Abstract
Core promoters are sites where transcriptional regulatory inputs of a gene are integrated to direct the assembly of the pre-initiation complex (PIC) and RNA polymerase II (Pol II) transcription output. Until now, core promoter functions have been investigated by distinct methods, including Pol II transcription initiation site mappings and structural characterization of PICs on distinct promoters. Here, we bring together these previously non-connected observations and hypothesize how, on metazoan TATA promoters, the precisely structured building up of TFIID-based PICs results in sharp transcription start site (TSS) selection; or, in contrast, how the less strictly controlled positioning of the TATA-less promoter DNA relative to TFIID-core PIC components results in alternative broad TSS selections by Pol II.
Keywords: RNA polymerase II (Pol II), transcription initiation, core promoter architecture, pre-initiation complex (PIC), TFIID, transcription start site selection (TSS)
RNA polymerase II transcription initiation, the disconnected ends
Eukaryotic RNA polymerase II (Pol II) gene transcription is a highly regulated process. One of the key steps in transcription initiation is the assembly of general transcription factors (GTFs; see Glossary) and Pol II into a pre-initiation complex (PIC) at core promoters [1-3]. TFIID, the first recruited factor, makes contacts with core promoter DNA elements (see below), promotes TATA-binding protein (TBP) loading on DNA upstream of the transcription start site (TSS), and works as a dynamic scaffold for the formation of Pol II PICs on all protein-coding genes ([4] and references therein).
Core promoters are crucial because they are the sites where transcriptional regulatory inputs of a gene are integrated to direct the rate of transcriptional output. Until now, transcription initiation events have been investigated from several, mostly disconnected directions: a) by determining the DNA sequence motifs that participate in Pol II transcription initiation, b) defining TSS positions genome-wide; or c) by solving the structure of PICs on distinct artificial or endogenous promoters (as discussed below).
However, despite decades of intensive research, the exact determinants of transcription initiation at core promoters remain elusive. As a result, it is not yet possible to predict transcription initiation patterns at a base resolution from DNA sequence alone. Here we bring together previously non-connected observations of human TSS usage and PIC formation to suggest a mechanism by which DNA sequence elements in core promoters may help to determine TSS usage. We hypothesize that core promoters containing a TATA box and a downstream promoter element (DPE) are characterized by precise loading of TFIID-based PICs, resulting in sharp TSS initiation; on the contrary, PIC assembly is less strictly controlled on TATA-less promoters, resulting in the alternative broad TSS selection by Pol II. As these mechanisms may differ in yeast or Drosophila, and due to the lack of TFIID-containing PIC structures from these model organisms, here we mainly discuss findings described at human core promoters.
Sequence motifs of mammalian core promoters
The core promoter was originally defined as the minimal DNA fragment sufficient to direct basal levels of transcription initiation by Pol II in vitro. These assays were performed on naked viral core promoter templates containing a TATA box and a well-defined TSS [5]. According to its definition, the core promoter typically extends approximately 50 bp up- and downstream of the TSS and can contain several distinct core promoter sequence elements. Bioinformatic studies of vertebrate core promoter sequences failed to identify one single core promoter element, or one unique combination of elements that would universally cluster close to the TSSs for the majority of core promoters [6].
Nevertheless, a series of individual core promoter elements has been shown to exist in smaller subsets of core promoters with positional constraints in relation to the TSS. These elements include the TATA box, the initiator (INR), the TCT initiator, DPE, Motif Ten Element (MTE), upstream of and downstream TFIIB recognition elements (BREu and BREd), and several others, which are known to interact with different components of the PIC (reviewed in [7-14]). Some of these elements can positively or negatively correlate with the presence of other core promoter sequence motifs; however, the regulatory significance of these correlations is still unclear. It should also be noted that the canonical TATA box, which was once believed to be a general feature of core promoters, is only present in less than 10% of all human Pol II promoters [15, 16].
Transcription initiation patterns define two core promoter types
Large scale sequencing approaches, such as Cap Analysis of Gene Expression (CAGE [17]), have allowed for 5’-ends of mRNAs to be accurately determined, and consequently TSS annotations genome wide [10, 13, 18]. Two main classes of promoter types were identified on the basis of the differential usage of TSSs [19]: i) sharp or focused promoters, which have a relatively tightly defined TSS position within a few base pairs (Figure 1A) and ii) broad or dispersed promoters, which show a relatively wide distribution of many TSSs in a 100-bp window (Figure 1B). This distinction between the two TSS patterns, referred to as promoter “shape” or “architecture”, may reflect different mechanisms of transcription initiation. Indeed, sharp promoters were found to be more likely associated with TATA box-containing promoters and to possess a higher frequency of other core promoter elements than broad promoters. Broad promoters are often TATA-less, overlapping with CpG-islands (or are simply CG-rich), and are often lacking other consensus core promoter elements (reviewed in [18]). Moreover, TATA box-associated sharp promoters are more involved in tight regulation of genes, which are often tissue-specific and/or developmentally regulated, while broad peak promoters are often associated with ubiquitously expressed genes, also called housekeeping genes [13, 19, 20].
