Abstract
To address the current lack of validated molecular standards for analytical ultracentrifugation (AUC), we investigated the suitability of double-stranded DNA molecules. We compared the hydrodynamic properties of linear and circular DNA as a function of temperature. Negatively supercoiled, nicked, and linearized 333 and 339 bp minicircles were studied. We quantified the hydrodynamic properties of these DNAs at five different temperatures, ranging from 4 to 37 °C. To enhance the precision of our measurements, each sample was globally fitted over triplicates and five rotor speeds. The exceptional stability of DNA allowed each sample to be sedimented repeatedly over the course of several months without aggregation or degradation, and with excellent reproducibility. The sedimentation and diffusion coefficients of linearized and nicked minicircle DNA demonstrated a highly homogeneous sample, and increased with temperature, indicating a decrease in friction. The sedimentation of linearized DNA was the slowest; supercoiled DNA sedimented the fastest. With increasing temperature, the supercoiled samples shifted to slower sedimentation, but sedimented faster than nicked minicircles. These results suggest that negatively supercoiled DNA becomes less compact at higher temperatures. The supercoiled minicircles, as purified from bacteria, displayed heterogeneity. Therefore, supercoiled DNA isolated from bacteria is unsuitable as a molecular standard. Linear and nicked samples are well suited as a molecular standard for AUC and have exceptional colloidal stability in an AUC cell. Even after sixty experiments at different speeds and temperatures, measured over the course of 4 months, all topological states of DNA remained colloidal, and their concentrations remained essentially unchanged.
Keywords: Analytical ultracentrifugation, Molecular standards, Double-stranded DNA, DNA minicircle, Hydrodynamics, Global analysis
Introduction
Analytical ultracentrifugation (AUC) is a widely used technique to study biological macromolecules in the solution phase (Hansen et al. 1994). Hydrodynamic analyses have provided important information about the composition, interactions, and structure of biological macromolecules (García de la Torre and Hernández Cifre 2020). AUC has been considered essential for characterizing macromolecules in solution since its invention by Theodor Svedberg (Harding 2017) nearly 100 years ago. A particular strength of AUC is the possibility to study molecules in a physiologically relevant solution environment, where size and shape distributions can be characterized as a function of temperature, sample concentration, and buffer conditions (Brookes and Demeler 2006). AUC data analysis is based on first principles to derive hydrodynamic properties such as sedimentation and diffusion coefficients, and partial concentration of multiple components in a mixture. The diffusion coefficient directly yields the frictional coefficient and the hydrodynamic radius of the molecule. If the partial specific volume (PSV) is known, the frictional ratio and molar mass can be determined as well. Experimental data can be modeled by finite element solutions of the Lamm equation (Cao and Demeler 2005, 2008) to yield the sedimentation and diffusion coefficients, and partial concentrations directly.
Unlike other solution-based composition characterization methods such as size exclusion chromatography, gel electrophoresis, or field flow fractionation, AUC does not require reference standards. This assertion is only true, however, if the instrument can be trusted to report accurate measurements, a question that has been investigated previously (Stoutjesdyk et al. 2020; Zhao et al. 2015). Failures by the instrument to accurately report temperature, radial positioning, rotor speed, time, or wavelength affect the reliability of all parameters determined in an AUC measurement, and could limit the value of the measurements produced by the instrument. To address this limitation, a well-characterized molecular standard should be used to confirm the accuracy of the instrument, giving the investigator the necessary confidence that the instrument can be trusted for measurements of other systems. The requirements for an AUC molecular standard are multifold:
Stability for an extended period;
Resistance to degradation or aggregation;
Predictable behavior over the temperature range accessible with the instrument;
Homogeneous in composition;
Precisely known molar mass;
No concentration-dependent non-ideality;
Ideal solution behavior at concentrations suitable for measurement in an AUC instrument;
Available at low cost to the scientific community;
High degree of purity;
Properties can be readily modulated to explore a range of size and shape factors; and
Available orthogonal method (e.g., mass spectrometry, electrophoresis, sequencing) to confirm molecular properties of the standard such as molar mass and composition.
Although commercial standards with characterized hydrodynamic properties do not exist, bovine serum albumin (BSA) has been used previously in a multi-laboratory study to establish reproducibility of AUC experiments (Zhao et al. 2015). BSA fits some of the criteria above. Importantly, in a suitable buffer it is stable at 4 °C for 1 year and, if sufficiently dilute, BSA can be observed under ideal solution conditions. Whereas BSA has a precisely known monomer mass, higher order oligomers occur that must be removed by extensive purification before a homogeneous preparation is obtained. Additionally, the stability of BSA at temperatures higher than 20 °C during AUC has not been explored. Finally, with only one size and one shape, BSA does not offer variability in size and shape factors. These factors limit the utility of BSA for use in the validation of AUC instruments.
Some or all of these limitations exist for other proteins as well. Many proteins are less stable than BSA, and more prone to aggregation and degradation over extended periods of time. In addition, many proteins form oligomeric states that vary depending on experimental conditions. Furthermore, the standard method used to assess proteins, SDS polyacrylamide gel electrophoresis, is unsuitable as an orthogonal quantification method because SDS modifies both the conformation and the oligomerization state of proteins in solution (Parker and Song 1992; Henrickson et al. 2023). We propose that double-stranded DNA (hereafter referred to as DNA, because we are not testing single-stranded DNA) satisfies the requirements for a molecular standard listed above, and here we investigate this proposition. Some of its unique additional benefits include:
High stability under many conditions;
Precisely known molar mass and homogenous when not synthesized chemically (contaminating incomplete products would be problematic);
High extinction coefficient at 260 nm for measurement under dilute and ideal solution conditions, and the ultraviolet (UV) extinction coefficient can be precisely calculated from the DNA sequence to support accurate quantification; and
Molar mass can be easily modulated by changing the length of the DNA molecule.
To investigate its suitability as a molecular standard, we chose two different lengths of DNAs (333 bp and 339 bp). Comparing DNA molecules of molar mass differing by less than 2% tests the precision that can be obtained from AUC. We compared different topological states (negatively supercoiled, nicked open circle, and linearized) of these DNA molecules to elucidate the hydrodynamic differences imposed by conformation and temperature.
Negatively supercoiled DNA can be readily extracted from bacterial cells in high quantities using standard molecular biology techniques or using commercially available kits. If it satisfies the other requirements listed above, it should be suitable as a molecular standard. Supercoiled DNA is, however, structurally diverse (Irobalieva et al. 2015) and likely fluctuates between different conformations. Although we are beginning to understand how to model the sedimentation of supercoiled DNA (Waszkiewicz et al. 2023), we do not yet fully understand how to account for the conformational heterogeneity. Nicked and linearized samples require additional processing steps, but removing supercoiling may yield useful molecular standards.
We investigated DNA over a range of temperatures. Temperature is important for DNA nanoparticle formation, stability, flexibility, and interactions with architectural proteins (Driessen et al. 2014). Understanding how the solution behavior of DNA changes with temperature should yield insight into DNA at the nanoscale. Yet, the effect of temperature on sedimentation and diffusion transport of DNA conformation and dynamics is a largely unexplored topic. To corroborate the AUC results with an orthogonal approach, we determined the composition of all DNA samples by polyacrylamide gel electrophoresis, which provides a very sensitive composition assessment (Waszkiewicz et al. 2023).
Materials and methods
Chemicals and reagents
100 bp DNA ladder was purchased from New England Biolabs (Ipswich, MA, USA). Calcium chloride and sodium chloride were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acrylamide was purchased from Fisher Scientific (Pittsburgh, PA, USA). SYBR Gold was purchased from ThermoFisher Scientific (Waltham, MA, USA). All other chemicals were purchased from VWR International (West Chester, PA, USA).
Generation and purification of minicircle DNA
Minicircle DNA was generated via λ-integrase mediated site-specific recombination as described previously (Fogg et al. 2006, 2021). Plasmids pMC333 (Fogg et al. 2021), and pMC339-BbvC1 (Fogg et al. 2006) were used to generate 333 bp and 339 bp minicircles, respectively. Supercoiled, linearized, and nicked samples of each minicircle were prepared as described in Waszkiewicz et al. (2023). Individual minicircle topoisomers (used as markers for electrophoresis) were prepared as described previously (Irobalieva et al. 2015).
