Abstract
Artificial intelligence (AI) and other data-driven technologies hold great promise to transform healthcare and confer the predictive power essential to precision medicine. However, the existing biomedical data, which are a vital resource and foundation for developing medical AI models, do not reflect the diversity of the human population. The low representation in biomedical data has become a significant health risk for non-European populations, and the growing application of AI opens a new pathway for this health risk to manifest and amplify. Here we review the current status of biomedical data inequality and present a conceptual framework for understanding its impacts on machine learning. We also discuss the recent advances in algorithmic interventions for mitigating health disparities arising from biomedical data inequality. Finally, we briefly discuss the newly identified disparity in data quality among ethnic groups and its potential impacts on machine learning.
Keywords: artificial intelligence, health equity, multiethnic machine learning, subpopulation shift, data inequality, transfer learning
INTRODUCTION
Biomedical sciences have become increasingly data driven. In the past two decades, we have witnessed revolutionary new technologies for generating and collecting biomedical data, exemplified by DNA and RNA sequencing and databases containing millions of electronic health records (EHRs). Genomic, transcriptomic, and other high-throughput technologies have become a primary driving force in discovering the molecular basis of disease. Large biobanks are systematically generating genomics and other biomedical data for the participants whose EHRs have also been collected (1-4). Such biomedical datasets provide essential training and testing data for machine learning model development and have become the foundation for building artificial intelligence (AI) capacity for precision medicine (5, 6). Researchers are developing AI models to utilize biomedical data for disease risk prediction and prognosis (7-13). However, the current data foundation for biomedical AI is biased, as most of the critical biomedical datasets were collected from cohorts of predominantly European ancestry (Figure 1). Recent statistics show that over 80% of the data from genome-wide association studies (GWAS) and clinical omics studies were collected from individuals of European ancestry, which constitute less than 20% of the world population (14-18).
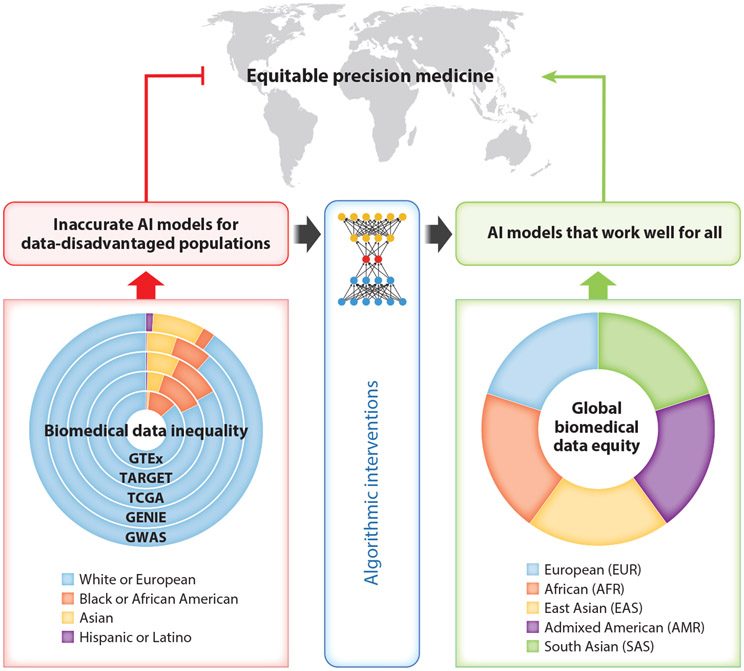
Figure 1.
Addressing the challenge of data inequality for AI-powered precision medicine. The path of the status quo (left) leads to data inequality, a significant obstacle to achieving equitable precision medicine for all. The ring graph on the left shows the ethnic/ancestry compositions of GWAS using data from the GWAS Diversity Monitor (https://gwasdiversitymonitor.com/) and several representative clinical omics studies, including GTEx (https://gtexportal.org/), TARGET (https://ocg.cancer.gov/programs/target), TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), and GENIE (https://www.aacr.org/professionals/research/aacr-project-genie/), using data from GTEx Portal and NCI Genomic Data Commons (https://portal.gdc.cancer.gov/). The ring graph on the right is a conceptual illustration of the goal of biomedical data equity for global populations, represented by the five super-populations defined by the 1000 Genomes Project (135). Algorithmic interventions can attenuate, but may not be able to eliminate, the negative impacts of data inequality. A new path (right) is essential to achieve equitable precision medicine that works well for all ethnic/ancestry groups. Abbreviations: GENIE, Genomics Evidence Neoplasia Information Exchange; GTEx, Genotype-Tissue Expression Project; GWAS, genome-wide association studies; NCI, National Cancer Institute; TARGET, Therapeutically Applicable Research to Generate Effective Treatments; TCGA, The Cancer Genome Atlas.
AI is revolutionizing biomedical research and healthcare, but in the meantime, it is opening a major pathway for data inequality to assert its negative impacts. The inadequate training data has resulted in inaccurate AI models for disease risk assessment, prognostic prediction, and medication usage for the data-disadvantaged populations (14, 19). The disparity in AI model performance is a significant impediment to equitable precision medicine (Figure 1). Precision medicine is poised to be less precise for most of the world’s population because of biomedical data inequality.
Recent studies show that multiethnic machine learning schemes differ significantly in their performance in the presence of data inequality and that transfer learning is an effective strategy to improve machine learning model performance on data-disadvantaged populations. In the following sections, we discuss the current status of biomedical data inequality among ethnic groups, the ongoing efforts to increase ethnic diversity in biomedical data, the impacts of data inequality and subpopulation shift on multiethnic machine learning, and the advances in machine learning strategies to mitigate the negative impacts of biomedical data inequality.
BIOMEDICAL DATA INEQUALITY
Biomedical data inequality has existed for a long time but has only recently been brought to wide attention (14-18). As biomedical research enters the era of big data, many large-scale datasets have been generated in recent years. These datasets provide unprecedented opportunities for data-driven knowledge discovery and enable the development of sophisticated AI models. However, severe data inequality widely exists in biomedical datasets. Table 1 shows examples of data inequality in some highly influential biomedical datasets, providing a snapshot of the degree of biomedical data inequality in a wide range of studies on health and disease. The data inequality is particularly severe in large-scale genomic, transcriptomic, proteomic, and other omic data (18). Statistics from the National Human Genome Research Institute (USA) provide an overview of the populations included in large-scale genomic studies: 87% European, 10% Asian, 8.5% unreported, 2% African, 1% Hispanic, and 0.5% others (20).
Table 1.
Examples of data inequality in biomedical datasets
| Dataset | Disease/phenotype | Data type | Ethnicity compositiona | Reference(s) | URL |
|---|---|---|---|---|---|
| ADNI-3 | Alzheimer’s disease | Genotype and image | Caucasian 86%, other 14% | 118, 119 | https://adni.loni.usc.edu/ |
| GENIE | Cancers | Genomic variation | White 87%, Black or African American 6%, Asian 5%, other 2% | 120 | https://www.aacr.org/professionals/research/aacr-project-genie/ |
| GTEx (v8) | Gene expression in normal tissues | Genotype and transcriptome | White 85%, African American 13%, Asian 1%, unknown 1% | 121, 122 | https://gtexportal.org/ |
| GWAS | Various | Genotype and phenotype | European 88%, Asian 8%, African, African American or Afro-Caribbean 2%, Hispanic or Latin American 1%, other/mixed 1% | 16 | https://gwasdiversitymonitor.com/ |
| Million Veteran Program | Various | Genotype and electronic health record | European 70%, African 19%, admixed American 9%, Asian 2% | 4, 123 | https://www.mvp.va.gov/pwa/ |
| MIMIC-IV | Various | Electronic health record | White 77%, Black or African American 10%, Asian 3%, Hispanic/Latino 4%, other 6% | 62, 124, 125 | https://doi.org/10.13026/07hj-2a80 |
| SHHS | Cardiovascular diseases related to sleep-disordered breathing | Electronic health record | White 86%, Black 9%, other 5% | 126, 127 | https://sleepdata.org/datasets/shhs |
| TARGET | Pediatric cancers | Multiomics | White 80%, Black or African American 13%, Asian 5%, other 2% | NAb | https://ocg.cancer.gov/programs/target |
| TCGA | Cancers | Multiomics | European ancestry 82%, African ancestry 6%, East Asian ancestry 6%, admixed ancestry 4%, other 2% | 33 | https://cancergenome.nih.gov/ |
| UK Biobank | Various | Genotype, genome sequence, and electronic health record | European ancestry 95%, African ancestry 2%, Central/South American ancestry 2%, East Asian ancestry 1% | 128 | https://www.ukbiobank.ac.uk/ |
The original terms from the information sources are used. The percentages were calculated using the patients with known race/ethnicity/ancestry information (as of August 2022).