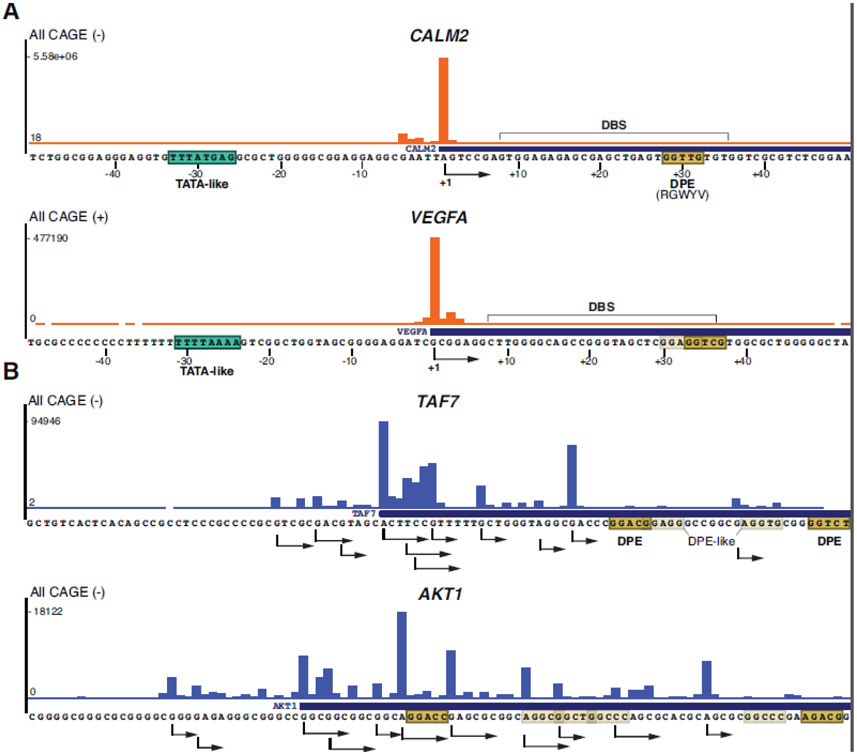
Figure 1. Sharp and broad promoter architectures.
UCSC browser genomic CAGE snapshots of the human TATA/DPE containing CALM2 and VEGFA promoters (A) and the TATA-less TAF7 and AKT1 promoters (B). CALM2 and TAF7 have been analyzed structurally by [31]. The CAGE mapped main TSS of the TATA-containing promoters (A) and the multiple mapped TSSs of the TATA-less promoters (B) are indicated with arrows showing the direction of Pol II transcription. The interval of All ENCODE CAGE tag densities (All CAGE) for each gene, on either forward (+) or reverse (−) strands, are indicated on the left. TATA or TATA-like elements (boxed in green), downstream promoter elements (DPEs, including its consensus sequence, are boxed in khaki) and alternative DPE-like sequences (transparent khaki boxes) are indicated. In A) DBS is highlighted (see text).
Although promoter shape is generally conserved between species, suggesting functional importance, sharp promoters are more evolutionary constrained, further reflecting different needs for the regulation of the corresponding genes [19, 21]. Importantly, sharp and broad promoters exhibit distinctive regulatory properties, as they respond differently toward activating pathways [22], and are regulated by distinct sets of coactivators [23]. Moreover, sharp and broad TSS patterns represent two end-points of a spectrum of multiple promoter shapes, supporting that these possible regulatory associations do not always define absolute rules at a genome-wide level. However, it has not yet been investigated whether or how these promoter architectures selectively result from regulated assembly of general transcription factors involved in the formation of the PIC for transcription.
Recent advances in the structural architecture of the TFIID complex and its role for PIC assembly
During transcription initiation, Pol II by itself cannot recognize core promoter sequences, and it requires the specific preassembly of a series of GTFs to guide Pol II to TSSs at core promoters to start transcription. According to the sequential PIC assembly model established using purified GTFs, TFIID, composed of the TBP and 13 TBP associated factors (TAFs) [24], is the first GTF to recognize and bind core promoter sequences. Promoter bound TFIID is then stabilized by the binding of TFIIA and subsequently by TFIIB. This promoter-TFIID-TFIIA-TFIIB complex further recruits TFIIF and Pol II to form the core PIC (cPIC). The holo PIC (hPIC) is assembled by the final arrival of TFIIE and TFIIH [25].