Gel electrophoresis
DNA samples were analyzed by gel electrophoresis through 5% polyacrylamide gels (acrylamide: bis-acrylamide = 29:1) in Tris–acetate buffer (pH 8.2) containing 150 mM NaCl and 10 mM CaCl2 at 40 V (~ 2 V/cm) for 25 h. Buffer was continuously recirculated during electrophoresis. Electrophoresis was performed using a PROTEAN II xi cell (Bio-Rad, Hercules, CA, USA). Gels were run at 4 °C (in a walk-in cold room), room temperature (~ 23 °C) (in the laboratory), or 37 °C. The electrophoretic apparatus (including the gel and running buffer) was pre-chilled (for 24 h) or preheated (for 5 h) and the temperature of the recirculating water bath water was verified before electrophoresis. For electrophoresis at 37 °C, the temperature was maintained by circulating 37 °C water from a recirculating water bath through the central core of the electrophoretic apparatus. When electrophoresis was complete, gels were subsequently stained with SYBR Gold then visualized using a FOTO/ANALYST Investigator imaging system and quantified using ImageQuant TL, version 8.1 (GE Healthcare Life Sciences, Marlborough, MA, USA). The identity of the topoisomers was determined by comparison to known purified topoisomer markers. The topoisomer distributions were determined by quantitation of the digital images of fluorescently stained gels using image analysis software as described previously (Waszkiewicz et al. 2023).
AUC experimental design
DNA samples were analyzed by sedimentation velocity analytical ultracentrifugation (SV AUC) in a Beckman Coulter Optima AUC™ at the Canadian Center for Hydrodynamics at the University of Lethbridge in Alberta, Canada. All studies were conducted using an AN50Ti rotor. Approximately 450 μl of each DNA sample (, see Fig. 1 (a) and (b)) in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 10 mM CaCl2 buffer was loaded into each sector of a 12 mm double-sector epon charcoal centerpiece (Beckman Coulter, Indianapolis, USA). Data were collected in intensity mode at 260 nm using the UV/Visible optics. Each sample was measured at five different temperatures of 4, 12, 20, 28, and 37 °C, in this order. At each of these temperatures, the samples were measured at four different rotor speeds of 15, 20, 26.7, and 35.6 krpm, in this order, and each of these experiments was performed in triplicate. After data collection at each temperature, speed, and replicate was completed, the AUC cells were shaken to redistribute the DNA samples thoroughly before the next run. The density and viscosity of the buffer at each temperature were calculated with UltraScan-III, version 4.0, release 6612 (Demeler and Gorbet 2016) (https://ultrascan.aucsolutions.com).
Fig. 1.
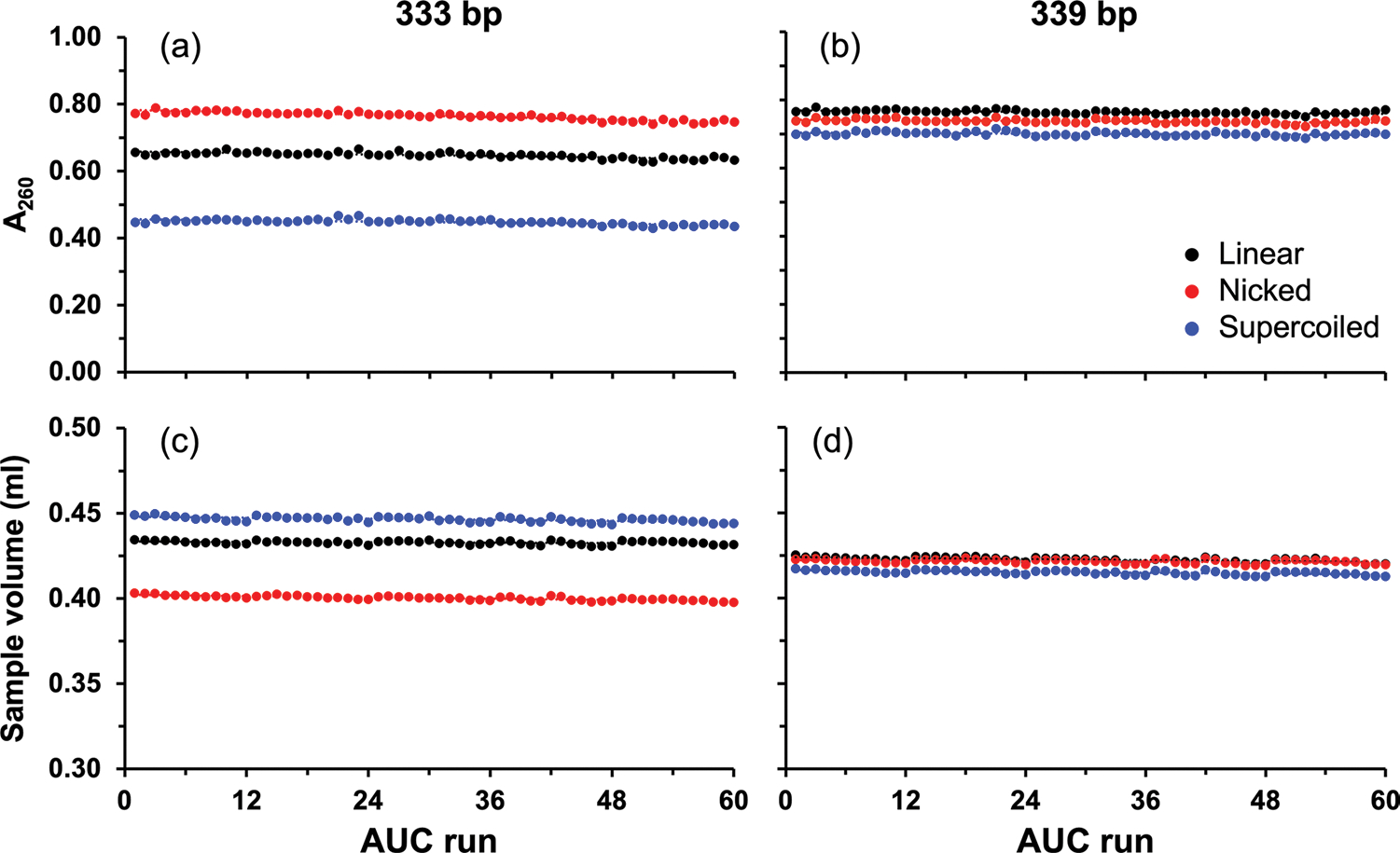
Effect of time on total concentration and volume of DNA. Change of total concentration (top, for a 333 bp and b 339 bp minicircle DNA) and volume (bottom, c 333 bp and d 339 bp minicircle DNA) for each topoisomer (black = linearized, red = nicked, blue = negatively supercoiled) over all experiments performed over a period of four months and 60 runs
AUC data analysis of individual datasets
All data were analyzed with UltraScan-III, version 4.0 (6612) (Demeler and Gorbet 2016) and were collected using the UltraScan data acquisition module (Savelyev et al. 2020). The data were first imported and converted into UltraScan-III OpenAUC format (Cölfen et al. 2010). Parallel distributed data analysis was performed on the UltraScan Science Gateway using XSEDE resources (Expanse, Bridges-2, Anvil) and on the Chinook cluster at the University of Lethbridge (Pierce et al. 2014). Two-dimensional spectrum analysis (2DSA) (Brookes et al. 2010) was used to remove time- and radially- invariant noise, and to fit the meniscus position. A final 2DSA refinement with up to ten iterations was obtained for each dataset. The models resulting from the final 2DSA fit were used to initialize a genetic algorithm analysis (GA) (Brookes and Demeler 2007), which was followed by a Monte Carlo analysis (GA-MC) (Demeler and Brookes 2008) to obtain 95% confidence intervals for the sedimentation and diffusion coefficients, the partial concentration of each solute in a dataset, and to quantify the effect of experimental stochastic noise on the obtained parameter values.
The DNA loading concentrations and volumes for each sample were tracked across all experiments. Loading concentrations were obtained directly from the fitted iterative 2DSA models. The sample volumes were calculated from the fitted meniscus positions and the positions at the cell bottom. All centerpieces used have a cell bottom position of 7.2 cm when at rest. Due to rotor stretch, which varies as a function of rotor speed, this position moves outward during the experiment. Using the rotor stretch calibrations performed in UltraScan (Stoutjesdyk et al. 2020), the stretch value was calculated and subtracted from the meniscus position to obtain the channel positions adjusted for a rotor at rest. The volume was then calculated according to Eq. 1, where is the sector angle (2.5°), is the pathlength (1.2 cm), the cell bottom at rest (7.2 cm), and is the speed-dependent rotor stretch adjusted meniscus position.
| (1) |
Global analysis
The signal generated by each sample for each of the fitted parameters must be considered when selecting a rotor speed for a sedimentation velocity (SV) experiment. The observed signals for the sedimentation and the diffusion transport are dependent on the rotor speed. At high rotor speeds, sedimentation resolution is favored. A fast-sedimenting particle, however, will spend less time diffusing before being pelleted, which limits the diffusion signal that can be observed (Gorbet et al. 2018; Williams et al. 2018). Hence, the diffusion signal is enhanced at lower rotor speeds. The optimal information from both transport parameters can be combined by performing a global analysis over both high-speed and low-speed experiments at the same temperature, and by including replicate measurements in the global fit to maximize precision. UltraScan supports the global fitting of datasets from multiple experimental conditions and can be used with either the 2DSA or GA analysis.
