Abstract
SARS-CoV-2 main protease () is a validated antiviral drug target of nirmatrelvir, the active ingredient in Pfizer’s oral drug Paxlovid. Drug-drug interactions limit the use of Paxlovid. In addition, drug-resistant mutants against nirmatrelvir have been identified from cell culture viral passage and naturally occurring variants. As such, there is a need for a second generation of inhibitors. In this study, we explored several reactive warheads in the design of inhibitors. We identified Jun11119R (sulfonamide warhead), Jun10221R (propiolamide warhead), Jun1112R (4-chlorobut-2-ynamide warhead), Jun10541R (nitrile warhead), and Jun10963R (dually activated nitrile warhead) as potent inhibitors. Jun10541R and Jun10963R also had potent antiviral activity against SARS-CoV-2 in Calu-3 cells with values of 2.92 and 6.47 μM, respectively. X-ray crystal structures of with Jun10541R and Jun10221 revealed covalent modification of Cys145. These inhibitors with diverse reactive warheads collectively represent promising candidates for further development.
Keywords: SARS-CoV-2, COVID-19, main protease, 3CL protease, antiviral
Graphical Abstract
1. Introduction
It has been more than three years since the beginning of the COVID-19 pandemic. As of March 10, 2023, more than 676 million positive cases have been reported, and over 6.8 million people have died [1]. The etiological agent of COVID-19 is SARS-CoV-2, an enveloped, non-segmented positive-sense, and single-stranded RNA virus.[2] SARS-CoV-2 belongs to the genus Betacoronavirus, which also includes SARS-CoV and MERS-CoV [2]. Built upon the knowledge gained in studying SARS-CoV and MERS-CoV, unprecedented progress has been made in vaccine and antiviral developments [3, 4]. FDA has approved several drugs for the prevention and treatment of SARS-CoV-2 infection. The viral RNA-dependent RNA polymerase (RdRp) inhibitors remdesivir, and molnupiravir are successful examples of drug repurposing. However, the use of remdesivir is limited by intravenous infusion and controversial clinical efficacy [5, 6]. Molnupiravir is a mutagen, and its use might cause mutation in the host [7]. SARS-CoV-2 encodes two viral proteases, the main protease () and papain-like protease (PLpro), both are high-profile validated antiviral drug targets [8, 9]. Starting from a lead compound PF-00835231 [10], developed in 2003 to target SARS-CoV , Pfizer designed the nitrile-containing nirmatrelvir [11, 12]. Given its low stability, nirmatrelvir is combined with metabolic enhancer ritonavir and is used in humans with the brand name Paxlovid [13]. Paxlovid is highly efficacious and reduces the risk of progression to severe COVID-19 by 89% compared to the placebo group [13]. However, the co-administration of ritonavir leads to multiple drug-drug interactions [14], which limit its use in certain patients with underlying diseases. Furthermore, like other RNA viruses, SARS-CoV-2 continues to evolve with or without drug selection pressure [15–17]. Specifically, multiple mutations in have been identified from naturally occurring viruses resistant to nirmatrelvir [15]. mutations can also evolve under drug selection pressure in cell culture [16, 18, 19]. Collectively, newer generations of inhibitors are needed to combat current and future coronavirus infections [8].
The SARS-CoV-2 Mpro active site’s malleability makes it feasible to accommodate structurally disparate inhibitors, including both covalent and noncovalent inhibitors [8, 20, 21]. is a cysteine protease, and the catalytic Cys145 is susceptible to covalent modification by reactive warheads from inhibitors. inhibitors with diverse reactive warheads have been reported (Figure 1), including nirmatrelvir (nitrile) [11], Jun9–62-2R (dichloroacetamide) [22], Jun9–88-6R (tribromoacetamide),[22] (R,R)-18 and YH-6 (chlorofluoroacetamide) [23, 24], YH-53 (benzothiazolyl ketone) [25], Y180 (ketoamide) [26], GD-9 (chloroacetamide) [27], 20a (nitrile), 23a (4-hydroxybut-2-ynamide), and 14f (vinyl sulfonamide) [28]. With our continuous interest in exploring reactive warheads for the inhibitor design[22] and the inspiration of previous work by others [28], we report the design, synthesis, enzymatic inhibition, antiviral activity, and X-ray crystal structures of covalent inhibitors with sulfonamide propiolamide, 4-chlorobut-2-ynamide, ketoamide, and nitrile warheads.
Figure 1.
SARS-CoV-2 inhibitors with diverse reactive warheads.
2. Results and Discussion
2.1. Chemistry
Synthesis of covalent inhibitors.
The inhibitors were synthesized using the one-pot Ugi four-component reaction (Ugi-4CR) as previously described (Figure 2).[22, 29] The diastereomers were separated by chiral HPLC for compounds with potent enzymatic inhibition. The absolute stereochemistry of Jun10541R was determined by X-ray crystallography, and the stereochemistry for the diastereomers of Jun11119, Jun10221, Jun1112, and Jun10963 was tentatively assigned based on their relative retention time in chiral HPLC.
Figure 2.
Synthesis of covalent SARS-CoV-2 inhibitors through Ugi-4CR. The R and S chirality refers to the chiral center at the pyridine substitution. w=warhead.
2.2. Exploring acrylamide and vinyl sulfonamide as the reactive warheads
We recently reported the design of covalent SARS-CoV-2 inhibitors with the acrylamide reactive warhead [22]. Interestingly, the unsubstituted acrylamide Jun9513 was inactive ( > 20 μM), while Jun10382 with the 2-chloroacrylamide warhead was active and had an of 4.22 μM (Figure 3). We, therefore, were motivated to explore other substituted acrylamides as novel reactive warheads. Compound Jun10542 with the 2-fluoroacrylamide was inactive ( > 20 μM). Jun10594 with the 2-bromoacrylamide ( = 4.26 μM) and Jun10943 with the 2-trifluoromethylacrylamide ( = 4.36 μM) had similar activity as Jun10382 ( = 4.22 μM), suggesting electron-withdrawing groups at the 2-position increase the electrophilicity of the acrylamide warhead. In contrast, compounds Jun10592 and Jun10945 with electron-donating substitutions (methyl and phenyl) at the 2-position were inactive ( > 20 μM). Compound 10905 with the 2-acetylamide substitution was also inactive ( > 20 μM). Next, we explored acrylamides with 3-substitutions. It was found that compounds with the 3-substituted acrylamides generally had reduced activity, and only four compounds, Jun10952 ( =12.39 μM), Jun1117 ( =3.97 μM), Jun10961 ( = 17.05 μM), and Jun10962 ( = 15.42 μM) had moderate inhibitory activity. Compounds Jun10946 and Jun10948 with the cyclized alkene were also inactive ( > 20 μM). Lastly, compounds Jun11119R ( = 0.31 μM) and Jun11119S ( = 6.49 μM) with the vinyl sulfonamide warhead had potent inhibitory activity. However, compound Jun11119R is moderately toxic in Vero E6 cells with a value of 18.4 μM. Collectively, our results showed that the electro-withdrawing substituents at the 2-position of acrylamide are preferred, while substitutions at the 3-position are generally not preferred. Electron-withdrawing substitutions at the 2-position likely increase the electrophilicity at the 3-position for the nucleophilic attack by the Cys145 thiolate. Vinyl sulfonamide is also a preferred warhead, but it leads to moderate cytotoxicity.
Figure 3.
SARS-CoV-2 inhibitors with substituted acrylamide warheads or vinyl sulfonamides. Results of Jun9513 and Jun10382 were reported.[22]
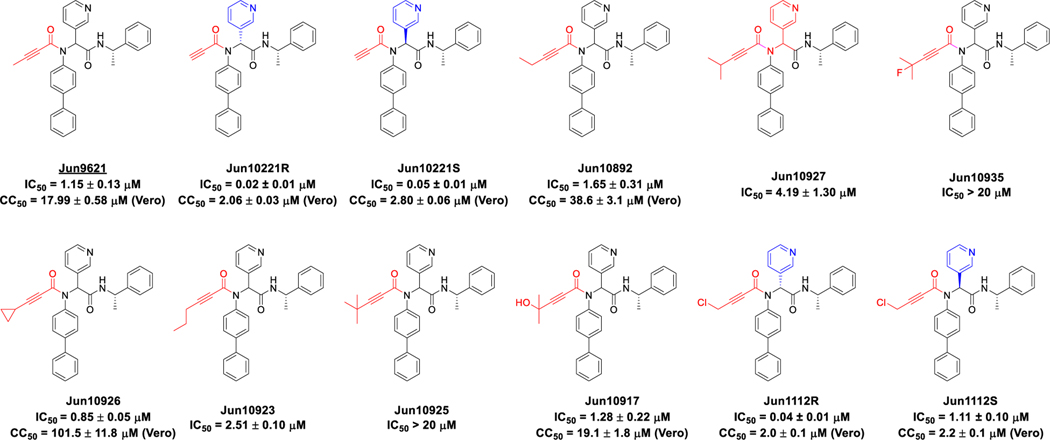
2.3. Exploring propiolamide as the reactive warhead
We recently reported the design of compound Jun9621 with the 2-butynamide warhead ( = 1.15 μM) [22], however, it was cytotoxic ( = 17.99 μM in Vero E6 cells). We, therefore, sought to explore unsubstituted and substituted propiolamides as warheads with reduced cytotoxicity (Figure 4). Both diastereomers Jun10221R ( = 0.02 μM) and Jun10221S ( = 0.05 μM) with the propiolamide warhead had potent inhibition (Figure 4). Unfortunately, both compounds were also highly cytotoxic ( < 3 μM). Adding hydrophobic substitutions at the 3-position of propiolamide led to compounds Jun10892 ( = 1.65 μM) and Jun10927 ( = 4.19 μM) with reduced enzymatic inhibitory activity. Compound Jun10935 with an additional fluoro-substitution at the 3-position of propiolamide was inactive ( > 20 μM). Unexpectedly, compound Jun10926 with the 3-cyclopropyl substitution had potent inhibition ( = 0.85 μM) while being non-cytotoxic ( = 101.5 μM). Compound Jun10923 with the 3-propyl substitution ( = 2.51 μM) was less active, while compound Jun10925 with the 3-tert-butyl substitution was not active ( > 20 μM). Notably, compound 10917 with the 4-hydroxy-4-methylpen-2-ynamide was active ( = 1.28 μM). The diastereomers Jun1112R ( = 0.04 μM) and Jun1112S ( = 1.11 μM) with the 4-chlorobut-2-ynamide warhead had potent inhibition. However, they were highly toxic, with values of 2.0 and 2.2 μM, respectively.
Figure 4.
SARS-CoV-2 inhibitors with substituted propiolamide warheads.
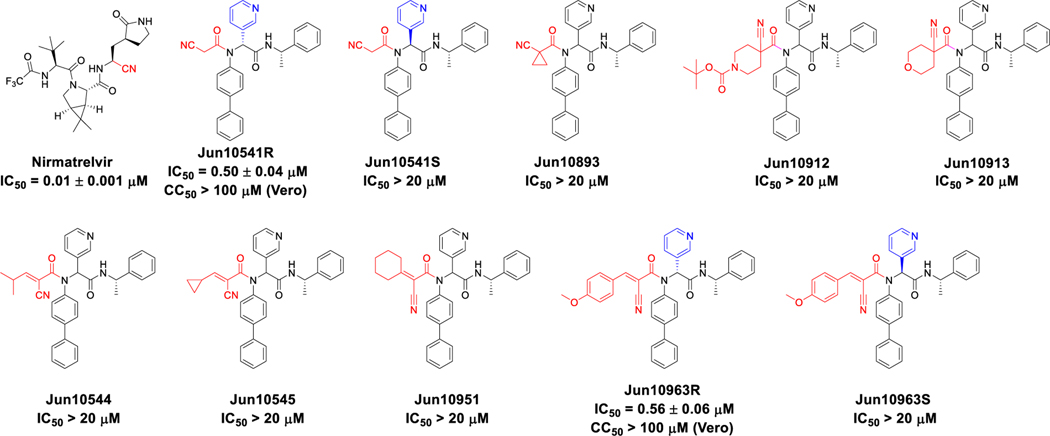
2.4. Exploring nitrile as the reactive group
Inspired by the high potency and selectivity of Pfizer’s oral inhibitor nirmatrelvir,[11] we were interested in exploring nitrile as a weakly reactive warhead in our scaffold. Gratifyingly, compound Jun10541R with the 2-cyanoacetamide warhead had potent inhibition with an of 0.50 μM (Figure 5). The corresponding diastereomer Jun10541S was inactive ( > 20 μM), consistent with our previous structure-activity relationship results [22, 29]. Adding substitutions at the 1-position led to compounds Jun10893, Jun10912, and Jun10913 with abolished inhibition ( > 20 μM). Next, we examined the dually activated 2-cyanoacrylamides as the Cys145 reactive warhead. Among the compounds tested (Jun10544, Jun10545, Jun10951, and Jun10963), compound Jun10963R had potent inhibition ( = 0.56 μM), and the corresponding diastereomer Jun10963S was inactive ( > 20 μM). Compound Jun10963R was also well tolerated in Vero E6 cells with a value over 100 μM. Compounds Jun10544, Jun10545, and Jun10951 with the aliphatic substitutions at the 3-position were inactive ( > 20 μM).