Several structural studies reported mammalian PIC assembly on TATA box promoters with only TBP [26-29]. However, the functions of the TFIID complex, which are essential for the transcription of almost all Pol II-transcribed genes [30], may not be fully recapitulated by TBP alone. The recent structures of the human TFIID-based PICs give a better appreciation of the contacts made amongst TFIID, several other PIC components, and the DNA sequence motifs [31-33]. In one of the studies, TFIID-based PICs were assembled on either TATA-containing or TATA-less promoters, providing an understanding of the role of TFIID for PIC assembly on various different core promoters [31]. These data, together with earlier EM structures, showed that TFIID is composed of three lobes, named A, B, and C, each playing distinct roles in PIC assembly [31, 34-39]. Lobe A is made of one copy of TAF5 and a histone octamer like structure, composed of four histone fold (HF)-containing TAF pairs interacting with TBP. TFIID-lobe C has been shown to bind first to a DPE-containing DNA fragment through multiple contacts including TAF1/TAF2, prior to loading of TBP [31, 37, 39-42]. In contrast with the rather static structural module made of lobes B and C, lobe A is dynamically attached to the rest of the complex, allowing TBP to be loaded onto the upstream core promoter region, termed TBP binding site (TBS), independently whether the core promoter contains a TATA box, a TATA-like motif or TATA-less sequences [31, 37, 41, 42]. During the loading phase, TFIID lobe A hands TBP over to TFIIA, which in turn is associated with TFIID lobe B through a flexible joint provided by the TAF4 stalk helix. The latter structures also provide an additional contact point with upstream core promoter sequences just 3’ from the TBS [41, 42]. In addition, the HMG box domain of TAF1, protruding from lobe C, was found in direct contact with the putative INR DNA region [31]. While these structural studies offer explanation to how GTFs assemble on promoters with distinct spatially constrained promoter motif composition, they have not been explored for their contribution to the transcription start site distributions observed in distinct promoter shapes, such as sharp and broad promoters.
Structural architectures of TFIID-based PIC assemblies at distinct classes of core promoters
The first TFIID structures were all assembled on TATA box-containing core promoters, either on an artificial so-called super core promoter (SCP [43]), or on natural yeast promoters [37, 39]. The more recent human TFIID-based PICs have been assembled not only on TATA box-containing (SCP or endogenous TATA-DPE or TATA-only), but also on TATA-less promoters (containing only a DPE element) [31]. Based on the cryo-EM PIC structures obtained either on TATA-DPE core promoters or TATA-less promoters, the authors proposed two distinct PIC assembly tracks.
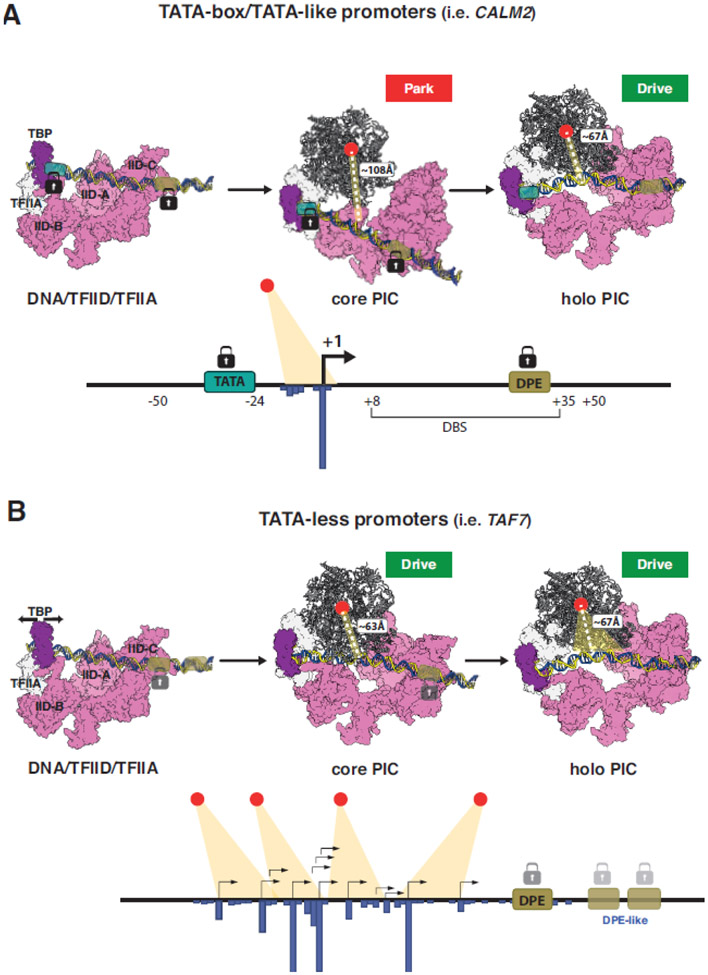
In the first track (Track I), on endogenous human HDM2 (MDM2), CALM2, and RPLP1 core promoters containing TATA and DPE elements, the authors describe a stepwise assembly. In the first step, TBP/TFIID, TFIIA, TFIIB, Pol II, and TFIIF bind around the TATA box and bend the DNA, while TFIID through its lobe C (TFIID-C) is well positioned on the downstream DPE sequences (Figure 2A). TBP/TFIIA bind around the TATA box and are separated by about 32 bp from the TFIID-C binding site (hereafter called DBS, which encompasses promoter sequences from about +8 to +35 relative to the start site; Figure 2A). The last base of the TBS is at position - 24 relative to the TSS, while the first base of the DBS is at position +8. Strong binding of TFIID-C to the DBS and of TFIID-lobe B to TFIIA and TBP leads to a stable assembly of TFIID on TATA-DPE core promoters with specific anchor points both upstream and downstream of the TSS (Figure 2A). The canonical 32 bp separation between lobes B and C was also visualized in TFIID/TFIIA complexes assembled on SCP DNA, in absence of Pol II and other GTFs [37, 38]. At this stage, the active site of Pol II is kept away from the promoter DNA, which is also the case when TFIIE is added. However, when TFIIH completes the hPIC, the path of the promoter DNA is changed to position the TATA-DPE promoter DNA close to the Pol II active site (Figure 2A). The different paths of the DNA in the cPIC and the hPIC were called Park (DNA far from Pol II active site) and Drive (DNA close to Pol II active site) conformations, respectively.
Figure 2. Structural models of TFIID-based PIC assemblies leading to either sharp or broad transcription start site selection on TATA/DPE or TATA-less promoters.
Structural models of TFIID-based PIC assemblies on TATA/DPE-containing promoters leading to sharp (A) or TATA-less promoters leading to broad (B) transcription start site selection are shown. The structural models [in A) and B) upper part] are based on PDB 7EG7, PDB 7EG9, PDB 7EG8, PDB 7EGA, PDB 7EGB [31]. Sharp and broad transcription architectures with a well-defined TSS (A) or many TSSs (B), respectively, are depicted [in the lower panels of A) and B)]. The active center of Pol II is shown with a red dot, and the distance of the active center from the TSSs is indicted with dotted lines in angstroms (Å). The TATA like box is indicated in green and the DPEs in khaki. The TFIID-C bound TFIID-binding site, DBS, is shown in (A). The well positioned binding of TBP/TFIID-lobe B on the TATA box, and TFIID lobe C on the DPE are represented by black padlocks (in A) and the less well positioned binding of TFIID on the DPE on the TATA-less promoter is indicated by gray padlocks (in B). TFIID is in pink, TBP is in magenta, TFIIA and TFIIB are in white. Pol II is shown in ribbon type representation. TFIIF, TFIIE and TFIIH were omitted for clarity. In B) sliding of TBP loading is indicated with a black two-headed arrow and the plasticity of TFIID-C binding to alternative DPE sequences is indicated by two-headed blue arrow.
During the transitions from promoter-TFIID-TFIIA complex to cPIC and from cPIC to TFIIE-bound PIC, TFIID is similarly anchored to the promoter DNA, through TBP and TFIID-C bound to their respective consensus sequences and separated by 32 bps. When TFIIH joins the assembly, it establishes contacts both with downstream promoter DNA and TFIID-C (through TAF2). This intermediate conformation (named by the authors pre-hPIC) resolves with the dissociation of TFIID-C from the DBS to form hPIC (Figure 3A). During this transition, the TAF1 HMG box is also displaced, allowing DNA relocation close to Pol II active site. Thus, TFIIH binding in hPIC induces a transition to the Drive conformation. In this track, many different steps are required to assemble a functional hPIC, providing several well-defined, potentially regulable, stable anchoring points to control the proper deposition of the PIC relative to promoter sequence elements, such as TATA, and DBS (or DPE). In agreement, the spacing between TBP and TFIID-C is kept fixed, preventing sliding of promoter DNA relative to cPIC components. Thus, we hypothesize that these TATA-DPE promoters may be characterized by precise, well-structured loading of Pol II and a regulable multistep process for promoter deposition into the active site, resulting in sharp initiation.
