Abstract
The simultaneous determination of carbaryl and its metabolite 1-naphthol is essential for risk assessment of pesticide exposure in agricultural and environmental samples. Herein, several bispecific nanobodies (BsNbs) with different lengths of hydrophilic linkers and junction sites were prepared and characterized for the simultaneous recognition of carbaryl and its metabolite 1-naphthol. It was found that the affinity of BsNbs to the analytes could be regulated by controlling linker length and linking terminal. Additionally, molecular simulation revealed that linker lengths affected the conformation of BsNbs, leading to alteration in sensitivity. The BsNb with G4S linker, named G4S-C-N-VHH, showing good thermal stability and sensitivity was used to develop a bispecific indirect competitive enzyme-linked immunosorbent assay (Bic-ELISA). The assay demonstrated a limit of detection of 0.8 ng/mL for carbaryl and 0.4 ng/mL for 1-naphthol in buffer system. Good recoveries from soil and rice samples were obtained, ranging from 80.0% to 112.7% (carbaryl) and 76.5% to 110.8% (1-naphthol), respectively. Taken together, this study firstly provided a BsNb with high sensitivity and efficiency against environmental pesticide and its metabolite, and firstly used molecular dynamics simulation to explore the influence of linker on recognition. The results are valuable for the application of immunoassay with high efficiency in the fields of environment and agriculture.
Capsule:
A highly sensitive bispecific nanobody against carbaryl and its metabolite 1-naphthol was development, and the recognition with the influence of linker was explored via molecular dynamics simulation and Bic-ELISA.
Keywords: Linker, Molecular dynamics simulation, Circular dichroism, Stability, Bic-ELISA
GRAPHICAL ABSTRACT
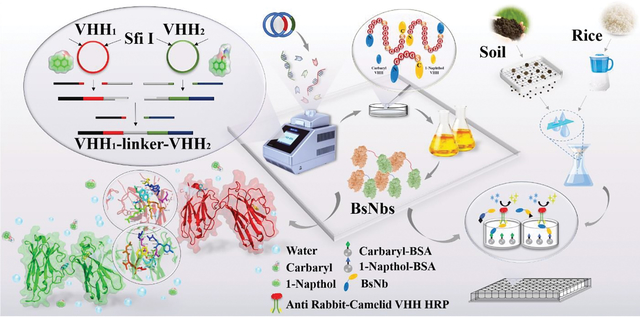
1. Introduction
Carbaryl is a water-soluble carbamate insecticide that has a long residual period in the soil, easily resulting in contamination of crops and food (Ćwieląg-Piasecka et al., 2021). It possesses systemic toxicity, causing damage to all organs of the human body upon ingestion (Ke et al., 2021). Compared to the 72.17% degradation rate in sterile soil, 59.0% carbaryl was eliminated in non-sterile soil (Li at al., 2019). It is worth noting that carbaryl can undergo photolysis or biodegradation by Micrococcus spp., resulting in the production of 1-naphthol and methylamine. 1-Naphthol is the main metabolites of carbaryl (Chen et al., 2020), which would cause significant harm to human body through contact or accidental ingestion (Chen et al., 2022). Currently, many countries and organizations have established strict maximum residue limits for carbaryl in crops or soil, including China (1 mg/kg in crops, No. 4.235) (GB2763–2021), USA (0.10 ppm, No. §180.169) (EPA, 2014), EU (0.5 mg/kg in barley, No. 0500010; 0.01 mg/kg in rice, No. 0500060) (EU, 2014) and IFS (1 mg/kg, No. 8) (CAC, 2004), while no specific maximum residue limits for 1-napthol had been set. Recently, some researches had revealed the presence of carbaryl and 1-naphthol residues ranging from 100 to 600 ng/g in soils and vegetables (Mei-Liang Chin-Chen et al., 2012). In addition, the real residues of carbaryl are approximately 4.77 mg/mL in fruits (Shahdost-fard at al., 2021) and 20 ng/g in other crops (Ding et al., 2023). Hence, it is necessary to tightly control the actual residue of carbaryl. Hence, it is necessary to tightly control the actual residue of carbaryl. However, since a single test of carbaryl cannot capture the true residue, a method that tests both the original pesticide and metabolites together is a desirable approach.
At present, the detection of carbaryl and its metabolites mainly relies on instruments, which are costly, require professional operators and have long turnaround times (Chen et al., 2021). An alternative method for rapid detection is immunoassay based on antibodies (Bhardwaj et al., 2020; Mills et al., 2022), which offers advantages of high sensitivity, specificity and cost-efficiency. In 1993, heavy-chain antibodies naturally lacking light chain were discovered firstly in camels (Hamers-Casterman et al., 1993). This discovery led to the subsequent identification of nanobody, (Nb), the variable domain of the heavy-chain of heavy-chain antibody (VHH) (Jia et al., 2020). Nbs possess several advantages, including small molecular weight, high expression, and ease of functional modification. As a result, it has been gradually applied in rapid detection of complex matrix samples (Bathula et al., 2021). Building upon the success of nanobodies, bispecific nanobody (BsNb) with two epitopes (Li et al., 2022) has been developed and can detect two different antigens simultaneously, thereby improving detection efficiency (Segovia-de Los Santos et al., 2022). Currently, bispecific antibody has been applied to the simultaneous detection, including bispecific Nb produced by He, at al. (2021) and Zhao at al. (2020), bispecific monoclonal antibody (BsmAb) constructed and used in ELISA by Wang, at al. (2016) and Hua, at al. (2013), and BsmAb used in colloidal gold immunochromatographic strip that established by Guo, at al. (2009). Nevertheless, they suffer from time consumption, high detection limits or poor stability. Moreover, they were unable to identify any potential effects of the linker on bispecific antibodies so that there is a lack of knowledge regarding the impact of linkers. To mitigate these challenges, we proposed that BsNbs produced by genetic engineering with linker could potentially overcome these defects and confer additional benefits.
With the aim to simultaneously determine carbaryl and its metabolite, 1-naphthol, we constructed a BsNb based on the monomer research conducted in our previous work. It turned out that the length of linker and its junction sites had a significant impact on the affinity of the BsNb. Consequently, the purpose of this study is to further study the comprehensive influence of the linker on affinity via combining experimental and molecular simulation methods, and then discuss the stability of BsNb. Finally, an ELISA-based simultaneous detection method was established to serve as a foundation for further studies involving the relevant BsNbs.
2. Materials and methods
2.1. Materials and Instruments
Coating antigen (1-naphthol-BSA and carbaryl-BSA) were previously prepared in our laboratory (Chen et al., 2022) and the laboratory of China Agricultural University, respectively (Figure S1). The pComb3xSS vector with ampicillin resistance and a lactose promoter, which dissolved in sterile water, was gifts from Prof. Bruce Hammock (UC Davis, California, USA). Absorbance was measured at wavelengths of 450 nm using a microplate reader (Multiskan FC, Thermo Fisher, Shanghai, China). The circular dichroism spectra were determined by Circular Dichroism (Applied Photophysics Ltd., UK). Other materials and instruments were summarized at Supplementary Materials (SM).
2.2. Construction of BsNb Plasmid Vector
The commonly used short peptide linker (G4S)3 was preferentially selected for the construction of two BsNbs, including (G4S)3-C-N-VHH and (G4S)3-N-C-VHH. The codon of linkers had undergone a retooling, redesign and optimization. To explore the influence on BsNbs, linkers with different lengths, including no linker, G4S, (G4S)2, (G4S)3 and (G4S)4, were designed as flexible linkers to construct the corresponding BsNbs. Next, the VHH genes of BsNbs, along with the specific linkers and SfiI restriction sites, were amplified by two rounds of PCR (Liu et al., 2022) with specific primers (Table S1). The first round of amplification was performed to obtain the gene sequences of carbaryl-VHH and 1-naphthol-VHH. Then the ultimate target fragment was amplified in the second round of PCR amplification by mixing the two target fragments. The optimum annealing temperature (53–65 °C) for this step was explored. The resulting PCR product, approximately 800 bp, was purified according to the DNA recovery verification. Finally, the purified PCR products were purified again after digesting with SfiI and subcloned into the pcomb3xSS using T4 DNA ligase (Fu et al., 2020). The reaction was allowed to proceed for at least 2 hours at 22 °C.
2.3. Expression and Purification of BsNb
After transforming the recombinant plasmid into E. coli DH5a, the colony PCR was performed and the positive clones were sent for sequencing. When obtaining a monoclonal strain with the correct sequence, the E. coli BL21 (DE3) was transformed with recombinant plasmid and the target protein was expressed in periplasmic cavity of E. coli BL21 via cultured in LB medium (seeing in SM). Then the protein in periplasmic lumen was extracted by cold osmotic shock method. Briefly, the bacterial liquid was centrifuged (4 °C, 10000 rpm, 3 min only) to isolate bacteria and was dissolved in sucrose solution. After being placed in −80 °C to freeze 2 h, the system was added distilled water to shaken for 1 h (4 °C, 250 rpm) and taken out for centrifugation (4 °C, 10000 rpm, 3 min only). The supernatant is the protein solution, which would be mixed with nickel filler at least 2 h to prepare for purification. Then purified with Ni-NTA resin by using 200 mmol/L imidazole in 0.01 mol/L PBS for elution. The specific procedures were shown in SM. Finally, the 12% SDS-PAGE and western blotting were both applied to identify the resulting BsNb fusion protein (Srinivasan et al., 2022).
