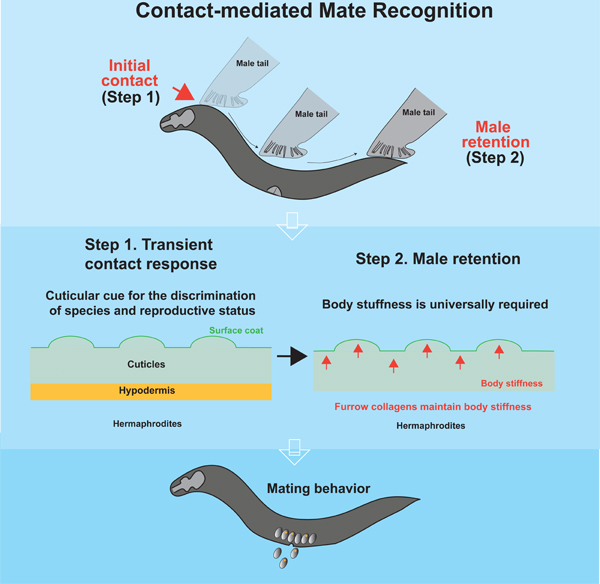
Summary
Physical contact is prevalent in the animal kingdom to recognize suitable mates by decoding information about sex, species, and maturity. Although chemical cues for mate recognition have been extensively studied, the role of mechanical cues remains elusive. Here we show that C. elegans males recognize conspecific and reproductive mates through short-range cues, and the attractiveness of potential mates depends on the sex and developmental stages of the hypodermis. We find that a particular group of cuticular collagens is required for mate attractiveness. These collagens maintain body stiffness to sustain mate attractiveness but do not affect the surface properties that evokes the initial step of mate recognition, suggesting that males utilize multiple sensory mechanisms to recognize suitable mates. Manipulations of body stiffness via physical interventions, chemical treatments, and 3D-printed bionic worms indicate that body stiffness is a mechanical property for mate recognition and increases mating efficiency. Our study thus extends the repertoire of sensory cues of mate recognition in C. elegans and provides a paradigm to study the important role of mechanosensory cues in social behaviors.
Keywords: Mate recognition, Mate searching, C. elegans, Collagen, Body stiffness
GRAPHICAL ABSTRACT:
Introduction
Mating is a social behavior conserved among metazoans that transfers genetic variation for phenotypic diversity within conspecific individuals. To maximize reproductive success and reduce the cost of energy for inappropriate mating, animals evolve different strategies to recognize conspecific and reproductive mates. Recognition of suitable mates requires accessing information from individuals through a broad array of sensory modalities 1–4. Animals then integrate sensory information to decode the chemical and physical properties of mates through complex neural networks and respond appropriately according to their location, species, sex, maturity, and even physiological conditions 3,5–7. Thus, identifying a complete set of sensory cues used in mate recognition is crucial to understanding the neural computation underlying social behaviors.
Studies of sensory cues for mate recognition mostly centered on chemical-based communication 1,7,8, in which animals leave chemical marks in the environment or on their bodies that instruct suitable mates. However, in the wild, the mixture of chemical cues from different species or sex may disturb the transmission and interpretation of the chemical signal, which likely leads to lower efficiency. Furthermore, chemical cues can be mimicked by predators, as with carnivorous fungi that secrete pheromone-like chemicals to attract their C. elegans prey 9. As a result, physical traits, such as colors, sounds, and body size, are used to enhance the specificity of mate recognition in many species 10–12. For instance, pigmentation of eyes and body are visual cues for mate selections and courtship behaviors in Drosophila 13,14. Males also use their forelegs to physically contact females to access cuticular pheromones for species and sex recognition 15–17. After contacting females, males display “love songs” by extending and vibrating a wing and finish copulation upon reception by females. Interestingly, cuticular extracts are insufficient to evoke mate recognition, suggesting the involvement of unidentified sensory cues to initiate courtship behaviors 17. These findings highlight that integrating multiple sensory cues is pivotal for the precision of mate recognition.
C. elegans is hermaphroditic nematode with two sexes, males and self-reproductive hermaphrodites. Hermaphrodites secrete pheromones as long-range cues in the environment to attract males remotely 2,6,18, and males move toward the source of pheromones sensed by olfactory and pheromone-sensing neurons 19. In addition to pheromonal cues, physical contact provides short-range signals for mate recognition in C. elegans 20,21. After contacting hermaphrodites, males restrict the locomotion area independent of pheromones, presumably increasing the chance of mating 20. Moreover, physical contact with hermaphrodites is essential for the sexual conditioning of males that reverses the learned aversive behaviors associated with starvation 20–22. Although physical contact is an essential step for contact-mediated mate recognition, Barrios et al. have shown that male-specific EF interneurons are dispensable for initial contact response to mates but are required for enduring changes of locomotion 21. Further studies speculated that persistent scanning of suitable mates provides additional information to discriminate mates 20,21. These findings highlight a complex neural basis with multiple steps in decoding mate information. However, the nature of contact-mediated sensory cue(s) for each step on hermaphrodites is unknown. Because physical contact is mainly controlled by male-specific ray sensory neurons, which are sensitive to mechanical stimulation 23, it raises the possibility that mechanical cues transmit the short-range signal for mate recognition. Indeed, mechanosensory proteins have been implicated in mating but what they sense is unknown 24.
In nematodes, a collagen-rich apical extracellular matrix, namely cuticle, maintains body shape and serves as a physical barrier to prevent pathogenic infection 25–27. The composition of collagens is crucial to determine surface structure 28,29, and the physical constraints of cuticles maintain body stiffness supported by internal organs and muscle tones 30–32. A recent study has shown that specific collagens in the cuticle are involved in maintaining body mechanical properties 32, but their role in mate recognition has not been explored.
Here we showed that the body stiffness of mates is a mechanical property that facilitates mate recognition. We demonstrated that C. elegans males physically contacted potential mates to access the information of species, sex, and reproductive stages. Through a candidate genetic screen, we discovered that the furrow collagens, including dpy-2, dpy-7, and dpy-10, were generally required for mates to attract males independent of their known roles in controlling body size and surface texture. Interestingly, these collagens were dispensable for the first step in mate recognition by evoking contact responses in males. Instead, furrow collagens maintain body stiffness as recently reported 32 in the second step. We found that cuticular cues, which evoke contact response, and a specific range of body stiffness were inseparable sensory cues, suggesting a two-step sensory mechanism for contact-mediated mate recognition. Lastly, we showed that mate attractiveness was associated with mating efficiency, indicating the biological significance of contact-mediated mate recognition in social interactions. We therefore provide evidence that the mechanical property conferred by furrow collagens facilitates mate recognition in nematodes. Since physical contact is prevalent in mate recognition across the animal kingdom, our study opens an avenue to study the roles of mechanical cues in social behaviors and the underlying neural mechanisms.
Results
C. elegans Males Utilize Contact-mediated Cues to Recognize Species, Sex, and Developmental Stages
To search for contact-mediated cues on hermaphrodites, we performed male retention assay that has been used to evaluate the sex drive of individual males with different ages, mating history, and nutritional status (Figure S1A) 20,21,33. We placed day 1 (D1) adult males with or without hermaphrodites on a food source and measured the sex drive of males by calculating the percentage of leaving males (See STAR Methods) 33. If alone, males had a strong tendency to leave the food source and explore the environment. In contrast, the presence of adult hermaphrodites fixed by paraformaldehyde for 30 min profoundly switched male locomotion patterns from exploration to local stay (Figure 1A and 1C). We used these behavioral changes as a readout to study mate recognition, as the presence of suitable mates retains males, while the loss of essential sensory information reduces male retention. Consistent with previous studies 6,7,21,34,35, we found that hermaphrodites utilized distinct sets of molecules apart from pheromonal, germline, and vulval signaling to retain males (Figure 1A, S1B, and S1C).
Figure 1. Sex and developmental stages of hypodermis are crucial for mate attractiveness.
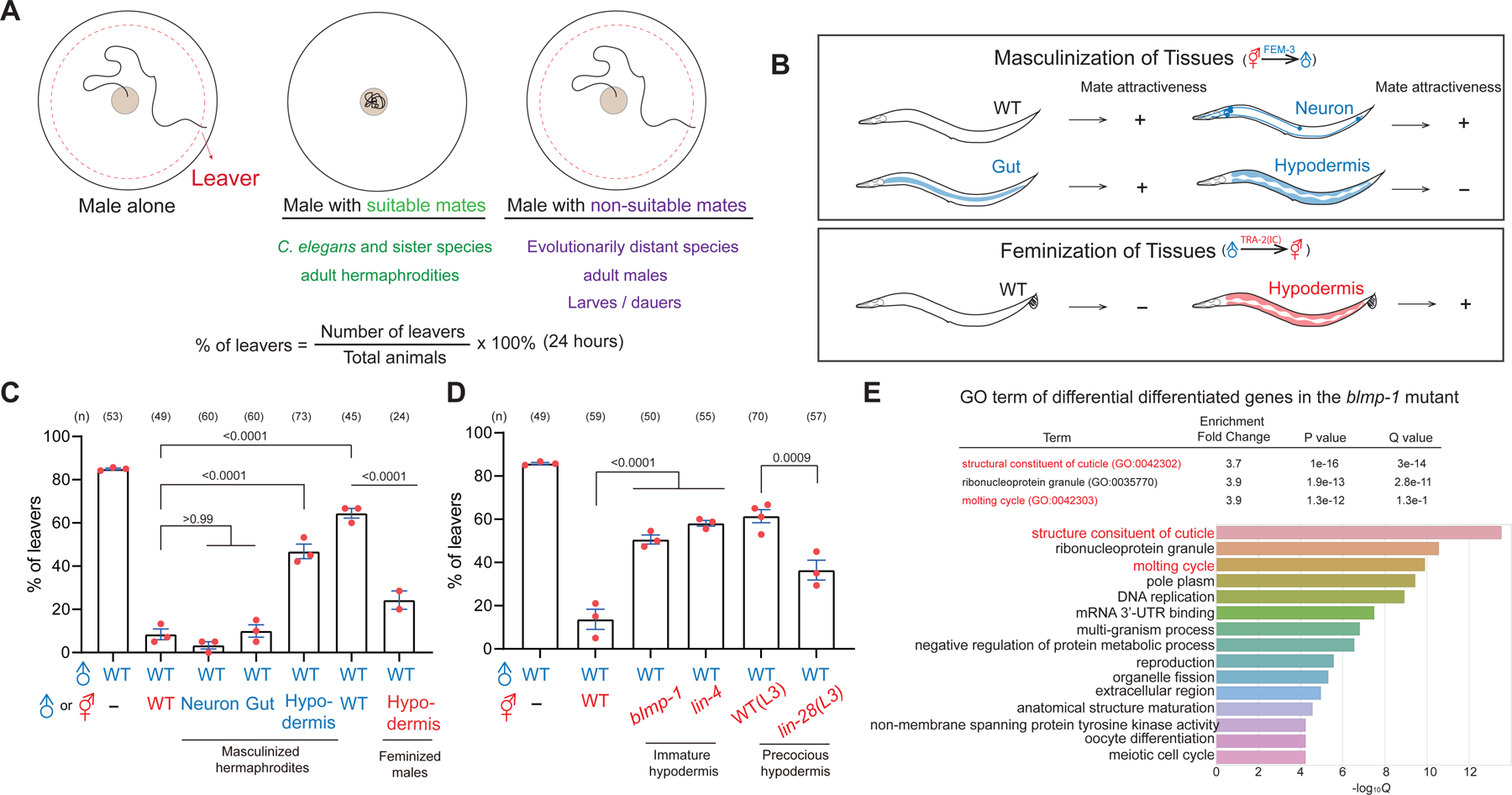
(A) Summary of figure S1 and a schematic diagram of male retention assay with the equation for quantification. (B) Schematic diagram of masculinized hermaphrodites and feminized males by expressing FEM-3 and TRA-2(IC). (C-D) Percentage of male leavers in male retention assay with transgenic and mutant hermaphrodites. In the subsequent figures, each dot represents the percentage of leavers in a population containing 15–20 males as one replica, and n indicates the number of total animals tested in all replicas. Error bar = S.E.M. One-way ANOVA with Bonferroni correction and p values are indicated. (E) Gene Ontology (GO) analysis of differentially expressed genes comparing the blmp-1 mutant and the wild-type.
We next wondered whether males are able to discriminate species, sex, and developmental stages of potential mates. In a previous study, Lipton et al. showed that males can discriminate the sexes through physical contact 20. Indeed, while the presence of hermaphrodites completely suppressed male exploration, the exploration behavior of males persisted in the presence of other males (Figure 1C) 20. Notably, we still observed a marginal but significant decrease in leaving tendency with male-male interaction compared to solitary males, suggesting that males still respond to the presence of conspecific males (Figure 1A and 1C).
