Summary
There has been growing research interest in developing methodology to evaluate healthcare centers’ performance with respect to patient outcomes. Conventional assessments can be conducted using fixed or random effects models, as seen in provider profiling. We propose a new method, using fusion penalty to cluster healthcare centers with respect to a survival outcome. Without any priori knowledge of the grouping information, the new method provides a desirable data-driven approach for automatically clustering healthcare centers into distinct groups based on their performance. An efficient alternating direction method of multipliers algorithm is developed to implement the proposed method. The validity of our approach is demonstrated through simulation studies, and its practical application is illustrated by analyzing data from the national kidney transplant registry.
Keywords: Latent class, Fixed effects, Random effects, Provider profiling, Fusion penalty
1 |. INTRODUCTION
Assessing the comparative performance of healthcare centers (e.g., hospitals, nursing homes, transplant centers, or dialysis facilities) has attracted significant interest over the past decades. The objective is to outline and compare the performance of these centers in order to facilitate improvements through accountability and feedback. This information can aid individuals in selecting the most suitable healthcare facility and also enable stakeholders and payers to identify areas requiring enhancements.
Our motivating example is the national kidney transplant registry data collected by the U.S. Organ Procurement and Transplantation Network (OPTN). Our goal is to evaluate transplant centers based on their five-year post-transplant graft survival rates. For patients with end-stage renal disease, kidney transplantation provides the best opportunity for survival. The Scientific Registry of Organ Recipients (SRTR) commonly utilizes the five-year post-transplant graft survival metric, defined as the time until either death or graft failure within five years following transplantation, for regulatory monitoring of transplant centers1,2. Consequently, we will employ the five-year post-transplant graft survival to assess the quality of care provided by transplant centers.
Traditionally, profiling methods have been developed to evaluate the quality of care provided by various healthcare centers, using multiple patient outcome quality measures, such as readmission, mortality, and hospitalization. Existing transplant center profiling approaches typically employ inference-based procedures and generate a three-tier system, indicating whether centers perform worse than expected, as expected, or better than expected. Random effects and fixed effects models are two prevalent analytical methods used in profiling3,4,5,6. However, both models have their drawbacks. For random effects models, healthcare centers on the tails of the distribution tend to have small sample sizes, leading to substantially shrunk estimates towards the population mean7,8. This may result in reduced sensitivity when classifying healthcare centers in the tail areas, causing the majority of healthcare centers to be classified as expected, despite noticeable heterogeneity9. Additionally, misspecification of the random effects distribution can pose challenges in both estimation and inference. In contrast, fixed effects models suffer from a loss of efficiency due to a large number of parameters. Moreover, the simultaneous testing of the null hypothesis for extensive healthcare center effects is computationally demanding.
In order to offer more comprehensive ratings for kidney transplantation, the SRTR has implemented a five-tier rating system10,11 that indicates whether a transplant center performs better than expected, somewhat better than expected, as expected, somewhat worse than expected, or worse than expected. However, concerns arise regarding the selection of appropriate cutoffs to categorize transplant centers into distinct groups. Furthermore, the decision regarding the total number of tiers is arbitrary.
To tackle the aforementioned challenges, we introduce a new fused effects model12 designed to automatically identify homogeneous groups of healthcare centers without requiring a priori classification knowledge. We employ Cox’s proportional hazards model13 with fusion penalty14 to cluster transplant centers based on the post-transplant graft survival outcome. Unlike random or fixed effects models, this new method offers a data-driven approach that does not rely on inference tests of statistical significance. Our model can also investigate risk factors associated with post-transplant graft survival. Our method can be considered as an alternative of the latent class model, where we use fusion penalty to “classify” providers into different latent groups.
We employ a local quadratic approximation to the partial likelihood and optimize the penalized partial likelihood with the fusion penalty. Prioritizing clustering accuracy, we opt for the smoothly clipped absolute deviation (SCAD)15 penalty function over the LASSO penalty16. Compared to the LASSO, the SCAD penalty is nearly unbiased in identifying groups and enforces a sparser solution more aggressively17. The alternating direction method of multipliers (ADMM) algorithm can be utilized to implement the estimation, ensuring rapid convergence18. Due to the information loss during computation, we perform refitting by maximizing the log partial likelihood with the grouped data to obtain accurate parameter estimates.
The remainder of this paper is structured as follows: in Section 2, we outline the penalized Cox’s regression model with fusion penalty for clustering healthcare centers. Section 3 evaluates the performance of our approach through Monte Carlo simulation studies. In Section 4, we demonstrate the proposed method using the kidney transplant data as a practical example. Finally, we summarize our methodology and discuss potential future directions in Section 5.
2 |. METHODS
2. 1 |. Model
We begin by introducing the required notations to formulate our model. For subject from healthcare center , we have data in the format , where the observed time is the minimum of the censoring time and the event time is the censoring indicator, and is a vector of predictors. The Cox’s proportional hazards model is
| (2.1) |
where is the hazard for patient of center at time is a baseline hazard function, is the center-specific effect, and is the vector of covariate coefficients. A constraint ensures that all parameters are identifiable. For estimation and inference, we often rely on the partial likelihood, where the unspecified baseline hazards can be canceled out. Patients are assumed to be independent within each healthcare center, i.e., we assume independence of given and . The partial likelihood for model (2.1) can be written as
| (2.2) |
where . We assume that belongs to one of groups , which are mutually exclusive partitions of ; and the number of groups is much smaller than that of centers, i.e., . Moreover, the number of groups and the group membership are unknown in advance.
We utilize the fused SCAD penalty to identify homogeneous center performance and then fuse them as shared parameters to classify groups of healthcare centers. Incorporating the fusion penalty into the partial likelihood (2.2) results in the following optimization problem:
| (2.3) |
where is the log of the partial likelihood, and is the SCAD penalty function15 defined as
with if and = 0 otherwise, is a tuning parameter, is a parameter that controls the concavity of the penalty functions. Following Ma et al.,17 we treat as a fixed constant.
