Abstract
Programmable RNA-guided DNA nucleases perform numerous roles in prokaryotes, but the extent of their spread outside prokaryotes is unclear. Fanzors, the eukaryotic homolog of prokaryotic TnpB proteins, have been detected in genomes of eukaryotes and large viruses, but their activity and functions in eukaryotes remain unknown. Here, we characterize Fanzors as RNA-programmable DNA endonucleases, using biochemical and cellular evidence. We found diverse Fanzors that frequently associate with various eukaryotic transposases. Reconstruction of Fanzors evolution revealed multiple radiations of RuvC-containing TnpB homologs in eukaryotes. Fanzor genes captured introns and proteins acquired nuclear localization signals, indicating extensive, long-term adaptation to functioning in eukaryotic cells. Fanzor nucleases contain a rearranged catalytic site of the RuvC domain, similar to a distinct subset of TnpBs, and lack collateral cleavage activity. We demonstrate that Fanzors can be harnessed for genome editing in human cells, highlighting the potential of these widespread eukaryotic RNA-guided nucleases for biotechnology applications.
Fanzor is an RNA-guided DNA endonuclease widespread in eukaryotes and their viruses.
INTRODUCTION
RNA-programmable DNA nucleases serve multiple roles in prokaryotes, including in mobile element defense and spread. These nucleases include argonaut, CRISPR, and the obligate mobile element–guided activity (OMEGA) systems, the latter of which include the TnpB, IscB, IsrB, and IshB nucleases. TnpB contains a RuvC-like nuclease domain [ribonuclease H (RNase H) fold] that is related to the nuclease domain of the type V CRISPR effector Cas12 (1, 2), specifically, Cas12f (3), suggesting a direct evolutionary path from TnpB enzymes to Cas12 (4–6). This relationship is supported by phylogenetic analysis of the RuvC-like domains, which indicates independent origins of Cas12s of different type V subtypes from distinct groups of TnpBs (6, 7). OMEGA systems encode the guide ωRNA adjacent to the nuclease gene, often overlapping the coding region, and biochemical and cellular experiments demonstrated that the ωRNA-TnpB complex is indeed an RNA-guided, programmable DNA endonuclease (4, 6).
RuvC domain–containing proteins are not limited to prokaryotes: A set of TnpB homologs, Fanzors, are present in eukaryotes (5). Mirroring the diversity of TnpBs in bacteria and archaea, Fanzors have been identified in diverse eukaryotic lineages, including metazoans, fungi, algae, amorphea, and some large double-stranded DNA (dsDNA) viruses. The identified Fanzors fall into two major groups: (i) Fanzor1 proteins are associated with eukaryotic transposons, including Mariners, IS4-like elements, Sola, Helitron, and MuDr, and occur predominantly in diverse eukaryotes; (ii) Fanzor2 proteins are found in IS607-like transposons and are present in dsDNA viral genomes. Despite the similarities between TnpB and Fanzors, Fanzors have not been surveyed comprehensively throughout eukaryotic diversity and have not been characterized experimentally.
Here, we report a comprehensive census of RNA-guided nucleases in eukaryotic and viral genomes, discovering a broad class of functional nucleases that have extensively spread within eukaryotes and their viruses. We examine the diversity of Fanzor systems in eukaryotes, perform a phylogenetic analysis to trace their evolution, and demonstrate their programmable, RNA-guided endonuclease activity biochemically and in cells, showcasing their utility as new genome editing tools.
RESULTS
Fanzor nucleases are TnpB homologs widespread in eukaryotes and viruses
We identified putative RNA-guided nucleases across 22,497 eukaryotic and viral assemblies from National Center for Biotechnology Information (NCBI) GenBank by searching for similarity to a multiple alignment of RuvC domains from known Fanzor1 and Fanzor2 proteins (5). We found 3655 putative nucleases with unique sequences (using a 70% similarity clustering threshold) that occurred across metazoans, fungi, choanoflagellates, algae, rhodophyta, diverse unicellular eukaryotes, and multiple viral families (Fig. 1, A and B), expanding the known diversity of eukaryotic RuvC homologs over 100-fold (Fig. 1A). These Fanzor homologs frequently occur in multiple copies across eukaryotic genomes, with some genomes carrying up to 122 copies. This widespread of the Fanzors is strongly suggestive of intragenomic mobility, similar to TnpBs (fig. S1A). Fanzor proteins also are typically substantially larger than TnpB, with a mean size of 620 residues, compared to 480 residues for TnpB proteins (Fig. 1C).
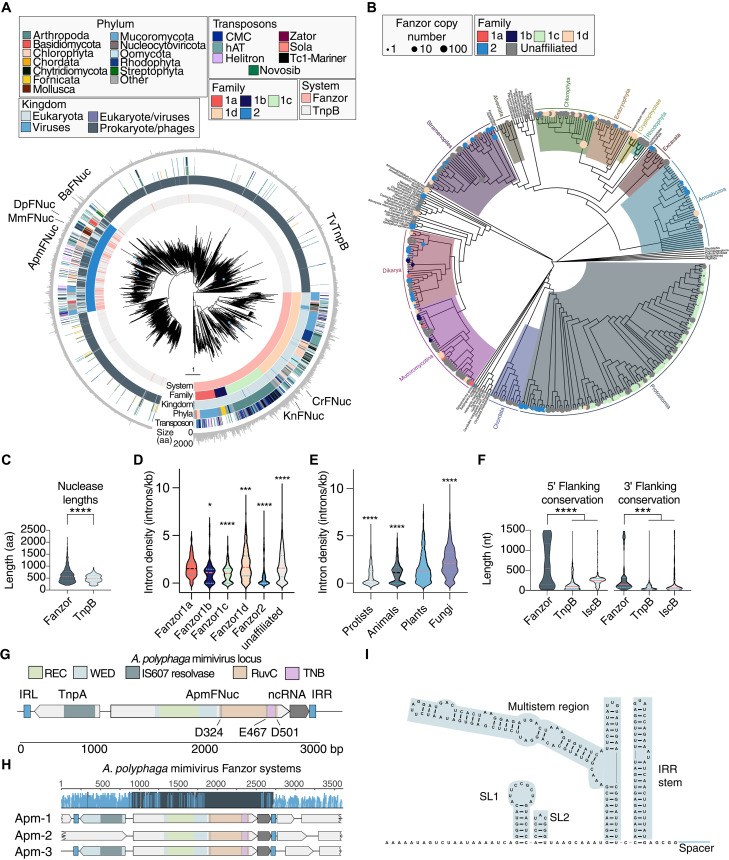
Fig. 1. Evolution of Fanzor nucleases and their association with noncoding fRNAs.
(A) Phylogenetic tree of representative Fanzor and TnpB proteins. The rings show protein system, Fanzor family designation, host superkingdom, phyla of their host species predicted associated transposons, and protein length from inside to outside. Proteins studied in this work are marked around the tree. Splits with bootstrap support less than 0.7 of 1 were collapsed, and the tree was rooted at the midpoint. (B) Fanzor systems projected onto the evolutionary tree of eukaryotes (33). Nodes and tips of the tree are marked with circles if there are Fanzors in the corresponding taxonomic group. Circle sizes are proportional to the Fanzor copy number and colored by family. (C) Comparison of protein lengths [amino acids (aa)] between Fanzor nucleases and TnpB nucleases (*P < 0.05; ****P < 0.0001, two-sided t test). (D) Intron density of Fanzor genes grouped by assigned families. A two-sided Student’s t test with multiple hypothesis correction is performed between each family against the rest (****P < 0.0001; ***P < 0.001). (E) Two-sided Student’s t test with multiple hypothesis correction is performed between each kingdom and the rest (***P < 0.001; ****P < 0.0001). (F) Comparison of predicted flanking noncoding conservation lengths at the 5′ end and 3′ end of the MGEs of IscB, TnpB, and Fanzor systems [****P < 0.0001, one-way analysis of variance (ANOVA)]. (G) Schematic of the A. polyphaga mimivirus (ApmFNuc) system, including the Fanzor ORF, associated IS607 TnpA, the noncoding RNA region, and the left and right inverted repeat elements (ILR and IRR). The WED, RuvC, and REC domains are annotated based on structural similarity with the Isdra2 TnpB structure (11). (H) Conservation of the three Fanzor loci in the A. polyphaga mimivirus genome, showing high conservation of the Fanzor protein-coding regions and the nearby noncoding regions. (I) RNA secondary structure of the conserved 3′ noncoding region from (H).
Phylogenetic analysis of our expanded set of Fanzor nucleases and a selection of closely related TnpBs revealed five distinct Fanzor clades supported by bootstrap analysis, with four Fanzor1 families (Fanzor1a to Fanzor1d) and a single Fanzor2 clade (Fig. 1A). In addition, there are a number of unaffiliated Fanzor systems that could not confidently be assigned to any Fanzor family based on phylogeny. Fanzors are each broadly represented in diverse eukaryotes, and Fanzor2 shows a pronounced enrichment of virus-encoded Fanzors (18.4%, P < 1017), including Phycodnaviridae, Ascoviridae, and Mimiviridae (Fig. 1A). Fanzor proteins often contain various domains, in addition to the RuvC-like nuclease domain; in particular, Fanzor2 members contain a helix-turn-helix domain, mimicking the domain architecture of the TnpBs (fig. S1B). Furthermore, direct comparison of specific Fanzors and their closest TnpBs further supports the close evolutionary relationship between these enzymes (fig. S1, C and D). In all families, Fanzors are interspersed with TnpBs, suggesting multiple acquisitions of TnpB during the evolution of eukaryotes. Moreover, TnpB-containing clades that include sparse Fanzors might reflect direct acquisitions from symbiotic bacteria (Fig. 1A).
Projecting Fanzor hosts onto the eukaryotic tree of life shows broad spread into amoebozoa, several other groups of unicellular eukaryotes, plants, fungi, and animals, including Chordata and Arthopoda (Fig. 1B). Notably, assimilation of Fanzors in eukaryotic genomes was accompanied by intron acquisition: Numerous Fanzor loci have intron densities similar to those in host genes, up to ~9.6 introns/kb (Fig. 1, D and E, and fig. S2).
