Summary
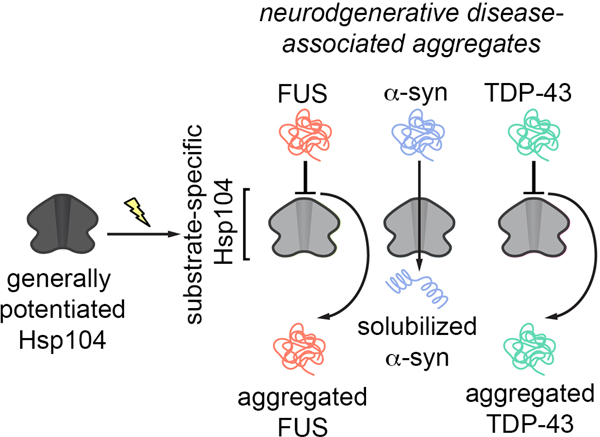
Hsp104 is an AAA+ protein disaggregase that solubilizes and reactivates proteins trapped in aggregated states. We have engineered potentiated Hsp104 variants to mitigate toxic misfolding of α-synuclein, TDP-43, and FUS implicated in fatal neurodegenerative disorders. Though potent disaggregases, these enhanced Hsp104 variants lack substrate specificity, and can have unfavorable off-target effects. Here, to lessen off-target effects, we engineer substrate-specific Hsp104 variants. By altering Hsp104 pore loops that engage substrate, we disambiguate Hsp104 variants that selectively suppress α-synuclein toxicity but not TDP-43 or FUS toxicity. Remarkably, α-synuclein-specific Hsp104 variants emerge that mitigate α-synuclein toxicity via distinct ATPase-dependent mechanisms, involving α-synuclein disaggregation or detoxification of soluble α-synuclein conformers. Importantly, both types of α-synuclein-specific Hsp104 variant reduce dopaminergic neurodegeneration in a C. elegans model of Parkinson’s disease more effectively than non-specific variants. We suggest that increasing the substrate specificity of enhanced disaggregases could be applied broadly to tailor therapeutics for neurodegenerative disease.
Graphical Abstract
eTOC blurb
In this paper, Mack et al. engineer Hsp104 variants with enhanced selectivity for α-synuclein that reduce dopaminergic neurodegeneration more effectively than non-specific Hsp104 variants. These findings suggest that increasing the substrate specificity of enhanced protein disaggregases could have broad applications in tailoring therapeutics for neurodegenerative disease.
Introduction
There are no effective therapies for several fatal neurodegenerative diseases including Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and frontotemporal dementia (FTD). PD is marked by misfolding of α-synuclein (α-syn), a protein normally found in presynaptic terminals, which may function in synaptic vesicle recycling.1,2 α-Syn misfolds into toxic oligomers and amyloid fibrils, accumulating in characteristic Lewy bodies in the cytoplasm of dopaminergic neurons that degenerate in PD.3–5 Indeed, protein misfolding and aggregation unite a spectrum of fatal neurodegenerative diseases.6 Thus, it is important to develop therapeutics that directly antagonize the underlying toxic protein-misfolding events in neurodegenerative disease. In this way, proteins can be restored to their native conformation and function, which may halt the debilitating trajectory of neurodegeneration.7–11
Hsp104, an asymmetric, hexameric AAA+ protein disaggregase from yeast, is an intriguing therapeutic agent to directly target toxic protein-misfolding events in neurodegenerative disease.12,13 Hsp104 uses energy from ATP binding and hydrolysis, and collaboration with Hsp70 and Hsp40, to reactivate proteins trapped in insoluble states.13–16 Hsp104 hexamers are dynamic and adopt open "lock-washer" spiral states and closed ring structures that translocate polypeptides across the central channel.14,17–20 During protein disaggregation, pore-loop tyrosines grip substrate and ATP hydrolysis-driven conformational changes at the spiral seam ratchet substrate either partially or completely through the channel.16,17,19,21–26 Thus, Hsp104 liberates individual polypeptides from soluble toxic oligomers, amorphous aggregates, stress-induced condensates, and amyloid fibrils, which can then regain their functional form.14–16,27 Curiously, Hsp104 is not found in metazoa, but is conserved in eubacteria, algae, fungi, protozoa, and plants.28 As humans lack an Hsp104 homolog, they have limited capacity to effectively counter overwhelming protein-misfolding events that underlie neurodegenerative disease.7,29 Introduction of Hsp104 into animal models (e.g. worm, fly, mouse, and rat) protects against deleterious protein misfolding and neurodegeneration connected to PD and polyglutamine-expansion disorders.27,30–33 Nonetheless, the ability of Hsp104 to counter the misfolding and toxicity of human disease-linked proteins can be limited, requiring high Hsp104 concentrations.14,27
To address this issue, we have engineered enhanced versions of Hsp104 that more effectively disaggregate various disease-linked proteins, including α-syn (linked to PD), and TDP-43 and FUS (linked to ALS/FTD) under conditions where wild-type (WT) Hsp104 is ineffective.34–42 Select potentiated variants reduce dopaminergic neurodegeneration in a C. elegans model of PD34 and reverse FUS aggregation and toxicity in mammalian cells.43 Typically, potentiated Hsp104 variants exhibit elevated ATPase activity, altered protomer cooperativity, altered substrate recognition, and prolonged substrate interactions, which enables more productive disaggregase activity.26,34,35,44 Though powerful disaggregases, these potentiated Hsp104 variants can exhibit off-target toxicity, which may limit their progression along the therapeutic pipeline through more complex model systems.34,45 This toxicity is likely caused by aberrant unfolding of essential substrates, resulting in unwanted off-target effects.35 Thus, enhanced substrate specificity is a desirable attribute for Hsp104 variants to effectively translate into higher organisms as therapeutics.46
Here, we hypothesized that specifically mutating Hsp104 residues known to contact substrate in a potentiated variant background would couple increased substrate-specificity to enhanced disaggregase activity. We found that specific alterations to pore-loop tyrosines that engage substrate directly endowed Hsp104 with the ability to selectively mitigate α-syn toxicity. Surprisingly, two classes of α-syn-specific Hsp104 variant emerged. The first class mitigated α-syn toxicity via ATPase-dependent disaggregation of α-syn inclusions. By contrast, an unanticipated second class mitigated α-syn toxicity via ATPase-dependent detoxification of soluble α-syn conformers without disaggregation of α-syn inclusions. Importantly, both types of α-syn-specific Hsp104 variant reduced dopaminergic neurodegeneration in a C. elegans model of PD more effectively than non-specific Hsp104 variants. Thus, we establish an important concept: specializing protein disaggregases against individual disease-associated substrates can improve their therapeutic utility. We anticipate that this concept can be applied broadly to diverse protein disaggregases and specific targets implicated in protein-misfolding disorders. In this way, specific toxic misfolding events could be remediated with tailor-made therapeutic disaggregases.
Results
Targeting Hsp104 pore-loop tyrosines via a rational engineering approach
Hsp104 consists of an N-terminal domain (NTD), nucleotide-binding domain 1 (NBD1), a middle domain (MD), nucleotide-binding domain 2 (NBD2), and a C-terminal domain (CTD) (Figure 1A).47 Hsp104 forms an asymmetric, ring-like hexamer, and threads substrate through its central channel, which is lined with substrate-binding pore loops (Figure 1B–D).17,18,21,22 Each NBD contains a highly-conserved tyrosine embedded within a pore-loop motif: ‘KYKG’ in NBD1 (residues 256-259) comprises pore-loop 1, and ‘GYVG’ in NBD2 (residues 661-664) comprises pore-loop 217,18,21,22 (Figure 1A–D).17,18,21,22 The pore-loop tyrosines, Y257 in pore-loop 1 and Y662 in pore-loop 2, are essential for disaggregation and substrate threading.14,16–18,21,22,24 The pore loops are arranged in the Hsp104 axial channel as a ‘spiral staircase’, allowing for substrate to be translocated through the channel (Figure 1B–D).17,18 Y257 and Y662 contact substrate directly.17 We reasoned that subtly changing the properties of these highly-conserved tyrosines would enable us to tune the substrate repertoire of Hsp104. We altered each pore-loop tyrosine to a series of hydrophobic, aromatic, or uncharged polar residues to preserve different features of the original tyrosine residue, which was found to be optimal for protein disaggregation.24,34 Using this rational engineering approach, we isolated α-syn-specific Hsp104 variants.
Figure 1. Pore-loop substitutions in Hsp104A503S do not generate substrate-specific Hsp104 variants.
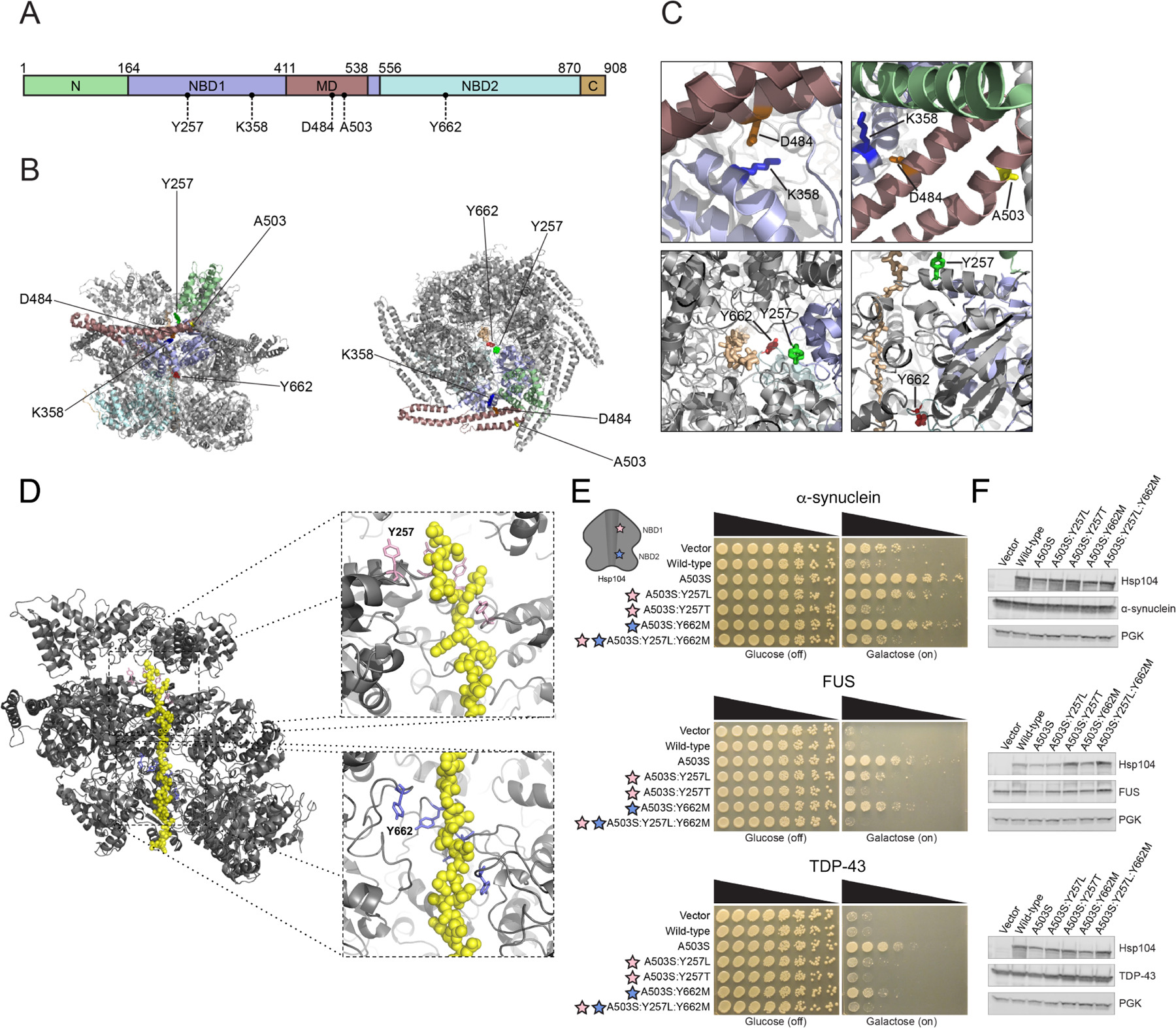
(A) Domain map of Hsp104 showing N-terminal domain (N; green), nucleotide-binding domain 1 (NBD1; purple), middle domain (MD; maroon), nucleotide-binding domain 2 (NBD2; light blue), and C-terminal domain (C, orange). Location of residues assessed in this work are shown. (B) Structure of Hsp104 hexamer (side-view; left, top-down view; right) with residues from this study indicated. Colors of domains in protomer correspond to (A). PDB: 5VY9. (C) Zooms into Hsp104 structure highlighting K358:D484 interaction, and location of these residues in relation to A503. Substrate-binding pore-loop tyrosine residues are shown (Y257 and Y662) relative to polypeptide (casein) substrate (tan) in the Hsp104 channel. PDB: 5VY9. (D) Structure of Hsp104 hexamer (gray) with two subunits omitted to reveal polypeptide (casein) substrate (yellow) in the Hsp104 channel. Pore loops (pink; Y257 in NBD1 and purple; Y662 in NBD2) line the channel of Hsp104 in a “spiral staircase” manner. Pore-loop residues are essential for substrate translocation, as they establish the main contacts between Hsp104 and substrate. PDB: 5VY9. (E) Δhsp104 yeast integrated with α-syn-YFP (top), FUS (middle), or TDP-43 (bottom) on a galactose-inducible promoter were transformed with Hsp104 variants or an empty vector control. Yeast were spotted onto glucose (uninducing, off) and galactose (inducing, on) media in a five-fold serial dilution. Stars indicate substitution to pore loop in NBD1 (pink), NBD2 (purple). (F) Integrated strains from (E) were induced in the presence of Hsp104 variants or empty vector control for 5 hours (FUS, TDP-43) or 8 hours (α-syn). Yeast were lysed and processed for Western blot. 3-Phosphoglycerate kinase (PGK) is a loading control.
Mutating pore-loop tyrosines in Hsp104A503S does not yield α-syn-specific variants
Mutating pore-loop tyrosines in WT Hsp104 does not enable suppression of α-syn, TDP-43, or FUS toxicity in yeast.34 Thus, we first set out to engineer substrate-specific Hsp104 variants using a generally potentiated Hsp104 variant, Hsp104A503S, as a starting scaffold.34 Our goal was to leverage the elevated activity of Hsp104A503S as a starting point to introduce substitutions that alter Hsp104 substrate selectivity. We assessed Hsp104 variant activity in powerful yeast models of α-syn, TDP-43, and FUS proteinopathy, which faithfully recapitulate several aspects of neurodegenerative disease, including protein aggregation and toxicity.48–50 Importantly, these valuable yeast models have enabled identification of genetic suppressors and drug candidates that mitigate neurodegeneration in C. elegans, fly, mouse, rat, and human patient-derived neuronal models of disease.34,51–57
Hsp104A503S potently suppressed α-syn, TDP-43, and FUS toxicity in yeast (Figure 1E). 34 In an effort to confer substrate specificity, we first introduced hydrophobic mutations to pore-loop 1 Y257 or pore-loop 2 Y662 to maintain the hydrophobic character of the tyrosine. Thus, we substituted a leucine at pore-loop 1 Y257. Although Y257 is highly conserved, leucine is also found rarely at this position in ~0.5% of Hsp104 homologues according to our Generative Regularized ModeLs of proteINs (GREMLIN) analysis of 5,812 Hsp104 species variants.58 Relative to Hsp104A503S, Hsp104A503S:Y257L displayed reduced activity, as it suppressed α-syn and FUS toxicity, but not TDP-43 toxicity in yeast (Figure 1E). Though substitution of a hydrophobic residue at pore-loop 1 Y257 reduced toxicity suppression, introducing a hydrophobic methionine residue at pore-loop 2 Y662 had a different effect. Hsp104A503S:Y662M suppressed toxicity of α-syn, TDP-43, and FUS, but not as strongly as Hsp104A503S (Figure 1E). Combining these pore-loop mutations in Hsp104A503S:Y257L:Y662M diminished any toxicity-suppression activity (Figure 1E).
Since hydrophobic substitutions at each pore loop did not lead to enhanced substrate specificity, we evaluated the effect of an uncharged, polar variant at Y257, and so introduced a threonine substitution. Hsp104A503S:Y257T was unable to suppress toxicity of α-syn, TDP-43, or FUS (Figure 1E). Expressing each pore-loop variant did not noticeably affect expression levels of Hsp104 or disease-associated substrates (Figure 1F). Altogether, slightly altering Hsp104 pore-loop tyrosines in the potentiated Hsp104A503S background did not generate any variants with the desired substrate specificity. Indeed, several pore-loop mutations diminished the ability of Hsp104A503S to mitigate proteotoxicity.
Hsp104K358D can be tailored via tuning pore loops for substrate specificity
Hsp104A503S is potentiated via disruption of interprotomer contacts between helix L1 and helix L3 of the MD17,26,40,59, but was not amenable for tuning substrate specificity (Figure 1E). Thus, we wondered whether substrate selectivity could be more effectively engineered in a Hsp104 variant potentiated by a different mechanism than Hsp104A503S. We first considered Hsp104 with a K358D mutation in NBD1 as a starting scaffold. Hsp104K358D has a mutation that disrupts interactions between D484 in the MD and K358 in NBD1 (Figure 1C) similar to the previously-studied Hsp104K358E.17,60 Hsp104K358E has elevated ATPase activity relative to Hsp104, but has been reported to be extremely toxic to yeast.60 By contrast, Hsp104K358D was not toxic to ∆hsp104 yeast at 30°C (Figure S1). Thus, we explored whether this Hsp104 variant could suppress toxicity of neurodegenerative disease-linked substrates, and whether we could now tune pore-loop residues to engender substrate specificity.
Hsp104K358D suppressed α-syn, FUS, and TDP-43 toxicity in yeast (Figure 2A). Thus, Hsp104K358D can antagonize proteotoxicity connected to PD and ALS/FTD. Using Hsp104K358D as a starting scaffold, we systematically altered the substrate-binding, pore-loop tyrosines, Y257 and Y662, as with Hsp104A503S. Interestingly, Hsp104K358D:Y257L suppressed toxicity of α-syn, FUS, and TDP-43 more effectively than Hsp104K358D (Figure 2A). Thus, a leucine at position 257 in pore-loop 1 is more productive than a tyrosine in the Hsp104K358D background in striking contrast to Hsp104A503S (Figure 1E, 2A). Interestingly, Hsp104K358D:Y662L, with a leucine at pore-loop 2 Y662, did not suppress FUS or TDP-43 toxicity, but mildly suppressed α-syn toxicity (Figure 2A). Thus, increased α-syn selectivity can originate by altering Y662 in the K358D background.
Figure 2. Hsp104K358D:Y662M selectively suppresses α-syn toxicity.