Ethnicity composition numbers for TARGET were derived from the NCI Genomic Data Commons (https://portal.gdc.cancer.gov/).
Abbreviations: ADNI, The Alzheimer’s Disease Neuroimaging Initiative; GENIE, Genomics Evidence Neoplasia Information Exchange; GTEx, The Genotype-Tissue Expression Project; GWAS, genome-wide association studies; MIMIC-IV, Medical Information Mart for Intensive Care, version IV; NA, not any; NCI, National Cancer Institute; SHHS, Sleep Heart Health Study; TARGET, Therapeutically Applicable Research to Generate Effective Treatments; TCGA, The Cancer Genome Atlas.
During the past decade, GWAS have become the most important source of knowledge on the genetic architecture of complex diseases (21). GWAS data also provide the basis for developing polygenic disease prediction models (22-29). Recent studies show that GWAS data inequality between the European and other ancestry populations is overwhelming (16, 18). The GWAS Diversity Monitor (https://gwasdiversitymonitor.com/) tracks the ancestral diversity of thousands of GWAS and shows that over 85% of GWAS data were collected from individuals of European descent, and the diversity has not improved in recent years (16). As we discuss more thoroughly in the following sections, such data inequality hinders equity in multiethnic machine learning and may lead to new health disparities.
SUBPOPULATION SHIFT
In machine learning, subpopulation shift refers to data distribution discrepancies among subpopulations. Here, we focus on subpopulations defined by ancestry or ethnicity. Researchers have observed ancestry- or ethnicity-associated differences in genetic and somatic DNA mutation (30-33), epigenetic modification (33-37), RNA and protein expression (33, 38-42), metabolic signatures (43-46), and microbiome profiles (47-49) in a wide range of biological processes critical to human health and diseases (32, 33, 38, 50-52). From the data science perspective, this indicates that the natural data generation mechanism may vary among populations of different ancestries. Such variations can lead to discrepancies in biomedical data distribution among ancestry groups, which has profound implications for multiethnic machine learning strategies.
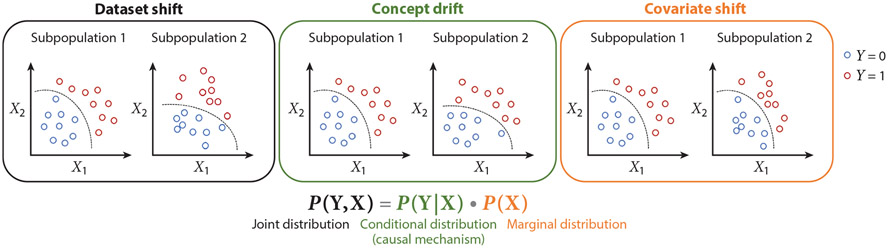
A machine learning problem consists of a domain and a learning task . The domain consists of a feature space and a probability distribution , where represents the input features. The learning task consists of a label space and a predictive function , learned from the feature–label pairs (, ). From the probabilistic perspective, can be written as , where represents the prediction targets. In machine learning, it is generally assumed that each (, ) is drawn from a single distribution . However, this assumption is violated due to the data distribution discrepancy across subpopulations. Given , both the marginal distribution and the conditional distribution may contribute to the joint distribution discrepancy. The marginal distribution and the conditional distribution correspondence to two types of dataset shift and have different implications for multiethnic machine learning. Here we consider a population consisting of two subpopulations. A covariate shift (53) is a scenario where but , while a concept drift (53, 54) is a scenario where but . A dataset shift (53, 55) is a more general scenario where at least one of the marginal or conditional distributions is different (Figure 2). Subpopulation shift is essentially a dataset shift (53, 55) caused by a data distribution discrepancy among subpopulations.
Figure 2.
A conceptual framework for elucidating the data distribution discrepancies among subpopulations and their implications for machine learning. We consider a population consisting of two subpopulations 1 and 2, where represents the input features for machine learning and represents the prediction target variable. From the machine learning perspective, the two subpopulations can be viewed as two domains. Covariate shift is the situation where the marginal distributions of the two domains are different while the conditional distributions of the two domains are the same. Concept drift is the situation where the conditional distributions of the two domains are different while the marginal distributions of the two domains are the same. Dataset shift is a more general situation where the joint distributions of the two domains are different because at least one of the conditional and marginal distributions is different. Given the relationship between the joint, conditional, and marginal distributions, covariate shift and concept drift are two special cases of dataset shift. The dashed curves represent the decision boundaries separating the two classes of the samples (Y = 0 and Y = 1). A decision boundary is determined by the conditional distribution that represents the causal mechanism (136) to generate from .
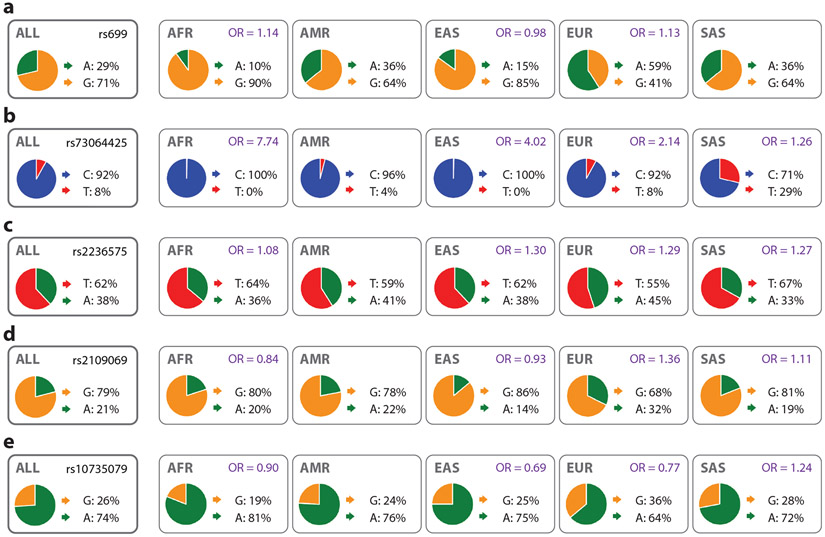
The genetic architectures of many diseases, mainly represented by the allele frequencies and effect sizes of the causal genetic variants, vary among ancestry groups (56-59). For instance, the allele frequency of rs699, a single-nucleotide variant (SNV) associated with hypertension, varies across different populations (Figure 3a). This SNV has two alleles: A (associated with lower arterial pressure) and G, with overall allele frequencies of 29% and 71%, respectively. However, the allele frequencies vary significantly among the five global super-populations defined by the 1000 Genomes Project: admixed American (AMR), African (AFR), East Asian (EAS), European (EUR), and South Asian (SAS). Allele A is the major allele in the European population with a frequency of 59% while being the minor allele in the non-European populations. The allele frequency also varies (to lesser extents) among the subpopulations of each of the five global ancestry populations. The effect size (odds ratio) of rs699 on preeclampsia, a severe pregnancy complication characterized by hypertension, varies among ancestry groups (60) (Figure 3a). The genetic architecture of COVID-19 also varies among ancestry groups (61). The allele frequencies and effect sizes of four genetic variants (rs73064425, rs2236575, rs2109069, and rs10735079) associated with the critical illness caused by COVID-19 vary significantly across the ancestry groups (Figure 3b-e).