Fanzor nucleases associate with diverse transposons
Fanzors commonly associate with different transposons (5). We performed a comprehensive transposon search (8) within 10 kb of Fanzors, analyzing the identity of the associated open reading frames (ORFs) by domain search (Fig. 1A; fig. S3, A and B; and table S1). Among eukaryotic transposons, we found both previously reported transposon families, including Mariner/Tc1, Helitron, and Sola, and families not previously known to associate with Fanzors, including hAT and CMC DNA transposons (fig. S3A and table S1). Fanzor-transposon associations included autonomous transposons encoding a transposase, such as in the Crypton and Mariner/Tc1 families, as well as non-autonomous transposons including only transposon ends, such as hAT, EnSpm, and Helitron families (fig. S3, A to D and table S1). Notably, the most frequent associations were with the DNA transposon hAT, suggesting that Fanzors might have some role with these transposons in the respective eukaryotic genomes. Fanzor1a, b, and d clades are most commonly associated with hAT, whereas Fanzor1c preferentially associated with LINE, CMC, and Mariner/Tc1 transposons (Fig. 1A and fig. S3, A to D). Fanzor2s associated with diverse transposons, including, Helitron, hAT, and IS607 (Fig. 1A and fig. S3, B to D). The IS607 transposons encode a TnpA-like transposase, further cementing the close relationship between Fanzor2 and TnpBs.
Fanzors are associated with conserved, structured noncoding RNAs
TnpB and IscB nucleases process the ends of the transposon-encoded RNA transcript into ωRNA, which complex with the respective nucleases to form a RNA-guided dsDNA endonuclease ribonucleoprotein (RNP) (4, 6, 9). We searched Fanzor loci for putative regions encoding OMEGA-like RNAs, based on conservation of noncoding sequence. We found conservation extending beyond the detectable Fanzor ORF on both 5′ and 3′ ends of the ORF, with the conserved regions substantially longer for some Fanzor families than those in TnpB and IscB loci, although some families like the viral-enriched Fanzor2 have noncoding lengths similar to those of TnpB systems (Fig. 1F and fig. S3, E and F). These conserved regions indicate either strong conservation within the transposon boundaries or longer guide RNAs associated with Fanzor enzymes.
To explore the potential activity and expression of these conserved regions, we selected the Fanzor2 from the Acanthamoeba polyphaga mimivirus (ApmFNuc) that is encoded within a IS607 transposon and contains a TnpA transposase and defined inverted terminal repeats (Fig. 1E). The A. polyphaga mimivirus genome contains three IS607 copies, which show strong sequence conservation, both within the protein-coding regions but also in the noncoding region at the 3′ ends of the IS607 MGE (Fig. 1, E and F). This noncoding sequence conservation extended 200 base nucleotides (nt) past the end of ApmFNuc ORF, ending upstream of the right inverted repeat (IRR), designating the right end (RE) of the MGE (Fig. 1G). In silico RNA secondary structure analysis predicted a stable fold (Fig. 1H and fig. S3E), suggesting that the transcript of this conserved region could function as a Fanzor-associated guide RNA, which we accordingly named Fanzor RNA (fRNA). In the alignment of ApmFNuc loci, the predicted fRNA structure was highly conserved, with the conservation extending upstream into the coding region of ApmFNuc, indicating possible cofolding with this portion of the coding region and potential RNA processing site (Fig. 1I and fig. S3G). This apparent RNA structure conservation is reminiscent of the OMEGA families, where both the IscB and TnpB families show limited structural variation (6), and processing of the upstream region of the mRNA releases functional guide RNAs (9).
Viral-encoded ApmFNuc is a fRNA-guided DNA endonuclease
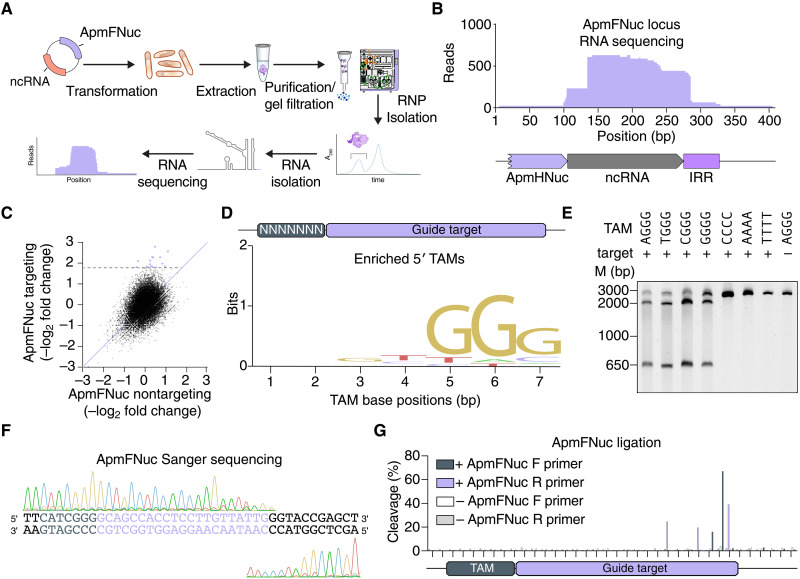
We hypothesized that the fRNA forms a complex with ApmFNuc and directs binding and DNA cleavage to a specific sequence in the target. To investigate potential fRNA-ApmFNuc binding, we coexpressed in Escherichia coli the A. polyphaga mimivirus Fanzor locus, containing the noncoding RNA region, and an E. coli codon-optimized ApmFNuc (Fig. 2A and table S2). Notably, ApmFNuc protein was unstable when expressed alone and required coexpression with its fRNA for protein stabilization and accumulation (fig. S4), similar to the instability of TnpB in the absence of ωRNA (4, 6). We purified the fRNA-ApmfNuc RNP and sequenced the RNA component of the complex. Small RNA sequencing revealed enriched coverage between the 3′ ends of the protein ORF and the IRR, in agreement with the evolutionary conservation across the region (Fig. 2B).
Fig. 2. Viral Fanzor RNPs can be programmed to cleave DNA targets in vitro.
(A) Schematic of the method used for identifying the ApmFNuc-associated noncoding RNA (ncRNA). The ApmFNuc protein is copurified with its noncoding RNA, allowing for the isolation of the noncoding RNA species and identification by small RNA sequencing. (B) RNA sequencing coverage of the ApmFNuc-1 noncoding RNA region showing robust expression of the noncoding RNA and its guide sequence extending past the IRR element. (C) Scatterplots of the fold change of individual TAM sequences in a 7N library plasmid relative to input plasmid library distribution with either ApmFNuc RNP with a targeting fRNA or a nontargeting fRNA. (D) Sequence motif of TAM preference computed from depleted TAMs, showing an NGGG-rich tam preference. (E) Biochemical validation of individual ApmFNuc TAM sequences including four preferred TAMs (TGGG, AGGG, CGGG, and GGGG) as well as 3 non-TAM sequences and 1 nontargeting sequence. ApmFNuc RNP is incubated with DNA targets containing each of these sequences, and cleavage is visualized by gel electrophoresis on 6% TBE gel. (F) Sanger sequencing traces of ApmFNuc RNP cleavage on the 5′ CGGG TAM target, showing cleavage downstream of the guide target. (G) NGS mapping of the TAM cleavage by ApmFNuc via NEB adaptor ligation. Cleavage products from in vitro cleavage reactions were prepared for sequencing via ligation of sequencing adaptors and PCR before NGS. Reads were aligned to the TAM target to map cleavage locations. Two separate reactions were ran in parallel with and without addition of ApmFNuc RNP. The cleavage products were amplified in both 5′ and 3′ directions with F denoting 3′ direction and R denoting the 5′ direction.
Testing RNP cleavage activity required both the engineering of a reprogrammed fRNA and the determination of any sequence preferences, akin to the target adjacent motif (TAM) in the case of TnpB and IscB (4, 6). We combined a 3′-terminal 21-nt targeting sequence with the fRNA scaffold determined through RNA profiling to engineer a synthetic fRNA, coexpressed the synthetic fRNA and ApmFNuc in E. coli, and isolated the reprogrammed RNP complex. To determine potential sequence preferences of ApmFNuc, we tested cleavage on a DNA target containing a randomized 7-nt TAM 5′ of a 21-nt target region complementary to the fRNA targeting sequence. We coincubated this TAM library with purified ApmFNuc RNPs containing either targeting or scrambled synthetic fRNA guide sequences and profiled the relative depletion of sequences with next-generation sequencing (NGS). TAM depletion analysis revealed a strong 5′ GGG motif adjacent to the target site (Fig. 2, C and D). We validated robust ApmFNuc activity on all possible NGGG TAMs, with no detectable cleavage of sequences lacking the TAM (Fig. 2E). In contrast to the G-rich ApmFNuc TAM, TnpB homologs of ApmFNuc universally prefer an A/T rich 5′ TAM (9). The GGG motif is present at the start of ApmFNuc MGE sequence and likely contributed to the TAM preference of ApmFNuc.
Cleavage locations of RNA-guided nucleases vary substantially, with cleavage sites located either upstream or downstream of the target sequence. To profile ApmFNuc cleavage patterns, we purified ApmFNuc reaction products and mapped the locations of the cleavage ends using Sanger sequencing. Cleavage occurred in the 3′ regions of the target sequence, with multiple nicks in both the target strand (TS) and the nontarget strand (NTS) (Fig. 2F). The cleavage behavior of ApmFNuc at the 3′ end of the target is similar to the cleavage patterns of Cas12 or TnpB nucleases and in general agreement with the properties of programmable RuvC domains (1, 4, 6). We sensitively quantified the relative preference for these different nicking sites using an NGS-based assay, finding that during dsDNA cleavage by ApmFNuc, the enzyme generated nicks in the NTS at positions 19 and 20 and in the TS at positions 15, 18, and 21 with all cleavage occurring inside the target region, indicating a slightly different cleavage pattern compared to TnpB nucleases (Fig. 2G).
