Abstract
A fundamental goal of population genetic studies is to identify historical biogeographic patterns and understand the processes that generate them. However, localized demographic events can skew population genetic inference. Assessing populations with multiple types of genetic markers, each with unique mutation rates and responses to changes in population size, can help to identify potentially confounding population-specific demographic processes. Here, we compared population structure and connectivity inferred from microsatellites and RAD loci among 17 populations of an arid-specialist lizard, the desert night lizard, Xantusia vigilis, in central California to test among historical processes structuring population genetic diversity. We found that both marker types yielded generally concordant insights into population genetic structure including a major phylogenetic break maintained between two populations separated by less than 10 kilometers, suggesting that either marker type could be used to understand generalized demographic patterns across the region for management purposes. However, we also found that the effects of demography on marker discordance could be used to elucidate population histories and distinguish among biogeographic hypotheses. Our results suggest that comparisons of within-population diversity across marker types provides powerful opportunities for leveraging marker discordance, particularly for understanding the creation and maintenance of contact zones among clades.
Introduction
Population-level processes are a major force in structuring species-level genetic diversity across space and time. Especially in the absence of dispersal barriers, comparative tests across multiple species offer important insight into the differential effects of historical and demographic processes on spatial signatures of genetic diversity (Avise, 2009; Knowles, 2009). These comparisons give rise to questions such as: why do geographic barriers affect different species or populations in different ways, even for taxa with similar dispersal capabilities and habitat usage (Charlesworth et al., 2003; Irwin, 2002)? What constitutes a barrier to dispersal, and by what mechanisms do barriers create and maintain these patterns in natural populations living in complex - or simple - habitats? Why do these potential geographic barriers seem to affect different species or even populations within a single species in different ways, even when the underlying biology of those species suggests similar dispersal capabilities and habitat usage (Myers et al., 2019)?
One powerful way to test among competing processes that structure standing genetic diversity is to compare marker types with different a) mutational properties and b) response to demographic history (Fischer et al., 2017; Miller et al., 2014). For example, microsatellite markers tend to overestimate within-population diversity due to their rapid mutation rates, large numbers of alleles, and the tendency for researchers to select highly polymorphic loci (Putman & Carbone, 2014; Queirós et al., 2015). Single nucleotide polymorphisms from RAD (Restriction site Associated DNA) markers are likely to be in slower-mutating sections of the genome, since many bioinformatic pipelines select fragments that appear in multiple individuals and finding homologous DNA fragments across many individuals or species relies on consistent enzyme cut site sequences motifs (Lowry et al., 2017). These differences allow within-population comparisons between the marker types, with the goal of identifying the relative timing of demographic events such as population isolation. Since microsatellites have a higher mutation rate than RAD loci, private microsatellite alleles should on average emerge in an isolated population before private RAD alleles. For example, comparing the two markers within a single population accounts for differences in effective population size and other demographic factors, we should be able to group timing of population isolation into three separate phases: 1) no private alleles, 2) relative excess of private microsatellite alleles, and 3) private alleles for both marker types. Using these three bins, we can tease apart demographic events that otherwise produce indistinguishable genetic signatures (Figure 1), even when both marker types reveal similar phylogeographic patterns in natural populations (DeFaveri et al., 2013; Gärke et al., 2012). Overall, leveraging variation in marker response to historical processes is a powerful, but underutilized, approach to population and evolutionary genetic analyses.
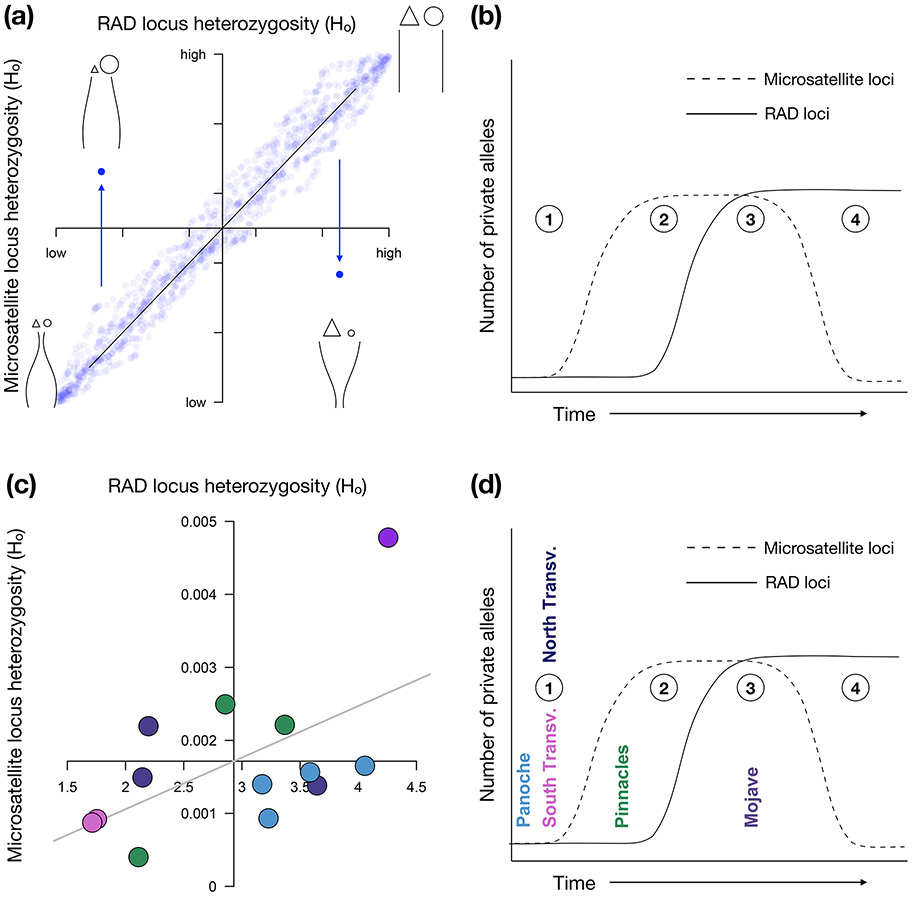
Figure 1. Predictions of genetic marker variation supporting different historical biogeographic scenarios.
(a) The concordance (or lack thereof) between values of observed heterozygosity (Ho) across RAD and microsatellite loci reflects different population histories (population size through time shown by curved lines in each quadrant, with time increasing towards the top). Most population histories will yield generally concordant levels of heterozygosity (blue points) across both marker types (lower left and upper right quadrants). Discordant heterozygosity values between the two marker types in which one marker is much higher than the other (lower right and upper left quadrants) suggest timing and strength of historical bottlenecks or founder events that will differentially affect these marker types. (b) The predictions from heterozygosity can be integrated with numbers and identity of private alleles across marker types to create a framework for testing among competing biogeographic hypotheses across populations (numbers in circles). Variation in correspondence between these values over time is due to the differential mutational and saturation properties of the two marker types. Recently isolated populations should have few private alleles at either marker type (1). Higher microsatellite mutation rates will generate microsatellite private alleles before RAD private alleles (2). Eventually private RAD alleles will occur (3), and finally random mutations in other populations could result in size homoplasy, rendering some microsatellite private alleles undetectable (4). (c) In our data, heterozygosity discordance shows two North Transverse populations in the upper left quadrant, showing signs of more recent bottlenecks, while Panoche populations occur in the lower right quadrant, indicating an older bottleneck followed by a population rebound. Axes are placed at mean heterozygosity values for each marker type, and the grey line represents the results from a linear regression of RAD heterozygosity agains microsatellite heterozygosity. (d) Discordance in private allele rates at our two marker types place the populations on a time-since-isolation axis. The Pinnacles populations show signatures of long-term isolation.
Retrieving information from different types of genetic markers with various characteristics, biases, and theoretical behaviors has a long history in population genetics. Notable marker types include electrophoresis polymorphisms in proteins and DNA fragment lengths, Sanger sequence data, microsatellites, and next-generation sequencing. In tandem with these approaches, theoretical developments helped to generate testable predictions, including the value of comparing predictions generated by the infinite sites and infinite alleles models (Kimura, 1971; Li, 1977; Griffiths & Tavaré, 1994; Tajima, 1996). Infinite sites models assume that the states a locus can take on are limited, but there are so many possible mutational loci that sample-wide homoplasy is unlikely. In contrast, the infinite alleles model assumes a restricted number of loci that can mutate, but that each locus can take on an infinite number of unique states. Neither of these theoretical constructs occurs in the real world, but rather markers behave on a spectrum between them (Ewens, 1974). Infinite alleles models generally capture the behavior of microsatellites, restriction length polymorphism data, and protein electrophoresis alleles, while the infinite sites models are often better descriptors of sequence and SNP data.
Marker comparisons specifically within geographically complex and biodiverse regions create the strongest tests of both biogeographic hypotheses and the relative contribution of different historical processes responsible for spatial patterns of standing diversity (Buonaccorsi et al., 2001; Portnoy et al., 2010). California, with its complex geological structure due to tectonic activity, has long been considered an engine responsible for biogeographic diversity, including high endemism, species richness, and complexity of population structure (Calsbeek et al., 2003; Feldman & Spicer, 2006; Gottscho, 2016; Lancaster & Kay, 2013; Wake, 2006). For central California, the most important abiotic factors affecting species distribution and population structure are a) topographic structure and history, particularly the uplift of the north-south Sierra Nevada and southern Coast Ranges and the tectonic-induced rotation of the east-west Transverse Ranges (Chatzimanolis & Caterino, 2007; Feldman & Spicer, 2006; Lapointe & Rissler, 2005) and b) rainfall gradients across this topography, especially the replicated rain shadow effects along eastern-facing mountain slopes (Hughes et al., 2009). The disjunct arid habitats in the central California rain shadows have high conservation importance and represent the northernmost distribution of many California desert species (Hill, 2003). These factors all combine to make central California biodiversity an ideal system for marker comparison studies.
In this study, we used an arid associated lizard species (the desert night lizard, Xantusia vigilis) to identify which of two competing scenarios about the historical distribution and connectivity of populations in central California’s xeric ecozones is most consistent with the available data. We then assessed contemporary genetic diversity to inform population management practices. Finally, we compared marker-specific patterns of diversity and allelic evolution to distinguish between two explanations for an unexpected combination of strong genetic affinities across large distances and a deep genetic break across a short geographic distance (<10 km). Together, these analyses help us understand how geography, habitat, and history interact to control barriers to migration among populations.