In a global analysis, multiple SV AUC datasets are globally fitted, which enhances signal-to-noise ratios and improves the confidence limits for the determined hydrodynamic parameters (Demeler and Gorbet 2016). This global fit generates models for datasets. The super-global model represents the best fit global model for all datasets. For each dataset, scaled and variable ratio models are also created. In a scaled model, the super-global model is scaled to match the total concentration of each individual dataset. The scaled model has the same set of solutes as the global model, as well as the same ratios between each solute as the best fit global model, but solute concentrations are scaled to match to total concentration observed in an individual sample. A variable ratio model allows the ratio between solutes to vary to account for micro-variation between solutes, as would be observed in a reversible association. In the variable ratio model, the solutes remain identical to those present in the super-global model.
For the homogeneous linearized and nicked minicircle DNA used in this study, global models were created for each sample at each temperature by combining corresponding GA-MC models for that sample performed at different speeds. This combined model was used to initialize a global GA fit. The resulting GA model was then used to initialize a global GA-MC analysis to obtain the sedimentation and diffusion coefficients for linearized and nicked minicircle DNA, and to determine the confidence intervals of each parameter. Heterogeneity was not observed in the SV experiments of linearized and nicked samples. The SV data from negatively supercoiled samples was not appropriate for a global GA-MC analysis due to heterogeneity in composition. Instead, for negatively supercoiled samples, a global 2DSA, followed by a 2DSA-MC (Brookes et al. 2010) was used to globally fit all replicates and speeds for a single temperature. In all cases, the super-global model results are reported. RMSD values were checked by inspecting the variable ratio models, to ensure the obtained fit reports random residuals for each individual dataset. By applying global analysis, the obtained model must fit many datasets under multiple conditions equally well, reducing the chance for over-fitting, and increasing the signal to noise ratio.
Hydrodynamic properties
SV experiments were fitted to the Lamm equation (Lamm 1929) to obtain sedimentation (, Eq. 2) and diffusion (, Eq. 3) coefficients. depends on the molar mass , the frictional coefficient , and the buoyancy of the particle, which is a function of its partial specific volume (PSV), , and solvent density . Both and are inversely proportional to the frictional coefficient of the solute.
| (2) |
| (3) |
where is Avogadro’s number, and is the universal gas constant. The molar masses are 205,721 Da and 209,429 Da for the 333 bp and 339 bp minicircles, respectively, as calculated from the DNA sequences (Fogg et al. 2021) using http://molbiotools.com/dnacalculator. To aid comparisons of AUC experiments performed at different temperatures and buffer conditions, and are reported after correction to standard conditions (20 °C in water, see Eq. 4 and Eq. 5).
| (4) |
| (5) |
correction requires knowledge of the PSV of DNA both at 20 °C in water, and at the experimental temperature in the buffer. Obtaining both values accurately requires instrumentation and sample amounts not available to us. For the purposes of this study, a constant PSV value was used for all temperatures, which is supported by earlier studies performed on DNA (Durchschlag 1986) that indicated a negligible variation of the PSV as a function of temperature. A PSV value for double-stranded DNA in water of 0.55 ml/g as reported earlier (Durchschlag 1986) was used here to correct to standard conditions. The Stokes–Einstein relationship relates the frictional coefficient, , of a solid sphere to the radius of the sphere in a solvent with viscosity (Eq. 6).
| (6) |
For any molecule, spherical or not, the hydrodynamic radius, , is the radius of a sphere consistent with the measured frictional coefficient (Eq. 7).
| (7) |
Substituting Eq. 3 into Eq. 7 yields the hydrodynamic radius, , directly from the diffusion coefficient (Eq. 8).
| (8) |
Results
Rationale
The overall goal of our study was to evaluate DNA as a suitable molecular standard for AUC instruments. To explore the effect of temperature, shape, and mass on the hydrodynamics of DNA, we measured two different lengths (333 bp and 339 bp) of linearized, nicked (open circle) and negatively supercoiled DNA by AUC over the temperature range accessible by the instrument (4–37 °C). Because AUC is sensitive to molar mass and conformation, the two sizes allow us to explore the impact of a small (six bp; ~ 0.6 helical turns) difference in length. Changes in topology and conformation will result in different frictional coefficients, which can also be measured by AUC.
Because AUC requires more material than other methods, such as electron cryotomography (cryoET), atomic force microscopy, or gel electrophoresis, we used negatively supercoiled minicircle DNA obtained from bacterial cells. Although it is possible to further modify the supercoiling post-extraction, and to purify individual topoisomers by preparative polyacrylamide gel electrophoresis, very few laboratories are equipped with the skills, experience, or equipment to do so, especially in the quantities required for AUC. If supercoiled DNA is to be used as an AUC molecular standard, it should be the same level of supercoiling as obtained from bacteria, without the need for additional manipulation.
The molecular mass of these minicircles (~ 206–209 kDa) falls in the middle of the range that can be interrogated using AUC, allowing meaningful data to be acquired over a wide range of rotor speeds, a benefit for a molecular standard. Previously, we performed AUC experiments at 20 °C on minicircles of a similar length (336 bp) to validate theoretical models capable of predicting their hydrodynamic properties (Waszkiewicz et al. 2023). Changing temperature changes DNA flexibility (Driessen et al. 2014) and alters the helical repeat (Kriegel et al. 2018; Duguet 1993), which will likely affect supercoiling-dependent conformations. Correcting for these effects may be non-trivial, possibly limiting the utility of supercoiled DNA as a molecular standard. Thus, we also tested linear and nicked (open circle) DNA samples that lacked supercoiling.
Composition analysis by polyacrylamide electrophoresis
Supercoiling in circular DNA molecules can be described by comparing the linking number , the total number of times the two single DNA strands coil about each other, to , the for a fully relaxed DNA molecule is equal to the number of base pairs divided by the helical repeat. The helical repeat is ~ 10.45 bp/turn at room temperature (~ 23 °C) under the conditions used for AUC (150 mM NaCl and 10 mM C aCl2) (Fogg and Zechiedrich, unpublished results). This helical repeat value translates to for the 333 bp minicircle and for the 339 bp minicircle under these conditions. For simplicity, we hereby refer to values rounded to the nearest integer. Actual (not rounded) values can be found in Supplementary Table 1.
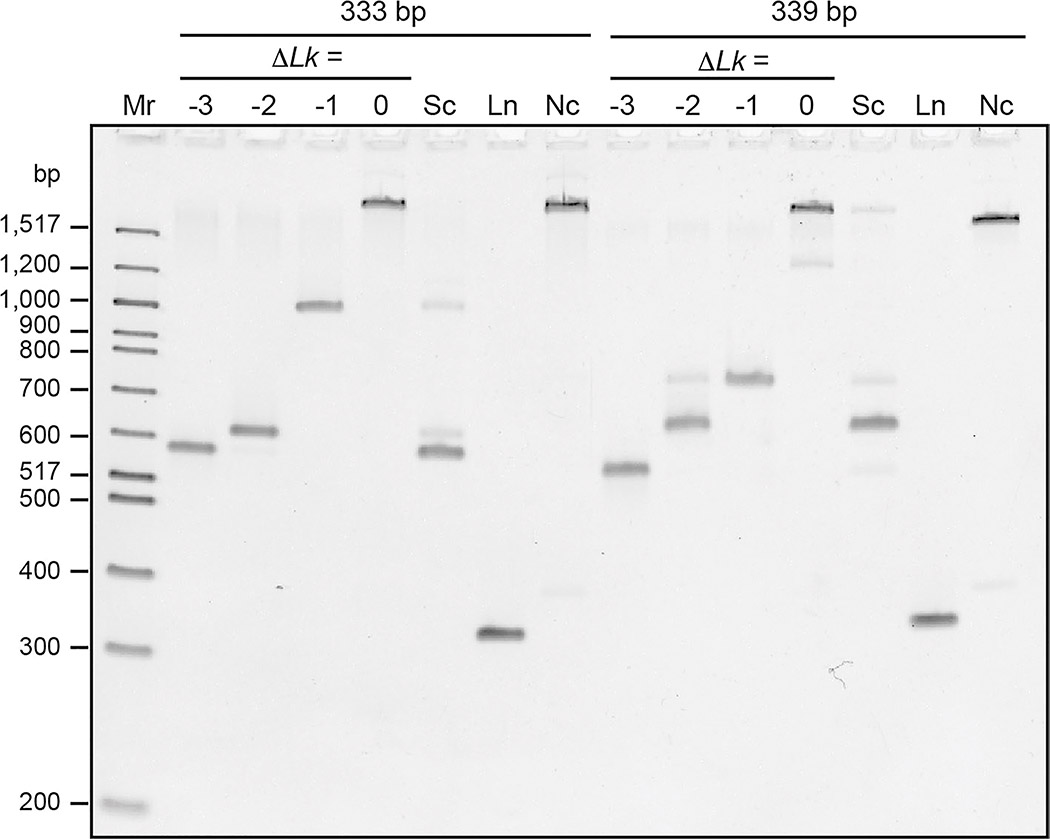
Because of their short lengths, 333 and 339 bp minicircles adopt a narrow distribution of topoisomers reflecting the steady-state level of supercoiling in bacteria (Fig. 2). The 333 bp topoisomer distribution was primarily , as well as and topoisomers. The negatively supercoiled 333 bp sample also contained a trace amount (1%) of supercoiled 666 bp minicircle (double-length minicircle). The 339 bp supercoiled sample contained primarily , and also , and topoisomers. Percentages reported for each species are the mean values ± standard deviation from three separate gels. Because the linking number cannot be changed without breaking one or more of the DNA strands, the topoisomer distribution does not change with temperature or over time.