2. 2 |. Estimation procedure
Note that the penalty function cannot be written in the form of addition of separate terms of and as in LASSO. Here, we introduce a new set of parameters , which are equivalent to the pairwise differences of healthcare center effects. Consequently, the above minimization problem (2.3) can be transformed into the following constraint optimization problem:
| (2.4) |
where . As a result, the alternating direction method of multipliers (ADMM) algorithm can be used to identify the groups in the objective function (2.4). The ADMM algorithm combines the strengths of dual decomposition and augmented Lagrangian methods for constrained optimization. The estimates of the parameters are obtained by the augmented Lagrangian
| (2.5) |
where are Lagrange multipliers, is the penalty parameter. We use ADMM to iteratively compute the estimates of . For given at step , the iterations can be specified as follows:
| (2.6) |
| (2.7) |
| (2.8) |
To update and , minimizing (2.6) is equivalent to minimizing
| (2.9) |
where is the constant independent of and . To solve the optimization problem (2.9), we approximate the nonlinear logpartial likelihood function using a two-term Taylor series expansion. At each iteration, we solve a reweighted least squares problem 19. Specifically, let with with with being the vector with ones, and . Let denote the gradient and Hessian of the log-partial likelihood with respect to , respectively. The log-partial likelihood can be approximated by the following quadratic form (see the supplementary material)
where with
where is the set of indices with (the times for which observation of the th subject in the th center is still at risk) and is the set of indices with (those at risk at time ). Since is a full matrix, it requires computation of elements, we instead replace by a diagonal matrix with the same diagonal elements as .20,21,19 This substitution works well because the diagonal elements of are much larger than the off-diagonal elements. Denote by and are computed based on and at iteration . Equation (2.9) can be rewritten as
| (2.10) |
where with being a vector whose th element is 1 and the remaining elements are 0. Thus, for given at the sth step, we set the derivatives and to obtain the following updates and :
| (2.11) |
where . We let mean to guarantee that the estimate satisfies the constraint , and
| (2.12) |
To update , we minimize the function
where . It is worth noting that by using the concave penalties, the objective function is no longer a convex function. However, it is convex with respect to each when for the SCAD penalty; consequently, given , the minimizer of with respect to is unique and has a closed-form solution as follows:
| (2.13) |
where is a groupwise soft thresholding operator. Finally, the Lagrange multiplier is updated by (2.8). This process is conducted iteratively until the convergence over a grid of values for . The iterative algorithm terminates when primal residuals and dual residuals are close to zero, i.e., and , where and are specified as suggested by Boyd et al.18 It is important to find appropriate initial values for the ADMM algorithm. In this paper, the initial values are obtained from the fixed effects Cox model, then we set and . If and are classified into the same group. As a result, we obtain estimated groups , and set to be the common value of ’s from the th group.
Due to the approximation in the computation, Equation (2.10) does not fully capture parameter information, and the iteration may also lead to a loss in efficiency. Once the groups have been identified, we conduct a refitting step to estimate and by maximizing the following log-partial likelihood with the grouped information:
| (2.14) |
where , and is the set of indices with (those at risk at time ). Here rather than the constraint (as for in Section 2.1), we assume that to simplify the interpretation, where . The above procedure can be conveniently implemented using R function coxph, which also provides standard errors and p-values of and .
After obtaining a path of solutions, it becomes essential to choose an optimal tuning parameter by minimizing the modified Bayesian Information Criterion (BIC) using a grid search.22. The BIC is given by
where and are the estimates of and at given , respectively. is the log-partial likelihood evaluated at and is the dimension of the parameter , and is a positive number depending on the total number of observations . If , the modified BIC reduces to the traditional BIC23 Following Wang et al22, .
We summarize our method in Algorithm 1 below.
Algorithm 1 ADMM algorithm for healthcare center clustering
| Require: Initialize and . |
| for s = 0,1,2,⋯do |
| Compute using (2.11) |
| Compute using (2.12) |
| Compute using (2.13) |
| Compute using (2.8) |
| if the convergence criterion is met, then |
| Stop and denote the last iteration by , |
| else |
| s = s + 1. |
| end if |
| end for |
| After identifying the groups, estimate using (2.14). |
| Ensure: Output |
3 |. SIMULATION
In this section, we evaluate the finite sample performance of the proposed methods through simulation studies. Conventionally, in Model (2.1), the healthcare center effect is treated as either a random effect or a fixed effect. We compare our fused effects model with SCAD penalty (SCAD) to the random effects model (RE) and fixed effects model (FE). Specifically, we examine the efficiency of estimation and the accuracy of healthcare center classification. We consider two different numbers of centers, and 100, and obtain all simulation results via 100 replicates.
Example 1. To assess the relative performance of different models, we mimic the censoring rate observed in the real-life application of kidney transplant centers in Section 4. We assume that among the center effects ’s, 10% are set to - 1, representing healthcare centers performing “better than expected,” 10% are set to 1, indicating centers performing “worse than expected,” and the remaining 80% are set to 0, representing centers performing “as expected.” The survival time is generated from the Weibull distribution with scale and shape parameters of 2 and 3, respectively. The censoring time is generated from a uniform distribution to achieve approximately a 70% censoring rate, which is close to the real data. The covariates are generated from the multivariate normal distribution with mean 0, variance 1 and an exchangeable correlation . The coefficient . We consider the number of patients in each center ~ Uniform (50,100).
