Abstract
Gnathostome adaptive immunity is defined by the antigen receptors, immunoglobulins (Ig) and T cell receptors (TCR), and the major histocompatibility complex (MHC). Cartilaginous fish are the oldest vertebrates with these adaptive hallmarks. We and others have unearthed non-rearranging antigen receptor-like genes in several vertebrates, some of them encoded in the MHC or in MHC paralogous regions. One of these genes, named UrIg, was detected in the class III region of the shark MHC that encodes a protein with typical Variable (V) and Constant (C) domains like those found in conventional Ig and TCR. As no transmembrane region was detected in gene models or cDNAs the protein does not appear to act as a receptor. Unlike some other shark Ig genes, the UrIg V region shows no evidence of RAG-mediated rearrangement, and thus is likely related to other V genes that predated the invasion of the RAG transposon. The UrIg gene is present in all elasmobranchs and evolves conservatively, unlike Ig and TCR. Also, unlike Ig/TCR, the gene is not expressed in secondary lymphoid tissues, but mainly in the liver. Recombinant forms of the molecule form disulfide-linked homodimers, which is the form also detected in many shark tissues by western blotting. mAbs specific for UrIg identify the protein in the extracellular matrix of several shark tissues by immunohistochemistry. We propose that UrIg is related to the V gene invaded by the RAG transposon, consistent with the speculation of emergence of Ig/TCR within the MHC or proto-MHC.
Keywords: MHC, Immunoglobulin, Liver, Extracellular Matrix, Shark
Introduction
Immunoglobulins (Ig) and T cell receptors (TCR) are the antigen receptors that coordinate adaptive immunity in jawed vertebrates (Gnathostomes) (1). Ig/TCR are generated by RAG-dependent gene rearrangement events during lymphocyte ontogeny. Igs and most γ/δ TCR recognize free antigen while α/β TCR recognize peptide or lipid antigens bound in the grooves of MHC class I and class II proteins (2). It is universally agreed that Ig and TCR were generated from a common ancestral antigen receptor (3–5), but speculation abounds on the nature of such a common ancestor since all living Gnathostomes have all three types of Ig-Superfamily (IgSF)-based antigen receptors and jawless fish and invertebrates lack such receptors.
Cartilaginous fishes are in the oldest vertebrate group with rearranging antigen receptors and the MHC (6). Other hallmarks of adaptive immunity such as primary (thymus) and secondary (spleen) lymphoid tissues, activation-induced cytidine deaminase (AID)-driven somatic hypermutation, diverse cytokine and chemokine networks, etc. also arose in cartilaginous fish. The only glaring omissions of shark adaptive immunity are class-switch recombination (classically arose in amphibians (7) although sharks also have a form of switch (8)), germinal centers (arose in reptiles/birds (9)), and lymph nodes (arose in mammals) (10). However, sharks have some primordial features of their adaptive immune system that were lost in most higher vertebrates (and all primates) such as the cluster (split) organization of Ig heavy (H) chain genes (11), linkage of beta2-microglobulin (β2m) to the MHC (12), somatic hypermutation of TCRα genes during thymic development (13), “germline-joining” of V, D, and J segments that can generate functional Ig genes in the genome (14–18), single-V domain-containing H chains that make dimers but do not associate with light (L) chains (15, 19, 20), and usage of IgH V regions in TCR (21, 22). Such ancestral features highlight the shark as an attractive model for immune studies.
Ig/TCR genes arose by an invasion of an IgSF exon by the RAG transposon early in Gnathostome evolution (23–25). The IgSF domain used by Ig/TCR is a so-called “VJ domain” in which the last (G) strand of the molecule bears a Gly-X-Gly motif in the N-terminal part of the strand involved in dimerization and a less conserved V(l)TVT motif in the C-terminal part (26, 27). There are not many genes besides Ig/TCR with this motif in the genome, most of them probably derived from an ancestral, non-rearranging VJ domain. A recent review compiled all of the reported animal VJ domains, most of which are involved in immunity (27). In addition, the C IgSF domains used by Ig/TCR are likewise special, so-called C1 domains that are the most compact IgSF domains also with distinguishing motifs: FYP in the B strand and C-V-H in the F strand (26, 28). Besides Ig/TCR, C1 domains are found in MHC class I, MHC class II, b2m and a few other molecules involved in immunity (29). Interestingly, several VJ and C1 domain genes are encoded in the MHC or in MHC paralogous regions, prompting the idea from several groups (including ours) that Ig/TCR and class I/II arose from a proto-MHC region (28, 30–32).
While examining the shark MHC we uncovered a non-rearranging Ig-like gene in the class III region that converges on mammalian IgG, near where other VJ exons are found (33). This was an unexpected result and not one based on hypothesis testing, but nevertheless consistent with an MHC origin of antigen receptors. Herein we report the basic features of the gene and molecule, which likely serves an innate-immune or structural role in sharks. We speculate on this molecule’s evolutionary significance in relation to antigen receptor emergence.
Materials and Methods
Animals
Wild-caught nurse sharks (Ginglymostoma cirratum) were maintained in artificial seawater at approx. 28°C in indoor tanks at the Institute of Marine and Environmental Technology, Baltimore, USA. Animals were anesthetized with MS222 (0.1%) before bleeds were harvested from the caudal vein with 1000 U/ml heparin reconstituted in shark-modified Phosphate-Buffered Saline (SPBS), then spun at 300g for 10 min to isolate blood plasma and buffy coat. We harvested major organs in SPBS and then euthanized animals according to protocol. All procedures were conducted in accordance with University of Maryland School of Medicine Institutional Animal Care and Use Committee (IACUC) protocols.
Database searches
While searching for MHC genes in databases from various cartilaginous fish genomes at the NCBI website (ncbi.nlm.nih.gov), we discovered the UrIg gene. The presence of UrIg was further confirmed by BLASTp, tblstn searches against various vertebrate species including the nurse shark transcriptomics database.
Phylogenetic tree analysis
The UrIg variable (V) and constant (C) IgSF domains were separately aligned against different sets of IgSF-containing genes (e.g., Ig, TCR) based on the predicted evolutionary origin and domain similarities using Clustal W. Phylogenetic trees were then constructed using the bootstrapping Neighbor-Joining method (34) with 500 runs.
rtPCR
First strand complementary DNA (cDNA) was made from 500ng of total RNA from various nurse shark “NJ” tissue using SuperScript IV First-Strand Synthesis kit (Invitrogen) following manufacturers protocol. Reverse transcription PCR (rtPCR) was performed using Go Taq Master Mix (Promega) with 35 cycles of denature (at 95°C) for 45 sec, annealing (see below for each gene) for 45 sec, and extension (at 72°C) for 60 sec, following 2 min of hot start at 95°C, and flowed by 5 min final extension at 72°C. The primers used to examine the gene expression wee: UrIg (C1-C3 IgSF domains) 5’- CCGGAAGAACATCTCGCTGCT-3’ & 5’- CCGGAAGAACATCTCGCTGCT-3’ (at 58°C annealing temperature); nucleoside-diphosphate kinase (NDPK: control) 5’-AACAAGGAACGAACCTTC-3’ & 5’-TCACTCATAGATCCAGTC-3’ (at 50°C annealing temperature). The PCR amplicons were visualized on 1% agarose gel.
UrIg recombinant expression and purification
The 4-domain UrIg molecule (V-C1-C2-C3) was cloned into the phCMV3 vector for mammalian expression, with an N-terminal murine Igκ signal peptide and C-terminal His tag. Protein was produced by transient transfection of expi293F cells (ThermoFisher) following the manufacturer’s standard protocols. Protein was purified from the medium by affinity chromatography with Ni Sepharose excel histidine-tagged protein purification resin (Cyvita). After binding, resin was washed with 20mM Tris, pH7.5, and then with 500 mM imidazole, 20mM Tris, 500mM NaCl, pH7.5 for elution. Protein was then dialyzed overnight at 4°C against 1X PBS/1mM EDTA. After dialysis NDSB-201 (MilliporeSigma) was added to a final concentration of 200mM and protein concentrated to approximately 1 mg/ml before size-exclusion chromatography on a Superdex 200 16/60 column (Cytiva) with running buffer Dulbecco’s PBS. Peak fractions were pooled, and samples were then concentrated and examined by SDS-PAGE, and for immunization of mice according to IACUC protocols.
Monoclonal antibody (mAb) production
mAbs were produced as previously described (35). Briefly, mice were immunized subcutaneously to 50 ug of UrIg protein emulsified in Incomplete Freund’s Adjuvant (IFA) and boosted three weeks later with 50 ug in IFA. Three days later, splenocytes were fused with myeloma line X63. Two weeks later, hybridomas were tested for reactivity by ELISA on 96 well plates coated with recombinant UrIg (1 ug/ml). Positive clones were expanded, and supernatants used for immunohistochemistry (IHC) and western blotting.
Cloning and production of trimeric CNA35
Collagen adhesin (CNA) 35 synthetic DNA with optimized codons for E. coli (Integrated DNA Technologies) was cloned in frame with the human collagen XVIII trimerization domain sequence (36) into pET23-His-Trx-thr vector using BamHI and EcoRI sites (37). The resultant construct encoded a thrombin-cleavable, His-tagged thioredoxin domain added to the N-terminus of collagen XVIII trimerization domain (18TD) fused with CNA35. The plasmid sequence was verified by Sanger sequencing.
The protein was expressed in BL21(DE3) strain of E. coli using 1mM IPTG induction for 3 hrs. at 37°C. Cells were collected by centrifugation and disrupted by ultrasonication in 20 mM TrisHCl pH8 on ice. Insoluble material was removed by centrifugation at 10,000 g at 4°C for 30 mins and supernatant was adjusted to include 50 mM sodium phosphate pH 8, 150 mM NaCl, and 5 mM imidazole. The His-tagged chimera protein was affinity purified using Ni-NTA column (Qiagen) by elution using the same buffer supplemented with increasing imidazole concentrations. The fractions with the protein of interest were pooled and dialyzed against 50 mM TrisHCl, 100 mM NaCl, pH8.3. 10mM CaCl2 and 0.01% sodium azide were added to the dialyzed protein sample to facilitate thrombin cleavage and prevent bacterial growth, respectively. Every week 4 U/ml of thrombin was added for a total time of 4 weeks to complete the thrombin cleavage at room temperature. His-tagged thioredoxin was separated from (18TD-CNA35)3 using Ni-NTA column (Qiagen), but this time the trimeric CNA35 was found in the flow through. The (18TD-CNA35)3 was dialyzed against 20 mM TrisHCl, pH8, loaded onto Q-Sepharose column (Cytiva) and eluted at ~10–20 mM of NaCl gradient. The fractions were pooled and run over the size-exclusion chromatography using the Superdex S200 Increase column (Cytiva) equilibrated with the coupling buffer (100 mM sodium carbonate, pH 8.8).
The conjugation with Alexa Fluor™ 488 NHS Ester (Thermo Fisher Scientific) was performed at 2 mg/ml protein concentration at 4 °C overnight using 1 mg dye/10 mg protein. The labeled protein was separated from unreacted dye by desalting into PBS buffer. The stock solution was adjusted to 1 mg/ml. The working dilution for staining was 1:1,000.
