SUMMARY
Cryptosporidiosis is a leading cause of life-threatening diarrhea in young children in resource-poor settings. To explore microbial influences on susceptibility, we screened 85 microbiota-associated metabolites for their effects on Cryptosporidium parvum growth in vitro. We identify eight inhibitory metabolites in three main classes: secondary bile salts/acids, a vitamin B6 precursor, and indoles. Growth restriction of C. parvum by indoles does not depend on the host aryl hydrocarbon receptor (AhR) pathway. Instead, treatment impairs host mitochondrial function and reduces total cellular ATP, as well as directly reducing the membrane potential in the parasite mitosome, a degenerate mitochondria. Oral administration of indoles, or reconstitution of the gut microbiota with indole-producing bacteria, delays life cycle progression of the parasite in vitro and reduces the severity of C. parvum infection in mice. Collectively, these findings indicate that microbiota metabolites impair mitochondrial function and contribute to colonization resistance to Cryptosporidium infection.
In brief
Funkhouser-Jones et al. show that metabolites produced by the microbiota inhibit growth of Cryptosporidium.Bacterially produced indoles impair energy production by host mitochondria and disrupt the membrane potential of the mitosome, a remnant parasite mitochondrion. These findings demonstrate the basis for resistance against enteric parasite infection provided by the microbiota.
Graphical Abstract
INTRODUCTION
Cryptosporidiosis, primarily caused by Cryptosporidium hominis or C. parvum (Cp) in humans, manifests as self-limiting diarrhea in healthy individuals and severe, chronic diarrhea in immunocompromised patients, particularly those with HIV/AIDS.1 Despite the parasite’s worldwide distribution,2 the true global burden of cryptosporidiosis was underestimated until the seminal Global Enteric Multicenter Study (GEMS) unexpectedly revealed that Cryptosporidium is second only to rotavirus as the cause of moderate to severe diarrhea in infants and toddlers (0–24 months) in resource-poor countries.3–5 Importantly, cryptosporidiosis in young children also increases their risk for severe malnutrition, long-term growth stunting, and death.3,6,7 However, for reasons not well understood, the incidence of cryptosporidiosis drops dramatically in children older than two years of age in these same populations.3,4 This inverse correlation between age and susceptibility to cryptosporidiosis is not specific to humans: other zoonotic hosts, including dairy cows,8 broiler chickens,9,10 and mice,11,12 are highly susceptible to Cryptosporidium as neonates but relatively resistant if infected as juveniles or adults. Although immune system maturation is an important factor in the development of resistance to Cryptosporidium infection,13,14 dramatic changes in the composition and diversity of the gut microbiome during early development also play a pivotal role in the incidence and severity of infection.11,15,16 For example, one prospective study of infants in Bangladesh found that the microbiota of infants that developed clinical cryptosporidiosis were less diverse one month prior to the onset of diarrheal symptoms than the microbiota of infected but asymptomatic infants.16 Additionally, studies in adult mice have consistently shown that loss of the microbiota either through antibiotic (Abx) treatment or gnotobiotic rearing sharply increases susceptibility of animals to Cryptosporidium infection.15,17
Although a diverse microbiota appears to protect against cryptosporidiosis, little is known about how the microbiota influences Cryptosporidium infection on a mechanistic level. One possibility is that the microbiota primes the immune system to respond to enteric pathogens. Indeed, one study in neonatal mice observed that stimulation of the intestinal immune response with poly(I:C) reduced Cp load but only when the gut microflora was present.18 A second possibility is that metabolites in the diet or produced by the microbiota, rather than the bacteria themselves, could affect Cryptosporidium growth. Precedent for this idea comes from a human challenge study that found high fecal indole levels (>2.5 mM), or indole-producing bacteria, were protective against the development of diarrhea in volunteers challenged with C. hominis or multiple strains of Cp.19 Additionally, prior studies have also shown that dietary supplementation with arginine can decrease severity of Cp infection in malnourished neonatal mice by stimulating the production of nitric oxide.20
We previously postulated that increased susceptibility to Cryptosporidium in neonates is partly explained by the presence of early-life gut metabolites that enhance parasite growth.11 Metabolites are known to undergo profound changes during development, concordant with alteration in the microbiota.11,21 Accordingly, we focused on the potential role gut metabolites that are common in adults may play in the development of age-dependent resistance to the parasite. We identified several classes of bacterial metabolites that inhibit parasite growth including indoles that act by impairing host mitochondrial function, leading to a reduction in total ATP levels in the host cell and reduced parasite growth in vitro and in vivo. Thus, high indole levels in the gut may decrease Cryptosporidium burden by depriving the parasite of essential host-derived nutrients or by inhibiting key parasite functions.
RESULTS
Microbial metabolite screen identifies inhibitors of Cp growth in vitro
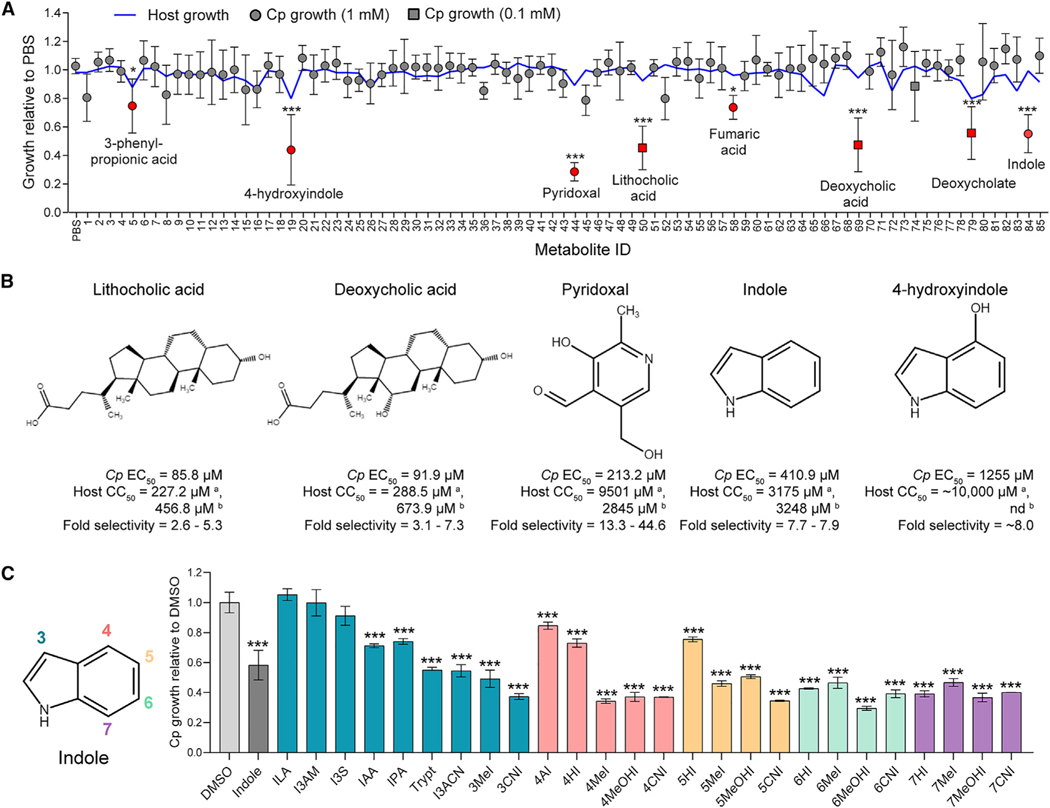
We screened a collection of 85 metabolites (Table S1) that are associated with the adult murine gut microbiota22 for their effects on Cp infection in a human ileocecal adenocarcinoma (HCT-8) cell line (Figure 1A). Cp oocysts were added simultaneously with individual metabolites to HCT-8 cell monolayers, and the numbers of parasites and host cells in each well were quantified 24 h post-infection (hpi) using an automated image-based assay.11 In total, we identified 8 metabolites that significantly inhibited Cp growth and none that enhanced infection (Figure 1A; Table S1). These inhibitory metabolites included indole and its derivative 4-hydroxyindole (4HI), three secondary bile acids or salts (deoxycholate, deoxycholic acid, and lithocholic acid), and pyridoxal hydrochloride, a precursor of the metabolically active form of vitamin B6, pyridoxal-5-phosphate. Dose-response curves indicated that secondary bile acids were the most potent against Cp (half maximal effective concentration [EC50] < 100 μM) but had low selectivity (2.5- to 3-fold) for the parasite because of host toxicity (Figure 1B). Pyridoxal hydrochloride was less potent against Cp (EC50 = 213.2 μM) than the secondary bile acids but had the highest fold selectivity (44.6-fold) for the parasite (Figure 1B). Indole and 4HI were the least potent (EC50 = 410.9 and 1,255 μM, respectively) but exhibited less host cell toxicity than secondary bile acids (Figure 1B). Indole is a well-known microbial metabolite that exhibits antimicrobial activity (e.g., Escherichia coli, Pseudomonas aeruginosa, Clostridium difficile).23–26 Moreover, fecal indole levels correlate with resistance to C. hominis or Cp in human challenge studies.19 Thus, we focused on defining the potential mechanism of action of indoles against Cp.
Figure 1. Gut metabolites, specifically secondary bile acids and indoles, inhibit C. parvum (Cp) infection in vitro.
(A) Effects of 85 intestinal metabolites at 1 mM (circles) or 0.1 mM (squares) on Cp infection in HCT-8 cells 24 hpi. Data plotted represent mean ± SD of Cp or mean host cell numbers (blue line) relative to PBS control for six independent experiments. Differences between Cp numbers for each metabolite and the PBS control were analyzed using a one-way ANOVA followed by Dunnett’s test for multiple comparisons. Metabolites that significantly inhibited Cp growth are indicated in red. *p < 0.05 and ***p < 0.001.
(B) Chemical structures of five inhibitory metabolites with their respective EC50 values for Cp and host cells and fold selectivity (host half maximal cytotoxic concentration [CC50] divided by Cp EC50). aHost CC50 values were calculated by counting Hoechst stained nuclei. CC50 values were calculated using a nonlinear regression curve fit with six replicates (three technical replicates from two independent experiments) per concentration. bHost CC50 values were calculated by Cell Titer-Glo Assay. nd, not done because of incompatibility with the assay. CC50 values were calculated using a nonlinear regression curve fit with nine replicates (three technical replicates from three independent experiments) per concentration.
(C) Screen of indole analogs (1 mM) modified at the 3-carbon (teal), 4-carbon (pink), 5-carbon (orange), 6-carbon (green), or 7-carbon (purple) positions and their effects on Cp infection in HCT-8 cells. Data plotted represent mean ± SD of six replicates (three technical replicates from two independent experiments). Differences between mean Cp numbers for each metabolite and the DMSO control were analyzed using a one-way ANOVA followed by Dunnett’s test for multiple comparisons. ***p < 0.001. ILA, indole-3-lactic acid; I3AM, indole-3-acetamide; I3S, indoxyl-3-sulfate; IAA, indole-3-acetic acid; IPA, indole-3-propionic acid; Trypt, tryptamine; I3ACN, indole-3-acetonitrile; MeI, methylindole; CNI, cyanoindole; AI, aminoindole; HI, hydroxyindole; MeOHI, methoxyindole.
Indole analogs modified at different carbon positions significantly inhibit Cp infection
Although both indole and 4HI significantly inhibited Cp growth, indole was 3X more potent than 4HI, while another indole analog included in the primary screen, 5-hydroxyindole-3-acetic acid, had no effect on Cp (Table S1). These results suggest that modifications at different carbon positions along the pyrrole or benzene rings of indole (Figure 1C) may change the potency of indole analogs. To examine the effect of modification position on indole efficacy against Cp, we tested a panel of 25 additional indole analogs modified at the 3-, 4-, 5-, 6-, and 7-carbon positions of indole using the in vitro growth assay (Figure 1C). Interestingly, nearly all the indole analogs (22 of 25) significantly inhibited Cp infection (Figure 1C; Table S2). The three exceptions were all modified at the 3-carbon position: indole-3-lactic acid (ILA), indole-3-acetamide (I3AM), and indoxyl-3-sulfate (I3S). However, other indoles modified at the 3-carbon position, such as indole-3-propionic acid (IPA) and 3-methylindole (also called skatole), did inhibit Cp, indicating that the side chain composition is more important than its position on the indole molecule. Indeed, certain side chains were more potent than others, particularly methyl, methoxy, and cyano groups. Of the 22 indoles that inhibited Cp, 11 were more potent than indole, and we chose 7-cyanoindole (7CNI) as the best analog for further study on the basis of its combined potency and selectivity.
Indole inhibition of Cp does not depend on the aryl hydrocarbon receptor pathway
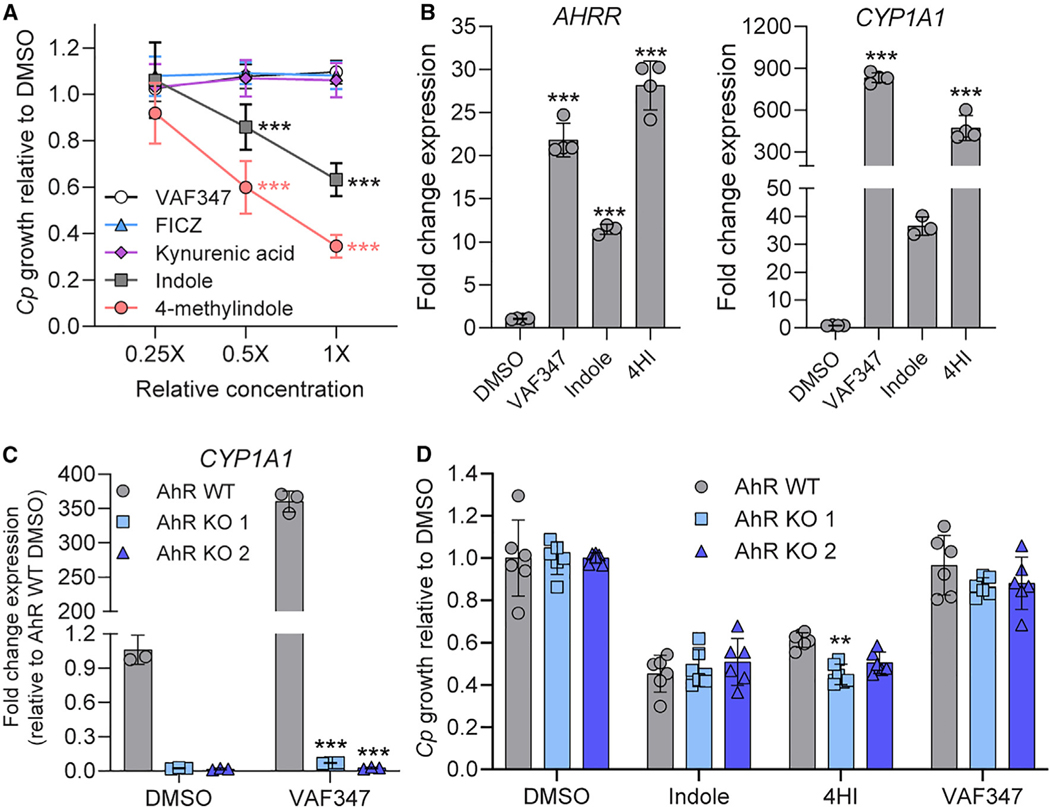
Many metabolites produced from microbial or host tryptophan metabolism including indole and its derivatives serve as endogenous ligands for the aryl hydrocarbon receptor (AhR) in animals.27 Furthermore, synthetic indoles such as 4-methylindole, 6-methylindole, and 7-methoxyindole have been shown to activate AhR signaling in vitro.28 To determine whether activation of the AhR pathway by indole analogs is sufficient to inhibit parasite growth, we treated Cp-infected cultures with agonists of AhR including tryptophan metabolites kynurenic acid29 and 6-formylindolo(3,2-b)carbazole FICZ30,31 as well as a highly specific synthetic AhR agonist, VAF347.32 Although indole and 4MeI significantly inhibited Cp in a dose-dependent manner (Figure 2A), none of the non-indole AhR agonists affected Cp growth (Figure 2A), even though the expression of AhR target genes AhRR and CYP1A1 were significantly upregulated in HCT-8 cells after 24 h treatment with AhR agonists (Figure 2B). To confirm that the AhR pathway is not involved in indole inhibition of Cp, we knocked out the AhR gene in HCT-8 cells using CRISPR-Cas9 to disrupt the first exon of the gene. We isolated two clonal lines, one with a 1 bp insertion (AhR knockout [KO] 1) and the other with an 11 bp deletion (AhR KO 2) (Figure S1). Both mutations are predicted to cause a frameshift with premature stop codons that result in truncated proteins of 29 and 25 amino acids, respectively (the full-length protein is 848 aa; Figure S1). Importantly, we confirmed that both AhR KO lines lost the ability to upregulate CYP1A1 gene expression upon VAF347 treatment (Figure 2C). However, Cp remained sensitive to indole and 4HI inhibition in AhR KO cells (Figure 2D), indicating that indoles do not act through the host AhR pathway to inhibit Cp.
