Abstract
An assay which combines the direct detection of Ureaplasma urealyticum with biovar determination was developed and applied to 618 urogenital specimens. U. urealyticum was detected by inhibitor-controlled PCR. A 429-bp fragment of the urease gene was amplified. The amplicons were labelled with digoxigenin during PCR. Biovar determination was performed by liquid hybridization with biotin-labelled biovar-specific probes, and the hybrids were detected with peroxidase-conjugated sheep anti-digoxigenin immunoglobulin G Fab fragments. Results of PCR and culture for 453 urogenital specimens from women and 105 urethral specimens from men could be compared. Among the specimens from women, 63% were PCR positive as well as culture positive, 0.9% were positive only by PCR, and 4% were positive only by culture. Among the specimens from men, 15% were PCR positive as well as culture positive, 1% were positive only by PCR, and 9% were positive only by culture. By using culture as the reference method, the PCR had a sensitivity of 94% and a specificity of 98% when applied to specimens from women and a sensitivity of 64% and a specificity of 99% when applied to specimens from men. Overall, 80% of the PCR-positive specimens contained biovar 1,13.5% contained biovar 2, and 6.5% contained both biovars.
The importance of ureaplasmas is obscured by the many asymptomatic persons from whom ureaplasmas can be isolated from urogenital specimens. However, male nongonococcal urethritis (4, 23–27) and chorioamnionitis may be caused by Ureaplasma urealyticum (10, 12). Patients suffering from clinical chorioamnionitis may deliver prematurely. U. urealyticum is isolated more often from preterm infants, stillborn fetuses, and spontaneously aborted fetuses than from fully developed neonates and fetuses from induced abortions. In premature neonates, particularly those with a birth weight of <1,500 g, U. urealyticum may cause respiratory distress syndrome, pneumonia, and meningitis (5).
In U. urealyticum 14 serotypes have been established by various serological methods (14, 18). On the basis of sensitivity to manganese, the polypeptide patterns, and percent homology by DNA-DNA hybridization experiments, the serotype standard strains can be divided into two groups or biovars (7, 15, 22). One biovar includes serotype standard strains 1, 3, 6, and 14 and is designated biovar 1. The other biovar comprises the remaining 10 of the established serotypes and is designated biovar 2 or T960; T960 is the name of the type strain of U. urealyticum. Biovar 1 has been called “parvo” because the genomes of these four serotype standard strains are much smaller than those of the serotype standard strains of biovar 2 (17).
Several investigators have suggested an association between certain serotypes and disease caused by the bacterium. Others have been unable to confirm that association (9, 16, 21, 30). An association between U. urealyticum and infection should probably be understood in terms of biovars rather than serotypes.
Robertson et al. (19) used minor differences found in the 16S rRNA gene sequences of the serotype standard strains 3 (biovar 1) and 8 (biovar 2) to provide primers for biovar-specific PCRs. The method is based on two separate PCRs with a common reverse primer and two different biovar-specific forward primers. Jacobs et al. (11) also used PCR for biovar determination. Their method is based on a single PCR with three different primers for the amplification of the 16S rRNA gene. Both methods described were applied to the serotype standard strains. Furthermore, Robertson et al. (19) applied their method to wild-type isolates which had previously been serotyped. The biovar determination was in agreement with that predicted by the serotyping results. Typing methods based on isolates are hampered by the fact that culture for U. urealyticum takes up to 7 days. We developed a method that allows the detection of ureaplasmas in clinical samples by PCR and biovar determination performed directly with the amplified samples within 1 day. The method is easy to apply to a large number of specimens in order to investigate a possible association between biovar and the role of ureaplasmas in disease.
MATERIALS AND METHODS
Strains.
U. urealyticum serotype standard strains 1 through 5, 6 (Pirillo), 7 (Cook), and 8 (T960) were obtained from F. Black, Institute of Medical Microbiology, University of Aarhus, Aarhus, Denmark. Serotype standard strains 9 (Vancouver) and 10 (Western) were obtained from Janet Robertson, Department of Medical Microbiology and Infectious Diseases, University of Alberta, Edmonton, Alberta, Canada. Serotype standard strains 11 through 14 were obtained from American Type Culture Collection (ATCC 33695, ATCC 33696, ATCC 33698, and ATCC 33697, respectively). For evaluation of the analytical specificity of the PCR, Ureaplasma diversum ATCC 43321, Ureaplasma gallorale ATCC 43346, Ureaplasma cati NCTC 11710, and strains of the following other urease-positive bacteria were used: Enterobacter cloacae, Proteus vulgaris, Proteus mirabilis, Yersinia enterocolitica, Klebsiella pneumoniae, Klebsiella oxytoca, Haemophilus influenzae, Haemophilus parainfluenzae, Morganella morganii, Streptococcus salivarius, Helicobacter pylori, and Bordetella parapertussis. These strains were obtained from the Department of Clinical Microbiology, Statens Serum Institut.
Clinical specimens.