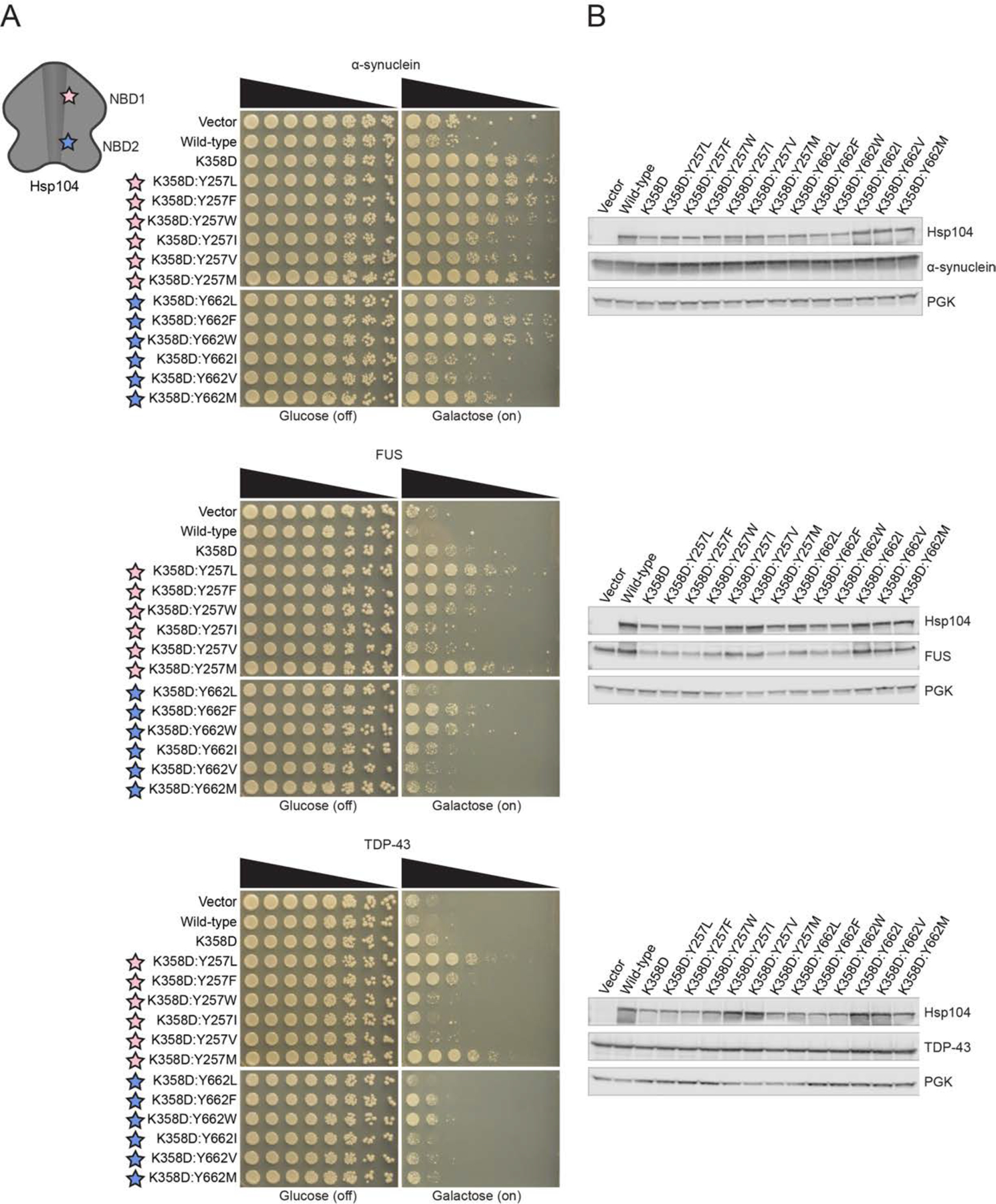
(A) Δhsp104 yeast integrated with α-syn-YFP (top), FUS (middle), or TDP-43 (bottom) on a galactose-inducible promoter were transformed with Hsp104 variants or an empty vector control. Yeast were spotted onto glucose (uninducing, off) and galactose (inducing, on) media in a five-fold serial dilution. Stars indicate substitution to pore loop in NBD1 (pink), NBD2 (purple). (B) Integrated strains from (A) were induced in the presence of Hsp104 variants or empty vector control for 5 hours (FUS, TDP-43) or 8 hours (α-syn). Yeast were lysed and lysates visualized via Western blot. 3-Phosphoglycerate kinase (PGK) is a loading control. See also Figure S1–S4.
We next explored hydrophobic, aromatic substitutions at pore loops 1 and 2. Hsp104K358D:Y257F/W and Hsp104K358D:Y662F/W suppressed toxicity of α-syn, FUS, and TDP-43 (Figure 2A). We also substituted hydrophobic residues isoleucine and valine at the pore-loop tyrosines. Hsp104K358D:Y257I/V slightly suppressed α-syn toxicity and did not suppress FUS or TDP-43 toxicity (Figure 2A). Thus, α-syn selectivity can also emerge by mutating Y257 in the K358D background. By contrast, Hsp104K358D:Y662I/V did not suppress α-syn, FUS, or TDP-43 toxicity (Figure 2A). Interestingly, even a minor change from leucine to isoleucine at position 662 in the K358D background resulted in diminished ability to suppress α-syn toxicity.
Generally, our pore-loop Hsp104 variants did not grossly affect α-syn or TDP-43 expression levels (Figure 2B). A subset of these variants mildly reduced FUS expression (Figure 2B), but we have shown before that this mild reduction in FUS levels is not required to suppress FUS toxicity by enhanced Hsp104 variants.35,40 Collectively, our data suggest that engineered pore-loop variants do not suppress disease-associated substrate toxicity by severely lowering substrate levels.
Given the changes in toxicity suppression when a single pore-loop tyrosine was mutated in the Hsp104K358D background, we next tested the effect of double pore-loop tyrosine mutations. As Hsp104K358D:Y257L showed robust suppression of α-syn, FUS, and TDP-43 toxicity, we used this variant as a starting scaffold to tune the second pore-loop tyrosine. Hsp104K358D:Y257L:Y662L suppressed toxicity of α-syn, but only slightly reduced FUS and TDP-43 toxicity (Figure 3A). Hsp104K358D:Y257L:Y662F/W suppressed toxicity of all three substrates, consistent with the finding that phenylalanine and tryptophan substitutions at either pore loop alone do not affect the activity of enhanced Hsp104 variants (Figure 3A).34 Interestingly, introducing polar, uncharged residues (S, T, Q, N) at pore-loop 2 Y662 ablated the toxicity suppression activity of Hsp104K358D:Y257L, again suggesting that pore-loop 2 Y662 is not as amenable to mutation as pore-loop 1 Y257 (Figure 3A). Overall, the double pore-loop Hsp104 variants were similar to those with single pore-loop mutations in that aside from a set of variants that mildly reduced FUS expression, the variants did not grossly affect disease-associated substrate levels (Figure 3B).
Figure 3. Hsp104K358D:Y257L:Y662M selectively suppresses α-syn toxicity.
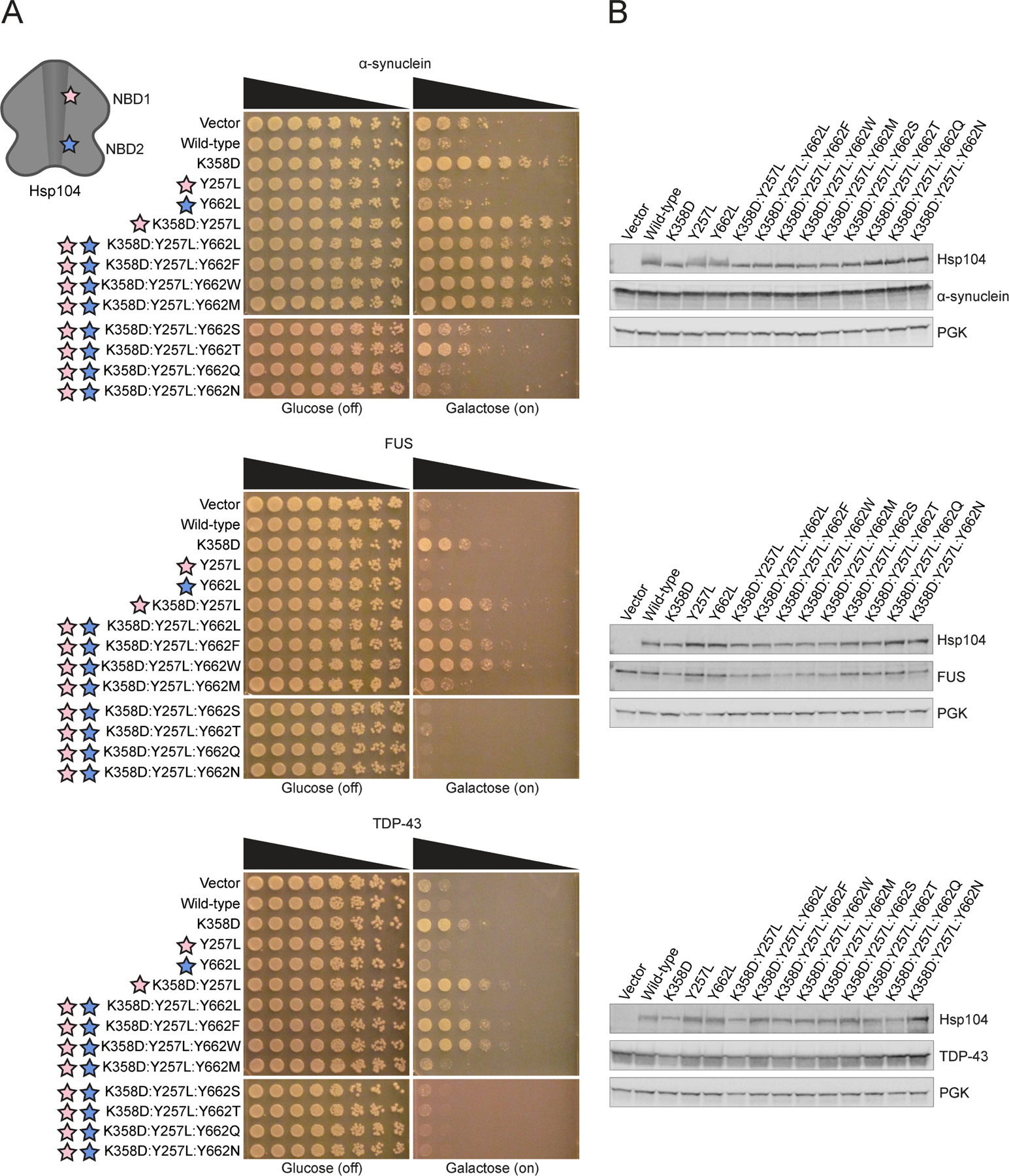
(A) Δhsp104 yeast integrated with α-syn-YFP (top), FUS (middle), or TDP-43 (bottom) on a galactose-inducible promoter were transformed with Hsp104 variants or an empty vector control. Yeast were spotted onto glucose (uninducing, off) and galactose (inducing, on) media in a five-fold serial dilution. Stars indicate substitution to pore loop in NBD1 (pink), NBD2 (purple). (B) Integrated strains from (A) were induced in the presence of Hsp104 variants or empty vector control for 5 hours (FUS, TDP-43) or 8 hours (α-syn). Yeast were lysed and lysates visualized via Western blot. 3-Phosphoglycerate kinase (PGK) is a loading control. See also Figure S3 and S4.
Our rational approach to altering the substrate repertoire of Hsp104 revealed several trends for pore-loop substitutions that are favorable for general potentiation. In the enhanced Hsp104K358D background, aromatic residues (W, F) at Y257 or Y662 maintained enhanced toxicity suppression activity (Figure 2A). Substituting aromatic residues for Y662 in the potentiated Hsp104K358D:Y257L background also maintained potentiated activity (Figure 3A). Hydrophobic residues (I, V) at Y257 in the Hsp104K358D background were not favorable for potentiated activity against TDP-43 or FUS but did yield more selective variants that mildly suppressed α-syn toxicity (Figure 2A). Additionally, uncharged, polar residues at Y662 in Hsp104K358D:Y257L eliminated potentiated activity (Figure 3A).
Y662M in pore-loop 2 of Hsp104K358D or Hsp104K358D:Y257L confers α-syn selectivity
We unearthed further α-syn-specific variants by substituting a Met residue at the NBD2 pore-loop tyrosine. Thus, a Y662M substitution at pore-loop 2 in the Hsp104K358D or Hsp104K358D:Y257L created variants that selectively suppressed α-syn toxicity (Figure 2A, 3A). Indeed, Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M suppressed α-syn toxicity but were ineffective against FUS or TDP-43 toxicity (Figure 2A, 3A). These variants did not grossly affect α-syn expression levels (Figure 2B, 3B). Interestingly, introducing a Met residue at pore-loop 1 (Y257) in the Hsp104K358D background did not confer substrate specificity, but rather strengthened suppression of α-syn, FUS, and TDP-43 toxicity (Figure 2A). Thus, replacing Tyr with Met in pore-loop 1 versus pore-loop 2 has distinct effects on the substrate selectivity of Hsp104.
Hsp104D484K can also be tailored via tuning pore loops for substrate specificity
In addition to Hsp104K358D, Hsp104D484K is another Hsp104 variant that disrupts MD-NBD1 interactions normally mediated by K358 in NBD1 and D484 in helix L2 of the MD (Figure 1C).60 To our surprise, Hsp104D484K was not toxic to yeast at 30°C (Figure S1). Thus, neither Hsp104K358D nor Hsp104D484K are overtly toxic to yeast at 30°C. We generated a set of pore-loop Hsp104D484K variants, which displayed very similar effects on α-syn, FUS, and TDP-43 toxicity and expression to those studied in the Hsp104K358D background (Figure S2A, B). For example, like Hsp104K358D:Y257L/M, Hsp104D484K:Y257L/M enhanced activity against α-syn, FUS, and TDP-43 (Figure S2A). Furthermore, mutation of Y257 to I or V, or Y662 to L or M yielded Hsp104D484K variants that selectively suppressed α-syn toxicity (Figure S2A). Thus, in potentiated backgrounds created by disrupting NBD1 and MD contacts (e.g., K358D and D484K) we establish general rules for: (a) mutating pore loops to enhance suppression of α-syn, FUS, and TDP-43 toxicity; and (b) mutating pore loops to selectively suppress α-syn toxicity. Specifically, enhanced activity is conferred by Y257L/M mutations and α-syn-selectivity is conferred by Y257I/V mutation or Y662L/M mutations.
Y257T/Q in pore-loop 1 of Hsp104K358D confer α-syn selectivity
We next introduced alternative uncharged, polar residues Thr, Gln, Ser, or Asn at pore-loop 1, Y257. As these substitutions still maintain the polar, uncharged character of tyrosine we reasoned they might tune the substrate repertoire of Hsp104. Remarkably, Hsp104K358D:Y257T/Q specifically suppressed α-syn toxicity and did not suppress FUS or TDP-43 toxicity (Figure 4A). Furthermore, Hsp104K358D:Y257T/Q did not reduce α-syn expression levels (Figure 4B). Interestingly, Hsp104K358D:Y257S/N did not effectively suppress α-syn, TDP-43, or FUS toxicity (Figure 4A). We focused on Hsp104K358D:Y257T as a representative among the uncharged, polar (Hsp104K358D:Y257T/Q) and hydrophobic (Hsp104K358D:Y257I/V) α-syn-specific variants. This variant suppressed α-syn toxicity to a lesser extent than Hsp104K358D:Y662M (Figure 2A, 4A), revealing that even among α-syn-specific variants, we can tune the degree of toxicity suppression.
Figure 4. Hsp104K358D:Y257T/Q selectively suppress α-syn toxicity.
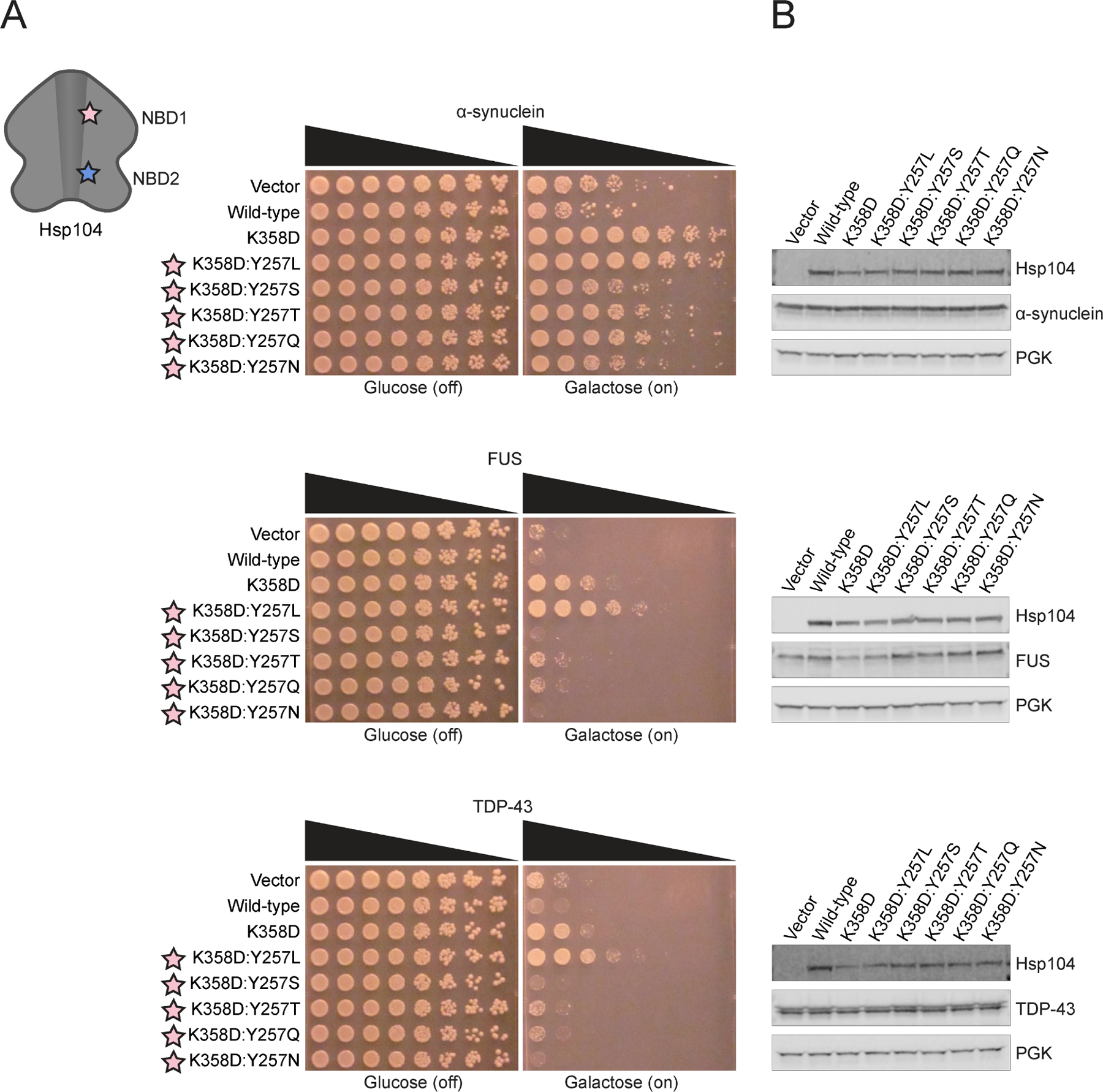
(A) Δhsp104 yeast integrated with α-syn-YFP (top), FUS (middle), or TDP-43 (bottom) on a galactose-inducible promoter were transformed with Hsp104 variants or an empty vector control. Yeast were spotted onto glucose (uninducing, off) and galactose (inducing, on) media in a five-fold serial dilution. Stars indicate substitution to pore loop in NBD1 (pink), NBD2 (purple). (B) Integrated strains from (A) were induced in the presence of Hsp104 variants or empty vector control for 5 hours (FUS, TDP-43) or 8 hours (α-syn). Yeast were lysed and lysates visualized via Western blot. 3-Phosphoglycerate kinase (PGK) is a loading control. See also Figure S3 and S4.