Figure 3.
Allele frequencies and effect sizes of genetic variants across five global super-populations defined by the 1000 Genomes Project: African (AFR), admixed American (AMR), East Asian (EAS), European (EUR), and South Asian (SAS). The odds ratio (OR) values represent the effect sizes of the genetic variants on (a) preeclampsia (60) and (b–e) COVID-19 (61) in different populations (where the data are available). Figure generated using the Ensembl Genome Browser (137) webpages, which show the allele frequencies of the five genetic variants (data from the 1000 Genomes Project phase 3).
In polygenic disease prediction, genotypes of the genetic variants associated with the disease are used as input features (), and the disease status is the prediction target (). The marginal distribution represents the allele frequencies of the causal genetic variants. The conditional distribution represents the dependency of the disease on the genotype of the causal genetic variants, which is mainly determined by their effect sizes on the disease. The allele frequencies and effect sizes of these causal genetic variants may vary among different subpopulations, leading to marginal and conditional distribution discrepancies. Similarly, the distribution of other molecular features (e.g., mRNA and protein expression) and their effects on the diseases may also vary among ancestry or ethnic groups (19), leading to subpopulation shift. Data inequality and subpopulation shift also exist in EHR datasets. For example, about 77% of patients with known ethnicities in MIMIC-IV (Medical Information Mart for Intensive Care, version IV) (62), the largest publicly available EHR dataset, are white (based on self-reported demographic data). For many clinical laboratory tests, there are significant value distribution differences among ancestry or ethnic groups (63, 64), suggesting the reference intervals (i.e., normal ranges) for these tests should be ethnicity dependent (65).
Making clinical decisions with AI models built using inadequate and incompatible data confers health risks for data-disadvantaged populations (66). Polygenic scores and medical AI models developed using data from cohorts of predominantly European ancestry show significantly lower performance on non-European populations (14, 19, 67-75). Despite the highly nonlinear genotype–phenotype relationship and nonadditive genetic interactions, linear polygenic models are widely used for disease risk prediction (29, 76). In the multiple linear regression framework, polygenic prediction for disadvantaged populations can be enhanced by calibrating parameters for genetic effect sizes or model sparsity patterns across ethnic groups (77-82). However, the linear polygenic models do not have the sufficient expressive capacity to learn and transfer complex representations across subpopulations with different genetic architectures. Recent studies indicate that the deep learning models capable of capturing complex nonlinear interactions generally outperform the linear disease prediction models (83-85).
MULTIETHNIC MACHINE LEARNING
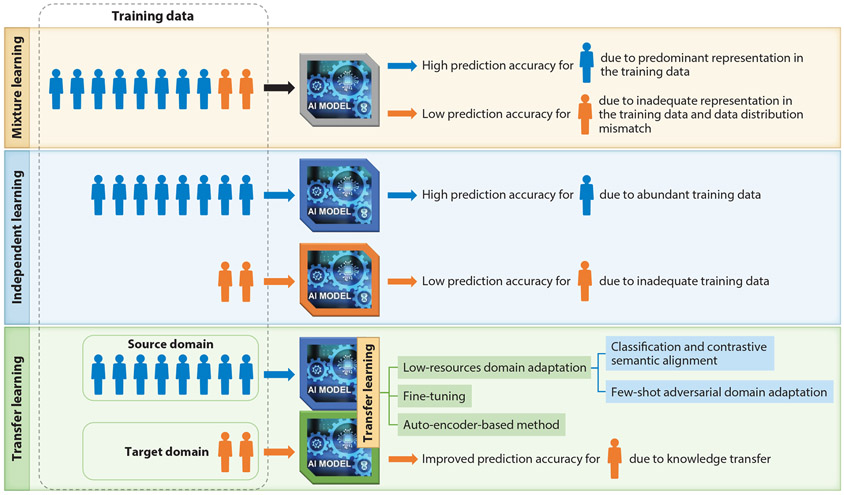
We have defined three categories for multiethnic machine learning schemes based on how they utilize the data from different subpopulations: mixture learning, independent learning, and transfer learning (19) (Figure 4). The mixture learning scheme indistinctly uses data from all subpopulations for model training. Currently, mixture learning is used as the standard machine learning scheme for multiethnic data. In the presence of data inequality, the performance of the mixture learning model on different subpopulations can be very different. The overall performance of the mixture learning model is mainly driven by its performance for the predominant subpopulation in the dataset. Its performance for the smaller subpopulations is often significantly lower due to inadequate representation in the training data and data distribution discrepancies with the predominant subpopulation. Another multiethnic machine learning scheme is independent learning, which uses data from different subpopulations separately to train an independent model for each subpopulation. The independent learning scheme also tends to generate machine learning models with low performance for the smaller subpopulations due to inadequate training data. In the transfer learning (86-92) scheme, knowledge learned from the data-rich subpopulation (source domain) is transferred to assist the learning task for the data-disadvantaged subpopulation (target domain).
Figure 4.
Multiethnic machine learning schemes. The mixture learning scheme indistinctly uses data from all subpopulations in model training. The independent learning scheme uses data from different subpopulations separately to train an independent model for each group. In the transfer learning scheme, knowledge learned from the data-rich subpopulation (source domain) is transferred to assist the learning task for the data-disadvantaged subpopulation (target domain).
The current prevalent machine learning scheme for multiethnic data, the mixture learning scheme, and its main alternative, the independent learning scheme, have major obstacles in training optimal machine learning models for data-disadvantaged subpopulations (19, 93-95). The two major challenges in multiethnic machine learning are data inequality and subpopulation shift. Both challenges can be addressed with transfer learning (Figure 5). In transfer learning, we consider a source domain with labeled samples and a target domain with labeled samples, where the represent features and the represent labels. For multiethnic machine learning tasks, data from an ethnic group with a larger sample size is designated as the source domain, and data from an ethnic group with a smaller sample size is designated as the target domain (Figure 4). The knowledge learned from the source domain can be transferred to assist in developing a machine learning model for the target domain.
Figure 5.
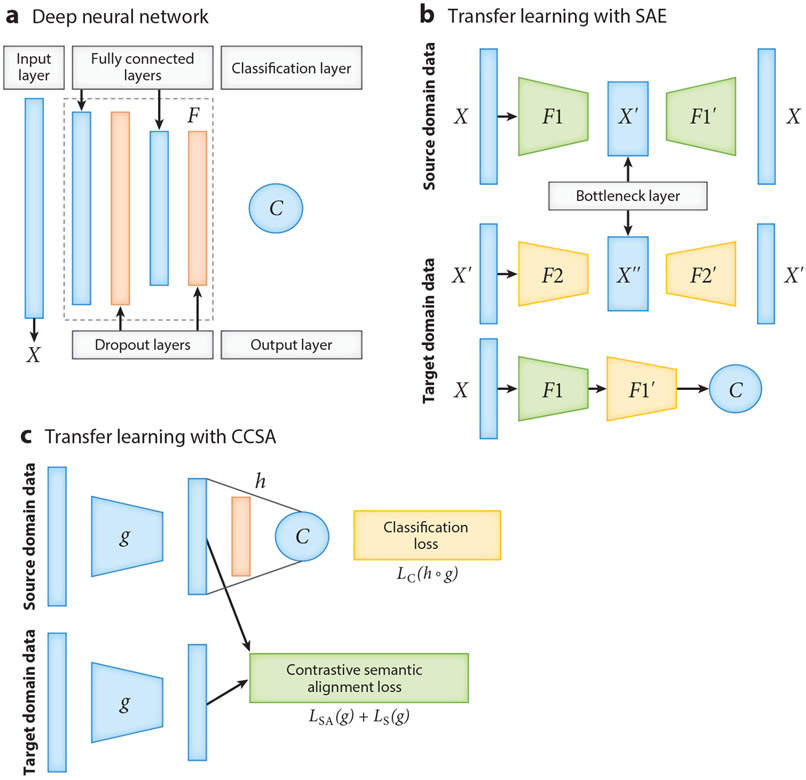
The neural network architectures for deep learning and deep transfer learning. (a) An example architecture of a deep neural network model, which includes an input layer; several hidden layers (marked as ), including fully connected layers and dropout layers; and one output layer . More fully connected layers can be added to the deep neural network model. (b) The neural network architecture of a stacked denoising auto-encoder (SAE) for transfer learning. (or ) is the encoder with two layers, including a fully connected layer and a dropout layer; (or ) is the decoder; the first and the second rows provide the structure of the first and second auto-encoders, respectively; and is a regression or classification layer. (c) The neural network architecture of classification and contrastive semantic alignment (CCSA) (105). CCSA minimizes the loss function , where represents the composition of a function that maps the input data to an embedding space and a function used to predict the output label from ; is a classification layer; is the classification loss; is the semantic alignment loss; is the separation loss; and is the weight used to balance the classification loss versus the contrastive semantic alignment loss .