A total of 618 randomly selected urogenital samples, 508 from women and 110 from men, received during the period from January to August 1996 with a request for culture for U. urealyticum, were analyzed by PCR. The only available information about the patients was age and sex. The median age for the men was 33 years (95% confidence interval, 31 to 35 years), and that for the women was 27 years (95% confidence interval, 26 to 28 years).
Culture of U. urealyticum and preparation of clinical specimens for PCR.
Specimens for culture were obtained on charcoal-impregnated cotton swabs and were inoculated first in 2 ml of Shepards 10C U+ medium. Immediately after expression of the contents of the swab, 50 μl of the broth was transferred to solid medium (U-agar plates) (20) and another 100 μl was mixed with 300 μl of a 20% slurry of Chelex-100 (Bio-Rad, Hercules, Calif.) in TE buffer (10 mM Tris [pH 8], 1 mM EDTA) and heated at 98°C for 10 min. The resin was pelleted by centrifugation at 20,000 × g for 10 min; 10 μl of the supernatant was subjected to PCR. All broth cultures were incubated at 37°C under atmospheric conditions, and all agar media were incubated at 37°C in 5% CO2. U. urealyticum was identified by colony formation on agar medium, and the quantitative estimate of growth was based on the change in color of a 10-fold dilution series of the inoculated broth. The estimation of U. urealyticum growth was expressed semiquantitatively as color-changing units (CCUs), and growth was considered to be negative or positive. The positive samples were assigned quantities of 101 to 105 CCUs. A CCU is the minimum inoculum required to produce growth that is indicated by a change in the color of the phenol red indicator.
Quantitation of urease-positive bacteria.
For evaluation of the analytical specificity the concentrations of U. diversum, U. cati, and U. gallorale were estimated by determination of the numbers of CCUs. The concentrations of the other urease-positive bacteria were determined by spectrophotometry at 600 nm (1 optical density (OD) unit corresponds to 1 × 109 bacteria/ml), and an equivalent of about 5 × 105 cells or CCUs was added to the PCR mixture in order to ensure that the amount exceeded the limit of detection.
PCR. (i) Primers.
The primers published by Blanchard et al. (3) were used for the PCR: primers U5 (forward; 5′-CAA TCT GCT CGT GAA GTA TTA C-3′) and U4 (reverse; 5′-ACG ACG TCC ATA AGC AAC T-3′). A 429-bp fragment of the urease gene was amplified with this primer set.
(ii) Construction of an internal process control for inhibition.
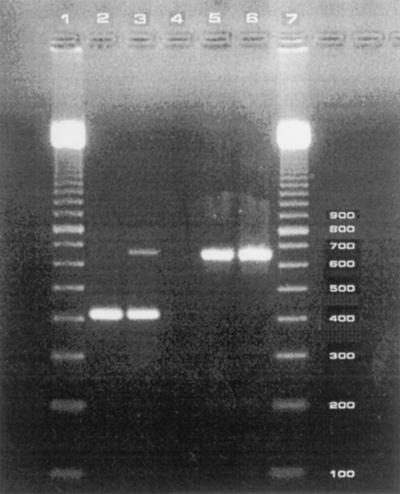
For the detection of the presence of Taq DNA polymerase inhibitors or suboptimal reaction conditions, an internal process control was constructed on the basis of the phage lambda genome. The lambda primers included the sequence of each of the primers U5 and U4 added to the 5′ ends of the corresponding lambda primers. Amplicons thus containing the binding sites of primers U5 and U4 were obtained by amplification of DNA from the lambda phage by the primers lambda-U5 and lambda-U4 (Table 1). After gel purification of the amplicons, the concentration producing no increase in the detection limit of purified U. urealyticum DNA was determined and was used as an internal process control in the assay. The electrophoretic position of the internal control is illustrated in Fig. 1. Preferential amplification of the shortest fragment occurred; consequently, the internal process control may be lacking in positive samples (Fig. 1).
TABLE 1.
Sequences of the oligonucleotide primers lambda-U5 and lambda-U4
| Primer | Sequencea | Use |
|---|---|---|
| Lamda-U5 | 5′-CAA TCT GCT CGT GAA GTA TTA Cct gac ggt ttc taa c-3′ | Forward |
| Lamda-U4 | 5′-ACG ACG TCC ATA AGC AAC Tga cat acg gaa ata g-3′ | Reverse |
The sequences of primers U5 and U4 are presented in capital letters.
FIG. 1.
Electrophoretic analysis of amplicons obtained by PCR with species-specific primers U5 and U4. Lower band (429 bp), specific for U. urealyticum; higher band (657 bp), internal process control. Lane 1, 100-bp marker; lane 2, positive sample lacking the internal process control; lane 3, positive sample with the internal process control; lane 4, inhibitory sample; lane 5, negative sample; lane 6, negative sample (water); lane 7, 100-bp marker.
(iii) Amplification.
