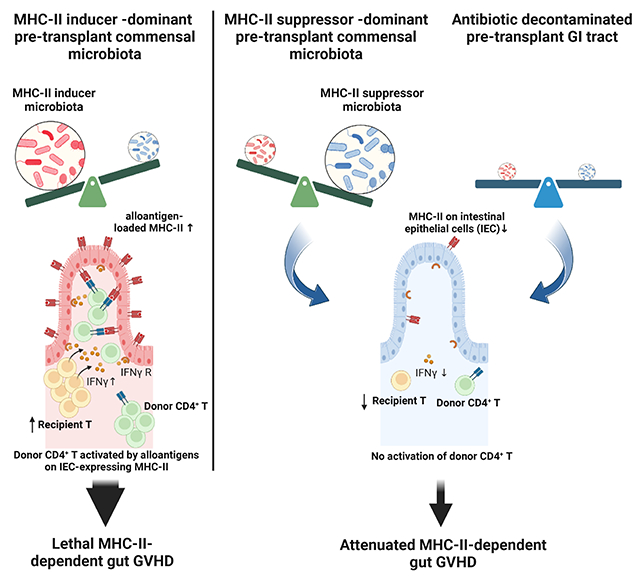
Summary
Acute graft-versus-host disease (GVHD) remains a major limitation of allogeneic stem cell transplantation (SCT) and severe intestinal manifestation is the major cause of early mortality. Intestinal microbiota control MHC class II (MHC-II) expression by ileal intestinal epithelial cells (IEC) that initiate GVHD. Here, we demonstrated that genetically identical mice of differing vendor origins had markedly different intestinal microbiota and ileal MHC-II expression, resulting in discordant GVHD severity. We utilized cohousing and antibiotic treatment to characterize the bacterial taxa positively and negatively associated with MHC-II expression. A large proportion of bacterial MHC-II inducers were vancomycin-sensitive, and peri-transplant oral vancomycin administration attenuated CD4+ T cell-mediated GVHD. We identified a similar relationship between pre-transplant microbes, HLA class II expression, and both GVHD and mortality in a large clinical SCT cohort. These data highlight therapeutically tractable mechanisms by which pre-transplant microbial taxa contribute to initiate GVHD independently of genetic disparity.
Keywords: Graft-versus-host disease (GVHD), microbiota, intestinal epithelial cells, MHC class II antigen presentation
Graphical Abstract
eTOC Blurb
Genetically identical recipient and donor pairs are associated with differential MHC-II expression on intestinal epithelial cells (IECs) and subsequent graft versus host disease, which is controlled by the composition of commensal bacteria. Koyama et al. identifiy taxa that positively (inducers) and negatively (suppressors) correlate with MHC-II expression on IECs, and show that they are regulated by IFNγ-secreting intestinal T cells.
Introduction
Allogeneic hematopoietic stem cell transplantation (SCT) is a curative therapy for hematopoietic malignancies and non-malignant diseases characterized by genomic aberrations that lead to immune deficiency or errors in metabolism 1,2 In the setting of malignancy, the major therapeutic benefit arises from donor T cells and NK cells which recognize host allogeneic and malignancy-specific disparity to eliminate malignant cells, termed graft-versus-leukemia (GVL) effects 3. However, these immune responses can also target healthy recipient tissue, resulting in acute and chronic graft-versus-host disease (aGVHD and cGVHD), which are the major complications of allogeneic SCT 4,5 Acute GVHD is mediated by donor T cells in the graft which respond to alloantigen presented by recipient antigen presenting cells (APC) and typically targets the skin, liver and gastrointestinal (GI) tract. Severe gut GVHD is a major determinant of transplant-related mortality (TRM) early after transplant 6,7 Severe aGVHD is also an important risk-factor for subsequent cGVHD 4.
The reason that aGVHD occurs in only a limited number of organs (skin, liver and GI tract) characterized by an environmental interface remains unclear. In the last decade, commensal microbiota and their derivatives (pathogen-associated molecular patterns (PAMPs) and metabolites) have been shown as pivotal in the modulation of GVHD 8. Innate inflammatory signals from the microbiota (PAMPs), damage to the mucus barrier by mucinolytic bacteria, and the depletion of protective metabolites (e.g. riboflavin- or bile acid-related metabolites and short-chain fatty acid (SCFA)) from relevant bacterial taxa, have been shown to influence transplant outcome in preclinical GVHD models 9. However, the immunological mechanisms through which distinct bacterial taxa influence disease pathology remain elusive.
Recently, we demonstrated that MHC class II (MHC-II) is expressed by intestinal epithelial cells (IEC) in the ileum under the influence of the microbiota and cytokines (interleukin-12 (IL-12) from macrophages and interferon-γ (IFN-γ) from type 1 innate lymphoid cells and T cells). Furthermore, transplanted mice whose IEC lack MHC-II expression are protected from CD4+ T cell-mediated GVHD lethality and gut pathology 10. In the current study, we showed that genetically identical mice of differing vendor origins had distinct compositions of commensal bacteria with prominently different MHC-II expression on IEC in the ileum at steady state, resulting in significantly discordant CD4+ T cell-mediated GVHD severity. We identified bacterial genera/species positively and negatively associated with MHC-II expression by IEC (hereinafter referred to as MHC-II inducers and suppressors respectively) from a series of experiments that involved cohousing to promote natural microbial transfer, and targeted alteration of bacterial composition by antibiotic treatment. Finally, we demonstrate HLA class II (HLA-II) expression by IEC in the ileum of patients and the clinical impact of bacterial taxa in the pre-transplant stool of individuals who subsequently developed grade 2-4 acute GVHD and/or transplant-related mortality (TRM). These data highlight the importance of microbiota composition for the initiation or suppression of immune-mediated pathology in the GI tract.
Results
Genetically identical mice of differing vendor origin exhibit prominent differences in IEC MHC-II expression and intestinal microbiota composition
To examine the influence of commensal microbiota on MHC-II expression by IEC at steady state, we obtained genetically identical sex- and age- matched mice from different vendors. We used cytometric analysis to compare MHC-II expression by ileal IEC between C57BL/6N (B6N JAX) and C57BL/6J (B6J JAX) mice derived from The Jackson Laboratory and mice from Taconic Biosciences (B6N TAC), Charles River Laboratories (B6N CR) or an Australian-derived colony maintained at Fred Hutch (referred to as B6N ADFH). MHC-II expression was absent on IEC in JAX B6N and B6J, while it was expressed at similar frequency and intensity among B6N from TAC, CR and ADFH (Figure 1 A – B). In addition, when we assessed other inbred strains of C57BL/6J, BALB/c and DBA/2 mice from JAX and TAC, we found that independently of genetic variation between the strains, JAX-derived mice did not express MHC-II on IEC (Figure 1B). Likewise, although C57Bl/6J (B6J) and B6J-derived B6N have minor genetic differences, both B6J and B6N strains from JAX lacked MHC-II expression (Figure 1 A – B). These results prompted us to investigate the intestinal microbiota in these differing vendor mice.
Figure 1. MHC II expression on intestinal epithelial cells (IEC) is dependent on intestinal microbiota rather than murine genetic variation.
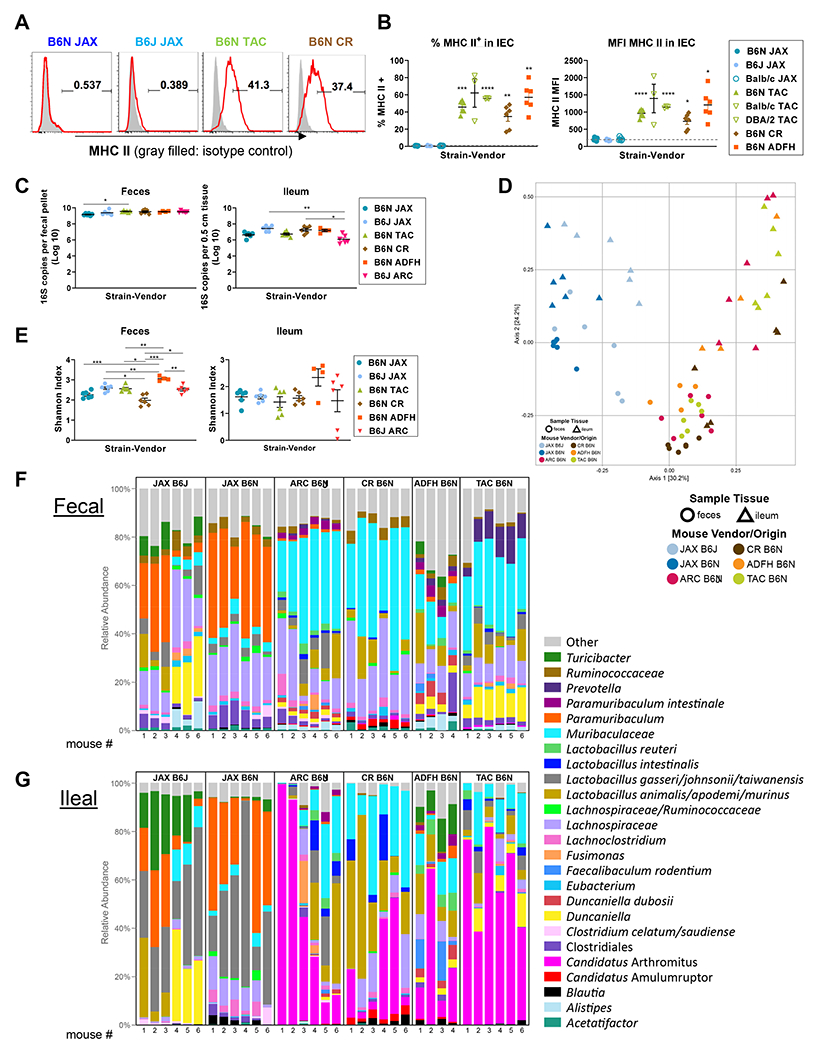
(A, B) Naïve B6J, B6N, BALB/c and DBA/2 mice derived from different vendors were compared for MHC-II expression on ileal IEC. (A) Representative flow plots and (B) quantification of positivity and MFI for MHC-II on IEC. Data of BALB/c and DBA/2 from Taconic are from 1 experiment, n = 3 per group. Remaining data are combined from 2 experiments, n = 6 per group. Brown-Forsythe and Welch ANOVA test with Dunnett’s T3 multiple comparisons against B6N JAX. (C-G) Fecal and ileal samples were collected from naive JAX B6J, JAX B6N, Taconic B6N, CR B6N, ADFH B6N and ARC B6J mice and underwent 16S rRNA gene sequencing. All data are combined from two independent litters except ARC B6J which was from one (n = 4 – 6 per group). (C) The total bacterial load in fecal and ileal samples was compared by Kruskal-Wallis test with Dunn’s multiple comparison test. (D) Multi-dimensional scaling (MDS) plot using the Bray-Curtis dissimilarity metric was used to visualize the fecal and ileal microbiota of mice from different vendors. Each point represents the intestinal microbiota in a single sample. (E) Shannon index shown by Welch’s t-test with p-values adjusted using Holm’s method. (F-G) Relative abundance of the top 25 bacterial taxa detected in fecal (F) and (G) ileal samples. (B, C, E) Show mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Given that gut bacterial decontamination with an antibiotic cocktail of cefoxitin, gentamycin, metronidazole and vancomycin abrogates MHC-II expression by ileal IEC 10, we anticipated that the microbiota responsible for MHC-II expression would be absent in JAX mice and present in mice derived from the other vendors. We thus conducted 16S rRNA gene sequencing on both fecal and ileal samples from these mice. In these analyses we also included samples collected from B6J mice derived from an Australian vendor (Animal Resources Centre (ARC) in Perth, Western Australia) which we predominantly used in our previous study 10. The bacterial loads within the feces and ileum of JAX mice (B6J and B6N) were similar to those of mice of differing vendor origin (Figure 1C). However, multidimensional scaling (MDS) by Bray-Curtis dissimilarity metric revealed that fecal and ileal samples obtained from JAX mice clustered separately to samples obtained from mice originating from other vendors. These data suggested that the intestinal bacterial composition in JAX mice, which lacked IEC MHC-II expression, differs from that of mice from other vendors (Figure 1D). Although the Shannon diversity index was not substantially altered across the different vendor mice (Figure 1E), the top 25 most abundant bacteria were distinct; JAX mice had high proportion of Bacteroidales (order), whereas the other vendor mice were enriched in Lactobacillus animalis/apodemi (species) and Clostridium (genus) (Figure 1F – G). Compared to fecal samples, ileal samples demonstrated lower bacterial diversity (mean Shannon diversity: 2.5 ± 0.4 vs. 1.6 ± 0.6, p < 0.001; Figure 1E) and the uniqueness of bacterial composition in JAX mice was particularly highlighted by analysis in the ileum (Figure 1D). Of note, the bacterial landscape was largely determined by the host origin (JAX B6J vs. ARC B6J or JAX B6N vs. CR/TAC/ADFH B6N) rather than minor genetic variation (JAX B6J vs. JAX B6N). In order to include rare bacterial species in the analyses of correlation with MHC-II expression, we expanded the target species to 99 and 61 taxa in fecal and ileal samples respectively; these were detected in more than 10 % of total samples (more than 3 specimens out of 28 samples per anatomic site). ARC mice were excluded from this analysis as we did not have MHC-II data paired to the microbiota samples from these mice. We considered four parameters: two for microbiota (a. the bacterial presence/absence and b. the bacterial abundance), and two for MHC-II expression by IEC (c. the percentage of MHC-II positive cells and d. the mean fluorescence intensity (MFI) of MHC-II). We calculated the correlation between bacterial species and MHC-II expression in four ways (a vs. c, a vs. d, b vs. c and b vs. d) and combined them statistically while accounting for the number of comparisons (see Statistical analysis for murine data in the Supplemental data) to define the bacterial species which positively and negatively correlate with MHC-II expression by IEC (hereinafter referred to as MHC-II inducers and suppressors respectively). We identified 7 MHC-II inducers and 12 MHC-II suppressors in fecal samples (Figure 2A) and 5 MHC-II inducers and 13 MHC-II suppressors in ileal samples (Figure 2B). A large proportion of species in the feces and ileum were congruent as either inducers or suppressors (100% of ileum inducers were found within the inducers in feces, whereas 46% of ileum suppressors were found in the feces and represented 50% of the total suppressors at this site). No species changed from a suppressor to an inducer, or vice versa. As expected, most of the inducers were significantly more abundant in non-JAX mice (TAC B6N, CR B6N, ADFH B6N and ARC B6J) compared to JAX mice, whereas the suppressors were significantly more abundant in JAX mice (JAX B6N and JAX B6J) (Figure 2C – D).
Figure 2. MHC II inducers are abundant in Non-JAX mice whereas MHC-II suppressors dominate in JAX mice.