Figure 2. Indoles do not inhibit C. parvum (Cp) through the host AhR pathway.
(A) Ratio of Cp relative to DMSO control in HCT-8 cells after 24 h treatment with serial dilutions of AhR agonists. Starting concentrations (1×) were 1 μM for VAF347 and FICZ and 1 mM for kynurenic acid, indole, and 4-methylindole. Data plotted represent mean ± SD of nine replicates (three technical replicates from three independent experiments). Differences between mean Cp ratio in 1×- or 0.5×-treated cultures versus 0.25×-treated cultures for each compound were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. ***p < 0.001.
(B) Fold change in gene expression of human AHRR or CYP1A1 genes (normalized to GAPDH) in uninfected HCT-8 cultures after 24 h treatment with VAF347 (250 nM), indole (1.5 mM), or 4-hydroxyindole (4HI; 2.5 mM) relative to 1% DMSO control. Data plotted represent mean ± SD of 3 or 4 technical replicates from a single experiment. For each gene, differences between fold change in gene expression for each treatment versus DMSO control were analyzed using a one-way ANOVA followed by Dunnett’s test for multiple comparisons. ***p < 0.001.
(C) Fold change in gene expression of human CYP1A1 genes (normalized to GAPDH) in uninfected HCT-8 AhR WT (gray) or KO cell lines (blue) after 24 h treatment with VAF347 (500 nM) relative to 1% DMSO control. Data plotted represent mean ± SD of three technical replicates from a single experiment. Differences between fold change in gene expression for each AhR KO versus AhR WT cell line for each treatment were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. ***p < 0.001.
(D) Ratio of Cp relative to DMSO control in HCT-8 AhR WT (gray) or KO cell lines (blue) after 24 h treatment with 0.5% DMSO, indole (1 mM), 4HI (1 mM), or VAF347 (500 nM). Data plotted represent mean ± SD of six replicates (three technical replicates from two independent experiments). Differences between Cp ratio in each AhR KO versus AhR WT cell line for each treatment were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. **p < 0.01.
See also Figure S1.
Indoles delay intracellular life cycle progression of Cp
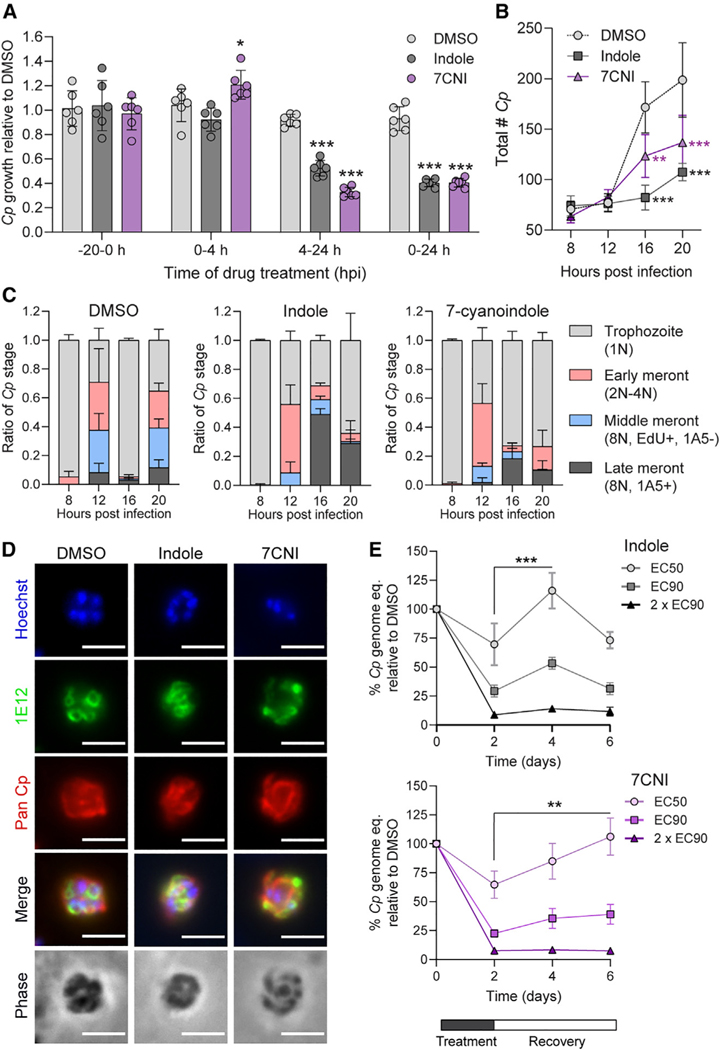
To test whether indoles inhibit parasite invasion and/or asexual replication, we quantified Cp growth in cultures treated with 90% maximal effective concentration (EC90) concentrations of either indole or 7CNI at time points corresponding to Cp invasion and parasitophorous vacuole formation (0–4 hpi), intracellular replication (4–24 hpi), or both processes (0–24 hpi). We also pre-treated host cells for 20 h before infection to see if indoles “prime” host cells against Cp infection. Pre-treatment of host cells did not affect subsequent Cp infection, and neither indole nor 7CNI inhibited Cp growth when cells were treated for the first four hours of infection only (Figure 3A). In contrast, when treatment was started 4 hpi, indole and 7CNI significantly inhibited Cp infection to nearly the same extent as in cultures treated for the full 24 h (Figure 3A). Taken together, these results indicate that (1) indoles must be present during infection to inhibit Cp; (2) indoles do not impair Cp excystation, invasion, or parasitophorous vacuole formation; and (3) indoles act at some point during the intracellular replicative stages of the parasite.
Figure 3. Indoles delay C. parvum (Cp) life cycle progression.
(A) Ratio of Cp numbers relative to DMSO control in HCT-8 cells after treatment with 1% DMSO or EC90 concentrations of indole (880 μM) or 7-cyanoindole (7CNI; 500 μM) for the indicated hours post-infection (hpi). Data plotted represent mean ± SD of six replicates (three technical replicates from two independent experiments). Differences between mean Cp ratio in indole or 7CNI-treated cultures vs in the DMSO control at each time point were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. *p < 0.05 and ***p < 0.001.
(B) Total number of Cp in HCT-8 cultures treated with 1% DMSO or EC90 concentrations of indole or 7CNI for the indicated hours post-infection. Data plotted represent mean ± SD of three independent experiments (same experiments as in C). Differences between mean Cp numbers in indole or 7CNI-treated cultures vs in the DMSO control at each time point were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. **p < 0.01 and ***p < 0.001.
(C) Ratio of the number of Cp in the trophozoite, early meront, middle meront, or late meront stages in infected HCT-8 cultures treated with 1% DMSO or EC90 concentrations of indole or 7CNI at the indicated hours post-infection. N, number of nuclei per parasite. Data plotted represent mean ± SD of three independent experiments.
(D) Immunofluorescence images of Cp in HCT-8 cultures treated with 1% DMSO or EC90 concentrations of indole or 7CNI 22 hpi. Parasites are labeled with membrane marker 1E12 (green) and a general Cp antibody, Pan Cp (red). Nuclei are stained with Hoechst. Scale bar, 3 μm.
(E) Washout experiments in Cp-infected air-liquid interface (ALI) cultures treated with 1% DMSO or indole at EC50 (577 μM), EC90 (1,894 μM), or 2 × EC90 (3,788 μM) or 7CNI at EC50 (379 μM), EC90 (688 μM), or 2 × EC90 (1,376 mM) for 48 h before washout. Cp genome equivalents were normalized to the DMSO control at each time point. Data plotted represent mean ± SD of six replicates (three technical replicates from two independent experiments). Differences between mean percentage Cp after washout versus mean percentage Cp at time of washout (2 dpi) for each indole concentration were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. **p < 0.01 and ***p < 0.001.
See also Figures S2 and S3.
After invasion, Cp begins its intracellular asexual cycle as a single nucleus trophozoite that undergoes three rounds of DNA replication with incomplete cytokinesis (early and middle meronts), followed by separation of the individual nuclei and membrane engulfment to form eight mature type I merozoites (late meront).33 The mature merozoites then egress from the host cell and invade a neighboring host cell to restart the asexual cycle. To more precisely define when indoles act during the intracellular stages of the parasite, we treated infected cultures with indole or 7CNI at EC90 concentrations while adding the thymidine analog EdU in 4 h pulses to label replicating parasite DNA and also staining for stage specific markers, as described previously.33–35 We then quantified the total number of parasites and the ratio of intracellular life cycle stages present at each time point (Figure S2). We found that the total number of Cp did not significantly differ between DMSO and indole or 7CNI-treated cultures until the start of the second round of merogony (16 hpi; Figure 3B), indicating that indoles inhibit parasite development. When the progression of specific replicative stages was examined over the first 24 hpi, it was evident that indole and 7CNI delayed progression through sequential life cycle stages, rather than stalling development at a specific time point (Figure 3C). To determine if indoles prevent merozoite maturation, egress, and/or re-invasion similar to the previously studied compound KDU691,33 we treated infected cultures with EC90 indole or 7CNI for 22 h before fixing and labeling with 1E12, a monoclonal antibody that labels the parasite sur face membrane.35 We did not observe any obvious morphological defects in mature merozoites in indole or 7CNI-treated cultures (Figure 3D), indicating indoles do not prevent formation of mature merozoites.
Indole inhibition is partly reversible after washout in a long-term culture system for Cp
To examine the reversibility of treatment, we used an air-liquid interface (ALI) Transwell culture system that supports both asexual and sexual development and allows long-term growth of Cp.34,36 We treated infected cultures with indole or 7CNI at EC50, EC90, and 2 × EC90 concentrations for two days before washing out the compounds and allowing the cultures to recover for 2–4 days before quantification of Cp and host genomes. After treatment with indole or 7CNI EC50 concentrations, there was significant recovery in Cp growth 2 days after indole washout and 4 days after 7CNI washout (Figure 3E). Some growth recovery (though not significant) occurred after washout of cultures treated with EC90 indole or 7CNI, while no recovery was observed in cultures treated with 2 × EC90 indole or 7CNI (Figure 3E). However, the lack of parasite recovery at the highest concentrations of indole and 7CNI was likely affected by the high level of host toxicity at these concentrations (Figure S3). Thus, inhibition of Cp growth by indoles is partially reversible at lower concentrations when host cell viability is not affected. These results suggest that, while indoles impair growth when present, there is rapid restoration of an environment that is favorable for parasite development upon removal.
Indole treatment upregulates host genes involved in endoplasmic reticulum stress and membrane transport
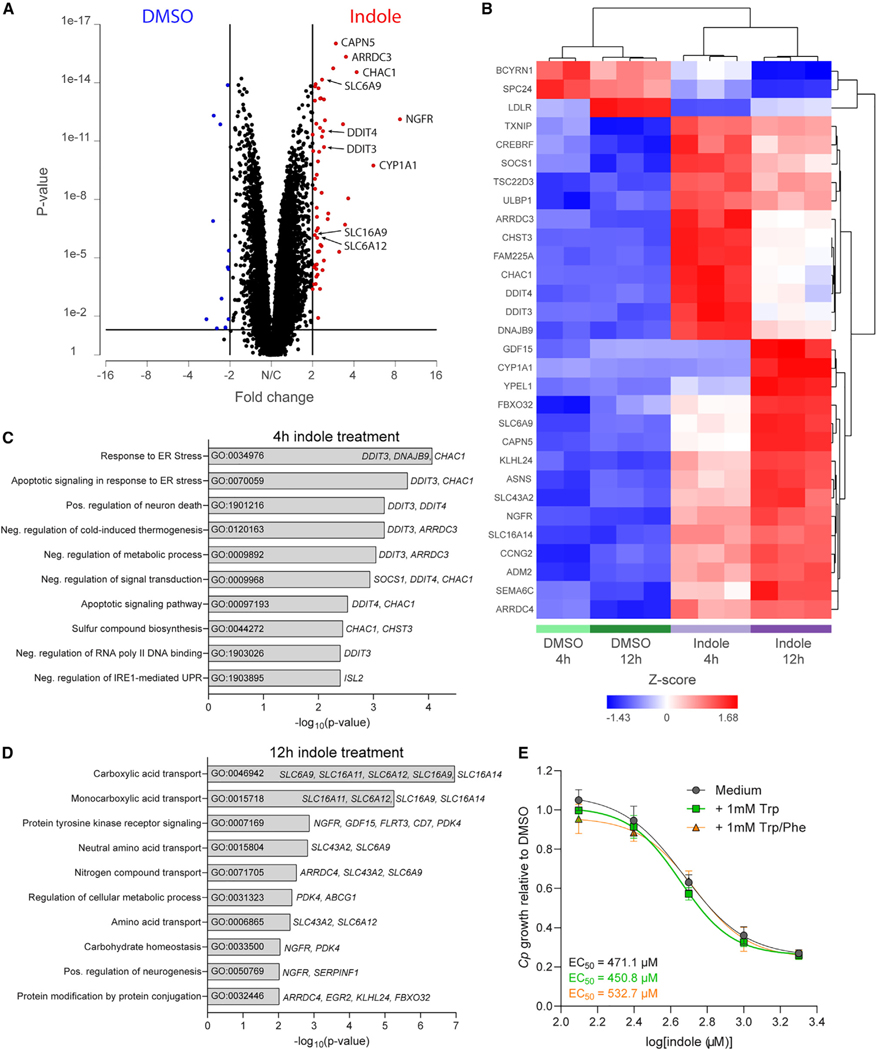
To determine if indoles alter host gene expression to create an unfavorable host cell environment for Cp growth, we performed RNA sequencing (RNA-seq) on HCT-8 cells treated with Cp EC90 concentration of indole or 1% DMSO for 4 or 12 h. When comparing all indole-treated (n = 6) with all DMSO-treated (n = 5) samples using length of treatment as a co-variable, we identified 68 genes that were significantly differentially expressed after indole treatment (false discovery rate [FDR] p value < 0.05, absolute fold change > 2): 57 upregulated genes (including AhR target gene CYP1A1) and 11 downregulated genes (Figure 4A, Table S3). Hierarchical clustering analysis of differentially expressed genes revealed that gene expression patterns separated into 3 main clusters: downregulated after indole treatment, highly upregulated after 4 h of indole treatment, and highly upregulated after 12 h of indole treatment (top 30 genes are shown in Figure 4B, all genes listed in Table S3). Gene Ontology (GO) process analysis performed in Enrichr on the subset of genes upregulated after 4 h of indole treatment found that the two most significant pathways were in response to endoplasmic reticulum (ER) stress (GO:0034976) and apoptotic signaling in response to ER stress (GO:0070059; Figure 4C). Interestingly, the expression of genes associated with these pathways, namely, DDIT3, DNAJB9, and CHAC1, goes down with longer exposure to indole (Figure 4B). In contrast, five out of the top 10 pathways upregulated after 12 h of indole treatment are involved in membrane transport of carboxylic acids (GO:0046942), monocarboxylic acids (GO:0015718), amino acids (GO:0015804 and GO:0006865), or nitrogen compounds (GO:0071705; Figure 4D). Taken together, these transcriptomics data suggest that short treatment with indole induces an ER stress response, which may cause the cells to upregulate transporters to restore imbalances in important nutrients. We reasoned that indole may be competing with the import of tryptophan or other aromatic amino acids like phenylalanine, both essential amino acids that cannot be made by the host cell. However, supplementation of cell culture medium with additional tryptophan or tryptophan plus phenylalanine did not rescue indole inhibition of Cp growth in HCT-8 cells (Figure 4E). Thus, indole inhibition of Cp is likely not due to a tryptophan deficiency in the host but may still result from a lack of another essential nutrient.
Figure 4. Indole induces ER stress and transporter upregulation in HCT-8 cells.
(A) Volcano plot of fold change vs p value after gene-specific analysis (GSA) of indole vs DMSO-treated HCT-8 cells highlighting genes significantly (p < 0.05) upregulated (red) or downregulated (blue) after indole treatment by >2-fold.
(B–D) Hierarchical clustering analysis of the 30 most differentially regulated genes (FDR-corrected p < 1 × 10−9) between indole and DMSO-treated HCT-8 cells. (C and D) Gene Ontology (GO) pathway analysis performed in Enrichr using genes significantly upregulated after (C) 4 h or (D) 12 h of indole treatment as input.
Upregulated genes associated with each pathway are listed to the right of the bar graph.