The PCR was performed in a Hybaid Thermal reactor with SuperTaq DNA polymerase (HT Biotechnology Ltd., Cambridge, England). The amplicons were labelled with digoxigenin (DIG) during PCR by the addition of digoxigenin-11-dUTP (DIG-11-dUTP) to the master mixture. Ten microliters of the Chelex-100-treated specimens (see above) was subjected to PCR in a final volume of 100 μl of reaction buffer (50 mM KCl, 10 mM Tris hydrochloride [pH 9.0] at 25°C, 2.5 mM MgCl2, 0.01% gelatin, 0.1% Triton X-100 [HT Biotechnology]) containing each of the primers at a concentration of 0.4 μM; dATP, dGTP, and dCTP each at a concentration of 62.5 μM; 125 μM dUTP; 0.4 μM DIG-11-dUTP; and 10 μl of the appropriate dilution of the internal process control. A hot-start procedure (6) was used by adding 1.0 U of SuperTaq DNA polymerase in 10 μl of reaction buffer (withheld from the master mixture). The thermal cycling parameters consisted of 50 cycles each of 94°C for 30 s and 56°C for 1 min.
Two positive controls for each biovar and two negative controls (sterile water handled as a clinical specimen) were included in each PCR experiment. The two positive controls for each biovar contained 10 and 100 times more DNA, respectively, than the limit of detection of the biovar.
(iv) Gel electrophoresis of amplified samples.
Twenty microliters of the amplified sample was analyzed on a composite gel consisting of 1% SeaKem agarose and 1% NuSieve agarose (both from FMC, Rockland, Maine) in 0.5× TBE buffer (Tris-borate-EDTA) containing 5 μg of ethidium bromide per ml. The DNA was visualized by UV fluorescence.
(v) Limit of detection.
A 10-fold dilution to 10−6 of 24-h-old culture of serotype standard strains 6 and 8 in 10C U − broth was made in Shepard’s 10C U+ broth. From each dilution 100 μl of the broth was treated with 300 μl of Chelex-100 as described above, and 10 μl of the supernatant was subjected to PCR. The remaining ureaplasma cultures were then incubated at 37°C, and when no further color changes in the 10C U+ medium occurred, the numbers of CCUs for both strains were read.
(vi) Competition during amplification of mixtures of biovar 1 to biovar 2 DNA at various ratios.
Biovar 1 and biovar 2 DNAs were mixed at various ratios before amplification, keeping the total amount of DNA constant. The minimum amount of DNA of each biovar used in the mixture and amplified alone resulted in an OD of ≥4.0 with the homologous probe and an OD of ≤0.03 with the heterologous probe.
Liquid hybridization. (i) Sequencing.
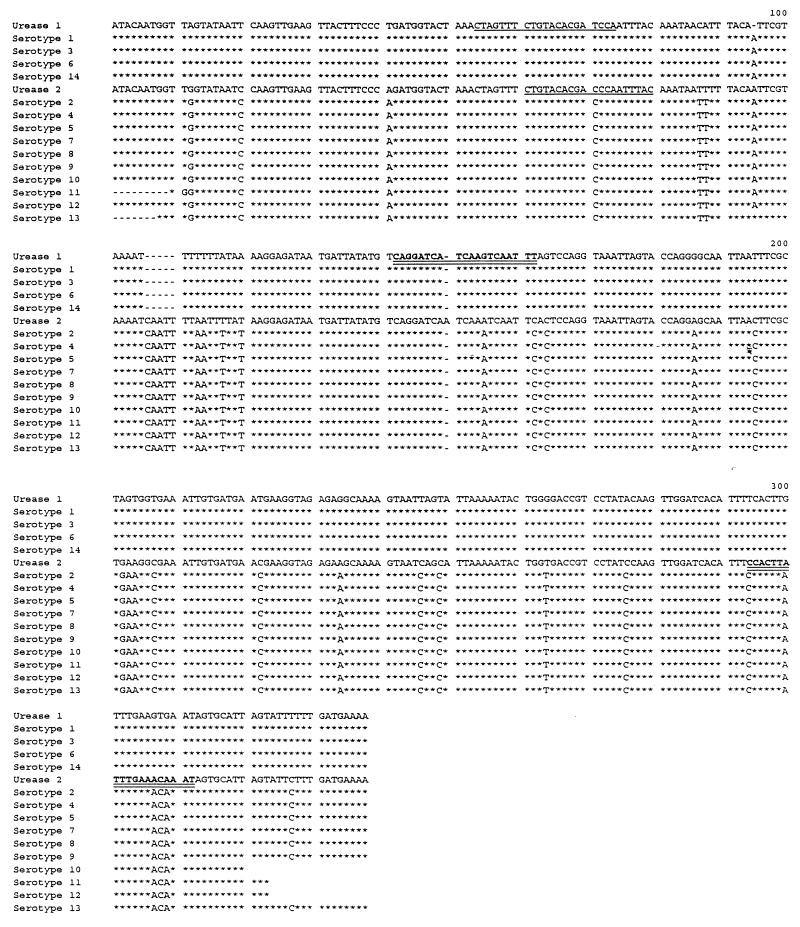
The urease gene amplicons from the 14 serotype standard strains were produced by PCR with the primers U5 and U4 and were subsequently sequenced by cycle sequencing. The Taq Dye Deoxy Terminator Cycle Sequencing kit from Applied Biosystems Inc. was used as described by the manufacturer. The sequencing products were analyzed on an Applied Biosystem 373A DNA sequencer. The alignment (see Fig. 3) showed differences between the two biovars, and different biovar-specific biotin-labelled oligonucleotides probes were synthesized. However, only one of the selected probes for each biovar showed biovar specificity by liquid hybridization. The sequence of the biovar 1-specific probe was 5′-AAA TTG ACT TGA TGA TCC TG-3′, and the sequence of the biovar 2-specific probe was 5′-ATT TGT TTC AAA TAA GTG G-3′. These are marked in Fig. 3.
