Abstract
Otopathogens in acute otitis media (AOM) have implications for care because the likelihood of resolution without antibiotics and optimal antibiotic agent varies by microorganism. We aimed to determine the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of nasopharyngeal (NP) qualitative polymerase chain reaction (PCR) for common bacterial otopathogens in children with AOM compared to NP culture. NP flocked swabs collected from enrolled children aged 6-35 months with uncomplicated AOM in Denver, CO were tested by culture and multiplex PCR. The sensitivity and NPV of PCR using culture as a reference were high (H. influenzae 93.3%, 98.0%; S. pneumoniae 94.2%, 95.1%; M. catarrhalis 92.3%, 86.4%); whereas the specificity and PPV were lower and varied by organism (54.2-84.1%, 55.1-69.2%, respectively). PCR detected 1.5 times more organisms than culture. NP PCR has a high predictive value for excluding otopathogens compared to culture and warrants exploration as a diagnostic tool.
Keywords: Pediatrics, Acute Otitis Media, Polymerase Chain Reaction, Otopathogens
1. Background:
Acute otitis media (AOM) affects over 60% of children by three years of age and is the most common reason children in the United States are prescribed an antibiotic[1,2,3]. Among children with AOM, Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are the predominant bacterial otopathogens. Up to 20% of children have exclusively viral AOM[4-7]. The associated pathogen has important implications for management because the severity of infection, risk of tympanic membrane rupture, and likelihood of resolution without an antibiotic differ between organisms[8-12]. Additionally, the optimal antibiotic agent varies by pathogen; nearly all M. catarrhalis isolates and a growing proportion of H. influenzae isolates produce beta-lactamase, rendering them resistant to amoxicillin[5,13,14].
Unfortunately, there are no clinical features that can distinguish between bacterial otopathogens[15], and testing is not conducted routinely for pathogens in clinical practice. Thus, national guidelines take a one-size-fits-most approach to management by recommending that immediate antibiotics be considered for nearly all children in the United States and that amoxicillin should be used as a first-line agent in most cases regardless of the associated organisms[16]. This results in significant antibiotic overuse because most AOM episodes resolve without treatment with an antibiotic[17]. One mechanism to reduce unnecessary antibiotic use for AOM is to recommend watchful waiting as first-line treatment for most children aged 2 years and older and select children younger than 2 years of age. Unfortunately, antibiotic prescribing remains high even in countries[18,19] that recommend watchful waiting for initial management; the use of watchful waiting has not increased in the United States over a decade after release of the 2013 American Academy of Pediatrics guidelines[20]. For other infections, such as pharyngitis, the use of rapid diagnostic testing (RDT) has substantially reduced unnecessary antibiotic use while assuring that the children likely to benefit from an antibiotic receive treatment with one [21]. We previously reported that the use of a RDT for AOM could potentially reduce unnecessary antibiotic use, including broad-spectrum use, by over 50% annually[22].
Tympanocentesis is not performed routinely in clinical practice, however, there is a strong correlation between the common bacterial otopathogens detected in the middle ear and nasopharynx (NP) during AOM. While the positive predictive value (PPV) of NP testing is variable, the negative predictive value (NPV) is high; based on the current prevalence of otopathogens, negative NP testing can effectively exclude the presence of bacterial otopathogens in the middle ear with over 92% accuracy[4,7]. Thus, NP testing could be an effective tool to reduce unnecessary antibiotic use and individualize care. Though culture is considered the gold standard for pathogen detection, the turnaround time for results and limited sensitivity reduce its utility in clinical practice. In contrast, polymerase chain reaction (PCR) may offer more timely results with increased sensitivity. We therefore aimed to determine the sensitivity, specificity, PPV and NPV for NP qualitative PCR for the most common bacterial otopathogens compared to NP culture among children 6-35 months of age with AOM.
