Renin cells are essential for survival. They control the composition and volume of our extracellular fluid, arterial blood pressure, tissue perfusion, and oxygen delivery. FoxD1+ cells are precursors for renin cells which in turn differentiate into smooth muscle cells (SMCs), mesangial cells (MCs), and pericytes (PCs). When arteriole assembly completes, mature renin cells are confined to the tip of the renal arterioles near the glomeruli, thus their name juxtaglomerular (JG) cells. The chromatin states and transcription factor (TF) repertoires that determine the differentiation trajectory of renin cells from their early progenitors to mature JG cells is unknown. We isolated single cells from FoxD1cre;mTmG mice at key developmental times in embryonic (E12: 15 embryos, E18: 17 embryos, and post-natal (P5: 12 mice 6F|6M, P30: 10 mice; 4F|6M) life and subjected them to integrated scRNA-seq and scATAC-seq. This approach allowed us to i) construct a single-cell atlas of chromatin accessibility and gene expression profiles, ii) establish the developmental trajectory that leads to the mosaic of cells that compose the kidney arterioles, and iii) identify transcription factors (TFs) that control the elusive, myo-endocrine adult renin-secreting JG cell.
We uncovered how successive changes in the transcriptome, accessibilome, and enriched TFs instruct FoxD1 progenitor’ cells to adopt the different cell fates that encompass the kidney vasculature (Figure A). As Ren1 expression is crucial to JG cell identity, we predicted which regulatory elements and TFs regulate Ren1 and identified MEF2 family members (Figure B). We confirmed the presence of Mef2c using immunohistochemistry (IHC) on mouse kidney sections throughout development in JG cells and SMCs (Figure C).
Figure: Chromatin and transcriptomic shifts during renin cell development.
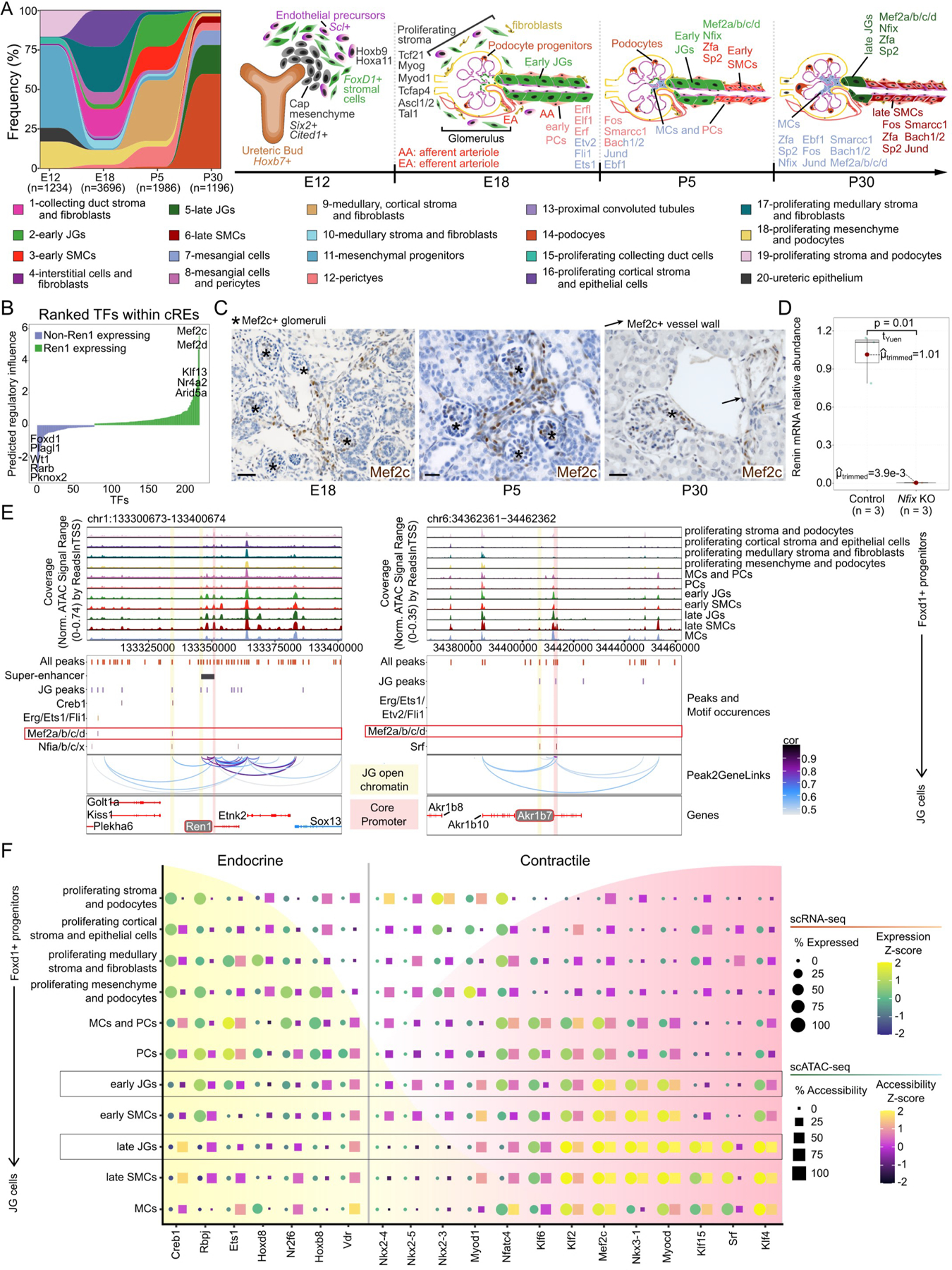
(A), Alluvial plot of cell frequency distribution (left) and individually enriched TF motifs (right) across developmental time points. Hoxb9 and Hoxa11 are enriched at E12. At E18, Tcf21, Myog, Myod1, Tcfap4, Ascl1 and Ascl2, and Tal1 are enriched in proliferating stroma. Between E18 and P5, Erfl, Elf1, Erf, Etv2, Fli1, Fos, Smarcc1, Bach1 and Bach2, Jund, and Ebf1 are enriched in early PCs and mixed MCs and PCs. In early JGs and SMCs, Mef2a/b/c/d, Nfix, Zfa, and Sp2 motifs are enriched. By P30, MCs are enriched for Fos, Smarcc1, Bach1 and Bach2, Jund, Ebf1, and to a lesser extent Mef2a/b/c/d, Nfix, Zfa, and Sp2. Late JGs are enriched for Mef2a/b/c/d, Nfix, Zfa, and Sp2 