(E) Ratio of C. parvum (Cp) relative to DMSO control in HCT-8 cells after 24 h treatment with a serial dilution of indole in growth medium supplemented with 1 mM tryptophan (Trp) or 1 mM Trp plus 1 mM phenylalanine (PHE). EC50 values were calculated for each medium using a nonlinear regression curve fit with six replicates (three technical replicates from two independent experiments) per indole concentration.
Indoles impair host mitochondrial function and reduce ATP production
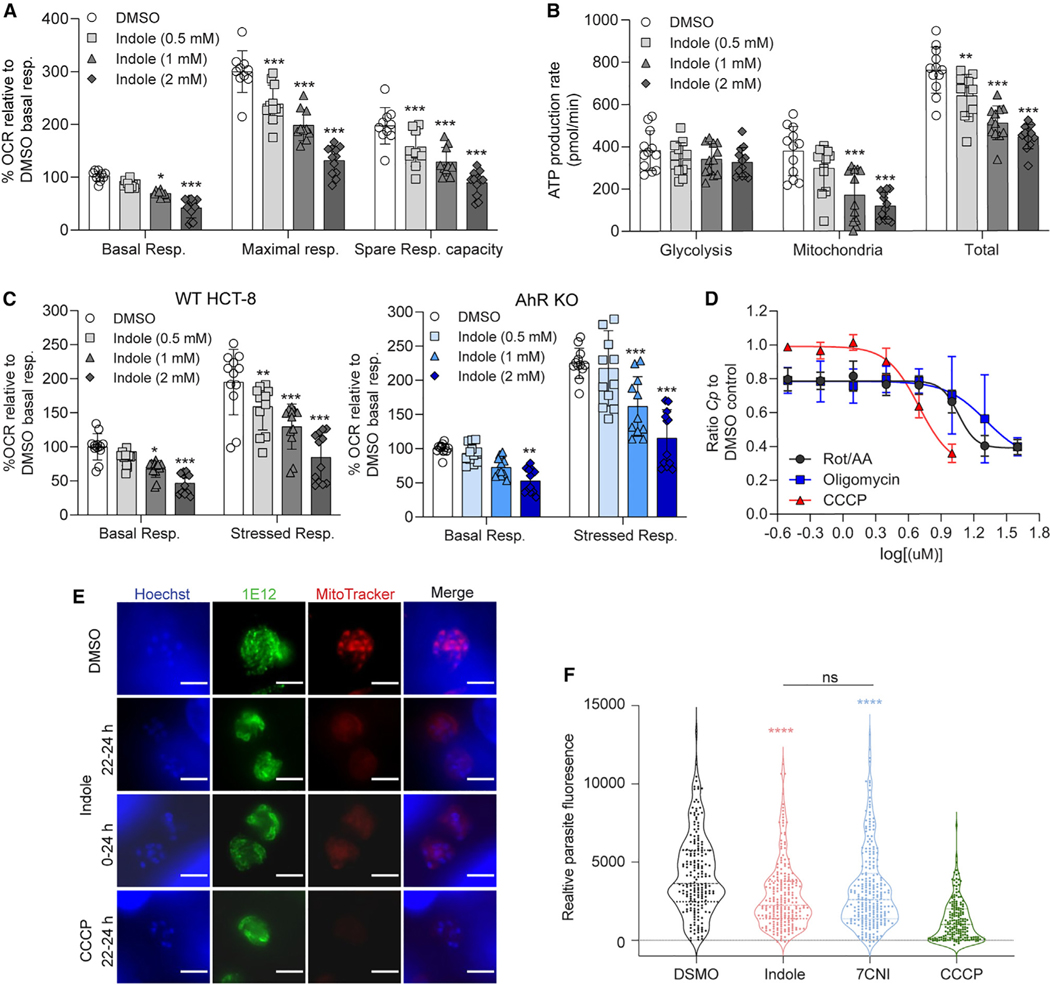
Previous studies have reported that indole can uncouple oxidative phosphorylation (OxPhos),37,38 inhibit the electron transport chain,37 and/or decrease ATP levels26,37,39 in P. putida,26 rat liver,37,38 and human enteroendocrine cells.39 To test whether indoles inhibit mitochondrial function in HCT-8 cells, we measured the oxygen consumption rate (OCR) and respiratory capacity of cells treated with indole or 7CNI. We found that indole and 7CNI significantly reduced both basal levels and maximal levels of mitochondrial respiration in HCT-8 cells in a dose-dependent manner (Figures 5A and S4). As a result, the spare respiratory capacity (maximal minus basal respiratory rate) of indole and 7CNI-treated cells was also diminished (Figures 5A and S4), indicating that the mitochondria in these cells are less able to respond to a sudden increase in energy demand (simulated by the addition of proton ionophore and OxPhos uncoupler FCCP in the assay). This dose-dependent impairment of mitochondrial function by indole and 7CNI translated into a significant reduction in ATP production rate by the mitochondria (Figures 5B and S4). Although ATP production rates by glycolysis remained similar between all treatments, there was a lower total ATP production rate in indole or 7CNI-treated cells (Figures 5B and S4).
Figure 5. Indole impairs host mitochondrial ATP production and affects C. parvum (Cp) mitosome potential.
(A) Metabolic analysis using the Seahorse XF Cell Mito Stress Test kit on HCT-8 cells treated for 18 h with 1% DMSO or indole (0.5, 1, or 2 mM). Data calculated asa percentage of the oxygen consumption rate (OCR) for each well relative to the mean basal OCR of DMSO control cells for that experiment. Spare respiratory capacity = maximal respiratory rate - basal respiratory rate for each well. Data plotted represent mean ± SD of 12 replicates (six technical replicates from two independent experiments). Differences between percentage OCR for each indole concentration vs the DMSO control for each measurement were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. *p < 0.05 and ***p < 0.001.
(B) Metabolic analysis using the Seahorse XF Real-Time ATP Rate assay on HCT-8 cells treated for 18 h with 1% DMSO or indole (0.5, 1, or 2 mM). Data plotted represent mean ± SD of ATP production rate (pmol/min) produced by glycolysis, the mitochondria, or total ATP (glycolysis + mitochondrial ATP rates) for 12 replicates (six technical replicates from two independent experiments). For each source of ATP, differences between ATP production rate for each indole concentration vs the DMSO control were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. **p < 0.01 and ***p < 0.001.
(C) Metabolic analysis using the Seahorse XF Cell Energy Phenotype Test kit on HCT-8 AhR WT cells (gray) or AhR KO cells (blue) treated for 18 h with 1% DMSO or indole (0.5, 1, or 2 mM). Data calculated as a percentage of OCR for each well relative to the mean basal OCR of DMSO control cells for that experiment. Data plotted represent mean ± SD of 12 replicates (six technical replicates from two independent experiments). For each cell line, differences between percentage OCR for each indole concentration vs the DMSO control were analyzed using a one-way ANOVA followed by Dunnett’s test for multiple comparisons. *p < 0.05, **p < 0.01, and ***p < 0.001.
(D) Ratio of Cp relative to DMSO control in HCT-8 cells after 24 h treatment with serial dilutions of mitochondrial complex I and III inhibitors rotenone and antimycin A, respectively (Rot/AA); ATP synthase inhibitor oligomycin; or proton gradient uncoupler carbonyl cyanide m-chlorophenyl hydrazone (CCCP). Inhibition curves were calculated for each compound using a nonlinear regression curve fit with six replicates (three technical replicates from two independent experiments) per concentration.
(E) Immunofluorescence images of Cp in HCT-8 cultures treated with 1% DMSO or 2 × EC90 concentrations of indole (1.76 mM) or 10 μM CCCP for the indicated hours post-infection. Parasites were labeled with membrane marker 1E12 (green), MitoTracker Red CMXRos (red), and nuclei were stained with Hoechst (blue). Scale bar, 3 μm.
(F) Distribution of MitoTracker intensity for indole-treated parasites. Cp in HCT-8 cultures treated with 1% DMSO or 2 × EC90 concentrations of indole (1.76 mM), 7-cyanoindole (7CNI) (1 mM), or 10 μM CCCP for 2 h starting 22 hpi. Parasite fluorescence intensity was measured on the basis of MitoTracker staining collected from at least 180 parasites from two independent experiments. Statistical analyses comparing each treatment group with control were performed using two-tailed Mann-Whitney U tests. ****p < 0.0001. ns, not significant. (DMSO compared with indole shown in red asterisk, DMSO compared with 7CNI shown in blue asterisk.).
See also Figure S4.
Although indole inhibition of Cp does not appear to act through the AhR pathway (Figure 2), AhR responsive genes were clearly upregulated in the RNA-seq experiments (Figure 4) raising the possibility that this pathway might affect mitochondrial function. To test this possibility, we examined mitochondrial function using the HCT-8 AhR KO 1 cell line. We found that mitochondrial basal respiratory rate and stressed respiratory rate (measured after the simultaneous addition of FCCP and oligomycin, an ATP synthase inhibitor) were significantly impaired in a dose-dependent manner in both HCT-8 wild-type (WT) and AhR KO cells (Figure 5C). Thus, impaired mitochondrial respiration is associated with inhibition of Cp growth in a pathway that does not depend on AhR. To determine whether inhibiting mitochondrial function was sufficient to inhibit Cp growth independently of indole, we performed the Cp growth assay in HCT-8 cells after treating with serial dilutions of OxPhos inhibitors oligomycin, carbonyl cyanide m-chlorophenyl hydrazone (CCCP; a proton ionophore structurally similar to FCCP), or a combination of rotenone and antimycin A (Rot/AA), inhibitors of complex I and complex III, respectively. All three treatments inhibited Cp growth in a dose-dependent manner with EC50 values of 11.5 μM for Rot/AA, 20.6 μM for oligomycin, and 5.0 μM for CCCP (Figure 5D), levels that were not due to host cell toxicity (Figure S4).
Unlike its host cell, Cp does not contain an intact mitochondrion but instead has a remnant mitosome organelle that lacks both a functional tricarboxylic acid (TCA) cycle and a cytochrome-dependent electron transport pathway and therefore does not directly contribute to ATP production.40 Despite lacking a conventional electron transport chain, Cp mitosomes do sequester cationic dyes traditionally used to stain live mitochondria, such as MitoTracker CMXRos, suggesting the mitosome has an alternative mechanism to generate a membrane potential.41 Here, we verified that a potent depolarizer of mitochondrial membrane potential, CCCP, also decreases MitoTracker staining in the Cp mitosome (Figures 5E and F). Similarly, treatment of Cp with indole or 7CNI, whether briefly (e.g., 2 h) or for an extended time period (e.g., 24 h), significantly decreased MitoTracker staining of Cp mitosomes, indicating indoles likely impair the membrane potential of this organelle (Figures 5E and F).
Oral treatment with 7CNI or introduction of indole-producing microbes temporarily suppresses Cp infection in vivo
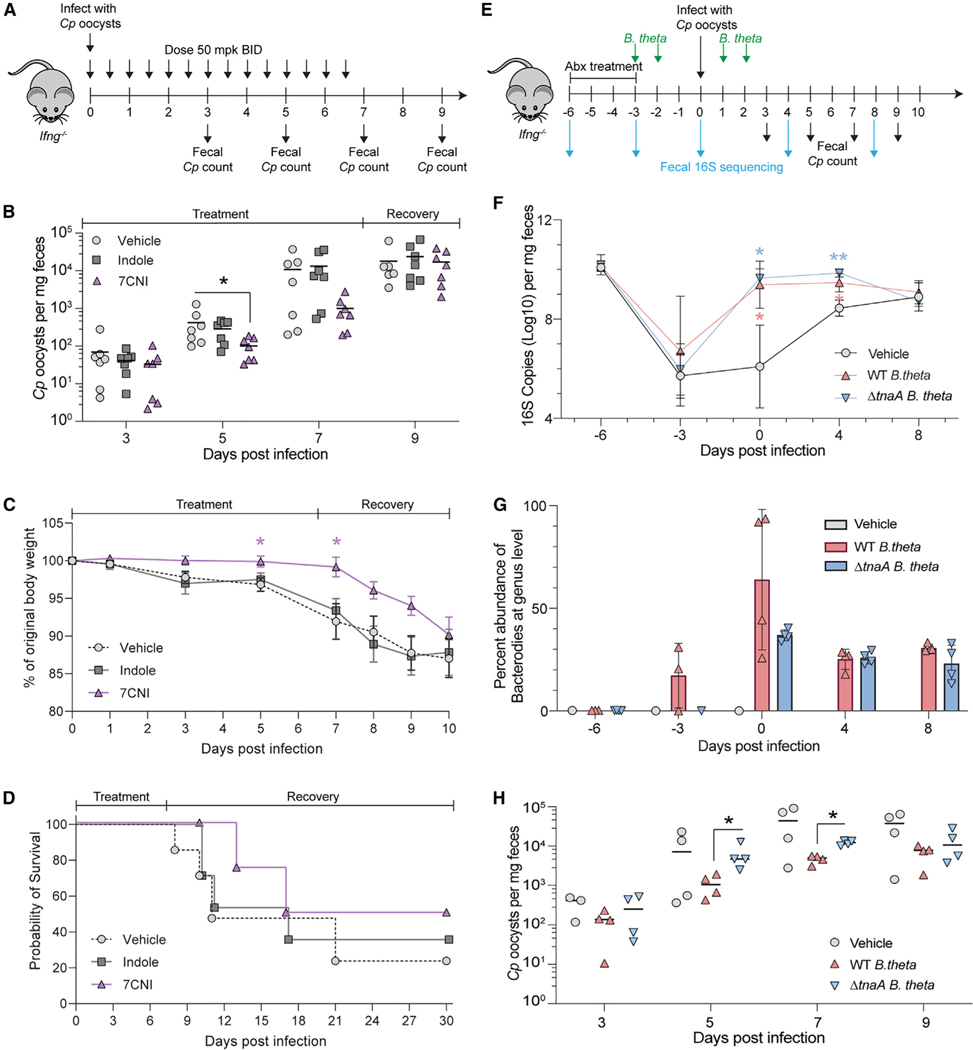
To determine whether oral treatment with indoles could alter infection in a mouse model of cryptosporidiosis, we treated Cp-infected interferon gamma KO (GKO) mice with 50 mg/kg indole or 7CNI by oral gavage twice daily for 7 days starting on the day of infection (Figure 6A). GKO mice are naturally susceptible to infection and when infected with the AUCP-1 strain of Cp used here, they do not require Abx treatment to result in a robust infection characterized by shedding of high numbers of oocysts. Although indole had no significant effect on Cp infection in vivo, 7CNI treatment significantly decreased the number of Cp oocysts shed in the feces of mice by 5 dpi (Figure 6B). The mean number of Cp oocysts per mg feces remained lower in 7CNI-treated mice at 7 dpi (the last day of treatment) but rebounded by 9 dpi (Figure 6B). Similarly, 7CNI-treated mice weighed significantly more than vehicle- or indole-treated mice 5 and 7 dpi but started to lose weight rapidly after treatment was terminated (Figure 6C). Although 7CNI treatment did not significantly increase survival rate, it did prolong the median survival time to 23.5 dpi compared with 11 or 17 dpi for vehicle or indole-treated mice, respectively (Figure 6D).
Figure 6. Exogenous indole treatment, or reconstitution with indole-producing bacteria, suppresses C. parvum (Cp) infection in GKO mice.
(A) GKO mice were treated twice daily by gavage with vehicle (10% DMSO in water) or 50 mg/kg indole or 7-cyanoindole (7CNI) for 7 days. Cp oocysts numbers were quantified from a single fecal pellet per mouse collected 3, 5, 7, and 9 dpi. All data plotted represent 7 mice per treatment group sampled over time from two independent experiments.
(B) Cp oocysts per mg feces for each mouse at the indicated days post-infection. Statistical analyses comparing each treatment group with vehicle control on individual days were performed using two-tailed Mann-Whitney U tests. *p < 0.05.
(C) Percent of original body weight plotted as mean ± SD. Statistical analysis performed using a mixed-effects model with a Geisser-Greenhouse correction for matched values, followed by Dunnett’s test for multiple comparisons. *p < 0.05.
(D) Combined survival curves of all 7 mice for the first 10 days then for the 4 mice from the second experiment for days 11–30.
(E) GKO mice were treated with antibiotic to suppress endogenous flora and then reconstituted with WT B. theta or ΔtnaA B. theta followed by challenge with Cp.
Oocysts numbers were quantified from fecal pellets collected 3, 5, 7, and 9 dpi. All data plotted represent 4 mice per treatment group sampled over time.
(F) Estimation of bacterial burdens in the gut by 16S rRNA qPCR. Mean ± SD. Statistical analyses comparing vehicle with WT B. theta group (shown in blue asterisk) or ΔtnaA B. theta group (shown in red asterisk) on individual days were analyzed using a two-way ANOVA followed by Dunnett’s test for multiple comparisons. *p < 0.05 and **p < 0.01.