Biogeographic hypotheses
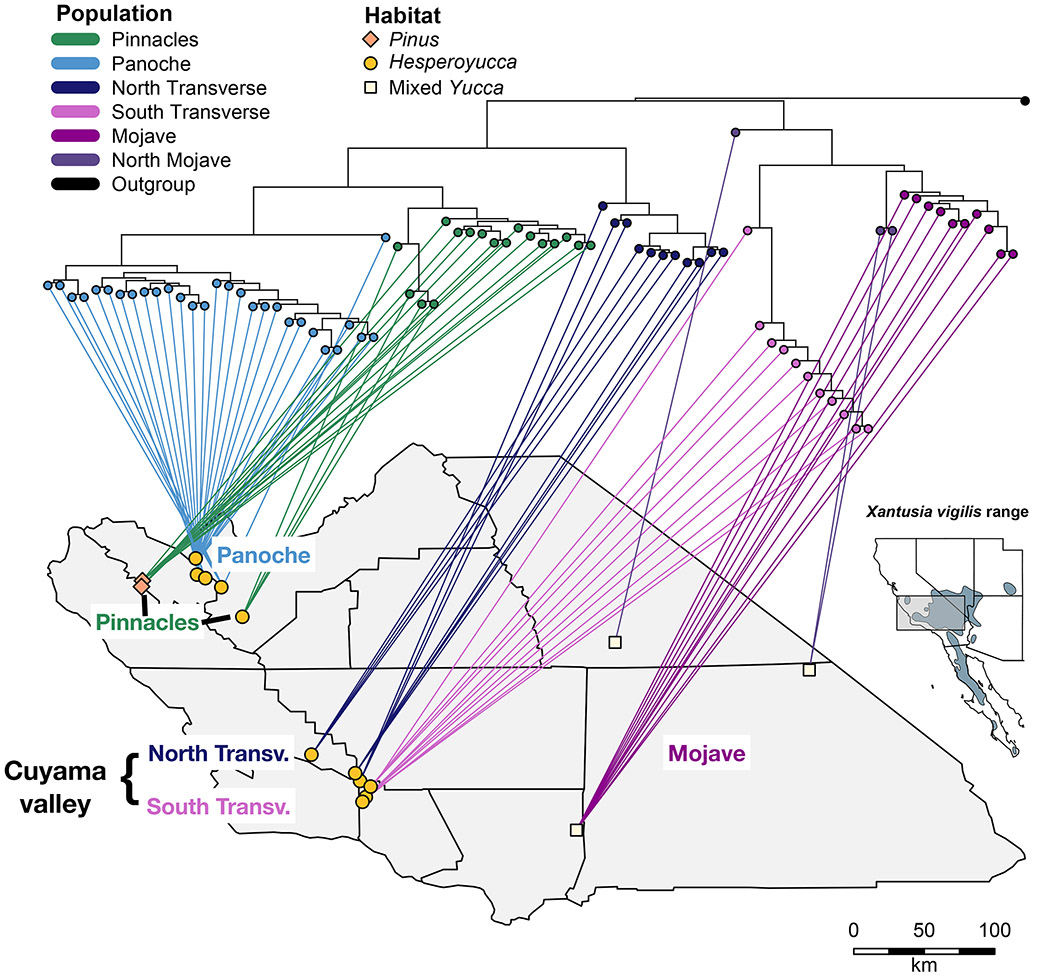
Xantusia vigilis is a very small (adult mass = 1.5g), secretive lizard commonly found throughout arid regions of the southwestern US and the Baja California peninsula of Mexico (see Figure 2 inset; Stebbins, 2003). Presumably due to limited dispersal rates and distances, low rates of inbreeding, and high effective population size (Davis et al., 2011), this species maintains genetic signatures of historical processes over long periods of time and boundaries between demes tend to be well-maintained (Sinclair et al., 2004). However, this species also shows unexpected and dramatic patterns of genetic similarity among non-neighboring populations (Leavitt et al., 2007). Xantusia vigilis is intimately tied to plant or rock cover objects, and several authors have suggested the importance of these specialized habitat associations in predicting historical distribution and resolving unexpected patterns in population connectivity (Noonan et al., 2013; Sinclair et al., 2004). These factors have also contributed to the presence of highly fragmented and disjunct populations across the northern range of X. vigilis, which includes isolated populations in central California and southern Utah (see Figure 2 inset). Here, we focus on the California populations, which consist of two isolated populations of X. vigilis about 150 miles northwest of the main range of the species, where it is associated with joshua trees (Yucca brevifolia) in the Mojave Desert: one regional population is found in isolated outcrops of chapparal yucca (Hesperoyucca whipplei) in the Panoche Hills region (comprising the Panoche, Ciervo, Tumey and Griswold Hill ranges) and another in gray pine (Pinus sabiniana) within Pinnacles National Park (Figure 2).
Figure 2: Collection locations, habitat, and phylogeographic relationships of seventeen sampled Xantusia vigilis populations.
Lizards in the northern part of the range shelter under the monocot shrub Hesperoyucca whippeli or under bark of fallen gray pine (Pinus sabiana) logs, unlike the mixed Yucca (brevifolia, baccata, schidigera) sheltering sites found in the Mojave desert. The full geographic range of X. vigilis range and the portion of the range sampled here (gray box) is indicated in the inset map. Note that the two main phylogenetic clades meet across a short geographic distance in the Cuyama valley in the Transverse Ranges, demonstrating a biogeographic break that does not follow habitat breaks. The northern and southern clades are reciprocally monophyletic, so no directionality of north-south colonization can be inferred from the tree.
California has the highest diversity in the world of unique and deeply divergent lineages of Xantusia. Although the membership within several distinct subclades of X. vigilis sensu stricto (also referred to as “Clade A” X. vigilis in Sinclair et al., 2004) across central California has been well supported with phylogenetic work using both mitochondrial and nuclear loci (Leavitt et al., 2007; Noonan et al., 2013), these previous studies have not resolved relationships among these subclades. The authors have suggested that the polytomy reflects range expansion followed by in situ diversification in the region. These relationships inform an important outstanding question in the historical biogeography of the system: the directionality and timing of range expansion along central California dispersal corridors. The two competing hypotheses about expansion from ancestral populations generally fall into the categories of “North-to-South” or “South-to-North,” and they have differing implications for both the drivers of expansion and conservation importance of the disjunct northern populations (Morafka & Banta, 1973). In the North-to-South scenario, the populations along the central Coast Range derive from an ancestral population near the northern range limit in the Pinnacles or Panoche area and are more distantly related to the populations in the Mojave Desert to the southwest. In this case, the main direction of population expansion and gene flow is from the northern populations to the southern populations, and the Pinnacles/Panoche populations would have important conservation value as repositories of ancestral genetic variation. In the South-to-North scenario, the ancestral population was centered in the main species range of the Mojave Desert, and the northernmost Pinnacles/Panoche populations are simply the most recent outpost of post-glacial range expansion into newly suitable habitat. Both North-to-South and South-to-North scenarios entail demographic processes of expansion, isolation, and population size changes that should be reflected in the contemporary genetic diversity and the distribution of alleles across the landscape.
Predictions of marker variance
Diversity metrics of the markers we use here, microsatellites and RADseq data, can depart from co-linearity in several specific scenarios (Figure 1a), which can improve our understanding of complex biogeographic scenarios. Due to their relatively fast mutation rate, microsatellites should have relatively higher allelic richness in populations that have fairly recently gone through an acute reduction in size, but have since rebounded demographically (Hoelzel, 1999; Martínez-Cruz et al., 2004). Alternatively, this pattern might be produced if a metapopulation with relatively low contemporary migration rates has recently received immigrants (Alexandri et al., 2017), although not all migration processes result in this pattern (Sunde et al., 2020; Zimmerman et al., 2020). In contrast, RAD data should have higher relative heterozygosity in very recently established or post-bottleneck populations compared to the same markers in non-bottlenecked populations. Rare alleles are disproportionately likely to drop out during founder events (Garza & Williamson, 2001). Since microsatellites can have more alleles per locus than sequence data, founder events should reduce the relative allelic richness of microsatellites more sharply than RAD loci (Maruyama & Fuerst, 1985), given that a relatively higher proportion of microsatellite alleles per locus should be in the under 10% frequency category that is most vulnerable to loss in a bottleneck (Luikart, 1998; Tajima, 1989). Low variance in both marker types can be produced by prolonged isolation at low population size, or by a recent and severe founder event (Nei et al., 1975).
The relative proportion of private alleles compared to shared alleles in the populations can distinguish between the scenarios posed above and provide information on the duration of population isolation (Figure 1b). The longer a population has been isolated, the more likely it is to have private alleles (Harpak et al., 2016). The proportion of private alleles should reflect dynamics in the population that occur post-bottleneck, while the allelic richness analyses should reflect processes that occur during the bottleneck. As such, these two approaches have more explanatory value combined than either does independently.
Due to the difference in mutation rates between our marker types, populations should go through four distinct phases following a demographic event that reduces population size, assuming no migration from surrounding populations. Immediately following the event, neither marker type will have a large number of private alleles, and any private alleles that are present should be due to random sampling events during population subdivision, rather than unique local mutations. In the second phase, new microsatellite alleles will emerge due to local mutations, but new private RAD alleles will still be rare due to their slower mutation rate (Lowry et al., 2017). In the third phase, both marker types will show many private alleles. In a theoretical fourth phase, the rapid mutation rate of microsatellites will lead to size homoplasy with alleles in other populations, rendering former private microsatellite alleles no longer identifiable as private. This pattern could emerge due to the stepwise nature of mutation in microsatellites that leads to re-emergences of some read lengths from different ancestral alleles and the size homoplasy that can arise due to interrupted microsatellite sequences (Estoup et al., 2002). Empirical data shows that size homoplasy is widespread in natural systems and increases in prevalence in populations separated by long time frames (Estoup et al., 2002; Lia et al., 2007). The combination of our two marker types should allow us to identify and order the relative occurrence time of demographic events to a greater degree of precision than either of our markers independently.
Using a combination of phylogeographic and demographic analyses, we reconstructed historical patterns of population structure and connectivity in X. vigilis across a complex geological and ecological landscape. In doing so, we leveraged the two marker types to differentiate potential migration patterns across a range of scenarios of patterns of historical connectivity between populations. By assessing biogeographic drivers structuring genetic marker discordance across populations, we provided a clear mechanism for testing among otherwise indistinguishable hypotheses that is useful for other systems with similarly intractable population histories.
Methods
Field collection and tissue acquisition
We collected tissue samples in the field from 354 X. vigilis between 2007 and 2014, which we augmented with five museum samples mainly from the northeastern Mojave desert (Table S1). We sampled across 17 populations in central California, loosely clustered into four regions (Pinnacles National Park, Panoche Hills, northwestern Transverse Ranges around the Cuyama Valley, and the Antelope Valley in the Mojave Desert, see Table 1) along a north-south latitudinal gradient (Figure 2, Figure S2a). Throughout the manuscript, we refer to our sampling populations by the names of local landmarks and include a numbered identifier referencing the regional genetic deme to which they belong (see Figure 2, Table 1). The Coast Range sites were selected by scanning Google Earth for patches of Hesperoyucca whipplei habitat, followed by an initial survey to check for lizard presence. There is a substantial gap in the lizard’s known range between the Transverse Ranges and the Pinnacles and Panoche area, corresponding to an area in which H. whipplei is present only in a few small stands (Figure S1). We surveyed available H. whipplei stands within that gap and found no evidence of X. vigilis (Figure S1), although the lizard is difficult to detect in some habitats and populations may be found in this area in the future. For most sites, we captured lizards by lifting, rolling, or opening decaying yucca trunks and rosettes (see also Davis et al., 2011). At the Pinnacles site, we captured lizards by first moving fallen Pinus logs onto a white sheet and prying off flakes of bark by hand, as well as flipping associated flakes of talus underneath the logs or on scree slopes. After capture, we took tail clip samples that were stored in 95% ethanol and kept at −20 C until analysis. For outgroup comparisons, we included a sample from X. wigginsi, Wiggin’s night lizard.