Figure 5.
SARS-CoV-2 inhibitors with substituted nitrile warheads.
2.5. Miscellaneous reactive warheads
In addition to the above conventional cysteine reactive warheads, we also explored a few potential cysteine reactive warheads, including oxoacetamide (Jun10938), diacetamide (Jun11812), un-activated alkyne (Jun1155, Jun1156, and Jun1191), and sulfonyl fluoride (Jun1191) (Figure 6). Alkynes were previously explored as a reactive warhead for the active-site cysteine in deubiquitinase [30, 31]. Among the compounds examined, Jun10938 with the oxoacetamide warhead and Jun1155 with the alkyne warhead inhibited with values of 3.81 and 4.63 μM, respectively.
Figure 6.
SARS-CoV-2 inhibitors with miscellaneous warheads.
2.6. Characterization of the mechanism of action
To characterize the mechanism of action, we chose four compounds with potent values, Jun10221R, Jun1112R, Jun10541R, and Jun10963R, and performed enzyme kinetic studies with different concentrations of inhibitors. The kinetic curves showed one liner phase (Figure S1), suggesting no enzyme depletion at the time scale of measurement (1 h). We therefore fit the curve with Morrison equation and obtained the inhibitory constant values [32]. is independent of concentration, and no preincubation with the inhibitor is required. Therefore, provides a more direct comparison of the inhibitor potency. The values for Jun10221R, Jun1112R, Jun10541R, and Jun10963R are 11.77 nM, 54.21 nM, 345.3 nM, and 573.3 nM, respectively (Figure 7). The results suggest that all four compounds are covalent reversible inhibitors of .
Figure 7.
plots for four potent inhibitors Jun10221R, Jun1112R, Jun10541R, and Jun10963R.
2.7. Antiviral activity of the inhibitors with novel warheads against SARS-CoV-2 replication in Calu-3 cells
We next selected compounds with potent inhibition ( < 1 μM) and low cellular cytotoxicity ( > 50 μM) for the SARS-CoV-2 antiviral assay. Compounds Jun10541R and Jun10963R met the criteria, and both compounds contain the nitrile warhead. The antiviral assay was performed with the infectious SARS-CoV-2 (isolate USA WA1/2020) in Calu-3 cells using the immunofluorescence assay [29]. Calu-3 is a human lung adenocarcinoma cell line that expresses TMPRSS2, rendering it a physiologically relevant cell line for SARS-CoV-2 infection, and it is widely used for antiviral testing [33]. Compounds Jun10541R and Jun10963R inhibited SARS-CoV-2 with values of 2.92 and 6.47 μM, respectively, and were not cytotoxic to Calu-3 cells at up to 100 μM (Figure 8). The value for positive control remdesivir was 0.17 μM.
Figure 8.
Antiviral activities of Mpro inhibitors Jun10541R and Jun10963R against SARS-CoV-2 in Calu-3 cells. The antiviral curve and the cytotoxicity curve were shown in blue and green, respectively.
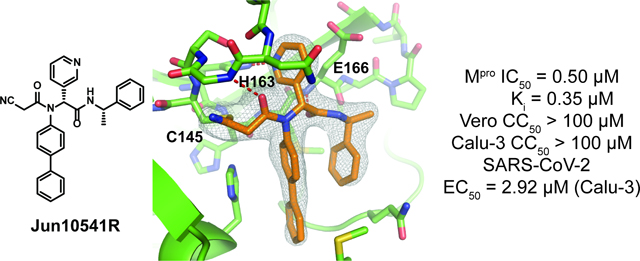
2.8. X-ray crystal structures of SARS-CoV-2 in complex with Jun10221 and Jun10541R
To characterize the binding poses of Jun10221 and Jun10541R with and illustrate interactions between the covalent warheads and catalytic cysteine, we determined their crystal structures at 2.50 Å (Jun10541R, PDB ID 8FIV; Figure. 9A) and 2.55 Å (Jun10221, PDB ID: 8FIW; Figure. 9B) resolution (Table S1). Jun10541R was crystalized with as a pure diastereomer, while Jun10221 was crystalized as a diastereomer mixture consisting of both the (R,S) and (S,S) diastereomers (the first chiral center refers to the -position of the S1 pyridine). Unlike many other cases in which the (R,S) diastereomers are significantly more active than the (S,S) diastereomers (Jun11119, Jun1112, Jun10541, Jun10963 and additional examples in references[22, 29]), the (R,S) and (S,S) diastereomers of Jun10221 had similar potency with values of 0.02 and 0.05 μM, respectively.
Figure 9.
Complex structures of SARS-CoV-2 with inhibitors Jun10541R (2.50 Å resolution) and 10221 (2.55 Å). (A) 2Fo-Fc map, shown in grey and contoured at 1 , illustrates the continuous electron density between the catalytic cysteine C145 and Jun10541R, which forms a covalent adduct through its nitrile warhead. (B) Jun10221 also forms a covalent adduct via its alkyne warhead, but has two conformations corresponding to both diastereomers. Here, the 2Fo-Fc map, shown in grey, is contoured at 0.5 to highlight the positioning of the -methylbenzene sidechain more clearly. (C) The conformations for both diastereomers of Jun10221 are shown from a different angle, offering an unobstructed view of the pyridyl stereocarbon, indicated in the figure, which causes the -methylbenzene sidechain to occupy either the S2 pocket for the (R, S) diastereomer (turquoise) or flip into the solvent exposed S3 site for the (S, S) diastereomer (pale blue). (D) 2Fo-Fc electron density map for inhibitor Jun10221. Sidechain density for the two diastereomers of Jun10221 is less defined at 1 σ (left panel), than 0.5 σ (right panel).
Jun10541R and Jun10221 bear electrophilic nitrile and alkyne warheads that covalently react with the catalytic Cys145, positioning the rest of the inhibitor two atoms from the thiol sulfur, where they occupy the active site in a similar fashion. The key, shared interactions include amide hydrogen bonds between the inhibitor and the peptide backbone of Gly143 and Glu166 and a hydrogen bond between the pyridyl nitrogen and the His163 sidechain of the S1 pocket (Figure 9A, B). The diphenyl and -methylphenyl also form hydrophobic interactions with the buried, nonpolar S2 pocket. Interestingly, in the Jun10221 complex structure, there is defined electron density corresponding to both the (R,S) and (S,S) diastereomers (Figure 9B, D). Although the electron density for (S,S)-Jun10221 diastereomer is slightly weaker than previous structures containing diastereomer pure (R,S) inhibitors, it is nonetheless apparent, especially when the 2Fo-Fc map is contoured at 0.5 σ (Figure 9B). Overall, the binding poses for both diastereomers of Jun10221 are very similar (Figure 9C), except that the pyridyl carbon in the (S,S) diastereomer is closer to the S1 pocket, which forces the amide group to rotate ~ 90° outwards and the -methylbenzene to flip out of the hydrophobic S2/S4 subpocket and into the solvent-exposed S3 site.
3. CONCLUSION
Cysteine proteases are challenging drug targets. Most cysteine protease inhibitors are covalent inhibitors, and it is difficult to achieve a delicate balance between potency and selectivity [34, 35]. No cysteine protease inhibitors had been approved by FDA before Pfizer’s Paxlovid, which contains the inhibitor nirmatrelvir and the metabolic enhancer ritonavir. Key to the success of nirmatrelvir is the choice of nitrile as the reactive warhead. Nitrile is a weakly reactive cysteine warhead compared to aldehyde, ketoamide, and ,-unsaturated ester [36]. Nirmatrelvir is highly selective compared to other inhibitors, including GC-376 and its analogs [33]. Several inhibitors have been desgiend with the nitrile warhead including nirmatrelvir [11, 12, 28]. This study explored acrylamide, vinyl suofonamide, propiolamide, nitrile, ketoamide, and alkyne as reactive warheads. Although compounds with the vinyl sulfonamide Jun11119R and the propiolamide Jun10221R/S had potent enzymatic inhibition, they were cytotoxic, which might be due to non-specific modification of host proteins. Similar compounds were reported earlier, but their cellular cytotoxicity and antiviral activity were not characterized [28]. Although Jun10221 with the propiolamide warhead was cytotoxic, the co-crystal structures revealed novel findings that are insightful for drug design. Specifically, both the R,S and S,S diasteromers of Jun10221 had similar inhibition with values of 0.02 and 0.05 μM, respectively. This starkly contradicts other compounds in the same series in which the R,S diastereomer is more potent than the S,S diastereomer [22, 29]. One explanation might be that the high reactivity of the propiolamide warhead dominates the potency. X-ray crystal structures revealed that both diastereomers bind to the S1 and S2 subsites with superimposable configurations. However, the -methylbenzene substitution binds in the flipped orientation with the R,S diastereomer facing towards the subsite between S2 and S4 pockets, while the S,S diastereomer projects away from the protein and occupies the solvent-exposed S3 site. The flipped orientations of the -methylbenzene in the two diastereomers of Jun10221 are reminiscent of the binding of GC-376 in , in which the benzyloxycarbonyl (Cbz) group adapts either the inward or outward orientations [32, 37, 38]. Notably, Jun10541R with the nitrile reactive warhead had potent enzymatic inhibition while being non-cytotoxic. Jun10541R also had potent antiviral activity against SARS-CoV-2 in Calu-3 cells with an of 2.92 μM. X-ray crystal structure showed a covalent modification of the Cys145 by the nitrile warhead. In summary, our results showed that nitrile is a promising warhead in designing inhibitors. Further optimization of Jun10541R might lead to drug candidates with in vivo animal efficacy. In addition, the co-crystal structure of with the S,S diasteromers of Jun10221 represent the first example in this compound series, reavealing a distinct binding pose that is informative in drug design.
4. EXPERIMENTAL SECTION
4.1. Cell lines and viruses.
VERO E6 cells (ATCC, CRL-1586) were cultured in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 5% heat-inactivated FBS in a 37°C incubator with 5% .
4.2. Protein expression and purification.
SARS CoV-2 main protease ( or 3CL) gene from strain BetaCoV/Wuhan/WIV04/2019 and SARS-CoV main protease from strain CDC#200301157 in the pET29a(+) vector with E. coli codon optimization were ordered from GenScript (Piscataway, NJ). The gene was then subcloned into pE-SUMO vector as described previously [32, 37]. The expression and purification of SARS-CoV-2 with unmodified N- and C-termini was detailed in our previous publication [37].
4.3. Enzymatic assays.
The main protease () enzymatic assays were carried out in reaction buffer containing 20 mM HEPES pH 6.5, 120 mM NaCl, 0.4 mM EDTA, 20% glycerol and 4 mM DTT. The percentage of inhibition and enzymatic values were calculated as previously described [32, 37]. The curves for selected compounds were shown in Figure S2. Briefly, the assay was performed in 96-well plates with 100 μL of 100 nM protein in the reaction buffer. Then 1 μL testing compound at various concentrations was added to each well and incubated at 30 °C for 30 min. The enzymatic reaction was initiated by adding 1 μL of 1 mM FRET substrate (the final substrate concentration is 10 μM). The reaction was monitored in a Cytation 5 image reader with filters for excitation at 360/40 nm and emission at 460/40 nm at 30 °C for 1 hr. The initial velocity of the enzymatic reaction with and without testing compounds was calculated by linear regression for the first 15 min of the kinetic progress curve.
For the Morrison plot, 10 μL 100 nM SARS-CoV-2 protein was added to 190 μl of reaction buffer containing testing compound and the FRET substrate, and the reaction was monitored for 2 hr. The final FRET substrate concentration in this assay is 20 μM. Detailed curve fitting and determination was described previously [32, 37]. The equation used for Mprrison plot curve fitting is: . Y: enzyme activity; : velocity in the absence of inhibitor; Et: enzyme concentration (0.005 μM); X: concentration of inhibitor (μM); : dissociation of inhibitor (μM); S: substrate concentration (20 μM); : the concentration of substrate which permits the enzyme to achieve half (35.25 μM).
4.4. Cytotoxicity measurement.
Evaluation of the cytotoxicity of compounds were carried out using the neutral red uptake assay [39, 40]. Briefly, 80,000 cells/mL of the tested cell lines were dispensed into 96-well cell culture plates at 100 μL/well. Twenty-four hours later, the growth medium was removed and washed with 150 μL PBS buffer. 200 μL fresh serum-free medium containing serial diluted compounds was added to each well. After incubating for 5 days at 37 °C, the medium was removed and replaced with 100 μL DMEM medium containing 40 μg/mL neutral red and incubated for 2–4 h at 37 °C. The amount of neutral red taken up was determined by measuring the absorbance at 540 nm using a Multiskan FC Microplate Photometer (Fisher Scientific). The values were calculated from best-fit dose response curves with variable slope in Prism 8.