Fig. 2.
Composition of DNA samples determined by electrophoresis. DNA samples were analyzed by polyacrylamide gel electrophoresis (5% polyacrylamide) in 150 mM NaCl and 10 mM CaCl2 (to replicate the conditions used in analytical ultracentrifugation). Mr: 100 bp DNA ladder (linear DNA, lengths in bp as indicated). 333 and 339 bp minicircle topoisomer markers ( as indicated) were also loaded. Negatively supercoiled (Sc), linear (Ln), and nicked (Nc) 333 bp and 339 bp DNA samples were analyzed. The identity of the topoisomers in the supercoiled samples was determined by comparison to the topoisomer markers
Although the linking number is unchangeable, the helical repeat of DNA depends on temperature (Kriegel et al. 2018; Duguet 1993). , determined by the helical repeat, and consequently , will both change with temperature. In particular, the negatively supercoiled samples become less supercoiled with increasing temperature. To allow comparison of different-sized molecules, was scaled to the DNA length to give the superhelical density (), which is equal to . The estimated values of at each temperature are listed in Supplementary Table 2. It should be emphasized that these values are estimates, not direct measurements. Over the full range of temperatures used in our study, we predict a small but significant temperature-dependent effect on , which should be taken into consideration when comparing data at different temperatures. For linear and nicked molecules, the DNA ends are not constrained, and the of these molecules can freely change without strand breakage. Thus, the effects of changes in helical repeat at different temperatures should be mitigated.
Electrophoresis experiments demonstrated that the linear and nicked samples consisted of single species with high purity (95–100%) (Fig. 2). Thus, a single enzymatic step (linearization or nicking) is sufficient to remove the heterogeneity and produce homogenous samples, which may be better as molecular standards for AUC.
To be suitable as a molecular standard for AUC, the composition and conformation of a molecule in question should be consistent across a wide range of temperatures. To determine whether this was the case for the DNA samples, gel electrophoresis was also performed at 4 °C and 37 °C (Supplementary Fig. 1). DNA samples migrated much more slowly at 4 °C than at higher temperatures. The distances migrated by the linear samples were measured and plotted against temperature (Supplementary Fig. 2 (a)), revealing a linear relationship. This temperature dependence results from buffer viscosity decreasing with increasing temperature (Seeton 2006), which allows for faster migration. To normalize for this generalized effect of temperature, distances migrated by the other samples were compared to the distance migrated by the linear samples. This normalization facilitated comparison among the gels run at different temperatures. Nicked samples migrated much more slowly than the linear and supercoiled samples at all temperatures. There were some subtle changes in the relative migration of the nicked minicircles at different temperatures (Supplementary Fig. 2 (b)), but these differences are relatively small in comparison to the difference between nicked, linear, and supercoiled samples. For the supercoiled samples, there was little change in the relative migration of the major species within each sample (Supplementary Fig. 2 (c) and (d)).
The electrophoresis results confirm that the composition of the DNA samples does not change with temperature. Any differences in relative electrophoretic migration were relatively subtle suggesting that the global conformation also does not significantly change, even for the supercoiled samples. The ability to use an orthogonal technique to verify composition and conformation meets another requirement for the suitability of DNA samples as a molecular standard for AUC.
Investigating the suitability of DNA as a molecular standard
As described above, a molecular standard for AUC must be stable for an extended period of time, produce meaningful data at a wide range of different rotor speeds, and exhibit predictable hydrodynamic behavior across a wide range of temperatures. We investigated whether DNA would meet these requirements by testing each DNA sample repeatedly at five different rotor speeds and five different temperatures. Each experiment was performed in triplicate and fitted globally over all replicates and rotor speeds. The DNA samples remained in the AUC cells throughout the course of these experiments and were resuspended by shaking the cell before each subsequent AUC run. To ascertain that DNA did not aggregate, degrade, or become lost due to irreversible pelleting, or sticking to cell windows or centerpiece surfaces, we monitored the sample composition, DNA concentration (Fig. 1 (a) and (b)), and sample volume (Fig. 1 (c) and (d)) over time. Linear fit coefficients and the percent sample loss as a function of concentration and volume for each sample over the course of all experiments are shown in Table 1. The fits indicated a negligible drift in the total concentration, which was < 5% for all samples over the course of the measurements, which included 60 runs with the same sample, collected over four months, five temperatures, four rotor speeds, and three replicates for each condition. Volume changes were negligible as well (Table 1). These data attest to the superior stability of double-stranded DNA of these lengths under these conditions in contrast to other biological macromolecules, such as RNA and protein, and suggest the suitability of DNA as a molecular standard for AUC experiments.
Table 1.
DNA sample and volume loss over four months
| Sample | Sample volume | ||||
|---|---|---|---|---|---|
|
| |||||
| Slope | Sample loss, % | Slope | Volume loss, % | ||
|
| |||||
| 333 bp | Linear | − 0.0004 | 3.58 | − 0.00002 | 0.27 |
| Nicked | − 0.0006 | 4.52 | − 0.00006 | 0.88 | |
| Supercoiled | − 0.0003 | 3.88 | − 0.00005 | 0.66 | |
| 339 bp | Linear | − 0.0002 | 1.53 | − 0.00005 | 0.70 |
| Nicked | − 0.0001 | 0.84 | − 0.00003 | 0.42 | |
| Supercoiled | − 0.0002 | 1.59 | − 0.00004 | 0.57 | |
Effect of temperature, circularity, and supercoiling on hydrodynamic parameters of DNA
The sedimentation coefficients, diffusion coefficients, and hydrodynamic radii of each linearized and nicked minicircle DNA sample are shown in Fig. 3 as a function of temperature. With increasing temperature, the sedimentation and diffusion coefficients of all linearized and nicked samples increased while the hydrodynamic radii decreased. The changes in the sedimentation and diffusion coefficients as a function of temperature appeared to follow a linear relationship. Fitting to a straight line resulted in values close to 1 for most of the fits (Table 2). The PSV of DNA does not change significantly as a function of temperature (Durchschlag 1986) or topology (Waszkiewicz et al. 2023). Because the PSV and the molar mass of the DNA samples remain constant (across the different forms—linearized, nicked, and supercoiled), the change in sedimentation and diffusion coefficients of each sample, when corrected to standard conditions, must be a consequence of changes in the frictional coefficient caused by changes in the conformation.
Fig. 3.
