Abstract
PURPOSE
Improved survival prediction and risk stratification in non–small-cell lung cancer (NSCLC) would lead to better prognosis counseling, adjuvant therapy selection, and clinical trial design. We propose the persistent homology (PHOM) score, the radiomic quantification of solid tumor topology, as a solution.
MATERIALS AND METHODS
Patients diagnosed with stage I or II NSCLC primarily treated with stereotactic body radiation therapy (SBRT) were selected (N = 554). The PHOM score was calculated for each patient's pretreatment computed tomography scan (October 2008-November 2019). PHOM score, age, sex, stage, Karnofsky Performance Status, Charlson Comorbidity Index, and post-SBRT chemotherapy were predictors in the Cox proportional hazards models for OS and cancer-specific survival. Patients were split into high– and low–PHOM score groups and compared using Kaplan-Meier curves for overall survival (OS) and cumulative incidence curves for cause-specific death. Finally, we generated a validated nomogram to predict OS, which is publicly available at Eashwarsoma.Shinyapps.
RESULTS
PHOM score was a significant predictor for OS (hazard ratio [HR], 1.17; 95% CI, 1.07 to 1.28) and was the only significant predictor for cancer-specific survival (1.31; 95% CI, 1.11 to 1.56) in the multivariable Cox model. The median survival for the high-PHOM group was 29.2 months (95% CI, 23.6 to 34.3), which was significantly worse compared with the low-PHOM group (45.4 months; 95% CI, 40.1 to 51.8; P < .001). The high-PHOM group had a significantly greater chance of cancer-specific death at post-treatment month 65 (0.244; 95% CI, 0.192 to 0.296) compared with the low-PHOM group (0.171; 95% CI, 0.123 to 0.218; P = .029).
CONCLUSION
The PHOM score is associated with cancer-specific survival and predictive of OS. Our developed nomogram can be used to inform clinical prognosis and assist in making post-SBRT treatment considerations.
Novel topology-based radiomics predicts OS for patients with NSCLC treated with SBRT in clinical nomogram.
INTRODUCTION
Approximately 230,000 cases of lung cancer are diagnosed, and 135,000 patients die from lung cancer each year in the United States.1 Proportionally, 85% of all lung cancer cases are non–small-cell lung cancer (NSCLC).2 Primary therapy has traditionally been surgical resection; however, stereotactic body radiation therapy (SBRT) is now an option for patients with inoperable tumors.3 For all cancers, clinical prognostication is essential for therapy selection. This is especially important in lung cancer, for which 5-year survival is highly variable in NSCLC ranging from nearly 70% for patients with stage I disease down to 10% for patients with metastatic disease.4 Although early stage has an overall positive prognosis, approximately 30% of patients do not survive at 5 years. This indicates a need to develop better risk-stratifying metrics.
CONTEXT
Key Objective
To validate a new topology-inspired radiomics metric (persistent homology [PHOM] score) for prediction of overall survival (OS) and risk stratification of patients with early-stage non–small-cell lung cancer treated with stereotactic body radiation therapy.
Knowledge Generated
Validated nomogram predicts OS using the PHOM score in conjunction with clinical variables. We demonstrate that the PHOM score provides unique information for OS prediction and can risk stratify across clinical subgroups.
Relevance
The nomogram tool can inform patients and providers about an individual's prognosis and can assist in making personal and medical decisions. Clinical trialists may find interest in risk stratification in trialing new therapies, and oncologists may use risk stratification provided by the PHOM score to assess the need of adjuvant therapy.
Risk calculators are clinical decision support tools that quantify prognosis. They typically use histopathologic and clinical covariates to predict outcomes. Risk calculators incorporating molecular and clinical information have been developed for lung cancer.5 Although clinical data are easily obtained, obtaining molecular data outside of tertiary medical centers is often challenging. By contrast, imaging data are far more accessible. Radiomics—the science of extracting and analyzing features from imaging data—has been increasingly used to predict clinical outcomes in cancer.6 Although radiomics already offers a wide toolset for image analysis, we propose a valuable extension with persistent homology (PHOM).
Persistent homology quantifies the global topology, or overall structure, of big data.7 Its high-level approach provides the benefit of discriminating signal from noise. Oncology already features structure analysis similar in spirit to PHOM. Glandular shape analysis through Gleason scoring is used to grade prostate cancer severity.8
We hypothesize that malignant and benign tissues exhibit distinct topology on clinical imaging. Malignant tumors are typically diffusely spread and heterogeneous in composition and may possess necrotic cavities. By contrast, benign tumors are well-circumscribed and likely to be homogeneous in appearance.9 Persistent homology quantifies these observations to permit their use in predictive models. Of note, PHOM has already been introduced to radiomics in the study of glioblastoma and liver cancer.10,11
Within lung cancer, PHOM has shown promise in predictive modeling via a metric named the PHOM score.12 In our previous study, we demonstrated that the PHOM score is associated with overall survival (OS) in patients with NSCLC treated with surgery or radiation. In this study, we use the advantage of using our institutional data to curate a cohort with greater resolution of clinical details. We selected patients treated with SBRT since their tumor margins on imaging are contoured for treatment purposes. The primary aim of this study was to assess our hypothesis that the PHOM score can predict OS in patients with NSCLC treated with SBRT. The secondary aim was to create clinical risk groups using the PHOM score. Here, we validate the PHOM score using a large prospectively collected, IRB-approved data set from our institution including 554 patients treated with SBRT for primary lung cancers from 2008 to 2019. We combine our PHOM score with clinical covariates to develop a prognostic nomogram that predicts duration of OS post-SBRT. This prognostic tool would assist providers and their patients in making both personal and medical decisions. We also demonstrate the PHOM score's ability to stratify patients into risk groups. Risk stratification would assist adjuvant therapy selection at the individual patient level and clinical trial design at a more global level.
