Structured Abstract
Objective:
External exposures, the host, and the microbiome interact in oncology. We aimed to investigate tumoral microbiomes in young-onset rectal cancers for profiles potentially correlative with disease etiology and biology.
Summary Background Data:
Young-onset rectal cancer is rapidly increasingly, with one in four new rectal cancer cases occurring under age 50. Its etiology is unknown.
Methods:
Young-onset (YO-; <50 years old) or later-onset rectal cancer (LORC, ≥50 years old) patients underwent pretreatment biopsied of tumor and tumor-adjacent normal tissue (TAN). After whole genome sequencing, metagenomic analysis quantified microbial communities comparing tumors vs. TANs and YO- vs. LORCs, controlling for multiple testing. Response to neoadjuvant therapy (NT) was categorized as major pathological response (MPR, ≤10% residual viable tumor) vs. non-MPR.
Results:
Our 107 tumor and 75 TANs from 37 (35%) YO- and 70 (65%) LORCs recapitulated bacterial species previously associated with colorectal cancers (all p<0.0001). YO and LORC tumoral microbiome signatures were distinct. After NT, 13 (12.4%) patients achieved complete pathologic response, while MPR, in 47(44%). Among YORCs, MPR was associated with Fusobacterium nucleaum, Bacteroides dorei, and Ruminococcus Bromii (all p<0.001), but MPR in LORC was associated with Ruminococcus Bromii (p<0.001). Network analysis of non-MPR tumors demonstrated a preponderance of oral bacteria not observed in MPR tumors.
Conclusions:
Microbial signatures were distinct between YORC and LORC. Failure to achieve an MPR was associated with oral bacteria in tumors. These findings urge furture studies to decipher correlative versus mechanistic associations but suggest a potential for microbial modulation to augment current treatments.
Mini-Abstract
One in every 4 cases of rectal cancer in the US is diagnosed in a young adult aged 18–50. We investigated the bacterial microbiome of young-onset vs. later-onset rectal cancers and respective adjacent normal tissues. We identified unique profiles and potential correlates with response to neoadjuvant therapy.
INTRODUCTION
Young adults aged between 18 and 50 represent the only population segment where the incidence of colorectal cancer (CRC) is persistently rising while the mortality rate has been stagnant1. Alarmingly, the most rapid rise is occurring among rectal cancers that are loco-regionally advanced and/or already metastatic2. These patients face often difficult multimodality therapies that span over long periods, complex pelvic operations with unfavorable bowel and sexual functional sequalae, the risk of permanent ostomy, and other challenges to the quality of life of a normal adulthood3,4,5. Therefore, YORC represents a unique phenotype of disease, and our current lack of understanding of disease etiology significantly hampers efforts in early detection and disease prevention6. Furthermore, it has been well established that response to neoadjuvant therapy benchmarks long term outcomes in patients with rectal cancer. Recently, novel neoadjuvant regimens have emerged and have improved the rates of complete pathologic response from 17% to up to 36%, while also increasing rates of complete clinical response and affording opportunities for non-operative management and rectal organ preservation7. Thus, toward the goal of improving mortality rates in these young adults, efforts to enhance response to neoadjuvant therapy, based on a better understanding of the disease biology, are needed.
While a patient’s microbiome consists of bacterial, viral, fungal, and archaeal species from a variety of sites throughout the body, tumoral microbes have recently been associated with both the development of CRC as well as the differential responses to cytotoxic therapies, perhaps in association with their ability to regulate immune infiltrates within tumors8,9,10,11 and modify chemotherapeutic drugs. For example, E coli isolates from CRC patients were shown to modify 5-fluoroacyl diminishing toxicity to CRC epithelial cells12. Inactivation of Oxaliplatin by Fusobacterium nucleatum was observed in CRC via autophagy13. Similarly, inactivation of Gemcitabine in pancreatic cancer has been noted to be secondary to enzymatic degradation from Gammaproteobacteria14. Recognizing reported associations among external exposures (the patient’s “exposome”), the host, and the microbiome in oncology, we hypothesized that tumoral microbiomes may reflect unique correlative and potentially causative factors in the development of YORC. Given the importance of these microbes in defining cancer outcomes, there is a pressing need to further define this interface in YORC.
Neoadjuvant therapy provides a unique platform to study disease biology with serial measurements both before and after intervention. We aimed to profile the differential and unique tumoral microbial profile of YORC versus later-onset rectal cancer (LORC), and to identify markers of response to therapy accounting for these differing disease phenotypes. We hypothesize that by differentiating YORC and LORC, we will be able to describe these microbial environments more precisely and identify biomarkers of response to therapy and generate hypotheses behind potential mechanisms explaining the increase in incidence and particularly aggressive nature of YORC. The identified microbial biomarker associations offer the potential to augment current standard of care therapeutics via microbial modulation therapies such as directed antibiosis or next generation targeted microbial modulation strategies.
METHODS
Under approval from the University of Texas MD Anderson Cancer Center (UTMDACC) Institutional Review Board, patients being treated for rectal adenocarcinoma with curative intent were offered enrollment into a prospective protocol as a part of the UTMDACC Young Onset Rectal Cancer Moonshot Program. Patients must be treatment-naïve at enrollment, and they received usual clinical care and cancer surveillance per their treating multidisciplinary teams. Patients were grouped as YORC (diagnosed between ages 19–50) vs. LORC (diagnosed between ages 51–80). The age groups were defined as consistent with literature and with the Young-Onset Colorectal Cancer Program at UTMDACC. Medical records were reviewed for clinicopathologic data, treatment details, and oncologic outcomes.
Patients with locally advanced rectal cancer (i.e. clinical stage T3, T4, or N+) were typically offered neoadjuvant therapy. Among these patients, endoscopic biopsy of tumors and adjacent grossly normal tissues (termed tumor associated normal, TAN) were collected prior to any therapy (Figure 1A). Patients were treated along standard of care pathways and resected specimens underwent pathologic examination with histologic grade and response to neoadjuvant therapy measured. Pretreatment biopsied tissue underwent whole genome sequencing by the MD Anderson Cancer Clinical Genomics Laboratory. Tumors were sequenced to 60x depth while paired TANs were sequenced to 30x depth. Metagenomic analysis was carried out with MetaPhlAn315 allowing taxonomical classification of sequencing reads. Alpha diversity metrics calculated included species richness, the Simpson diversity index, and the Shannon diversity index. Pathologic response was assessed in all patients who completed curative intent resection. All patients were followed to the last contact for survival and disease status.
Figure 1: Comparing the tumoral microbiome against paired tumor associated normal tissue.