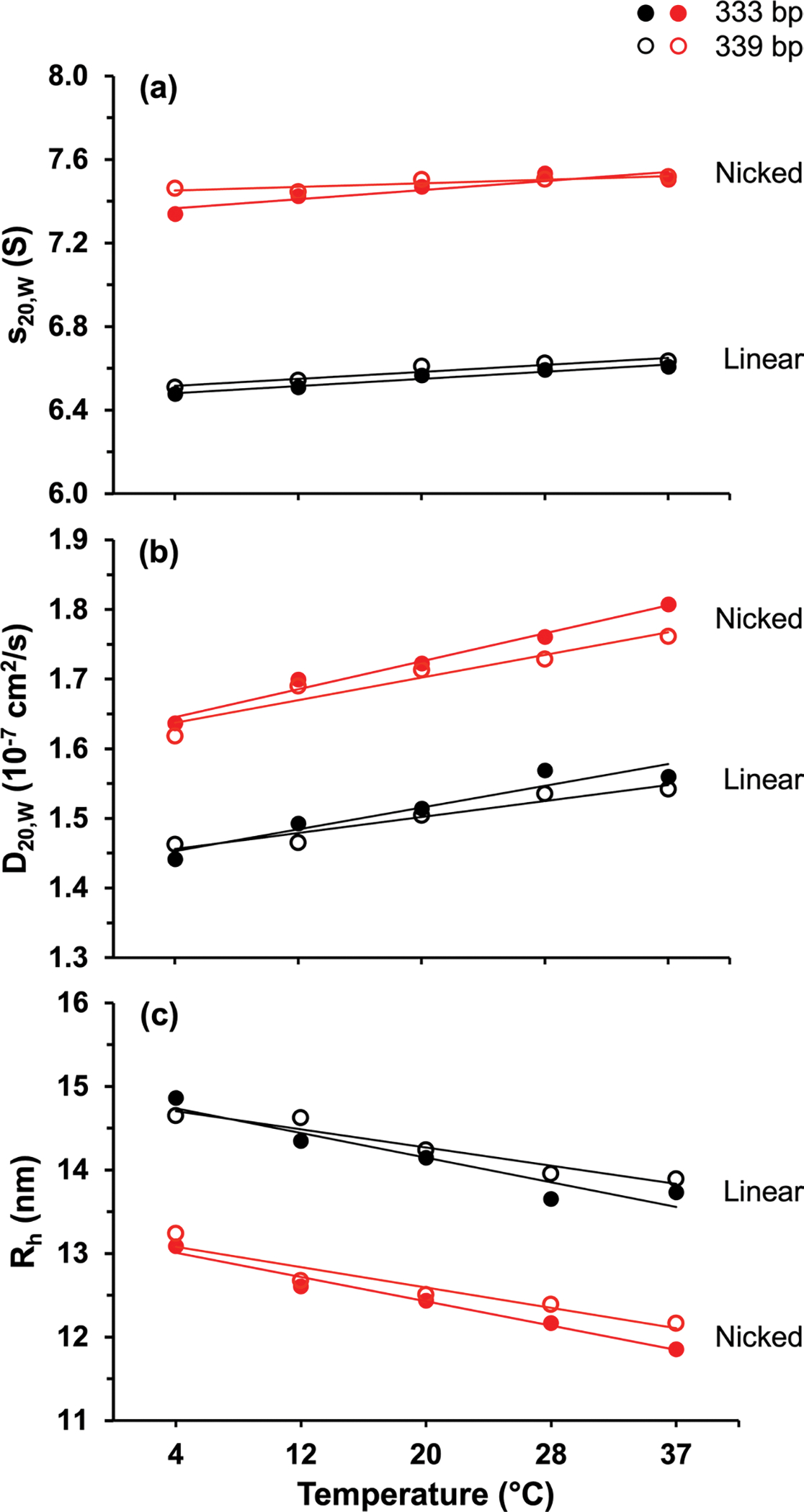
Hydrodynamic measurements of DNA. Hydrodynamic measurements of linearized (black) and nicked (red) minicircle DNA of 333 bp (filled circles) or 339 bp (open circles) length as a function of temperature for (top panel), (center panel), and (bottom panel)
Table 2.
Linear regression data for sedimentation and diffusion coefficients as a function of temperature
| 333 bp | 339 bp | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Linear | Nicked | Linear | Nicked | |||||
|
| ||||||||
| Slope | Slope | Slope | Slope | |||||
|
| ||||||||
| 0.034 s/°C | 0.96 | 0.044 s/°C | 0.83 | 0.033 s/°C | 0.91 | 0.017 s/°C | 0.74 | |
| 0.031 cm2/(s °C) | 0.91 | 0.040 cm2/(s °C) | 0.98 | 0.023 cm2/(s °C) | 0.93 | 0.033 cm2/(s °C) | 0.92 | |
| − 0.295 nm/°C | 0.90 | − 0.291 nm/°C | 0.98 | − 0.218 nm/°C | 0.93 | − 0.243 nm/°C | 0.90 | |
At each temperature, a clear difference was observed between linearized and nicked minicircles with the linearized DNA samples always sedimenting slower than the nicked. This result is consistent with earlier studies showing circular DNA has a reduced radius of gyration relative to linear DNA (Robertson et al. 2006). These observations suggest that linear and nicked DNA are more rigid and extended at lower temperatures and become more compact or flexible at higher temperatures. Previous research showed that increasing temperature decreases the persistence length of linear duplex DNA (Brunet et al. 2018), which is consistent with our observations. The persistence length is a measure of polymer stiffness, suggesting that the molecule becomes more flexible and less elongated with increasing temperature. Previous nuclear magnetic resonance experiments on 24 bp linear duplex DNA determined that the diffusion coefficient of short linear DNA increases with increasing temperature (Lapham et al. 1997), which is also consistent with our results.
We observed that the rate of change of sedimentation and diffusion coefficients as a function of temperature for the linear and nicked DNAs were greater for the shorter DNA length (333 bp). While these changes were subtle, they were significant (see Fig. 3). Although the molar masses of the two lengths of minicircles (333 and 339 bp) differ by less than 2%, their differences in hydrodynamic properties demonstrate the sensitivity of AUC.
Although heterogenous, the supercoiled samples consisted primarily (> 70%) of a single topoisomer (Fig. 2). Moreover, we previously showed that small differences in have only small effects on sedimentation (Waszkiewicz et al. 2023). At 4 °C, the first temperature evaluated, we observed a primarily homogeneous behavior (see Fig. 4). We, therefore, hoped that supercoiled DNA, as isolated out of bacteria, may have utility as a molecular standard. Overall, however, the sedimentation behavior of the supercoiled samples, and their response to temperature was much more complex than we had anticipated (Fig. 4, temperatures above 4 °C). The results reveal that supercoiled samples, when isolated directly from bacterial cells, are not suitable as a molecular standard. Nevertheless, the observations provide new insight into the behavior of supercoiled DNA.
Fig. 4.
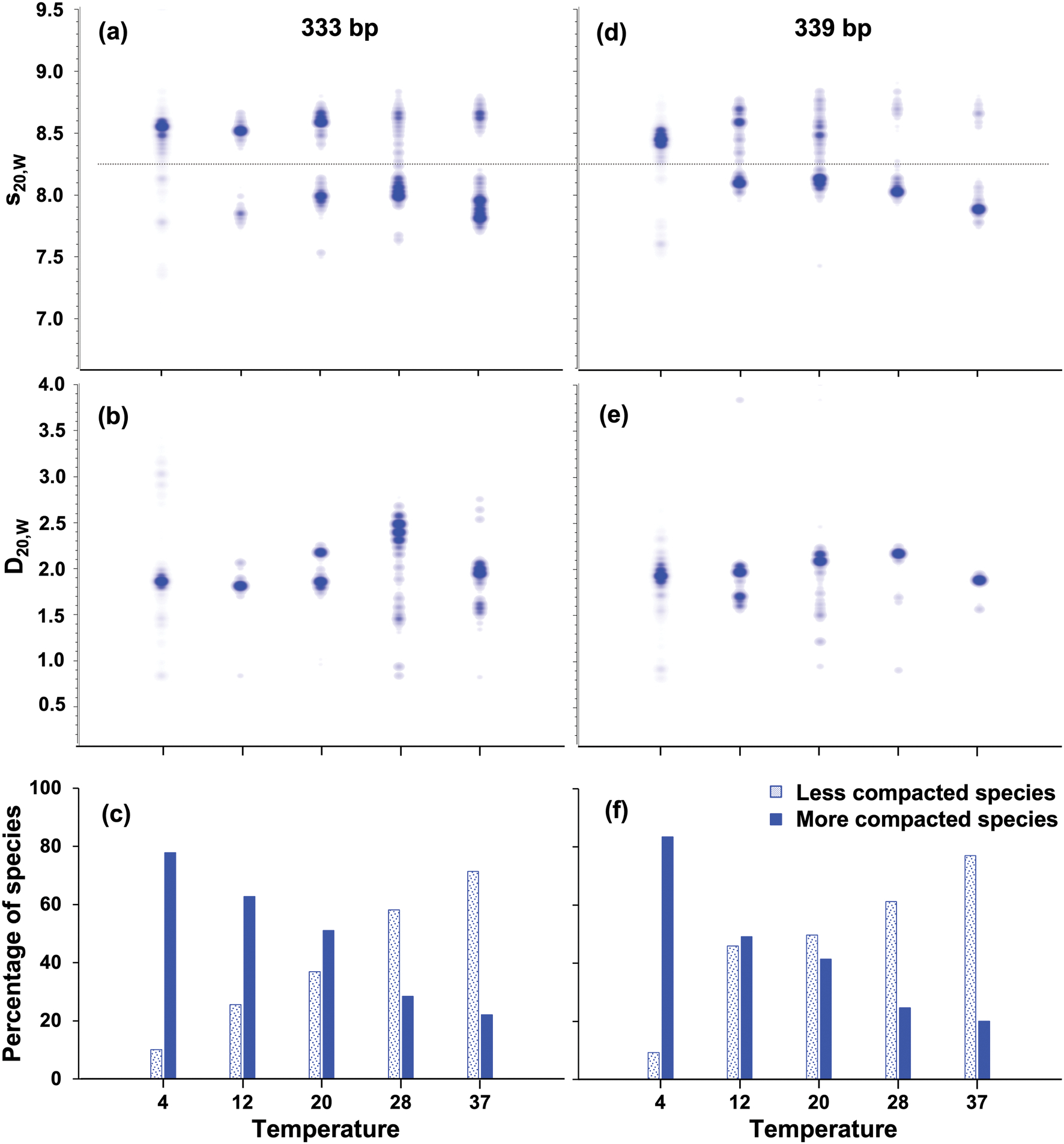
Sedimentation and diffusion coefficient distributions of DNA. Sedimentation coefficient distributions(top), diffusion coefficient distributions (center), and relative percentage of more compact (solid texture) and less compact (pattern texture) supercoiled minicircles. 333 bp minicircle results are shown in the left panels, and 339 bp supercoiled minicircle results are shown in the right panels. The horizontal line in panels (a) and (b) denotes the integration boundary for the integrated groups shown in (c) and (f)
Whereas the linearized and nicked minicircle DNA samples were sufficiently homogeneous to be analyzed by global GA analysis, the negatively supercoiled samples of multiple different topoisomers were best modeled by a global 2DSA, because their heterogeneity was too great to lend themselves to parsimonious regularization. The global 2DSA analysis revealed that multiple species with differing sedimentation coefficients were present in the supercoiled samples (Fig. 4). Two distinct peaks were observed in the sedimentation coefficient distributions (Fig. 5). One of the peaks with a higher sedimentation coefficient was predominant at lower temperatures and presumably represents a more compact conformation. With increasing temperature, a second peak with lower sedimentation coefficient emerged. Although this peak presumably represents a less compact conformation, its sedimentation coefficient is still higher than that for nicked DNA, suggesting that supercoiled DNA may shift to more open conformations at higher temperatures, but never becomes as open as nicked DNA. These two groups and this temperature effect were observed for both the 333 bp and the 339 bp minicircles. To quantify the relative change in concentration for each major peak shown in the supercoiled sample of Fig. 5, we integrated each peak integration limits: 7.063–8.225 s and 8.268–9.134 s). This integration boundary is shown as a dashed line in Fig. 4 (a) and (d). The relative concentration of each species changed from a predominantly fast-sedimenting species at low temperatures to a significantly more populated species with a lower sedimentation rate at higher temperatures (Fig. 4 (c) and (f)).