2.4. Effect of Linkers
The effect of C/N-terminal ligation on BsNb was explored firstly to realize linking terminal optimization. Then the better would be chosen to optimize the lengths of linkers. Different lengths of short peptide sequences were selected as linkers for construction. Their characteristics would be compared via enzyme-linked immunosorbent assay (ELISA) (Li et al., 2021), including preparation of coating hapten, incubation of BsNb, combination of secondary antibody and color development with TMB. The specific details were summarized at SM.
2.5. Model Building and Molecular Simulations
The structural model of the two BsNbs, G4S-C-N-VHH and (G4S)3-C-N-VHH, were built via AlphaFold2 (Chen et al., 2018). The credibility of the proposed models was evaluated using UCLA-DOE LAB-SAVES v6.0 (https://saves.mbi.ucla.edu/). To conduct docking simulations, the molecular structures of carbaryl and 1-naphthol were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The docking simulations were performed using leadit-2.3.2-Linux-x64 (http://www.biosolveit.de/LeadIT/) with the Enthalpy and Entropy (hybrid approach) ligand binding setting, while the other settings remained default. The interaction forces between BsNbs and ligands were analyzed using Open-Source PyMoL™ and LigPlus. Then, the semi-flexible docking complexed containing 1-naphthol (ligand) and BsNbs were analyzed via molecular dynamics (MD) simulations using GROMACS 2021.2 with the AMBER99SB-ILDN force field. MD carried out in a triclinic box filled with TIP3 water molecules and periodic bounding conditions were applied with a minimum distance of 1 Å, setting 100 ns of the simulation time and 310 K of simulation temperature.
2.6. Stability of BsNb
To assess different performances of BsNb, several stability performance indicators were tested and discussed, including thermal stability (37–100 °C), acid-base tolerance (pH 1.4–12.4) and organic solvent tolerance (10%, 20%, 30%, 40%, 50% of methanol, acetonitrile and acetone). Comprehensive analysis was performed via combining ELISA and circular dichroism for secondary structure determination in thermal stability identification (Fan et al., 2018). Herein, we further set up variable temperature program (37–90 °C) to research the detailed changes in sum of the valleys of secondary structure by circular dichroism, measured in every 1 °C rise.
2.7. Optimization of Bic-ELISA and Recovery Test
For different conditions of Bic-ELISA (Hu et al., 2022; Liang et al., 2022), the most critical parameters, including incubation temperature (4 °C, 25 °C, 37 °C and 50 °C) and concentrations of coating hapten (1 μg/mL, 500 ng/mL, 250 ng/mL and 125 ng/mL), was optimized in turns. The sensitivity of the assay was evaluated using the maximum absorption value (Amax), half-inhibitory concentration (IC50) and Amax/IC50. Then the optimal conditions were used for recovery verification of real samples, including soil sample (Gu et al., 2020) from farmland and grain rice. These samples were also evaluated by liquid chromatography tandem mass spectrometry (LC-MS/MS) to assess the accuracy and reliability of the proposed Bic-ELISA (Table S2). The specific details were shown in SM. Finally, the recovery (RR, %) of the sample additions and the coefficient of variation (CV, %) were calculated and analyzed. In a word, these protocols took at least one week to complete from the construction of the assay to simultaneous recognition, while only took 2 h from sample preparation to obtaining the detection results of carbaryl and 1-naphthol.
3. Results and discussion
3.1. Construction, Expression and Identification of BsNbs
Generally, the specific binding between antigens and antibodies relies on their compatible epitopes. For BsNb, when one epitope binds to an antigen, the other epitope remains available to bind another antigen. However, the successful implementation of this dual-recognition feature requires an appropriate connection method. Presently, several strategies for BsNb construction have been reported, including short peptide linking (Wichgers Schreur et al., 2020), natural hinge linking (Zhong et al., 2022), heavy chain transfer (Pekar et al., 2020), loop composing (Fernández-Quintero et al., 2022), and chemical linking (Chen et al., 2022). For the construction of BsNbs, the short peptide linking strategy (Bai et al., 2023) stands out due to its advantages of high stability, high solubility, high success rate, and natural conformation folding. Therefore, we opted to constructed BsNbs using this linking strategy. Considering that linker junction sites and lengths are critical factors affecting antibody activity, this study aimed to investigate the impact of different linker lengths on BsNbs. The detailed experimental process is outlined in Scheme 1.
Scheme 1.
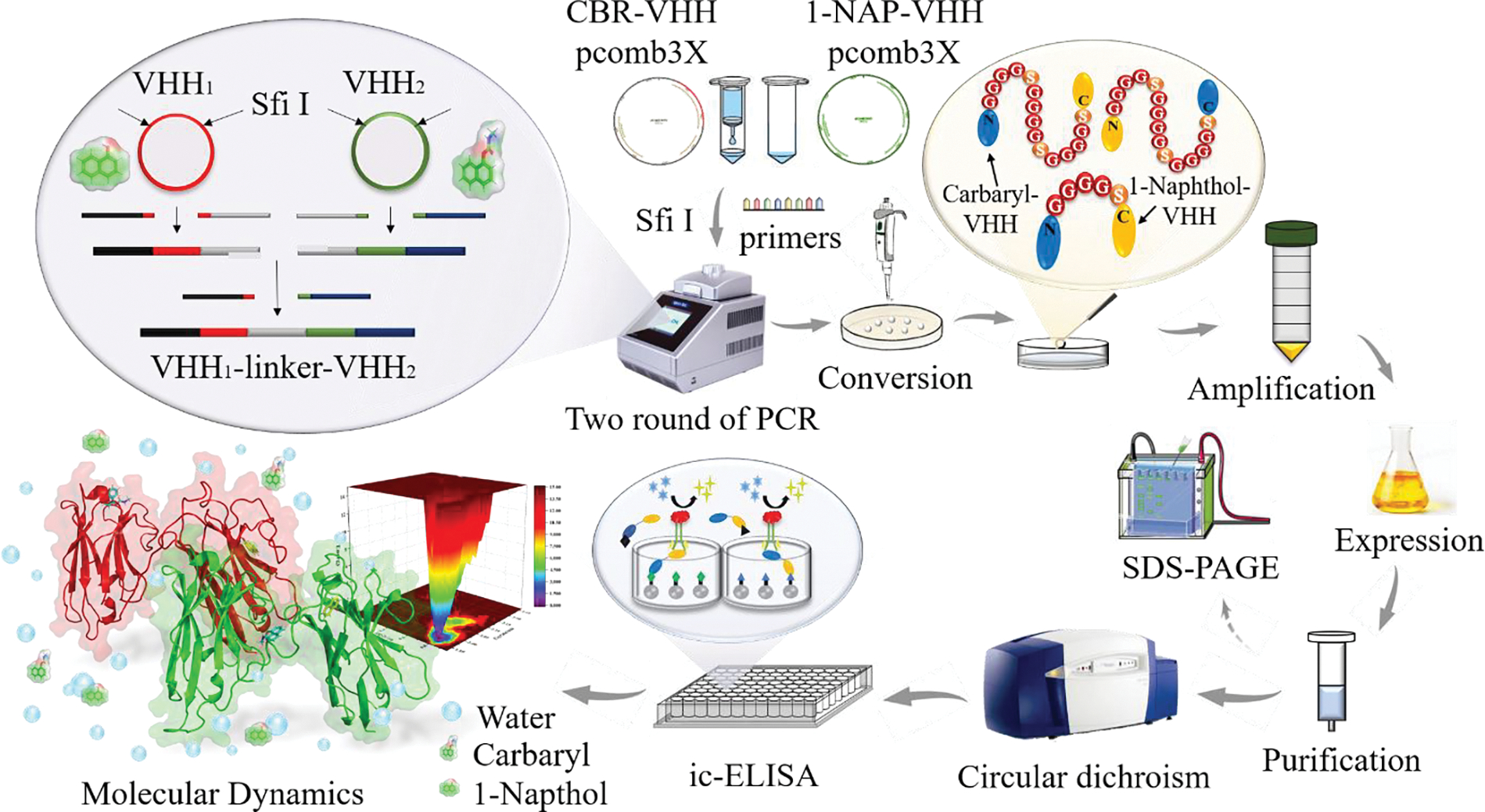
The diagram of BsNb construction and characterization.
The most commonly used short peptide linker (G4S)3 was preferentially selected to construct the recombinant plasmid of (G4S)3-C-N-VHH and (G4S)3-N-C-VHH. The codons of the linker used in this experiment contain a large number of G/C base pairs, which significantly contribute to improving stability and expression. Then, short peptides with different lengths, no linker/G4S/(G4S)2/(G4S)3/(G4S)4, were selected as flexible linkers for the construction of BsNbs. The monomer VHH genes were amplified to construct BsNb genes through overlap extension PCR and vector construction. The successful generation of the two VHH genes can be confirmed via RT-PCR and nucleic acid electrophoresis, where the molecular weight of the gene can be preliminarily determined based on the theoretical molecular weight. Additionally, the PCR product could be sent for sequencing and then aligned against a homologous sequence. Finally, antigen-antibody hybridization also can be used to detect whether the target gene is producing and translating into protein. For the first round of PCR, primers with different linkers were utilized to amplify the anti-carbaryl and anti-1-naphthol VHH, resulting in the generation of DNA bands measuring approximately 400 bp (Figure 1a, b). Subsequently, the obtained VHH genes were employed in an overlap extension process. The annealing temperature was optimized, and the results showed no non-specific amplification (Figure S2a). Thus, the usual annealing temperature of 60 °C was chosen for overlap extension PCR. This procedure yielded DNA bands of approximately 400 bp for the intermediate product and 800 bp for the overlap extension product (Figure 1c, d). The latter was purified for the subsequent vector construction.