We further asked whether C. elegans males can distinguish different species (Figure S1D and S1E). C. elegans males displayed a strong leaving tendency to four evolutionarily distant species, Steinernema carpocapsae, Rhabditis sp., Aunema tufa, and Pristionchus pacificus, but the leaving tendency of males toward sister species Caenorhabditis briggsae and remanei was also mildly increased without reaching statistical significance (Figure S1D and S1E). These data indicate that contact-mediated mate recognition of C. elegans is most prominent among different genera. Interestingly, we found that discrimination of sister strains was more discernable in C. remanei, a male-female species, suggesting that dioecious species have a stronger selection of different species (Figure S1F). These results demonstrate that Caenorhabditis males recognize different species through physical contact, and the degree of recognition is associated with the evolutionary distance (Figure 1A).
Lastly, we focused on the recognition of different developmental stages. C. elegans become reproductive adults after four larva stages in the well-fed condition. In unfavorable conditions, larvae switch off the reproductive cycle and enter the diapause stage called dauer. We found that the ability of hermaphrodites to retain males increased dramatically from the L3 larva stage and reached full capacity at the adult stage (Figure S1G). By contrast, dauer did not retain males (Figure S1G). We also showed that D1, D5, and D10 hermaphrodites were equally attractive, suggesting that age did not affect male retention (Figure S1H). Thus, males can discriminate developmental stages but not the ages (Figure 1A). Altogether, we conclude that C. elegans hermaphrodites present cues for males to recognize conspecific and reproductive mates.
Sexual State and Developmental Stages of Hypodermis Are Critical for Hermaphrodites to Retain Males
To gain insight into contact-mediated cues on hermaphrodites, we asked which tissue is essential for hermaphrodites to retain males. In C. elegans, the sex determination pathway is autonomously controlled by transcriptional programming. Masculinization of somatic tissues can be achieved by overexpressing the E3 ubiquitin ligase FEM-3, which degrades the transcription factor TRA-1 essential for hermaphroditic fate 36. We expressed FEM-3 in the hermaphrodites driven by tissue-specific promoters and subjected these masculinized hermaphrodites to male retention assay. Hypodermis-masculinized hermaphrodites significantly lost male retention, whereas masculinization of neurons and intestines had no effect (Figure 1B and 1C). Masculinization of hypodermis also led to the protrusive vulva and ray-like tail in hermaphrodites, supporting that the sexual identity was altered (Figure S2). Furthermore, hypodermis-feminized males with hermaphrodite-like tails enhanced male retention compared with wild-type males (Figure 1B, 1C, and S2). These data indicate the sexual identity of the hypodermis is critical for mates to retain males.
Since the sexual identity of hypodermis is crucial for retaining males, we wondered whether genes involved in the maturity of hypodermis affect male retention in hermaphrodites. lin-4 is a heterochronic gene involved in the maturation of neuronal and hypodermal cells 37. In the lin-4 mutant hermaphrodites, the development of hypodermal cells is retarded and cannot form adult-specific hypodermis. Adult lin-4 mutant hermaphrodites significantly lost the ability to retain males with immature hypodermis (Figure 1D). We also tested precocious mutant lin-28, whose adult-specific hypodermis are abnormally formed at L3 larva stage 38. In contrast to the lin-4 mutant, lin-28 mutant hermaphrodites retained males precociously at L3 larva stage (Figure 1D). To further corroborate the role of hypodermis, we tested an evolutionarily conserved transcription factor blmp-1 (B lymphocyte-induced maturation protein 1) that controls several aspects of the development of hypodermal cells 39–41. Consistently, loss of blmp-1 in hermaphrodites did not retain males effectively (Figure 1D). Altogether, these data suggest that the maturity of hypodermal cells is a determinant factor for mate recognition.
Epicuticular Components Play Minimal Roles in Retaining Males
To find the effectors in the hypodermis, we analyzed published RNA-seq data of blmp-1 mutant hermaphrodites from a previous study 40. GO analysis of 975 differentially expressed genes revealed an enrichment of cuticular constituents (Figure 1E; see Data S1 for more information). Cuticular constituents are secreted by the hypodermis, and the apical layer is covered by a surface coat, composed of lipids and glycoproteins (Figure 2A) 42,43. Several lines of evidence indicate that the surface coat is not required. First, adult hermaphrodites treated with organic solvents such as hexane or ethanol remained attractive to males, suggesting that lipids had a minimal effect (Figure S3A). Consistent with this, mutants of desaturases fat-5, fat-6, fat-7, or their double mutants did not alter male retention (Figure S3A). Furthermore, mutations in srf-2, srf-3, srf-5, bus-1, bus-4, bus-13, and bus-17 have been shown to reduce but do not completely abolish the contact response of males via regulating the expression of surface epitopes for defending pathogens 25,44–46. However, these mutants effectively retained males compared with that in the otherwise wild-type animals (Figure S3B). Altogether, these data support that the surface coat of cuticles does not play a crucial role in contact-mediated mate recognition.
Figure 2. Collagens are required and sufficient to promote mate attractiveness.
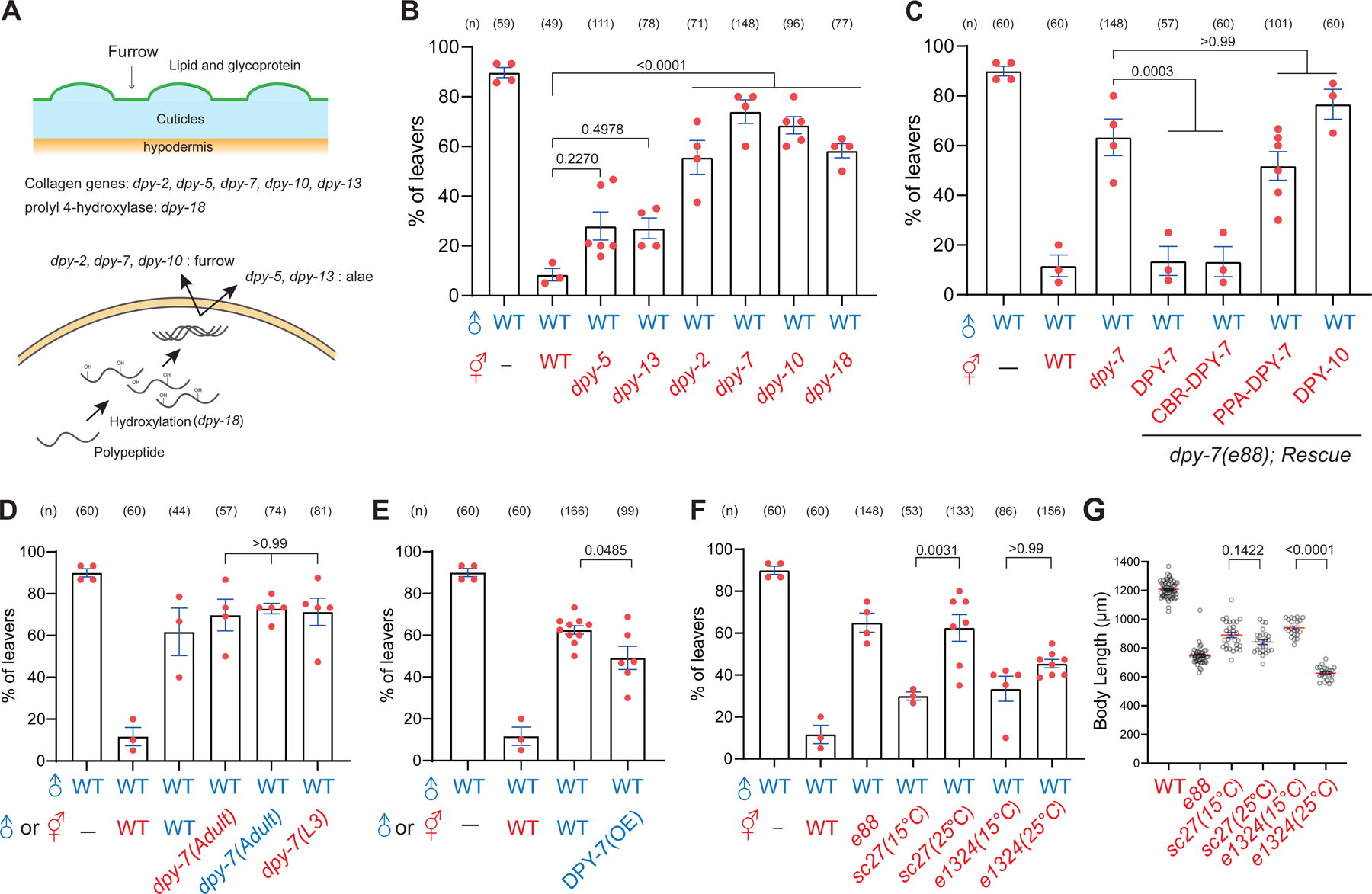
(A) (Top) A schematic diagram of C. elegans cuticles (blue) with its surface coat (green) and furrow (arrow). Mutant hermaphrodites tested for mate recognition are listed below. (Bottom) The schematic diagram of the biosynthesis pathway of collagens and related mutants. (B-F) Percentage of male leavers in male retention assay with mutant hermaphrodites in indicated genotypes. (G) Quantification of body length in the dpy-7 temperature-sensitive mutants.
Furrow Collagens Are Essential for Hermaphrodites to Retain Males
The cortical layer of the cuticle is mainly composed of collagens. We hypothesized that collagens or related proteins are essential to retaining males. Consistent with this hypothesis, we found that the mutation in dpy-18, a crucial enzyme for processing collagens 47, became less attractive to males (Figure 2A and 2B). To search for specific collagens involved in male retention, we focused on the downregulated genes encoding collagens or surface proteins in the blmp-1 mutant and selected candidates with gene expression at L4 and adult stages (See Data S1 for more information). When deletion mutant strains were unavailable, we constructed deleted mutants by introducing a STOP-IN cassette using the CRISPR-Cas9 method as previously described 48. While the locomotion behaviors and morphology of these mutants were grossly normal, mutant hermaphrodites exhibited a marginal decrease in male retention but did not reach statistical significance (Figure S4A). These observations prompted us to examine other collagen mutants with more robust phenotypes in body shape 41,49, including dpy-2, dpy-3, dpy-5, dpy-7, dpy-8, dpy-9, dpy-10, dpy-13, lon-2, sqt-1, sqt-3, and rol-6 mutants. Strikingly, we found that males were barely retained by hermaphrodites without furrow collagens, including dpy-2, dpy-3, dpy-7, dpy-8, dpy-9, and dpy-10 (Figure 2B, S4A, and S4B). Conversely, mutations in the rest of the genes remained attractive to males. It is noteworthy that alterations of body length in the Dpy and Lon mutants were not associated with the degree of male retention, suggesting that body length is not a determinant factor for mate recognition (Figure 2B, S4A, and S4C).
Because the dpy-7 mutant showed the most severe defects, we focused on dpy-7 and studied the underlying molecular mechanisms. Feeding RNAi of dpy-7 resulted in comparable defects of the e88 allele, confirming that DPY-7 activity is required for hermaphrodites to retain males (Figure S4D). Moreover, restoration of dpy-7 but not dpy-10 driven from a hypodermis-specific promoter rescued male retention in the dpy-7 mutant hermaphrodites, indicating that dpy-7 functions in the hypodermis and is not interchangeable quantitatively with other collagens (Figure 2C). Because dpy-7 is evolutionarily conserved among nematodes 50,51, we wondered whether the sequence variations in dpy-7 of different species confer the specificity of species recognition. To test this hypothesis, we cloned C. briggsae and P. pacificus dpy-7 orthologs and expressed them under the control of C. elegans hypodermis-specific promoter. The expression of CBR-dpy-7 significantly restored the male retention and body length in the C. elegans dpy-7 mutant, whereas the expression of PPA-dpy-7 had no discernible effects on both phenotypes (Figure 2C and S4E). These data indicate that the function of dpy-7 for retaining males is conserved in Caenorhabditis nematodes. Consistent with it, we generated a CBR-dpy-7 mutant by inserting a STOP-IN cassette in the dpy-7 ortholog of C. briggsae and showed that mutant hermaphrodites were unable to retain males (Figure S4F).
We next investigated whether dpy-7 is required to present sexual and reproductive signals on mates. dpy-7 mutant animals exhibited the same male retention regardless of their sexual identity. Likewise, the defects of the dpy-7 mutant were not further enhanced in juvenile larvae (L3) hermaphrodites (Figure 2D), indicating that dpy-7 is necessary to present mate information of sexual identity and developmental stages. Moreover, we overexpressed dpy-7 in C. elegans males and found that transgenic males became more attractive, suggesting that the expression level of dpy-7 is crucial for mates to retain males (Figure 2E).