4.5. Antiviral assay in Calu-3 cells.
Calu-3 cells (ATCC, HTB-55) grown in Minimal Eagles Medium supplemented with 1% non-essential amino acids, 1% penicillin/streptomycin, and 10% FBS are plated in 384 well plates. The next day, 50 nL of drug suspended in DMSO is added as an 8-pt dose response with three-fold dilutions between test concentrations in triplicate, starting at 40 μM final concentration. The negative control (DMSO, n=32) and positive control (10 μM Remdesivir, n=32) are included on each assay plate. Calu3 cells are pretreated with controls and test drugs (in triplicate) for 2 hours prior to infection. In BSL3 containment, SARS-CoV-2 (isolate USA-WA1/2020) diluted in serum free growth medium is added to plates to achieve an MOI=0.5. Cells are incubated continuously with drugs and SARS-CoV-2 for 48 hours. Cells are fixed and then immunstained with anti-dsRNA (J2) and nuclei are counterstained with Hoechst 33342 for automated microscopy. Automated image analysis quantifies the number of cells per well (toxicity) and the percentage of infected cells (dsRNA+ cells/cell number) per well. SARS-CoV-2 infection at each drug concentration was normalized to aggregated DMSO plate control wells and expressed as percentage-of-control (POC=% Infection sample/Avg % Infection DMSO cont). A non-linear regression curve fit analysis (GraphPad Prism 8) of POC Infection and cell viability versus the transformed concentration values to calculate values for Infection and values for cell viability. Selectivity index (SI) was calculated as a ratio of drug’s and values (SI = /).
4.6. crystallization and structure determination.
inhibitor was added to 20 mg/mL SARS-CoV-2 to a final concentration of 1.75 mM and incubated overnight at 4˚C. This mixture was then diluted four-fold with protein stock buffer (20 mM Tris pH 7.5, 200 mM NaCl, 1 mM DTT) then spun down at 13,000 × g for 1 min to remove precipitate. Crystals were grown by mixing the protein-inhibitor sample with an equal volume of crystallization buffer (20% PEG 3350, 0.2 M NaF) in a vapor diffusion, hanging drop apparatus. Crystals were then transferred to a drop with crystallization buffer containing 5 mM inhibitor for 1 h, followed by a brief soaking in a cryoprotectant solution of 30% PEG 3350 and 15% glycerol with 2 mM inhibitor. Crystals were then flash frozen in liquid nitrogen for X-ray diffraction.
X-ray diffraction data for the SARS-CoV-2 structures were collected on the SBC 19-ID beamline at the Advanced Photon Source (APS) in Argonne, IL, and processed with the HKL3000 software suite. The CCP4 versions of MOLREP[41] was used for molecular replacement using a previously solved SARS-CoV-2 structure, 6YB7. Structural refinement was performed using REFMAC5[42] and COOT [43]. The crystallographic statistics is shown in Supplementary Materials table S1. The complex structures for SARS-CoV-2 with inhibitors have been deposited in the Protein Data Bank with the accession IDs 8FIV (Jun10541R) and 8FIW (Jun10221).
4.7. General chemical methods.
All chemicals were purchased from commercial vendors and used without further purification unless otherwise noted. 1H and 13C NMR spectra were recorded on a Bruker-400 or −500 NMR spectrometer. Chemical shifts are reported in parts per million referenced with respect to residual solvent (CD3OD) 3.31 ppm, (DMSO-d6) 2.50 ppm, and (CDCl3) 7.26 ppm or from internal standard tetramethylsilane (TMS) 0.00 ppm. The following abbreviations were used in reporting spectra: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; dd, doublet of doublets; ddd, doublet of doublet of doublets. All reactions were carried out under N2 atmosphere, unless otherwise stated. HPLC-grade solvents were used for all reactions. Flash column chromatography was performed using silica gel (230–400 mesh, Merck). High resolution mass spectra were obtained using an OrbitrapTM for all the compounds, obtained in an Ion Cyclotron Resonance (ICR) spectrometer. The purity was assessed by using Shimadzu UPLC with Shimdazu C18-AQ column (4.6x150 mm P/N #227–30767-05) at a flow rate of 1 mL/min; and 220 nm; mobile phase A, 0.1% trifluoroacetic acid in , and mobile phase B, 0.1% trifluoroacetic acid in 90% CH3CN and 10% . The gradients are 0–2 mins 10% B, 2–15mins 10%-100% B, 15–18mins, 100% B, 18.1–20mins 10% B. All compounds submitted for testing were confirmed to be > 95.0% purity by HPLC traces. All final products were characterized by proton and carbon NMR, HPLC and HRMS.
General Procedure for Ugi-4CR Reaction:
Ug-4CR reaction was performed according to the reported procedure with modifications [44]. Amine (1.0 equiv) and aldehyde (1.0 equiv) were mixed in methanol (10 ml) and stirred at room temperature for 30 minutes. Then carboxylic acid (1.0 equiv) and isocyanide (1.0 equiv) were added sequentially and the resulting mixture was stirred at room temperature overnight. After that, the solvent was removed under reduced pressure and the crude product was purified with flash silica gel chromatography (methanol in dichloromethane 1–5% or acetone in hexane 30–80%).
N-([1,1’-biphenyl]-4-yl)-2-fluoro-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10542). White solid, 65% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.79–8.75 (t, J = 7.2 Hz, 1H), 8.43–8.30 (m, 2H), 7.62–7.09 (m, 16H), 6.24 (s, 0.5H), 6.21 (s, 0.5H), 5.20–4.96 (m, 3H), 1.39–1.37 (d, J = 6.9 Hz, 1.5H), 1.29–1.27 (d, J = 7.0 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.02, 167.87, 161.59, 161.53, 151.79, 151.74, 149.53, 144.70, 144.34, 139.70, 139.68, 139.16, 138.52, 138.50, 138.47, 138.45, 138.06, 131.16, 131.13, 130.81, 129.41, 128.72, 128.59, 128.23, 127.13, 126.72, 126.64, 126.52, 126.26, 126.02, 123.53, 123.24, 62.76, 48.91, 48.76, 22.71, 22.58. C30H26FN3O2, HRMS calculated for m/z [M+H]+: 480.2009 (calculated), 480.2004 (found).
N-([1,1’-biphenyl]-4-yl)-2-bromo-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10594). White solid, 70% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.74–8.71 (t, J = 11.9, 7.6 Hz, 1H), 8.41–8.28 (m, 2H), 7.60–7.07 (m, 16H), 6.22 (s, 0.5H), 6.19 (s, 0.5H), 5.18–4.95 (m, 2H), 1.38 (d, J = 6.8 Hz, 1.5H), 1.29 (d, J = 6.8 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 167.95, 167.82, 165.34, 165.30, 151.59, 151.54, 149.43, 144.72, 144.26, 139.61, 139.07, 138.62, 138.59, 137.95, 131.88, 131.83, 129.40, 128.72, 128.59, 128.25, 127.22, 127.14, 126.98, 126.66, 126.61, 126.27, 124.16, 121.58, 121.53, 62.43, 48.95, 48.75, 22.71, 22.58. C30H26BrN3O2, HRMS calculated for m/z [M+H]+: 540.1206 (calculated), 540.1200 (found).
N-([1,1’-biphenyl]-4-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)-2-(trifluoromethyl)acrylamide (Jun10943). White solid, 57% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.77–8.73(t, J = 11.1, 7.7 Hz, 1H), 8.44–8.31 (m, 2H), 7.62–7.58 (t, J = 7.4 Hz, 2H), 7.53–7.17 (m, 13H), 7.13–7.10 (t, 1H), 6.30 (s, 0.5H), 6.25 (s, 0.5H), 5.57–5.54 (m, 2H), 5.07–4.99 (m, 1H), 1.39–1.37 (d, J = 6.9 Hz, 1.5H), 1.30–1.28 (d, J = 6.8 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 171.45, 171.41, 168.54, 168.43, 151.58, 151.54, 149.20, 144.84, 144.41, 141.10, 141.05, 139.90, 139.87, 139.23, 139.00, 138.99, 137.86, 131.73, 131.58, 131.53, 131.38, 129.38, 128.69, 128.58, 128.11, 127.17, 127.11, 126.92, 126.67, 126.52, 126.31, 123.48, 123.19, 118.74, 61.94, 48.90, 48.73, 22.72, 22.64, 20.72. C31H26F3N3O2, HRMS calculated for m/z [M+H]+: 530.1965 (calculated), 530.1961 (found).
N-([1,1’-biphenyl]-4-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)methacrylamide (Jun10592). White solid, 80% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.72–8.68 (t, J = 8.9 Hz, 1H), 8.42–8.28 (m, 2H), 7.60–7.57 (t, J = 7.3 Hz, 2H), 7.46–7.19 (m, 12H), 7.14–7.10 (t, J = 7.4 Hz, 1H), 6.30 (s, 0.5H), 6.25 (s, 0.5H), 5.09–4.99 (m, 2H), 4.92–4.89 (d, J = 13.6 Hz, 1H), 1.74–1.71 (d, J = 11.0 Hz, 3H), 1.38–1.36 (d, J = 6.9 Hz, 1.5H), 1.31–1.29 (d, J = 6.9 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 171.44, 171.40, 168.54, 168.42, 151.58, 151.53, 149.20, 144.84, 144.41, 141.10, 141.04, 139.89, 139.87, 139.22, 138.98, 137.85, 131.58, 131.53, 129.38, 128.68, 128.58, 128.11, 127.17, 127.11, 126.91, 126.67, 126.52, 126.30, 123.48, 123.19, 118.73, 61.93, 48.89, 48.72, 22.72, 22.64, 20.72. C31H29N3O2, HRMS calculated for m/z [M+H]+: 476.2259 (calculated), 476.2255 (found).
N-([1,1’-biphenyl]-4-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)-2-phenylacrylamide (Jun10945). White solid, 55% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.76–8.74 (d, J = 7.5 Hz, 1H), 8.47–8.31 (m, 2H), 7.51–7.07 (m, 19H), 6.40 (s, 0.5H), 6.35 (s, 0.5H), 5.77 (s, 1H), 5.64 (s, 1H), 5.32–5.27 (d, J = 16.6 Hz, 1H), 5.11–5.06 (m, 1H), 1.42–1.40 (d, J = 7.0 Hz, 1.5H), 1.33–1.31 (d, J = 6.9 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 170.24, 168.72, 168.54, 151.76, 151.66, 144.84, 144.35, 139.16, 138.95, 131.99, 129.30, 128.75, 128.67, 128.58, 128.06, 127.14, 126.85, 126.68, 126.37, 126.32, 126.29, 126.13, 123.49, 123.18, 60.22, 48.95, 48.77, 22.81, 22.57. C36H31N3O2, HRMS calculated for m/z [M+H]+: 538.2415 (calculated), 538.2410 (found).
N-([1,1’-biphenyl]-4-yl)-2-acetamido-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10905). White solid, 70% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 9.79 (s, 1H), 8.80 (dd, J = 13.8, 7.9 Hz, 1H), 8.35 (d, J = 5.6 Hz, 1H), 7.59 (d, J = 7.6 Hz, 2H), 7.48–7.18 (m, 15H), 6.25 (s, 1H), 5.11–5.06 (m, 1H), 4.89 (d, J = 21.4 Hz, 1H), 4.63 (d, J = 38.1 Hz, 1H), 1.84 (s, 3H), 1.47 (d, J = 7.0 Hz, 1.5H), 1.40 (d, J = 7.0 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.62, 168.03, 166.48, 151.55, 151.44, 149.02, 148.98, 144.94, 144.64, 139.25, 139.10, 139.08, 138.48, 138.41, 138.10, 138.00, 131.99, 131.72, 130.66, 130.52, 129.38, 128.71, 128.63, 128.12, 127.20, 127.10, 126.93, 126.75, 126.72, 126.41, 123.37, 123.22, 55.37, 49.05, 49.01, 23.08, 23.04, 22.85, 22.61. C32H30N4O3, HRMS calculated for m/z [M+H]+:519.2396 (calculated), 519.2399 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-3-bromo-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10591). White solid, 43% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.78–8.74 (t, J = 7.6 Hz, 1H), 8.41–8.28 (m, 2H), 7.66–7.08 (m, 17H), 6.30–6.24 (t, J = 13.4 Hz, 2H), 5.05–4.98 (m, 1H), 1.40–1.38 (d, J = 6.8 Hz, 1.5H), 1.28–1.27 (d, J = 6.8 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.35, 168.21, 163.26, 151.67, 151.61, 149.39, 144.74, 144.41, 139.99, 139.04, 137.89, 131.86, 131.31, 130.91, 129.62, 129.58, 129.43, 128.72, 128.58, 128.34, 127.29, 127.19, 127.12, 127.08, 126.66, 126.25, 124.73, 124.70, 62.06, 48.87, 22.75, 22.62. C30H26BrN3O2, HRMS calculated for m/z [M+H]+: 540.1205 (calculated), 540.1199 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-3-chloro-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10894). White solid, 46% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.80–8.78 (t, J = 8.4 Hz, 1H), 8.47–8.34 (m, 2H), 7.65–7.55 (m, 5H), 7.49–7.43 (m, 4H), 7.33–7.12 (m, 8H), 6.86–6.82 (t, 1H), 6.35 (s, 0.5H), 6.31 (s, 0.5H), 5.10 (m, 1H), 1.45–1.43 (d, J = 6.8 Hz, 1.5H), 1.35–1.33 (d, J = 6.8 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.37, 168.24, 163.05, 151.62, 151.55, 149.33, 144.74, 144.42, 140.02, 139.08, 137.94, 134.93, 131.84, 129.42, 128.77, 128.71, 128.58, 128.32, 127.29, 127.08, 126.67, 126.26, 123.54, 123.25, 62.04, 48.87, 48.79, 22.73, 22.59. C30H26ClN3O2, HRMS calculated for m/z [M+H]+: 496.1747 (calculated), 496.1712 (found).