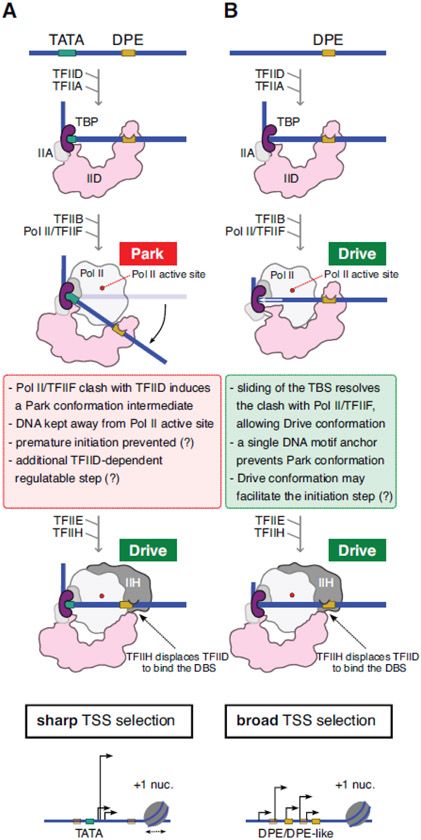
Figure 3. Step-wise assembly of PICs on core promoters resulting in either sharp or broad TSS selection.
TFIID-based PIC assembly steps on either TATA/DPE (A) or only DPE-containing (TATA-less) (B) core promoters resulting in either sharp (A) or broad (B) TSS selections are summarized. TATA boxes are shown with green boxes, DPEs and DPE-like sequences are shown in khaki boxes, where the DPE-like sequences are transparent. Pol II active sites are shown with a red dot. +1 nucleosomes (+1 nuc.) are depicted on the lower panels.
In the second series of structures obtained on endogenous human TATA-less PUMA (BBC3), TAF7, and POLB promoters (Track II) [31], the TBS-bound TBP and the DBS-bound TFIID-C were found to be separated by about 32 bps in the promoter-TFIID-TFIIA complex, similarly to observations made on TATA-DPE promoters. However, during the assembly of the cPIC on these TATA-less promoters, while the contact between TFIID-C and the DPE is unchanged, in the absence of TATA-like sequences TBP slides 10 bps upstream of the initial TBS when compared with the equivalent cPIC in the Track I (Figures 2A and 3A). This longer distance of 42 bps (instead of 32 bps), where the cPIC is fixed only on one side through TFIID-C/DBS (or DPE) contacts, prevents a steric clash between TFIID-B and the incoming Pol II/TFIIF complex, thus allowing the Pol II cleft to be positioned close to the promoter DNA. Therefore, in Track II all intermediate assemblies (cPIC, TFIIE-PIC and hPIC) directly adopt the Drive conformation [31, 42] (Figures 2B and 3B). The structure of a human TFIID-based PIC assembled on the SCP together with the human Mediator complex was recently solved and revealed similar dynamic interactions between TFIID, TFIIH, Pol II and promoter sequence elements [32], suggesting that the presence of Mediator has no major influence on DNA conformation in TFIID-based PICs.
How can the TFIID-based PIC structures be reconciled with sharp and broad transcription initiation?
When analyzing human CAGE-defined TSS profiles of the structurally characterized human promoters from the ENCODE database, we found that transcription initiation from the TATA/DPE core promoter (CALM2) was belonging to the sharp category, while transcription initiation from the TATA-less promoter (TAF7) showed a broad pattern (Figure 1A, 1B).
The above structural observations would suggest that when several strong TFIID positioning sequences are present upstream and downstream of the TSS in a given promoter, such as in the TATA-DPE category, these sequence elements can define a precise and stable cPIC in the Park conformation (Figure 3A). In this configuration, TBP and TFIID-C are stably positioned on their binding sequences and separated by ~32 bps, resulting in clashes between TFIIF-Pol II and TFIID-B, thereby keeping the promoter away from the active site. Further conformation changes are observed during transition from cPIC to TFIIE-PIC and to final assemble of the hPIC in the Drive conformation, now competent for transcription initiation at a well-defined sharp TSS (Figure 3A) [31].
In contrast, on TATA-less promoters we hypothesize that the anchoring of TFIID is less well defined, as TBP was found to slide 10 bp upstream during cPIC assembly, when compared to the Track I cPIC binding to the TATA sequences [31]. One can speculate that TFIID-C might also slide on the downstream DNA to interact with alternative DPEs, or DPE-like sequences (Figures 1B, 2B, and 3B), which have a rather weakly defined consensus (RGWYV) and can be found at several locations (Figure 1B, 2B). This would be further relaxed by the repositioning of TBP during the transition from promoter-TFIID-TFIIA complex to cPIC. Therefore, the positioning of the TATA-less promoter DNA relative to TFIID is not as strictly controlled and defined as for TATA-DPE promoters in Track I, thus, potentially allowing alternative TSS selections by Pol II.