Fig. 5.
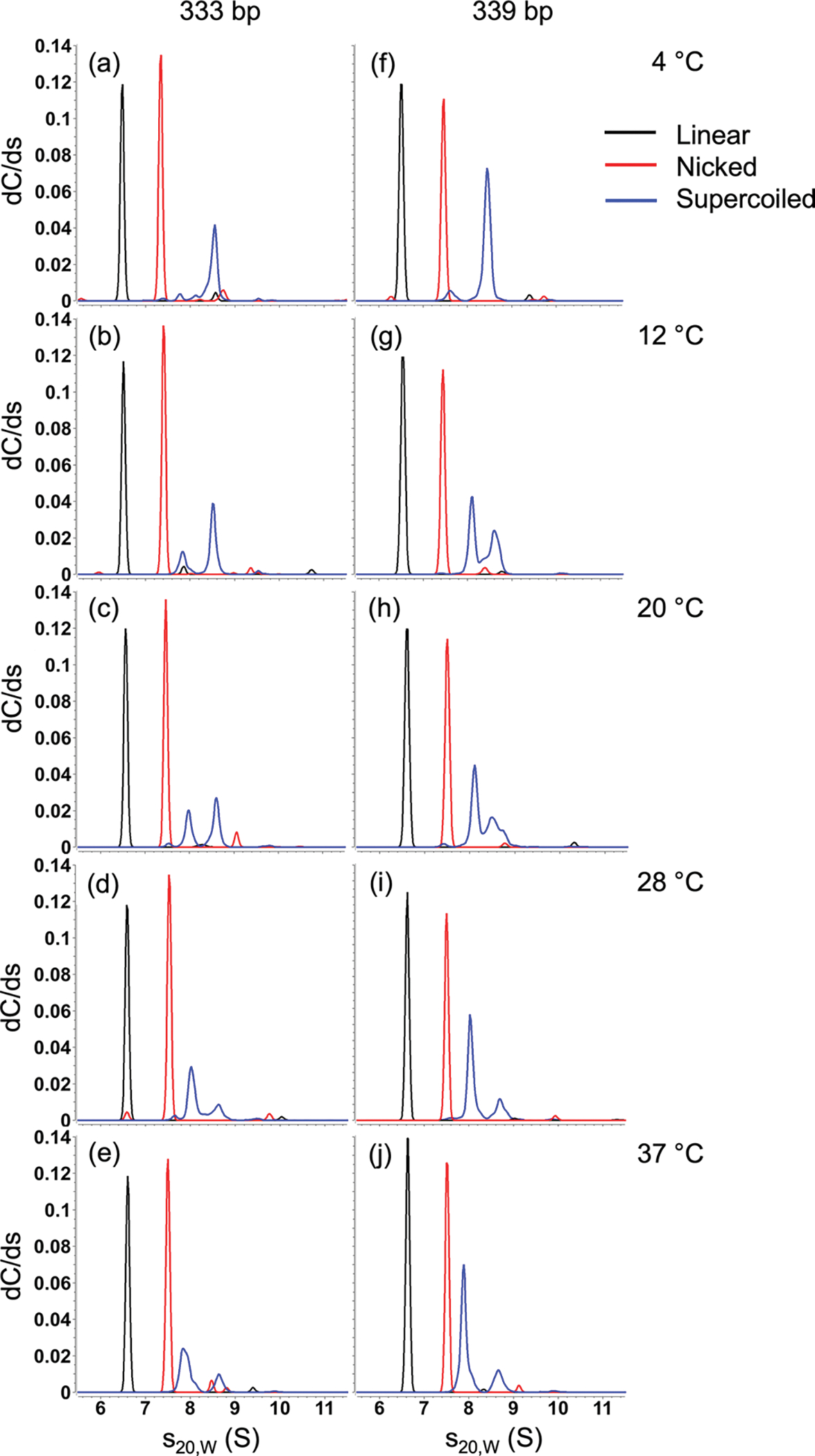
Effect of temperature on sedimentation coefficient distributions of DNA. Sedimentation coefficients of linearized (black), nicked (red) and supercoiled (blue) 333 bp (left) and 339 bp (right) DNA minicircles as a function of temperature
To facilitate comparison with the linear and nicked samples, weighted averages of the sedimentation coefficients of the supercoiled samples were plotted as a function of temperature (see Supplementary Fig. 3). Whereas the sedimentation coefficients of the linear and nicked samples increased with temperature, the opposite was the case for the supercoiled samples and the weighted average sedimentation coefficients decreased with temperature.
While the sedimentation coefficient results obtained for the negatively supercoiled minicircle samples provided a clear shifting pattern to slower sedimentation values as the temperature was increased, the diffusion coefficients (Fig. 4 (b) and (e)) were difficult to distinguish, and individual species (resolvable by gel electrophoresis) could not be resolved. Diffusion coefficients are derived from the fitted slope of the sedimenting boundary, which is cross-correlated with the heterogeneity in the boundary. Boundary slopes from multiple species tend to merge into boundaries with average slopes, decreasing the resolution of the diffusion signal in heterogeneous samples.
A complete list of the sedimentation and diffusion coefficient values for the linear, nicked, and supercoiled samples at each temperature can be found in Table 3. For the linear and nicked samples, there was very high precision in these measured values. For the supercoiling samples, the values reported with 95% confidence levels covered a broader range for each measurement.
Table 3.