fRNA-guided DNA endonucleases are present in diverse eukaryotic genomes
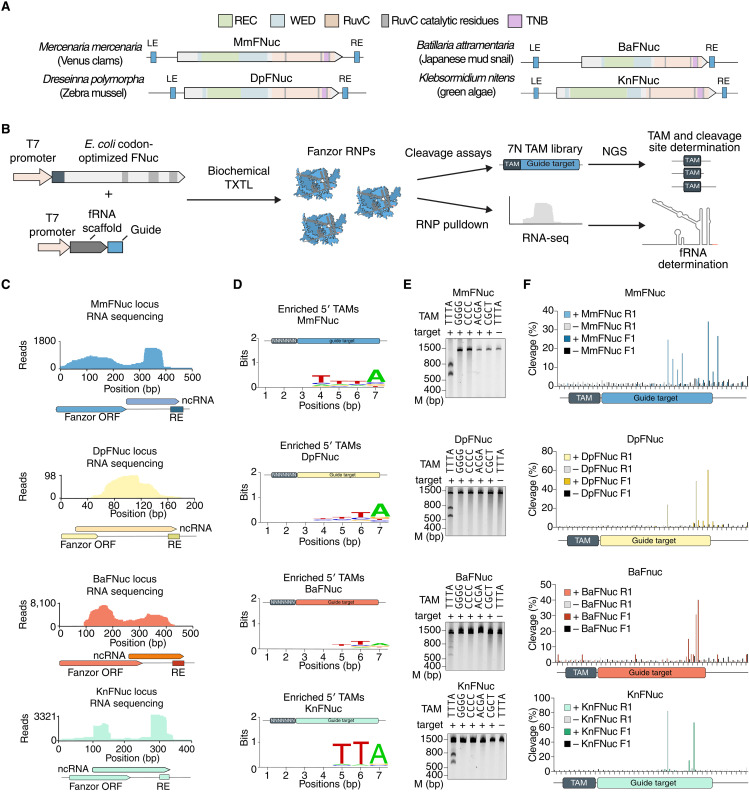
After demonstrating the activity of a viral Fanzor2, we sought to explore whether Fanzor2 proteins from diverse eukaryotes also are active RNA-guided nucleases. To this end, we chose three Fanzor2 representatives from three animals and a Fanzor 1 representative from a plant: (i) Fanzor2 from Mercenaria mercenaria (Venus clam; MmFNuc), (ii) Fanzor2 from Dreissena polymorpha (Zebra mussel; DpFNuc), (iii) Fanzor2 from Batillaria attramentaria (Japanese mud snail; BaFNuc), and (iv) Fanzor 1 from Klebsormidium nitens (freshwater green algae; KnFNuc) (Fig. 3A). MmFNuc, DpFnuc, BaFnuc, and KnFNuc are all represented by multiple copies in the respective organisms, with 7, 24, 5, and 5 copies per genome, respectively (Fig. 3A and fig. S5A), suggesting recent mobility of their associated transposons. We cloned constructs for coexpression of the fRNA and Fanzor nuclease in a cell-free transcription/translation system, allowing for isolation of the resulting RNPs to study their fRNA sequences and cleavage activity (Fig. 3B). We affinity-purified the RNPs and sequenced the bound fRNAs, demonstrating that all four Fanzors copurified with an RNA species derived from the 3′ noncoding region abutting the transposon RE (Fig. 3C). These fRNAs were highly structured with diverse structural motifs and domains (fig. S5B).
Fig. 3. Eukaryotic Fanzor orthologs are widespread across eukaryotic kingdoms, associate with fRNAs, and are RNA-guided nucleases.
(A) Locus schematics of four eukaryotic Fanzor systems from M. mercenaria, Dreseinna polymorpha, B. attramentaria, and K. nitens. WED, REC, and RuvC domains are identified by sequence and structural alignment with Isdra2 TnpB (11). (B) Schematic of screening for fRNA expression, TAM, activity, and cleavage locations via cell-free transcription/translation. RNA-seq, RNA sequencing. (C) Left: Small RNA sequencing of the MmFNuc locus showing expression of a noncoding RNA species extending outside the ORF. Middle: Weblogo visualization of the TAM sequence preference of MmFNuc identified by adaptor ligation assay on a 7N TAM library incubated with MmFNuc protein and fRNA. Right: Validation of MmFNuc cleavage by incubating the MmFNuc RNP with its correct TTTA TAM, four mutated TAMs, and a nontargeted plasmid. (D) Small RNA sequencing of the MmFNuc, DpFNuc, BaFNuc, KnFNuc, and loci showing expression of a noncoding RNA species extending outside the ORF. (E) Weblogo visualization of the TAM sequence preference of tested Fanzors identified by adaptor ligation assay on a 7N TAM library incubated with RNP. (E) Validation of Fanzor cleavage by incubation of RNP with their corresponding TAM, four mutated TAMs, and a nontargeted plasmid. (F) NGS mapping of the cleavage positions by MmFNuc, DpFNuc, and BaFNuc via NEB adaptor ligation of cleaved DNA targets that were incubated with the respective RNP complexes. Cleavage products from in vitro cleavage reactions were prepared for sequencing via ligation of sequencing adaptors and PCR before NGS. Reactions were performed with and without addition of each Fanzor RNP. The cleavage products were amplified in both 5′ and 3′ directions with F denoting 3′ direction (top) and R denoting the 5′ direction (bottom).
We next challenged a 7N TAM library with MmFNuc, DpFNuc, BaFNuc, and KnFNuc RNPs with fRNA guide sequences complementary to the library target, finding strong TAM selection corresponding to TTTA, TA, TTA, and TTA TAMs for MmFNuc, DpFNuc, BaFNuc, and KnFNuc, respectively (Fig. 3D). Incubation of RNPs with individual preferred TAMs showed robust cleavage, validating all four eukaryotic Fanzor enzymes as RNA-guided nucleases (Fig. 3E). As with ApmFNuc, these Fanzors generated multiple nicks in the top and bottom DNA strands near the 3′ end of the target (Fig. 3F). Specific cleavage sites showed diversity, with MmFNuc and KnFNuc nicking more upstream and downstream within the guide target sequence than DpFNuc or BaFNuc (Fig. 3F). KnFNuc produced highly focused nicks in both the top and the bottom strands rather than multiple nicks, suggesting mechanistic differences between Fanzor1 and Fanzor2 nucleases.
Given that ApmFNuc, MmFNuc, DpFNuc, BaFNuc, and KnFNuc all lack introns, we evaluated an intron-containing Fanzor1c from the unicellular green alga Chlamydomonas reinhardtii (CrFNuc) (fig. S6, A to C). There are six CrFNuc copies in the genome, and they are all associated with Helitron 2 transposons, which contain identifiable short target site duplications and asymmetrical terminal inverted repeats. Small RNA sequencing of a C. reinhardtii isolate showed strong enrichment of noncoding RNAs aligning to the 3′ untranslated region of the Cr-1 Fanzor mRNA (fig. S6D), which was strongly conserved across all six copies CrFNuc-1 (fig. S6, A and B). Computational secondary structure prediction for the CrFNuc-1 fRNA with the fRNAs of the other five loci revealed a conserved stable secondary structure with a conserved upstream region not present in the RNA sequencing trace, suggesting possible RNA processing of this region to serve as a guide RNA for CrFNuc-1 (fig. S6, E and F). Searches for similar sequences across the C. reinhardtii genome identified 20 additional distinct but highly conserved copies of the fRNA (fig. S6G). Coexpression of CrFNuc-1 either with its native fRNA on the 3′ end of the MGE or a scrambled RNA sequence produced stable RNP only when coexpressed with its fRNA, similar to ApmFNuc (fig. S6, H and I). However, when we coincubated the RNP with the 7N randomized TAM library plasmids, we did not detect cleavage, suggesting either failure to reconstitute the RNP activity under our experimental conditions or a lack of endonuclease activity of the native CrFNuc-1.
Fanzor nucleases contain a conserved rearranged catalytic site and lack collateral activity
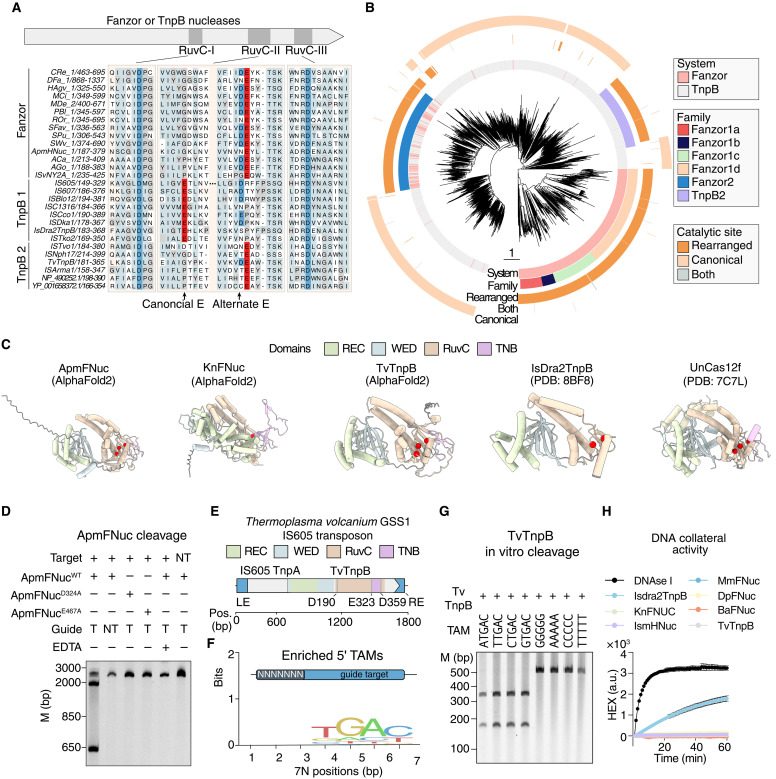
Alignment of Fanzor nucleases and TnpB members shows that, compared to the majority of TnpBs, Fanzor nucleases contain a substitution in the catalytic RuvC-II motif from a glutamate to a catalytically inert residue (proline or glycine) (Fig. 4A). To find TnpBs clades with this substitution, we searched for similarly modified RuvC nuclease domains among the TnpBs. We found a similar apparent inactivation of RuvC-II in TnpBs across multiple clades, including a monophyletic group, which we termed TnpB2, in contrast to canonical TnpB1 (Fig. 4, A and B). Given the demonstrated nuclease activity of ApmFNuc, we then searched for conserved acidic residues that could potentially compensate for the RuvC-II–inactivating mutations. All Fanzor proteins and TnpBs with a loss of the canonical glutamic acid in RuvC-II contained an alternative conserved glutamate approximately 45 residues away (Fig. 4, A and B).
Fig. 4. Rearranged RuvC catalytic residues enable Fanzor and TnpB on-target cleavage without collateral activity.