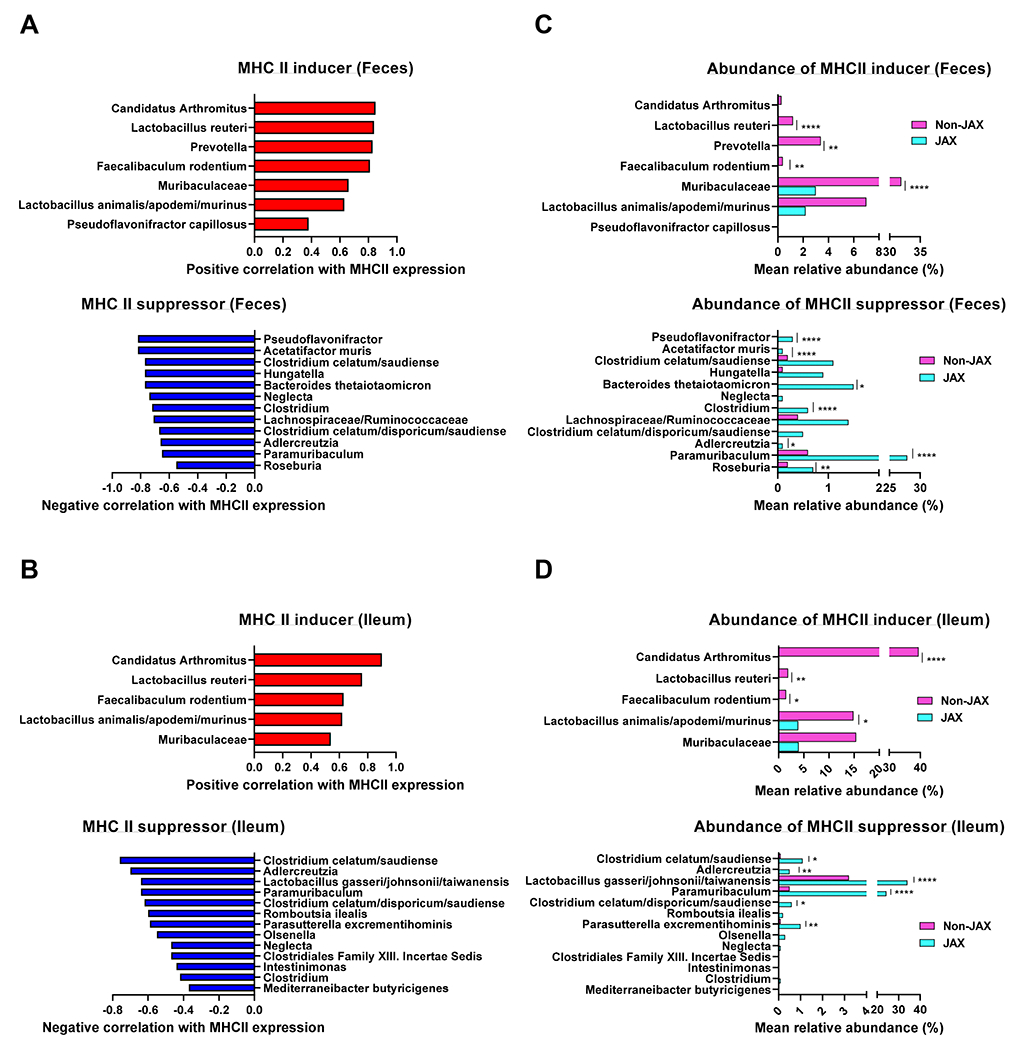
(A, C) Fecal and (B, D) ileal bacteria identified from naive JAX (B6J, B6N) or non-JAX (Taconic B6N, CR B6N, ADFH B6N and ARC B6J) mice were correlated with MHC-II expression by ileal IEC. Only taxa present in at least 3 specimens out of 34 specimens were included corresponding to 106 taxa in fecal samples and 67 taxa in ileal samples. (A, B) The taxa positively (red) or negatively (blue) correlated with MHC-II expression are referred to as MHC-II inducers and suppressors respectively. Due to the lack of MHC-II data paired to microbiota analyses, ARC B6J mice were excluded. (C, D) The relative abundance of taxa defined as MHC-II inducers or suppressors are shown in JAX (B6J and B6N) and non-JAX mice (Taconic B6N, CR B6N, ADFH B6N and ARC B6J mice). Wilcoxon Rank-sum test with p-values adjusted using Holm’s method. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Genetically identical mice from divergent vendors develop CD4+ T cell-mediated GVHD of differential severity that correlates with MHC II expression by IEC.
Next, we investigated whether mice with identical genetics but from different vendors exhibited differences in GVHD severity correlating with differences in MHC-II expression by IEC. To examine this in a GVHD model that is MHC-II-dependent, lethally irradiated male B6N mice from different vendors were transplanted with bone marrow (BM) from female B6 mice and Marilyn T cells (I-Ab-restricted male H-Y antigen-specific TCR transgenic T cells). Female B6N recipients served as non-GVHD controls. Male JAX recipients did not develop lethal acute GVHD and had similar survival to female recipients. In contrast, male CR and ADFH recipients developed significant GVHD lethality (Figure 3A). Clinical GVHD scores at day 7 post BMT were concordant with lethality (Figure 3A). Next, we validated these findings in a second system using polyclonal CD4+ T cells to induce GVHD. Lethally irradiated female B6N (H-2b) mice from different vendors were transplanted with BM and CD4+ T cells from MHC-mismatched female BALB/c (H-2d) mice. Recipients receiving T cell depleted (TCD) BM were used as non-GVHD controls. JAX recipients again developed less GVHD lethality and had lower clinical scores at day 7 compared to the recipients from other vendors (Figure 3B). Histopathological analysis of the ileum confirmed significantly increased GVHD in CR and ADFH recipients, compared to JAX recipients (Figure 3C). Next, we examined if the variance in microbiota and MHC-II expression by IEC affects the initial proliferation of donor T cells following BMT. Lethally irradiated male B6N recipients from JAX and CR were transplanted with Marilyn T cells, and alloantigen-specific donor CD4+ T cell proliferation was assessed early post-transplant (day 4). While high MHC-II expression by ileal IEC in CR-derived recipients was retained, alloreactive Marilyn T cell expansion in the GI tract was higher in CR recipients compared to JAX recipients, suggesting intestinal donor T cell priming was enhanced by MHC-II expressing IEC (Figure 3D). These data indicate that despite being genetically identical, recipients harboring MHC-II-inducers display more severe gut GVHD and expansion of donor T cells, compared to recipient harboring MHCII-suppressors. Together, these findings highlight the crucial role of pre-transplant intestinal microbial composition in determining transplant outcome.
Figure 3. Genetically identical mice from different vendors develop CD4+ T cell-mediated GVHD of varying severity that correlates with MHC-II expression by IEC.
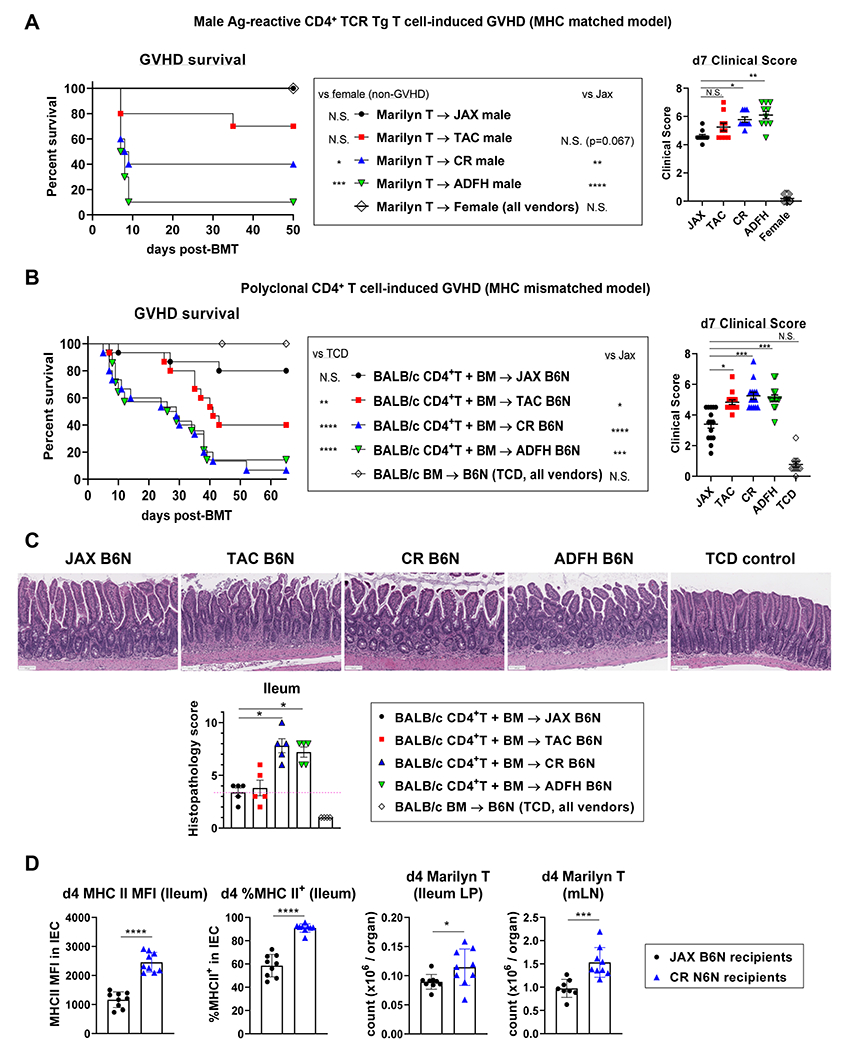
(A) Lethally irradiated B6N mice from the indicated vendors were transplanted with female B6 BM and 0.5 x 106 Marilyn T cells. Female recipients were used as negative controls. Survival by Kaplan-Meier analysis and clinical score on day 7 post BMT, combined from 2 independent experiments (n = 7 – 10). (B, C) Lethally irradiated female B6N mice were transplanted with BALB/c BM and 5 x 106 PC61-treated (regulatory T cell-depleted) CD4+ T cells. Recipients of TCD grafts are negative controls. (B) Survival by Kaplan-Meier analysis and clinical score on day 7, combined from 3 independent experiments (n = 15 – 11). (C) Intestinal histopathology scores and representative images on day 8 post BMT (n = 5 per T cell replete group, 4 per TCD from 1 experiment). (D) Lethally irradiated male B6N mice from JAX and CR vendors were transplanted with 1.0 x 106 Marilyn T cells and the mLN and ileum were analyzed on day 4 (n = 9 per group combined from 2 independent experiments) (A-C) Statistical analyses: Day 7: Kruskal-Wallis test (mean ± SEM). (D) t-test with Welch’s correction (mean ± SEM). *P < 0.05, ** P < 0.01, ***p < 0.001, ****p < 0.0001.
MHC-II-inducer but not suppressor microbiota transfer between CR and JAX mice during cohousing
To confirm that the distinct microbiota between JAX mice and other vendor mice determine MHC-II expression on IEC, we examined the effect of microbial transfer between two different vendor mice. After JAX B6N and CR B6N were cohoused for 4 weeks, MHC-II expression by ileal IEC and microbiota composition were examined. Age and shipment matched non-cohoused JAX and CR mice served as controls. After cohousing, JAX mice acquired MHC-II expression at comparable frequency and intensity to CR mice, while MHC-II expression remained absent in the non-cohoused JAX mice (Figure 4A). Fecal 16S rRNA sequencing demonstrated that cohousing amalgamated the microbiota in JAX and CR mice, while the microbiota in the non-cohoused JAX mice remained distinct (Figure 4B). The composition of microbiota was next examined in relation to MHC-II expression in 80 and 62 taxa from the feces and ileum, respectively. Many of the bacteria that correlated with MHC-II expression in the vendors comparison (Figure 2) reemerged and were congruent across the fecal and ileum analysis. Lactobacillus animalis/apodemi/murinus and Candidatus Arthromitus (species) were again seen as inducers, and Adlercreutzia (genus) and multiple species/genus in Clostridiales (order) were again noted as suppressors (Figure 4C – D). MHC-II inducers were significantly more abundant in cohoused JAX and co-housed/non-cohoused CR mice, whereas MHC-II suppressors were significantly more abundant in non-cohoused JAX mice compared to cohoused JAX and co-housed/non-cohoused CR mice (Figure 4C – D). Since IEC MHC-II expression in CR mice was not reduced following their cohousing with JAX mice (Figure 4A), we assessed the nature of the species that were successfully or unsuccessfully transferred during the cohousing process. MHC-II inducers were successfully transferred from CR to cohoused JAX mice, whereas most MHC-II suppressors failed to transfer (Figure S1A – B). We also found that the reduced GVHD lethality observed in JAX relative to CR recipients (Figure 3), was lost following co-housing (Figure S1C). Together, these data confirmed that the differential MHC-II expression by ileal IEC isolated from genetically identical mice of differing vendor origin is dependent on transferable intestinal microbiota. Notably, MHC-II suppressors, which are predominantly anaerobes, failed to transfer or successfully colonize after cohousing.
Figure 4. MHC II-inducing microbiota are transfered after cohousing.
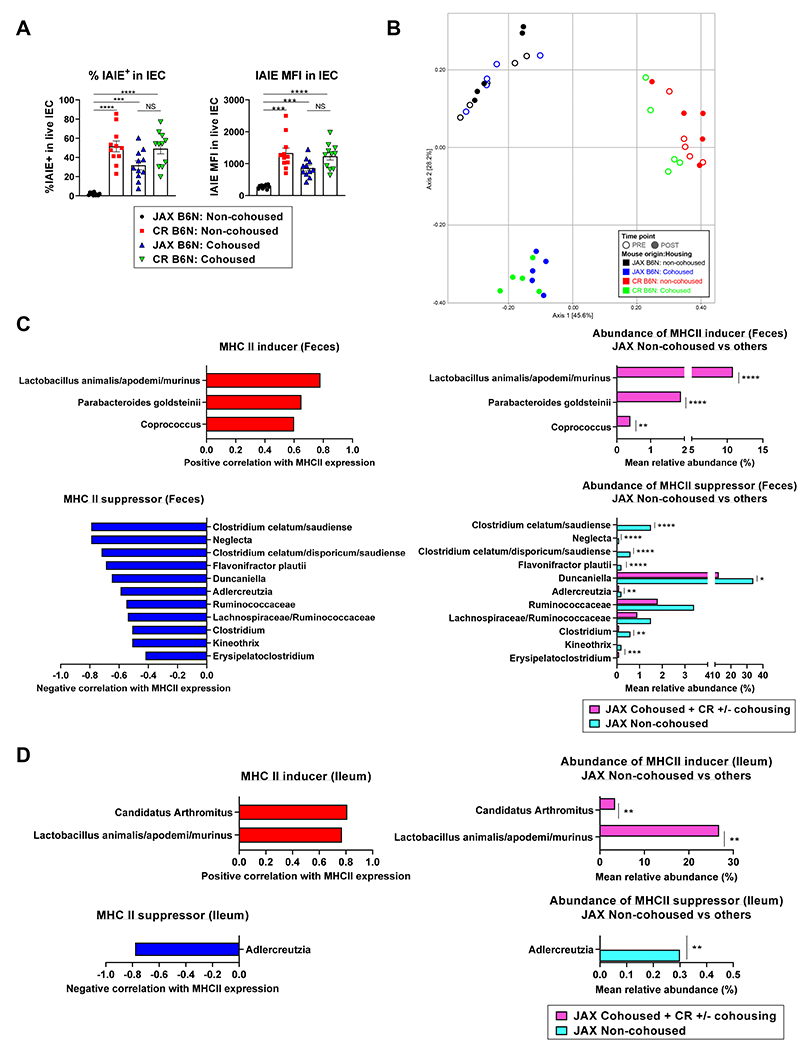
JAX B6N and CR B6N mice were housed separately (non-cohoused) or cohoused for 4 weeks. Fecal samples were collected before and after cohousing. Ileal MHC-II and microbiota samples were obtained after cohousing (n = 11 per group combined from 2 independent experiments except ileal microbiota where n = 5 per group from 1 experiment). Only taxa present in at least 3 specimens (out of 20 ileal samples or out of 44 mice with fecal samples) were analyzed equalling to 80 taxa in fecal samples and 62 taxa in ileal samples. (A) Quantification of MFI and % MHC-II expression by IEC (mean ± SEM). Statistical analysis by Brown-Forsythe and Welch ANOVA test with Dunnett’s T3 multiple comparisons. (B) MDS plot using the Bray-Curtis dissimilarity metric was used to visualize the fecal microbiota of mice from a representative experiment (n = 5 per group from 1 experiment) pre- and post-cohousing. Each point represents the fecal microbiota in a single sample. The fecal microbiota of JAX mice were similar to the microbiota of CR mice post-cohousing while the microbiota of control JAX mice that were not cohoused did not shift in composition post-cohousing. (C, D) MHC-II inducers (red) and MHC-II suppressors (blue) and their relative abundance in JAX non-cohoused mice (cyan) vs. others (JAX cohoused, CR-cohoused and CR non-cohoused mice: pink) in (C) feces and (D) ileum. Statistical analysis by Wilcoxon rank-sum test with p-values adjusted using Holm’s method. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S1.