Immunohistochemistry
Fresh frozen nurse shark tissues in Tissue-Tek O.C.T. compound (Sakura) were cryo-sectioned at 7μm thickness and briefly fixed in cold acetone. Slides could be stored at −80°C for several days. Slides were thawed, rehydrated in PBS for 5 min, and made permeable in PBS supplemented with 0.05% Tween 20 (PBS-T). Slides were blocked with 10% fetal calf serum (FCS) in PBS for 45 min at 4°C, then incubated with mAb sup for 5 min at 4°C. After washing with PBS-T, slides were incubated with the secondary antibody, goat anti-mouse IgG-Alexa 488 for 45 min at 4°C, washed, and mounted with ProLong gold with DAPI mounting media. Images were taken with Nikon Eclipse E800 and analyzed with Mosaic software.
For double immunofluorescence staining with CNA and mAb, kidney sections (7μm) were mounted on glass slides, air dried for 15–30 minutes at room temperature and fixed in acetone at −20°C for 10 minutes. Sections were washed for 5 minutes with 50 mM Tris 150mM NaCl 0.1% Tween-20 buffer (TBS-T). Blocking was performed with 10% goat serum for one hour (Invitrogen 50062Z) followed by incubation with primary anti-UrIg mAb O-1 (1:500 dilution in 1% goat serum TBS-T, Fig. 7) overnight in the cold room. Following 3×15 minutes washes with TBS-T, secondary antibody conjugated with Alexa568 and CNA conjugated with Alexa488 were incubated on sections for 1.5 hours at room temperature. Sections were then washed and mounted with anti-fade mounting solution with DAPI. Images were taken with Nikon Eclipse Ti microscope and analyzed with GIMP.
Fig. 7. UrIg monoclonal antibody specificity.
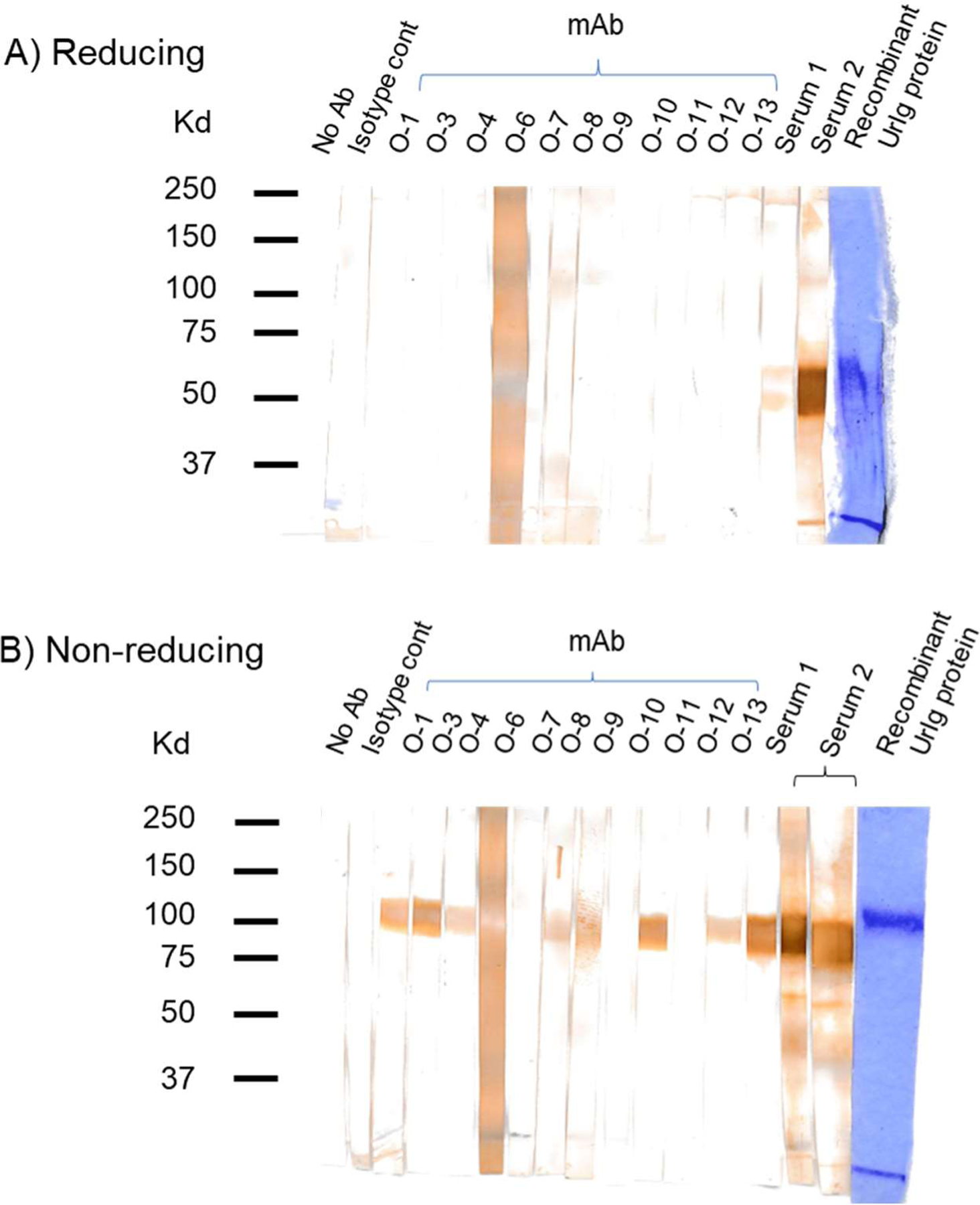
Western blotting test of UrIg-specific mAbs and antisera tested on recombinant UrIg protein under reducing (A) and nonreducing (B) conditions. The last lane on each blot is the UrIg protein stained by Coomassie Blue. All of the mAbs react with UrIg by ELISA (native), but only a subset (O-1, O-2, O-3, O-8, O-9, O-11, O-13) react with denatured and native UrIg. Antisera (from 2 mice) reacted with monomeric and dimeric UrIg, while the mAbs reacted only with the dimer.
Western blotting
Nurse shark tissues were dissected from animals in SPBS and minced into small pieces. Approximately 500 mg of tissue was dissociated with the frosted ends of microscope slides in 5 mls of 2% NP40/PBS lysis buffer containing protease inhibitors. Lysates were kept on ice for 30 minutes and then were cleared of nucleic acid and debris by centrifugation at 600 X g for 15 minutes. 30 ul of lysates were mixed with an equal volume of Laemmli Sample Buffer and subjected to SDS-PAGE. Gels were transferred to PVDF as described (38) and cut into strips for incubated with 500 ul mAb supernatant or antisera at 1/1000. Bands were revealed with the Vectastain ELITE ABC kit (Novus Biologicals), using precipitable diaminobenzidine (DAB) as substrate.
UrIg Modeling
AlphaFold-Multimer (39) was used to prepare a structural model for the full UrIg sequence. The relaxed predicted structure with highest confidence was then submitted to the GLYCAM-Web server (40) for addition of carbohydrate for N-linked glycan sites. Any modeled glycans with steric clashes were then adjusted manually to avoid clashing using Coot (41).
Results
UrIg is an MHC-linked Ig heavy chain gene
Cartilaginous fish databases were scanned for MHC-linked genes in NCBI. One gene in the MHC class III region of catshark (Scyliorhinus canicula) was annotated as a V region of Ig-kappa L chain (LOC119976294), but upon inspection it had a VJ exon followed by a C1 exon. The gene is encoded near the cluster of TNF genes, not far from other MHC-linked genes encoding VJ domains, the NK receptor NCR3 (33). Using this gene fragment, we searched other shark genomic and transcriptomic databases and found the ortholog in various shark genomes as well as full-length transcriptomes (GIWU01140354 and GIWU01140356) from a nurse shark liver database. In most cartilaginous fish species, the gene is composed of a leader segment, one VJ exon, followed by three constant domain exons (Fig. 1A–D). Upon examination of genomic sequences including nurse shark (position: 341176–354343 in JAHRHZ010000005 (Supplemental Fig. 1)), we found that the leader exon is “split” as is found for all Ig/TCR V genes (a short leader exon followed by an intron and then a short leader segment 5’ of the VJ exon, Fig. 1A), and each IgSF domain is encoded by a separate exon (Fig. 1B–D, Supplemental Fig. 1). No transmembrane exons were found in the genome, nor were any transcripts found with a TM region. Furthermore, there is no secretory tail at the end of the C3 exon and thus no identifiable cryptic splice site that would allow for alternative splicing, as is found for all vertebrate H chain mRNA (42). Besides the VC1C2C3 mRNA transcript, there is a shorter transcript in which the C2 domain exon is spliced out (Supplemental Fig. 2). In all species in which larger genomic contigs were available, the gene mapped to the class III region of MHC (Fig. 1E).
Fig. 1. Basic features of shark UrIg. A) UrIg is an ancient VJ domain.
