Abstract
Trichomonas vaginalis infection is the most prevalent nonviral sexually transmitted disease (STD) in the world. A PCR test using vaginal swab samples for the detection of T. vaginalis was developed to add T. vaginalis infection to the growing list of STDs that can be detected by DNA amplification techniques. A primer set, BTUB 9/2, was designed to target a well-conserved region in the beta-tubulin genes of T. vaginalis. All strains (15 of 15) of T. vaginalis tested were successfully detected by PCR giving a single predicted product of 112 bp in gel electrophoresis. No such targeted product was amplified with DNA from Trichomonas tenax, Trichomonas gallinae, Chlamydia trachomatis, Neisseria gonorrhoeae, Giardia lamblia, Chilomastix sulcatus, Dientamoeba fragilis, and Entamoeba histolytica. An optimal analytical sensitivity of one T. vaginalis organism per PCR was achieved. Culture, performed with the Inpouch TV culture system, was examined daily with a light microscope to identify T. vaginalis. Twenty-three of 350 (6.6%) vaginal swab samples from women attending an army medical clinic were culture positive for T. vaginalis. Of these culture positive specimens, PCR detected 22 of 23 (96%) with primer set BTUB 9/2, and wet preparation detected only 12 of 23 (52%). Seventeen specimens were BTUB 9/2-PCR positive and culture negative. Ten of these discordant specimens were determined to be as true positive by PCR using primer sets TVA 5-1/6 and/or AP65 A/B, which target different regions in the T. vaginalis genome, and seven were determined to be false positive. The sensitivity of BTUB 9/2-PCR was 97% and the specificity was 98%. The sensitivities of culture and wet preparation were 70 and 36%, respectively. The diagnosis of T. vaginalis infection by PCR is a sensitive and specific method that could be incorporated into a joint strategy for the screening of multiple STDs by using molecular amplification methods.
Trichomonas vaginalis infection is the most prevalent nonviral sexually transmitted disease in the world (18). In the United States, an estimated 3 million woman contract trichomoniasis every year (12). Symptomatic women with trichomoniasis usually complain of vaginal discharge, vulvovaginal soreness, and/or irritation. Dysuria and dyspareunia are also common (18, 21). T. vaginalis can be asymptomatic in 10 to 50% of women (9, 28). The organism can be recovered in 11% of men attending sexually transmitted disease (STD) clinics (13) and from 30 to 60% of male sexual partners of infected women, usually as a self-limited mild urethritis (9, 13). It can be transmitted to neonates during passage through an infected birth canal (2 to 17%), but the infection is usually asymptomatic and self limited (4). The incidence of trichomoniasis is highest in women with multiple partners and in groups with a high prevalence of other STDs (3). Whether trichomoniasis is a risk factor for human immunodeficiency virus transmission or just a marker for high-risk heterosexual activity remains unclear (15). An association of pelvic inflammatory disease, tubal infertility, and cervical cancer with previous episodes of trichomoniasis has been reported but may be explained by its association with other STDs (16, 17, 29). Complications of trichomonal vaginitis that have been reported include premature rupture of membranes, premature labor, low birth weight, and post-abortion or post-hysterectomy infection (2, 8, 9, 21, 22).
Traditionally physicians make the diagnosis based on clinical grounds, but in women, the characteristics of the vaginal discharge, including color and odor, are poor predictors of T. vaginalis (21, 23). Since no symptom alone or in combination is sufficient to diagnose T. vaginalis infection reliably, laboratory diagnosis is necessary (28). T. vaginalis may be identified in vaginal secretions by using a wet preparation, but this method is only 35 to 80% sensitive compared with culture (6, 14). The sensitivity of wet preparation is highly dependent on the expertise of the microscopist (9) and the prompt transport and laboratory processing of samples before the organism becomes lysed or loses motility. Other diagnostic techniques, such as fluorescent antibody (14), enzyme-linked immunosorbent assay (20), and a hybridization test (5), have been used to detect T. vaginalis and have had reported sensitivities between 70 and 90% (14). Culture in microaerophilic conditions is estimated to be 85 to 95% sensitive and has been considered the “gold standard” for diagnosis (9).
In this study, a PCR targeting the beta-tubulin genes of T. vaginalis was developed for the detection of the organism in vaginal swab samples. The targeted genes encode the amino acid sequence of beta-tubulin protein (11), a major component of the T. vaginalis cytoskeleton. Trichomonas PCR was compared with culture and wet preparation. Discrepant results were adjudicated by PCR using a previously described set of primers (19) and a set of primers targeting the adhesin genes of T. vaginalis. Since elevated pH of vaginal secretions in patients infected with T. vaginalis has been reported (21), its utility as a predictor was also explored in this study.