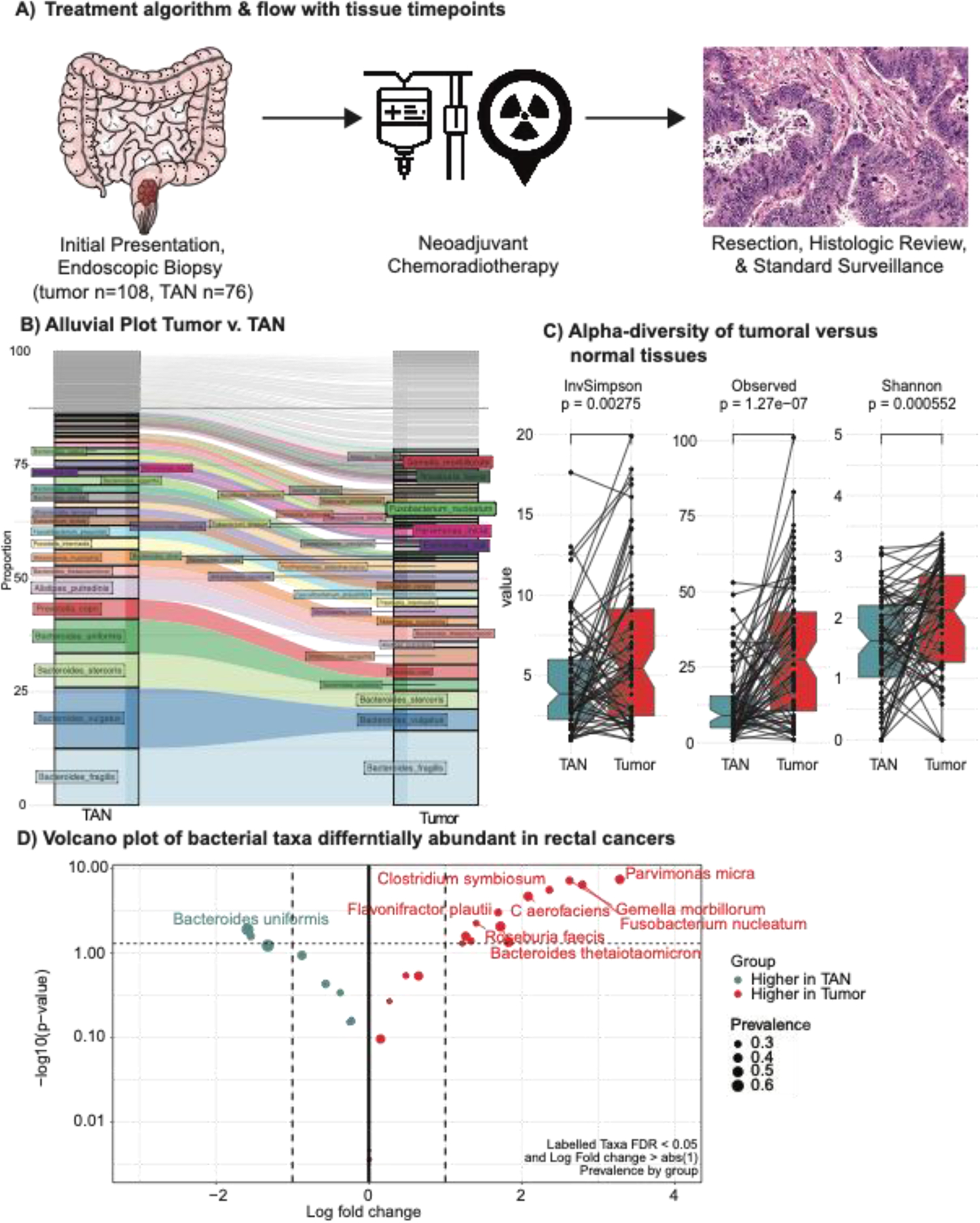
A) Treatment algorithm and tissue collection timepoint for young and late onset rectal cancer patients. B) Alluvial plot of bacterial species within tumor associated normal tissues and matched tumoral tissues. C) Alpha diversity plots of tumor associated normal tissues and tumoral tissues (inverse Simpson D) Volcano plot of differentially abundant bacteria in rectal cancers as compared to tumor adjacent normal tissues. Significantly associated bacteria are labeled and have an absolute log fold change of greater than 1 and p-value less than 0.05.
The primary descriptive aim was to compare the profile of matched tumor and TANs microbiome. In addition, we compared the tumor microbiome between YORC and LORC. Analysis of alpha diversity metrics comparing groups was carried out using Wilcoxon rank-sum or paired Wilcoxon rank-sum in case of matched tumor and TAN data. The Spearman rank correlation test was used to compare continuous variables. Beta diversity metrics were derived using Bray-Curtis dissimilarity and plotted on Principal Coordinate Analysis plots. Analysis of pairwise inter-sample distances to measure the association between the microbiome diversity and covariates of interest (e.g. environmental factors, clinical outcomes, treatment groups) was carried out with PERMANOVA16,17. Differential abundance analysis of identified taxonomies were performed with minimum of 20% prevalence using ANCOM-BC18. Network analysis was conducted on paired Tumor and TANs samples using secom distance function within ANCOM-BC with minimum of 20% prevalence, distance threshold of 0.60, 1000 replicates for p-value calculation and maximum p-value set at 0.05. Network communities were determined using edge betweenness score. Centrality scores were calculated using PageRank algorithm.
The primary outcome was pathologic response to neoadjuvant therapy. Major pathologic response was defined as <10% viable tumor cells. We additionally explored long-term survival using the Kaplan-Meier method. Hazard ratios and 95% confidence intervals will be calculated using univariate and multivariate Cox proportional hazards models to assess associations between time-to-outcome, clinical factors, and microbiome metrics. Adjustments for multiple comparisons will be done using the false-discovery rate (FDR) method at an α of 0.05.
Descriptive data were compared between two groups using Wilcoxon rank-sum test, t test, or Chi squared test where appropriate. Univariate analysis was performed on this data given the limited number of patients. Associations between bacterial taxa and survival were performed in a hypothesis testing fashion and therefore multiple testing was not corrected for in these survival analyses. Analyses were performed using STATA 13 (College Station, TX).
RESULTS
Clinical characteristics
Among 107 patients, 37 (35%) were YO- and 70 (65%) LORC. The median age of diagnosis in each age group was 42 and 61 respectively. Neoadjuvant therapy was administered in nearly all patients (37 YO and 69 LO; Table 1). Preoperative clinical assessment with conventional laboratory studies, imaging and endoscopy revealed complete clinical response as determined by the treating surgeon in 16 (15.0%).
Table 1:
Demographic and clinical details of patient cohort
| n=107 | |
|---|---|
| Age at diagnosis (median, IQR) | 53 (45, 64) |
| Race/Ethnicity | |
| Caucasian | 76 (71.0%) |
| Black or African American | 8 (7.5%) |
| Asian | 7 (6.5%) |
| Hispanic or Latino | 14 (13.1%) |
| Unknown/Other | 2 (1.9%) |
| Stage | |
| I/II | 10 (9.3%) |
| III | 86 (80.4%) |
| IV | 11 (10.3%) |
| Clinical Complete Response | 17 (15.9%) |
| Pathologic Complete Response | 12 (11.2%) |
| Major Pathologic Response | 47 (43.5%) |
Surgical pathology, available in 96 (89.7%) patients, revealed a complete pathologic response (ypT0N0) rate of 12.0% (13 patients). Major pathologic response was seen in 47 (43.9%) patients. After a median follow-up of 73 months (6.1 years) from diagnosis patients were followed post operatively and recurrence free survival, was measured and summarized by clinicopathologic factors in Table 2. Achievement of a MPR was associated with improved recurrence free survival (HR=0.08, 95% CI [0.01–0.61]; p=0.02).
