Abstract
The presence of endogenous amplification inhibitors in urine may produce false-negative results for the detection of Chlamydia trachomatis nucleic acids by tests such as PCR, ligase chain reaction (LCR), and transcription-mediated amplification (TMA). Consecutive urine specimens from 101 pregnant women and 287 nonpregnant women submitted for urinalysis were processed for C. trachomatis detection. Aliquots were spiked with the equivalent of one C. trachomatis elementary body and were tested by three commercial assays: AMPLICOR CT/NG, Chlamydia LCX, and Chlamydia TMA. The prevalence of inhibitors resulting in complete inhibition of amplification was 4.9% for PCR, 2.6% for LCR, and 7.5% for TMA. In addition, all three assays were partially inhibited by additional urine specimens. Only PCR was more often inhibited by urine from pregnant women than by urine from nonpregnant women (9.9 versus 3.1%; P = 0.011). A complete urinalysis including dipstick and a microscopic examination was performed. Logistic regression analysis revealed that the following substances were associated with amplification inhibition: beta-human chorionic gonadotropin (odds ratio [OR], 3.3) and crystals (OR, 3.3) for PCR, nitrites for LCR (OR, 14.4), and hemoglobin (OR, 3.3), nitrites (OR, 3.3), and crystals (OR, 3.3) for TMA. Aliquots of each inhibitory urine specimen were stored at 4 and −70°C overnight or were extracted with phenol-chloroform and then retested at dilutions of 1:1, 1:4, and 1:10. Most inhibition was removed by storage overnight at 4 or −70°C and a dilution of 1:10 (84% for PCR, 100% for LCR, and 92% for TMA). Five urine specimens (three for PCR and two for TMA) required phenol-chloroform extraction to remove inhibitors. The results indicate that the prevalence of nucleic acid amplification inhibitors in female urine is different for each technology, that this prevalence may be predicted by the presence of urinary factors, and that storage and dilution remove most of the inhibitors.
Nucleic acid amplification (NAA) techniques such as PCR, ligase chain reaction (LCR), and transcription-mediated amplification (TMA) have greatly improved our ability to diagnose Chlamydia trachomatis infections, and in recent years, they have been successfully applied to first-void urine specimens. NAA testing of first-void urine specimens has usually detected as many positive patients as testing of urethral or endocervical swabs by cell culture or antigen testing (1–3, 5, 6, 8, 10, 12, 15), but most of these studies have also revealed that none of the amplification tests is 100% sensitive. While the majority of processed specimens are amplifiable, some contain substances that inhibit NAA, thereby giving false-negative results, even if the specimen contains C. trachomatis nucleic acid. In a study comparing PCR and LCR testing of 767 female urine specimens, we observed 15 (1.9%) urine specimens which were positive by one test but not by the other, a discrepancy which could be explained by the presence of inhibitory substances (3). In a study of 200 urine specimens sent to a hospital clinical chemistry laboratory for routine urinalysis, we found that 9% of urine specimens from men and 18% of urine specimens from women contained PCR inhibitors (4). These observations provided the rationale for a prospective study to determine (i) the prevalence of inhibitors in urine specimens to be tested by PCR, LCR, and TMA; (ii) the urinary components associated with amplification inhibition; and (iii) treatment procedures which might remove inhibitors from urine.
MATERIALS AND METHODS
Specimens.
This laboratory-based study tested 388 freshly collected urine specimens submitted for routine urinalysis to clinical chemistry laboratories in three university teaching hospitals. Urine specimens (20 to 50 ml) were obtained from 101 pregnant women and 287 nonpregnant women between 15 and 40 years of age. The specimens were transported by courier at room temperature each morning, together with a printed copy of the urinalysis report, to the Regional Virology and Chlamydiology Laboratory at St. Joseph’s Hospital, aliquoted by one technologist, and tested blindly on the same day by three C. trachomatis nucleic acid detection assays.
Urinalysis.