(A) Alignment of the RuvC domains of Fanzor and TnpB nucleases (TnpB2) showing the alternative glutamate in RuvC-II versus the canonical glutamate that is typically observed in TnpB nucleases (TnpB1). (B) Phylogenetic tree of TnpB and Fanzor proteins, showing TnpBs and Fanzor nucleases with rearranged catalytic sites. (C) Predicted AlphaFold2 structure of ApmFNuc and TvTnpB compared with the solved structures of Isdra2TnpB and Uncas12f, showing that despite having a rearranged glutamate in the RuvC catalytic domain, the catalytic aspartates and glutamates form a putative active catalytic triad (red residues). Domains identified are highlighted in specific colors, and the disordered N-terminal region is colored dark gray. PDB, Protein Data Bank. (D) ApmFNuc RNP purified with either targeting (T) or nontargeting (NT) fRNAs as well as two catalytic dead ApmFNuc mutants (D324A and E467A) is tested on either a plasmid containing the correct target spacer DNA sequences, or a scrambled DNA sequence containing the 5′ TAM TGGG. EDTA is added in lane 5 to quench the cleavage reaction. (E) Schematic of the T. volcanium GSS1TnpB (TvTnpB) system, including the TnpB with a rearranged catalytic site, associated IS605 TnpA, and the LE and RE elements. (F) Sequence logo of the TAM for TvTnpB. (G) Biochemical validation of individual TAM preference by TvTnpB showing that the cleavage by TvTnpB is TAM (NTGAC) specific. TvTnpB RNP is incubated with targets containing different 5′ TAMs, and cleavage is visualized by gel electrophoresis. (H) ApmFNuc, TvTnpB, MmFNuc, DpFNuc, BaFNuc, and Isdra2TnpB DNA collateral cleavage activity are measured using an single-stranded DNA fluorescent reporter, showing a lack of collateral activity for nucleases with the rearranged glutamic acid in RuvC-II. Deoxyribonuclease I (DNase I) is used as a positive nuclease control for collateral cleavage activity. a.u., arbitrary units.
We compared AlphaFold2-generated structural models of ApmFNuc, MmFNuc, DpFNuc, BaFNuc, KnFNuc, and a TnpB from Thermoplasma volcanium GSS1 (TvTnpB) that both contain a rearranged catalytic site with the cryo–electron microscopy structures of TnpB from Deinococcus radiodurans R1 (Isdra2) and Cas12f from uncultured archaeon (UnCas12f) containing the canonical catalytic site (Fig. 4C and fig. S7A) (10, 11). This comparison showed that the alternative conserved glutamate of Fanzor nucleases and rearranged TnpB (E467 of ApmFNuc and E323 of TvTnpB) were in close proximity with the catalytic residues in the RuvC-I and RuvC-III motifs, suggesting that these alternative, conserved glutamates compensate for the mutation in RuvC-II (Fig. 4C and fig. S7A).
To test the predicted role of the conserved alternative glutamate in Fanzor activity, we purified two ApmFNuc RNP with mutations at predicted catalytic sites in RuvC-I (D324A) or the alternative glutamate in RuvC-II (E467A) (fig. S7, B to D). While the D324A mutant showed no change in the RNP stability during protein purification, we noticed a substantial decrease in the expression of the E467A mutant relative to the wild-type protein (fig. S3B). We compared the cleavage efficiencies of these mutants with that of the wild-type ApmFNuc and found, in agreement with the nuclease mechanism, that both RuvC-I and RuvC-II mutants abolished ApmFNuc cleavage activity (Fig. 4D). Thus, the alternative Fanzor glutamate is indeed essential for the nuclease activity. Activity required a temperature range of 30° and 40°C for optimal activity, similar to other mesophilic RuvC nucleases, needed complexing with magnesium or a compensatory metal ion, and was robust across a range of salt concentrations (fig. S7, E to G).
We profiled the activity of the TnpB2, TvTnpB, to determine whether these rearranged TnpBs were similarly active. We isolated TvTnpB RNPs by coexpressing the enzyme with its native locus in E. coli and profiled associated noncoding RNA by NGS (fig. S8). Expression of the noncoding RNA species mapped proximal to the RE element, similar to other TnpB systems (Fig. 4E and fig. S9A). Applying our TAM assay by coexpressing TvTnpB with a synthetic ωRNA containing a reprogrammed 21-nt spacer, incubating the RNP with a 7N TAM library plasmid, and sequencing the cleavage products, we found strong enrichment of a TGAC motif near the 5′ target spacer sequence (Fig. 4F). Notably, this TGAC motif is also present at the 5′ end of the left end (LE), marking the beginning of the TvTnpB-encoding transposon. Because T. volcanium is a thermophile, we optimized in vitro cleavage efficiency over a range of temperatures and determined the optimal temperature for cleavage at the TGAC TAM at 60°C (fig. S9B). We validated all four possible NTGAC TAM sequences along with four negative TAM sequences and found TAM-specific cleavage, similar to other Fanzors and TnpB nucleases (Fig. 4G). We profiled the ends of the cleavage products with NGS, mapping the cleavage position to position 22 in the nontargeting strand and positions 21 and 22 in the targeting strand (fig. S9C), with a similar cleavage pattern found by Sanger sequencing (fig. S9D).
Although the rearranged RuvC catalytic site of the Fanzors and TnpB2 did not affect on-target cleavage, we hypothesized that it could affect the collateral cleavage activity of the enzyme (8, 12). We profiled ApmFNuc, MmFNuc, DpFNuc, BaFNuc, TvTnpB, and the canonical TnpB Isdra2TnpB for either RNA or DNA collateral cleavage activity by coincubating the RNP complexes with their cognate targets along with either RNA or DNA cleavage reporters, single-stranded nucleic acid substrates functionalized with a quencher and fluorophore that become fluorescent upon nucleolytic cleavage. We found that, while all nucleases had similar on-target cleavage efficiencies (fig. S9E), the Fanzor orthologs and TvTnpB lacked detectable collateral DNA and RNA cleavage activity in contrast to the strong collateral cleavage activity Isdra2TnpB (Fig. 4H and fig. S9F) (4).
Fanzor nucleases contain nuclear localization signals and are functional for mammalian genome editing
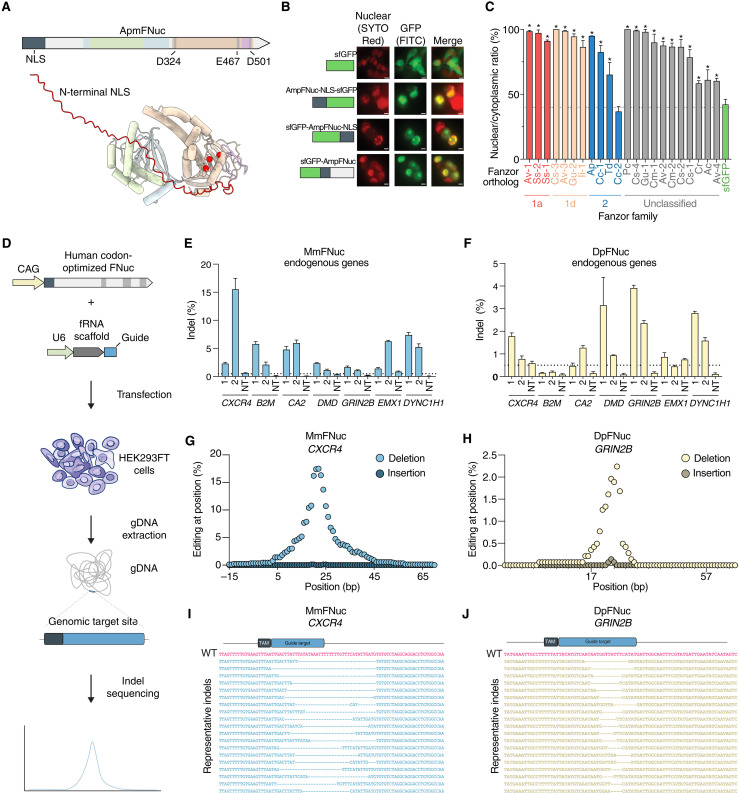
As eukaryotic RNA-guided endonucleases would need to enter the nucleus to access their genomic targets, we hypothesized that Fanzor nucleases might have harbor nuclear localization signals (NLS) to actively cross the nuclear membrane. In the AlphaFold2-predicted structures of ApmFNuc, we identified a disordered region of 64 amino acids at the N terminus (Fig. 5A). Computational prediction of the NLS identified a strong, positively charged NLS within the N-terminal region of ApmFNuc (fig. S10A).
Fig. 5. Fanzor nucleases contain NLS and have mammalian genome editing activity.
(A) Schematic of ApmFNuc showing the split RuvC domain and the predicted N-terminal NLS. NLS is colored in red, and the catalytic triad is shown as red space filling residues inside the cyan RuvC domain on the AF2-predicted ApmFNuc structure. (B) Confocal images of unmodified sfGFP, the predicted ApmFNuc NLS fused to sfGFP on either the N-terminal or C-terminal end, and sfGFP fused directly to the N terminus of ApmFNuc transfected into HEK293FT cells and stained with SYTO Red nuclear stain. Images display the nuclear stain (red), GFP signal (green), and a merged image. Scale bars, 10 μm. FITC, fluorescein isothiocyanate. (C) A quantitative analysis of 22 predicted Fanzor NLS sequences. Putative NLS sequences are fused to the N terminus of sfGFP, and the nuclear to cytoplasmic ratio of GFP fluorescence is quantitated (n = 3, *P < 0.01; one-way ANOVA with false discovery rate correction). (D) Schematic of Fanzor nucleases adapted for genome editing in mammalian cells. (E) The indel formation rates generated by MmFNuc across seven selected endogenous loci. For each locus, two fRNA guide sequences were tested, and a nontargeting guide is used as a negative control. (F) The indel formation rates generated by DpFNuc across seven selected endogenous loci. For each locus, two fRNA guide sequences were tested, and a nontargeting guide is used as a negative control. (G) Insertion and deletion rates at each base inside the quantification window generated by MmFNuc at the CXCR4 genomic locus. (H) Insertion and deletion rates at each base inside the quantification window generated by DpFNuc at the GRIN2b genomic locus. (I) Representative indel reads formed by MmFNuc at the CXCR4 genomic locus. (J) Representative indel reads formed by DpFNuc at the GRIN2b genomic locus. WT, wild type.