Figure 1.
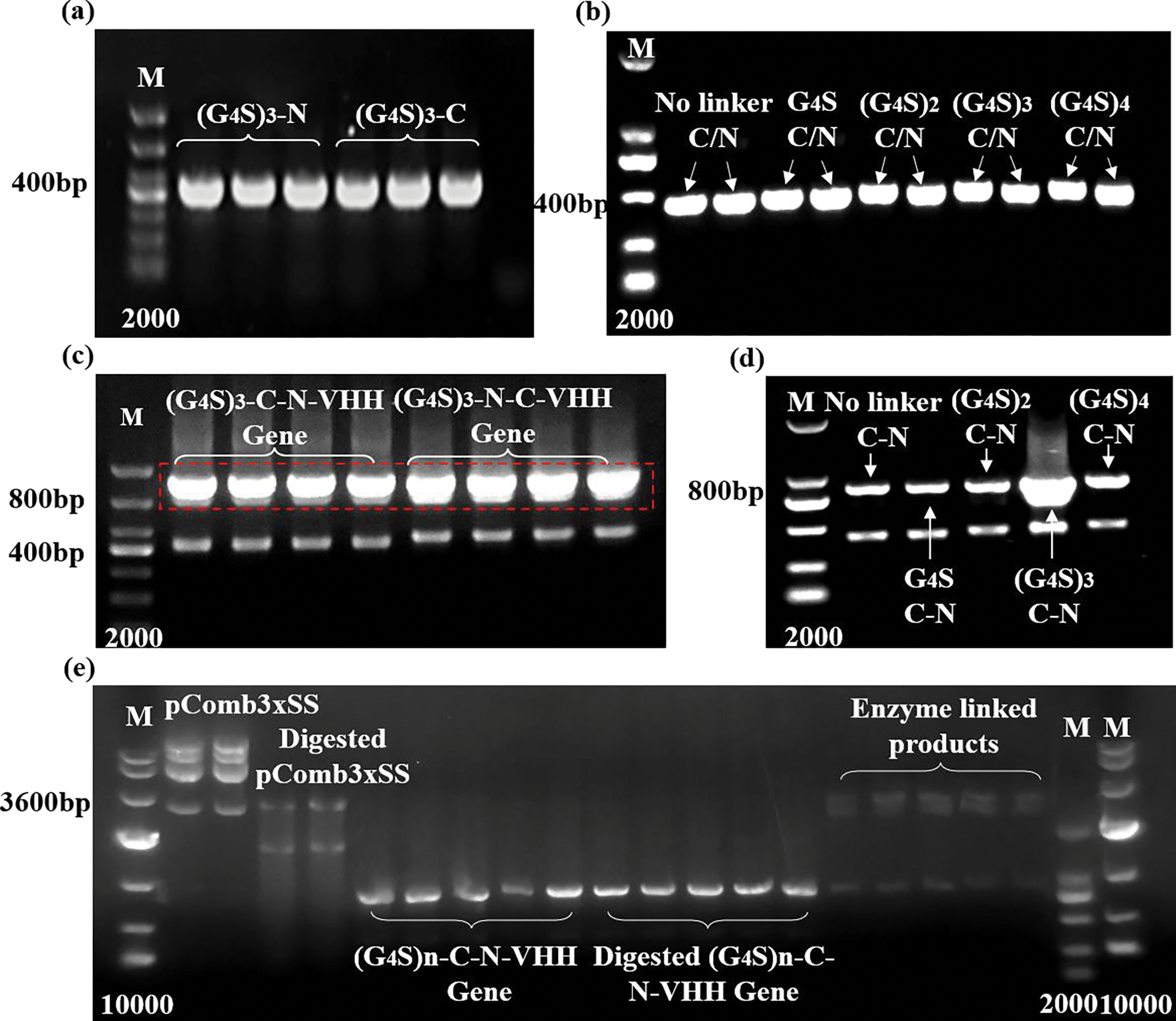
Vector construction. (a-b) The first round of PCR amplification of target fragments with (G4S)3-N/C genes and different linkers; (c-d) The second round of PCR amplification of recombinant gene fragments with different junction sites and linker lengths; (e) The results of enzyme digestion and ligation of empty plasmid and target genes.
Enzyme digestion of the empty plasmid pcomb3xSS was performed after PCR amplification and plasmid extraction. Gel purification yielded the target band of approximately 3600 bp (Figure 1e). Following enzyme digestion of the target fragments, no significant changes were observed due to the minimal difference in weight before and after digestion. After enzyme ligation, the chemically competent cells E. coli DH5a were transformed with recombinant plasmid. Both (G4S)3-C-N-VHH and (G4S)3-N-C-VHH were successfully obtained with the correct sequence (Figure S2b, c). On the LB plate, only strains containing the G4S and (G4S)3 linkers showed monoclonal growth, while strains without a linker or with (G4S)2 or (G4S)4 linkers exhibited difficulty in growth. Based on these results, the codons composing the peptides were re-optimized, followed by re-running the experiment. However, similar results were obtained, indicating the success of the enzyme digestion and ligation processes. The observed preference for growth in strains containing G4S or (G4S)3 linkers may be attributed to the correct folding of the translated proteins. Performing colony PCR on these monoclonal strains (Figure S2b), 5 samples showed an 800bp band (4 of G4S and 1 of (G4S)3), and only 2 samples showed the correct protein sequence upon sequencing. Notably, the protein fragment of G4S-C-N-VHH4 consisted of a total of 270 amino acids (Figure S3), while (G4S)3-C-N-VHH contained 280 amino acids (Figure S2d).
Following the acquisition of the target plasmid, the expression of BsNbs was achieved using the bacterial expression system. Subsequently, the expressed BsNbs were purified using a nickel column. The expression yields of G4S-C-N-VHH and (G4S)3-C-N-VHH were 10.13 mg/L and 8.45 mg/L, respectively (Figure S4a), which were higher than that of (G4S)3-N-C-VHH (1.29 mg/L). This difference in yield may be due to the fact that proteins with long linkers are easily influenced by proteases, leading to a reduction in actual yield, while proteins with short linkers can overcome this influence. The molecular weight of BsNb was approximately 35 kDa, as shown in Figure S4b. Whereas, there were several other molecular weights of proteins present, which may be attributed to the existence of other promoters. In general, the translation of proteins requires promoters. Since pComb3xSS contains an additional promoter, it can result in the production an unexpected protein with molecular weight of 25–30 kDa and fewer histidine (Figure S4b). When the low-concentration imidazole dosage used for purification was insufficient, it would lead to incomplete impurity removal. However, after washing impurities with the same amount of imidazole, it was found that the molecular weight of the residual G4S-C-N-VHH hybrid protein smaller compared to the others. This also indicated that the expression of this hybrid protein in G4S-pComb3xSS was relatively low, which may be affected by the length of the linker. Moreover, the expression of BsNb with a long linker was less than others, suggesting that other proteins with smaller molecular weight were dominant. As a result, the target protein band appeared slightly fuzzy. Nevertheless, the result of western blot in Figure S4c showed that only the 35 kDa band existed binding specificity among all the protein. This demonstrated the successful preparation of specific BsNb.
3.2. Effect of Junction Sites and Linker Length on Assay Sensitivity
Actually, the sequence order of fusion protein genes is very important. The N-terminal or C-terminal of Nb is always involved in the formation of active sites. It is necessary to avoid connecting these sites with other proteins to avoid affecting the function of the fusion protein after expression. In this section, the sensitivity of (G4S)3-C-N-VHH/(G4S)3-N-C-VHH was evaluated (Figure 2a–c), and different ligation sites resulted in different nanobody activities. The results in Figure 2b and c showed that (G4S)3-N-C-VHH had better antigen binding activity. Whereas, there was no significant improvement in sensitivity to carbaryl. In contrast, the IC50 values for (G4S)3-C-N-VHH were lower, indicating a 2-fold increase in sensitivity to 1-naphthol. Based on these findings, (G4S)3-C-N-VHH was selected to observe the effect of linker lengths.
Figure 2.
