Abstract
Shiga toxin (Stx)-producing Escherichia coli strains of serogroup O111 are the most frequently isolated non-O157 strains causing outbreaks of gastroenteritis with hemolytic-uremic syndrome. The O111 O-antigen gene cluster had been cloned and about half of it has been sequenced; we have now sequenced the remainder of the gene cluster, which is 12.5 kb in length and which comprises 11 genes. On the basis of sequence similarity, we have identified all the O-antigen genes expected, including five sugar biosynthetic pathway genes, three transferase genes, the O-unit flippase gene, and the O-antigen polymerase gene. By PCR testing with E. coli strains representing all 166 O-antigen forms, some randomly selected gram-negative bacteria, and Salmonella enterica serovar Adelaide, we showed that four O-antigen genes are highly specific to O111. This work provides the basis for a sensitive test for the rapid detection of E. coli O111. This is important both for decisions related to patient care, because early treatment may reduce the risk of life-threatening complications, and for the detection of sources of contamination.
Escherichia coli is a clonal species, with clones normally identified by their combination of O and H (and sometimes K) antigens. Some O antigens, such as O157 and O111, are characteristically found in pathogenic clones. An O111 clone was the first E. coli strain implicated as a cause of outbreaks of gastroenteritis (7, 19). E. coli O111:H2, O111:H8, O111:H12, and O111:nonmotile have been recognized as pathogenic clones. The O111 antigen has been classically associated with the serogroup of enteropathogenic E. coli and now has also been recognized as an O antigen of enterohemorrhagic and enteroaggregative E. coli (for a recent review, see reference 26). Since most laboratories do not screen stool samples for E. coli O111, the magnitude of the public health problem posed by these clones is probably underestimated. Nevertheless, there have been numerous reports of O111 strains as the cause of serious enteric disease, e.g., 28% of 50 outbreaks of infantile diarrhea in the United States from 1934 to 1987 (25), 33% of diarrhea cases in children in Brazil (33), an extensive outbreak involving more than 700 people in Finland (39), and more recently, documented outbreaks of hemolytic-uremic syndrome (HUS) in Australia (30) and Italy (8).
The O antigen, which contains many repeats of an oligosaccharide unit (O unit), is part of the lipopolysaccharide (LPS) present in the outer membrane of gram-negative bacteria. It contributes major antigenic variability to the cell surface, and on the basis of this variation, 166 O-antigen forms have been recognized in E. coli. The surface O antigen is subject to intense selection by the host immune system, which may account for the maintenance of many different O-antigen forms within species such as E. coli.
The O111 O antigen contains colitose, d-glucose, d-galactose, and N-acetyl-d-glucose. We have previously cloned the O111 O-antigen gene cluster (5) and sequenced about half of it and identified genes including colitose biosynthetic genes and the O-antigen flippase gene (4). In this paper we report the sequence of the remainder of this gene cluster. Analysis of the sequence revealed the third gene of the GDP-colitose pathway and good presumptive genes for the fourth and fifth steps, three presumptive sugar transferase genes for the synthesis of the O unit, and the O-antigen polymerase gene.
We have recently shown that the sequences of the E. coli O157 O-antigen transferase, flippase, and polymerase genes are O157 specific (40). In the study described here, we tested the O111 O-antigen transferase, flippase, and polymerase genes with representatives of all the 166 known E. coli O-antigen forms using PCR and found 4 of them to be specific to O111.
MATERIALS AND METHODS
Plasmids and bacterial strains.
Plasmids pPR1237, pPR1239, pPR1245, and pPR1246 are described elsewhere (5). Plasmids were maintained in E. coli K-12 strain JM109. Standard E. coli O group strains (23) were used (Table 1). The other strains used are also listed in Table 1, together with their sources.
TABLE 1.