(G) Relative abundance of Bacteroides at the genus level from each mouse at the indicated days post-infection.
(H) Cp oocysts per mg feces for each mouse at the indicated days post-infection. Statistical analyses comparing WT B. theta group with ΔtnaA B. theta group on individual days were performed using two-tailed Mann-Whitney U tests. *p < 0.05.
See Figures S5, S6, and S7.
To extend the studies on the role of indole in protection in vivo, we eliminated the endogenous microflora of mice by treatment with Abx and then reconstituted the microbiota with indole-producing bacteria before challenging with Cp. A variety of gram-negative bacteria express the tryptophanase gene (TnaA), which converts tryptophan to indole.42 Among these, members of the Bacteroidales and Clostridiales orders are among the most abundant taxa with the ability to generate indole within the gut.42 Hence, we chose Bacteroides thetaiotaomicron (B. theta) for reconstitution, as previous studies have shown that indoles produced by this species alter susceptibility to enterohemorrhagic E. coli infection by modulating virulence gene expression.43 For these experiments, mice were treated with a cocktail of Abx to suppress the endogenous flora and then reconstituted by oral gavage with WT B. theta that expresses TnaA or a mutant that lacks this biosynthetic pathway (ΔtnaA) (Figure 6E). Following Abx treatment, we confirmed the microbiota was depleted as assessed by qPCR analyses of 16S copy numbers (Figure 6F). Reconstitution with B. theta led to a rapid recovery of total bacterial levels, while vehicle-treated control mice returned more gradually to pre-Abx levels of gut bacteria (Figure 6F). The weight of the mice was not significantly different whether reconstituted with WT B. theta or the tnaA mutant (Figure S5). As expected, the richness of the endogenous bacterial communities was decreased by Abx treatment, and the number of observed species remained low in mice colonized with B. theta or administered vehicle (Figure S6). For all mice, the bacterial community structure, analyzed by weighted UniFrac distance, was disrupted by Abx, with the communities of mice administered WT or mutant B. theta converging by 0 dpi with Cp, while those of vehicle-treated mice remained disrupted and returned to a pre-Abx structure by 8 dpi (Figure S6). When analyzed taxonomically, reconstitution led to robust levels of Bacteroidales by 0 dpi in mice given WT or mutant B. theta but were absent in vehicle-treated mice (Figures S7). At the genus level, Bacteroides was generally undetected prior to Abx but became a dominant group in mice administered WT or mutant B. theta, with the only amplicon sequence variant identified for Bacteroides originating from B. theta (Figure 6G). In contrast, in mice given the vehicle control, Bacteroidales and Bacteroides were absent (Figures S7 and 6G). Having established that Abx treatment and reconstitution was effective in replacing the endogenous flora with WT B. theta capable of producing indole and the ΔtnaA mutant that lacks this ability, we tested the reconstituted mice for susceptibility to infection. Reconstitution with WT B. theta led to a significant reduction in oocyst shedding at 5 and 7 dpi compared with mice reconstituted with the ΔtnaA mutant, supporting a role for indole production in resistance to infection (Figure 6H). The difference in shedding was transient and the eventual convergence in susceptibility in mice given WT and mutant bacteria may be due to the emergence of other taxa, notably Clostridiales (Figure S7), members of which are also known to produce indoles.42
DISCUSSION
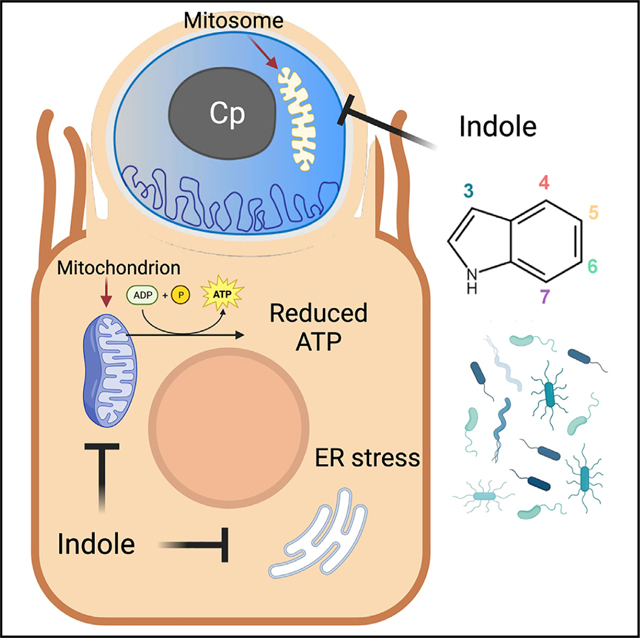
As an enteric pathogen, Cryptosporidium parasites are exposed to the billions of microbial inhabitants of the gut as well as the vast array of molecules they produce. Our screen of adult murine gut metabolites identified indole and its derivative, 4HI, as inhibitors of Cp growth in human cells. Further testing extended these findings to show that most indole analogs can inhibit the parasite regardless of modification position or side chain composition. Although indoles are known to signal through AhR, KO cell lines lacking AhR revealed that indole restriction of Cp is independent of this pathway. Instead, we found that indoles likely impact Cp growth indirectly by suppressing host mitochondrial respiration, leading to decreased ATP levels. Indole treatment also resulted in loss of membrane potential in the parasite mitosome, suggesting indoles can also impair function of this organelle. Indole treatment delayed progression of the parasite life cycle and was partially reversible upon washout, suggesting the effect is largely static. Treatment with indoles in vivo partially protected mice from Cp infection, as did reconstitution with indole-producing bacteria, suggesting that supplementation of indoles, or enhancement of microbial communities that produce them in large amounts, may be useful for reducing infection.
Cryptosporidium spp. have severely reduced metabolic capabilities and most lack both conventional mitochondria and the enzymes necessary for the TCA cycle and OxPhos.44 As such, Cp relies almost entirely upon host cells for their energy needs and prior studies have suggested that they may import phosphorylated nucleotides,45 including ATP. Furthermore, several metabolomic and proteomic studies have found that Cp and C. hominis infections induce host mitochondrial and glycolytic activity in cell lines (HCT-8 and COLO-680N) and experimental mouse models (C57BL/6J and BALB/c),46–49 with a concordant increase in cellular ATP levels.46,49 Our findings demonstrate that indoles inhibit host mitochondrial respiration in HCT-8 cells leading to a reduction in ATP production. Previous studies using intestinal enteroendocrine cells found that that indoles increased cellular NADH/NAD+ ratios, which would decrease the proton gradient across the inner mitochondrial membrane, reduce the OCR, and inhibit ATP synthesis.39,50 Consistent with this model, we found that treatment with known mitochondrial ETC inhibitors rotenone, antimycin A, oligomycin, and CCCP all had deleterious, dose-dependent effects on Cp growth. This mechanism of action is consistent with the observed developmental delay in Cp growth when cells were treated with indoles and with the observation that the effect was partially reversible upon washout of the compounds. Other studies have shown that Cp infection affects the metabolism of host cells,49 and future studies could examine the intersection of parasite infection with microbiota metabolite treatment, as these pathways likely interact.
Our findings also indicate that indoles directly affect the membrane potential of the mitosome, a degenerate organelle that lacks many features of normal mitochondria including an absence of DNA, complexes 2, 3, and 4 of the electron transport chain, and nearly all of the enzymes of the TCA cycle.40,51 As such, the mitosome does not contain a conventional system for generating ATP, although it maintains a membrane potential and consequently accumulates dyes like MitoTracker CMXRos. Previous studies have demonstrated that Fe-S cluster synthesis proteins are transported to the mitosome,52 and it is also likely the site of ubiquinone biosynthesis.40,51 The mitosome also contains an alternative oxidase that may function together with NAD(P) transhydrogenase and NDH2 as an alternative proton pump to establish a membrane gradient.40,51 Our studies demonstrate that treatment with indoles, or CCCP, results in decreased staining by MitoTracker, suggesting these agents disrupt the membrane potential and impair mitosome function, potentially contributing to the developmental delay of the parasite.
Our studies reveal that several classes of microbial metabolites impair growth of Cp in vitro including bile salts, indoles, and several short chain fatty acids (i.e., fumaric and propionic acids). Notably, the bile salt sodium taurocholate has been shown to enhance microneme secretion and increase the entry of Cp into host cells,53 although our studies conducted over longer time frames suggest the cumulative effect of secondary bile salts is inhibitory. Another recent study also demonstrated that growth of Cp is inhibited by short chain fatty acids,54 which are abundant metabolites produced by the microbiota. Notably, the active metabolites identified here are characteristic of the adult microbiota and are largely absent in neonatal mice, which are highly susceptible to cryptosporidiosis.11 Hence, the changes that occur in microbial metabolites with maturation of the microbiome likely contribute to colonization resistance seen in adults. We tested the role of indole derivatives in vivo and demonstrated that either direct administration or reconstitution with indole-producing bacteria provides partial protection against infection. The action of indoles in vivo could arise from the observed growth inhibitor properties we measured in vitro (AhR independent) but may also affect AhR signaling that increases barrier function and immunity in vivo.27,55,56 Consistent with a protective role for indole, previous studies have shown that healthy adults with high levels of fecal indole are resistant to challenge with Cryptosporidium.19 Indoles are produced from dietary tryptophan by a wide range of bacterial taxa that express the TnaA gene encoding tryptophanase including Clostridia, Bacteroidia, and Gammaproteobacteria.42 As such, exogenously delivered metabolites such as indoles, or enhancement of bacterial communities producing such entities, might provide an adjunct to Abx therapy in control of cryptosporidiosis.
Limitations of the study
Although our studies indicate that indoles can affect the membrane potential of the mitosome in Cp and also decrease ATP production from the mitochondria in human cells, they do not precisely define the extent to which either process contributes to parasite growth inhibition. The mitosome is poorly described and future studies could potentially elucidate key pathways in this organelle that depend on membrane potential. Oral administration of indoles, or reconstitution with indole-producing bacteria, demonstrates that these products contribute to resistance to Cp infection in mice, yet the effects of artificially manipulating these products were fairly modest. These limitations might reflect both rapid metabolism and potential excretion of indoles, as concentrations in the gut are expected to vary by location and over time. Additionally, our studies and those of others indicate that multiple metabolites contribute to resistance to Cp infection; hence, it is not unexpected that the sole manipulation of indole levels did not provide greater protection against Cp infection. Further studies using gnotobiotic mice could potentially better define a role for individual metabolites by reducing the contribution of other confounding factors.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, L. David Sibley (sibley@wustl.edu).
Materials availability
HCT-8 AhR knockout cell lines are available upon request.
Data and code availability
Raw RNA-seq reads and analyzed data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database and are publicly available as of the date of publication. 16S rRNA sequencing reads generated in this study have been deposited in the European Nucleotide Archive (ENA) and are publicly available as of the date of the publication. Accession numbers are listed in the key resources table. All other data reported in this paper will be shared by the lead author upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit polyclonal anti-RH | 35 | N/A |
| Rabbit polyclonal pan Cp | 35 | N/A |
| Mouse monoclonal 1A5 | 35 | N/A |
| Mouse monoclonal 1E12 | 35 | N/A |
| Alexa Fluor 488 goat anti-rabbit IgG (H + L) | Thermo Fisher Scientific | Cat#A11034; RRID: AB_2576217 |
| Alexa Fluor 488 goat anti-mouse IgG (H + L) | Thermo Fisher Scientific | Cat#A11029; RRID: AB_2534088 |
| Alexa Fluor 568 goat anti-mouse IgG (H + L) | Thermo Fisher Scientific | Cat#A11004; RRID: AB_2534072 |
| Alexa Fluor 594 goat anti-rabbit IgG (H + L) | Thermo Fisher Scientific | Cat#A11037; RRID: AB_2534095 |
| Alexa Fluor 647 goat anti-rabbit IgG (H + L) | Thermo Fisher Scientific | Cat#A21245; RRID: AB_2535813 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Bacteroides thetaiotaomicron: VPI-5482 strain | Laboratory of Vanessa Sperandio | N/A |
| Bacteroides thetaiotaomicron: VPI-5482 strain; ΔtnaA | Laboratory of Vanessa Sperandio | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Metabolites and indole analogs, see Table S4 | Sigma-Aldrich; AA Blocks, Inc. | N/A |
| Sodium taurocholate hydrate | Sigma-Aldrich | Cat#86339; CAS: 345,909–26-4 |
| Y-27632 dihydrochloride ROCK inhibitor | Tocris Biosciences | Cat#1254; CAS: 129,830–38-2 |
| VAF347 | Sigma-Aldrich | Cat#182690 |
| FICZ | AA Blocks, Inc. | Cat#AA007VKI; CAS: 172,922–91-7 |
| Kynurenic acid | Sigma-Aldrich | Cat#K3375; CAS: 492–27-3 |
| L-tryptophan | Sigma-Aldrich | Cat#T8941; Cas: 73–22-3 |
| L-phenylalanine | Sigma-Aldrich | Cat#P5482; Cas: 63–91-2 |
| Ampicillin | Sigma-Aldrich | Cat#A0166; Cas: 69–52-3 |
| Neomycin | Sigma-Aldrich | Cat#N1876; Cas: 1405–10-3 |
| Vancomycin | Sigma-Aldrich | Cat#V2002; Cas: 1404–93-9 |
| Puromycin | Sigma-Aldrich | Cat#P8833; Cas: 58–58-2 |
| Formaldehyde | Polysciences | Cat#04018; Cas: 50–00-0 |
| 2-mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| Triton X-100 | Thermo Fisher Scientific | Cat#BP151–100 |
| Lipofectamine 3000 Transfection Reagent |
Thermo Fisher Scientific | Cat#L3000001 |
| Hoechst 33,342 | Thermo Fisher Scientific | Cat#H3570 |
| ProLong Glass Antifade Mountant | Thermo Fisher Scientific | Cat#P36984 |
| Rotenone | Sigma-Aldrich | Cat#R8875; Cas: 83–79-4 |
| Antimycin A | Sigma-Aldrich | Cat#A8674; Cas: 1397–94-0 |
| Oligomycin | Sigma-Aldrich | Cat#O4876; Cas: 1404–19-9 |
| Carbonyl cyanide m-chlorophenylhydrazone (CCCP) | Sigma-Aldrich | Cat#C2759; Cas: 555–60-2 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Click-iT Plus EdU Alexa Fluor 488 Imaging kit | Thermo Fisher Scientific | Cat#C10637 |
| QIAamp DNA Mini kit | QIAGEN | Cat#51306 |
| QIAamp Powerfecal Pro DNA kit | QIAGEN | Cat#51804 |
| RNeasy Mini kit | QIAGEN | Cat#74104 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat#G7571 |
| RQ1 RNase-free DNase | Promega | Cat#M6101 |
| DNA-free DNA removal kit | Thermo Fisher Scientific | Cat#AM1906 |
| SuperScript VILO cDNA synthesis kit | Thermo Fisher Scientific | Cat#11754050 |
| TB Green Advantage qPCR premix | Takara Bio | Cat#639676 |
| Platinum Taq DNA Polymerase, High Fidelity | Thermo Fisher Scientific | Cat#11304011 |
| Seahorse XF Cell Energy Phenotype Test kit | Agilent | Cat#103325–100 |
| Seahorse XF Real-time ATP Rate Assay kit | Agilent | Cat#103592–100 |
| Seahorse XF Cell Mito Stress Test kit | Agilent | Cat#103015–100 |
| MitoTracker® Red CMXRos | Thermo Fisher Scientific | Cat# M7512 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE185652 |
| 16S rRNA reads | This paper | ENA: PRJEB58007 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse: NIH/3T3 | ATCC | CRL-1658 |
| Mouse: Ileal epithelial stem cells from C57BL/6 | Laboratory of Thaddeus Stappenbeck |
N/A |
| Human: Lenti-X 293T | Takara Bio | Cat#632180 |
| Human: HCT-8 | ATCC | CCL-244 |
| Human: HCT-8 AhR KO | This paper | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Cryptosporidium parvum AUCP-1 isolate | Laboratory of William Witola | N/A |
| Mouse: Infg KO (C57BL/6 background) | Jackson Laboratories | Cat#002287; RRID: IMSR_JAX:002,287 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primer: human CYP1A1 forward: ACATGCTGACCCTGGGAAAG |
PrimerBank | https://pga.mgh.harvard.edu/primerbank;PrimerBankID:189339226c2 |
| Primer: human CYP1A1 reverse: GGTGTGGAGCCAATTCGGAT |
PrimerBank | https://pga.mgh.harvard.edu/primerbank;PrimerBankID:189339226c2 |
| Primer: human AHRR forward: CTTAATGGCTTTGCTCTGGTCG |
61 | N/A |
| Primer: human AHRR reverse: TGCATTACATCCGTCTGATGGA |
61 | N/A |
| Primer: human GAPDH forward: TGAGTACGTCGTGGAGTCCA |
This paper | N/A |
| Primer: human GAPDH reverse: AGAGGGGGCAGAGATGATGA |
This paper | N/A |
| sgRNA for human AhR exon 1: TCACCTACGCCAGTCGCAAG | This paper | N/A |
| Primer: human AhR forward: GCACCATGAACAGCAGCAG |
This paper | N/A |
| Primer: human AhR reverse: TCCAAGTCCTCTGTCTCCCA |
This paper | N/A |
| Primer: C. parvum GAPDH forward: CGGATGGCCATACCTGTGAG |
34 | N/A |
| Primer: C. parvum GAPDH reverse: GAAGATGCGCTGGGAACAAC |
34 | N/A |
| Primer: mouse GAPDH forward: GCCATGAGTGGACCCTTCTT |
34 | N/A |
| Primer: mouse GAPDH reverse: GAAAACACGGGGGCAATGAG |
34 | N/A |
| Primer: V4 16S rRNA 515 forward: GTGCCAGCMGCCGCGGTAA |
71 | N/A |
| Primer: V4 16S rRNA 805 reverse: GACTACCAGGGTATCTAATCC |
71 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: lentiCRISPR v2 | 63 | Addgene Plasmid #52961 |
| Plasmid: lentiCRISPRv2-sgAhR | This paper | N/A |
| Plasmid: pMD2.g | Laboratory of Didier Trono | Addgene Plasmid #12259 |
| Plasmid: pMDLg/pRRE | 64 | Addgene Plasmid #12251 |
| Plasmid: pRSV-Rev | 64 | Addgene Plasmid #12253 |
|
| ||
| Software and algorithms | ||
|
| ||
| BioTek Gen5 v.3.02 | Agilent | https://www.agilent.com/en/product/cell-analysis/cell-imaging-microscopy/cell-imaging-microscopy-software/biotek-gen5-software-for-imagingmicroscopy-1623226 |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| QuantStudio Design & Analysis Software | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/global/forms/life-science/quantstudio-3–5-software.html |
| FIJI (ImageJ) | 65 | https://fiji.sc/ |
| Partek Flow | Partek, Inc. | https://www.partek.com/partek-flow/ |
| Enrichr | 70 | http://maayanlab.cloud/Enrichr/ |
| Wave v2.6.1 | Agilent | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolicanalysis/xf-software/seahorse-wavedesktop-software-740897 |
| R package: dada2 | 72 | https://benjjneb.github.io/dada2/ |
| R package: phyloseq | 74 | https://www.bioconductor.org/packages/release/bioc/html/phyloseq.html |
|
| ||
| Other | ||
|
| ||
| BioTek Cytation 3 cell imaging multimode reader | Agilent | https://www.biotek.com/products/detection-multi-mode-microplatereaders/cytation-hybrid-multimode-reader/ |
| Axioskop Mot Plus fluorescence microscope | Zeiss | N/A |
| QuantStudio 3 real-time PCR system | Applied Biosciences | Cat#A28137 |
| NovaSeq 6000 sequencing system | Illumina | https://www.illumina.com/systems/sequencing-platforms/novaseq.html |
| Seahorse XF96 Analyzer | Agilent | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolicanalysis/xf-analyzers |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cryptosporidium strain
C. parvum isolate AUCP-1 was maintained by repeated passage in male Holstein calves and purified from fecal material by sieve filtration, Sheather’s sugar flotation, and discontinuous sucrose gradient centrifugation as previously described.57,58 All calf procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Illinois at Urbana-Champaign. Purified oocysts were stored at 4°C in phosphate-buffered saline (PBS) plus 50 mM Tris and 10 mM EDTA (pH 7.2) for up to six months after fecal collection.