Sedimentation and diffusion coefficients of minicircle topoisomers with 95% confidence limits
| Sample | (°C) | (10−7 cm2 s− 1) | |||
|---|---|---|---|---|---|
|
| |||||
| 333 bp | Linear | 4 | 6.48 | 1.44 | |
| 12 | 6.51 | 1.49 | |||
| 20 | 6.57 | 1.51 | |||
| 28 | 6.59 | 1.57 | |||
| 37 | 6.61 | 1.56 | |||
| Nicked | 4 | 7.34 | 1.64 | ||
| 12 | 7.42 | 1.70 | |||
| 20 | 7.47 | 1.72 | |||
| 28 | 7.53 | 1.76 | |||
| 37 | 7.50 | 1.81 | |||
| Supercoiled | Less compact species | 4 | 7.84 ± 0.63 | 1.61 ± 1.76 | |
| 12 | 7.86 ± 0.39 | 1.95 ± 0.38 | |||
| 20 | 7.99 ± 0.29 | 2.17 ± 0.73 | |||
| 28 | 8.03 ± 0.27 | 2.40 ± 0.75 | |||
| 37 | 7.90 ± 0.41 | 1.97 ± 0.44 | |||
| More compact species | 4 | 8.52 ± 0.30 | 2.00 ± 1.28 | ||
| 12 | 8.52 ± 0.34 | 1.81 ± 0.90 | |||
| 20 | 8.60 ± 0.32 | 1.85 ± 0.52 | |||
| 28 | 8.56 ± 0.39 | 1.56 ± 0.61 | |||
| 37 | 8.65 ± 0.30 | 1.55 ± 0.29 | |||
| (°C) | (10−7 cm2 s− 1) | ||||
| 339 bp | Linear | 4 | 6.51 | 1.46 | |
| 12 | 6.54 | 1.47 | |||
| 20 | 6.61 | 1.50 | |||
| 28 | 6.62 | 1.54 | |||
| 37 | 6.63 | 1.54 | |||
| Nicked | 4 | 7.46 | 1.62 | ||
| 12 | 7.44 | 1.69 | |||
| 20 | 7.50 | 1.71 | |||
| 28 | 7.50 | 1.73 | |||
| 37 | 7.52 | 1.76 | |||
| Supercoiled | Less compact species | 4 | 7.71 ± 0.47 | 1.63 ± 0.50 | |
| 12 | 8.09 ± 0.34 | 1.99 ± 0.49 | |||
| 20 | 8.10 ± 0.33 | 2.10 ± 0.51 | |||
| 28 | 8.03 ± 0.31 | 2.16 ± 0.56 | |||
| 37 | 7.90 ± 0.39 | 1.89 ± 0.31 | |||
| More compact species | 4 | 8.45 ± 0.32 | 1.93 ± 0.93 | ||
| 12 | 8.57 ± 0.39 | 1.72 ± 0.42 | |||
| 20 | 8.57 ± 0.44 | 1.71 ± 0.60 | |||
| 28 | 8.68 ± 0.44 | 1.69 ± 0.36 | |||
| 37 | 8.68 ± 0.37 | 1.55 ± 0.34 | |||
Where confidence limits are not shown, the confidence limits were exactly zero as assessed by Monte Carlo analysis. The values of linear and nicked samples are obtained from global GA-MC models, whereas the values of supercoiled samples are obtained from global 2DSA-MC models
Discussion
The goal of this study was to assess the suitability of different isomeric forms of double-stranded DNA as molecular standards for validating AUC instruments. Linearized and nicked circular samples demonstrated a very high degree of homogeneity, with highly reproducible sedimentation and diffusion coefficients. The effect of temperature on sedimentation and diffusion of these samples appeared to follow a linear relationship. Global analysis included triplicate measurements at five different speeds, reducing the 95% confidence bands of each parameter value to near zero, producing very narrow sedimentation/diffusion coefficient distributions. Sedimentation and diffusion coefficients increased slightly with temperature, with distinct differences in both parameters between the two different minicircles. These parameter differences were greater at higher temperatures, and less noticeable at lower temperatures. Their stability in the AUC experiments, tested over four months on the same sample, resuspended by shaking the cell, and measured over 60 times at multiple rotor speeds, triplicate measurements, and temperatures ranging between 4 °C and 37 °C proved to be exceptional. No significant volume or concentration loss could be measured for any topoisomer, but linearized and nicked circles provided better homogeneity than supercoiled minicircles. Together, these results suggest that both linearized and open circular DNA molecules are quite suitable as a molecular standard, and satisfy all of the requirements.
Supercoiled samples isolated from bacterial cultures failed to satisfy the requirements for molecular standards. These heterogenous supercoiled samples yielded results that are complex and difficult to interpret. We had previously shown that purified supercoiled minicircle topoisomers with defined adopt multiple conformations (Irobalieva et al. 2015). This result suggested that individual negatively supercoiled minicircles may interconvert among the different conformations over time. The rate at which this possible conformational interchange takes place is not yet clear. Molecular dynamics revealed a possible pathway for interconversion, but could not provide accurate kinetics because of the way the solvent was simulated (Irobalieva et al. 2015). Future studies will be important in interpreting the species seen in AUC. The shift from a single major peak in the negatively supercoiled samples at lower temperatures to multiple peaks at higher temperatures suggests that the conformational equilibrium is shifted towards more open conformations. The fact that we observe differently sedimenting species suggests that the different conformations have sufficiently long lifetimes to be detected. If the interconversion is rapid, a bulk observation like AUC would report a weight averaged sedimentation profile for all interconverting conformations.
Electrophoresis and AUC are orthogonal approaches. The temperature-dependent effects observed in the AUC data were not reflected in the electrophoresis data. Electrophoresis and AUC measure different parameters. The separation of supercoiled topoisomers by electrophoresis is poorly understood but is thought to be closely correlated to their average writhe in solution (Vetcher et al. 2010; Zivanovic et al. 1986). Electrophoretic mobility is not a direct measure of hydrodynamic radius, especially when comparing different topologies. Sedimentation velocity, on the other hand, is inversely proportional to the hydrodynamic radius. The precise effect of temperature on supercoiled minicircles (and its reversibility) requires further study with purified supercoiled minicircles with a distinct .
Conclusions
Our results demonstrate how AUC can be used to distinguish between the solution behavior of 333 bp and 339 bp minicircle DNA and differentiate these DNAs according to topology (linear, nicked, and negatively supercoiled). Global fitting across multiple replicates and multiple rotor speeds provided superior precision, significantly reducing the confidence limits for each measured parameter compared to single AUC experiments as assessed by Monte Carlo analysis. Temperature affects the sedimentation and diffusion coefficients, as well as the hydrodynamic radius of all the tested forms of minicircle DNA in a systematic fashion, and, while subtle, these changes were readily measured and distinguished by AUC. Both linear and nicked DNA samples displayed highly homogeneous and well-defined sedimentation behavior across a wide range of temperatures and over time. There was a linear relationship among sedimentation, diffusion, and hydrodynamic radius as a function of temperature. A 2% molar mass difference in DNA molecules could be clearly resolved. These results illustrate the suitability of these DNAs for molecular standards over the range of temperatures accessible in AUC instruments. In addition, AUC revealed a significant change in sedimentation and diffusion transport due to conformational changes as a function of temperature, but was unable to resolve the topoisomer composition of supercoiled species isolated directly from bacterial cells. Our results further show that under conditions used for these experiments, DNA of different topological states is remarkably stable during AUC and does not degrade over four months of replicate experiments performed at different speeds and temperatures. Our results suggest that linear and nicked circular DNA have superior properties compared to other biological macromolecules and may be suitable as a molecular standard for AUC. Furthermore, the results revealed by this initial analysis have opened both a new method and a new line of investigation into the dynamics of supercoiled DNA at different temperatures. We suggest that the analysis for an eventually commercially available DNA molecular standard for AUC must be repeated on multiple calibrated instruments to confirm the results.
Supplementary Material
Acknowledgements
This work was funded by National Institutes of Health grant R35 GM141793 and National Science Foundation grant MCB 2107527 (to LZ), and the Biomolecular Interaction Technology Center, University of Delaware, the Canada 150 Research Chairs program C150–2017-00015, the National Institutes of Health grant 1R01GM120600, and the Canadian Natural Science and Engineering Research Council Discovery Grant DG-RGPIN-2019–05637 (to BD). The Canadian Center for Hydrodynamics is funded by the Canada Foundation for Innovation grant CFI-37589 (BD). UltraScan supercomputer calculations were supported through NSF/XSEDE grant TG-MCB070039N, and University of Texas grant TG457201 (BD).
Footnotes
Declarations
Conflict of interest Jonathan M. Fogg, Daniel J. Catanese, Jr., and Lynn Zechiedrich are co-inventors on issued and pending patents covering the supercoiled minicircle technology and uses and furthermore hold an equity stake in Twister Biotech, Inc. The other authors declare they have no competing financial interests.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00249-023-01671-y.
Data availability
The UltraScan software used to analyze the AUC data is open source and freely available from the Github repository (https://github.com/ehb54/ultrascan3). The AUC data are available in openAUC format upon request from the authors, and is stored in the UltraScan LIMS server at the Canadian Center for Hydrodynamics. Electrophoresis quantitation data is available upon request from the authors.