MATERIALS AND METHODS
T. vaginalis strains.
One strain from the American Type Culture Collection (Manassas, Va.) (ATCC SF-314 030001) and 14 strains of T. vaginalis (designated A to N) isolated by culture of vaginal secretions from patients attending an STD clinic were used to assess the sensitivity of the PCR primer sets. The specificity was evaluated by testing other related Trichomonas spp., flagellates, amoebae, or cervicovaginal pathogens (Trichomonas tenax ATCC 30207, Trichomonas gallinae ATCC 30002, Giardia lamblia ATCC SF-741 30888, Chilomastix sulcatus ATCC 50562, Dientamoeba fragilis ATCC 30948, Entamoeba histolytica ATCC SF-31-90015, Chlamydia trachomatis serovar E ATCC VR 3488, and Neisseria gonorrhoeae ATCC 19424).
DNA from T. vaginalis or the other microorganisms mentioned above was prepared from cultures by using the Chelex method (25). Mixtures of 50 μl of cultures with 200 μl of a 5% suspension of chelating resin (Chelex 100; Sigma, St. Louis, Mo.) in Tris buffer (0.01 M [pH 8.0]) were incubated at 56°C for 15 to 30 min. Preparations were mixed gently and then boiled for 8 to 10 min. After mixing again, preparations were centrifuged (12,000 × g) for 1 min in a microcentrifuge and stored at −70°C.
The analytical sensitivity was assayed with one clinical strain of T. vaginalis (strain M). Twofold dilutions of trichomonas organisms in culture media were initially prepared with 128 organisms (counted with a hemocytometer counting chamber) per PCR amplification, but the optimal analytical sensitivity of one organism per PCR amplification was achieved. Each dilution was processed separately by using the Chelex method to extract the DNA.
Clinical specimens.
Informed consent was obtained from all study participants. Before insertion of the speculum, clinicians collected vaginal swab samples (n = 350) from women attending the Epidemiology and Disease Control Clinic at the Womack Army Medical Center, Fort Bragg, N.C., from March to December 1997. The median age of the sampled population was 24 (range, 18 to 47), and 30% were Caucasian, 64% were African American, and 6% were of other races.
Vaginal swab samples were placed in 1 ml of a commercial PCR transport medium (AMPLICOR; Roche Diagnostic Systems, Branchburg, N.J.) and kept at 4°C until arrival at the laboratory within 4 days of collection. An equal volume of specimen diluent (AMPLICOR) was added to the sample, and the preparation was mixed, incubated at room temperature for 10 min, and stored at −70°C until tested.
A second vaginal swab sample was obtained after the insertion of the speculum. It was immediately touched to a glass slide together with a drop of normal saline for the microscopic (×100) wet examination of trichomonas in vaginal fluid. After the wet preparation was made, the swab was immediately inoculated into the Inpouch TV culture system (Biomed Diagnostics, Santa Clara, Calif.) (6). The Inpouch culture bag was sealed, incubated at 30°C, and examined daily with a light microscope to identify T. vaginalis. The pH of vaginal secretions was measured by using pH test strips (Sigma).
PCR primers.
A set of primers targeting a conserved region of the beta-tubulin genes of T. vaginalis (btub1, -2, and -3) was designed, synthesized, and tested. The sequences were as follows: for BTUB 9, 5′ CAT TGA TAA CGA AGC TCT TTA CGA T 3′ (positions 850 to 874); and for BTUB 2, 5′ GCA TGT TGT GCC GGA CAT AAC CAT 3′ (positions 961 to 938).
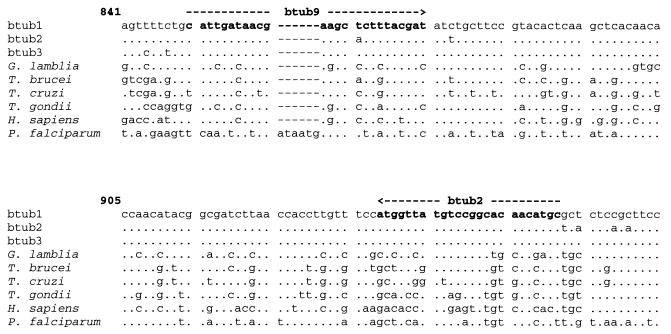
The DNA sequences for the primer set BTUB 9/2 were designed to target conserved regions of the three beta-tubulin genes of T. vaginalis to improve the analytical sensitivity (GenBank accession numbers: btub1, L05468; btub2, L05469; and btub3, L05470). In addition, to improve specificity, these DNA sequences were selected from the regions of the T. vaginalis beta-tubulin genes that differed substantially from the beta-tubulin gene sequences of humans and other microorganisms. (GenBank accession numbers: G. lamblia, X06748; Trypanosoma brucei subsp. rhodesiense, K02836; Trypanosoma cruzi, M97956; Toxoplasma gondii, M20025; Homo sapiens, V00598 J00317; and Plasmodium falciparum, G9981) (Fig. 1).