Separable Functions of dpy-7 for Body Shape and Male Retention
Our previous data indicated that body size is not a determinant factor for retaining males (Figure S4C), so we speculated that collagen/dpy-7 has two separate functions in male retention and body shape. To test this hypothesis, we analyzed two additional alleles of missense dpy-7 mutations. Specifically, sc27 and e1324 are missense temperature-sensitive mutations in the collagen domain with severe Dpy and Rol phenotypes that shorten body length at restrictive temperatures 52. At the permissive temperature of 15℃, both mutants retained males comparable to the otherwise wild-type hermaphrodites (Figure 2F). However, at the restrictive temperature of 25℃, the sc27 mutant allele was unable to retain males, whereas the e1324 mutant allele remained attractive (Figure 2F). By contrast, body length was drastically shortened in the e1324 mutant allele but not in the sc27 mutant allele (Figure 2G). Thus, the different expressivities of body shape and male retention in the sc27 and e1324 alleles support the notion that collagen/dpy-7 has separable functions for male retention and body shape.
The Purified Cuticles Are Not Sufficient for Male Retention
How do collagens carry the mate information on cuticles? Because physical contact with suitable mates is essential for males to restrict locomotion in the male retention assay 20,21, we first asked whether these collagens are required for the initial step by evoking the contact response of males. Adult C. elegans cuticles were then dissociated from interior organs by the SDS-containing buffer (See STAR Methods). Consistent with the prior studies 44, we found that purified cuticles from some Srf/Col/Bus mutants evoked slightly lower contact response (Figure S5A), suggesting that purified cuticles preserved features in live animals. Surprisingly, purified cuticles from wild-type and the dpy-7 mutant hermaphrodites evoked robust contact responses of males with reversals and turns (Figure 3A, 3B, and S5A), indicating that males can sense and respond to purified cuticles independent of dpy-7 collagen. Notably, purified cuticles from the wild-type and the dpy-7 mutant hermaphrodites evoked transient contact response but never sustained prolonged scanning essential for contact-mediated mate recognition, consistent with the observations that purified cuticles alone were unable to retain males regardless of the species, reproductive status, and sex (Figure 3C). Despite this partial effect, we found that such response to purified cuticles is species- and stage-specific because males rarely responded to P. pacificus and L3 larval cuticles, supporting the presence of conspecific cuticular cues in the adult stage for the recognition of species and developmental stages (Figure 3B).
Figure 3. The surface properties of cuticles are likely not involved in mate attractiveness.
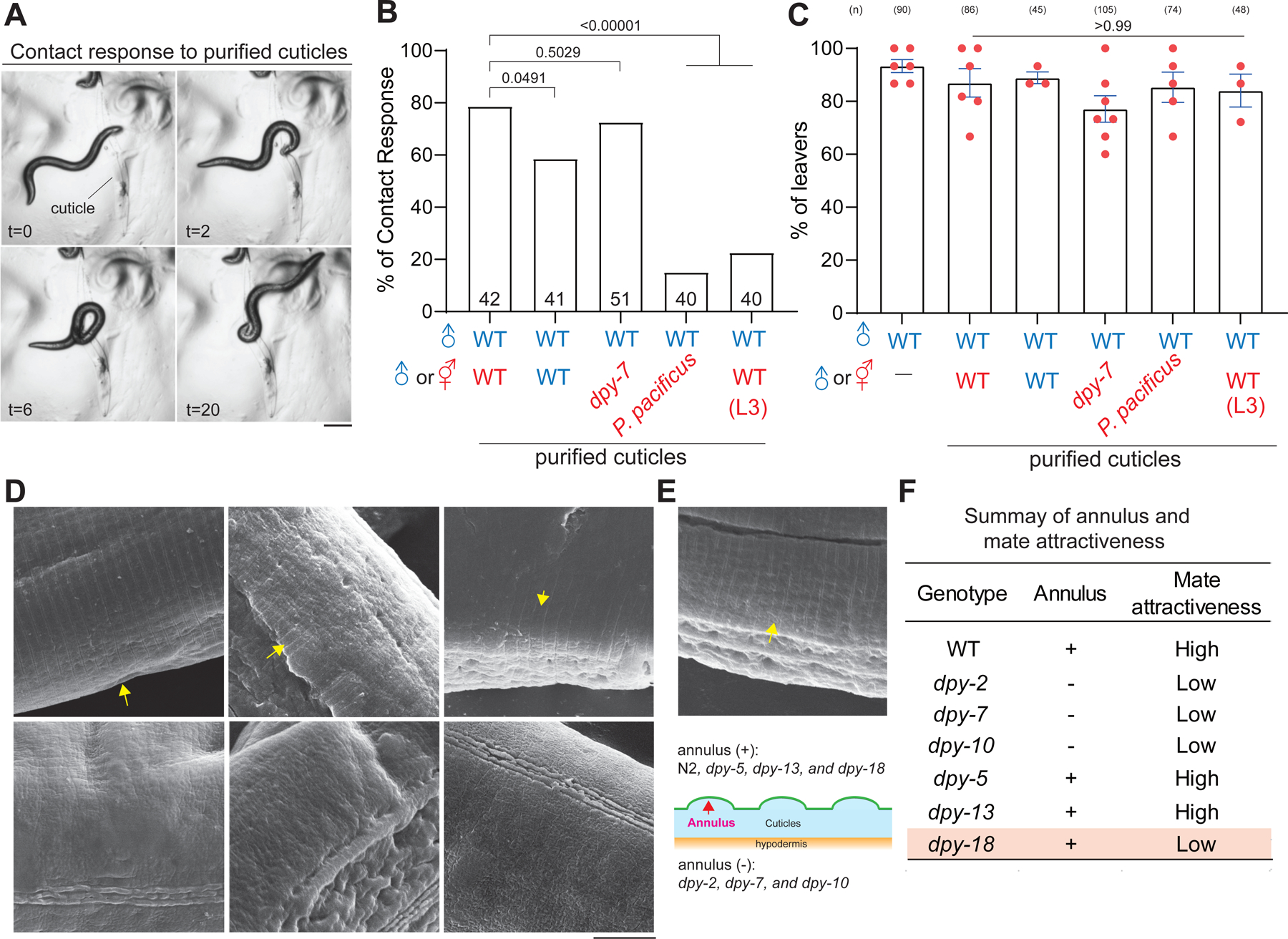
(A) Time series of bright field images of one C. elegans male contacting purified cuticles. Time in seconds. Scale bar = 0.25 mm. (B) Quantification of contact response with purified cuticles from indicated genotypes. The number of males tested with purified cuticles is indicated. Chi-square test with p values indicated. (C) Percentage of male leavers in male retention assay with purified cuticles from indicated species or genotypes. (D, E) Scanning electron microscope images of wild-type and indicated genotypes, and yellow arrows indicate the furrows. Scale bar = 5 μm. (F) Summary of results of male retention assay and SEM analysis in indicated genotypes.
We also asked whether the degree of contact response is associated with sexual discrimination. Sexual transformations of hypodermis did not significantly affect the transient contact responses of males to purified cuticles (Figure S5A), indicating the presence of additional cues for sexual discrimination during the prolonged scanning. These data support the previous speculations that multiple sensory cues are required for mate recognition 20,21. Together, we conclude that furrow collagens are likely involved in the subsequent step after male contact response.
Furrow Structures Are Not Required for Male Retention
dpy-2, dpy-3, dpy-7, dpy-8, dpy-9 and dpy-10 are necessary for forming circumferential furrow on cuticles 29,53,54. Previous studies have found that the furrow functions as a signaling hub to control innate immunity and physiology in C. elegans 49,55. We next asked whether the furrow structure provides a specific surface texture as a sensory cue. Using a scanning electron microscope (SEM) to visualize the ridges on cuticles, we confirmed that furrows were not formed in the dpy-2, dpy-7, and dpy-10 mutants but remained intact in the dpy-5 and dpy-13 mutants 29,32,49 (Figure 3D). However, furrows were detectable in the low attractive dpy-18 mutant, suggesting that the furrow might not contribute to the attractiveness of mates (Figure 3D–F).
Collagens Maintain Body Stiffness of Mates for Male Retention
In addition to the changes in surface texture, body stiffness measured by probing the cuticle with AFM (atomic force microscopy) is significantly reduced in the furrow mutants but not in other Dpy mutants like dpy-13 32. This observation raised the possibility that body stiffness is a mechanical cue for contact-mediated mate recognition. Interestingly, we observed that dpy-2, dpy-7, and dpy-10 furrow mutants but not dpy-5 and dpy-13 mutants were often ruptured from the vulva, arguably due to the changes of body stiffness (Figure 4A and S6A). Furthermore, we found that body stiffness was significantly increased in males measured by AFM, supporting the role of body stiffness in mate discrimination (Figure 4B). To directly test the role of body stiffness, we poked wild-type worms with microinjection needles to release the internal pressure and fixed ruptured worms with paraformaldehyde for male retention assay (Figure 4C and 4D). The release of internal pressure of wild-type hermaphrodites resulted in low male retention comparable to the furrow mutants (Figure 2B and 4D). We reasoned that the leaving of males was not due to aversive substances from the ruptured body because supernatant from ruptured worms did not affect male retention with intact hermaphrodites (Figure S6B and S6C). In addition to physical intervention, male attraction was also reduced with the gon-2 mutant hermaphrodites (Figure 4C and 4E), in which the body stiffness is significantly reduced due to the loss of internal pressure 30. We also manipulated the stiffness of hermaphrodites by treating animals with SDS-containing lysis buffer. We found that 30 minutes of fixation by paraformaldehyde could not prevent body rupture by the lysis buffer leading to softer body stiffness (Figure S6D), and the corpses were unable to retain males (Figure 4F and 4G). Conversely, hermaphrodites fixed with paraformaldehyde for 24 hours were resistant to the lysis buffer and preserved male retention (Figure 4F and 4G). These combined results suggest that body stiffness is essential for male retention. We conclude that collagens promote mate recognition by maintaining body stiffness.
Figure 4. Collagens maintain body stiffness for mate attractiveness.
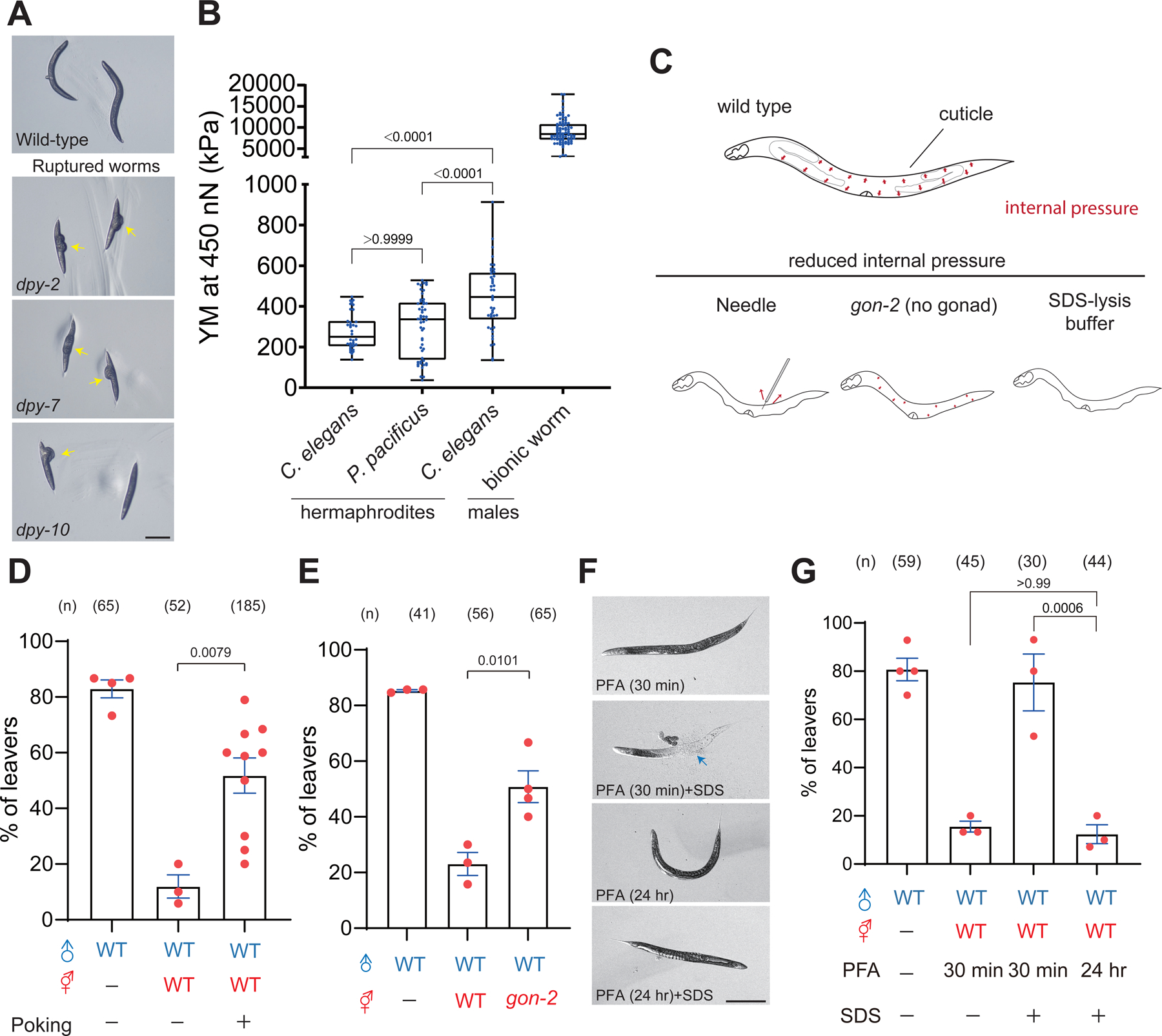
(A) Bright-field images of wild type and bursting hermaphrodites, including dpy-2, dpy-7, and dpy-10. Scale bar = 1 mm. (B) Young’s Modulus of C. elegans with indicated genotypes and treatments. Whisker and box plot with the median for stiffness measurement. Individual dots indicate the measurement of each trial. Whisker labels the range from maximum to minimum. (C) Schematic diagram of experimental procedures to release internal pressure by poking wild-type hermaphrodites or using the gon-2 mutant. (D, E) Percentage of male leavers in male retention assay with the mutant or the wild-type hermaphrodites under indicated treatments. (F) Bright-field images of wild-type hermaphrodites in different conditions. Scale bar = 1 mm. (G) Percentage of male leavers in male retention assay with the mutant or the wild-type hermaphrodites under indicated treatments.