Intestinal microbiota sensitive to oral vancomycin and gentamicin promote MHC-II expression on IEC
Intestinal decontamination by combinations of broad-spectrum antibiotics has been shown to abolish MHC-II expression on ileal IEC 10,11. However, the differential effects of individual antibiotics remain unclear. To generate an atlas of bacterial species which positively and negatively correlate with MHC-II expression by IEC (MHC-II inducers and suppressors), the comparison of bacterial taxa which perish or persist after various antibiotic treatments, and the relationship to MHC-II expression by IEC, represents an additional approach. ADFH B6N mice, which constitutively expressed MHC-II (Figure 1B) and exhibited high bacterial α-diversity among various vendor origin mice (Figure 1E) were treated with drinking water containing various antibiotics for two weeks. MHC-II expression by ileal IEC and bacterial microbiota from feces and ileum were subsequently examined. Compared to control water, a four-antibiotic cocktail (4AB; cefoxitin, gentamicin, metronidazole and vancomycin) which we had utilized in our previous study10, significantly decreased MHC-II expression. To a lesser extent, isolated treatment with gentamicin or vancomycin also significantly reduced MHC-II expression. Cefoxitin in isolation did not cause a significant reduction in MHC-II expression, although a clear trend was noted. In contrast, metronidazole had no effect on MHC-II expression. Indeed, a three-antibiotic cocktail excluding metronidazole (3AB; cefoxitin, gentamicin and vancomycin) had an equivalent effect on the reduction of MHC-II expression as the 4AB treatment (Figure 5A). These results suggest that species sensitive to gentamicin and vancomycin, but not metronidazole, are responsible for inducing MHC-II expression.
Figure 5. Peri-transplant oral vancomycin attenuates CD4+ T cell-mediated GVHD in an MHC-II and IEC dependent manner.
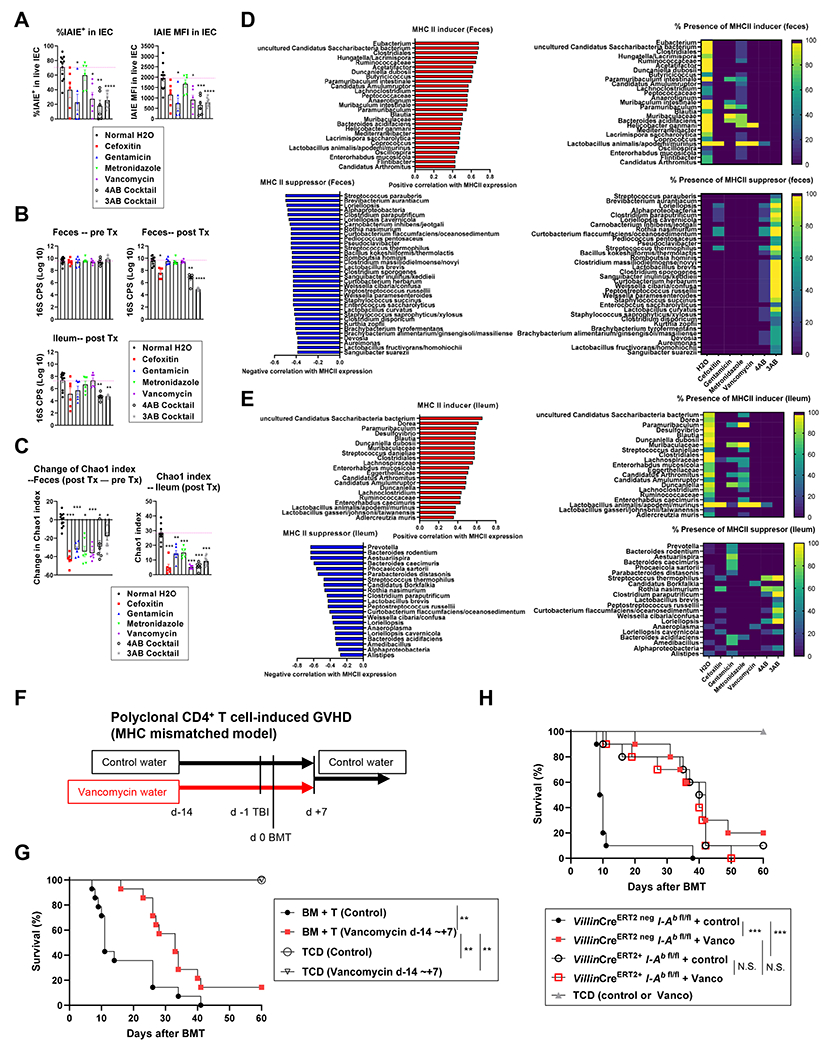
ADFH B6N mice were treated with drinking water containing various antibiotics as shown, for 2 weeks. Fecal samples were collected before and after treatment. Ileal MHC-II and microbiota were analyzed after treatment. Two sets of comparisons as 4AB cocktail, single antibiotics vs. normal H2O (n = 6 per group combined from 2 independent experiments) and 3AB cocktail vs. normal H2O (n = 6, 9 per group combined from 2 independent experiments) were combined and analyzed. Only taxa present in at least 3 specimens (out of 55 fecal or ileal samples) were analyzed representing 128 taxa in fecal samples and 61 taxa in ileal samples. (A) Quantification of MFI and % MHC-II expression by IEC. (B) Bacterial load presented as log10 (16S rRNA gene copies per sample as one pre- or post- treatment fecal pellet, or 0.5 cm Ileal tissue). (C) Chao1 index in post-treatment fecal (top) and ileal (bottom) samples. (D, E) MHC-II inducers (red) and MHC-II suppressors (blue) (left) and their frequency in each treatment group (right) of fecal (D) and ileal (E) samples. Statistical analysis by Brown-Forsythe ANOVA and Welch’s t-test with Dunnett’s T3 multiple comparison test (A), Kruskal-Wallis test with Dunn’s multiple comparison test against normal H2o (B), and Welch’s t-test with p-values adjusted using Holm’s method (C). Data are shown as mean ± SEM (A-C). (F - H) Female mice received either vancomycin water or normal drinking water from day −14 to day 7 after BMT, thereafter all mice received normal water. Mice were transplanted with BALB/c BM and 5 x 106 PC61-treated CD4+ T cells. (G) Recipients were CR B6N mice and (H) B6-background VillinERT2 neg I-Ab fl/fl or VillinERT2 + I-Ab fl/fl mice. Survival by Kaplan-Meier analysis combined from 2 independent experiments (G: n = 14 per T cell replete, n = 4 per TCD. H: n = 10 per T cell replete, n = 5 per TCD). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S2.
Of the single antibiotic treatments, only cefoxitin quantitatively reduced the total bacterial load in feces. In contrast, the cocktail treatments profoundly reduced the total bacterial load in both the feces and ileum (Figure 5B). Critically, single antibiotic treatments significantly reduced bacterial diversity, as determined by α-diversity (Chao1 richness [Figure 5C] and Shannon diversity [Figure S2 A–D]). 16S rRNA sequencing produced large lists of MHC-II inducer and suppressor species in the fecal and ileal samples (Figure 5D left – E left). When we estimated the detection frequency of inducers and suppressors after antibiotic treatment, cefoxitin and vancomycin, and to a lesser extent gentamicin depleted large fractions of inducers from the feces and ileum, whereas metronidazole retained the inducers (Figure 5D right – E right). In regard to the MHC-II suppressors, gentamicin retained multiple suppressors, particularly in the ileum. A comparison between the presence and absence of metronidazole within the cocktail treatments (4AB vs 3AB) demonstrated that metronidazole inhibited the expansion of suppressors when inducers were concurrently depleted by the combination of cefoxitin, vancomycin and gentamicin. Given that i) vancomycin in isolation significantly decreased MHC-II expression without increasing suppressors and ii) the 4AB cocktail blocked the blooming of suppressors but showed similar reductions in MHC-II expression on IEC to the 3AB cocktail where suppressors bloomed, the dominant determinants of MHC-II expression by IEC are inducers rather than suppressors (Figure 5D – E).
Qualitative change of microbiota by peri-transplant oral vancomycin treatment attenuates MHC-II-dependent GVHD
Since vancomycin in isolation abrogated most of the MHC-II inducers (Figure 5D right – E right), and this antibiotic is used clinically and is not absorbed, we next assessed MHC-II dependent (CD4+ T cell-mediated) GVHD in the presence of early peri-transplant oral vancomycin. Recipient mice treated with oral vancomycin from day −14 to day 7 after BMT (Figure 5F) had significantly reduced GVHD lethality compared to control recipients (Figure 5G). Next, we tested whether the protective effect of peri-transplant oral vancomycin is dependent on MHC-II expression by IEC. We thus compared the effects of vancomycin in recipients with or without IEC-specific MHC-II gene depletion (VillinCre-ERT2+ I-Ab-fl/fl or VillinCre-ERT2 neg I-Ab-fl/fl). Vancomycin treatment significantly reduced GVHD lethality in the presence of MHC-II expression by host IEC. In contrast, the protective effects of vancomycin were lost in the absence of MHC-II expression by recipient IEC (Figure 5H). These data confirm that peri-transplant oral vancomycin attenuates GVHD via a critical terminal pathway that is dependent on MHC-II expression by IEC. The late deaths in mice receiving early vancomycin were the result of severe skin GVHD, consistent with previous observations in mice where MHC II was conditionally deleted in IEC pre-transplant 10. Since control mice in which IEC express MHC-II succumb to rapid GVHD, we cannot distinguish whether this skin disease results from the possible redistribution of alloreactive T cells from the intestine (versus local cutaneous expansion), or simply reflects the natural course of GVHD in mice that survive long term.
The most clinically relevant off-target effect of the suppression of MHC-II in IEC is its influence on the protective graft-versus leukemia (GVL) effect. We thus examined the effect of oral antibiotics on GVL to determine how altering the intestinal microbiota influences immunity outside the GI tract. Lethally irradiated B6N CR mice were transplanted with BM and CD4+ T cells from BALB/c mice and a primary B cell acute lymphoblastic leukemia (C57Bl/6-background MHC-II+, Arf−/−MSCV-RCSD1-ABL2-ires-GFP+MSCV-IK6-ires-RFP) on day 0. The antibiotic-treated group received the 3AB cocktail that potently suppressed IEC MHC-II expression. As expected, antibiotic treatment profoundly attenuated GVHD, leading to significant improvement in overall survival. However, GVL was also attenuated (Figure S2 E–G). Thus, targeting MHC-II inducer bacteria with oral antibiotics during the peri-transplant period is an effective approach to prevent the initiation of GVHD but is associated with the downstream inhibition of MHC-II-dependent GVL.
Microbiota invoke IFNγ secretion in local lymphocytes to control MHC-II expression by IEC
To understand how microbiota control MHC-II expression by IEC, we examined intestinal and lymphoid organ immune subsets from CR mice harboring MHC-II inducer and JAX mice harboring MHC-II suppressor microbiota, respectively. As previously noted, IEC in CR mice had high MHC-II expression, but MHC-II expression by intestinal hematopoietic APCs (conventional dendritic cells (cDC) and macrophages) was marginally lower than in JAX mice (Figure S3 A–B). CR mice had increased frequencies and absolute numbers of conventional CD4+ and CD8+ T cells in the ileum compared to JAX mice (Figure S3 C–D). No differences were observed in the spleen (Figure S3 C–D). The T cell expansion observed in the ileum led us to investigate the function of these local T cells. IFNγ is the major driver of MHC-II expression on IEC, and IEC-specific IFNγ receptor deletion (in tamoxifen-treated villinCreERT2+ ifnγr fl/fl mice) profoundly decreased MHC-II expression relative to controls (Figure S4A). We therefore compared IFNγ production by T cells in genetically identical IFNγ-YFP reporter mice from different vendors (JAX GREAT and ADFH GREAT) (Figure S4B). As expected, ileal IEC from ADFH GREAT mice had high MHC-II expression at steady state (Figure S4 C–D). The expression of IFNγ receptor was similar in mice from the two vendors (Figure S4 C–D), consistent with the notion that differences in MHC-II expression in these mice (Figure 1B) are due to differences in local IFNγ. IFNγ expression by T cells (but not ILC1 or NK cells) from ADFH mice was increased on a per cell basis, as was the total number of IFNγ-expressing T cells in the ileal epithelial layer and lamina propria (Figure S4 E–F). These effects were localized to the GI tract and were not seen in the spleen (Figure S4F). The increase in IFNγ production by intestinal T cells in the ADFH GREAT mice persisted after TBI (Figure S4 G–I). We also evaluated fecal microbiota from naïve JAX and ADFH GREAT mice. IFNγ (YFP+) T cell numbers and MHC-II expression were positively and tightly correlated (Spearman’s correlation r = 0.92 – 0.93, p < 0.001) (Figure S5A). Fecal microbiota from JAX and ADFH GREAT mice were distinctively separate, and the bacterial species that correlated with MHC-II expression also strongly correlated with species associated with IFNγ-YFP+ T cells, both positively and negatively (Figure S5 B–D). When we compared these species to the MHC-II inducers and suppressors identified in our previous comparisons (vendor, cohousing and antibiotics), 12 species of MHC-II inducers and 3 species of MHC-II suppressors were detected in GREAT mice (Figure S5 E–F). These results are consistent with the notion that MHC-II inducers and suppressors control the size of the small intestine T cell compartment and their IFNγ production, resulting in the differential expression of MHC-II by IEC.