Bacterial strains and PCR pools
| Pool no. | Strains whose chromosomal DNA is included in the pool | Source |
|---|---|---|
| 1 | E. coli type strains for O serotypes 1, 2, 3, 4, 10, 16, 18, and 39 | IMVSa |
| 2 | E. coli type strains for O serotypes 40, 41, 48, 49, 71, 73, 88, and 100 | IMVS |
| 3 | E. coli type strains for O serotypes 102, 109, 119, 120, 121, 125, 126, and 137 | IMVS |
| 4 | E. coli type strains for O serotypes 138, 139, 149, 7, 5, 6, 11, and 12 | IMVS |
| 5 | E. coli type strains for O serotypes 13, 14, 15, 17, 19ab, 20, 21, and 22 | IMVS |
| 6 | E. coli type strains for O serotypes 23, 24, 25, 26, 27, 28, 29, and 30 | IMVS |
| 7 | E. coli type strains for O serotypes 32, 33, 34, 35, 36, 37, 38, and 42 | IMVS |
| 8 | E. coli type strains for O serotypes 43, 44, 45, 46, 50, 51, 52, and 53 | IMVS |
| 9 | E. coli type strains for O serotypes 54, 55, 56, 57, 58, 59, 60, and 61 | IMVS |
| 10 | E. coli type strains for O serotypes 62, 63, 64, 65, 66, 68, 69, and 70 | IMVS |
| 11 | E. coli type strains for O serotypes 74, 75, 76, 77, 78, 79, 80, and 81 | IMVS |
| 12 | E. coli type strains for O serotypes 82, 83, 84, 85, 86, 87, 89, and 90 | IMVS |
| 13 | E. coli type strains for O serotypes 91, 92, 95, 96, 97, 98, 99, and 101 | IMVS |
| 14 | E. coli type strains for O serotypes 103, 104, 105, 106, 107, 108, and 110 | IMVS |
| 15 | E. coli type strains for O serotypes 112, 162, 113, 114, 115, 116, 117, and 118 | IMVS |
| 16 | E. coli type strains for O serotypes 123, 165, 166, 167, 168, 169, 170, and 171 | —b |
| 17 | E. coli type strains for O serotypes 172, 173, 127, 128, 129, 130, 131, and 132 | —c |
| 18 | E. coli type strains for O serotypes 133, 134, 135, 136, 140, 141, 142, and 143 | IMVS |
| 19 | E. coli type strains for O serotypes 144, 145, 146, 147, 148, 150, 151, and 152 | IMVS |
| 20 | E. coli type strains for O serotypes 153, 154, 155, 156, 157, 158, 159, and 160 | IMVS |
| 21 | E. coli type strains for O serotypes 161, 163, 164, 8, 9, and 124 | IMVS |
| 22 | As for pool 21, plus E. coli O111 type strain Stoke W | IMVS |
| 23 | As for pool 21, plus E. coli O111:H2 strain C1250-1991 | —d |
| 24 | As for pool 21, plus E. coli O111:H12 strain C156-1989 | —e |
| 25 | As for pool 21, plus S. enterica serovar Adelaide | —f |
| 26 | Y. pseudotuberculosis strains of O groups IA, IIA, IIB, IIC, III, IVA, IVB, VA, VB, VI, and VII | —g |
| 27 | S. boydii strains of serogroups 1, 3, 4, 5, 6, 8, 9, 10, 11, 12, 14, and 15 | —h |
| 28 | S. enterica strains of serovars (each representing a different O group) Typhi, Montevideo, Ferruch, Jangwani, Raus, Hvittingfoss, Waycross, Dan, Dugbe, Basel, 65:i:e,n,z15, and 52:d:e,n,x,z15 | IMVS |
IVMS, Institute of Medical and Veterinary Science, Adelaide, Australia.
Serotype 123 was from IMVS; the rest were from Statens Serum Institut, Copenhagen, Denmark.
Serotypes 172 and 173 were from Statens Serum Institut; the rest were from IMVS.
C1250-1991 was from Statens Serum Institut.
C156-1989 was from Statens Serum Institut.
S. enterica serovar Adelaide was from IMVS.
S. Aleksic of Institute of Hygiene, Hamburg, Germany.
J. Lefebvre of Bacterial Identification Section, Laboratoire de Santé Publique du Québec, Ste-Anne-de-Bellevue, Quebec, Canada.
Sequencing and analysis.
The PCR DNA and double-stranded plasmid DNA used as templates for DNA sequencing were prepared with the Wizard PCR and DNA preparation kits (Promega), respectively. Thermocycle sequencing reactions based on the dideoxy termination method (35) were run in a Perkin-Elmer Cetus DNA thermal cycler by the procedure recommended by Perkin-Elmer Cetus. The reactions used primers labelled with the appropriate fluorescent dye and were run on an Applied Biosystems 377 automated DNA sequencer. Sequence data were assembled and analyzed by using the Australian National Genomic Information Service, which incorporates several sets of programs (34). Sequence data were assembled with the XDAP program (16). Sequence databases were searched with the National Center for Biotechnology Information BLAST network server (2). Analysis of open reading frames was carried out with the nucleotide interpretation program (37). Program BESTFIT (11) was used for pairwise sequence comparison. We used the algorithm described by Eisenberg et al. (13) to identify potential transmembrane segments from the amino acid sequence.