α-Syn-specific Hsp104 variants suppress α-syn toxicity through an ATPase-dependent mechanism
We next determined whether α-syn-specific Hsp104 variants suppressed α-syn toxicity via an ATP hydrolysis-dependent process as for other potentiated Hsp104 variants.34,39 Thus, we made double Walker A (DWA) mutations (K218T and K620T) that render Hsp104 unable to bind ATP at NBD1 and NBD2.14,39,61–64 The DWA mutations inhibited potentiated variants Hsp104A503S, Hsp104K358D, and Hsp104K358D:Y257L from suppressing toxicity of α-syn, FUS, and TDP-43 (Figure S3A). DWA mutations in Hsp104K358D:Y257T diminished suppression of α-syn toxicity (Figure S3A), suggesting Hsp104K358D:Y257T also employs an ATP hydrolysis-dependent mechanism. Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M were also inactivated by DWA mutations (Figure S3A). Thus, these α-syn-specific Hsp104 variants also work through a mechanism reliant on ATP hydrolysis. The reduction in therapeutic Hsp104 activity conferred by DWA mutations was not due to reduced Hsp104 levels (Figure S3B). Overall, these findings suggest that α-syn-specific Hsp104 variants suppress α-syn toxicity through an ATPase-dependent mechanism and not via a passive mechanism as with select Hsp104 species variants.65
α-Syn-specific Hsp104 variants do not suppress toxicity of PD-linked α-synE46K or α-synA53T
We next tested whether α-syn-specific Hsp104 variants could suppress the toxicity of α-syn variants, α-synE46K and α-synA53T, which cause rare familial forms of PD (Figure S4A).66,67 None of the Hsp104 variants grossly affected Hsp104 or α-syn expression (Figure S4B). As anticipated, Hsp104 was unable to mitigate toxicity of α-synE46K and α-synA53T, whereas Hsp104A503S reduced α-synE46K and α-synA53T toxicity (Figure S4B).35 By contrast, Hsp104K358D weakly reduced α-synE46K toxicity, but not α-synA53T toxicity, indicating that α-synA53T toxicity is more challenging to suppress (Figure S4B). Addition of the Y257L mutation to the Hsp104K358D background enabled suppression of α-synE46K and α-synA53T toxicity (Figure S4B). This finding provides further evidence that a leucine at position 257 in pore-loop 1 is more productive than a tyrosine for mitigating α-syn toxicity in the Hsp104K358D background. Interestingly, the α-syn-specific Hsp104 variants, Hsp104K358D:Y257T, Hsp104K358D:Y662M, and Hsp104K358D:Y257L:Y662M could not mitigate α-synE46K or α-synA53T toxicity (Figure S4B). Thus, the PD-linked E46K and A53T mutations in the amphipathic region of α-syn reduce the efficacy of Hsp104K358D:Y257T, Hsp104K358D:Y662M, and Hsp104K358D:Y257L:Y662M.
Determinants in the α-syn C-terminal acidic domain enable toxicity suppression by α-syn-selective Hsp104 variants
Hsp104-binding peptides can be enriched in aromatic residues, uncharged polar residues, as well as basic and acidic residues21. Hence, we wondered whether the acidic C-terminal domain of α-syn might enable toxicity mitigation by Hsp104 variants. Thus, we expressed α-syn constructs encompassing residues 1-95, 1-115, or 1-125 in yeast (Figure S4A). α-Syn1-95 lacks the entire C-terminal acidic region (amino acids 95-140), whereas α-syn1-115 and α-syn1-125 lack C-terminal portions of the acidic region and are naturally occurring fragments of α-syn that are more aggregation prone (Figure S4A).68 α-Syn1-95, α-syn1-115, and α-syn1-125 were robustly expressed and were toxic in yeast (Figure S4C). This toxicity could be mitigated by the generally potentiated Hsp104 variants, Hsp104A503S and Hsp104K358D, but not by Hsp104 (Figure S4D). By contrast, Hsp104K358D:Y257L reduced toxicity of α-syn1-115 and α-syn1-125 but was not effective against α-syn1-95 (Figure S4D). Thus, mutation of Y257 in the K358D background can reduce the ability to mitigate α-syn1-95 toxicity. Indeed, the α-syn-selective variants, Hsp104K358D:Y257T, Hsp104K358D:Y662M, and Hsp104K358D:Y257L:Y662M were unable to mitigate the toxicity of α-syn1-95 but could partially mitigate toxicity of α-syn1-115 and α-syn1-125 (Figure S4D). These findings indicate that determinants between residues 96-115 in the C-terminal acidic region of α-syn are critical for Hsp104K358D:Y257L, and the α-syn-selective Hsp104 variants to suppress α-syn toxicity. Thus, this region of α-syn acquires increased importance for toxicity mitigation by Hsp104K358D:Y257L, as well as the α-syn-selective Hsp104 variants Hsp104K358D:Y257T, Hsp104K358D:Y662M, and Hsp104K358D:Y257L:Y662M, but is less critical for Hsp104A503S and Hsp104K358D.
α-Syn-specific Hsp104 variants do not suppress FUS aggregation
We next evaluated the ability of the α-syn-specific Hsp104 variants to antagonize FUS aggregation in yeast. α-Syn-specific Hsp104 variants should not affect FUS aggregates in yeast, as FUS is no longer recognized as a substrate. In ALS/FTD and in yeast proteinopathy models, FUS mislocalizes to cytoplasmic aggregates50,69,70.50,69,70 Enhanced Hsp104 variants, Hsp104K358D and Hsp104K358D:Y257L, effectively suppressed FUS aggregation compared to Hsp104, which had no effect (Figure S5A, B). In agreement with our toxicity-suppression findings, α-syn-specific variants Hsp104K358D:Y257T, Hsp104K358D:Y662M, and Hsp104K358D:Y257L:Y662M did not antagonize FUS aggregation (Figure S5A, B). Thus, our α-syn-specific variants do not suppress FUS toxicity or aggregation in yeast, strongly advocating for tailored substrate specificity.
α-Syn-specific Hsp104 variants do not restore TDP-43 to the nucleus
As with FUS, we expect α-syn-specific Hsp104 variants should not suppress mislocalization of TDP-43, as these variants have an altered substrate repertoire. TDP-43 is normally localized to the nucleus in human cells, but mislocalizes to cytoplasmic aggregates in ALS/FTD pathology, which is recapitulated in yeast.48,51,71,72 Hsp104 was unable to return TDP-43 to the nucleus, whereas enhanced variants, Hsp104K358D and Hsp104K358D:Y257L, significantly restored nuclear localization to TDP-43, as ~39% and ~50% of cells respectively harbored nuclear TDP-43 (Figure S5C, D). Importantly, α-syn-specific variants Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M did not restore TDP-43 to the nucleus, and α-syn-specific variant Hsp104K358D:Y257T had only a slight effect on TDP-43 localization (Figure S5C, D). Thus, α-syn-specific Hsp104 variants do not suppress TDP-43 toxicity or restore TDP-43 to the nucleus, consistent with increased substrate selectivity.
α-Syn-specific Hsp104 variants differentially affect cytoplasmic α-syn foci
α-Syn initially localizes to the plasma membrane in yeast, but eventually forms toxic cytoplasmic inclusions that are detergent-insoluble, contain high molecular weight α-syn species, can react with Thioflavin-S (an amyloid-diagnostic dye), and cluster cytoplasmic vesicles reminiscent of aspects of Lewy pathology in PD.4,5,34,49,73–77 Hsp104 is unable to antagonize formation of cytoplasmic α-syn foci and return α-syn to the plasma membrane, whereas enhanced variants Hsp104K358D and Hsp104K358D:Y257L robustly clear α-syn foci (Figure 5A, B). Only ~18% cells had α-syn to the plasma membrane with Hsp104, whereas Hsp104K358D and Hsp104K358D:Y257L promoted α-syn plasma membrane localization in ~62% and ~48% of cells respectively (Figure 5B). α-Syn-specific variants Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M also eradicated α-syn foci (Figure 5A, B). Unexpectedly, α-syn-specific variant Hsp104K358D:Y257T did not significantly eliminate α-syn foci relative to Hsp104 (Figure 5A, B). Thus, Hsp104K358D:Y257T is likely not suppressing α-syn toxicity by resolving α-syn foci. The uncoupling of toxicity suppression and clearance of α-syn foci is a novel feature of Hsp104K358D:Y257T, as we have not previously engineered Hsp104 in this way. Thus, Hsp104K358D:Y257T emerges as a substrate-specific Hsp104 variant that is distinct from other α-syn-specific variants Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M.
Figure 5. α-Syn-specific Hsp104 variants differentially eradicate α-syn inclusions.
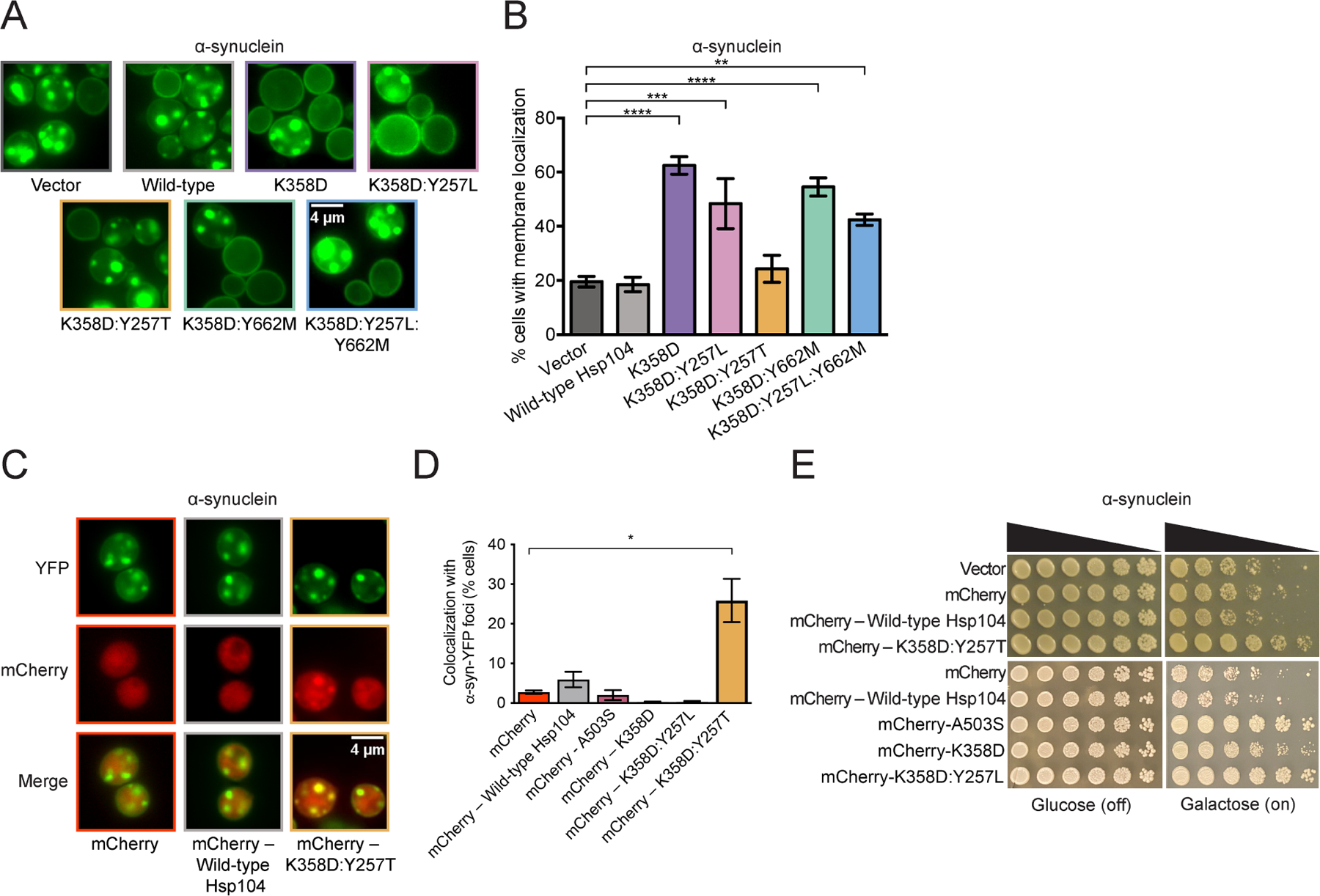
(A) Representative fluorescence microscopy images of Δhsp104 yeast integrated with α-syn-YFP and transformed with Hsp104 variants or an empty vector control. Bar, 4µm. (B) Localization of α-syn-YFP in yeast was quantified by counting the number of cells with plasma membrane-localized α-syn-YFP or α-syn-YFP foci in the cytoplasm. Values represent means ± SEM (n = 3–6). **P<0.01, ***P<0.001, ****P<0.0001; One-way ANOVA with Dunnett’s post-hoc test. (C) Representative fluorescence microscopy images of Δhsp104 yeast integrated with α-syn-YFP and transformed with mCherry – Hsp104 variants or mCherry alone as a control. Bar, 4µm. (D) Colocalization of mCherry–Hsp104 variants or mCherry alone with α-syn-YFP foci was quantified. Values represent means ± SEM (n = 2–6). *P<0.05; One-way ANOVA with Dunnett’s post-hoc test. (E) Δhsp104 yeast integrated with α-syn-YFP on a galactose-inducible promoter were transformed with mCherry – Hsp104 variants or mCherry alone. Yeast were spotted onto glucose (uninducing, off) and galactose (inducing, on) media in a five-fold serial dilution.
See also Figure S5.
Hsp104K358D:Y257T colocalizes with α-syn foci more frequently than Hsp104
As Hsp104K358D:Y257T suppressed α-syn toxicity in yeast but did not eliminate α-syn foci, we were curious if this variant colocalized with α-syn foci. One explanation for the uncoupling of toxicity suppression and clearance of α-syn foci is that Hsp104K358D:Y257T is still able to recognize and colocalize with α-syn foci to mitigate their toxicity. We tagged Hsp104 and Hsp104K358D:Y257T with mCherry to visualize their cellular localization in yeast expressing α-syn-YFP. C-terminally mCherry-tagged Hsp104K358D:Y257T more frequently colocalized with α-syn foci than mCherry-tagged Hsp104 or mCherry alone (Figure 5C, D). mCherry-Hsp104K358D:Y257T colocalized with α-syn foci in ~25% of cells (Figure 5D) and suppressed α-syn toxicity (Figure 5E). We also assessed whether the generally potentiated variants, mCherry-Hsp104K358D or mCherry-Hsp104K358D:Y257L, colocalized with α-syn foci. We find that cells with α-syn foci typically expressed very low levels of mCherry-Hsp104K358D, mCherry-Hsp104A503S, or mCherry-Hsp104K358D:Y257L, and we do not observe any colocalization with α-syn foci (Figure 5D). By contrast, cells that robustly express α-syn-YFP and mCherry-Hsp104K358D, mCherry-Hsp104A503S, or mCherry-Hsp104K358D:Y257L, do not contain α-syn-YFP foci and suppress α-syn toxicity (Figure 5E). Thus, the colocalization with α-syn foci is a specific feature of Hsp104K358D:Y257T, which we suggest contributes to the ability of Hsp104K358D:Y257T to mitigate α-syn toxicity. Hsp104K358D:Y257T may remodel α-syn conformers to a less toxic conformation or otherwise quench their toxicity (e.g., by shielding surfaces from interacting with other essential cell components, such as intracellular vesicles). Furthermore, Hsp104K358D:Y257T likely engages α-syn foci more efficiently than Hsp104, as it has enhanced substrate specificity.
α-Syn-specific Hsp104 variants have altered ATPase activity
To elucidate the underlying mechanism for substrate-specific toxicity suppression, we surveyed various biochemical properties of a representative set of Hsp104 variants. We first tested the ATPase activity to determine if α-syn-specific variants differ in ability to hydrolyze ATP relative to Hsp104 and potentiated variants. We purified the generally potentiated MD variant, Hsp104A503S, and the generally potentiated NBD1 variant, Hsp104K358D, both of which exhibit elevated ATPase activity compared to Hsp104 (Figure 6A).26,34,60 We also purified a generally potentiated pore-loop variant, Hsp104K358D:Y257L, and three α-syn specific variants, Hsp104K358D:Y257T, Hsp104K358D:Y662M, and Hsp104K358D:Y257L:Y662M. Interestingly, Hsp104K358D:Y257L exhibited reduced ATPase activity compared to Hsp104K358D, but slightly higher ATPase activity than Hsp104 (Figure 6A). Thus, Hsp104K358D:Y257L provides another example of a potentiated Hsp104 that does not have greatly elevated basal ATPase activity as with Hsp104D504C.34 Nonetheless, this effect was unexpected as mutating Y257 in WT Hsp104 does not typically reduce basal ATPase activity.14,21,39 This reduction in ATPase activity was even more pronounced for the α-syn-specific variant Hsp104K358D:Y257T, which had ATPase activity similar to Hsp104, but reduced ATPase activity compared to Hsp104K358D (Figure 6A). Thus, the K358D mutation sensitizes Hsp104 ATPase activity to Y257 mutations, perhaps due to destabilization of the small domain of NBD1 similar to another enhanced NBD1 variant, Hsp104E360R, which also bears a charge-reversing mutation in the vicinity of K35826,40.26,40
Figure 6. α-Syn-specific Hsp104 variants have distinct biochemical properties.