MHC-II suppressor bacteria can attenuate MHC-II-dependent GVHD
Although the bacterial landscape after antibiotic treatment suggests a dominant role for MHC-II inducers over suppressors for the control of epithelial MHC-II expression (Figure 5D right – E right), we sought to determine whether MHC-II suppressors can be leveraged to reduce MHC-II expression by IEC. We took the fecal contents from our 3AB-treated ADFH B6N mice, which include most MHC-II suppressors in expanded proportions (Figure 5D) and transferred these microbial communities enriched in MHC-II suppressor taxa into preconditioned recipients for 2 or 4 weeks. This transfer decreased ileal MHC-II expression in CR B6N mice and modulated microbial composition with an increase in MHC-II suppressors and a decrease in MHC-II inducers (Figure S6 A–D). We next selected two species (Clostridium sporogenes and Bacteroides thetaiotaomicron) from our MHC-II suppressor list (Figure S5F) that could be cultured and expanded in vitro. We inoculated these bacteria at a 1:1 ratio into B6N mice preconditioned with 3AB containing water. After 2-weeks of oral gavage with cultured bacteria or vehicle control, the mice were lethally irradiated and analyzed 24 hours later (Figure 6A). Mice that received suppressor bacteria had significantly decreased ileal MHC-II expression (Figure 6B). Both of the inoculated suppressor bacteria were detected in feces from suppressor-treated mice but not those from vehicle-treated mice (Figure 6C). We noted that Bacteroides thetaiotaomicron was the dominant transferred bacteria in these experiments (Figure 6D) and that this taxon was associated with MHC-II suppression across experiments (Figure 6E). When male B6N mice treated with these two suppressors were transplanted with BM and alloreactive Marilyn T cells, survival was significantly improved compared to the vehicle treated recipients (Figure 6F). These data confirm that following antibiotic mediated reduction of inducer species, MHC-II suppressors can maintain reduced MHC-II expression on IEC and modify the induction of MHC-II-dependent GVHD.
Figure 6. MHC-II suppressors attenuate MHC-II expression on IEC and GVHD.
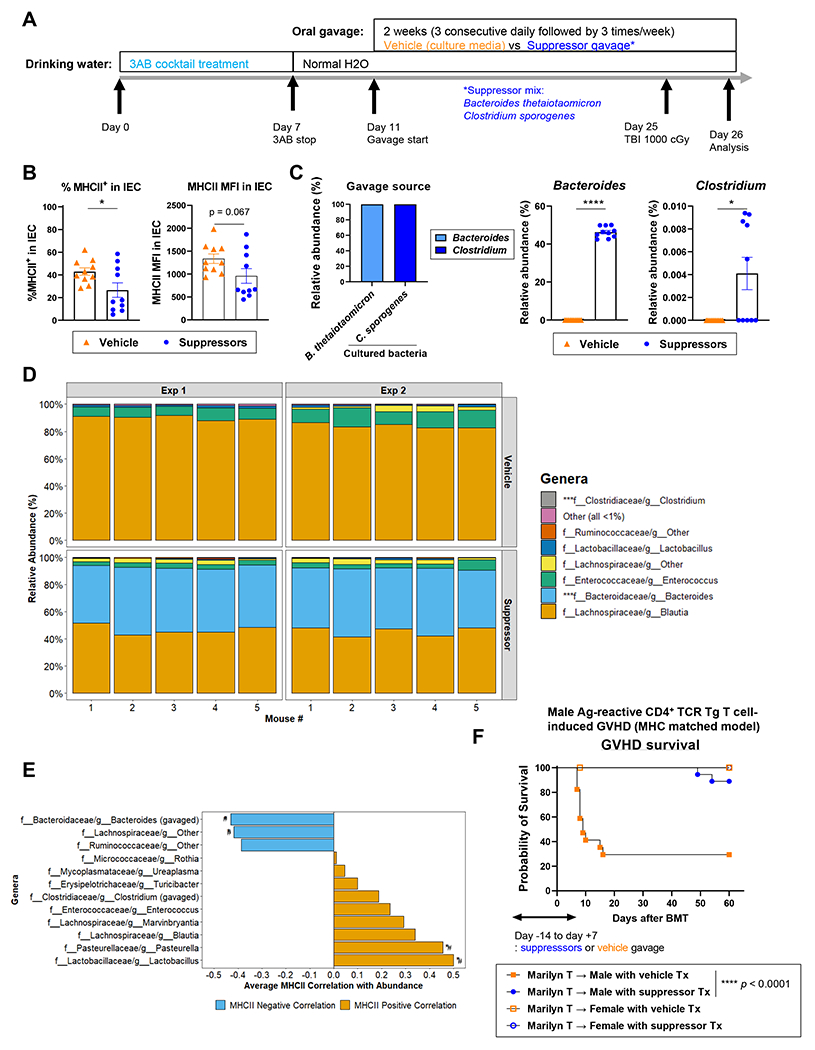
(A) Treatment schema. ADFH B6N mice received 3AB cocktail water for 7 days followed by normal drinking water. Four days later, mice were gavaged with MHC-II suppressor bacteria (1:1 mix of B. thetaiotaomicron and C. sporogenes) or vehicle (culture media) for 2 weeks. Mice were then lethally irradiated (1000 cGy) and analyzed 24 hours later. (B) Quantification of % MHC-II expression and MFI on ileal IEC (n = 10 per group, combined with 2 independent experiments. Statistical analysis by T-test with Welch’s correction [mean ± SEM]). (C) Quantification of relative abundance of Clostridium and Bacteroides (genus) in cultured bacteria and fecal samples (n = 10 per group, combined with 2 independent experiments. Statistical analysis by Mann-Whitney test [mean ± SEM]). (D) Genus-rank relative abundance of fecal bacteria. (E) Genera that negatively (blue) or positively (orange) correlated with MHC-II expression. #p < 0.1 (unadjusted). (F) ADFH B6N male and female mice were treated as described in (A), then transplanted with female B6 BM and 0.5 x 106 Marilyn T cells. Survival by Kaplan-Meier analysis (n = 17 - 18 per male group combined from 2 independent experiments). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S5 and S6.
Pre-transplant fecal microbiota from patients undergoing allogeneic unmodified peripheral blood stem cell transplantation correlate with aGVHD and TRM
Finally, we investigated whether pre-transplant fecal microbiota in patients correlated with the incidence of aGVHD and TRM. Pre-transplant stool samples were collected from a uniform cohort of 287 recipients of unmodified peripheral blood stem cell transplants (PBSCT) at MSKCC between January 2009 and December 2019, all of whom received Calcineurin inhibitor/Methotrexate-based GVHD prophylaxis (patient characteristics, selection criteria and antibiotic exposures, and the timing of sample collection are shown in Figure S7 A–C, Table S1, S2, S6). This cohort had an incidence of grade 2-4 GVHD of 40.8% and an incidence of TRM at 2-years of 15.7%. Pre-transplant fecal α-diversity was not significantly different among patients who developed grade 2-4 and those who developed grade 0-1 GVHD (Figure S7B). The microbial composition of patient samples is shown in Figure 7A. PERMANOVA testing demonstrated a statistically different overall composition between the pre-transplant samples from patients who went on to develop grade 2-4 vs grade 0-1 GVHD (p=0.047, R2=0.008) and between patients who succumbed to TRM within two years and those who did not (p=0.038, R2=0.009). We further assessed these compositional differences using linear discriminant analysis of effect size (LefSE), which demonstrated that patients who do not develop grades 2-4 GVHD have a higher abundance of taxa in the Clostridia class (p=0.02) in their pre-HCT gut microbiome (Figure 7B – C). Patients that later develop GVHD have a higher abundance of taxa in the Erysipelotrichia class, specifically in the genus of Erysipelatoclostridium (p=0.03) and Absiella (p=0.03), Flavonifractor genus (p=0.04) within the Oscillospiraceae family of the Clostridia class, and Negativicutes class (p=0.006). Upon interrogation of TRM within the 2 years after allo-HCT, patients with the highest TRM had pre-HCT microbiota with high abundance of members of the Bacilli class (p=0.04), specifically Lactobacillales (p=0.04), Enterococcus genus (p=0.005) and Enterococcaceae family (p=0.005). These results from LEfSe analysis were further validated with Wilcoxon rank-sum univariate testing, by comparing the abundance of these taxa in patients with GVHD grades 2-4 and TRM.
Figure 7. Pre-transplant Bacilli and Clostridia in stool correlate with grade 2-4 GVHD and TRM.
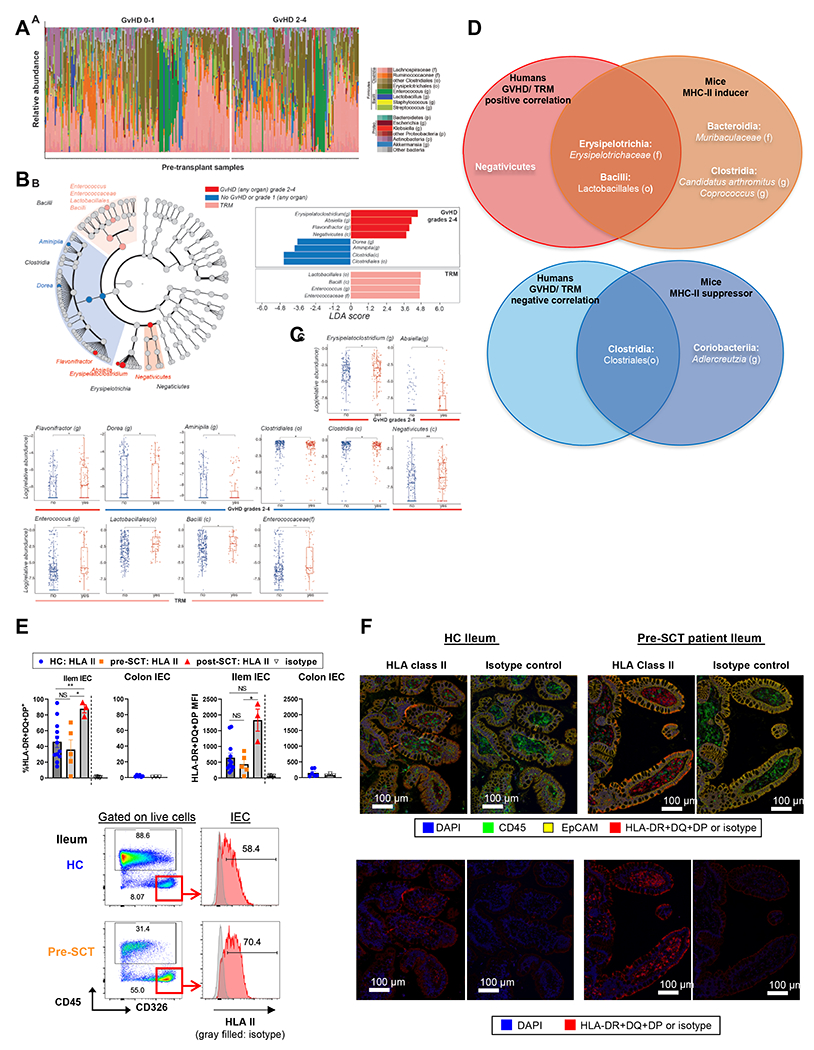
(A) Visualization of the composition of pre-HCT microbiota in 287 patients separated in patients who later develop GVHD grades 0-1 vs. 2-4. (B) LEfSe results are shown in a cladogram form (left) and bar plot (right). The cladogram illustrates the taxa associated with either TRM or GVHD grades 2-4 and their phylogenetic information. The bar plot illustrates the taxa and their LDA score. (C) Taxa-rank comparisons of patients who later develop GVHD grades 2-4 or not; and patients with TRM. The taxa were selected from LEfSe, as seen in panel (B). *p < 0.05, **p < 0.01. (D) Venn diagrams correlating inducers and suppressors across human and mouse. (E-F) Ileum and colon biopsies from healthy individuals and patients pre- or post-transplant were dissociated for flow cytometric analysis (E) or stained for immunofluorescent analysis (F). Representative flow plots with quantification of MFI and % HLA-II expression by IEC (E), and representative immunofluorescence images (F) are shown. (E) Ileum: n = 12, 5, 3 per healthy control (HC), pre-SCT, post-SCT. Colon: n = 6 per HC. Brown-Forsythe and Welch ANOVA test with Dunnett’s T3 multiple comparisons for % positivity of ileal HLA-II and Kruskal-Wallis test with Dunn’s multiple comparisons for ileal HLA-II MFI (mean ± SEM). (F) Ileum: n = 3, 3 per HC or pre-SCT. GVHD: graft versus host disease; TRM: transplant related mortality; LDA: linear discriminant analysis. NS: not significant. See also Figure S7, and Table S1–S3, S6.
By analyzing our combined data, we generated a summary of murine MHC-II inducer and suppressor bacteria at the genus-rank (Figure S7 D–E) and found clear overlap in the pre-transplant bacteria associated with GVHD between humans and mice (Figure 7D). We observed that Clostridia, which were less abundant in patients who developed grades 2–4 GVHD, were present in murine MHC-II suppressors (Lachnospiraceae [family], Clostridium [genus] and Neglecta [genus]) (Figure S7E). In humans, Erysipelotrichaceae (family) in the Erysipelotrichia class positively correlated with grade 2-4 aGVHD (Absiella [genus] and Erysipelatoclostridium [genus]). This finding is consistent with the observation that Faecalibaculum (genus) in Erysipelotrichaceae (family) was an MHC-II inducer in mice (Figure S7D). Additionally, we found that the Bacilli class correlated with TRM in humans, consistent with the potent MHC-II inducer Lactobacillus (genus) of the same Bacilli class being found in mice.
Finally, we confirmed that epithelial cells in the ileum of healthy individuals and patients being screened before BMT expressed HLA class II (HLA-II). In contrast, colonic epithelial cells from healthy individuals did not express HLA-II, consistent with our previous findings in mice 10 (Figure 7E – F). We obtained three ileum biopsy samples from post-transplant patients (range 2-6 months) and noted that IEC expressed significantly increased HLA-II relative to IEC pre-transplant (Figure 7E), similar to observations in our preclinical analyses. Thus, the expression of MHC-II by IEC is not limited to mice and is also likely active as a primary mechanism of GVHD in patients.
