Summary
Axonal transport is key to neuronal function. Efficient transport requires specific motor-cargo association in the soma, yet the mechanisms regulating this early step remain poorly understood. We found that EBP-1, the C. elegans ortholog of the canonical microtubule end binding protein EB1, promotes the specific association between kinesin-3/KIF1A/UNC-104 and Dense Core Vesicles (DCVs) prior to their axonal delivery. Using single-neuron, in vivo labelling of endogenous cargo and EBs, we observed reduced axonal abundance and reduced secretion of DCV cargo, but not other KIF1A/UNC-104 cargo, in ebp-1 mutants. This reduction could be traced back to fewer exit events from the cell body, where EBP-1 colocalized with the DCV sorting machinery at the trans Golgi, suggesting that this is the site of EBP-1 function. EBP-1 CH domain was required for directing microtubule growth on the Golgi, and mammalian EB1 interacted with KIF1A in an EBH domain dependent manner. Loss and gain of function experiments suggest a model in which both kinesin-3 binding and guidance of microtubule growth at the trans Golgi by EBP-1 promote motor-cargo association at sites of DCV biogenesis. In support of this model, tethering either EBP-1 or a kinesin-3/KIF1A/UNC-104 interacting domain from an unrelated protein to the Golgi restored the axonal abundance of DCV proteins in ebp-1 mutants. These results uncover an unexpected role for a microtubule associated protein and provide insight into how specific kinesin-3 cargo is delivered to the axon.
In brief
Kinesins must load cargo in the neuronal cell body prior to transporting it to synapses. Park et al. find that Golgi-enriched microtubule End Binding protein 1 interacts with kinesin-3 and orients microtubule growth on the Golgi to promote the association of the motor with nascent Dense Core Vesicles, thereby ensuring their axonal delivery.
Introduction
Axonal transport necessitates motor-cargo association and efficient cell body exit prior to long-range runs, but these early steps of transport remain poorly understood. The major axonal motor kinesin-3/KIF1A/UNC-104 is thought to be activated in the cell body following relief of autoinhibition and interaction of its PH domain with phospholipids on cargo vesicles.1–5 However, UNC-104/KIF1A can transport different cargo types, raising the question of whether additional mechanisms exist that could promote the transport of specific cargo.6–11
A case in point is synaptic vesicle precursors (SVPs) and dense core vesicles (DCVs), both UNC-104/KIF1A cargo, which differ from each other in membrane composition and biogenesis mechanisms in the cell body.12–19 Interestingly, recent work in C. elegans showed that the cargo adaptor complexes which sort DCVs and SVPs for axonal delivery (UNC-101/AP-1μ and APM-3/AP-3μ, respectively) are found on separate sub-domains of the trans Golgi.14, 20 This finding raises the possibility that the association of UNC-104/KIF1A with specific cargo could be regulated by proteins that reside at these spatially segregated biogenesis sites.
End Binding (EB) proteins are a conserved family of microtubule (MT) associated proteins (MAPs) that bind growing MT ends.21 EBs are characterized by an N-terminal calponin homology (CH) domain, which mediates MT end tracking, and a C terminal End-binding Homology (EBH) domain, which mediates interactions with EB binding proteins.22–25 EBs can recruit MTs to membranous organelles and in neurons they are thought to play important roles in controlling MT organization, either by regulating microtubule dynamics, mediating binding to structures such as the axon initial segment (AIS) or guiding MT growth along other MTs or the cortex.26–29 EBs also promote the initiation of retrograde transport from the distal axon by facilitating dynein recruitment and may compete with kinesin-3 for the MT GTP cap at presynapses.30–32 Unexpectedly, recent work revealed that cultured mammalian cells in which all three EBs were disrupted show relatively mild mitotic defects.29, 33 Similarly, worm embryos lacking all EBs are viable and develop normally.34 Hence, despite the extensive characterization of EBs at the structural, biochemical and cell biological levels, the full range of their functions in vivo remains to be elucidated.
Here, using single-neuron, endogenous labelling of cargo and EBs in vivo, we found a role for the C. elegans EBP-1 in promoting UNC-104/KIF1A dependent axonal delivery of DCVs but not SVPs or ATG-9 vesicles. EBP-1 is enriched near the trans Golgi with the DCV sorting machinery and ebp-1 mutants showed reduced numbers of DCVs exiting the cell body, suggesting that this is its relevant site of action. Consistently, DCV abundance in the axons of ebp-1 mutants could be restored by expression of a Golgi tethered fragment of EBP-1. We find that mammalian EB1 could rescue ebp-1 mutants – suggesting conservation of its function – and its EBH domain co-immunoprecipitated with kinesin-3/KIF1A. Loss and gain of function experiments indicate that EBP-1 promotes DCV transport through the microtubule binding CH domain, which is required for orienting microtubule growth on the Golgi, and the UNC-104/KIF1A binding EBH domain, which plays a more prominent role. We propose that through binding UNC-104, and through guiding MT ends on the Golgi, EBP-1 transiently enriches the motor near DCV sorting sites, thus promoting specific motor-cargo association and efficient transport. In support of this working model, EBP-1 function could be replaced by a Golgi-tethered UNC-104/KIF1A interacting domain from an unrelated protein, SYD-2.
Results
Loss of EBP-1 results in a specific loss of Dense Core Vesicle cargo in the axon
Recent in vitro studies highlighted the role of MAPs, including EBs, as regulators of molecular motors.30 To investigate the role of End Binding proteins in neuronal transport in vivo, we used the C. elegans cholinergic motor neuron DA9. The DA9 soma resides in the preanal ganglion, from where the axon grows posteriorly, traverses to the dorsal cord and grows anteriorly. Following an asynaptic region of ~45 μm in the dorsal cord, en passant presynaptic boutons form onto dorsal muscle at a stereotypic stretch of 70~90 μm (Figure 1A).35, 36 A DCV luminal cargo, INS-22::Emerald, expressed under control of a DA9 promoter, shows enrichment at the synaptic region and is also detected in asynaptic regions, consistent with previous reports (Figure 1C).37, 38
Figure 1: Loss of ebp-1 results in a specific loss of dense core vesicles from the axon.
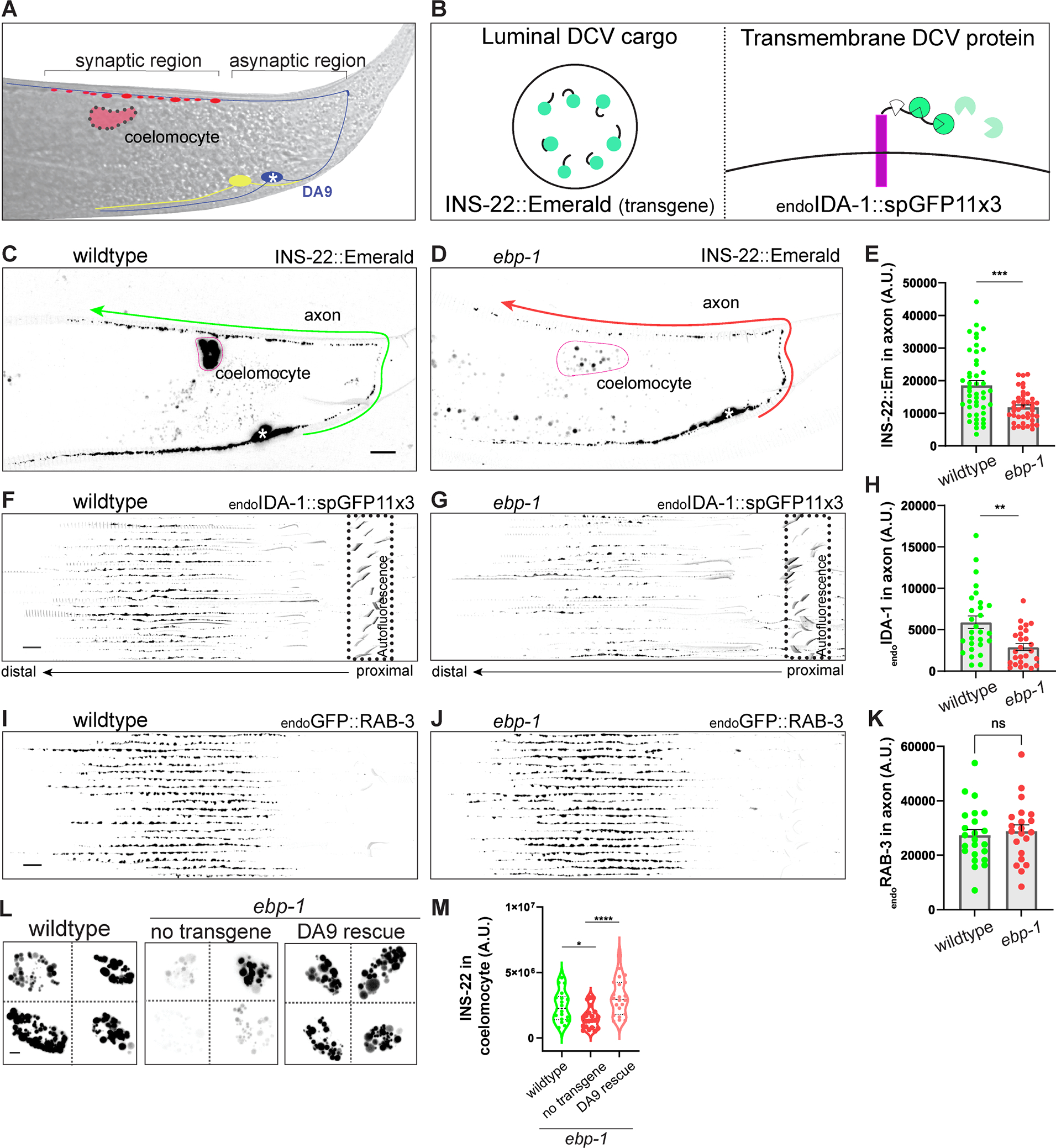
(A) Schematic of the DA9 motor neuron. DA9 axon traverses dorsally and forms en passant presynapses (red boutons). White asterisk indicates the soma in all figures. The ccDL coelomocyte, adjacent to DA9 synapses is in pink. VA12 neuron (yellow) synapses on the ventral side and is also labeled by Pmig-13 promoter. (B) Diagram of INS-22, luminal DCV cargo, transgene (left box) and splitGFP tagging strategy for labeling endogenous transmembrane DCV cargo, IDA-1 (endoIDA-1, right box). (C,D) Confocal images of wildtype (C) and ebp-1 mutant (D) expressing INS-22::Emerald. Green and red arrows points toward the more distal axon of DA9 and the straightened axon that spans the arrow line was quantified. Pink silhouette outlines the coelomocyte. Scale bar 10 μm. (E) Quantification of (C) and (D). (F-H) Twenty axons per genotype were straightened and aligned from cell body (proximal) to past the synaptic region (distal). Signal is from splitGFP tagged to endogenous IDA-1. Non-specific background fluorescence from the rectum (diagonal lines) is indicated by dotted box (F,G). Quantification of fluorescence in (H). Scale bar 10 μm. (I-K) Fluorescence of endogenously labeled RAB-3 vesicles in wildtype and ebp-1 mutant (I, J), quantified in (K). Scale bar 10 μm. See also Figure S1. (L-M) Representative images of INS-22::Emerald in coelomocyte in wildtype, ebp-1 mutant, and DA9 specific expression of EBP-1 in ebp-1 mutant. Scale bar 3 μm. Quantification of fluorescence in (M) n=21–46. See also Figure S3D for INS-22::Emerald in coelomocyte in unc-104/kinesin-3 mutant. ****p<0.0001; ***p<0.001; **p<0.01, *p<0.05 (t-test for (E), (H), (K) and one-way ANOVA for (M)). Each data point in the graphs represents a fluorescence measurement from a single animal. n number is counting each individual animal imaged. See also Table S1.
Examination of a previously described deletion allele of ebp-1 (he279) or a deletion allele generated for this study (wy1156, see methods), revealed a ~40% reduction of INS-22::Emerald in the axon of DA9 (Figure 1D and 1E), suggesting that EBP-1 is required for the normal abundance of DCVs in the axon (note that the deletions in the wy1156 and he279 alleles encompass both ebp-1 and the neighboring gene, ebp-3, which is likely a pseudogene.34 We therefore refer to ebp-1, ebp-3 mutants as ebp-1. All alleles used are null unless indicated otherwise; deletions of specific domains are noted by Δ).
To determine whether other DCV cargo requires EBP-1, we used CRISPR to tag the C terminus of IDA-1/phogrin, a transmembrane DCV protein, at its endogenous locus with spGFP11 (referred to as endoIDA-1) and visualized it in DA9 at native levels by expressing spGFP1–10 under Pmig-13 promoter (Figure 1B). endoIDA-1 was enriched at synapses compared to INS-22 in wildtype animals and showed a similar ~50% reduction in ebp-1 mutants (Figure 1F, 1G, 1H, S1I, and S1J). These results indicate that both luminal and transmembrane DCV cargo require EBP-1 for their axonal localization.