As the primary driving force of the recent AI advances, deep neural networks (DNNs) consisting of multiple layers of connected artificial neurons (Figure 5a) have outperformed traditional machine learning systems in a wide range of applications (96). DNNs are also particularly suitable for transfer learning, as they can learn transferable features that generalize well to novel tasks for domain adaptation (97). However, most deep learning and deep transfer learning algorithms were developed initially for visual recognition and language processing tasks, which provide rich algorithm resources but not an off-the-shelf solution that one can directly apply to tabular biomedical data. Machine learning experiments on genomic prediction of disease occurrence and omics-based disease prognosis have shown that transfer learning can significantly improve the predictive accuracy for data-disadvantaged subpopulations (19, 93-95, 98, 99). Here we discuss three transfer learning strategies that have been adapted and applied to mitigate the negative impacts of biomedical data inequality: a fine-tuning method, an auto-encoder-based method, and a domain adaptation method.
Fine-tuning is frequently used as a transfer learning method to improve DNN model performance and generalization (100). The general fine-tuning procedure involves (a) training a DNN on the source domain (a large subpopulation), (b) cutting off some layers of the network and replacing them with randomly initialized layers, and (c) tuning the network using backpropagation on the target domain (a smaller subpopulation) until the validation loss starts to increase. The key issue in the fine-tuning approach is the transferability of the layers. One can test the transferability of the layers along the DNNs by (a) changing the cutoff point in the network from where the bottom or top n layers will be frozen or fine-tuned and (b) setting different learning rates for each layer to find the optimal cutoff point and learning rate distribution for fine-tuning (100).
We developed an auto-encoder-based transfer learning strategy for improving cancer classification (101) and improving cancer prognosis prediction for data-disadvantaged ethnic groups (19). The method is based on stacked denoising auto-encoders (SAE) and uses unlabeled data from the source domain and labeled data from the target domain (Figure 5b). The basic idea is that using unlabeled data of the source domain to initialize the network parameters would improve the performance for the target domain. The SAE maps the input feature into different levels of representation and reconstructs it from the mapped space. During the training, the source domain data are used to pretrain an SAE, and then the model is fine-tuned using target domain data. The key parameters for this method include the number of SAE layers and their sizes.
Domain adaptation (102, 103) is a class of transfer learning methods that improve machine learning performance on the target domain by adjusting the distribution discrepancy across domains. The source domain and target domain are sampled from two different joint distributions, and , respectively. As discussed in the previous section, the difference between joint distributions may stem from the conditional distribution or the marginal distribution . Many domain adaptation methods can only handle marginal distribution adjustment (104). However, both marginal and conditional distributions may differ between subpopulations. It is essential to select domain adaptation methods that can simultaneously address the two significant challenges in multiethnic machine learning: the small sample size of the data-disadvantaged subpopulation (target domain) and the discrepancy of data distribution (both marginal and conditional distributions) between subpopulations. Low-resource domain adaptation methods such as classification and contrastive semantic alignment (CCSA) (105) are particularly suitable for addressing these challenges because (a) these methods can significantly improve target domain prediction accuracy by using very few labeled target samples in training and (b) these methods include semantic alignment in training and therefore can handle the domain discrepancy in both marginal and conditional distributions. The CCSA domain adaptation method (Figure 5c) utilizes a loss function comprising three terms: classification loss, semantic alignment loss, and separation loss. The semantic alignment loss is used to minimize the distance between samples of the same class but from different domains, the separation loss is used to maximize the distance between samples of different classes and domains, and the classification loss is used to maximize the prediction accuracy (105).
Subpopulation shift has been addressed by enforcing predictive performance parity on subpopulations (106). However, a fundamental challenge for machine learning fairness research (107-109) is the inherent trade-off between fairness and prediction accuracy (110, 111). The transfer learning scheme is not subject to this dilemma. In transfer learning, a machine learning model trained on a data-rich subpopulation (source domain) can aid in training a model for a data-disadvantaged subpopulation (target domain) without affecting its own prediction accuracy. Thus, transfer learning provides a Pareto improvement (112) for multiethnic machine learning (95). Pareto improvement is a generally desired scenario in which some parties are better off without negatively impacting other parties in the system.
In studies of the impacts of data inequality on machine learning (14, 19), the gap or ratio of the model performance metrics between groups is often used to measure the disparity of machine learning model performance across subpopulations. It should be noted that some performance metrics may not be suitable for evaluating machine learning model performance on data-disadvantaged populations with small sample sizes. As shown by Davis & Goadrich (113), the interpolation property of the precision-recall (PR) curve (114) may lead to inaccurate calculation of the area under the PR curve when the sample size is small. In contrast, the receiver operating characteristic curve (115) does not have this problem (113), thus providing a more stable performance metric for data-disadvantaged populations.
MACHINE LEARNING WITH MORE ANCESTRALLY BALANCED DATA
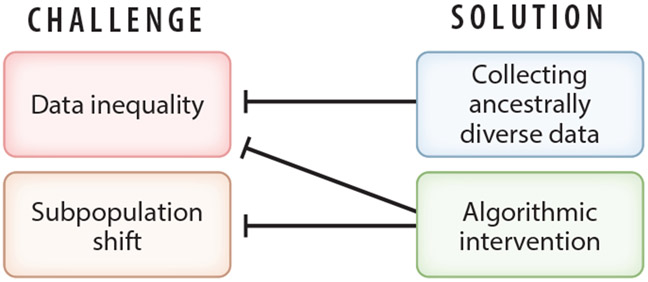
Machine learning experiments on synthetic data show that data inequality and subpopulation shift are the key factors underlying model performance disparities (19, 95). Currently, these challenges in multiethnic machine learning are being addressed on two fronts: data collection and algorithmic intervention (Figure 6). Large-scale efforts are underway to collect biomedical data from diverse populations (116). Table 2 lists some examples of current efforts to collect data from diverse or data-disadvantaged populations (this is by no means a complete list). Given the severe and ubiquitous biomedical data inequality that has accumulated for decades, there is a long way to go to achieve global biomedical data equity (Figure 1). As a result, medical AI faces a long-term challenge in attenuating the negative impacts of biomedical data inequality. However, we can expect the degree of data inequality to decrease gradually. Therefore, it is crucial to understand how the performance of different machine learning schemes changes as a function of the degree of data inequality. Recent experiments on synthetic data indicate that multiethnic machine learning schemes still perform differently even when data inequality is eliminated (i.e., different ancestry groups having the same sample size) because of different responses to the data distribution discrepancy among ancestry groups (subpopulation shift) (19).
Figure 6.