2. Methods:
2.1. Population:
Data were obtained as part of the larger NOTEARS AOM study[13], which took place at Denver Health and Hospital Authority (DHHA) in Denver, CO from 2019 to 2022 and enrolled children ages 6-35 months with uncomplicated AOM in primary care, urgent care, and emergency department settings. Children were included if they had an in-person visit, were diagnosed with AOM with a qualifying International Classification of Diseases version 10 code, had symptom onset within 10 days prior to enrollment, and were prescribed amoxicillin by their provider. Children were excluded if they had tympanic membrane rupture at diagnosis, a competing bacterial diagnosis (e.g., pneumonia), received more than 2 doses of systemic antibiotics in the prior 30 days, an underlying medical condition including immunocompromise, concurrent steroid use, underlying structural ear abnormality (e.g. cleft palate, Down syndrome, sensorineural hearing loss), or current tympanostomy tubes.
2.2. Sample collection:
Patients had an NP flocked swab in liquid Amies media (ESwab®, Copan Diagnostics) collected at enrollment by trained medical providers. Swabs were inserted through one nostril at a depth equivalent to the distance between the tip of the child’s nose and the tragus of the ear or the maximum depth as indicated on the swab and rotated for 10 seconds before removal. Children were enrolled whenever trained research staff were available (day or night, including weekends). Samples were transported to the clinical microbiology laboratory within one hour of collection and refrigerated for up to four hours prior to processing. Samples were vortexed for 20 seconds immediately prior to aliquoting and plating.
2.3. PCR Testing:
A 400 μL aliquot of specimen was frozen at −70°C for PCR testing. Specimens were batch-shipped to Quidel Laboratories (San Diego, CA) for PCR testing. Quidel staff were blinded to culture results and participant information. Nucleic acids were extracted using the NucliSENS® easyMAG® system (Quidel, San Diego, CA) per manufacturer’s instructions. Multiplex RT-PCR for S. pneumoniae, H. influenzae, and M. catarrhalis was completed on 5 μL of extracted nucleic acids using AnDiaTec® assay kits (Quidel, San Diego, CA). Nucleic acid amplification and detection were completed on the Applied Biosystems® (ABI) 7500 Fast Dx Real-Time PCR Instrument (ThermoFisher Scientific, Waltham, MA). Manufacturer recommendations for qualitative determination of ‘positive’ or ‘negative’ values were utilized using pre-defined cutoffs specified by the manufacturer[23,24]. The diagnostic sensitivity and specificity as reported by the manufacturer are as follows: S. pneumoniae (sensitivity 99.2%, specificity 98.4%); H. influenzae (sensitivity 100%, specificity 98.6%); and M. catarrhalis (sensitivity 100%, specificity 100%).
2.4. Culture Testing:
Specimens were cultured in the DHHA Clinical Microbiology Laboratory using standard laboratory techniques by inoculating 10 μL of specimen on blood agar, chocolate agar, and MacConkey agar, then streaking for isolation. Media were incubated at 35-37°C in 5-10% carbon dioxide and examined at 24, 48, and 72 hours for growth. Results were recorded semiquantitatively (i.e., rare, few, moderate, or many). Pathogens were identified using standard microbiologic methods, including biochemical and agglutination tests, supplemented, when necessary, by use of Vitek cards (Biomerieux, Durham, NC) and Microscan panels (Beckman Coulter, Brea, California).
2.5. Analysis:
The primary analysis included calculation of sensitivity, specificity, PPV, and NPV and associated 95% confidence intervals (CI) for S. pneumoniae, H. influenzae, and M. catarrhalis using culture as the reference method. Only children that had not received any doses of antibiotic at the time of specimen collection were included in the primary analysis. Secondary analyses included: 1) the number and percentage of children that tested positive for each microorganism based on the number of doses of antibiotics taken prior to specimen collection and 2) the proportion of children positive by PCR that were also positive by culture based on the number of doses of antibiotics taken prior to specimen collection. We did not evaluate the sensitivity and specificity for less common bacterial otopathogens (e.g., Streptococcus pyogenes) because we were unlikely to have a sufficient sample size for the analyses given their low frequency among young children with AOM[1,5]. All analyses were performed using SAS Enterprise Guide 7.1 (SAS Corporation, Cary, NC).