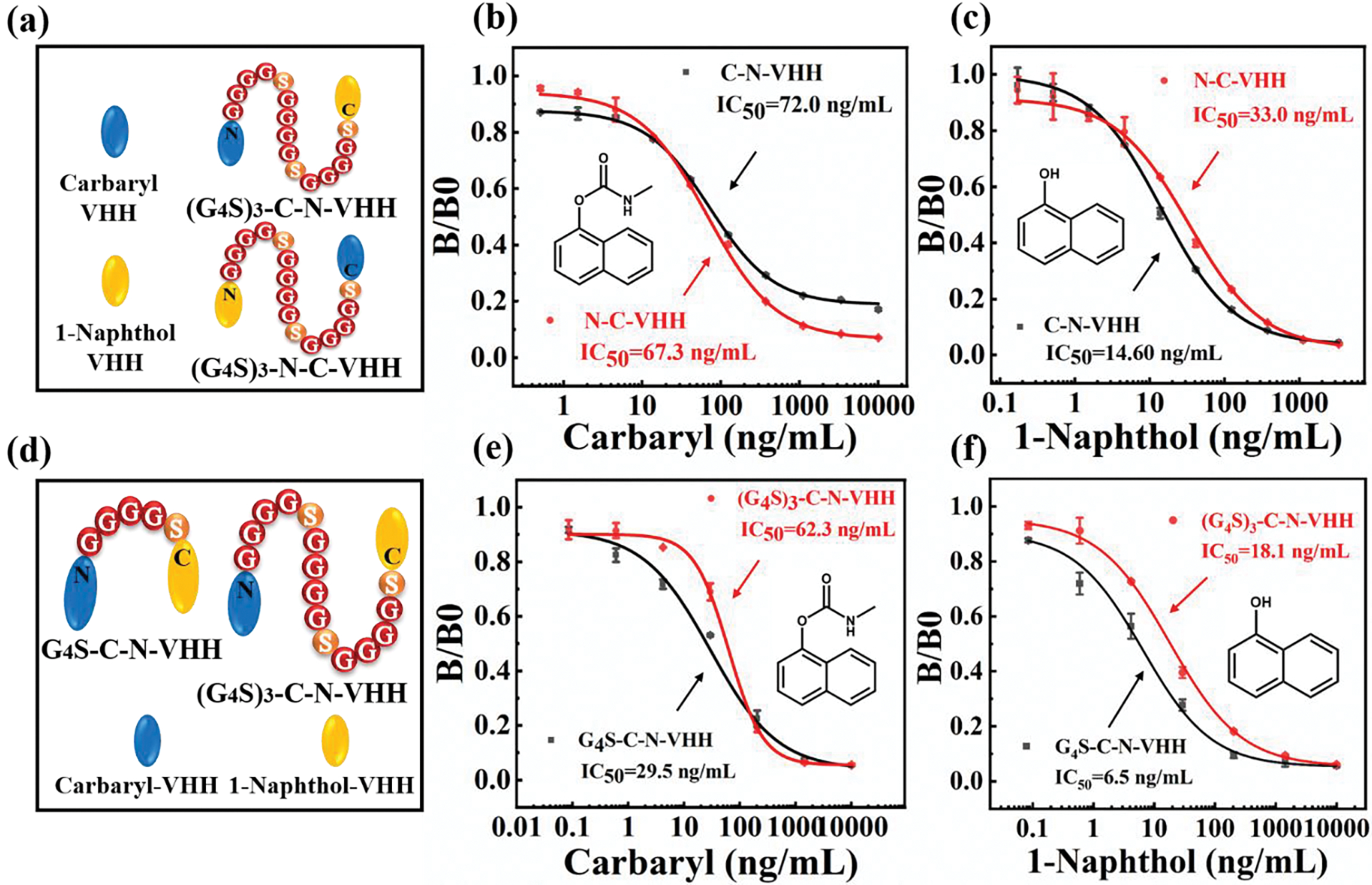
Effect of C/N junction sites and linker lengths on assay sensitivity. (a, d) Display of different junction sites and linker lengths; (b-c) The effect of linker junction sites for BsNbs against carbaryl and 1-naphthol; (e-f) The effect of different linker lengths for BsNbs against carbaryl and 1-naphthol.
Normally, two protein fragments need to be joined by a proper peptide in order to reduce steric hindrance. This may also be the main reason for the failure of plasmid construction without linkers. As a matter of fact, the activity and stability of BsNbs largely depends on the type/length of linkers, codons and metal ions. Among these factors, the amino acid composition of peptide chain will affect the flexibility and hydrophilicity of the linker, thereby affecting the functional activity. Notably, linkers with low electricity and hydrophobicity, (G4S)n, possess great flexibility and can effectively separate the two protein molecules. This allows the fusion protein to fold into its natural conformation and exhibit proper protein activity. Additionally, glycine and serine are ionizable in water with strong hydrophilicity, enabling the peptide segment to promote the stable existence of the protein in an aqueous solution. Nevertheless, if the linker is too short, the decrease in steric hindrance may not be significant, while if it is too long, the binding efficiency of BsNb to antigen may be affected due to conformational changes, or even lead to misfolding and susceptibility to protease hydrolysis. Hence, although the enzyme linkage results showed successful vector construction (Figure 1e), the strain could not grow when the plasmids without a linker/containing (G4S)2 and (G4S)4 were constructed in the same batch of recombinant plasmids. This suggests that these three linkers were not conducive to strain growth or protein expression. Thus, a further comparison of the binding activity was made between G4S-C-N-VHH and (G4S)3-C-N-VHH (Figure 2d–f). From the results of Figure 2e, f, G4S-C-N-VHH showed higher sensitivity to both carbaryl and 1-naphthol than (G4S)3-C-N-VHH. It had lower IC50 values of 29.5 ng/mL and 6.5 ng/mL in buffer system, respectively, and wider detection ranges. Based on this observation, we hypothesized that a five-residue length linker would be more suitable for construction of BsNbs. To further investigate the influence of linker length on the structure of BsNbs and the potential impact mechanism, molecular simulation was performed.
3.3. Model building and Molecular Simulations
To gain insights into the structural differences between G4S-C-N-VHH and (G4S)3-C-N-VHH, 3-D models of both BsNbs were generated using AlphaFold2 (Figure S5a and b). The models revealed that the C-chains of the two models largely overlapped, exhibiting only one difference arising in C-CDR2. However, significant differences were observed in the conformation of the N-chains. These conformational variances originated from the linker, which differed in length, and subsequently affected the conformation of the N-chains. Thus, it became evident that different linker lengths would influence the conformation of BsNbs, primarily affecting the conformation of N-chains. To validate the credibility of the two models, Ramachandran plot maps were obtained, showing that more than 95% of the residues located within the allowed region (Figure S5c and d). In addition, the 3D-1D scores for both BsNbs mostly exceeded 0.2, with 92.86%–94.07% residues (Figure S5e). These results affirmed the credibility of the models. Subsequently, molecular simulation was conducted to explore the location of the binding pocket in different BsNbs.
The flexible pockets of functional proteins are usually active binding areas. Due to their high flexibility, they can adapt to various insertion directions of ligands with different tightness, thereby resulting in various docking results and binding energies. The optimal binding conformation occurs when the small molecular ligand is in the mode that maximizes the binding energy, indicating the most stable binding conformation. Consequently, the docking model with the maximum binding energy was selected for analysis based on the scoring order by the software (Xiang et al., 2021). All the docking results above suggested that the affinity of BsNbs to ligands was related to the docking region and the depth of the docking pocket (Figure 3). Surprisingly, the caudal region of carbaryl inserted into the pocket on C-CDR2 region of G4S-C-N-VHH, therefore, forming 3 shorter hydrogen bonds with the Nitrogen and Oxygen atoms. The bond lengths of N-Ser52, O-Arg53 and O-Leu54 were 2.71 Å, 2.21 Å and 2.15 Å, respectively. However, the pocket formed by the C-CDR3 region of (G4S)3-C-N-VHH was notably larger and shallower. Theoretically, the expansion of the protein pocket often leads to a looser combination with ligands and a decrease in binding energy, resulting in extremely low affinity (Chen et al., 2018). Here, the docking results indeed showed that it just formed 2 hydrogen bonds with Asp106 and Thr108, the bond lengths of which were longer. Thus, G4S-C-N-VHH showed a stronger affinity to carbaryl than (G4S)3-C-N-VHH. As for 1-naphthol, the adsorption sites of 1-naphthol for BsNbs are mainly Oxygen atoms (Sun, 2023). Specifically, the Oxygen atom on benzene ring formed a strong hydrogen bond (~2.15 Å) with Cys175 in the N-CDR3 region of G4S-C-N-VHH. Additionally, there were 12 hydrophobic interactions with amino acids formed by 1-naphthol within a 4 Å range. In contrast, the docking pocket formed by N-CDR1 region of (G4S)3-C-N-VHH was noticeably shallower. This limits the interaction of 1-naphthol with the surface, resulting in a relatively weak interaction force indicated by a long hydrogen bond of approximately ~3.20 Å. It can be speculated that the difference in affinity for 1-naphthol between G4S-C-N-VHH and (G4S)3-C-N-VHH was influenced by the linker length affecting the conformation. To further determine the stronger binding of G4S-C-N-VHH to 1-naphthol, MD was performed, followed by calculation of the binding free energy.
Figure 3.