A complete urinalysis including dipstick and microscopic examination was performed for each urine specimen. Fresh urine specimens were tested for the presence of leukocytes, nitrites, protein, blood, ketone, and glucose, and their specific gravities and pHs were measured with the Multistix 8 SG dipstick (Bayer Inc., Etobicoke, Ontario, Canada). The dipstick was read, according to the manufacturer’s instructions, with an automated urine chemistry analyzer (Clinitex 200+; Bayer Corp.). Positive urinary protein results from the dipstick were confirmed by a semiquantitative sulfosalyic acid test, which simply involved the mixing of 1.0 ml of centrifuged urine to 3.0 ml of 3% sulfosalyic acid. After 5 min, the degree of turbidity caused by the precipitation of denatured protein was observed and was graded from negative to 4+. For microscopic examination of the urine, 10- to 15-ml aliquots were spun at 1,000 rpm for 5 min. A drop of the resulting sediment was transferred to a microscope slide. Examination for epithelial cells, erythrocytes and leukocytes, mucus, yeast, bacteria, casts, and various types of crystals (oxalate, phosphate, urate) was done under both low and high magnifications of the light microscope by trained technicians using standard criteria (17).
Detection of amplification inhibitors by spiking experiments.
C. trachomatis L2 434 was propagated in McCoy cells, and elementary bodies (EBs) were purified by differential centrifugation as described previously (3). EBs were resuspended in phosphate-buffered saline and counted by direct fluorescent-antibody (DFA) staining with monoclonal antibodies specific for major outer membrane proteins (Syva Microtrak, San Jose, Calif.). A serial dilution of C. trachomatis containing approximately one EB determined by DFA staining was tested in triplicate by AMPLICOR CT/NG (Roche Molecular Systems, Branchburg, N.J.), Chlamydia LCX (Abbott Diagnostics, North Chicago, Ill.), and Chlamydia TMA (Gen-Probe, San Diego, Calif.) to ensure that a spike of one EB would generate a positive signal in each test. This dilution gave positive readings in all three tests and was used to spike aliquots of urine specimens. Spiked and unspiked aliquots of urine were tested to detect inhibition of amplification. Complete inhibition was defined as a reduction in signal below the manufacturer’s cutoff. Partial inhibition was defined arbitrarily as a reduction in signal greater than 20% but less than 100%.
Removal of inhibitory activity.
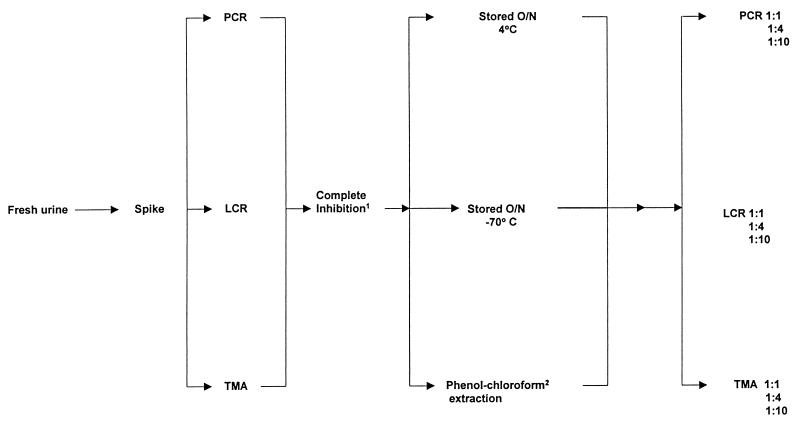
Only urine specimens showing complete inhibition were used to evaluate methods for removing inhibitory activity. Complete inhibition was defined as a signal dropping below the positive cutoff for each test. Urine specimens showing complete inhibition were retested by the algorithm shown in Fig. 1. Aliquots of urine were stored overnight at 4 and −70°C, while a third aliquot was extracted with phenol-chloroform and was allowed to precipitate overnight. Aliquots of urine and extracted nucleic acid were then retested undiluted and at dilutions of 1:4 and 1:10.
FIG. 1.