Specificity assay by PCR.
Chromosomal DNA was isolated with the Promega Genomic isolation kit. Each chromosomal DNA sample was checked by PCR of mdh or O-antigen genes. Twenty-eight pools were made, with 6 to 12 samples of DNA per pool (Table 1). Chromosomal DNAs from three E. coli O111 strains and Salmonella enterica serovar Adelaide were individually added to one pool containing another six samples to give pools 22 to 25. PCRs were carried out under the following conditions: denaturation at 94°C for 30 s, annealing at various temperatures (see Table 2) for 30 s, and extension at 72°C for 1 min for 30 cycles. The PCR was carried out in a volume of 25 μl for each pool. After the PCR, 10 μl of the PCR product from each pool was run on an agarose gel to check for amplified DNA.
TABLE 2.
PCR test specificity data obtained with O111 primers
| Gene | Base positions of the gene | Forward primer (base positions) | Reverse primer (base positions) | Length of the PCR fragment (bp) | No. of pools (pools 1 to 21 and 25 to 28) giving band(s) of correct size | Anealing temp (°C) of PCR |
|---|---|---|---|---|---|---|
| wbdH | 739–1932 | 866 (739–757) | 867 (1941–1924) | 1,203 | 0a | 60 |
| 976 (925–942) | 978 (1731–1714) | 807 | 0 | 60 | ||
| 976 (925–942) | 979 (1347–1330) | 423 | 0 | 60 | ||
| 977 (1165–1182) | 978 (1731–1714) | 567 | 0 | 60 | ||
| wzx | 8646–9911 | 969 (8646–8663) | 970 (9908–9891) | 1,263 | 0 | 55 |
| 1060 (8906–8923) | 1062 (9468–9451) | 563 | 0 | 60 | ||
| 1061 (9150–9167) | 1063 (9754–9737) | 605 | 0a | 50 | ||
| wzy | 9901–10953 | 900 (9976–9996) | 901 (10827–10807) | 852 | 0 | 60 |
| 980 (10113–10130) | 983 (10484–10467) | 372 | 0b | 61 | ||
| wbdL | 10931–11824 | 870 (10931–10949) | 871 (11824–11796) | 894 | 7 | 60 |
| wbdM | 11821–12945 | 868 (11821–11844) | 869 (12945–12924) | 1,125 | 0 | 60 |
| 984 (12042–12059) | 987 (12447–12430) | 406 | 0 | 60 | ||
| 985 (12258–12275) | 986 (12698–12681) | 441 | 0c | 65 |
One pool gave a band of the wrong size.
Two pools gave three bands of the wrong size, one pool gave two bands of the wrong size, and all other pools gave one band of the wrong size.
Twenty-two pools gave a band of the wrong size.
Nucleotide sequence accession number.
The DNA sequence of the E. coli O111 O-antigen gene cluster has been deposited in GenBank under accession no. AF078736.
RESULTS
Sequencing.
We have previously sequenced 6,962 bp covering the central region of the O111 gene cluster from O111 type strain Stoke W (4). Plasmids pPR1237, pPR1239, pPR1245, and pPR1246 (5) together cover the two unsequenced ends of the gene cluster. We first sequenced both ends of the inserts of these plasmids using the M13 forward and reverse primer sites located in the vector. Then, PCR walking was carried out to sequence further into each insert by using oligonucleotide primers based on the sequence data obtained and tagged with M13 forward or reverse primer sequences. This PCR walking procedure was repeated until the entire inserts were sequenced in the cases of pPR1237 and pPR1239. For pPR1246, we sequenced only from positions 12007 to 14516 after finding that the gnd gene starts at position 13124. The junctions between plasmid inserts were then sequenced by using primers based on the sequence obtained.
Sequences of 3,031 bp (positions 1 to 3031) and 4,536 bp (positions 9981 to 14516) were obtained for the 5′ and 3′ ends of the gene cluster, respectively (region 1 and 2, respectively, in Fig. 1). DNA from positions 1 to 273 encodes the C-terminal end of GalF; DNA from positions 13124 to 14516 encodes the N-terminal end of Gnd (Fig. 1). Thus, DNA from positions 274 to 13123 is the O111 O-antigen gene cluster with intergenic regions.