AFM analysis showed that P. pacificus hermaphrodites had a comparable range of stiffness to C. elegans hermaphrodites (Figure 4B), but P. pacificus was not able to retain C. elegans males (Figure S1E), indicating that body stiffness is not sufficient for male retention similar to purified cuticles. This is consistent with the notion that males utilize multiple sensory cues to verify the presence of suitable mates. We thus propose a two-step mechanism in which the first sensory cue existing on the cuticle evokes the contact response, and then body stiffness serves as the second sensory cue to sustain or substantiate the sensation. The combination of two sensory cues ensures the alternation of male behaviors in male retention assay. To test this hypothesis, we purified cuticles from the dpy-7 mutant to exclude the plausible mechanical stimulation by furrow and evoke the contact response of males (Figure 5A). Because we were only able to prepare 2–3 animals at a time, we compared the pooled data collected from several experiments. Consistent with our prediction, P. pacificus coated with purified dpy-7 cuticles evoked contact response and significantly restored male retention (Figure 5B and S5B), supporting a two-step sensory mechanism gated by cuticular cues and body stiffness for contact-mediated mate recognition.
Figure 5. A proper range of body stiffness of mates is likely essential to retain males.
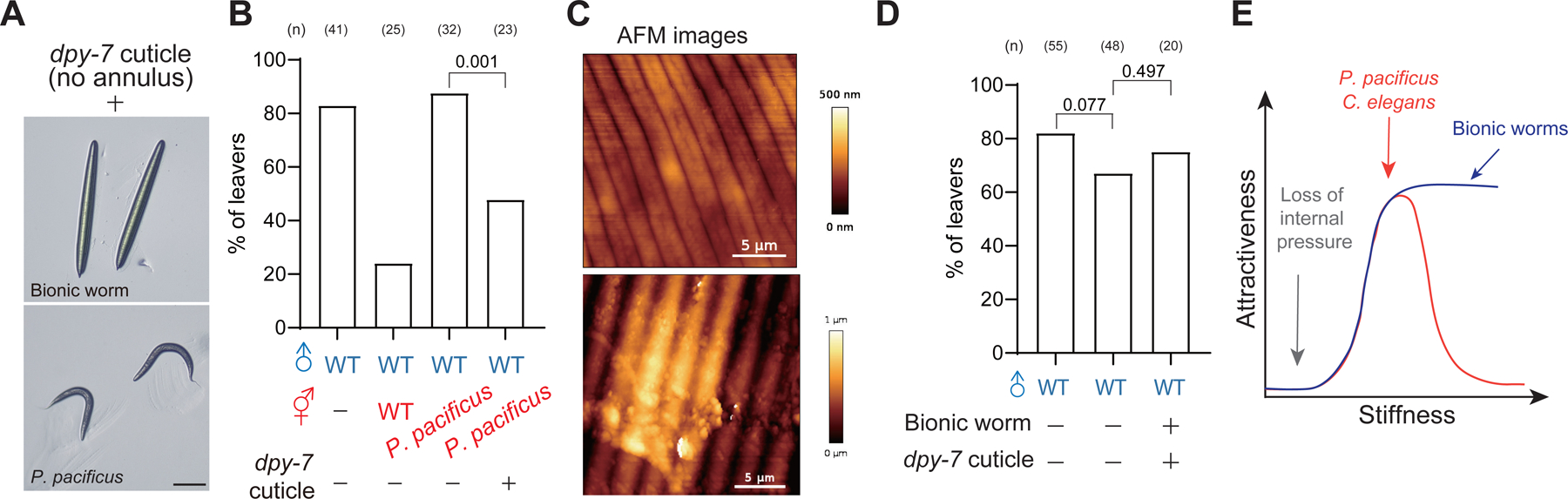
(A) Bright-field images of 3D-printed bionic worms and P. pacificus (Middle). Scale bar = 1mm. (B) Percentage of male leavers in male retention assay with the mutant or the wild-type hermaphrodites under indicated treatments. (C) AFM images of WT hermaphrodites and 3D-printed bionic worm. The scale bar is as indicated. (D) Percentage of male leavers in male retention assay with the 3D-printed bionic worm under indicated treatments. Chi-squared test and p values are indicated. (E) Two models of how stiffness controls mate attractiveness. The red line indicates a model where body stiffness falls in a range in different species that confers specific mechanical properties for mate recognition (model 1). The blue line indicates another model that mates become attractive once the body stiffness surpasses the threshold (model 2).
See also Figure S6.
To further understand whether mate recognition requires a specific range of body stiffness, we generated bionic worms made of resin through 3D printing that preserves the furrow structure on the surface, offering a much stiffer body shape (Figure 4B and 5C; see STAR Methods). 3D-printed bionic worms alone did not evoke contact responses, nor did they attract males in male retention assay (Figure 5D and S5B). In addition, we found that fake worms coated with purified dpy-7 cuticles evoked contact response but had a negligible effect on male retention, favoring that males likely sense a range of body stiffness for mate recognition (Figure 5D, 5E, and S5B). Together, our results suggest that cuticular cues evoke contact behaviors as the initial step, and then the body stiffness provides additional information for males to complete contact-mediated mate recognition.
Contact-mediated Mate Recognition Is Associated with Reproductive Success
Our results established a model whereby body stiffness controlled by furrow collagens is a mechanical property that retains males for contact-mediated mate recognition. However, it is unclear whether male retention contributes to reproductive success. To examine this issue, we paired 10 males with three wild-type, dpy-7, or dpy-13 mutant hermaphrodites, and counted the ratio of cross progeny after mating for 24 hours. Reproductive success was significantly reduced with the dpy-7 mutant hermaphrodite, compared to wild-type and dpy-13 mutant hermaphrodites (Figure 6A). Expression of DPY-7 in hypodermis restored mating success in the dpy-7 mutant hermaphrodites (Figure 6A). Male retention by contact-mediated mate recognition is thus important for reproductive success.
Figure 6. Loss of mate attractiveness is associated with lower mating efficiency.
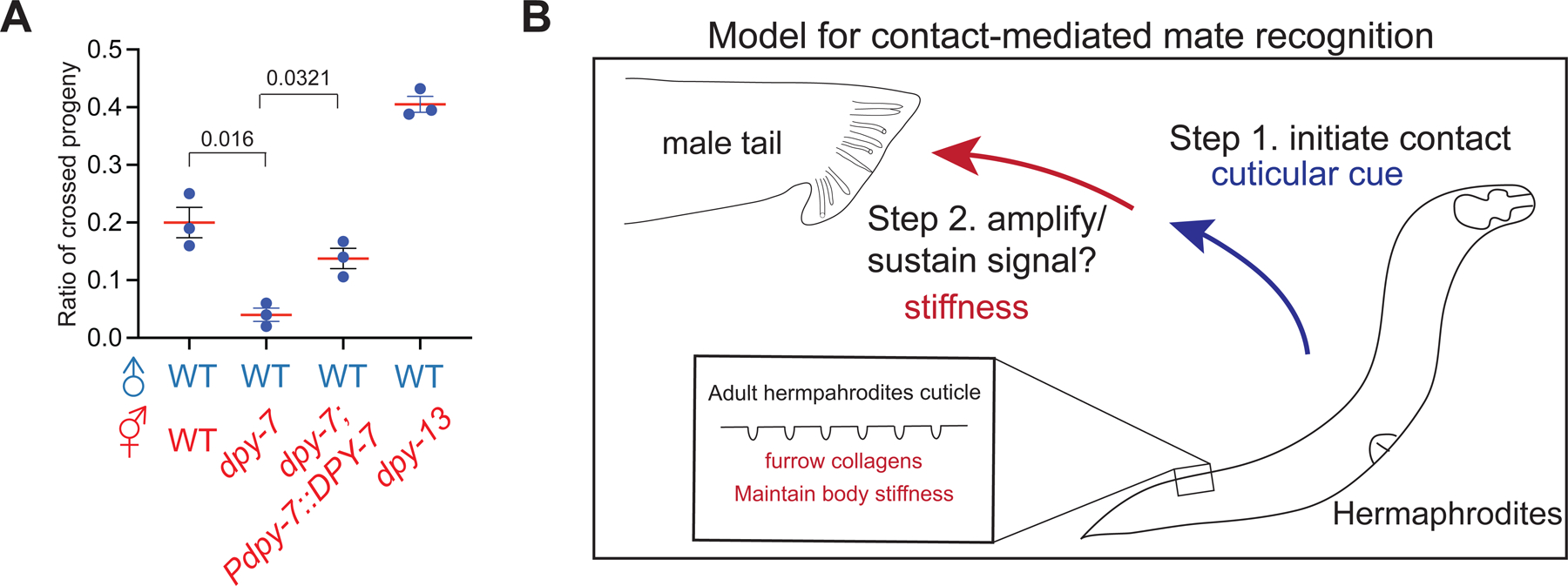
(A) Quantification of the ratio of crossed progeny with wild-type and indicated mutant hermaphrodites. (B) A proposed model for contact-mediated mate recognition in C. elegans. Surface-associated cues on cuticles evoke the contact response of C. elegans males as the first checkpoint. Mechanical signals conveyed by body stiffness then serve as the second signal for contact-mediated mate recognition.
DISCUSSION
Mate recognition is crucial to faithfully and efficiently exchange genetic materials within conspecific mates. In C. elegans, soluble ascaroside and as yet unknown volatile pheromones are secreted by hermaphrodites as long-range mate recognition cues to attract males 2,6,7,35. Here, we find that body stiffness is a mechanical property that allows males to validate the existence of potential mates. In addition to body stiffness, our results indicate that an unidentified cue(s) on cuticles serve as the first gated mechanism to evoke contact responses in males. The first gating mechanism offers effective discrimination between species and developmental stages, but not sexual identity. Unlike almost no contact response to different species and developmental stages, different sexual identities only slightly affected the transient contact response but had drastically different abilities to retain males. In addition, Srf mutant hermaphrodites evoked slightly lower contact responses but did not show discernible defects in male retention. These data suggest that male retention is sufficiently preserved with a certain level of contact responses, and the differences may not be revealed with the sensitivity of male retention assay. Once contact response is evoked, body stiffness of mates is universally required as a second step to sustain or amplify contact signals with the prolonged scanning that leads to long-term behavioral changes. Our data indicate that males were stiffer than hermaphrodites, but the contact responses were generally unchanged by sexual transformation, highlighting the role of body stiffness in sex discrimination. We note that our data do not completely exclude the role of sex-specific cuticular cues for sex discrimination with the sensitivity of our contact assay, because the sexual state of hypodermis is shown to be critical in mate-choice decisions through contact in a different experimental setting 56. Based on these observations, we propose that contact-mediated mate recognition requires the integration of multiple sensations through a two-step mechanism that accesses cuticular cues and body stiffness, respectively, to construct the representations of suitable mates in C. elegans (Figure 6B), thereby establishing a paradigm to study the neural computation of decision-making with well-annotated neural circuits in C. elegans males 57.