MATERIALS AND METHODS
Source of Data and Participants
We developed a data pipeline that calculated the PHOM score of computed tomography (CT) tumor scans to create validated survival regression models. Methodology to generate a PHOM score using tumor CT scans is described in our previous work.12 Only patients with early-stage (I or II) biopsy-proven primary NSCLC with no treatment history were eligible for this study. We used a single-institution prospectively collected data set of 803 patients receiving SBRT with pretreatment patient scans (scan dates ranging October 2008-November 2019) with corresponding clinical data. Clinical data including dates of diagnosis, last follow-up, and treatment were obtained from Epic chart review. Patients had uniform free-breathing CT parameters with a slice thickness of 3 mm.
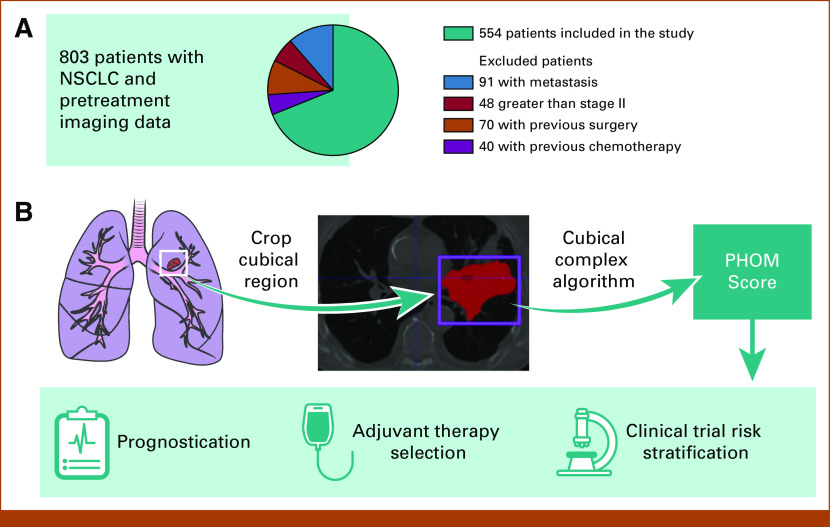
After excluding 249 patients with metastatic disease, nodal disease, and additional primary tumors and patients who received pretreatment, 554 patients met inclusion criteria (Fig 1A) All patients received SBRT as their primary treatment for local control.
FIG 1.
Conceptual overview of translating utility of PHOM score. (A) Eight hundred three patients were in our database with imaging and clinical data for NSCLC treated primarily with SBRT. Two hundred forty-nine patients were excluded on the basis of their stage or treatment history. (B) The PHOM score is algorithmically calculated from a tumor segmented by a clinician. Note that this particular cross-section demonstrates a large tumor greater than stage II but was selected for visualization purposes. Only stage II tumors and below were selected for analysis. (C) We envision the PHOM score to have utility in prognostication, therapy selection, and risk stratification. NSCLC, non–small-cell lung cancer; PHOM, persistent homology; SBRT, stereotactic body radiation therapy.
Model Development
Each patient's primary lung tumor had been segmented by a radiation oncologist. For each CT scan, we used the segmentation coordinates to crop scan to only include tumor tissue. The PHOM score was calculated from this data object (Fig 1B). As previously described, this metric reflects the number of disconnected foci of pixels with similar grayscale values in the segmented tumor object.12
We demonstrate the PHOM score's potential clinical utility through a Cox proportional hazards model and nomogram (Fig 1C). PHOM score, age, sex, Karnofsky Performance Status (KPS), Charlson Comorbidity Index (CCI), overall stage, and postradiation chemotherapy were included as covariates in a Cox proportional hazards model for OS and cancer-specific survival.
The multivariable Cox proportional hazards model for OS formed the basis of a nomogram to predict 1-, 2-, 5-, and 8-year survival. The nomogram preserves the underlying relationship between the coefficients in the multivariable Cox proportional hazards model and merely maps these coefficients to easily interpretable point values. Predicted median survival was calculated as a weighted sum of the individual survival predictions. This model was validated using bootstrap resampling of the original patient cohort. This validation method creates new Cox models by drawing patients from the original cohort with replacement (bootstrap sample). The performance of the bootstrap sample and original cohort against the bootstrap model is subtracted. This is repeated 1,000 times, and all the differences are averaged. This value is subtracted from the performance of the original model against the original cohort creating an optimism-corrected estimate. Alignment of the plotted optimism-corrected estimate with the plotted performance of the model on the original cohort is an indication of internal validity and lack of overfitting (Data Supplement [Fig 4]). Internal validation by bootstrap resampling has been shown to have stable performance estimates and low bias compared with other methods.13
To assess the relative contribution of each covariate to prediction ability of the OS multivariable Cox model, the drop in C index was calculated by omitting each covariate one at a time from the full model. The C index has been routinely used to evaluate the discriminating ability of a survival model.14
Risk Groups
The model development was the primary analysis of this study. Secondary analysis aimed to understand whether PHOM score might have potential to create risk groups. This cohort was divided into two groups by the median PHOM score of the entire cohort (PHOM = –0.0372). Kaplan-Meier curves were drawn to compare OS between the two groups, and log-rank statistics were used to compare the survival trends. Cumulative incidence curves were drawn to compare the cause of death between these two groups.