Given that DCVs are prominently lost in ebp-1 mutants from the dorsal axon where presynaptic sites lie, we tested whether their loss may reflect excessive secretion. We generated double mutants between ebp-1 and unc-31/CAPS, which is required for DCV secretion.37, 39, 40 If ebp-1 mutants lead to excessive secretion, we would expect unc-31 mutants to restore DCVs to the axon. However, ebp-1; unc-31 double mutants showed ~30% loss of the INS-22 signal compared to unc-31 mutants (Figure S1C and S1D), suggesting that loss of DCV cargo from the DA9 axon of ebp-1 mutants is caused by reduced delivery of DCVs, not over-secretion of their contents.
To examine how reduced axonal DCV abundance in ebp-1 mutants affects DCV cargo secretion, we measured INS-22 levels in coelomocytes, which are scavenger cells that endocytose secreted INS-22 (Pink outline; Figure 1A and 1C). Measuring the fluorescence of INS-22 endocytosed by coelomocytes serves as a proxy of secreted INS-22 over time and has been a replicable metric for DCV secretion in C. elegans.41 Quantification of INS-22::Emerald fluorescence in the closest coelomocyte to DA9 synapses revealed a 50% reduction in ebp-1 mutants compared to wildtype (Figure 1L). This effect could be rescued by cell specific expression of EBP-1 in DA9 (Figure 1L and 1M), indicating that the reduced INS-22 signal in coelomocytes is due to a reduction in INS-22 secreted from DA9 and not to a defect in coelomocyte function.
DCV cargos play important roles in the animal physiology. In C. elegans, neuropeptides both promote and inhibit egg-laying; specifically, ectopic expression of the neuropeptide NLP-3 was shown to induce premature egg-laying, leading to the retention of very few eggs in the animal’s uterus.42, 43 Consistent with a role for EBP-1 in promoting DCV delivery for secretion, we found that ebp-1 mutants suppressed the excessive egg-laying behavior of animals overexpressing NLP-3 (Figure S1A and S1B). Reintroducing ebp-1 cDNA under Ptph-1 promoter driving expression in egg-laying neural circuit was able to restore the excessive egg laying behavior, suggesting that the role of ebp-1 is important in the egg-laying neurons (Figure S1B). Collectively, these results indicate that loss of ebp-1 leads to reduced axonal abundance of DCVs and reduced secretion of DCV cargo.
DCVs are transported to the axon by kinesin-3/KIF1A/UNC-104, which also transports synaptic vesicle precursors (SVPs) and other synaptic cargo to the presynapses.6–8, 10, 11 To determine if the DCV phenotype of ebp-1 mutants reflects a general reduction in UNC-104 activity, we measured the axonal signal of RAB-3, a SVP marker, labelled at its endogenous locus with a FLP-ON GFP cassette and visualized with a DA9 FLP. Surprisingly, the endoRAB-3 signal was not decreased in ebp-1 mutants (Figure 1I, 1J, and 1K). The levels of other UNC-104 cargo, the transmembrane SVP protein SNG-1/synaptogyrin and the autophagic regulator ATG-9 were also unaffected in ebp-1 mutants (Figure S1E, S1F, S1G, and S1H). These results suggest that ebp-1 is specifically required for DCV trafficking.
We note that in ebp-1 mutants, the spacing between adjacent presynaptic boutons was reduced, a phenotype which could be best visualized with the active zone marker CLA-1 (Figure S2H andS2I).44 However, as described below in structure function analysis and rescue experiments, this phenotype stems from a separate function of EBP-1 that is not related to DCV delivery and is not pursued in this study.
The EBH domain is sufficient to restore axonal DCVs in ebp-1 mutants
To gain further insight into how EBP-1 functions in DCV trafficking, we assayed the requirement for different EB proteins and domains in DA9. We first examined the EBP-1 paralog, EBP-2, which is expressed in DA9 at similar levels to EBP-1 (Figure S4F). To assess the DCV loss phenotype, we measured overall fluorescence in the axon as well as the number of IDA-1 puncta in the synaptic region. ebp-2 (he278) deletion mutants showed only a minor reduction in axonal endoIDA-1 compared to wildtype, but that reduction was not on par with the phenotype observed in ebp-1 mutants (Figure S2A, S2B, and S2C). Furthermore, ebp-2 mutants did not enhance the phenotype of ebp-1 mutants (Figure S2B, S2C, S2D, and S2E). These results suggest that EBP-2 plays a relatively minor role in DCV trafficking.
Next, we used CRISPR to delete either the CH domain (ebp-1ΔCH) or the C terminus which contains the EBH domain and acidic tail (ebp-1ΔC; Figure 2A) from the ebp-1 genomic locus. In parallel, we tagged the resulting EBP-1 fragment at their endogenous loci with spGFP11 and confirmed that they were still expressed in DA9 with Pmig-13 driving spGFP1–10 (Figure S2F). Interestingly, both deletion mutants did not show a significant loss of endoIDA-1 from the axon (Figure 2C and 2D). In contrast, ebp-1ΔCH did lead to a reduced distance between synaptic boutons but ebp-1ΔC did not (Figure S2H and S2I), indicating that this phenotype is separate from the loss of axonal DCVs. Many EB binding proteins associate with EBs via a SxIP motif, which binds the EBH domain.23, 29 However, two point-mutations that are predicted to eliminate binding to SxIP motif proteins (Y264A and E272A) did not alter axonal DCV levels (Figure 2F and 2G). 23, 24 In contrast, deleting both the CH domain and the C-terminus phenocopied ebp-1 deletion (Figure 2E). A CH deletion with Y264A and E272A also phenocopied ebp-1, but the resulting protein was expressed at significantly lower levels (Figure 2F, 2G, and S2F), preventing us from drawing definitive conclusions about Y264A and E272A in DCV transport. Together, these results suggest that both MT binding and an association with an EBP-1 binding protein need to be eliminated to mimic ebp-1 deletion mutants.45–47
Figure 2: The EBH domain is sufficient for proper DCV trafficking.
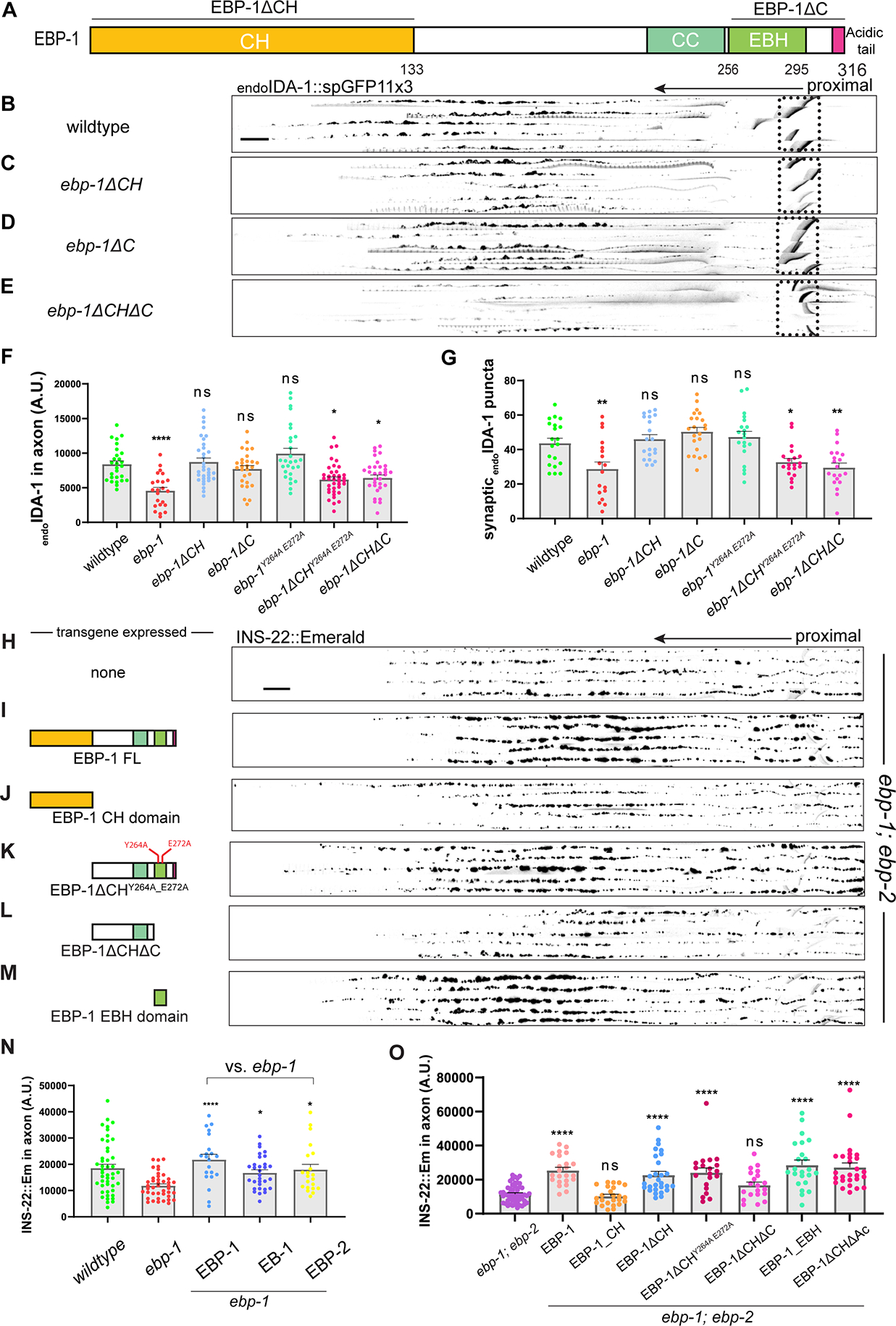
(A) Diagram of EBP-1 and its predicted domains. EBP-1ΔC truncates both EBH domain (lime green) and acidic tail (magenta). (B-E) Alignments of 5 representative axons per genotype showing endoIDA-1::GFP in DA9. Black arrow points distally. Autofluorescence from rectum in dotted box. Scale bar 10 μm. (F) Quantification of fluorescence of endoIDA-1. (G) Count of IDA-1 puncta in the synaptic region defined as 50–70 μm region that follows 45 μm asynaptic region from the turn of the commissure. See also Figure S2. (H-M) Alignments of 5 axons expressing INS-22::Emerald in ebp-1; ebp-2 double mutants per rescue transgene. (N) Quantification of INS-22::Emerald fluorescence in ebp-1 mutant with or without respective rescue transgene. (O) Quantification of INS-2::Emerald fluorescence in ebp-1; ebp-2 mutant with or without respective rescue transgene. n=18–38 ****p<0.0001; *p<0.05 (one-way ANOVA). CH, Calponin homology domain. CC, coiled-coil domain. EBH, End-binding Homology domain. FL, full length. Each data point in the graphs represents a fluorescence measurement or counted puncta from a single animal. n number is counting each individual animal imaged. See also Table S1.
To complement these loss of function experiments, we also tested the sufficiency of EBP-1 domains and EBP-1 homologs for rescuing ebp-1 mutants. DA9 specific expression of EBP-1 or its mammalian homolog EB1 fully rescued the loss of INS-22 labelled DCVs in ebp-1 mutants, indicating that EBP-1 functions cell autonomously and that EB1 function is conserved from invertebrates to vertebrates (Figure 2N). EBP-2 overexpression also rescued ebp-1 mutants (Figure 2N), suggesting that at high expression levels the functional differences between these proteins can be overcome. We next expressed either the Calponin Homology (CH) domain, which mediates MT tip tracking, or the remaining EBP-1 C-terminus comprising the EBH and acidic tail. These experiments were done in ebp-1; ebp-2 double mutants to prevent potential dimerization between EBP-1 fragments and native EBP-2.33 Interestingly, the CH domain could not rescue the double mutant phenotype, whereas the remaining C-terminus was sufficient to restore INS-22 levels to the axon (Figure 2J and 2K). The Y264A and E272A mutations which interfere with EB binding to SxIP motifs did not prevent the C-terminus from rescuing ebp-1; ebp-2 double mutants (Figure 2K). In contrast, a construct lacking both the EBH domain and acidic tail could not rescue the phenotype (Figure 2L). Finally, we found that the acidic tail was not required for rescuing activity and that expressing the EBH domain on its own could restore the INS-22 fluorescent signal to the axon of double mutants (Figure 2M and 2O). Taken together with the deletion of endogenous ebp-1 domains, these results suggest that both the CH and EBH domains contribute to DCV transport, but at high expression levels the EBH domain is sufficient to replace full length EBP-1. We speculate that EBH domain may be sufficient for a rescue at high overexpression since EB’s C terminus forms an a dimer that can bind other microtubule associated proteins and can form a phase to mediate non-stochiometric multiplex interactions in vitro.48–50 However, further mechanistic studies will be required to fully elucidate how highly overexpressed EBH domain can rescue the DCV transport defect in ebp-1 mutants.