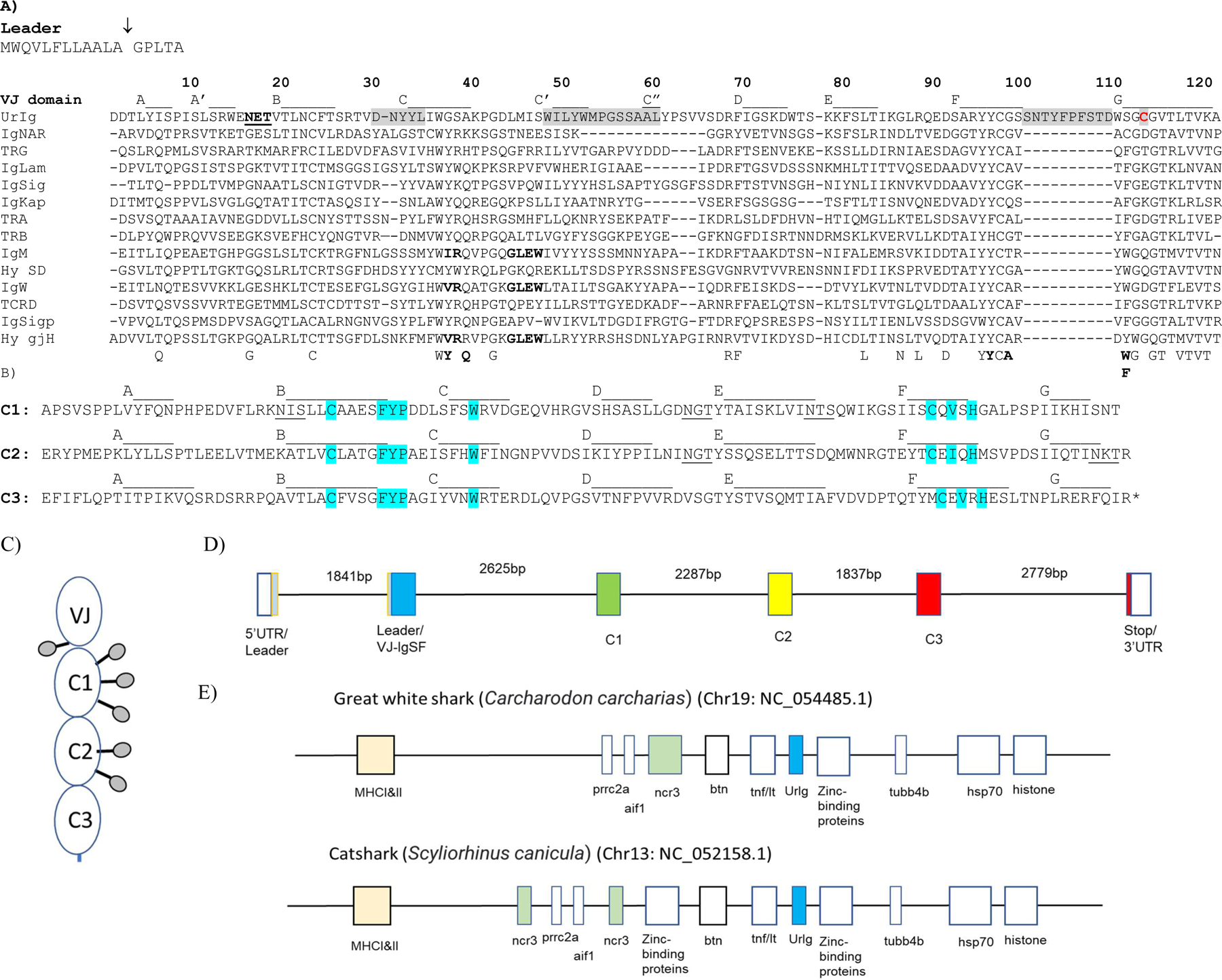
UrIg VJ domain aligned to nurse shark antigen receptor VJ domains: 4 types of TCR (α, β, γ, δ), IgM, IgW, IgNAR, and the 3 types of L chains (κ, λ, σ); the germline-joined Hydrolagus IgM V (Hyco-gjH) and single domain Hydrolagus IgM V (hyco-SD) are also aligned. Numbering and strand positions are based on the nurse shark UrIg mature protein sequence and AlphaFold prediction. Accession numbers for sequences are Nurse shark- UrIg (GIWU01140354.1); IgM (AAA50817); IgW (AAB08972): igNAR (AAB42621); IgLambda (sequence taken from; IgKappa (L16765); IgSigma (EF114759); TCRα (ADW95866); TCRβ (ADW95893): TCRδ (ADW95945); TCRγ (QNT23252). Spotted Ratfish (Hydrolagus colliei)- Hyco-SD (HHC1; seq taken from (63); Hyco-gjV (AAC12891). Conserved amino acids in VJ domains are shown below the figure. Residues that interact between V domains of bona fide Ig are bolded below the figure and in the IgM, IgW, and Hydrolagus V sequences. The potential glycosylation site in UrIg is underlined. CDR1, 2, and 3 are shaded. Only the CDR3 for UrIg is shown because the other CDR3s for Ig/TCR are somatically generated. The one free cysteine (Cys-113) in UrIg is shaded red. The arrow marking the leader segment indicates the intron of the “split leader.” Dashes indicate gaps in amino acid sequences. B) UrIg C domains bear classical C1 domain features found in Ig/TCR/MHC. The three C domains of UrIg are noted with the conserved residues in blue (esp. FYP and C-V-H). Strand positions are based on the AlphaFold prediction). Potential glycosylation sites are underlined. C. Schematic model of the UrIg monomer. Potential glycosylation sites shown as lollipops. D. Genomic (Intron/Exon) organization of nurse shark UrIg. Genomic DNA (position: 341176–354343 in JAHRHZ010000005) was aligned against a full-length transcript GIWU01140354 using splign. Each colored box is a coding region, and the untranslated regions (UTR) are in an open box. E. UrIg maps to the shark MHC in the class III region. UrIg, blue; the other VJ gene in the MHC, NCR3, in green; class I/II, light peach; other genes including class III genes are in open boxes. Mapping is not to scale, but simply shows relative locations. Wider boxes indicate clusters of multigenes.
The VJ domain bears the classic YYC in the F strand and GX(C)G and LTVK motifs in the G strand (Fig. 1A). Residues that interact between VH and VL are not well conserved in the UrIg V domain, suggesting that the V-UrIg may not form a closed dimer (bold residues below the alignment and in the IgM/IgW sequences in Fig. 1A). All three C domains are card-carrying C1 IgSF domains bearing the cardinal FYP and C-V-H motifs described in the Introduction (Fig. 1B). The overall 4-domain structure suggests that this molecule is a type of Ig and therefore we named it UrIg (Ig-original). Unlike Ig/TCR, there is no cysteine in the A strand of the first constant domain to make a disulfide bond with an L chain cysteine (also true of IgNAR, (43)), so if the molecule associates with L chains it must occur via noncovalent bonding. There is only one free cysteine in the entire molecule, found in the VJ domain GXG motif in the G strand, which may form a disulfide bond between UrIg H chains (Fig. 1A, Cys-113 in red). The molecule has 6 potential N-linked glycosylation sites (underlined in Fig.1A, B). From these sequence analyses we predicted that UrIg forms a disulfide-linked homodimer, perhaps with free V domains as for IgNAR and camelid IgG (discussed below) (44). This speculation is tested below with recombinant UrIg and with UrIg found in nurse shark tissues.
The entire UrIg protein sequence is only 30% identical to other shark IgH genes such as IgM (11, 14), IgNAR (19), and IgW (45–47), showing that UrIg diverged from other antigen receptors long ago. Domain-by-domain bioinformatic (e.g., BLAST) searches showed that the VJ domain is just as related to Vs of IgL chains, TCRs and IgH chains with no clear orthology, which is confirmed by the phylogenetic tree (Fig. 2A). The UrIg VJ domain is more like Ig/TCR V domains than the other VJ gene encoded in the class III region of the MHC, NKp30 (encoding gene: NCR3), which forms the root of the VJ tree (33, 48).
Fig. 2. Phylogenetic tree analyses of UrIg domains demonstrate UrIg’s ancient origin.
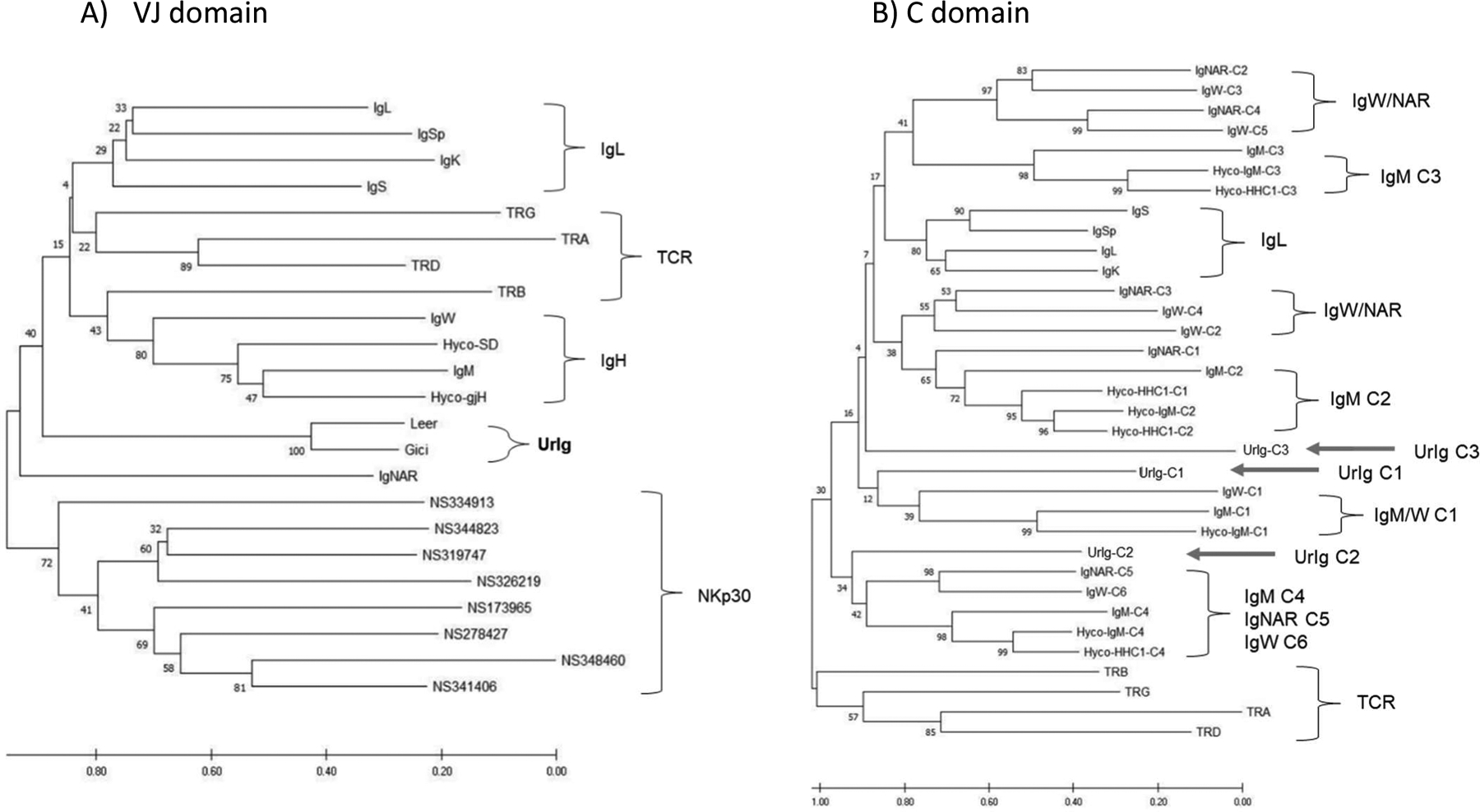
Neighbor-joining phylogenetic tree was performed with 500 bootstrap runs. The original tree is shown. Scale bars indicate the number of amino acid changes per site. A. Tree of VJ domains from nurse shark Ig/TCR and NKp30. UrIg VJ from two highly divergent Elasmobranchs nurse shark Ginglymostoma cirratum (Gici) and little skate Leucoraja erinacea (Leer) included. Ratfish Hydrolagus colliei IgM (Hyco-SD) and germline-joined IgM (Hyco-gH) included. CDR3 were removed from the sequences to construct the tree. B. Relationships of UrIg C domains to nurse shark Ig/TCR (and Hydrolagus IgM) C domains. Positions of the three UrIg C domains shown with arrows. Since clustering was more informative, we did not root these trees. Additional accession numbers for these analyses are: V- nurse shark IgS’ (Sp) (AAV34678); Nurse shark NCR3 genes (MT914154–914161); C- Hyco-IgM (AAC12891); TCRα (ADW95866; JAHRHZ010000115.1); TCRβ (ADW95903): TCRδ (ADW95942); TCRγ (ADW95913).