FIG. 3.
Alignment of the sequenced products of the 429-bp fragment of the urease gene. The sequences of serotype standard strains 1, 3, 6, and 14 (biovar 1) are compared to the published sequence of serotype standard strain 1 (13) designated Urease 1. The sequences of serotype standard strains 2, 4, 5, 7, 8, 9, 10, 11, and 13 (biovar 2) are compared to the published sequence of serotype standard strain 8 (2) designated Urease 2. The sequences of the probes that are not useful are marked by underlines, and the sequences of the useful probes are marked by double underlines.
(ii) Optimization of the liquid hybridization conditions.
The assay was performed at hybridization temperatures in the range of 40 to 55°C with the biovar 1-specific probe and at hybridization temperatures in the range of 45 to 60°C with the biovar 2-specific probe at 5°C intervals. At the optimal hybridization temperature, probe concentrations in the range 0.67 to 40 nM were evaluated.
(iii) Biovar determination by liquid hybridization.
Five microliters of the amplified products was added to wells of a heat-stable microtiter plate (Titergene; Biozyme). Each well contained 100 μl of hybridization solution (100 mM NaCl, 10 mM Tris-HCl [pH 8], 5 mM EDTA) and biotinylated biovar-specific probe. The plates were sealed with tape and subjected to a 2-min denaturation step at 95°C and a 10-min hybridization step at 40°C with the biovar 1-specific probe at 1.3 nM and 50°C with the biovar 2-specific probe at 1 nM in a Hybaid Thermal reactor. The hybrids were transferred to a microtiter plate (MaxiSorp; Nunc, Roskilde, Denmark) which had been coated overnight with 5 μg of streptavidin (Sigma) per ml in carbonate buffer (pH 9.6) at 4°C and blocked for 15 min with 1% blocking reagent (Boehringer Mannheim) (1:10 dilution of 10% blocking reagent dissolved in maleic acid buffer as described by the manufacturer) in phosphate-buffered saline (PBS; pH 7.4) with 0.05% Tween 20 (PBST), and the plates were incubated for 30 min at 37°C. After capture, three washes with PBST were performed and the bound hybrids were detected by incubation at 37°C for 30 min with 0.6 U of peroxidase-conjugated sheep anti-digoxigenin Fab fragments (Boehringer Mannheim) per ml in 1% blocking buffer. After three additional washes with PBST, the hybrids were visualized with ortho-phenylenediamine (KemEnTec, Copenhagen, Denmark) in citrate buffer (pH 5.0). The reaction was stopped after 30 min by the addition of H2SO4, and the A490 was read in an enzyme-linked immunosorbent assay reader (Molecular Devices, Menlo Park, Calif.). The cutoff OD value for the positive samples was three times the mean OD value for the negative control samples.
RESULTS
Detection of U. urealyticum. (i) Limit of detection.
The limit of detection for strains belonging to biovar 1 was calculated to be 0.012 to 0.12 CCU/10 μl of supernatant, corresponding to 2.4 × 100 to 2.4 × 101 CCUs/2 ml, and the limit of detection for strains belonging to biovar 2 was calculated to be 1.2 × 100 to 1.2 × 101 CCU/10 μl of supernatant, corresponding to 2.4 × 102 to 2.4 × 103 CCUs/2 ml. The experiment was repeated twice, with the same results obtained each time.
For twofold serial dilutions of amplicons obtained from the biovar 1 and biovar 2 strains, respectively, the results of agarose gel electrophoresis were compared with the results of the hybridization assay. It was shown that the hybridization assay was four times more sensitive than gel electrophoresis for the detection of amplicons.
(ii) Analytical specificity.
When DNAs from urease-producing walled bacteria (see Materials and Methods) were subjected to PCR, no amplicons were observed. However, amplicons of the expected size were observed when DNAs from U. gallorale and U. cati were subjected to the PCR.
PCR detection of U. urealyticum in clinical samples and comparison with culture results.
A total of 618 specimens were examined. In the culture assay 55 samples were overgrown with other bacteria; 5 samples were inhibitory to the PCR. Thus, results for 558 specimens were available for evaluation of the sensitivities and specificities of the two methods.