Bacteria lines
The wild type (WT) and tryptophanase knockout (ΔtnaA) Bacteroides thetaiotaomicron (B. theta) VPI-5482 strains were gifts from Vanessa Sperandio (University of Texas Southwestern Medical Center, Dallas, Texas, USA). Following growth on BHI-blood agar plates, single colonies were grown overnight at 37°C in anaerobic conditions in chopped meat media. The bacteria strains were aliquoted and frozen at −80°C in 20% glycerol until further processing. Colony forming units (CFUs) of frozen stocks were calculated based on serial dilution spotting on TSA +5% sheep’s blood plates (Thermo Fisher Scientific).
Cell lines
All cell lines were cultured at 37°C in a 5% CO2 incubator under normal atmospheric oxygen conditions. All cell lines were confirmed to be mycoplasma-free with the e-Myco plus Mycoplasma PCR detection kit (Boca Scientific). Human ileocecal adenocarcinoma cells (HCT-8; ATCC CCL-244) from a male patient and HCT-8 AhR knock-out cell lines (this study) were maintained in RPMI 1640 ATCC Modification medium (Thermo Fisher A14091–01) supplemented with 10% fetal bovine serum (Gibco).
Lenti-X 293T cells (Takara Bio, 632180), derived from the human kidney cell line HEK 293T, were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM high glucose; Sigma D6329) supplemented with 10% fetal bovine serum (Sigma).
NIH/3T3 embryonic mouse fibroblast cells (ATCC CRL-1658) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM high glucose; Sigma D6329) supplemented with 10% fetal bovine serum (Sigma).
Primary ileal intestinal epithelial stem cells (IECs) isolated from 8 to 10 week-old female C57BL/6 mice were expanded and maintained as 3D spheroid cultures in Matrigel (Corning) and 50% L-WRN conditioned medium (CM) containing 10 μM Y-27632 ROCK inhibitor (Torcis Biosciences), as previously described.59,60 The medium was changed every 2 days, and the cells were passaged every 3 days in a 1:6 split. For all experiments in this study, IECS were used between passages 4 and 26.
Mouse lines
Ifng knockout mice (referred to as GKO) were purchased from Jackson Laboratories (strain #002287, B6.129S7-Ifngtm1Ts/J) or bred in house in a specific-pathogen-free animal facility on a 12:12 light-dark cycle. Mice received irradiated laboratory rodent chow (Purina 5053) and autoclaved water ad libitum. Animals that became non-ambulatory during the course of infection were humanely euthanized in an SMARTBOX Auto CO2 euthanasia chamber. All mouse studies were approved by the Animal Studies Committee at the School of Medicine, Washington University in St. Louis.
For the first metabolite dosing experiment, nine GKO mice between 2 and 4 months old were split into 3 treatment groups: vehicle BID (1 male, 2 females), 50 mpk BID indole (1 male, 2 females) and 50 mpk BID 7CNI (2 males, 1 female). Within each treatment group, mice were co-housed with siblings but separated by sex. For the second metabolite dosing experiment, twelve GKO mice between 2 and 3 months old were split into the same 3 treatment groups as the first experiment (vehicle BID, 50 mpk BID indole, and 50 mpk BID 7CNI) with 2 males and 2 females per group. Within each treatment group, mice were co-housed with siblings but separated by sex. For the B. theta reconstitution experiment, twelve GKO mice between 8 and 16 weeks old were split into 3 treatment groups: vehicle (2 males, 2 females), WT B. theta (2 males, 2 females) and ΔtnaA B. theta (2 males, 2 females). Within each treatment group, mice were co-housed with siblings but separated by sex. All mice were given water containing 1 g/L ampicillin, 1 g/L neomycin and 0.5 g/L vancomycin (Sigma) for 3 days to deplete gut microbiota prior to the start of the experiment.
METHOD DETAILS
C. parvum oocyst preparation for infection
Before infection of cell culture or animal models, C. parvum oocysts were treated in a 40% bleach solution (commercial bleach containing 8.25% sodium hypochlorite) diluted in Dulbecco’s phosphate-buffered saline (DPBS; Corning) for 10 min on ice, then washed three times in DPBS containing 1% (wt/vol) BSA (BSA; Sigma). Bleached oocysts were stored for up to 1 week in DBPS plus 1% BSA at 4°C before infection. For most experiments, cells were infected with bleached oocysts added directly to the cell culture medium. For experiments requiring infection with sporozoites, bleached oocysts were excysted in 0.75% sodium taurocholate diluted in DPBS at 37°C for 1 h, then centrifuged at 1,250 x g for 3 min to pellet the sporozoites and remove the sodium taurocholate before resuspending in appropriate cell culture medium.
Initial screen of metabolites on C. parvum
Metabolites (Sigma-Aldrich, Table S4) were dissolved in appropriate solvent (Table S1) and screened at a final concentration of 1 mM. Metabolites that displayed significant host toxicity at 1 mM were re-screened at lower concentrations, specifically 0.1 mM for deoxycholic acid (DCA), sodium deoxycholate (SDC), and lithocholic acid (LCA) and 0.05 mM for prostaglandin E2. For the inhibition assay, HCT-8 cells were plated at 1.5 × 105 cells per well in black-sided, optically clear-bottomed 96-well plates (Greiner Bio-One) and grown for ~24h until confluent. Cells were then infected with 5 × 105 bleached C. parvum oocysts per well along with a single metabolite diluted in culture medium. After 24h of infection/treatment, cells were fixed in a 9:1 methanol:acetate solution for 5 min, washed twice with DPBS, permeabilized in 0.05% saponin in PBS for 10 min, and blocked in 0.05% saponin, 5% normal goat serum (NGS), and 5% fetal bovine serum (FBS) in PBS for 10 min before antibody staining. Parasites were labeled with an anti-RH antibody (rabbit polyclonal antibody raised against Toxoplasma strain RH that also recognizes all intracellular C. parvum stages35) diluted 1:1000 in PBS containing 1% NGS and 0.1% saponin, followed by Alexa Fluor goat anti-rabbit 594 secondary antibody (Thermo Fisher Scientific, diluted same as primary antibody). Host cells were stained with Hoechst 33342 (1 μg/mL, Thermo Fisher Scientific) for 10 min. Plates were imaged with a 103 objective on a BioTek Cytation 3 cell imager (9 images per well in a 3 × 3 grid). Gen5 software version 3.02 was used to quantify the total number of parasites (puncta in the Texas Red channel) and host cells (nuclei in the DAPI channel) per well. Each metabolite was screened in triplicate in six separate experiments. Relative parasite and host cell growth were calculated as a ratio of the number of C. parvum or host cells, respectively, in treated vs PBS negative control wells on each plate.
C. parvum growth assay
All C. parvum growth assays (aside from the initial metabolite screen) were performed as detailed here, with any modifications stated in the specific experimental sections. HCT-8 cells were plated at 1.5 × 105 cells per well in black-sided, optically clear-bottomed 96-well plates (Greiner BioOne) and grown for ~24h until confluent. Cells were infected with 1 × 105 bleached oocysts per well. After ~24h of infection/treatment, cells were fixed in 4% formaldehyde for 10 min, washed twice in DPBS, and then permeabilized and blocked in PBS containing 0.1% Triton X-100 and 1% BSA for 20 min before antibody staining. C. parvum parasites were labeled with Pan-Cp (rabbit polyclonal antibody raised against C. parvum that recognizes all stages of the parasite34) diluted 1:2,000 in PBS containing 0.1% Triton X-100 and 1% BSA, followed by Alexa Fluor goat anti-rabbit 488 secondary antibody (Thermo Fisher Scientific, diluted same as primary antibody). Host cell nuclei were stained with Hoechst 33342 (5 μg/mL, Thermo Fisher Scientific) for 20 min. Plates were imaged with a 10× objective on a BioTek Cytation 3 cell imager (9 images per well in a 3 × 3 grid). BioTek Gen5 software version 3.08 (Agilent) was used to quantify the total number of parasites (puncta in the GFP channel) and host cells (nuclei in the DAPI channel) per well.
Indole analog screen
Indole analogs (Tables S2 and S4) were ordered from Sigma-Aldrich (St. Louis, MO) or AA Blocks, Inc. (San Diego, CA) and reconstituted at 100 mM or 200 mM in DMSO. C. parvum growth assays were performed as described above with all analogs diluted to 1 mM in culture medium. Relative parasite number was calculated as a ratio of the number of C. parvum in treated wells vs the mean number of parasites in 1% DMSO negative control wells on each plate. Data plotted represents mean ± S.D. of six replicates (three technical replicates from two independent experiments).
Metabolite dose-response on C. parvum and host
To calculate metabolite EC50 values for C. parvum inhibition, metabolites were tested in a 7-point 1:2 serial dilution series starting at 200 mM (DCA, LCA and SDC), 4 mM (4HI and pyridoxal HCl), or 2 mM (indole and indole analogs). C. parvum growth assays were performed as described above, and relative parasite number was calculated as a ratio of the number of C. parvum in treated wells divided by the mean number of parasites in 1% DMSO negative control wells. To calculate CC50 values for host cytotoxicity, the growth assay was performed with the following modifications: HCT-8 cells were plated at a lower concentration (5 × 104 cells per well), cells were left uninfected, and the 7-point 1:2 serial dilution series started at 400 μM (DCA, LCA and SDC), 16 mM (4HI and pyridoxal HCl) or 8 mM (indole and indole analogs). Cells were fixed, permeabilized, stained with Hoechst and counted with the Cytation 3 as described above. Relative host toxicity was calculated as a ratio of the number of host nuclei in treated wells divided by the mean number of host nuclei in 1% DMSO negative control wells. We also monitored CC50 values for host cytotoxicity using HCT-8 cells that were plated at a lower concentration (1 × 104 cells per well). Cells were either left untreated or treated with an 8-point 1:2 serial dilution series starting at 400 μM (DCA, LCA), 16 mM (pyridoxal HCl) or 4 mM (indole). Cell viability was determined by Cell Titer-Glo Assay (Promega) based on quantification of ATP. The luminescence signal was measured by Cytation3 multi-mode plate imager (BioTek). Relative host toxicity was calculated as a ratio of luminescence value in treated wells divided by the mean number of luminescence value in 1% DMSO negative control wells. EC50 and EC90 values for C. parvum and host cells were calculated in GraphPad Prism 9 using a nonlinear regression curve fit with six replicates per data point (three technical replicates from two independent experiments). Fold selectivity of a metabolite for parasite vs host was calculated as host CC50 divided by C. parvum EC50.
C. parvum growth assay with AhR agonists
AhR agonists VAF347 (Sigma) and FICZ (AAblocks, Inc.) were reconstituted at 10 mM in DMSO, while indole, 4-methylindole (4MeI) and kynurenic acid (Sigma) were reconstituted at 100 mM in DMSO. Compounds were diluted in culture medium to a starting concentration of 1 mM (VAF347 and FICZ) or 1 μM (indole, 4MeI, and kynurenic acid), then serially diluted 1:2 twice. C. parvum growth assays were performed as described above, and relative parasite numbers were calculated as a ratio of the number of C. parvum in treated wells divided by the mean number in 1% DMSO negative control wells. Data plotted represents mean ± S.D. of 9 replicates (three technical replicates from three independent experiments).
Gene expression analysis of AhR target genes
HCT-8 cells were plated at 1 × 106 cells per well in 12-well culture plates and grown for ~24h until confluent. Medium in each well was then replaced with medium containing either 1% DMSO, VAF347 (250 nM), indole (1.5 mM) or 4-hydroxyindole (2.5 mM) and cultured for an additional 24h. Cells were then lysed in RLT buffer (QIAGEN) plus 1% β-mercaptoethanol and homogenized using a QIAshredder column (QIAGEN). RNA was extracted using the RNeasy Mini kit (QIAGEN), treated with RQ1 DNase (Promega) to remove DNA contamination, and converted to cDNA using the SuperScript VILO cDNA synthesis kit. Reverse transcription quantitative PCR (RT-qPCR) was run on a QuantStudio 3 real-time PCR system (Applied Biosciences) with TB Green Advantage qPCR premix (Takara Bio) and the following primers (5′ to 3′): human CYP1A1 (forward, ACATGCTGACCCTGGGAAAG; reverse, GGTG TGGAGCCAATTCGGAT; PrimerBank ID 189339226c2), human AHRR (forward, CTTAATGGCTTTGCTCTGGTCG; reverse, TGCA TTACATCCGTCTGATGGA61) and human GAPDH (forward, TGAGTACGTCGTGGAGTCCA; reverse, AGAGGGGGCAGAGAT GATGA; this study). Relative gene expression was calculated in QuantStudio Design and Analysis Software with the ΔΔCT method62 using human GAPDH as the reference gene and normalizing expression of each gene to its mean expression in the DMSO control samples. Data plotted represents mean ± S.D. of four technical replicates for all groups except for indole, which had 3 technical replicates due to RNA degradation in one of the samples.