Phylogenetic trees of the C domains (Fig. 2B) showed that the UrIg C1 domain weakly clusters with (and may be ancestral to) the IgM C1 domains of nurse shark and the Holocephalan Hydrolagus as well as the nurse shark IgW C1 domain. Interestingly, the UrIg C2 domain clustered weakly (and also may be ancestral to) the C-terminal domains of IgM (C4), IgNAR (C5), and IgW (C6). The UrIg C3 domain was not closely related to any IgH/IgL/TCR domain cluster. In summary, while the domains of UrIg are clearly VJ and C1 domains like those seen in antigen receptors, there is no high relatedness to any known Ig or TCR. Furthermore, while the bootstrap values are very low, a weak case can be made that UrIg domains are older than all other cartilaginous fish (and thus all other vertebrate) Ig/TCR domains.
We should briefly point out other features of the C domain tree, which were noted previously but now are definitive, perhaps reinforced with the addition of the UrIg domains. The relationships between the IgNAR, IgW, and IgM C domains (45, 47) as well as duplication by convergent evolution of the C1 domains from C2 domains in IgNAR (19) and the H-chain-only class of Hydrolagus IgM (15, 20), are robustly confirmed by this new tree.
UrIg is found in all Elasmobranchs and evolves conservatively
Nurse shark UrIg was used as bait to search for the gene in all other cartilaginous fish databases and was detected in all Elasmobranchs (sharks and rays) (Fig. 3). The alignment demonstrated that UrIg is highly conserved in all species (60–97% overall; 70–95% VJ exon), reaching back at least 300 million years. Note that the VJ domain is more conserved than the C domains, and that “CDR3” has been particularly well conserved over 300 million years of evolution. Such conservation suggests that UrIg is under strong negative selection, perhaps for binding to an evolutionarily conserved epitope. These characteristics are clearly unlike bona fide Igs and even other IgSF molecules and suggest that UrIg serves an innate immune or even a non-immune function in sharks.
Fig. 3. UrIg evolves conservatively among different cartilaginous fish.
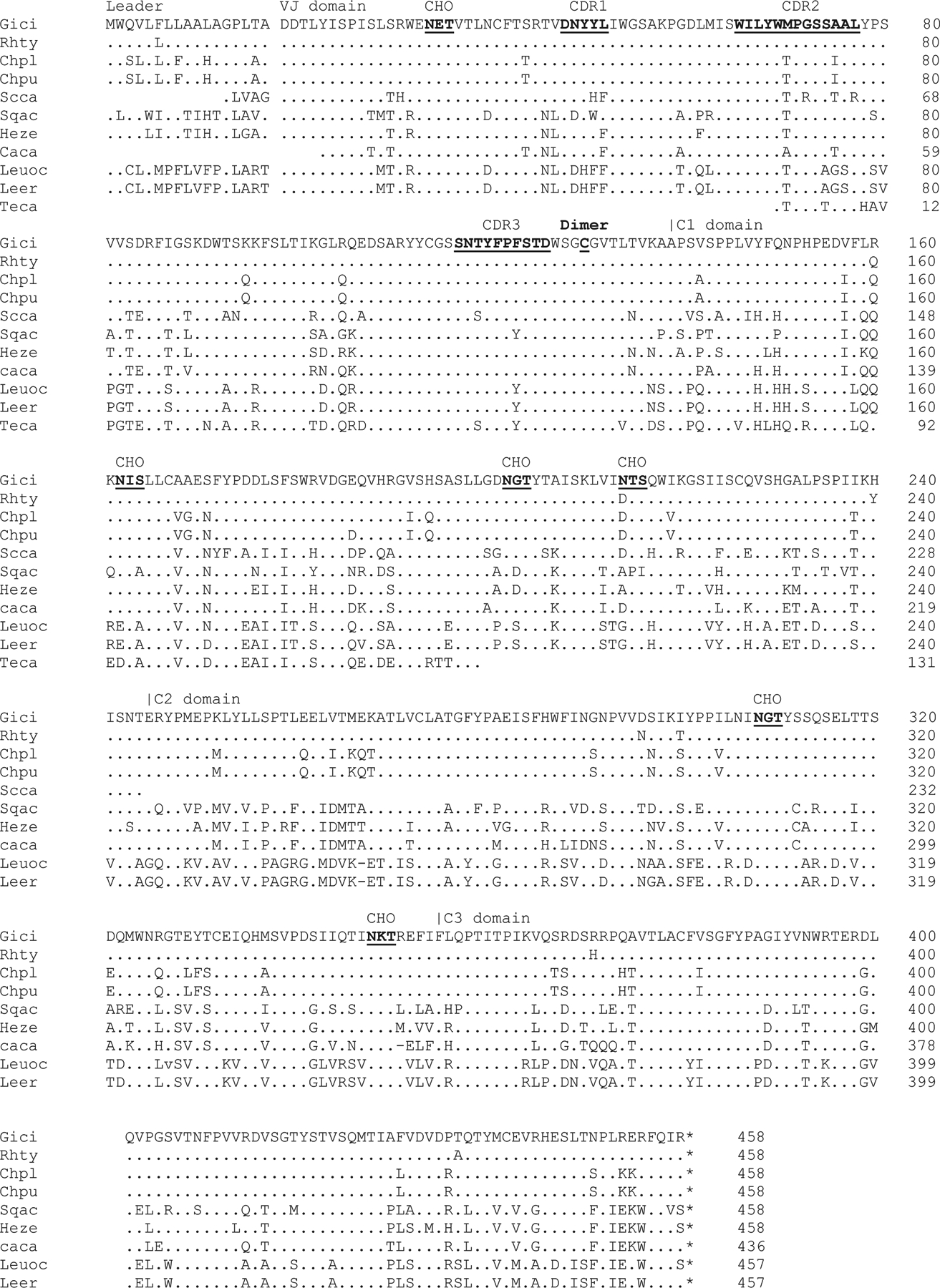
We identified cartilaginous fish UrIg from various databases. Although most of them are gene models (i.e., predicted genes), Clustal omega multiple sequence alignment (ebi.ac.uk) revealed high conservation to the nurse shark UrIg transcripts. Dashes and dots indicate absence and identical amino acid sequences compared to nurse shark UrIg, respectively. CDR, glycosylation sites, and cys-113 are underlined. Species abbreviation and accession numbers, when available)are: Leer: Little Skate: Leucoraja erinacea (JAMYXN010000071.1)*; Teca: Pacific electric ray: Tetronarce californica (GFBV01010368); Leuoc: Winter skate: Leucoraja ocellata (GEZH01007499+GEZH01041997); Sqac: spiny dogfish: Squalus acanthias (HAGU01063221.1); Caca: Great white shark: Carcharodon carcharias (NC_054485.1)*; Heze: Zebra bullhead shark: Heterodontus zebra (GGGL01499922.1)l Scca: small-spotted catshark Scyliorhinus canicula (NC_052158+CACTIS020018579)*; Rhty: Whale shark: Rhincodon typus (XP_048472529.1); Gici: Nurse shark: Ginglymostoma cirratum (GIWU01140354.1; GIWU01140356.1); Chpu: Brownbanded bamboo shark: Chiloscyllium punctatum (BEZZ01003013); Chpl: Whitespotted bamboo shark: Chiloscyllium plagiosum (GCC18650.1). Note that sequences with * indicate were manually curated from the genome assembly and/or transcripts databases.
UrIg is expressed in the liver and not primary or secondary lymphoid tissues
Igs are expressed at high levels in the spleen and epigonal of cartilaginous fish, and at lower levels in the gut, liver, and kidney (49, 50). TCRs are also expressed highly in the spleen and thymus, as in all other vertebrates. In contrast, UrIg is expressed at high levels in the liver and somewhat lower levels in the kidney with no or little mRNA expression in other tissues including immune organs (Fig. 4). It is not known whether the gene is expressed in hepatocytes or in infiltrating liver leukocytes.
Fig. 4. UrIg is expressed in the liver and kidney.
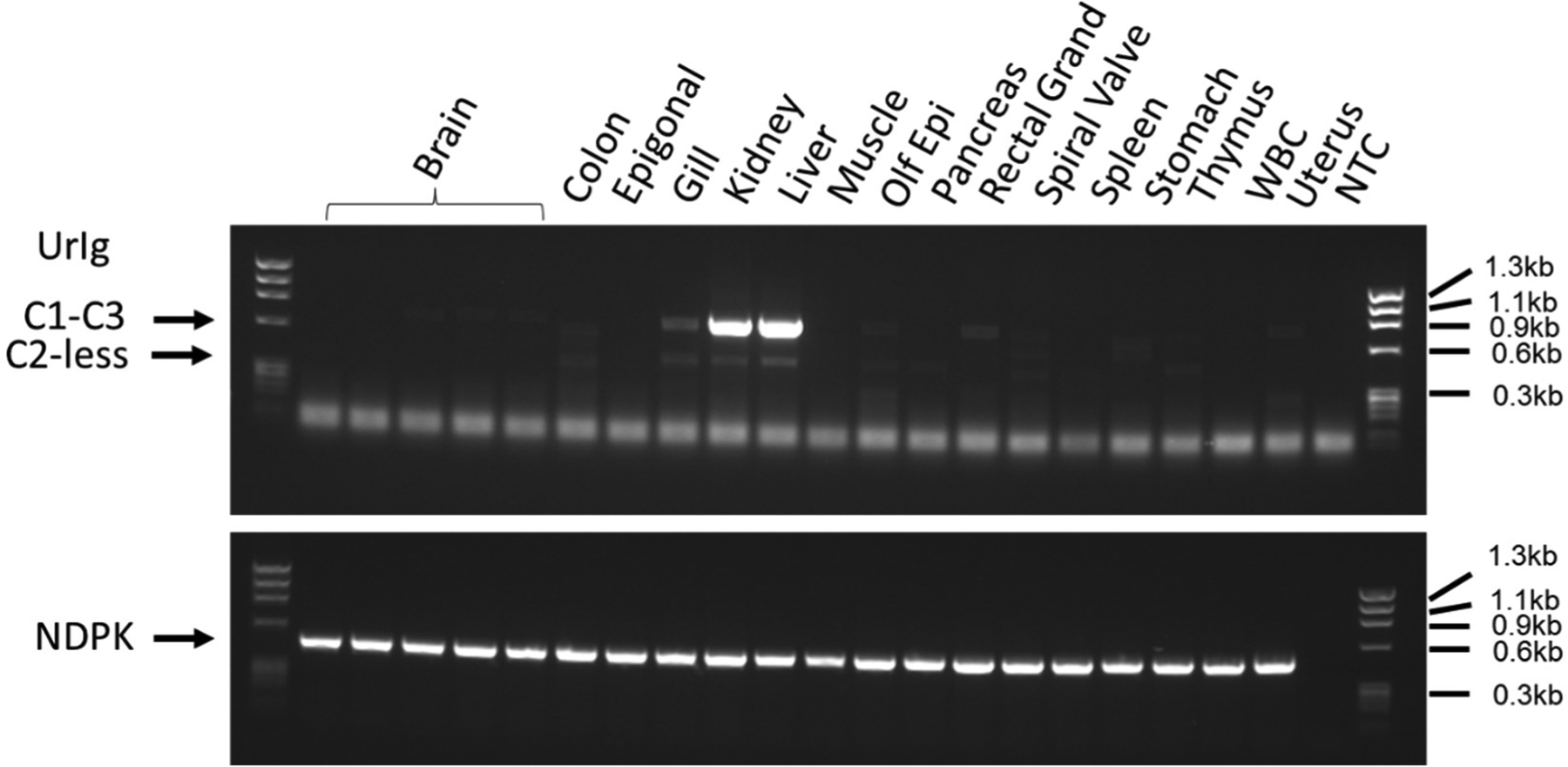
cDNA generated from nurse shark “G” total RNA was used for PCR amplification of UrIg. Primers were casted at the 5’ of the C1 and 3’ of the C3 domain to amplify the 792bp constant regions (arrow marked with C1-C3). These primers also amplify the C2-less short, spliced form (C2-less) expressed at very low levels. The uniform band at the bottom of the gel is presumably primer dimers which also appear in the control (NTC: non template control) lane. NDPK was used as a PCR control since it is nearly ubiquitously expressed. Size marker: phiX174 HaeIII.