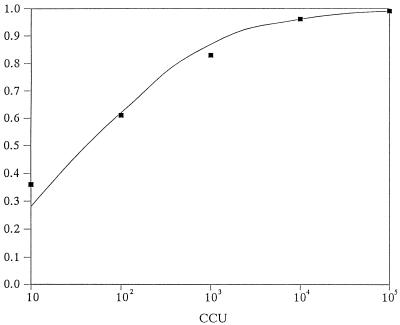
Among the specimens from women, 63% (287 of 453) were PCR positive as well as culture positive; 0.9% (4 of 453) were positive only by PCR, and 4% (20 of 453) were positive only by culture (Table 2). Among the specimens from men, 15% (16 of 105) were PCR positive as well as culture positive, 1% (1 of 105) were positive only by PCR, and 9% (9 of 105) were positive only by culture (Table 3). When culture was used as the reference method, the PCR method had a sensitivity of 94% (287 of 307) and a specificity of 98% (142 of 146) when applied to specimens from women (Table 2) and a sensitivity of 64% (16 of 25) and a specificity of 99% (79 of 80) when applied to specimens from men (Table 3). The PCR method was significantly more sensitive for the detection of U. urealyticum in specimens from women than in those from men. The predictive value of a positive test result was 99% (287 of 291) for specimens from women (Table 2) and 94% (16 of 17) for specimens from men (Table 3). The predictive value of a negative test result was 88% (142 of 162) for specimens from women (Table 2) and 90% (79 of 88) for specimens from men (Table 3). The sensitivity of PCR for specimens with different concentrations of U. urealyticum is as follows for women: 101 CCUs, 57%; 102 CCUs, 67%; 103 CCUs, 86%; 104 CCUs, 97%; and ≥105 CCUs, 99%. The sensitivity of PCR for specimens with different concentrations of U. urealyticum is as follows for men: 101 CCUs, 0%; 102 CCUs, 25%; 103 CCUs, 50%; 104 CCUs, 100%; and ≥105 CCUs, 100%. However, 48% (12 of 25) of the culture-positive specimens from men had ≤103 CCUs (Table 3), whereas 23% (71 of 307) of the culture-positive specimens from women had ≤103 CCUs (Table 2). On average, the culture-positive and PCR-negative specimens had fewer CCUs than the culture-positive and PCR-positive specimens (Table 2 and Table 3). Figure 2 illustrates the observed and estimated PCR-positive fractions as a function of the numbers of CCUs.
TABLE 2.
Detection of U. urealyticum in urogenital specimens from women by culture and PCR
| PCR result | No. of specimens
|
No. of culture-positive specimens with the following no. of CCUs:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total | Culture negative | Culture positive | 101 | 102 | 103 | 104 | 105 | |
| Negative | 162 | 142 | 20 | 3 | 9 | 5 | 2 | 1 |
| Positive | 291 | 4 | 287 | 4 | 18 | 32 | 56 | 177 |
| Total | 453 | 146 | 307 | 7 | 27 | 37 | 58 | 178 |
TABLE 3.
Detection of U. urealyticum in urethral specimens from men by culture and PCR
| PCR result | No. of specimens
|
No. of culture-positive specimens with the following no. of CCUs:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total | Culture negative | Culture positive | 101 | 102 | 103 | 104 | 105 | |
| Negative | 88 | 79 | 9 | 4 | 3 | 2 | 0 | 0 |
| Positive | 17 | 1 | 16 | 0 | 1 | 2 | 5 | 8 |
| Total | 105 | 80 | 25 | 4 | 4 | 4 | 5 | 8 |
FIG. 2.
Observed and estimated PCR-positive fraction as a function of the numbers of CCUs for the specimens.
Biovar determination. (i) Sequencing.
The alignment (Fig. 3) showed differences between the two biovars, and different biovar-specific biotin-labelled oligonucleotides probes were synthesized. However, only one of the selected probes for each biovar showed biovar specificity by liquid hybridization. The sequence of the biovar 1-specific probe was 5′-AAA TTG ACT TGA TGA TCC TG-3′, and the sequence of the biovar 2-specific probe was 5′-ATT TGT TTC AAA TAA GTG G-3′ (Fig. 3). The sequences of the relevant parts of the urease gene of serotype standard strain 1 and serotype standard strain 8 published by Neyrolles et al. (13) and Blanchard et al. (2) are included in Fig. 3. The useful and the useless probes are indicated in Fig. 1.
(ii) Optimization of the liquid hybridization conditions.
The optimal hybridization conditions with regard to temperature and probe concentration were determined. Under the conditions investigated, the optimal hybridization temperatures were 55°C with the biovar 1-specific probe and 45°C with the biovar 2-specific probe, and the optimal concentrations of the probes were 1.3 nM with the biovar 1-specific probe and 1 nM with the biovar 2-specific probe.
(iii) Analytical specificity.
The amplicons produced when DNAs from U. cati and U. gallorale were subjected to the PCR did not react with the biovar-specific probes.
(iv) Competition during amplification.
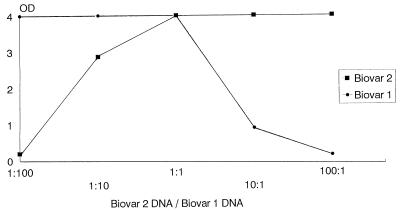
The ability to detect biovar 1 and biovar 2 by the PCR declined when the amount of heterologous DNA added to the mixture was increased (Fig. 4). The biovars could be detected with the homologous probe even when a 100-fold excess of the heterologous DNA was added to the reaction mixture. However, there was a decline in the OD value to 0.3 for the homologous probe.