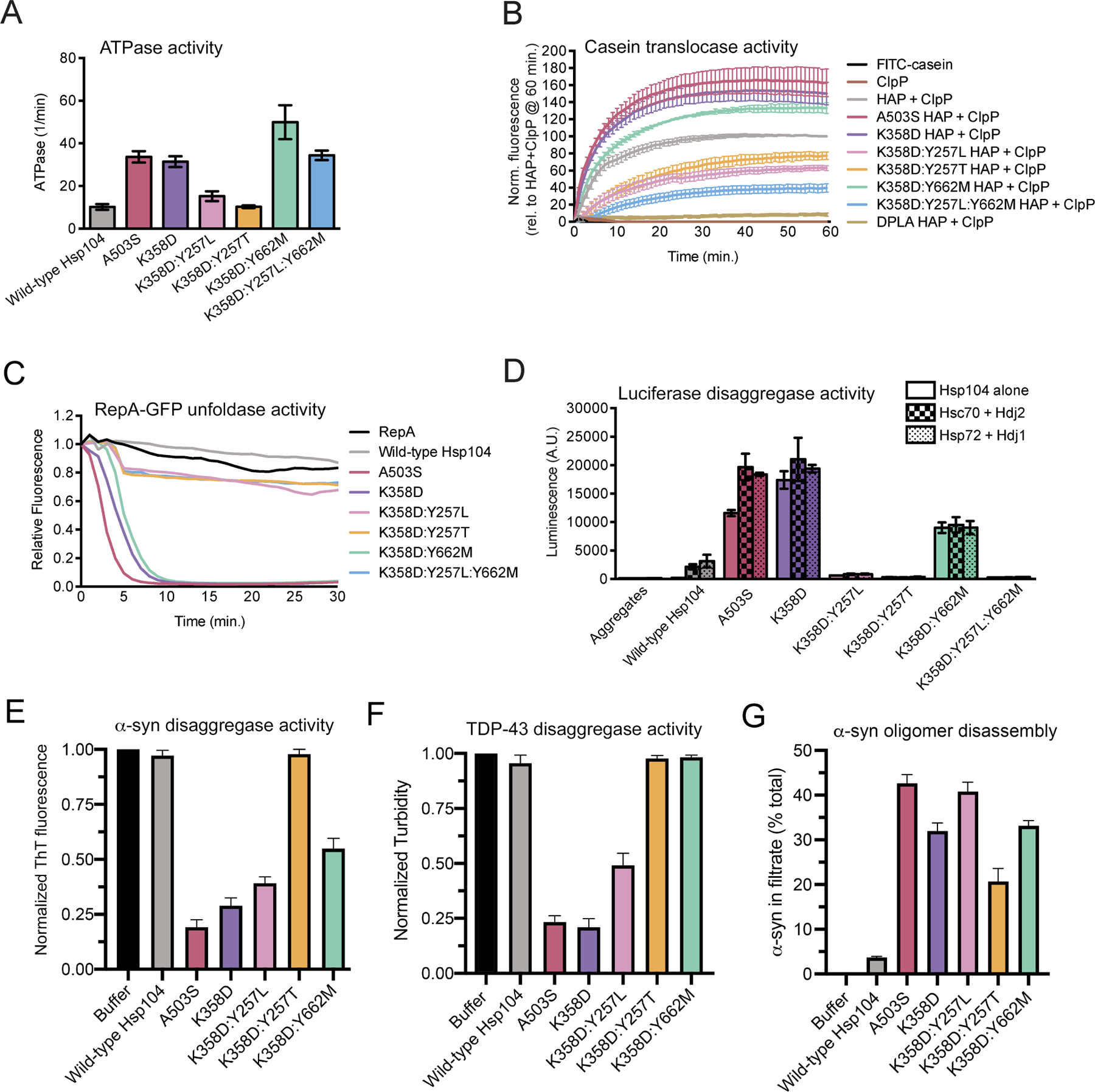
(A) ATPase activity of Hsp104 variants. Values represent means±SEM (n=4). (B) HAP variants (0.167μM hexamer) plus ClpP (21μM monomer) were incubated with FITC-casein, and FITC-casein degradation was measured by fluorescence. Negative controls FITC-casein alone (black) and ClpP alone control (dark brown) were included. Values were normalized to HAP plus ClpP at 60 minutes and represent means±SEM (n=3–6). (C) RepA1-25-GFP (0.7μM) was incubated with Hsp104 variants (2.1μM hexamer) plus GroELtrap (2.5μM tetradecamer), and RepA1-25-GFP unfolding was measured by fluorescence. Negative control of RepA1-25-GFP alone included. A representative trial is shown (n=3). (D) Hsp104 variants alone (1μM hexamer, solid bars) or with chaperone pairs Hsc70 and Hdj2 (0.167μM each, checkered bars), or Hsp72 and Hdj1 (0.167μM each, dotted bars), were incubated with urea-denatured luciferase aggregates. Reactivation activity was measured by luminescence. Negative control of luciferase aggregates alone included. Values represent means±SEM (n=2–8). (E) α-Syn fibrils (3µM monomer) were incubated without or with Hsp104, Hsp104A503S, Hsp104K358D, Hsp104K358D:Y257T or Hsp104K358D:Y662M (3µM) plus Hsc70 (3µM), Hdj1 (3µM), ATP (20 mM) and an ATP regeneration system (20mM creatine phosphate and 0.5µM creatine kinase) for 2h at 30°C. Disaggregation was assessed by Thioflavin-T (ThT) fluorescence. Values represent means±SEM (n=3). (F) TDP-43 fibrils (3µM monomer) were incubated without or with Hsp104, Hsp104A503S, Hsp104K358D, Hsp104K358D:Y257T or Hsp104K358D:Y662M (3µM) plus Hsc70 (3µM), Hdj1 (3µM), ATP (20mM) and an ATP regeneration system (20mM creatine phosphate and 0.5µM creatine kinase) for 2h at 30°C. Disaggregation was assessed by turbidity (absorbance at 350nm). Values represent means±SEM (n=3). (G) Soluble α-syn oligomers (0.5µM monomer) were incubated without or with Hsp104, Hsp104A503S, Hsp104K358D, Hsp104K358D:Y257L, Hsp104K358D:Y257T or Hsp104K358D:Y662M (1µM) plus ATP (20mM) and an ATP regeneration system (20mM creatine phosphate and 0.5µM creatine kinase) for 1h at 37°C. Reactions were then fractionated through a Microcon YM-100 (100-kDa molecular weight cut off) filter. The amount of α-syn in the filtrate fraction was then determined. Values represent means±SEM (n=3).
See also Figure S6.
The other α-syn-specific variants, Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M, exhibited ATPase activity equal to or greater than the activity of Hsp104A503S and Hsp104K358D (Figure 6A). Indeed, the Y662M mutation stimulated ATPase activity in the K358D background, and even counteracted the effect of the Y257L mutation such that Hsp104K358D:Y257L:Y662M had higher ATPase activity than Hsp104K358D:Y257L and similar ATPase activity to Hsp104K358D (Figure 6A). This result was also unanticipated as Y662 mutations do not typically affect WT Hsp104 ATPase activity,14,24,39 indicating that the K358D mutation sensitizes Hsp104 ATPase activity to Y662 mutations.
The difference in ATPase activity between α-syn-specific variants is a key biochemical distinction between them. These findings further suggest that α-syn-specific variants likely employ different toxicity-suppression mechanisms. Thus, the lower ATPase activity of Hsp104K358D:Y257T may limit clearance of α-syn foci in yeast, whereas elevated ATPase activity of Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M likely enables clearance of α-syn foci.
α-Syn-specific Hsp104 variants have altered translocase activity
Next, we assessed translocase activity of each Hsp104 variant. Hsp104 translocates substrates across its central channel during disaggregation.13,14 To assess substrate translocation, we evaluated each Hsp104 variant in the HAP background in vitro. HAP is an Hsp104 variant with three missense mutations (G739I:S740G:K741F) that enable interaction with the chambered protease, ClpP.22 Thus, HAP but not Hsp104 can translocate substrate into the proteolytic chamber of ClpP where it is degraded. We monitored translocation of a model unfolded substrate fluorescein isothiocyanate (FITC)-labeled casein through HAP via degradation by ClpP, which liberates FITC and increases fluorescence.14 Mutating each pore-loop Tyr to Ala (Y257A:Y662A), which ablates substrate translocation and disaggregation,14,21,22,39 resulted in the DPLA (‘double pore-loop alanine’) HAP variant that lacks translocation activity (Figure 6B). Thus, minimal passive translocation occurs in this system. We previously found that HAPA503V translocated substrate more rapidly than HAP34. Likewise, HAPA503S and HAPK358D translocated substrate more rapidly than HAP (Figure 6B). This accelerated substrate translocation may promote enhanced disaggregase activity.
Interestingly, α-syn-specific variants exhibited distinct activities. HAPK358D:Y662M translocated FITC-casein more effectively than HAP, whereas HAPK358D:Y257L:Y662M was less effective (Figure 6B). In the context of Hsp104, these variants have similar ATPase activity (Figure 6A). Thus, HAPK358D:Y257L:Y662M likely has reduced ability to grip FITC-casein resulting in decelerated translocation. HAPK358D:Y257L and HAPK358D:Y257T translocated FITC-casein at a similar rate but were less effective than HAP (Figure 6B). These findings suggest that interfering with Y257 of pore-loop 1 likely weakens grip of casein and thereby reduces translocase activity. Changes in substrate grip and translocase activity likely contribute to altered patterns of substrate specificity, as they could enable fine-tuning of the force used by Hsp104 to disaggregate different substrates. Indeed, Hsp104 variants with low translocase activity may only partially translocate substrate, thus changing the way a substrate is disaggregated.
α-Syn-specific Hsp104 variants have altered unfoldase activity
We next evaluated the unfoldase activity of each Hsp104 variant using RepA1-25-GFP as a model substrate. In these reactions, we included GroELtrap, which captures newly unfolded RepA1-25-GFP and prevents its refolding.78 Hsp104 is unable to unfold RepA1-25-GFP under these conditions, whereas potentiated Hsp104 variants, Hsp104A503S and Hsp104K358D, rapidly unfold RepA1-25-GFP34 (Figure 6C). Remarkably, α-syn-specific Hsp104 variants showed distinct unfoldase activity from one another. α-Syn-specific Hsp104K358D:Y662M unfolded RepA1-25-GFP almost as rapidly as Hsp104A503S and Hsp104K358D (Figure 6C). By contrast, α-syn-specific Hsp104K358D:Y257T and Hsp104K358D:Y257L:Y662M, as well as generally potentiated pore-loop variant Hsp104K358D:Y257L, only modestly unfolded RepA1-25-GFP (Figure 6C). Here too, it is likely that pore-loop 1 mutations weaken substrate gripping by Hsp104 and limit unfolding of the RepA1-25-GFP substrate. As Hsp104K358D:Y662M possessed strong unfoldase activity similar to generally potentiated Hsp104 variants, enhanced substrate recognition or unfolding power may contribute to its mechanism of toxicity suppression. The stark difference in unfoldase activity between α-syn-specific Hsp104 variants is another distinction in their biochemical properties that could contribute to different modes of α-syn toxicity suppression in yeast.
α-Syn-specific Hsp104 variants have altered luciferase disaggregase activity
Next, we assessed disaggregase activity of each Hsp104 variant against aggregated luciferase. Differences in the ability to reactivate this model substrate could help to illuminate general disaggregase properties of substrate-selective Hsp104 variants. Hsp104 was unable to reactivate disordered luciferase aggregates on its own, and required Hsp70 and Hsp40 (Figure 6D).15,34 Generally potentiated variants, Hsp104A503S and Hsp104K358D, exhibited similar activities, and had high luciferase reactivation activity even in the absence of Hsp70 and Hsp40 (Figure 6D). Interestingly, the generally potentiated pore-loop variant Hsp104K358D:Y257L was less active than Hsp104, indicating that Y257L mutation is inhibitory with respect to specifically disaggregating luciferase (Figure 6D). Indeed, it appears that Y257L alters activity against model substrates in vitro, but not against disease substrates in yeast. Of the three α-syn specific Hsp104 variants, only Hsp104K358D:Y662M had luciferase reactivation activity (Figure 6D). Hsp104K358D:Y662M was more active than Hsp104 and displayed comparable activity in the absence and presence of Hsp70 and Hsp40 (Figure 6D). Notably, each Hsp104 variant showed similar activity with two different human Hsp70 and Hsp40 pairs, Hsc70/Hdj2, and Hsp72/Hdj1, suggesting this activity is not dependent on the presence of specific Hsp70 or Hsp40 variants (Figure 6D). The distinct luciferase reactivation activity of Hsp104K358D:Y662M and lack of activity of Hsp104K358D:Y257L, Hsp104K358D:Y257T, and Hsp104K358D:Y257L:Y662M, likely result from a specific effect on luciferase from perturbing pore-loop 1. Altering pore-loop 1 in the context of the enhanced Hsp104K358D background must interfere with disaggregation of disordered luciferase aggregates. Indeed, these variants may no longer recognize luciferase effectively as a result of altered substrate-specificity.
Hsp104K358D:Y662M disaggregates α-syn fibrils but not TDP-43 fibrils
Next, we assessed the ability of Hsp104 variants to disaggregate preformed α-syn and TDP-43 fibrils.34 Hsp104 displayed limited ability to dissociate α-syn or TDP-43 fibrils under these conditions, whereas Hsp104A503S and Hsp104K358D effectively dismantled α-syn and TDP-43 fibrils (Figure 6E, F). Hsp104K358D:Y257L also effectively disaggregated α-syn and TDP-43 fibrils (Figure 6E, F). Thus, although ineffective against luciferase (Figure 6D), Hsp104K358D:Y257L was active against α-syn and TDP-43 fibrils (Figure 6E, F). By contrast, Hsp104K358D:Y257T was unable to dissolve α-syn or TDP-43 fibrils (Figure 6E, F). Thus, the ability of Hsp104K358D:Y257T to reduce α-syn toxicity is separated from α-syn disaggregation. Finally, Hsp104K358D:Y662M could disassemble α-syn fibrils, but not TDP-43 fibrils (Figure 6E, F), indicating a change in substrate specificity that explains the ability to reduce α-syn toxicity, but not TDP-43 toxicity.
Hsp104K358D:Y257T and Hsp104K358D:Y662M disassemble soluble α-syn oligomers
We next assessed the ability of Hsp104 variants to disassemble preformed, soluble α-syn oligomers, which are a toxic species that accumulate in yeast.76,79 Under our conditions, Hsp104 displayed limited ability to dissociate soluble α-syn oligomers, whereas Hsp104A503S, Hsp104K358D, and Hsp104K358D:Y257L effectively disassembled soluble α-syn oligomers (Figure 6G). Importantly, the α-syn-selective variants, Hsp104K358D:Y257T and Hsp104K358D:Y662M, also effectively disassembled soluble α-syn oligomers (Figure 6G). Thus, these α-syn-selective variants directly remodel toxic α-syn conformers, which may enable mitigation of α-syn toxicity in yeast.
α-Syn-specific Hsp104 variants show distinct toxicity phenotypes at 37°C
As the α-syn-specific Hsp104 variants had differing biochemical profiles, we wondered if these variants showed differences in their off-target toxicity to yeast. Thus, we evaluated their toxicity at 37°C in the absence of neurodegenerative disease-associated substrate. Yeast are mildly stressed at 37°C as essential proteins are slightly unfolded. Potentiated Hsp104 variants can nonspecifically target key proteins that are slightly unfolded, resulting in a strong toxic phenotype when expressed in yeast.35 Thus, this assay can be used to uncover Hsp104 variants that exhibit less off-target toxicity that may be advanced to more complex model systems. Consistent with previous results,34 the potentiated variants Hsp104A503V and Hsp104K358D were toxic to yeast relative to Hsp104 at 37°C but not 30°C (Figure S6A). Interestingly, generally potentiated Hsp104K358D:Y257L and α-syn-specific Hsp104K358D:Y257T virtually eliminated this toxic phenotype (Figure S6A). Low toxicity to yeast could result from reduced targeting of essential, unfolded substrates in yeast and enhanced recognition of toxic substrates. It appears that the Y257L mutation reduces Hsp104 activity against off-target yeast substrates, but not against disease substrates. The ability to suppress toxicity of neurodegenerative disease-associated substrates, coupled with very low inherent toxicity to yeast, are desirable attributes for translating these disaggregases into effective therapeutic agents. Surprisingly, however, both Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M were noticeably toxic to yeast at 37°C (Figure S6A). We suggest these variants could be targeting a substrate under mild stress conditions that is essential for yeast. It seems the Y662M mutation reduces Hsp104 activity specifically against TDP-43 and FUS, but not against α-syn, or off-target yeast substrates.
α-Syn-specific Hsp104 variants confer yeast thermotolerance to different extents
Next, we assessed the ability of α-syn-specific Hsp104 variants to confer thermotolerance in yeast. One of the central functions of Hsp104 is to help yeast survive heat shock by resolubilizing proteins that aggregate during stress.62,80–84 We hypothesized that substrate-specific Hsp104 variants should be defective in conferring thermotolerance, in that they are no longer able to recognize a broad array of substrates for solubilization. We evaluated each Hsp104 variant under the native heat shock element (HSE) Hsp104 promoter relative to Hsp104 in their ability to confer thermotolerance to yeast. We first induced Hsp104 expression at 37°C, then heat-shocked yeast at 50°C (Figure S6B). All Hsp104 variants were expressed at similar levels (Figure S6C). Empty vector conferred no thermotolerance (very few yeast survive a 30-minute heat shock at 50°C), whereas Hsp104 transformed into a Δhsp104 strain effectively complemented the yeast survival observed in yeast expressing Hsp104 endogenously (Figure S6B). Potentiated variants Hsp104A503V and Hsp104K358D did not retain full Hsp104 activity in conferring thermotolerance (Figure S6B). Likewise, Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M also conferred reduced thermotolerance (Figure S6B). This reduced activity is perhaps due to off-target toxicity at the 37°C pretreatment step (Figure S6A). Interestingly, Hsp104K358D:Y257L and Hsp104K358D:Y257T conferred similar levels of reduced thermotolerance (Figure S6B). For these variants, which are not toxic at 37°C (Figure S6A), the reduced thermotolerance indicates that they do not disaggregate the complete repertoire of Hsp104 substrates (Figure S6B).
Several α-syn-specific Hsp104 variants prevent α-syn-induced neurodegeneration in C. elegans
We next tested whether the Hsp104 variants that selectively suppressed α-syn toxicity in yeast can prevent neurodegeneration in a C. elegans model of PD, which has successfully validated PD-relevant modifiers of α-syn toxicity.34,55,56,85,86 Preventing α-syn-induced neurodegeneration in this model is a key step in translating our findings in yeast to a full metazoan nervous system. We tested α-syn specific Hsp104K358D:Y257T, Hsp104K358D:Y662M, and Hsp104K358D:Y257L:Y662M, as well as generally potentiated Hsp104K358D and Hsp104K358D:Y257L. In this transgenic model, expression of human α-syn and Hsp104 variants is driven selectively in the dopaminergic neurons of worms using the endogenous promoter of the dopamine transporter gene, Pdat-1. When α-syn is expressed alone, ~23% of worms retained the full complement of dopaminergic neurons at day 7, and ~10% at day 10 post hatching (Figure 7A), which recapitulates the progressive α-syn neurotoxicity in human α-synucleinopathies. Hsp104 was unable to protect against α-syn-induced neurodegeneration (Figure 7A).34 Surprisingly, generally potentiated Hsp104K358D and Hsp104K358D:Y257L also did not significantly protect against neurodegeneration (Figure 7A). Moreover, the α-syn-specific Hsp104K358D:Y257L:Y662M did not protect against neurodegeneration, suggesting that this variant also did not translate from yeast to worm.
Figure 7. Several α-syn-specific Hsp104 variants prevent dopaminergic neuron degeneration in a C. elegans model of PD.