This analysis was repeated with optimized tertiles. Tertiles were calculated by selecting the two PHOM score cutoff values that resulted in the maximal chi-square value of the Kaplan-Meier curve. The low-risk group (n = 211) was defined as below –0.460, medium-risk group (n = 172) as between –0.460 and 0.668, and high-risk group (n = 171) as above 0.668. Both Kaplan-Meier curves and cumulative incidence curves were drawn for these three groups. Benjamini-Hochberg correction was used to correct for multiple comparisons for Kaplan-Meier pairwise comparisons. Post hoc Kaplan-Meier curves were drawn in a similar method to assess PHOM risk stratification in clinical subgroupings by KPS, CCI, and overall stage.
Data Availability
All data were processed and analyzed using R (v3.6.1) and Python (v3.7.6).15-17 Anonymized data may be requested from the authors. Code with explanation for this data pipeline can be found at GitHub.18
RESULTS
Patient Characteristics
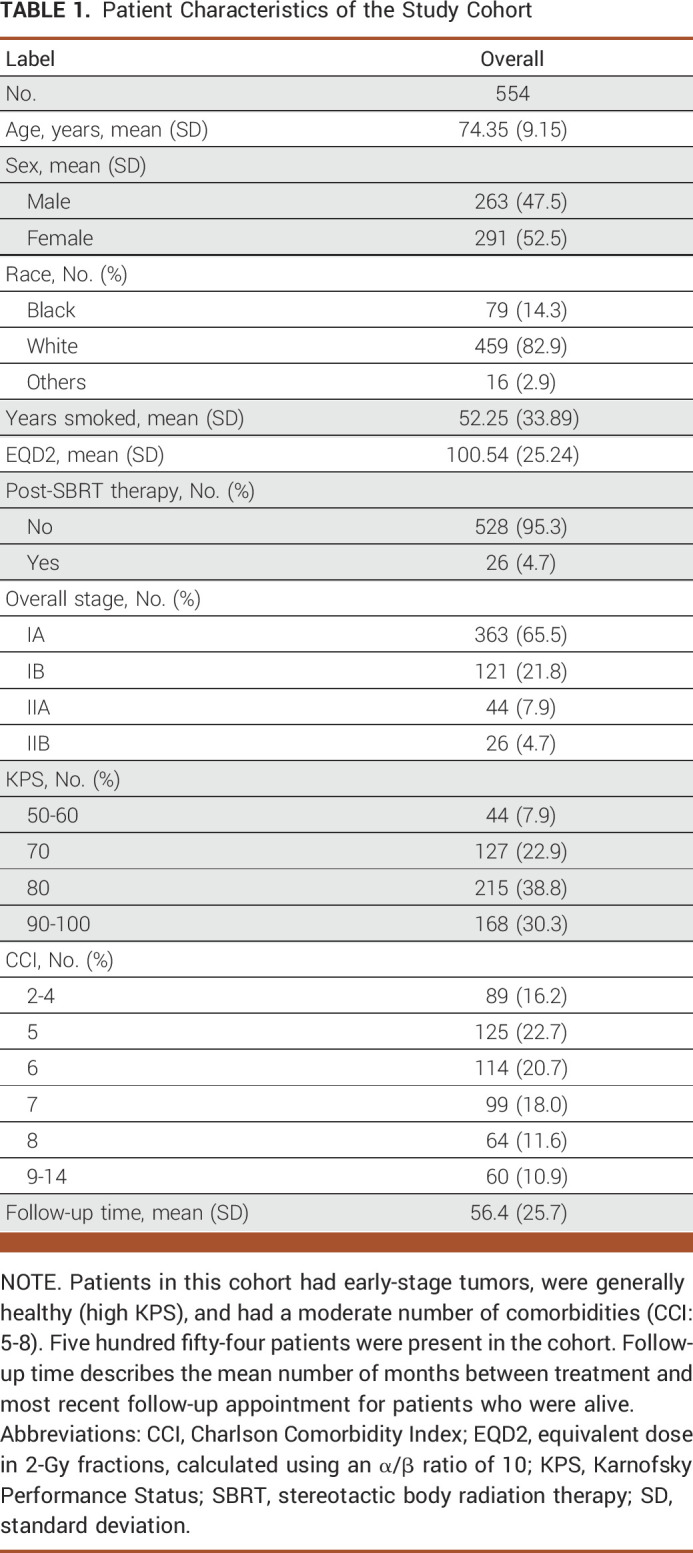
The patients in this study were generally able-bodied individuals with early-stage NSCLC and a moderate number of comorbidities. Table 1 shows a descriptive overview of the patient population. All patients had stage I or II primary NSCLC (none had nodal disease). No patients in this cohort had treatment before SBRT, but 26 patients (4.7%) had some form of post-SBRT chemotherapy. The KPS in this study ranged from 50 to 100. Most patients had baseline independent functional status corresponding to the KPS of 80 (215 patients) and 90-100 (168 patients). Using an α/β ratio of 10, the mean dose received over treatment course was 100.54 Gy (standard deviation [SD], 25.24 Gy) equivalent dose in 2-Gy fractions. Eighty-nine patients had minimal comorbidity corresponding to a CCI of 2-4. Most patients had additional commodities with scores ranging from 5 to 8. Sixty patients had severe comorbidity (CCI > 8). One hundred fifteen (20.8%) patients died of cancer, and 323 (58.3%) patients died of other causes. One hundred sixteen patients (20.9%) were alive at last follow-up, and the median follow-up time for those alive was approximately 4.7 years (SD, 2.14 years).
TABLE 1.