To evaluate the localization of ApmFNuc and its NLS, we fused superfolder green fluorescent protein (sfGFP) to the N terminus of ApmFNuc and attached the N-terminal portion of ApmFNuc containing the NLS to either the N terminus or C terminus of sfGFP. We visualized sfGFP localization via fluorescent microscopy, finding that sfGFP with the NLS from ApmFNuc fused to either terminus had strong nuclear localization (Fig. 5B). Fusion of sfGFP with the complete ApmFNuc also caused strong nuclear localization of sfGFP (Fig. 5B). These results suggest that ApmFNuc indeed contains a functional NLS, likely acquired after the capture of TnpBs by eukaryotes.
We next performed a broad search for Fanzor-encoded NLS sequences by analyzing each Fanzor ORF for a predicted NLS. We found that across all Fanzor families, ~60% of ORFs had readily identifiable NLS sequences, on par with the prediction accuracy of a validated set of NLS-containing proteins (13) and substantially greater than the fraction of NLS sequences predicted for cytosolic human proteins (fig. S10, B to D). We selected a subset of 22 Fanzors across Fanzor1 and Fanzor2 families with predicted N-terminal NLS sequences and screened these proteins by fusing the N-terminal 100 amino acids of each Fanzor ortholog to sfGFP, transfecting this panel into human embryonic kidney (HEK) 293FT cells, and visualizing sfGFP distribution. We found that 21 of 22 predicted N-terminal NLS sequences were functional for nuclear localization in mammalian cells, with varying nuclear localization efficiencies (Fig. 5C and fig. S10E). This experimental validation of the predicted NLS domains shows that Fanzor nucleases acquired mechanisms for nuclear import to access the genome and perform their genomic functions.
We next tested whether Fanzor nucleases could be adopted for mammalian genome editing by codon-optimizing ApmFNuc, DpFNuc, MmFNuc, and BaFNuc for mammalian expression and engineering their fRNA guide scaffolds for optimal U6-based expression in mammalian cells by removing poly-U stretches (fig. S11). We designed a reporter plasmid carrying the 21-nt target matching the fRNA guide and evaluated editing by NGS of generated insertions and deletions (indels). DpFNuc, MmFNuc, and ApmFNuc with engineered fRNAs had detectable editing activity, with DpFNuc and MmFNuc, achieving ~0.5 to 1% editing on plasmids inside human cells (fig. S12, A to D). We analyzed the indel patterns of DpFnuc and MmFNuc and found 2- to 35-bp deletions near the 3′ end of the target site (fig. S12, E and F), similar to the indel cleavage patterns of other programmable RuvC containing nucleases, such as Cas12 or TnpB (1, 4, 6). Because DpFNuc and MmFNuc displayed the highest levels of plasmid editing, we designed a panel of guides against seven endogenous genomic targets (Fig. 5D) and found varying levels of editing, from ~0.5 to 15% (Fig. 5, E and F), validating Fanzors as RNA-guided nucleases with activity in mammalian cells. As with plasmid editing, editing outcomes were primarily large deletions, ranging in size from 1 to 25 bp (Fig. 5, G to J). To evaluate whether Fanzor1 orthologs are also functional for genome editing, we also tested KnFNuc’s editing efficiency and found editing up to 2% across multiple endogenous genomic targets (fig. S13), showing that both Fanzor1 and Fanzor2 nucleases can be reprogrammed for human genome editing.
DISCUSSION
RNA-guided DNA endonucleases are prominent in prokaryotes including roles in innate immunity mediated by prokaryotic Argonautes (14), adaptive immunity by CRISPR systems (15–17), RNA-guided transposition by CRISPR-associated transposases (18, 19), and still uncharacterized functions of OMEGA nucleases in transposon life cycles (4, 6). In eukaryotes, whereas RNA-guided cleavage of RNA is the cornerstone of the RNA-interference defense machinery and posttranscriptional regulation (20, 21), RNA-guided cleavage of genomic DNA has not been demonstrated, to our knowledge. We show here that the Fanzors, previously uncharacterized eukaryotic homologs (5) of the OMEGA effector nucleaseTnpB (6), are RNA-guided, programmable DNA nucleases. Saito et al., have characterized Fanzor nucleases biochemically, solved the structure of Fanzor1, and engineered the nucleases for mammalian genome editing (22). We also extensively searched diverse genomes of eukaryotes and their viruses to discover thousands of additional RuvC-containing Fanzor nucleases, providing the starting point for further exploration of this family of proteins.
Phylogenetic analysis of the Fanzors together with their closest TnpB relatives revealed five major Fanzor families, which all contain Fanzor nucleases interspersed with prokaryotic TnpBs, suggesting that TnpBs entered the eukaryotic genomes on multiple, independent occasions. Considering the high abundance of TnpBs in bacteria and archaea, and their mobility, along with the exposure of unicellular eukaryotes to bacteria, this apparent history of multiple jumps into eukaryotic genomes does not appear unexpected. Furthermore, given the widespread of Fanzors in eukaryotes, together with the near ubiquity of TnpBs in bacteria and archaea, it appears likely that TnpBs were originally inherited from both archaeal and bacterial partners in the original endosymbiosis that triggered eukaryogenesis (23). Subsequent events of TnpB capture by eukaryotes could occur via additional endosymbioses as well as sporadic contacts with bacterial DNA. Notably, however, the high intron density in many Fanzors implies their long evolution in many groups of eukaryotes. The history of Fanzor2, however, is quite distinct from the four Fanzor1 families. This variety of Fanzors are enriched in viruses and in IS607 transposons and are far more closely similar to TnpB than members of other Fanzor families, suggesting likely origin from phagocytosis of TnpB-containing bacteria by amoeba and subsequent spread via amoeba-trophic giant viruses (24).
Association of Fanzor nucleases with transposases suggests a role for their RNA-guided nuclease activity in transposition similarly to the case of TnpB. The exact nature of that role, however, remains unknown. TnpB has been reported to boost the persistence of the associated transposons in bacterial populations (25, 26). TnpB and Fanzors potentially could perform different mechanistic roles in transposon maintenance. In particular, these RNA-guided nucleases could target sites from which a transposon was excised, initiating homology-directed repair through a transposon-containing locus, restoring the transposon in the original site and thus serving as an alternate mechanism of transposon propagation (26). The association of TnpBs and Fanzors with diverse types of transposases suggests that the function(s) of the RNA-guided nucleases do not strictly depend on the transposition mechanism.
Our biochemical characterization of both viral and eukaryotic Fanzor nucleases revealed both similarities with the homologous TnpB and Cas12 RNA-guided nucleases and several notable distinctions. Like TnpB and Cas12, Fanzor nucleases generate double-stranded breaks through a single RuvC domain and cleave the target DNA near the 3′ end of the target. However, unlike canonical TnpB and Cas12 enzymes, which have strong collateral activity against free single-stranded DNA, Fanzor nucleases and a subset of related TnpBs contain rearranged catalytic sites that are not conducive to collateral activity. In contrast to the T-rich TAMs of TnpB and PAMs of Cas12, the Fanzor TAM preference is diverse, with a GC preference observed for the viral ApmFNuc and A/T rich preferences for the eukaryotic MmFNuc, DpFNuc, and BaFNuc. In some cases, the TAM preference agrees with the insertion site sequence, which is compatible with the role of Fanzors in transposition. Last, the fRNA of Fanzors overlaps with the transposon IRR and terminal inverted repeat (TIR), much like TnpB’s ωRNA, but extends farther downstream of the Fanzor ORF, in contrast to the ωRNAs that ends near the 3′ regions of the TnpB ORF. Furthermore, although the Fanzor nucleases originated from TnpB, some features of these eukaryotic RNA-guided nucleases notably differ from those of the prokaryotic ones, reflecting their adaptation functioning in eukaryotic cells, such as the acquisition of introns and functional NLS sequences for nuclear localization.
We demonstrate that Fanzor nucleases can be applied for efficient genome editing with detectable cleavage and indel generation activity in human cells. While the Fanzor nucleases are compact (~600 amino acids), which could facilitate delivery, and their eukaryotic origins might help to mitigate the immunogenicity of these nucleases in humans, additional engineering is needed to further improve the activity of these systems in human cells, as has been accomplished for other miniature RNA-guided nucleases such as Cas12f (27–30). The broad distribution of Fanzor nucleases among diverse eukaryotic lineages and associated viruses suggests that many more currently unknown RNA-guided systems could exist in eukaryotes, serving as a rich resource for future characterization and development of new biotechnologies.
MATERIALS AND METHODS
Computational discovery of Fanzor systems
A profile of the Fanzor RuvC domain (Fanzor profile) was constructed by aligning the previously discovered Fanzor proteins (seed sequences) with MUSCLE v5 (-align), extracting the RuvC domain, and building a profile HMM with hmmbuild (default options) from the HMMER v3 suite of programs. An initial set of putative Fanzor proteins was gathered by searching all annotated proteins and translated ORFs (stop codon to stop codon) longer than 100 residues in NCBI eukaryotic and viral assemblies (one assembly per species) as well as all full-length proteins annotated on eukaryotic and viral sequences in GenBank (hmmsearch -E 0.001 -Z 61295632). To predict introns in Fanzor ORFs, AUGUSTUS v3.5.0 and Spaln v2.4.13f were applied to the genomic region containing the ORF (10 kb upstream/downstream). AUGUSTUS was used for ab initio gene prediction when there was an available parameter set of the same class as the target species. Tantan was used to soft-mask the genome before gene prediction using an “-r” parameter of 0.01 if the genome AT fraction was less than 0.8 and 0.02 otherwise (with the suggested scoring matrix for AT-rich genomes). Spaln was used to splice-align Fanzor proteins to the Fanzor ORFs (default options). The protein query set for Spaln was generated by searching UniClust90 and GenBank eukaryotic proteins with the Fanzor profile. The Fanzor profile was iteratively refined by repeatedly searching the initial set of proteins (hmmsearch -E 0.0001 –domE 1000 -Z 69000000), extracting the RuvC domain, clustering with MMseq2 (--min-seq-id 0.5 -c 0.9), aligning the cluster representatives with the profile seed sequences, manually refining the alignment, building a new profile, and using the new profile for the next round. Three rounds of refinement were completed. The refined profile was used for a final round of searches, and clusters that would have been included in the profile were kept for the subsequent filtering steps. To reduce the likelihood of including genome assembly contaminants in downstream analysis, all Fanzor proteins from NCBI assemblies marked as contig level completeness or those originating from contigs shorter than 50 kb (only from assemblies) were discarded. The remaining sequences were clustered using a combination of Diamond v2.1.6 (--evalue 0.0001 --id 70 --query-cover 90 --subject-cover 90 --max-target-seqs 500 --comp-based-stats 3) and MCL (-I 4.0). Each cluster was aligned with MUSCLE, and a consensus sequence was computed using a custom Python script. The RuvC domains were extracted from each consensus sequence, and all aligned with MUSCLE. The alignment was manually inspected and filtered to yield a final set of Fanzor sequences.
