SUMMARY
How the Venus flytrap (Dionaea muscipula) evolved the remarkable ability to sense, capture and digest animal prey for nutrients has long puzzled the scientific community1. Recent genome and transcriptome sequencing studies have provided clues to the genes thought to play a role in these tasks2–5. However, proving a causal link between these and any aspect of the plant’s hunting behavior has been challenging due to the genetic intractability of this non-model organism. Here, we use CRISPR/Cas9 methods to generate targeted modifications in the Venus flytrap genome. The plant detects prey using touch-sensitive trigger hairs located on its bilobed leaves6. Upon bending, these hairs convert mechanical touch signals into changes in the membrane potential of sensory cells, leading to rapid closure of the leaf lobes to ensnare the animal7. Here, we generate mutations in trigger hair-expressed MscS-like (MSL)-family mechanosensitive ion channel genes FLYCATCHER1 (FLYC1) and FLYCATCHER2 (FLYC2)5, and find that double mutant plants have a reduced leaf-closing response to mechanical ultrasound stimulation. While we cannot exclude off-target effects of the CRISPR/Cas9 system, our genetic analysis is consistent with these and other functionally-redundant mechanosensitive ion channels acting together to generate the sensory system necessary for prey detection.
Graphical Abstract
eTOC Blurb
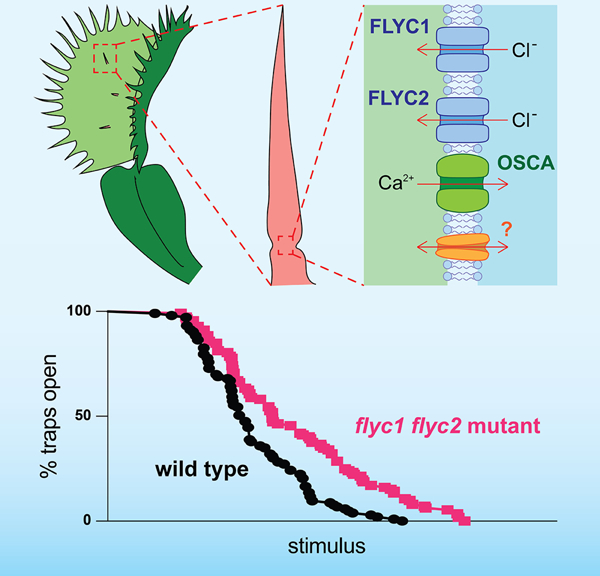
Ion channel genes FLYC1 and FLYC2 are expressed in touch-sensing structures of the carnivorous Venus flytrap plant. Procko et al. show that when these genes are mutated, Venus flytrap leaves have a reduced response to mechanical ultrasound stimulation. Thus, these channels may function redundantly with others for sensing mechanical stimuli.
RESULTS AND DISCUSSION
Bending of a Venus flytrap trigger hair initiates an action potential that is propagated across the leaf8. When the hair or another is bent a second time in short succession, the second action potential causes the two leaf lobes to close9,10. To find genes involved with this touch sensitivity, transcriptional analyses have identified at least three putative mechanosensitive ion channels that are differentially expressed in trigger hairs: FLYC1 and FLYC2, and a member of the OSCA family of membrane stretch-activated calcium-permeable channels3,5. It has been proposed that the opening of these channels in trigger hair sensory cells in response to membrane deformation is required for cell membrane depolarization, thereby initiating electrical signaling. However, to our knowledge, targeted mutations in the genome of any carnivorous plant have not yet been reported, and only two studies have demonstrated successful Venus flytrap transformation11,12. As such, challenges remain as to conclusively prove that any given gene is involved in a particular process in carnivorous plants.
Using CRISPR/Cas9 to generate targeted mutations in the Venus flytrap genome
In recent years, the CRISPR/Cas9 system has proven to be a powerful method for targeted genome modifications of model and agriculturally-important plant species13. Here, we bombarded Venus flytrap callus generated from a clonal wild-type line with gold particles coated with plasmid DNA containing components of the CRISPR/Cas9 system. In addition to Cas9, our plasmid included four guide RNAs (gRNAs) targeting two different sites in each of FLYC1 and FLYC2 (Figure S1A; genomic sites g1 through g4). The plasmid further contained 35S sequences driving expression of the fluorescent protein mCitrine (mCit) for visual identification of transformed tissue. Using this approach, we regenerated and selected a single plantlet carrying the DNA construct and exhibiting mosaic mCit fluorescence (Figure S1B). Subsequent splitting of the plant material and propagation in tissue culture was used to isolate a clonal line with ubiquitous mCit expression (Figure 1A).
Figure 1. Generation of flyc1 mutant Venus flytrap plants.
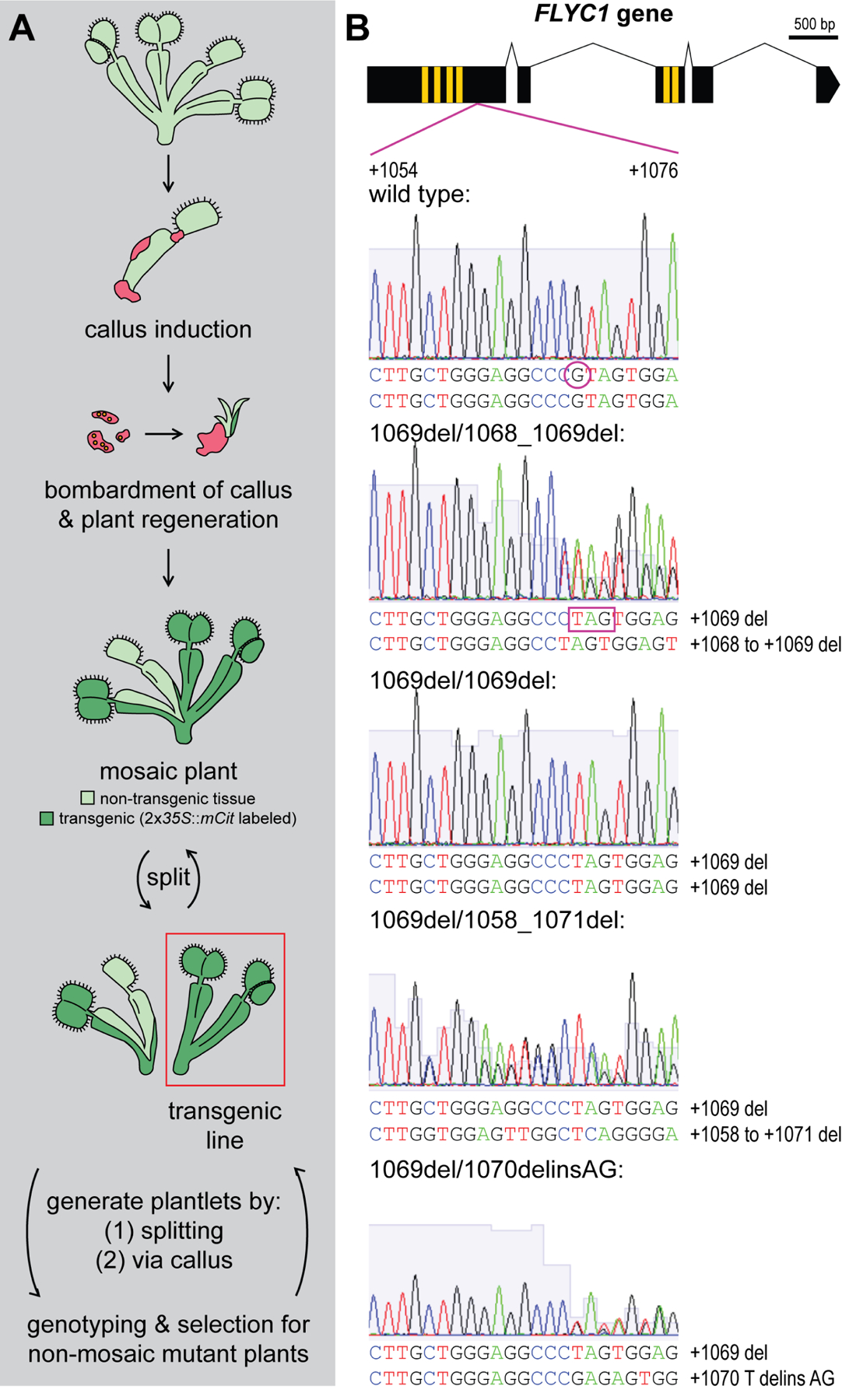
(A) Schematic of the method used to generate mutant Venus flytrap plants. Leaves were cut from a clonal wild-type Venus flytrap line in tissue culture and callus (red) generated. The callus was removed from the plant and bombarded with gold particles coated in plasmid DNA. Following bombardment, a mosaic transgenic plant was regenerated and identified by the presence of fluorescent mCitrine (mCit) protein expression under the 35S promoter (dark green). Further splitting of the mosaic plant at the rhizome was used to isolate a transgenic line with ubiquitous mCit expression (red box). From this, plantlets for genotyping were generated either by (1) splitting at the rhizome or (2) through leaf dissection, callus generation and plant regeneration, which in principle generates new plants from a smaller pool of cells more likely to be genetically uniform. Plantlets showing mosaicism for one or more mutations were positively selected for further rounds of splitting and regeneration until plants exhibiting evidence of non-mosaic, deleterious mutations on both FLYC1 (or FLYC2) alleles were found. These were clonally propagated in culture before moving to soil.