EBP-1 localizes near AP-1μ positive Golgi carriers and is required for cell body exit of DCVs
Since loss of DCVs from the axon in ebp-1 mutants was not due to increased secretion, we next tested whether it reflected reduced cargo delivery from the cell body to the axon. For this, we measured how many endoIDA-1/Phogrin positive vesicles exited the cell body after photobleaching the residual signal of endoIDA-1 in the proximal axon (Figure 3A). In ebp-1 mutants, we observed a much higher incidence of zero exit events (no DCV exiting the cell body during an imaging window) over multiple experimental replicates (Figure 3D and 3E), suggesting that EBP-1 promotes cell body exit of DCVs. The velocity of endoIDA-1 puncta exiting the cell body was similar between wildtype and mutants (Figure 3B and 3C), indicating that EBP-1 is not required for UNC-104/KIF1A motility in the proximal axon. The number of cell body exit events for a non-DCV cargo of UNC-104, the SVP protein SNG-1/synaptogyrin, was not affected in ebp-1 mutants (Figure 3E). These results raise the hypothesis that reduced cell-body exit underlies the reduced levels of axonal DCVs in ebp-1 mutants.
Figure 3: ebp-1 is required for efficient cell body exit of DCVs.
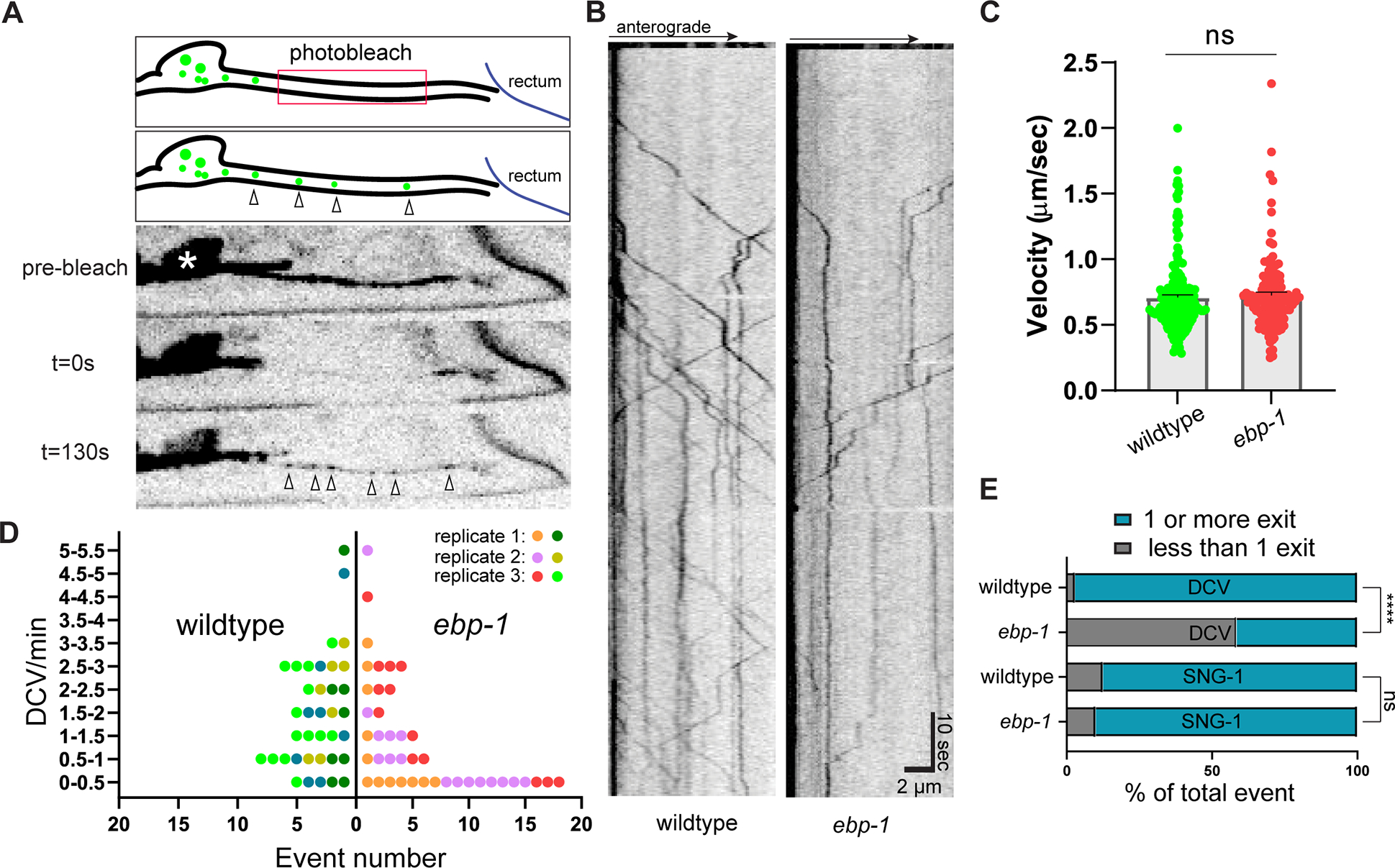
(A) Three frame montage of photobleaching experiment with diagram. Proximal axon outside the soma (white asterisk) is bleached to eliminate background from residual DCVs and allow visualization of DCVs exiting the cell body (indicated with empty arrow heads). Cell body exit events of endoIDA-1 positive DCV are imaged for 120–180 seconds at three frames/second. (B) Kymograph of endogenous DCV exiting cell body in wildtype and ebp-1 mutant. Scale bar is ten seconds vertical and 2 μm horizontal. (C) Velocity of DCV exiting the cell body in wildtype and ebp-1 mutant. n=133–183 events in 27–31 animals. t-test (D) “Superplot” of exit event frequency over multiple replicates.65 Each dot indicates an animal and each color indicates different experimental replicates. Wildtype is shown in green color palette and ebp-1 mutant in red. n=37–41. (E) Proportion of zero event (less than one vesicle exiting during an imaging window) in wildtype and ebp-1 mutant. See also Figure S3 for comparison with DCV and SVP transport defect in unc-104/kinesin-3 mutant. **p<0.01 (n=37–41 for DCV and n=7–10 for SNG-1, Chi-square test). See also Table S1.
The finding that ebp-1 is required for cell body exit of DCVs is suggestive of a transport function. However, we were surprised not to observe an accumulation of DCV cargo in the somatodendritic compartment, as would be expected when axonal delivery is reduced. We therefore examined DCVs in a strong, bona fide transport mutant, unc-104/kinesin-3. unc-104 mutants show a near complete loss of DCVs from the axon (Figure S3A), and we observed reduced INS-22 in coelomocytes in these mutants (Figure S3D and S3E), suggesting that in DA9 neuron failure in DCV transport does not lead to excessive secretion from the cell body or dendrite. Similar to ebp-1 mutants, unc-104 mutants showed only minor accumulation of DCV cargo in the somatodendritic compartment (Figure S3C), an observation that contrasts with the massive accumulation of another UNC-104 cargo, SVPs, in the dendrite of unc-104 mutants (Figure S3B and S3C). These results suggest that failure of DCV cargo leads to cargo degradation rather than misaccumulation. Consistently, western blot analysis of worm lysates showed reduced INS-22 levels in both ebp-1 and unc-104 mutants (Figure S3F and S3G). We conclude that the phenotypes of ebp-1 mutants are consistent with transport defects.
To examine the potential function of EBP-1 in the cell body, we examined the subcellular localization of endogenously tagged EBP-1 and EBP-2. We tagged both proteins at the C terminus and confirmed that they were functional by MT tip tracking (Figure S4G), consistent with previous studies using tagged EBs.51 Both EBP-1 and EBP-2 were uniformly distributed throughout the neurons as expected for a cytosolic protein (Figure S4A and S4B). Interestingly, whereas EBP-2 tracked MT tips more efficiently (Figure S4G, S4H, and S4I), in the cell body, a local accumulation of EBP-1 was observed in two or three punctate structures (Figure 4B). This localization was not unique to DA9 but could be observed across many neuronal cell bodies in the ventral cord (Figure 4A). The EBP-1 punctate signal recovered quickly but incompletely following photobleaching, suggesting that at least some of the EBP-1 molecules in the cell body puncta can exchange freely with the cytosol (Figure S4F). EBP-2 did not show such clear localization to cell body puncta, either in a wildtype background or in ebp-1 mutants (Figure 4B and S4C).
Figure 4: Endogenous EBP-1 is enriched in the vicinity of late Golgi carriers.
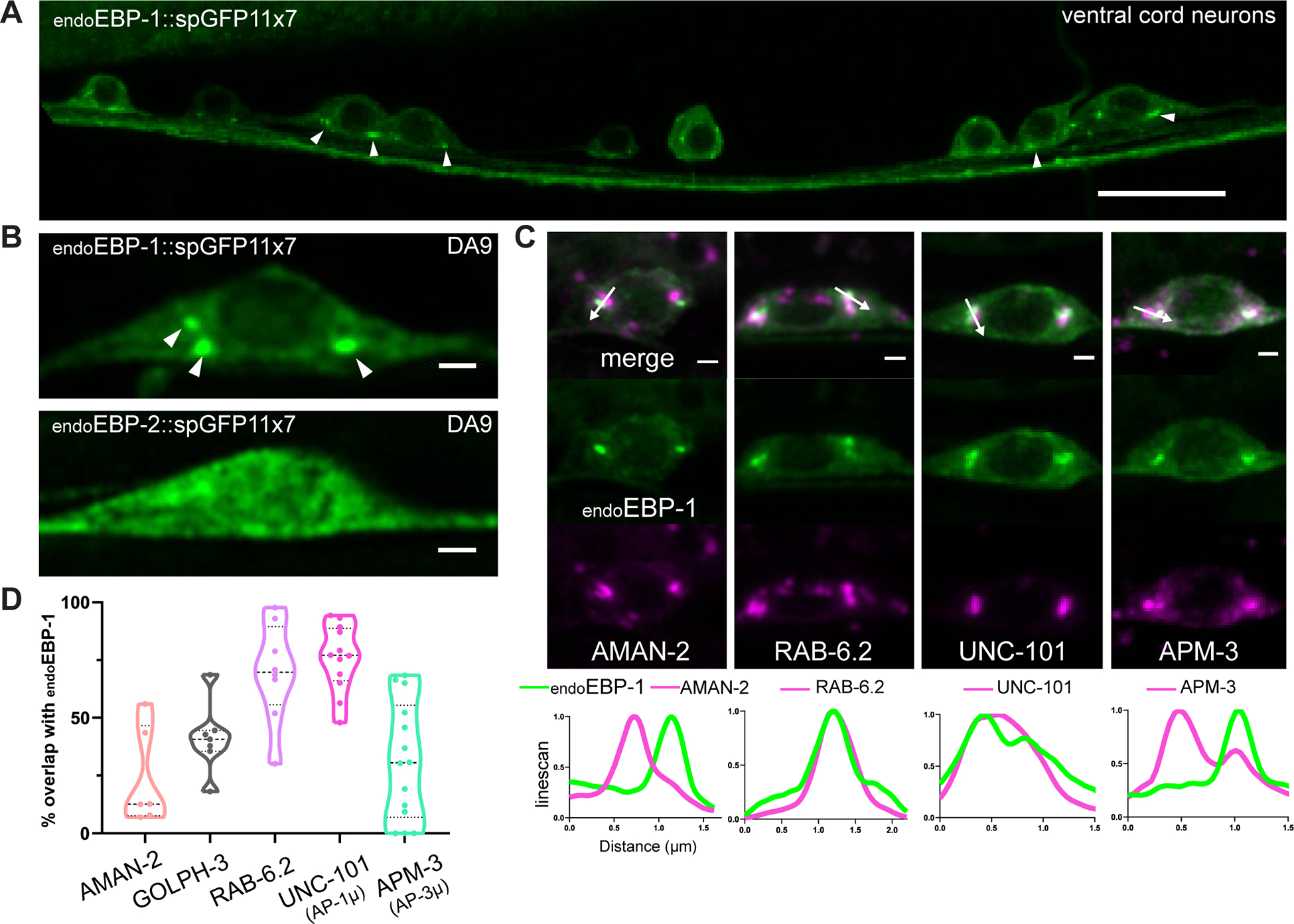
(A) Airyscan image of endogenous EBP-1 in ventral neurons that express Pmig-13::spGFP1–10. Note consistent enrichment of EBP-1 in 2–3 puncta per neuronal cell body indicated by white arrowheads. Scale bar 10 μm. See also Figure S4A and S4B. (B) Airyscan images of endogenous EBP-1 and EBP-2 in the DA9 cell body. White arrowheads pointing EBP-1 cell body structure. Scale bar 1 μm. See also Figure S4C and S4F. (C) Endogenous EBP-1 in green and various secretory pathway markers in magenta, imaged in DA9 cell body. A linescan is drawn in the direction of the white arrow. Scale bar 1 μm. See also Figure S4D and S4E for unc-101/AP-1μ involvement in DCV biogenesis. (D) Quantification of precent area overlap between endogenous EBP-1 and each Golgi marker in maximum projection image of DA9 cell body z-stack. Each data point in the graphs represents a fluorescence measurement from a single animal. n number is counting each individual animal imaged. See also Table S1.