Patient Characteristics of the Study Cohort
Survival Models
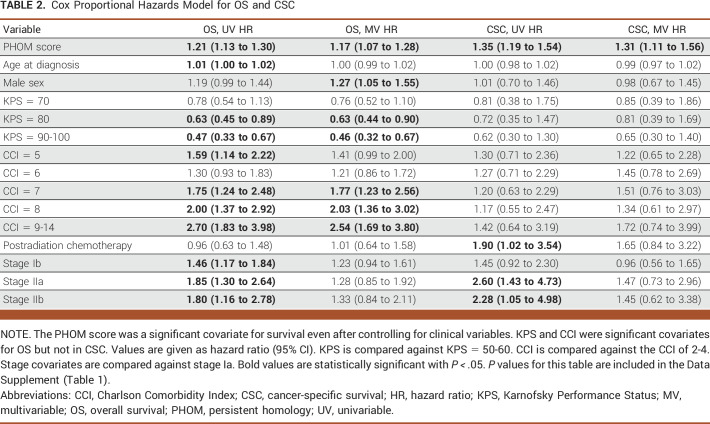
PHOM score was a significant predictor of both OS (hazard ratio [HR], 1.17; 95% CI, 1.07 to 1.28; P < .001) and cancer-specific survival (HR, 1.31; 95% CI, 1.11 to 1.56; P = .0014) in their respective multivariable Cox models. Table 2 lists the exact hazard ratios for all Cox models, and the Data Supplement ([Fig 1]) shows a forest plot visualization of the data. The Data Supplement ([Table 1]) shows the associated P values. For OS, the KPS of 90-100 was associated with improved survival (HR, 0.46; 95% CI, 0.32 to 0.67; P < .001) and increased comorbidity was associated with poor OS (HR, 2.54; 95% CI, 1.69 to 3.80; P < .001).
TABLE 2.
Cox Proportional Hazards Model for OS and CSC
We drew nomograms, which are user-friendly visual tools that represent the underlying survival regression models. Nomograms were calibrated for 1-, 2-, 5-, and 8-year OS as shown in the Data Supplement ([Fig 5]). The model was internally validated using bootstrap resampling, and calibration curves are shown in the Data Supplement ([Fig 4]). The apparent and optimism bias–corrected curves showed relatively strong alignment, with the ideal curve indicating that our model is internally valid and not overfitted. The Data Supplement ([Table 2]) demonstrates the relative contribution of each covariate to the predictive ability of the multivariable Cox model with a relative drop in C index. The Data Supplement ([Fig 3]) shows the proportion chi square contributed by each variable in the multivariable Cox model. Both analyses revealed that PHOM score, KPS, and CCI were the most important variables in the model and that each of these variables contributed unique information to the predictive ability of our model.
Each covariate from the multivariable Cox proportional hazards model is associated with a certain point value, and summing the point values for a given patient's parameter generates a score that can be used to assign 1-, 2-, 5-, and 8-year OS chance. The weighted sum of these survival chances gives the predicted median survival. An online interactive version of this nomogram is available at Eashwarsoma.Shinyapps.19 We provide the exact risk point values associated with each clinical variable in this nomogram as a supplemental CSV file: Predictors_Points_Survival.csv.
Risk Stratification
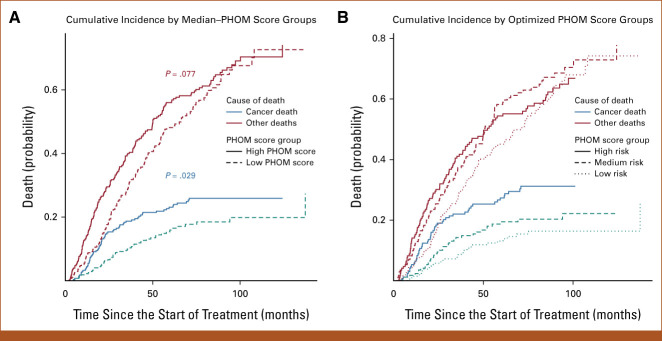
The PHOM score produced well-defined risk categories for OS and cancer death incidence when stratified by median and optimized cutoff values. Figure 2A shows that patients with PHOM scores below the cohort median had a median survival of 45.4 months (95% CI, 40.1 to 51.8), which was significantly higher compared with patients in the high–PHOM score group (median, 29.2 months; 95% CI, 23.6 to 34.3; P < .001). Figure 2B shows survival differences predicted by optimized PHOM score cutoffs. The median survival for the high-PHOM risk group was 25.7 months (95% CI, 20.9 to 31.0). The median survival for the medium- and low-risk groups was 34.8 months (95% CI, 30.0 to 44.9) and 47.0 months (95% CI, 40.9 to 58.1), respectively. These groups had significant differences in OS (P < .001); however, the P value is somewhat moot as the cutoffs were calculated by optimizing the chi-square value of the Kaplan-Meier curve. We applied these cutoffs to clinical groups stratified by KPS, CCI, or overall stage as shown in the Data Supplement ([Fig 2]). This post hoc analysis demonstrated that higher PHOM risk score cutoffs were still associated with poorer survival in all clinical strata except for patients in stage IB, IIA, and IIB where the trend was similar but did not reach statistical significance.
FIG 2.