Algorithm showing procedures used to study specimens containing inhibitors of PCR, LCR, and TMA. Complete Inhibition1, signal below the cutoff of positivity for the test; Phenol-chloroform2, nucleic acid precipitated overnight.
PCR.
Aliquots of urine unspiked and spiked with C. trachomatis were tested by the AMPLICOR CT/NG test. The spike contained one EB of C. trachomatis, equivalent to approximately 10 target molecules for PCR. AMPLICOR CT/NG was performed according to the manufacturer’s instructions. Briefly, 0.5 ml of urine was centrifuged at 16,000 × g for 5 min at room temperature. The pellet was resuspended in 0.25 ml of lysis buffer, vortexed, and incubated for 15 min at room temperature. An equal volume of specimen diluent buffer was added, and the tubes were vortexed and centrifuged at 16,000 × g for 10 min. A 50-μl aliquot of the supernatant was used for amplification. Urine specimens that demonstrated complete inhibition were retested following overnight storage at 4 and −70°C and after extraction with phenol-chloroform according to the algorithm (Fig. 1).
LCR.
The Chlamydia LCX assay was performed according to the manufacturer’s instructions. One milliliter of urine was centrifuged at 16,000 × g for 15 min, and the pellet was resuspended in 1 ml of urine resuspension buffer. The tubes were placed in a heating block at 95°C for 15 min. After cooling to room temperature, 0.2 ml was added to the unit dose and the samples were amplified and read on the LCX instrument.
TMA.
The Gen-Probe Chlamydia TMA assay was performed according to the manufacturer’s instructions. Briefly, 1.5 ml of urine was incubated for 10 min at 37°C followed by centrifugation at 8,000 × g for 5 min. The supernatant was decanted, and the pellet was resuspended in 0.2 ml of specimen diluent buffer. Twenty five microliters of amplification reagent was added to separate tubes, followed by the addition of 200 μl of oil reagent. Fifty microliters of processed specimen was pipetted under the oil, and the tubes were incubated for 10 min at 95°C in a heating block. The tubes were cooled to 42°C, and 25 μl of enzyme reagent was added and the mixture was incubated for 1 h at 42°C. Twenty microliters of termination reagent was added, and the mixture was incubated for 10 min at 42°C. The probe was added and the tubes were incubated for 15 min at 60°C. The selection reagent was added and the tubes were incubated for 10 min at 60°C prior to the hybridization protection assay, which was performed according to the instructions in the manufacturer’s package insert.
Data analysis.
Variables and outcome predictors were recoded prior to analysis to indicate the absence or presence of urinary components. Both univariate and multivariate analyses were performed to determine which urinary components were associated with complete amplification inhibition. Univariate analysis was carried out by the continuity-corrected chi-square test. For multivariate analysis, forward logistic regression modeling was performed with SPSS software (version 7.0). Type I (alpha) error rate was set at 0.05 (two tailed for all analyses).
RESULTS
A total of 388 urine specimens were examined in the study; of these, 101 were positive for beta-human chorionic gonadotropin (beta-HCG). The number of urine specimens containing inhibitors for the three assays is shown in Table 1. A total of 27 urine specimens (7.0%) had inhibitors for PCR, 15 (3.9%) had inhibitors for LCR, and 46 (11.9%) had inhibitors for TMA. For PCR, 19 urine specimens had complete inhibitory activity, as defined by a reduction in the signal to a level below the manufacturer’s cutoff, and 8 additional urine specimens had detectable or partial inhibitory activity, as defined by at least a 20% reduction in the signal. Ten urine specimens completely inhibited the LCR and 5 additional urine specimens partially inhibited the LCR. For TMA, 29 urine specimens had complete inhibitory activity and 17 specimens had partial inhibitory activity. The percentage of urine specimens from pregnant women with complete inhibitory activity was slightly lower than the percentage from nonpregnant women for both LCR (1.0 versus 3.1%) and TMA (6.9 versus 7.7%). For PCR, 9.9 and 3.1% of urine specimens from pregnant and nonpregnant women, respectively (P = 0.011), had complete inhibitory activity; for any detectable inhibitors of PCR, the values were 11.9 and 5.2%, respectively (P = 0.038). Inhibition appeared to be test specific because 11 of 77 urine specimens were inhibitory for two amplification tests, and none were inhibitory for all three tests.