EBP-1 puncta in the cell body were highly reminiscent of Golgi stacks in C. elegans neurons, prompting us to test colocalization with Golgi markers.15, 52 Using Airyscan microscopy, we observed that AMAN-2, a cis-medial Golgi marker, did not show high overlap with EBP-1, whereas the trans Golgi marker GOLPH-3 had a higher overlap (Figure 4C and 4D). Interestingly, EBP-1 showed 69.7% overlap with a trans Golgi/late Golgi vesicular carrier marker, RAB-6.2. This suggests that EBP-1 localizes in the vicinity of the trans Golgi and post Golgi carriers. Previous work has shown that different subdomains of the trans Golgi show a differential enrichment of the cargo adaptor complexes AP-1 and AP-3.14 AP-3μ /APM-3 is important for sorting SVPs to the axon while AP-1μ/UNC-101 is required for sorting DCV cargo, which we confirmed in DA9 (Figure S4D and S4E).14, 20 We found that EBP-1 had an average of 75.4% overlap in area with UNC-101, while it only overlapped 31.2% with APM-3 on average (Figure 4C and 4D). Together, these results indicate that EBP-1 is required for cell body exit of endoIDA-1 and is enriched in cell body puncta in the vicinity of trans Golgi carriers and the DCV sorting machinery AP-1μ/UNC-101.
EBP-1 functions in the neuronal soma to promote DCV trafficking to the axon
The enrichment of EBP-1 in cell body puncta and its role in promoting cell body exit of DCVs suggested that the neuronal soma might be the site where it functions in DCV trafficking. We tested whether EBP-1 co-traffics with endoIDA-1 or INS-22 but could not detect co-movement events in the axon (not shown), even though in the cell body EBP-1 did show colocalization with IDA-1 (Figure S5A). These results suggest that any interaction with exiting DCVs may be transient or that EBP-1 may function prior to DCVs entering the axon.
EB1 is required for Golgi integrity in HeLa and RPE cells.29 In DA9, UNC-101::RFP was not mislocalized in ebp-1 mutants (not shown). Since in various cell-types the Golgi has been shown to nucleate microtubules,53, 54 we tested if regulation of Golgi organization and Golgi-associated microtubules might underlie EBP-1 function in DCV transport in DA9. We initially tested the localization of the minus-end associated protein PTRN-1/CAMSAP but did not observe an enrichment at the sites of the EBP1- puncta (Figure S5B). Furthermore, RFP::PTRN-1 localization and abundance were unaffected in ebp-1 mutants (Figure S5D and S5E), suggesting that EBP-1 is not involved in Golgi organization of microtubule minus-end tethering in DA9. In addition, ebp-1 mutants did not affect microtubule stability or sensitivity to Colchicine (Figure S5C), consistent with the mild microtubule organization defects we previously reported in the axons of ebp-1 mutants.28 To test whether EBP-1 specifically regulates microtubules at the Golgi, we imaged endogenous EBP-2 comets in the cell body (video S1). In wildtype animals, microtubules grew in various orientations in the cell body, with an enrichment (32.1%) of comets associated with UNC-101::RFP at the Golgi. Interestingly, although overall comet numbers were not changed in ebp-1 mutants, we observed a clear and specific reduction in Golgi-associated comets (13.9%), which could also be observed in the ebp-1ΔCH mutants (Figure 5A–D). Loss of ebp-2 did not lead to a reduction of Golgi-associated growing MTs (as measured with EBP-1 as a plus-end marker), consistent with the observation that EBP-2 is not enriched at the Golgi (Figure S5F and S5G). Since the total number of EBP-2 comets was not reduced in ebp-1 mutants, we speculated that Golgi-localized EBP-1 is required to orient microtubule growth on the Golgi rather than to nucleate polymers at this site. To further distinguish whether Golgi-associated EBP-2 comets represent microtubule growth or nucleation events, we visualized the universal microtubule nucleator gamma tubulin/TBG-1 and its binding partner GIP-1 (both labelled at their endogenous loci) and did not observe a significant enrichment at the Golgi (Figure 5E and 5F). Taken together, these data suggest that ebp-1 is required to direct the growth of cell body microtubules at the Golgi.
Figure 5: Loss of ebp-1 disrupts Golgi associated microtubule growth in neuronal soma.
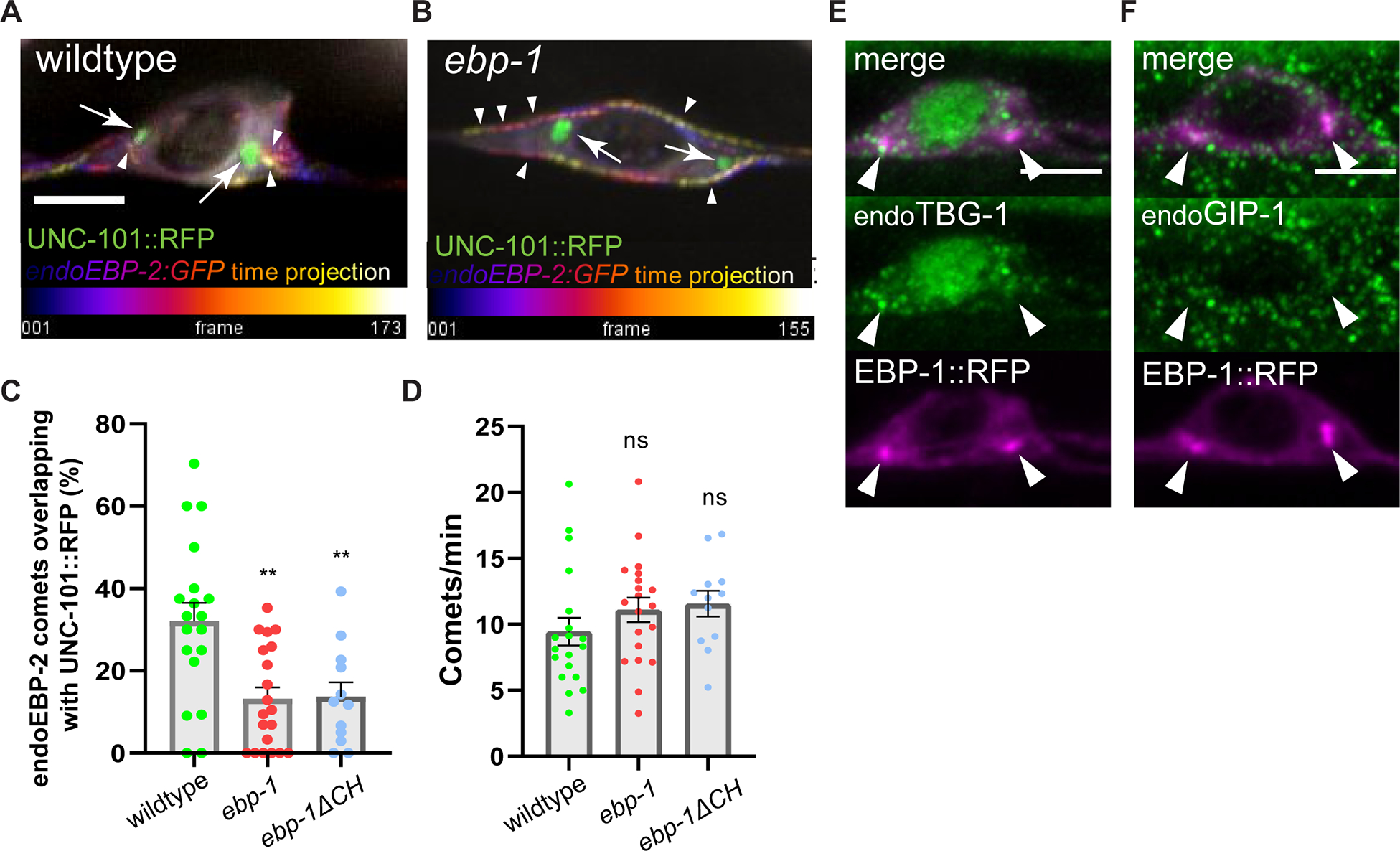
(A, B) Temporal projection of endogenous EBP-2 comet trajectory. Time is pseudo color-coded from purple to yellow, indicated by white arrowhead. UNC-101::RFP is shown in green, indicated by white arrow. Scale bar 3 μm. See also Video S1. (C, D) Quantification of percentage of endoEBP-2 comets overlapping with UNC-101::RFP out of total EBP-2 comets observed (C) and the number of comets per minute (D) for each condition. n=12–20. (E, F) Representative image of endogenously labeled TBG-1 (E) and GIP-1 (F) with EBP-1::RFP. Scale bar 3 μm. See also Figure S5. **p<0.01 (one-way ANOVA for (C) and (D)). Each data point in the graphs represents a count or an average comet number from a single animal. n number is counting each individual animal imaged. See also Table S1 and Video S1.
Since the ebp-1ΔCH mutant mimicked the Golgi microtubules defects but not the DCV transport defects of a full ebp-1 deletion, we speculated that directing microtubule growth at the Golgi is redundant with another function of EBP-1. This would be consistent with the need to eliminate both the CH and EBH domains to mimic an ebp-1 deletion. To test whether EBP-1 has additional functions in the cell body prior to DCV exit, we sought to restrict its location by fusing it to the integral trans Golgi proteins BRE-4 or GOLGIN-84. For this chimera, we used a version of EBP-1 lacking the CH domain to prevent interactions with MTs from affecting the localization of the chimeric protein or from influencing microtubules around the Golgi, and since expression of EBP-1 lacking this domain could rescue ebp-1; ebp-2 mutants (Figure 2K). Both RFP::EBP-1ΔCH::BRE-4 and RFP::EBP-1ΔCH::GOLGIN-84 chimeras showed punctate localization in the cell body and could not be detected in the axon (Figure 6D’ and 6D”). Interestingly, this chimeric protein was sufficient to restore endoIDA-1 to the axon in ebp-1 mutants (Figure 6E). Control constructs RFP::BRE-4 or RFP::EBP-1ΔCHΔC::BRE-4 did not rescue axonal DCVs, consistent with the inability of overexpressed EBP-1ΔCHΔC to rescue ebp-1; ebp-2 mutants (Figure 6F, 6H, and 6I). Conversely, RFP::EBP-1::BRE-4 did not rescue the reduced spacing between synaptic boutons in ebp-1 ΔCH mutants, suggesting that reduced synaptic spacing is a separate phenotype that is not due to EBP-1 function in the Golgi (Figure S6B). These data support the conclusion that EBP-1 promotes DCV trafficking to the axon by acting in the vicinity of the Golgi in the cell body.
Figure 6: EB1 interacts with KIF1A and EBP-1 function in neuronal soma to promote DCV trafficking.