It is conceivable that on TATA-less promoters, TFIID is only positioned by downstream sequence elements, which are rather loosely defined. Thus, a more poorly defined interaction of TFIID with upstream and downstream promoter motifs would position the Pol II active site close to several putative TSSs (Drive conformation), creating a broad transcription initiation pattern (Figures 2B and 3B). On these broad peak promoters several scenarios are possible: i) only one PIC assembles at a time and the same PIC may slide to different DPEs in the same region for separate initiation events; (ii) distinct consecutively arriving TFIID-based PICs bind to slightly different DPE sites; or (iii) the distinct TSSs represent single PICs specific to each cell, but they appear as multiple TSSs in the CAGE data obtained from a cell population. Variation in promoter shape is not expected to rely exclusively on the differential presence of a TATA-box or a DPE sequence. Differently from the Saccharomyces cerevisiae shooting gallery model [44], mammalian Pol II is not believed to perform a directional promoter scanning to assess candidate TSSs. Instead, separate PICs would independently assemble around each TSS [45]. Yet, Chou et al. (2022) inferred a rather short DNA window (~20 bp) over which mammalian Pol II could select alternative initiation sites once assembled in the PIC [46]. The authors interpreted the data as Pol II itself moving by stochastic motion after DNA melting and template strand loading before TSS selection. According to our model, this short range of “Brownian” motion along the DNA is consistent with the TFIID-dependent dynamic repositioning of the PIC on closely-spaced alternative motifs, especially for TATA-less promoters.
Concluding remarks
Future structural studies will be required to increase the resolution and the numbers of hPIC structures at several distinct core promoters to further dissect the functional differences by which Pol II can initiate transcription from one major location (sharp TSS selection) or from several less defined sites (broad TSS selection). Such structures could also be carried out in the presence of certain nucleotide analogues that would allow Pol II to initiate transcription synthesis.
As broad peak promoter range was also suggested to be influenced by the positioning of the +1 nucleosome, which may reside at +30/+60 distance from the dominant TSS [47], another key question is how nucleosome positioning will affect Pol II initiation on the different core promoter types with distinct TSS architecture. In this regard, the structure of a TFIID-based hPIC in presence of the reconstituted +1 nucleosome, with its edge positioned at +40, was recently described [33]. As expected, at this position the nucleosome does not cover the DBS, and the overall architecture of the PIC resembles the one on naked DNA, apart from defined direct contacts of Mediator/TFIIH with the nucleosome [33]. Notably, the repositioning the nucleosomes further apart (+50/+70) was not mirrored by the repositioning of the PIC, suggesting that the underlying DNA motifs remain the major drivers of PIC localization [33]. Whether the same holds true also for TATA-less promoters remains to be assessed. A second recent study reported the structure of a TBP-based PIC with the +1 nucleosome edge positioned progressively closer to the TSS (+18/+10) [48]. In this configuration TFIID-C and the nucleosome would compete for DBS binding, suggesting that the nucleosome could impair TFIID mediated PIC stabilization, in accordance with the low transcriptional activity of promoters partially invaded by nucleosomes. How the +1 nucleosome might contribute to the fine tuning of TSS selection on different promoter classes remains to be defined. The tremendous advances made by cryo-EM techniques should make it possible to answer these and other outstanding questions (see Outstanding questions).
Outstanding questions.
Will it be possible to visualize transcription initiation at endogenous promoters in cells by cryo-electron tomography (cryo-ET), or related approaches?
Is it possible to resolve by cryo-EM a more representative number of endogenous TFIID-based PIC structures at core promoters containing several combinations of DNA elements to get the underlying grammar of Pol II initiation?
Does the TAF1 HMG domain-INR interaction(s) play a role in regulating TSS selection by Pol II?
Are other factors recognizing TSSs and influence promoter shape selection?
Are there other promoter recognition complexes than TFIID (containing TBPL1 or TBPL2), playing a role in core promoter type recognition and consequent initiation of Pol II transcription, TSS selection?
How are +1 nucleosomes interacting with TFIID-based PICs on TATA/DPE or TATA-less promoters? Are +1 nucleosomes influencing sharp and broad TSS selection?
Do distinct TSSs represent single PICs specific to single cells in a cell population, or even in one given cell several TSS selection can occur?
Can partial TFIID-based PICs regulate differently TSS selection?
Highlights.
Core promoter elements, such as the TATA box and downstream promoter elements (DPEs), participate in the determination of RNA polymerase II (Pol II) transcription initiation
Metazoan Pol II starts mRNA transcription either from one major site, called sharp (or focused) transcription start site (TSS) selection, or a broad region, called broad (or dispersed) TSS selection
Recently, several cryo-EM structures of human TFIID-containing preinitiation complexes (PICs) have been determined
Based on these structures, we propose a model for how the presence or absence of core promoter elements in conjunction with TFIID-based PICs could define sharp or broad TSS selection
Acknowledgements
We are grateful to S. Bour for help in making Figure 2. This work was financially supported by Agence Nationale de la Recherche (ANR) ANR-19-CE11-0003-02, ANR-20-CE12-0017-03, ANR-22-CE11-0013-01_ACT; Fondation pour la Recherche Médicale (EQU-2021-03012631); NIH MIRA (R35GM139564); and NSF (Award Number: 1933344) grants. A.B. has been supported by the Fondation ARC pour la recherche sur le cancer (ARCPOST- DOC2021080004113). This work, as part of the ITI 2021-2028 program of the University of Strasbourg, was also supported by IdEx Unistra (ANR-10-IDEX-0002), and by SFRI-STRAT’US project (ANR 20-SFRI-0012) and EUR IMCBio (ANR-17-EURE-0023) under the framework of the French Investments for the Future Program.