FIG. 4.
Competition during amplification of various ratios of biovar 1 DNA and biovar 2 DNA. Biovar 1 and biovar 2 DNAs were mixed at various ratios before amplification, but the total amount of DNA was kept constant.
(v) Clinical samples.
The result of a liquid hybridization assay is illustrated in Fig. 5. The hybridization assay did not detect ureaplasma DNA in any specimens negative by gel electrophoresis. Among the samples received with a request for culture for U. urealyticum, 80% (246 of 308) of the positive specimens contained biovar 1, 13.5% (41 of 308) contained biovar 2, and 6.5% (21 of 308) contained both biovars. There was no significant difference in the biovar distribution among the specimens from men and women. However, only a small number of positive specimens from men were available for the analysis.
FIG. 5.
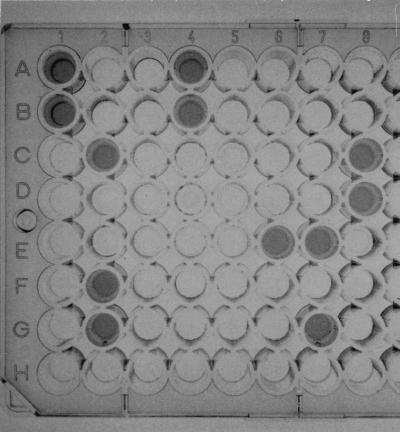
Liquid hybridization assay for assignment of U. urealyticum biovars, with the following results: hybridization with the biovar 1-specific probe, 1A to 3H and 4A to 4F; positive specimens containing biovar 1, 1A, 1B, 2C, 2F, and 2G; positive controls of biovar 1, 4A and 4B; positive controls of biovar 2, 4C and 4D; negative controls, 4E and 4F; hybridization with the biovar 2-specific probe, 5A to 7H and 8A to 8F. Positive specimens containing biovar 2, 6E, 7E, and 7G; positive controls of biovar 1, 8A and 8B; positive controls of biovar 2, 8C and 8D; negative controls, 8E and 8F.
DISCUSSION
Development of the assay.
Detection of PCR products by liquid hybridization has several advantages. The method yields objective results compared to those obtained by gel electrophoresis, and the hybridization step adds specificity and may lower the limit of detection. The method developed allows detection of ureaplasmas in clinical samples by PCR and biovar determination performed directly with the amplified samples within 1 day.
In the commercially available PCR enzyme-linked immunosorbent assay (DIG detection) from Boehringer Mannheim a ratio of labelled/unlabelled nucleotides of 1:19 is used. However, labelled dUTP is very expensive, and experience with other PCRs has shown that the limit of detection remained satisfactory even when the ratio was decreased to 1:312. Therefore, this concentration was chosen.
When the hybridization temperature exceeded the optimum for the biovar 1-specific probe (55°C) and the biovar 2-specific probe (45°C), the hybrids remained denatured, resulting in a decline in the OD value.
Other liquid hybridization assays in our laboratory were run at 50 and 40°C. From a practical point of view, these temperatures were chosen since unspecific binding was not observed.
We showed that with detection of U. urealyticum by PCR and biovar determination by liquid hybridization it was possible to detect a mixed infection (Fig. 4). However, competition during amplification did occur. When the ratio of biovar 1 DNA/biovar 2 DNA exceeded 1, the OD value for the biovar 2-specific probe declined, and when the ratio fell below 1, the OD value for the biovar 1-specific probe declined. However, the assay was sufficiently robust to detect both biovars in the presence of 100-fold excess of one of the biovars.
The diagnostic PCR.
The analytical specificities of primers U4 and U5 were high. They allowed the amplification of a product of the expected size from ureaplasma strains but not from other urease-producing bacteria. However, when DNAs from U. gallorale and U. cati were subjected to the PCR, amplicons of the expected size were observed. None of the amplicons hybridized with the biovar-specific probes, and since U. gallorale and U. cati are believed not to be pathogenic for humans, it was considered to be without importance for investigation of clinical specimens. When the assay was applied to clinical specimens and culture was used as the “gold standard,” the sensitivity was calculated to be 94% for specimens from women and 64% for specimens from men. The hybridization assay did not identify any additional U. urealyticum-positive specimens, although this detection method was found to be at least four times more sensitive than gel electrophoresis. The low sensitivity for specimens from men was found to be correlated to the smaller amount of microorganisms present in these samples than that present in samples from women. Blanchard et al. (3) observed a sensitivity of 92% when they subjected specimens from the urogenital tract to this PCR.