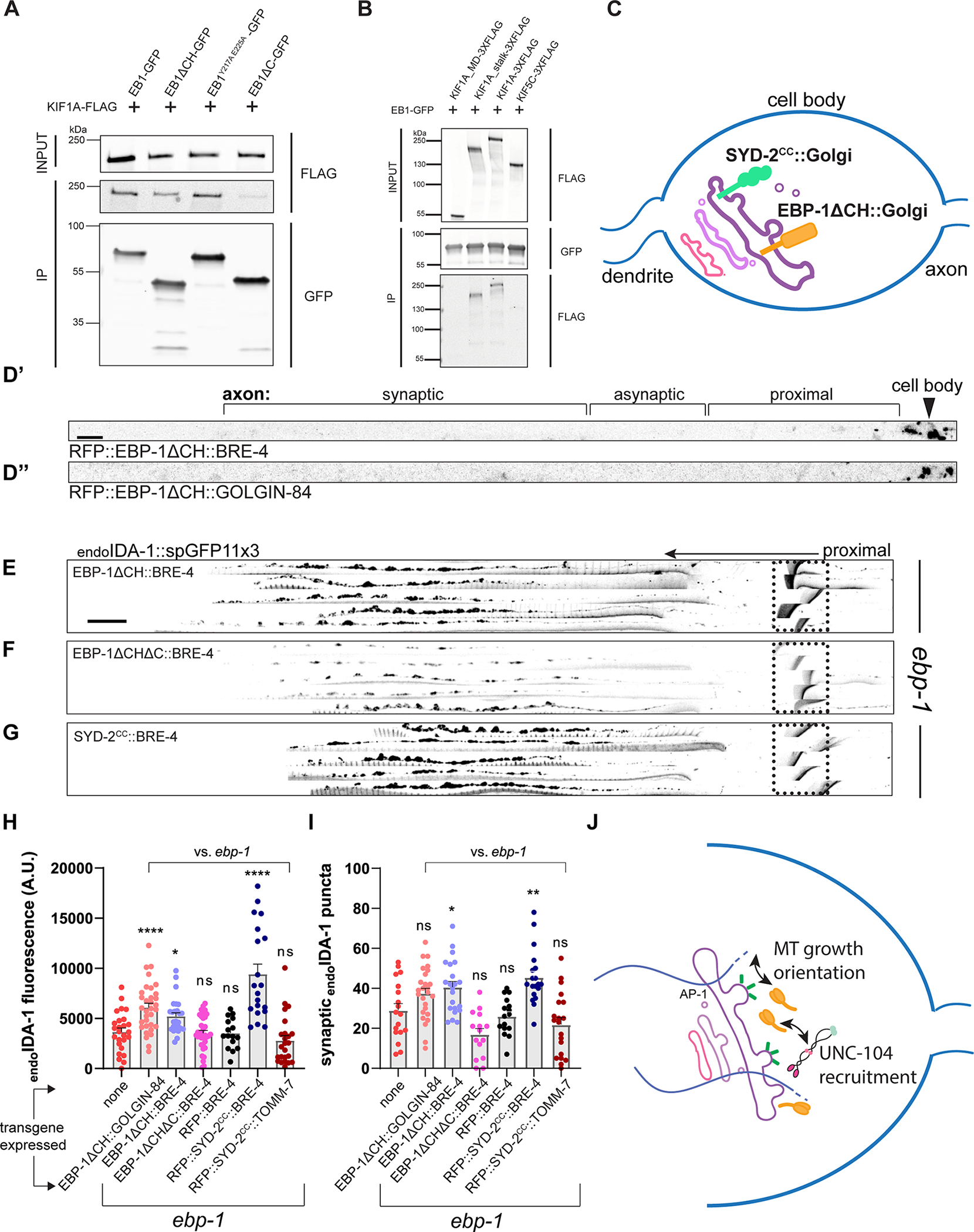
(A) co-IP between EB1 fragments and KIF1A. EB1 fragments stained with anti-GFP and KIF1A with anti-FLAG. (B) co-IP between EB1 and KIF1A motor domain, KIF1A stalk, KIF1A, and KIF5C. See also Figure S6A. (C) Diagram of Golgi protein chimera with EBP-1ΔCH and SYD-2CC. (D) Representative images of RFP::EBP-1ΔCH::BRE-4 (D’) and RFP::EBP-1ΔCH::GOLGIN-84 (D”) showing that the chimeric proteins are restricted to the DA9 cell body. Black arrowhead indicates DA9 cell body. Scale bar 10 μm. (E-G) Alignments of 5 representative axons per rescue showing endoIDA-1::GFP in DA9. Black arrow points distally. Autofluorescence from rectum in dotted box. Scale bar 10 μm. (H) Quantification of fluorescence of endoIDA-1 for each rescue condition. n=16–34. (I) Count of endoIDA-1 puncta in the synaptic region. n=15–26. (J) Model diagram showing EBP-1 biasing kinesin-3 localization by interaction and directing microtubule growth near Golgi. ****p<0.0001; **p<0.01, *p<0.05 (one-way ANOVA for (F) and (G). t-test for (H) and (I)). Each data point in the graphs represents a fluorescence measurement or counted puncta from a single animal. n number is counting each individual animal imaged. See also Table S1.
EB1 interacts with KIF1A and a KIF1A/UNC-104 interacting domain can replace EBP-1 in the soma
Since the DCV phenotype of ebp-1 mutants was suggestive of transport defects, we tested whether it could involve UNC-104/kinesin-3, which is the motor for DCVs. Previously, EBs have been shown to physically interact with other kinesin motors and with dynactin, either directly or indirectly.27, 55–59 However, an interaction between KIF1A/UNC-104 and EB has not been described. C. elegans UNC-104 formed aggregates when expressed in mammalian cell culture (not shown), so we tested its highly conserved mammalian homolog KIF1A and the mammalian EB1, which could robustly rescue the DCV phenotype of ebp-1 mutants in C. elegans (Figure 2N). FLAG tagged KIF1A and GFP tagged EB1 co-immunoprecipitated from HEK293T cells, while the neuronally enriched kinesin-1/KIF5C did not co-immunoprecipitate with EB1 (Figure 6B). EB1ΔCH was also able to pull down full length KIF1A (Figure 6A and S6A), albeit not as efficiently as full length EB1, suggesting that MT tip localization might facilitate the interaction by promoting encounters of EB1 and KIF1A at MT ends. Interestingly, the EB1ΔC construct minimally co-immunoprecipitated with KIF1A but EB1Y217A E225A -GFP still pulled down KIF1A (Figure 6A third and fourth lane). We also found that the motor domain of KIF1A was dispensable for this interaction and the stalk region alone could co-immunoprecipitate with EB1-GFP (Figure 6B), consistent with previous reports of EBs interacting with other kinesins via their stalk regions.55, 57–59
We noted that the domains that mediated the EB1-KIF1A interaction reflected the ability of DA9-expressed EBP-1 fragments to rescue the DCV phenotype of ebp-1; ebp-2 mutants (Figure 2K and 2O), suggesting that an interaction between EBP-1 and UNC-104/KIF1A might be important for DCV trafficking. Since we found that EBP-1 functions in the cell body near Golgi-derived carriers, we asked whether it could act by transiently recruiting UNC-104 to these sites. For this, we tested whether ebp-1 mutants can be rescued by artificially recruiting UNC-104/KIF1A to the Golgi with a chimera between the Golgi-resident protein BRE-4 and a UNC-104/KIF1A binding domain from a protein not related to EBs. We used the UNC-104 interacting coiled-coil domain from SYD-2/liprin-α (SYD-2coiled-coil), which was shown to directly interact with kinesin-3 in worms and mammals.60, 61 RFP::SYD-2coiled-coil::BRE-4 expression in DA9 was able to fully rescue the loss of endoIDA-1 from the axon of ebp-1 mutants (Figure 6G, 6H, and 6I). In contrast, RFP::SYD-2coiled-coil::TOMM-7, which localized to mitochondria in the cell body and axon, could not rescue (Figure 6H and 6I). This result suggests that transiently localizing UNC-104 to the vicinity of Golgi carriers is sufficient to compensate for EBP-1 function in promoting axonal delivery of DCVs. Collectively, our results support a working model in which Golgi-localized EBP-1 promotes axonal DCV transport by facilitating motor-cargo association through its interactions with kinesin-3/UNC-104 and with microtubules (Figure 6J).
Discussion
How specific motor-cargo associations are regulated in the cell body prior to transport in the axon is unclear. We report a role for EBP-1 in promoting the initial steps of transport of one KIF1A/UNC-104 cargoes – DCV. Loss of EBP-1 leads to a specific reduction in axonal DCVs, while other cargoes (ATG-9, RAB-3, SNG-1) remain unperturbed. This reduction can be traced to a reduced number of DCVs exiting the cell body, suggesting that EBP-1 functions in neuronal soma. Consistently, EBP-1 (but not its paralog EBP-2), is enriched in the vicinity of AP-1μ which sorts DCVs at the trans Golgi. EBP-1 is required to direct MT growth on the Golgi in a CH domain dependent manner, and mammalian EB1 co-immunoprecipitates with KIF1A in an EBH domain dependent manner. These results support a working model in which directing MTs to the trans Golgi and interacting with the motor in this location are two EBP-1 functions that promote the early steps of DCV movement, with the latter function playing a more prominent role. In agreement, tethering a MT-binding deficient version of EBP-1 to the Golgi or anchoring to this organelle a KIF1A/UNC-104 interacting domain from an unrelated protein, SYD-2, can restore DCVs to ebp-1 mutant axons. These results provide insight into how the specificity of motor-cargo association in the cell body is generated and reveal an unexpected role of a MT associated protein.
Coupling cargo sorting with motor-association in the neuronal soma for efficient axonal transport
SVP and DCV biogenesis involves complex sorting steps, followed by an association between the transport packet and a motor, but how these processes are coordinated is unclear. 15, 62, 63 In cases where the same motor carries different cargo, specific mechanisms that promote association with given cargos should exist. The KIF1A/UNC-104 Pleckstrin Homology domain binds PI(4,5)P2 and PI(4)P and is thought to be the main determinant in cargo recognition.3–5, 17 Deletion or a point mutation in the PH domain abolishes trafficking of DCVs, SVPs, lysosomes, and ATG-9 vesicles.3, 5, 17 However, it is unclear if membrane recognition by a PH domain is sufficient to explain the transport of vesicles that likely differ in size and membrane properties.
We found that EBP-1 specifically promotes the cell body exit of DCVs transported by UNC-104. EBP-1 is specifically enriched near the adaptor complex that sorts DCVs, but not SVPs, for axonal delivery. Thus, we propose that spatial segregation between these adaptor complexes could underlie the ability of EBP-1 to specifically promote association between UNC-104 and DCVs. We propose that two EBP-1 functions operate redundantly at DCV biogenesis sites: interactions with UNC-104 and steering microtubule growth, with the latter indirectly enriching kinesin-3 at the trans Golgi. The redundancy between these functions is supported by the need to eliminate both CH and EBH domains to mimic a full ebp-1 deletion. The sufficiency of the EBH domain to rescue DCV transport upon overexpression may be because EBH domains bind microtubule associated proteins.23, 51 Exactly how EBP-1 interacts with UNC-104 at the Golgi remains to be determined: we did not observe a direct interaction between EB1 and KIF1A in a yeast two hybrid assay (not shown) but the two proteins share many potential interactors (data not shown).51, 64
Neuronal function of EBs
EBs are highly conserved, and while they are routinely used as markers of growing MT ends, the full range of their physiological roles remains to be discovered. This observation is underscored by the surprisingly mild phenotype in mammalian cells where all 3 EBs are disrupted or in worm embryos with a triple EBP deletion.29, 33, 34 In neurons, EBs were proposed to organize neuronal MTs by guiding polymer growth along other MTs, linking MTs to actin-based structures or to the AIS, or controlling polymer dynamicity.26–28, 31, 32. We show that in addition to these cytoskeletal functions, which mostly take place along neurites, EBP-1 has a specific role in promoting the transport of DCVs from the cell body. This EB function is conserved since EB1 could rescue ebp-1 mutants and interacted with KIF1A.
The localization of EBs to MT ends makes them ideal for regulating molecular motors, which often associate with the plus ends, or fall from it, at the end of a run.30 Consistently, EBs were shown to interact with several kinesins and to help load dynactin/dynein on MTs at the distal axon.32, 55, 57–59. In non-neuronal cells, EBs function in microtubule organization at the Golgi.29 In agreement with these functions, we found that Golgi-localized EBP-1 directs growing plus-ends to the trans Golgi, which is consistent with a role in promoting the association between kinesin-3 /UNC-104 with nascent DCVs.
How EBP-1, but not EBP-2 is recruited to the Golgi is unclear. Functional differences between mammalian EB1 and EB2 have also been reported,29, 33 as well as different tip-tracking propensities, similar to our observations in C. elegans. In non-neuronal cell culture, a complex of GM130 / AKAP450 / Myomegalin promotes EB1/3 localization to Golgi.29 These proteins are not conserved in C. elegans, and examination of mutants that could mediate Golgi localization of EBP-1 (dynein/dhc-1, dynactin/dnc-1, CAMSAP/ptrn-1, CLASP/cls-2, cls-3, EB2/ebp-2, KIF1A/unc-104, AP-1μ/unc-101, ninein/noca-1) did not reveal EBP-1 mislocaization (not shown). Hence, future studies will be required to precisely determine the sequences that confer specific subcellular localizations to EBPs or that dictate their tip-tracking efficiency, which should also lead to a better understanding of their physiological roles.
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents should be directed to the lead contact (shaul.yogev@yale.edu).
Materials Availability
All transgenic C. elegans strains and recombinant plasmids generated for this study are available from the lead contact upon request.
Data and Code Availability
Raw microscopy data and western blots reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Strains and maintenance
All C. elegans strains were grown on nematode growth medium plates seeded with E. coli OP50 as previously described.66 The N2 Bristol strain was used as wildtype and for outcrosses. All animals were grown at 20°C for experiments/injection.
METHOD DETAILS
Cloning and constructs
Genes of interest were PCR cloned from either cDNA or gDNA of N2 Bristol strain using PfuUltra II Fusion HS DNA polymerase (Agilent). Amplicon was ligated into pSM vector backbone for expression in C. elegans and EGFP-C1 (Clontech) for mammalian cells. All plasmid ligation step was done using In-fusion seamless cloning (Takara Bio). Appropriate mutagenesis was done using QuikChange Lightning Multi Site-Directed mutagenesis kit (Agilent). Final product plasmids were confirmed by Sanger sequencing. All plasmids are freely available upon request.