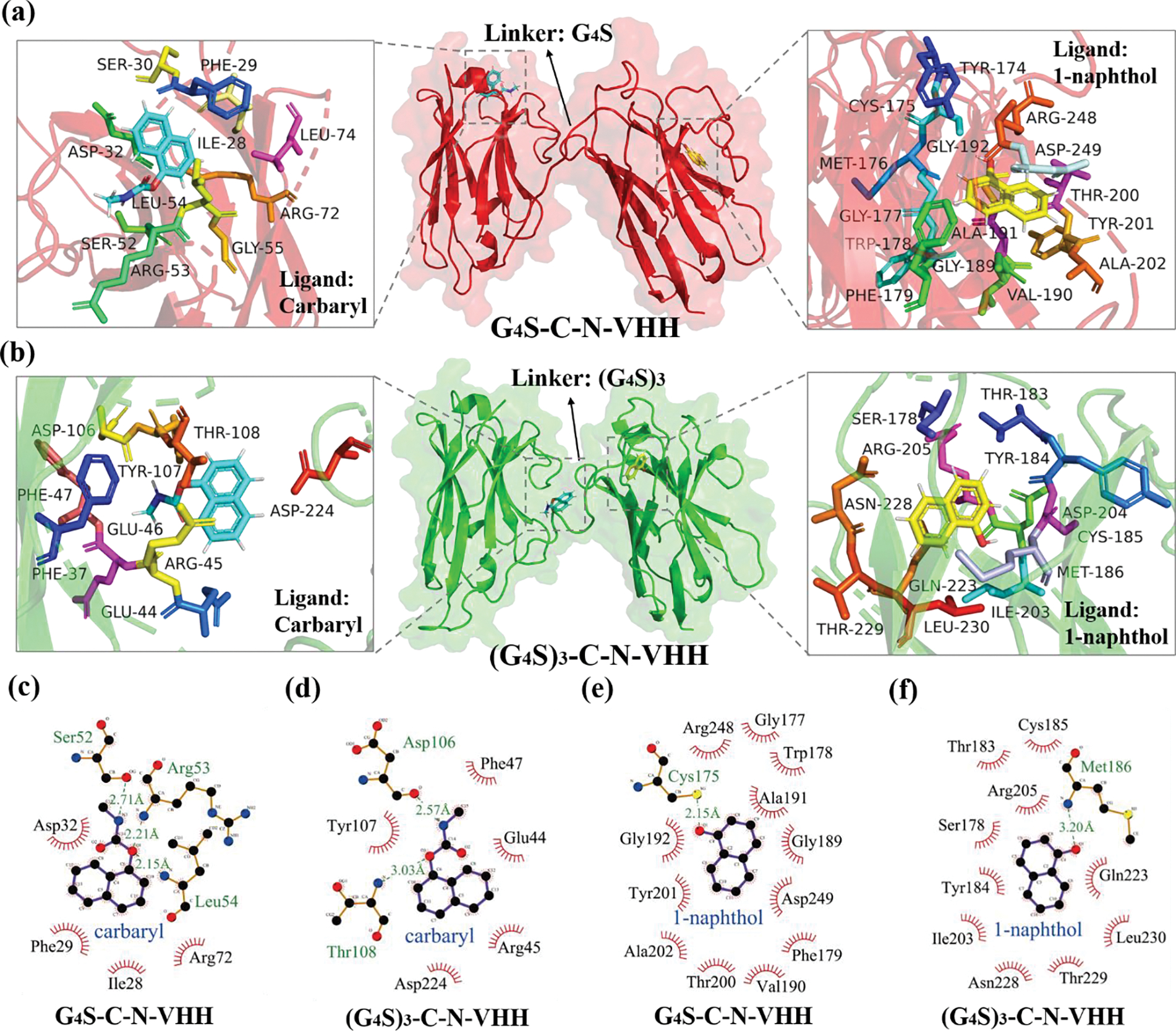
Molecular docking and interaction force analysis of G4S-C-N-VHH and (G4S)3-C-N-VHH with carbaryl and 1-naphthol. (a) Molecular docking of G4S-C-N-VHH; (b) Molecular docking of (G4S)3-C-N-VHH; (c) Interaction analysis between G4S-C-N-VHH and carbaryl; (c) Interaction force between G4S-C-N-VHH and 1-naphthol; (e) Interaction force analysis of (G4S)3-C-N-VHH.
As shown in Figure S6, the ligand-receptor complexes reached an equilibrium state after 10 ns (G4S-C-N-VHH)/50 ns ((G4S)3-C-N-VHH) of MD simulation. Herein, the calculated root-mean-square deviation (RMSD) values on binding region had fluctuated within the range of 2 Å, indicating a stable binding and dynamic equilibrium (Xiang et al., 2021). The overall root-mean-square fluctuation (RMSF) value of G4S-C-N-VHH complex was slightly lower than that of the (G4S)3-C-N-VHH complex (Figure S6b), suggesting that G4S-C-N-VHH complex was more stable and had lower degrees of freedom (Xiang et al., 2021). Furthermore, the smaller radius of gyration of the G4S-C-N-VHH (Figure S6c) complex indicated a tighter binding between G4S-C-N-VHH and the ligand (Yang et al., 2019). The free energy landscapes with the lowest energy point (the purple region) demonstrated that the system had reached an equilibrium steady state (Figure S6e–f), indicating the credibility of the simulation under this stability condition. Finally, the enthalpy change (ΔH) and Gibbs free energy variation (ΔG) were further calculated from the MD simulation results (Table 1).
Table 1.
Gibbs free energy parameters via MD.
| System | △H (kJ/mol) | T△S (kJ/mol) | △G (kJ/mol) | Ki (nM) | Affinity |
|---|---|---|---|---|---|
|
| |||||
| G4S+1-naphthol | −72.179 | −4.226 | −67.953 | 1.245×10−3 | Very strong |
| (G4S)3+1-naphthol | −64.395 | −20.211 | −44.184 | 18.17 | Strong |
The (ΔG) of G4S-C-N-VHH complex with 1-naphthol was −67.953 kJ/mol, lower than that of the (G4S)3-C-N-VHH complex (−44.184 kJ/mol), confirming that G4S-C-N-VHH binds more tightly to the ligand and exhibits greater stability. Meanwhile, the lower Ki value of G4S-C-N-VHH indicated a stronger affinity for 1-naphthol. In conclusion, these results suggested that G4S-C-N-VHH and 1-naphthol possess functional groups with stronger interactions. Different linker lengths indirectly influence the affinity of BsNbs by affecting their conformation. The greater stability and sensitivity of G4S-C-N-VHH were further demonstrated, justifying its selection for stability testing.
3.4. Stability of BsNb
Thermal stability of Nb is an important performance index storage stability. High thermal stability of Nb also means that they are less affected by temperature changes, especially temperature rise. Thus, improving thermal stability of nanobodies can prolong their storage time at room temperature or higher temperature. It can also be used in samples that heat treatment without the need for cooling, effectively maintaining their activity and reducing processing time, thereby improving the utilization value. Inspired by the impact of incubation temperature on the sensitivity of BsNb, further exploration was conducted to understand the influence of different incubation time at different incubation temperatures.
As shown in Figure 4a, b, the A450 nm of BsNb obtained at 50 °C was close to those obtained at low temperature, indicating that BsNb had a certain tolerance to high temperature. Interestingly, it only took 20 minutes to reach equilibrium at 50 °C, albeit with a slight decrease in sensitivity, whereas it took over 40 min at 4 °C to reach the binding equilibrium. These results suggested a positive correlation between temperature and binding affinity within 50 °C with a moderate increase in temperature promoting the binding rate of nanobodies. Additionally, it was observed that the equilibrium time required for incubation at higher temperature was shorter within a certain temperature range. However, when the incubation temperature exceeded 30 min, the binding affinity decreased to some extent. To further understand the temperature tolerance of BsNb, the protein solutions were heated at different temperatures for 10 min, and the effect of temperature on their secondary structure was explored using Circular Dichroism (Figure 4c). The circular dichroism spectra showed a trough signal at 216 nm, indicating that the secondary structure of BsNb remained unaffected when the temperature was below 50 °C. However, when the temperature rose above 75 °C, the wave valley exhibited a slight upward shift, suggesting a subtle change in the secondary structure with a decrease in the content of α-helix and an increase in the content of anti-parallel chains. Notably, when the samples were treated at 100 °C, a visible leftward shift in the trough occurred, accompanied by the appearance of a signal at 215 nm, indicating significant structural changes in the protein. Furthermore, we observed the detailed changes in the secondary structure at 216 nm with increasing temperature to compare the thermal stability of each monomer with BsNb (Figure 4d). Surprisingly, it was found that the secondary structure of BsNb remained relatively stable when the temperature was increased up to 90 °C. In contrast, the monomers showed a significant decrease in stability below 70 °C. The denaturation temperature of the carbaryl nanobody was determined to be 60 °C, while that of 1-naphthol nanobody was 65 °C. These findings indicated that the BsNb obtained in this work exhibited higher thermal stability, maintaining its secondary structure up to 90 °C, which can be attributed to the crosslinking of nanobodies by the linker, increasing the overall structure stability (Sun, 2023).
Figure 4.