Discussion
Minimizing GVHD whilst maintaining/maximizing GVL represents the single largest unmet need in allogeneic SCT. In contrast to CD8+ T cell-mediated (MHC class I-dependent) GVHD in which recipient hematopoietic APC are primarily required for the induction of GVHD 12,13, CD4+ T cell-mediated (MHC class II-dependent) GVHD is induced by both recipient hematopoietic APC and non-hematopoietic APC 14,15. Recently we demonstrated that MHC-II expression on intestinal IEC is necessary and sufficient for lethal CD4+ T cell-mediated GVHD and the presence of intestinal microbiota drives MHC-II expression on IEC via local cytokine production (IL-12 and IFN-γ) that consequently promotes lethal gut GVHD 10. In the current study we demonstrate that specific bacterial taxa control MHC-II expression on IEC, independent of genetic donor-host disparities.
Whilst several studies have reported associations between specific bacterial species and GVHD outcomes, the mechanisms driving these effects were unknown. Several clinical studies have demonstrated that dominance of Enterococcus genus and the loss of Clostridia class after SCT are associated with an increased risk of GVHD 16–19. The presence of Enterococcus in mixed taxa inoculation into gnotobiotic mice increases colonic Th17 differentiation and systemic IFN-γ 18. The protective effects of Clostridia on GVHD are thought to lie in the production of SCFAs, including butyrate, that promote intestinal barrier integrity 20 and regulatory T cell generation 21–23. Recently Tuganbaev et al defined bacterial MHC-II inducers and suppressors utilizing comparisons between different antibiotic treatments in naïve mice11 . They reported Lactobacillus murinus to be an MHC-II suppressor. By contrast, we identified Lactobacillus/animalis/apodemi/murinus species as a universal MHC-II inducer. In our study these taxa were not separated, thus the three sub-species, which have different glycoside hydrolase gene expression, likely also have different effects on MHC-II expression 24. Among other MHC-II inducers identified across our multiple experiments, Candidatus Arthromitus, a segmented filamentous bacteria (SFB), and Muribaculaceae were found as universal MHC-II inducers in our analyses. SFB are potent Th17 inducers due to their ability to adhere to epithelium and stimulate the production of RegIIIγ and IFN-γ 25,26. SFB have also been shown to induce MHC-II expression in gnotobiotic mice 11,27. We have previously shown that the presence of Muribaculaceae (previously known as S24-7) in the pre-transplant period correlates with subsequent GVHD 28,29; here we confirm these bacteria as inducers of MHC-II. Vancomycin treatment before BMT eradicated most MHC-II inducers and reduced CD4+ T cell-dependent GVHD in an MHC-II and IEC dependent manner. Of note, antibiotic decontamination of the gut profoundly mitigated GVHD, but also attenuated GVL, albeit less prominently. The impact of T cell priming and expansion induced by IEC on subsequent T cell migration to the other organs, such as the bone marrow (where GVL activity occurs) early after BMT, thus needs consideration and further investigation is required.
Regarding MHC-II suppressors, we found many taxa in the Clostridia class that negatively correlate with MHC-II expression by IEC. In addition, reductions in MHC-II could be invoked by elimination of MHC-II inducers, even in the absence of MHC-II suppressors, suggesting that inducers mediate the dominant biological effect. Bacteroides thetaiotaomicron was consistently found as an MHC-II suppressor. Recently this species has been reported to augment colonic GVHD via mucus degradation 30. However, when we orally inoculated recipient mice with B. thetaiotaomicron and C. sporogenes prior to transplant, MHC-II expression was reduced and GVHD survival was improved. Although the two bacteria were mixed evenly in the gavage, B. thetaiotaomicron dominated relative to C. sporogenes in the inoculated mice. B. thetaiotaomicron may thus influence GVHD via multiple mechanisms, that are both temporal (before versus after BMT) and site (ileum versus colon) specific. In murine colitis models, B. thetaiotaomicron also exerts protective effects 31,32. B. thetaiotaomicron-derived outer membrane vesicles stimulate PBMC-derived DC from healthy individuals, but not from inflammatory bowel disease patients, to secrete IL-10 33. This axis has been shown to mediate cytoprotection in a DSS colitis model 34.
It is important to note that our pre-transplant microbial modifications result in a different outcome to interventions after BMT and during GVHD. We have demonstrated potent positive and negative associations between bacterial species that promote MHC-II expression by IEC in the ileum and their ability to modulate IFN-γ production by T cells. Thus, T cells in the ileum that secrete IFN-γ were expanded in mice from vendors with high MHC-II-expression at this site. B. thetaiotaomicron negatively correlated with both MHC-II expression and IFN-γ production. Critically, we show that the critical pathway that regulates GVHD is IFN-γ mediated MHC-II expression by IEC - supported by the fact that GVHD protection afforded by vancomycin treatment relies on the depletion of MHC-II inducers and reduction of MHC-II expression by IEC. Furthermore we show that IFNγ signaling directly in IEC is essential for MHC-II expression.
A challenge in preclinical studies is the disparity of microbes harbored in humans and mice 35,36. For example, within the Lactobacillales order, Lactobacillus genus (Lactobacillaceae family) is abundant in mouse but not in human, whereas the Enterococcus genus (Enterococcaceae family) is more abundant in human than mouse 36. It is well known that Candidatus Arthromitus is rarely detected in adult human feces but it is present in early life, principally within the ileum 37,38, raising the potential importance of anatomical considerations. We demonstrated that pre-transplant human microbes indeed correlated with GVHD and TRM within the wide taxonomic classification of MHC-II inducers and suppressors identified in our murine systems. A large number of Clostridium species and other bacteria within the Eubacteriales order within the Clostridia class correlated with human GVHD and fell into the murine MHC-II suppressor category. The Lactobacillales order is shared by the Enterococcus genus and correlated with clinical TRM after transplant, consistent with the Lactobacillus genus that includes Lactobacillus animalis apodemi/murinus and Lactobacillus reuteri being found as MHC-II inducers in mice. The Erysipelotrichia class, which includes the Erysipelatoclostridium and Absiella genus, was associated with clinical GVHD, a finding consistent with the identification of Faecalibaculum rodentium as an MHC-II inducer in mice. Notably, our analyses demonstrated that humans, both healthy and patients with hematological malignancies pre-transplant, express HLA class II on their IEC in the ileum, and expression is further enhanced after transplant.
In conclusion, our findings highlight the complexity of the microbial ecosystem that controls IFN-γ production by local T cell subsets, antigen presentation by IEC and the initiation of subsequent tissue-specific immune pathology. These studies provide a broad framework that defines potential microbiota-focused strategies to prevent the initiation of severe acute GVHD and demonstrate the significant impact of non-genetic, environmental determinants to transplant outcome.
Limitation of the study
We acknowledge that human and murine commensal microbial taxa are different and many species and genus rank do not overlap. However, our data demonstrate that common taxa positively or negatively correlate with MHC-II expression by IEC and subsequent disease. In this study, we did not identify the bacteria-derived molecules that are differentially expressed by MHC-II inducers and suppressers to control the production of IFNγ by T cells in the ileum.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Geoffrey R Hill (grhill@fredhutch.org).
Material availability
This study did not generate new, unique reagents.
Dada and code availability
16S RNA gene seq data have been deposited at GEO and are publicly available as of the date of publication. For murine data, the Bioproject accession numbers : Bioproject PRJNA980199 https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA980199. For human patients data, the Bioproject ID numbers to access the raw and processed files of stool 16S sequencing are listed in Table S6. Flow cytometry gating strategies are included in the supplemental figures.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice.
Sex- and age-matched (7-8 week old) C57BL/6J (B6J, H-2b, CD45.2+), C57BL/6N (B6N, H-2b, CD45.2+), BALB/c (H-2d, CD45.2+) and DBA/2 (H-2d, CD45.2+) were purchased from Jackson Laboratory (JAX; Bar Harbor, ME, USA), Taconic Biosciences (TAC; Albany, NY, USA) and Charles River Laboratories (CR; Wilmington, MA, USA) and the Animal Resources Centre (ARC; Perth, WA, Australia). C57BL/6N, B6.Ifnγ-YFP (Stock 017581, B6.129S4-Ifngtm3.1Lky/J) and B6.Ifnγr−/− (Stock 003288, B6.129S7-Ifngr1tm1Agt/J) mice which were maintained in QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia and subsequently bred at the Fred Hutchinson Cancer Center (Australian-derived Fred Hutch maintained colony, referred to as ADFH B6N, ADFH GREAT and ADFH Ifnγr−/. I-Ab-fl/fl (Stock 013181, B6.129X1-H2-Ab1tm1Koni/J) 39 or Ifnγr1fl/fl (Stock 025394, C57BL/6N-Ifngr1tm1.1Rds/J) and Villin-Cre (Stock 004586, B6. SJL-Tg(Vil-cre)997Gum/J) were intercrossed to generate VillinCre-ERT2I-Ab-fl/fl or VillinCre-ERT2 Ifnγr1fl/fl, respectively. Marilyn Tg (Rag2−/− background) mice on a B6 background originated from Dr P Matzinger, NIH, Bethesda, MD, USA 40 and were backcrossed onto a B6 β-actin-luciferase background (Marilynluc+, CD90.1+ CD45.2+). For cohousing experiments, mice were housed as described previously 28. Mice were housed in sterilized microisolator cages and received acidified autoclaved water (pH 2.5) at the Fred Hutch animal facility. Experiments were performed with sex and age-matched animals using littermates where possible. All animal procedures were undertaken using protocols approved by FHCC IACUC (51055 and 51074).
Human samples.
Patients receiving alloSCT at Memorial Sloan Kettering Cancer Center (MSKCC) were screened, and pre-transplant stool samples were collected from a uniform cohort of 287 recipients of unmodified peripheral blood stem cell transplants (PBSCT) at MSKCC between January 2009 and December 2019 (Figure S7A, Table S1–2). All analysis with human stool samples were conducted with institutional IRB approval from MSKCC (16-834).
Ileal (12 from healthy individuals, 5 from pre-transplant patients and 3 from post-transplant patients) and colonic (6 from healthy individuals) biopsy samples were obtained by the endoscopic procedure. Healthy individuals were participants through colorectal cancer screening. One piece of biopsy tissue per participant was collagenase-digested and analyzed by flow cytometry. Among these participants, formalin-fixed paraffin-embedded ileal tissue blocks from 3 healthy individuals and 3 pre-transplant patients were sectioned and stained for immunohistochemistry. All analysis with human ileal and colonic samples were conducted with FHCC IRB approval (RG1004585 and RG0815001).
METHOD DETAILS
Stem Cell Transplantation.
B6N mice were transplanted as described previously 10 with 1100 cGy total body irradiation (TBI, 137Cs source at 84 cGy/min) on day −1.. For MHC-matched male antigen-specific GVHD model, male or female B6N mice were transplanted with 5 x 106 B6 bone marrow (BM) and 0.5 x 106 luciferase-expressing Marilyn cells (Rag2−/−) on day 0. BM cells were depleted of T cells by antibody and complement as previously described (T cell depletion = TCD). Marilyn CD4+ T cells (Rag2Rag2−/− background) were enriched using CD4 MACS system (Miltenyi). For MHC-mismatched BMT utilizing WT polyclonal CD4+ T cells, lethally irradiated female B6N mice were transplanted with 5 x 106 BM and 5 x 106 CD4+ T cells (MACS purified) from Treg-depleted BALB/c donor mice (PC-61.5.3 (Bio X Cell, Lebanon, MH USA); 500μg/ animal, day - 3 and −1, i.p.). TCD BM grafts were utilized as GVHD negative controls. Animal procedures were undertaken using protocols approved by the institutional animal ethics committee (FHCC IACUC #51055). The severity of systemic GVHD was assessed by scoring as previously described (maximum index = 10) 41. For survival experiments, transplanted mice were monitored daily and those with GVHD clinical scores ≥ 6 were sacrificed and the date of death registered as the next day, in accordance with institutional guidelines. In some experiments, recipient mice received either vancomycin water or normal drinking water from day −14 to day 7 after BMT, thereafter all mice were treated with normal water. Unless explicitly stated, mice did not receive antibiotics during transplantation.
For GVL experiments, lehally irradiated (1000 cGy) female ADFH B6N mice were transplanted with 10 x 106 BM and 0.5 x 106 CD4+ T cells (MACS purified) from BALB/c donor mice with 0.5 x 106 primary B-progenitor acute lymphoblastic leukemia (Arf−/− MSCV-RCSD1-ABL2-ires-GFP + MSCV-IK6-ires-RFP) 42 which was kindly provided by Dr. Charles Mullighan (St Judes, Menphis). The recipients were injected with anti-NK1.1 mAb (clone: PK136) 500 μg/dose and anti-CD8b mAb (clone: 53-5.8) 150 μg/dose intraperitoneally on day −1 and +1 to enable complete and rapid engraftment. Recipients were treated with an antibiotic cocktail consisting of vancomycin, cefoxitin and gentamicin (3AB) added to the drinking water from day −7 to day 28 after BMT. The control mice received control water without antibiotics. Leukemia deaths were defined as animals requiring sacrifice in the presence of >50% GFP+ RFP+ leukemia cells in blood or marrow and GVHD clinical scores <5 in the preceding week. GVHD deaths were defined as those in mice with clinical scores ≥5 and leukemia <10% in blood and marrow. Analysis utilized competing risk models using R software whereby GVHD death is treated as a competing risk for leukemia death as published 43.
Cell isolation from intestine.
Longitudinally sectioned pieces of the murine terminal ileum (3 cm next to the cecum) were processed for epithelial layer preparation (Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer’s protocol. Dithiothreitol was excluded from the entire process. For human biopsy samples, defrosted tissue was incubated in 9 ml digestion buffer (0.01% DNase I, 150 U/ml collagenase type I, 5% FBS, 25 mM HEPES, Penicillin 100 IU/ml, streptomycin 100 ug/ml, 2 mM L-glutamine, 1.1 mM MgCl2, 1.1 mM CaCl2 in RPMI 1640) for 30 minutes at 37 degrees, then washed and filtered through 70 μm filter for flow cytometric analysis.
Oral antibiotic treatment.
For microbiota depletion, an antibiotic cocktail (termed four antibiotic cocktail: 4AB) consisting of vancomycin, metronidazole, cefoxitin and gentamicin (final concentrations 1 mg/ml each) was added to the drinking water for 14 days before analysis 44. The antibiotic cocktail consisting of vancomycin, cefoxitin and gentamicin (termed three antibiotic cocktail: 3AB) or each single antibiotic in isolation was added to the drinking water for 14 days before analysis. The control mice received control water without antibiotics.
Bacterial culture and gavage.
Bacteroides thetaiotaomicron (VPI 5482) and Clostridium sporogenes (ATCC 15579) were culture in Mega media (Table S5) in an anaerobic chamber. These bacteria were mixed at the adjusted concentration (OD600 0.5 to 0.7 per dose) for in vivo treatment (termed suppressor mix). ADFH B6N mice were pre-conditioned by 3AB (1 mg/ml each antibiotic) for 7 days, then switched to normal drinking water. 4 days later, they were treated with oral gavage of suppressor mix or culture media (vehicle control) 200 μl/dose for 3 consecutive days followed by twice a week treatment for a total of 2 weeks for ileal MHC-II analysis or 3 weeks for transplant (day −14 to day 7 after BMT).
Antibiotic enriched suppressor collection and gavage.