TABLE 1.
Urine specimens from pregnant and nonpregnant women showing complete or partial inhibition for PCR, LCR, or TMA tests
| Group | No. (%) of inhibitorya urine specimens
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| PCR
|
LCR
|
TMA
|
|||||||
| Complete | Partial | Total | Complete | Partial | Total | Complete | Partial | Total | |
| Pregnant (n = 101) | 10 (9.9) | 2 (1.9) | 12 (11.9) | 1 (1.0) | 2 (2.0) | 3 (2.9) | 7 (6.9) | 5 (5.0) | 12 (11.9) |
| Nonpregnant (n = 287) | 9 (3.1) | 6 (2.1) | 15 (5.2) | 9 (3.1) | 3 (1.0) | 12 (4.2) | 22 (7.7) | 12 (4.1) | 34 (11.8) |
| Total (n = 388) | 19 (4.9) | 8 (2.1) | 27 (7.0) | 10 (2.6) | 5 (1.3) | 15 (3.9) | 29 (7.5) | 17 (4.4) | 46 (11.9) |
Complete inhibition was defined as a reduction in the signal to a level below the cutoff for positivity in each assay; partial inhibition was defined as greater than 20% but less than 100% of the signal achieved by the positive control spike.
The results of a univariate analysis of urine parameters associated with complete inhibition of PCR, LCR, and TMA are presented in Table 2. PCR inhibition was significantly associated (P < 0.05) with the presence of hemoglobin, beta-HCG, crystals, and bacteria. The presence of glucose and nitrites was significantly associated (P < 0.05) with LCR inhibition, while the presence of hemoglobin, protein, ketones, nitrites, crystals, and bacteria was associated (P < 0.05) with TMA inhibition.
TABLE 2.
Univariate analysis of urinary substances associated with complete inhibition for PCR, LCR, or TMA
| Inhibitory substancea | PCR
|
LCR
|
TMA
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of specimens with complete inhibitory activity/total no. tested
|
Pb | No. of specimens with complete inhibitory activity/total no. tested
|
Pb | No. of specimens with complete inhibitory activity/total no. tested
|
Pb | ||||
| Present | Absent | Present | Absent | Present | Absent | ||||
| Glucose | 0/19 | 20/369 | 0.297 | 2/10 | 18/378 | <0.05* | 2/29 | 18/359 | 0.660 |
| Hemoglobin | 1/19 | 98/369 | <0.05* | 5/10 | 94/378 | 0.072 | 10/29 | 83/359 | <0.001*** |
| Protein | 4/19 | 81/369 | 0.926 | 4/10 | 81/378 | 0.161 | 11/29 | 74/359 | <0.05* |
| Ketone | 2/19 | 30/369 | 0.711 | 3/10 | 29/378 | 0.113 | 6/29 | 26/359 | <0.05* |
| Nitrites | 2/19 | 23/309 | 0.457 | 5/10 | 20/378 | <0.001*** | 6/29 | 19/359 | <0.01** |
| beta-HCG | 12/19 | 89/369 | <0.001*** | 1/10 | 100/378 | 0.242 | 7/29 | 94/359 | 0.809 |
| Crystals | 12/19 | 116/369 | <0.01** | 4/10 | 124/378 | 0.633 | 17/29 | 111/359 | <0.01** |
| Bacteria | 15/19 | 206/369 | <0.05* | 6/10 | 215/378 | 0.844 | 24/29 | 197/359 | <0.01** |
Six other substances not statistically related were leukocytes, urates, oxalates, phosphates, casts, and yeasts.