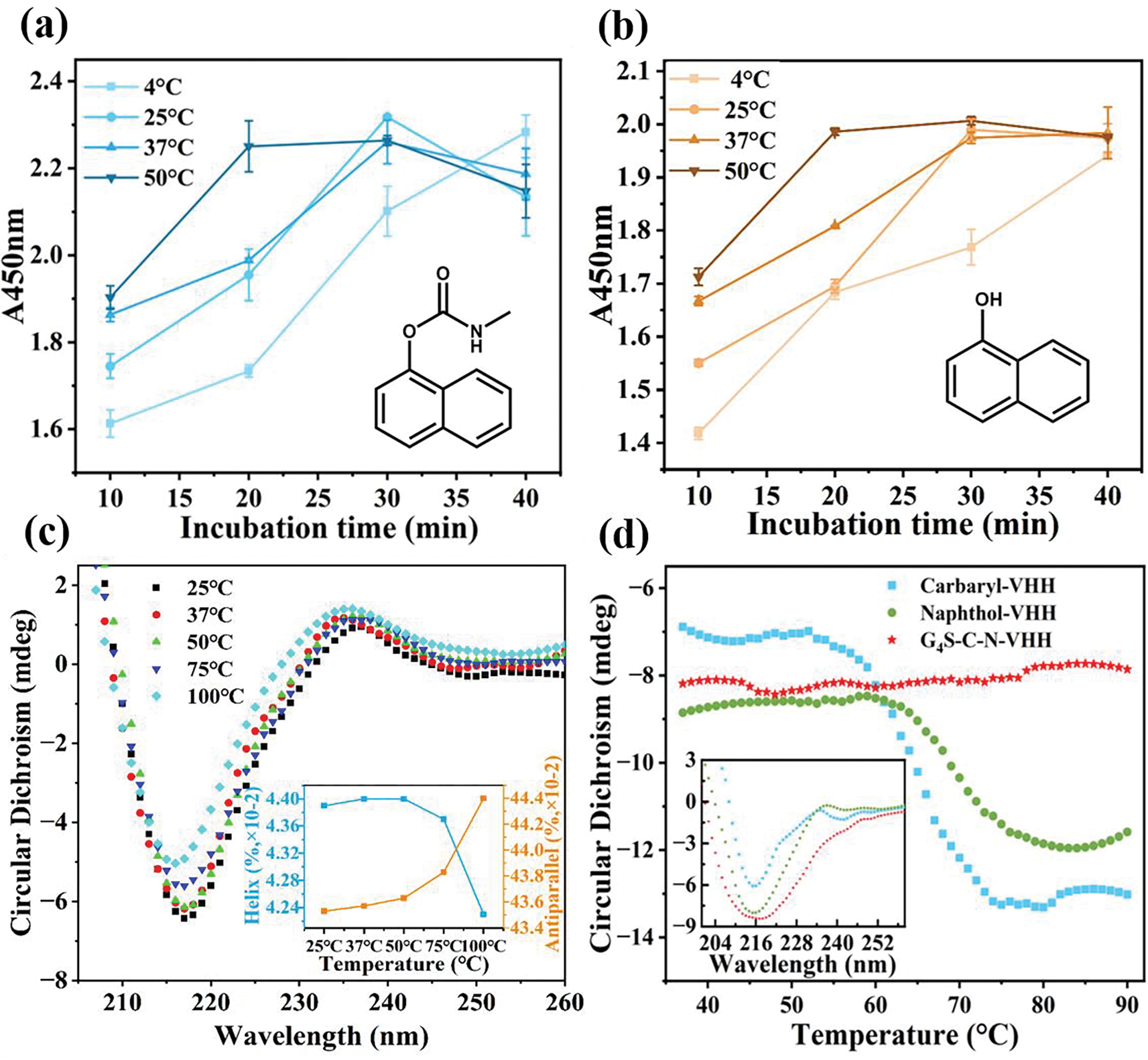
Thermal stability of BsNb. (a-b) Equilibrium time of BsNb binding to antigen at different temperatures; (c-d) Circular Dichroism results for thermal stability of BsNb.
Through the organic stability test (Figure S7), it was observed that BsNb maintained its binding activity in solvents containing no more than 10% methanol, 20% acetonitrile or 10% acetone solvent. Consequently, in sample pretreatment, it is important to combine with nitrogen drying to minimize the concentration of organic solvents. Regarding the effect of pH values (Figure S8), different pH values had varying influences on the drug sensitivity of BsNb. In most cases, the sensitivity of the Nbs tends to decrease under acidic or alkaline conditions. However, in this experiment, the BsNb exhibited some advantages under acidic or alkaline conditions, particularly in the detection of carbaryl or 1-naphthol, where its sensitivity remained relatively stable under alkaline conditions (Figure S8). The specific folding mode of BsNb may contribute to this phenomenon, though further investigation is required to fully understand its underlying mechanism.
3.5. Optimization of Bic-ELISA and Sample Validation
Herein, we initially investigated the optimization of incubation temperature to enhance sensitivity. The results (Figure S9) demonstrated a decrease in the antigen binding capacity with the decreased in temperature, while the sensitivity to carbaryl/1-naphthol increased. This may be due to the slowed binding speed of BsNb at lower temperatures, increasing the opportunity for small-molecule drugs to bind. Additionally, the pocket cavity within BsNb’s binding site may slightly shrink at lower temperatures, allowing small-molecule drugs to more easily enter and bind to hapten with BSA. Notably, the BsNb maintained its monomer binding activity with the IC50 values of 18.8 ng/g for carbaryl and 6.3 ng/g for 1-naphthol at an incubation temperature of 25 °C. Considering cost-effectiveness, room temperature was chosen for subsequent reactions. Regarding the optimization of coating hapten concentration (Figure S10), the obtained Amax/IC50 values of different concentrations suggested that the optimal reaction concentration was 250 ng/mL for both carbaryl or 1-naphthol.
Subsequently, soil samples from farmland and grain rice were selected for the detection of carbaryl and 1-naphthol pollutants. The results of matrix effects (Figure S11) showed that a 1:20 dilution with PBS for carbaryl and 1:40 dilution with PBS for 1-naphthol analysis could effectively eliminate the matrix effects. The spiked sample extraction was then measured by ic-ELISA under the established conditions. Moreover, the same spiked sample extraction for carbaryl and 1-naphthol was also analyzed using HPLC-MS/MS (Table S3). Finally, the LC-MS/MS detection limits obtained were 1.7 ng/g for carbaryl and 1.3 ng/g for 1-naphthol. The RR of carbaryl and 1-naphthol in soil and rice using ELISA ranged from 80.0 to 112.7% and from 76.5 to 110.8%, respectively, with a CV of less than 15%. Additionally, the analysis results of the multianalyte ic-ELISA were in good agreement with that from HPLC-MS/MS (Table S4).
Compared with other detection methods (Table 2), our BsNb demonstrated the ability to recognize two antigens simultaneously while maintaining sensitivity. Although the polyclonal antibody obtained by Sun (Sun et al., 2010) achieved a sensitivity of 0.3 ng/g for carbaryl, its narrow detection limit restricts its practical applications. Moreover, its cross-reactivity with 1-naphthol, with an IC50 of 600 ng/g, indicates that its specificity is not superior to other methods. Regarding 1-naphthol detection, the multianalyte ic-ELISA developed in this study exhibited a lower detection limit, a wider linear range and excellent thermal stability compared to the pAb-ELISA developed by Kramer (Kramer et al., 1994), the fluorescent sensor by Qin and Yan (Qin and Yan, 2018), the mAb-ELISA by Chen (Chen et al., 2020) and the Nb-ELISA by Chen (Chen et al., 2022). Overall, this work provides valuable insights for the detection and related research of small molecules and multiple analytes. However, further improvements are needed in terms of sensitivity and simultaneous detection methods although the whole construction process is fast and easy to operate. In the future, new detection methods of BsNb will continue to be researched to fully exploit its advantages in molecular technology, especially the development of dual-channel mix-and-read detection method like dual-channel fluorescence resonance energy transfer. Furthermore, BsNb may potentially be used in chromatographic column to eliminate the cross-interference from other drugs or for sample pretreatment, leveraging its high specificity and stability.
Table 2.
The comparison of ELISA detection methods for carbaryl or 1-naphthol.
| Ab | Substrate | Sensitive antigen | IC50 (ng/mL) | Linear Range (ng/mL) | LOD (ng/mL) | Thermal stability (°C) | Reference |
|---|---|---|---|---|---|---|---|
|
| |||||||
| pAb | TMB | carbaryl | ND | ND | 10 | ND | (Wang et al., 2005) |
| pAb | TMB | carbaryl/2% of cross reactivity to 1-naphthol | 0.3/600 | 0.1–2 | 0.015 | ND | (Sun et al., 2010) |
| ScFv-AP | PNPP | carbaryl | 15 | 3.5–76 | 1.6 | ND | (He et al., 2019) |
| Nb | TMB | carbaryl | 5.4 | 0.8–38 | 0.3 | 60 | (Liu et al., 2019) |
| pAb | TMB | 1-naphthol | 72 | 10–1000 | 10 | ND | (Kriimer et al., 1994) |
| mAb | TMB | 1-naphthol | 11.2 | 4.0–31.4 | 2.2 | ND | (Chen et al., 2020) |
| Nb | TMB | 1-naphthol | 8.3 | 2.7–25.6 | 1.4 | 65 | (Chen et al., 2022) |
| ALP-Nb | pNPP | 1-naphthol | 7.6 | 0.5–107.8 | 0.1 | ND | (Chen et al., 2022) |
| BsNb | TMB | carbaryl/1-naphthol | 18.8/6.3 |
2.1–270.9
1.1–112.0 |
0.8/0.4 | 90 | This work |
4. Conclusions
In this work, we successfully demonstrated a reliable ic-ELISA based on the anti-carbaryl and anti-1-naphthol BsNb fusion protein connected by a short peptide for the simultaneous detection of carbaryl and 1-naphthol in soil and rice samples. The comprehensive effect of the BsNb constructed with G4S was better than that of BsNbs constructed with other lengths of short peptides. Molecular docking analysis revealed that linker lengths affected the conformation of BsNbs, thereby influencing sensitivity. MD further confirmed the lower binding free energy and higher affinity of G4S-C-N-VHH. Consequently, the BsNb with G4S was chosen for performance identification and was found that retained better antigen-binding capacity, showing good specificity under both acidic or alkaline conditions. Additionally, the BsNb maintained comparable binding activity in organic solvents that did not exceed 10% methanol, 20% acetonitrile or 10% acetone. In terms of thermal stability, the BsNb showed high-temperature tolerance, with a denaturation temperature increased by at least 20 °C compared to the monomers. Moreover, as the temperature decreased, the antigen-binding activity of the BsNb showed no significant change, while the sensitivity increased. Based on these results, the multianalyte ic-ELISA was successfully used to analyze carbaryl and its metabolite 1-naphthol in real samples. The assay demonstrated appropriate specificity, recovery, and accuracy in different seasons with varying temperature. The results of proposed method showed a good correlation with those obtained by LC-MS/MS.