ADFH B6N mice were treated with 3AB drinking water for 2 weeks and cecal contents and feces were subsequently collected in PBS and filtered through a 100 μm filter. The flow-through was aliquoted as a large frozen batch (termed “antibiotic enriched fecal suppressor feed”). Naïve CR B6N mice were administered the antibiotic enriched fecal suppressor feed or PBS (200 μl/dose) by oral gavage daily for 2 or 4 weeks. In some experiments, mice were subjected to a bowel cleansing with PEG solution (200 μl/dose, gavaged at 30 min intervals x 6) prior to the 1st day of fecal feed or PBS administration. PEG solution contained PEG 3350 (77.5 g/L), sodium chloride (1.9 g/L), sodium sulfate (7.4 g/L), potassium chloride (0.98 g/L) and sodium bicarbonate (2.2 g/L) diluted in sterile tap water 45.
Flow cytometry.
For surface staining, cell suspensions were incubated with anti-CD16/CD32 (2.4G2) followed by staining with antibodies against CD45.2, MHC class II (I-A/I-E), CD326 (EpCAM), Rat IgG2b isotype control (all BioLegend); CD45 (BD Biosciences). 7AAD (Sigma) was added before cell acquisition. For hematopoietic subsets analysis, after the incubation with Fixable Viability Stain 440UV and anti-CD16/CD32, cells were stained with antibodies as shown in STAR methods and Figure S4B. Samples were acquired with a BD Symphony A3 (BD) and analysis was performed with FlowJo v10 (Tree Star) software.
Histologic analysis.
H&E stained sections of formalin-fixed tissue were coded and examined in a blinded fashion by A.D.C. using a semi-quantitative scoring system, as previously described 15. Images of GVHD target tissues were acquired using an Aperio AT Turbo slide scanner (Leica Biosystems, Wetzlar, Germany) and analyzed using Aperio ImageScope software (Leica).
Bacterial DNA extraction of murine intestinal samples, broad-range PCR, sequencing and processing of sequence reads.
Fecal (1 pellet) and 0.5 cm terminal ileum samples were collected from mice and stored frozen at −80°C until processing. DNA was extracted using the BiOstic Bacteremia DNA Isolation Kit (Qiagen Germantown, MD) with the following modifications. Frozen samples (fecal or ileum) were directly added to the bead tubes with 200 μL lysis (MBL) buffer. A plastic micro-pestle was used to grind tissue or fecal material until sufficiently suspended in the buffer. An additional 250 μL lysis buffer was added and the extraction continued according to the manufacturer’s protocol. DNA was eluted in 150 μL buffer for fecal samples and 120 μL for ileum samples. Negative extraction controls (buffer controls) using blank swabs were included for every 15-20 samples to monitor reagent contamination. Air samples were collected in the mouse facility by unsheathing the swab, moving through the air, and re-sheathing. These samples help inform regarding environmental contamination during sample collection. Total bacterial DNA was quantified with a TaqMan-based quantitative PCR (qPCR) assay targeting the 16S rRNA gene as previously described 46. No-template PCR controls (sterile water added to PCR) were also included to monitor contamination during PCR amplification and library preparation. Broad-range 16S rRNA gene PCR with Illumina sequencing was performed on mouse fecal and ileum tissue samples as previously described, as well as all control samples 47,48. Briefly, broad-range PCR targeting the V3-V4 hypervariable region of the 16S rRNA gene was performed with equimolar concentrations of each sample being barcoded, pooled and sequenced on the Illumina MiSeq instrument (Illumina, San Diego, CA) using the v3- 600 cycle paired end reads (2 x 300) MiSeq Kit. A mock community purchased from ATCC (https://www.atcc.org/products/msa-1003) with known bacterial composition was used as a positive control to evaluate if bacterial taxa present in this community were detected with our laboratory processes and bioinformatics pipeline (Table S4 A–E).
Raw sequence reads were demultiplexed using Illumina MiSeq’s onboard software. Demultiplexed reads were processed using barcodecop v0.41 (Hoffman NG. barcodecop. 2019. Available at: https://github.com/nhoffman/barcodecop). The DADA2 package (PMID 27214047) was used for error correction, dereplication, paired-end assembly, and chimera removal and a list of unique sequence variants (SVs) were generated. Sequence reads have been deposited in the NCBI Short Read Archive (Bioproject Accession: In Progress). A custom reference set was created to assign taxonomy to individual SVs as previously described 48. The list of bacterial taxa used for the creation of the mouse gut reference set used for our analyses is provided as Table S4 F. Taxonomic assignments were made as described previously 48. Briefly, a multiple sequence alignment of both query and reference sequences was created using cmalign 49 and query sequences were placed on the phylogenetic tree using pplacer 50. Taxonomy was assigned to each unique SV based on location on the tree. Bacterial taxa represented by fewer than 25 reads in a sample were excluded from that sample to minimize environmental contaminant sequences from being included in the final dataset. Bacterial taxa that were judged to be contaminant sequences were removed from the final dataset (see list of bacterial taxa filtered from the mouse gut dataset prior to statistical analyses, in the supplementary material). Results with read counts and taxonomic outputs for positive and negative controls are also provided in the supplementary material.
For the bacterial suppressor inoculation experiments, fecal microbioal DNA was extracted using PowerFecal Pro DNA Kit (Qiagen Germantown, MD) and cleaned up using DNeasy PowerClean Pro Cleanup Kit (Qiagen). Extracted DNA was 16S amplicon sequenced and taxonomy was assigned by Zymo Research (Irvine, CA).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis for murine data.
Data were analysed using GraphPad Prism (ver. 9.4.0) and R (ver. 4.0.3). All hypothesis tests were two-sided unless otherwise specified, with statistical significance defined as p<0.05. Survival curves were plotted using Kaplan-Meier estimates and compared by log-rank test. If the data was normally distributed, Brown-Forsythe ANOVA test and Welch’s t-test were used to compare groups. P-values were adjusted to account for multiple pairwise comparisons using Dunnett’s procedure or Holm’s method. For data that was not normally distributed, the Kruskal-Wallis test and Wilcoxon rank-sum test were used to compared groups. P-values were adjusted to account for multiple pairwise comparisons using Dunn’s multiple comparion procedure or Holm’s method. The relative abundance of each taxon was centered log-ratio (CLR) transformed prior to statistical comparisons 51. CLR-transformed relative abundance of each taxon was compared between groups using the Wilcoxon rank-sum test with p-values adjusted using Holm’s method. MHC-II expression (measured as the percentage of MHC-II positive cells and the the mean fluorescence intensity (MFI) of MHC-II) was compared between the presence/absence of each taxon using Somers’ D rank correlation and the Wilcoxon rank-sum test. MHC-II expression was also correlated with CLR-transformed relative abundance using Spearman’s rank correlation coefficient. This produced four comparisons per taxon (two measures of MHC-II expression x either presence/absence or relative abundance). The four corresponding p-values were combined into a single p-value using the adjusted harmonic mean p-value method to account for these four separate and non-independent comparisons 52. These combined p-values (one per taxon) were further adjusted using Holm’s method to accounting for the number of taxa tested. The two Somers’ D rank correlation estimates and two Spearman’s rank correlation estimates were averaged per taxon. Taxa with statistically significant associations between MHC-II expression (after the p-value adjustments) were classified as MHC-II inducers or suppressors if the corresponding average correlations with MHC-II expression were positive or negative, respectively.
Patient selection and stool sample sequencing.
Patients that underwent allo-HCT at Memorial Sloan Kettering Cancer Center between September 2009 and November 2019 were screened for inclusion. Included patients all underwent first allo-HCT with peripheral blood stem cell graft (PBSC) and received calcineurin inhibitor/methotrexate GVHD prophylaxis. Stool samples between days −30 and 0 (relative to HCT) were analyzed. For patients with multiple stool samples, the one closer to the HCT day was used. The consort diagram is shown in Figure S7 and the patient characteristics are shown in Table S1 - S2. Patients provided written consent to an IRB-approved biospecimen-collection protocol.
After processing the stool samples, the bacterial cell walls were lysed with silica bead-beating; nucleic acids were isolated and the 16S ribosomal-RNA gene V4-V5 variable region was amplified and sequenced on the Illumina MiSeq platform, as previously described 16. The DADA2 pipeline was used to identify the amplicon sequence variants (ASVs) present 53 which were then mapped to the NCBI 16S rRNA sequence database using BLAST. 54
Statistical analysis and comparison of microbial compositions in patient samples
Patients were divided in groups based on developing GVHD (specifically grades 2-4) or transplant related mortality within the first 2 years after allo-HCT. The microbiome composition of the patients was first visualized (Fig. 7 A) and further assessed by permutational multivariate analysis of variance (PERMANOVA) testing and linear discriminant analysis of effect size (LEfSe). PERMANOVA testing is a multivariate non-parametric statistical test used to compare the microbiome of patients with and without GVHD (grades 2-4) and TRM (within the first 2 years), using the differences of the composition’s centroids and dispersion. 55 LEfSe is a computational method that combines tests for statistical significance and biological relevance to identify the taxa most likely to explain differences between two groups. 56 The observed differences from LEfSe analysis were further validated with Wilcoxon rank-sum univariate testing, by comparing the abundance of these taxa in patients with GVHD grades 2-4 and TRM. Taxa (at the genus level) present at a median relative abundance of ≥2x10−3 and present in at least 10% of the samples were included for assessment for LEfSe and PERMANOVA testing.
Immunofluorescence microscopy.
We obtained ileal biopsy samples from 12 healthy individuals, 5 pre-transplant patients and 1 post-transplant patient, and colonic biopsy samples from 6 healthy individuals. The use of volunteer and patient speciments was approved by the Fred Hutchinson Cancer Center IRB. Formalin-fixed paraffin embedded samples were stained as previously described 57. In brief, human biopsy tissues were fixed with 10% formalin, then placed in 70% ethanol before paraffin-embedding. Sections (4 μm) were baked for 1 hour at 60°C. The slides were then dewaxed and stained on a Leica BOND Rx stainer (Leica, Buffalo Grove, IL) using Leica Bond reagents for dewaxing (Dewax Solution), antigen retrieval/antibody stripping (Epitope Retrieval Solution 2), and rinsing after each step (Bond Wash Solution). Antigen retrieval and antibody stripping steps were performed at 100°C with all other steps at ambient temperature. Endogenous peroxidase was blocked with 3% H2O2 for 5 minutes followed by protein blocking in TCT buffer (0.05M Tris, 0.15M NaCl, 0.25% Casein, 0.1% Tween 20, 0.05% ProClin300 pH 7.6) for 10 minutes. The first primary antibody (position 1) was applied for 60 minutes followed by the secondary antibody application for 20 minutes and the application of the tertiary TSA-amplification reagent (PerkinElmer OPAL fluor) for 20 minutes. A wash was performed after the secondary and tertiary applications using high-salt TBST solution (0.05M Tris, 0.3M NaCl, and 0.1% Tween-20, pH 7.2-7.6). Species-specific Polymer HRP was used for all secondary applications, either Leica’s PowerVision (PV) Poly-HRP anti-mouse/Rabbit IgG Detection or Opal Polymer HRP Ms + Rb. The primary and secondary antibodies were stripped with retrieval solution for 20 minutes before repeating the process with the second primary antibody (position 2) starting with a new application of 3% H2O2. The process was repeated until three positions were completed. The three utilized primary and secondary antibodies with fluorophores are shown in Table S3. Slides were then stained with DAPI for 5 minutes, rinsed and coverslipped with Prolong Gold Antifade reagent (Invitrogen/Life Technologies, Grand Island, NY). Slides were cured overnight at room temperature, then whole slide images were acquired on the Vectra Polaris Quantitative Pathology Imaging System (Akoya Biosciences, Marlborough, MA). The entire tissue was selected for processing using Phenochart and the images were spectrally unmixed using inForm software and exported as multi-image TIF files, which were analyzed with HALO Link image analysis software (Indica Labs, Cooales, NM).
Supplementary Material
Table S4. Murine microbiota controls and gut reference set, related to Figure 1–5. (A) Murine Microbiota Dataset – Mock bacterial community controls. (B) Murine Microbiota Dataset - DNA Extraction Controls. (C) Murine Microbiota Dataset - PCR no-template controls (water in PCR). (D) Murine Microbiota Dataset - Environmental controls. (E) List of bacterial taxa filtered from murine dataset prior to statistical analyses. (F) The mouse gut reference set.
Table S6. Human microbiota data deposition accession number, related to Figure 7.