Body Stiffness is A Mechanical Property Used for Mate Recognition
In Drosophila, conspecific courtship behaviors require species-specific contact-dependent pheromones on cuticles for courtship behaviors 8,16,17,58. Furthermore, males and females possess different surface pheromones to prevent homosexual courtship behaviors 17,59. The specific distribution of surface-coated molecules, such as glycoproteins, could even convey spatial information that facilitates mating behaviors 60. Our study showed that collagens provide a mechanical signal as a short-range sensory cue to validate the presence of suitable mates. How does body stiffness facilitate mate recognition? Our data suggest that surface texture is negligible as males could recognize P. pacificus coating cuticles without furrow structure, and the 3D-printed bionic worms did not retain males even with the furrow structure. In addition, body stiffness is insufficient for mate recognition because males do not display contact response to 3D-printed bionic worms and P. pacificus. Thus, our data favor the model that body stiffness provides a second checkpoint to confirm the physical presence of mates. Studying the precise range of the mechanical property is needed to comprehensively understand the neural basis of contact-mediated mate recognition, which is a future direction to uncover the complex sensory mechanisms at cellular and molecular levels.
We showed that the furrow collagen had a conserved function for mate recognition in Caenorhabditis species, and three mutant alleles of dpy-7 differentially affected mate attractiveness and body length. dpy-7 encodes 318 amino acids, and e88, sc27, and e1324 alleles are missense mutations on residue 101 (G to R), 156 (G to R), and 189 (G to Y), respectively 52. One speculation is that the N-terminal region of dpy-7 is crucial for body stiffness, whereas the C-terminal region mainly controls body length. Although our data suggest that overexpression of dpy-7 in males enhanced male retention, it is still unknown whether dpy-7 is sufficient to modulate proper body stiffness. Further studies are needed to address these unsolved questions.
Unknown Cuticular Cues for Contact-mediated Mate Recognition
Our study also supports the hypothesis that unknown surface cues on cuticles initiate the contact response of males. Our genetic analysis of lipid- and glycan-related surface genes revealed that purified cuticles from some surface mutants evoke less contact response of males, suggesting that, similar to Drosophila and Rotifer, lipid- and glycan-derivatives may have a minor role in the first step of mate recognition in C. elegans. In addition to lipid- and glycan-derivatives, self-1, a small peptide secreted from hypodermis, has been reported for kin recognition in P. pacificus 61. Even though self-1 has no apparent homolog in C. elegans, other peptides are known to be secreted by the epidermis 27,62. Therefore, it is tempting to speculate that peptidergic signaling may be involved in mate recognition. Although our genetic analysis favors a model that collagens mainly contribute to body stiffness for mate attractiveness, we do not exclude the possibility that collagens are also components of cuticular cues for mate recognition, given that the dpy-18 mutant is distinct from furrow mutants with a lower occurrence of body ruptured. Further studies are needed to identify the surface cues and clarify these missing gaps for contact-mediated mate recognition.
What Is the Possible Sensory Mechanism in C. elegans Males?
C. elegans males integrate the sensory information from multiple cues associated with collagens and decide appropriate behaviors to increase the chance of mating. What is the neural mechanism in males? We reason that the role of body stiffness in mate recognition is to sustain the contact between males and hermaphrodites for decoding mate information. One possibility is that body stiffness provides specific mechanical signals (stiffness) sensed by males. In this model, sensory neurons such as ray neurons and post-cloacal sensilla may be directly activated by body stiffness or indirectly activated by the stretch with the opening and closing of rays in males63. Another possibility is that body stiffness is required to build the scaffold of the body shape where cuticular cues can be fully exposed to males for persistent scanning. Although our data with stiffer 3D-printed bionic worms favor the first model that proper body stiffness is a mechanical cue, further research is needed to identify the mechanosensory receptors and dissect neural mechanisms for contact-mediated mate recognition. Our understanding of contact-dependent cues on hermaphrodites is thus crucial to dissect the neural networks. Because the neural mechanisms of contact-mediated mate recognition are largely unknown, our study provides a genetic framework to understand the neural mechanisms and neural computation of decision-making in social behaviors.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Chun-Hao Chen (chunhaochen@ntu.edu.tw)
Material availability
C. elegans strains and DNA constructs generated in this study are available from the Lead Contact upon request.
Data and Code Availability
All the primary data used in this study have been deposited at Figshare and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this study will be shared by the lead contact upon request.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| E. coli: Strain OP50 | Caenorhabditis Genetics Center (CGC) | WormBase: OP50 |
| Experimental Models: C. elegans Strains | ||
| Caenorhabditis elegans: Strain N2: wild isolate | CGC | WormBase: N2 |
| Caenorhabditis remanei: Strain PB4641: wild isolate | CGC | WormBase: C. remanei |
| Caenorhabditis briggsae: Strain AF16: wild isolate | CGC | WormBase: C. briggsae |
| Auanema. sp: Strain PS8402: wild isolate | Paul Sternberg Lab | WormBase: Auanema. sp. |
| Rhabditis. n. sp.: Strain PS1130: wild isolate | Paul Sternberg Lab | WormBase: Rhabditis. n. sp. |
| Pristionchus pacificus: Strain PS312: wild isolate | Ray Hong Lab | WormBase: P. pacificus |
| Steinernema carpocapsae: wild isolate | Paul Sternberg Lab | N/A |
| C. elegans: Strain MT4009: lin-39(n1760)/ dpy-17(e164) unc-32(e189) | CGC | WormBase: lin-39 |
| C. elegans: Strain DR476: daf-22(m130) | Chun-Liang Pan Lab | WormBase: daf-22 |
| C. elegans: Strain PS867: unc-32(e189) glp-1(q231) | Paul Sternberg Lab | WormBase: glp-1 |
| C. elegans: Strain CB5414: srd-1(eh1) | CGC | WormBase: srd-1 |
| C. elegans: Strain BC119: blmp-1(s71) | CGC | WormBase: blmp-1 |
| C. elegans: Strain CT8: lin-4 (ma104) | Chih-Peng Chan Lab | WormBase: lin-4 |
| C. elegans: Strain MT1524: lin-28 (n719) | Chih-Peng Chan Lab | WormBase: lin-14 |
| C. elegans: Strain VC1463: gon-2(ok465) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) | CGC | WormBase: gon-2 |
| C. elegans: Strain AT6: srf-2(yj262) | CGC | WormBase: srf-2 |
| C. elegans: Strain CB6627: srf-3(e2689) | CGC | WormBase: srf-3 |
| C. elegans: Strain CL261: him-5(e1490); srf-5(ct115) | CGC | WormBase: srf-5 |
| C. elegans: Strain CB5609: bus-1(e2678) | CGC | WormBase: bus-1 |
| C. elegans: Strain CB5443: bus-4(e2693) | CGC | WormBase: bus-4 |
| C. elegans: Strain CB5635: bus-13(e2710) | CGC | WormBase: bus-13 |
| C. elegans: Strain CB6081: bus-17(e2800) | CGC | WormBase: bus-17 |
| C. elegans: Strain CB3584: dpy-5(e61); him-5(e1490) | Paul Sternberg Lab | WormBase: dpy-5 |
| C. elegans: Strain BE93: dpy-2 (e8) | CGC | WormBase: dpy-2 |
| C. elegans: Strain CB88: dpy-7(e88) | CGC | WormBase: dpy-7 |
| C. elegans: Strain BE27: dpy-7(sc27) | CGC | WormBase: dpy-7 |
| C. elegans: Strain CB1324: dpy-7(e1324) | CGC | WormBase: dpy-7 |
| C. elegans: Strain CB128: dpy-10(e128) | CGC | WormBase: dpy-10 |
| C. elegans: Strain PS424: dpy-13(e184) | Paul Sternberg Lab | WormBase: dpy-13 |
| C. elegans: Strain IG1685: frIs7(Pnlp-29::GFP, Pcol-19::DsRed); dpy-3(e27) | Nathalie Pujol Lab | WormBase: dpy-3 |
| C. elegans: Strain IG1699: frIs7(Pnlp-29::GFP, Pcol-19::DsRed); dpy-8(e130) | Nathalie Pujol Lab | WormBase: dpy-8 |
| C. elegans: Strain IG466: frIs7(Pnlp-29::GFP, Pcol-19::DsRed) dpy-9(e12) | Nathalie Pujol Lab | WormBase: dpy-9 |
| C. elegans: Strain NTU27: dpy-18(e364) | CGC | WormBase: dpy-18 |
| C. elegans: Strain CB1350: sqt-1(e1350) | CGC | WormBase: sqt-1 |
| C. elegans: Strain BE8: sqt-3(sc8) | CGC | WormBase: sqt-3 |
| C. elegans: Strain HE1006: rol-6(su1006) | CGC | WormBase: rol-6 |
| C. elegans: Strain CB769: bli-1(e769) | CGC | WormBase: bli-1 |
| C. elegans: Strain CB768: bli-2(e768) | CGC | WormBase: bli-2 |
| C. elegans: Strain CB678: lon-2(e678) | CGC | WormBase: lon-2 |
| C. elegans: Strain PS8825: clec-47(sy1532) | In this paper | WormBase: clec-47 |
| C. elegans: Strain PS8544: clec-87(sy1396) | In this paper | WormBase: clec-87 |
| C. elegans: Strain PS8578: clec-88(sy1409) | In this paper | WormBase: clec-88 |
| C. elegans: Strain PS8584: clec-91(sy1415) | In this paper | WormBase: clec-91 |
| C. elegans: Strain PS8787: col-81(sy1520) | In this paper | WormBase: col-81 |
| C. elegans: Strain PS8780: col-88(sy1513) | In this paper | WormBase: col-88 |
| C. elegans: Strain PS8819: col-120(sy1526) | In this paper | WormBase: col-120 |
| C. elegans: Strain PS8821: col-129(sy1528) | In this paper | WormBase: col-129 |
| C. elegans: Strain PS8634: col-135(sy1435) | In this paper | WormBase: col-135 |
| C. elegans: Strain PS8822: col-137(sy1529) | In this paper | WormBase: col-137 |
| C. elegans: Strain PS8827: col-139(sy1534) | In this paper | WormBase: col-139 |
| C. elegans: Strain PS8829: col-156(sy1536) | In this paper | WormBase: col-156 |
| C. briggsae: Strain PS8520: dpy-10(sy1387) | In this paper | N/A |
| C. briggsae: Strain: NTU6: dpy-7(chc1) | In this paper | N/A |
| C. elegans: Strain NTU95: chcEx045 Pelt-2::FEM-3::SL2::mCherry | In this paper | N/A |
| C. elegans: Strain NTU125: Pdpy-7::FEM-3::SL2::mCherry | In this paper | N/A |
| C. elegans: Strain NTU94: chcEx044(Pdpy-7::TRA-2(IC)::SL2::mCherry) | In this paper | N/A |
| C. elegans: Strain NTU183: dpy-7(e88); chcEx99(Pdpy-7::Ppa-dpy-7) | In this paper | N/A |
| C. elegans: Strain NTU177: dpy-7(e88); chcEx96(Pdpy-7::dpy-10) | In this paper | N/A |
| C. elegans: Strain NTU3: dpy-7(e88); Pdpy-7::chcEx1(Pdpy-7::cbr-dpy-7) | In this paper | N/A |
| C. elegans: Strain NTU7: chcEx2(Pdpy-7::dpy-7, Pblmp-1::GFP) | In this paper | N/A |
| C. elegans: Strain NTU5: dpy-7(e88); chcEx2(Pdpy-7::dpy-7, Pblmp-1::GFP) | In this paper | N/A |
| C. elegans: Strain NTU90: chcEx043(Prab-3::fem-3::SL2::mcherry) | In this paper | N/A |
| C. elegans: Strain NTU89: chcEx042 (Prab-3::tra-2(IC)::SL2::mcherry) | In this paper | N/A |
| C. elegans: Strain BX156: fat-6(tm331); fat-7(wa36) | CGC | WormBase: fat-6; fat-7 |
| C. elegans: Strain BX160: fat-7(wa36) fat-5(tm420) | CGC | WormBase: fat-7; fat-5 |
| C. elegans: Strain BX52: fat-4(wa14) fat-1(wa9) | CGC | WormBase: fat-4; fat-1 |
| C. elegans: Strain BX107: fat-5(tm420) | CGC | WormBase: fat-5 |
| C. elegans: Strain BX153: fat-7(wa36) | CGC | WormBase: fat-7 |
| Chemicals | ||
| Hexane | Merck | Catalog number: 139386 |
| Ethanol | Merck | Catalog number: 51976 |
| Paraformaldehyde | Agar Scientific | Catalog number: AGR1026 |
| Software and algorithms | ||
| Primary datasets | Figshare | DOI:10.6084/m9.figshare.23641011 |
| Others | ||
| 3D-printed worms | BMF Nano Materials Technology Co., Ltd | N/A |
This study does not use original code for data analysis.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
C. elegans strains were cultured and maintained as previously described 64. In brief, animals were cultivated on nematode growth medium (NGM) plates seeded with E. coli OP50 bacteria at 20℃, except for the temperature-sensitive strains. A list of mutant alleles and transgenic strains used in this study is available in the key resources table.