Statistically significant differences are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
When multivariate analysis was performed to determine which substances were associated with inhibition, logistic regression modeling revealed that hemoglobin (odds ratio [OR], 3.29; P = 0.004), nitrites (OR, 3.57; P = 0.025), and crystals (OR, 3.18; P = 0.005) were associated with inhibition of TMA (Table 3). For PCR, beta-HCG (OR, 3.43; P = 0.038) and crystals (OR, 3.59; P = 0.025) were associated with inhibition, and for LCR only nitrites were significantly associated with inhibition (OR, 14.36; P = 0.011).
TABLE 3.
Multivariate analysis of urinary substances associated with complete inhibition of amplification by PCR, LCR, and TMA assays
| Assay | Inhibitory substancea | OR | P value |
|---|---|---|---|
| TMA | Hemoglobin | 3.29 | 0.004 |
| Nitrites | 3.57 | 0.025 | |
| Crystals | 3.18 | 0.005 | |
| PCR | beta-HCG | 3.43 | 0.038 |
| Crystals | 3.59 | 0.025 | |
| LCR | Nitrites | 14.36 | 0.011 |
Substances associated with inhibition were determined by logistic regression analysis.
The effects of storage at different temperatures and dilution of the urine on the inhibitory activity are indicated in Table 4. An insufficient volume of 5 urine specimens for TMA and 1 urine specimen for LCR left 24 and 9 urine specimens, respectively, for inhibitor removal studies. Storage overnight at 4°C removed inhibitory substances from 8 of 19 (42.1%) specimens for PCR, 5 of 9 (55.5%) specimens for LCR, and 19 of 24 (79.1%) specimens for TMA. Storage at −70°C similarly removed inhibitory substances from 11 of 19 (57.9%) specimens for PCR, 4 of 9 (44.4%) specimens for LCR, and 19 of 24 (79.1%) specimens for TMA. PCR inhibitors were removed further by dilution to 1:4 (after storage at 4 or −70°C) for 73.6% (14 of 19) of the specimens, and further dilution to 1:10 increased the proportion to 84.2% (16 of 19). Thus, 3 of 19 specimens resisted PCR inhibitor removal after storage and dilution. Dilution of specimens with inhibitors of TMA increased the numbers rendered noninhibitory from 19 to 22, leaving 2 of 24 persistently inhibitory urine specimens. Dilutions removed all inhibitory activity for LCR. Phenol-chloroform extraction removed the inhibitors from all urine specimens.
TABLE 4.
Removal of amplification inhibitors by various treatments
| Treatment | Dilution | No. of urine specimens rendered noninhibitory/total no. tested (%) for the following:
|
||
|---|---|---|---|---|
| PCR | LCR | TMA | ||
| 4°C overnight | 1:1 | 8/19 (42.1) | 5/9 (55.5) | 19/24 (79.1) |
| 1:4 | 14/19 (73.6) | 7/9 (77.7) | 22/24 (91.6) | |
| 1:10 | 16/19 (84.2) | 9/9 (100) | 22/24 (91.6) | |
| −70°C overnight | 1:1 | 11/19 (57.9) | 4/9 (44.4) | 19/24 (79.1) |
| 1:4 | 14/19 (73.6) | 9/9 (100) | 22/24 (91.6) | |
| 1:10 | 16/19 (84.2) | 9/9 (100) | 22/24 (91.6) | |
| Phenol-chloroform | 1:1 | 19/19 (100) | 9/9 (100) | 24/24 (100) |
| 1:4 | 19/19 (100) | 9/9 (100) | 24/24 (100) | |
| 1:10 | 19/19 (100) | 9/9 (100) | 24/24 (100) | |
Because the AMPLICOR CT/NG had its own internal control, we were able to compare its ability to identify urine specimens containing PCR inhibitors. The AMPLICOR internal control detected 13 (3.4%) urine specimens that had inhibitory activity, while the exogenous DNA spike method detected 19 (4.9%) specimens that had completely inhibitory activity and an additional 8 specimens that had partial inhibitory activity.