The study was approved by the Colorado Multiple Institute Review Board and informed consent was obtained from the parents of all participants.
3. Results:
In total, 215 children were enrolled. Twelve were withdrawn from the study, however, the parent of one child who was withdrawn allowed us to use the child’s laboratory data, leaving 204 children for the analysis. Of the 203 with children with culture results available included, at the time of enrollment, 148 (72.9%) had not taken any doses of antibiotics prior to NP specimen collection at enrollment, 19 (9.3%), 31 (15.3%), and 5 (2.5%) had taken 1, 2, and an unknown number of doses respectively. Table 1 shows the demographic and clinical features of children included in the primary analysis. Of children who had not taken any antibiotics before enrollment, 123 (83.1%) had one or more bacterial otopathogens detected by NP culture and 131 (88.5%) had one or more detected by NP PCR. M. catarrhalis was the most frequently isolated organism by culture (78, 52.7%) followed by S. pneumoniae (51, 34.5%), and H. influenzae (30, 20.3%). A minority of children (25, 16.9%) had no bacterial otopathogens isolated and 55 (37.2%) had multiple bacterial otopathogens isolated.
Table 1:
Demographic and clinical features of children included in the primary analysis.
| Characteristic | N (%) N=148 |
|---|---|
| Age (n (%), months) | 18.1 months |
| 6-11 | 47 (31.8) |
| 12-23 | 62 (41.9) |
| 24-35 | 39 (26.3) |
| Sex (n (%)) | |
| Female | 86 (58.1) |
| Male | 62 (41.9) |
| Race (n (%)) | |
| White | 83 (56.1) |
| Unknown or Not Reported | 49 (33.1) |
| African American/Black | 11 (7.4) |
| Other or Multiracial | 5 (3.4) |
| Ethnicity (n (%)) | |
| Hispanic/Latinx | 113 (76.3) |
| Not Hispanic/Latinx | 33 (22.3) |
| Unknown or Not Reported | 2 (1.4) |
| Language Preference1 (n (%)) | |
| English | 112 (75.7) |
| Spanish | 36 (24.3) |
| Attends school or daycare (n (%)) | 39 (26.4) |
| Diagnostic Setting (n (%)) | |
| Emergency Department or Urgent Care | 106 (71.6) |
| Pediatric primary care | 18 (12.2) |
| Family medicine primary care | 24 (16.2) |
| Time period1 (n (%)) | |
| Pre COVID-19 | 81 (54.7) |
| Peri COVID-19 | 67 (45.3) |
Pre COVID-19 includes patients enrolled before 03/01/2020 and Peri COVID-19 includes patients enrolled on or after 03/01/2020.