Microbe strains
The OP50 E. coli strain was used as the food for the worms.
METHOD DETAILS
Germline Transformation
Germline transformation was performed by microinjecting plasmids in gonads as described 65. In brief, gonads of D1 adult worms were visualized by an inverted microscope (Zeiss, Axio Observer) at 40X objective, and the DNA mixture with co-injection markers was injected through a microinjector (Eppendorf FemtoJet 4X).
Molecular Biology
Molecular cloning was mostly conducted by recombination-based cloning method with GenBuilder DNA Assembly kit (GenScript, Catalog number: L00744). cDNA of fem-3 and tra-2(IC) was amplified from the cDNA library prepared by SuperScript III First-Strand Synthesis System (ThermoFisher, Catalog number: 18080051) and cloned into pPD95.75 based plasmid containing either hypodermis (dpy-7)-, pan-neuronal (rab-3)-, or intestine (elt-3)-specific promoter and SL2::mCherry sequence. cDNA for dpy-7, CBR-dpy-7, and PPA-dpy-7 prepared as mentioned above were cloned into the plasmid with a hypodermis-specific promoter for the rescue experiments. For feeding RNAi experiments, ~0.8 kb of dpy-7 genomic DNA was cloned using primers described below: dpy-7-F (5’-CCACGTGGCAAAAGCCACCG-3’), dpy-7-R (5’-CCACCACGTGCTGGCTTCTC-3’. Sequences were then cloned into L4440 and transformed with E. coli HT115.
Male Retention Assay
The male retention assay was conducted as previously described 21. The assay was performed in a 9 cm plate filled with 10 ml nematode growth medium (NGM) agar, and each plate had 18 μl OP50 bacteria (OD600=0.4-0.6) spotted (~1 cm2) on the center overnight. Before the assay, mates with indicated sex, developmental stages, or species were fixed by 8% paraformaldehyde for 30 min at room temperature unless noted specifically. Fixed worms were washed twice with M9 buffer before being placed on the bacteria patch. Next, one male was placed with two mates on the food patch of each assay plate, and a population of 15–20 virgin D1 males was assayed for one experimental set. A male is considered a leaver when the moving track reaches 0.5 cm away from the plate edge. Plates were kept at 20℃, and the proportion of males was scored at 24 hours.
To release the internal pressure of worms, animals were poked by a microinjection needle 3–5 times until the worm body was ruptured. Then, ruptured worms were immediately fixed with 8% paraformaldehyde for 30 minutes. Cuticles were coated by gently placing purified cuticles on fixed P. pacificus or 3D-printed bionic worms.
Quantification of the percentage of leaving males in male retention assay
We define the leaver as the locomotion track reaching 0.5 cm away from the plate edge in male retention assay at 24 hours 33. The percentage of leaving is calculated by dividing the number of leavers by the number of total tested animals, as shown below.
The value of the percentage of leaving males is pooled across replicas to estimate the standard error of mean (S.E.M.).
Gene Ontology (GO) and Enrichment Analysis
The N2 and blmp-1 transcriptome comparison was performed using the published dataset from Gene Expression Omnibus (Access number: GSE179411) 40. In brief, differential expression between wild-type and blmp-1 mutant strains was determined with DESeq2 (v1.22.0; padj<0.05 and FDR>2) 66. 883 upregulated and 653 downregulated genes were subjected to gene ontology (GO) and enrichment analysis by Gene Enrichment Analysis in WormBase with q value threshold of 0.1 (Data S1) 67,68. 975 genes were included in the GO analysis.
RNA Interference
Feeding RNAi experiments were conducted as described with the E. coli HT115 bacterial strain69. In brief, liquid RNAi bacterial cultures were diluted 1:100 and incubated at 37 ℃ for 3–4 hours until the OD600 reached 0.4–0.6, followed by RNAi production with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 1 hour at room temperature. 100 μl bacteria culture was seeded on nematode growth medium (NGM) plates with 1 mM IPTG and ampicillin. Synchronized L1 (P0) larvae were reared on RNAi plates, and adult F1 animals were transferred to a new RNAi plate. Quantification was done with F2 animals in all feeding RNAi experiments.
CRISPR/Cas9 Mutagenesis
CRISPR-Cas9 stop-in method is performed as described with modifications 48. In brief, all crRNA and the universal tracrRNA were synthesized from IDT, Coralville, IA. The guide RNA sequences for all deletion mutants generated by the CRISPR stop-in method are listed in Table S1. crRNA and tarcrRNA were dissolved in RNAase-free water to make 100 μM working stocks. Purified Cas9 protein (IDT, Catalog number: 1081060) was used with the solution buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 10% Glycerol, and 1 mM DTT). The CRISPR-Cas9 complex was made in sequential steps. First, 5 μl of tracrRNA (0.4 μg/μl) and 2.8 μl of crRNA (0.4 μg/μl) were mixed with 0.5 μl of the Cas9 protein (10 μg/μl) and then were incubated at 37 ℃ for 10 minutes followed by cooling down at room temperature. 2.2 μl of single-strand DNA donor (1 μg/μl) and pRF4(rol-6(su1006)) and RNase-free water were then added to the mixture to reach a final volume of 20 μl. The CRISPR-Cas9 complex was used for microinjection with N2 (C. elegans) or AF16 (C. briggsae) strain.
Scanning Electron Microscopy (SEM) of C. elegans
SEM was conducted as previously described 70. Roughly a hundred synchronized animals in the adult stage were collected in a 1.5 mL Eppendorf and pre-fixed in 0.1M phosphate buffer with 2.5% glutaraldehyde overnight. After fixation, animals were washed with 0.1 M phosphate buffer three times and subsequently incubated with 1% osmium tetroxide in 0.12M phosphate buffer for 30 minutes at 4 °. After washing animals with 0.1M phosphate buffer three times, samples were dehydrated through a sequential gradient of ethanol from 30%−100% and were completely dehydrated by 100% acetone. Then, animals were dried by critical point dehydration (CPD) by a critical point dryer (Hitachi HCP-2), mounted on stubs, and coated with gold using an ion coater (Eiko Engineering IB-2). Cuticle surface structures from at least 10 animals in indicated genotypes were observed by scanning electron microscope (FEI Inspect S, 15kV).
Microscopy and Quantification of Images
Images were taken by ZEN Blue 3.3 software using 10X objective in a Zeiss Imager Z2 microscope with an Axiocam 506 Mono camera. For imaging, worms were anesthetized in 1% sodium azide and mounted on 2 % agarose pads. In order to measure the body length, worms were anesthetized as mentioned above, and the DIC images under 10X objective were taken and analyzed by ZEN Blue 3.3 software. Bright-field images of ruptured worms, cuticles, and 3D-printed bionic worms were taken with a C-mount camera on a dissecting microscope.
Purification of Nematode Cuticles
The purification method is modified from the previous study to purify intact cuticles 71. Synchronized animals at preferred stages were collected in a 1.5 ml Eppendorf tube and washed three times in M9 by centrifugation at 1000 rpm. We discarded the supernatant and resuspended worm pellet with M9 buffer to reach the density of 50–100 worms/μl. 1 μl of worm was dropped on a glass slide and 50 μl of SDS-DTT solution (0.25% SDS, 200 mM DTT, 20mM HEPES, pH 8.0, 3% sucrose, stored at −20°) was added for 30 minutes at room temperature until tissues were transparent. A microinjection needle was used to transfer purified cuticles on an NGM plate, and purified cuticles were washed three times with M9 buffer. Two intact cuticles with indicated genotypes or species were used for male retention assay and 7–10 purified cuticles were used for contact response assay.
Quantification of Male Contact Response to Purified Cuticles
All the experiments were done with day 1 (D1) adult virgin males. Purified cuticles with indicated genotypes, sex, or developmental stages were placed on NGM plates seeded with E. coli OP50. A single male was placed on the cuticle-containing NGM plate (7–10 purified cuticles), and the behavioral response of first contact with purified cuticles was scored. Contact response was defined as backward movements for at least 0.5 body length with curling tail postures of males when moving across a purified cuticle. Each male was scored once and removed from the plate after scoring. Over 30 males were tested in each condition.
Fabrication of 3D-printed Bionic Worms
3D-printed worms were fabricated by a projecting microstereolithography-based 3D printing technique with a customized acrylic photosensitive resin, HTL (BMF Nano Materials Technology Co., Ltd), with a tensile strength of around 71500 kPa. A 3D model of bionic worms including furrows was generated by AutoCAD (AutoDesk, Inc) and was imported into the printer for 3D printing. 3D printing was conducted by curing the resin layer-by-layer exposed to 405 nm UV light (30 mW / cm2), and the resolution was 2–10 μm. The printed products were cleaned, vacuum-dried, and polished before use in experiments.
Quantification of Mating Efficiency
10 D1 adult males and 3 D1 hermaphrodites were placed on a NGM plate with a small food patch (~7 μL OP50 bacteria) on the center for 24 hours. Then, males and hermaphrodites were removed, and the fertilized eggs cultured for 3 days until the animals reached the L4 larval stage. The number of total animals and males (cross progeny) were counted, and the mating efficiency calculated as (# of males/ # of total animals).
Atomic Force Microscopy
AFM was conducted as previously described 32,54. D1 young adult worms were fixed in 4% paraformaldehyde (PFA) for 30 min and then rinsed several times in 50 mM NaCl or M9 buffer. The worms were then transferred as described before to a ~2 mm thick 4 % agarose bed in a Petri dish (30 mm). Heads and tails were fixed with the tissue glue (Dermabond, Ethicon), and the dish was filled with 50 mM NaCl or M9 buffer. AFM data of worms were obtained using a NanoWizard IV or III (JPK, Bruker, Santa Barbara, CA, USA) coupled with an inverted optical microscope Ti-Eclipse (Nikon, Japan). NSC12 tipless cantilevers (7.5 N/m; MikroMash) with a 10 μm diameter borosilicate bead attached (produced by sQUBE www.sQUBE.de) were used. The cantilever sensitivity and stiffness were calibrated without contact based on the thermal spectrum in liquid. The tip was positioned above the worm, using the optical image to ensure to measure glue-free area below the pharynx and/or in the tail. Force curves were generated in force spectroscopy mode, applying a setpoint of 450 nN while the cantilever speed was set at 0.5 or 1.0 μm/s, reaching an indentation of approximately 0.5 to 2 μm with at least 5 curves recorded in a few microns separated areas. AFM data were analyzed using the JPK analysis software. All force curves were processed to set the baseline to determine the tip-sample contact point and to subtract cantilever bending. Young’s Modulus was calculated within the software by fitting the Hertz/Sneddon model.
QUANTIFICATION AND STATISTICAL ANALYSIS
One-way ANOVA and two-way ANOVA with Bonferroni correction and Chi-squared test are conducted by Prism (Version 9.3.1) as indicated in Figure Legends. Error bars in bar graphs represent the standard error of means (S.E.M.).
Supplementary Material
Data S1. List of Differentially Expressed Genes In the blmp-1 Mutant for Gene Ontology Analysis. Related to STAR Methods and Figure 2.
(A) Raw data of RNAseq analysis of N2 and blmp-1 mutant. (B) Upregulated genes in the RNAseq analysis. (C) Downregulated genes in the RNAseq analysis. (D) List of genes used for gene ontology analysis.
Table S1. Sequence Information of Strains Generated by the CRISPR STOP-IN Method. Related to STAR Methods and Figure S4.
Bullet Points.
C. elegans males recognize species, sex, and reproductive stages of their mates.
Cuticular cues on adult hermaphrodites evoke transient contact response of males.
Proper Body stiffness is a ubiquitous mechanical factor for mates to retain males.
Cuticular cues and body stiffness are inseparable for mate recognition by contact.
Weng et al. identify two inseparable sensory cues by which C. elegans males identify suitable mating partners. A cuticular signal evokes contact behaviors of males, and then body stiffness maintained by collagens ensures that males stay in contact with their partners. This study reveals a vital role of mechanical signal for social communications.