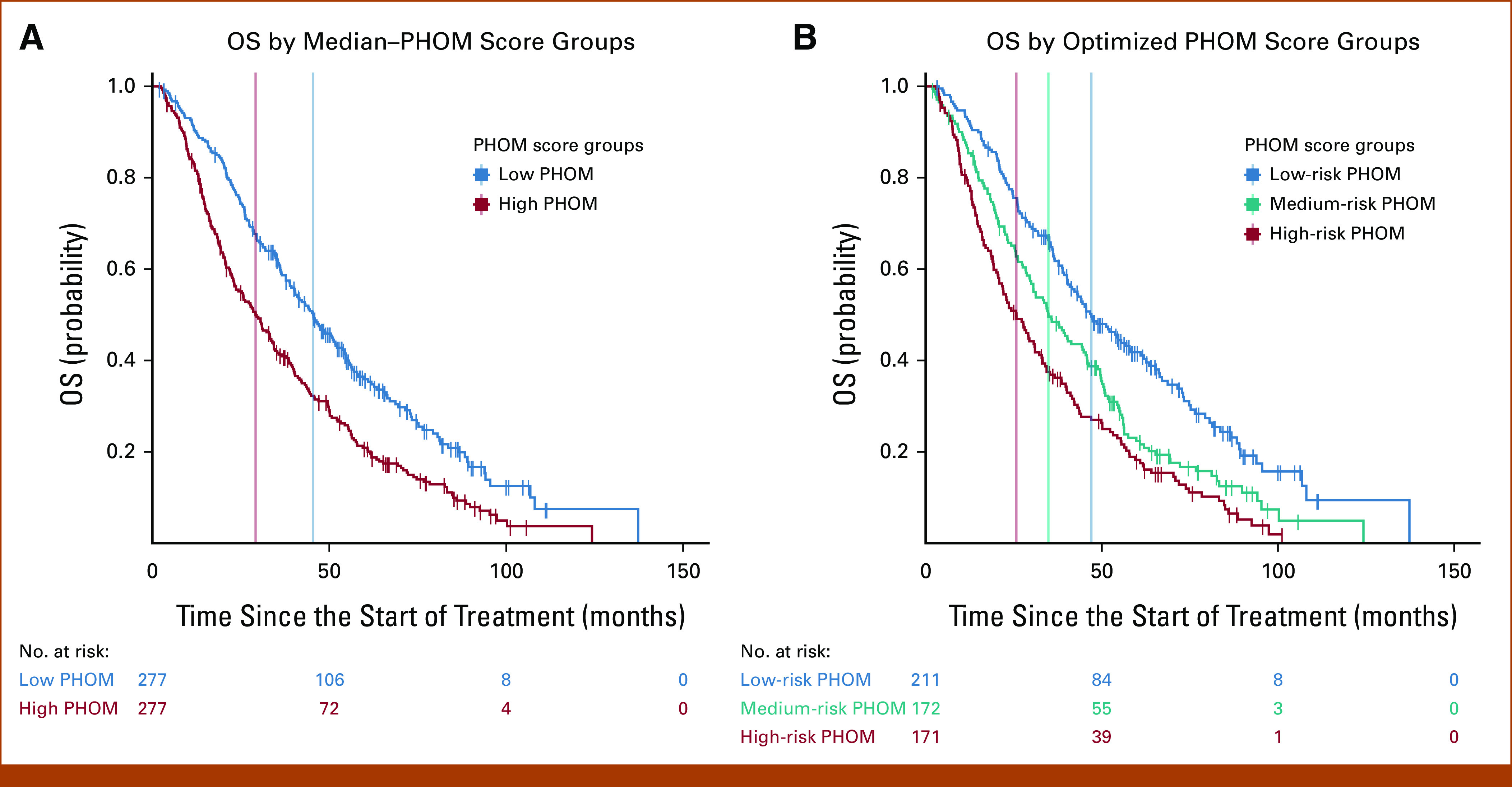
Kaplan-Meier OS by PHOM score. Stratifying the cohort by median PHOM score groups and optimized cutoffs produced well-demarcated survival curves. (A) The median survival for the high-PHOM group was 29.2 months (95% CI, 23.6 to 34.3). The median survival for the low-PHOM group was significantly higher at 45.4 months (95% CI, 40.1 to 51.8; P < .001). (B) The median survival for the high-PHOM risk group was 25.7 months (95% CI, 20.9 to 31.0). This was significantly less than that for both the medium- (P = .020) and low-risk PHOM score groups (P < .001). The median survival for the medium-risk group was 34.8 months (95% CI, 29.9 to 44.9). This was significantly less than that of the low-risk PHOM score group (P < .001). The median survival for the low-risk group was 47.0 months (95% CI, 40.9 to 58.1). However, it is important to note that these P values are somewhat moot since the tertiles were calculated to maximize the chi-square value of the KM curve. KM, Kaplan-Meier; OS, overall survival; PHOM, persistent homology.
Figure 3A compares cause-specific death between PHOM score groups divided by cohort median. Having an above median PHOM score was associated with a 0.244 (95% CI, 0.192 to 0.296) chance of cancer-specific death by month 65, which was significantly higher compared with the below median PHOM score chance of death of 0.171 (95% CI, 0.123 to 0.218; P = .029). However, for other causes of death, a high versus low PHOM score corresponded to a nonsignificant 0.582 (95% CI, 0.521 to 0.642) versus 0.499 (95% CI, 0.436 to 0.563) chance of death by month 65. Figure 3B compares cause-specific death among PHOM score groups divided by optimized PHOM score risk groups. For low-, medium-, and high-risk PHOM score groups, the chance of cancer death by month 65 was 0.146 (95% CI, 0.0950 to 0.197), 0.194 (95% CI, 0.133 to 0.255), and 0.296 (95% CI, 0.224 to 0.366), respectively (P = .0013). For low-, medium-, and high-risk PHOM score groups, the chance of death from other causes by month 65 was 0.474 (95% CI, 0.401 to 0.546), 0.612 (95% CI, 0.535 to 0.689), and 0.551 (95% CI, 0.474 to 0.629), respectively.