Data inequality and subpopulation shift are the two key challenges in multiethnic machine learning. These challenges are being addressed on two fronts. Collecting more ancestrally diverse data will gradually reduce the degree of data inequality, and algorithmic intervention (e.g., transfer learning) can mitigate the impacts of data inequality and subpopulation shift on multiethnic machine learning.
Table 2.
Examples of ongoing efforts to collect more ancestrally diverse biomedical data
| Project | Disease/ phenotype |
Data type | Populationsa | Reference(s) | URL |
|---|---|---|---|---|---|
| All of Us | Various | Genome sequence, omics data, and EHR | Black, African American or African; Asian; Hispanic Latino or Spanish; and White | 2, 129 | https://allofus.nih.gov/ |
| Ancestry Networks for the Human Cell Atlas | Healthy human cell | Single-cell multiomics | African, African American, Afro-Caribbean, Asian, Latinx, and Middle Eastern populations | 130 | https://chanzuckerberg.com/rfa/ancestry-networks-human-cell-atlas/ |
| Global Biobank Meta-Analysis Initiative | Various | Genotype and EHR | Global populations | 131 | https://www.globalbiobankmeta.org/ |
| H3 Africa | Various | Genotype and EHR | African | 132 | https://h3africa.org/ |
| The PAGE Study | Various | Genotype and phenotype | African American, Asian, Hispanic/Latino, Native American, Native Hawaiian | 133 | https://www.pagestudy.org/ |
| TOPMed | Heart, lung, blood, and sleep disorders | Multiomics | African, Asian, European, and Hispanic/Latino ancestry populations | 134 | https://topmed.nhlbi.nih.gov/ |
| WW-ADNI | Alzheimer’s disease | Genotype and image | Argentina, Australia, Canada, China, Japan, Korea, Mexico, North America, and Taiwan | 118 | https://www.alz.org/research/for_researchers/partnerships/wwadni |
The original terms from the information sources are used.
The Global Biobank Meta-Analysis Initiative includes 24 biobanks with more than 2.2 million genotyped samples (as of January 2023) from different origins and ancestries (https://www.globalbiobankmeta.org/). Of the 24 biobanks, 9 are in North America, 8 are in Europe, 5 are in Asia, 1 is in Africa, and 1 is in Australia.
Abbreviations: EHR, electronic health record; H3 Africa, Human Heredity and Health in Africa; PAGE, Population Architecture using Genomics and Epidemiology; TOPMed, Trans-Omics for Precision Medicine; WW-ADNI, Worldwide Alzheimer’s Disease Neuroimaging Initiative.
Understating the influence of data inequality on machine learning has important implications for resource allocation in biomedical data collection and generation. For example, proportional representation is widely accepted and implemented as a criterion for equity in resource allocation. However, although the population of the United States is more ancestrally diverse than most developed countries, proportional representation in the United States means that only about 27% of the data will be collected from all non-European ancestry groups combined, which can still lead to significant disparities in AI model performance. Therefore, using proportional representation in the developed countries where most biomedical studies are conducted is not adequate for achieving health equity from a machine learning perspective. Collecting approximately equal amounts of biomedical data from all ancestry groups is essential to achieving equitable AI-empowered precision medicine.
DATA QUALITY DISPARITY
Current research on biomedical data inequality almost exclusively focuses on the disparity in data quantity. However, recent research provides evidence of significant disparity in data quality between ancestry groups (117). Wickland et al. (117) found that exome sequencing coverage is lower for patients of African ancestry in data from The Cancer Genome Atlas, which may hamper the detection of the African-specific DNA variants. At this point, it is unclear whether there is a widespread data quality disparity among ancestry or ethnic groups in biomedical datasets. The data quality disparity is a serious issue that can broadly impact biomedical research and healthcare, and it warrants a thorough investigation. The data quality disparity can also exacerbate the existing disparity in multiethnic machine learning because low-quality data from the disadvantaged populations provide weaker and noisier signals that are more difficult for machine learning models to capture and utilize. In light of the discovery of biomedical data quality disparity, the concept of data inequality can be expanded to include not only disparity in data quantity but also disparity in data quality.
SUMMARY POINTS.
Biomedical data inequality confers a significant health risk for people of non-European ancestry, which constitute over 80% of the world’s population.
Artificial intelligence (AI) greatly empowers precision medicine, but in the meantime, it opens a major pathway for biomedical data inequality to manifest and amplify its health risks to data-disadvantaged groups.
AI-empowered precision medicine is set to be less precise for data-disadvantaged populations, which can generate new health disparities.
These new health disparities can impact any disease where data inequality exists, so the negative impacts would be broad.
Algorithmic interventions such as using transfer learning can mitigate the negative impacts of data inequality.
In many cases, transfer learning provides a generally desired Pareto improvement in multiethnic machine learning, and it is not subject to the dilemma between fairness and prediction accuracy.
There is an urgent need to improve the ethnic (or ancestral) diversity in biomedical data, and proportion representation is insufficient to build the data foundation for equitable AI-empowered precision medicine in developed countries.
Even as the ethnic (or ancestral) diversity in biomedical data increases, the subpopulation shift will remain a significant challenge for multiethnic machine learning, which can be addressed with algorithms such as transfer learning.
ACKNOWLEDGMENTS
This work was supported by the US National Cancer Institute grant R01CA262296.
Glossary
- Biomedical data inequality
the significant disparity in the amount of data collected from populations of different ancestries or ethnicities
- Multiethnic machine learning
machine learning using data from a population consisting of multiple subpopulations of different ancestries or ethnicities
- Polygenic disease prediction
predicting disease risk or occurrence using the genotype data of multiple genetic variants associated with the disease
- Effect size
the effect size of a causal genetic variant on a disease represents the strength of its influence on the phenotype or disease and is usually expressed as an odds ratio in GWAS
- Causal genetic variant
the DNA variation responsible for the variation of a phenotype or disease in a population
- Artificial neuron
the basic computing unit in artificial neural networks that transforms the input signals into an output signal using an activation function
- Auto-encoder
a type of neural network for unsupervised learning of efficient data representations through a process of encoding and decoding for reconstructing the input data
- Loss function
the difference between estimated and true outputs of the machine learning model; during training and validation, it is used to optimize the model parameters for high prediction accuracy
- Pareto improvement
a change in a system that results in a new situation where some parties in the system are better off, and no party is worse off
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Halldorsson BV, Eggertsson HP, Moore KHS, Hauswedell H, Eiriksson O, et al. 2022. The sequences of 150,119 genomes in the UK Biobank. Nature 607:732–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.All Us Res. Progr. Investig. 2019. The “All of Us” Research Program. N. Engl. J. Med 381:668–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karczewski KJ, Snyder MP. 2018. Integrative omics for health and disease. Nat. Rev. Genet 19:299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, et al. 2016. Million Veteran Program: a megabiobank to study genetic influences on health and disease. J. Clin. Epidemiol 70:214–23 [DOI] [PubMed] [Google Scholar]