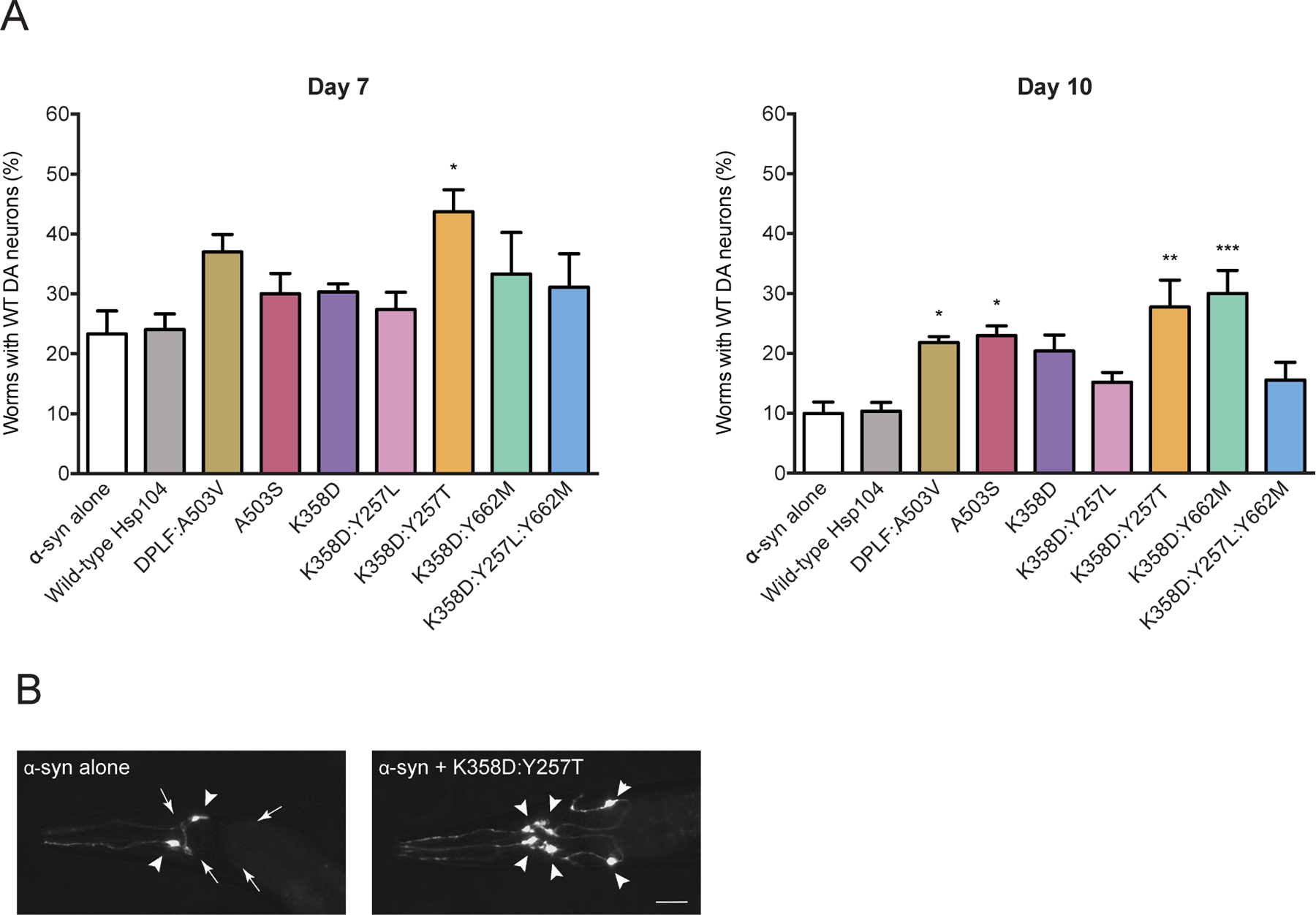
(A) Hsp104 variants and α-syn were co-expressed in the dopaminergic (DA) neurons of C. elegans. Animals expressing Hsp104K358D:Y257T are significantly protected against α-syn-induced DA neuron degeneration at day 7 post-hatching (left). DA neurodegeneration was exacerbated at day 10 post-hatching (right). Though not protective at day 7 post-hatching, overexpression of Hsp104 variants Hsp104DPLF:A503V, Hsp104A503S, and Hsp104K358D:Y662M protected against α-syn-induced DA neurodegeneration at day 10 post-hatching (right). Notably, the Hsp104K358D:Y257T variant consistently exhibits neuroprotection at both the earlier and later timepoints. Values represent means±SEM, n=30 per replicate, three independent experiments were performed/variant, and three distinct worm stable lines were generated for each Hsp104 variant. *P<0.05, **P<0.01, ***P<0.001; One-way ANOVA with Dunnett’s post-hoc test (compared to α-syn alone control). (B) A representative image of C. elegans DA neurons in worms expressing α-syn alone (left) or α-syn + Hsp104K358D-Y257T (right). Nematodes have six anterior DA neurons (4 CEP and 2 ADE), which were scored at day 7 and 10 post-hatching. Left, the worm has only two normal neurons (2 CEP), where the other four neurons have degenerated. Right, the full complement of six anterior DA neurons expressing Hsp104K358D-Y257T + α-syn, indicating a protective activity against α-syn toxicity. Triangles show normal neurons while arrows depict regions where there are degenerating or missing neurons. Bar, 10µm.
See also Figure S7.
Remarkably, α-syn-specific variant, Hsp104K358D:Y257T, strongly protected against α-syn-induced neurodegeneration at days 7 and 10 (Figure 7A, B). Despite only moderate α-syn toxicity suppression in yeast, Hsp104K358D:Y257T displayed the strongest protection in the context of a metazoan nervous system, as ~43% of worms retained the full complement of DA neurons at day 7, and ~27% at day 10 (Figure 7A, B). Hsp104K358D:Y257T is more effective in the C. elegans PD model than previously engineered potentiated variants Hsp104A503S and Hsp104DPLF:A503V 34 (Figure 7A, B). Hsp104K358D:Y257T expressed lower mRNA levels than generally potentiated variants in transgenic C. elegans and did not affect α-syn expression levels (Figure S7). This finding suggested that the reduced neurodegeneration is due to the enhanced activity of this α-syn-specific Hsp104 variant.
Interestingly, another α-syn-specific variant, Hsp104K358D:Y662M, conferred neuroprotection but was only significantly protective against neurodegeneration at a later timepoint (day 10), where ~30% of worms retained the full complement of WT neurons (Figure 7A). As Hsp104K358D:Y662M did not prevent neurodegeneration at an earlier time point (day 7), this variant did not appear able to delay disease onset, but instead limited the progression of α-syn neurotoxicity at a threshold of greater severity. Delayed prevention of α-syn-induced neurodegeneration could stem from a unique remodeling activity of Hsp104K358D:Y662M. Using an Hsp104 variant that stops disease progression earlier (Hsp104K358D:Y257T), followed by Hsp104K358D:Y662M which works more effectively later in disease, may be an advantageous combination strategy for mitigating α-syn toxicity in dopaminergic neurons.
Discussion
Here, we engineered Hsp104 variants that selectively suppress toxicity of α-syn. Unexpectedly, the Hsp104 background used to engineer enhanced substrate specificity was critical. For example, we found that Hsp104A503S, a potentiated Hsp104 variant that breaks MD-MD contacts,26,40 was not amenable for introducing substrate specificity via mutation of substrate-binding, pore-loop tyrosines. By contrast, α-syn-selective Hsp104 variants could be isolated by mutating pore-loop tyrosines in enhanced backgrounds that instead break NBD1-MD contacts, e.g. Hsp104K358D or Hsp104D484K.60 Thus, the precise nature of the underlying potentiation mechanism is an important aspect to consider when tuning Hsp104 variants26.
By making subtle changes to the substrate-binding, pore-loop tyrosines that line the axial channel of Hsp104, we re-wired enhanced Hsp104 variants (Hsp104K358D or Hsp104D484K) into substrate-specific variants. Thus, we shifted the substrate repertoire of Hsp104 such that α-syn toxicity could be mitigated, whereas TDP-43 or FUS toxicity could not. α-Syn-selectivity could be conferred by Y257I/V/T/Q mutations in pore-loop 1 or by Y662L/M mutations in pore-loop 2 in the K358D background. Surprisingly, two classes of α-syn-specific Hsp104 variant emerged that reduced α-syn toxicity via distinct mechanisms.
The first class, which includes Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M, mitigated α-syn toxicity via ATPase-dependent disaggregation of α-syn inclusions. Similar to our previously engineered, potentiated Hsp104 variants, Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M exhibited elevated ATPase activity compared to Hsp104. Hsp104K358D:Y662M and Hsp104K358D:Y257L:Y662M cleared α-syn foci in yeast but did not affect TDP-43 or FUS aggregation. Importantly, Hsp104K358D:Y662M dissolved α-syn fibrils but not TDP-43 fibrils in vitro. Thus, these Hsp104 variants have altered substrate specificity, which reduces disaggregase activity against some substrates (e.g., TDP-43) but permits effective α-syn disaggregation.
The second class, which includes Hsp104K358D:Y257T, mitigated α-syn toxicity via ATPase-dependent detoxification of α-syn conformers without disaggregation of α-syn inclusions. Intriguingly, Hsp104K358D:Y257T exhibited similar ATPase activity to WT Hsp104. However, Hsp104K358D:Y257T was unable to disaggregate luciferase in vitro, but could confer some level of thermotolerance in vivo indicating that it retains some disaggregase activity. Unexpectedly, Hsp104K358D:Y257T neither eliminated α-syn foci in yeast nor disaggregated α-syn fibrils in vitro. Importantly, Hsp104K358D:Y257T was more effective than WT Hsp104 in disassembling soluble α-syn oligomers in vitro, which are a neurotoxic species76,79,87,88. Moreover, unlike WT Hsp104, Hsp104K358D:Y257T colocalized with α-syn foci. Hsp104K358D:Y257T may partially remodel α-syn inclusions into less toxic structures. For example, Hsp104K358D:Y257T could selectively disassemble soluble α-syn oligomers throughout the cell, including any associated with α-syn inclusions. In addition, Hsp104K358D:Y257T might extract essential proteins trapped in α-syn foci, or mitigate toxic interactions between α-syn and intracellular vesicles or organelles.5,73,74,89
Hsp104 variants from each class (Hsp104K358D:Y662M and Hsp104K358D:Y257T) prevented dopaminergic neuron degeneration in a C. elegans model of PD more effectively than previous non-α-syn-specific, enhanced Hsp104 variants. Hsp104K358D:Y257T protected against neurodegeneration throughout the course of disease in this model, whereas Hsp104K358D:Y662M protection was restricted to the later time point. These data highlight the utility of the transgenic worm model in enabling temporal distinctions in neuromodulation to be parsed in vivo. Thus, substrate-specific Hsp104 variants effectively suppress neurodegeneration in the context of an intact metazoan nervous system, thereby translating the therapeutic benefits of these variants from yeast to metazoa. These findings bode well for subsequent validation, given the proven translational efficacy of the C. elegans α-syn model used in these studies, which has reproducibly demonstrated a predictive capacity to yield outcomes representative of mammalian models of PD.90
We have also established that the level of off-target toxicity in yeast inherent to potentiated Hsp104 variants does not necessarily predict whether these variants will be neuroprotective in C. elegans. For example, Hsp104K358D:Y662M exhibited off-target toxicity in yeast, but mitigated α-syn-induced dopaminergic neurodegeneration at later time points in C. elegans. Hsp104K358D:Y662M could target essential yeast proteins for unfolding, but these proteins may not be as crucial or may be absent from C. elegans. By contrast, Hsp104K358D:Y257L:Y662M exhibited off-target toxicity in yeast but did not prevent neurodegeneration in C. elegans. The challenge of translating our findings from yeast to worm likely reflects key differences between the two model systems. One important distinction is that the proteome of dopaminergic neurons in C. elegans likely has crucial differences from the yeast proteome. For example, dopamine modulates the propensity of α-syn to oligomerize and impacts neurodegeneration in mice, dopaminergic neuron culture, and C. elegans.91,92 Thus, the absence of dopamine in yeast, as well different sets of Hsp104-interacting proteins in yeast versus worm renders the direct translation of Hsp104 variants to metazoa challenging. Nevertheless, our efforts to engineer α-syn-specific Hsp104 variants have yielded Hsp104K358D:Y662M and Hsp104K358D:Y257T, which outperform prior potentiated variants. It will be important to advance these Hsp104 variants to mammalian models of α-synucleinopathies.
It will also be of great interest to engineer substrate-specific Hsp104 variants to specifically target a range of other misfolded proteins in neurodegenerative disease, including TDP-43 and FUS. At a minimum, our findings suggest that engineering substrate-specific disaggregases can improve their ability to confer neuroprotection. We envision that increasing the substrate specificity of enhanced disaggregases could be applied broadly to tailor therapeutics for neurodegenerative disease. Beyond Hsp104, we anticipate that fine-tuning disaggregases found in humans, such as Hsp110, Hsp70, and Hsp40, nuclear-import receptors, Skd3, TRIMs, and polyD/E proteins like DAXX will also be immensely valuable.7,29,46,93–97 We suggest that highly tuned, specific disaggregases that reverse targeted toxic misfolding events represent an exciting avenue for therapeutic agents in neurodegenerative disease.7,11,46
Limitations of the Study
We have not assessed the substrate selectivity of α-synuclein-specific Hsp104 variants in a proteome-wide manner, which will be important to address in future studies. Moreover, we have not assessed α-synuclein-specific Hsp104 variants in mammalian models. In future studies, it will be important to advance the α-synuclein-specific Hsp104 variants to mammalian models of α-synucleinopathies, including human neurons in culture and mouse models.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James Shorter (jshorter@pennmedicine.upenn.edu).
Materials availability
Plasmids generated in this study will be made readily available to the scientific community. We will honor requests in a timely fashion. Material transfers will be made with no more restrictive terms than in the Simple Letter Agreement or the Uniform Biological Materials Transfer Agreement and without reach through requirements.
Data and code availability
All raw images of yeast spotting, western blots, and C. elegans experiments are available in Mendeley Data. All data are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
All custom code has been deposited to GitHub and Zenodo. DOI are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Hsp104 polyclonal | Enzo Life Sciences | Cat#ADI-SPA-1040-F; RRID:AB_2039208 |
| Rabbit anti-FUS polyclonal | Bethyl Laboratories | Cat# A300-302A; RRID:AB_309445 |
| Rabbit anti-TDP-43 polyclonal | Proteintech | Cat#10782; RRID: AB_615042 |
| Rabbit anti-GFP polyclonal | Sigma-Aldrich | Cat# G1544; RRID: AB_439690 |
| Mouse anti-α-Synuclein monoclonal (Syn303) | Kelvin Luk (University of Pennsylvania) | RRID:AB_2315395 |
| Mouse anti-PGK1 monoclonal | Thermo Fisher | Cat#459250; RRID:AB_2532235 |
| IRDye 800CW Goat anti-Mouse IgG secondary antibody | LI-COR | Cat# 926–32210; RRID:AB_621842 |
| IRDye 680RD Goat anti-Rabbit IgG secondary antibody | LI-COR | Cat# 926–68071; RRID:AB_10956166 |
| Bacterial and virus strains | ||
| Escherichia coli DH5α competent cells | Thermo Fisher | Cat#18265017 |
| Escherichia coli BL21-CodonPlus (DE3)-RIL competent cells | Agilent | Cat#230245 |
| Chemicals, peptides, and recombinant proteins | ||
| Creatine phosphate | Roche | Cat#10621722001 |
| Adenosine 5’-triphosphate disodium salt hydrate (ATP) | Sigma-Aldrich | Cat#A3377 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#4693159001 |
| Casein fluorescein isothiocyanate from bovine milk (FITC-Casein) | Sigma-Aldrich | Cat#C0528 |
| Creatine kinase | Roche | Cat#10127566001 |
| Firefly luciferase | Sigma-Aldrich | Cat#L9506 |
| Lysozyme | Sigma-Aldrich | Cat#L6876 |
| His-TEV protease | Cupo and Shorter98 | N/A |
| Hsp104 | Jackrel et al.34 | N/A |
| Hsp104A503S | Jackrel et al.34 | N/A |
| Hsp104K358D | This paper | N/A |
| Hsp104K358D:Y257L | This paper | N/A |
| Hsp104K358D:Y257T | This paper | N/A |
| Hsp104K358D:Y662M | This paper | N/A |
| Hsp104K358D:Y257L:Y662M | This paper | N/A |
| HAP | Jackrel et al.34 | N/A |
| HAPA503S | This paper | N/A |
| HAPK358D | This paper | N/A |
| HAPK358D:Y257L | This paper | N/A |
| HAPK358D:Y257T | This paper | N/A |
| HAPK358D:Y662M | This paper | N/A |
| HAPK358D:Y257L:Y662M | This paper | N/A |
| GroELtrap | Jackrel et al.34 | N/A |
| RepA1-25-GFP | Lopez et al.99 | N/A |
| ClpP | Jackrel et al.34 | N/A |
| Hsc70 | Enzo Life Sciences | Cat#ADI-SPP-751-F |
| Hdj2 | Enzo Life Sciences | Cat#ADI-SPP-405-F |
| Hsp72 | Michalska et al.20 | N/A |
| Hdj1 | Michalska et al.20 | N/A |
| α-Syn | Jackrel et al.34 | N/A |
| TDP-43 | Jackrel et al.34 | N/A |
| Critical commercial assays | ||
| ATPase Activity Kit (Colorimetric) | Innova Biosciences | Cat#601–0120 |
| Luciferase Assay Reagent | Promega | Cat#E1483 |
| Human α-Synuclein ELISA Kit | Thermo Fisher | Cat#KHB0061 |
| QuikChange Site-Directed Mutagenesis Kit | Agilent | Cat# 200518 |
| Deposited data | ||
| Raw images of yeast spotting, western blots, and C. elegans experiments | Mendeley data | doi: 10.17632/t7k9w8txzk.1 |
| Experimental models: Organisms/strains | ||
| S. cerevisiae W303a (MATa, can1-100, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, ade2-1) | Jackrel et al.34 | N/A |
| S. cerevisiae W303a∆hsp104 (MATa, can1-100, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, ade2-1, hsp104::KanMX) | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-α-syn-YFP-pAG304GAL-α-syn-YFP | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-FUS | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-TDP-43 | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-TDP-43-GFPS11-pAG305GAL-GFPS1-10 | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-FUS-GFP | Jackrel et al.34 | N/A |
| C. elegans UA44 (baln11 [Pdat-1::α-syn, Pdat-1: :GFP]) | Cao et al.85 | N/A |
| C. elegans UA367 (baEx198 a,b,c [Pdat-1::HSP104-K358D, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA368 (baEx199 a,b,c [Pdat-1::HSP104-K358D:Y257L, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA369 (baEx200 a,b,c [Pdat-1::HSP104-K358D:Y257T, rol-6];baln11 [Pdat-1::α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA370 (baEx201 a,b,c [Pdat-1::HSP104-K358D:Y257L:Y662M, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1::GFP]) | This study | N/A |
| C. elegans UA371 (baEx202 a,b,c [Pdat-1::HSP104-K358D: Y662M, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA256 (baEx147 a,b,c [Pdat-1::Hsp104 WT, Pmyo-2::mCherry]; baIn11 [Pdat-1:: α-syn, Pdat-1::GFP]) | Jackrel et al.34 | N/A |
| C. elegans UA259 (baEx150 a,b,c [Pdat-1::Hsp104-A503S, Pmyo-2::mCherry]; baIn11 [Pdat-1::α-syn, Pdat-1::GFP]) | Jackrel et al.34 | N/A |
| C. elegans UA262 (baEx153 a,b,c [Pdat-1::Hsp104-DPLF:A503V, Pmyo-2::mCherry]; baIn11[Pdat-1::α-syn, Pdat-1::GFP]) | Jackrel et al.34 | N/A |
| Oligonucleotides | ||
| See Table S1 | ||
| Recombinant DNA | ||
| pRS416GAL | Jackrel et al.34 | N/A |
| pRS416GAL-Hsp104 | Jackrel et al.34 | N/A |
| pRS416GAL-Hsp104A503S | Jackrel et al.34 | N/A |
| pRS416GAL-Hsp104A503S:Y257L | This study | N/A |
| pRS416GAL-Hsp104A503S:Y257T | This study | N/A |
| pRS416GAL-Hsp104A503S:Y662M | This study | N/A |
| pRS416GAL-Hsp104A503S:Y257L:Y662M | This study | N/A |
| pRS416GAL-Hsp104K358D | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257F | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257W | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257I | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257V | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257M | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662F | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662W | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662I | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662V | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662M | This study | N/A |
| pRS416GAL-Hsp104D484K | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257L | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257F | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257W | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257I | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257V | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257M | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662L | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662F | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662W | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662I | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662V | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662M | This study | N/A |
| pRS416GAL-Hsp104Y257L | This study | N/A |
| pRS416GAL-Hsp104Y662L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662F | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662W | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662M | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662S | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662T | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662Q | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662N | This study | N/A |
| pRS416GAL-Hsp104K218T:K620T | Torrente et al.39 | N/A |
| pRS416GAL-Hsp104K218T:A503S:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:K358D:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:Y257L:K358D:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:Y257T:K358D:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:K358D:K620T:Y662M | This study | N/A |
| pRS416GAL-Hsp104K218T:Y257L:K358D:K620T:Y662M | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257S | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257T | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257Q | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257N | This study | N/A |
| pRS416GAL-mCherry | This study | N/A |
| pRS416GAL-Hsp104-mCherry | This study | N/A |
| pRS416GAL-Hsp104K358D-mCherry | This study | N/A |
| pRS416GAL-Hsp104A503S-mCherry | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257T-mCherry | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L-mCherry | This study | N/A |
| pRS416HSE-Hsp104 | Jackrel et al.34 | N/A |
| pRS416HSE-Hsp104A503V | Jackrel et al.34 | N/A |
| pRS416HSE-Hsp104K358D | This study | N/A |
| pRS416HSE-Hsp104K358D:Y257L | This study | N/A |
| pRS416HSE-Hsp104K358D:Y257T | This study | N/A |
| pRS416HSE-Hsp104K358D:Y662M | This study | N/A |
| pRS416HSE-Hsp104K358D:Y257L:Y622M | This study | N/A |
| pAG413GAL | Alberti et al.100 | N/A |
| pAG413GAL-α-syn | This study | N/A |
| pAG413GAL-α-synE46K | This study | N/A |
| pAG413GAL-α-synA53T | This study | N/A |
| pAG413GAL-α-syn1-95 | This study | N/A |
| pAG413GAL-α-syn1-115 | This study | N/A |
| pAG413GAL-α-syn1-125 | This study | N/A |
| pAG415GAL | Alberti et al.100 | N/A |
| pAG415GAL-α-syn | This study | N/A |
| pAG415GAL-α-synE46K | This study | N/A |
| pAG415GAL-α-synA53T | This study | N/A |
| pAG415GAL-α-syn1-95 | This study | N/A |
| pAG415GAL-α-syn1-115 | This study | N/A |
| pAG415GAL-α-syn1-125 | This study | N/A |
| pNOTAG-Hsp104 | Jackrel et al.34 | N/A |
| pNOTAG-Hsp104A503S | Jackrel et al.34 | N/A |
| pNOTAG-Hsp104K358D | This study | N/A |
| pNOTAG-Hsp104K358D:Y257L | This study | N/A |
| pNOTAG-Hsp104K358D:Y257T | This study | N/A |
| pNOTAG-Hsp104K358D:Y662M | This study | N/A |
| pNOTAG-Hsp104K358D:Y257L:Y662M | This study | N/A |
| pNOTAG-HAP | Jackrel et al.34 | N/A |
| pNOTAG-HAPA503S | This study | N/A |
| pNOTAG-HAPK358D | This study | N/A |
| pNOTAG-HAPK358D:Y257L | This study | N/A |
| pNOTAG-HAPK358D:Y257T | This study | N/A |
| pNOTAG-HAPK358D:Y662M | This study | N/A |
| pNOTAG-HAPK358D:Y257L:Y662M | This study | N/A |
| pTrc99A-GroELtrap | Jackrel et al.34 | N/A |
| pBAD-RepA1-25-GFP | Lopez et al.99 | N/A |
| pET28b-ClpP-his | Jackrel et al.34 | N/A |
| pE-SUMO-Hsp72 | This study | N/A |
| pE-SUMO-Hdj1 | Michalska et al.20 | N/A |
| pHis-TEV | Cupo and Shorter98 | N/A |
| pT7-7 α-syn | Lo Bianco et al.27 | N/A |
| pDUET-GST-TEV-TDP-43 | Johnson et al.71 | N/A |
| Software and algorithms | ||
| Prism 9 | GraphPad | N/A |
| ImageJ | Rueden et al.101 | N/A |
| Other | ||
| Custom code for a semi-automated procedure was used to quantify colocalization between the red and green images. | Zenodo | doi: 10.5281/zenodo.8185164 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Yeast strains
Yeast strains used were wild-type W303a (MATa, can1-100, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, ade2-1) or the isogenic strain W303aΔhsp104.34 The yeast strains W303aΔhsp104-pAG303GAL-α-syn-YFP-pAG304GAL-α-syn-YFP, W303aΔhsp104-pAG303GAL-FUS, W303aΔhsp104-pAG303GAL-TDP-43, W303aΔhsp104-pAG303GAL-TDP-43-GFPS11-pAG305GAL-GFPS1-10, and W303aΔhsp104-pAG303GAL-FUS-GFP have been described previously.34,35,102 Yeast were grown in rich medium (YPD) or in synthetic media without amino acids used for selection. 2% sugar (dextrose, raffinose, or galactose) was added to synthetic media.