The Rand Index (RI)24 is employed to assess the level of agreement between the estimated partitions and the true partitions. Each pair of observations and falls to one of four categories: (i) true positive (TP) where and from the same group are assigned to the same cluster; (ii) true negative (TN) where and from different groups are assigned to different clusters; (iii) false negative (FN) where and from different groups are assigned to the same cluster; (iv) false positive (FP) where and from the same group are assigned to different clusters. The Rand Index is given by
Intuitively, TP and TN represent agreement between the true group and the estimated cluster, while FP and FN indicate disagreement between the true group and the estimated cluster. RI ranges from 0 to 1, with a larger value indicating a higher degree of agreement.
The top panel of Table 1 presents the mean, median, and standard deviation (SD) of the estimated number of groups , the percentage of equal to the true number of groups (per), and the RI for evaluating the classification accuracy. As expected, the RI and the percentages of correctly classifying centers in comparison to the reference are very close to 1 and improve as increases. To visually display the distribution of , the histograms of are depicted in Figure 1
TABLE 1.
The mean, median, and standard deviation (SD) of , the percentage (per) of equal to the true number of subgroups, and the Rand Index (RI) value by our method with in Examples 1 to 3, respectively.
| m | mean | median | SD | per | RI | |
|---|---|---|---|---|---|---|
| Example 1 | 50 | 3.08 | 3.00 | 0.339 | 0.91 | 0.960 |
| 100 | 3.05 | 3.00 | 0.261 | 0.93 | 0.969 | |
| Example 2 | 50 | 8.23 | 8.00 | 1.362 | ||
| 100 | 9.43 | 9.00 | 1.653 | |||
| Example 3 | 50 | 4.06 | 4.00 | 0.343 | 0.91 | 0.963 |
| 100 | 4.04 | 4.00 | 0.281 | 0.92 | 0.970 |
FIGURE 1.
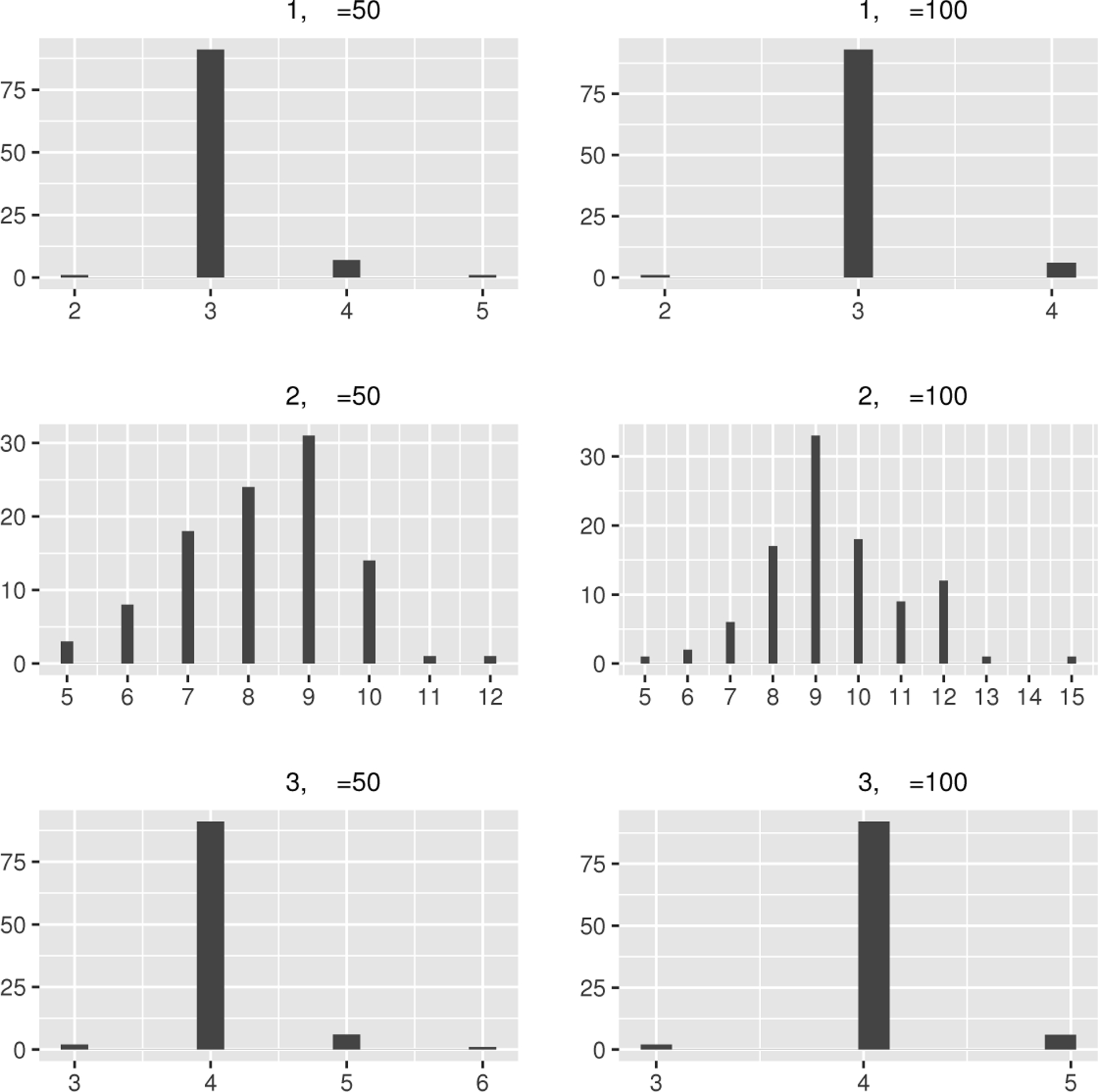
The histograms of in Example 1–3.