motifs, and late SMCs for Fos, Smarcc1, Bach1 and Bach2, Jund, Ebf1, Zfa, and Sp2. (B), TFs ranked by predicted regulatory score. (C), Mef2c IHC in kidney sections at E18, P5, and P30. At E18, Mef2c is found along AAs, interlobular arterioles, and MCs while at P5, it is found inside glomeruli, MCs, AAs, interlobular arterioles, and at the JGA. Scale bars: 50μm. (D), Deletion of Nfix in cultured As4.1 cells decreases Ren1 mRNA (n=3, p=0.01, Yuen-Welch’s test). Three gRNAs were designed for Nfix knockout with 92% transfection efficiency. (E), Browser tracks identify enriched open chromatin and TF motif sites at Ren1 (left) and Akr1b7 (right). (F), Dot plot for markers of endocrine or contractile phenotypes throughout JG cell differentiation.
We found the TF, nuclear factor I X (Nfix), was enriched in JG cells (Figure A, right), and confirmed co-localization with renin from E18 to P30 by RNAscope (not shown). We knocked out Nfix in renin-expressing As4.1 cells using CRISPR-Cas9 and showed a significant decrease (99.6% drop) in Ren1 mRNA (Figure D), indicating the functional relevance of Nfix. Ongoing in vivo studies will define whether Nfix affects the development of JG cells, SMCs and/or PCs and their ability to switch on-off the renin phenotype in responses to homeostatic challenges.
We identified enriched TF binding sites at Ren1 and Akr1b7, two independent markers of JG cells (Figure E). We observed significant motif occurrences for MEF2 members and Nfix in a region unique to JGs with co-accessibility to the Ren1 core promoter. (Figure E). We also identify overrepresented MEF2 motif occurrences within open chromatin of JG populations at the Akr1b7 locus (Figure E).
Past work highlighted the bivalent nature of renin-expressing JG cells between endocrine and contractile phenotypes1. Mef2c has an essential role in skeletal muscle growth and differentiation, and we observed significant increases in transcript abundance (FWelch’s and Games-Howell post-hoc tests: early JGs versus: early SMCs (pHolm-adj = 3.27×10−9) or PCs (pHolm-adj = 0.03)) and accessibility (early JGs versus: early SMCs (pHolm-adj = 0.00) or PCs (pHolm-adj = 8.99×10−5)) for Mef2c by the formation of early JGs (Figure F). Furthermore, we observed increased expression of the Mef2c paralog, Srf, in P30 populations of JGs (late JGs versus: PCs (pHolm-adj = 0.00), early JGs (pHolm-adj = 2.52×10−10) or late SMCs (pHolm-adj = 1.87×10−13)) which is involved in SMC development and maintenance and likely contributes to the plasticity of cells able to express renin1 (Figure F).