(Z)-N-([1,1’-biphenyl]-4-yl)-3-iodo-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10895). White solid, 51% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.77–8.75 (d, J = 7.7 Hz, 1H), 8.43–8.30 (m, 2H), 7.62–7.51 (m, 5H), 7.46–7.36 (m, 5H), 7.29–7.20 (m, 5H), 7.13–7.10 (dd, J = 11.4, 7.0 Hz, 2H), 6.82–6.78 (t, 1H), 6.32 (s, 0.5H), 6.28 (s, 0.5H), 5.06–4.99 (m, 1H), 1.41–1.40 (d, J = 6.8 Hz, 1.5H), 1.31–1.30 (d, J = 6.8 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.37, 168.24, 163.05, 151.62, 151.55, 149.33, 144.74, 144.42, 140.02, 139.08, 137.94, 134.93, 131.84, 129.42, 128.77, 128.71, 128.58, 128.32, 127.29, 127.08, 126.67, 126.26, 123.54, 123.25, 62.04, 48.87, 48.79, 22.73, 22.59. C30H26IN3O2, HRMS calculated for m/z [M+H]+: 588.1066 (calculated), 588.1060 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-2-methyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)but-2-enamide (Jun10906). White solid, 77% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.71–8.66 (t, J = 8.7 Hz, 1H), 8.41–8.28 (m, 2H), 7.60 (s, 2H), 7.46–7.35 (m, 7H), 7.29–7.14 (m, 6H), 6.29 (s, 0.5H), 6.27 (s, 0.5H), 5.79 (s, 1H), 5.66–5.60 (dd, J = 14.9, 7.6 Hz, 1H), 5.08–4.99 (m, 1H), 1.57 (d, J = 6.9 Hz, 1.5H), 1.54 (d, J = 6.9 Hz, 1.5H), 1.42–1.31 (m, 6H). 13C NMR (101 MHz, DMSO-d6) 172.53, 168.66, 168.56, 151.47, 144.85, 144.45, 140.30, 139.25, 138.52, 138.49, 137.80, 132.91, 132.86, 131.37, 131.30, 129.37, 129.27, 129.21, 128.67, 128.58, 128.06, 127.15, 127.11, 126.87, 126.67, 126.38, 126.33, 123.47, 123.20, 62.09, 48.87, 48.71, 22.70, 22.64, 14.43, 13.38. C32H31N3O2, HRMS calculated for m/z [M+H]+: 490.2415 (calculated), 490.2411(found).
N-([1,1’-biphenyl]-4-yl)-3-methyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)but-2-enamide (Jun10911). White solid, 83% yield, dr = 1:1.1H NMR (400 MHz, DMSO-d6) 8.67 (s, 1H), 8.36–8.25 (m, 2H), 7.60–7.10 (m, 16H), 6.27 (s, 0.5H), 6.26 (s, 0.5H), 5.39 (s, 1H), 5.00 (s, 1H), 2.08–2.05 (d, J = 10.5 Hz, 3H), 1.61 (s, 3H), 1.38 (s, 1.5H), 1.29 (s, 1.5H). 13C NMR (101 MHz, DMSO-d6) 169.05, 168.82, 166.49, 151.60, 151.53, 149.14, 139.49, 139.25, 137.81, 131.82, 129.41, 128.61, 128.56, 128.17, 127.09, 126.96, 126.91, 126.70, 126.30, 123.45, 117.99, 61.60, 48.86, 48.75, 27.34, 22.79, 22.65, 20.44. C32H31N3O2, HRMS calculated for m/z [M+H]+: 490.2410 (calculated), 490.2413 (found).
(E)-4-([1,1’-biphenyl]-4-yl(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)amino)-4-oxobut-2-enoic acid (Jun10952). White solid, 69% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 12.98 (s, 1H), 8.78–8.74 (t, J = 8.1 Hz, 1H), 8.40–8.27 (m, 2H), 7.63–7.55 (m, 5H), 7.48–7.39 (m, 5H), 7.27–7.7.01 (m, 6H), 6.63–6.54 (m, 2H), 6.29 (s, 0.5H), 6.27 (s, 0.5H), 5.05–4.97 (m, 1H), 1.38–1.36 (d, J = 7.1 Hz, 1.5H), 1.27–1.25 (d, J = 7.0 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.29, 168.14, 166.52, 166.49, 151.64, 149.41, 144.39, 140.03, 139.00, 137.90, 134.24, 132.16, 131.76, 129.45, 128.72, 128.58, 128.36, 127.27, 127.20, 127.12, 127.07, 126.68, 126.27, 123.53, 123.24, 62.38, 48.92, 48.83, 22.74, 22.61. C31H27N3O4, HRMS calculated for m/z [M+H]+: 506.2000 (calculated), 506.1998 (found).
Methyl (E)-4-([1,1’-biphenyl]-4-yl(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)amino)-4-oxobut-2-enoate (Jun1117). White solid, 80% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.81 (t, J = 8.4 Hz, 1H), 8.32 (s, 1H), 7.64–7.19 (m, 16H), 7.11 (d, J = 7.6 Hz, 1H), 6.73–6.62 (m, 2H), 6.34 (s, 0.5H), 6.31 (s, 0.5H), 5.09–4.98 (m, 1H), 3.61 (d, J = 4.7 Hz, 3H), 1.39 (d, J = 7.0 Hz, 1.5H), 1.28 (d, J = 6.9 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.25, 168.10, 165.64, 165.62, 163.54, 163.50, 151.74,127.20, 127.10, 126.93 151.66, 149.44, 144.72, 144.36, 140.09, 138.96, 137.91, 137.89, 134.83, 131.74, 131.16, 130.76, 130.55, 129.44, 129.42, 128.71, 128.57, 128.36, 127.30, 127.20, 127.12, 127.05, 126.67, 126.26, 123.52, 123.22, 62.43, 52.51, 48.94, 48.85, 22.72, 22.59. C32H29N3O4, HRMS calculated for m/z [M+H]+:520.2236 (calculated), 520.2239 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-4-oxo-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)pent-2-enamide (Jun10961). White solid, 66% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.79–8.75 (t, J = 7.1 Hz, 1H), 8.41–8.29 (m, 2H), 7.63–7.60 (t, J = 7.2 Hz, 2H), 7.57–7.55 (d, J = 8.1 Hz, 2H), 7.45–7.40 (dd, J = 10.2, 5.1 Hz, 4H), 7.38–7.34 (t, J = 6.3 Hz, 3H), 7.28–7.17 (m, 4H), 7.11–7.09 (d, J = 7.5 Hz, 1H), 6.97–6.90 (dd, J = 15.2, 11.0 Hz, 1H), 6.48–6.46 (dd, J = 15.7, 8.4 Hz, 1H), 6.32 (s, 0.5H), 6.29 (s, 0.5H), 5.04–4.98 (m, 1H), 2.15–2.13 (dd, J = 6.8, 2.2 Hz, 3H), 1.39–1.37 (d, J = 7.1 Hz, 1.5H), 1.27 (d, J = 7.1 Hz, 1.5H).13C NMR (101 MHz, DMSO-d6) 198.13, 198.10, 168.30, 168.14, 151.68, 151.62, 149.42, 144.70, 144.40, 139.99, 139.02, 137.89, 137.72, 131.73, 129.44, 128.71, 128.59, 128.35, 127.22, 127.13, 127.05, 126.67, 126.28, 123.54, 123.26, 62.42, 48.91, 48.80, 29.22, 22.73, 22.58. C32H29N3O3, HRMS calculated for m/z [M+H]+: 504.2209 (calculated), 504.2208 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-3-(furan-2-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10962). White solid, 75% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.77–8.73 (t, J = 8.5 Hz, 1H), 8.42–8.30 (m, 2H), 7.68–7.58 (m, 4H), 7.51–7.36 (m, 7H), 7.30–7.18 (m, 3H), 7.13–7.11 (m, 1H), 6.83–6.76 (dd, J = 6.8, 3.4 Hz, 1H), 6.55–6.54 (d, J = 4.3 Hz, 1H), 6.37 (s, 0.5H), 6.35 (s, 0.5H), 6.11–6.01 (m, 1H), 5.06–5.00 (m, 1H), 1.41–1.40 (d, J = 7.1 Hz, 1.5H), 1.30–1.28 (d, J = 7.0 Hz, 1.5H).13C NMR (101 MHz, DMSO-d6) 168.74, 168.60, 165.38, 165.34, 151.66, 151.60, 151.14, 149.27, 145.68, 144.83, 144.50, 139.60, 139.08, 137.88, 132.02, 129.45, 128.95, 128.70, 128.57, 128.29, 127.16, 127.12, 127.09, 127.02, 126.70, 126.27, 123.50, 123.21, 116.38, 116.34, 115.73, 112.96, 62.09, 48.85, 48.75, 22.78, 22.63. C34H29N3O3, HRMS calculated for m/z [M+H]+: 528.2218 (calculated), 528.2216 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-3-methoxy-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10436). White solid, 81% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.65–8.61 (t, J = 9.2 Hz, 1H), 8.37–8.25 (m, 2H), 7.62–7.33 (m, 13H), 7.27–7.15 (m, 4H), 6.32–6.29 (d, J = 11.2 Hz, 1H), 5.03–4.89 (m, 2H), 3.45–3.43 (d, J = 6.4 Hz, 3H), 1.38–1.36 (d, J = 7.0 Hz, 1H), 1.27–1.25 (d, J = 7.2 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) 168.92, 168.82, 166.37, 166.32, 162.00, 151.56, 151.49, 149.13, 144.88, 144.53, 139.45, 139.28, 137.82, 137.79, 132.04, 132.00, 129.40, 128.68, 128.55, 128.18, 127.13, 127.00, 126.97, 126.72, 126.27, 123.46, 123.18, 97.73, 61.54, 58.40, 48.70, 22.76, 22.60. C31H29N3O3, HRMS calculated for m/z [M+H]+: 492.2220 (calculated), 492.2218 (found).
N-([1,1’-biphenyl]-4-yl)-3-oxo-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)cyclopent-1-ene-1-carboxamide (Jun10946). White solid, 82% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.78 (t, J = 8.4 Hz, 1H), 8.46 – 8.30 (m, 2H), 7.59–7.19 (m, 16H), 7.10 (d, J = 7.8 Hz, 1H), 6.32 (s, 0.5H), 6.28 (s, 0.5H), 5.71 (d, J = 15.2 Hz, 1H), 5.09–4.97 (m, 1H), 2.72–2.62 (m, 2H), 2.18 (d, J = 5.0 Hz, 2H), 1.38 (d, J = 7.0 Hz, 1.5H), 1.29 (d, J = 6.9 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 208.51, 170.09, 170.06, 168.16, 168.02, 151.73, 151.68, 149.48, 144.70, 144.33, 139.81, 138.91, 138.26, 138.21, 137.93, 132.88, 131.70, 131.66, 130.94, 130.56, 129.41, 128.73, 128.61, 128.31, 127.23, 127.17, 126.97, 126.81, 126.65, 126.29, 123.57, 123.27, 48.99, 48.83, 34.46, 34.43, 29.94, 22.70, 22.63. C34H29N3O3, HRMS calculated for m/z [M+H]+:528.2287 (calculated), 528.2297 (found).