Let and be the average estimates of center effects from “better than expected”, “as expected”, and “worse than expected” groups, with true values being −1,0 and 1, respectively. The top panel of Table 2 presents the bias, the standard deviation (SD), the standard error (SE), and the coverage probability of 95% confidence intervals (CP) of the estimators and . The estimator serves as a reference, such as the national norm. The Oracle estimators are obtained with a priori knowledge of the true grouping information. As not all replications are clustered into three groups by SCAD, we only use the replications with the estimated number of groups equal to three to compute the bias, SD, SE, and CP of and . For the Oracle, the measures are calculated based on all 100 replications. From Table 2. We notice that our method performs very closely to the Oracle, as it can accurately recover the group structure. Evidently, our estimators and align well with the corresponding true values on average for all cases. The inference is also adequately precise, with a strong correspondence between SD and SE, and the coverage probabilities are near the nominal level of 0.95. All the original estimates of , and without the refitting step are reported in Table S1 of the supplementary material.
TABLE 2.
The bias, standard deviation (SD), standard error (SE), and the coverage probability of 95% confidence intervals (CP) of the estimators in Example 1 and Example 3.
| m=50 | m=100 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| m | Method | Bias | SD | SE | CP | Bias | SD | SE | CP | |
| Example 1 | SCAD | 0.014 | 0.090 | 0.113 | 98.9 | 0.008 | 0.070 | 0.078 | 97.8 | |
| Oracle | 0.009 | 0.094 | 0.105 | 96.0 | 0.018 | 0.078 | 0.074 | 94.0 | ||
| SCAD | 0.028 | 0.094 | 0.086 | 91.5 | 0.010 | 0.055 | 0.061 | 97.8 | ||
| Oracle | 0.006 | 0.094 | 0.084 | 92.0 | 0.006 | 0.055 | 0.060 | 98.0 | ||
| Example 3 | SCAD | 0.009 | 0.086 | 0.087 | 93.4 | 0.004 | 0.063 | 0.062 | 97.4 | |
| Oracle | 0.004 | 0.082 | 0.087 | 95.0 | 0.002 | 0.061 | 0.062 | 98.0 | ||
| SCAD | 0.005 | 0.069 | 0.089 | 97.8 | 0.019 | 0.058 | 0.062 | 94.8 | ||
| Oracle | 0.004 | 0.084 | 0.086 | 96.0 | 0.004 | 0.059 | 0.061 | 94.0 | ||
| SCAD | 0.006 | 0.088 | 0.085 | 92.3 | 0.008 | 0.057 | 0.060 | 93.5 | ||
| Oracle | 0.009 | 0.087 | 0.085 | 93.0 | 0.006 | 0.055 | 0.060 | 96.0 | ||
We compute the mean squared error (MSE) of using the formula for each replicated dataset. The top panel of Table 3 presents the MSE of the estimator , the bias, the standard deviation (SD), the standard error (SE), and the coverage probability of 95% confidence intervals (CP) for the estimators and . It is worth mentioning that, in contrast to Table 2, which only displays results for replications with an estimated group count of three, Table 3 includes results for all replicates, regardless of whether the estimated number of groups is three or not. The MSEs of from our fused effects model are much smaller than those from the random effects and fixed effects models in all cases. As a result, our method not only accurately classifies centers but also obtains precise estimates of these centers. To graphically visualize the numerical results of Table 3, the boxplots of the MSE of are depicted in Figure 2 For the estimators , our method and the random effects model exhibit satisfactory performance, while the fixed effects model yields larger bias.
TABLE 3.
The bias, standard deviation (SD), standard error (SE), and the coverage probability of 95% confidence intervals (CP) of the estimators in Examples 1 to 3.
| Method | MSE | Bias | SD | SE | CP | Bias | SD | SE | CP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Example 1 | 50 | SCAD | 0.022 | 0.011 | 0.043 | 0.045 | 95.0 | 0.012 | 0.043 | 0.045 | 95.0 |
| FE | 0.078 | 0.029 | 0.045 | 0.047 | 91.0 | 0.026 | 0.045 | 0.047 | 94.0 | ||
| RE | 0.189 | 0.005 | 0.044 | 0.046 | 97.0 | 0.008 | 0.044 | 0.046 | 95.0 | ||
| Oracle | 0.002 | 0.003 | 0.043 | 0.046 | 97.0 | 0.005 | 0.043 | 0.046 | 94.0 | ||
| 100 | SCAD | 0.017 | 0.002 | 0.034 | 0.032 | 93.0 | 0.004 | 0.033 | 0.032 | 94.0 | |
| FE | 0.074 | 0.034 | 0.032 | 0.033 | 88.0 | 0.032 | 0.034 | 0.033 | 87.0 | ||
| RE | 0.034 | 0.001 | 0.032 | 0.033 | 95.0 | 0.001 | 0.033 | 0.033 | 94.0 | ||
| Oracle | 0.001 | 0.002 | 0.032 | 0.032 | 95.0 | 0.001 | 0.033 | 0.032 | 93.0 | ||
| Example 2 | 50 | SCAD | 0.321 | 0.001 | 0.032 | 0.030 | 93.0 | 0.002 | 0.031 | 0.030 | 94.0 |
| FE | 1.183 | 0.002 | 0.033 | 0.030 | 93.0 | 0.002 | 0.031 | 0.030 | 94.0 | ||
| RE | 1.810 | 4e-4 | 0.032 | 0.030 | 94.0 | 0.003 | 0.031 | 0.030 | 94.0 | ||
| 100 | SCAD | 0.242 | 0.002 | 0.019 | 0.021 | 97.0 | 0.002 | 0.022 | 0.021 | 93.0 | |
| FE | 1.080 | 0.001 | 0.020 | 0.021 | 98.0 | 0.003 | 0.022 | 0.021 | 93.0 | ||
| RE | 0.060 | 0.001 | 0.019 | 0.021 | 98.0 | 0.001 | 0.022 | 0.021 | 93.0 | ||
| Example 3 | 50 | SCAD | 0.016 | 0.004 | 0.047 | 0.046 | 94.0 | 0.007 | 0.044 | 0.046 | 97.0 |
| FE | 0.079 | 0.031 | 0.046 | 0.047 | 90.0 | 0.027 | 0.045 | 0.047 | 92.0 | ||
| RE | 0.847 | 0.003 | 0.045 | 0.046 | 96.0 | 0.006 | 0.044 | 0.046 | 96.0 | ||
| Oracle | 0.003 | 9e-5 | 0.044 | 0.046 | 96.0 | 0.003 | 0.042 | 0.046 | 97.0 | ||
| 100 | SCAD | 0.012 | 0.001 | 0.033 | 0.032 | 94.0 | 1e-4 | 0.032 | 0.032 | 92.0 | |
| FE | 0.077 | 0.035 | 0.032 | 0.033 | 88.0 | 0.034 | 0.033 | 0.032 | 89.0 | ||
| RE | 0.039 | 0.003 | 0.032 | 0.033 | 94.0 | 0.001 | 0.033 | 0.033 | 92.0 | ||
| Oracle | 0.001 | 0.003 | 0.032 | 0.032 | 94.0 | 0.002 | 0.033 | 0.032 | 92.0 | ||
FIGURE 2.