DISCUSSION
In this study of 388 urine specimens from women, the prevalence of urine specimens containing detectable inhibitors was 7.0% for PCR, 3.9% for LCR, and 11.9% for TMA. When complete inhibition was considered, the prevalence of inhibitory urine specimens was 4.9% for PCR, 2.6% for LCR, and 7.5% for TMA. Inhibition rates were similar for urine specimens from pregnant and nonpregnant women for LCR (4.2 versus 2.9%) and TMA (11.8 versus 11.9%). For PCR, however, the proportion of inhibitory urine specimens from pregnant women was 11.9%, whereas the proportion from nonpregnant women was 5.2% (P = 0.038). The mechanism of PCR inhibition associated with beta-HCG is unknown.
In a letter recently published by Jensen et al. (9), the investigators reported that 15 of 1,136 urine specimens (1.3%) from pregnant patients possibly had plasmid LCR inhibitors because these samples which were negative by the LCX test had positive results by other tests such as DFA analysis, enzyme immunoassay, major outer membrane protein or LCR, but they did not perform inhibitor studies. As described in another letter (13), the investigators assumed a rate of 3% inhibition for cervical swabs from prostitutes because of differences seen between NAA testing and other assays, but they did not perform studies for LCR inhibitors. Berg et al. (1) used DNA spiking and reported the presence of LCR inhibitors in 10 of 382 (2.6%) urine specimens from men attending a sexually transmitted disease clinic. Our LCR inhibition rate of 3.9%, which was achieved with a DNA spike equivalent to 1 EB, was similar to the previously reported rates presented above and particularly the 2.6% rate reported by Berg et al. (1), who used the LCX-positive control DNA equivalent to 50 inclusion-forming units.
PCR has been shown by others to have reduced sensitivity due to inhibitors (7, 11, 18, 19). Those studies have shown that inhibitors could be removed by dilution, freezing and thawing, heating, or prolonged storage of the sample. Roche Molecular Systems has developed an internal amplification control to identify specimens containing PCR inhibitors. The internal control can be incorporated into both the manual AMPLICOR CT/NG test and the semiautomated COBAS AMPLICOR system (14). Specimens yielding a negative result for the internal control are interpreted as inhibitory or nonamplifiable and thus are not reported as negative, thereby reducing the number of false-negative results. In our study the internal control detected 13 inhibitory specimens, while our exogenous DNA spike method detected 19 inhibitory specimens. The discrepancies observed between the kit’s internal control and our DNA spike for female urine specimens may be due to weakly inhibitory specimens for which the results were below the limit of detection for the Roche internal control. The kit internal control has a slightly higher number of target molecules (20 copies of plasmid) compared with the one EB that we used in our spike. In this study 15 specimens were positive for C. trachomatis by either PCR, LCR, or TMA, but none of these urine specimens contained inhibitors by either the DNA spike method or the Roche internal control method. For this reason, the efficiency of the internal control for identifying specimens with false-negative results could not be determined. Further studies with larger numbers of specimens will be required to determine if the use of an internal amplification control is a cost-effective approach for monitoring amplification inhibition.
To our knowledge this is the first study to attempt to correlate inhibition of NAA with substances found in urine. Nitrites were associated with LCR inhibition, beta-HCG and crystals were associated with PCR inhibition, and hemoglobin, nitrites, and crystals were associated with TMA inhibition. These results, however, do not establish a direct causal relationship between the implicated urinary substance and inhibition since a small number of urine specimens (n = 22) which had lost their inhibitory activity after storage were found on subsequent urinalysis to have retained the implicated inhibitory substance. Not all urine specimens were retested by urinalysis. For the 11 urine specimens that were inhibitory in two NAA assays, we attempted to determine whether one assay was more sensitive to a single substance, i.e., nitrites. One urine specimen containing nitrites and beta-HCG was inhibitory for TMA and PCR but not for LCR. Since nitrites were associated with inhibition for TMA (OR, 3.57) and LCR (OR, 14.36) by multivariate logistic regression analysis and this urine specimen was not inhibitory for LCR, the existence of a complex interaction between urinary substances resulting in amplification inhibition is suggested.