FIG. 1.
O-antigen gene cluster of E. coli O111. The regions sequenced in this study are labelled regions 1 and 2. Both gene names and open reading frame (orf) numbers are given.
O111 O-antigen genes.
Figure 1 shows the structure of the O111 gene cluster. Two (wbdH and gmd) and three (wzy, wbdL, and wbdM) genes were predicted from the sequence in regions 1 and 2, respectively. With the 6 genes located in the previously sequenced segment, there are 11 genes in the O111 gene cluster; all have the same transcriptional direction from galF to gnd. The nucleotide and amino acid sequences were used to search available databases for an indication of possible function.
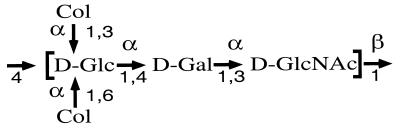
The structure of the O111 O unit is known (Fig. 2) (21). We expected to find in the gene cluster transferase genes for galactose, glucose, and colitose; genes for the synthesis of GDP-colitose; an O-antigen flippase gene (wzx); and an O-antigen polymerase gene (wzy). The gene encoding the transferase for the addition of GlcNac as the first sugar is located outside of the O-antigen gene cluster because O111 strains are wecA dependent (1).
FIG. 2.
Structure of the O111 O antigen. Col, colitose; Glc, glucose; Gal, galactose; GlcNAc, N-acetylglucosamine.
The deduced amino acid sequence of Orf-1 shares about 64% similarity with that of the WbbP gene (17) of Shigella dysenteriae. Both WbbP and Orf-1 have similar predicted hydrophobic profiles of four transmembrane segments. WbbP is a galactosyl transferase involved in the synthesis of the LPS core, and it is likely that orf-1, which we have named wbdH, is the galactosyl transferase gene.
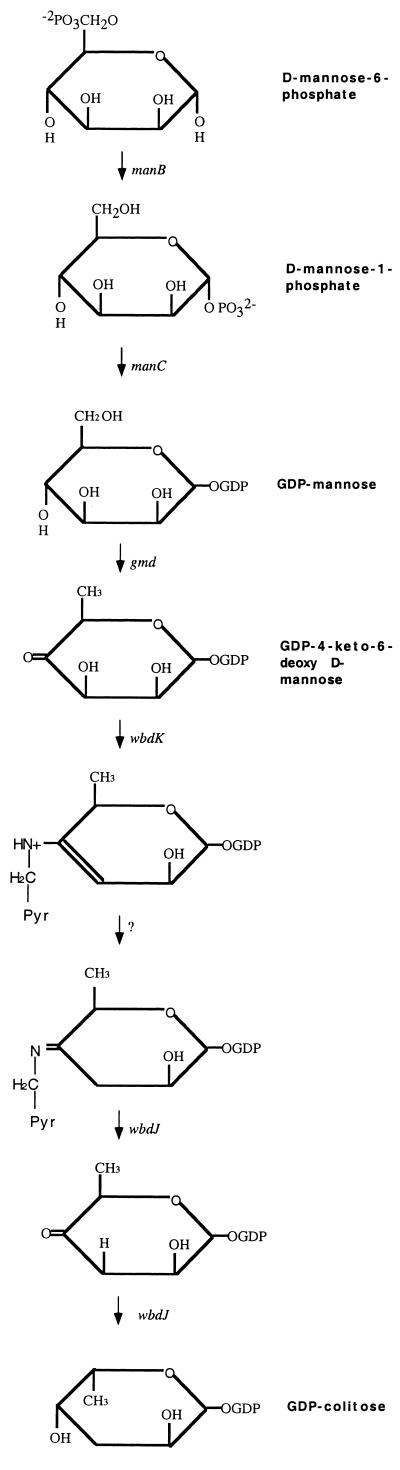
Of the five genes orf-2, orf-4, orf-5, orf-6, and orf-7 orf-2 was newly sequenced. Figure 3 shows the potential GDP-colitose biosynthetic pathway (4). orf-4 and orf-5 were named manC and manB, respectively, on the basis of their high levels of similarity to other manC and manB genes (4). orf-2 has 85.7% identity at the amino acid level to the gmd gene recently identified in the E. coli K-12 colanic acid gene cluster (38) and is deduced to be the expected gmd gene. The next step is a pyridoxamine 5-phosphate-dependent dehydrase reaction (Fig. 3). orf-7 shows similarity (53% similarity at the amino acid level) with ddhC, encoding a CDP-4-keto-6-deoxy-d-glucose-3-dehydrase in the abequose pathway in Yersinia pseudotuberculosis (22), and may be the expected dehydrase. It has been named wbdK. orf-6 has 37.2% identity at the amino acid level with fcl, encoding the fucose synthetase of E. coli colanic acid (3), and was named wbdJ. Fcl is a bifunctional enzyme, converting GDP-4-keto-6-deoxymannose through GDP-4-keto-6-deoxygalactose to GDP-l-fucose by steps similar to the last two steps of the proposed GDP-colitose pathway. Thus, we propose that WbdJ carries out the last two reactions (Fig. 3).