Table 1:
Population-level information, genetic diversity, and allele privacy.
| Population | Code | Latitude | Longitude | Number of individuals: RAD |
Number of individuals: microsatellites |
RAD Ho |
Microsatellite richness |
RAD private alleles |
Microsatellite private alleles |
|---|---|---|---|---|---|---|---|---|---|
| Panoche Region | 29 | 210 | 0.0015 | 3.857 | 0.165 | 0.334 | |||
| Panoche | PAN4 | 36.66068 | −120.75175 | 14 | 83 | 0.0017 | 4.074 | 0.052 | 0.112 |
| Griswold | PAN3 | 36.53129 | −120.74115 | 8 | 57 | 0.0014 | 3.189 | 0.027 | 0.067 |
| Tumey | PAN2 | 36.50301 | −120.67454 | 2 | 19 | 0.0016 | 3.594 | - | 0.050 |
| Ciervo | PAN1 | 36.43077 | −120.54703 | 5 | 51 | 0.0009 | 3.246 | 0.016 | 0.058 |
| Pinnacles Region | 17 | 16 | 0.0019 | 3.243 | 0.171 | 0.463 | |||
| Pinnacles | PINN3 | 36.48287 | −121.17629 | 9 | 9 | 0.0022 | 3.226 | 0.112 | 0.255 |
| South Chalone | PINN2 | 36.43547 | −121.18401 | 4 | 3 | 0.0025 | 2.920 | 0.050 | 0.076 |
| Curry Mountain | PINN1 | 36.19482 | −120.37915 | 4 | 4 | 0.0004 | 2.109 | 0.040 | 0.048 |
| North Transverse Region | 11 | 15 | 0.0016 | 3.260 | 0.171 | 0.394 | |||
| Caliente Ridge | NT3 | 35.09398 | −119.8280 | 4 | 6 | 0.0014 | 3.600 | 0.069 | 0.225 |
| Cuyama | NT2 | 34.94589 | −119.47849 | 4 | 5 | 0.0015 | 2.177 | 0.078 | 0.034 |
| Ballinger | NT1 | 34.88359 | −119.43983 | 3 | 4 | 0.0022 | 2.177 | - | 0.090 |
| South Transverse Region | 11 | 16 | 0.0013 | 1.811 | 0.176 | 0.313 | |||
| Quatal | ST3 | 34.83917 | −119.35552 | 3 | 2 | 0.0022 | - | - | - |
| Apache Canyon | ST2 | 34.75505 | −119.39395 | 6 | 11 | 0.0009 | 1.697 | 0.217 | 0.024 |
| Dry Canyon | ST1 | 34.71718 | −119.42048 | 2 | 3 | 0.0009 | 1.714 | - | 0.052 |
| Mojave Region | 14 | 61 | 0.0044 | 4.069 | 0.301 | 0.386 | |||
| East Mojave | MOJ1 | 35.76989 | −115.85529 | 2 | - | - | - | - | - |
| NE Mojave | MOJ2 | 35.99031 | −117.40484 | 1 | - | - | - | - | - |
| Antelope Valley | MOJ3 | 34.49113 | −117.71298 | 11 | 61 | 0.0048 | 4.178 | 0.311 | 0.252 |
Habitat classifications
We collected samples from three major habitat types (Figure 2). North of the Transverse Ranges, Hesperoyucca whipplei is distributed into discrete patches on sandstone formations, within which it is the dominant woody shrub species, and therefore all the collection sites were straightforwardly characterized as Hesperoyucca mesohabitat. In the disjunct populations at Pinnacles National Park, the habitat has been defined as gray pine (Pinus sabiniana)-blue oak (Quercus douglasii) woodland (Sawyer & Keeler-Wolf, 2009) and lizards also occurred in gray pine-manzanita (Arctostaphylos sp.) associations at higher elevations as well as under loose rocks several meters from vegetation. In the Transverse Range collection sites, lizards were also collected in H. whipplei, but here the yucca tended to be interspersed with woody shrubs such as desert tea (Ephedra californica) and common sagebrush (Artemisia tridentata) as well as at least two tree species, California juniper (Juniperus californica) and single leaf pinyon (Pinus monophyla). Our Mojave sites were dominated by creosote bush (Larrea tridentata) and lizards were found mostly under joshua trees (Yucca brevifolia), Mojave yucca (Y. schidigera), and banana yucca (Y. baccata).
DNA Extraction and Microsatellite Genotyping
We extracted DNA from tissue samples using a standard Qiagen Blood and Tissue spin column kit or a Chelex-based protocol and amplified eight microsatellite loci using a Qiagen Multiplex PCR kit under standard amplification conditions (see Davis et al., 2011 for primer information). We visualized fluorescently-labelled products on ABI 3170XL machines at the University of California, Berkeley and on ABI 3730XL machines at University of Michigan, and we scored genotypes in GeneMapper v4.0 using allele panels created from previous analysis of 1,140 X. vigilis from the Antelope Valley (MOJ3, see Table 1) population (Davis, 2012; Davis et al., 2011; Davis Rabosky et al., 2012). We discarded any individual from further analysis that we could not confidently genotype at five or more loci. We checked our microsatellite data for null alleles using the null.all function summary two mean value from the R package ‘PopGenReport’ (Adamack & Gruber, 2014) and Hardy-Weinberg equilibrium using the R package ‘pegas’ (Paradis, 2010). One locus was dropped from further analysis due to a frequency of null alleles over 22 percent. The allele calls for the microsatellites are available in a Structure formatted file in our Dryad repository (DOI:10.5061/dryad.31zcrjdht).
Next-Generation Sequencing and Data Processing
We performed double digest Restriction site Associated DNA (ddRAD) sequencing on a subset of individuals (N = 104 X. vigilis, plus 10 outgroup samples) following the protocol developed by Peterson, Weber, Kay, Fisher, & Hoekstra (2012). We restricted total genomic DNA using the enzymes EcoR1 and Msp1 and then used a QIAquick gel extraction kit to size select fragments between 100 and 200 base pairs. We used 24 unique barcodes and four unique indices (following Peterson et al., 2012) to individually mark genomic DNA from 96 individuals per multiplexed lane. We sequenced individuals across three runs on an Illumina HiSeq 2500 at the University of Michigan Sequencing Core with 200 base pair paired end reads.
After preliminary analysis, we removed individuals that aligned poorly with the remaining dataset. We retained 81 X. vigilis individuals and one X. wigginsi sample as an outgroup. We uploaded fastq files to NCBI’s short read archive under BioProject PRJNA649707. We used FastQC to assess the quality of our sequences (Andrews et al., 2011). The initial results showed adapter contamination in some of our sequences. We processed our RAD results using the ipyrad pipeline (Eaton & Overcast, 2020) using the default settings. The first step in ipyrad includes adapter trimming. After it ran, we reran the cleaned sequences in FastQC to check that the adapter contamination had been successfully removed. The results showed that the adapter contamination had been removed, but the nucleotide ratios in the first 10 nucleotides were not even, and in some individuals the per tile sequence quality scores in the final 10 nucleotides were lower than average. We continued the ipyrad run with a cutoff of 20 individuals per locus sequenced in the output. We used a custom R script to parse the gphocs output file from ipyrad. We first removed all fragments without an X. wigginsi outgroup sequence. We then removed all fragments that were not variable within the X. vigilis samples. For each fragment, we trimmed any basepairs that were uncalled in some individuals along the edges of the sequences, then removed individuals with indels called in the remaining central portion of the sequence. We retained fragments that were longer than 50 bp in length after trimming. We removed fragments with a minor allele frequency below 0.05, and any fragment where one allele was represented only in heterozygotes. We wrote out separate fasta files for each locus, which we used to format program-specific input files for later analysis. These per-locus fasta files, and the code used to make them, are available in our Dryad repository (DOI:10.5061/dryad.31zcrjdht).
Locus diversity and characteristics
We calculated within-population and within-region allelic richness using the R package ‘heirfstat’ for microsatellites (Goudet & Jombart, 2022). We removed populations with fewer than three microsatellite genotypes (east Mojave (MOJ1), northeast Mojave (MOJ2), South Chalone (PINN2), Curry Mountain (PINN1), and Quatal (ST3)). We rarefied populations to three individuals and found the allelic richness for each locus. We found the mean value for the richness for each of our seven loci in each population. We repeated the rarefaction 50 times and then found the mean of the 50 mean values for each population. We did the same with our regional designations, which follow our phylogeographic tree (Figure 2). The regional groups are the Mojave, the southern Transverse Range populations, the northern Transverse Range populations, the Pinnacles populations, and the Panoche populations. For RAD data, we used a custom R script that calculated the average proportion of heterozygote calls per base pair in each individual (Singhal et al., 2017). We then found the mean of the individual heterozygosity values for each population and region.
To identify the proportion of private alleles in each population and region, we used a custom R script for both microsatellites and RAD data. For each locus, we first rarefied the number of individuals to three per population. For each population in turn, we then identified the alleles found in the population and the alleles found in all other populations. We calculated the proportion of the alleles in the focal population that were unique to that population. For example, a population with one allele that occurred only in that population and one that occurred elsewhere would have a privacy proportion of 0.5. We found the mean privacy proportion across all alleles for each population or region. We repeated the rarefaction 50 times and found the mean across results. The scripts for this analysis are available in our Dryad repository (DOI:10.5061/dryad.31zcrjdht).
Phylogeography
To reconstruct the phylogenetic relationships and major genetic splits within our focal X. vigilis populations, we used the programs iqtree2 1.6.12 (Nguyen et al., 2015) and astral 5.7.1 (Zhang et al., 2018) to construct a single coalescent tree from trees based on the individual RAD fragments output by our bioinformatics pipeline. This approach is statistically more robust to incomplete lineage sorting, which could be common in our individual-level dataset. We generated trees for each fragment using iqtree2 with 1,000 bootstraps each, setting the X. wigginsi sequence as an outgroup. We collected the output tree files into a single file, then used astral to estimate a single species tree from a randomly selected subset of 1,000 gene trees. We used iqtree2 to quantify concordance between our gene trees and the astral-derived species tree. To visually assess the relationship between phylogeny and geography, we mapped each sample from its collection point to its location on the phylogenetic tree using phytools (Revell, 2012).
We used Structure v.2.3 to identify population-level deme groupings and find evidence of admixture in our RAD and microsatellite data (Pritchard et al., 2000). For the RAD Structure input, we randomly selected one SNP per fragment using a custom R script, avoiding selecting from the first or last 10 bp of each sequence due to the FastQC results. We ran each marker type at K values from one to seven. For microsatellites, we performed 2,000,000 steps and 1,000,000 burnin steps. For the RAD data, we did 200,000 steps with 100,000 burnin steps. We performed 10 runs at each K value for our RAD data and 30 for our microsatellite data and used the Evanno delta K method to select the value of K that best fit the data (Evanno et al., 2005) using the program StructureHarvester web v.0.6.94 (Earl & vonHoldt, 2012). Both marker types identified two demes as the best supported value of K. For both marker types, the location of the split matched the major division in our phylogeographic tree, with Mojave and South Transverse in one group and Pinnacles, Panoche, and North Transverse in another. Within each of these groups, we repeated Structure runs from K=1 to K=7 for both marker types, with 50 runs at each K value for microsatellites and 10 for RAD data. Other values were the same as our initial runs. We repeated the Evanno delta K method for both regional groups in both marker types.