CRISPR/Cas9 KO of AHR in HCT-8 cells
A short guide RNA (5′-TCACCTACGCCAGTCGCAAG-3′) targeting the first exon of the human AHR gene (NM_001621) was cloned into BsmBI-digested Cas9 plasmid lentiCRISPR v2 (Addgene #52961).63 The resulting plasmid (pLentiCRISPRv2-sgAhR) was transfected along with support plasmids pMD2.g (Laboratory of Didier Trono, Addgene #12259), pMDLg/pRRE (Addgene #12251),64 and pRSV-Rev (Addgene #12253)64 in equimolar concentrations into Lenti-X 293T cells (Takara Bio) using Lipofectamine 3000 reagent (Thermo Fisher Scientific). Lentivirus-containing supernatant was collected 72h post-transfection, passed through a 0.45 μM polyethersulfone (PES) filter, and diluted 1:2 in cell culture medium containing 10 μg/mL polybrene (Sigma) before adding to HCT-8 cells. Stable transgenic cells were selected for using puromycin (16 mg/mL) for 12 days starting 48 h after virus addition, with puromycin medium changes every 2–3 days. The transgenic cell population was serially diluted to isolate single cells, which were amplified and sequenced (Genewiz, Inc.) with primers flanking the targeted region (forward: 5′-GCACCATGAACAGCAGCAG-3′; reverse: 5′-TCCAAGTCCTCTGTCTCCCA-3′) to identify clonal lines with identical deleterious mutations in both alleles of AHR. Loss of AhR function was confirmed by treating AhR WT and KO cell lines with VAF347 (500 nM) or 1% DMSO for 24 h before harvesting RNA and analyzing CYP1A1 gene expression as described above. Data plotted represents mean ± S.D. of three technical replicates per treatment group from one experiment. Cp growth assay in HCT-8 WT vs AhR KO cell lines was performed as described above, treating with indole (1 mM), 4HI (1 mM), VAF347 (500 nM) or 0.5% DMSO. Data plotted represents mean ± S.D. of six replicates (three technical replicates from two independent experiments).
Sliding window treatment with indoles
C. parvum growth assays were performed as described above with the following modifications: after allowing the HCT-8 cells to adhere to the bottom of the wells (4 h post-seeding), medium in “pre-treatment” wells was replaced with EC90 concentrations of indole (877 μM) or 7-cyanoindole (500 μM) in 0.87% DMSO medium. Medium in all other wells was replaced with 0.87% DMSO medium. After an additional 20h of culture, all wells were washed twice with medium and then infected with 1 × 105 bleached oocysts. EC90 concentrations of indole or 7-cyanoindole in 0.87% DMSO medium were added to 0–4 h and 0–24 h treatment wells, with the remaining wells receiving 0.87% DMSO medium. All wells were washed twice with medium 4 hpi, and medium was replaced with EC90 concentrations of indole or 7-cyanoindole in 0.87% DMSO medium for the 4–24 h and 0–24 h treatment wells or 0.87% DMSO in medium for all other wells. All cells were fixed at 24 hpi and stained and imaged as described above. Relative parasite numbers were calculated as a ratio of the number of C. parvum in treated wells divided by the mean number in DMSO negative control wells. Data plotted represents mean ± S.D. of six replicates (three technical replicates from two independent experiments).
Antibody and EdU labeling of C. parvum
HCT-8 cells were plated at 4.5 × 105 cells per 12-mm-diameter glass coverslip (Thermo Fisher Scientific) in 24-well tissue culture plates and incubated until confluency (~24 h). Monolayers were infected with ~1 × 106 excysted sporozoites, washed twice with DPBS at 4 hpi, then treated at EC90 concentrations of indole (880 μM) or 7-cyanoindole (500 μM) in 1% DMSO medium. For EdU pulse labeling, one set of two coverslips per treatment group was incubated with 10 μM EdU for 4 h, then fixed in 4% formaldehyde. EdU was then added to another set of two coverslips per treatment group for 4h before fixing, and this cycle was repeated until 20 hpi. All coverslips were permeabilized/blocked in 0.1% Triton X-100 + 1% BSA in DPBS for 20 min, then treated with the Click-iT Plus EdU 488 imaging kit (Thermo Fisher Scientific) for 30 min to label EdU. Parasites were labeled with mouse monoclonal antibody 1A5 and rabbit polyclonal antibody Pan Cp, followed by anti-mouse Alexa Fluor 568 (Thermo Fisher Scientific) and anti-rabbit Alexa Fluor 647 (Thermo Fisher Scientific) and Hoechst nuclear stain. Coverslips were mounted on glass slides using ProLong Glass antifade mountant (Thermo Fisher Scientific) and sealed with nail polish. The number of parasites at each life stage was counted from 10 fields using a 100× oil immersion objective on a Zeiss Axioskop Mot Plus fluorescence microscope. The sum of parasites at each life stage at each time point was divided by the total number of C. parvum for that time point. Ratios were averaged across three independent experiments.
For membrane labeling of C. parvum merozoites, HCT-8 cells were cultured on coverslips as described above and infected with 5 × 105 C parvum oocysts. Cells were washed twice 4 hpi with DPBS, and EC90 concentrations of indole (880 mM) or 7-cyanoindole (500 μM) in 1% DMSO medium were added. Monolayers were fixed and stained 22 hpi with mouse monoclonal antibody 1E12 and rabbit polyclonal antibody Pan Cp, followed by anti-mouse Alexa Fluor 488 (Thermo Fisher Scientific) and anti-rabbit Alexa Fluor 568 (Thermo Fisher Scientific) and Hoechst nuclear stain. Coverslips were mounted on glass slides using ProLong Glass antifade mountant (Thermo Fisher Scientific) and sealed with nail polish. Images were acquired on a Zeiss Axioskop Mot Plus fluorescence microscope with a 100× oil immersion objective using AxioVision software (Carl Zeiss, Inc.) and processed in ImageJ (https://fiji.sc/).65
ALI cultures and washout assays
Air-liquid interface (ALI) cultures for long-term C. parvum growth were generated as previously described.34,36 Briefly, irradiated mouse 3T3 fibroblasts (i3T3, ATCC CRL-1658) cells were plated on transwells (polyester membrane, 0.4-μm pore, 6.5 mm insert; Corning Costar) coated with 10% Matrigel (Corning) at a density of 8 × 104 cells per transwell. Cells were cultured for 24 h at 37°C in DMEM high glucose medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 0.1 mg/mL streptomycin. Mouse intestinal epithelial cells (mIEC) spheroids were trypsinized and plated on the i3T3 feeder layer at 5 × 104 mIECs per transwell and cultured in 50% L-WRN conditioned medium (CM) supplemented with 10 μM Y-27632 (ROCK inhibitor, RI), as defined previously,60,66 with 200 μL and 400 μL medium in the top and bottom compartments of the transwell, respectively. Medium was changed every 2 to 3 days, and top medium was removed after 7 days to initiate the air-liquid interface.
To determine the indole and 7CNI EC50 and EC90 concentrations for C. parvum in ALI cultures, C. parvum oocysts (5 × 104 per transwell) were added to the top of ALI cultures 3 days post top medium removal. After ~4h of infection, the tops of the transwells were washed twice with DPBS to remove unexcysted oocysts, and the bottom medium was changed to medium containing indole or 7CNI in an 8 point serial dilution series. Approx. 48 hpi, cells were scraped from the transwell membranes, and DNA extraction was performed using the QIAamp DNA mini kit (QIAGEN). C. parvum and host genome equivalents were quantified on a QuantStudio 3 real-time PCR system (Applied Biosystems) with primers for their respective GAPDH genes, as previously described.34 EC50 and EC90 values were calculated using a nonlinear regression curve fit with seven replicates per data point (2–3 technical replicates from 3 independent experiments).
For indole and 7CNI washout experiments, C. parvum oocysts (5 × 104 per transwell) were added to the top of ALI cultures 3 days post top medium removal. After ~4h of infection, the tops of the transwells were washed twice with DPBS to remove unexcysted oocysts, and three transwells were scaped for DNA extraction (D0). The bottom medium in the remaining transwells was changed to medium containing 1% DMSO; indole at EC50 (577 μM), EC90 (1894 μM) or 2 x EC90 (3788 μM); or 7CNI at EC50 (379 μM), EC90 (688 μM) or 2 x EC90 (1376 μM). After ~48 h, the indoles were washed out by transferring transwells to clean wells with normal growth medium in the bottom compartments. A subset of transwells at each indole/7CNI concentration were scraped and collected for DNA at D2 (day of wash out), D4 (two days of recovery) and D6 (four days of recovery) post infection. C. parvum and host genome equivalents were quantified from DNA samples by qPCR as described above. Values from experimental samples were normalized to the mean of the DMSO control samples at each time point. Data plotted represents mean ± S.D. of six replicates (three technical replicates from two independent experiments).
Transcriptomics of indole-treated HCT-8 cells
HCT-8 cells were plated at 9 × 105 cells per well in 12-well culture plates and grown for ~24h until confluent. Cells were infected with ~4 × 106 excysted Cp sporozoites per well and incubated for 4 h before washing 2X with DPBS to remove extracellular parasites. Cells were then treated with either indole (880 μM) or 1% DMSO in cell culture medium for an additional 4 h or 12 h before harvesting RNA. Cells were lysed in RLT buffer (QIAGEN) plus 1% β-mercaptoethanol (Sigma) and homogenized using a QIAshredder column (QIAGEN). RNA was extracted using the RNeasy Mini kit (QIAGEN) and treated with the DNA-free DNA Removal kit (Thermo Fisher Scientific). Total RNA from three technical replicates per treatment condition was submitted to the Genome Access Technology Center (Washington University School of Medicine) for library prep and sequencing. RNA integrity was determined using an Agilent Bioanalyzer (all samples had an RIN of 10) and library preparation was performed with 5–10 μg of total RNA per sample. Ribosomal RNA was removed by poly-A selection using Oligo-dT beads (mRNA Direct kit, Life Technologies). mRNA was then fragmented in reverse transcriptase buffer by heating to 94°C for 8 min. mRNA was reverse transcribed using SuperScript III RT enzyme (Life Technologies) and random hexamers, per manufacturer’s instructions. A second strand reaction was performed to yield ds-cDNA, and then cDNA was blunt-ended before adding an A base to the 3′ ends and ligating Illumina sequencing adapters to both ends. Ligated fragments were amplified for 12–15 cycles using primers incorporating unique dual index tags, then sequenced on an Illumina NovaSeq 6000 to generate 150 bp, paired-end reads.
Demultiplexed fastq files were imported into Partek Flow (Partek, Inc.) and mapped to the Homo sapiens hg38 genome build (NCBI GenBank assembly ID GCA_000001405.15) using the STAR 2.7.8a aligner with default parameters.67 The number of reads mapping to each gene was then quantified based on the human Ensembl Transcripts release 100. At this point, we removed one of the replicates treated with DMSO for 4 h from further analysis due to most of its reads mapping to intergenic regions (indicative of DNA contamination). For the remaining 11 samples, gene expression values were normalized across samples by dividing the number of reads per gene by the total number of reads per sample to obtain counts per million (CPM) values. A pairwise comparison of indole-treated (6 total) and DMSO-treated samples (5 total) was performed using the Partek gene-specific analysis (GSA) algorithm using default parameters. Volcano plots were generated in Partek Flow. Genes were considered significantly differentially expressed if the FDR-corrected p value was less than 0.05 and the absolute fold change was greater than 2. Hierarchical clustering analysis of significant genes was performed in Partek Flow using a Euclidean point distance matrix and average linkage for the cluster distance metric. Gene Ontology (GO) pathway analyses were performed in Enrichr (http://maayanlab.cloud/Enrichr/)68–70 with the GO Biological Process 2021 database.
C. parvum growth with amino acid supplementation
HCT-8 cell culture medium was supplemented with 1 mM L-tryptophan (Sigma) with or without 1 mM L-phenylalanine (Sigma), and then filter sterilized. Indole was tested in a 5-point 1:2 serial dilution starting at 2 mM in all three media. C. parvum growth assays were performed as described above, and relative parasite number was calculated as a ratio of the number of C. parvum in each well divided by the mean number of parasites in the 1% DMSO, non-supplemented medium control wells. Indole EC50 values on Cp in each medium type were calculated using a nonlinear regression curve fit (with six replicates per data point (three technical replicates from two independent experiments).
Metabolic assays using Seahorse
For all metabolic assays, mitochondrial oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured using a Seahorse XF96 Analyzer (Agilent). HCT-8 cells or HCT-8 AhR KO cells were plated at 2 × 104 cells per well in a 96-well Seahorse XF96 cell culture microplate (Agilent) and grown for ~24h. Medium was replaced with fresh medium containing either 1% DMSO or 1:2 serial dilutions of indole (starting at 2 mM) or 7CNI (starting at 1 mM) and cultured for an additional 18h (cell confluency 90%). A Seahorse XFe96 extracellular flux sensor cartridge (Agilent) was hydrated overnight in distilled water in a non-CO2, 37°C incubator. The water was then replaced with XF Calibrant (Agilent), and the sensor cartridge was incubated for an additional ~1h in a non-CO2, 37°C incubator. Assay medium was prepared by adding 1 mM pyruvate (Agilent), 2 mM glutamine (Agilent) and 10 mM glucose (Agilent) to Seahorse XF DMEM medium, pH 7.4 (Agilent).
For assays run with the Seahorse XF Cell Energy Phenotype kit (Agilent), cells were washed once with assay medium before incubating in assay medium in a non-CO2, 37°C incubator for 1h. OCR and ECAR levels were measured for five cycles of mixing (2 min) and measuring (3 min) before and after supplementation via injection ports with a mix of FCCP (1 μM) and oligomycin (1 μM).
For assays run with the Seahorse XF Real-time ATP Rate Assay kit (Agilent), cell growth medium was replaced with assay medium and incubated in a non-CO2, 37°C incubator for 1h. Assay medium was then replaced with fresh assay medium prior to running the assay. OCR and ECAR levels were measured for four cycles of mixing (2 min) and measuring (3 min) before and after sequential supplementation via injection ports with oligomycin (1.5 mM), followed by a mix of rotenone (0.5 μM) and antimycin A (0.5 μM).
For assays run with the Seahorse XF Cell Mito Stress Test Kit (Agilent), cells were washed once with assay medium before incubating in assay medium in a non-CO2, 37°C incubator for 1h. OCR and ECAR levels were measured for four cycles of mixing (2 min) and measuring (3 min) before and after sequential supplementation via injection ports with oligomycin (1.5 μM), followed by FCCP (1 mM), and then a mix of rotenone (0.5 μM) and antimycin A (0.5 μM).
Results were initially analyzed in Wave v2.6.1 (Agilent) before importing into Graphpad Prism 9. All assays were performed twice with six technical replicates per sample per plate. Wells were removed from analysis if the OCR or ECAR values were negative or identified as an outlier by a ROUT test with Q = 1% performed in GraphPad Prism 9. Data plotted represents mean ± S.D. of 10–12 replicates (5–6 technical replicates from two independent experiments).
Mitotracker staining of indole treated C. parvum
HCT-8 cells were cultured on coverslips and infected with 5 × 105 C parvum oocysts. Cells were washed twice with DPBS and medium was supplemented with 2 x EC90 concentrations of indole (1.76 mM), 7-cyanoindole (1 mM) or 10 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP, Sigma) in 1% DMSO media. Infected cells were treated for 2 h (22–24 hpi) or continuously for 24 hpi vs. 1% DMSO control. MitoTracker Red CMXRos (50 nM final concentration) was added to the culture 45 min before fixation. Monolayers were fixed and stained at 24 hpi with mouse monoclonal antibody 1E12 followed by anti-mouse Alexa Fluor 488 (Thermo Fisher Scientific) and Hoechst nuclear stain. Coverslips were mounted on glass slides using ProLong Glass antifade mountant (Thermo Fisher Scientific) and sealed with nail polish. Images were acquired on a Zeiss Axioskop Mot Plus fluorescence microscope with a 1003 oil immersion objective using AxioVision software (Carl Zeiss, Inc.) and the relative fluorescence intensity was processed in ImageJ (https://fiji.sc/).65 Example parasites were chosen from four replicates (two technical replicates from two independent experiments).
C. parvum growth assay with mitochondrial inhibitors
Rotenone (Sigma), antimycin A (Sigma), oligomycin (Sigma) and carbonyl cyanide m-chlorophenyl hydrazone (CCCP, Sigma) were reconstituted at 20 mM in DMSO. Compounds were diluted in DMSO to 4 mM each with rotenone and antimycin-A mixed together, each at 4 mM. Compounds were serially diluted 1:2 7 times in DMSO, then diluted in cell culture medium to 1% DMSO with the highest concentration of each compound starting at 40 μM. C. parvum growth assays were performed as described above, and relative parasite numbers were calculated as a ratio of the number of C. parvum in treated wells divided by the mean number in 1% DMSO negative control wells. Data plotted represents mean ± S.D. of 4 replicates (two technical replicates from two independent experiments). Data for CCCP at 40 μM and 20 μM were not included due to observed host toxicity at those concentrations.