Glossary:
- Cap Analysis of Gene Expression (CAGE):
an approach to identify and monitor the activity (transcription initiation frequency) of TSSs at single base-pair resolution across the genome. CAGE allows high-throughput gene expression profiling with simultaneous identification of the TSSs, including promoter usage analysis.
- Core Promoter:
genomic DNA sequence where the PIC assembles and that encompasses the site of transcription initiation and extends both upstream and downstream for ~30-50 bp.
- Downstream core promoter element (DPE):
a core promoter element with a consensus sequence of RGWYV, located about 28–33 nucleotides downstream of the TSS.
- Genaral Transcription Factors (GTFs):
also known as basal transcriptional factors, are TFs that bind to specific sites at core promoters and are necessary to recruit RNA Polymerase II (Pol II) and to initiate mRNA synthesis at the correct position. Pol II GTFs are: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH.
- Initiator (INR):
a DNA sequence element that overlaps a transcription start site. It has a loose consensus sequence of YYA+1NWYY.
- Pre-initiation complex (PIC):
composed of six GTFs and Pol II assembled on the core promoter and required for the transcription of protein-coding genes in eukaryotes. The preinitiation complex positions Pol II at transcription start sites, opens the core promoter DNA, and positions the template strand in the RNA polymerase II active site for starting transcription.
- TATA binding protein (TBP):
a subunit of the Pol II GTF, TFIID. The C-terminal core of TBP is highly conserved and contains two repeats that produce a saddle-shaped structure which binds to the TATA box. When TBP binds to a TATA box DNA, it distorts the DNA by inserting amino acid side-chains between base pairs, partially unwinding the helix, and bending it by almost ~90°. Note that TBP is also involved in transcription initiation by RNA polymerases I and III.
- TATA box:
a DNA sequence motif found about 30 base pairs upstream of the TSS in a subset of metazoan core promoters. The TATA box is named after its conserved DNA sequence, characterized by repetitive T and A base pairs, and it has a consensus sequence of TATAWAW.
- TBP associated factors (TAFs):
identified biochemically in stable complexes with TBP, composing the TFIID GTF complex. Comparison of yeast, plant, Drosophila, human TFIID compositions indicated a set of 13 TAFs that are conserved across many eukaryotic species. These 13 evolutionarily conserved TAFs have been designated from TAF1 to TAF13.
- Transcription factor IID (TFIID):
a large multi-protein GTF complex formed by the TATA box binding protein (TBP) and 13 (in metazoans and 14 in yeast) TBP-associated protein factors (TAFs). Among the TAFs, TAF4, TAF5, TAF6, TAF9, TAF10, TAF12 are present in the complex in two copies, generating a 20-subunit complex with a molecular weight of ~1.3 MDa. TFIID plays an important role in core promoter recognition, TBP loading onto promoter, and in nucleating the PIC assembly at almost all Pol II promoters.
- Transcription start site (TSS):
the location in a core promoter where transcription begins and corresponds to the first nucleotide (+1) incorporated into the mRNA molecule.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roeder RG (1996) The role of general initiation factors in transcription by RNA polymerase II. Trends.Biochem.Sci 21, 327–335. [PubMed] [Google Scholar]
- 2.Orphanides G and Reinberg D (2002) A unified theory of gene expression. Cell 108 (4), 439–51. [DOI] [PubMed] [Google Scholar]
- 3.Thomas MC and Chiang CM (2006) The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41 (3), 105–78. [DOI] [PubMed] [Google Scholar]
- 4.Bhuiyan T and Timmers HTM (2019) Promoter Recognition: Putting TFIID on the Spot. Trends Cell Biol 29 (9), 752–763. [DOI] [PubMed] [Google Scholar]
- 5.Weil PA et al. (1979) Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell 18, 469–484. [DOI] [PubMed] [Google Scholar]
- 6.Kadonaga JT (2012) Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol 1 (1), 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vo Ngoc L et al. (2017) The punctilious RNA polymerase II core promoter. Genes Dev 31 (13), 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vo Ngoc L et al. (2019) The RNA Polymerase II Core Promoter in Drosophila. Genetics 212 (1), 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juven-Gershon T et al. (2006) Perspectives on the RNA polymerase II core promoter. Biochem Soc Trans 34 (Pt 6), 1047–50. [DOI] [PubMed] [Google Scholar]
- 10.Sandelin A. et al. (2007) Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet 8 (6), 424–36. [DOI] [PubMed] [Google Scholar]
- 11.Muller F et al. (2007) New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem 282 (20), 14685–9. [DOI] [PubMed] [Google Scholar]