Table 2:
Clinical Associations with Recurrence Free, Cancer Specific Survival, and Overall Survival
| Recurrence Free Survival | |||
|---|---|---|---|
| HR | 95% CI | p | |
| Age at diagnosis | 1.00 | 0.97–1.03 | 0.95 |
| Young Onset | 0.87 | 0.35–2.13 | 0.76 |
| Stage | |||
| I/II | Ref | Ref | - |
| III | 1.61 | 0.21–12.27 | 0.64 |
| IV | 9.06 | 1.11–73.79 | 0.04 |
| Neoadjuvant rectal (NAR) score | |||
| High | Ref | Ref | - |
| Low/Intermediate | cong | cong | - |
| Clinical Response | 1.25 | 0.42–3.70 | 0.69 |
| Pathologic Complete Response | colinear | colinear | - |
| Major Path Response | 0.08 | 0.01–0.61 | 0.02 |
| Adjuvant Treatment | 0.93 | 0.49–1.76 | 0.82 |
Ref - Reference value
Bacterial communities in tumor vs. normal tissue
To profile bacterial communities within these tumors, non-human WGS reads that mapped to bacterial entities were identified and compared between matched tumor and tumor adjacent normal tissues. While the general bacterial communities and composition of matched TAN and tumoral tissues are largely equivalent, notable changes in mean bacterial abundances of select bacterial taxa and composition between normal tissues and tumoral tissues were as demonstrated in the alluvial plot (Figure 1B). Significantly, there was an increase in bacterial diversity in tumoral tissue as compared to matched TAN. Changes in alpha diversity, measures of overall bacterial diversity within tumor tissues, are demonstrated Figure 1C (inverse Simpson p=0.003, observed p<0.001, and Shannon p<0.001). A predominance of bacterial species previously reported to be associated with CRC noted in our cohort, including Fusobacterium Nucleatum9,19–23, Parvimonas micra21,24,25, Clostridium symbiosum24,26. We also identified species that have been reported to be associated with CRC but previously found in gut microbiome Collinsella aerofaciens 27,28, and in plasma circulating microbiome, Gemella morbillorum 25(Figure 1D, Supplementary Table 1, Supplemental Digital Content 1, http://links.lww.com/SLA/E740).
Bacterial communities of tumor vs. normal tissue in YO vs LORC
When we compare the tumor bacterial communities in YO- vs. LORCs, we noted that both alpha and beta diversity were equivalent between the two cohorts (alpha p=0.602, beta p=0.425, Figure 2A & B). However, when comparing bacterial composition of tumoral tissues in YORC against matched adjacent normal, a number mutually exclusive bacteria taxa showed higher abundances in tumors as compared to matched normal tissues: significantly higher prevalence of populations of Escherichia coli, Clostridium symbiosum, Parvimonas micra, Bacteroides thetaiotaomicron and Collinsella aerofaciens were seen in YORC tumoral as compared to normal tissues (Figure 2C, Supplementary Table 2, Supplemental Digital Content 1, http://links.lww.com/SLA/E740). In contrast, the LORC cohort demonstrated higher prevalence in a wide array of species including Fusobacterium nucleatum, Parvimonas micra, Clostridium symbiosum, and Gemella morbillorum in tumoral vs. normal tissues. We also observed new significant associations with Dorea longicatena, and Hungatella nathewayi showing more prevalent populations in the LORC tumors as compared to matched TAN (Figure 2D, Supplementary Table 3, Supplemental Digital Content 1, http://links.lww.com/SLA/E740).
Figure 2: Comparing the tumoral microbiome of young versus late onset rectal cancer.
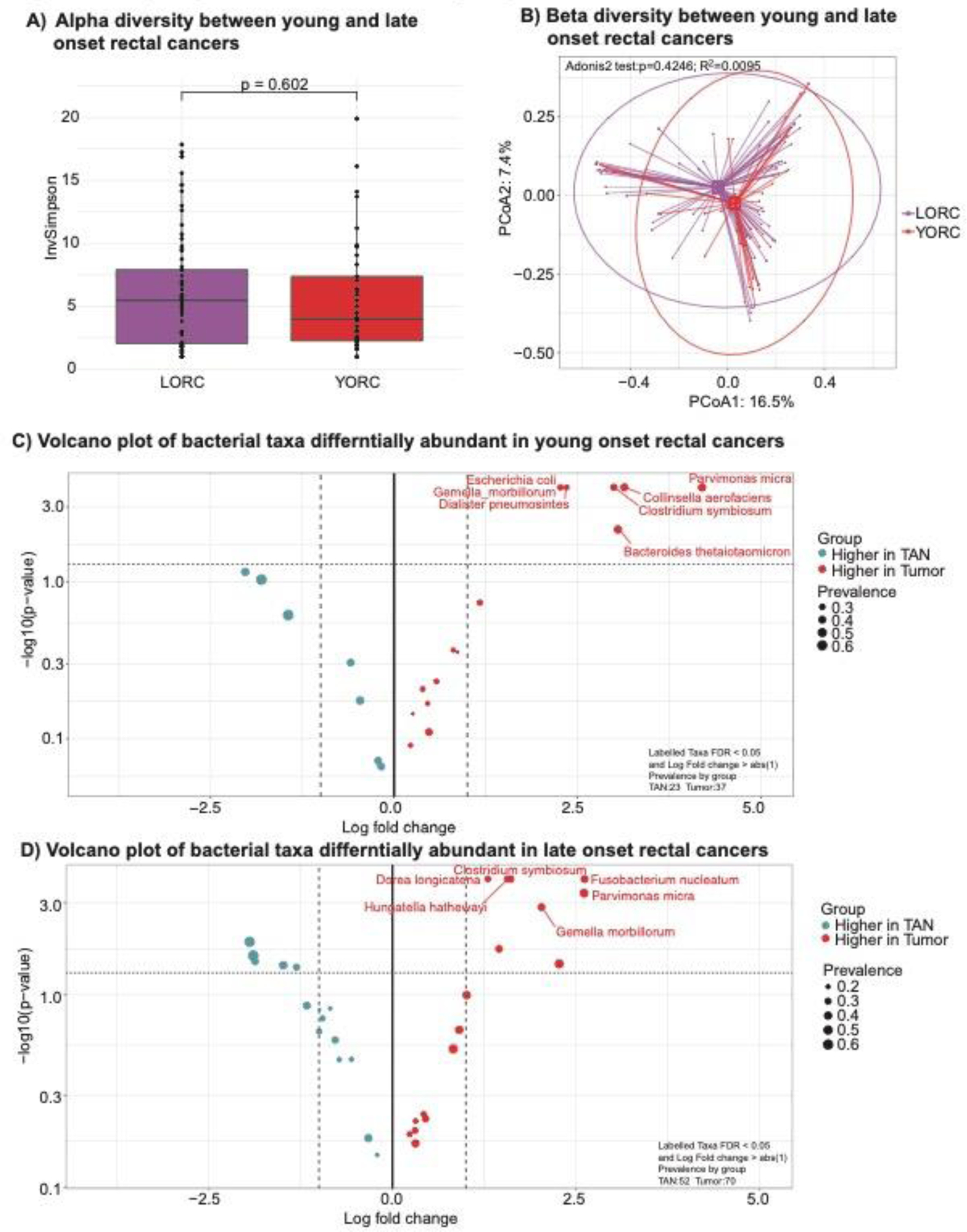
A) Alpha diversity between young and late onset rectal cancers (p=0.761). B) Beta diversity between young and later onset rectal cancers (p=0.406). C) Volcano plot of differentially abundant bacteria in rectal cancers as compared to tumor adjacent normal tissues in young onset patients (age < 50 years old). Significantly associated bacteria are labeled and have an absolute log fold change of greater than 1 and p-value less than 0.05. D) Volcano plot of differentially abundant bacteria in rectal cancers as compared to tumor adjacent normal tissues in late onset patients (age ≥ 50 years old). Significantly associated bacteria are labeled and have an absolute log fold change of greater than 1 and p-value less than 0.05.