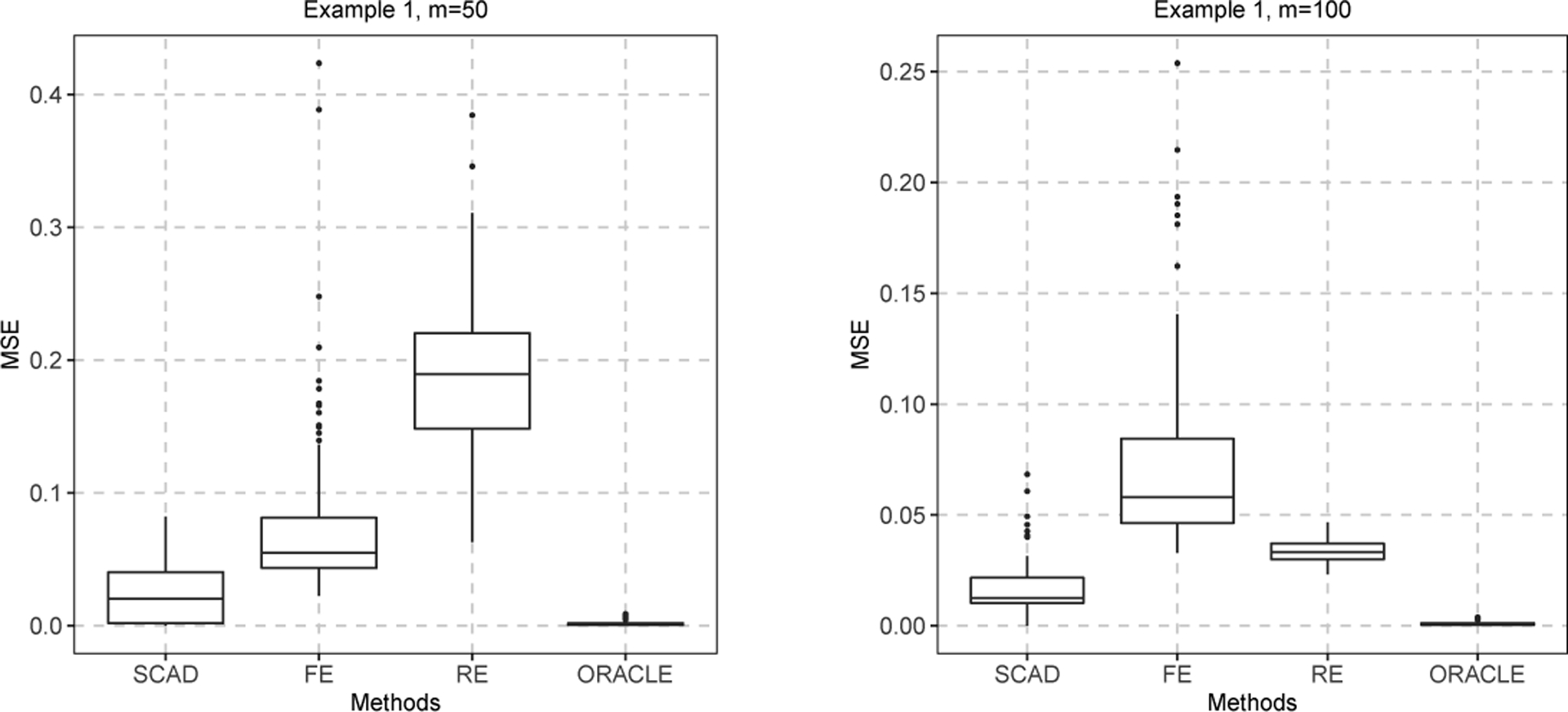
The boxplots of the MSE of with and in Example 1.
Example 2. In this example, we generate the covariates from the multivariate normal distribution with mean 0, variance 1, and an exchangeable correlation . The survival time is generated from the Weibull distribution with scale and shape parameters being 2 and 3, respectively. The censoring time is generated from a uniform distribution to achieve about a 70% censoring rate. We set the coefficient , and the number of patients in each center ~ Uniform (50, 100). To demonstrate the robustness of our method, is simulated from the standard normal distribution . Consequently, the random effects model is the correct model.