(B) Examples of mutations detected in the FLYC1 gene at the g1 site. Top, schematic of the FLYC1 gene, showing exons (black boxes), transmembrane-coding regions (yellow boxes), and region overlapping the g1 gRNA-targeted site (purple lines). Below, Sanger sequencing results of PCR products from genomic DNA template showing deleterious mutations at the g1 site in the FLYC1 gene of the diploid Venus flytrap genome for different mutant lines. The region covering +1054 to +1076 of the gene is shown (relative to the +1 ATG start site). del indicates deletion, delins indicates deletion-insertion. All alleles result in frameshifts coding for predicted truncated forms of the FLYC1 protein lacking the pore domain (transmembrane region 6). For example, a deletion of a single G residue at position +1069 (purple circle in wild-type allele) causes a frameshift mutation and premature stop codon (for example, purple box in mutant line 1069del/1068_1069del).
See also Figure S1.
To generate plants with deleterious mutations in FLYC1 and FLYC2, we repeatedly split and/or regenerated the transgenic plant material. New plantlets were subjected to PCR-based Sanger sequencing and genotyping (Figure 1A and S1C–D). Those showing mosaicism for one or more mutations were positively selected for further rounds of splitting and genotyping. Because we observed greater activity of Cas9 at the FLYC1 locus, we initially selected for flyc1 single mutants. Following this process of positive selection, we identified 56 of 276 transgenic plantlets tested as containing deleterious frameshift mutations in both FLYC1 alleles. Specifically, at the first gRNA site (g1) in the FLYC1 gene, we identified four different genotypes (Figure 1B). These mutations led to predicted truncation of the protein, including loss of the pore domain at the sixth transmembrane region14. The plantlets were then genotyped at the FLYC2 locus, and three flyc1 single mutant lines chosen for further analysis (Figure S1E). One of these lines, 1069del/1068_1069del, was heterozygous for two different mutations at the g1 site, while the other lines (1069del/1069del lines #1 and #2) showed homozygosity for a single base deletion at position +1069 of the g1 site. Despite sharing the same mutations, these two 1069del/1069del lines were isolated from separate culturing plates >1 month apart generated by splitting different parental plants, and therefore are only distantly related and considered here as independent. All three flyc1 single mutant lines displayed wild-type sequences at the g3 and g4 sites of the FLYC2 gene (Figure S1E). Single mutant plants were then clonally propagated in tissue culture before moving wild-type control and mutants simultaneously to soil for phenotypic analysis.
Leaves of flyc1 single mutants are likely normal
On soil, flyc1 mutants appeared similar to wild-type (Figure 2A). To better understand how mutations in FLYC1 might affect trap function, we analyzed the 1069del/1068_1069del single mutant line. The sensory cells responsible for detecting touch are localized to an indentation zone at the trigger hair base where maximum flexure of the hair occurs6,15. We observed no apparent difference in the morphology of these cells (Figure 2B).
Figure 2. flyc1 single mutants resemble wild-type plants.
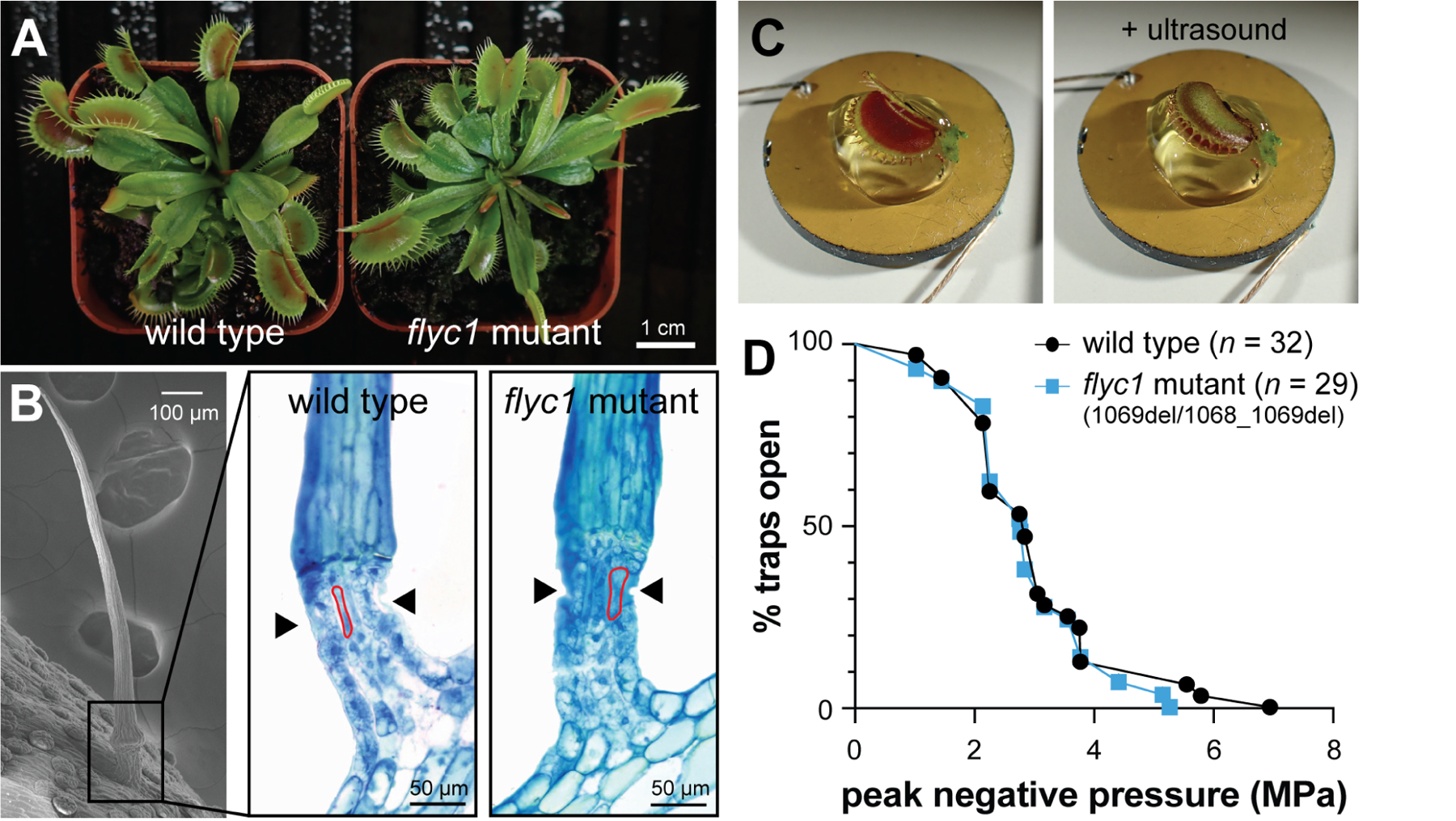
(A) Examples of wild-type and a flyc1 mutant plant on soil.