C. elegans
Nematodes were grown and maintained using well-established procedures.103 Plasmid constructs were injected into worms to generate transgenic animals using previously described methods.104 Strains UA367 (baEx198 a,b,c [Pdat-1::HSP104-K358D, rol-6];baln11 [Pdat-1::α-syn, Pdat-1::GFP]), UA368 (baEx199 a,b,c [Pdat-1::HSP104-K358D:Y257L, rol-6];baln11 [Pdat-1::α-syn, Pdat-1::GFP]), UA369 (baEx200 a,b,c [Pdat-1::HSP104-K358D:Y257T, rol-6];baln11 [Pdat-1::α-syn, Pdat-1::GFP]), UA370 (baEx201 a,b,c [Pdat-1::HSP104-K358D:Y257L:Y662M, rol-6];baln11 [Pdat-1::α-syn, Pdat-1::GFP]), and UA371 (baEx202 a,b,c [Pdat-1::HSP104-K358D:Y662M, rol-6];baln11 [Pdat-1::α-syn, Pdat-1::GFP]) were generated by injecting 50 ng/μl of corresponding plasmid construct into UA44 (baln11 [Pdat-1::α-syn, Pdat-1::GFP]) with phenotypic marker (rol-6, 50 ng/μl, for roller expression). Three independent stable lines were created for each group (a, b, c). Strain UA256 (baEx147 a,b,c [Pdat-1::Hsp104 WT, Pmyo-2::mCherry]; baIn11 [Pdat-1::α-syn, Pdat-1::GFP]), UA259 (baEx150 a,b,c [Pdat-1::Hsp104-A503S, Pmyo-2::mCherry]; baIn11 [Pdat-1::α-syn, Pdat-1::GFP]), and UA262 (baEx153 a,b,c [Pdat-1::Hsp104-DPLF:A503V, Pmyo-2::mCherry]; baIn11[Pdat-1:: α-syn, Pdat-1::GFP]) were previously generated.34
METHOD DETAILS
Yeast plasmids
Hsp104 variants were under control of a galactose-inducible promoter on pRS416GAL plasmids, except for in thermotolerance assays where they were under control of the HSE promoter on pRS416HSE plasmids. For mCherry-tagged Hsp104 variants, mCherry is located at the C-terminal end of Hsp104, separated by a glycine-serine linker (also on pRS416GAL plasmids). For experiments in Figure S4, α-syn variants (α-syn, α-synE46K, α-synA53T, α, α-syn1-115, or α-syn1-125) were under control of a galactose-inducible promoter on pAG413GAL and pAG415GAL plasmids.
Site-directed mutagenesis
Mutations were introduced into Hsp104 through QuikChange site-directed mutagenesis (Agilent) and confirmed by DNA sequencing.
Yeast transformation and spotting assays
Plasmids containing Hsp104 variants were transformed into yeast using a standard lithium acetate and polyethylene glycol procedure.105 For spotting assays, yeast cultures were grown to saturation overnight at 30°C in dropout media containing raffinose. Raffinose cultures were then normalized according to OD600, five-fold serial diluted, and spotted onto glucose and galactose plates using a 96-bolt replicator tool. Plates were grown at 30°C for 2–3 days and imaged.
Toxicity spotting assay
pRS416GAL plasmids containing Hsp104 variants were transformed into W303aΔhsp104 yeast. Yeast cultures were grown to saturation overnight at 30°C in dropout media containing raffinose. Raffinose cultures were then normalized according to OD600 and five-fold serial diluted. The cultures were spotted onto two sets of glucose and galactose plates using a 96-bolt replicator tool. One set of plates was grown at 30°C, and the other at 37°C, for 2–3 days and subsequently imaged.
Western blotting
Hsp104 variants transformed into appropriate yeast strains were grown to saturation overnight at 30°C in dropout media containing raffinose. Cultures were normalized to OD600 = 0.3 and grown in galactose dropout media at 30°C to induce Hsp104 and disease substrate expression (α-syn cultures induced for 8h, TDP-43 and FUS cultures induced for 5h). Galactose cultures were then normalized according to OD600 and the equivalent of 6ml culture with an OD600 = 0.6 were harvested by centrifugation. Media was aspirated, and the cell pellets were resuspended in 0.1M NaOH and incubated at room temperature for 5min. Cells were pelleted again by centrifugation, supernatant removed, and pellet was resuspended in 100µL 1X SDS sample buffer and boiled for 4–5min. Samples were separated via SDS-PAGE (4–20% gradient, Bio-Rad) and transferred to a PVDF membrane (Millipore) using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Membranes were blocked for at least 1h at room temperature and then incubated with primary antibodies (rabbit anti-Hsp104 polyclonal (Enzo Life Sciences); rabbit anti-FUS polyclonal (Bethyl Laboratories); rabbit anti-TDP-43 polyclonal (Proteintech); rabbit anti-GFP polyclonal (Sigma-Aldrich); mouse anti-PGK1 monoclonal (Thermo Fisher)) at 4°C overnight. Membranes were washed multiple times with PBS-T, incubated with secondary antibodies (goat anti-mouse and goat anti-rabbit, LI-COR) for 1h at room temperature, and washed again multiple times with PBS-T (final wash with PBS). Membranes were imaged using a LI-COR Odyssey FC Imaging system.
Thermotolerance assay
Hsp104 variants under the HSE promoter were transformed into W303aΔhsp104 yeast. Yeast cultures were grown to saturation overnight at 30°C in glucose dropout media. Cultures were normalized to OD600 = 0.3 and grown in glucose dropout media at 30°C for at least 4h, after which the equivalent of 6 ml culture with an OD600 = 0.6 was grown at 37°C for 30 min (if assessing Hsp104 expression, samples would be harvested at this stage for western blot as described above). Cultures were then heat-shocked at 50°C in 1.5ml Eppendorf tubes in an Eppendorf Thermomixer for 30min and incubated on ice for 2min. Cultures were diluted appropriately, plated on glucose dropout media, and incubated at 30°C. After 2–3 days, colonies were counted using an aCOLyte colony counter and software (Synbiosis). W303a yeast were transformed with an empty pRS416Gal vector and treated as above for endogenous Hsp104 control.
Fluorescence microscopy
Yeast strains for fluorescence microscopy were as in Jackrel et al.34 Hsp104 variants on pRS416GAL plasmids were transformed into each yeast strain (α-syn-YFP, FUS-GFP, or TDP43-GFPS11 from Jackrel et al.34) and grown to saturation overnight at 30°C in dropout media containing raffinose. Cultures were normalized to OD600 = 0.3 and grown at 30°C in galactose dropout media to induce Hsp104 and disease substrate expression (α-syn cultures induced for 8h, TDP-43 and FUS cultures induced for 5h). For α-syn and FUS cultures, cells were harvested and processed for microscopy, and imaged as live cells. For TDP-43 cultures, cells were harvested and fixed by spinning cells down, resuspending in 1ml cold 70% ethanol, and immediately pelleting cells again. Cells were washed 3 times with cold PBS and resuspended in Vectashield mounting medium with 4’,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Cells were imaged at 100X magnification using a Leica DM IRBE microscope. Cells were analyzed using ImageJ software, and a minimum of 200 cells were quantified per sample in at least three independent trials. One-way ANOVA with Dunnett’s post hoc test was performed using GraphPad Prism Software.
For fluorescence microscopy to assess mCherry-GFP localization, Hsp104 variants with a C-terminal mCherry tag, or mCherry alone (on pRS416GAL plasmids) were transformed into the α-syn-YFP strain described in Jackrel et al.34 Cultures were treated as α-syn cultures described above. After induction, cells were harvested, stained with Hoechst dye (channel not shown in Figure 5C) and imaged at 100X magnification using a Leica DM IRBE microscope. For analysis of mCherry colocalization with GFP, cells were imaged in red (TX2) and green (L5) channels. A semi-automated procedure was used to quantify colocalization between the red and green images (code available at https://github.com/bensondaled/mack-cell-annotation). Cells containing α-syn-YFP foci were manually identified and selected using a custom interface. A colocalization metric for each cell was then defined as the Pearson correlation between the pixels in the red and green channels. A cell was defined to exhibit colocalization if this measure exceeded 0.85, a uniform threshold applied to all images and chosen for its agreement with manual colocalization assessments. Alternatively, colocalization was determined manually. A minimum of 200 cells were quantified per sample in at least three independent trials. One-way ANOVA with Dunnett’s post hoc test was performed using GraphPad Prism Software.
C. elegans dopaminergic neurodegeneration
For dopaminergic neurodegeneration analyses, the transgenic animals were scored as described previously.106 Briefly, on the day of analysis, the six anterior dopaminergic neurons [four CEP (cephalic) and two ADE (anterior deirid)] were examined in 30 animals randomly. Each nematode was considered normal when all six anterior DA neurons were present. However, if a worm exhibited any degenerative phenotype, such as a missing dendritic process, cell body loss, or a blebbing neuronal process, it was scored as degenerating. Three independent transgenic worm lines were analyzed per genetic background for a total of ninety animals per transgenic line and an average of total percentage of worms with normal neurons was reported in the study. A one-way ANOVA, followed by a Dunnett’s post hoc test, was performed for statistical analysis using GraphPad Prism Software.
Quantitative Real-Time PCR in nematodes
RNA isolation and qRT-PCR were performed on worms using previously published methods.106,107 Briefly, total RNA was isolated from 100 young adult (day 4 post hatching) nematodes from corresponding transgenic line using TRI reagent (Molecular Research Center). 1μg of RNA was used for cDNA synthesis using the iScript Reverse Transcription Supermix for qRT-PCR (Bio-Rad, Hercules, CA, USA). qRT-PCR was performed using IQ-SYBR Green Supermix (Bio-Rad) with the Bio-Rad CFX96 Real-Time System. Hsp104 expression levels were normalized to two reference genes (snb-1 and y45). α-syn expression levels were normalized to three reference genes (ama-1, cdc-42, and tba-1). The reference target stability was analyzed by GeNorm and passed for all reference genes listed above. Three technical replicates with three biological replicates were performed in this study. Each primer pair was confirmed for at least 90–100% efficiency by standard curve analyses. The following primers were used for the assays:
hsp104 Forward: GGCCATGAAGAATTGACACAAG
hsp104 Reverse: CCTTAAATCAGCGGCGGTAG
α-syn Forward: ATGTAGGCTCCAAAACCAAGG
α-syn Reverse: ACTGCTCCTCCAACATTTGTC
snb-1 Forward: CCGGATAAGACCATCTTGACG
snb-1 Reverse: GACGACTTCATCAACCTGAGC
y45 Forward: GTCGCTTCAAATCAGTTCAGC
y45 Reverse: GTTCTTGTCAAGTGATCCGACA
ama-1 Forward: TCCTACGATGTATCGAGGCAA
ama-1 Reverse: CTCCCTCCGGTGTAATAATGA
cdc-42 Forward: CCGAGAAAAATGGGTGCCTG
cdc-42 Reverse: TTCTCGAGCATTCCTGGATCAT
tba-1 Forward: ATCTCTGCTGACAAGGCTTAC
tba-1 Reverse: GTACAAGAGGCAAACAGCCAT
Protein purification
Hsp104 and HAP
Hsp104 and HAP proteins were purified as described34 with the following modifications. Eluate from Affi-Gel Blue Gel was equilibrated to a low-salt buffer Q (~100mM NaCl, 20mM TRIS pH 8.0, 5mM MgCl2, 0.5mM EDTA) and purified via ResourceQ anion exchange chromatography. Buffer Q (20mM TRIS pH 8.0, 50mM NaCl, 5mM MgCl2, 0.5mM EDTA) was used as running buffer, and the protein was eluted with a linear gradient of buffer Q+ (20mM TRIS pH 8.0, 1M NaCl, 5mM MgCl2, 0.5mM EDTA). The eluted protein was buffer-exchanged into high-salt storage buffer (40mM HEPES-KOH pH 7.4, 500mM KCl, 20mM MgCl2) plus 10% glycerol and 1mM DTT and snap-frozen.
GroELtrap
pTrc99A-GroELtrap was transformed into DH5α competent E. coli cells (Thermo Fisher). Cells were grown in 2xYT medium with appropriate antibiotics at 37°C with shaking until OD600 reached ~ 0.4 – 0.6. Protein overexpression was induced with 1mM IPTG, and cells were grown at 37°C until OD600 ~ 2.0. Cells were harvested by spinning (5000g, 4°C, 15 min) and pellet was resuspended in 50mM sodium phosphate buffer and centrifuged (5000g, 4°C, 15min). Pellet was resuspended in low-salt buffer (50mM Tris-HCl pH 7.5, 1mM EDTA, 1mM DTT, 50mM NaCl) and 10mg lysozyme per g cell pellet. Sample stirred gently for 5min, lysed through sonication, and centrifuged (17,000 g, 4°C, 30min). Clarified lysate loaded onto HiTrap Q HP column (GE Healthcare) and eluted through salt gradient using low-salt buffer (as described above) and high-salt buffer (50mM Tris-HCl pH 7.5, 1mM EDTA, 1mM DTT, 500mM NaCl108). Collected fractions were exchanged into the following TKME-100 buffer: 20mM TRIS-HCl pH 7.5, 100mM KCl, 10mM MgCl2, 0.1mM EDTA, 5mM DTT, 10% glycerol, 0.005% Triton X-100 and snap-frozen.
RepA1-25-GFP
pBAD-RepA1-25-GFP was transformed into BL21 (DE3)-RIL cells. Cells were inoculated in 2xYT medium with appropriate antibiotics at 37°C with shaking until OD600 reached ~ 0.6 – 0.8. Protein overexpression was induced with 1mM IPTG, and cells were grown at 30°C for 4h. Cells were harvested by spinning (4000rpm, 25min) and pellet was resuspended in 40mM HEPES-KOH pH 7.4 plus 2mM 2-Mercaptoethanol (BME) and EDTA-free protease inhibitors. Cells were lysed using a sonicator and centrifuged (16,000rpm, 20min). The resulting pellet was washed twice with HM buffer (40mM HEPES-KOH pH 7.4, 20mM MgCl2) plus 2mM BME. After each wash, cells were centrifuged (16,000 rpm, 20min). Pellet was then resuspended in buffer containing 8M urea, 40mM Tris-HCl pH 6.8, 500mM NaCl, 10% glycerol (v/v) and agitated slowly overnight at 25°C. Sample was centrifuged (16,000rpm, 20min) and supernatant was kept. Supernatant was incubated with Ni-NTA beads (HisPur™ Ni-NTA Resin, Thermo Scientific) pre-equilibrated in buffer containing 8M urea, 40mM Tris pH 6.8, 500mM NaCl, 10% glycerol (v/v) for 2h on a spinning wheel at 25°C at the lowest speed. Sample washed 5 times with buffer containing 8M urea, 40mM Tris-HCl pH 6.8, 500mM NaCl, 20mM imidazole, 10% glycerol (v/v). Sample then washed 5 times with buffer containing 8M urea, 40mM Tris-HCl pH 6.8, 500mM NaCl, 40mM imidazole, 10% glycerol (v/v). Sample eluted with buffer containing 8 M urea, 40mM Tris-HCl pH 6.8, 500mM NaCl, 500mM imidazole, 10% glycerol (v/v). Sample was dialyzed overnight into buffer containing 40mM HEPES-KOH pH 7.4, 20mM imidazole, 150mM KCl, 2mM BME, 10% glycerol (v/v) at 4°C and re-loaded onto a Ni-NTA column (HisTrap™ HP, GE Healthcare). An imidazole gradient was applied (from 20mM to 500mM) over 20CV in buffer containing 40mM HEPES-KOH pH 7.4, 20mM imidazole, 150mM KCl, 2mM BME, 10% glycerol (v/v). The purity of eluted fractions was assessed using SDS-PAGE. Collected fractions were buffer-exchanged into HKM-150 buffer (40mM HEPES-KOH pH 7.4, 150mM KCl, 20mM MgCl2) plus 2 mM BME and 10% glycerol (v/v) and snap-frozen.
ClpP
C-terminally His-tagged ClpP was purified as described34.