Modeling shows UrIg to converge on mammalian IgG
Bona fide Igs are dimers of IgH/IgL dimers. The H chains are disulfide-bonded to each other and to L chains. There is, however, only one free cysteine in UrIg, in the G strand of the VJ domain (Fig. 1A, Cys-113), suggesting that H chain dimers form but there is no disulfide-bonded L chain. AlphaFold relaxed predicted structure of UrIg with highest confidence placed the V-V domains so that a disulfide bond would be formed between the Cys residues 113 and 113’ (marked red in Fig. 5A). In contrast, if the V-V dimer domains were modeled as a standard Fab or L chain dimer of V domains, the free Cys would be on the distal sides of that dimer so it is unlikely that the V-V will pair like a typical Fab. The AlphaFold model of the C2-C3 region predicts an Fc-like (CH2-CH3) arrangement of the domains, and the N-linked glycan site at position 291 in UrIg (penultimate sugar in Fig. 1C) is very close to that of position 297 in a human IgG Fc, where the equivalent glycan is attached. In mammals this glycan is crucial for IgG effector functions (51). Thus, we predict that the UrIg C2-C3 domains will pair like mammalian IgG C2-C3. The other glycans (underlined in Fig. 1A, B) are modeled to be exposed to the solvent, most conspicuously at the unique hinge region between the VJ and C1 domain; this sugar likely protects UrIg from protease attack—note that UrIg has a hinge-like region at the N-terminus of the C1 domain (APSVSPPL).
Fig. 5. UrIg AlphaFold Protein Models. A. Complete dimeric molecule.
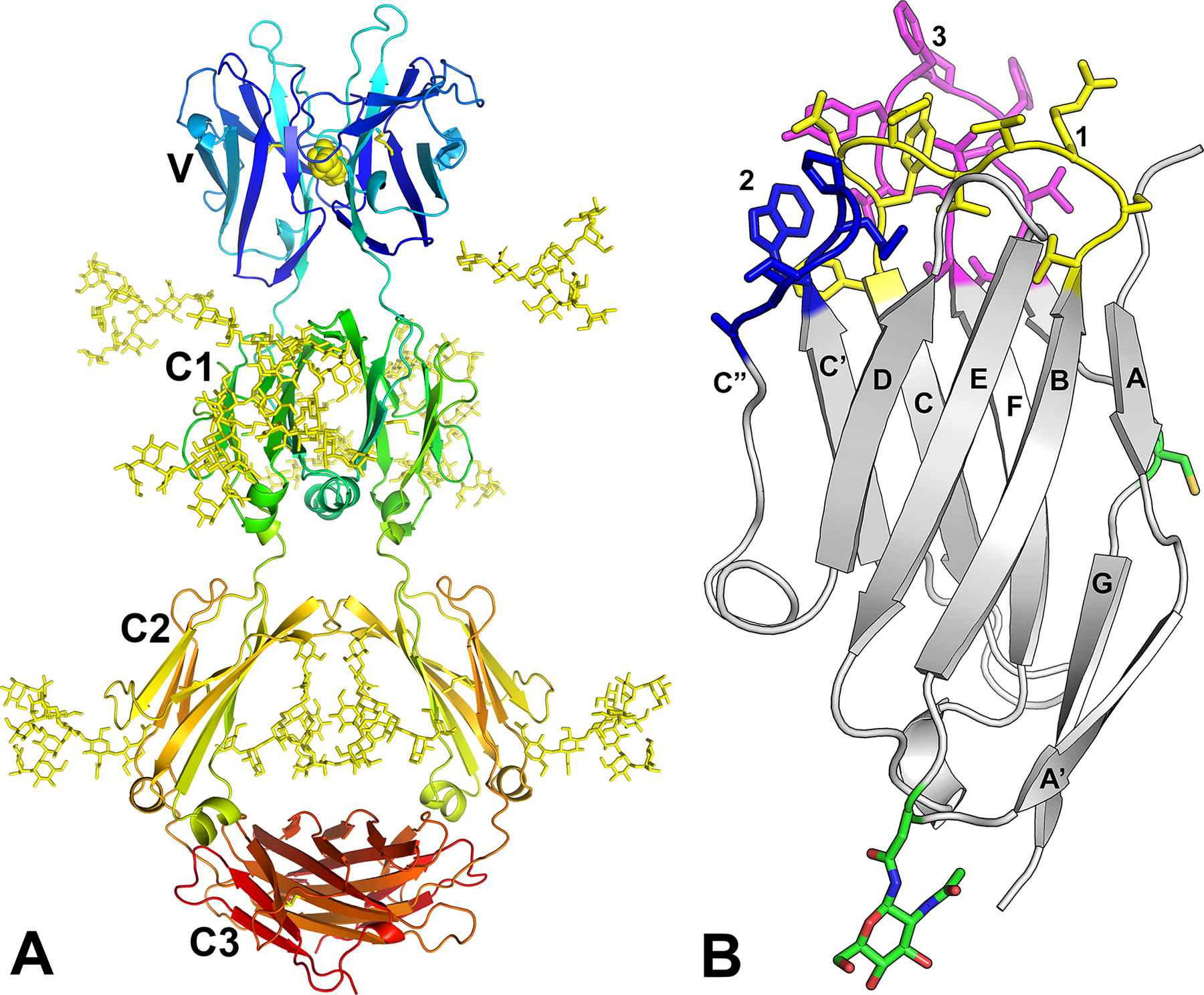
Glycans were added to the model with GLYCAM-Web followed by manual adjustments to avoid steric clashes. The predicted disulfide bond at position Cys-113 in the V domain is shown with yellow schematic. B. V domain with CDR sidechains indicated. The model of the UrIg V domain is shown with CDR-like loops colored yellow (CDR1), blue (CDR2) and magenta (CDR3). Side chains are shown for the unpaired Cys in the G strand and putative N-linked Asn in the AB loop.
The V domain was modeled as well, with amino acid side chains on the CDR (Fig. 5B). Note the large number of aromatic residues in the loops that are modeled to interact (2 Phe’s in CDR3, Trp in CDR2, 3 Tyr’s). We emphasize that this is a model and we are not certain whether the CDR3 points up or folds over and perhaps buries those aromatic residues.
Recombinant UrIg forms a secreted disulfide-linked dimer
The UrIg 4-domain H chain, with a C-terminal His-tag, was produced in mammalian expi293F cells. The fact that UrIg was secreted readily into the supernatant suggested that it folded correctly and likely does not have to associate with another partner, e.g., Ig L chains. The calculated protein molecular weight for this construct was 50,730 g/mol. There are 6 potential N-linked glycosylation sites per monomer (one in V, three in C1, two in C2, Fig. 1A–C), suggesting an approximate molecular weight increase between 9,000 and 12,500, depending on the occupancy and type of carbohydrate at each site. Thus, we expected a monomer to have a molecular weight of ~60,000–63,500 and dimer of ~120,000–127,000. As mentioned, there is one Cys in the V domain (residue 113, sequential numbering) that modeling predicts to be surface-exposed, thus the possibility of dimerization via this residue. SDS-PAGE of purified UrIg showed a band slightly larger than 100,000 for the non-reduced sample and between 50,000–75,000 for the reduced sample (Fig. 6A). The non-reduced sample eluted in size-exclusion chromatography close to the molecular weight standard γ-globulin (~158,000) (Fig. 6B). Thus, the secreted, recombinant UrIg forms a disulfide-linked dimer.
Fig. 6. Recombinant UrIg forms a secreted dimer.
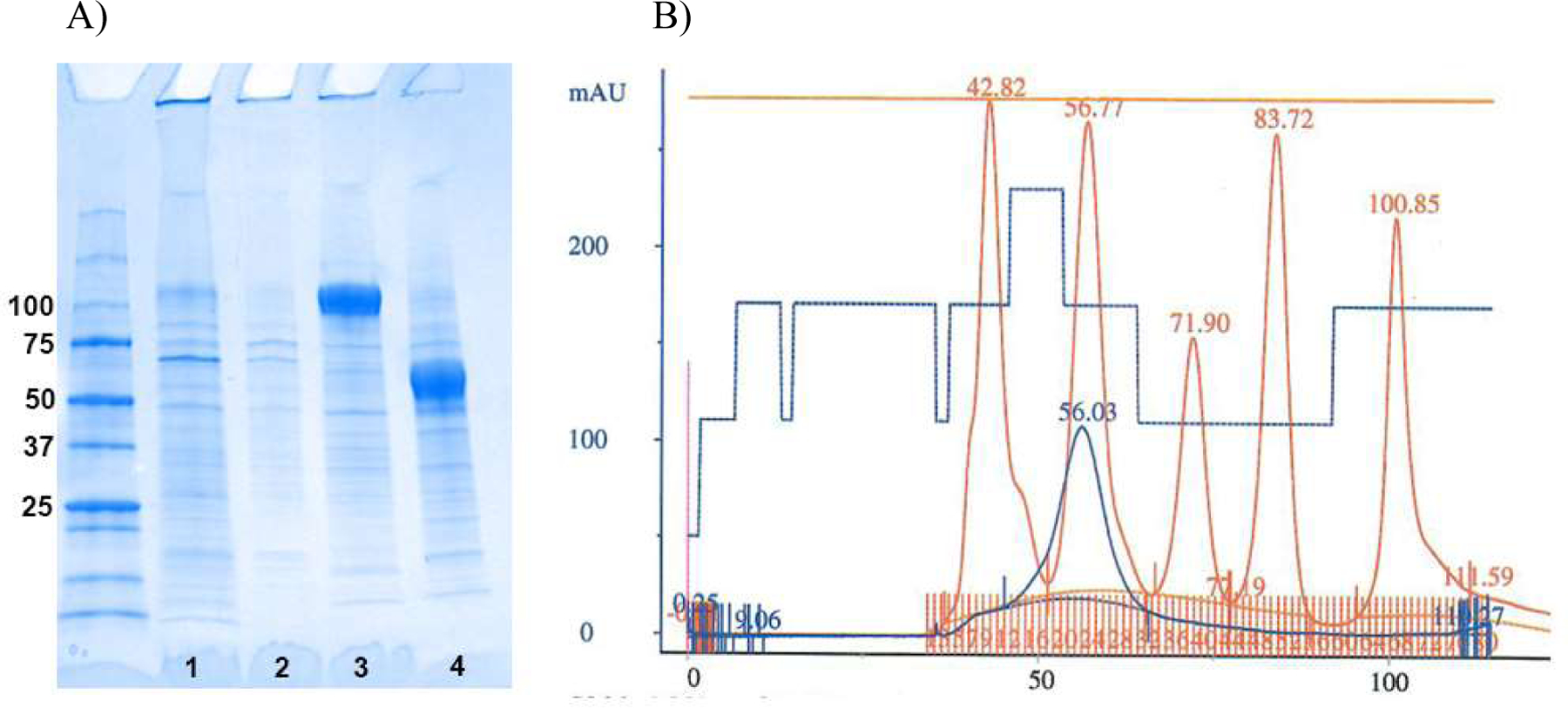
A. Coomassie-stained gel with MW markers (BioRad, Precision Plus dual color), followed by: Lane 1-media after depletion by Ni excel resin (non-reducing), Lane 2-high salt wash of Ni excel resin before elution (non-reducing), Lane 3-eluted UrIg (non-reducing), Lane 4-eluted UrIg (reducing). B. Size exclusion chromatography (Superdex 200, 16/60) of UrIg after affinity purification. UrIg: blue, MW standards: red, with ml on the x-axis and mAu on the y axis. Elution positions (ml) are shown at the top of each standard peak. UrIg runs at the approximate weight expected for a dimer. MW standards (BioRad) are: vitamin B12, 100.9ml, 1,350 dal; myoglobin, 83.7ml, 17,000 dal; ovalbumin, 71.9ml, 44,000 dal; γ-globulin, 56.8ml, 158,000 dal; thyroglobulin, 42.8mlm.