(B) Left, scanning electron micrograph of a trigger hair, and right, representative toluidine blue-stained cross-sections at the base of the trigger hair (boxed) of wild-type and flyc1 (1069del/1068_1069del) mutant plants (n = 3, both genotypes), showing the indentation zone (arrowheads) and sensory cells (examples outlined in red).
(C) Left, an open Venus flytrap leaf mounted on ultrasound gel atop a 2 cm diameter ultrasound transducer, and right, a closed leaf after ultrasound stimulation.
(D) The peak negative pressure (PNP) following pulsed ultrasound stimulation required for trap closure of wild-type and flyc1 (1069del/1068_1069del) mutant plants (not significant, logrank test).
See also Figures S1–S3.
We next tested the ability of flyc1 (1069del/1068_1069del) mutant leaves to respond to a harsh touch stimulus. For this, we bent the trigger hair (~5–30°) using a fire-polished Pasteur pipette attached to a manually-controlled micromanipulator. Using this assay, we observed no defect in the ability of mutant leaves to respond to this strong touch stimulus. Specifically, 53 of 55 wild-type and 33 of 33 flyc1 (1069del/1068_1069del) leaves closed after one or two touches of a trigger hair at 10 s intervals.
The lack of a detectable difference in flyc1 mutant leaves in our harsh touch-based assay may be due to the deflection of the trigger hairs being large enough to overcome any subtle defects in sensory loss. Indeed, the trigger is extremely sensitive and can detect an angular deflection of just a few degrees16. As such, to more finely assess the ability of the leaf to respond to mechanical stimulation, we developed a novel ultrasound-based assay. Detached, open traps were placed with one leaf lobe resting on ultrasound gel atop a 2 cm diameter transducer. Mechanical stimulation was then applied in the form of pulsed, mechanical ultrasound waves of increasing peak negative pressure (PNP) until the trap closed (100 ms stimuli pulsed at 10 s intervals; Figure 2C). Because there was some minor variation between wild-type leaves scored on different days (Figure S2A), we ensured that mutant plants were always scored alongside equivalent age-matched wild-type controls on any given day. This variation was not due to our experimental equipment (Figure S2B), but rather likely represents biological differences in our greenhouse-grown plants over time. For example, plant growth and trap function is known to be affected seasonally and by temperature9.
To induce trap closure, the ultrasound waves may open mechanosensitive channels in the leaf by: (1) causing mechanical perturbations directly on cell membranes17, (2) generating small deflections in the trigger hair as the leaf moves during pulsing (Figure S2C–F), which in turn deforms the membranes of the sensory cells, or (3) some combination of these two mechanisms or other effects18,19. Strikingly, we found that despite FLYC1 being highly up-regulated in sensory cells5, there was no quantitative difference between flyc1 (1069del/1068_1069del) mutants and wild-type traps in their response to ultrasound stimulation (Figure 2D).
As further confirmation of our findings, we tested the two additional flyc1 (1069del/1069del) single mutant lines. Surprisingly, while one of these lines also appeared wild-type, the other had a slight but significant increase in trap sensitivity to ultrasound (Figure S3A and B), despite the two lines sharing the same genotype at the FLYC1 and FLYC2 loci. This apparent defect was not due to differences in the size of the traps scored respective to wild-type plants, and nor was it due to a defect in trigger hair morphology (Figure S3C and D). Thus, our findings suggest that line effects may be present that are independent of the CRISPR/Cas-9 targeted mutations at the FLYC1 and FLYC2 genes. We hypothesize that these differences might be caused by an unknown off-target effect of the CRISPR/Cas9 system, despite the system generally considered to have few off-target effects in plants20–22. Alternatively, line differences may be caused by somaclonal variation as a result of tissue culture propagation23.
Together, our results are most consistent with mutations in FLYC1 not affecting Venus flytrap leaf closure in response to mechanical stimulation (2 of 3 lines). Thus, FLYC1 alone cannot be responsible for prey touch detection in Venus flytrap.
flyc1 flyc2 double mutants have defects in trap closure
Our inability to measure a consistent quantitative difference in the Venus flytrap’s response to mechanical stimulation in flyc1 single mutants suggests that there might be functional redundancy with other mechanosensitive ion channels expressed in the sensory cells. To test this, we proceeded to isolate a flyc1 flyc2 double mutant. Of the 56 flyc1 mutant plantlets we identified above, two of these also carried a deleterious frameshift mutation in one of the two FLYC2 alleles. Further splitting and positive selection recovered two plantlets split from the same parental pool—and thus most likely recently clonal and treated here as a single line—from 864 tested which harbored identical deleterious mutations in both FLYC1 and both FLYC2 alleles (Figure 3A). Due to the occurrence of chimeric PCR products during genotyping, we were unable to match the mutations shown at the g3 site of FLYC2 with one of the particular mutations at the g4 site to determine the two complete flyc2 alleles in the double mutant; however, all combinations result in premature stop codons and a predicted loss of the pore domain. These plants were clonally propagated, and a selection of progeny (n = 3 for each isolate) confirmed by Sanger sequencing before moving plants to soil. On soil, we observed typical variation in plant size (Figures 3B and S4A–G; see STAR Methods).
Figure 3. flyc1 flyc2 double mutant leaves have wild-type morphology.