FIG. 1.
PCR primers BTUB 9 and BTUB 2 for the detection of T. vaginalis with the beta-tubulin genes. Primers were selected from well-conserved and specific regions of the genes (btub1, -2, and -3). Sequences of the beta-tubulin genes of T. vaginalis were compared to the beta-tubulin gene sequences of humans and to those of other pathogens. A dot indicates the same base, and a letter indicates a different base compared to the T. vaginalis btub1 gene sequence.
Discrepant results between the BTUB 9/2 PCR, wet preparation, and culture were adjudicated by PCR with primer set TVA 5/6, which has been previously described (19), and with primer set AP65 A/B, which is designed to target the adhesin genes of T. vaginalis (1). Primer TVA 5 was slightly modified by removing four bases at the 5′ end to avoid primer dimer formation and by including four more bases of the targeted sequence (102-bp clone A6p) at the 3′ end (our TVA 5-1), as follows: for TVA 5, 5′ GATC ATG TTC TAT CTT TTC A 3′ (positions 1 to 20); for TVA 5-1, 5′ ATG TTC TAT CTT TTC A TTGT 3′ (positions 5 to 24); and for TVA 6, 5′ GAT CAC CAC CTT AGT TTA CA 3′ (positions 102 to 83). Primer set AP65 A/B was designed to target a conserved region in the ap65 adhesin genes (GenBank accession numbers U18346 and U18347). Genes ap65-1 and -2 encode the AP65 protein that mediates T. vaginalis cytoadherence to the vaginal epithelium (1). The sequences of the primers were as follows: for AP65 A, 5′ GAT TCC TCT TCA CAC ACC CAC CAG 3′ (positions 304 to 327); and for AP65 B, 5′ AAT ACG GCC AGC ATC TGT AAC GAC 3′ (positions 512 to 489).
PCR.
PCR was performed with 10 μl of cultures (including all T. vaginalis strains and the other microorganisms mentioned above) processed by the Chelex method, or with 50 μl of vaginal swab samples processed with AMPLICOR specimen buffers. PCRs were performed in an automated thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.). The final reaction mixture (100 μl) contained 25 pmol of each primer, 2.5 mM deoxynucleoside triphosphate, 1× PCR buffer (10 mM Tris HCl [pH 8.4], 50 mM KCl), and 2 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Cetus) overlaid with 2 drops of mineral oil. Since 1.0 mM MgCl2 is a component of the AMPLICOR specimen buffers, 1.5 mM MgCl2 was added to the reaction mixture; otherwise, 2.5 mM MgCl2 was used when cultures of microorganisms processed with Chelex were tested.
A touchdown method for thermal cycling was used. Cycling times were 75 s at 95°C followed by 60 cycles of denaturation temperature 95°C for 45 s, annealing temperature beginning at 62°C and ending at 52°C for 45 s, and extension temperature of 72°C for 1 min. The annealing temperature was lowered one degree every four cycles until reaching 52°C, and this annealing temperature was then kept until the end of the cycling process.
To avoid product carryover, PCRs were set up in a room separate from all activities involving amplified target sequences, including thermocycling areas, PCR product storage, and gel electrophoresis. A separate set of pipetting devices was devoted to the setup of PCRs, and aerosol barrier pipette tips were used. Negative controls, including uninoculated transport media, were used throughout the specimen preparation and PCR process. A low copy number (n = 50/PCR) of trichomonas as positive control was included in every PCR run.
Detection of amplified targets.
The primer set BTUB 9/2 was designed to amplify a DNA product of 112 bp from the three beta-tubulin genes (Fig. 1). Primer set TVA 5-1/6 amplified a DNA product of 98 bp from a segment of the T. vaginalis genome (102-bp clone A6p). Primer set AP65 A/B amplified a DNA product of 209 bp from both ap65-1 and -2 adhesin genes of T. vaginalis (sequence numbers 304 to 512).
Twenty microliters of amplified product was electrophoresed at 120 V in 12% polyacrylamide gels in Tris-borate-EDTA buffer and stained with ethidium bromide (0.01 mM). The sizes of the amplified products were assessed by comparison with a commercial 50-bp weight marker (XIII; Boehringer Mannheim, Indianapolis, Ind.).
Adjudication of discrepant results and statistical analysis.