The grouping results of using our method are presented in the second panel of Table 1 The median of the estimated number of groups is 8 with and 9 with . The fusion penalty generally tends to select fewer groups for and more groups for . As the number of parameters increases with , the median and the standard deviation of also grow. To graphically visualize the distribution of , the histograms of are shown in Figure 1
The second panel of Table 3 presents the mean squared error (MSE) of the estimator , the bias, the standard deviation (SD), the standard error (SE), and the coverage probability of 95% confidence intervals (CP) for the estimators and . From Table 3. we observe that the MSE values of using our method are smaller than those in the fixed effects model. Moreover, our method outperforms the random effects model when the number of centers is small , likely due to the shrinkage of the predicted values of ’s in the random effects model. As the number of centers increases, the random effects model performs the best, but our method’s performance is only slightly worse than the random effects model and still much better than the fixed effects model. All methods exhibit similar performance in estimating and .
Example 3. In this example, we evaluate the performance of the proposed model with four groups. The covariates , and the censoring time are generated from the same distributions as in Example 1. We assume that among the center effects ’s, 10% are set to 1 and 10% are set to 2, indicating centers performing at different levels of “worse than expected”; 20% are set to −1.5, signifying centers performing “better than expected”; the remaining 60% are set to 0, indicating centers performing “as expected”.
The grouping results of using our method are presented in the bottom panel of Table 1, where the medians of over the 100 replicates are 4, which is the true number of subgroups, and the mean values are very close to 4. Furthermore, the RI values and the percentage of correctly selecting the number of subgroups approach 1. Therefore, our methods perform well in cases with an even number of groups. To graphically visualize the distribution of , the histograms of are shown in Figure 1.
Let , and be the average estimates for the ’s from four groups where the true values are −1.5,0,1, and 2, respectively. The bottom panel of Table 2 presents the bias, the standard deviation (SD), the standard error (SE), and the coverage probability of 95% confidence intervals (CP) for the estimators , and . The estimator , which has the smallest absolute value, is set as the reference. From the bottom panel of Table 2. we can see that the means of , and are close to the true values and the Oracle estimators. We also observe that the SDs of , and are close to the corresponding SEs, resulting in valid coverage probabilities.
The bottom panel of Table 3 reports the MSE of the estimator , the bias, the standard deviation (SD), the standard error (SE), and the coverage probability of 95% confidence intervals (CP) for the estimators and . From the bottom panel of Table 3 we observe that the MSE values of using SCAD are smaller than those of the random effects and fixed effects models. For the estimators , our method and the random effects model exhibit satisfactory performance, while the fixed effects model produces larger bias. These results indicate that the proposed method performs well with an even number of groups.
4 ।. APPLICATION
To demonstrate the proposed methods, we conduct an evaluation of kidney transplant centers using the national kidney transplant registry data obtained from the U.S. Organ Procurement and Transplantation Network (https://optn.transplant.hrsa.gov/data/). We limit the study cohort to adult kidney transplant recipients (age ≥ 18) who received a transplant between January 1, 2007, and December 31, 2007. The analysis cohort includes 4198 patients from 60 centers, with the number of patients per center ranging from 50 to 100. In our dataset, out of 4198 patients who received their kidney transplant in 2007, 1137 (27.1%) either died or experienced graft failure within five years after receiving a kidney transplant. All others were censored at five years of follow-up. In our survival analysis, the failure time (termed “graft survival”) is defined as the duration (in years) from transplantation to graft failure or death, whichever occurred first. The censoring time is at the end of the five-year period after the transplantation. Thus, the observed time to event is , with a 72.9% censoring rate. We apply our method to investigate the risk factors of five-year graft survival using Cox’s proportional hazards model and assess the performance of centers concerning their graft outcomes.
The study cohort included 15 baseline characteristics of donors and recipients. The characteristics of the study population are as follows: Time on end-stage renal disease (ESRD, reference: <1 years), donor age (reference: 30–45 years old), donor gender (male = 1, female=0), donor body mass index (BMI, reference: normal), donor race (reference: white), donor history of hypertension (DON-HTN, yes=1, no=0), donor meeting expanded criteria (DON-EC, yes=1, no=0), recipient gender (male = 1, female=0), recipient race (reference: white), recipient insulin dependent diabetes (REC-DIAB Type I, yes=1, no=0), recipient non-insulin dependent diabetes (REC-DIAB Type II, yes=1, no=0), recipient age at transplant (REC-AGE, reference: 50–60), recipient body mass index (reference: normal), recipient previous kidney transplant (REC-PREV-KI, yes=1, no=0), recipient total cold ischemia time (REC-COLD-ISCH, > 20 hours = 1,< 20 hours =0).
We use the Akaike Information Criteria (AIC) to assess the performance of three methods. A smaller value of AIC indicates better performance. When treating the center effect as a random intercept, we obtain an AIC of 18501.43. For the fixed effects model, the AIC value is 18535.64. If is estimated by our fused effects model, the 60 centers are classified into 3 groups with an AIC of 18451.24. Our method leads to a significant improvement in model fitting.
Table 4 reports the estimate (Est.), standard error (SE), and p-value of for testing the significance of the coefficients by our fused effects model with the SCAD penalty (SCAD), the fixed effects model (FE) and the random effects model (RE). Longer time spent in end-stage renal disease, donor age over 60, black donor, donor history of hypertension, black recipient, recipient aged 60 or older at transplant, recipient with non-insulin dependent diabetes, and donor with low body mass index all have a significantly worse effect on five-year graft survival according to all three methods, with p-values less than 0.05. Asian recipients, on the other hand, tended to experience better survival outcomes.
TABLE 4.