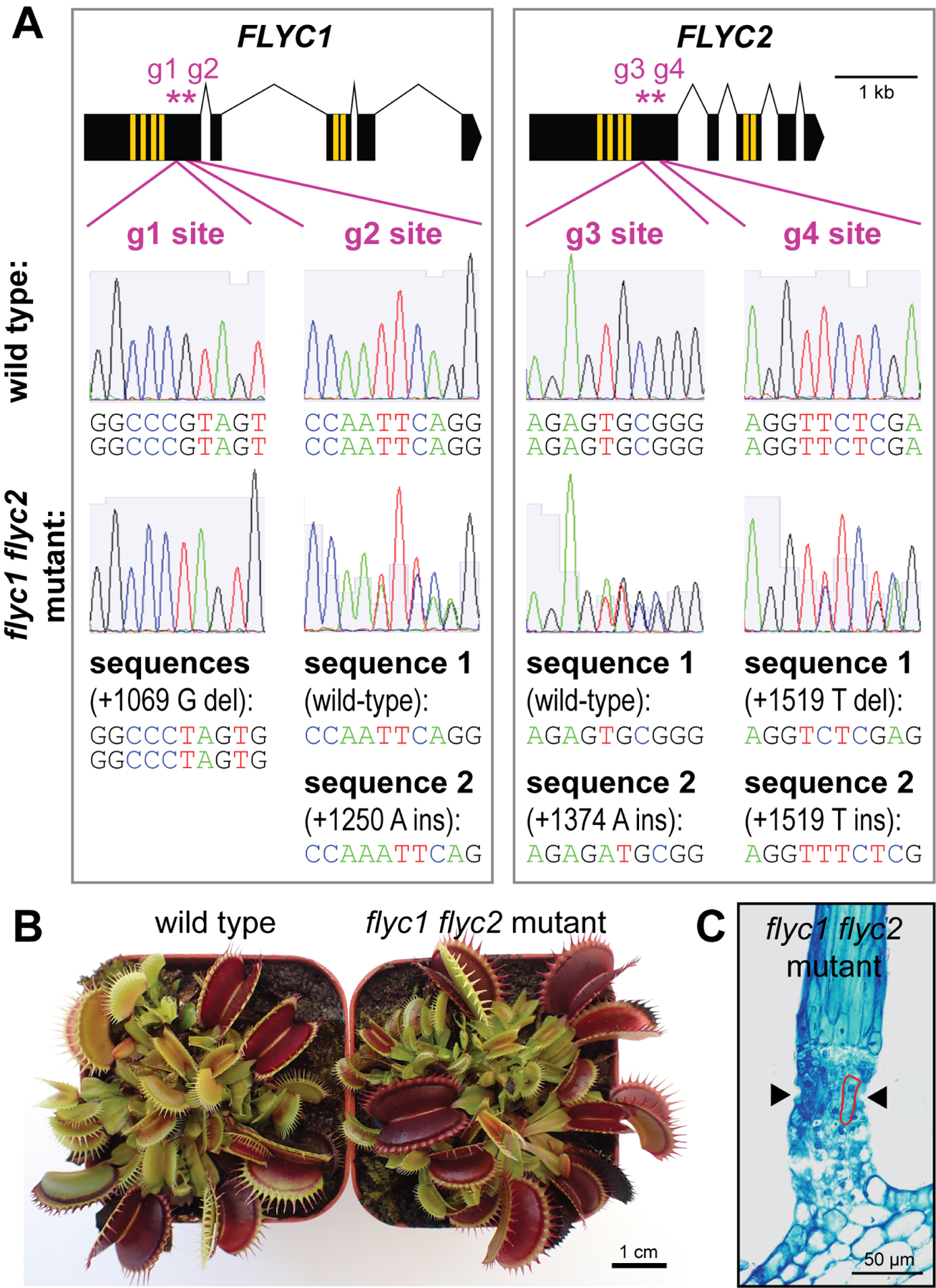
(A) Mutations in the double mutant. Top, schematic representations of the FLYC1 (see Figure 1B) and FLYC2 genes (black boxes, exons; yellow, transmembrane regions). Purple asterisks mark the locations of the gRNA targeted sites, g1 through g4. Below, Sanger sequencing of PCR products from genomic DNA template of wild-type and double mutant plants showing mutations on the two homologous chromosomes in the diploid genome. Residue positions are in the coding sequences relative to the +1 ATG sites. Deletion, del; insertion, ins.
(B) Examples of wild-type and flyc1 flyc2 mutant plants on soil.
(C) Toluidine blue-stained cross-section of the base of a flyc1 flyc2 mutant trigger hair (see Figure 2B; n = 3).
See also Figures S1 and S4.
Similar to flyc1 single mutants, our flyc1 flyc2 double mutant had normal trigger hair morphology at the sensory zone (Figure 3C). This suggests that any possible sensory defect, if present, is unlikely to be due to morphological abnormalities. In addition, we saw no defect in their response to a strong touch stimulus using a Pasteur pipet-mounted micromanipulator (36 of 40 wild-type and 38 of 40 mutant leaves closed after one or two touches of a trigger hair at 10 s intervals).
To more finely assess the ability of the double mutant to respond to mechanical stimulation, we again subjugated the plants to our quantitative ultrasound-based assay. Here, we found that flyc1 flyc2 mutant leaves showed a significant defect, requiring a greater PNP for trap closure compared to wild-type (Figure 4A). This defect was apparent for plants generated from both double mutant isolates (Figure S4H). Furthermore, we saw a slight correlation between trap size and the PNP required for closure in both wild-type and mutants, with larger traps requiring greater stimulation for both genotypes (Figures 4B and 4C). This is consistent with previous findings that larger traps require greater force on the trigger hair to initiate action potential firing16, and thus supports our ultrasound-based assay as a method to probe Venus flytrap leaf function.
Figure 4. flyc1 flyc2 double mutants have a reduced leaf response to mechanical ultrasound stimulation.
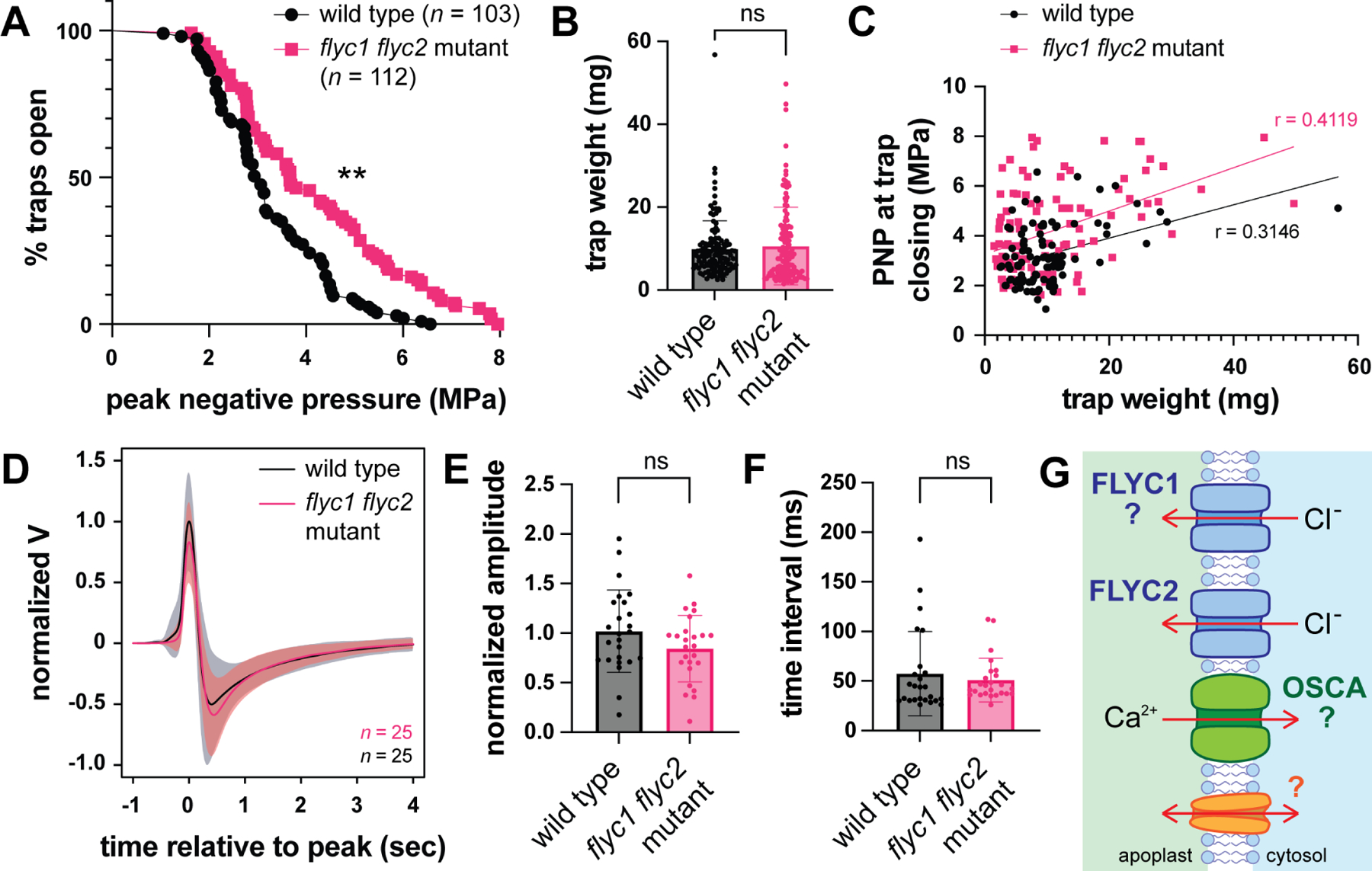
(A) The peak negative pressure (PNP) following pulsed ultrasound stimulation required for trap closure in wild-type and flyc1 flyc2 mutant plants (**p < 0.0001, logrank test).