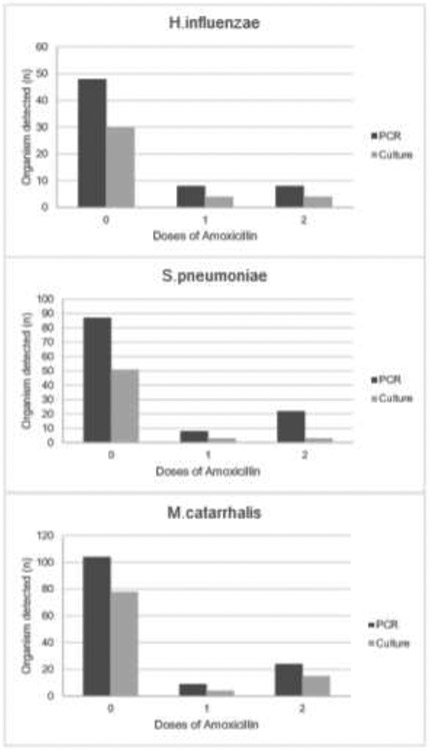
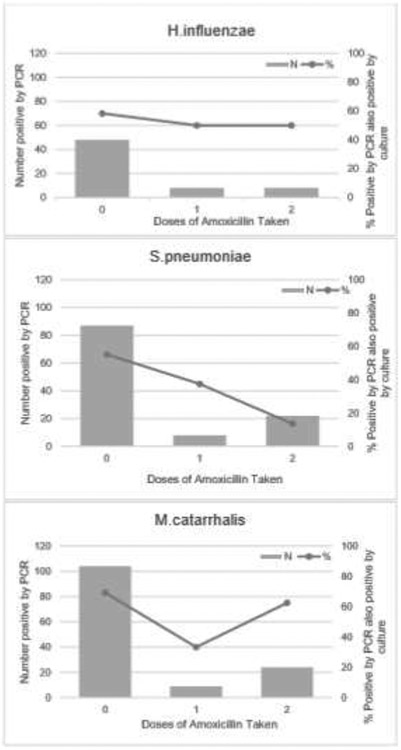
The sensitivity and NPV of NP PCR compared to NP culture was high for all organisms (H. influenzae 93.3%, 98.0%; S. pneumoniae 94.2%, 95.1%; M. catarrhalis 92.3%, 86.4%, respectively). The specificity was highly variable ranging from 54.2% (95% CI: 42.0, 66.1) for S. pneumoniae to 83.1% (95% CI: 74.8, 98.8) for M. catarrhalis with correspondingly low positive predictive values (Table 2). For all organisms NP PCR was substantially more likely to detect pathogens than NP culture (Figure 1). Patients who had taken amoxicillin were substantially less likely to have S. pneumoniae and H. influenzae isolated by culture, whereas isolation of M. catarrhalis was largely unaffected by amoxicillin consumption (Figure 2).
Table 2:
Sensitivity, specificity, negative and positive predictive values of nasopharyngeal PCR compared to nasopharyngeal culture.
| Organism | Na N=148 |
Prevalence % (95%CI) |
TPb (n) |
FP c (n) |
TN d (n) |
FN e (n) |
Sensitivity % (95%CI) |
Specificity % (95%CI) |
Positive Predictive Value % (95%CI) |
Negative Predictive Value % (95%CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| H.influenzae | 30 | 20.3 (14.3, 27.8) | 28 | 20 | 98 | 2 | 93.3 (76.5, 98.8) | 83.1 (74.8, 98.8) | 58.3 (43.3, 72.1) | 98.0 (92.3, 99.7) |
| S.pneumoniae | 51 | 34.5 (27.0, 42.8) | 48 | 39 | 58 | 3 | 94.2 (82.8, 98.5) | 59.8 (49.3, 69.5) | 55.1 (44.2, 65.7) | 95.1 (85.4, 98.7) |
| M.catarrhalis | 78 | 52.7 (44.3, 60.1) | 72 | 32 | 38 | 6 | 92.3 (83.4, 96.8) | 54.2 (42.0, 66.1) | 69.2 (59.3, 77.7) | 86.4 (72.0, 94.3) |
Children may have had more than one organism detected.
True positive
False positive
True negative
False negative
Figure 1:
Number of children for whom organisms were detected by PCR and culture by doses of amoxicillin taken.
Figure 2:
Proportion of children positive by PCR and culture by doses of amoxicillin taken at enrollment.
4. Discussion:
For young children with AOM, NP PCR had high sensitivity and NPV compared to NP culture for common bacterial otopathogens. Here we demonstrate that NP PCR in children with AOM detected more pathogens compared to culture and was able to detect pathogens in children who had taken antibiotics.