ACKNOWLEDGMENTS
We thank members of the Chen and the Sternberg laboratories for insightful discussions. We also thank Ray Hong, Chun-Liang Pan, Yi-Chun Wu, and Shih-Peng Chan for sharing reagents and strains. We thank WormBase for providing organized genetic and genomic information. Some strains were provided by the C. elegans Genetics Center, funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We are grateful to the technical assistance of Technology Commons, College of Life Science, National Taiwan University with scanning electron microscopy (SEM) and confocal microscopy (CLSM). We thank the C. elegans Core Facility of the National Core Facility for Biopharmaceuticals, Ministry of Science of Technology in Taiwan. This study was funded by the Ministry of Science and Technology, Taiwan, to C.-H.C. (MOST 109-2636-B-002-016; MOST 110-2636-B-002-013, NSTC 111-2636-B-002-027) and N. I. H. to P.W.S. (R01NS113119 and R240D023041). The study is also funded by the French National Research Agency ANR-22-CE13-0037-01, by the “Investissements d’Avenir” French Government program (ANR-16-CONV-0001), and from Excellence Initiative of Aix-Marseille University -A*MIDEX and institutional grants from CNRS, Aix Marseille University, National Institute of Health and Medical Research (Inserm) to the CIML. The NanoWizard III (JPK) was funded by the Ministry of Science, Research and the Arts of Baden-Württemberg (Az: 33-7532.20) and the University of Freiburg (‘Strategiefonds’), granted to Winfried Römer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no conflict of interest.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- 1.Asaba A, Hattori T, Mogi K, and Kikusui T (2014). Sexual attractiveness of male chemicals and vocalizations in mice. Front Neurosci 8, 231. 10.3389/fnins.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leighton DH, and Sternberg PW (2016). Mating pheromones of Nematoda: olfactory signaling with physiological consequences. Curr Opin Neurobiol 38, 119–124. 10.1016/j.conb.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto D, and Koganezawa M (2013). Genes and circuits of courtship behaviour in Drosophila males. Nature reviews. Neuroscience 14, 681–692. 10.1038/nrn3567. [DOI] [PubMed] [Google Scholar]
- 4.Ellingsen DM, Leknes S, Loseth G, Wessberg J, and Olausson H (2015). The Neurobiology Shaping Affective Touch: Expectation, Motivation, and Meaning in the Multisensory Context. Front Psychol 6, 1986. 10.3389/fpsyg.2015.01986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrero DM, Moeller LM, Osakada T, Horio N, Li Q, Roy DS, Cichy A, Spehr M, Touhara K, and Liberles SD (2013). A juvenile mouse pheromone inhibits sexual behaviour through the vomeronasal system. Nature 502, 368–371. 10.1038/nature12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, Malik RU, Edison AS, Sternberg PW, and Schroeder FC (2008). A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454, 1115–1118. 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chasnov JR, So WK, Chan CM, and Chow KL (2007). The species, sex, and stage specificity of a Caenorhabditis sex pheromone. Proceedings of the National Academy of Sciences of the United States of America 104, 6730–6735. 10.1073/pnas.0608050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billeter JC, Atallah J, Krupp JJ, Millar JG, and Levine JD (2009). Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature 461, 987–991. 10.1038/nature08495. [DOI] [PubMed] [Google Scholar]
- 9.Hsueh YP, Gronquist MR, Schwarz EM, Nath RD, Lee CH, Gharib S, Schroeder FC, and Sternberg PW (2017). Nematophagous fungus Arthrobotrys oligospora mimics olfactory cues of sex and food to lure its nematode prey. eLife 6. 10.7554/eLife.20023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Richards-Zawacki CL, Devar A, and Dugas MB (2016). Poison frog color morphs express assortative mate preferences in allopatry but not sympatry. Evolution 70, 2778–2788. 10.1111/evo.13079. [DOI] [PubMed] [Google Scholar]
- 11.Baker CA, Clemens J, and Murthy M (2019). Acoustic Pattern Recognition and Courtship Songs: Insights from Insects. Annual review of neuroscience 42, 129–147. 10.1146/annurev-neuro-080317-061839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conte GL, and Schluter D (2013). Experimental confirmation that body size determines mate preference via phenotype matching in a stickleback species pair. Evolution 67, 1477–1484. 10.1111/evo.12041. [DOI] [PubMed] [Google Scholar]
- 13.Connolly K, Burnet B, and Sewell D (1969). Selective Mating and Eye Pigmentation: An Analysis of the Visual Component in the Courtship Behavior of Drosophila Melanogaster. Evolution 23, 548–559. 10.1111/j.1558-5646.1969.tb03540.x. [DOI] [PubMed] [Google Scholar]
- 14.Kopp A, Duncan I, Godt D, and Carroll SB (2000). Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553–559. 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- 15.Sato K, and Yamamoto D (2020). Contact-Chemosensory Evolution Underlying Reproductive Isolation in Drosophila Species. Front Behav Neurosci 14, 597428. 10.3389/fnbeh.2020.597428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohatsu S, Koganezawa M, and Yamamoto D (2011). Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron 69, 498–508. 10.1016/j.neuron.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 17.Thistle R, Cameron P, Ghorayshi A, Dennison L, and Scott K (2012). Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell 149, 1140–1151. 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon JM, and Sternberg PW (2002). Evidence of a mate-finding cue in the hermaphrodite nematode Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 99, 1598–1603. 10.1073/pnas.032225799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, and Jorgensen EM (2007). The sensory circuitry for sexual attraction in C. elegans males. Curr Biol 17, 1847–1857. 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Lipton J, Kleemann G, Ghosh R, Lints R, and Emmons SW (2004). Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. J Neurosci 24, 7427–7434. 10.1523/JNEUROSCI.1746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrios A, Nurrish S, and Emmons SW (2008). Sensory regulation of C. elegans male mate-searching behavior. Current biology : CB 18, 1865–1871. 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakai N, Iwata R, Yokoi S, Butcher RA, Clardy J, Tomioka M, and Iino Y (2013). A sexually conditioned switch of chemosensory behavior in C. elegans. PLoS One 8, e68676. 10.1371/journal.pone.0068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H, Yue X, Cheng H, Zhang X, Cai Y, Zou W, Huang G, Cheng L, Ye F, and Kang L (2018). OSM-9 and an amiloride-sensitive channel, but not PKD-2, are involved in mechanosensation in C. elegans male ray neurons. Sci Rep 8, 7192. 10.1038/s41598-018-25542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brugman KI, Susoy V, Whittaker AJ, Palma W, Nava S, Samuel ADT, and Sternberg PW (2022). PEZO-1 and TRP-4 mechanosensors are involved in mating behavior in Caenorhabditis elegans. PNAS Nexus 1, pgac213. 10.1093/pnasnexus/pgac213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouger V, Bordet G, Couillault C, Monneret S, Mailfert S, Ewbank JJ, Pujol N, and Marguet D (2014). Independent synchronized control and visualization of interactions between living cells and organisms. Biophys J 106, 2096–2104. 10.1016/j.bpj.2014.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martineau CN, Kirienko NV, and Pujol N (2021). Innate immunity in C. elegans. Curr Top Dev Biol 144, 309–351. 10.1016/bs.ctdb.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, and Ewbank JJ (2008). Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog 4, e1000105. 10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SJ, Goodman MB, and Pruitt BL (2007). Analysis of nematode mechanics by piezoresistive displacement clamp. Proceedings of the National Academy of Sciences of the United States of America 104, 17376–17381. 10.1073/pnas.0702138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McMahon L, Muriel JM, Roberts B, Quinn M, and Johnstone IL (2003). Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol Biol Cell 14, 1366–1378. 10.1091/mbc.e02-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilpin W, Uppaluri S, and Brangwynne CP (2015). Worms under Pressure: Bulk Mechanical Properties of C. elegans Are Independent of the Cuticle. Biophys J 108, 1887–1898. 10.1016/j.bpj.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petzold BC, Park SJ, Ponce P, Roozeboom C, Powell C, Goodman MB, and Pruitt BL (2011). Caenorhabditis elegans body mechanics are regulated by body wall muscle tone. Biophys J 100, 1977–1985. 10.1016/j.bpj.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggad D, Brouilly N, Omi S, Essmann CL, Dehapiot B, Savage-Dunn C, Richard F, Cazevieille C, Politi KA, Hall DH, et al. (2023). Meisosomes, folded membrane microdomains between the apical extracellular matrix and epidermis. eLife 12. 10.7554/eLife.75906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wexler LR, Miller RM, and Portman DS (2020). C. elegans Males Integrate Food Signals and Biological Sex to Modulate State-Dependent Chemosensation and Behavioral Prioritization. Current biology : CB 30, 2695–2706 e2694. 10.1016/j.cub.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leighton DH, Choe A, Wu SY, and Sternberg PW (2014). Communication between oocytes and somatic cells regulates volatile pheromone production in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 111, 17905–17910. 10.1073/pnas.1420439111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan X, Zhou Y, Chan CM, Yang H, Yeung C, and Chow KL (2019). SRD-1 in AWA neurons is the receptor for female volatile sex pheromones in C. elegans males. EMBO Rep 20. 10.15252/embr.201846288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barton MK, Schedl TB, and Kimble J (1987). Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee RC, Feinbaum RL, and Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 38.Ambros V (1989). A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell 57, 49–57. 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 39.Kaletsky R, Yao V, Williams A, Runnels AM, Tadych A, Zhou S, Troyanskaya OG, and Murphy CT (2018). Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS genetics 14, e1007559. 10.1371/journal.pgen.1007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu YZ, Jiang HS, Han HF, Li PH, Lu MR, Tsai IJ, and Wu YC (2022). C. elegans BLMP-1 controls apical epidermal cell morphology by repressing expression of mannosyltransferase bus-8 and molting signal mlt-8. Developmental biology 10.1016/j.ydbio.2022.03.011. [DOI] [PubMed] [Google Scholar]
- 41.Sandhu A, Badal D, Sheokand R, Tyagi S, and Singh V (2021). Specific collagens maintain the cuticle permeability barrier in Caenorhabditis elegans. Genetics 217. 10.1093/genetics/iyaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blaxter ML (1993). Cuticle surface proteins of wild type and mutant Caenorhabditis elegans. The Journal of biological chemistry 268, 6600–6609. [PubMed] [Google Scholar]
- 43.Page AP, and Johnstone IL (2007). The cuticle. WormBook, 1–15. 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gravato-Nobre MJ, Stroud D, O’Rourke D, Darby C, and Hodgkin J (2011). Glycosylation genes expressed in seam cells determine complex surface properties and bacterial adhesion to the cuticle of Caenorhabditis elegans. Genetics 187, 141–155. 10.1534/genetics.110.122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blaxter ML, Page AP, Rudin W, and Maizels RM (1992). Nematode surface coats: actively evading immunity. Parasitol Today 8, 243–247. 10.1016/0169-4758(92)90126-m. [DOI] [PubMed] [Google Scholar]
- 46.Darby C, Chakraborti A, Politz SM, Daniels CC, Tan L, and Drace K (2007). Caenorhabditis elegans mutants resistant to attachment of Yersinia biofilms. Genetics 176, 221–230. 10.1534/genetics.106.067496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill KL, Harfe BD, Dobbins CA, and L’Hernault SW (2000). dpy-18 encodes an alpha-subunit of prolyl-4-hydroxylase in caenorhabditis elegans. Genetics 155, 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H, Park H, Liu J, and Sternberg PW (2018). An Efficient Genome Editing Strategy To Generate Putative Null Mutants in Caenorhabditis elegans Using CRISPR/Cas9. G3 8, 3607–3616. 10.1534/g3.118.200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dodd W, Tang L, Lone JC, Wimberly K, Wu CW, Consalvo C, Wright JE, Pujol N, and Choe KP (2018). A Damage Sensor Associated with the Cuticle Coordinates Three Core Environmental Stress Responses in Caenorhabditis elegans. Genetics 208, 1467–1482. 10.1534/genetics.118.300827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratnappan R, Vadnal J, Keaney M, Eleftherianos I, O’Halloran D, and Hawdon JM (2016). RNAi-mediated gene knockdown by microinjection in the model entomopathogenic nematode Heterorhabditis bacteriophora. Parasit Vectors 9, 160. 10.1186/s13071-016-1442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilleard JS, Barry JD, and Johnstone IL (1997). cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Molecular and cellular biology 17, 2301–2311. 10.1128/MCB.17.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnstone IL, Shafi Y, and Barry JD (1992). Molecular analysis of mutations in the Caenorhabditis elegans collagen gene dpy-7. The EMBO journal 11, 3857–3863. 10.1002/j.1460-2075.1992.tb05478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thein MC, McCormack G, Winter AD, Johnstone IL, Shoemaker CB, and Page AP (2003). Caenorhabditis elegans exoskeleton collagen COL-19: an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Developmental dynamics : an official publication of the American Association of Anatomists 226, 523–539. 10.1002/dvdy.10259. [DOI] [PubMed] [Google Scholar]
- 54.Essmann CL, Elmi M, Shaw M, Anand GM, Pawar VM, and Srinivasan MA (2017). In-vivo high resolution AFM topographic imaging of Caenorhabditis elegans reveals previously unreported surface structures of cuticle mutants. Nanomedicine 13, 183–189. 10.1016/j.nano.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Taffoni C, and Pujol N (2015). Mechanisms of innate immunity in C. elegans epidermis. Tissue Barriers 3, e1078432. 10.1080/21688370.2015.1078432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo J, Barrios A, and Portman DS (2023). C. elegans males optimize mate-choice decisions via sex-specific responses to multimodal sensory cues. bioRxiv 10.1101/2023.04.08.536021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LT, Bayer EA, Duerr JS, et al. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71. 10.1038/s41586-019-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Billeter JC, and Levine JD (2013). Who is he and what is he to you? Recognition in Drosophila melanogaster. Curr Opin Neurobiol 23, 17–23. 10.1016/j.conb.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Moon SJ, Lee Y, Jiao Y, and Montell C (2009). A Drosophila gustatory receptor essential for aversive taste and inhibiting male-to-male courtship. Current biology : CB 19, 1623–1627. 10.1016/j.cub.2009.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snell TW, Shearer TL, Smith HA, Kubanek J, Gribble KE, and Welch DB (2009). Genetic determinants of mate recognition in Brachionus manjavacas (Rotifera). BMC Biol 7, 60. 10.1186/1741-7007-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lightfoot JW, Wilecki M, Rodelsperger C, Moreno E, Susoy V, Witte H, and Sommer RJ (2019). Small peptide-mediated self-recognition prevents cannibalism in predatory nematodes. Science 364, 86–89. 10.1126/science.aav9856. [DOI] [PubMed] [Google Scholar]
- 62.Lee SH, Omi S, Thakur N, Taffoni C, Belougne J, Engelmann I, Ewbank JJ, and Pujol N (2018). Modulatory upregulation of an insulin peptide gene by different pathogens in C. elegans. Virulence 9, 648–658. 10.1080/21505594.2018.1433969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu KS, and Sternberg PW (1995). Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron 14, 79–89. [DOI] [PubMed] [Google Scholar]
- 64.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mello CC, Kramer JM, Stinchcomb D, and Ambros V (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Angeles-Albores D, Lee R, Chan J, and Sternberg P (2018). Two new functions in the WormBase Enrichment Suite. MicroPubl Biol 2018. 10.17912/W25Q2N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis P, Zarowiecki M, Arnaboldi V, Becerra A, Cain S, Chan J, Chen WJ, Cho J, da Veiga Beltrame E, Diamantakis S, et al. (2022). WormBase in 2022-data, processes, and tools for analyzing Caenorhabditis elegans. Genetics 220. 10.1093/genetics/iyac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, and Ahringer J (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2, RESEARCH0002. 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peixoto CA, de Melo JV, Kramer JM, and de Souza W (1998). Ultrastructural analyses of the Caenorhabditis elegans rol-6 (su1006) mutant, which produces abnormal cuticle collagen. J Parasitol 84, 45–49. [PubMed] [Google Scholar]
- 71.Zhang S, Banerjee D, and Kuhn JR (2011). Isolation and culture of larval cells from C. elegans. PLoS One 6, e19505. 10.1371/journal.pone.0019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. List of Differentially Expressed Genes In the blmp-1 Mutant for Gene Ontology Analysis. Related to STAR Methods and Figure 2.