Hsp70 and Hsp40
Hsc70 and Hdj2 were from Enzo Life Sciences. Hdj1 was purified as in Michalska et al.20 Hsp72 was purified as for Hsc70 in Michalska et al.20
α-Syn and TDP-43
α-Syn and TDP-43 were purified as described.34
ATPase assay
Hsp104 (0.25µM monomer in Hsp104 buffer, see Hsp104 purification) was incubated with ATP (1 mM) for 5min at 25°C in luciferase-refolding buffer (LRB: 25mM HEPES-KOH pH 7.4, 150mM KAOc, 10mM MgAOc, 10mM DTT). The final reaction buffer contained 6.3% of HKM-500 buffer. ATPase activity was evaluated by the release of inorganic phosphate, which was measured using a malachite green phosphate detection kit (Innova Biosciences). Background hydrolysis at time zero was subtracted.
FITC-casein degradation assay
FITC-casein (0.1µM) was incubated with HAP or HAP variants (1µM monomer), ClpP (21µM monomer), ATP (5mM), and an ATP regeneration system (1mM creatine phosphate and 0.25µM creatine kinase). ClpP was first buffer-exchanged into luciferase-refolding buffer (LRB: 25mM HEPES-KOH pH 7.4, 150mM KAOc, 10mM MgAOc, 10mM DTT) at 25°C. Reaction mixtures were assembled 25°C in LRB and degradation of FITC-casein was monitored by measuring fluorescence (excitation 490nm, emission 520nm) using a Tecan Infinite M1000 plate reader.
RepA1-25-GFP unfoldase assay
RepA1-25-GFP (0.7µM) was incubated with Hsp104 or Hsp104 variants (2.1µM hexamer), ATP (4mM), ARS (20mM creatine phosphate, 0.06µg/µl creatine kinase). GroELtrap (2.5µM tetradecamer) was included to prevent refolding of unfolded RepA1-25-GFP. Hsp104 variants were buffer-exchanged into TKME-100 buffer at 25°C. Reactions were assembled on ice in TKME-100 buffer plus 20µg/ml BSA. RepA1-25-GFP unfolding was measured by fluorescence (excitation 395nm, emission 510nm) using a Tecan Safire2, which was heated to 30°C prior to reading.
Luciferase reactivation assay
Aggregated luciferase (100nM) was incubated with Hsp104 or Hsp104 variants (1µM monomer), ATP (5mM), and an ATP regeneration system (10mM creatine phosphate, 0.25µM creatine kinase) in the presence or absence of co-chaperones Hsp70 (Hsc70 or Hsp72, 0.167µM) and Hsp40 (Hdj2 or Hdj1, 0.167µM) for 90min at 25°C in LRB. The final reaction buffer contained 8.6% of HKM-500 buffer. After 90min, luciferase activity was measured with a luciferase assay reagent (Promega). Recovered luminescence was measured using a Tecan Infinite M1000 plate reader.
α-Syn and TDP-43 fibril disaggregation
α-Syn (80µM) was assembled into fibrils via incubation in 40mM HEPES-KOH (pH 7.4), 150 mM KCl, 20 mM MgCl2, and 1 mM dithiothreitol for 48h at 37°C with agitation. α-Syn fibrils (3µM monomer) were incubated without or with Hsp104, Hsp104A503S, Hsp104K358D, Hsp104K358D:Y257L, Hsp104K358D:Y257T or Hsp104K358D:Y662M (3µM) plus Hsc70 (3µM), Hdj1 (3µM), ATP (20mM) and an ATP regeneration system (20mM creatine phosphate and 0.5µM creatine kinase) for 2h at 30°C. Disaggregation was assessed by Thioflavin-T fluorescence. 27 To generate TDP-43 fibrils, GST-TEV-TDP-43 (6µM) was incubated with TEV protease in 40mM HEPES-KOH (pH 7.4), 150mM KCl, 20mM MgCl2, and 1mM dithiothreitol for 16h at 25°C with agitation, by which time all the TDP-43 had aggregated. TDP-43 fibrils (3µM monomer) were incubated without or with Hsp104, Hsp104A503S, Hsp104K358D, Hsp104K358D:Y257L, Hsp104K358D:Y257T or Hsp104K358D:Y662M (3µM) plus Hsc70 (3µM), Hdj1 (3µM), ATP (20 mM) and an ATP regeneration system (20mM creatine phosphate and 0.5µM creatine kinase) for 2h at 30°C. Disaggregation was assessed by turbidity (absorbance at 350nm).
Soluble α-syn oligomer disassembly
Soluble α-syn oligomers were purified by gel filtration as described.109 For disassembly experiments, soluble α-syn oligomers (0.5 μM monomer) were incubated without or with Hsp104, Hsp104A503S, Hsp104K358D, Hsp104K358D:Y257L, Hsp104K358D:Y257T or Hsp104K358D:Y662M (1µM) in KHM buffer (40 mM HEPES-KOH, pH 7.4, 150 mM KCl, 20 mM MgCl2, 1 mM DTT) plus ATP (20 mM) and an ATP regeneration system (20mM creatine phosphate and 0.5µM creatine kinase) for 1 hour at 37°C. Reactions were then fractionated through a Microcon YM-100 (100-kDa molecular weight cut off) filter (Milllipore). Soluble α-syn oligomers are trapped by the filter, and only α-syn monomers enter the filtrate fraction.27 The amount of α-syn in the filtrate fraction was then determined using α-syn ELISA Kit (Thermofisher).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification is as described in the figure legends. Statistical analyses were performed using GraphPad Prism (GraphPad Software, Inc.; La Jolla, CA, USA) as described in figure legends.
Supplementary Material
Highlights.
Tuning Hsp104 specificity to selectively detoxify α-synuclein (α-syn)
Hsp104 variants with enhanced selectivity for α-syn oligomers and fibrils
Hsp104 variants with enhanced selectivity for soluble α-syn oligomers
α-syn-specific Hsp104 variants confer increased neuroprotection in C. elegans
Acknowledgements
We thank Ben Deverett for assistance with analysis of microscopy and Kelvin Luk for Syn303 antibody. K.L.M. was supported by an NSF graduate research fellowship (DGE-1321851). E.B.M. was supported by a Milton Safenowitz Post-Doctoral Fellowship from the ALS Association and NIH grants F32NS108598 and K99AG075242. J.L. was supported by an Alzheimer's Association Research Fellowship and a Warren Alpert Distinguished Scholars Fellowship Award. E.C. was supported by NIH grant T32GM008076 and a Blavatnik Family Fellowship in Biomedical Research. M.E.J. was supported by NIH grant R35GM128772. R.R.C. was supported by NIH grants T32GM008275, F31AG060672, and a Blavatnik Family Fellowship in Biomedical Research. L.M.C. was supported by an NSF graduate research fellowship (DGE-0822). K.A.C. and G.A.C. were supported by NIH grants R15NS106460 and R15NS104857, respectively. J.S. was supported by NIH grants (DP2OD002177, R01GM099836), a Muscular Dystrophy Association Research Award (MDA277268), an ALS Association Award, the Life Extension Foundation, a Linda Montague Pechenik Research Award, the Packard Center for ALS Research at Johns Hopkins University, and Target ALS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest
J.S. is a consultant for Dewpoint Therapeutics, Neurmora, and ADRx. J.S. is an advisor and a shareholder for Confluence Therapeutics. All other authors declare no competing interests.
References
- 1.Abeliovich A, and Gitler AD (2016). Defects in trafficking bridge Parkinson's disease pathology and genetics. Nature 539, 207–216. 10.1038/nature20414. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Wang L, Bao H, Premi S, Das U, Chapman ER, and Roy S (2019). Functional cooperation of alpha-synuclein and VAMP2 in synaptic vesicle recycling. Proc Natl Acad Sci U S A 116, 11113–11115. 10.1073/pnas.1903049116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson MX, Trojanowski JQ, and Lee VM (2019). alpha-Synuclein Pathology in Parkinson's Disease and Related alpha-Synucleinopathies. Neurosci Lett, 134316. 10.1016/j.neulet.2019.134316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki K, Yagi N, Aoyama K, Choong CJ, Hayakawa H, Fujimura H, Nagai Y, Goto Y, and Mochizuki H (2019). Parkinson's disease is a type of amyloidosis featuring accumulation of amyloid fibrils of alpha-synuclein. Proc Natl Acad Sci U S A 116, 17963–17969. 10.1073/pnas.1906124116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shahmoradian SH, Lewis AJ, Genoud C, Hench J, Moors TE, Navarro PP, Castano-Diez D, Schweighauser G, Graff-Meyer A, Goldie KN, et al. (2019). Lewy pathology in Parkinson's disease consists of crowded organelles and lipid membranes. Nat Neurosci 22, 1099–1109. 10.1038/s41593-019-0423-2. [DOI] [PubMed] [Google Scholar]
- 6.Chuang E, Hori AM, Hesketh CD, and Shorter J (2018). Amyloid assembly and disassembly. J Cell Sci 131. 10.1242/jcs.189928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shorter J (2017). Designer protein disaggregases to counter neurodegenerative disease. Curr Opin Genet Dev 44, 1–8. 10.1016/j.gde.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shorter J (2016). Engineering therapeutic protein disaggregases. Mol Biol Cell 27, 1556–1560. 10.1091/mbc.E15-10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shorter J (2008). Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals 16, 63–74. 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 10.March ZM, Mack KL, and Shorter J (2019). AAA+ Protein-Based Technologies to Counter Neurodegenerative Disease. Biophys J 116, 1380–1385. 10.1016/j.bpj.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fare CM, and Shorter J (2021). (Dis)Solving the problem of aberrant protein states. Dis Model Mech 14. 10.1242/dmm.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackrel ME, and Shorter J (2015). Engineering enhanced protein disaggregases for neurodegenerative disease. Prion 9, 90–109. 10.1080/19336896.2015.1020277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shorter J, and Southworth DR (2019). Spiraling in Control: Structures and Mechanisms of the Hsp104 Disaggregase. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a034033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, and Shorter J (2012). Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell 151, 778–793. 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glover JR, and Lindquist S (1998). Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82. [DOI] [PubMed] [Google Scholar]
- 16.Yoo H, Bard JAM, Pilipenko EV, and Drummond DA (2022). Chaperones directly and efficiently disperse stress-triggered biomolecular condensates. Mol Cell 82, 741–755 e711. 10.1016/j.molcel.2022.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gates SN, Yokom AL, Lin J, Jackrel ME, Rizo AN, Kendsersky NM, Buell CE, Sweeny EA, Mack KL, Chuang E, et al. (2017). Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokom AL, Gates SN, Jackrel ME, Mack KL, Su M, Shorter J, and Southworth DR (2016). Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat Struct Mol Biol 23, 830–837. 10.1038/nsmb.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye X, Lin J, Mayne L, Shorter J, and Englander SW (2019). Hydrogen exchange reveals Hsp104 architecture, structural dynamics, and energetics in physiological solution. Proc Natl Acad Sci U S A 116, 7333–7342. 10.1073/pnas.1816184116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michalska K, Zhang K, March ZM, Hatzos-Skintges C, Pintilie G, Bigelow L, Castellano LM, Miles LJ, Jackrel ME, Chuang E, et al. (2019). Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events. Structure 27, 449–463 e447. 10.1016/j.str.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lum R, Niggemann M, and Glover JR (2008). Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J Biol Chem 283, 30139–30150. 10.1074/jbc.M804849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tessarz P, Mogk A, and Bukau B (2008). Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol 68, 87–97. 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 23.Castellano LM, Bart SM, Holmes VM, Weissman D, and Shorter J (2015). Repurposing Hsp104 to Antagonize Seminal Amyloid and Counter HIV Infection. Chem Biol 22, 1074–1086. 10.1016/j.chembiol.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lum R, Tkach JM, Vierling E, and Glover JR (2004). Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279, 29139–29146. 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 25.Sweeny EA, Jackrel ME, Go MS, Sochor MA, Razzo BM, DeSantis ME, Gupta K, and Shorter J (2015). The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol Cell 57, 836–849. 10.1016/j.molcel.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye X, Lin J, Mayne L, Shorter J, and Englander SW (2020). Structural and kinetic basis for the regulation and potentiation of Hsp104 function. Proc Natl Acad Sci U S A 10.1073/pnas.1921968117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo Bianco C, Shorter J, Regulier E, Lashuel H, Iwatsubo T, Lindquist S, and Aebischer P (2008). Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest 118, 3087–3097. 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erives AJ, and Fassler JS (2015). Metabolic and chaperone gene loss marks the origin of animals: evidence for hsp104 and hsp78 chaperones sharing mitochondrial enzymes as clients. PLoS One 10, e0117192. 10.1371/journal.pone.0117192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shorter J (2011). The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 6, e26319. 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, and Morimoto RI (2000). Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A 97, 5750–5755. 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cushman-Nick M, Bonini NM, and Shorter J (2013). Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet 9, e1003781. 10.1371/journal.pgen.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vacher C, Garcia-Oroz L, and Rubinsztein DC (2005). Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum Mol Genet 14, 3425–3433. 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 33.Perrin V, Regulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, Luthi-Carter R, and Deglon N (2007). Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington's disease. Mol Ther 15, 903–911. 10.1038/mt.sj.6300141. [DOI] [PubMed] [Google Scholar]
- 34.Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, and Shorter J (2014). Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell 156, 170–182. 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackrel ME, and Shorter J (2014). Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis Model Mech 7, 1175–1184. 10.1242/dmm.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackrel ME, Yee K, Tariq A, Chen AI, and Shorter J (2015). Disparate Mutations Confer Therapeutic Gain of Hsp104 Function. ACS Chem Biol 10, 2672–2679. 10.1021/acschembio.5b00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryan JJ, Sprunger ML, Holthaus K, Shorter J, and Jackrel ME (2019). Engineered protein disaggregases mitigate toxicity of aberrant prion-like fusion proteins underlying sarcoma. J Biol Chem 294, 11286–11296. 10.1074/jbc.RA119.009494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tariq A, Lin J, Noll MM, Torrente MP, Mack KL, Murillo OH, Jackrel ME, and Shorter J (2018). Potentiating Hsp104 activity via phosphomimetic mutations in the middle domain. FEMS Yeast Res 18. 10.1093/femsyr/foy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torrente MP, Chuang E, Noll MM, Jackrel ME, Go MS, and Shorter J (2016). Mechanistic Insights into Hsp104 Potentiation. J Biol Chem 291, 5101–5115. 10.1074/jbc.M115.707976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tariq A, Lin J, Jackrel ME, Hesketh CD, Carman PJ, Mack KL, Weitzman R, Gambogi C, Hernandez Murillo OA, Sweeny EA, et al. (2019). Mining Disaggregase Sequence Space to Safely Counter TDP-43, FUS, and alpha-Synuclein Proteotoxicity. Cell Rep 28, 2080–2095 e2086. 10.1016/j.celrep.2019.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard MK, Miller KR, Sohn BS, Ryan JJ, Xu A, and Jackrel ME (2023). Probing the drivers of Staphylococcus aureus biofilm protein amyloidogenesis and disrupting biofilms with engineered protein disaggregases. mBio, e0058723. 10.1128/mbio.00587-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan JJ, Bao A, Bell B, Ling C, and Jackrel ME (2021). Drivers of Hsp104 potentiation revealed by scanning mutagenesis of the middle domain. Protein Sci 30, 1667–1685. 10.1002/pro.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda K, Clatterbuck-Soper SF, Jackrel ME, Shorter J, and Mili S (2017). FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination. J Cell Biol 216, 1015–1034. 10.1083/jcb.201608022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durie CL, Lin J, Scull NW, Mack KL, Jackrel ME, Sweeny EA, Castellano LM, Shorter J, and Lucius AL (2019). Hsp104 and Potentiated Variants Can Operate as Distinct Nonprocessive Translocases. Biophys J 116, 1856–1872. 10.1016/j.bpj.2019.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackrel ME, and Shorter J (2014). Reversing deleterious protein aggregation with re-engineered protein disaggregases. Cell Cycle 13, 1379–1383. 10.4161/cc.28709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mack KL, and Shorter J (2016). Engineering and Evolution of Molecular Chaperones and Protein Disaggregases with Enhanced Activity. Front Mol Biosci 3, 8. 10.3389/fmolb.2016.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sweeny EA, and Shorter J (2016). Mechanistic and Structural Insights into the Prion-Disaggregase Activity of Hsp104. J Mol Biol 428, 1870–1885. 10.1016/j.jmb.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson BS, McCaffery JM, Lindquist S, and Gitler AD (2008). A yeast TDP-43 proteinopathy model: Exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc Natl Acad Sci U S A 105, 6439–6444. 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Outeiro TF, and Lindquist S (2003). Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science 302, 1772–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, and Gitler AD (2011). Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 9, e1000614. 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, et al. (2017). Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371. 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jackson KL, Dayton RD, Orchard EA, Ju S, Ringe D, Petsko GA, Maquat LE, and Klein RL (2015). Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther 22, 20–28. 10.1038/gt.2014.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, et al. (2013). Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science 342, 983–987. 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, et al. (2013). Yeast reveal a "druggable" Rsp5/Nedd4 network that ameliorates alpha-synuclein toxicity in neurons. Science 342, 979–983. 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, et al. (2006). Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson's models. Science 313, 324–328. 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khurana V, Peng J, Chung CY, Auluck PK, Fanning S, Tardiff DF, Bartels T, Koeva M, Eichhorn SW, Benyamini H, et al. (2017). Genome-Scale Networks Link Neurodegenerative Disease Genes to alpha-Synuclein through Specific Molecular Pathways. Cell Syst 4, 157–170 e114. 10.1016/j.cels.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ovchinnikov S, Kamisetty H, and Baker D (2014). Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife 3, e02030. 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heuck A, Schitter-Sollner S, Suskiewicz MJ, Kurzbauer R, Kley J, Schleiffer A, Rombaut P, Herzog F, and Clausen T (2016). Structural basis for the disaggregase activity and regulation of Hsp104. Elife 5. 10.7554/eLife.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lipinska N, Zietkiewicz S, Sobczak A, Jurczyk A, Potocki W, Morawiec E, Wawrzycka A, Gumowski K, Slusarz M, Rodziewicz-Motowidlo S, et al. (2013). Disruption of ionic interactions between the nucleotide binding domain 1 (NBD1) and middle (M) domain in Hsp100 disaggregase unleashes toxic hyperactivity and partial independence from Hsp70. J Biol Chem 288, 2857–2869. 10.1074/jbc.M112.387589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franzmann TM, Czekalla A, and Walter SG (2011). Regulatory circuits of the AAA+ disaggregase Hsp104. J Biol Chem 286, 17992–18001. 10.1074/jbc.M110.216176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parsell DA, Sanchez Y, Stitzel JD, and Lindquist S (1991). Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature 353, 270–273. [DOI] [PubMed] [Google Scholar]
- 63.Schaupp A, Marcinowski M, Grimminger V, Bosl B, and Walter S (2007). Processing of proteins by the molecular chaperone Hsp104. J Mol Biol 370, 674–686. 10.1016/j.jmb.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 64.Schirmer EC, Queitsch C, Kowal AS, Parsell DA, and Lindquist S (1998). The ATPase activity of Hsp104, effects of environmental conditions and mutations. J Biol Chem 273, 15546–15552. 10.1074/jbc.273.25.15546. [DOI] [PubMed] [Google Scholar]
