 ± 0.6 | 1.6 ± 0.8 | 0.5 ± 0.2 | 1.9 ± 0.6 | 0.7 ± 0.2 | 0.5 ± 0.3 |
| 1.0 | 1.7 | 0.9 | 0.8 | 0.7 | 0.4 | 1.1 | 1.1 | 0.4 | 2.1 | 0.8 | 0.4 | |
| [0.9-2.8] | [0.8-4.6] | [0.5-1.3] | [0.5-1.1] | [0.5-1.0] | [0.3-0.4] | [0.8-1.9] | [1.0-2.4] | [0.4-0.7] | [1.4-2.2] | [0.5-0.8] | [0.3-0.8] | |
| Ampk | 1.4 ± 1.1 | 2.2 ± 2.6 | 0.9 ± 0.3 | 0.9 ± 0.4 | 1.8 ± 0.5 | 0.5 ± 0.1 | 1.2 ± 1.4 | 0.8 ± 0.9 | 0.7 ± 0.3 | 0.7 ± 0.4 | 0.6 ± 0.2 | 0.6 ± 0.2 |
| 1.0 | 1.3 | 0.9 | 0.9 | 1.6 | 0.5 | 0.6 | 0.5 | 0.7 | 0.6 | 0.6 | 0.6 | |
| [0.7-2.0] | [0.7-3.4] | [0.5-1.2] | [0.5-1.3] | [1.4-2.3] | [0.4-0.6] | [0.5-1.6] | [0.4-1.5] | [0.6-0.7] | [0.4-1.1] | [0.5-0.8] | [0.5-0.8] | |
| Nampt | 1.0 ± 0.4 | 3.7 ± 4.0 | 5.1 ± 2.6 | 1.5 ± 0.5 | 0.8 ± 0.3 | 0.9 ± 0.1 | 2.0 ± 1.5 | 1.9 ± 0.7 | 2.0 ± 0.4 | 3.3 ± 1.8 | 2.1 ± 0.9 | 1.6 ± 0.8 |
| 1.0 | 2.3 | 4.2 | 1.3 | 1.0 | 0.9 | 1.5 | 1.6 | 2.1 | 2.4 | 2.2 | 1.4 | |
| [0.8-1.4] | [1.7-4.7] | [3.2-8.1] | [1.1 -2.1] | [0.5-1.0] | [0.8-1.1] | [1.1-2.6] | [1.4-2.5] | [1.7-2.2] | [1.4-5.1] | [1.5-2.4] | [1.2-1.9] | |
| Pparg | 0.9 ± 0.3 | 1.8 ± 0.8 | 0.9 ± 0.3 | 1.2 ± 0.6 | 0.8 ± 0.3 | 0.5 ± 0.1 | 1.8 ± 1.6 | 1.9 ± 0.5 | 0.5 ± 0.3 | 1.9 ± 0.8 | 1.1 ± 0.6 | 0.7 ± 0.4 |
| 1.0 | 1.9 | 0.9 | 1.1 | 0.8 | 0.5 | 1.7 | 2.2 | 0.6 | 1.6 | 1.0 | 0.5 | |
| [0.5-1.3] | [1.0-2.6] | [0.8-1.1] | [0.7-1.8] | [0.6-1.0] | [0.4-0.6] | [0.6-2.5] | [1.5-2.3] | [0.2-0.9] | [1.4-2.3] | [0.6-1.7] | [0.4-1.0] | |
| Pgc1a | 1.1 ± 0.7 | 1.3 ± 1.0 | 0.5 ± 0.2 | 0.7 ± 0.2 | 1.2 ± 0.4 | 0.5 ± 0.2 | 1.8 ± 1.4 | 1.4 ± 0.8 | 1.5 ± 2.4 | 2.3 ± 2.3 | 1.9 ± 1.6 | 0.5 ± 0.2 |
| 1.0 | 1.2 | 0.6 | 0.7 | 1.3 | 0.5 | 1.2 | 1.5 | 0.7 | 2.1 | 1.2 | 0.4 | |
| [0.8-1.6] | [0.4-2.0] | [0.4-0.6] | [0.6-0.9] | [0.9-1.5] | [0.3-0.6] | [0.8-3.0] | [0.7-2.1] | [0.4-1.0] | [0.7-2.5] | [0.9-2.2] | [0.3-0.8] | |
| Ucp2 | 1.1 ± 0.5 | 1.3 ± 0.6 | 0.1 ± 0.1 | 0.8 ± 0.4 | 0.7 ± 0.3 | 0.2 ± 0.1 | 1.1 ± 1.0 | 0.7 ± 0.2 | 0.4 ± 0.6 | 1.0 ± 0.4 | 0.6 ± 0.4 | 0.1 ± 0.1 |
| 1.0 | 1.1 | 0.04 | 0.7 | 0.8 | 0.2 | 0.6 | 0.8 | 0.2 | 1.0 | 0.4 | 0.05 | |
| [0.6-1.6] | [0.8-1.7] | [0.02-0.2] | [0.5-1.2] | [0.5-1.0] | [0.1-0.3] | [0.4-2.3] | [0.5-0.9] | [0.1-0.9] | [0.6-1.4] | [0.3-0.7] | [0.02-0.2] | |
Data (2-ΔΔCt) are shown as mean ± standard deviation, median and [interquartile interval]. CG: control group, DF: day-fed, RF-n: restricted night-fed, and RF-d: restricted day-fed. ZT3, ZT11 and ZT17: Zeitgeber times 3, 11 and 17, corresponding to 0900, 1700, and 2300h, respectively.
Funding Statement
Funding: this work was supported by Sao Paulo Research Foundation (Fapesp): grants n° 2007/58365-3 and n° 2012/02234-6 and the National Council of Technological and Scientific Development (CNPq): grant no. 150569/2015-7.
Footnotes
Funding: this work was supported by Sao Paulo Research Foundation (Fapesp): grants n° 2007/58365-3 and n° 2012/02234-6 and the National Council of Technological and Scientific Development (CNPq): grant no. 150569/2015-7.
REFERENCES