Natural nurse shark UrIg also forms a dimer and is present in many tissues
The purified recombinant UrIg was used to immunize mice to produce antisera and mAbs. A panel of mAbs was generated and tested on the recombinant protein by ELISA. The antisera and several of the mAbs recognized the recombinant UrIg molecule at ~120kd under non-reducing conditions by western blotting, and the antisera, but not the mAbs, also recognized the reduced UrIg at 60kd (Fig. 7A, B).
Tissue lysates from all nurse shark tissues were prepared and tested for the presence of UrIg. Although mRNA expression is mostly limited to the liver (Fig. 4 and data not shown), we were able to detect UrIg protein in many tissues under non-reducing conditions at 120kd (Fig. 8). Interestingly, in the five brain tissues, telencephalon (cerebrum), diencephalon (optic lobes), mesencephalon (midbrain), metencephalon (cerebellum), and myelencephalon (medulla oblongata) UrIg was found predominantly as a monomer.
Fig. 8. UrIg is present in many nurse shark tissues.
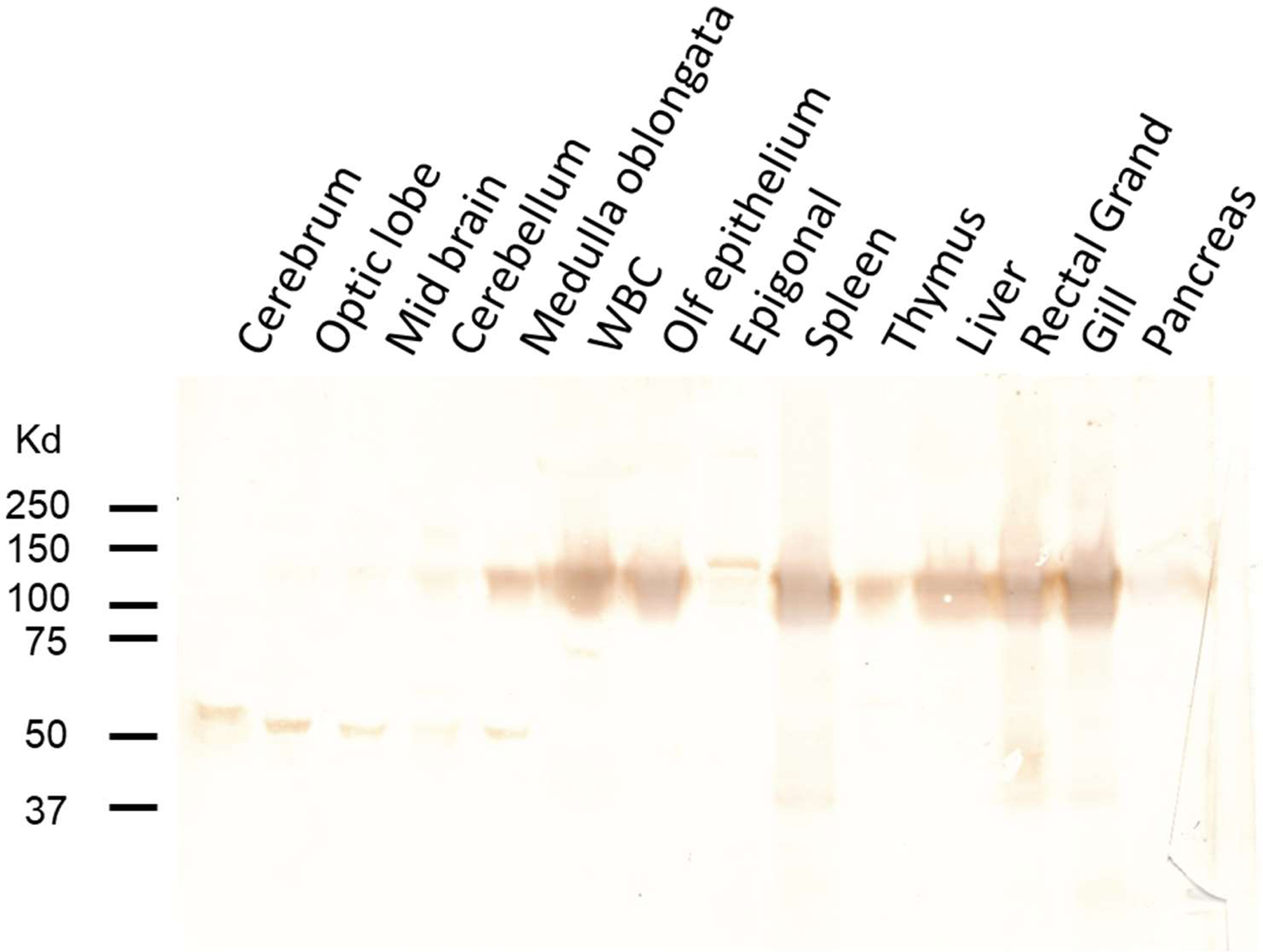
Western blotting analysis with mouse antisera on14 shark tissues demonstrated UrIg protein is found as a dimer in most tissues. Gel of denatured shark lysates was run under non-reducing conditions. Note that UrIg is predominantly a monomer in the five regions of shark brain: Cerebrum, Optic lobe, Midbrain, Cerebellum, Medulla oblongata. WBC (white blood cells) were from the buffy coat of centrifuged blood.
UrIg is present in the extracellular matrix of many shark tissues
It was a puzzle that UrIg is expressed as mRNA by the liver, but the protein can easily be detected in many tissues by western blotting (Fig. 8). We tested the exact tissue localization by immunohistochemistry of the spleen and found that the mAbs and antisera stained in a “stick-like” pattern around the splenic white pulp and around the vasculature, expected of staining of the extracellular matrix (ECM)(Fig. 9A). A mAb specific for IgNAR was used to show the staining of lymphocytes in the white pulp (Fig. 9B). Staining of other tissues on the western blot such as gill (Fig 9D) and others (not shown) also showed “stick-like” staining of defined areas. To test whether the mAbs truly stained ECM, we co-stained kidney sections with an UrIg-specific mAb O-1 (in red, see western blot for mAb O-1 reactivity, Fig. 7) and a reagent that detects collagen (CNA in green).
Fig. 9. UrIg is found in the extracellular matrix of nurse shark tissues. A-C. Staining of spleen.
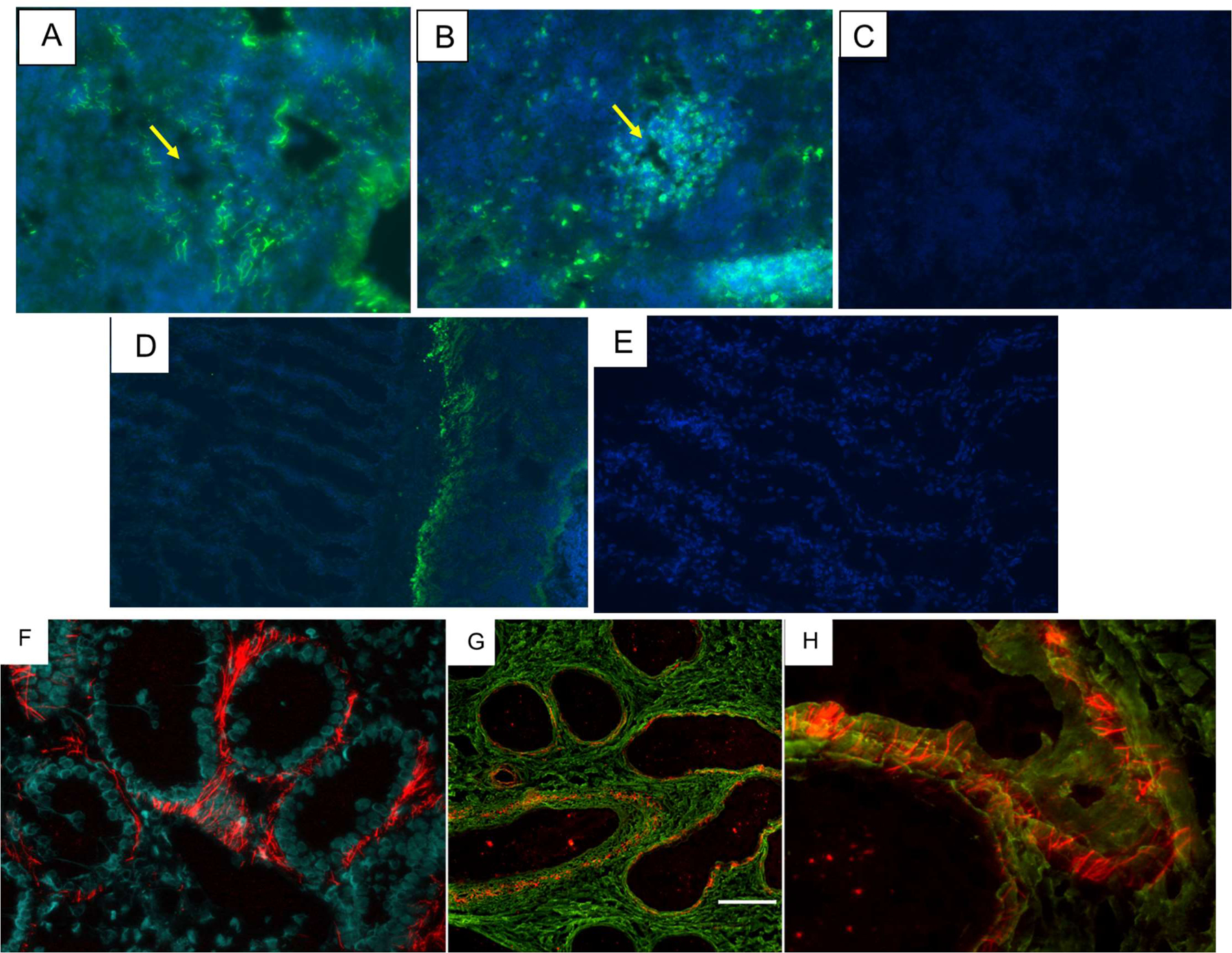
A. Immunofluorescent staining of nurse shark spleen with the UrIg-specific mAb O-1 staining (in green) extracellular matrix surrounding the white pulp in a “stick-like” fashion. Central arteriole of the white pulp is shown with an arrow. Merged images of mAb staining (green) and DAPI (blue). B. IgNAR-specific mAb showing staining of lymphocytes in the splenic white pulp and scattered plasma cells in the red pulp. C. Spleen with no primary antibody. Merged images of mAb staining (green) and DAPI (blue). Arrows point out to central arteries, D-E. Staining of gill with UrIg-specific mAb O-1 (D), and no primary antibody (E). UrIg staining is mainly at the insertion sites of the gill filaments and weak staining of the filaments themselves. Merged images of mAb staining (green) and DAPI (blue). F-H. Immunofluorescent staining of renal sections. F. mAb UrIg staining alone shows extracellular localization of UrIg outside of tubules and blood vessels. Blue DAPI staining shows tubular nuclei. G/H. UrIg/collagen double-stains. To visualize collagen matrix (18TD-CNA35)3 conjugated with Alexa 488 that was used which binds to any triple collagen helix. Double immunofluorescent staining for UrIg (red) and collagen (green). UrIg rod-like parallel structures are cross linking collagen layers. Magnifications are 600x (F, G) and 800x (H). Scale bar in (G) indicates 50 mm.