NP testing for children with AOM could be a useful tool to reduce unnecessary antibiotic exposure. Optimally, children should be diagnosed with AOM using stringent criteria and initial observation should be used for most children. However, in clinical practice AOM is often incorrectly diagnosed (30-50%)[13,16,25-28], and yet 95% of children are prescribed an immediate antibiotic. Of those, 40% are prescribed a broad-spectrum antibiotic[29,30]. Given the high NPV, a negative NP PCR during AOM indicates that the organism is unlikely to be present. NP PCR could reduce or exclude the need for an antibiotic in children without a bacterial pathogen or who exclusively have a pathogen that is likely to resolve without an antibiotic (e.g., M. catarrhalis). Similarly, it may be a useful tool to prevent unnecessary prescribing of broad-spectrum antibiotics. Genes associated with beta-lactamase production by H. influenzae are well described and could be incorporated into a PCR panel. In cases where an antibiotic might be warranted, a negative NP PCR for M. catarrhalis or beta-lactamase producing H. influenzae indicates that a child may be unlikely to benefit from a broader spectrum antibiotic. This might be particularly helpful for children with chronic or recurrent AOM, although we did not assess performance characteristics in these populations. We recently reported that the routine use of rapid PCR testing for common bacterial otopathogens during AOM episodes, coupled with use of a clinical decision algorithm to guide management (watchful waiting versus immediate antibiotic and antibiotic agent), is likely to reduce unnecessary antibiotic use[21]. We anticipate that results could be available within several hours in most health settings including outpatient clinics given the wide-spread use of rapid PCR for other conditions such as Group A streptococcal pharyngitis and SARS-CoV-2 infection. Thus, like pharyngitis, clinicians could provide recommendations for analgesics and antipyretics and follow-up with families the same or next day with test results. Importantly, the correlation between NP and middle ear fluid bacterial otopathogens during AOM is imperfect[4,7]. There are several reasons for this: discordance including high carriage rates of bacterial otopathogens in the nasopharynx, the presence of biofilms in the nasopharynx, and the timing of sample collection during illness. For example, early in the course of infection organisms may be more likely to be detected in the NP than in the middle ear fluid. Nearly all of these scenarios result in more otopathogens detected in the NP than in middle ear fluid. Fortunately, the absence of a bacterial otopathogen in the nasopharynx indicates it is unlikely to be in the middle ear fluid in most cases. Thus, the risk of a false negative from NP sampling is low.
Conversely, the use of NP PCR would still result in some antibiotic overprescribing because the presence of an otopathogen in the NP does not always indicate that the same organism is also present in the middle ear. For example, some children may carry S. pneumoniae, H. influenzae, or M. catarrhalis in the nasopharynx (even without AOM). Nevertheless, we previously estimated that use of an NP PCR based RDT for AOM in routine clinical practice could substantially reduce overall antibiotic prescribing and broad-spectrum antibiotic prescribing [21]. Thus, testing via NP PCR may be considered a potential tactic to reduce unnecessary antibiotic use for AOM, but should be used in conjunction with other antimicrobial stewardship interventions including those to improve diagnostic accuracy. Additionally, PCR is likely to be more sensitive than culture and could reliably detect organisms when culture results are falsely negative. Patients who had potential otopathogens detected by PCR, but not culture, were classified as false positives. However, it is more likely that PCR was simply more sensitive than culture. This is particularly relevant for S. pneumoniae which is fastidious and may not grow well aerobically. The sensitivity of PCR compared to culture could enhance its use in clinical practice. However, one potential challenge with the use of PCR is the inability to get most antimicrobial susceptibility testing.