Discrepant specimens (those found to be culture negative and PCR positive) were determined to be T. vaginalis positive by performing PCR with other primer sets (TVA 5-1/6 and AP65 A/B). The gold standard was considered to be either culture positivity or PCR positivity with primer set BTUB 9/2 and at least one other primer set (TVA 5-1/6 or AP65 A/B). Sensitivity and specificity were calculated by using the defined gold standard. Confidence intervals for sensitivity and specificity were determined based on a normal approximation to the binomial distribution. Agreement between PCR and culture was assessed by using the kappa test (7).
RESULTS
Primer set BTUB 9/2 amplified the predicted 112-bp product in all 15 T. vaginalis strains tested. The analytical sensitivity of PCR with primer set BTUB 9/2 on the twofold dilutions of T. vaginalis organisms was the amplification of the DNA of one organism per PCR (see Fig. 3). No targeted PCR products were amplified when DNAs from other vaginal pathogens or protozoa were tested with the BTUB 9/2 primer set. However, a slightly bigger product (125 bp) was obtained (in gel electrophoresis) when DNA from the oral T. tenax was tested.
FIG. 3.
Analytical sensitivity testing of T. vaginalis PCR with primer set BTUB 9/2 (A) and primer set TVA 5-1/6 (B). Twofold dilutions of strain M of T. vaginalis were processed separately to extract the DNA and were tested by PCR.
PCR primer sets TVA 5-1/6 and AP65 A/B, which were used to resolve discrepant results in our clinical study, amplified the predicted 98-bp product in 14 of 15 strains of T. vaginalis and the predicted 209-bp product in 15 of 15 strains of T. vaginalis, respectively. Neither primer set TVA 5-1/6 (Fig. 2) nor primer set TVA 5/6 (not modified) amplified one particular clinical strain of T. vaginalis (strain L). None of the other vaginal pathogens or protozoa tested was detected by primer sets TVA 5-1/6 or AP65 A/B. The DNA corresponding to one copy of T. vaginalis (strain M) was amplified by both sets of primers TVA 5-1/6 (Fig. 3) and AP65 A/B (data not shown).
FIG. 2.
Detection of T. vaginalis strains F to N by PCR with primer sets BTUB 9/2 (A) and primer set TVA 5-1/6 (B). Strains F to N are representative of 14 strains of T. vaginalis (designated A to N) isolated from vaginal secretions of patients attending a city STD clinic in Baltimore, Md.
Twenty-three of 350 vaginal swab samples (6.6%) were culture positive for T. vaginalis. Wet preparation detected 52% (12 of 23), and BTUB 9/2 PCR detected 96% (22 of 23) of these positive cultures. Seventeen specimens were BTUB 9/2 PCR positive and culture negative. Ten of these discrepant specimens were determined to be true positives by PCR with primer sets TVA 5-1/6 and/or AP65 A/B targeting different regions in the T. vaginalis genome, and seven could not be confirmed as positives and were considered false positives (Table 1). Primer set TVA5-1/6 detected 61% (13 of 23) of specimens positive by culture. Primer set AP65 A/B detected 4 of 9 (44%) positive cultures that were not detected by PCR with TVA 5-1/6, and 6 of 10 (60%) negative cultures detected positive by two or more PCR primer sets (Table 1). Three specimens that were found to be positive by wet preparation could not be confirmed positive by culture or PCR with any of the primer sets tested and were deemed false positives.
TABLE 1.
Resolution of discrepant results between culture and PCR for the detection of T. vaginalis in 350 vaginal swab samples
| No. of samples | Result by culturea | Result with PCR primer set:
|
||
|---|---|---|---|---|
| BTUB 9/2 | TVA 5-1/6 | AP65 A/B | ||
| 14 | + | + | + | NTb |
| 4 | + | + | − | + |
| 4 | + | + | − | − |
| 1 | + | − | − | − |
| 5 | − | + | + | + |
| 4 | − | + | + | − |
| 1 | − | + | − | + |
| 7 | − | + | − | − |
| 310 | − | − | − | NT |
As determined the Trichomonas Inpouch TV culture system (Biomed Diagnostics, Santa Clara, Calif.).
NT, not tested.
After resolving discrepant results that were culture negative and PCR positive, the sensitivity of PCR with the primer set BTUB 9/2 was 97% (32 of 33; 95% confidence interval [CI], 95.2 to 98.8), and the specificity was 98% (310 of 317; 95% CI, 96.3 to 99.3) (Table 2). The sensitivity of culture was 70% (23 of 33; 95% CI, 64.9 to 74.5) and the specificity was 100% (317 of 317). The sensitivity of wet preparation was 36% (12 of 33; 95% CI, 31.3 to 41.4) and the specificity was 99% (314 of 317; 95% CI, 98 to 100). Although culture and the PCR with primer set TVA 5-1/6 had the same sensitivity and specificity (Table 2), their agreement was only moderate (κ = 0.58). In comparison, there was substantial agreement between culture and PCR with primer set BTUB 9/2 (κ = 0.78). The prevalence of T. vaginalis in this STD population was 9.4% (33 of 350) after the adjudication of discrepant results.