We used the Clumpak web server to cluster results across runs at the best supported values of K within the two regional groupings (Kopelman et al., 2015). Since our microsatellite dataset had highly variable sample sizes across populations, we repeated our full-population analysis on a randomly sampled subset of ten individuals per populations. For populations with fewer samples, we used all individuals. All relevant input and output files, and the code used to make them, are available in our Dryad repository (DOI:10.5061/dryad.31zcrjdht).
For comparison with our Structure results, we mapped the size range between our largest and smallest microsatellite alleles in each population. This visual analysis allowed us to determine whether our Structure analyses were capturing changes in allelic states, allele frequencies, or overall reductions in allelic richness due to historical bottlenecks. We also found the average pairwise number of dissimilarities between populations in our RAD data. For each RAD fragment, we calculated the proportion of pairwise dissimilarities between two randomly chosen individuals for each pair of populations and within each population. We subsampled the number of pairwise dissimilarities to equal the lowest value in the set, then found the mean value. We plotted the results as a heatmap between pairs of populations. The code for this analysis is available in our Dryad repository (DOI: 10.5061/dryad.31zcrjdht). This analysis allowed us to contextualize the Structure results in relation to the underlying variance characteristics of the data.
Historical movement corridors
To identify historical patterns of population splitting and migration between our three regional groupings (Mojave + South Transverse, North Transverse, and Pinnacles + Panoche), we performed an Approximate Bayesian analysis with Random Forest model selection in the programs diyabc v.1.1.27 and abcranger v.1.16.23 (Collin et al., 2021). We subsampled the data such that each group was represented by an equal number of individuals. To generate the input file, we retained only SNPs from our Structure input that were represented by at least one called individual in each regional deme. We assigned an equal sex ratio to our populations. We set our minimum allele frequency threshold to 0.05. With these constraints, we retained 4,782 loci. We set broad priors for population sizes from 0 to 1,000,000 individuals, and priors for time since each split between populations between 0 and 1,000,000 generations. We based our model selection on the 50 default summary statistics generated by the diyabc platform. We tested all possible topologies of three regional groupings resulting in nine total models. We generated 12,000 simulated datasets, then used abcranger to select the topology that best fit the data based on 2,000 random forest trees. For the best-fit topology, we estimated the number of generations before present that the populations divided using abcranger. To best use our data around regional patterns of uncalled alleles, we created a dataset of only the northern populations (North Transverse, Pinnacles, and Panoche). Using the same approach as the full dataset, we tested every possible topology between these three populations and estimated the number of generations since divergence for the best supported topology.
Finally, we assessed the direction of migration between the major population clusters found in the iqtree tree. In this analysis, we followed the philosophy of rangeExpansion package (Peter & Slatkin, 2013) which relies on the observation that one-time, directional population movements often increase the proportion of rare alleles in the expanding population (Hallatschek et al., 2007; Klopfstein et al., 2006). Since rare alleles are more likely to be derived than ancestral, the expected outcome of a range expansion event is to increase the relative proportion of derived alleles in the newly established population (Slatkin & Excoffier, 2012). However, some of our populations have been separated for many generations, leading to many loci differing between populations but being fixed within populations, preventing us from using allele frequencies due to few alleles being variable in pairs of populations.
To best use the available data, we created a simple pairwise directionality metric between populations. For each RAD locus, we found the putatively ancestral allelic state using a designated outgroup animal, our X. wigginsi sample. If an allele from the X. vigilis sample matched the outgroup allele, we considered that allele to be ancestral. We then found all derived alleles from the X. vigilis individuals in the sample. In cases in which one population had only derived alleles while a second population contained both ancestral and derived alleles, we considered the ancestral+derived allele population to be the source population. For each pair of populations, we resampled sets of three individuals per population one hundred times each. For each set, we recorded the detectable direction, if one could be identified. We found the sum of the directionality estimates for that fragment, then summed again across shared fragments. Finally, we normalized the directionality measures by the number of shared fragments between each pair of populations. We followed the groupings of populations in iqtree output, first finding directionality between the major north and south geographical areas, then between the regional demes nested within each area.
Results
Locus characteristics, diversity, and private alleles
The observed frequency of null alleles in the microsatellite loci retained for analysis ranged from 0.04 to 0.21 per locus. No locus was significantly (p < 0.05) out of Hardy-Weinberg equilibrium in more than three populations. We found per-population allelic richness levels ranging from 1.71 in Dry Canyon (ST1) to 4.16 in Antelope Valley (MOJ3) microsatellites. Per-region allelic richness was highest in the Mojave region (4.17) and lowest in the South Transverse region (1.8). Private allele frequency per population ranged from 0.020 in Apache Canyon (ST2) to 0.263 in the Pinnacles population (PINN3), while private allele frequency per region ranged from 0.347 in the Panoche region to 0.441 in the Pinnacles region (Table 1). We found that many microsatellite alleles were region-specific rather than population-specific, so we concentrated on patterns of regional privacy going forward.
The ipyrad infile started with 58,359 loci, which we aggressively filtered down to a final dataset of 8,596 loci. The number of uncalled loci per individual ranged from 812 to 5,702. For RAD data, the proportion of heterozygote calls per base pair ranged from 0.0004 at Curry Mountain (PINN1) to 0.005 in Antelope Valley (MOJ3). Per-region heterozygosity was again highest for the Mojave (0.004) and lowest for the South Transverse region (0.0013). We found that the regional per-fragment frequency of private alleles ranged from 0.164 in the Panoche region to 0.301 in the Mojave region. At the population level, the frequency of private alleles ranged from 0.016 in the Ciervo population (PANO1) to 0.310 in Antelope Valley (MOJ3) (see Table 1 for full results).
Phylogeography
Our iqtree analysis recovered two major clades of X. vigilis across central California: one composed of samples from the Mojave and extending into the southern Transverse Ranges, and another spanning from the northern Transverse ranges to the central Coast Ranges (Figure 2, Figure S2a for population names). The regional groupings were supported by high bootstrap values, but all splits had low gene concordance values (Figure S2b). The west Mojave X. vigilis (MOJ) were nested within the east Mojave samples. Also nested in that clade, sister to the west Mojave samples, were individuals from three populations in the southern Transverse Ranges: Quatal Canyon, Dry Canyon, and Apache Canyon (ST3, ST2, ST1). The central Coast Range clade contains individuals from the Transverse Ranges from Ballinger Canyon (NT1) and Cuyama Valley (NT2). Slightly farther north are the Caliente Ridge (NT3) samples, which also cluster in this clade. The central Coast Range samples are split between localized Panoche and Pinnacles clades, with the further south Curry Mountain (PINN1) clustering with Pinnacles.
For both RAD data and microsatellites, the Evanno delta K method identified Structure runs at K=2 as the value that best fit the data (Figure S3a, S3c). The Evanno delta K method identified K=2 as the best supported value for both marker types in the southern population (Figure S3b). Both marker types separated the Mojave population and the South Transverse populations (Figure 3a, 3b). In the northern population, the best K value for microsatellites was two and the best K value for RADseq data was three (Figure S3b). In the RAD data, the North Transverse, Pinnacles, and Panoche populations all form demes (Figure 3a). The microsatellites show a regional deme in the Panoche area, with all other populations sharing a second deme (Figure 3b). The regional deme signature was especially strong in the southern, smaller and more isolated Panoche populations (PANO1, PANO2, and PANO3). The largest Panoche population (PANO4) retained significant signal of the wider northern microsatellite deme.
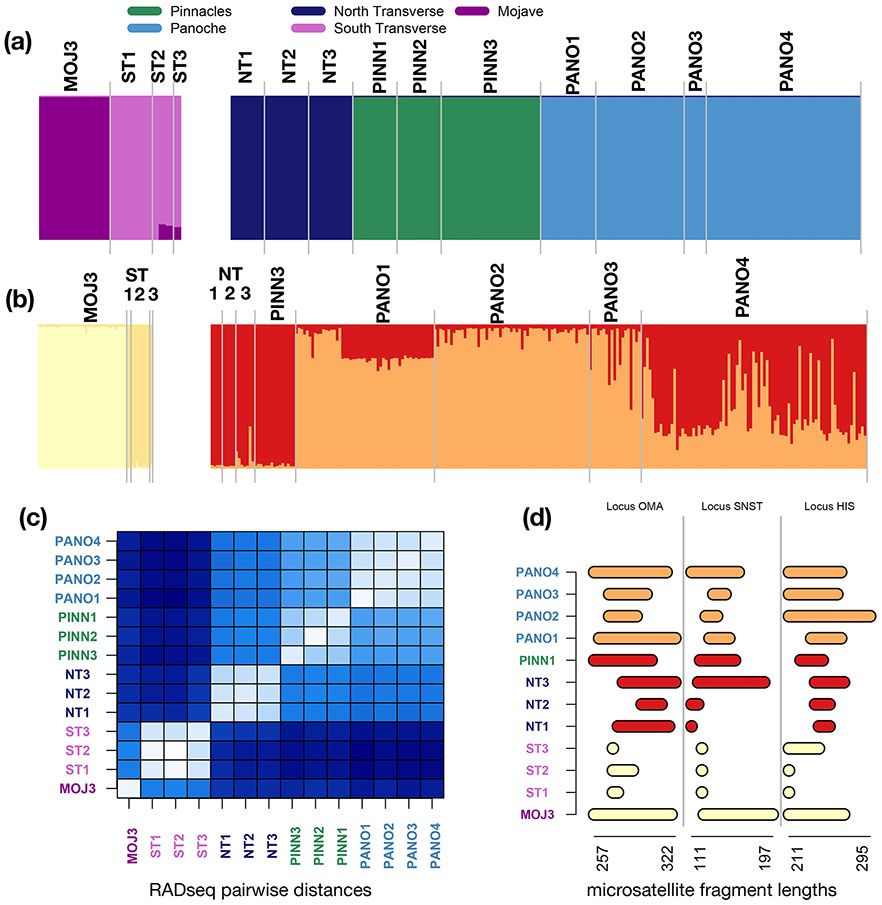
Figure 3. Structure demes and marker-specific patterns of diversity.
(a) RADseq Structure results for northern and southern population groups (b) Microsatellite Structure results. While patterns are broadly congruent, microsatellites distinguish the smaller populations in the Panoche region (PANO1-3) from the other populations in the northern region. (c) Average pairwise number of SNP differences between each pair of individuals in each population. This measure recapitulates the Structure demes. The three South Transverse populations are the most distinct from any other regional deme (dark bands), while the two larger Pinnacles (PINN2 and PINN3) populations are the most similar to every other group (lighter bands). (d) Range between the largest and smallest allele in each population for three microsatellite loci. Populations are colored by their RADseq structure deme. North and South Transverse populations are observably different in allele size range, while allele size range seems to shift more smoothly between the north Transverse, Pinnacles, and Panoche regions.