There are numerous additional advantages to NP PCR compared to culture or middle ear fluid testing. First, PCR has a relatively fast turn-around and many health systems now have access to PCR on site. Second, PCR based assays coupled with testing for influenza, SARS-CoV-2, and respiratory syncytial virus could provide an efficient and practical panel for testing. Such a panel would minimize NP sampling for children and provide additional comprehensive information to guide treatment and isolation recommendations. Third, the ability of PCR to identify pathogens in children who have taken some doses of an antibiotic could be useful to guide antibiotic choice for children who fail initial antibiotic treatment. Finally, nearly all clinicians and clinical staff can reliably collect an NP specimen, though few clinicians are trained or competent in tympanocentesis[31]. It remains to be seen whether mid-turbinate or anterior nasal swabs could also be used for detection of otopathogens, which could further simplify sample collection given that they are widely used due to their introduction during the COVID-19 pandemic. A prior study indicated moderate to good correlation between mid-turbinate and nasopharyngeal results for these organisms[31].
This evaluation has several strengths. We were able to evaluate the test performance among a prospective cohort of children with uncomplicated AOM diagnosed in a typical practice environment. Inclusion of children who had taken varying doses of amoxicillin at the time of specimen collection allowed us to compare the detection of pathogens by culture and PCR by antibiotic exposure. This is potentially important clinically for children who are failing antibiotic therapy and methodologically for collection of microbiologic data in pragmatic clinical trials that might enroll children after they have started an antibiotic.
The study also has limitations. First, although other authors have previously described an association between NP and middle ear fluid otopathogens, we did not test for otopathogens in middle ear fluid to confirm this association in our population [4,7,32]. Second, the reliability of all tests is subject to the technique used for sample collection. All specimens were collected by practicing clinicians who were trained in NP collection; however, we did not directly observe or validate sample collection techniques. Third, patients were recruited across only one health system in Colorado and only younger children with uncomplicated infections were included. Thus, results may not be generalizable to other geographic areas with different pathogen prevalence or among different patient populations. Fourth, we did not assess the potential clinical impact of use of the RDT in this study. We previously estimated that use of an RDT would reduce adverse drug events and disutility among children[22]. However, a future study is needed to assess clinical outcomes directly. Finally, the sample size was small for some organisms, which may have resulted in broader precision estimates than if more children had been included.
In conclusion, NP PCR has relatively high negative predictive value compared to culture for excluding common bacterial otopathogens in young children with uncomplicated AOM and warrants further exploration as a diagnostic tool to individualize care and reduce unnecessary antibiotic use.
Funding/Support (if any):
Funding was provided by The Gerber Foundation (Award Number: N/A). HF received salary support from the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number K23HD099925. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Role of Funder/Sponsor (if any):
The funder had no role in the design or interpretation of the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: HF serves as a senior scientific advisor for QuidelOrtho and holds a patent for diagnosing and treating otitis media #63/335,801. QuidoOrtho had no role in the design or interpretation of the study and Quidel staff were blinded to culture results. SD serves as a consultant and receives grant support from Biofire, serves as a consultant for Karius, and receives grant support from Pfizer for projects not pertaining to this project.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Prior Presentation of Work: Interim results from this study were presented at the IDWeek (Infectious Disease Society of America) conference in 2021 and the study was awarded the Top Abstract in Diagnostics.