Exploratory analyses for neoadjuvant therapy response
Clinical factors associated with MPR were investigated in a univariate fashion (Table 3). When bacterial associations with achieving an MPR was investigated, patients with Bacteroides xylanisolvens had a lower chance of achieving an MPR. (Figure 3A, Supplementary Table 4, Supplemental Digital Content 1, http://links.lww.com/SLA/E740). Again performing subset analyses of tumors in YORC, Alistipes finegoldii, Bacteroides dorei , Bacteroides xylanisolvens ,Blautia wexlerae, Campylobacter ureolyticus, Coprococcus comes, Dialister pneumosintes, Dorea longicatena, Flavonifractor plautii, Fusobacterium nucleatum, Odoribacter splanchnicus, Parabacteroides distasonis, Parabacteroides merdae, Peptostreptococcus stomatis, Roseburia faecis, and Ruminococcus bromii showed significantly higher populations in those not achieving a MPR (Figure 3B, Supplementary Table 5, Supplemental Digital Content 1, http://links.lww.com/SLA/E740). Conversely, LORCs showed associations with associations with Bacteroides cellulosilyticus, Bacteroides dorei, Bacteroides xylanisolvens, Barnesiella intestinihominis, Bifidobacterium adolescentis, Blautia obeum, Dialister pneumosintes, Dorea formicigenerans, Eggerthella lenta, Firmicutes bacterium CAG 83, Fretibacterium fastidiosum, Peptostreptococcus stomatis, Roseburia inulinivorans, Ruminococcus bromii, Solobacterium moorei, and Streptococcus sanguinis and having a less than MPR (Figure 3C, Supplementary Table 6, Supplemental Digital Content 1, http://links.lww.com/SLA/E740).
Table 3:
Univariate logistic regression of factors associated with major pathologic response (MPR)
| OR (95% CI) | p | |
|---|---|---|
| Age at diagnosis | 1.01 (0.98–1.04) | 0.61 |
| Young onset | 1.59 (0.69–3.66) | 0.27 |
| Stage | ||
| I/II | Ref | - |
| III | 1.35 (0.34–5.40) | 0.67 |
| IV | null | - |
Ref - Reference value
Figure 3: Microbial markers of response to neoadjuvant therapy.
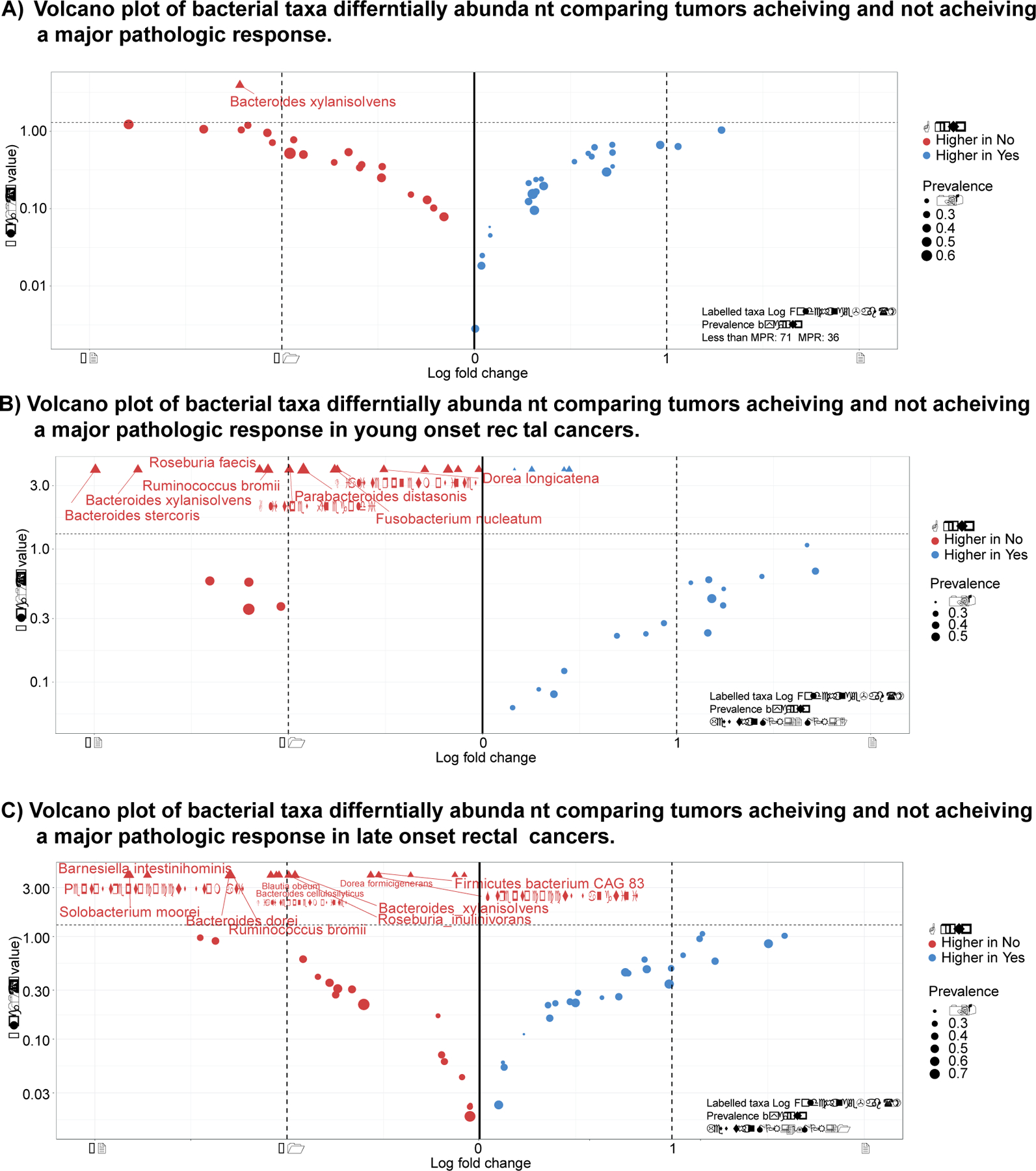
A) Volcano plot of differentially abundant bacteria in rectal cancers achieving a major pathologic response (<10% viable cells) as compared to rectal cancers not achieving a major pathologic response. Significantly associated bacteria are labeled and have an absolute log fold change of greater than 1 and p-value less than 0.05. B) Volcano plot of differentially abundant bacteria in rectal cancers in young onset patients achieving a major pathologic response (<10% viable cells) as compared to rectal cancers not achieving a major pathologic response. Significantly associated bacteria are labeled and have an absolute log fold change of greater than 1 and p-value less than 0.05. C) Volcano plot of differentially abundant bacteria in rectal cancers in late onset patients achieving a major pathologic response (<10% viable cells) as compared to rectal cancers not achieving a major pathologic response. Significantly associated bacteria are labeled and have an absolute log fold change of greater than 1 and p-value less than 0.05.