Microsatellite allele ranges show that the Northern and Southern Transverse Ranges had both become fixed for a single allele at some loci, but those loci differed between the two demes (Figure 3c). For most loci, the Mojave and Panoche populations had the largest size ranges, with other populations intermediate between them. The RAD pairwise difference heat map showed lower pairwise distances for within-deme population pairs than pairs from different demes, demonstrating that the Structure demes reflect raw genetic dissimilarities in the data (Figure 3d). Together, the iqtree, Structure, and locus characteristic analyses pointed to a strong biogeographic break between northern and southern groups in the Cuyama Valley in the Transverse Ranges.
Historical movement corridors
Our full dataset diyabc analysis supported a topology with the Mojave as the ancestral population, with the Panoche population splitting from the North Transverse population with a posterior probability of 0.760. This topology loosely supports the ‘south up’ hypothesis. However, the time estimates show that the splits occurred in the distant past, consistent with the reciprocally monophyletic iqtree results. Parameter estimation set the split between the Mojave and the North Transverse + Panoche populations at 752,153 generations before the present with 0.05 quantile ranges from 392,486 to 989,211 generations. The split between North Transverse and Pinnacles/Panoche populations occurred around 745,751 [ 406,884, 989,054] generations ago. The similar split time estimates are consistent with previous work showing a polytomy in the A-clade X. vigilis (Leavitt et al., 2007). In the north-only dataset, the model with the highest support (0.691 posterior probability score) set Pinnacles as the ancestral population, with the North Transverse population splitting first, followed by the Panoche population. The model put the Pinnacles-North Transverse split at 764,848 [ 495,269 to 986,880] generations before the present and the Panoche-North Transverse split at 506,592 [313,737 to 756,122] generations before present.
Our RAD allele-based directionality analysis shows that the southern populations (Mojave and South Transverse) are a source of migrant alleles to the northern populations (Panoche, Pinnacles, and North Transverse). Within the southern populations, the Mojave is a source for the South Transverse populations. The Pinnacles and North Transverse populations are a source for the Panoche populations.
Locus comparison
Our comparison between RAD and microsatellite heterozygosity showed populations occupying each of our four quadrants (Figure 1c), which we set using the mean values for heterozygosity for both marker types. Antelope Valley (MOJ3) had the highest heterozygosity for both markers, while the South Transverse populations had low values for both markers. Pinnacles and North Transverse populations showed variable levels of deviation from our regression line. The Panoche populations all clustered in the lower right quadrant, which we previously designated as indicating post-bottleneck rebound. No other regional deme showed a consistent pattern of clustering within a single quadrant. Private allele discordance indicates that the Panoche, North Transverse, and South Transverse populations may have experienced a relatively more recent bottleneck, while private allele patterns show the Pinnacles populations in a position consistent with recovery from an older bottleneck (Table 1, Figure 1d). The Antelope Valley (MOJ3) populations have been large and stable.
Discussion
In this paper, we leveraged patterns of variation between two types of genetic markers across populations of the desert night lizard, Xantusia vigilis, to test among biogeographic hypotheses that could not be resolved using conventional approaches. Despite a history of range expansion across physical barriers such as the Transverse Ranges in our study area, we surprisingly found that the most extreme phylogeographic break appears without any clear geographic barriers to gene flow. By incorporating population-specific discordance in heterozygosity and allele privacy across our marker types, our methods for leveraging genetic marker type discordance to our analytical advantage can be applied across other systems with similarly intractable population histories.
Despite the inherent biases associated with both of our marker types (Arnold et al., 2013; Putman & Carbone, 2014), they recover highly concordant results for both phylogeographic structure (Figure 3) and within-population diversity (Figure 1). Our paired analysis (Figure 3) generally agreed on population groupings and on the strong differentiation between the north and south Transverse Range populations. We also found several areas of disagreement between marker types, for which the nature, magnitude, and directionality of discordance were informative for both reconstructing population histories and understanding the creation and maintenance of contact zones among clades. Our study area contains many complex biogeographic patterns which have been influenced by geological and climatic history. To demonstrate how our analytic approaches helped to discriminate among demographic hypotheses and how these methods could be applied to organisms with similar ecologies or geological histories, we discuss four inferences below: one wholistic bioegeographic perspective and three regional case studies within our broader study area.
Tests of biogeographic hypotheses: Expansions and boundaries
One of the unresolved biogeographic questions we tested is whether the northern range-limit populations of X. vigilis represent a historical refugium for the species (North-to-South hypothesis), or whether they are a recent offshoot of the main, Mojave Desert population (South-to-North hypothesis; Morafka & Banta, 1973). We found support for the South-to-North hypothesis, although the expansion process most likely occurred many generations in the past. Our phylogeographic tree showed reciprocal monophyly between the two regions, rather than showing one nested in the other (Figure 2). To determine directionality, we instead relied on the evolutionary history of individual RAD fragments (Figure 4). Our results indicate that the Mojave populations are the source of the northern populations, but that the expansion event happened so long ago that the central Coast Range X. vigilis represent a valuable and unique genetic resource within the broader species. This result is consistent with previous mitochondrial trees that showed an expansion of the A-clade X. vigilis approximately 1.5 mya (Leavitt et al., 2007).
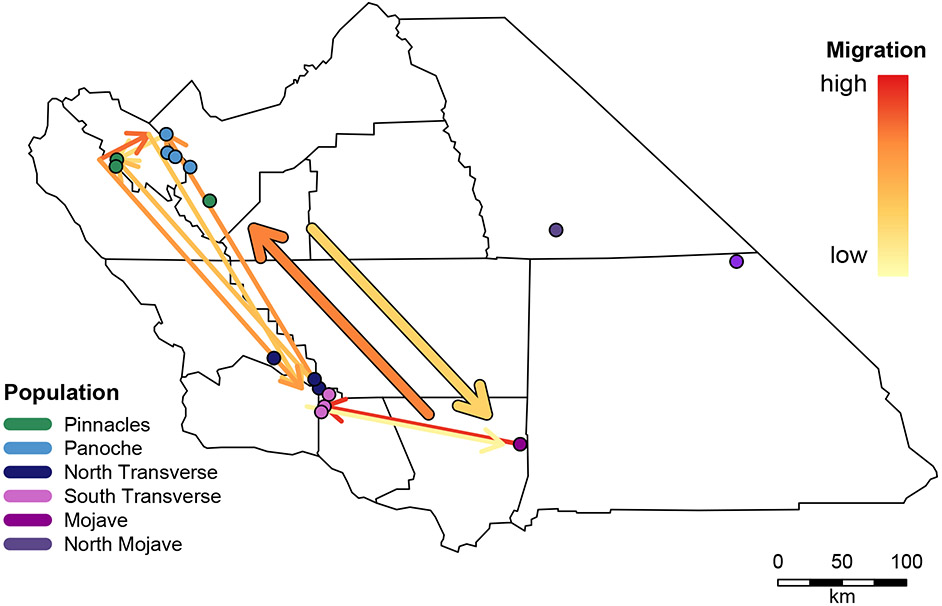
Figure 4.
Patterns of regional connectivity and migration directionality in RAD data. Historical signatures of expansion between the major clades recovered in our iqtree analysis using RAD loci. We follow the nested structure of the phylogeographic tree, first testing directionality between the northern and southern groups, and then between major divisions within the groups. Our analysis shows expansion from south to north (thick arrows). Within the two major groups, expansion from the Mojave populations to the South Transverse populations, and from Pinnacles toward Panoche.
Our study also resolved the geographic location of a major phylogenetic break in X. vigilis to the Cuyama valley in California’s Transverse Ranges (Figure 2), where the Mojave clade meets the Coast Range clade. In this broad pattern, X. vigilis is similar to many other California species or species groups that show biogeographic breaks in the Transverse Ranges (Chatzimanolis & Caterino, 2007; Gottscho, 2016). However, the break we detect falls in between the edges of Transverse Range-specific biogeographic units detected by comparative phylogeography (Chatzimanolis & Caterino, 2007). The phylogeographic patterns used to identify the Transverse Range biogeographic regions may be old enough that they were formed during the Transverse Range uplift, which occurred between five and three million years ago (Nicholson et al., 1994). In contrast, X. vigilis likely entered the area 1.5 million years ago, dispersing over the Transverse Ranges rather than being divided by their uplift (Leavitt et al., 2007). Our study populations show a secondary subdivision between the northern Transverse range populations and the Panoche-Pinnacles clade, which could correspond to the glacial-lake barrier shown in other Central Valley lizards (Papenfuss & Parham, 2013; Richmond et al., 2017). However, caution should be exercised in this interpretation due to the sampling gap between the North Transverse and Panoche/Pinnacles populations. Although our surveys in the area did not locate any X. vigilis or their preferred habitat (Figure S1), future sampling in the area could rule in or rule out the glacial lake hypothesis.
Many other animal species show similar phylogeographic breaks in the Cuyama valley area, including species with very different ecology and contemporary distributions compared to X. vigilis. The blunt-nosed leopard lizard (Gambelia sila) is a Central Valley species that reaches its southern range limit in the Cuyama valley, where it introgresses with the long-nosed leopard lizard (Gambelia wislizenii). The leopard lizard is a diurnal active hunter that prefers warm, open habitats (Lortie et al., 2020) and occurs throughout lowland the San Joaquin Valley (Germano et al., 2011; Richmond et al., 2017). Despite these differences compared to the natural history of night lizards, the two species share remarkably similar phylogeographic patterns in the region. Other species showing similar patterns in the area include pond turtles Actinemys (Spinks et al., 2010, 2014), wood rats Neotoma (Matocq, 2002), and silk moths Calosaturnia (Rubinoff et al., 2021). The Transverse Ranges in general and the Cuyama valley in particular are hotspots of phylogeographic lineage breaks across California herpetofauna (Rissler et al., 2006).
The precise paleoclimatic events that facilitated expansion by the A-clade X. vigilis are likely to remain elusive. However, looking at modern vegetation could provide some clues. Both marker types show lower genomic diversity in populations that shelter under Hesperoyucca whipplei relative to those that use other sheltering habitat (Figures 1-2, Table 1). We hypothesize that this pattern reflects the isolation and small size of Hesperoyucca patches in our study area. Yucca stands in the Mojave and pine stands and rocky habitat in the Pinnacles area seem to provide better opportunities for dispersal and gene flow. From this evidence, we might hypothesize that a more uniform region-wide distribution of shelter plant species similar to either the modern Mojave or Pinnacles could have facilitated the expansion of X. vigilis into their current range. In addition, our observed correlations between habitat type, genomic diversity, and subpopulation isolation indicate that habitat destruction in Hesperoyucca areas will have proportionally higher regional genomic diversity consequences for X. vigilis than a similar scale of habitat destruction in other regions.
Transverse Ranges populations: A phylogeographic museum
The Cuyama valley, a small area in the northern Transverse Ranges, holds populations from the two major phylogeographic lineages of California A-clade X. vigilis (Figures 2 and 3). Our directionality analysis shows a strong signal of the Mojave population being a source of migrants for the South Transverse populations, and while the North Transverse regions share genetic identity with Panoche and Pinnacles (Figure 4). However, both the South and North Transverse populations are monophyletic rather than nested within a source population, indicating that they have been in place for many generations and/or have received very few migrants (Figure 2). Our Structure results reflect a similar pattern, with even the two geographically closest populations, Quatal (ST3) and Ballinger (NT1), consistently assigned to opposite demes with little evidence of admixture (Figure 3a,b). Given their apparent long residency and geographic proximity, why do we observe so little admixture between these two regional demes?