- 65.March ZM, Sweeney K, Kim H, Yan X, Castellano LM, Jackrel ME, Lin J, Chuang E, Gomes E, Willicott CW, et al. (2020). Therapeutic genetic variation revealed in diverse Hsp104 homologs. Elife 9. 10.7554/eLife.57457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. (1997). Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science 276, 2045–2047. 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 67.Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, et al. (2004). The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 55, 164–173. 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 68.Sorrentino ZA, Vijayaraghavan N, Gorion KM, Riffe CJ, Strang KH, Caldwell J, and Giasson BI (2018). Physiological C-terminal truncation of alpha-synuclein potentiates the prion-like formation of pathological inclusions. J Biol Chem 293, 18914–18932. 10.1074/jbc.RA118.005603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE, Bosco DA, Hayward LJ, Brown RHJ, Lindquist S, et al. (2011). A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol 9. 10.1371/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. (2011). A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A 108, 20881–20890. 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, and Gitler AD (2009). TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem 284, 20329–20339. 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Figley MD, and Gitler AD (2013). Yeast genetic screen reveals novel therapeutic strategy for ALS. Rare Dis 1, e24420. 10.4161/rdis.24420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, et al. (2008). The Parkinson's disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci U S A 105, 145–150. 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soper JH, Roy S, Stieber A, Lee E, Wilson RB, Trojanowski JQ, Burd CG, and Lee VM (2008). Alpha-synuclein-induced aggregation of cytoplasmic vesicles in Saccharomyces cerevisiae. Mol Biol Cell 19, 1093–1103. 10.1091/mbc.E07-08-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zabrocki P, Pellens K, Vanhelmont T, Vandebroek T, Griffioen G, Wera S, Van Leuven F, and Winderickx J (2005). Characterization of alpha-synuclein aggregation and synergistic toxicity with protein tau in yeast. FEBS J 272, 1386–1400. 10.1111/j.1742-4658.2005.04571.x. [DOI] [PubMed] [Google Scholar]
- 76.Tenreiro S, Reimao-Pinto MM, Antas P, Rino J, Wawrzycka D, Macedo D, Rosado-Ramos R, Amen T, Waiss M, Magalhaes F, et al. (2014). Phosphorylation modulates clearance of alpha-synuclein inclusions in a yeast model of Parkinson's disease. PLoS Genet 10, e1004302. 10.1371/journal.pgen.1004302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franssens V, Boelen E, Anandhakumar J, Vanhelmont T, Buttner S, and Winderickx J (2010). Yeast unfolds the road map toward alpha-synuclein-induced cell death. Cell Death Differ 17, 746–753. 10.1038/cdd.2009.203. [DOI] [PubMed] [Google Scholar]
- 78.Weber-Ban EU, Reid BG, Miranker AD, and Horwich AL (1999). Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401, 90–93. 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 79.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, and Glabe CG (2003). Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489. 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 80.Sanchez Y, and Lindquist SL (1990). HSP104 required for induced thermotolerance. Science 248, 1112–1115. 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 81.Lindquist S, and Kim G (1996). Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci U S A 93, 5301–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanchez Y, and Taulien JB, Lindquist KA, S. (1992). Hsp104 is required for tolerance to many forms of stress. EMBO J 11, 2357–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parsell DA, Kowal AS, Singer MA, and Lindquist S (1994). Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372, 475–478. [DOI] [PubMed] [Google Scholar]
- 84.Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. (2015). Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell 162, 1286–1298. 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao S, Gelwix CC, Caldwell KA, and Caldwell GA (2005). Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J Neurosci 25, 3801–3812. 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harrington AJ, Hamamichi S, Caldwell GA, and Caldwell KA (2010). C. elegans as a model organism to investigate molecular pathways involved with Parkinson's disease. Dev Dyn 239, 1282–1295. 10.1002/dvdy.22231. [DOI] [PubMed] [Google Scholar]
- 87.Winner B, Jappelli R, Maji SK, Desplats PA, Boyer L, Aigner S, Hetzer C, Loher T, Vilar M, Campioni S, et al. (2011). In vivo demonstration that alpha-synuclein oligomers are toxic. Proc Natl Acad Sci U S A 108, 4194–4199. 10.1073/pnas.1100976108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Diógenes MJ, Dias RB, Rombo DM, Vicente Miranda H, Maiolino F, Guerreiro P, Näsström T, Franquelim HG, Oliveira LM, Castanho MA, et al. (2012). Extracellular alpha-synuclein oligomers modulate synaptic transmission and impair LTP via NMDA-receptor activation. J Neurosci 32, 11750–11762. 10.1523/jneurosci.0234-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mahul-Mellier AL, Burtscher J, Maharjan N, Weerens L, Croisier M, Kuttler F, Leleu M, Knott GW, and Lashuel HA (2020). The process of Lewy body formation, rather than simply alpha-synuclein fibrillization, is one of the major drivers of neurodegeneration. Proc Natl Acad Sci U S A 117, 4971–4982. 10.1073/pnas.1913904117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gaeta AL, Caldwell KA, and Caldwell GA (2019). Found in Translation: The Utility of C. elegans Alpha-Synuclein Models of Parkinson's Disease. Brain Sci 9. 10.3390/brainsci9040073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mor DE, Tsika E, Mazzulli JR, Gould NS, Kim H, Daniels MJ, Doshi S, Gupta P, Grossman JL, Tan VX, et al. (2017). Dopamine induces soluble alpha-synuclein oligomers and nigrostriatal degeneration. Nat Neurosci 20, 1560–1568. 10.1038/nn.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Conway KA, Rochet JC, Bieganski RM, and Lansbury PT Jr. (2001). Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science 294, 1346–1349. 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 93.Guo L, Fare CM, and Shorter J (2019). Therapeutic Dissolution of Aberrant Phases by Nuclear-Import Receptors. Trends Cell Biol 29, 308–322. 10.1016/j.tcb.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O'Donovan K, Fare CM, Diaz Z, Singh N, et al. (2018). Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 173, 677–692 e620. 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cupo RR, and Shorter J (2020). Skd3 (human ClpB) is a potent mitochondrial protein disaggregase that is inactivated by 3-methylglutaconic aciduria-linked mutations. Elife 9. 10.7554/eLife.55279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang L, Agrawal T, Zhu G, Yu S, Tao L, Lin J, Marmorstein R, Shorter J, and Yang X (2021). DAXX represents a new type of protein-folding enabler. Nature 597, 132–137. 10.1038/s41586-021-03824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu G, Harischandra DS, Ghaisas S, Zhang P, Prall W, Huang L, Maghames C, Guo L, Luna E, Mack KL, et al. (2020). TRIM11 Prevents and Reverses Protein Aggregation and Rescues a Mouse Model of Parkinson's Disease. Cell Rep 33, 108418. 10.1016/j.celrep.2020.108418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cupo RR, and Shorter J (2020). Expression and Purification of Recombinant Skd3 (Human ClpB) Protein and Tobacco Etch Virus (TEV) Protease from Escherichia coli. Bio Protoc 10, e3858. 10.21769/BioProtoc.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lopez KE, Rizo AN, Tse E, Lin J, Scull NW, Thwin AC, Lucius AL, Shorter J, and Southworth DR (2020). Conformational plasticity of the ClpAP AAA+ protease couples protein unfolding and proteolysis. Nat Struct Mol Biol 27, 406–416. 10.1038/s41594-020-0409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Alberti S, Gitler AD, and Lindquist S (2007). A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919. 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jackrel ME, Tariq A, Yee K, Weitzman R, and Shorter J (2014). Isolating potentiated Hsp104 variants using yeast proteinopathy models. J Vis Exp, e52089. 10.3791/52089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berkowitz LA, Knight AL, Caldwell GA, and Caldwell KA (2008). Generation of stable transgenic C. elegans using microinjection. J Vis Exp 10.3791/833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gietz RD, and Schiestl RH (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2, 31–34. 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 106.Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, and Caldwell GA (2008). Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc Natl Acad Sci U S A 105, 728–733. 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim H, Perentis RJ, Caldwell GA, and Caldwell KA (2018). Gene-by-environment interactions that disrupt mitochondrial homeostasis cause neurodegeneration in C. elegans Parkinson's models. Cell Death Dis 9, 555. 10.1038/s41419-018-0619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, and Sigler PB (1994). The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 371, 578–586. 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 109.Kumar ST, Donzelli S, Chiki A, Syed MMK, and Lashuel HA (2020). A simple, versatile and robust centrifugation-based filtration protocol for the isolation and quantification of α-synuclein monomers, oligomers and fibrils: Towards improving experimental reproducibility in α-synuclein research. J Neurochem 153, 103–119. 10.1111/jnc.14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw images of yeast spotting, western blots, and C. elegans experiments are available in Mendeley Data. All data are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
All custom code has been deposited to GitHub and Zenodo. DOI are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Hsp104 polyclonal | Enzo Life Sciences | Cat#ADI-SPA-1040-F; RRID:AB_2039208 |
| Rabbit anti-FUS polyclonal | Bethyl Laboratories | Cat# A300-302A; RRID:AB_309445 |
| Rabbit anti-TDP-43 polyclonal | Proteintech | Cat#10782; RRID: AB_615042 |
| Rabbit anti-GFP polyclonal | Sigma-Aldrich | Cat# G1544; RRID: AB_439690 |
| Mouse anti-α-Synuclein monoclonal (Syn303) | Kelvin Luk (University of Pennsylvania) | RRID:AB_2315395 |
| Mouse anti-PGK1 monoclonal | Thermo Fisher | Cat#459250; RRID:AB_2532235 |
| IRDye 800CW Goat anti-Mouse IgG secondary antibody | LI-COR | Cat# 926–32210; RRID:AB_621842 |
| IRDye 680RD Goat anti-Rabbit IgG secondary antibody | LI-COR | Cat# 926–68071; RRID:AB_10956166 |
| Bacterial and virus strains | ||
| Escherichia coli DH5α competent cells | Thermo Fisher | Cat#18265017 |
| Escherichia coli BL21-CodonPlus (DE3)-RIL competent cells | Agilent | Cat#230245 |
| Chemicals, peptides, and recombinant proteins | ||
| Creatine phosphate | Roche | Cat#10621722001 |
| Adenosine 5’-triphosphate disodium salt hydrate (ATP) | Sigma-Aldrich | Cat#A3377 |
| cOmplete, Mini, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#4693159001 |
| Casein fluorescein isothiocyanate from bovine milk (FITC-Casein) | Sigma-Aldrich | Cat#C0528 |
| Creatine kinase | Roche | Cat#10127566001 |
| Firefly luciferase | Sigma-Aldrich | Cat#L9506 |
| Lysozyme | Sigma-Aldrich | Cat#L6876 |
| His-TEV protease | Cupo and Shorter98 | N/A |
| Hsp104 | Jackrel et al.34 | N/A |
| Hsp104A503S | Jackrel et al.34 | N/A |
| Hsp104K358D | This paper | N/A |
| Hsp104K358D:Y257L | This paper | N/A |
| Hsp104K358D:Y257T | This paper | N/A |
| Hsp104K358D:Y662M | This paper | N/A |
| Hsp104K358D:Y257L:Y662M | This paper | N/A |
| HAP | Jackrel et al.34 | N/A |
| HAPA503S | This paper | N/A |
| HAPK358D | This paper | N/A |
| HAPK358D:Y257L | This paper | N/A |
| HAPK358D:Y257T | This paper | N/A |
| HAPK358D:Y662M | This paper | N/A |
| HAPK358D:Y257L:Y662M | This paper | N/A |
| GroELtrap | Jackrel et al.34 | N/A |
| RepA1-25-GFP | Lopez et al.99 | N/A |
| ClpP | Jackrel et al.34 | N/A |
| Hsc70 | Enzo Life Sciences | Cat#ADI-SPP-751-F |
| Hdj2 | Enzo Life Sciences | Cat#ADI-SPP-405-F |
| Hsp72 | Michalska et al.20 | N/A |
| Hdj1 | Michalska et al.20 | N/A |
| α-Syn | Jackrel et al.34 | N/A |
| TDP-43 | Jackrel et al.34 | N/A |
| Critical commercial assays | ||
| ATPase Activity Kit (Colorimetric) | Innova Biosciences | Cat#601–0120 |
| Luciferase Assay Reagent | Promega | Cat#E1483 |
| Human α-Synuclein ELISA Kit | Thermo Fisher | Cat#KHB0061 |
| QuikChange Site-Directed Mutagenesis Kit | Agilent | Cat# 200518 |
| Deposited data | ||
| Raw images of yeast spotting, western blots, and C. elegans experiments | Mendeley data | doi: 10.17632/t7k9w8txzk.1 |
| Experimental models: Organisms/strains | ||
| S. cerevisiae W303a (MATa, can1-100, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, ade2-1) | Jackrel et al.34 | N/A |
| S. cerevisiae W303a∆hsp104 (MATa, can1-100, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, ade2-1, hsp104::KanMX) | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-α-syn-YFP-pAG304GAL-α-syn-YFP | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-FUS | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-TDP-43 | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-TDP-43-GFPS11-pAG305GAL-GFPS1-10 | Jackrel et al.34 | N/A |
| S. cerevisiae W303aΔhsp104-pAG303GAL-FUS-GFP | Jackrel et al.34 | N/A |
| C. elegans UA44 (baln11 [Pdat-1::α-syn, Pdat-1: :GFP]) | Cao et al.85 | N/A |
| C. elegans UA367 (baEx198 a,b,c [Pdat-1::HSP104-K358D, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA368 (baEx199 a,b,c [Pdat-1::HSP104-K358D:Y257L, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA369 (baEx200 a,b,c [Pdat-1::HSP104-K358D:Y257T, rol-6];baln11 [Pdat-1::α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA370 (baEx201 a,b,c [Pdat-1::HSP104-K358D:Y257L:Y662M, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1::GFP]) | This study | N/A |
| C. elegans UA371 (baEx202 a,b,c [Pdat-1::HSP104-K358D: Y662M, rol-6];baln11 [Pdat-1:: α-syn, Pdat-1 ::GFP]) | This study | N/A |
| C. elegans UA256 (baEx147 a,b,c [Pdat-1::Hsp104 WT, Pmyo-2::mCherry]; baIn11 [Pdat-1:: α-syn, Pdat-1::GFP]) | Jackrel et al.34 | N/A |
| C. elegans UA259 (baEx150 a,b,c [Pdat-1::Hsp104-A503S, Pmyo-2::mCherry]; baIn11 [Pdat-1::α-syn, Pdat-1::GFP]) | Jackrel et al.34 | N/A |
| C. elegans UA262 (baEx153 a,b,c [Pdat-1::Hsp104-DPLF:A503V, Pmyo-2::mCherry]; baIn11[Pdat-1::α-syn, Pdat-1::GFP]) | Jackrel et al.34 | N/A |
| Oligonucleotides | ||
| See Table S1 | ||
| Recombinant DNA | ||
| pRS416GAL | Jackrel et al.34 | N/A |
| pRS416GAL-Hsp104 | Jackrel et al.34 | N/A |
| pRS416GAL-Hsp104A503S | Jackrel et al.34 | N/A |
| pRS416GAL-Hsp104A503S:Y257L | This study | N/A |
| pRS416GAL-Hsp104A503S:Y257T | This study | N/A |
| pRS416GAL-Hsp104A503S:Y662M | This study | N/A |
| pRS416GAL-Hsp104A503S:Y257L:Y662M | This study | N/A |
| pRS416GAL-Hsp104K358D | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257F | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257W | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257I | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257V | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257M | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662F | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662W | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662I | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662V | This study | N/A |
| pRS416GAL-Hsp104K358D:Y662M | This study | N/A |
| pRS416GAL-Hsp104D484K | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257L | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257F | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257W | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257I | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257V | This study | N/A |
| pRS416GAL-Hsp104D484K:Y257M | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662L | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662F | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662W | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662I | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662V | This study | N/A |
| pRS416GAL-Hsp104D484K:Y662M | This study | N/A |
| pRS416GAL-Hsp104Y257L | This study | N/A |
| pRS416GAL-Hsp104Y662L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662L | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662F | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662W | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662M | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662S | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662T | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662Q | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L:Y662N | This study | N/A |
| pRS416GAL-Hsp104K218T:K620T | Torrente et al.39 | N/A |
| pRS416GAL-Hsp104K218T:A503S:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:K358D:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:Y257L:K358D:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:Y257T:K358D:K620T | This study | N/A |
| pRS416GAL-Hsp104K218T:K358D:K620T:Y662M | This study | N/A |
| pRS416GAL-Hsp104K218T:Y257L:K358D:K620T:Y662M | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257S | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257T | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257Q | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257N | This study | N/A |
| pRS416GAL-mCherry | This study | N/A |
| pRS416GAL-Hsp104-mCherry | This study | N/A |
| pRS416GAL-Hsp104K358D-mCherry | This study | N/A |
| pRS416GAL-Hsp104A503S-mCherry | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257T-mCherry | This study | N/A |
| pRS416GAL-Hsp104K358D:Y257L-mCherry | This study | N/A |
| pRS416HSE-Hsp104 | Jackrel et al.34 | N/A |
| pRS416HSE-Hsp104A503V | Jackrel et al.34 | N/A |
| pRS416HSE-Hsp104K358D | This study | N/A |
| pRS416HSE-Hsp104K358D:Y257L | This study | N/A |
| pRS416HSE-Hsp104K358D:Y257T | This study | N/A |
| pRS416HSE-Hsp104K358D:Y662M | This study | N/A |
| pRS416HSE-Hsp104K358D:Y257L:Y622M | This study | N/A |
| pAG413GAL | Alberti et al.100 | N/A |
| pAG413GAL-α-syn | This study | N/A |
| pAG413GAL-α-synE46K | This study | N/A |
| pAG413GAL-α-synA53T | This study | N/A |
| pAG413GAL-α-syn1-95 | This study | N/A |
| pAG413GAL-α-syn1-115 | This study | N/A |
| pAG413GAL-α-syn1-125 | This study | N/A |
| pAG415GAL | Alberti et al.100 | N/A |
| pAG415GAL-α-syn | This study | N/A |
| pAG415GAL-α-synE46K | This study | N/A |
| pAG415GAL-α-synA53T | This study | N/A |
| pAG415GAL-α-syn1-95 | This study | N/A |
| pAG415GAL-α-syn1-115 | This study | N/A |
| pAG415GAL-α-syn1-125 | This study | N/A |
| pNOTAG-Hsp104 | Jackrel et al.34 | N/A |
| pNOTAG-Hsp104A503S | Jackrel et al.34 | N/A |
| pNOTAG-Hsp104K358D | This study | N/A |
| pNOTAG-Hsp104K358D:Y257L | This study | N/A |
| pNOTAG-Hsp104K358D:Y257T | This study | N/A |
| pNOTAG-Hsp104K358D:Y662M | This study | N/A |
| pNOTAG-Hsp104K358D:Y257L:Y662M | This study | N/A |
| pNOTAG-HAP | Jackrel et al.34 | N/A |
| pNOTAG-HAPA503S | This study | N/A |
| pNOTAG-HAPK358D | This study | N/A |
| pNOTAG-HAPK358D:Y257L | This study | N/A |
| pNOTAG-HAPK358D:Y257T | This study | N/A |
| pNOTAG-HAPK358D:Y662M | This study | N/A |
| pNOTAG-HAPK358D:Y257L:Y662M | This study | N/A |
| pTrc99A-GroELtrap | Jackrel et al.34 | N/A |
| pBAD-RepA1-25-GFP | Lopez et al.99 | N/A |
| pET28b-ClpP-his | Jackrel et al.34 | N/A |
| pE-SUMO-Hsp72 | This study | N/A |
| pE-SUMO-Hdj1 | Michalska et al.20 | N/A |
| pHis-TEV | Cupo and Shorter98 | N/A |
| pT7-7 α-syn | Lo Bianco et al.27 | N/A |
| pDUET-GST-TEV-TDP-43 | Johnson et al.71 | N/A |
| Software and algorithms | ||
| Prism 9 | GraphPad | N/A |
| ImageJ | Rueden et al.101 | N/A |
| Other | ||
| Custom code for a semi-automated procedure was used to quantify colocalization between the red and green images. | Zenodo | doi: 10.5281/zenodo.8185164 |