FIG. 3.
Potential biosynthetic pathway of GDP-colitose (4) with gene names. The pathway commences with d-mannose-6-phosphate, which is converted from fructose-6-phosphate by the phosphomannose isomerase encoded by manA. The manA gene is located outside the O-antigen gene cluster (6). Genes assigned to each step are based on sequence similarity described in the text. Pyr, pyridoxamine.
The orf-8 gene was sequenced previously and was shown by sequence comparison to be wzx, now known to be the O-unit flippase gene (4). orf-8 is predicted to encode an integral inner membrane protein with 12 transmembrane segments and shows similarity to many Wzx proteins, including the presence of the conserved motif found within a 50-amino-acid segment near the amino-terminal end of Wzx proteins (38). This gene has been named wzx.
The orf-9 gene encodes a protein with 10 predicted transmembrane segments with a large cytoplasmic loop (70 amino acids). This inner membrane topology is a characteristic feature for all known O-antigen polymerases (24), and we believe that orf-9 is the O-antigen polymerase gene, wzy.
The deduced product of the orf-10 gene shares a low level of similarity (46%) with that of lsi-2 of Neisseria gonorrhoeae. Lsi-2 is responsible for adding GlcNac to galactose in the synthesis of the lipooligosaccharide of N. gonorrhoeae (10). We have named this gene wbdL and suggest that it is the glucose or colitose transferase gene.
The orf-11 gene shares good similarity at both the nucleotide (about 56% identity) and the amino acid (about 65% similarity) levels with TrsE. TrsE is a putative sugar (Gal or GalNAc) transferase in the synthesis of the LPS outer core of Yersinia enterocolitica (36). It has been named wbdM, and we suggest that it encodes the colitose or glucose transferase.
The orf-3 gene shows a high level of similarity to WcaH and WbdQ of the E. coli colanic acid and O157 gene clusters, respectively. The functions of these genes are unknown. We have named it wbdI.
In summary, three putative transferase genes, five identified or putative GDP-colitose synthesis genes, an O-antigen polymerase gene, and a flippase gene were located in the O111 O-antigen gene cluster. There is one gene less than might have been expected because there are two colitose residues with different linkages; however, they account for all the genes expected if a single transferase is responsible for adding both colitose residues to the O unit.
Identification of O111-specific genes.
O-antigen gene clusters generally contain about 8 to 20 genes that fall into three general classes: (i) genes for the synthesis of nucleotide sugar precursors such as dTDP-rhamnose or GDP-N-acetyl-perosamine; (ii) genes for the transfer of sugars to build the O unit; and (iii) genes which carry out specific assembly or processing steps in conversion of the O unit to the O antigen as part of the complete LPS (see the reviews by Reeves [31, 32] and Whitfield [41]). Genes of the first class are commonly present in many O-antigen clusters, and sequence similarity is usually sufficient to identify these genes in database searches and may give cross-reactions in DNA-based assays. Genes of the second class are often group specific because they are specific for both sugars of the linkage. Genes of the third class encode proteins such as the O-antigen polymerase and the flippase; these are most easily identified on the basis of predicted transmembrane segments rather than the sequence per se and may also be O-antigen specific. We have shown that genes of the second and third classes are frequently group specific in S. enterica (unpublished data) and that oligonucleotide primers based on the E. coli O157 O-antigen transferase, wzx, and wzy genes are O157 specific (40).