(B) Weight of wild-type and mutant leaf traps scored by ultrasound stimulation (ns, not significant, t-test).
(C) PNP required for trap closing as a function of trap size (weight). Note a weak correlation between size and the required PNP for both mutant and wild-type traps (0.25 < r < 0.5).
(D) Change in extracellular potential of the Venus flytrap leaf as a result of trigger hair bending, normalized to the wild-type peak amplitude. Solid black and red lines show average change in extracellular potential, and gray and light red clouds show standard deviation for wild-type and flyc1 flyc2 mutant leaves, respectively.
(E) The peak amplitude of the change in leaf extracellular potential normalized to wild-type following trigger hair bending (ns, not significant, t test).
(F) The time interval between the peak and trough of the leaf action potential following trigger hair bending (ns, not significant, t test).
(D) A possible model showing the plasma membrane of a trigger hair sensory cell expressing redundantly-acting mechanosensitive channels required for membrane depolarization following a touch stimulus. These include FLYC2 and possibly FLYC1 (indicated by question mark), as well as OSCA and possibly other as yet undescribed channels. Arrows indicate the direction of ion movement following membrane deformation and channel opening.
See also Figures S2–S4.
While we cannot rule out the possibility of off-target effects of CRISPR/Cas9 on genes other than FLYC1 and FLYC2 (see above), a likely explanation for our results is that loss of function of these two genes leads to decreased sensitivity of the Venus flytrap to mechanical stimulation. The differential expression of these two genes in trigger hairs5 suggests that this defect is likely at the level of mechanoperception and not due to defects in downstream processes; for example, the all-or-nothing action potential that propagates across the leaf blade following trigger hair bending7. Thus, we hypothesized that leaf action potentials following mechanical stimulation would appear wild-type. While we were unable to measure action potentials resulting from ultrasound stimulation due to likely artifacts in the recordings (see STAR Methods), we observed that action potentials of double mutant leaves following manual trigger hair bending were not significantly different in amplitude and duration (Figure 4D–F). Likewise, the time taken for trap closure following ultrasound stimulation was similar between wild-type and mutant plants (Figure S4I). Together, these results are consistent with a sensory defect in flyc1 flyc2 double mutants in their trap closing response to mechanical ultrasound stimulation.
Conclusions
Here, we provide genetic evidence that redundancy between FLYC2, possibly FLYC1, and other mechanosensory ion channels in the trigger hair likely underlies prey touch sensation in the Venus flytrap (Figure 4G). These other channels most probably include an OSCA family member, for which homologs in Arabidopsis thaliana are mechanosensitive24,25 and for which in the Venus flytrap at least one member is differentially expressed in trigger hairs5. However, a limitation of our mutant analysis is the possibility of non-specific mutant line effects, perhaps due to CRISPR/Cas9-mediated off-target mutations or somaclonal variation (e.g., see Figure S3). Future work analyzing additional mutant lines or higher order mutants with other candidate mechanosensitive ion channels is needed to fully resolve these issues.
To test for a defect in trap function we developed a novel ultrasound-based assay. This was necessitated due to the fact that both our flyc1 single and flyc1 flyc2 double mutant plants appeared to respond normally to manual trigger hair bending. While this might argue that these genes have only a minor role, if any, in touch sensation, in light of possible redundancy with other mechanosensitive ion channels5 this result is perhaps not so surprising. The sensory cells can detect trigger hair deflection of just a few degrees16, and manual bending may be sufficiently large to overcome any subtle sensory defect. Pulsing with mechanical ultrasound waves provides an alternative means to mechanically stimulate the leaf and provide a quantitative readout. While ultrasound stimulation causes sudden movement of the plant tissue (Figure S2C–F), which may cause trigger hair deflection, it is also possible that ultrasound deforms the sensory cell membranes directly or the membranes of other cells in the leaf that also contribute to trap closing17,26. Alternatively, ultrasound may have other unrelated effects on the leaf, such as changes in temperature18,19. Despite these limitations, the assay provides a new method for quantitatively investigating subtle differences in trap physiology, which are not revealed by harsh trigger hair touch alone.
Why would the Venus flytrap require multiple mechanosensory ion channels for prey touch detection? Perhaps high levels of redundancy between mechanosensory channels is important for generating a robust sensory system necessary for prey capture. This system is important to support nutrient acquisition in the nutrient-poor soils in which the plant grows. Indeed, the trigger hair is exquisitely sensitive and can respond to the force of very small prey, such as ants16.
Finally, the testing of hypotheses regarding the genes required for Venus flytrap feeding behavior has traditionally been constrained by the seeming inability to validate gene function using mutant analysis. Here, our application of the CRISPR/Cas9 system to these non-model plants provides a new framework for functional gene discovery in plant carnivores.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carl Procko (procko@salk.edu).
Materials availability
There are no restrictions on materials generated for this manuscript.
Data Availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Plant materials and growth conditions
Venus flytraps (strain CP01) were propagated in tissue culture (1/3 × Murashige and Skoog salts and vitamins [Caisson Labs], 3% sucrose and 4.3 g/L gellan gum [Caisson Labs]) as previously described5. For phenotyping, plants were moved to soil (sphagnum peat moss, Premier Tech Horticulture, Canada) and grown in a greenhouse (25–30°C; mixed natural and artificial lighting; ~16 h day/8 h night cycles). Soil was kept constantly moist using purified water. Plants were hardened on soil > 6 months prior to ultrasound-based phenotyping.
METHOD DETAILS
Molecular cloning
To find regions of the FLYC1 and FLYC2 genes to target using the CRISPR/Cas9 system, we used CasOT-1.027 to locate gRNA targeting sites. These were designed within a 381 bp region of the first exon of FLYC1 (+1023 to +1403, relative to the +1 ATG start site of the coding sequence) and a 448 bp region of the FLYC2 gene for which we had confirmed the absence of any introns (+1237 to +1684, relative to the coding sequence). To minimize off-target effects, we checked potential gRNA targeting sequences against the transcriptome of our clonal strain5 and proceeded with those having the greatest number of mismatches against potential off targets, with at least one mismatch in the seed region.
gRNAs, plant kanamycin selection cassette and CRISPR/Cas9 expression cassettes were assembled into a plant binary vector using the Golden Gate/modular cloning system28. The gRNA scaffold was amplified from pICH86966_AtU6p_sgRNA_PDS (Addgene # 46966) 21 using forward primers incorporating the gRNA targeting sequence (g1 site: 5’ TGTGGTCTCAATTGCTTGCTGGGAGGCCCGTAG GTTTTAGAGCTAGAAATAGCAAG 3’; g2 site: 5’ TGTGGTCTCAATTGTCGATGTGGTATCCAATTC GTTTTAGAGCTAGAAATAGCAAG 3’; g3 site: 5’ TGTGGTCTCAATT GTCCAAAAGCTTCAGAGTGC GTTTTAGAGCTAGAAATAGCAAG 3’; g4 site: 5’ TGTGGTCTCAATT GGCATCGCCCTCGAGAACCT GTTTTAGAGCTAGAAATAGCAAG 3’) with reverse primer 5’ TGTGGTCTCAAGCGTAATGCCAACTTTGTAC 3’. Level 1 (L1) assembly was then performed using the g1, g2, g3 and g4 PCR products, the Arabidopsis thaliana U6 promoter (pICSL01009_AtU6pro; Addgene #46968) 21, and L1 destination vectors28 pICH47751 (Addgene #48002), pICH47761 (Addgene #48003), pICH47781 (Addgene #48005), and pICH47772 (Addgene #48004), respectively. Next, Level 2 (L2) assembly was performed using the four L1 gRNA plasmids, pICH47732::NOSp-NPTII-OCST (Addgene #51144; a gift from Jonathan D Jones), pICH47742::2x35S-5’UTR-hCas9(STOP)-NOST (Addgene #49771) 29, pICH41822 (Addgene #48021) 28, and the L2 destination vector pAGM4723 (Addgene #48015) 28.