Most UrIg staining was observed in the anterior and middle portions of kidneys surrounding tubules (Fig. 9F). No staining was detected in the glomeruli (not shown). Staining was extracellular surrounding tubules and apparently forming rod-like structures with an orientation perpendicular to the tubular cell membrane. To visualize collagen matrix, we developed a trimeric form of a natural collagen binding protein (CNA35) conjugated to a fluorescent dye (see Materials and Methods). CNA35 monomer itself was demonstrated to have much better specific binding to collagen than existing fluorescent techniques currently used for collagen visualization (52, 53). Generation of multivalent CNA35 dendrimers (2–4-mers) by native chemical ligation remarkably enhanced the affinity and attenuated the dissociation kinetics (54). Inspired by these multivalent CNA35 probes we designed our own system for recombinant production of trimeric CNA35 in bacteria. The CNA35 sequence was genetically fused to a trimerization domain of collagen XVIII (18TD), which has a picomolar trimerization potential (55). The resulting hybrid protein molecule (18TD-CNA35)3 was conjugated to Alexa 488 and used for co-staining of tissues. The UrIg (red) is seen «stitching» layered collagen structures in shark kidneys (Fig. 9F–H). Note that in all tissues the UrIg is found associated with collagen, but in defined areas that require further study.
The data, taken together, suggest that the UrIg expressed by the liver is transported by the blood or by other cells (Note that PBL (WBC) were western blot-positive, Fig. 8) throughout the body and deposited in the ECM.
Discussion
UrIg converges on IgG and likely predates Ig/TCR emergence
The UrIg sequence suggested that the molecule is an IgH chain that made a disulfide-linked dimer via a unique S-S bond and not associated with Ig L chains. This prediction was born out in the production of the recombinant molecule and by western blotting of shark tissues. An AlphaFold model predicts that the three C1 domains form a structure highly reminiscent of IgG, even at the level of the canonical glycan in the UrIg (and IgGH) C2 domain that provides flexibility to the Fc region and is vital for effector function (51).
As mentioned in the Introduction, cartilaginous fish IgH genes are in the so-called “cluster organization” with multiple V-D-D-J-C genes that undergo rearrangement within a cluster during B cell ontogeny (11, 56). A phenomenon called “germline-joining” is the apparent consequence of RAG activity in germ cells that can initiate rearrangement within these clusters (14–17). Most of the clusters having such germline joining that have been detected result in pseudogenes, but there are two examples of functional IgH and IgL genes generated by this mechanism in the nurse shark, and one example in Hydrolagus. In all cases, it is clear that the process was “recent” as the joined genes are closely related to non-germline-joined clusters; furthermore, in the two nurse shark joins there are few bases in the CDR3-encoding regions suggestive of little TdT activity in the germline-joined events in germ cells. UrIg, in contrast, is highly conserved in all Elasmobranchs, going back 300 million years (Fig. 3). The “CDR3” of UrIg is quite long and relatively hydrophobic. Furthermore, the C domains are not closely related to any of the shark IgC regions of IgM, IgNAR, or IgW, unlike the three germline-joined cartilaginous fish Ig genes (Fig. 2). So, while it is possible that there was a germline-joining event prior to the emergence of cartilaginous fish (or Elasmobranchs), it is more likely that UrIg emergence preceded the development of rearranging Ig/TCR; this hypothesis is also suggested by the trees, in which the UrIg C domains seem ancestral. Using the same logic, it is unlikely that UrIg resulted from a reverse-transcribed IgH gene (retroposon) that inserted into the genome (also the transcript would have had to retain all of the introns).
While the modeling of the C domains was relatively straightforward, this was not true of the VJ domains. We think it most likely that the cysteine-113 in the center of the Gly-X-Gly motif of the G strand, almost unique among VJ domains (27), provides the disulfide bond covalently linking the UrIg chains. However, do the UrIg V domains join together like VH/VL to make a conventional binding site or do the 2 domains bind antigen independently, like IgNAR (43)? The conserved amino acid residues known to interact between VH/VL and TCRα/β to facilitate dimer formation are not well conserved in the UrIg VJ, but the AlphaFold model does suggest a V-V association. Further studies of the natural UrIg will allow discrimination of these two possibilities.
What is UrIg’s function?
UrIg is transcribed in the liver yet is found as protein in ECM throughout the shark body (Fig. 8, 9). First, is UrIg produced by hepatocytes or infiltrating hematopoietic cells? We should be able to address that question with in situ hybridization studies in combination with IHC using existing mAbs. So far, in situ hybridization has not been sensitive enough to detect the UrIg-expressing cells in the liver, perhaps because of overall low expression by hepatocytes. Second, it will be a challenge to understand how UrIg is transported to the ECM throughout the body, yet with existing reagents we should be able to address that question in the future as well. Lastly, and most importantly, what is UrIg’s function in the ECM? Does it perform an innate immune function such as acting as a secreted pattern recognition receptor, or could it have an ECM supporting function? Based on its MHC-linkage in the class III region we propose that it does have an innate immune function; such a function would also make sense if UrIg (or an UrIg-like molecule) were co-opted to function in the adaptive immune system after the RAG transposon invasion.
UrIg is encoded in the MHC and might be related to the Ig/TCR precursor
Based on the presence of VJ and C1 domains in the MHC and MHC-paralogous regions, Du Pasquier suggested that Ig/TCR emerged from the MHC or MHC precursor (26). Subsequently, other groups including our own also provided evidence for such a scenario (27, 30–32). UrIg is encoded in the class III region of the MHC, near to the TNF gene cluster (57) and close to other VJ single-exon genes like NCR3 (Fig. 1E). The MHC class III region is well known for the presence of genes involved in innate immunity and inflammatory responses, and immune genes in the class III region clearly were present in the proto MHC prior to the emergence of adaptive immunity (28, 58, 59). Thus, we think it plausible that UrIg is related to the VJ gene that was invaded by the RAG transposon, perhaps most related. While UrIg is clearly Ig-like in overall structure, the VJ domain is not more related to VH, VL, TCRα, or TCRβ, suggesting that it is either highly derived, or it emerged before the split of Ig and TCR. To date, we have not detected UrIg in agnathans or deuterostomes that arose from ancestors prior to the genome-wide duplications that occurred in vertebrates (30). There are, however, VJ domains present in agnathans and invertebrates, yet little is known about their genetic history or functions (26, 60–62). Studies of these molecules, and further studies of the structure and ligand identification of UrIg, will be crucial in piecing together the origins of adaptive immunity.
Supplementary Material
Key points.
Discovery of a non-rearranging immunoglobulin gene, UrIg, that maps to the shark MHC
UrIg is a 4-domain dimer expressed by the liver, and the protein is found in the ECM
UrIg may predate invasion of an immunoglobulin superfamily gene by the RAG transposon
Acknowledgements
We thank Erik Cruz for technical support and Marc Elslinger for help with the AlphaFold modeling.
This work was supported by National Institutes of Health grants R01AI140326 and R01AI170844.
Footnotes
Disclosure
The authors have no financial conflicts of interest.