Table S7. Details on human participant profile, related to Figure 7. (A) Participants with stool samples. (B) Participants with endoscopic biopsy samples.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BUV395 anti-mouse CD45 | BD Biosciences | Cat# 564279, RRID AB_2651134 |
| BV785 anti-mouse CD326 (EpCAM) | BioLegend | Cat# 118245, RRID AB_2860639 |
| Pacific Blue anti-mouse I-A/I-E | BioLegend | Cat# 107620, RRID AB_493527 |
| Pacific Blue Rat IgG2b, κ Isotype control | BioLegend | Cat# 400627, RRID AB_493561 |
| Brilliant Violet 650 rat anti-mouse CD19 | BD Bioscience | Cat# 563235, RRID AB_2738085 |
| PE/Cyanine5 anti-mouse CD19 Antibody | BioLegend | Cat# 115510, RRIDAB_313645 |
| PE/Dazzle™ 594 anti-mouse CD3ε Antibody | BioLegend | Cat# 100348, RRID AB_2564029 |
| Brilliant Violet 510™ anti-mouse CD90.2 (Thy-1.2) Antibody | BioLegend | Cat# 140319, RRID AB_2561395 |
| BUV805 Mouse Anti-Rat CD90/Mouse CD90.1 | BD Bioscience | Cat# 741974, RRID AB_2871278 |
| BUV496 Rat Anti-Mouse CD4 | BD Bioscience | Cat# 612952 |
| Brilliant Violet 785™ anti-mouse CD4 Antibody | BioLegend | Cat# 100453, RRID AB_2565843 |
| BUV805 Rat Anti-Mouse CD8a | BD Bioscience | Cat# 612898, RRID AB_2870186 |
| Brilliant Violet 650™ anti-mouse CD8a Antibody | BioLegend | Cat# 100742, RRID AB_2563056 |
| BUV661 Rat Anti-Mouse CD8b | BD Bioscience | Cat# 741585, RRID AB_2870998 |
| Brilliant Violet 711™ anti-mouse CD317 (BST2, PDCA-1) Antibody | BioLegend | Cat# 127039, RRID AB_2832459 |
| BUV737 Rat Anti-Mouse Siglec-H | BD Bioscience | Cat# 748293, RRID AB_2872719 |
| Biotin Rat Anti-Mouse CD119 | BD Bioscience | Cat# 558771, RRID AB_397115 |
| Biotin Rat IgG2a, κ Isotype Ctrl Antibody | Biolegend | Cat# 400503, RRID AB_2783537 |
| PE Streptavidin | Biolegend | Cat# 405203 |
| Foxp3 PE-Cyanine5 | eBioscence | Cat# 15-5773-82, RRID AB_468806 |
| Brilliant Violet 711™ anti-mouse Ly-6G Antibody | BioLegend | Cat# 127643, RRID AB_2565971 |
| APC/Cy7 anti-mouse Ly-6G | BioLegend | Cat# 127624, RRID AB_10640819 |
| Brilliant Violet 605 anti-mouse CD11c | BioLegend | Cat# 117334, RRID AB_2562415 |
| PE/Cy7 anti-mouse CD64 (FcγRI) | BioLegend | Cat# 139314, RRID AB_2563904 |
| PerCP/Cyanine5.5 anti-mouse/human CD11b | BioLegend | Cat# 101228, RRID AB_893232 |
| CD200 Receptor Monoclonal Antibody (OX110), APC | eBioscience | Cat# 17-5201-82, RRID AB_10717289 |
| APC Rat IgG2a, κ Isotype Ctrl Antibody | BioLegend | Cat# 400512, RRID AB_2814702 |
| FITC Rat Anti-Mouse Vβ 6 T-Cell Receptor | BD Biosciences | Cat# 553193, RRID AB_394700 |
| Alexa Fluor® 700 anti-mouse Ly-6C | BD Biosciences | Cat# 128024, RRID AB_10643270 |
| BUV563 Hamster Anti-Mouse CD49b | BD Bioscience | Cat# 741280, RRID AB_2870819 |
| FITC anti-mouse NK-1.1 Antibody | BioLegend | Cat# 108706, RRID AB_313393 |
| APC/Cyanine7 anti-mouse NK-1.1 Antibody | BioLegend | Cat# 108724, RRID AB_830871 |
| Purified Rat Anti-Mouse CD16/CD32 (Mouse BD Fc Block™) | BD Bioscience | Cat# 553142, RRID AB_394656 |
| Alexa Fluor® 488 anti-human CD326 (EpCAM) Antibody | BioLegend | Cat# 324210, RRID AB_756084 |
| Brilliant Violet 510™ anti-human CD45 Antibody | BioLegend | Cat# 304036, RRID AB_2561940 |
| PE Anti-HLA DR + DP + DQ antibody [WR18] | Abcam | Cat# ab23901 |
| Purified anti-human CD326 (EpCAM) Antibody | BioLegend | Cat# 32402, RRID AB_756076 |
| CD45, Leucocyte Common Antigen (Concentrate) | DAKO | PartNumber: M070101-2 Code Number: M0701 |
| Anti-HLA DR + DP + DQ antibody [CR3/43] | Abcam | Cat# ab7856 |
| Mouse IgG1 Isotype Control | R&D systems | Cat# MAB002 |
| PowerVision+ Poly-HRP IHC Detection System | Leica Biosystems | Cat# PV6107 |
| 1X OPAL ANTI-MS + RB HRP (Formerly Opal Polymer HRP Ms + Rb) | Akoya Bioscences | SKU: ARH1001EA |
| Akoya Opal 570 reagent | Akoya Bioscences | Cat# FP1488001KT |
| Akoya Opal 520 reagent Pack | Akoya Bioscences | Cat# FP1487001KT |
| Akoya Opal 690 reagent | Akoya Bioscences | Cat$ FP1497001KT |
| Purified CD25 (clone PC-61.5.3) | BioXcell | Cat# BE0012 |
| Bacterial and virus strains | ||
| Bacteroides thetaiotaomicron (VPI 5482) | ATCC | Cat# 29148 |
| Clostridium sporogenes | ATCC | Cat# 15579 |
| Biological samples | ||
| Human ileal and colonic biopsy tissues from healthy donors and pre, post transplant patients | Fred Hutchinson Cancer Center (FHCC) | N/A |
| Human fecal samples | Memorial Sloan Kettering Cancer Center | N/A |
| Chemicals, reagents, peptides, and recombinant proteins | ||
| 7-Aminoactinomycin D (7AAD) | Sigma Aldrich | Cat# A9400-5MG |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma Aldrich | Cat#P1585-1MG |
| Ionomycin | Sigma Aldrich | Cat#I0634-1MG |
| BD Horizon™ Fixable Viability Stain 440UV | BD Biosciences | Cat# 566332, RRID AB_2869748 |
| Tamoxifen | MP Biomedicals | CAS#10540-29-1 |
| NaHCO3 | Sigma | Cat# S4019 |
| MgSO4.7H2O | Sigma | Cat# M63-500 |
| NaCl | Sigma | Cat# S3014 |
| CaCl2 | Sigma | Cat# C7902 |
| FeSO4 | Sigma | Cat# F8048 |
| BBL™ Trypticase Peptone | BD Biosciences | Cat# 211921 |
| Bacto Yeast extract | Gibco | Cat# 212750 |
| Glucose | Sigma | Cat# G8270 |
| Cysteine (free base) | Sigma | Cat# C1276 |
| KH2PO4 | Sigma | Cat# P0662 |
| K2HPO4 | Sigma | Cat# P3786 |
| meat extract | Sigma | Cat# 70164 |
| Menadione (Vitamin K) | Sigma | Cat# M5625 |
| Hematin | Sigma | Cat# H3281 |
| ATCC Viotamin mix | ATCC | Cat# MD-VS |
| ATCC Mineral mix | ATCC | Cat# MD-TMS |
| Tween 80 | Sigma | Cat# P1754 |
| cellobiose | Sigma | Cat# C7252 |
| maltose | Sigma | Cat# M5885 |
| fructose | Sigma | Cat# F0127 |
| sodium acetate | Fisher (Fluka) | Cat# BP333-500 |
| sodium sulfate | Sigma | Cat# S9627 |
| malic acid | Sigma | Cat# M7937 |
| Collagenase type I | Gibco | Cat# 17100017 |
| Deoxyribonuclease I from bovine pancreas | Sigma | SKU: DN25-1G |
| HEPES 1M | Gibco | Cat# 15630080 |
| Pen Strep | Gibco | Cat# 15140122 |
| L-glutamine | Gibco | Cat# 25030081 |
| MgCl2 | Sigma | SKU: M8266-100G |
| CaCl2 | Sigma | SKU: C1016-100G |
| TaqMan 1000RXN Gold/Buffer Pkg Kit; custom order | ThermoFisher Scientific | Cat# A26028 |
| AmpliTaq Gold DNA polymerase, LD | ThermoFisher Scientific | Cat# 4338856 |
| Custom Taqman Probe | ThermoFisher Scientific | Cat# 450024 |
| Nextera® XT Index Kit v2 Set A (96 Indices, 384 Samples) | Illumina | Cat# FC-131-2001 |
| Nextera® XT Index Kit v2 Set B (96 Indices, 384 Samples) | Illumina | Cat# FC-131-2002 |
| Nextera® XT Index Kit v2 Set C (96 Indices, 384 Samples) | Illumina | Cat# FC-131-2003 |
| Nextera® XT Index Kit v2 Set D (96 Indices, 384 Samples) | Illumina | Cat# FC-131-2004 |
| KAPA HotStart ReadyMix, 500 x 25 μL reactions | Roche | Cat# KK2602 |
| Magnetic Bead, Agencourt® AMPure® XP, 5 mL | Beckman Coulter | Cat# A63880 |
| PhiX Control v3 | Illumina | Cat# FC-110-3001 |
| AccuPrime™ Taq DNA Polymerase High Fidelity | ThermoFisher Scientific | Cat# 12346086 |
| Quant-iT™ dsDNA Assay Kit, high sensitivity | ThermoFisher Scientific | Cat# Q33120 |
| MiSeq Reagent Kit v3 | Illumina | Cat# MS-102-3003 |
| ATCC 20 Strain Staggered Mix Genomic Material | ATCC | Cat# MSA-1003 |
| Critical commercial assays | ||
| eBioscience™ Foxp3 / Transcription Factor Staining Buffer Set | eBioscience | Cat# 00-5523-00 |
| Cytometric Bead Array; IFNγ, IL-6, TNF, IL-17A | BD Biosciences | Cat#560485 |
| Lamina Propria Dissociation Kit, mouse | MACS Miltenyi Biotec | Cat#130-097-410 |
| QIAamp BiOstic Bacteremia DNA Isolation Kit | QIAGEN | Cat#12240-50 |
| MiSeq Reagent Kit v3 (600-cycle) | Illumina | Cat# MS-102-3003 |
| QIAamp PowerFecal Pro DNA Kit | QIAGEN | Cat# 51804 |
| DNeasy PowerClean Pro Cleanup Kit | QIAGEN | Cat# 12997-50 |
| Deposited data | ||
| Murine fecal and ileal 16S RNA gene sequence | This article | Accession: Bioproject PRJNA980199 |
| Human pre-transplant patients fecal 16S RNA gene sequence | MSKCC | Accession: Listed in Table S6 |
| Experimental models: Cell lines | ||
| (Bacteria described in ”Bacterial and virus strains”) | ||
| Experimental models: Organisms/strains | ||
| Mouse: B6J ARC: C57BL/6JArc | The Animal Resources Centre (ARC), Perth, AU | Cat# 000664 |
| Mouse: B6J JAX: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: B6N JAX: C57BL/6NJ | The Jackson Laboratory | RRID:IMSR_JAX:005304 |
| Mouse: B6N CR: C57BL/6NCrl | Charles River | Strain Code: 027 |
| Mouse: B6N TAC: C57BL/6NTac | Taconic | Model#: B6-F, B6-M |
| Mouse: B6N ADFH: C57BL/6N | Maintained at FHCC | N/A |
| Mouse: BALB/c: BALB/cJ | The Jackson Laboratory | RRID:IMSR_JAX:000651 |
| Mouse: B6.I-Ab−/−: H2-Ab−/− | Australian National University, Canberra, AU | Grusby M.J., et al. 1991 |
| Mouse: Villin Cre-ERT2 | Dr R Blumberg, Harvard Medical School, Boston, MA, USA | El Marjou F., et al. 2004 |
| Mouse: I-Ab-fl/fl: B6.129X1-H2-Ab1tm1Koni/J | The Jackson Laboratory | RRID:IMSR_JAX:013181 |
| Mouse: B6.Ifnγ-YFP: B6.129S4-Ifngtm3.1Lky/J (GREAT) | The Jackson Laboratory | RRID:IMSR_JAX:017581 |
| Mouse: B6.Ifnγ-YFP: B6.129S4-Ifngtm3.1Lky/J (GREAT) | Maintained at QIMR Berghofer, then FHCC | RRID:IMSR_JAX:017581 |
| Mouse: B6.Ifngr−/− : B6.129S7-Ifngr1tm1Agt/J | The Jackson Laboratory | RRID:IMSR_JAX:003288 |
| Mouse: C57BL/6N-Ifngr1tm1.1Rds/J | The Jackson Laboratory | RRID:IMSR_JAX:025394 |
| Mouse: Marilynluc+: Rag2−/− Marilyn mice were backcrossed onto a B6 β-actin-luciferase background | QIMR Berghofer, Brisbane, AU | N/A |
| Villin-Cre-ERT2 strain and I-Ab -fl/fl or Ifngr1 l/fl strains were intercrossed to generate: | N/A | N/A |
| Mouse: Villin-Cre-ERT2 I-Ab-fl/fl | FHCC | N/A |
| Mouse: Villin-Cre-ERT2 Ifngr1 fl/fl | FHCC | N/A |
| Oligonucleotides | ||
| N/A | ||
| Recombinant DNA | ||
| N/A | ||
| Software and algorithms | ||
| BD FACSDiva software version 9.1 | BD Bioscience | https://www.bdbiosciences.com/in/instruments/software/facsdiva/ |
| FlowJo v10 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism (ver. 9.4.0) | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| R (ver. 4.0.3) | R Foundation for Statistical Computing | https://www.r-project.org/ |
| HALO Link image analysis software | Indica Labs | https://indicalab.com/halo/ |
| barcodecop v0.41 | Dr. Noah Hoffman | https://github.com/nhoffman/barcodecop |
| DADA2 package | Callahan et al. Nat. Methods. PMID 27214047 | https://benjjneb.github.io/dada2/index.html |
| ya16sdb pipeline | Dr. Noah Hoffman | https://github.com/nhoffman/ya16sdb. |
| cmalign | Nawrocki and Eddy. PMID 24008419 | http://eddylab.org/infernal/ |
| pplacer | Matsen et al. BMC Bioinform. PMID 21034504 | https://matsen.fhcrc.org/pplacer/ |
| Other | ||
| BD Symphony A3 | BD Biosciences | https://www.bdbiosciences.com/en-us/products/instruments/flow-cytometers/research-cell-analyzers/bd-facsymphony-a3 |
| Vectra Polaris Quantitative Pathology Imaging System | Akoya Biosciences | https://www.akoyabio.com/wp-content/uploads/2021/11/Vectra_Polaris_Product_Note_with_MOTiF_Akoya.pdf |
Highlights.
MHC-II inducer taxa preferentially control MHC-II expression by intestinal epithelia.
Pre-transplant vancomycin depletes MHC-II inducers and attenuates GVHD.
Pre-transplant commensal bacterial taxa control CD4+ T-mediated GVHD.
Pre-transplant taxa in the GI tract correalate with clinical transplant outcomes.
Acknowledgments
We thank S. J. Weaver and A. L. Koehne in Experimental Histopathology for expert preparation of histology samples. We also thank all physicians, nurses, pharmacists and support personnel for their care of the patients in this study.