Following Golden Gate assembly, the CRISPR vector was modified further to include two additional selection cassettes. First, a hygromycin resistance cassette was PCR amplified from Gateway plasmid pH7m34GW (Invitrogen) using forward primer 5’ acagatatcACTAGTatcatacatgagaattaagggagtcacgttatgacc 3’, which contains flanking EcoRV and SpeI restriction enzyme sites, and reverse primer 5’ acagtttaaacatcagcttgcatgccggtcga 3’, which contains a flanking PmeI site. The PCR product was digested with PmeI and EcoRV, and inserted into the Cas9/CRISPR construct at the single PmeI site. Clones were tested with SpeI/PmeI digest and Sanger sequencing. Second, a 2x35S::mCitrine::35S terminator cassette was PCR amplified using primers 5’ acagtttaaacGTTGCTCGCGGCCAACATGG 3’ and 5’ acagtttaaacggtcgacctgcaggtcactgg 3’, each containing a flanking PmeI site. Template consisted of a Gateway plasmid (pJZ1) generated by recombining a 2x35S promoter sequence in pDONR P4-P1R, an mCitrine (mCit) sequence in pDONR221, a short “mock” sequence in pDONR P2R-P330, with destination vector pK7m34GW. The PCR product was digested and inserted into the CRISPR construct at the PmeI site to create the final plasmid pCP.CRSP7, which was used for transformation (Figure S1A). Orientation of the mCit expression cassette was determined by Sanger sequencing.
Transformation
Plant transformation methods were similar to those previously reported for the related carnivorous plant species, Drosera spatulata31. Briefly, callus was generated from the clonal Venus flytrap line by cutting petioles, rhizome and trap tissue and placing the explants onto growth medium supplemented with hormones (1 mg/L 6-benzylaminopurine [Sigma] and 0.5 mg/L kinetin [Sigma]). Tissue was cultivated in the dark at 25°C over a 6–12 month period to generate callus for multiple rounds of bombardments. Successful transformation was achieved using freshly cut pieces of callus bombarded with DNA-coated 1 μm gold particles at 1,350 psi using a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad). Coating was performed by adding 5 μg of pCP.CRSP7 plasmid DNA to 3 mg of sterile gold particles in 50 μL H2O. To this, 20 μL 0.1 M spermidine (Sigma) and 50 μL 2.5 M CaCl2 was added with constant agitation and vortexed for 10 min at room temperature. Gold particles were then washed with ethanol and resuspended in 50 μL ice-cold ethanol. 8 μL aliquots were added to macrocarriers for biolistic delivery. Following bombardment, callus was grown in the dark on hormone-supplemented medium for 7 d before transferring to light conditions (16 h light/8 h dark cycles; 30 μmol/m2/s). At 11 d post-bombardment, callus was moved to growth medium supplemented with hormones and 5 μg/mL hygromycin. ~1 month post-bombardment a small sector of mCit positive cells on a single piece of callus was observed using fluorescence microscopy. At ~2 months, the mCit-positive callus was transferred to medium containing hormones and 10 μg/mL hygromycin. After ~3 months, a small shoot was observed exhibited mosaic expression of mCit (Figure S1B). As other mCit positive shoots appeared from the same clump of callus, these were cut from the callus and moved to growth medium with 10 μg/mL hygromycin but without hormones, so as to further promote shoot regeneration. Following further splitting of plants at the rhizome and continual propagation, a plant was ultimately selected and propagated which displayed mCit fluorescence in all tissues (Figure 1A).
Genotyping and selection of mutant plant lines
Transgenic plants were subjected to multiple rounds of splitting and genotyping to positively select for flyc1 single and flyc1 flyc2 double mutants (Figures 1A and S1C–D). Genomic DNA template for genotyping was extracted from 3–6 leaves of a given plantlet using a CTAB-based method5. PCR amplification of the FLYC1 region was performed using primers CP1229 (5’ CAGTGGCTATTGCTTGCTCCTCG 3’) and CP0996 (5’ GATCACTGTGGTTCTTGCCACTTGG 3’). For FLYC2, primers used were CP1036 (5’ TGGCCATGTCGTTCCATGTGAGC 3’) and CP1038 (5’ GCAATGAGGGCTCATCCTCGATGC 3’). Sanger sequencing of the PCR products was performed using these same primers, or with internal primers CP1670 (5’ CCCGACGTAAATCAACCTCATCAGACG 3’) and CP1672 (5’ AGGGGATTAGTCGGACGGAAAGTC 3’) for FLYC1, or CP1671 (5’ CTTTGACCGGATTCAGGAGGCATTG 3’) or CP1005 (5’ GCGGGGTTGTCTATTCCAGCAGAG 3’) for FLYC2 (see Figure S1C and D). The most likely allele sequences with the minimum number of mutations were resolved from the Sanger sequencing chromatograms. For single mutants, plantlets which had the same genotype and were collected from the same culturing plate on the same day were generally considered to have arisen from the same CRISPR/Cas9-mediated mutagenesis events, and were thus pooled and treated as single lines. Following clonal propagation of the single mutant lines, n = 3, 4 and 3 soil-grown plants for 1069del/1068_1069del, and 1069del/1069del lines #1 and #2, respectively, were further genotyped to confirm the presence of the expected mutations. One of the four 1069del/1069del line #1 plants retained a small residual amount of mosaicism for the wild-type allele. In tissue culture, double mutant plants generally appeared slightly smaller than wild-type, although on soil typical size variation was seen (Figure S4A–G).
Microscopy
Toluidine blue staining and light microscopy of Venus flytrap trigger hair cross sections was performed as previously described5. Images of the trigger hair during ultrasound stimulation were taken with a ZEISS Axio Zoom.V16 Microscope with Objective Plan Z 1.0x/0.25 FWD 60mm mounted with a Zeiss Axiocam 705 mono camera, and displacement in the x-y plane measured using Zeiss Zen 3.4 (Blue edition) software. To measure the time of trap closure, time-lapse movies were taken on the same instrument and similarly analyzed with Zeiss Zen 3.4 software. For scanning electron microscopy (SEM) of the trigger hair, plants were fixed in 2.5% glutaraldehyde, 4% formaldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) under microwaves for 1 min, with additional 20 min at room temperature. Then, samples were washed in buffer 3x 15 min, post-fixed in buffered 1% osmium tetroxide for 40 min, washed in buffer, dehydrated in a gradient series of ethanol until absolute (3x), critical-point-dried (Leica CPD300), mounted on carbon tape, gold-sputtered with a 4 nm layer, and observed on a Zeiss Sigma VP operated at 5 kV using SE detector.