Abele-Horn et al. (1) have also evaluated the diagnostic PCR of Blanchard et al. (3). The efficiency of the PCR was compared with that of culture for detection of U. urealyticum in 468 clinical specimens (gynecological specimens, urological specimens, and specimens from newborn infants). The sensitivity of PCR versus culture was 95%. Willoughby et al. (29) used other primers to amplify a 459-bp fragment from nucleotides 542 to 999 of the ureasegene of U. urealyticum for diagnostic purposes. Teng et al. (28) evaluated this PCR. By culture, 5 of the 50 specimens were positive for U. urealyticum and 4 were doubtfully positive (growth in liquid medium but no positive identification on solid medium). PCR showed positive test results for 12 specimens which included the positive and doubtfully positive culture specimens. These results indicated that this PCR was more sensitive than culture for detection of U. urealyticum but could be explained by the less optimal culturing technique (medium, lack of staining of the colonies by manganese sulfate, and handling of the specimens between collection and the initiation of culture).
Biovar determination.
Among the samples received with a request for culture for U. urealyticum, 80% of the positive specimens contained biovar 1, 13.5% contained biovar 2, and 6.5% contained both biovars. Clerc et al. (8) applied the PCRs of Robertson et al. (19) to 350 noncloned U. urealyticum isolates obtained from 1,100 clinical specimens. The specimens were urethral and endocervical swab, semen, urine, and a few endotracheal swab specimens. There was no information about the patients. Of 350 U. urealyticum-positive specimens, 76% contained the parvo biovar (biovar 1), 17% contained the T960 biovar (biovar 2), and 7% contained both biovars. This is the same biovar distribution found in the present study for specimens received with a request for culture for U. urealyticum.
Robertson et al. (19) and Jacobs et al. (11) used PCR to amplify the 16S rRNA genes of ureaplasma isolates. Both methods were applied to the serotype standard strains. Furthermore, Robertson et al. (19) applied their method to wild-type isolates which had previously been serotyped. The biovar determination was in agreement with that predicted by the serotyping results. Typing methods based on isolates of U. urealyticum are hampered by the fact that culture of U. urealyticum takes up to 7 days. We developed a method that allows the detection of ureaplasmas in clinical samples by PCR and biovar determination performed directly with the amplified samples within 1 day.
ACKNOWLEDGMENT
We thank Henrik Wachman, Statistical Department, Statens Serum Institut, for statistical analyses.
REFERENCES
- 1.Abele-Horn M, Wolf C, Dressel P, Pfaff F, Ruckdeschel G. Polymerase chain reaction versus culture for detection of Ureaplasma urealyticum and Mycoplasma hominis in the urogenital tract of adults and the respiratory tract of newborns. Eur J Clin Microbiol Infect Dis. 1996;7:595–598. doi: 10.1007/BF01709369. [DOI] [PubMed] [Google Scholar]
- 2.Blanchard A. Urea plasma urealyticum urease genes; use of a UGA tryptophan codon. Mol Microbiol. 1990;4:669–676. doi: 10.1111/j.1365-2958.1990.tb00636.x. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard A, Hentschel J, Duffy L, Baldus K, Cassell G H. Detection of Ureaplasma urealyticum by polymerase chain reaction in the urogenital tract of adults, in amniotic fluid, and the respiratory tract of newborns. Clin Infect Dis. 1993;17:S148–S153. doi: 10.1093/clinids/17.supplement_1.s148. [DOI] [PubMed] [Google Scholar]
- 4.Brown M B, Cassell G H, Taylor-Robinson D, Shepard M C. Measurement of antibody to Ureaplasma urealyticum by an enzyme-linked immunosorbent assay and detection of antibody responses in patients with nongonococcal urethritis. J Clin Microbiol. 1983;17:288–295. doi: 10.1128/jcm.17.2.288-295.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassell G H, Waites K B, Watson H L, Crouse D T, Harasawa R. Ureaplasma urealyticum intrauterine infection: role in prematurity and disease in newborns. Clin Microbiol Rev. 1993;6:69–87. doi: 10.1128/cmr.6.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou Q, Russell M, Birch D E, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen C, Black F T, Freundt E A. Hybridization experiments with deoxyribonucleic acids from Ureaplasma urealyticum serovars I to VIII. Int J Syst Bacteriol. 1981;31:259–262. [Google Scholar]
- 8.Clerc M T, Renaudin H, de Barbeyrac B, Robertson J A, Stemke G W, Bébéar C. Proceedings of the 10th International Congress of the International Organization for Microplasmatology, Bordeaux, France. 1994. PCR, a useful tool for biovar determination in Ureaplasma urealyticum; p. 326. [Google Scholar]
- 9.Cracea E, Constantinescu S, Lazar M. Serotypes of Ureaplasma urealyticum isolated from patients with nongonococcal urethritis and gonorrhea and from asymptomatic urethral carriers. Sex Transm Dis. 1985;12:219–223. doi: 10.1097/00007435-198510000-00010. [DOI] [PubMed] [Google Scholar]