Given the high number of bacterial species associated with MPR and non-MPR, we performed cluster analysis to examine for commonalities among these bacterial species. Species associated with non-MPR were noted to have a significant subset of primarily oral bacteria, similar to Fusobacterium nucleatum and Parvimonas micra. Studying bacterial networks of those communities with a greater than 20% prevalence in these tumors, comparing tumors achieving an MPR, a majority of colonic type bacterial networks are noted (Figure 4A). Similar network analysis of those not achieving a MPR, however, showed a network with a majority of oral bacterium present, distinct from those achieving a MPR (Figure 4B).
Figure 4: Associations with major pathologic response, oral bacteria and Survival.
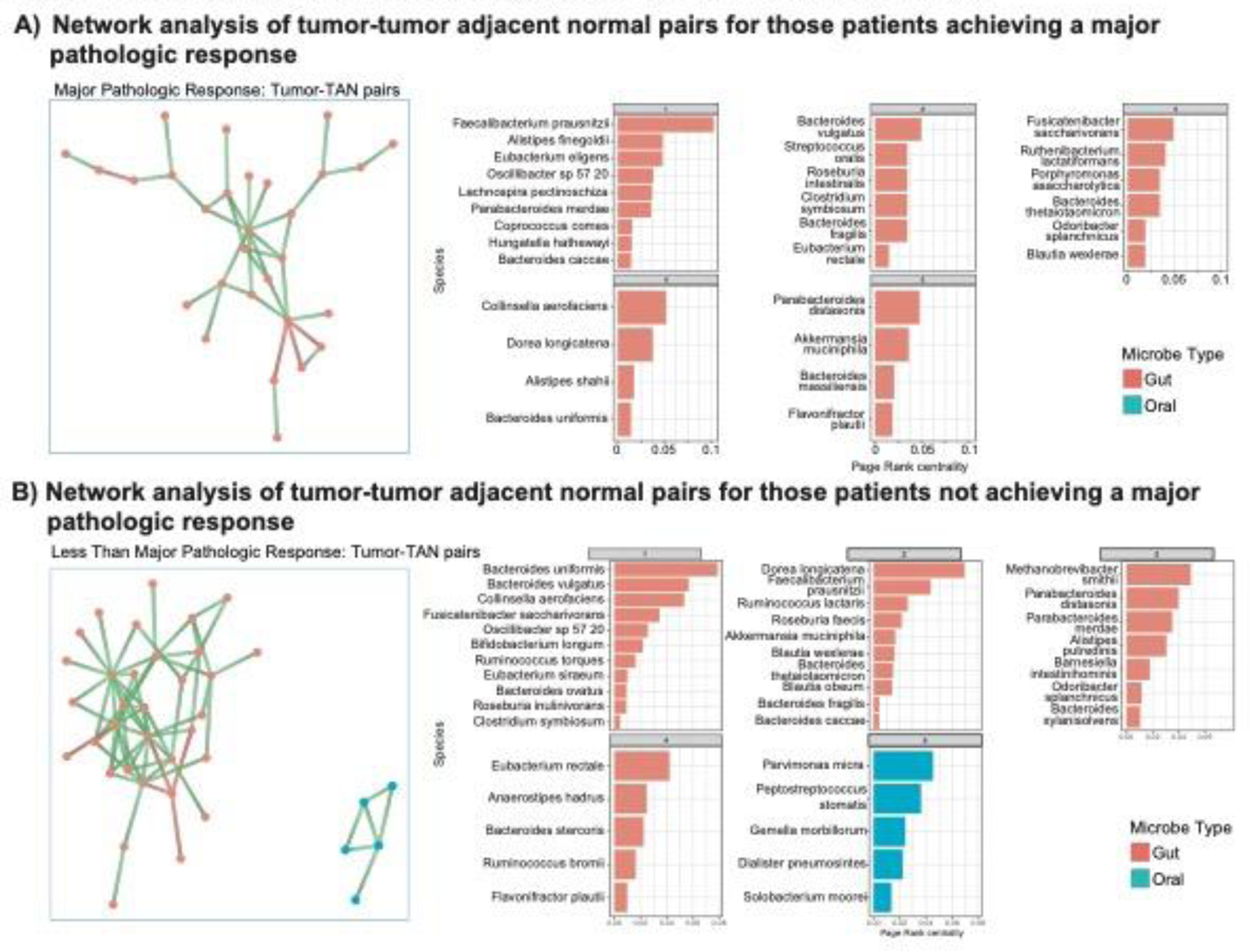
A) Network analysis of tumor-tumor adjacent normal pairs for those patients achieving a major pathologic response. B) Network analysis of tumor-tumor adjacent normal pairs for those patients not achieving a major pathologic response. C) Kaplan-Meier recurrence free survival curve of those patients with and without Escherichia coli present within their tumors (HR=0.14, p=0.06). D) Kaplan-Meier overall survival curve of those patients with and without Ruminococcus torques present within their tumors (HR=3.42, p=0.03).
Finally, given these associations with response to neoadjuvant therapy we queried bacterial associations with survival. Hypothesis driven univariate and multivariate survival analysis by those bacteria significantly associated with response to neoadjuvant therapy in either the YORC or LORC cohorts was then performed. Of these associations, listed in Supplementary Table 7, Supplemental Digital Content 1, http://links.lww.com/SLA/E740, none were significant.
Discussion
The alarming epidemiologic trends for young onset CRC and in particular YORC demand a deeper investigation towards disease etiology and biology. Given the growing understanding of the role of microbiome in linking exposome and human disease, we aimed to define the tumoral microbial profile of YO- vs. LORCs and to explore potential markers for treatment response. We found that the tumoral bacterial signatures were distinct for YO- vs. LORCs. Signatures associated with response to neoadjuvant therapy also differed among YO- and LORCs. These observational findings urge further studies to decipher correlative versus mechanistic associations, but hint at the potential to utilize microbial modulation tools to augment standard of care treatment regimens.
Our primary aim was to describe the tumoral microbial environments in YORC more precisely, in reference to LORC. First, we identified several species in tumor microbiome which had been previously associated with CRC, thereby validating previous studies, and establishing correlations between tumoral bacterial species and rectal cancer specifically. Second, we intriguingly found that unique subsets of specific bacteria were associated with rectal cancers as compared to TANs. The presence of these species in tumors is further reflected in the increase in microbial diversity of rectal cancers. These findings demonstrate the statistical importance of including matched normal tissues in microbiome studies.
Most importantly, we found little overlap in the tumor microbial signatures of YO vs LORCs. Indeed, only a small number of specific bacterial species were associated with YORC, while a larger array of species was associated with LORC. Few common bacterial taxa between these groups were noted, including Gemella morbillorum, and Parvimonas micra. Interestingly, Clostridium symbiosum, which had been previously demonstrated as a marker of CRC29. While P micra has been shown to be associated with tumorigenesis, immune response, and a worse prognosis30–32. Furthermore, Escherichia coli, commonly associated with CRC development and hypothesized to lead to CRC development through both the expression of polyketide synthase33 and the production of the genotoxin colibactin34, was differentially abundant only in the YORC cohort. Moreover, Escherichia coli has also been associated with the development of liver metastases by promoting vascular invasion and aiding in the development of pre-metastatic niches within the liver suggesting a potential link with the high rate of young patients presenting with metastatic disease35. In addition, Collinsella aerofaciens, associated with previous appendectomy demonstrated the highest proportion of differential abundance within YORC compared to matched normal tissues27. Distinct from these findings of YORC, the LORC cohort contains what might be considered more “typical” bacteria entities including species such as Fusobacterium nucleatum and Parvinomas micra. This potentially suggests that previous CRC-related work had included mostly LORC patients, and likely underlines again the need to design studies enriching for YORC to allow in-depth study of it as a distinct entity. The unique microbial pathways and alterations between YORC and LORC cohorts identified herein warrants further corroboration and study.