Ultrasound stimulation of Venus flytrap traps
For ultrasound-based phenotyping, we used a slightly modified setup to that previously described32. Briefly, a 2 MHz lithium niobate transducer (Boston Piezo-Optics Inc, Bellingham, MA) was attached to a waveform generator (Keysight 33600 A Series) to control stimulus duration and frequency. Pressures were controlled using a 300-W amplifier (VTC2057574, Vox Technologies, Richardson, TX). The peak negative pressure (PNP) measurements were determined using a Precision Acoustics Fiber-Optic Hydrophone connected to a Tektronix TBS 1052B Oscilloscope by testing every % gain used in the experiments (e.g. see Figure S2B). Fly trap heads from small-leaf plants (Figure S4D–G) were carefully detached and placed on top of ultrasound gel (Aquasonic, Clinton Township, MI) that was used to couple the transducer to the trap heads. A 100 ms stimulus was pulsed every 10 s, starting from 0 MPa to 8.5 MPa in small increments until the flytrap head closed. The experimenter was blinded to whether the traps were wild-type or mutant. Non-functional traps which failed to close during pulsing and subsequently in response to even a harsh manual touch stimulus following the ultrasound assay were excluded from analysis. These leaves may be undergoing senescence or showing other aspects of Venus flytrap biology, such as dormancy. For single mutant assays shown in Figure 2, three plants of both wild-type and mutant were scored (n ≥ 10 traps from each; two mutant leaves did not respond to even a harsh touch stimulus, and were excluded from analysis). For Figure 4, traps were collected from 9 mutant and 10 wild-type plants (n = 10–20 leaves scored from each plant). 26/129 wild-type and 19/131 flyc1 flyc2 mutant leaves failed to respond and were excluded from downstream analysis. Following stimulation, traps were scored as either open or having closed (a binary event), and a logrank statistical test applied. Stimulation was stopped once the initial rapid closure occurred, and does not include hermetical sealing of the trap.
Leaf action potential measurements
Initially, we attempted to measure action potentials of leaves stimulated with ultrasound. The ultrasound transducer was set up as described above. Venus flytrap leaves (open or closed) were cut at the petiole and the traps placed on top of ultrasound gel. A silver wire ground electrode was placed on the petiole, while the recording electrode was coiled slightly at its tip and placed on the exposed outer surface of one of the trap lobes. Electrical connection was enhanced with a drop of electrode gel between the end coil of the electrode and the leaf (Signa Gel, Parker Laboratories, Inc, USA). Electrical signals were first amplified (100×) by a differential amplifier (DP-311, Warner Instruments), and data were further amplified (10×) and sampled at 10 kHz using a Multiclamp 700B amplifier, Digidata 1550B, and pClamp 10.7 acquisition software (all from Molecular Devices, LLC). Multiple ultrasound pulses (100 ms duration) were delivered at ≥ 20 s intervals. Recordings were carried out in response to peak to peak pressures ranging from 0.5–8 MPa. Using this experimental setup, upon ultrasound stimulation we observed perpendicular, step-like changes in the leaf surface potential that appeared non-biological and for which the measured voltage change correlated in size with the stimulus intensity. These changes may be a result of the sudden movement of the plant tissue against the recording electrode when ultrasound was applied, and thus are likely artifacts of the experimental method.
For recordings of action potentials in Figure 4, we measured the extracellular potential of the leaf traps using surface electrodes. For ease of manipulation, recordings were taken from traps > 1.5 cm in length. The measuring electrode (0.25 mm diameter silver wire, Thermo Fisher, USA) with an end loop of 1 mm was placed onto the outer surface of one of the two trap lobes of a leaf on an intact plant. The electrical connection was enhanced with a drop of electrode gel between the end loop of the electrode and the leaf (Signa Gel, Parker Laboratories, Inc, USA). The reference electrode was inserted into the wet soil. A single trigger hair on the same lobe as the recording electrode was then bent, and the electrical signal recorded using the Spike Recorder software and Plant Spiker Box (Backyard Brains, USA) per the manufacturer’s instructions. Recordings were imported into R33 using the tuneR package34 and trimmed to include 1 s prior and 4 s following the peak amplitude. For wild-type and mutant plants, electrical spikes were recorded from n = 25 leaves from 11 and 10 plants, respectively. In addition, a further 6 wild-type and 1 mutant leaves were either non-responsive to touch or displayed an atypical electrical behavior (a large downward change in the extracellular field potential only) and were excluded from the analysis. The change in electrical potential is relative to the starting potential at 1 s prior to the peak change, and normalized to the wild-type average peak amplitude.
QUANTIFICATION AND STATISTICAL ANALYSIS
For comparing wild-type to mutant leaves following ultrasound stimulation, a logrank comparison (Mantel-Cox test) was performed using GraphPad Prism version 9.0.0 (GraphPad Software, San Diego, CA, USA). For comparing the PNP at closing to leaf weight, a nonlinear regression was performed using GraphPad Prism version 9.0.0. Leaf action potentials were analyzed in R (see above), and all other statistical tests performed with GraphPad Prism version 9.0.0.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| 6-benzylaminopurine | Sigma | Cat#B3408 |
| kinetin | Sigma | Cat#K0753 |
| ultrasound gel | Aquasonic | N/A |
| Critical commercial assays | ||
| Gateway LR clonase II enzyme mix | Invitrogen | Cat#11791–020 |
| Plant Spiker Box | Backyard Brains | N/A |
| Deposited data | ||
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| Dionaea muscipula: strain CP01 | Procko et al., 2021 | N/A |
| Dionaea muscipula: mutant strains | this manuscript | N/A |
| Oligonucleotides | ||
| see Method Details for primers used to generate constructs and for genotyping | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Plasmid: pICH86966_AtU6p_sgRNA_PDS | Addgene | 46966 |
| Plasmid: pICSL01009_AtU6pro | Addgene | 46968 |
| Plasmid: pICH47751 | Addgene | 48002 |
| Plasmid: pICH47761 | Addgene | 48003 |
| Plasmid: pICH47781 | Addgene | 48005 |
| Plasmid: pICH47772 | Addgene | 48004 |
| Plasmid: pICH47732::NOSp-NPTII-OCST | Addgene | 51144 |
| Plasmid: ppICH47742::2x35S-5’UTR-hCas9(STOP)-NOST | Addgene | 49771 |
| Plasmid: pICH41822 | Addgene | 48021 |
| Plasmid: pAGM4723 | Addgene | 48015 |
| Plasmid: pH7m34GW | Invitrogen | N/A |
| Plasmid: pJZ1 | this manuscript | N/A |
| Plasmid: pCP.CRSP7 | this manuscript | N/A |
| Software and algorithms | ||
| GraphPad Prism v 9.0.0 | GraphPad Software | N/A |
| CasOT v 1.0 | Xiao et al, 2014 | N/A |
| R | R Core Team, 2021 | N/A |
| tuneR | Ligges et al, 2023 | N/A |
| Other | ||
| PDS-1000/He Biolistic Particle Delivery System | Bio-Rad | N/A |
| 2 MHz lithium niobate ultrasound transducer | Boston Piezo-Optics Inc | N/A |
| Keysight 33600 A Series Waveform generator | Keysight Technologies | N/A |
| 300 W amplifier | Vox Technologies | VTC2057574 |
Highlights.
The CRISPR/Cas9 system can be used to genetically modify Venus flytrap plants
Loss of the mechanosensitive ion channel FLYC1 does not impair leaf function
flyc1 flyc2 double mutants have a reduced response to mechanical stimulation
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health (NIH) awards 1F32GM101876 (C.P.), 1R01NS115591 (S.H.C.) and 5R35GM122604 (J.C.); Howard Hughes Medical Institute (J.C.); a Salk Innovation Grant (C.P., J.C., S.H.C.); a Salk Women & Science Research Award (W.M.W.); the Kenneth G. and Elaine A. Langone Fellowship of the Damon Runyon Cancer Research Foundation DRG-2476–22 (W.M.W.); and a University of San Diego Faculty Research Grant (L.B.). SEM was performed at the Waitt Advanced Biophotonics Core with L. Andrade and U. Manor, supported by the Waitt Foundation and Core Grant applications NCI CCSG (CA014195) and NINDS Neuroscience Center (NS072031). We thank A. Patapoutian, O. Yarishkin, Y. Burko, E. Daniels, S. McDowell, M. Anderson, A. Chakraborty, E. Edsinger and members of the Chory, Chalasani, Patapoutian and Baird laboratories for assistance and helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
A patent application regarding proteins reported in this publication has been filed (PCT/US2021/058708).