- 1.Cox KH, Takahashi JS. Circadian clock genes and the transcriptional architecture of the clock mechanism. J Mol Endocrinol. 2019;63(4):R93–R102. doi: 10.1530/JME-19-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 3.Delezie J, Dumont S, Dardente H, Oudart H, Grechez-Cassiau A, Klosen P, et al. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26(8):3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 4.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaix A, Lin T, Le HD, Chang MW, Panda S. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab. 2019;29(2):303.e4–319.e4. doi: 10.1016/j.cmet.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stenvers DJ, Scheer FAJL, Schrauwen P, la Fleur SE, Kalsbeek A. Circadian clocks and insulin resistance. Nat Rev Endocrinol. 2019;15(2):75–89. doi: 10.1038/s41574-018-0122-1. [DOI] [PubMed] [Google Scholar]
- 8.Opperhuizen AL, van Kerkhof LW, Proper KI, Rodenburg W, Kalsbeek A. Rodent models to study the metabolic effects of shiftwork in humans. Front Pharmacol. 2015;6:50–50. doi: 10.3389/fphar.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 10.Morbiato, Frigato, Dinarello, Maradonna, Facchinello, Argenton et al. Feeding Entrainment of the Zebrafish Circadian Clock Is Regulated by the Glucocorticoid Receptor. Cells. 2019;8(11):1342–1342. doi: 10.3390/cells8111342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20(24):7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan SD, Lamia KA. AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol. 2013;366(2):163–169. doi: 10.1016/j.mce.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Araujo LD, Roa SL, Bueno AC, Coeli-Lacchini FB, Martins CS, Uchoa ET, et al. Restricted Feeding Schedules Modulate in a Different Manner the Expression of Clock Genes in Rat Hypothalamic Nuclei. Front Neurosci. 2016;10:567–567. doi: 10.3389/fnins.2016.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laguna-Abreu MT, Koenigkam-Santos M, Colleta AM, Elias PC, Moreira AC, Antunes-Rodrigues J, et al. Time course of vasopressin and oxytocin secretion after stress in adrenalectomized rats. Horm Metab Res. 2005;37(2):84–88. doi: 10.1055/s-2005-861159. [DOI] [PubMed] [Google Scholar]
- 18.Satoh Y, Kawai H, Kudo N, Kawashima Y, Mitsumoto A. Time-restricted feeding entrains daily rhythms of energy metabolism in mice. Am J Physiol Regul Integr Comp Physiol. 2006;290(5):R1276–R1283. doi: 10.1152/ajpregu.00775.2005. [DOI] [PubMed] [Google Scholar]
- 19.Sabath E, Salgado-Delgado R, Guerrero-Vargas NN, Guzman-Ruiz MA, del Carmen Basualdo M, Escobar C, et al. Food entrains clock genes but not metabolic genes in the liver of suprachiasmatic nucleus lesioned rats. FEBS Lett. 2014;588(17):3104–3110. doi: 10.1016/j.febslet.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 20.Guan D, Xiong Y, Trinh TM, Xiao Y, Hu W, Jiang C, et al. The hepatocyte clock and feeding control chronophysiology of multiple liver cell types. Science. 2020;369(6509):1388–1394. doi: 10.1126/science.aba8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zvonic S, Ptitsyn AA, Conrad SA, Scott LK, Floyd ZE, Kilroy G, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55(4):962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 22.Guo B, Chatterjee S, Li L, Kim JM, Lee J, Yechoor VK, et al. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012;26(8):3453–3463. doi: 10.1096/fj.12-205781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam D, Guo B, Chatterjee S, Chen MH, Nelson D, Yechoor VK, et al. The adipocyte clock controls brown adipogenesis through the TGF-beta and BMP signaling pathways. J Cell Sci. 2015;128(9):1835–1847. doi: 10.1242/jcs.167643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Zhang Y, Li T, Han J, Wang Y. The tight junction protein TJP1 regulates the feeding-modulated hepatic circadian clock. Nat Commun. 2020;11(1):589–589. doi: 10.1038/s41467-020-14470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Báez-Ruiz A, Escobar C, Aguilar-Roblero R, Vázquez-Martínez O, Díaz-Muñoz M. Metabolic adaptations of liver mitochondria during restricted feeding schedules. Am J Physiol Gastrointest Liver Physiol. 2005;289(6):G1015–G1023. doi: 10.1152/ajpgi.00488.2004. [DOI] [PubMed] [Google Scholar]
- 26.Barnea M, Haviv L, Gutman R, Chapnik N, Madar Z, Froy O. Metformin affects the circadian clock and metabolic rhythms in a tissue-specific manner. Biochim Biophys Acta. 2012;1822(11):1796–1806. doi: 10.1016/j.bbadis.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Zhu Z, Xie M, Xue J. Involvement of adenosine monophosphate-activated protein kinase in the influence of timed high-fat evening diet on the hepatic clock and lipogenic gene expression in mice. Nutr Res. 2015;35(9):792–799. doi: 10.1016/j.nutres.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Lomb DJ, Laurent G, Haigis MC. Sirtuins regulate key aspects of lipid metabolism. Biochim Biophys Acta. 2010;1804(8):1652–1657. doi: 10.1016/j.bbapap.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 29.Noriega LG, Feige JN, Canto C, Yamamoto H, Yu J, Herman MA, et al. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 2011;12(10):1069–1076. doi: 10.1038/embor.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rappou E, Jukarainen S, Rinnankoski-Tuikka R, Kaye S, Heinonen S, Hakkarainen A, et al. Weight loss is associated with increased NAD(+)/SIRT1 expression but reduced PARP activity in white adipose tissue. J Clin Endocrinol Metab. 2016;101(3):1263–1273. doi: 10.1210/jc.2015-3054. [DOI] [PubMed] [Google Scholar]
- 31.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150(3):620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberdi G, Rodriguez VM, Macarulla MT, Miranda J, Churruca I, Portillo MP. Hepatic lipid metabolic pathways modified by resveratrol in rats fed an obesogenic diet. Nutrition. 2013;29(3):562–567. doi: 10.1016/j.nut.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9(4):327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104(31):12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohamed JS, Hajira A, Pardo PS, Boriek AM. MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT-1/PGC-1α network in skeletal muscle. Diabetes. 2014;63(5):1546–1559. doi: 10.2337/db13-1364. [DOI] [PubMed] [Google Scholar]