Thirteen pairs of oligonucleotide primers that bind to the transferase, wzy, and wzx genes of O111 (Table 2) were tested by PCR with each of 28 DNA pools. Each of the 12 pairs that bound to wbdH, wzx, wzy, and wbdM produced a band of the predicted size with the pools containing O111 DNA (pools 22 to 24) and no band of the predicted size with any other pool (Table 2). The pools include DNA from strains representing the 166 known E. coli, 11 O-antigen forms and Y. pseudotuberculosis, 12 Shigella boydii, and 13 S. enterica O-antigen forms. Because pool 21 included DNA from all strains present in pools 22 to 24 other than the O111 strain DNA, we conclude that the 12 pairs of primers all give a positive PCR test result with each of three unrelated O111 strains but not with any of the other strains tested. One pair of primers for wbdL was tested and was found to be nonspecific (Table 2), so no further test was carried out. Thus, two of the three transferase genes and the wzx and wzy genes are highly specific for the O111 gene cluster.
DISCUSSION
We now have the sequence of the entire E. coli O111 O-antigen gene cluster and have identified with various degrees of precision all genes required for synthesis of the O antigen. All genes have low G+C contents (Fig. 1), as observed for other O-antigen gene clusters, and this indicates that the O111 O-antigen gene cluster was acquired by transfer from another species.
We have identified four genes specific to O111. They are highly specific, not being detected by PCR in strains with any of the 165 other known E. coli O-antigen forms or any other bacteria tested, including S. enterica serovar Adelaide, which has the same O antigen as E. coli O111 (21), and lend themselves to PCR-based methods for the identification of O111 strains to replace time-consuming plating and serotyping methods.
Of the 166 reported E. coli O antigens (14, 27, 28, 36), for only about 4 has the gene cluster been fully sequenced. In a 1989 review (12) only 23 structures were listed. Some new structures have been determined more recently, but the majority are still not known, and it remains quite possible that there will be other O antigens with linkages between sugars similar to those in O111. Thus, it is not surprising to find that one of the potential O111 transferase genes (wbdL) exists in another gene cluster. The E. coli serotyping scheme is not yet fully comprehensive, and there are almost certainly other as yet unidentified O antigens. For this reason field strains and conditions need to be tested to confirm the specificity, although we believe that all or most of these genes will be specific to O111 strains. Further specificity can be gained by use of a combination of these genes, perhaps by PCR with primers that bind to adjacent genes. Some of the primers produced bands of the wrong size in some or all of the sample pools. We believe that this is due to chance priming elsewhere on the chromosome. This problem can be avoided by using other primer pairs for those genes.
We previously sequenced the O157 gene cluster of E. coli and identified O157-specific genes (40). In the study described in this paper, we did the same for E. coli O111. The O157:H7 clone has been shown to be an important agent of food-borne disease in humans worldwide and has received particular attention. However, an increasing number of non-O157:H7 Shiga toxin (Stx)-producing E. coli strains including O111 strains have been isolated from humans suffering from HUS and diarrhea, and it is important for laboratories and public health surveillance systems to have the ability to detect and monitor all of these serotypes (18). Because of the very low infective dose of O157:H7 and O111 strains (20, 30), bacteria entering the human food chain can still pose a health problem after enormous dilution, making surveillance difficult. Great efforts have been made to develop a method for the timely and accurate detection of the O157:H7 strain (9, 15, 20, 29) by PCR-based methods, which are particularly useful for the rapid detection of organisms present at low concentrations. PCRs for the detection of O157:H7 strains with probes based on the stx and eaeA genes and a plasmid have been developed, but each probe cross-reacted with other E. coli strains even when only a small number of strains was tested (15). In a recent study of an O111 outbreak in Australia, Paton et al. (30) noticed that probes based on the stx gene not only picked up the O111 strain which was the cause of the outbreak but also picked up many other strains. Thus, the O111- and O157-specific genes identified in this and our previous studies provide much more specific probes for the detection of these two serotypes. Such a test could be a PCR-based test to check foodstuffs, animal feces, and human feces for the presence of O157 and O111 strains.
The currently accepted methods for the detection of Stx-positive O157 strains consist of assays for the detection of Stx, either directly or by PCR, coupled with plating on selective medium, followed by serotyping (O157 O-antigen determination). Most attention has been directed toward detection of the O157:H7 clone, and this has probably led to a higher frequency of detection of small outbreaks caused by this clone. The ability to use β-glucuronidase substrates, cefixime, and telluride for enrichment has made it much easier to detect O157:H7 clones than other Stx-producing E. coli clones causing HUS. In the absence of a specific enrichment, specific PCR tests are of even greater importance for the detection of non-O157:H7 organisms. The use of O157- or O111-specific PCR allows one to use PCR tests for the detection of both stx and the O antigen, avoiding the need to include serological testing and making enrichment less critical.