TABLE 2.
Comparison of PCR performed with primer set BTUB 9/2 and TVA 5-1/6, culture, and wet preparation for the detection of T. vaginalis in 350 vaginal swab samples
| Method | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|
| PCR | ||||
| With BTUB 9/2 | 97.0 | 97.8 | 82.1 | 99.7 |
| With TVA 5-1/2 | 69.7 | 100 | 100 | 96.9 |
| Culturea | 69.7 | 100 | 100 | 96.9 |
| Wet preparation | 36.4 | 99.1 | 80.0 | 93.7 |
Values obtained with the Trichomonas Inpouch TV culture system.
A similar proportion of confirmed trichomonas-positive patients was symptomatic (24 of 33 [72.7%]) compared to patients who were negative for trichomonas (230/317 [72.6%]) (P = 0.9). Sixty-seven percent (22 of 33) of confirmed T. vaginalis-positive patients had vaginal secretions of pH of >4.5 compared to 43% (136 of 317) of negative patients (P = 0.009). The odds ratio of having a trichomonas infection was 2.7 (95% CI, 1.3 to 5.6) when pH was >4.5. In this study, treatment for trichomoniasis was provided according to clinic standard methods on the basis of wet preparation results. In a retrospective analysis, 52% (17 of 33) of patients with confirmed trichomonas infections received appropriate treatment with metronidazole at the clinic site, five of them because bacterial vaginosis was suspected. In addition, no diagnosis or treatment was given to 15% (5 of 33) of infected patients even though three had genitourinary symptoms (Table 3).
TABLE 3.
Clinical diagnosis and treatment received by patients with PCR-confirmed infections with trichomonasa
| Clinical diagnosis | No. of patients | Treatment | No. of genitourinary symptoms |
|---|---|---|---|
| Vaginitis (trichomoniasis) | 12 | Metronidazole | 10 |
| None | 5 | None | 3 |
| Vaginitis (bacterial vaginosis) | 5 | Metronidazole | 4 |
| Vaginitis (unspecified) | 5 | Doxycycline | 3 |
| Vulvovaginal candidiasis | 2 | Antifungal | 1 |
| Genital herpes | 2 | Acyclovir | 2 |
| Chlamydia | 1 | Doxycycline | 1 |
| Condylomata acuminata | 1 | Not specified | 1 |
n=33. Treatment for trichomoniasis at the clinic was given on the basis of wet preparation results.
DISCUSSION
Trichomonas PCR with primer set BTUB 9/2 was 97% sensitive and 98% specific. It was more sensitive than wet preparation or culture. One culture-positive specimen could not be detected by any of the three PCR primer sets and was assumed to be a true positive by definition. The number of confirmed T. vaginalis-positive samples was increased by 39% by PCR compared to culture (32 of 350 [9.1%] versus 23 of 350 [6.6%]). PCR with primer set BTUB 9/2 for the detection of T. vaginalis had good analytical sensitivity and was able to amplify one trichomonas organism per PCR. The predicted DNA product (112 bp) in the targeted beta-tubulin gene was amplified with all T. vaginalis strains tested (15 of 15). The analytical specificity of primer set BTUB 9/2 was optimal, since no targeted DNA products were detected with other protozoa or vaginal pathogens.
In the T. vaginalis genome, there are several copies of the three genes encoding beta-tubulin proteins (beta-tubulin 1, 2, and 3) (11). Primer set BTUB 9/2 was designed to target a well-conserved region in all three beta-tubulin genes, thus improving sensitivity because of increased number of DNA target copies available for amplification. This region appears to be moderately conserved across other trichomonas species, because a slightly bigger PCR product of 125 bp was amplified by primer set BTUB 9/2 when DNA from the oral T. tenax was tested but not with DNA from T. gallinae. Although the 125-bp product detected with DNA of T. gallinae is similar in size to the product of T. vaginalis (112 bp), which could be difficult to differentiate by standard gel electrophoresis, specificity should not be affected since each Trichomonas species appears to be limited to its specific site of colonization (T. tenax inhabits the mouth and T. vaginalis only the genital and urinary tracts) (13).
The specificity of the primers depends on the annealing temperature, with higher temperatures favoring higher specificity. The use of a touchdown protocol increased the specificity of the PCR by favoring the amplification of targeted copies of DNA amplified during early cycles at higher annealing temperatures and eliminating spurious products. The DNA polymerase used (AmpliTaq Gold) is not active until heated to 95°C, thus simulating a hot-start PCR technique. The use of this enzyme also avoided erroneous amplification of DNA products due to nonspecific annealing of primers at lower temperatures.