As YORCs are disproportionately already stage III and IV at diagnosis, neoadjuvant therapy represents mainstay treatment pathway for the majority of the YORC cases. Given multiple proposed mechanisms of intratumoral bacteria altering or inactivating cytotoxic chemotherapies or response to radiation therapy13,14,36, we explored microbial markers associated with MPR to neoadjuvant therapy as species showing significant abundance. Bacterial associations with response and LORC were limited to only Bacteroides xylanisolvens, despite being a larger cohort studied. While a variety of bacterial species in the YORC cohort were associated with either attaining or failing to attain MPR. When studying those bacteria associated with a MPR, typical colonic bacteria were largely identified. While those not achieving a MPR were shown to have a greater proportion of oral bacterial networks, suggesting a potential association with either chemotherapy inactivation and/or immune exclusionary mechanisms at play. Indeed, prior studies have described alterations in oral bacteria such as Fusobacterium nucleatum in association with changes in the rates of response to neoadjuvant therapy13,20.
This observational work must be viewed considering its limitations. The most significantly of which is the inherent underpowering of these studies given the need for multiple testing corrections inherent in exploratory microbiome studies. In our limited single institutional series, alternative comparative analyses such as examining more extreme age groups using lower or higher age cutoffs, defining more than two age groups, or treating age as a continuous variable, suffered from loss in power in our unsupervised exploratory analyses. Nevertheless, recapitulation of a number of previous observations suggests that these bacterial signals and validated and reproduced across cohorts. Secondly, it should be emphasized that the current study has explored for associations between microbial species and differential age groups with CRC. The study design did not investigate mechanisms for oncogenesis to understand specific causation of disease. Future studies that utilize germ-free animal models to isolate impact of specific microbiome would be needed. Finally, our patients were all treated under standard of care at our referral cancer center which introduces inherent variation in care and therefore the generalizability of these findings requires further validation. This is perhaps best demonstrated in the high rates of MPR in the stage IV cohort who received more aggressive pre-treatment than those with localized disease. Finally, this work requires further functional validation in pre-clinical models as well as early studies of bacterial modulation in these populations.
In conclusion, we demonstrated previously established as well as novel associations between intra-tumoral bacterial species and the development of rectal cancer and response to neoadjuvant therapies. Interestingly, these populations are nearly mutually exclusive when comparing YO- vs. LORC. Bacterial species most highly associated with tumor tissues were rarely found in normal tissues, and oral microbial species persisting in tumor microbiome appeared to be associated with treatment response. Further work is needed to understand the dissemination of these bacteria through the digestive tract versus systemic immune circulation. Given demonstrated mechanisms of chemotherapy inactivation and immune exclusion, populations of these microbes offer plausible targets to augment standard of care therapies to maximize response, particularly in YORCs.
Supplementary Material
AMERICAN SURGICAL ASSOCIATION 2023 FRIDAY, APRIL 21.
SCIENTIFIC SESSION III
Discussion
Young-Onset Rectal Cancer: Unique Tumoral Microbiome and Correlation with Response to Neoadjuvant Therapy
*Michael White1, *Ashish Damania2, *Jumanah Y. Alshenaifi3, *Pranoti Sahasrabhojane2, *Oliver Peacock1, *Brian Bednarski1, *Jillian Losh2, *Matthew C. Wong2, *Zuzana Berkova3, *Sa Nguyen1, *Neal Bhutiani4, George Chang1, Jennifer A. Wargo4, *Nadim Ajami2, *Scott Kopetz3, Y. Nancy You1
1 Colon and Rectal Surgery, University of Texas MD Anderson Cancer, Houston, TX. 2 Platform for Innovative Microbiome and Translational research, Department of Genomic Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX. 3 Department of Gastrointestinal Medical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX. 4 Surgical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX
Dr. Matt Kalady (Columbus, OH): Good morning. Thank you. Matt Kalady from Columbus, Ohio. No relevant disclosures.
First of all, congratulations to you and your group from MD Anderson on some very nice work. As we’ve heard, the incidence of young-onset colorectal cancer is on the rise, but there is still a significant knowledge gap regarding the etiology, and further studies clearly need to be done.
As the authors have presented, the microbiome is a potential contributing and also modifiable factor. Bacterial species have been shown to be associated with cancer development and response to therapy in many cancers including colorectal cancer. The current paper provides some new information regarding microbial signatures in young-onset rectal cancer compared to older patients and also correlations with response to neoadjuvant therapy.
As with all exploratory and hypothesis-generating work, the next steps defining correlation versus causation will be critical, which brings me to my first question. What’s the next step towards the translation of these findings? What models will you use to try to sort out all these different bacteria to figure out what is causative and what’s just correlative.
My second question relates on how you determined your age cutoff. Defining young versus old has always been debated, and it’s likely that there’s a spectrum or a gradient of changes in the colorectal microbiome as people age, which I think provides a gray area around where the cutoff age is chosen. With all of your studies, you have tons and tons of data to data-mine. Have you considered examining more of the extremes of age without the gray zone, say for patients less than 40 versus those greater than 60? This might make the analysis and data a bit cleaner to see if you can identify some more causative factors that way.
And then lastly, if I interpret this correctly, your data shows associations with the increased abundance of bacteria but not necessarily where things decrease. Bacteria secrete several factors to create a local metabolomic environment. Some of the metabolites might be beneficial and provide anti-cancer effects or improve treatment sensitivity and losing those could be harmful for patients. Have you looked at the data where there is a decrease in the bacteria in any of these populations as opposed to just abundance?
Again, congratulations.
Response From Michael White: Absolutely, thank you very much for those comments and excellent points. Regarding your first point, looking for mechanistic associations, I think that’s really where we as an oncologic microbial research community need to go. There have been a plethora of correlative studies recently, and as far as preclinical models, the most common way that this has been studied and the way that we’ve studied it currently is to essentially grow sterile tumor cells that can be injected either into the rectums or the flanks of mice, and compare those to those grown in culture with say Parvimonas or Fusobacterium and then to compare the two tumors that are grown in those two mice.
Your third point actually informs the problem with that model really perfectly because those are bacteria that are under a lot less stress than those bacteria within a patient’s tumor that are competing with other bacterial species and other microbial metabolites for all of those resources, and we know that that stress and pressure induces phenotypic changes within those bacteria, so with that being said, the limitations of those models are resulting in the development of newer models with more complex bacterial consortia within the tumors themselves.
And then finally, I think it speaks to the need of translational studies so that we can look at microbial alterations within the same patients and negate all of the multiple factors and all of the noise when we compare between patients, and so our ongoing studies using either fecal microbiota transplant (FMT) or starting to look at treatment with standard antimicrobials is I think a path forward there.
Regarding your second question, the age cutoff used, it’s a point of debate in I think almost every lab meeting that we have. We regularly discuss how to analyze this data and define a cutpoint. I agree that it certainly is unlikely to be, you know, a log 10 binary variable. That being said, with all of the limitations of data analysis of these microbial studies, we have to at least start there, but now that we have some bacteria of interest, we can begin to ask more targeted questions where the multiple testing parameters can go away and you can look at single bacterial species within the extremes of age, and especially in our genetic analyses, we’re beginning to look at early-, middle-, and late-onset rectal cancers and find different ways to model that.