The estimates (Est.), standard error (SE), and P-value of in the national kidney transplant study.
| Variable | Fused | FE | RE | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Est | SE | P-value | Est | SE | P-value | Est | SE | P-value | |
| Time on ESRD | |||||||||
| < 1 years | Ref | ||||||||
| 1–5 years | 0.093 | 0.041 | 0.022 | 0.100 | 0.041 | 0.015 | 0.086 | 0.041 | 0.035 |
| > 5 years | 0.150 | 0.040 | 0.000 | 0.142 | 0.042 | 0.001 | 0.143 | 0.041 | 0.001 |
| Donor age | |||||||||
| < 15 | −0.063 | 0.044 | 0.149 | −0.073 | 0.044 | 0.098 | −0.069 | 0.044 | 0.114 |
| 15–30 | −0.066 | 0.042 | 0.113 | −0.067 | 0.042 | 0.112 | −0.075 | 0.042 | 0.073 |
| 30–45 | Ref | ||||||||
| 45–60 | 0.073 | 0.040 | 0.068 | 0.076 | 0.040 | 0.061 | 0.075 | 0.040 | 0.062 |
| > 60 | 0.113 | 0.043 | 0.009 | 0.115 | 0.044 | 0.009 | 0.114 | 0.043 | 0.009 |
| Donor race | |||||||||
| White | Ref | ||||||||
| Black | 0.066 | 0.028 | 0.021 | 0.072 | 0.029 | 0.015 | 0.071 | 0.029 | 0.013 |
| Asian | 0.001 | 0.030 | 0.987 | 0.004 | 0.031 | 0.908 | 0.001 | 0.030 | 0.969 |
| DON-HTN | 0.090 | 0.035 | 0.011 | 0.088 | 0.036 | 0.014 | 0.087 | 0.035 | 0.014 |
| DON-EC | −0.003 | 0.045 | 0.953 | −0.002 | 0.046 | 0.973 | −0.003 | 0.046 | 0.947 |
| Recipient gender | 0.048 | 0.030 | 0.115 | 0.053 | 0.031 | 0.086 | 0.053 | 0.031 | 0.083 |
| Recipient race | |||||||||
| White | Ref | ||||||||
| Black | 0.079 | 0.030 | 0.009 | 0.090 | 0.034 | 0.008 | 0.091 | 0.032 | 0.004 |
| Asian | −0.100 | 0.037 | 0.007 | −0.105 | 0.038 | 0.005 | −0.099 | 0.037 | 0.008 |
| Recipient BMI | |||||||||
| Normal | Ref | ||||||||
| Under | 0.054 | 0.029 | 0.068 | 0.050 | 0.030 | 0.093 | 0.054 | 0.030 | 0.070 |
| Over | 0.005 | 0.037 | 0.896 | 0.003 | 0.038 | 0.934 | −0.001 | 0.037 | 0.971 |
| Obesity | 0.026 | 0.037 | 0.484 | 0.024 | 0.038 | 0.530 | 0.027 | 0.037 | 0.472 |
| REC-PREV-KI | 0.046 | 0.030 | 0.127 | 0.038 | 0.031 | 0.220 | 0.053 | 0.030 | 0.084 |
| REC-COLD-ISCH | 0.006 | 0.030 | 0.837 | −0.004 | 0.033 | 0.895 | 0.014 | 0.031 | 0.662 |
| REC-AGE | |||||||||
| 18–35 | 0.050 | 0.034 | 0.150 | 0.055 | 0.035 | 0.116 | 0.045 | 0.035 | 0.189 |
| 35–50 | −0.029 | 0.038 | 0.439 | −0.025 | 0.039 | 0.520 | −0.029 | 0.038 | 0.449 |
| 50–60 | Ref | ||||||||
| 60–70 | 0.112 | 0.034 | 0.001 | 0.110 | 0.035 | 0.001 | 0.114 | 0.034 | 0.001 |
| > 70 | 0.082 | 0.030 | 0.006 | 0.081 | 0.030 | 0.007 | 0.095 | 0.030 | 0.001 |
| REC-DIAB | |||||||||
| Type I | 0.056 | 0.029 | 0.054 | 0.061 | 0.030 | 0.042 | 0.057 | 0.030 | 0.055 |
| Type II | 0.075 | 0.030 | 0.012 | 0.071 | 0.031 | 0.021 | 0.075 | 0.030 | 0.014 |
| Donor gender | 0.005 | 0.030 | 0.872 | 0.007 | 0.030 | 0.818 | 0.005 | 0.030 | 0.863 |
| Donor BMI | |||||||||
| Normal | Ref | ||||||||
| Under | 0.104 | 0.037 | 0.005 | 0.108 | 0.037 | 0.004 | 0.103 | 0.037 | 0.005 |
| Over | 0.037 | 0.035 | 0.292 | 0.036 | 0.035 | 0.300 | 0.034 | 0.035 | 0.329 |
| Obesity | −0.008 | 0.035 | 0.815 | −0.009 | 0.035 | 0.798 | −0.003 | 0.035 | 0.927 |
Next we utilize the standardized mortality ratio (SMR) as an evaluation measure to assess center-specific survival, defined as the ratio of the observed number of deaths at a given center to the number expected if the center had mortality equal to the population average25,4. An SMR greater or smaller than 1 indicates that the center’s observed mortality ratio is under-performing or over-performing relative to the population norm, respectively. Let be the common value of estimators ’s in group with . Table 5 reports the estimates and relative to a reference and the number of elements (num.) in each group, standard error (SE), p-value and SMR by our fused effects method. We identify 75.0% (45) of centers as the reference, 10.0% (6) of centers better than the reference and 15.0% (9) of centers worse than the reference. The estimated SMR of better and worse centers are 0.574 and 1.636, respectively.
TABLE 5.
Results of in the national kidney transplant study.
| Est | −0.556 | ref | 0.492 |
| num | 6 | 45 | 9 |
| SE | 0.129 | ref | 0.073 |
| P-value | <0.001 | ref | <0.001 |
| SMR | 0.574 | 1.000 | 1.636 |
In our national kidney transplant dataset, we only have a center-specific factor - the numbers of patients within a center (denoted by center size ). Since center-level confounder, e.g., is not identifiable with the center-specific effect , we cannot include directly in our model. Along the lines of He et al.4, we instead fit the model , where is the estimated center specific effect. The p-value is 0.915, suggesting a minimal correlation between the center-specific effects and the center size. This observation is further supported by the boxplots in Figure 3.