In our study, PCR with primer set TVA 5-1/6 detected 61% (14 of 23) of culture-positive specimens. In previous studies, when compared with culture, PCR with primer set TVA 5/6 detected 90% (44 of 49) of culture-positive distal vaginal swab samples from an STD clinic in Pittsburgh, Pa. (10), and detected 91% (41 of 45) of culture-positive tampon specimens collected from a population of the Torres Strait Island (24). The target sequence of primer set TVA 5/6 is the full length of the 102-bp A6p clone isolated by Riley et al. from the T. vaginalis genome with the restriction enzyme Sau3A (recognition sequence, ↓GATC [the arrow indicates the cleavage site]) (19). Since both TVA5 and TVA6 primers included this recognition sequence at the 5′ end, we designed the primer TVA 5-1, which excluded the GATC sequence, to avoid the formation of primer dimers during amplification. This slight modification is unlikely to explain the difference in the proportion of positive cultures detected by PCR with primer set TVA 5-1/6 compared to previous studies with primer set TVA 5/6 (not modified). A possible explanation for this difference is that a genetically more diverse group of T. vaginalis strains were represented in the population of our study, which differed in the targeted region of primer set TVA 5/6 and were not detected. This conclusion is supported by the failure of primer sets TVA 5-1/6 or TVA 5/6 to detect T. vaginalis strain L.
The Chelex method for DNA preparation from cultures was simple, inexpensive, and easy to perform and prevented contamination as a result of manipulation of specimens used in other DNA preparation methods. As chelating resin, Chelex 100 has been used to extract heavy metals from solutions and, when used for DNA extraction, may also eliminate PCR inhibitors present in the samples and/or prevent the disruption of the genomic DNA as suggested by Walsh et al. (25).
Vaginal swab samples for the PCR screening of T. vaginalis are adequate and are comparable to posterior vaginal vault specimens collected with a speculum during a pelvic examination (27). In our study, the vaginal swab samples were collected by the clinicians, but self-administered vaginal swab samples, which are practical, rapid, and easy to obtain, can also be used for screening. Other studies have demonstrated that self-administered vaginal swab samples are comparable to physician-administered samples for the PCR detection of trichomonal infection (26).
An increased pH of the vaginal secretions in patients infected with T. vaginalis has been previously reported (21). In our study, two-thirds of the patients with confirmed positive T. vaginalis infection had vaginal secretion pHs of >4.5. Nevertheless, these patients represented only 16% of the total number of patients with elevated pHs, making pH an inadequate predictor for trichomonas infection.
Retrospectively, only 36% of patients with confirmed T. vaginalis infection were diagnosed by wet preparation, only 52% received appropriate treatment with metronidazole at the clinic, and no clinical diagnosis or treatment was given to 15% of the infected patients. These findings suggest the need for a more accurate diagnostic test for trichomonas, such as PCR, to prevent complications, transmission to sexual partners, and asymptomatic carriers, and to decrease possible transmission of HIV (2, 8, 9, 15, 21, 22).
Although wet preparation had minimal cost, its sensitivity is highly dependent on the expertise of the microscopist (9), prompt transport, and laboratory processing before the organism lyses or loses motility. In our study, even when the wet preparation was performed at the collection site, its sensitivity (36%) was suboptimal compared to PCR. Culture had a better sensitivity (70%) than wet mount examination but required more time for laboratory turnaround since cultures are held for 1 week. PCR results are available in 2 to 3 days and provided the highest sensitivity. The cost of PCR testing comes mainly from the cost of reagents, which is about two dollars per specimen, approximately equivalent to the cost of an Inpouch TV culture bag. The technician time is probably equivalent since cultures require daily examination by light microscopy. Although trichomonas PCR requires more technical skill, molecular amplification techniques are currently in use in many laboratories for the detection of C. trachomatis and N. gonorrhoeae infections. Thus, trichomonas PCR could easily be incorporated into the work flow of other diagnostic amplification procedures.
Vaginal swab samples for trichomonas PCR were transported in and processed with AMPLICOR specimen buffers, which is the transport and processing method for the C. trachomatis-N. gonorrhoeae PCR test (AMPLICOR, Roche Diagnostic System) currently in research use in our laboratory. Preliminary results indicate that 2 sucrose phosphate, which is a common chlamydia culture transport medium, is also an adequate transport media for this trichomonas PCR method, as well as for the C. trachomatis-N. gonorrhoeae PCR test, as suggested by the manufacturer. Thus, specimens transported in 2 sucrose phosphate media can be subsequently processed by the Chelex method. Incorporating PCR with BTUB 9/2 for T. vaginalis into the routine laboratory diagnostic methods using the same processed sample used for the PCR detection of C. trachomatis and N. gonorrhoeae could be a cost-effective strategy when screening programs for multiple STDs are implemented.