Overnight storage at either 4 or −70°C removed 42.1 and 55.5% of the inhibitors for PCR and LCR, respectively, and 79.1% of the inhibitors for TMA. Storage at either 4 or −70°C combined with dilution to 1:10 removed all of the inhibitors for LCR and most of the inhibitors for PCR (16 of 19) and TMA (22 of 24). Phenol-chloroform extraction removed inhibitors from all urine specimens. The mechanism(s) involved in the removal of amplification inhibitors following storage at 4 or −70°C and dilution remain unknown. Dilution alone may remove inhibitory activity. Alternatively, freezing and thawing may destroy labile inhibitory molecules or release additional target DNA molecules by disrupting microbial cells more efficiently. This might account for the increased sensitivity observed with the AMPLICOR CT/NG test when frozen specimens are assayed (16). Alternatively, freezing and thawing may act directly on labile proteinaceous inhibitors, inducing conformational changes and the subsequent loss of inhibitory activity. Salts, especially those containing divalent cations, particularly Mg2+ ions, which have a significant effect on the activity of Taq polymerase, could explain the inhibitory activity of urinary crystals for PCR and LCR.
In summary, we have demonstrated a variable rate of inhibition for C. trachomatis NAA assays ranging from 3.9 to 11.9%. An elevated rate of inhibition related to pregnancy was found for PCR but not LCR or TMA. In this study none of the inhibitory urine specimens were from infected patients, and the 15 urine specimens with endogenous C. trachomatis nucleic acid had no inhibitors. Most but not all of the inhibition was removed by storage and dilution, and only five urine specimens (three for PCR and two for TMA) required phenol-chloroform extraction to remove the inhibitors. The use of storage and dilution of urine specimens should be considered to reduce the inhibitory effects of urine specimens, especially those to be tested by PCR or TMA. Alternatively, tests most significantly affected by urinary inhibitors may benefit from the use of an internal positive control which gives a signal just above the positive cutoff value. More studies are warranted to determine whether urinary factors could be predictive of NAA inhibition.
ACKNOWLEDGMENTS
We thank Roche Molecular Systems, Abbott Diagnostics, and GenProbe for supplying the Chlamydia kits and Michael St. Pierre, Loraine Vaillancount, and Janet Spenser for collecting urine specimens for this study.
REFERENCES
- 1.Berg E S, Anestad G, Moi H, Storvold G, Skaug K. False-negative results of a ligase chain reaction assay to detect Chlamydia trachomatis due to inhibitors in urine. Eur J Clin Microbiol Infect Dis. 1997;16:727–731. doi: 10.1007/BF01709252. [DOI] [PubMed] [Google Scholar]
- 2.Chernesky M A, Jang D, Lee H, Burczak J D, Hu H, Sellors J, Tomazic-Allen S J, Mahony J B. Diagnosis of Chlamydia trachomatis infections in men and women by testing first-void urine by ligase chain reaction. J Clin Microbiol. 1994;32:2682–2685. doi: 10.1128/jcm.32.11.2682-2685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chernesky M A, Jang D, Sellors J, Luinstra K, Chong S, Castriciano S, Mahony J B. Urinary inhibitors of polymerase chain reaction and ligase chain reaction and testing of multiple specimens may contribute to lower assay sensitivities for diagnosing Chlamydia trachomatis infected women. Mol Cell Probes. 1997;11:243–249. doi: 10.1006/mcpr.1997.0109. [DOI] [PubMed] [Google Scholar]
- 4.Chong S, Mahony J B, Jang D, Luinstra K, Chernesky M. 64th Conjoint Meeting on Infectious Diseases. 1996. Inhibitors of Chlamydia trachomatis PCR in urine, abstr. D3. [Google Scholar]