Transgenic C. elegans strain generation
Transgenic animals were generated via injecting plasmids or CRISPR reagents directly into the gonads of young adult animal following the standard injection method.67 Resulting F1s transmitted the injected expression arrays to its progenies were singled. Animals with preferred transmission rate were mounted on slide glass and were further visually screened for desired expression level of extrachromosomal array on an inverted microscope. Lastly, for genetically modified animals, they were PCR genotyped and the PCR product was Sanger sequenced to verify proper insertion.
CRISPR/Cas9 genome editing
All CRISPR strains were generated following the published protocol,67 with the exception of the wy1156 deletion that was generated using another protocol described previously.68 wy1156 starts 12 amino acids into ebp-1 and ends 39 base pairs before the stop codon of the pseudogene ebp-3. It also includes a 14bp insertion, such that the remaining ebp-3 sequence is not in frame. Custom designed crRNA, trRNA, and Cas9 are from IDT. Details of crRNA sites and repair template used are in the Table S1.
Fluorescence microscopy and sample preparation
L4 larvae animals were grown at 20°C, 20–24 hours prior to imaging. One day adult animals were used mainly for quantification and assessment of phenotype. For endogenous EBP-1/EBP-2 comet imaging, adult animals were bleached and synchronized L1 larvae were isolated to image L3 worms. For general imaging, worms were immobilized on agar pads with 10 mM Levamisole dissolved in M9 buffer. For time-lapse imaging, one day adult animals were incubated in 0.5 mM levamisole at room temperature for 6 min 30 seconds (4 min 30 seconds for L3) prior to mounting on 10% agarose pad in M9 without additional paralytics. For Colchicine drug treatment, Colchicine was dissolved in 0.5 mM Levamisole at 30 mg/mL. Animals were incubated in Colchicine dissolved solution for 30 min and subsequently moved to fresh M9 without paralytics for live imaging. Andor Dragonfly spinning-disk confocal microscope equipped with a plan apochromat objective (63x, 1.4 NA, oil) and a Zyla scientific CMOS camera was used for standard fluorescence imaging. Live images in the cell body were acquired through an HC PL APO 63x/1.40NA OIL CS2 objective or a PL APO 100x/1.40NA CS2 objective built on a Laser Safe DMi8 inverted microscope (Leica) equipped with a VT-iSIM system (BioVision). Images were captured with an ORCA-Flash4.0 camera (Hamamatsu) and controlled by MetaMorph Advanced Confocal Acquisition Software Package. The same imaging settings (laser power, exposure time, and gain) were used throughout the study.
In photobleaching experiments, region of interest was photobleached using 405 nm laser with a Perkin-Elmer Photokinesis FRAP unit. Pre-bleach and post-bleach time lapse was imaged at 3 Hz using an Olympus BX61 microscope equipped with a Hamamatsu ORCA-Flash4.0 LT camera. For high resolution imaging, Airyscan function of Carl Zeiss LSM880 confocal laser scanning microscope (63x, 1.4 NA, plan-apochromat, oil) with Airyscan detector was used. Raw Airyscan images were processed using ZEN imaging software (Zeiss).
Egg laying quantification
Quantification of unlaid egg was performed using adult animals 30 hours after staging late L4 larvae. The adult animals were bleached to dissolve the body and impermeable eggs were counted on a 96-well plate. Precise protocol is detailed in a previous work.42.
Western blot with whole worm lysate
Adult animals were bleached and remaining eggs were moved to a new nematode growth media (NGM) plate to hatch. From this stage synchronized animals, eight L4 larvae were moved to new plates to expand in 20°C. Per genetic condition, a dozen 60 mm NGM plates with OP50 spots were used for the expansion step. After 50~70 hours, each plate was washed with M9 to collect the adult animals. Collected worms were washed with fresh M9 every hour on a spinning wheel at room temperature. After 4~6 washes, when OP50 was sufficiently cleaned out, the worms were spun down to rid the supernatant and were rapidly frozen with liquid nitrogen for storage.
Each frozen sample was thawed on ice. With liquid nitrogen-cooled mortar and pestle, the samples were ground into powder form. Pulverized samples were then resuspended in buffer with 1% Triton X-100 and incubated for 20 minutes at 4°C for lysis. The lysate was spun down and the supernatant was dosed for concentration using Pierce BCA Protein Assay. After the protein concentration of each sample was adjusted, the loading samples were boiled with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer at 95°C for 10 minutes and then samples were run on 4~20% Mini-PROTEAN TGX Stain-Free gel and transferred to 0.2 μm nitrocellulose membrane (both Bio Rad). After blocking with 5% milk, the membrane was incubated with anti-αTubulin (Sigma-Aldrich) and anti-GFP (Abcam) primary antibody overnight. After washing out the primary antibodies, the membrane was stained with Li-COR fluorescence secondary antibodies for quantification.
Cell culture, transfection, and co-IP
HEK293T cells (American Type Culture Collection) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% Fetal Bovine Serum, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin, and 100 mg/mL streptomycin (Gibco) at 37 °C and 5% CO2. HEK293T cells were grown in 6-well plate for all IP experiments. For transfection, equimolar amount of EB1-GFP, EB1 fragment-GFP, and KIF1A-3XFLAG were transiently transfected using FuGene HD (Promega)/Lipofectamine 2000 (Thermo Fisher). 48 hours after transfection, cells were lysed with a buffer containing 1% Triton X-100 for 20 minutes. The lysate was spun down and the supernatant was incubated with anti-GFP magnetic beads (Chromotek) for 2 hours in 0.33% Triton X-100 solution. After incubation, beads were washed and the samples were eluted with SDS-PAGE sample buffer by 10 minutes incubation at 95°C. Samples were then run on 4~20% Mini-PROTEAN TGX Stain-Free gel and transferred to 0.2 μm nitrocellulose membrane. Membrane was incubated with M2 anti-FLAG (Sigma-Aldrich) and anti-GFP (Abcam) overnight in 4 °C. After the overnight incubation, the membrane was washed with TBS five times andwas stained with Li-COR fluorescence secondary antibodies (Li-COR) for two hours in room temperature. After three washes with TBST (0.1% triton) followed by two washes with TBS, the membrane was imaged on Li-COR Odyssey imager.
QUANTIFICATION AND STATISTICAL ANALYSIS
Fluorescence quantification
Raw imaging files were imported to ImageJ for further analysis. Puncta number was counted using FindFoci plugin using similar settings for all images analyzed.69 Kymographs were generated with ImageJ Multi Kymograph plugin where resulting traces were analyzed manually in Microsoft Excel. Movement correction was done with StackReg plugin for time-lapse movies with minor drift before kymograph analysis.
Statistical analysis
Statistical analysis was performed on Prism 9 (GraphPad) and Microsoft Excel. Three experimental replicates were included in each data set and data were considered significant at p ≤ 0.050 with statistical test indicated in each figure legend.
Supplementary Material
Video S1. Golgi-associated EBP-2 comets are reduced in ebp-1 mutant. Related to Figure 5.
Representative time-lapse movie of endogenously labeled EBP-2 (green) and RFP labeled UNC-101/AP-1μ (magenta) in DA9 neuron cell body. A number of EBP-2 comets are associated with UNC-101 in wildtype. In ebp-1 mutants, the number of comets associated with UNC-101::RFP is reduced. To avoid ambiguity, comet signals that completely overlap with UNC-101::RFP were counted as associated.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-FLAG M2 | Sigma-Aldrich | F3165 |
| Anti-GFP | Abcam | Ab290 |
| Anti-α Tubulin | Sigma-Aldrich | T5168 |
| GFP-Trap | Chromotek | Gtmak-20 |
| Bacterial and virus strains | ||
| Escherichia coli: OP50 Strain | Caenorhabditis Genetics Center | OP50 |
| Stellar Competent Cells | Takara Bio | Cat# 636766 |
| Chemicals, peptides, and recombinant proteins | ||
| Alt-R S.p.Cas9 Nuclease V3 | IDT | Cat# 1081059 |
| Critical commercial assays | ||
| Takara Bio In-Fusion seamless cloning | Takara Bio | Cat# 638949 |
| QuikChange Lightning Multi Site-Directed mutagenesis kit | Agilent | Cat# 210513 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher | Cat# 23225 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | CB_12022001 |
| Experimental models: Organisms/strains | ||
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA] | ceIS308 from Miller lab | KG5082 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (wy1156) | ebp-1 (wy1156) from Shen lab | MTS97 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS1308 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279) | ebp-1 (he279) from van den Heuvel lab | MTS1342 |
| rab-3 (ox699), shyIS43 [Pglr-4::FLP::NLSx2, Podr-1::rfp] | rab-3 (ox699) from Jorgensen lab | MTS1161 |
| rab-3 (ox699), shyIS43 [Pglr-4::FLP::NLSx2, Podr-1::rfp]; ebp-1 (he279) | This Study | MTS1234 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]/tmC9 [F36H1.2 (tmIS1221); Pmyo-2::Venus]; shyEx187 [Punc-122::rfp] | tmC9 from Mitani lab | MTS1623 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]/tmC9 [F36H1.2 (tmIS1221); Pmyo-2::Venus]; shyEx187 [Punc-122::rfp]; ebp-1 (wy1156) | This Study | MTS1624 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]tmC9 [F36H1.2 (tmIS1221); Pmyo-2::Venus]; shyEx187 [Punc-122::rfp]; ebp-1 (wy1156); shyEx438 [Pmig-13::ebp-1] | This Study | MTS1797 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; unc-31 (e928) | unc-31 (e928) from Hodgkin lab | MTS1656 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; unc-31 (e928); ebp-1 (wy1156) | This Study | MTS1644 |
| shyEx271 [Pmig-13::atg-9::gfp, Podr-1::rfp] | This Study | MTS1063 |
| shyEx271 [Pmig-13::atg-9::gfp, Podr-1::rfp]; ebp-1 (he279) | This Study | MTS1132 |
| wyIS401 [Pmig-13::STOP::sng-1::gfp]; wySi324 [Punc-4C::CRE] | wyIS401 and wySi324 from Shen lab | MTS1312 |