ACKNOWLEDGMENTS
This study was supported by grant #DAMD17-96-1-6309, U.S. Army Medical Research and Material Command, Fort Detrick, Md. 21702.
We thank Marie Tenant and the clinicians at the EDC for their collaboration in specimen collection; Jessie Chou, Kimberly Crotchfelt, and Anna Sierra-Honinman for laboratory assistance; and Katherine Kacena, Anita Kacena, M. Atenas, and Diana Perkins for assistance with preparation of the manuscript.
REFERENCES
- 1.Alderete J F, O’Brien J L, Arroyo R, Engbring J A, Musatovova O, Lopez O, Lauriano C, Nguyen J. Cloning and molecular characterization of two genes encoding adhesion proteins involved in Trichomonas vaginalis cytoadherence. Mol Microbiol. 1995;17:69–83. doi: 10.1111/j.1365-2958.1995.mmi_17010069.x. [DOI] [PubMed] [Google Scholar]
- 2.Coth M F, Pastorek J G, Nugent R P, Hiller S H, Gibbs R S, Martin D H, Eschebach D A, Edelman R, Carey J H, Regan J A, Krohn M A, Klebanoff M A, Vijaya Rao A, Rhoads G G the Vaginal Infections and Prematurity Study Group. Trichomonas vaginalis associated with low birth weight and preterm delivery. Sex Transm Dis. 1997;24:353–360. doi: 10.1097/00007435-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Coth M F, Pastorek J G, Nugent R P, Yerg D E, Martin D H, Eschenbach D A, et al. Demographic and behavioral predictors of Trichomonas vaginalis infection among pregnant women. Obstet Gynecol. 1991;78:1087–1092. [PubMed] [Google Scholar]
- 4.Danesh I S, Stephen J M, Gorbach J. Neonatal Trichomonas vaginalis infection. J Emerg Med. 1995;13:51–54. doi: 10.1016/0736-4679(94)00112-x. [DOI] [PubMed] [Google Scholar]
- 5.DeMeo L R, Draper D L, McGregor J A, Moore D F, Peter C R, Kapernick P S, McCormack W M. Evaluation of a deoxyribonucleic acid probe for the detection of Trichomonas vaginalis in vaginal secretions. Am J Obstet Gynecol. 1996;174:1339–1342. doi: 10.1016/s0002-9378(96)70682-8. [DOI] [PubMed] [Google Scholar]
- 6.Draper D, Parker R, Patterson E, Jones W, Beutz M, French J, Borchardt K, McGregor J. Detection of Trichomonas vaginalis in pregnant women with the InPouch TV culture system. J Clin Microbiol. 1993;31:1016–1018. doi: 10.1128/jcm.31.4.1016-1018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleiss J L. Statistical methods for rates and proportions. 2nd ed. New York, N.Y: John Wiley and Sons; 1981. [Google Scholar]
- 8.Hardy P H, Hardy J B, Nell E E, Graham D A, Spence M R, Rosenbaum R C. Prevalence of six sexually transmitted disease agents among pregnant inner-city adolescents and pregnancy outcome. Lancet. 1984;ii:333–337. doi: 10.1016/s0140-6736(84)92698-9. [DOI] [PubMed] [Google Scholar]
- 9.Heine P, McGregor J A. Trichomonas vaginalis: a reemerging pathogen. Clin Obstet Gynecol. 1993;36:137–144. doi: 10.1097/00003081-199303000-00019. [DOI] [PubMed] [Google Scholar]
- 10.Heine R P, Wiesenfeld H C, Sweet R L, Witkin S S. Polymerase chain reaction analysis of distal vaginal specimens: a less invasive strategy for detection of Trichomonas vaginalis. Clin Infect Dis. 1997;24:985–987. doi: 10.1093/clinids/24.5.985. [DOI] [PubMed] [Google Scholar]
- 11.Katiyar S K, Edlind T D. β-tubulin genes of Trichomonas vaginalis. Mol Biochem Parasitol. 1994;64:33–42. doi: 10.1016/0166-6851(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 12.Kent H L. Epidemiology of vaginitis. Am J Obstet Gynecol. 1991;165:1168–1176. doi: 10.1016/s0002-9378(12)90722-x. [DOI] [PubMed] [Google Scholar]
- 13.Krieger J N. Trichomoniasis in men: old issues and new data. Sex Transm Dis. 1995;22:83–96. [PubMed] [Google Scholar]
- 14.Krieger J N, Tam M R, Stevens C E, Nielsen I O, Hale J H, Kiviat N B, Holmes K K. Diagnosis of trichomoniasis: comparison of conventional wet-mount examination with cytologic studies, cultures, and monoclonal antibody staining of direct specimens. JAMA. 1988;259:1223–1227. doi: 10.1001/jama.259.8.1223. [DOI] [PubMed] [Google Scholar]