Computational discovery of TnpBs
A profile HMM was constructed from a multiple sequence alignment of subsets of Fanzor and used to query a custom database of prokaryotic and metagenomic assemblies using HMMER (-E 0.0001 -Z 61295632). Sequences identical to another sequence were discarded, and the remaining were clustered with MMseqs2 (--min-seq-id 0.7 -c 0.9 -s 7). The split-RuvC domain was extracted from each cluster representative and further clustered with MMseqs2 (--min-seq-id 0.5 -c 0.9 -s 7) for a two-step clustering process. These split-RuvC domain cluster representatives were aligned with MUSCLE, and sequences without alignment to the conserved DED motif were discarded.
Phylogenetic analysis of Fanzors and TnpBs
To make a phylogenetic tree of TnpB and Fanzor sequences, the split-RuvC domain was extracted from every Fanzor consensus sequence and aligned to the split-RuvC domain of a 3k random subset of the two-step clustered TnpB representatives using MUSCLE (-super5). Sequences appearing to be fragments were discarded from the alignment, and the remaining sequences were realigned. An approximately maximum-likelihood phylogenetic tree was constructed with FastTree2 (-lg -gamma). All branches with a local support value (as computed by FastTree) less than 0.7 were collapsed, and the tree rooted at the midpoint. The subsequent tree was visualized with R and the ggtree suite of packages.
Prediction of NLS in Fanzors
NLStradamus was used with default threshold at 0.6 and model option 2 (four-state bipartite model) to predict NLS domains. For background false-positive rate determination, a comprehensive search on UniProt is performed by looking for homo sapiens cytosolic proteins (with reviewed status), and a total of 1126 proteins are pulled out for analysis. For on-target false-negative rate determination, the original set of training sequences that include known NLS containing proteins from NLStradamus is used (13). NLS sequences cloned for experimental testing are listed in table S3.
Prediction of transposon associations with Fanzor systems
A random forest selective binary classifier (RFSB) transposon classifier (31) was used to classify Fanzor-transposon associations by inputting the surrounding 10-kb genomic sequence around the Fanzor protein. The classify mode was used with default parameters to make the prediction. Afterward, all predicted DNA transposons were mapped back to the phylogenetic tree. For all Fanzor nucleases that were classified with transposons, cd-hit was used to cluster these sets of Fanzor proteins with default parameters to find any clusters with two or more sequences for multiple sequence alignments. Then, these clusters containing (>2 Fanzor systems) were blasted against all Repbase-documented transposons (32). LE and RE elements, TIR, and their associated transposons are then determined by either protein homology to known transposons in Repbase or high similarity of TIR/LE/RE element to known transposon profiles.
Prediction of Fanzor-associated noncoding RNA
Fanzors that were not simply ORF translations were clustered along their entire length at 70% sequence identity and 95% coverage with MMseqs2 (--min-seq-id 0.7 -c 0.95). Each cluster with at least two sequences was subject to noncoding RNA prediction. For each cluster, the 5′ region of the first exon plus 1.5 kb upstream bases and 3′ region of the last exon plus 1.5 kb downstream bases were cut from sequence. The 5′ and 3′ regions were aligned separately with MAFFT (default options). Each column of the alignment was scored for conservation, and the change point in conservation scores was predicted with the R changepoint package to detect a drop in conservation. If the predicted change point was found to be at least 13 bases outside of the exon boundary of every sequence in the alignment, then the conserved portion of the exon, plus 11 bases past the change point, was folded with RNAalifold from the ViennaRNA software suite.
Fanzor and TnpB protein purification
To purify Fanzor or TnpB protein, Rosetta2 DE3 pLys cells were transformed with a twin-strep-sumo tag fused to the N terminus of a Fanzor or TnpB construct along with the predicted fRNA/ωRNA driven by a separate vector. Following transformation, single colonies were picked from the agar plate containing antibiotics and picked into a starter culture of 10 ml for overnight incubation at 37°C. The starter culture was transferred to 2 liters of TB with the designated antibiotics and grown until the optical density reached between 0.6 and 0.8. The culture was moved to 4°C for 30 min before induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside induction. The cultures were then grown at 16°C overnight and harvested by centrifugation the next day. The pellet is then flash-frozen at −80°C and subsequently homogenized in lysis buffer [0.02 M tris-HCl (pH 8.0), 0.5 M NaCl, 1 mM dithiothreitol (DTT), and 0.1 M cOmplete, EDTA-free Protease Inhibitor Cocktail (Merck Millipore)] with high-pressure sonication for 15 min. The homogenized lysates are then centrifuged at 14,000 rpm for 30 min at 4°C. The clarified supernatant is isolated from the subsequent bacterial pellet and incubated with Strep-TactinXT 4Flow high-capacity resin (catalog no. 2-5030-010) for 1 hour. Following incubation, the crude solution is loaded onto a Glass Econo-Column Column for gravity flow chromatography and washed three times with the previously described lysis buffer. To elute tagged protein, 10 U of sumo protease is then added onto the column for on-column cleavage overnight at 4°C. The next day, the eluent is collected and concentrated through an Amicon Ultra-15 Centrifugal Filter (catalog no. UFC9030) before continuing to fast protein liquid chromatography (FPLC). To purify desired protein from added sumo protease, the concentrated eluent is loaded onto a Superdex 200 Increase 10/300 GL gel filtration column (GE Healthcare). The column was equilibrated with running buffer [10 mM Hepes (pH 7.0 at 25°C), 1 M NaCl, 5 mM MgCl2, and 2 mM DTT]. The Peak fractions containing RNP are pulled and analyzed by SDS–polyacrylamide gel electrophoresis. Correct fractions are concentrated again with Amicon filter tubes, and subsequently, buffer is exchanged into storage buffer [0.02 M tris HCL (pH 8), 0.25 M NaCl, 50% glycerol, and 2 mM DTT] and stored at −20 for further use. TnpB proteins follow the same purification procedure with the following modifications: T7 express [New England Biolabs (NEB)] pLys strain is used for transformation and subsequent culture.
Cell-free transcription/translation TAM screen
Fanzor protein sequences were E. coli codon–optimized using the Integrated DNA Technologies (IDT) codon optimization tool, and fRNA scaffolds were synthesized by IDT eBlock gene fragments. Cell-free transcription/translation reactions were carried out using the PURExpress In Vitro Protein Synthesis Kit (NEB) as per the manufacturer’s protocol with half-volume reactions, using 75 ng of template for the protein of interest, 125 ng of template for the corresponding fRNA or ωRNA with a guide targeting the TAM library, and 30 ng of TAM library plasmid. Reactions were incubated at 37°C for 4 hours and then quenched by heating up to 95°C for 15 min and cooling down to 4°C. Ten micrograms of RNase A (Qiagen) is added followed by a 15-min incubation at 50°C. DNA was extracted by polymerase chain reaction (PCR) purification, and adaptors were ligated using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB) using the NEBNext Adaptor for Illumina (NEB) as per the manufacturer’s protocol. Following adaptor ligation, cleaved products were amplified specifically using one primer specific to the TAM library backbone and one primer specific to the NEBNext adaptor with a 10-cycle PCR using NEBNext High Fidelity 2X PCR Master Mix (NEB) with an annealing temperature of 65°C, followed by a second 12-cycle round of PCR to further add the Illumina i5 adaptor. Amplified libraries were gel extracted, quantified by qubit (Invitrogen), and subjected to paired-end sequencing on an Illumina MiSeq with Read 1 (200 cycles), Index 1 (8 cycles), Index 2 (8 cycles), and Read 2 (80 cycles). TAMs were extracted, position weight matrix based on the enrichment score was generated, and Weblogos were visualized on the basis of this position weight matrix using a custom Python script. All sequencing primers used are listed in table S4.
In vitro biochemical TAM screen
Purified RNP (1 μM) and 100 ng of the 7N TAM library were incubated at 37°C in NEB buffer 3 for 3 hours. Subsequently, reaction is purified and analyzed following the same procedure as cell-free transcription/translation TAM screen. TAM library sequence and guides used are listed in table S5.
Cell-free transcription/translation cleavage assays
Cell-free transcription/translation reactions were carried out using the PURExpress In Vitro Protein Synthesis Kit (NEB) as per the manufacturer’s protocol with half-volume reactions using 75 ng of template for the protein of interest and a 100 ng of fRNA or ωRNA. Reactions were incubated at 37°C for 4 hours to allow for RNP formation and then placed on ice to quench in vitro transcription/translation. Fifty to 100 ng of target substrate was then added, and the reactions were incubated at the specified temperature for 1 additional hour. Reactions were then quenched by heating up to 95° for 15 min and cooling back down to 50°C for addition of 10 μg of RNase A (Qiagen) for 10-min incubation. DNA was extracted by PCR purification using minElute columns (Qiagen) and run on 6% Novex Tris-Borate-EDTA (TBE) gels (Thermo Fisher Scientific) as per the manufacturer’s protocols, as specified in figures. Gels were stained with 1× SYBR Gold (Thermo Fisher Scientific) for 10 to 15 min and imaged on a ChemiDoc imager (Bio-Rad) with optimal exposure settings. Each condition was performed twice for replicability.
In vitro cleavage assays
dsDNA substrates were produced by PCR amplification of pUC19 plasmids containing the target sites and the TAM sequences. All ωRNA and fRNA used in the biochemical assays was in vitro–transcribed using the HiScribe T7 Quick High Yield RNA Synthesis kit (NEB) from the DNA templates purchased from IDT. Target cleavage assays performed with Fanzor orthologs contained 10 nM DNA substrate, 1 μM protein, and 4 μM fRNA in a final 1× reaction buffer of NEB buffer 3. Assays were allowed to proceed at 37°C for 2 hours, then briefly shifted to 50°C for 5 min, and immediately placed on ice to help relax the RNA structure before RNA digestion. Reactions were then treated with RNase A (Qiagen) and Proteinase K (NEB) and purified using a PCR cleanup kit (Qiagen). DNA was resolved by gel electrophoresis on Novex 6% TBE polyacrylamide gels (Thermo Fisher Scientific).