(A) Raw data of RNAseq analysis of N2 and blmp-1 mutant. (B) Upregulated genes in the RNAseq analysis. (C) Downregulated genes in the RNAseq analysis. (D) List of genes used for gene ontology analysis.
Table S1. Sequence Information of Strains Generated by the CRISPR STOP-IN Method. Related to STAR Methods and Figure S4.
Data Availability Statement
All the primary data used in this study have been deposited at Figshare and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this study will be shared by the lead contact upon request.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| E. coli: Strain OP50 | Caenorhabditis Genetics Center (CGC) | WormBase: OP50 |
| Experimental Models: C. elegans Strains | ||
| Caenorhabditis elegans: Strain N2: wild isolate | CGC | WormBase: N2 |
| Caenorhabditis remanei: Strain PB4641: wild isolate | CGC | WormBase: C. remanei |
| Caenorhabditis briggsae: Strain AF16: wild isolate | CGC | WormBase: C. briggsae |
| Auanema. sp: Strain PS8402: wild isolate | Paul Sternberg Lab | WormBase: Auanema. sp. |
| Rhabditis. n. sp.: Strain PS1130: wild isolate | Paul Sternberg Lab | WormBase: Rhabditis. n. sp. |
| Pristionchus pacificus: Strain PS312: wild isolate | Ray Hong Lab | WormBase: P. pacificus |
| Steinernema carpocapsae: wild isolate | Paul Sternberg Lab | N/A |
| C. elegans: Strain MT4009: lin-39(n1760)/ dpy-17(e164) unc-32(e189) | CGC | WormBase: lin-39 |
| C. elegans: Strain DR476: daf-22(m130) | Chun-Liang Pan Lab | WormBase: daf-22 |
| C. elegans: Strain PS867: unc-32(e189) glp-1(q231) | Paul Sternberg Lab | WormBase: glp-1 |
| C. elegans: Strain CB5414: srd-1(eh1) | CGC | WormBase: srd-1 |
| C. elegans: Strain BC119: blmp-1(s71) | CGC | WormBase: blmp-1 |
| C. elegans: Strain CT8: lin-4 (ma104) | Chih-Peng Chan Lab | WormBase: lin-4 |
| C. elegans: Strain MT1524: lin-28 (n719) | Chih-Peng Chan Lab | WormBase: lin-14 |
| C. elegans: Strain VC1463: gon-2(ok465) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) | CGC | WormBase: gon-2 |
| C. elegans: Strain AT6: srf-2(yj262) | CGC | WormBase: srf-2 |
| C. elegans: Strain CB6627: srf-3(e2689) | CGC | WormBase: srf-3 |
| C. elegans: Strain CL261: him-5(e1490); srf-5(ct115) | CGC | WormBase: srf-5 |
| C. elegans: Strain CB5609: bus-1(e2678) | CGC | WormBase: bus-1 |
| C. elegans: Strain CB5443: bus-4(e2693) | CGC | WormBase: bus-4 |
| C. elegans: Strain CB5635: bus-13(e2710) | CGC | WormBase: bus-13 |
| C. elegans: Strain CB6081: bus-17(e2800) | CGC | WormBase: bus-17 |
| C. elegans: Strain CB3584: dpy-5(e61); him-5(e1490) | Paul Sternberg Lab | WormBase: dpy-5 |
| C. elegans: Strain BE93: dpy-2 (e8) | CGC | WormBase: dpy-2 |
| C. elegans: Strain CB88: dpy-7(e88) | CGC | WormBase: dpy-7 |
| C. elegans: Strain BE27: dpy-7(sc27) | CGC | WormBase: dpy-7 |
| C. elegans: Strain CB1324: dpy-7(e1324) | CGC | WormBase: dpy-7 |
| C. elegans: Strain CB128: dpy-10(e128) | CGC | WormBase: dpy-10 |
| C. elegans: Strain PS424: dpy-13(e184) | Paul Sternberg Lab | WormBase: dpy-13 |
| C. elegans: Strain IG1685: frIs7(Pnlp-29::GFP, Pcol-19::DsRed); dpy-3(e27) | Nathalie Pujol Lab | WormBase: dpy-3 |
| C. elegans: Strain IG1699: frIs7(Pnlp-29::GFP, Pcol-19::DsRed); dpy-8(e130) | Nathalie Pujol Lab | WormBase: dpy-8 |
| C. elegans: Strain IG466: frIs7(Pnlp-29::GFP, Pcol-19::DsRed) dpy-9(e12) | Nathalie Pujol Lab | WormBase: dpy-9 |
| C. elegans: Strain NTU27: dpy-18(e364) | CGC | WormBase: dpy-18 |
| C. elegans: Strain CB1350: sqt-1(e1350) | CGC | WormBase: sqt-1 |
| C. elegans: Strain BE8: sqt-3(sc8) | CGC | WormBase: sqt-3 |
| C. elegans: Strain HE1006: rol-6(su1006) | CGC | WormBase: rol-6 |
| C. elegans: Strain CB769: bli-1(e769) | CGC | WormBase: bli-1 |
| C. elegans: Strain CB768: bli-2(e768) | CGC | WormBase: bli-2 |
| C. elegans: Strain CB678: lon-2(e678) | CGC | WormBase: lon-2 |
| C. elegans: Strain PS8825: clec-47(sy1532) | In this paper | WormBase: clec-47 |
| C. elegans: Strain PS8544: clec-87(sy1396) | In this paper | WormBase: clec-87 |
| C. elegans: Strain PS8578: clec-88(sy1409) | In this paper | WormBase: clec-88 |
| C. elegans: Strain PS8584: clec-91(sy1415) | In this paper | WormBase: clec-91 |
| C. elegans: Strain PS8787: col-81(sy1520) | In this paper | WormBase: col-81 |
| C. elegans: Strain PS8780: col-88(sy1513) | In this paper | WormBase: col-88 |
| C. elegans: Strain PS8819: col-120(sy1526) | In this paper | WormBase: col-120 |
| C. elegans: Strain PS8821: col-129(sy1528) | In this paper | WormBase: col-129 |
| C. elegans: Strain PS8634: col-135(sy1435) | In this paper | WormBase: col-135 |
| C. elegans: Strain PS8822: col-137(sy1529) | In this paper | WormBase: col-137 |
| C. elegans: Strain PS8827: col-139(sy1534) | In this paper | WormBase: col-139 |
| C. elegans: Strain PS8829: col-156(sy1536) | In this paper | WormBase: col-156 |
| C. briggsae: Strain PS8520: dpy-10(sy1387) | In this paper | N/A |
| C. briggsae: Strain: NTU6: dpy-7(chc1) | In this paper | N/A |
| C. elegans: Strain NTU95: chcEx045 Pelt-2::FEM-3::SL2::mCherry | In this paper | N/A |
| C. elegans: Strain NTU125: Pdpy-7::FEM-3::SL2::mCherry | In this paper | N/A |
| C. elegans: Strain NTU94: chcEx044(Pdpy-7::TRA-2(IC)::SL2::mCherry) | In this paper | N/A |
| C. elegans: Strain NTU183: dpy-7(e88); chcEx99(Pdpy-7::Ppa-dpy-7) | In this paper | N/A |
| C. elegans: Strain NTU177: dpy-7(e88); chcEx96(Pdpy-7::dpy-10) | In this paper | N/A |
| C. elegans: Strain NTU3: dpy-7(e88); Pdpy-7::chcEx1(Pdpy-7::cbr-dpy-7) | In this paper | N/A |
| C. elegans: Strain NTU7: chcEx2(Pdpy-7::dpy-7, Pblmp-1::GFP) | In this paper | N/A |
| C. elegans: Strain NTU5: dpy-7(e88); chcEx2(Pdpy-7::dpy-7, Pblmp-1::GFP) | In this paper | N/A |
| C. elegans: Strain NTU90: chcEx043(Prab-3::fem-3::SL2::mcherry) | In this paper | N/A |
| C. elegans: Strain NTU89: chcEx042 (Prab-3::tra-2(IC)::SL2::mcherry) | In this paper | N/A |
| C. elegans: Strain BX156: fat-6(tm331); fat-7(wa36) | CGC | WormBase: fat-6; fat-7 |
| C. elegans: Strain BX160: fat-7(wa36) fat-5(tm420) | CGC | WormBase: fat-7; fat-5 |
| C. elegans: Strain BX52: fat-4(wa14) fat-1(wa9) | CGC | WormBase: fat-4; fat-1 |
| C. elegans: Strain BX107: fat-5(tm420) | CGC | WormBase: fat-5 |
| C. elegans: Strain BX153: fat-7(wa36) | CGC | WormBase: fat-7 |
| Chemicals | ||
| Hexane | Merck | Catalog number: 139386 |
| Ethanol | Merck | Catalog number: 51976 |
| Paraformaldehyde | Agar Scientific | Catalog number: AGR1026 |
| Software and algorithms | ||
| Primary datasets | Figshare | DOI:10.6084/m9.figshare.23641011 |
| Others | ||
| 3D-printed worms | BMF Nano Materials Technology Co., Ltd | N/A |
This study does not use original code for data analysis.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