Supplementary Material
Highlights.
Carbaryl and its metabolite 1-naphthol are harmful to environment and human health.
Bispecific nanobodies were constructed to simultaneously recognize them.
The linker effect was firstly revealed via molecular simulation and ELISA.
It is valuable for immunoassay application in environment and agriculture.
Acknowledgments
This work was supported by the National Key Research and Development of China (2019YFE0116600), Guangdong Provincial Science and Technology Project, China (2022A0505050061), Guangdong Basic and Applied Basic Research Foundation (2021A1515110513) and the Guangdong Special Support Program, China (2019TX05N052). Partial support was provided by NIH-NIEHS (RIVER Award) R35 ES030443–01 and NIH-NIEHS (Superfund Award) P42 ES004699.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Materials
Several relative materials and methods; the UV wavelength of carbaryl-BSA/1-naphthol-BSA; primers design; affinity constant checkerboard assay curves; verification of target vector construction; structure models of BsNbs; results of MD; identification of organic tolerance and pH tolerance; effects of coating hapten; optimization of coating hapten concentrations for ELISA; parameters of LC-MS/MS. Supplementary data to this article can be found online at xxx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bai Z, Wang J, Li J, Yuan H, Wang P, Zhang M, Feng Y, Cao X, Cao X, Kang G, de Marco A, Huang H 2023. Design of nanobody-based bispecific constructs by in silico affinity maturation and umbrella sampling simulations. Comput. Struct. Biotechnol. J. 21, 601–13. 10.1016/j.csbj.2022.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathula NV, Bommadevara H, Hayes JM 2021. Nanobodies: the future of antibody-based immune therapeutics. Cancer Biother. Radio. 36(2), 109–22. 10.1089/cbr.2020.3941. [DOI] [PubMed] [Google Scholar]
- Bhardwaj J, Kim M, Jang J 2020. Rapid airborne influenza virus quantification using an antibody-based electrochemical paper sensor and electrostatic particle concentrator. Environ. Sci. Technol. 54(17), 10700–12. 10.1021/acs.est.0c00441. [DOI] [PubMed] [Google Scholar]
- Chen J, Huang C, Zhao W, Ren J, Ji F, Jia L 2022. Snoop ligase enables highly efficient generation of C-C-linked bispecific nanobodies targeting TNF-α and IL-17A. Bioconjugate Chem. 33, 1446–55. 10.1021/acs.bioconjchem.2c00143. [DOI] [PubMed] [Google Scholar]
- Chen Z, Huang Z, Huang S, Zhao J, Sun Y, Xu Z, Liu J 2021. Effect of proteins on the oxidase-like activity of CeO2 nanozymes for immunoassays. The Analyst. 146(3), 864–73. 10.1039/D0AN01755H. [DOI] [PubMed] [Google Scholar]
- Chen Z, Liu X, Xiao Z, Fu H, Huang Y, Huang S, Shen Y, He F, Yang X, Hammoc k BD, Xu Z 2020. Production of a specific monoclonal antibody for 1-naphthol based on novel hapten strategy and development of an easy-to-use ELISA in urine samples. Ecotox. Environ. Safe. 196, 110533. 10.1016/j.ecoenv.2020.110533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wu H, Shen Y, Wang H, Zhang Y, Hammock BD, Li Z, Luo L, Lei H, Xu Z 2022. Phosphate-triggered ratiometric fluoroimmunoassay based on nanobody-alkaline phosphatase fusion for sensitive detection of 1-naphthol for the exposure assessment of pesticide carbaryl. J. Hazard. Mater. 424(Pt C), 127411. 10.1016/j.jhazmat.2021.127411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang X, Wang B, Rao M, Wang H, Lei H, Liu H, Zhang Y, Sun Y, Xu Z 2018. Production of antigen-binding fragment against O, O-diethyl organophosphorus pesticides and molecular dynamics simulations of antibody recognition. Int. J. Mol. Sci. 19(5), 1381. 10.3390/ijms19051381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhang Y, Chen J, Lin Z, Wu M, Shen Y, Luo L, Wang H, Wen X, Hammock BD, Lei H, Xu Z 2022. Production and characterization of biotinylated anti-fenitrothion nanobodies and development of sensitive fluoroimmunoassay. J. Agr. Food Chem. 70(13), 4102–11. 10.1021/acs.jafc.2c00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Hao Y, Bao F, Zhang H, Liu Y, Ao K, Fu S, Wu Q, Wang Z 2022. Preparation and identification of a single domain antibody specific for adenovirus vectors and its application to the immunoaffinity purification of adenoviruses. AMB Express. 12(1). 10.1186/s13568-022-01422-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-Chen M, Rambla-Alegre M, Durgbanshi A, Bose D, Mourya SK, Esteve-Romero J, Carda-Broch S 2012. Micellar liquid chromatographic determination of carbaryl and 1-naphthol in water, soil, and vegetables. Int. J. Anal. Chem. 2012,1–7. 10.1155/2012/809513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ćwieląg-Piasecka I, Debicka M, Medyńska-Juraszek A 2021. Effectiveness of carbaryl, carbofuran and metolachlor retention in soils under the influence of different colloid. Minerals. 11(9),924. 10.3390/min11090924. [DOI] [Google Scholar]
- Codex Alimentarius Commission pesticide index. 2004. No. 8. http://www.fao.org/fao-who-codexalimentarius/codex-texts/dbs/pestres/pesticides/en/.2023/04/28.
- Ding Y, Song X, Chen J 2019. Analysis of pesticide residue in tomatoes by carbon nanotubes/β-cyclodextrin nanocomposite reinforced hollow fiber coupled with HPLC. J. Food Sci. 84(6),1651–9. 10.1111/1750-3841.14640. [DOI] [PubMed] [Google Scholar]
- European Commission (EU). 2014. Carbaryl. No. 1096/2014. https://ec.europa.eu/food/plant/pesticides/eu-pesticides-database/start/screen/mrls/searchpr. 2023/06/26. [Google Scholar]
- States Environmental Protection Agency (EPA). 2014. Code of Federal Regulations. Protection of Environment. 24(Pt 180), No. §180.169, 169–171. [Google Scholar]
- Fan K, Jiang B, Guan Z, He J, Yang D, Xie N, Nie G, Xie C, Yan X 2018. Fenobody: a ferritin-displayed nanobody with high apparent affinity and half-life extension. Anal. Chem. 90(9), 5671–7. 10.1021/acs.analchem.7b05217. [DOI] [PubMed] [Google Scholar]
- Fernández-Quintero ML, Kroell KB, Grunewald LJ, Fischer AM, Riccabona JR, Lied l KR 2022. CDR loop interactions can determine heavy and light chain pairing preferences in bispecific antibodies. mAbs. 14(1),2024118. 10.1080/19420862.2021.2024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Wang Y, Xiao Z, Wang H, Li Z, Shen Y, Lei H, Sun Y, Xu Z, Hammock BD 2020. A rapid and simple fluorescence enzyme-linked immunosorbent assay for tetrabromobisphenol A in soil samples based on a bifunctional fusion protein. Ecotox. Environ. Safe. 188, 109904. 10.1016/j.ecoenv.2019.109904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GB2763–2021[S]. 2021. National food safety standard-Maximum residue limits for pesticides in food. No. 4.235. http://down.foodmate.net/standard/sort/3/97819.html. 2023/06/26 [Google Scholar]
- GB/T 25782–2010 1-Naphthol[S]. 2010. National Standards of the People’s Republic of China. https://std.samr.gov.cn/gb. 2023/04/28. [Google Scholar]
- Gu L, Zou Y, Li Y, Zeng K, Zhu N, Zhu F, Gyimah E, Yakubu S, Meng H, Zhang Z 2020. High-throughput chemiluminescence immunoassay based on Co2+/hemin synergistic catalysis for sensitive detection tetrabromobisphenol A bis (2-hydroxyethyl) ether in the environments. Sci. Total Environ. 714, 136880. 10.1016/j.scitotenv.2020.136880. [DOI] [PubMed] [Google Scholar]
- Guo Y; Liu S; Gui W; Zhu G 2009. Gold immunochromatographic assay for simultaneous detection of carbofuran and triazophos in water samples. Anal. Biochem. 389 (1), 32–39. 10.1016/j.ab.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R 1993. Naturally occurring antibodies devoid of light chain s. Nature. 363, 446–448. 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- He J, Tao X, Wang K, Ding G, Li J, Li Q, Gee SJ, Hammock BD, Xu T 2019. One-step immunoassay for the insecticide carbaryl using a chicken single-chain variable fragment (scFv) fused to alkaline phosphatase. Anal. Biochem. 572, 9–15. 10.1016/j.ab.2019.02.022. [DOI] [PubMed] [Google Scholar]
- He J, Chen X, Shi S, Tang F, Huo N, Gu S 2021. Multivalent nanobody as capture antibody-based enzyme linked immunosorbent assay for detection of 3-phenoxybenzoic acids in urine. Anal. Biochem. 632, 114390. 10.1016/j.ab.2021.114390. [DOI] [PubMed] [Google Scholar]
- HJ960–2018[S]. 2018. National Environmental Protection Standards of the People’s Republic of China. https://www.mee.gov.cn/.2023/04/28.