3-([1,1’-biphenyl]-4-yl)-4-(cyclohex-1-en-1-yl)-4-oxo-N-((S)-1-phenylethyl)-2-(pyridin-3-yl)butanamide (Jun10948). White solid, 82% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.66 (t, J = 8.4 Hz, 1H), 8.32 (dd, J = 11.4, 4.7 Hz, 1H), 7.57 (t, J = 6.9 Hz, 2H), 7.45–7.37 (m, 7H), 7.33 (t, J = 7.8 Hz, 3H), 7.22–7.15 (m, 5H), 6.28 (s, 0.5H), 6.24 (s, 0.5H), 5.75 (s, 1H), 5.70 (d, J = 15.2 Hz, 1H), 5.04–4.95 (m, 1H), 1.92 (d, J = 14.2 Hz, 2H), 1.78–1.72 (m, 2H), 1.34 (d, J = 6.6 Hz, 3H), 1.28 (d, J = 7.1 Hz, 3H). 13C NMR (101 MHz, DMSO-d6) 171.84, 171.79, 168.63, 168.54, 151.47, 151.44, 149.11, 144.83, 144.44, 140.28, 140.25, 139.29, 138.57, 138.54, 137.79, 134.89, 134.84, 131.98, 131.36, 131.29, 131.26, 129.38, 128.67, 128.58, 128.05, 127.15, 127.10, 126.86, 126.67, 126.32, 126.28, 123.48, 123.21, 61.98, 55.38, 48.86, 48.70, 26.10, 24.66, 22.69, 22.64, 22.04, 22.02, 21.48. C34H33N3O2, HRMS calculated for m/z [M+H]+:516.2651 (calculated), 516.2655 (found).
(R)-2-(N-([1,1’-biphenyl]-4-yl)vinylsulfonamido)-N-((S)-1-phenylethyl)-2-(pyridin-3-yl)acetamide (Jun 11119R). White solid, 39% yield. 1H NMR (400 MHz, DMSO-d6) 8.85 (dd, J = 20.8, 7.8 Hz, 1H), 8.45 (d, J = 16.2 Hz, 2H), 7.63–7.25 (m, 17H), 6.94 (dd, J = 16.4, 9.8 Hz, 1H), 6.15–5.96 (m, 2H), 5.10–5.01 (m, J = 7.2 Hz, 1H), 1.31 (d, J = 7.0 Hz, 3H).13C NMR (101 MHz, DMSO-d6) 168.26, 151.08, 149.71, 144.33, 140.03, 139.28, 137.50, 136.25, 135.96, 133.44, 133.38, 131.17, 129.39, 128.73, 128.22, 127.29, 127.23, 127.06, 126.89, 126.75, 126.57, 126.43, 123.72, 63.55, 48.92, 48.83, 46.59, 22.50, 22.36. C29H27N3O3S, HRMS calculated for m/z [M+H]+: 498.1851 (calculated), 498.1862 (found).
(S)-2-(N-([1,1’-biphenyl]-4-yl)vinylsulfonamido)-N-((S)-1-phenylethyl)-2-(pyridin-3-yl)acetamide (Jun 11119S). White solid, 40% yield. 1H NMR (400 MHz, DMSO-d6) 8.82 (t, J = 8.6 Hz, 1H), 8.36 (dd, J = 21.7, 5.1 Hz, 2H), 7.63–7.13 (m, 18H), 6.18–6.03 (m, 2H), 5.10–5.01 (m, J = 7.1 Hz, 1H), 1.44 (d, J = 7.1 Hz, 3H).13C NMR (101 MHz, DMSO-d6) 168.49, 151.16, 149.67, 144.40, 144.29, 139.25, 137.58, 136.48, 133.46, 133.40, 130.91, 129.39, 129.37, 128.64, 128.62, 128.24, 127.24, 127.20, 127.05, 126.92, 126.77, 126.28, 126.26, 123.46, 63.71, 63.53, 48.96, 48.91, 46.78, 46.15, 22.68, 22.60. C29H27N3O3S, HRMS calculated for m/z [M+H]+: 498.1851 (calculated), 498.1862 (found).
N-([1,1’-biphenyl]-4-yl)-N-((S)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)propiolamide (Jun10221R). White solid, 31% yield. 1H NMR (400 MHz, DMSO-d6) 8.78–8.72 (dd, J = 16.3, 7.1 Hz, 1H), 8.42–8.29 (m, 2H), 7.65–6.88 (m, 16H), 6.21–6.12 (m, 2H), 5.08–4.99 (m, J = 13.9, 6.7 Hz, 1H), 1.41–1.39 (d, J = 6.7 Hz, 1.5H), 1.28–1.27 (d, J = 6.4 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 167.98, 151.60, 149.53, 144.37, 139.16, 137.87, 131.98, 129.43, 128.71, 128.60, 128.27, 127.19, 127.02, 126.80, 126.60, 123.56, 83.92, 77.14, 61.93, 48.83, 22.55, 21.36. C30H25N3O2, HRMS calculated for m/z [M+H]+: 460.1948 (calculated), 460.1945 (found).
N-([1,1’-biphenyl]-4-yl)-N-((R)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)propiolamide (Jun10221S). White solid, 30% yield. 1H NMR (400 MHz, DMSO-d6) 8.78–8.76 (d, J = 7.8 Hz, 1H), 8.41–8.27 (m, 2H), 7.62–7.07 (m, 17H), 6.18 (s, 1H), 5.01 (m, 1H), 1.27 (s, 1.5H), 1.25 (s, 1.5H). 13C NMR (101 MHz, DMSO-d6) 13C NMR (101 MHz, DMSO) 170.79, 167.97, 153.13, 151.59, 149.51, 144.35, 140.01, 139.13, 138.25, 137.86, 131.97, 130.70, 129.40, 128.69, 128.58, 128.25, 127.18, 127.00, 126.79, 126.58, 126.41, 126.26, 123.54, 83.91, 77.12, 61.91, 60.21, 48.96, 48.82, 22.54, 21.22, 14.55. C30H25N3O2, HRMS calculated for m/z [M+H]+: 460.1948 (calculated), 460.1945 (found).
N-([1,1’-biphenyl]-4-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)pent-2-ynamide (Jun10892). White solid, 81% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.75–8.73 (d, J = 7.7 Hz, 1H), 8.40–8.29 (m, 2H), 7.58–7.47 (m, 5H), 7.43–7.30 (m, 8H), 7.20–7.15 (t, J = 6.3 Hz, 2H), 7.09–7.07 (d, J = 7.4 Hz, 1H), 6.18 (s, 0.5H), 6.16 (s, 0.5H), 5.03–4.93 (m, 1H), 2.05–1.97 (m, 2H), 1.36–1.35 (d, J = 7.0 Hz, 1.5H), 1.26–1.24 (d, J = 6.9 Hz, 1.5H), 0.68–0.64 (t, J = 7.5 Hz, 3H). 13C NMR (101 MHz, DMSO-d6) 168.22, 167.99, 154.09, 154.04, 151.60, 151.53, 149.43, 144.41, 139.40, 138.84, 137.87, 137.84, 132.00, 129.41, 128.69, 128.59, 128.17, 127.16, 126.99, 126.66, 126.60, 126.28, 123.58, 123.28, 96.01, 75.17, 55.39, 48.93, 48.79, 22.74, 22.57, 12.78, 11.95. C32H29N3O2, HRMS calculated for m/z [M+H]+: 488.2261 (calculated), 488.2259 (found).
N-([1,1’-biphenyl]-4-yl)-4-methyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)pent-2-ynamide (Jun10927). White solid, 77% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.78–8.76 (d, J = 7.7 Hz, 1H), 8.44–8.31 (m, 2H), 7.60–7.51 (m, 5H), 7.46–7.27 (m, 9H), 7.18–7.10 (m, 2H), 6.21 (s, 0.5H), 6.19 (s, 0.5H), 5.02 (m, 1H), 2.45–2.37 (m, 1H), 1.40–1.38 (d, J = 7.1 Hz, 1.5H), 1.29–1.27 (d, J = 7.0 Hz, 1.5H), 0.77 (dd, J = 7.9, 4.3 Hz, 6H). 13C NMR (101 MHz, DMSO-d6) 168.23, 167.99, 154.17, 154.12, 151.60, 151.54, 149.47, 144.66, 144.43, 139.98, 139.95, 139.51, 138.96, 137.89, 137.86, 132.06, 131.06, 130.76, 129.42, 128.69, 128.59, 128.15, 127.15, 127.00, 126.67, 126.60, 126.28, 123.60, 123.31, 99.24, 74.97, 61.67, 48.93, 48.79, 22.73, 22.56, 21.76, 21.75, 19.99. C33H31N3O2, HRMS calculated for m/z [M+H]+: 502.2399 (calculated), 502.2402 (found).
N-([1,1’-biphenyl]-4-yl)-4-fluoro-4-methyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)pent-2-ynamide (Jun10935). White solid, 70% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.63–8.57 (dd, J = 14.5, 7.7 Hz, 1H), 8.37–8.25 (m, 2H), 7.59–7.01 (m, 16H), 6.15 (s, 0.5H), 6.11 (s, 0.5H), 5.07–4.98 (m, 1H), 1.56–1.43 (m, 6H), 1.38–1.36 (d, J = 7.0 Hz, 1.5H), 1.26–1.24 (d, J = 6.9 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 171.65, 171.45, 168.52, 168.36, 152.01, 151.91, 149.24, 149.22, 144.92, 144.38, 139.42, 139.11, 138.13, 138.09, 131.10, 130.65, 129.37, 129.35, 128.64, 128.52, 128.06, 127.13, 127.03, 126.95, 126.92, 126.67, 126.20, 126.03, 123.30, 122.98, 97.27, 95.42, 63.71, 63.68, 60.22, 55.38, 48.81, 48.64, 27.12, 26.88, 22.79, 22.64, 21.23, 14.56. C33H30FN3O2, HRMS calculated for m/z [M+H]+: 520.2355 (calculated), 520.2349 (found).
N-([1,1’-biphenyl]-4-yl)-3-cyclopropyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)propiolamide (Jun10926). White solid, 83% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.74–8.72 (d, J = 7.7 Hz, 1H), 8.41–8.27 (m, 2H), 7.60–7.57 (t, J = 7.3 Hz, 2H), 7.53–7.32 (m, 10H), 7.27–7.25 (d, J = 8.0 Hz, 3H), 6.18 (s, 0.5H), 6.15 (s, 0.5H), 5.03–4.96 (m, 1H), 1.37–1.36 (d, J = 7.0 Hz, 1.5H), 1.27–1.25 (d, J = 7.0 Hz, 1.5H), 0.69–0.66 (dd, J = 8.0, 3.9 Hz, 2H), 0.18–0.14 (dd, J = 8.5, 4.8 Hz, 2H). 13C NMR (101 MHz, DMSO-d6) 168.55, 168.33, 154.37, 154.32, 151.91, 151.85, 149.78, 144.75, 140.28, 139.78, 139.28, 138.16, 132.40, 129.77, 129.02, 128.92, 128.53, 127.49, 127.36, 126.98, 126.94, 126.61, 123.92, 123.63, 98.92, 70.86, 61.90, 61.85, 49.25, 49.12, 23.06, 22.89, 14.90, 9.61, 9.55. C33H29N3O2, HRMS calculated for m/z [M+H]+:500.2265 (calculated), 500.2259 (found).
N-([1,1’-biphenyl]-4-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)hex-2-ynamide (Jun10923). White solid, 79% yield, dr = 1:1. 1H (400 MHz, DMSO-d6) 8.77–8.75 (d, J = 7.6 Hz, 1H), 8.43–8.30 (m, 2H), 7.59–7.33 (m, 13H), 7.23–7.18 (m, 2H), 7.12–7.10 (t, J = 8.0 Hz, 1H), 6.22 (s, 0.5H), 6.19 (s, 0.5H), 5.08–4.97 (m, 1H), 2.09–2.04(dd, J = 6.4 Hz, 2H), 1.40–1.38 (d, J = 7.0 Hz, 1.5H), 1.29–1.28 (d, J = 6.9 Hz, 1.5H), 1.14–1.05 (m, 2H), 0.54–0.50 (m, 3H). 13C NMR (101 MHz, DMSO-d6) 168.22, 168.00, 154.09, 154.04, 151.61, 151.54, 149.43, 144.68, 144.41, 139.98, 139.51, 138.84, 137.88, 137.85, 132.03, 129.40, 128.69, 128.59, 128.14, 127.16, 127.13, 127.00, 126.75, 126.61, 126.28, 123.56, 123.27, 94.71, 75.89, 61.67, 48.94, 48.79, 22.73, 22.56, 20.90, 20.14, 13.07. C33H31N3O2, HRMS calculated for m/z [M+H]+: 502.2420 (calculated), 502.2413 (found).
N-([1,1’-biphenyl]-4-yl)-4,4-dimethyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)pent-2-ynamide (Jun10925). White solid, 76% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.78–8.75 (dd, J = 7.7, 4.0 Hz, 1H), 8.45–8.31 (m, 2H), 7.59–7.27 (m, 14H), 7.18–7.10 (m, J = 7.2 Hz, 2H), 6.21 (s, 0.5H), 6.19 (s, 0.5H), 5.01 (m, 1H), 1.40–1.38 (d, J = 6.8 Hz, 1.5H), 1.29–1.27 (d, J = 7.1 Hz, 1.5H), 0.87–0.85 (t, J = 6.8 Hz, 9H). 13C NMR (101 MHz, DMSO-d6) 168.26, 168.00, 154.24, 154.19, 151.61, 151.54, 149.49, 149.46, 144.64, 144.45, 140.06, 140.03, 139.58, 139.04, 137.90, 137.86, 132.12, 129.43, 128.68, 128.59, 128.14, 127.15, 127.00, 126.69, 126.59, 126.28, 123.61, 123.32, 101.46, 74.33, 74.30, 61.63, 61.57, 48.93, 48.79, 29.65, 29.62, 27.33, 22.73, 22.56. C34H33N3O2, HRMS calculated for m/z [M+H]+: 516.2581 (calculated), 516.2573 (found).