Metabolite treatment of C. parvum-infected GKO mice
In the first set of in vivo metabolite experiments, indole and 7-cyanoindole solutions were prepared at 5 mg/mL in vehicle (10% DMSO and Millipore-filtered water), aliquoted and stored at −20°C. A total of 9 GKO mice (Jackson Laboratories #002287) between 2 and 4 months old were split into 3 treatment groups: vehicle BID (1 male, 2 females), 50 mpk BID indole (1 male, 2 females) and 50 mpk BID 7CNI (2 males, 1 female). Within each treatment group, mice were co-housed with siblings but separated by sex. On the first day of the experiment (0 dpi), all mice were infected by oral gavage with 2 × 104 Cp oocysts mixed in 200 μL of their respective treatment solution. Mice received an additional 200 μL gavage of their treatment solution approx. every 12 h for a total of 7 days. Mice were weighed every 24–48 hpi and survival was recorded. A single fecal pellet was collected per mouse 3, 5, 7, and 9 dpi. Fecal pellets were stored at −80°C until further processing. The experiment was terminated 10 dpi.
In the second set of in vivo metabolite experiments, indole and 7-cyanoindole solutions were prepared at 10 mg/mL in vehicle (10% DMSO and Millipore-filtered water), aliquoted and stored at −20°C. A total of 12 GKO mice between 2 and 3 months old were split into the same 3 treatment groups as previously (vehicle BID, 50 mpk BID indole, and 50 mpk BID 7CNI) with 2 males and 2 females per group. Within each treatment group, mice were co-housed with siblings but separated by sex. On the first day of the experiment (0 dpi), all mice were infected by oral gavage with 2 × 104 Cp oocysts in 100 μl PBS. Approx. 30 min later, all mice received 100 μL of their respective treatment solution via oral gavage. Mice received an additional 100 μL gavage of their treatment solution approx. every 12 h for a total of 7 days. Mice were weighed every 24–48 hpi and survival was recorded. A single fecal pellet was collected per mouse 3, 5, 7, and 9 dpi. Fecal pellets were stored at −80°C until further processing. The experiment was terminated at 30 dpi.
DNA was extracted from fecal pellets using the QIAamp Powerfecal Pro DNA kit (QIAGEN) following the manufacturer’s protocol. The number of Cp oocysts per fecal pellet was calculated using qPCR for the Cp GAPDH gene as described above and then divided by the weight of the pellet in mg to obtain the number of Cp per mg feces.
B. theta reconstitution of C. parvum-infected GKO mice
GKO mice between 8 and 16 weeks old were separated by sex and split into 3 treatment groups: vehicle (2 males, 2 females), WT B. theta (2 males, 2 females) and ΔtnaA B. theta (2 males, 2 females). Within each treatment group, mice were co-housed with siblings but separated by sex. All mice were given water (Abx water) containing 1 g/L ampicillin, 1 g/L neomycin and 0.5 g/L vancomycin (Sigma) for 3 days to deplete gut microbiota. Following Abx treatment (−3 dpi), groups of mice were inoculated with WT B. theta or with ΔtnaA B. theta (1 × 108 CFU) while control mice were orally administered vehicle (10% glycerol in Millipore-filtered water). Mice received three additional treatments at −3, −2, and 1, 2 dpi to reconstitute gut microbiota. On the first day of the experiment (0 dpi), all mice were infected by oral gavage with 2 × 104 Cp oocysts in 100 μL PBS. Fecal pellets were collected at 3, 5, 7, and 9 dpi for measuring Cp oocysts in feces, and at −6, −3, 0, 4, 8 dpi for analyzing gut microbiota. Fecal pellets were stored at −80°C until further processing. The experiment was terminated at 16 dpi and mice were humanely sacrificed. DNA was extracted from fecal pellets using the QIAamp Powerfecal Pro DNA kit (QIAGEN) following the manufacturer’s protocol. The number of Cp oocysts per fecal pellet was calculated using qPCR for the Cp GAPDH gene as described above and then divided by the weight of the pellet in mg to obtain the number of Cp per mg feces.
16S rRNA analyses
For quantification of bacterial levels, the 16S rRNA gene was amplified by qPCR and quantified using SYBR Green. Primers for qPCR consisted of 515 F and 805 R to detect the V4 hypervariable region of the 16S rRNA gene: (forward, 5′-GTGCCAGCMGCCGCGG TAA-3′; reverse, 5′-GACTACCAGGGTATCTAATCC-3′). For 16S rRNA sequencing, PCR amplification of the V4 region was performed as described previously.71 Briefly, each sample was amplified in triplicate with Golay-barcoded primers specific for the V4 region (F515/R805) and confirmed by gel electrophoresis. Platinum High Fidelity Taq (Invitrogen) was used for PCR. Amplicons were pooled and purified with 0.6x Agencourt Ampure XP beads (Beckman-Coulter) according to the manufacturer’s instructions. The final pooled samples, along with aliquots of the three sequencing primers, were sequenced using a 2 × 250 bp Illumina protocol by the Center for Genome Sciences (Washington University School of Medicine). Read quality control and the resolution of amplicon sequence variants (ASVs) were performed with the dada2 R package.72 ASVs that were not assigned to the kingdom Bacteria were filtered out. The remaining reads were assigned taxonomy using the Ribosomal Database Project (RDP trainset 16/release 11.5) 16S rRNA gene sequence database.73 Ecological analyses, such as alpha-diversity (richness, Shannon diversity) and beta-diversity analyses (unweighted and weighted UniFrac distances), were performed using PhyloSeq and additional R packages.74
QUANTIFICATION AND STATISTICAL ANALYSES
All statistical analyses were performed in GraphPad Prism 9 unless otherwise specified. A one-way ANOVA (single variable) or two-way ANOVA (multiple variables) followed by a Dunnett’s multiple comparisons test was used to compare each experimental group to a control group (defined in each figure legend). EC50 and EC90 values were calculated using a nonlinear regression curve fit (log inhibitor vs normalized response – variable slope). For mouse experiments, each treatment group was compared to control group on individual days using a two-tailed Mann-Whitney U test. Comparisons of body weight between treatment groups were analyzed using a mixed-effects model with a Geisser-Greenhouse correction for matched values, followed by a Dunnett’s test for multiple comparisons. Statistical parameters for each experiment including statistical test used, technical replicates (n), independent biological replicates (N) and standard error are reported in the figure legends and associated method details.
Supplementary Material
Highlights.
The microbiota provides resistance to infection by the protozoan parasite Cryptosporidium
Indole metabolites produced by adult microbiota inhibit Cryptosporidium growth in vitro
Indoles inhibit oxidative phosphorylation and decrease ATP levels in host cells
Indoles also depolarize the mitosome in the parasite, thus likely inhibiting vital functions
ACKNOWLEDGMENTS
We thank William Witola (University of Illinois at Urbana-Champaign) for providing the Cp oocysts and Vanessa Sperandio for providing the B. theta strains used here, as well as Lynn Foster for technical assistance. Support was provided in part by grants from the NIH (AI 145496 to L.D.S. and DK109081 to K.L.V.). We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University School of Medicine for help with genomic analysis. The center is partially supported by NCI Cancer Center Support Grant (P30 CA91842) to the Siteman Cancer Center and by Institute of Clinical and Translational Sciences/Clinical and Translational Science Award (ICTS/CTSA) grant UL1TR002345 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. This publication is solely the responsibility of the authors and does not necessarily represent the official view of the NCRR or NIH.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112680.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSTIY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research.
REFERENCES
- 1.O’Connor R,M, Shaffie R, Kang G, and Ward HD (2011). Cryptosporidiosis in patients with HIV/AIDS. AIDS 25, 549–560. 10.1097/QAD.0b013e3283437e88. [DOI] [PubMed] [Google Scholar]
- 2.Putignani L, and Menichella D. (2010). Global distribution, public health and clinical impact of the protozoan pathogen cryptosporidium. Interdiscip. Perspect. Infect. Dis. 2010, 753512. 10.1155/2010/753512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222. 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.Kotloff KL, Nasrin D, Blackwelder WC, Wu Y, Farag T, Panchalingham S, Sow SO, Sur D, Zaidi AKM, Faruque ASG, et al. (2019). The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS). Lancet Global Health 7, e568–e584. 10.1016/S2214-109X(19)30076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine MM, Nasrin D, Acácio S, Bassat Q, Powell H, Tennant SM, Sow SO, Sur D, Zaidi AKM, Faruque ASG, et al. (2020). Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Global Health 8, e204–e214. 10.1016/S2214-109X(19)30541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Checkley W, Epstein LD, Gilman RH, Black RE, Cabrera L, and Sterling CR (1998). Effects of Cryptosporidium parvum infection in Peruvian children: growth faltering and subsequent catch-up growth. Am. J. Epidemiol. 148, 497–506. 10.1093/oxfordjournals.aje.a009675. [DOI] [PubMed] [Google Scholar]
- 7.Mølbak K, Andersen M, Aaby P, Højlyng N, Jakobsen M, Sodemann M, and da Silva AP (1997). Cryptosporidium infection in infancy as a cause of malnutrition: a community study from Guinea-Bissau, west Africa. Am. J. Clin. Nutr. 65, 149–152. 10.1093/ajcn/65.1.149. [DOI] [PubMed] [Google Scholar]
- 8.Zambriski JA, Nydam DV, Bowman DD, Bellosa ML, Burton AJ, Linden TC, Liotta JL, Ollivett TL, Tondello-Martins L, and Mohammed HO (2013). Description of fecal shedding of Cryptosporidium parvum oocysts in experimentally challenged dairy calves. Parasitol. Res. 112, 1247–1254. 10.1007/s00436-012-3258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsay DS, Blagburn BL, Sundermann CA, and Giambrone JJ (1988). Effect of broiler chicken age on susceptibility to experimentally induced Cryptosporidium baileyi infection. Am. J. Vet. Res. 49, 1412–1414. [PubMed] [Google Scholar]
- 10.Hatkin JM, Giambrone JJ, and Blagburn BL (1993). Kinetics of serumantibody responses in broiler chicks against Cryptosporidium baileyi. Avian Pathol. 22, 525–532. 10.1080/03079459308418940. [DOI] [PubMed] [Google Scholar]
- 11.VanDussen KL, Funkhouser-Jones LJ, Akey ME, Schaefer DA,Ackman K, Riggs MW, Stappenbeck TS, and Sibley LD (2020). Neonatal mouse gut metabolites influence cryptosporidium parvum infection in intestinal epithelial cells. mBio 11, e02582–20. 10.1128/mBio.02582-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherwood D, Angus KW, Snodgrass DR, and Tzipori S. (1982). Experimental cryptosporidiosis in laboratory mice. Infect. Immun. 38, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lantier L, Lacroix-Lamandé S, Potiron L, Metton C, Drouet F, Guesdon W, Gnahoui-David A, Le Vern Y, Deriaud E, Fenis A, et al. (2013). Intestinal CD103+ dendritic cells are key players in the innate immune control of Cryptosporidium parvum infection in neonatal mice. PLoS Pathog. 9, e1003801. 10.1371/journal.ppat.1003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potiron L, Lacroix-Lamandé S, Marquis M, Levern Y, Fort G, Franceschini I, and Laurent F. (2019). Batf3-Dependent intestinal dendritic cells play a critical role in the control of cryptosporidium parvum infection. J. Infect. Dis. 219, 925–935. 10.1093/infdis/jiy528. [DOI] [PubMed] [Google Scholar]
- 15.Charania R, Wade BE, McNair NN, and Mead JR (2020). Changes in the microbiome of cryptosporidium-infected mice correlate to differences in susceptibility and infection levels. Microorganisms 8, 879. 10.3390/microorganisms8060879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey MA, Medlock GL, Alam M, Kabir M, Uddin MJ, Nayak U,Papin J, Faruque ASG, Haque R, Petri WA, and Gilchrist CA (2021). Megasphaera in the stool microbiota is negatively associated with diarrheal cryptosporidiosis. Clin. Infect. Dis. 73, e1242–e1251. 10.1093/cid/ciab207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harp JA, Wannemuehler MW, Woodmansee DB, and Moon HW (1988). Susceptibility of germfree or antibiotic-treated adult mice to Cryptosporidium parvum. Infect. Immun. 56, 2006–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lantier L, Drouet F, Guesdon W, Mancassola R, Metton C, Lo-Man R, Werts C, Laurent F, and Lacroix-Lamandé S. (2014). Poly(I:C)-induced protection of neonatal mice against intestinal Cryptosporidium parvum infection requires an additional TLR5 signal provided by the gut flora. J. Infect. Dis. 209, 457–467. 10.1093/infdis/jit432. [DOI] [PubMed] [Google Scholar]
- 19.Chappell CL, Darkoh C, Shimmin L, Farhana N, Kim DK, Okhuysen PC, and Hixson J. (2016). Fecal indole as a biomarker of susceptibility to cryptosporidium infection. Infect. Immun. 84, 2299–2306. 10.1128/IAI.00336-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro IC, Oliveira BB, Slowikowski JJ, Coutinho BP, Siqueira FJWS, Costa LB, Sevilleja JE, Almeida CA, Lima AAM, Warren CA, et al. (2012). Arginine decreases Cryptosporidium parvum infection in undernourished suckling mice involving nitric oxide synthase and arginase. Nutrition 28, 678–685. 10.1016/j.nut.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laursen MF, Sakanaka M, von Burg N, Mö rbe U, Andersen D, Moll JM, Pekmez CT, Rivollier A, Michaelsen KF, Mølgaard C, et al. (2021). Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 6, 1367–1382. 10.1038/s41564-021-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E,Koga Y, and Benno Y. (2012). Impact of intestinal microbiota on intestinal luminal metabolome. Sci. Rep. 2, 233. 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chimerel C, Field CM, Piñ ero-Fernandez S, Keyser UF, and Summers DK (2012). Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim. Biophys. Acta 1818, 1590–1594. 10.1016/j.bbamem.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darkoh C, Plants-Paris K, Bishoff D, and DuPont HL (2019). Clostridium difficile modulates the gut microbiota by inducing the production of indole, an interkingdom signaling and antimicrobial molecule. mSystems 4. 10.1128/mSystems.00346-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garbe TR, Kobayashi M, and Yukawa H. (2000). Indole-inducible proteins in bacteria suggest membrane and oxidant toxicity. Arch. Microbiol. 173, 78–82. 10.1007/s002030050012. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Hong H, Heo A, and Park W. (2013). Indole toxicity involves theinhibition of adenosine triphosphate production and protein folding in Pseudomonas putida. FEMS Microbiol. Lett. 343, 89–99. 10.1111/1574-6968.12135. [DOI] [PubMed] [Google Scholar]
- 27.Hubbard TD, Murray IA, and Perdew GH (2015). Indole and tryptophan metabolism: endogenous and dietary routes to ah receptor activation. Drug Metab. Dispos. 43, 1522–1535. 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stepankova M, Bartonkova I, Jiskrova E, Vrzal R, Mani S, Kortagere S, and Dvorak Z. (2018). Methylindoles and methoxyindoles are agonists and antagonists of human aryl hydrocarbon receptor. Mol. Pharmacol. 93, 631–644. 10.1124/mol.118.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiNatale BC, Murray IA, Schroeder JC, Flaveny CA, Lahoti TS,Laurenzana EM, Omiecinski CJ, and Perdew GH (2010). Kynurenic acid is a potent endogenous aryl hydrocarbon receptor ligand that synergistically induces interleukin-6 in the presence of inflammatory signaling. Toxicol. Sci. 115, 89–97. 10.1093/toxsci/kfq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rannug A, Rannug U, Rosenkranz HS, Winqvist L, Westerholm R, Agurell E, and Grafströ m AK (1987). Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J. Biol. Chem. 262, 15422–15427. [PubMed] [Google Scholar]