- Hu Y, Wang Y, Nie L, Lin J, Wu S, Li S, Wu J, Ji X, Lv H, Muyldermans S, Wang S 2022. Exploration of specific nanobodies as immunological reagents to detect milk allergen of β-lactoglobulin without interference of hydrolytic peptides. J. Agr. Food Chem. 70(48), 15271–82. 10.1021/acs.jafc.2c06175. [DOI] [PubMed] [Google Scholar]
- Hua X; Wang L; Li G; Fang Q; Wang M; Liu F 2013. Multi-analyte enzyme-linked immunosorbent assay for organophosphorus pesticides and neonicotinoid insecticides using a bispecific monoclonal antibody. Anal. Methods 5 (6), 1556–1563. 10.1039/c3ay26398c. [DOI] [Google Scholar]
- Jia M, E Z, Zhai F, Bing X. 2020. Rapid multi-residue detection methods for pesticides an d veterinary drugs. Molecules. 25(16), 3590. 10.3390/molecules25163590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z, Zhu Q, Jiang W, Zhou Y, Zhang M, Jiang M, Hong Q 2021. Heterologous expression and exploration of the enzymatic properties of the carbaryl hydrolase CarH from a newly isolated carbaryl-degrading strain. Ecotox. Environ. Safe. 224, 112666. 10.1016/j.ecoenv.2021.112666. [DOI] [PubMed] [Google Scholar]
- Kramer PM, Marco M, Hammock BD 1994. Development of a selective enzyme-linked immunosorbent assay for l-naphthol-the major metabolite of carbaryl (l-naphthyl N-Methylcar bamate). J. Agr. Food Chem. 42, 934–43. 10.1021/jf00040a019. [DOI] [Google Scholar]
- Li B, Qin X, Mi L 2022. Nanobodies: from structure to applications in non-injectable and bispecific biotherapeutic development. Nanoscale. 14(19), 7110–22. 10.1039/d2nr00306f. [DOI] [PubMed] [Google Scholar]
- Li F, Di L, Liu Y, Xiao Q, Zhang X, Ma F, Yu H 2019. Carbaryl biodegradation by Xylaria sp. BNL1 and its metabolic pathway. Ecotox. Environ. Safe. 167,331–7. 10.1016/j.ecoenv.2018.10.051. [DOI] [PubMed] [Google Scholar]
- Li Z, Dong J, Vasylieva N, Cui Y, Wan D, Hua X, Huo J, Yang D, Gee SJ, Hammock BD 2021. Highly specific nanobody against herbicide 2,4-dichlorophenoxyacetic acid for monitoring of its contamination in environmental water. Sci. Total Environ. 753, 141950. 10.1016/j.scitotenv.2020.141950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Zeng Y, Luo L, Xu Z, Shen Y, Wang H, Hammock BD 2022. Detection of acrylamide in foodstuffs by nanobody-based immunoassays. J. Agr. Food Chem. 70(29), 9179–86. 10.1021/acs.jafc.2c01872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sui J, Li C, Peng X, Wang Q, Jiang N, Xu Q, Wang L, Lin J, Zhao G 2022. Preparation of a nanobody specific to dectin 1 and is anti-inflammatory effects on fungal keratitis. Int. J. Nanomed. 17, 537–51. 10.2147/IJN.S338974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang K, Wu S, Wang Z, Ding G, Hao X, Li Q, Li J, Gee SJ, Hammock BD, Xu T 2019. Development of an immunoassay for the detection of carbaryl in cereals based on a camelid variable heavy-chain antibody domain. J. Sci. Food Agr. 99(9), 4383–90. 10.1002/jsfa.9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C, Dillon MJ, Kulabhusan PK, Senovilla-Herrero D, Campbell K 2022. Multiplex lateral flow assay and the sample preparation method for the simultaneous detection of three marine toxins. Environ. Sci. Technol. 56(17), 12210–7. 10.1021/acs.est.2c02339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekar L, Busch M, Valldorf B, Hinz SC, Toleikis L, Krah S, Zielonka S 2020. Biophysical and biochemical characterization of a VHH-based IgG-like bi- and trispecific antibody platform. mAbs. 12(1), 1812210. 10.1080/19420862.2020.1812210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Yan B 2018. A facile indicator box based on Eu3+ functionalized MOF hybrid for the determination of 1-naphthol, a biomarker for carbaryl in urine. Sens. Actuators B Chem. 259, 125–32. 10.1016/j.snb.2017.12.060. [DOI] [Google Scholar]
- Segovia-de Los Santos P, Quintero-Campos P, Morais S, Echaides C, Maquieira Á, Lassabe G, Gonzalez-Sapienza G 2022. Bispecific single-domain antibodies as highly standardized synthetic calibrators for immunodiagnosis. Anal. Chem. 94(2), 1342–9. 10.1021/acs.analchem.1c04603. [DOI] [PubMed] [Google Scholar]
- Shahdost-fard F, Fahimi-Kashani N, Hormozi-nezhad MR 2021. A ratiometric fluorescence nanoprobe using CdTe QDs for fast detection of carbaryl insecticide in apple. Talanta. 221,121467. 10.1016/j.talanta.2020.121467. [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Alzogaray V, Selvakumar D, Nathan S, Yoder JB, Wright KM, Klinke S, Nwafor JN, Labanda MS, Goldbaum FA, Schön A, Freire E, Tomaselli GF, Amzel LM, Ben-Johny M, Gabelli SB 2022. Development of high-affinity nanobodies specific for NaV1.4 and NaV1.5 voltage-gated sodium channel isoforms. J. Biol. Chem. 298(4), 101763. 10.1016/j.jbc.2022.101763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang Y, Wang S 2010. Development of chemiluminescence enzyme-linked immunosorbent assay for the screening of metolcarb and carbaryl in orange juice, cabbage and cucumber. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 27(3), 338–46. 10.1080/19440040903403016. [DOI] [PubMed] [Google Scholar]
- Sun Y, Leng R, Ma X, Zhang J, Han B, Zhao G, Ai Y, Hu B, Ji Z, Wang X 2023. Economical amidoxime-functionalized non-porous β-cyclodextrin polymer for selective detection and extraction of uranium. Chem. Eng. J. 459,141687. 10.1016/j.cej.2023.141687. [DOI] [Google Scholar]
- Wang F, Wang H, Shen Y, Li Y, Dong J, Xu Z, Yang J, Sun Y, Xiao Z 2016. Bispecific monoclonal antibody-based multianalyte ELISA for furaltadone metabolite, malachite green, and leucomalachite green in aquatic products. J. Agric. Food Chem. 64, 8054–8061. 10.1021/acs.jafc.6b03233. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang C, Zhang Y 2005. Development of a flow-through enzyme-linked immunosorbent assay and a dipstick assay for the rapid detection of the insecticide carbaryl. Anal. Chim. Acta. 535(1–2), 219–25. 10.1016/j.aca.2004.12.009. [DOI] [Google Scholar]
- Wichgers Schreur PJ, van de Water S, Harmsen M, Bermúdez-Méndez E, Drabek D, Grosveld F, Wernike K, Beer M, Aebischer A, Daramola O, Rodriguez Conde S, Brennan K, Kozub D, Søndergaard Kristiansen M, Mistry KK, Deng Z, Hellert J, Guardado-Calvo P, Rey FA, van Keulen L, Kortekaas J 2020. Multimeric single-domain antibody complexes protect against bunyavirus infections. eLife. 9. 10.7554/eLife.52716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H, Sun Q, Wang W, Li S, Xiang X, Li Z, Liao X, Li H 2021. Study of conformational and functional changes caused by binding of environmental pollutant tonalide to human serum albumin. Chemosphere. 270, 129431. 10.1016/j.chemosphere.2020.129431. [DOI] [PubMed] [Google Scholar]
- Yang J, Chen Y, Liu Z, Yang L, Tang J, Miao M, Gan N, Li H 2019. Differences between the binding modes of enantiomers S/R-nicotine to acetylcholinesterase. RSC Adv. 9(3),1428–40. 10.1039/c8ra09963d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li Y, Wu X, Li L, Liu J, Wang Y, Liu Y, Li Q, Wang Z 2020. Identification of anti-CD16a single domain antibodies and their application in bispecific antibodies. Cancer Biol. Ther. 21(1), 72–80. 10.1080/15384047.2019.1665953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Zhang M, Ning Y, Mao G, Li X, Deng Q, Chen X, Zuo D, Zhao X, Xie E, Wang H, Guo L, Li B, Xiao K, He X 2022. Development of a bispecific antibody targeting PD-L1 and TIGIT with optimal cytotoxicity. Sci. Rep. 12(1). 10.1038/s41598-022-22975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