| wyIS401 [Pmig-13::STOP::sng-1::gfp]; wySi324 [Punc-4C::CRE]; ebp-1 (he279) | This Study | MTS1400 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy99[ebp-1_134–316aa]) | This Study | MTS1416 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy116[ebp-1_1–256aa]) | This Study | MTS1730 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy147[ebp-1_134–256aa]) | This Study | NTS1866 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy102[ebp-1_Y264A_E272A]) | This Study | MTS1522 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy136[ebp-1_134–316aa_Y264A_E272A]) | This Study | MTS1827 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278) | ebp-2 (he278) from van den Heuvel lab | MTS929 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278); shyEx444 [Pmig-13::ebp-1] | This Study | MTS1817 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278); shyEx304 [Pmig-13::ebp-1 (1–133aa)] | This Study | MTS1045 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278); shyEx243 [Pmig-13::ebp-1 (134–316aa)] | This Study | MTS944 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278); shyEx408 [Pmig-13::ebp-1 (134–316aa, Y264A, E272A)] | This Study | MTS1568 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278); shyEx407 [Pmig-13::ebp-1 (134–256aa)] | This Study | MTS1567 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278); shyEx421 [Pmig-13::ebp-1 (257–316aa)] | This Study | MTS1616 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (wy1156); shyEx222 [Pmig-13::ebp-1] | This Study | MTS862 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (wy1156); shyEx227 [Pmig-13::EB1 (mus musculus)] | This Study | MTS867 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (wy1156); shyEx224 [Pmig-13::ebp-2] | This Study | MTS864 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; ebp-1 (he279); ebp-2 (he278); shyEx405 [Pmig-13::ebp-1 (134–313aa)] | This Study | MTS1565 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-2 (he278) | This Study | MTS1347 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279); ebp-2 (he278) | This Study | MTS1343 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy102[ebp-1_Y264A_E272A]); ebp-2 (he278) | This Study | MTS1829 |
| ida-1 (shy96[ida-1::GFP11x3]);; shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy116[ebp-1_1–256aa]); ebp-2 (he278) | This Study | MTS1869 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (shy99[ebp-1_134–316aa]); ebp-2 (he278) | This Study | MTS1940 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS1000 |
| ebp-1 (shy98[ebp-1_134–316aa::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS1321 |
| ebp-1 (shy103[ebp-1_Y264A_E272A::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS1643 |
| ebp-1 (shy121[ebp-1_1–256aa::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS1701 |
| ebp-1 (shy153[ebp-1_134–256aa::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS1870 |
| ebp-1 (shy154[ebp-1_134–316aa_Y264A_E272A::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS1892 |
| ebp-2 (shy70[ebp-2::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp] | This Study | MTS959 |
| rab-3 (ox699), shyIS43 [Pglr-4::FLP::NLSx2, Podr-1::rfp]; ebp-1 (shy147[ebp-1_134–256aa]) | This Study | MTS1856 |
| wyIs685 [Pmig-13::cla-1::gfp, Pmig-13::rab-3::tdTomato, Podr-1::gfp] | wyIs685 from Shen lab | TV18675 |
| wyIs685 [Pmig-13::cla-1::gfp, Pmig-13::rab-3::tdTomato, Podr-1::gfp]; ebp-1 (wy1156) | This Study | MTS35 |
| wyIs685 [Pmig-13::cla-1::gfp, Pmig-13::rab-3::tdTomato, Podr-1::gfp]; ebp-1 (shy99[ebp-1_134–316aa]) | This Study | MTS1975 |
| wyIs685 [Pmig-13::cla-1::gfp, Pmig-13::rab-3::tdTomato, Podr-1::gfp]; ebp-1 (shy116[ebp-1_1–256aa]) | This Study | MTS1976 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyIS49 [Prab-3::GFP1–10, Pmyo-2::mCherry] | This Study | MTS1934 |
| ebp-2 (shy70[ebp-2::GFP11x7]); shyIS49 [Prab-3::GFP1–10, Pmyo-2::mCherry] | This Study | MTS1939 |
| ebp-2 (shy70[ebp-2::GFP11x7]); shyIS49 [Prab-3::GFP1–10, Pmyo-2::mCherry]; ebp-1 (he279) | This Study | MTS1158 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx340 [Pmig-13::apm-3::rfp, Podr-1::gfp] | This Study | MTS1272 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx288 [Pmig-13::aman-2::rfp, Podr-1::gfp] | This Study | MTS1273 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx341 [Pmig-13::rfp::rab-6.2, Podr-1::gfp] | This Study | MTS1280 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx354 [Pmig-13::unc-101::rfp, Podr-1::gfp] | This Study | MTS1310 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx376 [Pmig-13::rfp::golph-3, Podr-1::gfp] | This Study | MTS1417 |
| shyEx430 [Pmig-13::rfp::ebp-1 (134–316aa)::bre-4, Podr-1::gfp] | This Study | MTS1857 |
| shyEx427 [Pmig-13::rfp::ebp-1 (134–316aa)::golgin-84, Podr-1::gfp] | This Study | MTS1899 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279); shyEx430 [Pmig-13::rfp::ebp-1 (134–316aa)::bre-4, Podr-1::gfp] | This Study | MTS1736 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279); shyEx427 [Pmig-13::rfp::ebp-1 (134–316aa)::golgin-84, Podr-1::gfp] | This Study | NTS1683 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279); shyEx489 [Pmig-13::rfp::ebp-1 (134–256aa)::bre-4, Podr-1::gfp] | This Study | MTS1960 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279); shyEx490 [Pmig-13::rfp::bre-4] | This Study | MTS1963 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279); shyEx491 [Pmig-13::rfp::syd-2 (2–699aa)::bre-4, Podr-1::gfp] | This Study | MTS1978 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS36 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279); shyEx492 [Pmig-13::rfp::syd-2 (2–699aa)::tomm-7, Podr-1::gfp] | This Study | MTS1979 |
| wyIs685 [Pmig-13::cla-1::gfp, Pmig-13::rab-3::tdTomato, Podr-1::gfp]; ebp-1 (shy99[ebp-1_134–316aa]); shyEx515 [Pmig-13::ebp-1::bre-4] | This Study | MTS2079 |
| vsIS75 [Pnlp-3::nlp-3, Pmyo-2::mCherry] | vsIS75 from Koelle lab | LX2519 |
| vsIS75 [Pnlp-3::nlp-3, Pmyo-2::mCherry]; ebp-1 (he279) | This Study | MTS2158 |
| vsIS75 [Pnlp-3::nlp-3, Pmyo-2::mCherry]; ebp-1 (he279); shyEx543 [Ptph-1::ebp-1, Podr-1::GFP] | This Study | MTS2215 |
| ida-1 (shy96[ida-1::GFP11x3]); shyIS40 [Pitr-1::gfp1–10, Podr-1::rfp]; shyEx300 [Pmig-13::ebp-1::rfp, Podr-1::gfp] | This Study | MTS1559 |
| gip-1(wow3[gfp::gip-1]); shyEx300 [Pmig-13::ebp-1::rfp, Podr-1::gfp] | gip-1 (wow3) from Feldman lab | MTS1527 |
| tbg-1 (shy152); shyIS35 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx300 [Pmig-13::ebp-1::rfp, Podr-1::gfp] | This Study | MTS2098 |
| ebp-2 (shy70[ebp-2::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx354 [Pmig-13::unc-101::rfp, Podr-1::gfp]; ebp-1 (he279) | This Study | MTS1373 |
| ebp-2 (shy70[ebp-2::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx354 [Pmig-13::unc-101::rfp, Podr-1::gfp] | This Study | MTS2372 |
| wyIS463 [Pmig-13::STOP::gfp::tba-1, Podr-1::rfp]; wySi265 [Punc-4C::cre, Punc-122::RFP], tba-1 (ok1135); shyEx300 [Pmig-13::ebp-1::rfp, Podr-1::gfp] | This Study | MTS2257 |
| wyIS463 [Pmig-13::STOP::gfp::tba-1, Podr-1::rfp]; wySi265 [Punc-4C::cre, Punc-122::RFP], tba-1 (ok1135); shyEx300 [Pmig-13::ebp-1::rfp, Podr-1::gfp]; ebp-1 (he279) | This Study | MTS2262 |
| wyIS463 [Pmig-13::STOP::gfp::tba-1, Podr-1::rfp], wyIS720 [Pmig-13::STOP::RFP::PTRN-1]; wySi265 [Punc-4C::cre, Punc-122::RFP], tba-1 (ok1135); shyEx300 [Pmig-13::ebp-1::rfp, Podr-1::gfp]; ebp-1 (he279) | This Study | MTS1005 |
| ebp-1 (shy68[ebp-1::GFP11x7]); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; shyEx413 [Pmig-13::STOP::tagRFP::PTRN-1_396–1110aa]; wySi265 [Punc-4C::cre, Punc-122::RFP] | This Study | MTS2500 |
| wyIS463 [Pmig-13::STOP::gfp::tba-1, Podr-1::rfp]; wySi265 [Punc-4C::cre], tba-1 (ok1135); wyIS363 [Pmig-13::STOP::tdTomato::RAB-3] | wyIS463, wyIS363, and wySi265 from Shen lab | TV18649 |
| wyIS463 [Pmig-13::STOP::gfp::tba-1, Podr-1::rfp]; wySi265 [Punc-4C::cre], tba-1 (ok1135); wyIS363 [Pmig-13::STOP::tdTomato::RAB-3]; unc-104 (e1265) | This Study | MTS2499 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; unc-104 (e1265) | This Study | MTS1203 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]/tmC9 [F36H1.2 (tmIS1221); Pmyo-2::Venus]; unc-104 (e1265) | This Study | MTS2283 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; unc-101 (m1) | This Study | MTS873 |
| ceIS308 [Pmig-13::ins-22::Em, Pmig-13::RFPA]; apb-3 (ok429) | This Study | MTS874 |
| ebp-2 (shy70); shyIS32 [Pmig-13::gfp1–10, Podr-1::rfp]; ebp-1 (he279) | This Study | MTS1158 |
| wyIS463 [Pmig-13::STOP::gfp::tba-1, Podr-1::rfp], wyIS720 [Pmig-13::STOP::RFP::PTRN-1]; wySi265 [Punc-4C::cre], tba-1 (ok1135) | wyIS463, wyIS720, and wySi265 from Shen lab | TV19492 |
| Oligonucleotides | ||
| Alt-R® CRISPR-Cas9 tracrRNA | IDT | Cat# 1072533 |
| Alt-R CRISPR-Cas9 crRNAs | IDT | See Table S1 |
| CRISPR HDR repair templates | Sigma-Aldrich | See Table S1 |
| Recombinant DNA | ||
| Pmig-13::ebp-1 | This Study | pJHP59 |
| Pmig-13::ebp-2 | This Study | pJHP70 |
| Pmig-13::ebp-1ΔCH | This Study | pJHP75 |
| Pmig-13::ebp-1_CH | This Study | pJHP76 |
| Pmig-13::ebp-1ΔCH_Y264A_E272A | This Study | pJHP105 |
| Pmig-13::ebp-1_EBH | This Study | pJHP162 |
| Pmig-13::EB1 (MAPRE1 of mus musculus) | This Study | pJHP72 |
| Pmig-13::EB2 (MAPRE2 of homo sapiens) | This Study | pJHP73 |
| Pmig-13::EB3 (MAPRE3 of homo sapiens) | This Study | pJHP74 |
| Pmig-13::ebp-1::RFP | This Study | pJHP163 |
| Pmig-13::RFP::rab-6.2 | This Study | pJHP87 |
| Pmig-13::unc-101::RFP | This Study | pJHP88 |
| Pmig-13::apm-3::RFP | This Study | pJHP89 |
| Pmig-13::RFP::golph-3 | This Study | pJHP91 |
| Pmig-13::aman-2_1–99::RFP | This Study | pJHP71 |
| Pmig-13::RFP::ebp-1ΔCH::bre-4 | This Study | pJHP95 |
| Pmig-13::RFP::ebp-1ΔCH::golgin-84 | This Study | pJHP96 |
| Pmig-13::RFP::ebp-1ΔCHΔC::bre-4 | This Study | pJHP119 |
| Pmig-13::RFP::ebp-1ΔCH_Y264A_E272A::bre-4 | This Study | pJHP120 |
| Pmig-13::RFP::bre-4 | This Study | pJHP121 |
| Pmig-13::RFP::syd-2_CC::tomm-7 | This Study | pJHP129 |
| Pmig-13::RFP::syd-2_CC::bre-4 | This Study | pJHP161 |
| Pmig-13::ATG-9::GFP | This Study | pJHP63 |
| Ptph-1::ebp-1 | This Study | pJHP145 |
| PCMV-EB1-GFP | This Study | pJHP164 |
| PCMV-EB1ΔCH-EGFP | This Study | pJHP165 |
| PCMV-EB1ΔC-EGFP | This Study | pJHP166 |
| PCMV-EB1_Y217A_E225A-EGFP | This Study | pJHP167 |
| PCMV-KIF1A-3XFLAG | This Study | pJHP168 |
| PCMV-KIF1A_MD-3XFLAG | This Study | pJHP169 |
| PCMV-KIF1A_stalk-3XFLAG | This Study | pJHP170 |
| PCMV-KIF5C-3XFLAG | This Study | pJHP171 |
| pET22b-mEB2-eGFP-6xHis | Addgene | Plasmid #130982 |
| EB3-tdTomato | Addgene | Plasmid #50708 |
| tdTomato-KIF1A | Addgene | Plasmid #102850 |
| pGFP-Kif5c | Addgene | Plasmid #71853 |
| Software and algorithms | ||
| Image J 1.49v | NIH | https://fiji.sc/ |
| MATHLAB 2022a | MathWorks | https://www.mathworks.com/products/matlab.html |
| Illustrator 27.0 | Adobe | https://www.adobe.com/de/products/illustrator.html |
| Prism 9 | Graphpad | https://www.graphpad.com/features |
Highlights.
EBP-1/EB1 is enriched near DCV biogenesis sites in C. elegans neurons.
ebp-1 mutants have reduced DCV delivery from cell body to axon.
EB1 interacts with kinesin-3 and EBP-1 orients microtubule growth on the Golgi.
Recruiting kinesin-3 to the Golgi restores DCVs to the axon in ebp-1 mutants.