References
- Brookes E, Demeler B (2006) Genetic algorithm optimization for obtaining accurate molecular weight distributions from sedimentation velocity experiments. Prog Colloid Polym Sci 131:78–82. [Google Scholar]
- Brookes E, Cao W, Demeler B (2010) A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J 39(3):405–414. 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- Brookes E, Demeler B. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments. GECCO ‘07: Proceedings of the 9th annual conference on Genetic and evolutionary computation, London, July 7–11, 2007, 361–368, CM 978–1–59593–697–4/07/0007, 10.1145/1276958.1277035. [DOI] [Google Scholar]
- Brunet A, Salomé L, Rousseau P, Destainville N, Manghi M, Tardin C (2018) How does temperature impact the conformation of single DNA molecules below melting temperature? Nucleic Acids Res 46(4):2074–2081. 10.1093/nar/gkx1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Demeler B (2005) Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution of the Lamm equation. Biophys J 89(3):1589–1602. 10.1529/biophysj.105.061135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Demeler B (2008) Modeling analytical ultracentrifugation experiments with an adaptive space-time finite element solution for multicomponent reacting systems. Biophys J 95(1):54–65. 10.1529/biophysj.107.123950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cölfen H, Laue TM, Wohlleben W, Schilling K, Karabudak E, Langhorst BW, Brookes E, Dubbs B, Zollars D, Rocco M, Demeler B (2010) The open AUC project. Eur Biophys J EBJ 39(3):347–359. 10.1007/s00249-009-0438-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeler B, Brookes E (2008) Monte Carlo analysis of sedimentation experiments. Colloid Polym Sci 286(2):129–137. 10.1007/s00396-007-1714-9 [DOI] [Google Scholar]
- Demeler B, Gorbet GE (2016) Analytical ultracentrifugation data analysis with UltraScan-III. In: Uchiyama S, Arisaka F, Stafford W, Laue T (eds) Analytical ultracentrifugation. Springer, Tokyo, pp 119–143. 10.1007/978-4-431-55985-6_8 [DOI] [Google Scholar]
- Driessen RP, Sitters G, Laurens N, Moolenaar GF, Wuite GJ, Goosen N, Dame RT (2014) Effect of temperature on the intrinsic flexibility of DNA and its interaction with architectural proteins. Biochemistry 53(41):6430–6438. 10.1021/bi500344j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguet M (1993) The helical repeat of DNA at high temperature. Nucleic Acids Res 21(3):463–468. 10.1093/nar/21.3.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durchschlag H (1986) Specific volumes of biological macromolecules and some other molecules of biological interest. In: Hinz HJ (ed) Thermodynamic data for biochemistry and biotechnology. Springer, Berlin, Heidelberg, pp 45–128. 10.1007/978-3-642-71114-5_3 [DOI] [Google Scholar]
- Fogg JM, Kolmakova N, Rees I, Magonov S, Hansma H, Perona JJ, Zechiedrich EL (2006) Exploring writhe in supercoiled minicircle DNA. J Phys Condens Matter 18(14):S-145–S−159. 10.1088/0953-8984/18/14/S01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg JM, Judge AK, Stricker E, Chan HL, Zechiedrich L (2021) Supercoiling and looping promote DNA base accessibility and coordination among distant sites. Nat Commun 12(1):5683. 10.1038/s41467-021-25936-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García de la Torre J, Hernández Cifre JG (2020) Hydrodynamic properties of biomacromolecules and macromolecular complexes: concepts and methods a tutorial mini-review. J Mol Biol 432(9):2930–2948. 10.1016/j.jmb.2019.12.027 [DOI] [PubMed] [Google Scholar]
- Gorbet GE, Mohapatra S, Demeler B (2018) Multi-speed sedimentation velocity implementation in UltraScan-III. Eur Biophys J 47(7):825–835. 10.1007/s00249-018-1297-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC, Lebowitz J, Demeler B (1994) Analytical ultracentrifugation of complex macromolecular systems. Biochemistry 33(45):13155–13163. 10.1021/bi00249a001 [DOI] [PubMed] [Google Scholar]
- Harding SE (2017) From nano to micro: the huge dynamic range of the analytical ultracentrifuge for characterising the sizes, shapes and interactions of molecules and assemblies in Biochemistry and polymer science. Eur Biophys J 47(7):697–707. 10.1007/s00249-018-1321-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson A, Montina T, Hazendonk P, Lomonte B, Neves-Ferreira AG, Demeler B (2023) SDS-induced hexameric oligomerization of myotoxin-II from Bothrops asper assessed by sedimentation velocity and nuclear magnetic resonance. Eur Biophys J. 10.1007/s00249-023-01641-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irobalieva RN, Fogg JM, Catanese DJ, Sutthibutpong T, Chen M, Barker AK, Ludtke SJ, Harris SA, Schmid MF, Chiu W, Zechiedrich L (2015) Structural diversity of supercoiled DNA. Nat Commun 12(6):8440. 10.1038/ncomms9440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel F, Matek C, Dršata T, Kulenkampff K, Tschirpke S, Zacharias M, Lankaš F, Lipfert J (2018) The temperature dependence of the helical twist of DNA. Nucleic Acids Res 46(15):7998–8009. 10.1093/nar/gky599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm O (1929) Die Differentialgleichung der Ultrazentrifugierung. Ark Mat Astr Fys 21B:1–4. [Google Scholar]
- Lapham J, Rife JP, Moore PB, Crothers DM (1997) Measurement of diffusion constants for nucleic acids by NMR. J Biomol NMR 10(3):255–262. 10.1023/a:1018310702909 [DOI] [PubMed] [Google Scholar]
- Parker W, Song PS (1992) Protein structures in SDS micelle-protein complexes. Biophys J 61(5):1435–1439. 10.1016/S0006-3495(92)81949-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce MS. Marru BD, Singh R, Gorbet G (2014) The Apache Airavata Application Programming Interface: Overview and Evaluation with the UltraScan Science Gateway. In: Proceedings of the 9th Gateway Computing Environments Workshop (GCE ‘14). IEEE Press, Piscataway, NJ, USA, 25–29. 10.1109/GCE.2014.15 [DOI] [Google Scholar]
- Robertson RM, Laib S, Smith DE (2006) Diffusion of isolated DNA molecules: dependence on length and topology. Proc Natl Acad Sci USA 103(19):7310–7314. 10.1073/pnas.0601903103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelyev A, Gorbet GE, Henrickson A, Demeler B (2020) Moving analytical ultracentrifugation software to a good manufacturing practices (GMP) environment. PLoS Comput Biol 16(6):e1007942. 10.1371/journal.pcbi.1007942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeton CJ (2006) Viscosity temperature correlation for liquids. Tribol Lett 22:67–78. [Google Scholar]
- Stoutjesdyk M, Henrickson A, Minors G, Demeler B (2020) A calibration disk for the correction of radial errors from chromatic aberration and rotor stretch in the Optima AUC™ analytical ultracentrifuge. Eur Biophys J 49(8):701–709. 10.1007/s00249-020-01434-z [DOI] [PubMed] [Google Scholar]
- Vetcher AA, McEwen AE, Abujarour R, Hanke A, Levene SD (2010) Gel mobilities of linking-number topoisomers and their dependence on DNA helical repeat and elasticity. Biophys Chem 148(1–3):104–111. 10.1016/j.bpc.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszkiewicz R, Ranasinghe M, Fogg JM, Catanese DJ, Ekiel-Jeżewska ML, Lisicki M, Demeler B, Zechiedrich L, Szymczak P (2023) DNA supercoiling-induced shapes alter minicircle hydrodynamic properties. Nucleic Acids Res 51:4027–4042. 10.1093/nar/gkad183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TL, Gorbet GE, Demeler B (2018) Multi-speed sedimentation velocity simulations with UltraScan-III. Eur Biophys J 47(7):815–823. 10.1007/s00249-018-1308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ghirlando R, Alfonso C, Arisaka F, Attali I, Bain DL, Bakhtina MM, Becker DF, Bedwell GJ, Bekdemir A, Besong TM, Birck C, Brautigam CA, Brennerman W, Byron O, Bzowska A, Chaires JB, Chaton CT, Cölfen H, Connaghan KD, Crowley KA, Curth U, Daviter T, Dean WL, Díez AI, Ebel C, Eckert DM, Eisele LE, Eisenstein E, England P, Escalante C, Fagan JA, Fairman R, Finn RM, Fischle W, de la Torre JG, Gor J, Gustafsson H, Hall D, Harding SE, Cifre JG, Herr AB, Howell EE, Isaac RS, Jao SC, Jose D, Kim SJ, Kokona B, Kornblatt JA, Kosek D, Krayukhina E, Krzizike D, Kusznir EA, Kwon H, Larson A, Laue TM, Le Roy A, Leech AP, Lilie H, Luger K, Luque-Ortega JR, Ma J, May CA, Maynard EL, Modrak-Wojcik A, Mok YF, Mücke N, Nagel-Steger L, Narlikar GJ, Noda M, Nourse A, Obsil T, Park CK, Park JK, Pawelek PD, Perdue EE, Perkins SJ, Perugini MA, Peterson CL, Peverelli MG, Piszczek G, Prag G, Prevelige PE, Raynal BD, Rezabkova L, Richter K, Ringel AE, Rosenberg R, Rowe AJ, Rufer AC, Scott DJ, Seravalli JG, Solovyova AS, Song R, Staunton D, Stoddard C, Stott K, Strauss HM, Streicher WW, Sumida JP, Swygert SG, Szczepanowski RH, Tessmer I, Toth RT 4th, Tripathy A, Uchiyama S, Uebel SF, Unzai S, Gruber AV, von Hippel PH, Wandrey C, Wang SH, Weitzel SE, Wielgus-Kutrowska B, Wolberger C, Wolff M, Wright E, Wu YS, Wubben JM, Schuck P (2015) A multilaboratory comparison of calibration accuracy and the performance of external references in analytical ultracentrifugation. PLoS ONE 10(5):e0126420. 10.1371/journal.pone.0126420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanovic Y, Goulet I, Prunell A (1986) Properties of supercoiled DNA in gel electrophoresis The V-like dependence of mobility on topological constraint DNA-matrix interactions. J Mol Biol 192(3):645–660. 10.1016/0022-2836(86)90282-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The UltraScan software used to analyze the AUC data is open source and freely available from the Github repository (https://github.com/ehb54/ultrascan3). The AUC data are available in openAUC format upon request from the authors, and is stored in the UltraScan LIMS server at the Canadian Center for Hydrodynamics. Electrophoresis quantitation data is available upon request from the authors.