ACKNOWLEDGMENT
This investigation was supported by Bioproperties (Australia) Pty Ltd.
REFERENCES
- 1.Alexander D C, Valvano M A. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J Bacteriol. 1994;176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul A F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Andrianopoulos K, Wang L, Reeves P R. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998;180:998–1001. doi: 10.1128/jb.180.4.998-1001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bastin D A, Reeves P R. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 5.Bastin D A, Romana L K, Reeves P R. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol Microbiol. 1991;5:2223–2231. doi: 10.1111/j.1365-2958.1991.tb02152.x. [DOI] [PubMed] [Google Scholar]
- 6.Berlyn M K B, Low K B, Rudd K E. Linkage map of Escherichia coli K-12. In: Neidhardt F D, Curtiss III R, Ingraham J L, Low K B, Lin E C C, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: American Society for Microbiology; 1996. pp. 1715–1902. [Google Scholar]
- 7.Bray J. Isolation of antigenically homogeneous strains of Bact. coli neapolitanum from summer diarrhea of infants. J Pathol Bacteriol. 1945;57:239–247. [Google Scholar]
- 8.Caprioli A, Luzzi I, Rosmini F, Resti C, Edefonti A, Perfumo F, Farina C, Goglio A, Gianviti A, Rizzoni G. Community-wide outbreak of hemolytic-uremic syndrome associated with non-O157 verocytotoxin-producing Escherichia coli. J Infect Dis. 1994;169:208–211. doi: 10.1093/infdis/169.1.208. [DOI] [PubMed] [Google Scholar]
- 9.Cubbon M D, Coia J E, Hanson M F, Thomson-Carter F M. A comparison of immunomagnetic separation, direct culture and polymerase chain reaction for the detection of verocytotoxin-producing Escherichia coli O157 in human faeces. J Med Microbiol. 1996;44:219–222. doi: 10.1099/00222615-44-3-219. [DOI] [PubMed] [Google Scholar]
- 10.Danaher R J, Levin J C, Arking D, Burch C L, Sandlin R, Stein D C. Genetic basis of Neisseria gonorrhoeae lipooligosaccharide antigenic variation. J Bacteriol. 1995;177:7275–7279. doi: 10.1128/jb.177.24.7275-7279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutton G G S, Parolis L A S. Polysaccharide antigens of Escherichia coli. In: Dea I G M, editor. Biomedical and biotechnological advances in industrial polysaccharides. Proceedings of the 3rd International Workshop on Recent Developments in Industrial Polysaccharides. New York, N.Y. 1989. [Google Scholar]
- 13.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 14.Ewing W H. Edwards and Ewing’s identification of the Enterobacteriaceae. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. [Google Scholar]
- 15.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gleeson T J, Staden R. An X windows and UNIX implementation of our sequence analysis package. Comput Appl Biosci. 1991;7:398. doi: 10.1093/bioinformatics/7.3.398. [DOI] [PubMed] [Google Scholar]
- 17.Gohmann S, Manning P A, Alpert C A, Walker M J, Timmis K N. Lipopolysaccharide O-antigen biosynthesis in Shigella dysenteriae serotype 1: analysis of the plasmid-carried rfp determinant. Microb Pathog. 1994;16:53–64. doi: 10.1006/mpat.1994.1005. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R P, Clarke R C, Wilson J B, Read S C, Rahn K, Renwick S A, Sandhu K A, Alves D, Karmali M A, Lior H, Mcewen S A, Spika J S, Gyles C L. Growing concerns and recent outbreaks involving non-O157:H7 serotypes of verotoxigenic Escherichia coli. J Food Prot. 1996;59:1112–1122. doi: 10.4315/0362-028X-59.10.1112. [DOI] [PubMed] [Google Scholar]
- 19.Kauffmann F, Dupont A. Escherichia strains from infantile epidemic gastroenteritis. Acta Pathol Microbiol Scand. 1950;27:552–563. doi: 10.1111/j.1699-0463.1950.tb04927.x. [DOI] [PubMed] [Google Scholar]
- 20.Keene W E, McAnulty J M, Hoesly F C, Williams L P, Hedberg K, Oxman G L, Barrett T J, Pfaller M A, Fleming D W. A swimming-associated outbreak of hemorrhagic colitis caused by Escherichia coli O157:H7 and Shigella sonnei. N Engl J Med. 1994;331:579–584. doi: 10.1056/NEJM199409013310904. [DOI] [PubMed] [Google Scholar]