Small RNA sequencing
Heterologous expression in E. coli
Rosetta2 chemically competent E. coli were transformed with plasmids containing the locus of interest. A single colony was used to seed a 5-ml overnight culture. Following overnight growth, cultures were spun down, resuspended in 750 μl of TRI reagent (Zymo), and incubated for 5 min at room temperature. Zirconia/silica beads (0.5 mm; BioSpec Products) were added, and the culture was vortexed for approximately 1 min to mechanically lyse cells. Two hundred microliters of chloroform (Sigma-Aldrich) was then added, and culture was inverted gently to mix and incubated at room temperature for 3 min, followed by spinning at 12000g at 4°C for 15 min. The aqueous phase was used as input for RNA extraction using a Direct-zol RNA miniprep plus kit (Zymo). Extracted RNA was treated with 10 U of deoxyribonuclease I (DNase I; NEB) for 30 min at 37°C to remove residual DNA and purified again with an RNA Clean & Concentrator-25 kit (Zymo). Ribosomal RNA (rRNA) was removed using the RiboMinus Transcriptome Isolation Kit for bacteria (Thermo Fisher Scientific) as per the manufacturer’s protocol using half-volume reactions. The purified sample was then treated with 20 U of T4 polynucleotide kinase (NEB) for 6 hours at 37°C and purified again with an RNA Clean & Concentrator-25 (Zymo) kit. The purified RNA was treated with 20 U of 5′ RNA phosphatase (Lucigen) for 30 min at 37°C and purified again using an RNA Clean & Concentrator-5 kit (Zymo). Purified RNA was used as input to an NEBNext Small RNA Library Prep for Illumina (NEB) as per the manufacturer’s protocol with an extension time of 60 s and 16 cycles in the final PCR. Amplified libraries were gel extracted, quantified by quantitative PCR (qPCR) using the KAPA Library Quantification Kit for Illumina (Roche) on a StepOne Plus machine (Applied Biosystems/Thermo Fisher Scientific), and sequenced on an Illumina NextSeq with Read 1 (42 cycles), Read 2 (42 cycles), and Index 1 (6 cycles). Adapters were trimmed using CutAdapt and mapped to loci of interest using BWA-align. Reads were visualized using Genious.
Ribonucleoprotein
RNPs were purified as described. One hundred microliters of concentrated RNP was used as input. The above protocol was followed with the following modifications: Three hundred microliters of TRI reagent (Zymo) and 60 μl of chloroform (Sigma-Aldrich) were used for RNA extraction.
PureExpress RNPs
Seventy five nanograms of plasmid encoding the fanzor ORF and 125 ng of the plasmid containing the locus were incubated in 1 U of PURExpress reactions for 4 hours at 37°C. Afterward, the RNP is affinity-purified using the protocol described above for heterologous Rosetta cell protein production and subjected to the same pipeline for small RNA sequencing.
C. reinhardtii was obtained from the University of Minnesota (CRC). The algae was lysed in TRIzol with glass beads vigorously shaken for 2 hours at room temperature. Then, the above protocol was followed with the following modifications: rRNA was removed using a plant specific ribominus rRNA depletion kits as per the manufacturer’s protocol, and the rRNA-depleted sample was purified using Agencourt RNAClean XP beads (Beckman Coulter) before T4 Polynucleotide Kinase (PNK) treatment. T4 PNK treatment was performed for 1.5 hours and purified with an RNA Clean & Concentrator-5 kit (Zymo). Final PCR in the small RNA library prep contained 10 cycles.
Collateral activity testing
DNase alert and RNase alert were purchased from IDT. RNP (1 μM) or 10 μl of PureExpress generated RNP and 10 nM DNA target containing either the target spacer or a scramble spacer are diluted in 1× DNase/RNase alert reaction buffer into 50-μl reactions. The solution is mixed well in the reaction test tube and subsequently aliquoted into 384-well plates. The plates are loaded onto applied biosystems qPCR machines, and reactions were ran at 37°C for ApmFNuc2, DrpFNuc2, BaaFNuc2, MemFNuc2, and Isdra2 TnpB and 60°C for TvoTnpB. The SYBR and HEX channel fluorescence intensity is recorded every minute for a duration of 60 min. The intensity is normalized by subtracting the nontarget DNA sequence from the target DNA sequence group. A positive control DNAse (2 μl) and RNAse (2 μl) is ran along with the HEREMES/TnpB group as a positive control to monitor the assay.
Cloning PAM/TAM libraries
Target sequences with 7N degenerate flanking sequences were synthesized by IDT and amplified by PCR with NEBNext High Fidelity 2× Master Mix (NEB). Backbone plasmid was digested with restriction enzymes (pUC19: Kpn I and Hind III, Thermo Fisher Scientific) and treated with FastAP alkaline phosphatase (Thermo Fisher Scientific). The amplified library fragment was inserted into the backbone plasmid by Gibson assembly at 50°C for 1 hour using 2× Gibson Assembly Master Mix (NEB) with an 8:1 molar ratio of insert:vector. The Gibson assembly reaction was then isopropanol precipitated by the addition of an equal volume of isopropanol (Sigma-Aldrich), the final concentration of 50 mM NaCl, and 1 μl of GlycoBlue nucleic acid coprecipitant (Thermo Fisher Scientific). After a 15-min incubation at room temperature, the solution was spun down at max speed at 4°C for 15 min, then the supernatant was pipetted off, and the pelleted DNA has resuspended in 12 μl Tris-EDTA (TE) and incubated at 50°C for 10 min to dissolve. Two microliters was then transformed by electroporation into Endura Electrocompetent E. coli (Lucigen) as per the manufacturer’s instructions, recovered by shaking at 37°C for 1 hour, then plated across five 22.7 cm by 22.7 cm BioAssay plates with the appropriate antibiotic resistance. After 12 to 16 hours of growth at 37°C, cells were scraped from the plates and midi- or maxi-prepped using a NucleoBond Midi- or Maxi-prep kit (Machery Nagel). The sequence for TAM libraries and guides used are provided in table S5.
Cleavage position mapping by NGS
RNP (1 μM) and 100 ng of the target plasmid were incubated at 37° for 3 hours in NEB buffer 3. Reactions were quenched by placing at 4°C or on ice and adding 10 μg of RNase A (Qiagen) and 8 U Proteinase K (NEB) each followed by a 5-min incubation at 37°C. DNA was extracted by PCR purification, and adaptors were ligated using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB) using the NEBNext Adaptor for Illumina (NEB) as per the manufacturer’s protocol. Following adaptor ligation, cleaved products were amplified specifically using one primer specific to the target plasmid (one on 5′ side of the cleavage and one on 3′ side of the cleavage) and one primer specific to the NEBNext adaptor with a 12-cycle PCR using NEBNext High Fidelity 2× PCR Master Mix (NEB) with an annealing temperature of 63°C, followed by a second 20-cycle round of PCR to further add the Illumina i5 adaptor. Amplified libraries were gel extracted, quantified by qubit dsDNA kit (Invitrogen), and subject to single-end sequencing on an Illumina MiSeq with Read 1 (100 cycles), Index 1 (8 cycles), and Index 2 (8 cycles). All sequencing primers are listed in table S4.
Confocal images of nuclear localization
The N-terminal predicted NLS sequences of fanzor are cloned onto N-terminal of sfGFP by Gibson assembly into a pCMV promoter backbone (NLS sequences cloned are listed in table S3). Twenty four hours before transfection, 15,000 HEK293FT cells were plated onto a glass bottom 96-well plates precoated with poly-d-lysine. One hundred nanograms of NLS-sfGFP construct is transfected into HEK293FT cells using Lipofectamine 3000, and 24 hours after transfection, cells were fixed and permeabilized using a Fix and Perm kit (Thermo Fisher Scientific) and subsequently stained by either DAPI or SYTO-Red nuclear stain (Thermo Fisher Scientific). All wells were measured via confocal microscopy at room temperature. Cells were focused in the 488-nm channel on the basis of the sfGFP protein. For each well, a 2 by 2 field of view image at ×20 magnification was collected under the following settings and stitched around the center point. Images were collected in 488 nm (32.8% power, 100-ms exposure), 359 nm (35.2% power, 100-ms exposure), and 633 nm (80% power, 100-ms exposure).
Mammalian cell culture and transfection
Mammalian cell culture experiments were performed in the HEK293FT line (Thermo Fisher Scientific) grown in Dulbecco’s modified Eagle’s medium with high glucose, sodium pyruvate, and GlutaMAX (Thermo Fisher Scientific), additionally supplemented with 1× penicillin-streptomycin (Thermo Fisher Scientific), 10 mM Hepes (Thermo Fisher Scientific), and 10% fetal bovine serum (VWR Seradigm). All cells were maintained at confluency below 80%.
All transfections were performed with Lipofectamine 3000 (Thermo Fisher Scientific). Cells were plated 16 to 20 hours before transfection to ensure 90% confluency at the time of transfection. For 96-well plates, cells were plated at 20,000 cells per well. For each well on the plate, transfection plasmids were combined with OptiMEM I Reduced Serum Medium (Thermo Fisher Scientific) to a total of 10 μl.
Mammalian genome editing
fRNA scaffold backbones were cloned into a pUC19-based human U6 expression backbone, and human codon-optimized Fanzor proteins were cloned into pCAG-based destination vector by Gibson Assembly. Then, 50 ng of protein expression construct, 50 ng of the corresponding guide construct, and an optionally 20 ng of luciferase reporter were transfected in one well of a 96-well plate using Lipofectamine 3000 transfection reagent. After 48 hours, reporter DNA was harvested by washing the cells once in 1× Dulbecco's phosphate-buffered saline (DPBS) (Sigma-Aldrich) and resuspended in 50 μl of QuickExtract DNA Extraction Solution (Lucigen) and cycled at 65°C for 15 min, 68°C for 15 min and then 95°C for 10 min to lyse cells. Lysed cells (2.5 μl) were used as input into each PCR reaction. For library amplification, target reporter regions were amplified with a 12-cycle PCR using NEBNext High Fidelity 2X PCR Master Mix (NEB) with an annealing temperature of 63°C for 15 s, followed by a second 18-cycle round of PCR to add Illumina adapters and barcodes. The libraries were gel-extracted and subject to single-end sequencing on an Illumina MiSeq with Read 1 (220 cycles), Index 1 (8 cycles), Index 2 (8 cycles), and Read 2 (80 cycles). Insertion/deletion (indel) frequency was analyzed using CRISPResso2. All sequencing primers are listed in table S4. Guides used for genomic target are listed in table S5.