And then finally, your final question on how we’re beginning to translate this into the clinic, I’ll say at MD Anderson, we have an ongoing trial looking at fecal transplantation for MSI-high checkpoint-blockade-refractory patients that has had some promising results and obviously yields quite a few biospecimens that can be looked at in translational labs, and then we have a number of other trials in preparation looking at bacterial consortia as well as treatment with antibiotic therapies, that hopefully can give us more targeted answers to these questions similar to how the FMT trials in melanoma have resulted in very similar outcomes.
Dr. David Shibata (Memphis, TN): David Shibata, Memphis. Congratulations to you and your team and particularly for Dr. You’s leadership in this space. Early onset rectal cancer is a very important and critical issue that we are all struggling with right now. So a couple things. I do re-emphasize Dr. Kalady’s point about the age cutoffs. It’s very hard to tease out what a difference between a 49-year-old and a 50-year-old would be based on these cutoffs. But I do sympathize that with the assigned lower-age cutoff as you start losing sample size if you become too stringent.
Could you clarify that you did in fact exclude MSI-high and hereditary cases in this situation, because MSI-high cases certainly have their own immune microenvironment issues that may impact the microbiome?
And my last point on this would be just from a technical standpoint, and congratulations on doing this prospectively, were these in fact frozen versus FFP samples?
And actually I do have an additional question for you on this. Philosophically, what does your group think about the differences between the fecal microbiome, the normal tumor microbiome versus the tumor microbiome, and in your work, did you see correlations? Which one is most relevant remains a very complicated question.
Response From Michael White: Absolutely, so thank you for those points. I think you’re absolutely correct on the age cutoff question and would agree with you there. We hope to with further analyses tease that out as a nonbinary variable. These were all frozen samples that were sequenced, that were analyzed, and then I apologize, the third question?
Dr. Diana Farmer (Sacramento, CA): Fecal microbiome.
Response From Michael White: We did collect fecal microbiome on a significant subset of these patients. We do not see very strong correlatives, and actually there are sequencing difficulties with oral microbes in the gut microbiome that are leading us to now collect oral swabs on these patients to see if the oral microbiome actually informs the tumoral microbiome similarly to the gut microbiome itself.
Dr. Andreas Kaiser (Duarte, CA): Thank you for your great presentation, obviously a hot topic. By the way, I’m Andreas Kaiser from City of Hope, and I have a few questions. One is regarding the protocol in terms of you had 14% of metastatic cancer, and how did the radiation affect your results, and when was actually the control taken again?
And the second is more like a general question. What is really there first, the chicken or the egg? So how does our microbiome change with aging in general without having a cancer?
And third, is it really a fair comparison to look at the nasty, necrotic tumor sitting there that secondarily grows a lot of bacteria anyway because it’s dead tissue as opposed to then having a response and normalization of the microbiome?
Response From Michael White: Thank you for those questions. The answer to one and three is somewhat interrelated. We took these biopsies pre-treatment to hopefully negating the effects of response to therapy but potentially informing future outcomes of how their microbial populations could affect their response to therapy. Patients that had metastatic disease at presentation similarly were treatment-naïve and underwent a biopsy of normal and tumoral tissue before and at the end of their treatment, and all of those patients were treated with curative intent, primary resection as well as metastasectomy.
As far as the effects of aging on our gut microbiome as well as our global microbiome, we certainly see changes throughout one’s lifetime, and the quantification of that is certainly outside the scope of my expertise, but it’s something that we see that the aging microbiome is very distinct and unique in those patients and potentially something that we can look at here as we begin to collect more of these patients, but we do know that there are significant alterations with just the presence of a colorectal cancer itself.
Dr. Andreas Kaiser (Duarte, CA): I mean the question is the control group, right?
Response From Michael White: Yes the control group is inherently difficult, but luckily there are some standard cadaveric sequencing publicly available as well as benign biopsies, and other groups have collected data that is publicly available that we’ll look at.
Dr. Nathalie Johnson (Portland, OR): Nathalie Johnson, Portland, Oregon. Really fascinating work. Congratulations. This is next level. Would like to piggyback off of one of the questions Dr. Shibata asked and that is to clarify how clearly how you looked at microsatellite instability in this patient group. Of course in younger people, you would expect to see more MSI, and it does impact the density of tumor-infiltrating lymphocytes, response or nonresponse to chemotherapy, and maybe somehow impacts the tumor microenvironment and the immune response with bacteria. Would you please clarify so we might understand if it is age versus other genomic changes like microsatellite instability that might separate these groups out.
Response From Michael White: Yes, absolutely, and I apologize for overlooking that with my previous response. The samples analyzed here were all MSI stable colorectal cancers.
Dr. Susan Galandiuk (Louisville, KY): I very much enjoyed your paper. There are some differences in microbiome within the layers of the bowel wall. For those rectal cancer patients who did not need neoadjuvant treatment, did you look at differences in the microbiome within layers of the bowel wall in the resected specimen?
Response From Michael White: So that is an excellent point. We’re in the process now of looking at the spatial distribution of these microbes using genomics and COSMICs with an overlaid RNA scope sequencing so that we can understand that distribution because certainly the mucosal surface has a very unique microbial community informed by the fecal microbiome as compared to the muscular layers of the abdominal wall, which we would expect to be much more reflective of the systemic microbial community, and yeah, ongoing work, and hopefully we’ll have some answers for you soon.
Dr. Diana Farmer (Sacramento, CA): And our final question from Dr. Strong.
Dr. Vivian Strong (New York, NY): Thank you very much. Vivian Strong from New York. First of all, I did want to congratulate you also for your excellent work in examining how the tumor microbiome affects outcomes for your cancer patients.
In our own work looking at the effects of the microbiome for gastric cancer, we have been learning, as you have, that this process is not just a unidimensional process. It’s a complex interaction of the microbiome with the immune system with tumor-infiltrating lymphocytes, T-cell exhaustion, and so forth, so that really leads me to my two questions. What are your plans in terms of looking at how these microbial differences and the outcomes that you’ve seen? How does that interact or how do you think this will interact with the T-cell environment with T-cell exhaustion with metabolomics and other factors of that nature?
And then the second question is in light of a recent New England Journal of Medicine article that came out this month looking at H. pylori infection in gastric cancer patients and how the ability for that infection to predispose to gastric cancer depending on the genomic pathogenic germline mutations has an effect, do you think that there could be similar factors at play in this cohort that you’ve looked at?
Response From Michael White: Yes, absolutely. To answer your last question first, some of the bacterial species that were seen are very similar to H. pylori, like Helicobacter hepaticus is coming up, and induced immune response. We’re beginning to see other groups have shown that the canonical molecular subtype categorization actually correlates with the type of microbial species that they’re seeing, and so not only in the development but the development of the immune environment of these tumors seems very dependent, at least correlatively, on the bacterial species present at that point in time.
Dr. Diana Farmer (Sacramento, CA): Thank you very much for this provocative conversation.