REFERENCES
- 1.Darwin CR (1875). Insectivorous Plants (John Murray)
- 2.Bemm F, Becker D, Larisch C, Kreuzer I, Escalante-Perez M, Schulze WX, Ankenbrand M, Van de Weyer AL, Krol E, Al-Rasheid KA, et al. (2016). Venus flytrap carnivorous lifestyle builds on herbivore defense strategies. Genome Res 26, 812–825. 10.1101/gr.202200.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iosip AL, Bohm J, Scherzer S, Al-Rasheid KAS, Dreyer I, Schultz J, Becker D, Kreuzer I, and Hedrich R (2020). The Venus flytrap trigger hair-specific potassium channel KDM1 can reestablish the K+ gradient required for hapto-electric signaling. PLoS Biol 18, e3000964. 10.1371/journal.pbio.3000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palfalvi G, Hackl T, Terhoeven N, Shibata TF, Nishiyama T, Ankenbrand M, Becker D, Förster F, Freund M, Iosip A, et al. (2020). Genomes of the Venus Flytrap and Close Relatives Unveil the Roots of Plant Carnivory. Current Biology 30, 2312–2320.e2315. 10.1016/j.cub.2020.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Procko C, Murthy S, Keenan WT, Mousavi SAR, Dabi T, Coombs A, Procko E, Baird L, Patapoutian A, and Chory J (2021). Stretch-activated ion channels identified in the touch-sensitive structures of carnivorous Droseraceae plants. Elife 10. 10.7554/eLife.64250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd FE (1942). The Carnivorous Plants (Chronica Botanica Company; ). [Google Scholar]
- 7.Hedrich R, and Neher E (2018). Venus Flytrap: How an Excitable, Carnivorous Plant Works. Trends in Plant Science 23, 220–234. 10.1016/j.tplants.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Sanderson JB (1873). I. Note on the electrical phenomena which accompany irritation of the leaf of Dionæa muscipula. Proceedings of the Royal Society of London 21, 495–496. 10.1098/rspl.1872.0092. [DOI] [Google Scholar]
- 9.Brown WH, and Sharp LW (1910). The Closing Response in Dionaea. Botanical Gazette 49, 290–302. 10.1086/330177. [DOI] [Google Scholar]
- 10.Forterre Y, Skotheim JM, Dumais J, and Mahadevan L (2005). How the Venus flytrap snaps. Nature 433, 421–425. 10.1038/nature03185. [DOI] [PubMed] [Google Scholar]
- 11.Suda H, Mano H, Toyota M, Fukushima K, Mimura T, Tsutsui I, Hedrich R, Tamada Y, and Hasebe M (2020). Calcium dynamics during trap closure visualized in transgenic Venus flytrap. Nature Plants 6, 1219–1224. 10.1038/s41477-020-00773-1. [DOI] [PubMed] [Google Scholar]
- 12.Makowski W, Krolicka A, Nowicka A, Zwyrtkova J, Tokarz B, Pecinka A, Banasiuk R, and Tokarz KM (2021). Transformed tissue of Dionaea muscipula J. Ellis as a source of biologically active phenolic compounds with bactericidal properties. Appl Microbiol Biot 105, 1215–1226. 10.1007/s00253-021-11101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma X, Zhu Q, Chen Y, and Liu Y-G (2016). CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Molecular Plant 9, 961–974. 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Jojoa-Cruz S, Saotome K, Tsui CCA, Lee WH, Sansom MSP, Murthy SE, Patapoutian A, and Ward AB (2022). Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1. Nature Communications 13. ARTN 850 10.1038/s41467-022-28511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benolken RM, and Jacobson SL (1970). Response Properties of a Sensory Hair Excised from Venus’s Flytrap. The Journal of General Physiology 56, 64–82. 10.1085/jgp.56.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherzer S, Federle W, Al-Rasheid KAS, and Hedrich R (2019). Venus flytrap trigger hairs are micronewton mechano-sensors that can detect small insect prey. Nature Plants 5, 670–675. 10.1038/s41477-019-0465-1. [DOI] [PubMed] [Google Scholar]
- 17.Vasan A, Orosco J, Magaram U, Duque M, Weiss C, Tufail Y, Chalasani SH, and Friend J (2021). Ultrasound Mediated Cellular Deflection Results in Cellular Depolarization. Advanced Science 9 10.1002/advs.202101950. [DOI] [PMC free article] [PubMed]
- 18.O’Brien WD Jr. (2007). Ultrasound-biophysics mechanisms. Prog Biophys Mol Biol 93, 212–255. 10.1016/j.pbiomolbio.2006.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyler WJ (2011). Noninvasive neuromodulation with ultrasound? A continuum mechanics hypothesis. Neuroscientist 17, 25–36. 10.1177/1073858409348066. [DOI] [PubMed] [Google Scholar]
- 20.Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang D-L, Wang Z, Zhang Z, Zheng R, Yang L, et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proceedings of the National Academy of Sciences 111, 4632–4637. 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nekrasov V, Staskawicz B, Weigel D, Jones JDG, and Kamoun S (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nature Biotechnology 31, 691–693. 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Peterson BA, Haak DC, Nishimura MT, Teixeira PJPL, James SR, Dangl JL, and Nimchuk ZL (2016). Genome-Wide Assessment of Efficiency and Specificity in CRISPR/Cas9 Mediated Multiple Site Targeting in Arabidopsis. Plos One 11. 10.1371/journal.pone.0162169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miguel C, and Marum L (2011). An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. Journal of Experimental Botany 62, 3713–3725. 10.1093/jxb/err155. [DOI] [PubMed] [Google Scholar]
- 24.Murthy SE, Dubin AE, Whitwam T, Jojoa-Cruz S, Cahalan SM, Mousavi SAR, Ward AB, and Patapoutian A (2018). OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife 7. 10.7554/eLife.41844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jojoa-Cruz S, Saotome K, Murthy SE, Tsui CCA, Sansom MSP, Patapoutian A, and Ward AB (2018). Cryo-EM structure of the mechanically activated ion channel OSCA1.2. eLife 7. 10.7554/eLife.41845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DiPalma JR, McMichael R, and DiPalma M (1966). Touch Receptor of Venus Flytrap, Dionaea muscipula. Science 152, 539–540. 10.1126/science.152.3721.539. [DOI] [PubMed] [Google Scholar]
- 27.Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, and Zhang B (2014). CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180–1182. 10.1093/bioinformatics/btt764. [DOI] [PubMed] [Google Scholar]
- 28.Weber E, Engler C, Gruetzner R, Werner S, and Marillonnet S (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, e16765. 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belhaj K, Chaparro-Garcia A, Kamoun S, and Nekrasov V (2013). Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9. Artn 39 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, and Chory J (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30, 1529–1541. 10.1101/gad.283234.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Procko C, Radin I, Hou C, Richardson RA, Haswell ES, and Chory J (2022). Dynamic calcium signals mediate the feeding response of the carnivorous sundew plant. Proc Natl Acad Sci U S A 119, e2206433119. 10.1073/pnas.2206433119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duque M, Lee-Kubli CA, Tufail Y, Magaram U, Patel J, Chakraborty A, Lopez JM, Edsinger E, Vasan A, Shiao R, et al. (2022). Sonogenetic control of mammalian cells using exogenous Transient Receptor Potential A1 channels. Nature Communications 13. ARTN 600 10.1038/s41467-022-28205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. (2021). R: A language and environment for statistical computing R Foundation for Statistical Computing; (https://www.R-project.org/). [Google Scholar]
- 34.Ligges U, S.K., Mersmann O and Schnackenberg S (2023). tuneR: Analysis of Music and Speech
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