Acknowledgments
We would like to thank D. Weston and E. Boyden for MiSeq instrumentation support; G. Feng, G. Choi and D. Wang for gel imager support; L. Villiger for guide engineering strategies; K. Chung for FPLC access; F. Chen and Y. Zhang for confocal microscopy help; and Z. Tang, R. Desimone, and J. Crittenden for support and helpful discussions. We thank the members of the Abudayyeh-Gootenberg labs for support and advice.
Funding: J.S.G. and O.O.A. are supported by NIH grants 1R21-AI149694, R01-EB031957, R01-AG074932, and R56-HG011857; The McGovern Institute Neurotechnology (MINT) program; the K. Lisa Yang and Hock E. Tan Center for Molecular Therapeutics in Neuroscience; G. Harold and Leila Y. Mathers Charitable Foundation; NHGRI Technology Development Coordinating Center Opportunity Fund; MIT John W. Jarve (1978) Seed Fund for Science Innovation; Impetus Grants; Cystic Fibrosis Foundation Pioneer Grant #GOOTEN21XX2; Google Ventures; FastGrants; Harvey Family Foundation; Winston Fu; and the McGovern Institute. E.V.K. is supported through the NIH Intramural Research Program (National Library of Medicine).
Author contributions: O.O.A., J.S.G., and E.V.K. conceived the study and participated in the design, execution, and analysis of experiments. K.J. designed and performed the experiments in this study, performed computational analyses, and analyzed the data. J.L. developed computational pipelines for Fanzor discovery and performed computational analyses. S.S. and M.T. purified proteins and participated in experiments. A.K. assisted with confocal microscopy experiments. N.Y. performed computational analyses related to Fanzor discovery, alignment, tree building, and domain discovery. W.B. assisted with neighboring ORF and transposon analysis. K.K. assisted with structural analyses and drawings. K.J., J.L., E.V.K., O.O.A., and J.S.G wrote the manuscript with help from all authors.
Competing interests: A patent application has been filed by MIT related to this work with O.O.A., J.S.G., K.J., and J.L. as co-inventors. J.S.G. and O.O.A. are co-founders of Sherlock Biosciences, Proof Diagnostics, and Tome Biosciences. The other authors declare that they have no other competing interests.
Data and materials availability: Sequencing data will be available at Sequence Read Archive (PRJNA1002369). Expression plasmids are available from Addgene under UBMTA; support information and computational tools are available at https://abugootlab.org/. Plasmids can also be provided by MIT pending scientific review and a completed material transfer agreement; requests for the plasmids should be submitted to the corresponding authors. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S5
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.B. Zetsche, J. S. Gootenberg, O. O. Abudayyeh, I. M. Slaymaker, K. S. Makarova, P. Essletzbichler, S. E. Volz, J. Joung, J. van der Oost, A. Regev, E. V. Koonin, F. Zhang, Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.I. Fonfara, H. Richter, M. Bratovǐ, A. Le Rhun, E. Charpentier, The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). [DOI] [PubMed] [Google Scholar]
- 3.L. B. Harrington, D. Burstein, J. S. Chen, D. Paez-Espino, E. Ma, I. P. Witte, J. C. Cofsky, N. C. Kyrpides, J. F. Banfield, J. A. Doudna, Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.T. Karvelis, G. Druteika, G. Bigelyte, K. Budre, R. Zedaveinyte, A. Silanskas, D. Kazlauskas, Č. Venclovas, V. Siksnys, Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.W. Bao, J. Jurka, Homologues of bacterial TnpB_IS605 are widespread in diverse eukaryotic transposable elements. Mob. DNA. 4, 12–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.H. Altae-Tran, S. Kannan, F. E. Demircioglu, R. Oshiro, S. P. Nety, L. J. McKay, M. Dlakić, W. P. Inskeep, K. S. Makarova, R. K. Macrae, E. V. Koonin, F. Zhang, The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Shmakov, A. Smargon, D. Scott, D. Cox, N. Pyzocha, W. Yan, O. O. Abudayyeh, J. S. Gootenberg, K. S. Makarova, Y. I. Wolf, K. Severinov, F. Zhang, E. V. Koonin, Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky, J. A. Doudna, CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. P. Nety, H. Altae-Tran, S. Kannan, F. E. Demircioglu, G. Faure, S. Hirano, K. Mears, Y. Zhang, R. K. Macrae, F. Zhang, The transposon-encoded protein TnpB processes its own mRNA into ωRNA for guided nuclease activity. CRISPR J. 6, 232–242 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. N. Takeda, R. Nakagawa, S. Okazaki, H. Hirano, K. Kobayashi, T. Kusakizako, T. Nishizawa, K. Yamashita, H. Nishimasu, O. Nureki, Structure of the miniature type V-F CRISPR-Cas effector enzyme. Mol. Cell 81, 558–570.e3 (2021). [DOI] [PubMed] [Google Scholar]
- 11.R. Nakagawa, H. Hirano, S. N. Omura, S. Nety, S. Kannan, H. Altae-Tran, X. Yao, Y. Sakaguchi, T. Ohira, W. Y. Wu, H. Nakayama, Y. Shuto, T. Tanaka, F. K. Sano, T. Kusakizako, Y. Kise, Y. Itoh, N. Dohmae, J. van der Oost, T. Suzuki, F. Zhang, O. Nureki, Cryo-EM structure of the transposon-associated TnpB enzyme. Nature 616, 390–397 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O. O. Abudayyeh, J. S. Gootenberg, S. Konermann, J. Joung, I. M. Slaymaker, D. B. T. Cox, S. Shmakov, K. S. Makarova, E. Semenova, L. Minakhin, K. Severinov, A. Regev, E. S. Lander, E. V. Koonin, F. Zhang, C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. N. Nguyen Ba, A. Pogoutse, N. Provart, A. M. Moses, NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10, 202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. C. Swarts, K. Makarova, Y. Wang, K. Nakanishi, R. F. Ketting, E. V. Koonin, D. J. Patel, J. van der Oost, The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 21, 743–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.P. D. Hsu, E. S. Lander, F. Zhang, Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.F. Hille, H. Richter, S. P. Wong, M. Bratovǐ, S. Ressel, E. Charpentier, The biology of CRISPR-Cas: Backward and forward. Cell 172, 1239–1259 (2018). [DOI] [PubMed] [Google Scholar]
- 17.J. A. Doudna, E. Charpentier, The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 18.J. Strecker, A. Ladha, Z. Gardner, J. L. Schmid-Burgk, K. S. Makarova, E. V. Koonin, F. Zhang, RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. E. Klompe, P. L. H. Vo, T. S. Halpin-Healy, S. H. Sternberg, Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). [DOI] [PubMed] [Google Scholar]
- 20.G. J. Hannon, RNA interference. Nature 418, 244–251 (2002). [DOI] [PubMed] [Google Scholar]
- 21.G. Hutvagner, M. J. Simard, Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32 (2008). [DOI] [PubMed] [Google Scholar]
- 22.M. Saito, P. Xu, G. Faure, S. Maguire, S. Kannan, H. Altae-Tran, S. Vo, A. Desimone, R. K. Macrae, F. Zhang, Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature 620, 660–668 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.P. López-García, D. Moreira, The symbiotic origin of the eukaryotic cell. C. R. Biol. 346, 55–73 (2023). [DOI] [PubMed] [Google Scholar]
- 24.M. Boyer, N. Yutin, I. Pagnier, L. Barrassi, G. Fournous, L. Espinosa, C. Robert, S. Azza, S. Sun, M. G. Rossmann, M. Suzan-Monti, B. La Scola, E. V. Koonin, D. Raoult, Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. U.S.A. 106, 21848–21853 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C. Pasternak, R. Dulermo, B. Ton-Hoang, R. Debuchy, P. Siguier, G. Coste, M. Chandler, S. Sommer, ISDra2 transposition in Deinococcus radiodurans is downregulated by TnpB. Mol. Microbiol. 88, 443–455 (2013). [DOI] [PubMed] [Google Scholar]
- 26.C. Meers, H. Le, S. R. Pesari, F. T. Hoffmann, M. W. G. Walker, J. Gezelle, S. H. Sternberg, Transposon-encoded nucleases use guide RNAs to selfishly bias their inheritance. bioRxiv 10.1101/2023.03.14.532601 (29 March 2023). 10.1101/2023.03.14.532601. [DOI] [PubMed]
- 27.G. Bigelyte, J. K. Young, T. Karvelis, K. Budre, R. Zedaveinyte, V. Djukanovic, E. Van Ginkel, S. Paulraj, S. Gasior, S. Jones, L. Feigenbutz, G. S. Clair, P. Barone, J. Bohn, A. Acharya, G. Zastrow-Hayes, S. Henkel-Heinecke, A. Silanskas, R. Seidel, V. Siksnys, Miniature type V-F CRISPR-Cas nucleases enable targeted DNA modification in cells. Nat. Commun. 12, 6191 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Z. Wu, Y. Zhang, H. Yu, D. Pan, Y. Wang, Y. Wang, F. Li, C. Liu, H. Nan, W. Chen, Q. Ji, Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat. Chem. Biol. 17, 1132–1138 (2021). [DOI] [PubMed] [Google Scholar]
- 29.X. Xu, A. Chemparathy, L. Zeng, H. R. Kempton, S. Shang, M. Nakamura, L. S. Qi, Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol. Cell 81, 4333–4345.e4 (2021). [DOI] [PubMed] [Google Scholar]
- 30.D. Y. Kim, J. M. Lee, S. B. Moon, H. J. Chin, S. Park, Y. Lim, D. Kim, T. Koo, J.-H. Ko, Y.-S. Kim, Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat. Biotechnol. 40, 94–102 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.K. Riehl, C. Riccio, E. A. Miska, M. Hemberg, TransposonUltimate: Software for transposon classification, annotation and detection. Nucleic Acids Res. 50, e64 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.W. Bao, K. K. Kojima, O. Kohany, Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. A. Rees, K. Cranston, Automated assembly of a reference taxonomy for phylogenetic data synthesis. Biodivers Data J. 5, e12581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S5
Legend for data S1
Data S1