Acknowledgments
The authors thank the CGC and NBRP Mitani lab for various strains. We also thank Kang Shen for strains and Sander van den Heuvel for SV1877 strain. We would also like to thank Michael Koelle for LX2519 strain as well as sharing resources for egg-laying behavior and Selim Çetinkaya for the initial worm husbandry for the project. We thank the members of the Yogev, De Camilli, and Hammarlund labs for discussions and technical assistance. This work was supported by NIH R35-GM133573 and DK45735.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Niwa S, Lipton DM, Morikawa M, Zhao C, Hirokawa N, Lu H, and Shen K (2016). Autoinhibition of a Neuronal Kinesin UNC-104/KIF1A Regulates the Size and Density of Synapses. Cell Rep 16, 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, Meyhofer E, and Verhey KJ (2009). Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol 7, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klopfenstein DR, Tomishige M, Stuurman N, and Vale RD (2002). Role of Phosphatidylinositol(4,5)bisphosphate Organization in Membrane Transport by the Unc104 Kinesin Motor. Cell 109, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar J, Choudhary BC, Metpally R, Zheng Q, Nonet ML, Ramanathan S, Klopfenstein DR, and Koushika SP (2010). The Caenorhabditis elegans Kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. PLoS Genet 6, e1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klopfenstein DR, and Vale RD (2004). The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol Biol Cell 15, 3729–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall DH, and Hedgecock EM (1991). Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65, 837–847. [DOI] [PubMed] [Google Scholar]
- 7.Stavoe AK, Hill SE, Hall DH, and Colon-Ramos DA (2016). KIF1A/UNC-104 Transports ATG-9 to Regulate Neurodevelopment and Autophagy at Synapses. Dev Cell 38, 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahn TR, Angleson JK, MacMorris MA, Domke E, Hutton JF, Schwartz C, and Hutton JC (2004). Dense core vesicle dynamics in Caenorhabditis elegans neurons and the role of kinesin UNC-104. Traffic 5, 544–559. [DOI] [PubMed] [Google Scholar]
- 9.Lo KY, Kuzmin A, Unger SM, Petersen JD, and Silverman MA (2011). KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett 491, 168–173. [DOI] [PubMed] [Google Scholar]
- 10.Yonekawa Y, Harada A, Okada Y, Funakoshi T, Kanai Y, Takei Y, Terada S, Noda T, and Hirokawa N (1998). Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol 141, 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pack-Chung E, Kurshan PT, Dickman DK, and Schwarz TL (2007). A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat Neurosci 10, 980–989. [DOI] [PubMed] [Google Scholar]
- 12.Gondre-Lewis MC, Park JJ, and Loh YP (2012). Cellular mechanisms for the biogenesis and transport of synaptic and dense-core vesicles. Int Rev Cell Mol Biol 299, 27–115. [DOI] [PubMed] [Google Scholar]
- 13.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, et al. (2006). Molecular anatomy of a trafficking organelle. Cell 127, 831–846. [DOI] [PubMed] [Google Scholar]
- 14.Li P, Merrill SA, Jorgensen EM, and Shen K (2016). Two Clathrin Adaptor Protein Complexes Instruct Axon-Dendrite Polarity. Neuron 90, 564–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ailion M, Hannemann M, Dalton S, Pappas A, Watanabe S, Hegermann J, Liu Q, Han HF, Gu M, Goulding MQ, et al. (2014). Two Rab2 interactors regulate dense-core vesicle maturation. Neuron 82, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sumakovic M, Hegermann J, Luo L, Husson SJ, Schwarze K, Olendrowitz C, Schoofs L, Richmond J, and Eimer S (2009). UNC-108/RAB-2 and its effector RIC-19 are involved in dense core vesicle maturation in Caenorhabditis elegans. J Cell Biol 186, 897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hummel JJA, and Hoogenraad CC (2021). Specific KIF1A-adaptor interactions control selective cargo recognition. J Cell Biol 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emperador-Melero J, Huson V, van Weering J, Bollmann C, Fischer von Mollard G, Toonen RF, and Verhage M (2018). Vti1a/b regulate synaptic vesicle and dense core vesicle secretion via protein sorting at the Golgi. Nat Commun 9, 3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Camilli P, and Jahn R (1990). Pathways to regulated exocytosis in neurons. Annu Rev Physiol 52, 625–645. [DOI] [PubMed] [Google Scholar]
- 20.Burgess J, Jauregui M, Tan J, Rollins J, Lallet S, Leventis PA, Boulianne GL, Chang HC, Le Borgne R, Kramer H, et al. (2011). AP-1 and clathrin are essential for secretory granule biogenesis in Drosophila. Mol Biol Cell 22, 2094–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akhmanova A, and Steinmetz MO (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9, 309–322. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi I, and Ikura M (2003). Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J Biol Chem 278, 36430–36434. [DOI] [PubMed] [Google Scholar]
- 23.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366–376. [DOI] [PubMed] [Google Scholar]
- 24.Honnappa S, John CM, Kostrewa D, Winkler FK, and Steinmetz MO (2005). Structural insights into the EB1-APC interaction. EMBO J 24, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maurer SP, Fourniol FJ, Bohner G, Moores CA, and Surrey T (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leterrier C, Vacher H, Fache MP, d’Ortoli SA, Castets F, Autillo-Touati A, and Dargent B (2011). End-binding proteins EB3 and EB1 link microtubules to ankyrin G in the axon initial segment. Proc Natl Acad Sci U S A 108, 8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, and Rolls MM (2010). Directed microtubule growth, +TIPs, and kinesin-2 are required for uniform microtubule polarity in dendrites. Curr Biol 20, 2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yogev S, Cooper R, Fetter R, Horowitz M, and Shen K (2016). Microtubule Organization Determines Axonal Transport Dynamics. Neuron 92, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang C, Wu J, de Heus C, Grigoriev I, Liv N, Yao Y, Smal I, Meijering E, Klumperman J, Qi RZ, et al. (2017). EB1 and EB3 regulate microtubule minus end organization and Golgi morphology. J Cell Biol 216, 3179–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guedes-Dias P, Nirschl JJ, Abreu N, Tokito MK, Janke C, Magiera MM, and Holzbaur ELF (2019). Kinesin-3 Responds to Local Microtubule Dynamics to Target Synaptic Cargo Delivery to the Presynapse. Curr Biol 29, 268–282 e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alves-Silva J, Sanchez-Soriano N, Beaven R, Klein M, Parkin J, Millard TH, Bellen HJ, Venken KJ, Ballestrem C, Kammerer RA, et al. (2012). Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins). J Neurosci 32, 9143–9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moughamian AJ, Osborn GE, Lazarus JE, Maday S, and Holzbaur EL (2013). Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J Neurosci 33, 13190–13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. (2009). Mammalian end binding proteins control persistent microtubule growth. J Cell Biol 184, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt R, Fielmich LE, Grigoriev I, Katrukha EA, Akhmanova A, and van den Heuvel S (2017). Two populations of cytoplasmic dynein contribute to spindle positioning in C. elegans embryos. J Cell Biol 216, 2777–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klassen MP, and Shen K (2007). Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell 130, 704–716. [DOI] [PubMed] [Google Scholar]
- 36.White JG, Southgate E, Thomson JN, and Brenner S (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314, 1–340. [DOI] [PubMed] [Google Scholar]
- 37.Hammarlund M, Watanabe S, Schuske K, and Jorgensen EM (2008). CAPS and syntaxin dock dense core vesicles to the plasma membrane in neurons. J Cell Biol 180, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison LM, Edwards SL, Manning L, Stec N, Richmond JE, and Miller KG (2018). Sentryn and SAD Kinase Link the Guided Transport and Capture of Dense Core Vesicles in Caenorhabditis elegans. Genetics 210, 925–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speese S, Petrie M, Schuske K, Ailion M, Ann K, Iwasaki K, Jorgensen EM, and Martin TF (2007). UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci 27, 6150–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Keimpema L, Kooistra R, Toonen RF, and Verhage M (2017). CAPS-1 requires its C2, PH, MHD1 and DCV domains for dense core vesicle exocytosis in mammalian CNS neurons. Sci Rep 7, 10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fares H, and Greenwald I (2001). Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brewer JC, Olson AC, Collins KM, and Koelle MR (2019). Serotonin and neuropeptides are both released by the HSN command neuron to initiate Caenorhabditis elegans egg laying. PLoS Genet 15, e1007896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee N, Bhattacharya R, Gorczyca M, Collins KM, and Francis MM (2017). Local neuropeptide signaling modulates serotonergic transmission to shape the temporal organization of C. elegans egg-laying behavior. PLoS Genet 13, e1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xuan Z, Manning L, Nelson J, Richmond JE, Colon-Ramos DA, Shen K, and Kurshan PT (2017). Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, et al. (2001). CLASPs Are CLIP-115 and −170 Associating Proteins Involved in the Regional Regulation of Microtubule Dynamics in Motile Fibroblasts. Cell 104, 923–935. [DOI] [PubMed] [Google Scholar]
- 46.Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, and Medema RH (2011). A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol 21, 1356–1365. [DOI] [PubMed] [Google Scholar]
- 47.van der Vaart B, Manatschal C, Grigoriev I, Olieric V, Gouveia SM, Bjelic S, Demmers J, Vorobjev I, Hoogenraad CC, Steinmetz MO, et al. (2011). SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J Cell Biol 193, 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sen I, Veprintsev D, Akhmanova A, and Steinmetz MO (2013). End binding proteins are obligatory dimers. PLoS One 8, e74448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song X, Yang F, Yang T, Wang Y, Ding M, Li L, Xu P, Liu S, Dai M, Chi C, et al. (2023). Phase separation of EB1 guides microtubule plus-end dynamics. Nat Cell Biol 25, 79–91. [DOI] [PubMed] [Google Scholar]
- 50.Maan R, Reese L, Volkov VA, King MR, van der Sluis EO, Andrea N, Evers WH, Jakobi AJ, and Dogterom M (2023). Multivalent interactions facilitate motor-dependent protein accumulation at growing microtubule plus-ends. Nat Cell Biol 25, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang K, Toedt G, Montenegro Gouveia S, Davey NE, Hua S, van der Vaart B, Grigoriev I, Larsen J, Pedersen LB, Bezstarosti K, et al. (2012). A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol 22, 1800–1807. [DOI] [PubMed] [Google Scholar]
- 52.Luo L, Hannemann M, Koenig S, Hegermann J, Ailion M, Cho MK, Sasidharan N, Zweckstetter M, Rensing SA, and Eimer S (2011). The Caenorhabditis elegans GARP complex contains the conserved Vps51 subunit and is required to maintain lysosomal morphology. Mol Biol Cell 22, 2564–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. (2007). Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell 12, 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, de Heus C, Liu Q, Bouchet BP, Noordstra I, Jiang K, Hua S, Martin M, Yang C, Grigoriev I, et al. (2016). Molecular Pathway of Microtubule Organization at the Golgi Apparatus. Dev Cell 39, 44–60. [DOI] [PubMed] [Google Scholar]
- 55.Jaulin F, and Kreitzer G (2010). KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J Cell Biol 190, 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ligon LA, Shelly SS, Tokito M, and Holzbaur EL (2003). The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol Biol Cell 14, 1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kornakov N, Mollers B, and Westermann S (2020). The EB1-Kinesin-14 complex is required for efficient metaphase spindle assembly and kinetochore bi-orientation. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris EJ, Nader GP, Ramalingam N, Bartolini F, and Gundersen GG (2014). Kif4 interacts with EB1 and stabilizes microtubules downstream of Rho-mDia in migrating fibroblasts. PLoS One 9, e91568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, and Walczak CE (2011). Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell 22, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin H, Wyszynski M, Huh KH, Valtschanoff JG, Lee JR, Ko J, Streuli M, Weinberg RJ, Sheng M, and Kim E (2003). Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J Biol Chem 278, 11393–11401. [DOI] [PubMed] [Google Scholar]
- 61.Wagner OI, Esposito A, Kohler B, Chen CW, Shen CP, Wu GH, Butkevich E, Mandalapu S, Wenzel D, Wouters FS, et al. (2009). Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans. Proc Natl Acad Sci U S A 106, 19605–19610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gotz TWB, Puchkov D, Lysiuk V, Lutzkendorf J, Nikonenko AG, Quentin C, Lehmann M, Sigrist SJ, and Petzoldt AG (2021). Rab2 regulates presynaptic precursor vesicle biogenesis at the trans-Golgi. J Cell Biol 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rizalar FS, Roosen DA, and Haucke V (2021). A Presynaptic Perspective on Transport and Assembly Mechanisms for Synapse Formation. Neuron 109, 27–41. [DOI] [PubMed] [Google Scholar]
- 64.Stucchi R, Plucinska G, Hummel JJA, Zahavi EE, Guerra San Juan I, Klykov O, Scheltema RA, Altelaar AFM, and Hoogenraad CC (2018). Regulation of KIF1A-Driven Dense Core Vesicle Transport: Ca(2+)/CaM Controls DCV Binding and Liprin-alpha/TANC2 Recruits DCVs to Postsynaptic Sites. Cell Rep 24, 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lord SJ, Velle KB, Mullins RD, and Fritz-Laylin LK (2020). SuperPlots: Communicating reproducibility and variability in cell biology. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dokshin GA, Ghanta KS, Piscopo KM, and Mello CC (2018). Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics 210, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, and Fire AZ (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herbert AD, Carr AM, and Hoffmann E (2014). FindFoci: a focus detection algorithm with automated parameter training that closely matches human assignments, reduces human inconsistencies and increases speed of analysis. PLoS One 9, e114749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Golgi-associated EBP-2 comets are reduced in ebp-1 mutant. Related to Figure 5.
Representative time-lapse movie of endogenously labeled EBP-2 (green) and RFP labeled UNC-101/AP-1μ (magenta) in DA9 neuron cell body. A number of EBP-2 comets are associated with UNC-101 in wildtype. In ebp-1 mutants, the number of comets associated with UNC-101::RFP is reduced. To avoid ambiguity, comet signals that completely overlap with UNC-101::RFP were counted as associated.
Data Availability Statement
Raw microscopy data and western blots reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