FIG 3.
Cumulative incidence of death by specific cause. Stratifying the cohort by median–PHOM score groups and optimized cutoffs produced well-demarcated cumulative incidence curves for cancer-specific death but not death because of other causes. (A) Having a high median PHOM score was associated with a 0.244 (95% CI, 0.192 to 0.296) chance of cancer-specific death by month 65. A low median PHOM score was associated with a significantly less chance of death by month 65 at 0.171 (95% CI, 0.123 to 0.218; P = .029). However, for other causes of death, a high versus low PHOM score corresponds to a nonsignificant 0.582 (95% CI, 0.521 to 0.642) versus 0.499 (95% CI, 0.436 to 0.563) chance of death by month 65. (B) For low-, medium-, and high-risk PHOM score groups, the chance of cancer death by month 65 was 0.146 (95% CI, 0.0950 to 0.197), 0.194 (95% CI, 0.133 to 0.255), and 0.296 (95% CI, 0.224 to 0.366), respectively. For low-, medium-, and high-risk PHOM score groups, the chance of death from other causes by month 65 was 0.474 (95% CI, 0.401 to 0.546), 0.612 (95% CI, 0.535 to 0.689), and 0.551 (95% CI, 0.474 to 0.629), respectively. Similarly, the difference among incidence of death at month 65 was significant when assessing cancer death (P = .0013) but not other deaths. One hundred fifteen patients in this cohort died of cancer, of which 80 were due to nodal and distal progression and the remaining 35 were due to new primary, local, or lobar progression. The P values are moot similar to the Kaplan-Meier risk stratification analysis since we maximized over the chi-square value of the KM curve. KM, Kaplan-Meier; PHOM, persistent homology.
DISCUSSION
Our novel topologically inspired radiomics variable predicts survival and risk stratifies patients with NSCLC. To our knowledge, there is currently no similar topology-based imaging metric that has demonstrated survival prediction in patients with NSCLC. Stratifying PHOM score by the cohort median reveals worse OS and cancer-specific survival in the high–PHOM score group. PHOM score was also the only significant predictor in the multivariable cancer-specific Cox model. In addition, it differentiated cancer-specific death from death by other causes, suggesting that the PHOM score is truly measuring cancer severity (Figs 2 and 3).
Interestingly, our post hoc analysis in the Data Supplement ([Fig 2]) revealed that PHOM score continued to stratify survival across all clinical subgroupings of KPS and CCI. This is expected as PHOM score is a measure of the tumor itself, whereas KPS and CCI are measures of overall health. Stage did not contribute to predicting survival in the multivariable models, possibly because of high local control rates when early-stage tumors are treated with SBRT.20 However, in the Data Supplement ([Fig 2G]), the PHOM score identified high-risk patients within the Stage IA group. Although Stage IA patients are thought to be at low risk overall, approximately 30% of patients with this stage do not survive over a 5-year interval.21 We need better risk stratification, and we offer the PHOM score tertiles as a possibility. Risk-stratifying early-stage NSCLC cancers can signal a need for adjuvant systemic therapy and assist clinical trialists in creating treatment groups on the basis of risk. However, further study is needed as this was a post hoc analysis. Although stages IB, IIA, and IIB had similar trends when grouping by PHOM risk scores, they did not reach statistical significance.
The PHOM score's greatest strength is its ability to predict OS. The C-index demonstrates the predictive ability of a model. When omitting variables one at a time from the model, we found that KPS had the greatest drop in C-index score and CCI and PHOM score resulted in similar C-index drops. This reflects the importance and unique information that each covariate brings to the model (Data Supplement [Table 2]). The calibration plot demonstrates strong internal validity of our nomogram's predictive ability (Data Supplement [Fig 4]).
To our knowledge, this is the first article that uses PHOM to predict OS in NSCLC. Our previous work was limited to demonstrating statistical association with OS and did not include important covariates such as KPS and CCI because of limitations in available data.12 We speculate our metric measures underlying structural tissue heterogeneity. Increased tissue heterogeneity may indicate aggressive tumor growth providing an explanation of why our metric correlates well with cancer-specific survival and predicts OS. Direct pathology data and tumor marker data were not available in this study but should be assessed in future studies to define the biologic significance of the PHOM score. Our new imaging PHOM metric adds to the other metrics that have been developed for glioblastoma and breast and prostate cancer clinical outcomes research.8,10,22
Persistent homology has opened up a new suite of tools for image analysis. We have shown that the PHOM score predicts survival in NSCLC even after controlling for clinical metrics. How the PHOM score could interact with traditional radiomic measures remains to be answered. In addition, the PHOM score could be incorporated with nonimaging data. Genomic models such as the genomically adjusted radiation dose predict individualized response to different radiation doses as demonstrated in a meta-analysis.23,24 We foresee future clinical models that incorporate genomic, imaging, and other data modalities to more precisely guide therapy.