With the available evidence, we hypothesize that at least one of the major regional lineages arrived in the Cuyama valley just prior to some paleoclimatic event that dramatically reduced migration rates to their low modern levels. Considering our observed differences in genetic diversity between cover-vegetation types, a reasonable scenario could involve a drying event that replaced contiguous vegetation cover with more xeric vegetation, with fragmented stands of appropriate cover plants interspersed in an inhospitable matrix. This scenario would fit with past work on the species showing patterns of an expansion followed by long-term stasis in the species (Leavitt et al., 2007).
Fault movements are biogeographically significant throughout our study area. The Cuyama valley region in particular is highly geologically active, with both rotation and subduction occurring (DeLong et al., 2007; Luyendyk et al., 1980; Prothero et al., 2008). The complex geological history of the region has likely contributed to the many species and clade boundaries that occur in the area (Chatzimanolis & Caterino, 2007). Crust movements in the Cuyama valley have not been mapped finely enough for us to identify how they might have impacted the historical locations of our sampled populations. However, we cannot rule out that their geographical separation could have been considerably different within the recent past.
Paleoclimatic reconstruction shows that the Cuyama valley had a relatively wet climate during the dry periods of the last glacial maximum and the Younger Dryas (DeLong et al., 2007). As such, it may function as a phylogeographic museum, preserving historical patterns of reticulate population identity formerly common to the broader area, rather than being an example of conditions that create absolute barriers to expansion in X. vigilis. If further sampling in the region identifies nearby populations, this scenario would be supported if they too reflected a complex mixture of affinities to the regional populations. However, our surveys, data on the distribution of X. vigilis from publicly available sources, and data on the distribution of their favored cover plants indicate that the Cuyama valley populations may be relatively isolated (Figure S1).
Worldwide, many other areas served as climatically-stable refugia during the last glacial maximum, including areas of the Amazon (Bonaccorso et al., 2006), the Eastern Afromontane Biodiversity Hotspot (Demos et al., 2014), and southern Australia (Byrne, 2008). These locations may also preserve a disproportionate amount of phylogeographic diversity, particularly if conditions have since changed to reduce migration rates of the species concerned. If this mechanism is widespread, high lineage diversity of a variety of low-dispersal organisms, such as snails, plants that spread mostly through vegetative mechanisms, or tropical-forest understory specialist birds, might be preserved in historically stable habitat patches. As in the case of Cuyama valley for X. vigilis, these phylogeographic hot spots may not be readily distinguishable from surrounding habitat in the modern day.
Panoche Hills: Recovery from old bottleneck results in microsatellite - RAD data conflict
Movement of the San Andreas fault near the Central Valley may help explain another unexpected pattern of deme affiliation. Our Panoche (PANO1-4) and Curry Mountain (PINN1) samples are on the eastern (stationary) side of the fault, while the Pinnacles populations (PINN2 and PINN3) are on the western (moving) side. In the time lag since the putative expansion of X. vigilis 1.5 mya (Leavitt et al. 2007), the fault has moved approximately 70 km (Argus & Gordon, 2001, Greg Middleton pers. comm.). Several of our analyses point to this movement structuring the patterns of connectivity that are observable today. The Pinnacles populations group with North Transverse in the microsatellite Structure results (Figure 3a, b). Our directionality analysis shows Pinnacles as a source of migrants to both Panoche and North Transverse populations (Figure 4). Our diyabc analysis also shows Pinnacles as ancestral, with North Transverse branching before Panoche. A further discordance between marker types occurs within the Panoche population. The microsatellite Structure results detect a regional deme in the southern Panoche populations (PANO1, 2, and 3), which is absent from the RADseq results (Figure 3a,b).
Insights about the demographic histories of our populations from discordance in allelic diversity between our marker types can resolve these observations. The Panoche populations, clustered in the lower right quadrant in Figure 1c, all show a signature of an old bottleneck followed by a rebound (Figure 1a). This signature is particularly strong in the more southern populations which border the present-day sampling gap between Panoche and the Transverse Range populations. A rebound in population size after such a bottleneck could account for the local demes within the Panoche populations in the microsatellite Structure results, particularly in the Ciervo Hills (PANO1) samples (Figure 3a,b), which could have emerged due to stochastic changes in relative allele frequency during the bottleneck and rebound process. Under this scenario, Pinnacles (PINN2 and 3), Curry Mountain (PINN1) and North Transverse (NT1-3) share a regional ancestral microsatellite allelic signature. This interpretation is strengthened by the observation that the microsatellite signatures in the larger, more dense Panoche (PANO4) proper populations show significant signatures of admixture with the Pinnacles/North Transverse deme, while the lower-diversity Griswold (PANO3), Ciervo (PANO1), and Tumey Hills (PANO2) populations carry the signatures of the location-specific deme (Figure 2, Figure 3b). Future work surveying for X. vigilis in the gap between the Panoche-Pinnacles area and the Cuyama valley may alter this conjecture, revealing instead a south to north stepping-stone process.
Similar mechanisms likely act worldwide, particularly in areas that experienced suboptimal climate conditions in recent paleoclimatic history. The Panoche deme’s location in a rain shadow may be particularly relevant here. Such rain shadow habitats might be optimal for survival of dry-adapted organisms during cold and wet climatic conditions, but vulnerable to bottlenecking due to drought when global conditions change. Rain shadow-driven arid areas of global biological importance include but are not limited to the Atacama Desert on the western coast of South America (Rech et al., 2010), the Eastern Arc mountains in Tanzania and Kenya (Burgess et al., 2007; Lovett, 1996), and the Central Asian high plateau north of the Himalayas (Tewari & Kapoor, 2013).
Curry Mountain: Tectonic drift separates populations that retain strong co-ancestry
Another seemingly paradoxical result we found that may be explained by San Andreas fault movement was the relationship between the samples taken from the main areas of Pinnacles and Panoche versus Curry Mountain (PINN1), which is approximately 70 km south of both of the larger populations (Figure 2). The Curry Mountain (PINN1) samples cluster with the Pinnacles lizards (PINN2 and PINN3) in both marker types, despite Panoche being geographically closer and more similar in habitat. However, at the likely time of the expansion of the A-clade X. vigilis (Leavitt et al. 2007), Pinnacles would have been geographically very close to Curry Mountain (PINN1), explaining the ongoing genetic similarity between the populations. Movement of the Pacific plate along the San Andreas fault carried the PINN2 and PINN3 populations approximate 70 km north since the estimated time of population establishment (Argus & Gordon, 2001, Greg Middleton pers. comm.). This type of strike-slip faulting displacement occurs elsewhere throughout the world. The rotating Pacific plate, which drives the movement of the San Andreas fault, also causes faulting throughout the Pacific rim. Locations along the North American west coast (Brothers et al., 2020), in Japan (Hosoi et al., 2020), and in New Zealand (Michailos et al., 2020) could experience similar displacement.
Conclusions
Our phylogeographic results show that X. vigilis has the ability to maintain biogeographic breaks between two closely adjacent demes over large timescales. In an apparently contradictory pattern, they also show close population co-ancestry over large geographic distances. When combined with previous work on this species, we hypothesize that the biogeographic history of X. vigilis is largely composed of long periods of population stasis, with little dispersal of any kind. Throughout the species’ history in their current range, there have been substantial geographic expansions in which individuals successfully established new populations. For similar scenarios of punctuated dispersal events across long time periods, our work demonstrates the utility of comparing phylogeographic signal in marker types with different mutational properties to successfully resolve complex histories of migration and demographic change. We propose that our approach is applicable to organisms in similarly tectonically active and paleoclimatically complex habitats worldwide.
Supplementary Material
Table S1: Individual-level data, including sample name, collection latitude and longitude, the regional population to which the collection location belonged, and the marker types at which the sample had been sequenced.
Figure S1: sampling effort for X. vigilis in the Coast Ranges between our Cuyama Valley and Pinnacles/Panoche populations.
Figure S2: map with sample names and bootstrap and gene concordance values for the iqtree analysis
Figure S3: Structure supplemental analysis, including Evanno delta K results, full population structure results, and equal sample size microsatellite Structure plot
Acknowledgements
We thank California Department of Fish and Game, the U.S. Fish and Wildlife Service, and Pinnacles National Park for scientific collection permits. We thank many people for assistance in field collection of samples: Ammon Corl, Dean Leavitt, Heather Mostman Liwanag, William Mautz, Theodore J. Papenfuss, Amy Patten, Michael Powers, Richard Seymour, Joseph Belli, and Barry Sinervo. We thank Ryan O’Dell and Greg Middleton for help in understanding the flora and geology of Central California. We thank the Natural History Museum of Los Angeles County, Monte L. Bean Museum at Brigham Young University, and the Museum of Vertebrate Zoology at the University of California, Berkeley for loaning tissue samples. Genetic work was done at the University of Michigan Biodiversity Laboratory. We also thank three anonymous reviewers for helpful comments in the preparation of this manuscript. This study was supported by startup funds from the University of Michigan to ARDR and from the U.S. Bureau of Land Management to MFW and ARDR, and the National Institutes of Health and National Institute of Allergy and Infectious Diseases Award T32Il45821 to IAH. The authors declare no competing conflicts of interest.
Data Accessibility Statement
RADseq data are available on NCBI’s Short Read Archive, as BioProject PRJNA649707. Microsatellite repeat data, scripts for analysis and figures, and intermediate output files are available on DataDryad as dataset https://doi.org/10.5061/dryad.31zcrjdht.