N-([1,1’-biphenyl]-4-yl)-4-hydroxy-4-methyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)pent-2-ynamide (Jun10917). White solid, 68% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.79–8.76 (m, 1H), 8.45–8.31 (m, 2H), 7.59–7.34 (m, 13H), 7.24–7.20 (m, 2H), 7.12–7.10 (d, J = 7.2 Hz, 1H), 6.21 (s, 0.5H), 6.19 (s, 0.5H), 5.41–5.40 (d, J = 7.7 Hz, 1H), 5.06–4.99 (m, 1H), 1.40–1.39 (d, J = 7.0 Hz, 1.5H), 1.30–1.28 (d, J = 6.8 Hz, 1.5H), 1.00–0.98 (m, 6H). 13C NMR (101 MHz, DMSO-d6) 168.18, 167.92, 154.04, 153.99, 151.62, 151.55, 149.52, 149.48, 144.64, 144.42, 140.02, 139.99, 139.51, 138.73, 137.91, 137.87, 131.99, 130.94, 130.65, 129.43, 128.69, 128.61, 128.17, 127.16, 127.01, 126.73, 126.71, 126.59, 126.29, 123.62, 123.33, 98.28, 75.25, 75.22, 63.41, 63.39, 61.71, 61.66, 60.23, 55.39, 48.96, 48.82, 30.75, 30.67, 22.74, 22.58, 21.24, 14.57. C33H31N3O3, HRMS calculated for m/z [M+H]+: 518.2366 (calculated), 518.2354 (found).
N-([1,1’-biphenyl]-4-yl)-4-chloro-N-((S)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)but-2-ynamide (Jun1112R). White solid, 35% yield. 1H NMR (400 MHz, DMSO-d6) 8.91 (s, 1H), 8.76–8.74 (d, J = 7.9 Hz, 1H), 8.51 (s, 2H), 7.55–7.48 (m, 4H), 7.39–7.27 (m, 10H), 7.22–7.18 (m, 1H), 6.13 (s, 1H), 4.94 (m, 1H), 4.31 (s, 2H), 1.23 (s, 1.5H), 1.21 (s, 1.5H). 13C NMR (101 MHz, DMSO-d6) 167.02, 158.47, 158.19, 152.97, 144.13, 140.46, 139.19, 137.83, 131.63, 129.42, 129.18, 128.74, 128.32, 127.27, 127.23, 127.07, 126.57, 88.26, 79.41, 60.15, 48.97, 30.02, 22.41. C31H26ClN3O2, HRMS calculated for m/z [M+H]+: 508.1722 (calculated), 508.1720 (found).
N-([1,1’-biphenyl]-4-yl)-4-chloro-N-((R)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)but-2-ynamide (Jun1112S). White solid, 35% yield. 1H NMR (400 MHz, DMSO-d6) 8.87 (s, 1H), 8.79–8.77 (d, J = 7.6 Hz, 1H), 8.31 (s, 2H), 7.53–7.50 (dd, J = 10.9, 8.4 Hz, 3H), 7.37–7.26 (m, 7H), 7.18–7.11 (m, 4H), 7.05–7.02 (m, 2H), 6.10 (s, 1H), 4.91 (m, 1H), 4.32 (s, 2H), 1.31 (s, 1.5H), 1.30 (s, 1.5H). 13C NMR (101 MHz, DMSO-d6) 166.83, 158.45, 158.13, 153.01, 144.49, 140.48, 139.19, 137.80, 131.66, 129.41, 128.84, 128.68, 128.31, 127.27, 127.24, 127.06, 126.23, 88.25, 79.44, 60.08, 49.18, 30.04, 22.59. C31H26ClN3O2, HRMS calculated for m/z [M+H]+: 508.1722 (calculated), 508.1720 (found).
N-([1,1’-biphenyl]-4-yl)-2-cyano-N-((S)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acetamide (Jun10541R). White solid, 38% yield. 1H NMR (400 MHz, DMSO-d6) 8.67–8.65 (d, J = 7.9 Hz, 1H), 8.30–8.26 (m, 2H), 7.54–7.52 (d, J = 7.4 Hz, 2H), 7.47–7.45 (d, J = 8.0 Hz, 2H), 7.38–7.18 (m, 11H), 7.12–7.09 (dd, J = 7.9, 4.8 Hz, 1H), 6.12 (s, 1H), 4.97–4.90 (m, 1H), 3.52–3.51 (d, J = 4.1 Hz, 2H), 1.20 (s, 1.5H), 1.19 (s, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.18, 163.43, 151.54, 149.44, 144.31, 140.24, 139.05, 137.85, 137.81, 131.67, 131.06, 129.43, 128.72, 128.34, 127.32, 127.22, 127.03, 126.64, 123.51, 116.06, 62.36, 48.78, 26.95, 22.49. C30H26N4O2, HRMS calculated for m/z [M+H]+: 475.2080 (calculated), 475.2065 (found).
N-([1,1’-biphenyl]-4-yl)-2-cyano-N-((R)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acetamide (Jun10541S). White solid, 37% yield. 1H NMR (400 MHz, DMSO-d6) 8.67–8.65 (d, J = 7.9 Hz, 1H), 8.30–8.26 (m, 2H), 7.54–7.52 (d, J = 7.5 Hz, 2H), 7.47–7.44 (m, 2H), 7.38–7.18 (m, 11H), 7.12–7.09 (dd, J = 7.9, 4.8 Hz, 1H), 6.12 (s, 1H), 4.94 (m, 1H), 3.52–3.51 (d, J = 4.3 Hz, 2H), 1.20 (s, 1.5H), 1.19 (s, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.18, 163.43, 151.54, 149.44, 144.31, 140.24, 139.05, 137.85, 137.81, 131.67, 131.06, 129.43, 128.72, 128.34, 127.32, 127.22, 127.03, 126.63, 123.51, 116.07, 62.36, 48.78, 26.95, 22.49. C30H26N4O2, HRMS calculated for m/z [M+H]+: 475.2080 (calculated), 475.2065 (found).
N-([1,1’-biphenyl]-4-yl)-1-cyano-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)cyclopropane-1-carboxamide (Jun10893). White solid, 66% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.71–8.67 (t, J = 8.6 Hz, 1H), 8.40–8.27 (m, 2H), 7.63–7.33 (m, 12H), 7.21–7.17 (m, 2H), 7.09–7.07 (d, J = 7.3 Hz, 2H), 6.19 (s, 0.5H), 6.15 (s, 0.5H), 5.06–4.95 (m, 1H), 1.79–1.71 (m, 1H), 1.64–1.59 (m, 1H), 1.45–1.40 (m, 2H), 1.36–1.34 (d, J = 7.1 Hz, 1.5H), 1.27–1.25 (d, J = 6.5 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.09, 167.98, 164.73, 164.69, 151.84, 151.78, 149.43, 144.73, 144.32, 140.06, 139.13, 138.01, 137.50, 137.41, 132.66, 132.60, 131.06, 130.64, 129.40, 128.70, 128.57, 128.27, 127.20, 127.12, 126.97, 126.78, 126.64, 126.26, 123.47, 123.17, 119.34, 63.46, 63.41, 48.90, 48.75, 22.68, 22.61, 18.72, 18.52, 14.84. C32H28N4O2, HRMS calculated for m/z [M+H]+: 501.2223 (calculated), 501.2219 (found).
tert-butyl 4-([1,1’-biphenyl]-4-yl(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)carbamoyl)-4-cyanopiperidine-1-carboxylate (Jun10912). White solid, 70% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.65 (s, 1H), 8.38–8.25 (m, 2H), 7.61–7.33 (m, 11H), 7.19–7.02 (m, 3H), 6.14 (s, 0.5H), 6.09 (s, 0.5H), 5.75 (s, 1H), 5.02–4.95 (m, 1H), 3.89–3.79 (m, J = 14.2 Hz, 2H), 2.80 (s, 2H), 1.93–1.82 (m, 3H), 1.38 (s, 12H). 13C NMR (101 MHz, DMSO-d6) 168.33, 168.09, 166.24, 154.00, 152.10, 152.00, 149.44, 144.81, 144.30, 140.78, 138.96, 138.13, 136.49, 136.42, 129.41, 128.62, 128.54, 128.40, 127.13, 127.03, 126.64, 126.21, 79.69, 79.67, 64.49, 55.38, 48.91, 48.74, 43.50, 43.38, 33.36, 28.47, 22.75, 22.56. C39H41N5O4, HRMS calculated for m/z [M+H]+: 644.3160 (calculated), 644.3150 (found).
N-([1,1’-biphenyl]-4-yl)-4-cyano-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)tetrahydro-2H-pyran-4-carboxamide (Jun10913). White solid, 68% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.69–8.65 (t, J = 8.5 Hz, 1H), 8.40–8.27 (m, 2H), 7.64–6.93 (m, 16H), 6.17 (s, 0.5H), 6.13 (s, 0.5H), 5.07–4.98 (m, 1H), 3.85–3.77 (q, J = 9.0, 5.7 Hz, 2H), 3.39–3.33 (t, J = 11.1 Hz, 2H), 2.23–2.14 (m, 1H), 2.04–1.97 (m, 1H), 1.87–1.78 (dd, J = 24.0, 14.0 Hz, 2H), 1.39–1.37 (d, J = 7.0 Hz, 1.5H), 1.27–1.25 (d, J = 6.8 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.32, 168.11, 166.21, 152.07, 151.97, 149.42, 144.82, 144.30, 140.70, 138.93, 138.11, 136.43, 136.36, 129.42, 129.40, 128.66, 128.54, 128.40, 127.17, 127.07, 127.02, 127.00, 126.65, 126.21, 118.89, 64.47, 63.38, 63.32, 55.38, 33.98, 33.92, 33.88, 33.83, 22.77, 22.59. C34H32N4O3, HRMS calculated for m/z [M+H]+: 545.2475 (calculated), 545.2470 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-2-cyano-4-methyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)pent-2-enamide (Jun10544). White solid, 52% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.76–8.74 (d, J = 7.7 Hz, 1H), 8.43–8.30 (m, 2H), 7.59–7.10 (m, 17H), 6.20 (s, 0.5H), 6.18 (s, 0.5H), 5.05–4.98 (m, J = 7.4 Hz, 1H), 2.45–2.36 (m, J = 6.8 Hz, 1H), 1.39–1.38 (d, J = 7.0 Hz, 1.5H), 1.29–1.27 (d, J = 7.0 Hz, 1.5H), 0.78–0.77 (d, J = 5.9 Hz, 6H). 13C NMR (101 MHz, DMSO-d6) 168.01, 164.58, 162.58, 151.72, 149.54, 144.31, 140.02, 139.15, 138.42, 138.04, 132.00, 131.97, 131.02, 129.42, 128.73, 128.60, 128.27, 127.23, 127.00, 126.95, 126.65, 126.59, 126.30, 123.57, 114.02, 109.61, 62.75, 60.22, 48.81, 31.39, 22.59, 21.22, 21.04, 21.01, 14.56. C34H32N4O2, HRMS calculated for m/z [M+H]+: 529.2525 (calculated), 529.2524 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-2-cyano-3-cyclopropyl-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10545). White solid, 44% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.80–8.75 (t, J = 8.1 Hz, 1H), 8.45–8.31 (m, 2H), 7.59–7.10 (m, 17H), 6.32 (s, 0.5H), 6.28 (s, 0.5H), 5.73–5.70 (d, J = 14.4 Hz, 1H), 5.08–4.99 (m, 1H), 2.74–2.68 (m, 2H), 2.26–2.16 (m, 2H), 1.38–1.37 (d, J = 7.1 Hz, 1.5H), 1.29–1.28 (d, J = 7.0 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.10, 168.01, 166.99, 162.36, 151.74, 149.49, 144.70, 144.36, 139.96, 139.14, 138.19, 138.04, 132.22, 132.17, 129.42, 129.20, 128.78, 128.73, 128.59, 128.28, 127.20, 127.13, 127.00, 126.87, 126.65, 126.34, 126.28, 126.03, 114.67, 114.13, 107.63, 63.05, 55.37, 48.88, 22.66, 22.58, 15.75, 10.76. C34H30N4O2, HRMS calculated for m/z [M+H]+: 527.2370 (calculated), 527.2375 (found).