While further experimental evidence of the importance of MEF2 members at the renin locus is ongoing, an accumulation of data supports their relevance. MEF2 family members are transcriptional effectors of the Notch pathway, and MEF2 activation has been linked to stimulation by p3002, which is critical to remodeling chromatin at the renin locus3. Additionally, myocardin (Myocd) family factors are direct targets of MEF2 members4, and Myocd expression is significantly increased (early JGs versus: early SMCs (pHolm-adj = 1.43×10−9) or PCs (pHolm-adj = 9.95×10−12); late JGs versus: late SMCs (pHolm-adj = 0.00) or MCs (pHolm-adj = 1.39×10−9)) in JG populations (Figure F). Clinical support for Mef2c relevancy is seen in Rett, Angelman, Pitt-Hopkins syndromes, CDKL5 deficiency disorder, and the observation of duplex kidneys in pediatric populations with Mef2c haploinsufficiency5. The evidence points to MEF2 members as providing a pioneering and sustaining role in guiding renin cells through specific developmental stages to form the classical JG cell.
We provide the first developmental trajectory of renin cells in a single-cell atlas of kidney vascular development enriched for the renin lineage (Figure A). Specifically, we identified the MEF2 and NFI TF families as enriched and likely to control renin cell fate (Figure A, F). It will be exciting to define whether other factors and chromatin domains identified in this trajectory atlas control the development and plasticity of renin cells. Ultimately, determining the epigenomic-transcriptomic landscape of the rare JG cell is necessary to understand and predict responses in cardiovascular and kidney pathologies3 and in the design of pharmaceutical compounds to treat hypertension.
Acknowledgments
The authors gratefully acknowledge the technical and analytical help of Minghong Li, Xiuyin Liang, Fang Xu, Devon Farrar and Thomas Wagamon. We thank Vidya Nagalakshmi for preliminary protocols of nuclei isolation.
Sources of Funding
This work was supported by NIH grants DK 096373 and DK 116718 (to RAG), and DK 116196, DK 096373, and HL 148044 (to MLSSL).
Non-standard Abbreviations and Acronyms
- E12
embryonic day 12
- E18
embryonic day 18
- FBs
fibroblasts
- JGA
juxtaglomerular apparatus
- JGs
juxtaglomerular cells
- MCs
mesangial cells
- MEF
myocyte enhancer factor
- P5
five days old
- P30
one month old
- PCs
pericytes
- SE
super-enhancer
- SMCs
smooth muscle cells
Footnotes
Data and Code Availability
• The single cell omics data have been uploaded to the Gene Expression Omnibus (GEO) with the accession IDs GEO: GSE218570.
• Code to reproduce analyses is available at https://github.com/jpsmith5/renin_analysis
• Any additional information is available from the Lead Contact(s), R. Ariel Gomez, upon request.
Disclosures
None.
References
- 1.Brunskill EW, Sequeira-Lopez MLS, Pentz ES, Lin E, Yu J, Aronow BJ, Potter SS, Gomez RA. Genes that confer the identity of the renin cell. Journal of the American Society of Nephrology : JASN 2011;22:2213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacilotto N, Chouliaras KM, Nikitenko LL, Lu YW, Fritzsche M, Wallace MD, Nornes S, García-Moreno F, Payne S, Bridges E, et al. MEF2 transcription factors are key regulators of sprouting angiogenesis. Genes & development 2016;30:2297–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sequeira-Lopez MLS, Gomez RA. Renin cells, the kidney, and hypertension. Circulation research 2021;128:887–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, tead and foxo proteins during cardiovascular development. Development (Cambridge, England) 2006;133:4245–56. [DOI] [PubMed] [Google Scholar]
- 5.Vrečar I, Innes J, Jones EA, Kingston H, Reardon W, Kerr B, Clayton-Smith J, Douzgou S. Further clinical delineation of the MEF2C haploinsufficiency syndrome: Report on new cases and literature review of severe neurodevelopmental disorders presenting with seizures, absent speech, and involuntary movements. Journal of pediatric genetics 2017;6:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]