- 15.Laga M, Manoka A, Kivuvu M, Malele B, Tuliza M, Nzila N, Goeman J, Behets F, Batter V, Alary M, Heyward W L, Rider R W, Piot P. Non-ulcerative sexually transmitted diseases as risk factors for HIV-1 transmission in women: results from a cohort study. AIDS. 1993;7:95–102. doi: 10.1097/00002030-199301000-00015. [DOI] [PubMed] [Google Scholar]
- 16.Okonofua F E, Ako-Nai K A, Dighitoghi M D. Lower genital tract infection in infertile Nigerian women compared with controls. Genitourin Med. 1995;71:163–168. doi: 10.1136/sti.71.3.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paisarntantiwong R, Brockmann S, Clarke L, Landesman S, Feldman J, Minkoff H. The relationship of vaginal trichomoniasis and pelvic inflammatory disease among women colonized with Chlamydia trachomatis. Sex Transm Dis. 1995;22:344–347. doi: 10.1097/00007435-199511000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Rein M F. Trichomonas vaginalis. In: Mandell G L, Bennet J E, Dolin R, editors. Principles and practices of infection disease. New York, N.Y: Churchill Livingston; 1995. pp. 2493–2497. [Google Scholar]
- 19.Riley D E, Roberts M C, Takayama T, Krieger J N. Development of a polymerase chain reaction-based diagnosis of Trichomonas vaginalis. J Clin Microbiol. 1992;30:465–472. doi: 10.1128/jcm.30.2.465-472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma P, Malla N, Gupta I, Ganguly N K, Mahajan R C. A comparison of wet mount, culture and enzyme linked immunosorbent assay for the diagnosis of trichomoniasis in women. Trop Geogr Med. 1991;43:257–260. [PubMed] [Google Scholar]
- 21.Sobel J D. Vaginitis. N Engl J Med. 1996;337:1896–1903. doi: 10.1056/NEJM199712253372607. [DOI] [PubMed] [Google Scholar]
- 22.Soper D E, Bump R C, Hurt W G. Bacterial vaginosis and trichomoniasis vaginitis are risk factors for cuff cellulitis after abdominal hysterectomy. Am J Obstet Gynecol. 1990;163:1016–1021. doi: 10.1016/0002-9378(90)91115-s. [DOI] [PubMed] [Google Scholar]
- 23.Spence M R, Hollander D H, Smith J, McCaig L, Sewel D, Brockman M. The clinical and laboratory diagnosis of Trichomonas vaginalis infection. Sex Transm Dis. 1980;7:168–171. doi: 10.1097/00007435-198010000-00004. [DOI] [PubMed] [Google Scholar]
- 24.Tabrizi S N, Paterson B, Fairley C K, Bowden F J, Garland S M. A self-administered technique for the detection of sexually transmitted diseases in remote communities. J Infect Dis. 1997;176:289–292. doi: 10.1086/517269. [DOI] [PubMed] [Google Scholar]
- 25.Walsh P S, Metzger D A, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 26.Wawer M J, McNairn D, Wabwire-Mangen F, Paxton L, Gray R H, Kiwanuka N. Self-administered vaginal swabs for population-based assessment of Trichomonas vaginalis prevalence. Lancet. 1995;345:131–132. doi: 10.1016/s0140-6736(95)90100-0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 27.Witkin S S, Inglis S R, Polaneczky M. Detection of Chlamydia trachomatis and Trichomonas vaginalis by polymerase chain reaction in introital specimens from pregnant women. Am J Obstet Gynecol. 1996;175:165–167. doi: 10.1016/s0002-9378(96)70268-5. [DOI] [PubMed] [Google Scholar]
- 28.Wolner-Hanssen P, Krieger J N, Steven C E, Kiviat N B, Koustsky L, Mcritchlow C, De Roven T, Hillier S, Holmes K K. Clinical manifestation of vaginal trichomoniasis. JAMA. 1989;264:571–576. doi: 10.1001/jama.1989.03420040109029. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z F, Graham S, Yu S Z, et al. Trichomonas vaginalis and cervical cancer: a prospective study in China. Ann Epidemiol. 1995;5:325–332. doi: 10.1016/1047-2797(94)00101-x. [DOI] [PubMed] [Google Scholar]