References
- 1.Flajnik MF 2018. A cold-blooded view of adaptive immunity. Nat Rev Immunol 18: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pishesha N, Harmand TJ, and Ploegh HL. 2022. A guide to antigen processing and presentation. Nat Rev Immunol 22: 751–764. [DOI] [PubMed] [Google Scholar]
- 3.Davis MM, and Bjorkman PJ. 1988. T-cell antigen receptor genes and T-cell recognition. Nature 334: 395–402. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman J 2018. Unfinished Business: Evolution of the MHC and the Adaptive Immune System of Jawed Vertebrates. Annu Rev Immunol 36: 383–409. [DOI] [PubMed] [Google Scholar]
- 5.Litman GW, Rast JP, and Fugmann SD. 2010. The origins of vertebrate adaptive immunity. Nat Rev Immunol 10: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flajnik MF 2014. Re-evaluation of the immunological Big Bang. Curr Biol 24: R1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Good RA, and Papermaster BW. 1964. Ontogeny and Phylogeny of Adaptive Immunity. Adv Immunol 27: 1–115. [DOI] [PubMed] [Google Scholar]
- 8.Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, and Hsu E. 2012. Origin of immunoglobulin isotype switching. Curr Biol 22: 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flajnik MF 2002. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol 2: 688–698. [DOI] [PubMed] [Google Scholar]
- 10.Neely HR, and Flajnik MF. 2016. Emergence and Evolution of Secondary Lymphoid Organs. Annu Rev Cell Dev Biol 32: 693–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinds KR, and Litman GW. 1986. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature 320: 546–549. [DOI] [PubMed] [Google Scholar]
- 12.Ohta Y, Shiina T, Lohr RL, Hosomichi K, Pollin TI, Heist EJ, Suzuki S, Inoko H, and Flajnik MF. 2011. Primordial linkage of β2-microglobulin to the MHC. J Immunol 186: 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ott JA, Castro CD, Deiss TC, Ohta Y, Flajnik MF, and Criscitiello MF. 2018. Somatic hypermutation of T cell receptor α chain contributes to selection in nurse shark thymus. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kokubu F, Hinds K, Litman R, Shamblott MJ, and Litman GW. 1988. Complete structure and organization of immunoglobulin heavy chain constant region genes in a phylogenetically primitive vertebrate. Embo j 7: 1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rast JP, Amemiya CT, Litman RT, Strong SJ, and Litman GW. 1998. Distinct patterns of IgH structure and organization in a divergent lineage of chrondrichthyan fishes. Immunogenetics 47: 234–245. [DOI] [PubMed] [Google Scholar]
- 16.Lee SS, Fitch D, Flajnik MF, and Hsu E. 2000. Rearrangement of immunoglobulin genes in shark germ cells. J Exp Med 191: 1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, and Flajnik MF. 2001. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci U S A 98: 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokubu F, Litman R, Shamblott MJ, Hinds K, and Litman GW. 1988. Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J 7: 3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, and Flajnik MF. 1995. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374: 168–173. [DOI] [PubMed] [Google Scholar]
- 20.Anumukonda K, Francis M, Currie P, Tulenko F, and Hsu E. 2022. Heavy chain-only antibody genes in fish evolved to generate unique CDR3 repertoire. Eur J Immunol 52: 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Criscitiello MF, Saltis M, and Flajnik MF. 2006. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci U S A 103: 5036–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ott JA, Ohta Y, Flajnik MF, and Criscitiello MF. 2021. Lost structural and functional inter-relationships between Ig and TCR loci in mammals revealed in sharks. Immunogenetics 73: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakano H, Huppi K, Heinrich G, and Tonegawa S. 1979. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280: 288–294. [DOI] [PubMed] [Google Scholar]
- 24.Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, Zhang Y, Yu W, Pontarotti P, Escriva H, Le Petillon Y, Liu X, Chen S, Schatz DG, and Xu A. 2016. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell 166: 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flajnik MF 2016. Evidence of G.O.D.’s Miracle: Unearthing a RAG Transposon. Cell 166: 11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du Pasquier L, Zucchetti I, and De Santis R. 2004. Immunoglobulin superfamily receptors in protochordates: before RAG time. Immunol Rev 198: 233–248. [DOI] [PubMed] [Google Scholar]
- 27.Dornburg A, and Yoder JA. 2022. On the relationship between extant innate immune receptors and the evolutionary origins of jawed vertebrate adaptive immunity. Immunogenetics 74: 111–128. [DOI] [PubMed] [Google Scholar]
- 28.Zucchetti I, De Santis R, Grusea S, Pontarotti P, and Du Pasquier L. 2009. Origin and evolution of the vertebrate leukocyte receptors: the lesson from tunicates. Immunogenetics 61: 463–481. [DOI] [PubMed] [Google Scholar]
- 29.Williams AF, and Barclay AN. 1988. The immunoglobulin superfamily--domains for cell surface recognition. Annu Rev Immunol 6: 381–405. [DOI] [PubMed] [Google Scholar]
- 30.Flajnik MF, and Kasahara M. 2010. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, Yang Z, Huang J, Cheng X, Wang X, Yang S, Ren L, Lian Z, Han H, and Zhao Y. 2019. Identification of Two Nonrearranging IgSF Genes in Chicken Reveals a Novel Family of Putative Remnants of an Antigen Receptor Precursor. J Immunol 202: 1992–2004. [DOI] [PubMed] [Google Scholar]
- 32.Ohta Y, Kasahara M, O’Connor TD, and Flajnik MF. 2019. Inferring the “Primordial Immune Complex”: Origins of MHC Class I and Antigen Receptors Revealed by Comparative Genomics. J Immunol 203: 1882–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinlein A, Janes ME, Kincer J, Almeida T, Matz H, Sui J, Criscitiello MF, Flajnik MF, and Ohta Y. 2021. Analysis of shark NCR3 family genes reveals primordial features of vertebrate NKp30. Immunogenetics 73: 333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Stecher G, Li M, Knyaz C, and Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flajnik MF, Ferrone S, Cohen N, and Du Pasquier L. 1990. Evolution of the MHC: antigenicity and unusual tissue distribution of Xenopus (frog) class II molecules. Mol Immunol 27: 451–462. [DOI] [PubMed] [Google Scholar]
- 36.Boudko SP, Sasaki T, Engel J, Lerch TF, Nix J, Chapman MS, and Bachinger HP. 2009. Crystal structure of human collagen XVIII trimerization domain: A novel collagen trimerization Fold. J Mol Biol 392: 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boudko SP, Zientek KD, Vance J, Hacker JL, Engel J, and Bachinger HP. 2010. The NC2 domain of collagen IX provides chain selection and heterotrimerization. J Biol Chem 285: 23721–23731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flajnik MF, Taylor E, Canel C, Grossberger D, and Du Pasquier L. 2015. Reagents Specific for MHC Class I Antigens of Xenopus1. American Zoologist 31: 580–591. [Google Scholar]
- 39.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, Ronneberger O, Bodenstein S, Zielinski M, Bridgland A, Potapenko A, Cowie A, Tunyasuvunakool K, Jain R, Clancy E, Kohli P, Jumper J, and Hassabis D. 2022. Protein complex prediction with AlphaFold-Multimer. bioRxiv: 2021.2010.2004.463034. [Google Scholar]
- 40.Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, and Woods RJ. 2008. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J Comput Chem 29: 622–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, and Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, and Hood L. 1980. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell 20: 313–319. [DOI] [PubMed] [Google Scholar]
- 43.Roux KH, Greenberg AS, Greene L, Strelets L, Avila D, McKinney EC, and Flajnik MF. 1998. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual mammalian immunoglobulins. Proc Natl Acad Sci U S A 95: 11804–11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flajnik MF, Deschacht N, and Muyldermans S. 2011. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol 9: e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greenberg AS, Hughes AL, Guo J, Avila D, McKinney EC, and Flajnik MF. 1996. A novel “chimeric” antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur J Immunol 26: 1123–1129. [DOI] [PubMed] [Google Scholar]
- 46.Berstein RM, Schluter SF, Shen S, and Marchalonis JJ. 1996. A new high molecular weight immunoglobulin class from the carcharhine shark: implications for the properties of the primordial immunoglobulin. Proc Natl Acad Sci U S A 93: 3289–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson MK, Strong SJ, Litman RT, Luer CA, Amemiya CT, Rast JP, and Litman GW. 1999. A long form of the skate IgX gene exhibits a striking resemblance to the new shark IgW and IgNARC genes. Immunogenetics 49: 56–67. [DOI] [PubMed] [Google Scholar]
- 48.Flajnik MF, Tlapakova T, Criscitiello MF, Krylov V, and Ohta Y. 2012. Evolution of the B7 family: co-evolution of B7H6 and NKp30, identification of a new B7 family member, B7H7, and of B7’s historical relationship with the MHC. Immunogenetics 64: 571–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rumfelt LL, Diaz M, Lohr RL, Mochon E, and Flajnik MF. 2004. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J Immunol 173: 1129–1139. [DOI] [PubMed] [Google Scholar]
- 50.Miracle AL, Anderson MK, Litman RT, Walsh CJ, Luer CA, Rothenberg EV, and Litman GW. 2001. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int Immunol 13: 567–580. [DOI] [PubMed] [Google Scholar]
- 51.Bournazos S, and Ravetch JV. 2017. Fcgamma Receptor Function and the Design of Vaccination Strategies. Immunity 47: 224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krahn KN, Bouten CV, van Tuijl S, van Zandvoort MA, and Merkx M. 2006. Fluorescently labeled collagen binding proteins allow specific visualization of collagen in tissues and live cell culture. Anal Biochem 350: 177–185. [DOI] [PubMed] [Google Scholar]
- 53.Boerboom RA, Krahn KN, Megens RT, van Zandvoort MA, Merkx M, and Bouten CV. 2007. High resolution imaging of collagen organisation and synthesis using a versatile collagen specific probe. J Struct Biol 159: 392–399. [DOI] [PubMed] [Google Scholar]
- 54.Breurken M, Lempens EH, Meijer EW, and Merkx M. 2011. Semi-synthesis of a protease-activatable collagen targeting probe. Chem Commun (Camb) 47: 7998–8000. [DOI] [PubMed] [Google Scholar]
- 55.Annunen S, Korkko J, Czarny M, Warman ML, Brunner HG, Kaariainen H, Mulliken JB, Tranebjaerg L, Brooks DG, Cox GF, Cruysberg JR, Curtis MA, Davenport SL, Friedrich CA, Kaitila I, Krawczynski MR, Latos-Bielenska A, Mukai S, Olsen BR, Shinno N, Somer M, Vikkula M, Zlotogora J, Prockop DJ, and Ala-Kokko L. 1999. Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome, but other mutations cause overlapping Marshall/Stickler phenotypes. Am J Hum Genet 65: 974–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malecek K, Lee V, Feng W, Huang JL, Flajnik MF, Ohta Y, and Hsu E. 2008. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol 6: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redmond AK, Pettinello R, Bakke FK, and Dooley H. 2022. Sharks Provide Evidence for a Highly Complex TNFSF Repertoire in the Jawed Vertebrate Ancestor. J Immunol 209: 1713–1723. [DOI] [PubMed] [Google Scholar]
- 58.Abi-Rached L, Gilles A, Shiina T, Pontarotti P, and Inoko H. 2002. Evidence of en bloc duplication in vertebrate genomes. Nat Genet 31: 100–105. [DOI] [PubMed] [Google Scholar]
- 59.Suurvali J, Jouneau L, Thepot D, Grusea S, Pontarotti P, Du Pasquier L, Ruutel Boudinot S, and Boudinot P. 2014. The proto-MHC of placozoans, a region specialized in cellular stress and ubiquitination/proteasome pathways. J Immunol 193: 2891–2901. [DOI] [PubMed] [Google Scholar]
- 60.Chen R, Zhang L, Qi J, Zhang N, Zhang L, Yao S, Wu Y, Jiang B, Wang Z, Yuan H, Zhang Q, and Xia C. 2018. Discovery and Analysis of Invertebrate IgV(J)-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily. J Immunol 200: 2869–2881. [DOI] [PubMed] [Google Scholar]
- 61.Pancer Z, Mayer WE, Klein J, and Cooper MD. 2004. Prototypic T cell receptor and CD4-like coreceptor are expressed by lymphocytes in the agnathan sea lamprey. Proc Natl Acad Sci U S A 101: 13273–13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haruta C, Suzuki T, and Kasahara M. 2006. Variable domains in hagfish: NICIR is a polymorphic multigene family expressed preferentially in leukocytes and is related to lamprey TCR-like. Immunogenetics 58: 216–225. [DOI] [PubMed] [Google Scholar]
- 63.Lee SS, Tranchina D, Ohta Y, Flajnik MF, and Hsu E. 2002. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 16: 571–582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