References
- Adamack AT, & Gruber B (2014). P op G en R eport: Simplifying basic population genetic analyses in R. Methods in Ecology and Evolution, 5(4), 384–387. 10.1111/2041-210X.12158 [DOI] [Google Scholar]
- Alexandri P, Megens H, Crooijmans RPMA, Groenen MAM, Goedbloed DJ, Herrero-Medrano JM, Rund LA, Schook LB, Chatzinikos E, Triantaphyllidis C, & Triantafyllidis A (2017). Distinguishing migration events of different timing for wild boar in the Balkans. Journal of Biogeography, 44(2), 259–270. 10.1111/jbi.12861 [DOI] [Google Scholar]
- Andrews S, Lindenbaum P, Howard B, & Ewels P (2011). FastQC: A quality control tool for high throughput sequence data (0.11.9). www.bioinformatics.babraham.ac.uk/projects/ [Google Scholar]
- Argus DF, & Gordon RG (2001). Present tectonic motion across the Coast Ranges and San Andreas fault system in central California. GSA Bulletin, 113(113), 1580–1592. [Google Scholar]
- Arnold B, Corbett-Detig RB, Hartl D, & Bomblies K (2013). RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Molecular Ecology, 22(11), 3179–3190. 10.1111/mec.12276 [DOI] [PubMed] [Google Scholar]
- Avise JC (2009). Phylogeography: Retrospect and prospect. Journal of Biogeography, 36(1), 3–15. 10.1111/j.1365-2699.2008.02032.x [DOI] [Google Scholar]
- Bonaccorso E, Koch I, & Peterson AT (2006). Pleistocene fragmentation of Amazon species’ ranges. Diversity <html_ent Glyph="@amp;" Ascii="&"/> Distributions, 12(2), 157–164. 10.1111/j.1366-9516.2005.00212.x [DOI] [Google Scholar]
- Brothers DS, Miller NC, Barrie JV, Haeussler PJ, Greene HG, Andrews BD, Zielke O, Watt J, & Dartnell P (2020). Plate boundary localization, slip-rates and rupture segmentation of the Queen Charlotte Fault based on submarine tectonic geomorphology. Earth and Planetary Science Letters, 530, 115882. 10.1016/j.epsl.2019.115882 [DOI] [Google Scholar]
- Buonaccorsi VP, Mcdowell JR, & Graves JE (2001). Reconciling patterns of inter-ocean molecular variance from four classes of molecular markers in blue marlin (Makaira nigricans). Molecular Ecology, 10(5), 1179–1196. 10.1046/j.1365-294X.2001.01270.x [DOI] [PubMed] [Google Scholar]
- Burgess ND, Butynski TM, Cordeiro NJ, Doggart NH, Fjeldså J, Howell KM, Kilahama FB, Loader SP, Lovett JC, Mbilinyi B, Menegon M, Moyer DC, Nashanda E, Perkin A, Rovero F, Stanley WT, & Stuart SN (2007). The biological importance of the Eastern Arc Mountains of Tanzania and Kenya. Biological Conservation, 134(2), 209–231. 10.1016/j.biocon.2006.08.015 [DOI] [Google Scholar]
- Byrne M (2008). Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quaternary Science Reviews, 27(27–28), 2576–2585. 10.1016/j.quascirev.2008.08.032 [DOI] [Google Scholar]
- Calsbeek R, Thompson JN, & Richardson JE (2003). Patterns of molecular evolution and diversification in a biodiversity hotspot: The California Floristic Province. Molecular Ecology, 12(4), 1021–1029. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D, & Barton NH (2003). The effects of genetic and geographic structure on neutral variation. Annual Review of Ecology, Evolution, and Systematics, 34(1), 99–125. 10.1146/annurev.ecolsys.34.011802.132359 [DOI] [Google Scholar]
- Chatzimanolis S, & Caterino MS (2007). Toward a Better Understanding of the “Transverse Range Break”: Lineage Diversificationin Southern California. Evolution, 61(9), 2127–2141. https://doi.org/doi: 10.1111/j.1558-5646 [DOI] [PubMed] [Google Scholar]
- Collin F, Durif G, Raynal L, Lombaert E, Gautier M, Vitalis R, Marin J, & Estoup A (2021). Extending approximate Bayesian computation with supervised machine learning to infer demographic history from genetic polymorphisms using DIYABC Random Forest. Molecular Ecology Resources, 21(8), 2598–2613. 10.1111/1755-0998.13413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR (2012). Kin presence drives philopatry and social aggregation in juvenile Desert Night Lizards (Xantusia vigilis). Behavioral Ecology, 23(1), 18–24. 10.1093/beheco/arr144 [DOI] [Google Scholar]
- Davis AR, Corl A, Surget-Groba Y, & Sinervo B (2011). Convergent evolution of kin-based sociality in a lizard. Proceedings of the Royal Society B: Biological Sciences, 278(1711), 1507–1514. 10.1098/rspb.2010.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis Rabosky AR, Corl A, Liwanag HEM, Surget-Groba Y, & Sinervo B (2012). Direct fitness correlates and thermal consequences of facultative aggregation in a desert lizard. PLoS ONE, 7(7), e40866. 10.1371/journal.pone.0040866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFaveri J, Viitaniemi H, Leder E, & Merilä J (2013). Characterizing genic and nongenic molecular markers: Comparison of microsatellites and SNPs. Molecular Ecology Resources, 13(3), 377–392. 10.1111/1755-0998.12071 [DOI] [PubMed] [Google Scholar]
- DeLong SB, Minor SA, & Arnold LJ (2007). Late Quaternary alluviation and offset along the eastern Big Pine fault, southern California. Geomorphology, 90(1–2), 1–10. 10.1016/j.geomorph.2007.01.018 [DOI] [Google Scholar]
- Demos TC, Kerbis Peterhans JC, Agwanda B, & Hickerson MJ (2014). Uncovering cryptic diversity and refugial persistence among small mammal lineages across the Eastern Afromontane biodiversity hotspot. Molecular Phylogenetics and Evolution, 71, 41–54. 10.1016/j.ympev.2013.10.014 [DOI] [PubMed] [Google Scholar]
- Earl DA, & vonHoldt BM (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Eaton DAR, & Overcast I (2020). ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics, 36(8), 2592–2594. 10.1093/bioinformatics/btz966 [DOI] [PubMed] [Google Scholar]
- Estoup A, Jarne P, & Cornuet J-M (2002). Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Molecular Ecology, 11(9), 1591–1604. 10.1046/j.1365-294X.2002.01576.x [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, & Goudet J (2005). Detecting the number of clusters of individuals using the software structure: A simulation study. Molecular Ecology, 14(8), 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Ewens WJ (1974). A note on the sampling theory for infinite alleles and infinite sites models. Theoretical Population Biology, 6(2), 143–148. 10.1016/0040-5809(74)90020-3 [DOI] [PubMed] [Google Scholar]
- Feldman CR, & Spicer GS (2006). Comparative phylogeography of woodland reptiles in California: Repeated patterns of cladogenesis and population expansion. Molecular Ecology, 15(8), 2201–2222. 10.1111/j.1365-294X.2006.02930.x [DOI] [PubMed] [Google Scholar]
- Fischer MC, Rellstab C, Leuzinger M, Roumet M, Gugerli F, Shimizu KK, Holderegger R, & Widmer A (2017). Estimating genomic diversity and population differentiation – an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genomics, 18(1). 10.1186/s12864-016-3459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärke C, Ytournel F, Bed’hom B, Gut I, Lathrop M, Weigend S, & Simianer H (2012). Comparison of SNPs and microsatellites for assessing the genetic structure of chicken populations: Population differentiation: SNPs vs SSRs. Animal Genetics, 43(4), 419–428. 10.1111/j.1365-2052.2011.02284.x [DOI] [PubMed] [Google Scholar]
- Garza JC, & Williamson EG (2001). Detection of reduction in population size using data from microsatellite loci. Molecular Ecology, 10(2), 305–318. 10.1046/j.1365-294x.2001.01190.x [DOI] [PubMed] [Google Scholar]
- Germano DJ, Rathbun GB, Saslaw LR, Cypher BL, Cypher EA, & Vredenburgh LM (2011). The San Joaquin Desert of California: Ecologically Misunderstood and Overlooked. Natural Areas Journal, 31(2), 138–147. 10.3375/043.031.0206 [DOI] [Google Scholar]
- Gottscho AD (2016). Zoogeography of the San Andreas Fault system: Great Pacific Fracture Zones correspond with spatially concordant phylogeographic boundaries in western North America: Zoogeography of the San Andreas Fault system. Biological Reviews, 91(1), 235–254. 10.1111/brv.12167 [DOI] [PubMed] [Google Scholar]
- Goudet J, & Jombart T (2022). hierfstat: Estimation and Tests of Hierarchical F-Statistics (0.5-11) [R]. https://CRAN.R-project.org/package=hierfstat [Google Scholar]
- Griffiths RC, & Tavaré S (1994). Sampling theory for neutral alleles in a varying environment. Philosophical Transactions of the Royal Society B. 344(1310):403–410. 10.1098/rstb.1994.0079 [DOI] [PubMed] [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, & Nelson DR (2007). Genetic drift at expanding frontiers promotes gene segregation. Proceedings of the National Academy of Sciences, 104(50), 19926–19930. 10.1073/pnas.0710150104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpak A, Bhaskar A, & Pritchard JK (2016). Mutation rate variation is a primary determinant of the distribution of allele frequencies in humans. PLOS Genetics, 12(12), e1006489. 10.1371/journal.pgen.1006489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K (2003). Richness, Rarity, and Endemism (Reptiles). In Atlas of the Biodiversity of California (pp. 30–31). California Department of Fish and Game. [Google Scholar]
- Hoelzel AR (1999). Impact of population bottlenecks on genetic variation and the importance of life-history; a case study of the northern elephant seal. Biological Journal of the Linnean Society, 68(1–2), 23–39. 10.1111/j.1095-8312.1999.tb01156.x [DOI] [Google Scholar]
- Hosoi J, Danhara T, Iwano H, Matsubara N, Amano K, & Hirata T (2020). Development of the Tanakura strike-slip basin in Japan during the opening of the Sea of Japan: Constraints from zircon U–Pb and fission-track ages. Journal of Asian Earth Sciences, 190, 104157. 10.1016/j.jseaes.2019.104157 [DOI] [Google Scholar]
- Hughes M, Hall A, & Fovell RG (2009). Blocking in areas of complex topography, and its influence on rainfall distribution. Journal of the Atmospheric Sciences, 66(2), 508–518. 10.1175/2008JAS2689.1 [DOI] [Google Scholar]
- Irwin DE (2002). Phylogeographic breaks without geographic barriers to gene flow. Evolution, 56(12), 2383–2394. [DOI] [PubMed] [Google Scholar]
- Kimura M (1971). Theoretical foundation of population genetics at the molecular level. Theoretical Population Biology, 2(2), 174–208. [DOI] [PubMed] [Google Scholar]
- Klopfstein S, Currat M, & Excoffier L (2006). The fate of mutations surfing on the wave of a range expansion. Molecular Biology and Evolution, 23(3), 482–490. 10.1093/molbev/msj057 [DOI] [PubMed] [Google Scholar]
- Knowles LL (2009). Statistical Phylogeography. Annual Review of Ecology, Evolution, and Systematics, 40, 593–612. [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, & Mayrose I (2015). Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources, 15(5), 1179–1191. 10.1111/1755-0998.12387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster LT, & Kay KM (2013). Origin and diversification of the California flora: Re-examining classic hypotheses with molecular phylogenies. Evolution, 67(4), 1041–1054. 10.1111/evo.12016 [DOI] [PubMed] [Google Scholar]
- Lapointe F, & Rissler LJ (2005). Congruence, consensus, and the comparative phylogeography of codistributed species in California. The American Naturalist, 166(2), 290–299. 10.1086/431283 [DOI] [PubMed] [Google Scholar]
- Leavitt DH, Bezy RL, Crandall KA, & Sites JW Jr (2007). Multi-locus DNA sequence data reveal a history of deep cryptic vicariance and habitat-driven convergence in the desert night lizard Xantusia vigilis species complex (Squamata: Xantusiidae). Molecular Ecology, 16(21), 4455–4481. 10.1111/j.1365-294X.2007.03496.x [DOI] [PubMed] [Google Scholar]