N-([1,1’-biphenyl]-4-yl)-2-cyano-2-cyclohexylidene-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acetamide (Jun10951). White solid, 36% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.78–8.76 (d, J = 7.7 Hz, 1H), 8.45–8.29 (m, 2H), 7.60–7.06 (m, 17H), 6.25 (s, 0.5H), 6.21 (s, 0.5H), 5.76–5.75 (d, J = 2.2 Hz, 1H), 5.06–4.98 (m, 1H), 2.40–2.38 (m, 2H), 2.16 (s, 2H), 1.44–1.42 (m, 4H), 1.37–1.35 (d, J = 6.8 Hz, 1.5H), 1.29–1.27 (d, J = 6.8 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.19, 167.96, 165.85, 162.85, 151.76, 151.65, 149.56, 149.51, 144.73, 144.18, 139.93, 138.94, 138.05, 137.75, 137.73, 131.51, 131.48, 130.46, 129.43, 129.41, 128.65, 128.59, 128.32, 127.15, 126.98, 126.69, 126.60, 126.27, 123.58, 123.26, 115.67, 115.62, 62.56, 60.22, 55.37, 49.04, 48.81, 33.57, 31.79, 27.53, 27.05, 22.78, 22.51. C36H34N4O2, HRMS calculated for m/z [M+H]+: 555.2690 (calculated), 555.2688 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-2-cyano-3-(4-methoxyphenyl)-N-((S)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10963R). White solid, 38% yield. 1H NMR (400 MHz, DMSO-d6) 8.73–8.71 (d, J = 7.7 Hz, 1H), 8.30–8.24 (dd, J = 18.6, 3.5 Hz, 2H), 7.78 (s, 1H), 7.71–7.68zz (d, J = 8.9 Hz, 2H), 7.51–7.49 (d, J = 7.1 Hz, 2H), 7.44–7.42 (d, J = 8.4 Hz, 2H), 7.32–7.30 (d, J = 7.9 Hz, 2H), 7.27–7.20 (m, 3H), 7.18–7.00 (m, 8H), 6.96–6.93 (dd, J = 8.7, 3.8 Hz, 2H), 6.21 (s, 1H), 4.93 (m, 1H), 3.71 (s, 3H), 1.31 (s, 1.5H), 1.30 (s, 1.5H). 13C NMR (101 MHz, DMSO-d6) 167.95, 163.82, 162.94, 152.09, 151.70, 149.52, 144.68, 139.68, 138.95, 138.22, 138.05, 132.43, 132.14, 130.77, 129.38, 128.59, 128.26, 127.14, 126.92, 126.76, 126.27, 125.02, 123.30, 116.00, 115.18, 103.80, 63.16, 56.03, 48.93, 22.67. C38H32N4O3, HRMS calculated for m/z [M+H]+: 593.2485 (calculated), 593.2466 (found).
(E)-N-([1,1’-biphenyl]-4-yl)-2-cyano-3-(4-methoxyphenyl)-N-((R)-2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acrylamide (Jun10963S). White solid, 38% yield. 1H NMR (400 MHz, DMSO-d6) 8.72–8.70 (m, 1H), 8.32–8.25 (m, 2H), 7.70–7.68 (d, J = 8.5 Hz, 2H), 7.51–6.94 (m, 19H), 6.21 (s, 1H), 4.97–4.90 (m, 1H), 3.71 (s, 3H), 1.31–1.29 (d, J = 7.1Hz 3H). 13C NMR (101 MHz, DMSO-d6) 167.95, 163.82, 162.94, 152.09, 151.70, 149.52, 144.68, 139.68, 138.95, 138.22, 138.06, 132.43, 132.14, 130.77, 129.38, 128.59, 128.26, 127.14, 126.92, 126.76, 126.27, 125.02, 123.30, 116.00, 115.18, 103.80, 63.16, 56.03, 48.93, 22.67. C38H32N4O3, HRMS calculated for m/z [M+H]+: 593.2485 (calculated), 593.2466 (found).
N-([1,1’-biphenyl]-4-yl)-2-oxo-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)butanamide (Jun10938). White solid, 65% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.41 (s, 1H), 8.35–8.33 (d, J = 4.6 Hz, 1H), 8.25–8.20 (m, 1H), 7.73–6.89 (m, 16H), 6.15 (s, 1H), 6.08 (s, 0.5H), 6.05 (s, 0.5H), 3.84 (m, 1H), 2.64 (m, J = 7.1 Hz, 1H), 2.34–2.30 (t, J = 7.1 Hz, 1H), 1.73–1.55 (m, 2H), 1.14–1.12 (d, J = 6.0 Hz, 1.5H), 0.98–0.96 (d, J = 6.1 Hz, 1.5H).13C NMR (101 MHz, DMSO-d6) 172.60, 172.56, 168.67, 168.57, 151.51, 151.47, 149.13, 144.85, 144.46, 140.32, 139.28, 138.55, 138.52, 137.81, 132.94, 132.88, 131.38, 131.32, 129.38, 129.29, 129.22, 128.68, 128.59, 128.06, 127.16, 127.12, 126.88, 126.69, 126.39, 126.35, 123.48, 123.21, 62.12, 48.88, 48.72, 22.70, 22.63, 14.44, 13.41, 13.38. C31H29N3O3, HRMS calculated for m/z [M+H]+: 492.2219 (calculated), 492.2215 (found).
N-([1,1’-biphenyl]-4-yl)-2-(4-methylpiperazin-1-yl)-2-oxo-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)acetamide (Jun11812). White solid, 46% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.85 (d, J = 7.7 Hz, 1H), 8.30 (s, 1H), 7.58–7.12 (m, 18H), 6.26 (s, 0.5H), 6.21 (s, 0.5H), 5.12–5.02 (m, 1H), 3.50–3.36 (m, 3H), 3.23–3.16 (m, 2H), 3.18 (d, J = 14.5 Hz, 1H), 3.11–3.04 (m, 1H), 2.36–2.23 (m, 1H), 2.12 (dt, J = 12.9, 6.7 Hz, 1H), 2.06 (s, 3H), 1.39 (d, J = 7.0 Hz, 1.5H), 1.32 (d, J = 7.0 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.20, 167.96, 165.37, 165.33, 162.28, 162.23, 151.69, 151.58, 149.54, 149.50, 144.67, 144.13, 140.13, 139.08, 138.05, 138.00, 136.37, 136.29, 131.57, 130.48, 130.17, 129.40, 129.39, 128.65, 128.62, 128.27, 127.20, 127.16, 126.99, 126.97, 126.67, 126.60, 126.31, 123.61, 123.29, 61.80, 61.78, 54.48, 54.08, 49.13, 48.93, 45.99, 45.21, 22.78, 22.49. C34H35N5O3, HRMS calculated for m/z [M+H]+: 567.2818 (calculated), 567.2820 (found).
N-([1,1’-biphenyl]-4-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)but-3-ynamide (Jun1155). White solid, 75% yield, dr = 1:1. 1H NMR (400 MHz, CDCl3) 8.45 (s, 2H), 7.51–7.26 (m, 18H), 6.72 (s, 0.5H), 6.22 (s, 0.5H), 5.18–5.11 (m,1H), 3.06 (s, 2H), 1.48 (s, 1.5H), 1.46 (s, 1.5H). 13C NMR (101 MHz, CDCl3) 168.00, 167.57, 151.53, 149.88, 142.95, 141.89, 139.50, 138.21, 137.30, 130.64, 129.93, 128.88, 128.75, 127.93, 127.44, 127.05, 126.29, 122.94, 76.65, 71.84, 61.78, 60.35, 49.54, 26.91, 21.70, 14.19. C31H27N3O2, HRMS calculated for m/z [M+H]+: 474.2182 (calculated), 474.2186 (found).
N-([1,1’-biphenyl]-4-yl)-N-(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)but-3-ynamide (Jun1156). White solid, 70% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.68 (dd, J = 12.6, 7.7 Hz, 1H), 8.38–8.26 (m, 2H), 7.62–7.08 (m, 18H), 6.25 (s, 0.5H), 6.22 (s, 0.5H), 5.04–4.95 (m, 1H), 2.72 (d, J = 8.4 Hz, 1H), 2.38–2.14 (m, 4H), 1.38 (d, J = 7.0 Hz, 1.5H), 1.26 (d, J = 6.9 Hz, 1.5H). 13C NMR (101 MHz, DMSO-d6) 168.70, 168.57, 151.61, 151.53, 149.23, 144.84, 144.42, 139.78, 139.23, 138.96, 138.90, 137.78, 137.75, 131.88, 131.70, 131.31, 129.38, 128.69, 128.54, 128.21, 127.21, 127.05, 126.68, 126.23, 123.45, 123.15, 84.17, 84.13, 71.76, 61.81, 48.82, 48.71, 34.11, 22.77, 22.60, 14.31. C32H29N3O2, HRMS calculated for m/z [M+H]+: 488.2338 (calculated), 488.2342 (found).
2-([1,1’-biphenyl]-4-yl(prop-2-yn-1-yl)amino)-N-((S)-1-phenylethyl)-2-(pyridin-3-yl)acetamide (Jun10924). White solid, 70% yield, dr = 1:1. 1H NMR (400 MHz, CDCl3) 8.64 (dd, J = 32.3, 2.3 Hz, 1H), 8.54 (ddd, J = 16.7, 4.9, 1.6 Hz, 1H), 7.72 (ddt, J = 38.8, 8.0, 2.0 Hz, 1H), 7.57–7.50 (m, 4H), 7.42 (td, J = 7.6, 4.9 Hz, 2H), 7.30–7.04 (m, 9H), 6.91 (dd, J = 6.6, 3.0 Hz, 1H), 5.40 (s, 0.5H), 5.34 (s, 0.5H), 5.12–5.01 (m, 1H), 3.97–3.75 (m, 2H), 2.18 (dt, J = 26.8, 2.4 Hz, 1H), 1.39 (d, J = 6.9 Hz, 1.5H), 1.27 (d, J = 6.9 Hz, 1.5H). 13C NMR (101 MHz, CDCl3) 168.75, 168.69, 150.52, 150.46, 149.73, 149.58, 146.69, 146.48, 142.70, 142.52, 140.42, 136.73, 136.70, 134.65, 134.29, 131.60, 131.47, 128.86, 128.84, 128.67, 128.51, 128.06, 127.98, 127.44, 127.23, 126.95, 126.91, 126.67, 126.64, 126.20, 125.93, 123.61, 123.52, 118.31, 117.80, 79.09, 78.88, 74.09, 73.93, 66.81, 66.74, 49.06, 48.88, 40.79, 40.26, 21.70, 21.27. C30H27N3O, HRMS calculated for m/z [M+H]+: 446.2236 (calculated), 446.2217 (found).
2-([1,1’-biphenyl]-4-yl(2-oxo-2-(((S)-1-phenylethyl)amino)-1-(pyridin-3-yl)ethyl)amino)-1,1-difluoro-2-oxoethane-1-sulfonyl fluoride (Jun1191). Yellow solid, 60% yield, dr = 1:1. 1H NMR (400 MHz, DMSO-d6) 8.76 (t, J = 7.6 Hz, 1H), 8.41–8.28 (m, 2H), 7.63–7.09 (m, 16H), 6.27 (s, 0.5H), 6.24 (s, 0.5H), 5.06–4.96 (m, 1H), 1.39 (d, J = 6.8 Hz, 1.5H), 1.28 (d, J = 7.2 Hz, 1.5H).13C NMR (101 MHz, CDCl3) 171.32, 149.42, 149.09, 144.74, 140.88, 134.80, 133.64, 131.61, 128.85, 128.82, 128.67, 128.04, 126.98, 126.36, 126.34, 123.95, 121.42, 115.44, 113.83, 58.54, 53.21. C29H24F3N3O4S, HRMS calculated for m/z [M+H]+: 568.1518 (calculated), 568.1523 (found).
Supplementary Material
Diverse reactive warheads were explored in targeting SARS-CoV-2 main protease.
Jun10541R with the nitrile warhead had an of 0.50 μM against .
Jun10963R with a dually activated nitrile warhead had an of 0.56 μM.
Jun10541R and Jun10963 inhibited SARS-CoV-2 with values of 2.92 and 6.47 μM.
X-ray crystals showed covalent conjugation with nitrile and alkyne warheads.
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health (NIH) grant AI158775 to J. W. The antiviral assay in Calu-3 cells was conducted through the NIAID preclinical service under a non-clinical evaluation agreement. We thank Michael Kemp for assistance with crystallization and X-ray diffraction data collection. We also thank the staff members of the Advanced Photon Source of Argonne National Laboratory, particularly those at the Structural Biology Center (SBC), with X-ray diffraction data collection. SBC-CAT is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02–06CH11357.
Jun Wang reports financial support was provided by National Institute of Allergy and Infectious Diseases. Jun Wang has patent #WO2022119756A1 pending to University of Arizona.
ABBREVIATIONS
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- Ugi-4CR
Ugi four-component reaction
main protease
- 3CLpro
3-chymotrypsin-like protease
Footnotes
ASSOCIATED CONTENT
Supporting information: Tables S1-S2; HNMR and CNMR spectra for final products; Molecular Formula Strings.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of competing interest
Jun Wang is an inventor of a filed patent claiming the use of Jun10541R and related compounds as potential COVID-19 antiviral drugs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].https://coronavirus.jhu.edu/map.html Accessed on July 15th, 2023.