- 12.Ohler U and Wassarman DA (2010) Promoting developmental transcription. Development 137 (1), 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haberle V and Stark A (2018) Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol 19 (10), 621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vo Ngoc L. et al. (2020) Identification of the human DPR core promoter element using machine learning. Nature 585 (7825), 459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.FitzGerald PC et al. (2004) Clustering of DNA sequences in human promoters. Genome Res 14 (8), 1562–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bajic VB et al. (2006) Mice and men: their promoter properties. PLoS Genet 2 (4), e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiraki T. et al. (2003) Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A 100 (26), 15776–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lenhard B. et al. (2010) Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat Rev Genet 13 (4), 233–45. [DOI] [PubMed] [Google Scholar]
- 19.Carninci P. et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38 (6), 626–35. [DOI] [PubMed] [Google Scholar]
- 20.Schug J. et al. (2005) Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6 (4), R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schor IE et al. (2017) Promoter shape varies across populations and affects promoter evolution and expression noise. Nat Genet 49 (4), 550–558. [DOI] [PubMed] [Google Scholar]
- 22.Zabidi MA et al. (2015) Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518 (7540), 556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haberle V. et al. (2019) Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570 (7759), 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tora L (2002) A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev 16 (6), 673–5. [DOI] [PubMed] [Google Scholar]
- 25.Buratowski S. et al. (1989) Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–561. [DOI] [PubMed] [Google Scholar]
- 26.He Y. et al. (2016) Near-atomic resolution visualization of human transcription promoter opening. Nature 533 (7603), 359–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aibara S. et al. (2021) Structures of mammalian RNA polymerase II pre-initiation complexes. Nature 594 (7861), 124–128. [DOI] [PubMed] [Google Scholar]
- 28.Abdella R. et al. (2021) Structure of the human Mediator-bound transcription preinitiation complex. Science 372 (6537), 52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rengachari S. et al. (2021) Structure of the human Mediator-RNA polymerase II pre-initiation complex. Nature 594 (7861), 129–133. [DOI] [PubMed] [Google Scholar]
- 30.Warfield L. et al. (2017) Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID. Mol Cell 68 (1), 118–129 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X. et al. (2021) Structural insights into preinitiation complex assembly on core promoters. Science 372, eaba8490. [DOI] [PubMed] [Google Scholar]
- 32.Chen X. et al. (2021) Structures of the human Mediator and Mediator-bound preinitiation complex. Science 372 (6546), eabg0635. [DOI] [PubMed] [Google Scholar]
- 33.Chen X. et al. (2022) Structures of +1 nucleosome-bound PIC-Mediator complex. Science 378 (6615), 62–68. [DOI] [PubMed] [Google Scholar]
- 34.Brand M. et al. (1999) Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286 (5447), 2151–3. [DOI] [PubMed] [Google Scholar]
- 35.Andel F. et al. (1999) Three-dimensional structure of the human TFIID-IIA-IIB complex. Science 286, 2153–2156. [DOI] [PubMed] [Google Scholar]
- 36.Cianfrocco MA et al. (2013) Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 152 (1-2), 120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel AB et al. (2018) Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362 (6421), eaau8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louder RK et al. (2016) Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531 (7596), 604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolesnikova O. et al. (2018) Molecular structure of promoter-bound yeast TFIID. Nat Commun 9 (1), 4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nogales E. et al. (2017) Towards a mechanistic understanding of core promoter recognition from cryo-EM studies of human TFIID. Curr Opin Struct Biol 47, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel AB et al. (2020) Recent insights into the structure of TFIID, its assembly, and its binding to core promoter. Curr Opin Struct Biol 61, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X and Xu Y (2022) Structural insights into assembly of transcription preinitiation complex. Curr Opin Struct Biol 75, 102404. [DOI] [PubMed] [Google Scholar]
- 43.Juven-Gershon T. et al. (2006) Rational design of a super core promoter that enhances gene expression. Nat Methods 3 (11), 917–22. [DOI] [PubMed] [Google Scholar]
- 44.Qiu C. et al. (2020) Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae. Genome Biol 21 (1), 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luse DS et al. (2020) A unified view of the sequence and functional organization of the human RNA polymerase II promoter. Nucleic Acids Res 48 (14), 7767–7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou SP et al. (2022) Genetic dissection of the RNA polymerase II transcription cycle. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haberle V. et al. (2014) Two independent transcription initiation codes overlap on vertebrate core promoters. Nature 507 (7492), 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abril-Garrido J. et al. (2023) Structural basis of transcription reduction by a promoter-proximal +1 nucleosome. Mol Cell 83 (11), 1798–1809 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]