- 5.Li R, Chen Y, Ritchie MD, Moore JH. 2020. Electronic health records and polygenic risk scores for predicting disease risk. Nat. Rev. Genet 21:493–502 [DOI] [PubMed] [Google Scholar]
- 6.Zhang A, Xing L, Zou J, Wu JC. 2022. Shifting machine learning for healthcare from development to deployment and from models to data. Nat. Biomed. Eng 6:1330–45 [DOI] [PubMed] [Google Scholar]
- 7.Uddin S, Khan A, Hossain ME, Moni MA. 2019. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inform. Decis. Making 19:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho DSW, Schierding W, Wake M, Saffery R, O’Sullivan J. 2019. Machine learning SNP based prediction for precision medicine. Front. Genet 10:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Cui Y. 2022. Clinical time-to-event prediction enhanced by incorporating compatible related outcomes. PLOS Digital Health 1(5):e0000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ching T, Zhu X, Garmire LX. 2018. Cox-nnet: An artificial neural network method for prognosis prediction of high-throughput omics data. PLOS Comput. Biol 14(4):e1006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajkomar A, Dean J, Kohane I. 2019. Machine learning in medicine. N. Engl. J. Med 380:1347–58 [DOI] [PubMed] [Google Scholar]
- 12.Cheerla A, Gevaert O. 2019. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics 35:i446–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leist AK, Klee M, Kim JH, Rehkopf DH, Bordas SPA, et al. 2022. Mapping of machine learning approaches for description, prediction, and causal inference in the social and health sciences. Sci. Adv 8:eabk1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. 2019. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51:584–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerrero S, López-Cortés A, Indacochea A, García-Cárdenas JM, Zambrano AK, et al. 2018. Analysis of racial/ethnic representation in select basic and applied cancer research studies. Sci. Rep 8:13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills MC, Rahal C. 2020. The GWAS Diversity Monitor tracks diversity by disease in real time. Nat. Genet 52:242–43 [DOI] [PubMed] [Google Scholar]
- 17.Gurdasani D, Barroso I, Zeggini E, Sandhu MS. 2019. Genomics of disease risk in globally diverse populations. Nat. Rev. Genet 20:520–35 [DOI] [PubMed] [Google Scholar]
- 18.Sirugo G, Williams SM, Tishkoff SA. 2019. The missing diversity in human genetic studies. Cell 177:26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y, Cui Y. 2020. Deep transfer learning for reducing health care disparities arising from biomedical data inequality. Nat. Commun 11:5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natl. Hum. Genome Res. Inst. 2021. Diversity in genomic research. Fact Sheet, Natl. Hum. Genome Res. Inst., Bethesda, MD. https://www.genome.gov/about-genomics/fact-sheets/Diversity-in-Genomic-Research [Google Scholar]
- 21.Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, et al. 2021. Genome-wide association studies. Nat. Rev. Methods Primers 1:59 [Google Scholar]
- 22.Torkamani A, Wineinger NE, Topol EJ. 2018. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet 19:581–90 [DOI] [PubMed] [Google Scholar]
- 23.Lambert SA, Abraham G, Inouye M. 2019. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet 28:R133–42 [DOI] [PubMed] [Google Scholar]
- 24.Lewis CM, Vassos E. 2020. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SW, Mak TS-H, O’Reilly PF. 2020. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15:2759–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wray NR, Lin T, Austin J, McGrath JJ, Hickie IB, et al. 2021. From basic science to clinical application of polygenic risk scores: a primer. JAMA Psychiatry 78:101–09 [DOI] [PubMed] [Google Scholar]
- 27.Polygenic Risk Score Task Force Int. Common Dis. Alliance. 2021. Responsible use of polygenic risk scores in the clinic: potential benefits, risks and gaps. Nat. Med 27:1876–84 [DOI] [PubMed] [Google Scholar]
- 28.Kullo IJ, Lewis CM, Inouye M, Martin AR, Ripatti S, Chatterjee N. 2022. Polygenic scores in biomedical research. Nat. Rev. Genet 23:524–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Zhou X. 2021. Genetic prediction of complex traits with polygenic scores: a statistical review. Trends Genet. 37:995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Özdemir BC, Dotto G-P. 2017. Racial differences in cancer susceptibility and survival: more than the color of the skin? Trends Cancer 3:181–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oak N, Cherniack AD, Mashl RJ, Hirsch FR, Ding L, et al. 2020. Ancestry-specific predisposing germline variants in cancer. Genome Med. 12:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, et al. 2018. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell 34:549–60.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrot-Zhang J, Chambwe N, Damrauer JS, Knijnenburg TA, Robertson AG, et al. 2020. Comprehensive analysis of genetic ancestry and its molecular correlates in cancer. Cancer Cell 37:639–54.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmad A, Azim S, Zubair H, Khan MA, Singh S, et al. 2017. Epigenetic basis of cancer health disparities: looking beyond genetic differences. Biochim. Biophys. Acta 1868:16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia Y-Y, Ding Y-B, Liu X-Q, Chen X-M, Cheng S-Q, et al. 2014. Racial/ethnic disparities in human DNA methylation. Biochim. Biophys. Acta 1846:258–62 [DOI] [PubMed] [Google Scholar]
- 36.Galanter JM, Gignoux CR, Oh SS, Torgerson D, Pino-Yanes M, et al. 2017. Differential methylation between ethnic sub-groups reflects the effect of genetic ancestry and environmental exposures. eLife 6:e20532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahmani E, Shenhav L, Schweiger R, Yousefi P, Huen K, et al. 2017. Genome-wide methylation data mirror ancestry information. Epigenet. Chromatin 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bisogno LS, Yang J, Bennett BD, Ward JM, Mackey LC, et al. 2020. Ancestry-dependent gene expression correlates with reprogramming to pluripotency and multiple dynamic biological processes. Sci. Adv 6:eabc3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park CS, De T, Xu Y, Zhong Y, Smithberger E, et al. 2019. Hepatocyte gene expression and DNA methylation as ancestry-dependent mechanisms in African Americans. NPJ Genom. Med 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sjaarda J, Gerstein HC, Kutalik Z, Mohammadi-Shemirani P, Pigeyre M, et al. 2020. Influence of genetic ancestry on human serum proteome. Am. J. Hum. Genet 106:303–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mogil LS, Andaleon A, Badalamenti A, Dickinson SP, Guo X, et al. 2018. Genetic architecture of gene expression traits across diverse populations. PLOS Genet. 14:e1007586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gay NR, Gloudemans M, Antonio ML, Abell NS, Balliu B, et al. 2020. Impact of admixture and ancestry on eQTL analysis and GWAS colocalization in GTEx. Genome Biol. 21:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J, Yao J, Deng S, Balasubramanian R, Jimenez MC, et al. 2022. Differences in metabolomic profiles between black and white women and risk of coronary heart disease: an observational study of women from four US cohorts. Circ. Res 131:601–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasishta S, Ganesh K, Umakanth S, Joshi MB. 2022. Ethnic disparities attributed to the manifestation in and response to type 2 diabetes: insights from metabolomics. Metabolomics 18:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel MJ, Batch BC, Svetkey LP, Bain JR, Turer CB, et al. 2013. Race and sex differences in small-molecule metabolites and metabolic hormones in overweight and obese adults. OMICS 17:627–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Valkengoed IGM, Argmann C, Ghauharali-van der Vlugt K, Aerts JMFG, Brewster LM, et al. 2017. Ethnic differences in metabolite signatures and type 2 diabetes: a nested case–control analysis among people of South Asian, African and European origin. Nutr. Diabetes 7:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brooks AW, Priya S, Blekhman R, Bordenstein SR. 2018. Gut microbiota diversity across ethnicities in the United States. PLOS Biol. 16:e2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ang QY, Alba DL, Upadhyay V, Bisanz JE, Cai J, et al. 2021. The East Asian gut microbiome is distinct from colocalized White subjects and connected to metabolic health. eLife 10:e70349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, et al. 2018. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat. Med 24:1526–31 [DOI] [PubMed] [Google Scholar]