- [2].Hu B, Guo H, Zhou P, Shi ZL, Characteristics of SARS-CoV-2 and COVID-19, Nat. Rev. Microbiol. 19 (2021) 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tregoning JS, Flight KE, Higham SL, Wang Z, Pierce BF, Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape, Nat. Rev. Immunol. 21 (2021) 626–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Tao K, Tzou PL, Nouhin J, Bonilla H, Jagannathan P, Shafer RW, SARS-CoV-2 Antiviral Therapy, Clin. Microbiol. Rev. 34 (2021) e0010921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].WHO Solidarity Trial Consortium. Remdesivir and three other drugs for hospitalised patients with COVID-19: final results of the WHO Solidarity randomised trial and updated meta-analyses, Lancet, 399 (2022) 1941–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ader F, Bouscambert-Duchamp M, Hites M, Peiffer-Smadja N, Poissy J, Belhadi D, Diallo A, Le MP, Peytavin G, Staub T, Greil R, Guedj J, Paiva JA, Costagliola D, Yazdanpanah Y, Burdet C, Mentre F, DisCoVeRy Study G, Remdesivir plus standard of care versus standard of care alone for the treatment of patients admitted to hospital with COVID-19 (DisCoVeRy): a phase 3, randomised, controlled, open-label trial, Lancet Infect. Dis. 22 (2022) 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhou S, Hill CS, Sarkar S, Tse LV, Woodburn BMD, Schinazi RF, Sheahan TP, Baric RS, Heise MT, Swanstrom R, beta-d-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells, J. Infect. Dis. 224 (2021) 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tan B, Joyce R, Tan H, Hu Y, Wang J, SARS-CoV-2 Main Protease Drug Design, Assay Development, and Drug Resistance Studies, Acc. Chem. Res. 56 (2023) 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tan H, Hu Y, Jadhav P, Tan B, Wang J, Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease, J. Med. Chem. 65 (2022) 7561–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Boras B, Jones RM, Anson BJ, Arenson D, Aschenbrenner L, Bakowski MA, Beutler N, Binder J, Chen E, Eng H, Hammond H, Hammond J, Haupt RE, Hoffman R, Kadar EP, Kania R, Kimoto E, Kirkpatrick MG, Lanyon L, Lendy EK, Lillis JR, Logue J, Luthra SA, Ma C, Mason SW, McGrath ME, Noell S, Obach RS, MN OB, O’Connor R, Ogilvie K, Owen D, Pettersson M, Reese MR, Rogers TF, Rosales R, Rossulek MI, Sathish JG, Shirai N, Steppan C, Ticehurst M, Updyke LW, Weston S, Zhu Y, White KM, Garcia-Sastre A, Wang J, Chatterjee AK, Mesecar AD, Frieman MB, Anderson AS, Allerton C, Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19, Nat. Commun. 12 (2021) 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, Dantonio A, Di L, Eng H, Ferre R, Gajiwala KS, Gibson SA, Greasley SE, Hurst BL, Kadar EP, Kalgutkar AS, Lee JC, Lee J, Liu W, Mason SW, Noell S, Novak JJ, Obach RS, Ogilvie K, Patel NC, Pettersson M, Rai DK, Reese MR, Sammons MF, Sathish JG, Singh RSP, Steppan CM, Stewart AE, Tuttle JB, Updyke L, Verhoest PR, Wei L, Yang Q, Zhu Y, An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19, Science, 374 (2021) 1586–1593. [DOI] [PubMed] [Google Scholar]
- [12].Joyce RP, Hu VW, Wang J, The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations, Med. Res. Rev. 31 (2022) 1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hammond J, Leister-Tebbe H, Gardner A, Abreu P, Bao W, Wisemandle W, Baniecki M, Hendrick VM, Damle B, Simon-Campos A, Pypstra R, Rusnak JM, Investigators E-H, Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19, N. Engl. J. Med. 386 (2022) 1397–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].www.fda.gov/media/158165/download. Accessed on July 15th, 2023.
- [15].Hu Y, Lewandowski EM, Tan H, Zhang X, Morgan RT, Zhang X, Jacobs LMC, Butler SG, Gongora MV, Choy J, Deng X, Chen Y, Wang J, Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir, ACS Cent. Sci. (2023) 10.1021/acscentsci.3c00538. [DOI] [PMC free article] [PubMed]
- [16].Iketani S, Mohri H, Culbertson B, Hong SJ, Duan Y, Luck MI, Annavajhala MK, Guo Y, Sheng Z, Uhlemann AC, Goff SP, Sabo Y, Yang H, Chavez A, Ho DD, Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, Nature, 613 (2022) 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yang KS, Leeuwon SZ, Xu S, Liu WR, Evolutionary and Structural Insights about Potential SARS-CoV-2 Evasion of Nirmatrelvir, J. Med. Chem. 65 (2022) 8686–8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jochmans D, Liu C, Donckers K, Stoycheva A, Boland S, Stevens SK, De Vita C, Vanmechelen B, Maes P, Trüeb B, Ebert N, Thiel V, De Jonghe S, Vangeel L, Bardiot D, Jekle A, Blatt LM, Beigelman L, Symons JA, Raboisson P, Chaltin P, Marchand A, Neyts J, Deval J, Vandyck K, The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir, mBio, (2023) e0281522. [DOI] [PMC free article] [PubMed]
- [19].Zhou Y, Gammeltoft KA, Ryberg LA, Pham LV, Tjørnelund HD, Binderup A, Duarte Hernandez CR, Fernandez-Antunez C, Offersgaard A, Fahnøe U, Peters GHJ, Ramirez S, Bukh J, Gottwein JM, Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system, Sci. Adv. 8 (2022) eadd7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ghosh AK, Mishevich JL, Mesecar A, Mitsuya H, Recent Drug Development and Medicinal Chemistry Approaches for the Treatment of SARS-CoV-2 Infection and COVID-19, ChemMedChem, 17 (2022) e202200440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ghosh AK, Brindisi M, Shahabi D, Chapman ME, Mesecar AD, Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and Covid-19 Therapeutics, ChemMedChem, 15 (2020) 907–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ma C, Xia Z, Sacco MD, Hu Y, Townsend JA, Meng X, Choza J, Tan H, Jang J, Gongora MV, Zhang X, Zhang F, Xiang Y, Marty MT, Chen Y, Wang J, Discovery of Di-and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity, J. Am. Chem. Soc.143 (2021) 20697–20709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yamane D, Onitsuka S, Re S, Isogai H, Hamada R, Hiramoto T, Kawanishi E, Mizuguchi K, Shindo N, Ojida A, Selective covalent targeting of SARS-CoV-2 main protease by enantiopure chlorofluoroacetamide, Chem Sci. 13 (2022) 3027–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hirose Y, Shindo N, Mori M, Onitsuka S, Isogai H, Hamada R, Hiramoto T, Ochi J, Takahashi D, Ueda T, Caaveiro JMM, Yoshida Y, Ohdo S, Matsunaga N, Toba S, Sasaki M, Orba Y, Sawa H, Sato A, Kawanishi E, Ojida A, Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease, J. Med. Chem. 65 (2022) 13852–13865. [DOI] [PubMed] [Google Scholar]
- [25].Konno S, Kobayashi K, Senda M, Funai Y, Seki Y, Tamai I, Schäkel L, Sakata K, Pillaiyar T, Taguchi A, Taniguchi A, Gütschow M, Müller CE, Takeuchi K, Hirohama M, Kawaguchi A, Kojima M, Senda T, Shirasaka Y, Kamitani W, Hayashi Y, 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents, J. Med. Chem. 65 (2022) 2926–2939. [DOI] [PubMed] [Google Scholar]
- [26].Quan BX, Shuai H, Xia AJ, Hou Y, Zeng R, Liu XL, Lin GF, Qiao JX, Li WP, Wang FL, Wang K, Zhou RJ, Yuen TT, Chen MX, Yoon C, Wu M, Zhang SY, Huang C, Wang YF, Yang W, Tian C, Li WM, Wei YQ, Yuen KY, Chan JF, Lei J, Chu H, Yang S, An orally available M(pro) inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron, Nat. Microbiol. 7 (2022) 716–725. [DOI] [PubMed] [Google Scholar]
- [27].Gao S, Song L, Claff T, Woodson M, Sylvester K, Jing L, Weiße RH, Cheng Y, Sträter N, Schäkel L, Gütschow M, Ye B, Yang M, Zhang T, Kang D, Toth K, Tavis J, Tollefson AE, Müller CE, Zhan P, Liu X, Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors, J. Med. Chem. 65 (2022) 16902–16917. [DOI] [PubMed] [Google Scholar]
- [28].Stille JK, Tjutrins J, Wang G, Venegas FA, Hennecker C, Rueda AM, Sharon I, Blaine N, Miron CE, Pinus S, Labarre A, Plescia J, Burai Patrascu M, Zhang X, Wahba AS, Vlaho D, Huot MJ, Schmeing TM, Mittermaier AK, Moitessier N, Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CLpro covalent inhibitors, European Journal of Medicinal Chemistry, 229 (2022) 114046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kitamura N, Sacco MD, Ma C, Hu Y, Townsend JA, Meng X, Zhang F, Zhang X, Ba M, Szeto T, Kukuljac A, Marty MT, Schultz D, Cherry S, Xiang Y, Chen Y, Wang J, Expedited Approach toward the Rational Design of Noncovalent SARS-CoV-2 Main Protease Inhibitors, J. Med. Chem. 65 (2022) 2848–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ekkebus R, van Kasteren SI, Kulathu Y, Scholten A, Berlin I, Geurink PP, de Jong A, Goerdayal S, Neefjes J, Heck AJ, Komander D, Ovaa H, On terminal alkynes that can react with active-site cysteine nucleophiles in proteases, J. Am. Chem. Soc. 135 (2013) 2867–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mons E, Kim RQ, van Doodewaerd BR, van Veelen PA, Mulder MPC, Ovaa H, Exploring the Versatility of the Covalent Thiol-Alkyne Reaction with Substituted Propargyl Warheads: A Deciding Role for the Cysteine Protease, J. Am. Chem. Soc. 143 (2021) 6423–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T, Zhang X, Tarbet B, Marty MT, Chen Y, Wang J, Boceprevir GC -376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease, Cell Res. 30 (2020) 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hu Y, Ma C, Szeto T, Hurst B, Tarbet B, Wang J, Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity against Coronaviruses, ACS Infect. Dis. 7 (2021) 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Siklos M, BenAissa M, Thatcher GR, Cysteine proteases as therapeutic targets: does selectivity matter? A systematic review of calpain and cathepsin inhibitors, Acta Pharm Sin B, 5 (2015) 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gehringer M, Laufer SA, Emerging and Re-Emerging Warheads for Targeted Covalent Inhibitors: Applications in Medicinal Chemistry and Chemical Biology, J. Med. Chem. 62 (2019) 5673–5724. [DOI] [PubMed] [Google Scholar]
- [36].Brogi S, Ibba R, Rossi S, Butini S, Calderone V, Gemma S, Campiani G, Covalent Reversible Inhibitors of Cysteine Proteases Containing the Nitrile Warhead: Recent Advancement in the Field of Viral and Parasitic Diseases, Molecules, 27 (2022) 2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sacco MD, Ma C, Lagarias P, Gao A, Townsend JA, Meng X, Dube P, Zhang X, Hu Y, Kitamura N, Hurst B, Tarbet B, Marty MT, Kolocouris A, Xiang Y, Chen Y, Wang J, Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L, Sci. Adv. 6 (2020) eabe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Xia Z, Sacco M, Hu Y, Ma C, Meng X, Zhang F, Szeto T, Xiang Y, Chen Y, Wang J, Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir, ACS Pharmacol. Transl. Sci. 4 (2021) 1408–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Repetto G, del Peso A, Zurita JL, Neutral red uptake assay for the estimation of cell viability/cytotoxicity, Nat. Protoc. 3 (2008) 1125–1131. [DOI] [PubMed] [Google Scholar]
- [40].Ma C, Zhang J, Wang J, Pharmacological Characterization of the Spectrum of Antiviral Activity and Genetic Barrier to Drug Resistance of M2-S31N Channel Blockers, Mol. Pharmacol. 90 (2016) 188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Vagin A, Teplyakov A, MOLREP: an automated program for molecular replacement, J. Appl. Crystallogr. 30 (1997) 1022–1025. [Google Scholar]
- [42].Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA, REFMAC5 for the refinement of macromolecular crystal structures, Acta Crystallogr. D Biol. Crystallogr. 67 (2011) 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallogr. D Biol. Crystallogr. 60 (2004) 2126–2132. [DOI] [PubMed] [Google Scholar]
- [44].Jacobs J, Grum-Tokars V, Zhou Y, Turlington M, Saldanha SA, Chase P, Eggler A, Dawson ES, Baez-Santos YM, Tomar S, Mielech AM, Baker SC, Lindsley CW, Hodder P, Mesecar A, Stauffer SR, Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease, J. Med. Chem. 56 (2013) 534–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.