Funding:
G.R.H. and M.K. are supported by NIH R01 HL148164. T.W.R. and D.S.H. are supported by the Biostatistics Shared Resource of the Fred Hutch/University of Washington/Seattle Children’s Cancer Consortium (P30 CA015704). Experimental histopathology at Fred Hutch was supported by NIH P30 CA015704. Scientific Computing Infrastructure at Fred Hutch was supported by ORIP grant S10 OD028685. M.R.M.B. was supported by National Cancer Institute award numbers, R01-CA228358, R01-CA228308, P30 CA008748 MSK Cancer Center Support Grant/Core Grant and P01-CA023766 ; NHLBI award number R01-HL123340 and R01-HL147584; National Institute on Aging award number P01-AG052359, and Tri Institutional Stem Cell Initiative, and additional funding was received from The Lymphoma Foundation, The Susan and Peter Solomon Family Fund, The Solomon Microbiome Nutrition and Cancer Program, Cycle for Survival, Parker Institute for Cancer Immunotherapy, Paula and Rodger Riney Multiple Myeloma Research Initiative, Starr Cancer Consortium, and Seres Therapeutics. O.M. was supported by the American Society of Clinical Oncology Young Investigator Award, a Hyundai Hope on Wheels Young Investigator Award, a Cycle for Survival Equinox Innovation Award, a Collaborative Pediatric Cancer Research Program Award, Michael Goldberg Fellowship and Tow Center for Developmental Oncology Career Development Award. N.H. is supported by Inter disciplinary Training Grant in Cancer T32 CA080416 to NGH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Declaration of Interests
W.M.G. is a scientific advisory board member for Freenome, Guardant Health, and SEngine and consultant for DiaCarta, Natera, Guidepoint and GLG. He is an investigator in a clinical trial sponsored by Janssen Pharmaceuticals and receives research support from Tempus and LucidDx. M.R.M.B. has received research support and stock options from Seres Therapeutics and stock options from Notch Therapeutics and Pluto Therapeutics; he has received royalties from Wolters Kluwer; has consulted, received honorarium from or participated in advisory boards for Seres Therapeutics, Vor Biopharma, Rheos Medicines, Frazier Healthcare Partners, Nektar Therapeutics, Notch Therapeutics, Ceramedix, Lygenesis, Pluto Therapeutics, GlaskoSmithKline, Da Volterra, Thymofox, Garuda, Novartis (Spouse), Synthekine (Spouse), Beigene (Spouse), Kite (Spouse); he has IP Licensing with Seres Therapeutics and Juno Therapeutics; and holds a fiduciary role on the Foundation Board of DKMS (a nonprofit organization). MSK has institutional financial interests relative to Seres Therapeutics. K.A.M. serves in an advisory role and holds stock in PostBiotics Plus Research and has consulted for Incyte. D.N.F. and T.L.F. have financial relationships with BD for licensure of molecular diagnosis of bacterial vaginosis, unrelated to the research presented in this article. G.R.H. has consulted for Generon Corporation, NapaJen Pharma, iTeos Therapeutics, Neoleukin Therapeutics and receives research funding from Compass Therapeutics, Syndax Pharmaceuticals, Applied Molecular Transport, Serplus Technology, Heat Biologics, Laevoroc Oncology,iTeos Pharmaceuticals and Genentech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.D’Souza A, Fretham C, Lee SJ, Arora M, Brunner J, Chhabra S, Devine S, Eapen M, Hamadani M, Hari P, et al. (2020). Current Use of and Trends in Hematopoietic Cell Transplantation in the United States. Biol Blood Marrow Transplant 26, e177–e182. 10.1016/j.bbmt.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Witte T, Bowen D, Robin M, Malcovati L, Niederwieser D, Yakoub-Agha I, Mufti GJ, Fenaux P, Sanz G, Martino R, et al. (2017). Allogeneic hematopoietic stem cell transplantation for MDS and CMML: recommendations from an international expert panel. Blood 129, 1753–1762. 10.1182/blood-2016-06-724500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiden PL, Flournoy N, Thomas ED, Prentice R, Fefer A, Buckner CD, and Storb R (1979). Antileukemic effect of graft-versus-host disease in human recipients of allogeneic-marrow grafts. N Engl J Med 300, 1068–1073. 10.1056/NEJM197905103001902. [DOI] [PubMed] [Google Scholar]
- 4.Zeiser R, and Blazar BR (2017). Pathophysiology of Chronic Graft-versus-Host Disease and Therapeutic Targets. N Engl J Med 377, 2565–2579. 10.1056/NEJMra1703472. [DOI] [PubMed] [Google Scholar]
- 5.Zeiser R, and Blazar BR (2017). Acute Graft-versus-Host Disease - Biologic Process, Prevention, and Therapy. N Engl J Med 377, 2167–2179. 10.1056/NEJMra1609337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacMillan ML, Robin M, Harris AC, DeFor TE, Martin PJ, Alousi A, Ho VT, Bolanos-Meade J, Ferrara JL, Jones R, et al. (2015). A refined risk score for acute graft-versus-host disease that predicts response to initial therapy, survival, and transplant-related mortality. Biol Blood Marrow Transplant 21, 761–767. 10.1016/j.bbmt.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naymagon S, Naymagon L, Wong SY, Ko HM, Renteria A, Levine J, Colombel JF, and Ferrara J (2017). Acute graft-versus-host disease of the gut: considerations for the gastroenterologist. Nat Rev Gastroenterol Hepatol 14, 711–726. 10.1038/nrgastro.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyama M, and Hill GR (2019). The primacy of gastrointestinal tract antigen-presenting cells in lethal graft-versus-host disease. Blood 134, 2139–2148. 10.1182/blood.2019000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fredricks DN (2019). The gut microbiota and graft-versus-host disease. J Clin Invest 129, 1808–1817. 10.1172/JCI125797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koyama M, Mukhopadhyay P, Schuster IS, Henden AS, Hulsdunker J, Varelias A, Vetizou M, Kuns RD, Robb RJ, Zhang P, et al. (2019). MHC Class II Antigen Presentation by the Intestinal Epithelium Initiates Graft-versus-Host Disease and Is Influenced by the Microbiota. Immunity. 10.1016/j.immuni.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuganbaev T, Mor U, Bashiardes S, Liwinski T, Nobs SP, Leshem A, Dori-Bachash M, Thaiss CA, Pinker EY, Ratiner K, et al. (2020). Diet Diurnally Regulates Small Intestinal Microbiome-Epithelial-Immune Homeostasis and Enteritis. Cell 182, 1441–1459 e1421. 10.1016/j.cell.2020.08.027. [DOI] [PubMed] [Google Scholar]
- 12.Shlomchik WD, Couzens MS, Tang CB, McNiff J, Robert ME, Liu J, Shlomchik MJ, and Emerson SG (1999). Prevention of graft versus host disease by inactivation of host antigen-presenting cells. Science 285, 412–415. [DOI] [PubMed] [Google Scholar]
- 13.Toubai T, Tawara I, Sun Y, Liu C, Nieves E, Evers R, Friedman T, Korngold R, and Reddy P (2012). Induction of acute GVHD by sex-mismatched H-Y antigens in the absence of functional radiosensitive host hematopoietic-derived antigen-presenting cells. Blood 119, 3844–3853. 10.1182/blood-2011-10-384057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Demetris AJ, McNiff J, Matte-Martone C, Tan HS, Rothstein DM, Lakkis FG, and Shlomchik WD (2012). Profound depletion of host conventional dendritic cells, plasmacytoid dendritic cells, and B cells does not prevent graft-versus-host disease induction. Journal of immunology 188, 3804–3811. 10.4049/jimmunol.1102795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koyama M, Kuns RD, Olver SD, Raffelt NC, Wilson YA, Don AL, Lineburg KE, Cheong M, Robb RJ, Markey KA, et al. (2012). Recipient nonhematopoietic antigen-presenting cells are sufficient to induce lethal acute graft-versus-host disease. Nat Med 18, 135–142. 10.1038/nm.2597. [DOI] [PubMed] [Google Scholar]
- 16.Peled JU, Gomes ALC, Devlin SM, Littmann ER, Taur Y, Sung AD, Weber D, Hashimoto D, Slingerland AE, Slingerland JB, et al. (2020). Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med 382, 822–834. 10.1056/NEJMoa1900623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, et al. (2014). Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant 20, 640–645. 10.1016/j.bbmt.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, Clurman AG, Armijo G, Gomes ALC, Shono Y, et al. (2019). Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science 366, 1143–1149. 10.1126/science.aax3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC, et al. (2015). Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol Blood Marrow Transplant 21, 1373–1383. 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, Oravecz-Wilson K, Wu SR, Sun Y, Rossi C, et al. (2016). Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nature immunology 17, 505–513. 10.1038/ni.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 22.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, and Rudensky AY (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236. 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 24.Sun Z, Harris HM, McCann A, Guo C, Argimon S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, et al. (2015). Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6, 8322. 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keilbaugh SA, Shin ME, Banchereau RF, McVay LD, Boyko N, Artis D, Cebra JJ, and Wu GD (2005). Activation of RegIIIbeta/gamma and interferon gamma expression in the intestinal tract of SCID mice: an innate response to bacterial colonisation of the gut. Gut 54, 623–629. 10.1136/gut.2004.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, and Setoyama H (1995). Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol 39, 555–562. 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 28.Varelias A, Ormerod KL, Bunting MD, Koyama M, Gartlan KH, Kuns RD, Lachner N, Locke KR, Lim CY, Henden AS, et al. (2017). Acute graft-versus-host disease is regulated by an IL-17-sensitive microbiome. Blood 129, 2172–2185. 10.1182/blood-2016-08-732628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowerman KL, Varelias A, Lachner N, Kuns RD, Hill GR, and Hugenholtz P (2020). Continuous pre- and post-transplant exposure to a disease-associated gut microbiome promotes hyper-acute graft-versus-host disease in wild-type mice. Gut Microbes 11, 754–770. 10.1080/19490976.2019.1705729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayase E, Hayase T, Jamal MA, Miyama T, Chang CC, Ortega MR, Ahmed SS, Karmouch JL, Sanchez CA, Brown AN, et al. (2022). Mucus-degrading Bacteroides link carbapenems to aggravated graft-versus-host disease. Cell 185, 3705–3719 e3714. 10.1016/j.cell.2022.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li K, Hao Z, Du J, Gao Y, Yang S, and Zhou Y (2021). Bacteroides thetaiotaomicron relieves colon inflammation by activating aryl hydrocarbon receptor and modulating CD4(+)T cell homeostasis. Int Immunopharmacol 90, 107183. 10.1016/j.intimp.2020.107183. [DOI] [PubMed] [Google Scholar]
- 32.Delday M, Mulder I, Logan ET, and Grant G (2019). Bacteroides thetaiotaomicron Ameliorates Colon Inflammation in Preclinical Models of Crohn’s Disease. Inflamm Bowel Dis 25, 85–96. 10.1093/ibd/izy281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durant L, Stentz R, Noble A, Brooks J, Gicheva N, Reddi D, O’Connor MJ, Hoyles L, McCartney AL, Man R, et al. (2020). Bacteroides thetaiotaomicron-derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome 8, 88. 10.1186/s40168-020-00868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonseca S, Carvalho AL, Miquel-Clopes A, Jones EJ, Juodeikis R, Stentz R, and Carding SR (2022). Extracellular vesicles produced by the human gut commensal bacterium Bacteroides thetaiotaomicron elicit anti-inflammatory responses from innate immune cells. Front Microbiol 13, 1050271. 10.3389/fmicb.2022.1050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hugenholtz F, and de Vos WM (2018). Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci 75, 149–160. 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, Bresciani A, Martinez I, Just S, Ziegler C, et al. (2016). The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol 1, 16131. 10.1038/nmicrobiol.2016.131. [DOI] [PubMed] [Google Scholar]
- 37.Yin YS, Wang Y, Zhu LY, Liu W, Liao NB, Jiang MZ, Zhu BL, Yu HWD, Xiang C, and Wang X (2013). Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. Isme Journal 7, 615–621. 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimoto K, Joshi SK, and Koni PA (2002). A conditional null allele of the major histocompatibility IA-beta chain gene. Genesis 32, 152–153. [DOI] [PubMed] [Google Scholar]
- 40.Lantz O, Grandjean I, Matzinger P, and Di Santo JP (2000). Gamma chain required for naive CD4+ T cell survival but not for antigen proliferation. Nature immunology 1, 54–58. 10.1038/76917. [DOI] [PubMed] [Google Scholar]
- 41.Cooke KR, Kobzik L, Martin TR, Brewer J, Delmonte J, Crawford JM, and Ferrara JLM (1996). An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation. I. The roles of minor H antigens and endotoxin. Blood 88, 3230–3239. [PubMed] [Google Scholar]
- 42.Churchman ML, Low J, Qu C, Paietta EM, Kasper LH, Chang Y, Payne-Turner D, Althoff MJ, Song G, Chen SC, et al. (2015). Efficacy of Retinoids in IKZF1-Mutated BCR-ABL1 Acute Lymphoblastic Leukemia. Cancer Cell 28, 343–356. 10.1016/j.ccell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang P, Tey SK, Koyama M, Kuns RD, Olver SD, Lineburg KE, Lor M, Teal BE, Raffelt NC, Raju J, et al. (2013). Induced regulatory T cells promote tolerance when stabilized by rapamycin and IL-2 in vivo. Journal of immunology 191, 5291–5303. 10.4049/jimmunol.1301181. [DOI] [PubMed] [Google Scholar]
- 44.Hulsdunker J, Ottmuller KJ, Neeff HP, Koyama M, Gao Z, Thomas OS, Follo M , Al-Ahmad A, Prinz G, Duquesne S, et al. (2018). Neutrophils provide cellular communication between ileum and mesenteric lymph nodes at graft-versus-host disease onset. Blood 131, 1858–1869. 10.1182/blood-2017-10-812891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Roy T, Debedat J, Marquet F, Da-Cunha C, Ichou F, Guerre-Millo M, Kapel N , Aron-Wisnewsky J, and Clement K (2018). Comparative Evaluation of Microbiota Engraftment Following Fecal Microbiota Transfer in Mice Models: Age, Kinetic and Microbial Status Matter. Front Microbiol 9, 3289. 10.3389/fmicb.2018.03289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, and Fredricks DN (2012). Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PloS one 7, e37818. 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golob JL, Pergam SA, Srinivasan S, Fiedler TL, Liu C, Garcia K, Mielcarek M, Ko D, Aker S, Marquis S, et al. (2017). Stool Microbiota at Neutrophil Recovery Is Predictive for Severe Acute Graft vs Host Disease After Hematopoietic Cell Transplantation. Clin Infect Dis 65, 1984–1991. 10.1093/cid/cix699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivasan S, Chambers LC, Tapia KA, Hoffman NG, Munch MM, Morgan JL, Domogala D, Lowens MS, Proll S, Huang ML, et al. (2020). Urethral microbiota in men: Association of Haemophilus influenzae and Mycoplasma penetrans with nongonococcal urethritis. Clin Infect Dis. 10.1093/cid/ciaa1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nawrocki EP, and Eddy SR (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsen FA, Kodner RB, and Armbrust EV (2010). pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11, 538. 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gloor GB, and Reid G (2016). Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol 62, 692–703. 10.1139/cjm-2015-0821. [DOI] [PubMed] [Google Scholar]
- 52.Wilson DJ (2019). The harmonic mean p-value for combining dependent tests. Proceedings of the National Academy of Sciences of the United States of America 116, 1195–1200. 10.1073/pnas.1814092116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callahan BJ, McMurdie PJ, and Holmes SP (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11, 2639–2643. 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, and Holmes SP (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McArdle BH, and Anderson MJ (2001). Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 82, 290–297. Doi 10.1890/0012-9658(2001)082[0290:Fmmtcd]2.0.Co;2. [DOI] [Google Scholar]
- 56.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12, R60. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koyama M, Samson L, Ensbey KS, Takahashi S, Clouston AD, Martin PJ, and Hill GR (2023). Lithium attenuates graft-versus-host disease via effects on the intestinal stem cell niche. Blood 141, 315–319. 10.1182/blood.2022015808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. Murine microbiota controls and gut reference set, related to Figure 1–5. (A) Murine Microbiota Dataset – Mock bacterial community controls. (B) Murine Microbiota Dataset - DNA Extraction Controls. (C) Murine Microbiota Dataset - PCR no-template controls (water in PCR). (D) Murine Microbiota Dataset - Environmental controls. (E) List of bacterial taxa filtered from murine dataset prior to statistical analyses. (F) The mouse gut reference set.
Table S6. Human microbiota data deposition accession number, related to Figure 7.
Table S7. Details on human participant profile, related to Figure 7. (A) Participants with stool samples. (B) Participants with endoscopic biopsy samples.