- 50.Nédélec Y, Sanz J, Baharian G, Szpiech ZA, Pacis A, et al. 2016. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167:657–69.e21 [DOI] [PubMed] [Google Scholar]
- 51.Yang HC, Chen CW, Lin YT, Chu SK. 2021. Genetic ancestry plays a central role in population pharmacogenomics. Commun. Biol 4:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulford AJ, Wing C, Dolan ME, Wheeler HE. 2021. Genetically regulated expression underlies cellular sensitivity to chemotherapy in diverse populations. Hum. Mol. Genet 30(3–4):305–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moreno-Torres JG, Raeder T, Alaiz-RodríGuez R, Chawla NV, Herrera F. 2012. A unifying view on dataset shift in classification. Pattern Recognit. 45:521–30 [Google Scholar]
- 54.Gama J, Žliobaitė I, Bifet A, Pechenizkiy M, Bouchachia A. 2014. A survey on concept drift adaptation. ACM Comput. Surveys 46:1–37 [Google Scholar]
- 55.Quionero-Candela J, Sugiyama M, Schwaighofer A, Lawrence ND. 2009. Dataset Shift in Machine Learning. Cambridge, MA: MIT [Google Scholar]
- 56.Lam M, Chen CY, Li Z, Martin AR, Bryois J, et al. 2019. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet 51:1670–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graham SE, Clarke SL, Wu KH, Kanoni S, Zajac GJM, et al. 2021. The power of genetic diversity in genome-wide association studies of lipids. Nature 600:675–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galinsky KJ, Reshef YA, Finucane HK, Loh PR, Zaitlen N, et al. 2019. Estimating cross-population genetic correlations of causal effect sizes. Genet. Epidemiol 43:180–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown BC, Asian Genet. Epidemiol. Netw. Type 2 Diabetes Consort., Ye CJ, Price AL, Zaitlen N. 2016. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet 99:76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang G, Zhao J, Yi J, Luan Y, Wang Q. 2016. Association between gene polymorphisms on chromosome 1 and susceptibility to pre-eclampsia: an updated meta-analysis. Med. Sci. Monit 22:2202–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, et al. 2021. Genetic mechanisms of critical illness in COVID-19. Nature 591:92–98 [DOI] [PubMed] [Google Scholar]
- 62.Johnson A, Bulgarelli L, Pollard T, Horng S, Celi LA, Mark R. 2022. MIMIC-IV (version 2.0). PhysioNet. 10.13026/7vcr-e114 [DOI] [Google Scholar]
- 63.Lim E, Miyamura J, Chen JJ. 2015. Racial/ethnic-specific reference intervals for common laboratory tests: a comparison among Asians, Blacks, Hispanics, and White. Hawai’i J. Medic. Public Health 74:302–10 [PMC free article] [PubMed] [Google Scholar]
- 64.Rappoport N, Paik H, Oskotsky B, Tor R, Ziv E, et al. 2019. Comparing ethnicity-specific reference intervals for clinical laboratory tests from EHR data. J. Appl. Lab. Med 3:366–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manrai AK, Patel CJ, Ioannidis JPA. 2018. In the era of precision medicine and big data, who is normal? JAMA 319:1981–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, et al. 2016. Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med 375:655–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prive F, Aschard H, Carmi S, Folkersen L, Hoggart C, et al. 2022. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet 109:12–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, et al. 2017. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet 100:635–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duncan L, Shen H, Gelaye B, Meijsen J, Ressler K, et al. 2019. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun 10:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen M-H, Raffield LM, Mousas A, Sakaue S, Huffman JE, et al. 2020. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182:1198–213.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou W, Kanai M, Wu K-HH, Rasheed H, Tsuo K, et al. 2021. Global Biobank Meta-analysis Initiative: powering genetic discovery across human diseases. Cell Genom. 2(10):100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Guo J, Ni G, Yang J, Visscher PM, Yengo L. 2020. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat. Commun 11:3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conti DV, Darst BF, Moss LC, Saunders EJ, Sheng X, et al. 2021. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet 53:65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cavazos TB, Witte JS. 2021. Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. Hum. Genet. Genom. Adv 2:100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li J, Bzdok D, Chen J, Tam A, Ooi LQR, et al. 2022. Cross-ethnicity/race generalization failure of behavioral prediction from resting-state functional connectivity. Sci. Adv 8:eabj1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dai Z, Long N, Huang W. 2020. Influence of genetic interactions on polygenic prediction. G3 10:109–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruan Y, Lin YF, Feng YA, Chen CY, Lam M, et al. 2022. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet. 54:573–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai M, Xiao J, Zhang S, Wan X, Zhao H, et al. 2021. A unified framework for cross-population trait prediction by leveraging the genetic correlation of polygenic traits. Am. J. Hum. Genet 108:632–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang H, Zhan J, Jin J, Zhang J, Ahearn TU, et al. 2022. Novel methods for multi-ancestry polygenic prediction and their evaluations in 3.7 million individuals of diverse ancestry. bioRxiv 2022.03.24.485519. 10.1101/2022.03.24.485519 [DOI] [Google Scholar]
- 80.Coram MA, Fang H, Candille SI, Assimes TL, Tang H. 2017. Leveraging multi-ethnic evidence for risk assessment of quantitative traits in minority populations. Am. J. Hum. Genet 101:218–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao J, Cai M, Hu X, Wan X, Chen G, Yang C. 2022. XPXP: improving polygenic prediction by cross-population and cross-phenotype analysis. Bioinformatics 38:1947–55 [DOI] [PubMed] [Google Scholar]
- 82.Weissbrod O, Kanai M, Shi H, Gazal S, Peyrot WJ, et al. 2022. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet 54:450–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou X, Chen Y, Ip F, Jiang Y, Cao H, et al. 2021. Deep learning methods improve polygenic risk analysis and prediction for Alzheimer’s disease. Res. Sq rs.3.rs-818364/v1. 10.21203/rs.3.rs-818364/v1 [DOI] [Google Scholar]
- 84.Muneeb M, Feng S, Henschel A. 2022. An empirical comparison between polygenic risk scores and machine learning for case/control classification. Res. Sq rs.3.rs-1298372/v1. 10.21203/rs.3.rs-1298372/v1 [DOI] [Google Scholar]
- 85.Badré A, Zhang L, Muchero W, Reynolds JC, Pan C. 2021. Deep neural network improves the estimation of polygenic risk scores for breast cancer. J. Hum. Genet 66:359–69 [DOI] [PubMed] [Google Scholar]
- 86.Yang Q, Zhang Y, Dai W, Pan SJ. 2020. Transfer Learning. Cambridge, UK: Cambridge Univ. Press [Google Scholar]
- 87.Pan SJ, Yang Q. 2010. A survey on transfer learning. IEEE Trans. Knowl. Data Eng 22:1345–59 [Google Scholar]
- 88.Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C. 2018. A survey on deep transfer learning. Paper presented at the 27th International Conference on Artificial Neural Networks (ICANN; 2018), Rhodes, Greece, Oct. 4–7 [Google Scholar]
- 89.Weiss K, Khoshgoftaar TM, Wang D. 2016. A survey of transfer learning. J. Big Data 3:9 [Google Scholar]
- 90.Sevakula RK, Singh V, Verma NK, Kumar C, Cui Y. 2019. Transfer learning for molecular cancer classification using deep neural networks. IEEE/ACM Trans. Comput. Biol. Bioinform 16:2089–100 [DOI] [PubMed] [Google Scholar]
- 91.Ebbehoj A, Thunbo Mø, Andersen OE, Glindtvad MV, Hulman A. 2022. Transfer learning for nonimage data in clinical research: a scoping review. PLOS Digit. Health 1:e0000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhuang F, Qi Z, Duan K, Xi D, Zhu Y, et al. 2021. A comprehensive survey on transfer learning. Proc. IEEE 109:43–76 [Google Scholar]