Although our results are promising for NSCLC, some limitations must be considered. Our nomogram is only validated for patients with early-stage NSCLC who receive SBRT therapy as primary treatment. Although we do not see a reason for this metric to not work for surgical patients, we cannot formally say that our nomogram is validated for this population. All the patients in this cohort received care at a tertiary health care center and sometimes became candidates for SBRT because of comorbidities contraindicating surgery. As such, they are more likely to be sicker than the average patient with early-stage NSCLC, which is reflected in our OS rates being less than the typical median survival of over 5 years.25 CT standards vary among institution, so it is not clear whether sources of imaging variance have an impact on PHOM score. However, our previous work assessed scans with varied parameters different from this institution's parameters and was still successful, suggesting a potential resilience of this score to this particular confounder.12
We could not generate a cancer-specific survival curve because of poor calibration curves. We believe this to be a result of the limited number of cancer-specific deaths—likely because of successful treatment of early-stage tumors. For completeness, we include these attempted curves in the Data Supplement ([Fig 6]).
In conclusion, publicizing this nomogram, recruiting multi-institutional data, and conducting prospective studies could address these limitations in future studies. In addition, future work could analyze how the PHOM score interacts with traditional radiomics. The widespread imaging capability of modern hospitals makes imaging metrics such as the PHOM score practical for use in patient care. In cohort studies or randomized controlled trials, the PHOM score could be used to risk stratify patients to investigate who might benefit from adjuvant therapy. In the long term, we envision the creation of a risk calculator that extracts the most useful features of imaging data to both prognosticate and guide clinical decisions.
ACKNOWLEDGMENT
The authors thank members of Theory Division for their constructive feedback and valuable input in this project. We thank Jessica Scarborough, PhD, for advice and code to generate optimal PHOM score tertiles. J.G.S. was supported by NIH grant R37 CA244613, NIH grant U54 (Radiation Oncology Biology Integration Network), and the American Cancer Society through their Research Scholar Grant. E.S. and R.R.W. were supported by the Case Comprehensive Cancer Center Summer Training Grant for medical students. K.Y. was supported by the Computational Genomic Epidemiology of Cancer (CoGEC) Program at the Case Comprehensive Cancer Center (T32CA094186), Young Investigator Award from ASCO Conquer Cancer Foundation, and RSNA Research Resident Grant.
Adam Litzler
Employment: Epic Systems
Stock and Other Ownership Interests: Epic Systems
Travel, Accommodations, Expenses: Epic Systems
Peng Qi
Research Funding: Varian Medical Systems
Nathan A. Pennell
Consulting or Advisory Role: Lilly, Merck, Genentech, Pfizer, Mirati Therapeutics, Janssen Oncology, Sanofi/Regeneron, resistanceBio, Takeda, Novartis, Vial, Bayer, AnHeart Therapeutics, Summit Therapeutics
Research Funding: AstraZeneca (Inst), Merck (Inst), Loxo (Inst), Spectrum Pharmaceuticals (Inst), Bristol Myers Squibb (Inst), Mirati Therapeutics (Inst), Sanofi (Inst), AnHeart Therapeutics (Inst), Navire (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/204570/summary
Daniel Raymond
Stock and Other Ownership Interests: Bristol Myers Squibb, Zimmer BioMet
Honoraria: KLS Martin
Consulting or Advisory Role: KLS Martin
Research Funding: Pacira Pharmaceuticals
Michael W. Kattan
Consulting or Advisory Role: Novartis, GlaxoSmithKline, Exosome Diagnostics, Merck, Stratify Genomics, Renalytix AI, SkinCure Oncology, Verici Dx
Research Funding: Novo Nordisk (Inst), Celgene (Inst), Otsuka (Inst), Bayer (Inst)
Jacob G. Scott
Stock and Other Ownership Interests: Cvergenx, Empyrean Medical Systems
Consulting or Advisory Role: Cvergenx, Empyrean Medical Systems, SkinCure Oncology, ResistanceBio, Vironexis
Patents, Royalties, Other Intellectual Property: I am a coinventor on IP licensed to Cvergenx and Moffitt Cancer Center for personalized radiation dosing
No other potential conflicts of interest were reported.
SUPPORT
J.G.S. was supported by NIH grant R37 CA244613, NIH grant U54 CA274513, and American Cancer Society. E.S. and R.R.W. were supported by the Case Comprehensive Cancer Center Summer Training Grant for medical students. K.Y. was supported by the Computational Genomic Epidemiology of Cancer (CoGEC) Program at the Case Comprehensive Cancer Center (T32CA094186), Young Investigator Award from ASCO Conquer Cancer Foundation, and RSNA Research Resident Grant.