- 31.Rannug U, Rannug A, Sjöberg U, Li H, Westerholm R, and Bergman J. (1995). Structure elucidation of two tryptophan-derived, high affinity Ahreceptor ligands. Chem. Biol. 2, 841–845. 10.1016/1074-5521(95)90090-x. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence BP, Denison MS, Novak H, Vorderstrasse BA, Harrer N,Neruda W, Reichel C, and Woisetschläger M. (2008). Activation of the aryl hydrocarbon receptor is essential for mediating the anti-inflammatory effects of a novel low-molecular-weight compound. Blood 112, 1158–1165. 10.1182/blood-2007-08-109645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Funkhouser-Jones LJ, Ravindran S, and Sibley LD (2020). Defining stage-specific activity of potent new inhibitors of cryptosporidium parvum growth in vitro. mBio 11, e00052–20. 10.1128/mBio.00052-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, Baldridge MT, VanDussen KL, Shen B, Kuhlenschmidt MS, et al. (2019). A stem-cell-derived platform enables complete cryptosporidium development in vitro and genetic tractability. Cell Host Microbe 26, 123–134.e8. 10.1016/j.chom.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilke G, Ravindran S, Funkhouser-Jones L, Barks J, Wang Q, VanDussen KL, Stappenbeck TS, Kuhlenschmidt TB, Kuhlenschmidt MS, and Sibley LD (2018). Monoclonal antibodies to intracellular stages of cryptosporidium parvum define life cycle progression in vitro. mSphere 3, e00124–18. 10.1128/mSphere.00124-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilke G, Wang Y, Ravindran S, Stappenbeck TS, Witola WH, and Sibley LD (2020). In vitro culture of Cryptosporidium parvum using stem cell-derived intestinal epithelial monolayers. In Cryptosporidium Methods in Molecular Biology, Mead Jand Arrowood M, eds. (Humana Press; ), pp. 351–372. [DOI] [PubMed] [Google Scholar]
- 37.Sakai M, Tohyama K, and Mutai M. (1982). Effect of indole on adenylateenergy charge and mitochondrial phosphorylative activity of rat liver. Int. J. Biochem. 14, 569–572. 10.1016/0020-711x(82)90037-4. [DOI] [PubMed] [Google Scholar]
- 38.Chimerel C, Murray AJ, Oldewurtel ER, Summers DK, and Keyser UF (2013). The effect of bacterial signal indole on the electrical properties of lipid membranes. ChemPhysChem 14, 417–423. 10.1002/cphc.201200793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, and Reimann F. (2014). Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 9, 1202–1208. 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Roellig DM, Guo Y, Li N, Frace MA, Tang K, Zhang L,Feng Y, and Xiao L. (2016). Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genom. 17, 1006. 10.1186/s12864-016-3343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts CW, Roberts F, Henriquez FL, Akiyoshi D, Samuel BU, Richards TA, Milhous W, Kyle D, McIntosh L, Hill GC, et al. (2004). Evidence for mitochondrial-derived alternative oxidase in the apicomplexan parasite Cryptosporidium parvum: a potential anti-microbial agent target. Int. J. Parasitol. 34, 297–308. 10.1016/j.ijpara.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Boya BR, Kumar P, Lee JH, and Lee J. (2021). Diversity of the tryptophanase gene and its evolutionary implications in living organisms. Microorganisms 9, 2156. 10.3390/microorganisms9102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar A, and Sperandio V. (2019). Indole signaling at the host-microbiota-pathogen interface. mBio 10, e01031–19. 10.1128/mBio.01031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, et al. (2004). The genome of Cryptosporidium hominis. Nature 431, 1107–1112. 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 45.Pawlowic MC, Somepalli M, Sateriale A, Herbert GT, Gibson AR, Cuny GD, Hedstrom L, and Striepen B. (2019). Genetic ablation of purine salvage in Cryptosporidium parvum reveals nucleotide uptake from the host cell. Proc. Natl. Acad. Sci. USA 116, 21160–21165. 10.1073/pnas.1908239116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vélez J, Velasquez Z, Silva LMR, Gärtner U, Failing K, Daugschies A, Mazurek S, Hermosilla C, and Taubert A. (2021). Metabolic signatures of cryptosporidium parvum-infected HCT-8 cells and impact of selected metabolic inhibitors on C. Parvum infection under physioxia and hyperoxia. Biology 10, 60. 10.3390/biology10010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T, Liu H, Jiang N, Wang Y, Wang Y, Zhang J, Shen Y, and Cao J. (2021). Comparative proteomics reveals Cryptosporidium parvum manipulation of the host cell molecular expression and immune response. PLoS Neglected Trop. Dis. 15, e0009949. 10.1371/journal.pntd.0009949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karpe AV, Hutton ML, Mileto SJ, James ML, Evans C, Shah RM, Ghodke AB, Hillyer KE, Metcalfe SS, Liu JW, et al. (2021). Cryptosporidiosis modulates the gut microbiome and metabolism in a murine infection model. Metabolites 11, 380. 10.3390/metabo11060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller CN, Panagos CG, Mosedale WRT, Kvá c M, Howard MJ, and Tsaousis AD (2019). NMR metabolomics reveals effects of Cryptosporidium infections on host cell metabolome. Gut Pathog. 11, 13. 10.1186/s13099-019-0293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Mir MY, Nogueira V, Fontaine E, Avé ret N, Rigoulet M, and Leverve X. (2000). Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 275, 223–228. 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 51.Salomaki ED, Terpis KX, Rueckert S, Kotyk M, Varadínová ZK, Cepi cka I, Lane CE, and Kolisko M. (2021). Gregarine single-cell transcriptomics reveals differential mitochondrial remodeling and adaptation in apicomplexans. BMC Biol. 19, 77. 10.1186/s12915021-01007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller CN, Jossé L, and Tsaousis AD (2018). Localization of Fe-S biosynthesis machinery in cryptosporidium parvum mitosome. J. Eukaryot. Microbiol. 65, 913–922. 10.1111/jeu.12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng H, Nie W, Sheoran A, Zhang Q, and Tzipori S. (2006). Bile acidsenhance invasiveness of Cryptosporidium spp. into cultured cells. Infect. Immun. 74, 3342–3346. 10.1128/IAI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keelaghan AP, Charania R, and Mead JR (2022). The effect of short-chain fatty acids on growth of cryptosporidium parvum in vitro. Microorganisms 10, 1822. 10.3390/microorganisms10091822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, Li T, and Yin Y. (2018). Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 8, 13. 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roager HM, and Licht TR (2018). Microbial tryptophan catabolites inhealth and disease. Nat. Commun. 9, 3294. 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arrowood MJ, and Sterling CR (1987). Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J. Parasitol. 73, 314–319. [PubMed] [Google Scholar]
- 58.Current WL (1990). Techniques and laboratory maintenance of Cryptosporidium. In Cryptosporidiosis of Man and Animals, Dubey JP, Speer CA, and Fayer R, eds. (CRC Press; ), pp. 44–77. [Google Scholar]
- 59.Miyoshi H, Ajima R, Luo CT, Yamaguchi TP, and Stappenbeck TS(2012). Wnt5a potentiates TGF-beta signaling to promote colonic crypt regeneration after tissue injury. Science 338, 108–113. 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyoshi H, and Stappenbeck TS (2013). In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li YF, Wang DD, Zhao BW, Wang W, Yuan SQ, Huang CY, Chen YM, Zheng Y, Keshari RP, Xia JC, and Zhou ZW (2012). Poor prognosis of gastric adenocarcinoma with decreased expression of AHRR. PLoS One 7, e43555. 10.1371/journal.pone.0043555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 63.Sanjana NE, Shalem O, and Zhang F. (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784. 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, and Naldini L. (1998). A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471. 10.1128/JVI.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M,Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.VanDussen KL, Sonnek NM, and Stappenbeck TS (2019). L-WRN conditioned medium for gastrointestinal epithelial stem cell culture shows replicable batch-to-batch activity levels across multiple research teams. Stem Cell Res. 37, 101430. 10.1016/j.scr.2019.101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128. 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q,Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. (2021). Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90. 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108, 4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA,and Holmes SP (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. 10.1038/Nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, and Tiedje JM (2014). Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McMurdie PJ, and Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-seq reads and analyzed data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database and are publicly available as of the date of publication. 16S rRNA sequencing reads generated in this study have been deposited in the European Nucleotide Archive (ENA) and are publicly available as of the date of the publication. Accession numbers are listed in the key resources table. All other data reported in this paper will be shared by the lead author upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit polyclonal anti-RH | 35 | N/A |
| Rabbit polyclonal pan Cp | 35 | N/A |
| Mouse monoclonal 1A5 | 35 | N/A |
| Mouse monoclonal 1E12 | 35 | N/A |
| Alexa Fluor 488 goat anti-rabbit IgG (H + L) | Thermo Fisher Scientific | Cat#A11034; RRID: AB_2576217 |
| Alexa Fluor 488 goat anti-mouse IgG (H + L) | Thermo Fisher Scientific | Cat#A11029; RRID: AB_2534088 |
| Alexa Fluor 568 goat anti-mouse IgG (H + L) | Thermo Fisher Scientific | Cat#A11004; RRID: AB_2534072 |
| Alexa Fluor 594 goat anti-rabbit IgG (H + L) | Thermo Fisher Scientific | Cat#A11037; RRID: AB_2534095 |
| Alexa Fluor 647 goat anti-rabbit IgG (H + L) | Thermo Fisher Scientific | Cat#A21245; RRID: AB_2535813 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Bacteroides thetaiotaomicron: VPI-5482 strain | Laboratory of Vanessa Sperandio | N/A |
| Bacteroides thetaiotaomicron: VPI-5482 strain; ΔtnaA | Laboratory of Vanessa Sperandio | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Metabolites and indole analogs, see Table S4 | Sigma-Aldrich; AA Blocks, Inc. | N/A |
| Sodium taurocholate hydrate | Sigma-Aldrich | Cat#86339; CAS: 345,909–26-4 |
| Y-27632 dihydrochloride ROCK inhibitor | Tocris Biosciences | Cat#1254; CAS: 129,830–38-2 |
| VAF347 | Sigma-Aldrich | Cat#182690 |
| FICZ | AA Blocks, Inc. | Cat#AA007VKI; CAS: 172,922–91-7 |
| Kynurenic acid | Sigma-Aldrich | Cat#K3375; CAS: 492–27-3 |
| L-tryptophan | Sigma-Aldrich | Cat#T8941; Cas: 73–22-3 |
| L-phenylalanine | Sigma-Aldrich | Cat#P5482; Cas: 63–91-2 |
| Ampicillin | Sigma-Aldrich | Cat#A0166; Cas: 69–52-3 |
| Neomycin | Sigma-Aldrich | Cat#N1876; Cas: 1405–10-3 |
| Vancomycin | Sigma-Aldrich | Cat#V2002; Cas: 1404–93-9 |
| Puromycin | Sigma-Aldrich | Cat#P8833; Cas: 58–58-2 |
| Formaldehyde | Polysciences | Cat#04018; Cas: 50–00-0 |
| 2-mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| Triton X-100 | Thermo Fisher Scientific | Cat#BP151–100 |
| Lipofectamine 3000 Transfection Reagent |
Thermo Fisher Scientific | Cat#L3000001 |
| Hoechst 33,342 | Thermo Fisher Scientific | Cat#H3570 |
| ProLong Glass Antifade Mountant | Thermo Fisher Scientific | Cat#P36984 |
| Rotenone | Sigma-Aldrich | Cat#R8875; Cas: 83–79-4 |
| Antimycin A | Sigma-Aldrich | Cat#A8674; Cas: 1397–94-0 |
| Oligomycin | Sigma-Aldrich | Cat#O4876; Cas: 1404–19-9 |
| Carbonyl cyanide m-chlorophenylhydrazone (CCCP) | Sigma-Aldrich | Cat#C2759; Cas: 555–60-2 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Click-iT Plus EdU Alexa Fluor 488 Imaging kit | Thermo Fisher Scientific | Cat#C10637 |
| QIAamp DNA Mini kit | QIAGEN | Cat#51306 |
| QIAamp Powerfecal Pro DNA kit | QIAGEN | Cat#51804 |
| RNeasy Mini kit | QIAGEN | Cat#74104 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat#G7571 |
| RQ1 RNase-free DNase | Promega | Cat#M6101 |
| DNA-free DNA removal kit | Thermo Fisher Scientific | Cat#AM1906 |
| SuperScript VILO cDNA synthesis kit | Thermo Fisher Scientific | Cat#11754050 |
| TB Green Advantage qPCR premix | Takara Bio | Cat#639676 |
| Platinum Taq DNA Polymerase, High Fidelity | Thermo Fisher Scientific | Cat#11304011 |
| Seahorse XF Cell Energy Phenotype Test kit | Agilent | Cat#103325–100 |
| Seahorse XF Real-time ATP Rate Assay kit | Agilent | Cat#103592–100 |
| Seahorse XF Cell Mito Stress Test kit | Agilent | Cat#103015–100 |
| MitoTracker® Red CMXRos | Thermo Fisher Scientific | Cat# M7512 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE185652 |
| 16S rRNA reads | This paper | ENA: PRJEB58007 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Mouse: NIH/3T3 | ATCC | CRL-1658 |
| Mouse: Ileal epithelial stem cells from C57BL/6 | Laboratory of Thaddeus Stappenbeck |
N/A |
| Human: Lenti-X 293T | Takara Bio | Cat#632180 |
| Human: HCT-8 | ATCC | CCL-244 |
| Human: HCT-8 AhR KO | This paper | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Cryptosporidium parvum AUCP-1 isolate | Laboratory of William Witola | N/A |
| Mouse: Infg KO (C57BL/6 background) | Jackson Laboratories | Cat#002287; RRID: IMSR_JAX:002,287 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primer: human CYP1A1 forward: ACATGCTGACCCTGGGAAAG |
PrimerBank | https://pga.mgh.harvard.edu/primerbank;PrimerBankID:189339226c2 |
| Primer: human CYP1A1 reverse: GGTGTGGAGCCAATTCGGAT |
PrimerBank | https://pga.mgh.harvard.edu/primerbank;PrimerBankID:189339226c2 |
| Primer: human AHRR forward: CTTAATGGCTTTGCTCTGGTCG |
61 | N/A |
| Primer: human AHRR reverse: TGCATTACATCCGTCTGATGGA |
61 | N/A |
| Primer: human GAPDH forward: TGAGTACGTCGTGGAGTCCA |
This paper | N/A |
| Primer: human GAPDH reverse: AGAGGGGGCAGAGATGATGA |
This paper | N/A |
| sgRNA for human AhR exon 1: TCACCTACGCCAGTCGCAAG | This paper | N/A |
| Primer: human AhR forward: GCACCATGAACAGCAGCAG |
This paper | N/A |
| Primer: human AhR reverse: TCCAAGTCCTCTGTCTCCCA |
This paper | N/A |
| Primer: C. parvum GAPDH forward: CGGATGGCCATACCTGTGAG |
34 | N/A |
| Primer: C. parvum GAPDH reverse: GAAGATGCGCTGGGAACAAC |
34 | N/A |
| Primer: mouse GAPDH forward: GCCATGAGTGGACCCTTCTT |
34 | N/A |
| Primer: mouse GAPDH reverse: GAAAACACGGGGGCAATGAG |
34 | N/A |
| Primer: V4 16S rRNA 515 forward: GTGCCAGCMGCCGCGGTAA |
71 | N/A |
| Primer: V4 16S rRNA 805 reverse: GACTACCAGGGTATCTAATCC |
71 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: lentiCRISPR v2 | 63 | Addgene Plasmid #52961 |
| Plasmid: lentiCRISPRv2-sgAhR | This paper | N/A |
| Plasmid: pMD2.g | Laboratory of Didier Trono | Addgene Plasmid #12259 |
| Plasmid: pMDLg/pRRE | 64 | Addgene Plasmid #12251 |
| Plasmid: pRSV-Rev | 64 | Addgene Plasmid #12253 |
|
| ||
| Software and algorithms | ||
|
| ||
| BioTek Gen5 v.3.02 | Agilent | https://www.agilent.com/en/product/cell-analysis/cell-imaging-microscopy/cell-imaging-microscopy-software/biotek-gen5-software-for-imagingmicroscopy-1623226 |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/ |
| QuantStudio Design & Analysis Software | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/global/forms/life-science/quantstudio-3–5-software.html |
| FIJI (ImageJ) | 65 | https://fiji.sc/ |
| Partek Flow | Partek, Inc. | https://www.partek.com/partek-flow/ |
| Enrichr | 70 | http://maayanlab.cloud/Enrichr/ |
| Wave v2.6.1 | Agilent | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolicanalysis/xf-software/seahorse-wavedesktop-software-740897 |
| R package: dada2 | 72 | https://benjjneb.github.io/dada2/ |
| R package: phyloseq | 74 | https://www.bioconductor.org/packages/release/bioc/html/phyloseq.html |
|
| ||
| Other | ||
|
| ||
| BioTek Cytation 3 cell imaging multimode reader | Agilent | https://www.biotek.com/products/detection-multi-mode-microplatereaders/cytation-hybrid-multimode-reader/ |
| Axioskop Mot Plus fluorescence microscope | Zeiss | N/A |
| QuantStudio 3 real-time PCR system | Applied Biosciences | Cat#A28137 |
| NovaSeq 6000 sequencing system | Illumina | https://www.illumina.com/systems/sequencing-platforms/novaseq.html |
| Seahorse XF96 Analyzer | Agilent | https://www.agilent.com/en/product/cell-analysis/real-time-cell-metabolicanalysis/xf-analyzers |