- 93.Gao Y, Cui Y. 2021. Multi-ethnic survival analysis: transfer learning with cox neural networks. Proc. Mach. Learn. Res 146:252–57 [Google Scholar]
- 94.Toseef M, Li X, Wong K-C. 2022. Reducing healthcare disparities using multiple multiethnic data distributions with fine-tuning of transfer learning. Brief. Bioinform 23(3):bbac078. [DOI] [PubMed] [Google Scholar]
- 95.Gao Y, Cui Y. 2022. Deep transfer learning provides a Pareto improvement for multiethnic genomic prediction of diseases. bioRxiv 2022.09.22.509055. 10.1101/2022.09.22.509055 [DOI] [Google Scholar]
- 96.LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521:436–44 [DOI] [PubMed] [Google Scholar]
- 97.Long M, Cao Y, Wang J, Jordan MI. 2015. Learning transferable features with deep adaptation networks. Proc. Mach. Learn. Res 37:97–105 [Google Scholar]
- 98.Zhao Z, Fritsche LG, Smith JA, Mukherjee B, Lee S. 2022. The construction of cross-population polygenic risk scores using transfer learning. Am. J. Hum. Genet 109:1998–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tian P, Chan TH, Wang Y-F, Yang W, Yin G, Zhang YD. 2022. Multiethnic polygenic risk prediction in diverse populations through transfer learning. Front. Genet 13:906965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yosinski J, Clune J, Bengio Y, Lipson H. 2014. How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst 27:3320–28 [Google Scholar]
- 101.Sevakula RK, Singh V, Verma NK, Kumar C, Cui Y. 2018. Transfer learning for molecular cancer classification using deep neural networks. IEEE/ACM Trans. Comput. Biol. Bioinform 16(6):2089–100 [DOI] [PubMed] [Google Scholar]
- 102.Csurka G. 2017. Domain Adaptation in Computer Vision Applications. Cham, Switz.: Springer [Google Scholar]
- 103.Guan H, Liu M. 2021. Domain adaptation for medical image analysis: a survey. IEEE Trans. Biomed. Eng 69:1173–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Long M, Zhu H, Wang J, Jordan MI. 2017. Deep transfer learning with joint adaptation networks. Proc. Mach. Learn. Res 70:2208–17 [Google Scholar]
- 105.Motiian S, Piccirilli M, Adjeroh DA, Doretto G. 2017. Unified deep supervised domain adaptation and generalization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV 2017), pp. 5716–26. Los Alamitos, CA: IEEE Comput. Soc. [Google Scholar]
- 106.Maity S, Mukherjee D, Yurochkin M, Sun Y. 2021. Does enforcing fairness mitigate biases caused by subpopulation shift? Adv. Neural Inf. Process. Syst 34:25773–84 [Google Scholar]
- 107.Rajkomar A, Hardt M, Howell MD, Corrado G, Chin MH. 2018. Ensuring fairness in machine learning to advance health equity. Ann. Intern. Med 169:866–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen IY, Pierson E, Rose S, Joshi S, Ferryman K, Ghassemi M. 2021. Ethical machine learning in healthcare. Annu. Rev. Biomed. Data Sci 4:123–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mehrabi N, Morstatter F, Saxena N, Lerman K, Galstyan A. 2021. A survey on bias and fairness in machine learning. ACM Comput. Surveys 54:1–35 [Google Scholar]
- 110.Zhao H, Gordon G. 2019. Inherent tradeoffs in learning fair representations. Adv. Neural Inf. Process. Syst 32:15675–85 [Google Scholar]
- 111.Menon AK, Williamson RC. 2018. The cost of fairness in binary classification. Proc. Mach. Learn. Res 81:107–18 [Google Scholar]
- 112.Chatterjee DK. 2011. Encyclopedia of Global Justice. Dordrecht, Neth.: Springer Sci. Bus. Media [Google Scholar]
- 113.Davis J, Goadrich M. 2006. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning (ICML ’06), pp. 233–40. New York: Assoc. Comput. Mach. [Google Scholar]
- 114.He H, Garcia EA. 2009. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng 21:1263–84 [Google Scholar]
- 115.Hanley JA, McNeil BJ. 1982. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143:29–36 [DOI] [PubMed] [Google Scholar]
- 116.Fatumo S, Chikowore T, Choudhury A, Ayub M, Martin AR, Kuchenbaecker K. 2022. A roadmap to increase diversity in genomic studies. Nat. Med 28:243–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wickland DP, Sherman ME, Radisky DC, Mansfield AS, Asmann YW. 2022. Lower exome sequencing coverage of ancestrally African patients in The Cancer Genome Atlas. J. Natl. Cancer Inst 114:1192–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weber CJ, Carrillo MC, Jagust W, Jack CR Jr., Shaw LM, et al. 2021. The Worldwide Alzheimer’s Disease Neuroimaging Initiative: ADNI-3 updates and global perspectives. Alzheimer’s Dement. 7(1):e12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, et al. 2017. The Alzheimer’s Disease Neuroimaging Initiative 3: continued innovation for clinical trial improvement. Alzheimer’s Dement. 13:561–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pugh TJ, Bell JL, Bruce JP, Doherty GJ, Galvin M, et al. 2022. AACR Project GENIE: 100,000 cases and beyond. Cancer Discov. 12(9):2044–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.GTEx (Genotype-Tissue Expression) Consort. 2020. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369:1318–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.GTEx (Genotype-Tissue Expression) Consort. 2022. Data set summary of analysis samples. GTEx Analysis Release; v8, accessed on Oct. 8, 2022. https://gtexportal.org/home/tissueSummaryPage [Google Scholar]
- 123.Wendt FR, Pathak GA, Vahey J, Qin X, Koller D, et al. 2022. Modeling the longitudinal changes of ancestry diversity in the Million Veteran Program. bioRxiv 2022.01.24.477583. 10.1101/2022.01.24.477583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Meng C, Trinh L, Xu N, Enouen J, Liu Y. 2021. Interpretability and fairness evaluation of deep learning models on MIMIC-IV dataset. Sci. Rep 12:7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johnson AEW, Bulgarelli L, Shen L, Gayless A, Shammout A, et al. 2023. MIMIC-IV, a freely accessible electronic health record dataset. Sci. Data 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sleep Heart Health Study. 2022. Sleep Mean Health Study dataset: race. Natl. Sleep Res. Resour, accessed on Oct. 8, 2022. https://sleepdata.org/datasets/shhs/variables/race [Google Scholar]
- 127.Zhang G-Q, Cui L, Mueller R, Tao S, Kim M, et al. 2018. The National Sleep Research Resource: towards a sleep data commons. J. Am. Med. Inform. Assoc 25:1351–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pan-UK Biobank. 2022. Overview: pan-ancestry genetic analysis of the UK Biobank. Web Resour., Pan-UK Biobank, accessed on Oct. 8, 2022. https://pan.ukbb.broadinstitute.org/docs/technical-overview [Google Scholar]
- 129.All Us Res. Progr. 2023. Data snapshots. Web Resour, All Us Res. Progr., Natl. Inst. Health, Bethesda, MD. https://www.researchallofus.org/data-tools/data-snapshots/ [Google Scholar]
- 130.Chan-Zuckerberg Initiat. 2023. Ancestry networks for the Human Cell Atlas. Web Resour., Chan-Zuckerberg Initiat., San Francisco. https://chanzuckerberg.com/science/programs-resources/single-cell-biology/ancestry-networks [Google Scholar]
- 131.Zhou W, Kanai M, Wu K-HH, Rasheed H, Tsuo K, et al. 2022. Global Biobank Meta-analysis Initiative: powering genetic discovery across human disease. Cell Genom. 2:100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mulder N, Abimiku A, Adebamowo SN, de Vries J, Matimba A, et al. 2018. H3Africa: current perspectives. Pharmgenom. Pers. Med 11:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, et al. 2019. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.TOPMed (Trans-Omics Precis. Med.). 2022. About TOPMed. Web Resour., TOPMed, Natl. Heart, Lung Blood Inst. Bethesda, MD. https://www.nhlbiwgs.org/ [Google Scholar]
- 135.Auton A, Abecasis GR, Altshuler DM, Durbin RM, Abecasis GR, et al. 2015. A global reference for human genetic variation. Nature 526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schölkopf B, Janzing D, Peters J, Sgouritsa E, Zhang K, Mooij J. 2012. On causal and anticausal learning. In Proceedings of the 29th International Coference on International Conference on Machine Learning, pp. 459–66. New York: Assoc. Comput. Mach. [Google Scholar]
- 137.Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, et al. 2022. Ensembl 2022. Nucleic Acids Res. 50:D988–95 [DOI] [PMC free article] [PubMed] [Google Scholar]