- 5.Crotchfelt K A, Pare B, Gaydos C, Quinn T C. Detection of Chlamydia trachomatis by the Gen-Probe AMPLIFIED Chlamydia trachomatis Assay (AMP-CT) in urine specimens from men and women and endocervical specimens from women. J Clin Microbiol. 1998;36:391–394. doi: 10.1128/jcm.36.2.391-394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Barbeyrac B, Rodriguez P, Dutilh B, Le Roux P, Bebear C. Detection of Chlamydia trachomatis by ligase chain reaction and cell culture in urogenital specimens. Genitourin Med. 1995;71:382–386. doi: 10.1136/sti.71.6.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forbes B A, Hicks K E. Substances interfering with direct detection of Mycobacterium tuberculosis in clinical specimens by PCR: effects of bovine serum albumin. J Clin Microbiol. 1996;34:2125–2128. doi: 10.1128/jcm.34.9.2125-2128.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaydos C A, Ngeow Y F, Lee H H, Canavaggio M, Welsh L E, Johanson J, Quinn T C. Urine as a diagnostic specimen for the detection of Chlamydia trachomatis in Malaysia by ligase chain reaction. Sex Transm Dis. 1995;23:402–406. doi: 10.1097/00007435-199609000-00010. [DOI] [PubMed] [Google Scholar]
- 9.Jensen I P, Thorsen P, Moller B R. Sensitivity of ligase chain reaction assay of urine from pregnant women for Chlamydia trachomatis. Lancet. 1997;349:329–330. doi: 10.1016/s0140-6736(05)62829-2. [DOI] [PubMed] [Google Scholar]
- 10.Lee H H, Chernesky M A, Schachter J, Burczak J D, Andrews W W, Muldoon S, Leckie G, Stamm W E. Diagnosis of Chlamydia trachomatis genitourinary infection in women by ligase chain reaction assay of urine. Lancet. 1995;345:213–216. doi: 10.1016/s0140-6736(95)90221-x. [DOI] [PubMed] [Google Scholar]
- 11.Miettinen A, Vuorinen P, Varis T, Hallstrom O. Comparison of enzyme immunoassay antigen detection, nucleic acid hybridization and PCR assay in the diagnosis of Chlamydia trachomatis infection. Eur J Clin Microbiol Infect Dis. 1995;14:546–549. doi: 10.1007/BF02113438. [DOI] [PubMed] [Google Scholar]
- 12.Mouton J W, Verkooyen R, Meijden W I, van Rijsoort-Vos T H, Goessens W H F, Kluytmans J A J W, Deelen S D A, Luijendijk A, Verbrugh H A. Detection of Chlamydia trachomatis in male and female urine specimens by using the amplified Chlamydia trachomatis test. J Clin Microbiol. 1997;35:1369–1372. doi: 10.1128/jcm.35.6.1369-1372.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabenau H, Berger A, Doerr H W, Weber B. Testing for Chlamydia trachomatis in urine. Lancet. 1997;349:1024–1025. doi: 10.1016/S0140-6736(05)62923-6. [DOI] [PubMed] [Google Scholar]
- 14.Rosenstraus M, Wang Z, Chang S-Y, DeBonville D, Spadoro J. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J Clin Microbiol. 1998;36:191–197. doi: 10.1128/jcm.36.1.191-197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schepetiuk S, Kok T, Martin L, Waddell R, Higgins G. Detection of Chlamydia trachomatis in urine samples by nucleic acid tests: comparison with culture and enzyme immunoassay of genital swab specimens. J Clin Microbiol. 1997;35:3355–3357. doi: 10.1128/jcm.35.12.3355-3357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stary A, Choueiri B, Horting-Muller I. Detection of Chlamydia trachomatis in urethral and urine samples from symptomatic and asymptomatic male patients by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1996;15:465–471. doi: 10.1007/BF01691313. [DOI] [PubMed] [Google Scholar]
- 17.Strasinger S K. Urinalysis and body fluids. Philadelphia, Pa: F. A. Davis Co.; 1985. pp. 205–208. [Google Scholar]
- 18.Verkooyen R P, Luijendijk A, Huisman W M, Goessens W H F, Kluytmans J A J W, van Rijsoort-Vos J H, Verbrugh H A. Detection of PCR inhibitors in cervical specimens by using the AMPLICOR Chlamydia trachomatis assay. J Clin Microbiol. 1996;34:3072–3074. doi: 10.1128/jcm.34.12.3072-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiedbrauk D L, Werner J C, Drevon A M. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]