AUTHOR CONTRIBUTIONS
Conception and design: Eashwar Somasundaram, Raoul R. Wadhwa, Adam Litzler, Rowan Barker-Clarke, Kevin Stephans, Daniel Raymond, Jacob G. Scott
Administrative support: Jacob G. Scott
Provision of study materials or patients: Gregory Videtic, Kevin Stephans
Collection and assembly of data: Eashwar Somasundaram, Adam Litzler, Peng Qi, Gregory Videtic, Kevin Stephans
Data analysis and interpretation: Eashwar Somasundaram, Raoul R. Wadhwa, Adam Litzler, Kevin Stephans, Nathan A. Pennell, Daniel Raymond, Kailin Yang, Michael W. Kattan
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Adam Litzler
Employment: Epic Systems
Stock and Other Ownership Interests: Epic Systems
Travel, Accommodations, Expenses: Epic Systems
Peng Qi
Research Funding: Varian Medical Systems
Nathan A. Pennell
Consulting or Advisory Role: Lilly, Merck, Genentech, Pfizer, Mirati Therapeutics, Janssen Oncology, Sanofi/Regeneron, resistanceBio, Takeda, Novartis, Vial, Bayer, AnHeart Therapeutics, Summit Therapeutics
Research Funding: AstraZeneca (Inst), Merck (Inst), Loxo (Inst), Spectrum Pharmaceuticals (Inst), Bristol Myers Squibb (Inst), Mirati Therapeutics (Inst), Sanofi (Inst), AnHeart Therapeutics (Inst), Navire (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/204570/summary
Daniel Raymond
Stock and Other Ownership Interests: Bristol Myers Squibb, Zimmer BioMet
Honoraria: KLS Martin
Consulting or Advisory Role: KLS Martin
Research Funding: Pacira Pharmaceuticals
Michael W. Kattan
Consulting or Advisory Role: Novartis, GlaxoSmithKline, Exosome Diagnostics, Merck, Stratify Genomics, Renalytix AI, SkinCure Oncology, Verici Dx
Research Funding: Novo Nordisk (Inst), Celgene (Inst), Otsuka (Inst), Bayer (Inst)
Jacob G. Scott
Stock and Other Ownership Interests: Cvergenx, Empyrean Medical Systems
Consulting or Advisory Role: Cvergenx, Empyrean Medical Systems, SkinCure Oncology, ResistanceBio, Vironexis
Patents, Royalties, Other Intellectual Property: I am a coinventor on IP licensed to Cvergenx and Moffitt Cancer Center for personalized radiation dosing
No other potential conflicts of interest were reported.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2020. CA Cancer J Clin 70:7-30, 2020 [DOI] [PubMed] [Google Scholar]
- 2.Duma N, Santana-Davila R, Molina JR: Non–small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc 94:1623-1640, 2019 [DOI] [PubMed] [Google Scholar]
- 3.Chang JY, Mehran RJ, Feng L, et al. : Stereotactic ablative radiotherapy for operable stage i non-small-cell lung cancer (revised STARS): Long-term results of a single-arm, prospective trial with prespecified comparison to surgery. Lancet Oncol 22:1448-1457, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstraw P, Chansky K, Crowley J, et al. : The IASLC lung cancer staging project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol 11:39-51, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Sperduto PW, Yang TJ, Beal K, et al. : Estimating survival in patients with lung cancer and brain metastases. JAMA Oncol 3:827, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillies RJ, Kinahan PE, Hricak H: Radiomics: Images are more than pictures, they are data. Radiology 278:563-577, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlsson G: Topology and data. Bull Am Math Soc 46:255-308, 2009 [Google Scholar]
- 8.Gordetsky J, Epstein J: Grading of prostatic adenocarcinoma: Current state and prognostic implications. Diagn Pathol 11:25, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba AI, Câtoi C: Comparative Oncology. Bucharest, The Publishing House of the Romanian Academy, 2007 [PubMed] [Google Scholar]
- 10.Crawford L, Monod A, Chen AX, et al. : Predicting clinical outcomes in glioblastoma: An application of topological and functional data analysis. J Am Stat Assoc 115:1-12, 2019. 34012183 [Google Scholar]
- 11.Oyama A, Hiraoka Y, Obayashi I, et al. : Hepatic tumor classification using texture and topology analysis of non-contrast-enhanced threedimensional t1-weighted MR images with a radiomics approach. Sci Rep 9:8764, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somasundaram E, Litzler A, Wadhwa R, et al. : Persistent homology of tumor CT scans is associated with survival in lung cancer. Med Phys 48:7043-7051, 2021 [DOI] [PubMed] [Google Scholar]
- 13.Steyerberg EW, Harrell FE: Prediction models need appropriate internal, internal–external, and external validation. J Clin Epidemiol 69:245-247, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pencina MJ, D'Agostino RB: Overall C as a measure of discrimination in survival analysis: Model specific population value and confidence interval estimation. Stat Med 23:2109-2123, 2004 [DOI] [PubMed] [Google Scholar]
- 15.R Core Team : R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2012 [Google Scholar]
- 16.RStudio Team : RStudio: Integrated Development Environment for R. Boston, MA, RStudio, PBC, 2020 [Google Scholar]
- 17.Van Rossum G, Drake FL: Python 3 Reference Manual. Scotts Valley, CA, CreateSpace, 2009 [Google Scholar]
- 18.Somasundaram E: PHOM LUNG CA CCF. Github. https://github.com/eashwarsoma/phom-lungca-ccf
- 19.Somasundaram E: Expected survival properties. R Shiny Apps. https://eashwarsoma.shinyapps.io/LungCancerTDATest/
- 20.Sebastian NT, Xu-Welliver M, Williams TM: Stereotactic body radiation therapy (SBRT) for early stage non-small cell lung cancer (NSCLC): Contemporary insights and advances. J Thorac Dis 10:S2451-S2464, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Port JL, Kent MS, Korst RJ, et al. : Tumor size predicts survival within stage IA non-small cell lung cancer. Chest 124:1828-1833, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Nicolau M, Levine AJ, Carlsson G: Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci 108:7265-7270, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott JG, Berglund A, Schell MJ, et al. : A genome-based model for adjusting radiotherapy dose (GARD): A retrospective, cohort-based study. Lancet Oncol 18:202-211, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott JG, Sedor G, Ellsworth P, et al. : Pan-cancer prediction of radiotherapy benefit using genomic-adjusted radiation dose (gard): A cohort-based pooled analysis. Lancet Oncol 22:1221-1229, 2021 [DOI] [PubMed] [Google Scholar]
- 25.Nesbitt JC, Putnam JB, Walsh GL, et al. : Survival in early-stage non-small cell lung cancer. Ann Thorac Surg 60:466-472, 1995 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data were processed and analyzed using R (v3.6.1) and Python (v3.7.6).15-17 Anonymized data may be requested from the authors. Code with explanation for this data pipeline can be found at GitHub.18