- 10.Gray D J, Robinson H B, Malone J, Thomson R B. Adverse outcome in pregnancy following amniotic fluid isolation of Ureaplasma urealyticum. Prenat Diagn. 1992;12:111–117. doi: 10.1002/pd.1970120206. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs E, Vonski M, Stemke G W, Robertson J A. Identification of ureaplasma biotypes. Med Microbiol Lett. 1994;3:31–35. [Google Scholar]
- 12.Kundsin R B, Leviton A, Allred E N, Poulin S A. Ureaplasma urealyticum infection of the placenta in pregnancies that ended prematurely. Obstet Gynecol. 1996;87:122–127. doi: 10.1016/0029-7844(95)00376-2. [DOI] [PubMed] [Google Scholar]
- 13.Neyrolles O, Ferris S, Behbahani N, Montagnier L, Blanchard A. Organization of Ureaplasma urealyticum gene cluster and expression in a suppressor strain of Escherichia coli. J Bacteriol. 1996;178:647–655. doi: 10.1128/jb.178.3.647-655.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn P A, Arshoff L U, Li H C. Serotyping of Ureaplasma urealyticum by immunoperoxidase assay. J Clin Microbiol. 1981;13:670–676. doi: 10.1128/jcm.13.4.670-676.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson J A, Chen M H. Effects of manganese on the growth and morphology of Ureaplasma urealyticum. J Clin Microbiol. 1984;19:857–864. doi: 10.1128/jcm.19.6.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson J A, Honore L H, Stemke G W. Serotypes of Ureaplasma urealyticum in spontaneous abortion. Pediatr Infect Dis. 1986;5:S270–S272. doi: 10.1097/00006454-198611010-00014. [DOI] [PubMed] [Google Scholar]
- 17.Robertson J A, Pyle L E, Stemke G W, Finch L R. Human ureaplasmas show diverse genome sizes by pulsed-field electrophoresis. Nucleic Acids Res. 1990;18:1451–1455. doi: 10.1093/nar/18.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson J A, Stemke G W. Expanded serotyping scheme for Ureaplasma urealyticum strains isolated from humans. J Clin Microbiol. 1982;15:873–878. doi: 10.1128/jcm.15.5.873-878.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson J A, Vekris A, Bébéar C, Stemke G W. Polymerase chain reaction using 16S rRNA gene sequences distinguishes the two biovars of Ureaplasma urealyticum. J Clin Microbiol. 1993;31:824–830. doi: 10.1128/jcm.31.4.824-830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shepard M C. Culture media for ureaplasmas. In: Razin S, Tully J G, editors. Methods in mycoplasmology. London, Untied Kingdom: Academic Press; 1983. pp. 137–146. [Google Scholar]
- 21.Shepard M C, Lunceford C D. Serological typing of Ureaplasma urealyticum isolates from urethritis patients by an agar growth inhibition method. J Clin Microbiol. 1978;8:566–574. doi: 10.1128/jcm.8.5.566-574.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swenson C E, Van Hamont J, Dunbar B S. Specific protein differences among strains of Ureaplasma urealyticum as determined by two-dimensional gel electrophoresis and a sensitive silver strain. Int J Syst Bacteriol. 1983;33:417–421. [Google Scholar]
- 23.Taylor-Robinson D. Possible role of ureaplasmas in nongonococcal urethritis. In: Hobson D, Holmes K K, editors. Nongonococcal urethritis. Washington, D.C: American Society for Microbiology; 1977. pp. 30–37. [Google Scholar]
- 24.Taylor-Robinson D. The history of nongonococcal urethritis. Sex Transm Dis. 1996;23:86–91. doi: 10.1097/00007435-199601000-00020. [DOI] [PubMed] [Google Scholar]
- 25.Taylor-Robinson D, Csonka G W. Laboratory and clinical aspects of mycoplasma infection of the human genitourinary tract. In: Morton R S, Harris J R W, editors. Recent advances in sexually transmitted diseases. Edinburgh, United Kingdom: Churchill Livingstone; 1981. pp. 151–185. [Google Scholar]
- 26.Taylor-Robinson D, Csonka G W, Prentice M J. Human intra-urethral inoculation of ureaplasmas. Q J Med. 1977;46:309–326. [PubMed] [Google Scholar]
- 27.Taylor-Robinson D, Purcell R H, London W T, Sly D L. Urethral infection of chimpanzees by Ureaplasma urealyticum. J Med Microbiol. 1978;11:197–201. doi: 10.1099/00222615-11-2-197. [DOI] [PubMed] [Google Scholar]
- 28.Teng K, Li M, Yu W, Li H, Shen D, Liu D. Comparison of PCR with culture for detection of Ureaplasma urealyticum in clinical samples from patients with urogenital infections. J Clin Microbiol. 1994;32:2232–2234. doi: 10.1128/jcm.32.9.2232-2234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willoughby J J, Russell W C, Thirkell D, Burdon M G. Isolation and detection of urease genes in Ureaplasma urealyticum. Infect Immun. 1991;59:2463–2469. doi: 10.1128/iai.59.7.2463-2469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng X, Watson H L, Waites K B, Cassell G H. Serotype diversity and antigen variation among invasive isolates of Ureaplasma urealyticum from neonates. Infect Immun. 1992;60:3472–3474. doi: 10.1128/iai.60.8.3472-3474.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]