References
- 1.Kaur R, Morris M, Pichichero ME. Epidemiology of Acute Otitis Media in the Postpneumococcal Conjugate Vaccine Era. Pediatrics 2018;140(3):e20170101. doi: 10.1542/peds.2017-4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poole NM, Shapiro DJ, Fleming-Dutra KE, Hicks LA, Hersh AL, Kronman MP. Antibiotic Prescribing for Children in United States Emergency Departments: 2009-2014. Pediatrics 2019;143(2):e20181056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler AM,et al. Association of Inappropriate Outpatient Pediatric Antibiotic Prescriptions With Adverse Drug Events and Health Care Expenditures. JAMA Netw Open 2022;5(5):e2214153. doi: 10.1001/jamanetworkopen.2022.14153. Erratum in: JAMA Netw Open. 2022 Jun 1;5(6):e2221479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaur R, Czup K, Casey JR, Pichichero ME. Correlation of nasopharyngeal cultures prior to and at onset of acute otitis media with middle ear fluid cultures. BMC Infect Dis 14 2014;640. doi: 10.1186/s12879-014-0640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaur R, Fuji N, Pichichero ME. Dynamic changes in otopathogens colonizing the nasopharynx and causing acute otitis media in children after 13-valent (PCV13) pneumococcal conjugate vaccination during 2015-2019. Eur J Clin Microbiol Infect Dis 2021;41:37–44.doi: 10.1007/s10096-021-04324-0. [DOI] [PubMed] [Google Scholar]
- 6.Casey JR, Adlowitz DG, Pichichero ME. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. Pediatr Infect Dis J 2010;29:304–9. doi: 10.1097/INF.0b013e3181c1bc48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsyshina S, et al. Detection of respiratory pathogens in pediatric acute otitis media by PCR and comparison of findings in the middle ear and nasopharynx. Diagn Microbiol Infect Dis 2016;85:125–30. doi: 10.1016/j.diagmicrobio.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frost HM, Hersh AL. Rethinking Our Approach to Management of Acute Otitis Media. JAMA Pediatr 2022;176:439–440. doi: 10.1001/jamapediatrics.2021.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broides A, Dagan R, Greenberg D, Givon-Lavi N, Leibovitz E. Acute otitis media caused by Moraxella catarrhalis: epidemiologic and clinical characteristics. Clin Infect Dis 2009;49:1641–7. doi: 10.1086/647933. [DOI] [PubMed] [Google Scholar]
- 10.Liu K, Kaur R, Almudevar A, Pichichero ME. Higher serum levels of interleukin 10 occur at onset of acute otitis media caused by Streptococcus pneumoniae compared to Haemophilus influenzae and Moraxella catarrhalis. Laryngoscope 2013;123:1500–5. doi: 10.1002/lary.23973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein JO. Otitis media. Clin Infect Dis 1994;19:823–33. doi: 10.1093/clinids/19.5.823. [DOI] [PubMed] [Google Scholar]
- 12.Howie VM, Ploussard JH. Efficacy of fixed combination antibiotics versus separate components in otitis media. Effectiveness of erythromycin estrolate, triple sulfonamide, ampicillin, erythromycin estolate- triple sulfonamide, and placebo in 280 patients with acute otitis media under two and one-half years of age. Clin Pediatr (Phila) 1972;11:205–14. doi: 10.1177/000992287201100407. [DOI] [PubMed] [Google Scholar]
- 13.Frost HM, Dominguez S, Parker S, Byars A, Michelson S, Keith A, Jenkins TC. Clinical failure rates of amoxicillin for the treatment of acute otitis media in young children. Open Forum Infect Dis 2020;7(Suppl 1):S682–3. doi: 10.1093/ofid/ofaa439.1524. [DOI] [Google Scholar]
- 14.Yamada S et al. β-Lactamase-non-producing ampicillin-resistant Haemophilus influenzae is acquiring multidrug resistance. J Infect Public Health 2020;13:497–501. doi: 10.1016/j.jiph.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Leibovitz E,et al. Can acute otitis media caused by Haemophilus influenzae be distinguished from that caused by Streptococcus pneumoniae? Pediatr Infect Dis J 2003;22:509–15. doi: 10.1097/01.inf.0000069759.79176.e1. [DOI] [PubMed] [Google Scholar]