- Li W (1977). Distribution of nucleotide differences between two randomly chosen cistrons in a finite population. Genetics, 85(2), 331–337. 10.1093/genetics/85.2.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lia VV, Bracco M, Gottlieb AM, Poggio L, & Confalonieri VA (2007). Complex mutational patterns and size homoplasy at maize microsatellite loci. Theoretical and Applied Genetics, 115(7), 981–991. 10.1007/s00122-007-0625-y [DOI] [PubMed] [Google Scholar]
- Lortie CJ, Braun J, Westphal M, Noble T, Zuliani M, Nix E, Ghazian N, Owen M, & Scott Butterfield H (2020). Shrub and vegetation cover predict resource selection use by an endangered species of desert lizard. Scientific Reports, 10(1), 4884. 10.1038/s41598-020-61880-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett JC (1996). Elevational and latitudinal changes in tree associations and diversity in the Eastern Arc mountains of Tanzania. Journal of Tropical Ecology, 12(5), 629–650. 10.1017/S0266467400009846 [DOI] [Google Scholar]
- Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, & Storfer A (2017). Breaking RAD: An evaluation of the utility of restriction site-associated DNA sequencing for genome scans of adaptation. Molecular Ecology Resources, 17(2), 142–152. 10.1111/1755-0998.12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G (1998). Distortion of allele frequency distributions provides a test for recent population bottlenecks. Journal of Heredity, 89(3), 238–247. 10.1093/jhered/89.3.238 [DOI] [PubMed] [Google Scholar]
- Luyendyk B, Kamerling M, & Terres R (1980). Geometric model for Neogene crustal rotations in southern California. GSA Bulletin, 91(4), 211–217. [Google Scholar]
- Martínez-Cruz B, Godoy JA, & Negro JJ (2004). Population genetics after fragmentation: The case of the endangered Spanish imperial eagle (Aquila adalberti). Molecular Ecology, 13(8), 2243–2255. 10.1111/j.1365-294X.2004.02220.x [DOI] [PubMed] [Google Scholar]
- Maruyama T, & Fuerst P (1985). Population bottlenecks and nonequilibrium models in population genetics. Genetics, 111, 675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matocq MD (2002). Phylogeographical structure and regional history of the dusky-footed woodrat, Neotoma fuscipes. Molecular Ecology, 11(2), 229–242. 10.1046/j.0962-1083.2001.01430.x [DOI] [PubMed] [Google Scholar]
- Michailos K, Warren-Smith E, Savage MK, & Townend J (2020). Detailed spatiotemporal analysis of the tectonic stress regime near the central Alpine Fault, New Zealand. Tectonophysics, 775, 228205. 10.1016/j.tecto.2019.228205 [DOI] [Google Scholar]
- Miller JM, Malenfant RM, David P, Davis CS, Poissant J, Hogg JT, Festa-Bianchet M, & Coltman DW (2014). Estimating genome-wide heterozygosity: Effects of demographic history and marker type. Heredity, 112(3), 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morafka DJ, & Banta BH (1973). The Distribution and Microhabitat of Xantusia vigilis (Reptilia: Lacertilia) in the Pinnacles National Monument, San Benito and Monterey Counties, California. Journal of Herpetology, 7(2), 97. 10.2307/1563207 [DOI] [Google Scholar]
- Myers EA, Xue AT, Gehara M, Cox CL, Davis Rabosky AR, Lemos-Espinal J, Martínez-Gómez JE, & Burbrink FT (2019). Environmental heterogeneity and not vicariant biogeographic barriers generate community-wide population structure in desert-adapted snakes. Molecular Ecology, 28(20), 4535–4548. 10.1111/mec.15182 [DOI] [PubMed] [Google Scholar]
- Nei M, Maruyama T, & Chakraborty R (1975). The bottleneck effect and genetic variability in populations. Evolution, 29(1), 1–10. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Schmidt H, von Haeseler A, & Minh B (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274. https://doi.org/doi.org/ 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson C, Sorlien CC, Atwater T, Crowell JC, & Luyendyk BP (1994). Microplate capture, rotation of the western Transverse Ranges, and initiation of the San Andreas transform as a low-angle fault system. 5. [Google Scholar]
- Noonan BP, Pramuk JB, Bezy RL, Sinclair EA, de Queiroz K, & Sites JW (2013). Phylogenetic relationships within the lizard clade Xantusiidae: Using trees and divergence times to address evolutionary questions at multiple levels. Molecular Phylogenetics and Evolution, 69(1), 109–122. 10.1016/j.ympev.2013.05.017 [DOI] [PubMed] [Google Scholar]
- Papenfuss T, & Parham J (2013). Four new species of California legless lizards (Anniella). Brevoria, 536(1), 1–17. https://doi.org/doi.org/ 10.3099/MCZ10.1 [DOI] [Google Scholar]
- Paradis E (2010). pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics, 26, 419–420. [DOI] [PubMed] [Google Scholar]
- Peter BM, & Slatkin M (2013). Detecting range expansions from genetic data. Evolution, 67(11), 3274–3289. https://doi.org/doi: 10.1111/evo.12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, & Hoekstra HE (2012). Double Digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE, 7(5), e37135. 10.1371/journal.pone.0037135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy DS, Mcdowell JR, Heist EJ, Musick JA, & Graves JE (2010). World phylogeography and male-mediated gene flow in the sandbar shark, Carcharhinus plumbeus. Molecular Ecology, 19(10), 1994–2010. 10.1111/j.1365-294X.2010.04626.x [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, & Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prothero DR, Kelly TS, Mccardel KJ, & Wilson EL (2008). Magnetostratigraphy, biostratigraphy, and tectonic rotation of the Miocene Caliente formation, Ventura County, California. New Mexico Museum of Natural History and Science Bulletin, 44, 255–272. [Google Scholar]
- Putman AI, & Carbone I (2014). Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecology and Evolution, n/a-n/a. 10.1002/ece3.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queirós J, Godinho R, Lopes S, Gortazar C, de la Fuente J, & Alves PC (2015). Effect of microsatellite selection on individual and population genetic inferences: An empirical study using cross-specific and species-specific amplifications. Molecular Ecology Resources, 15(4), 747–760. 10.1111/1755-0998.12349 [DOI] [PubMed] [Google Scholar]
- Rech JA, Currie BS, Shullenberger ED, Dunagan SP, Jordan TE, Blanco N, Tomlinson AJ, Rowe HD, & Houston J (2010). Evidence for the development of the Andean rain shadow from a Neogene isotopic record in the Atacama Desert, Chile. Earth and Planetary Science Letters, 292(3–4), 371–382. 10.1016/j.epsl.2010.02.004 [DOI] [Google Scholar]
- Revell LJ (2012). phytools: An R package for phylogenetic comparative biology (and other things): phytools: R package. Methods in Ecology and Evolution, 3(2), 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- Richmond JQ, Wood DA, Westphal MF, Vandergast AG, Leaché AD, Saslaw LR, Butterfield HS, & Fisher RN (2017). Persistence of historical population structure in an endangered species despite near-complete biome conversion in California’s San Joaquin Desert. Molecular Ecology, 26(14), 3618–3635. 10.1111/mec.14125 [DOI] [PubMed] [Google Scholar]
- Rissler LJ, Hijmans RJ, Graham CH, Moritz C, & Wake DB (2006). Phylogeographic lineages and species comparisons in conservation analyses: A case study of California herpetofauna. The American Naturalist, 167(5), 655–666. [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Doorenweerd C, McElfresh JS, & Millar JG (2021). Phylogeography of an endemic California silkmoth genus suggests the importance of an unheralded central California province in generating regional endemic biodiversity. Molecular Phylogenetics and Evolution, 164, 107256. 10.1016/j.ympev.2021.107256 [DOI] [PubMed] [Google Scholar]
- Sawyer JO, & Keeler-Wolf T (2009). A Manual of California Vegetation (2nd ed.). California Native Plant Society. [Google Scholar]
- Sinclair EA, Bezy RL, Bolles K, Camarillo JLR, Crandall KA, & Sites JW Jr (2004). Testing Species Boundaries in an Ancient Species Complex with Deep Phylogeographic History: Genus Xantusia (Squamata: Xantusiidae). The American Naturalist, 164(3), 396–413. [DOI] [PubMed] [Google Scholar]
- Singhal S, Huang H, Title PO, Donnellan SC, Holmes I, & Rabosky DL (2017). Genetic diversity is largely unpredictable but scales with museum occurrences in a species-rich clade of Australian lizards. Proceedings of the Royal Society B: Biological Sciences, 284(1854), 20162588. 10.1098/rspb.2016.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, & Excoffier L (2012). Serial founder effects during range expansion: A spatial analog of genetic drift. Genetics, 191(1), 171–181. 10.1534/genetics.112.139022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinks PQ, Thomson RC, & Bradley Shaffer H (2010). Nuclear gene phylogeography reveals the historical legacy of an ancient inland sea on lineages of the western pond turtle, Emys marmorata in California. Molecular Ecology, 19(3), 542–556. 10.1111/j.1365-294X.2009.04451.x [DOI] [PubMed] [Google Scholar]
- Spinks PQ, Thomson RC, & Shaffer HB (2014). The advantages of going large: Genome-wide SNPs clarify the complex population history and systematics of the threatened western pond turtle. Molecular Ecology, 23(9), 2228–2241. 10.1111/mec.12736 [DOI] [PubMed] [Google Scholar]
- Stebbins R (2003). A field guide to western reptiles and amphibians. Houghton Mifflin Publishing Company. [Google Scholar]
- Sunde J, Yıldırım Y, Tibblin P, & Forsman A (2020). Comparing the performance of microsatellites and RADseq in population genetic studies: Analysis of data for pike (Esox lucius) and a synthesis of previous studies. Frontiers in Genetics, 11, 218. 10.3389/fgene.2020.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1989). The effect of change in population size on DNA polymorphism. Genetics, 123(3), 597–601. 10.1093/genetics/123.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1996). Infinite-allele model and infinite-site model in population genetics. Journal of Genetics, 75(1), 27–31. 10.1007/BF02931749 [DOI] [Google Scholar]
- Tewari VP, & Kapoor KS (2013). Western Himalayan cold deserts: Biodiversity, eco-restoration, ecological concerns and securities. Annals of Arid Zones, 52(3 & 4), 225–232. [Google Scholar]
- Wake DB (2006). Problems with species: Patterns and processes of species formation in salamanders. Annals of the Missouri Botanical Garden, 93(1), 8–23. 10.3417/0026-6493(2006)93[8:PWSPAP]2.0.CO;2 [DOI] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, & Mirarab S (2018). ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics, 19(S6). 10.1186/s12859-018-2129-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SJ, Aldridge CL, & Oyler-McCance SJ (2020). An empirical comparison of population genetic analyses using microsatellite and SNP data for a species of conservation concern. BMC Genomics, 21(1), 382. 10.1186/s12864-020-06783-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Individual-level data, including sample name, collection latitude and longitude, the regional population to which the collection location belonged, and the marker types at which the sample had been sequenced.
Figure S1: sampling effort for X. vigilis in the Coast Ranges between our Cuyama Valley and Pinnacles/Panoche populations.
Figure S2: map with sample names and bootstrap and gene concordance values for the iqtree analysis
Figure S3: Structure supplemental analysis, including Evanno delta K results, full population structure results, and equal sample size microsatellite Structure plot
Data Availability Statement
RADseq data are available on NCBI’s Short Read Archive, as BioProject PRJNA649707. Microsatellite repeat data, scripts for analysis and figures, and intermediate output files are available on DataDryad as dataset https://doi.org/10.5061/dryad.31zcrjdht.