- 21.Kenne L, Lindberg B, Soderholm E, Bundle D R, Griffith D W. Structural studies of the O-antigens from Salmonella greenside and Salmonella adelaide. Carbohydr Res. 1983;111:289–296. doi: 10.1016/0008-6215(83)88313-x. [DOI] [PubMed] [Google Scholar]
- 22.Kessler A, Haase A, Reeves P R. Molecular analysis of the 3,6-dideoxyhexose pathway genes of Yersinia pseudotuberculosis serogroup IIa. J Bacteriol. 1993;175:1412–1422. doi: 10.1128/jb.175.5.1412-1422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lior H. Classification of Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 31–72. [Google Scholar]
- 24.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moyenuddin M, Wachsmuth I K, Moseley S L, Bopp C A, Blake P A. Serotype, antimicrobial resistance, and adherence properties of Escherichia coli strains associated with outbreaks of diarrheal illness in children in the United States. J Clin Microbiol. 1989;27:2234–2239. doi: 10.1128/jcm.27.10.2234-2239.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ørskov I, Ørskov F, Rowe B. Six new E. coli O groups: O165, O166, O167, O168, O169 and O170. APMIS. 1984;92:189–193. doi: 10.1111/j.1699-0463.1984.tb02819.x. [DOI] [PubMed] [Google Scholar]
- 28.Ørskov I, Wachsmuth K, Taylor D N, Echeverria P, Rowe B, Sakazaki R, Ørskov F. Two new Escherichia coli O groups: O172 from ≪Shiga-like≫toxin II-producing strains (EHEC) and O173 from enteroinvasive E. coli (EIEC) APMIS. 1991;99:30–32. doi: 10.1111/j.1699-0463.1991.tb05114.x. [DOI] [PubMed] [Google Scholar]
- 29.Park C H, Vandel N M, Hixon D L. Rapid immunoassay for detection of Escherichia coli O157 directly from stool specimens. J Clin Microbiol. 1996;34:988–990. doi: 10.1128/jcm.34.4.988-990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paton A W, Ratcliff R M, Doyle R M, Seymour-Murray J, Davos D, Lanser J A, Paton J C. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J Clin Microbiol. 1996;34:1622–1627. doi: 10.1128/jcm.34.7.1622-1627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reeves P R. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 32.Reeves P R. Biosynthesis and assembly of lipopolysaccharide. In: Neuberger A, van Deenen L L M, editors. Bacterial cell wall, new comprehensive biochemistry. Vol. 27. Amsterdam, The Netherlands: Elsevier Science Publishers; 1994. pp. 281–314. [Google Scholar]
- 33.Regua A H, Bravo V L, Leal M C, Lobo Leite M E. Epidemiological survey of the enteropathogenic Escherichia coli isolated from children with diarrhoea. J Trop Pediatr. 1990;36:176–179. doi: 10.1093/tropej/36.4.176. [DOI] [PubMed] [Google Scholar]
- 34.Reisner A H, Bucholtz C A, Smelt J, McNeil S. Proceedings of the Twenty-Sixth Annual Hawaii International Conference on Systems Science. Vol. 1. 1993. Australia’s National Genomic Information Service; pp. 595–602. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain- terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skurnik M, Venho R, Toivanen P, Alhendy A. A novel locus of Yersinia enterocolitica serotype O-3 involved in lipopolysaccharide outer core biosynthesis. Mol Microbiol. 1995;17:575–594. doi: 10.1111/j.1365-2958.1995.mmi_17030575.x. [DOI] [PubMed] [Google Scholar]
- 37.Staden R. Graphic methods to determine the function of nucleic acid sequences. A summary of ANALYSEQ options. Nucleic Acids Res. 1984;12:521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson G, Andrianopoulos K, Hobbs H, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viljanen M K, Peltola T, Junnila S Y, Olkkonen L, Jarvinen H, Kuistila M, Huovinen P. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet. 1990;336:831–834. doi: 10.1016/0140-6736(90)92337-h. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Reeves P R. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]