- 16.Lieberthal AS,et al. The diagnosis and management of acute otitis media. Pediatrics 2013;131:e964–99. 10.1542/peds.2012-3488 [DOI] [PubMed] [Google Scholar]
- 17.Venekamp RP, Sanders SL, Glasziou PP, Del Mar CB, Rovers MM. Antibiotics for acute otitis media in children. Cochrane Database Syst Rev 2013; (1):CD000219. doi: 10.1002/14651858.CD000219.pub3 [DOI] [PubMed] [Google Scholar]
- 18.Hansen MP, et al. Treatment of acute otitis media in general practice: quality variations across countries. Fam Pract 2012;29(1):63–8. 10.1136/bmjopen-2019-035343. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki HG, Dewez JE, Nijman RG, Yeung S. Clinical practice guidelines for acute otitis media in children: a systematic review and appraisal of European national guidelines. BMJ Open 2020;10(5):e035343. doi: 10.1136/bmjopen-2019-035343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smolinski NE, Antonelli PJ, Winterstein AG. Watchful Waiting for Acute Otitis Media. Pediatrics 2022;150(1): e2021055613. 10.1542/peds.2021-055613 [DOI] [PubMed] [Google Scholar]
- 21.Maltezou HC, et al. Evaluation of a rapid antigen detection test in the diagnosis of streptococcal pharyngitis in children and its impact on antibiotic prescription. J Antimicrob Chemother 2008;62:1407–12. doi: 10.1093/jac/dkn376. [DOI] [PubMed] [Google Scholar]
- 22.Sebastian T, Toseef M, Kurtz M, & Frost H Nasopharyngeal rapid diagnostic testing to reduce unnecessary antibiotic use and individualize management of acute otitis media. Antimicrobial Stewardship & Healthcare Epidemiology 2023;3(1), E49. doi: 10.1017/ash.2023.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andiatec. Streptococcus pneumoniae real time PCR Kit with Extraction Control. Ref 940200 2016;1–3. [Google Scholar]
- 24.Andiatec. Haemophilus influenzae real time PCR Kit with Extraction Control. Ref 950200 2016;1–3. [Google Scholar]
- 25.Shaikh N, Stone MK, Kurs-Lasky M, Hoberman A. Interpretation of tympanic membrane findings varies according to level of experience. Paediatr Child Health 2016; 21:196–8. doi: 10.1093/pch/21.4.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf RM, Langford KT, Patterson BL. Improving Adherence to AAP Acute Otitis Media Guidelines in an Academic Pediatrics Practice through a Quality Improvement Project. Pediatr Qual Saf 2022;7:e553. doi: 10.1097/pq9.0000000000000553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pichichero ME, Poole MD. Assessing diagnostic accuracy and tympanocentesis skills in the management of otitis media. Arch Pediatr Adolesc Med 2001;155:1137–42. doi: 10.1001/archpedi.155.10.1137. [DOI] [PubMed] [Google Scholar]
- 28.Tähtinen PA, Laine MK, Huovinen P, Jalava J, Ruuskanen O, Ruohola A. A placebo-controlled trial of antimicrobial treatment for acute otitis media. N Engl J Med 2011;364:116–26. doi: 10.1056/NEJMoa1007174. [DOI] [PubMed] [Google Scholar]
- 29.Frost HM, Bizune D, Gerber JS, Hersh AL, Hicks LA, Tsay SV. Amoxicillin Versus Other Antibiotic Agents for the Treatment of Acute Otitis Media in Children. J Pediatr 2022; 251:98–104.e5. doi: 10.1016/j.jpeds.2022.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGrath LJ, et al. Utilization of nonguideline concordant antibiotic treatment following acute otitis media in children in the United States. Pharmacoepidemiol Drug Saf 2023;32:256–265. doi: 10.1002/pds.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin JM, Simonian A, Muniz G, Green M, Shaikh N. Comparison of Bacterial Culture Results Obtained from Nasopharyngeal and Midturbinate Swabs in Children With Upper Respiratory Tract Infection Symptoms. Pediatr Infect Dis J 2018;37(11):e275–e277. doi: 10.1097/INF.0000000000002027. [DOI] [PubMed] [Google Scholar]
- 32.van Dongen TM, van der Heijden GJ, van Zon A, Bogaert D, Sanders EA, Schilder AG. Evaluation of concordance between the microorganisms detected in the nasopharynx and middle ear of children with otitis media. Pediatr Infect Dis J 2013;32(5):549–52. doi: 10.1097/INF.0b013e318280ab45. [DOI] [PubMed] [Google Scholar]