Acknowledgments
This work was supported in part by the generous philanthropic contributions to The University of Texas MD Anderson Cancer Center Moon Shots Program™, the Colorectal Cancer Alliance Chris4Life Early Career Investigator Award (to MGW), and the University of Texas Anderson Cancer Center Core Support Grant (P30 CA016672) from the National Institute of Health.
Footnotes
This work was presented as a podium presentation at the 143rd Annual Meeting of the American Surgical Association, April 2023, Toronto, Canada
REFERENCES
- 1.Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin Mar 1 2023;doi: 10.3322/caac.21772 [DOI] [PubMed]
- 2.Bailey CE, Hu CY, You YN, et al. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA Surg Jan 2015;150(1):17–22. doi: 10.1001/jamasurg.2014.1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You YN, Lee LD, Deschner BW, Shibata D. Colorectal Cancer in the Adolescent and Young Adult Population. JCO Oncol Pract Jan 2020;16(1):19–27. doi: 10.1200/JOP.19.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey CE, Tran Cao HS, Hu CY, et al. Functional deficits and symptoms of long-term survivors of colorectal cancer treated by multimodality therapy differ by age at diagnosis. J Gastrointest Surg Jan 2015;19(1):180–8; discussio 188. doi: 10.1007/s11605-014-2645-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rooney MK, De B, Corrigan K, et al. Patient-reported Bowel Function and Bowel-related Quality of Life After Pelvic Radiation for Rectal Adenocarcinoma: The Impact of Radiation Fractionation and Surgical Resection. Clin Colorectal Cancer Feb 15 2023;doi: 10.1016/j.clcc.2023.02.003 [DOI] [PMC free article] [PubMed]
- 6.Cheng J, Lao YJ, Wang Q, et al. Predicting Distant Metastasis in Young-Onset Colorectal Cancer After Surgery: A Retrospective Study. Front Oncol 2022;12:804038. doi: 10.3389/fonc.2022.804038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Aguilar J, Chow OS, Smith DD, et al. Effect of adding mFOLFOX6 after neoadjuvant chemoradiation in locally advanced rectal cancer: a multicentre, phase 2 trial. Lancet Oncol Aug 2015;16(8):957–66. doi: 10.1016/S1470-2045(15)00004-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overacre-Delgoffe AE, Bumgarner HJ, Cillo AR, et al. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity Dec 14 2021;54(12):2812–2824 e4. doi: 10.1016/j.immuni.2021.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galeano Nino JL, Wu H, LaCourse KD, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature Nov 2022;611(7937):810–817. doi: 10.1038/s41586-022-05435-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freitas JA, Gullo I, Garcia D, et al. The Adaptive Immune Landscape of the Colorectal Adenoma-Carcinoma Sequence. Int J Mol Sci Sep 10 2021;22(18)doi: 10.3390/ijms22189791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acosta-Gonzalez G, Ouseph M, Lombardo K, Lu S, Glickman J, Resnick MB. Immune environment in serrated lesions of the colon: intraepithelial lymphocyte density, PD-1, and PD-L1 expression correlate with serrated neoplasia pathway progression. Hum Pathol Jan 2019;83:115–123. doi: 10.1016/j.humpath.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaCourse KD, Zepeda-Rivera M, Kempchinsky AG, et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep Nov 15 2022;41(7):111625. doi: 10.1016/j.celrep.2022.111625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell Jul 27 2017;170(3):548–563 e16. doi: 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science Sep 15 2017;357(6356):1156–1160. doi: 10.1126/science.aah5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods Jun 10 2012;9(8):811–4. doi: 10.1038/nmeth.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly BJ, Gross R, Bittinger K, et al. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics Aug 1 2015;31(15):2461–8. doi: 10.1093/bioinformatics/btv183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang ZZ, Chen G, Alekseyenko AV. PERMANOVA-S: association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics Sep 1 2016;32(17):2618–25. doi: 10.1093/bioinformatics/btw311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin H, Peddada SD. Analysis of compositions of microbiomes with bias correction. Nat Commun Jul 14 2020;11(1):3514. doi: 10.1038/s41467-020-17041-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salvucci M, Crawford N, Stott K, Bullman S, Longley DB, Prehn JHM. Patients with mesenchymal tumours and high Fusobacteriales prevalence have worse prognosis in colorectal cancer (CRC). Gut Aug 2022;71(8):1600–1612. doi: 10.1136/gutjnl-2021-325193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serna G, Ruiz-Pace F, Hernando J, et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann Oncol Oct 2020;31(10):1366–1375. doi: 10.1016/j.annonc.2020.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allali I, Delgado S, Marron PI, et al. Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 2015;6(3):161–72. doi: 10.1080/19490976.2015.1039223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res Feb 2012;22(2):299–306. doi: 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res Feb 2012;22(2):292–8. doi: 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, He Y, Xia L, et al. Expansion of Colorectal Cancer Biomarkers Based on Gut Bacteria and Viruses. Cancers (Basel) Sep 25 2022;14(19)doi: 10.3390/cancers14194662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong TNY, Wang X, Nakatsu G, et al. Association Between Bacteremia From Specific Microbes and Subsequent Diagnosis of Colorectal Cancer. Gastroenterology Aug 2018;155(2):383–390 e8. doi: 10.1053/j.gastro.2018.04.028 [DOI] [PubMed] [Google Scholar]
- 26.Xie YH, Gao QY, Cai GX, et al. Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies. EBioMedicine Nov 2017;25:32–40. doi: 10.1016/j.ebiom.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi F, Liu G, Lin Y, et al. Altered gut microbiome composition by appendectomy contributes to colorectal cancer. Oncogene Feb 2023;42(7):530–540. doi: 10.1038/s41388-022-02569-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Du L, Shi D, et al. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat Commun Nov 19 2021;12(1):6757. doi: 10.1038/s41467-021-27112-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avuthu N, Guda C. Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer. Microbiol Spectr Aug 31 2022;10(4):e0001322. doi: 10.1128/spectrum.00013-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowenmark T, Li X, Lofgren-Burstrom A, et al. Parvimonas micra is associated with tumour immune profiles in molecular subtypes of colorectal cancer. Cancer Immunol Immunother Oct 2022;71(10):2565–2575. doi: 10.1007/s00262-022-03179-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowenmark T, Lofgren-Burstrom A, Zingmark C, et al. Tumour Colonisation of Parvimonas micra Is Associated with Decreased Survival in Colorectal Cancer Patients. Cancers Nov 30 2022;14(23)doi: 10.3390/cancers14235937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Zhang X, Zhou Y, et al. Parvimonas micra promotes colorectal tumorigenesis and is associated with prognosis of colorectal cancer patients. Oncogene Sep 2022;41(36):4200–4210. doi: 10.1038/s41388-022-02395-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ardito CM, Gruner BM, Takeuchi KK, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell Sep 11 2012;22(3):304–17. doi: 10.1016/j.ccr.2012.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navas C, Hernandez-Porras I, Schuhmacher AJ, Sibilia M, Guerra C, Barbacid M. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell Sep 11 2012;22(3):318–30. doi: 10.1016/j.ccr.2012.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertocchi A, Carloni S, Ravenda PS, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell May 10 2021;39(5):708–724 e11. doi: 10.1016/j.ccell.2021.03.004 [DOI] [PubMed] [Google Scholar]
- 36.Shiao SL, Kershaw KM, Limon JJ, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell Sep 13 2021;39(9):1202–1213 e6. doi: 10.1016/j.ccell.2021.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