Figure 3.
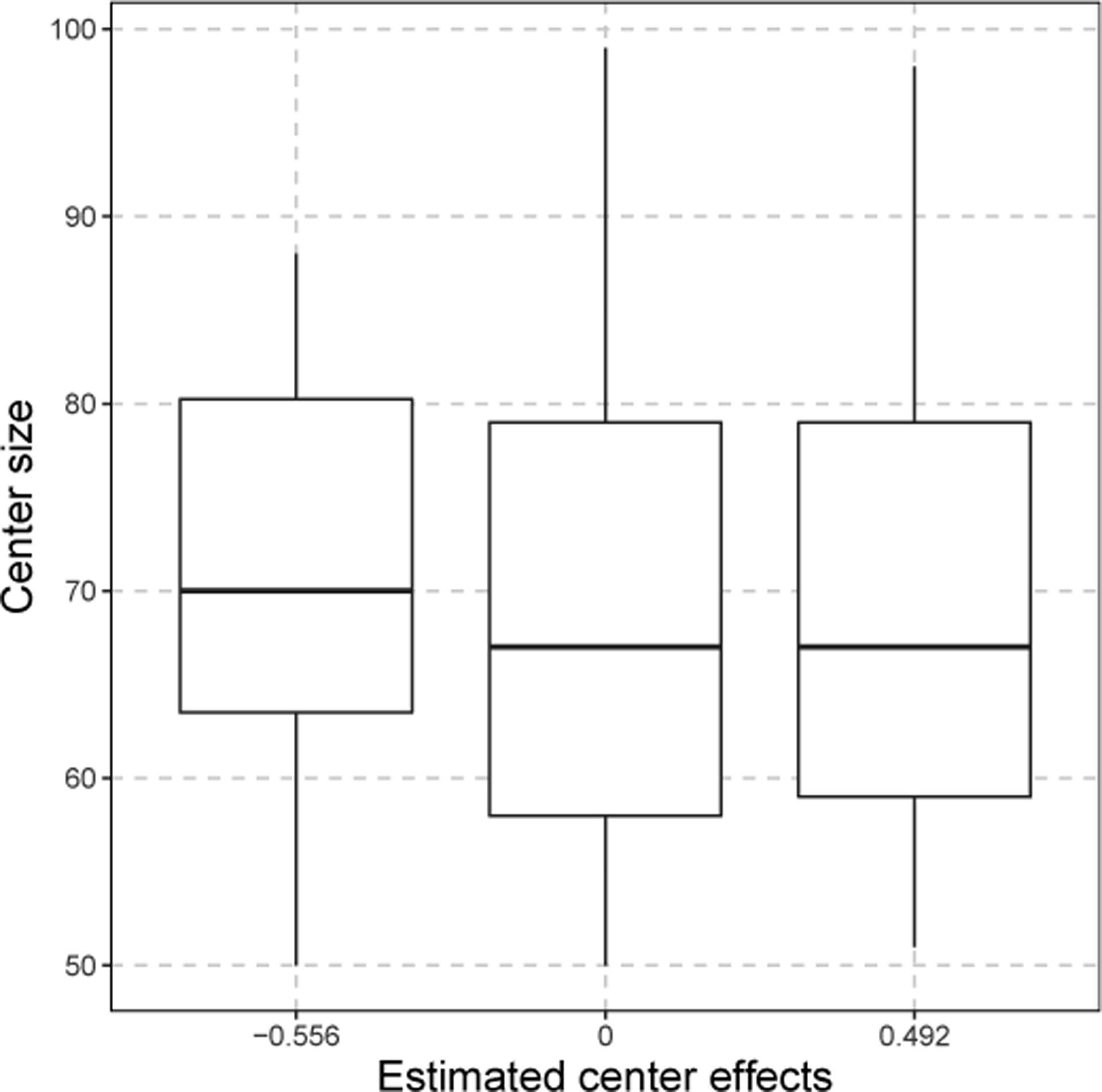
The boxplots of center size from three groups in the national kidney transplant study.
5 |. DISCUSSION
In this study, we proposed an innovative fusion method to assess healthcare centers concerning survival outcomes. Our method proves more efficient than the fixed effects model by using fewer parameters for healthcare centers. Additionally, it outperforms random effects models in identifying healthcare centers with better classification accuracy and lower bias. Through simulation studies, we demonstrated that our method surpasses existing approaches in performance. Our model can be thought as an alternative way to latent class models, as we use fusion penalty to classify different healthcare centers into latent subgroups.
There are several potential extensions for our method. First, when the number of centers is very large, fitting the pairwise fusion penalized Cox model directly becomes computationally demanding. In such cases, we could employ the divide-andconquer strategy26, which typically involves dividing the full sample into multiple subsets, solving the optimization problem for each subset, and combining the subset-specific estimates into a single estimate. Second, our method can be applied to cluster regression coefficients in the Cox’s proportional hazards model when parameters are partially heterogeneous across subgroups. For instance, grouping treatment heterogeneity can enable the provision of precise medical treatments to diverse patient subgroups27.17.
Finally, it is of interest to consider more sophisticated hierarchical data, e.g., patients clustered within practitioners which are clustered within practices. For this case, a three-level Cox’s proportional hazards model with two fusion effects can be proposed. Let denote the covariates for patient nested within doctor in center , the three-level Cox model can be written as
where is the center-specific effect, is the practitioner-specific effect, is the hazard for patient treated by practitioner in center at time , and is a baseline hazard function. We can cluster and by using two fusion penalties. Estimation and inference of this model is of future interest.
Supplementary Material
ACKNOWLEDGEMENT
This research is partly supported by NIH grants R21 EY031884, R21 EY033518, R21 AG063370, UL1 TR002345, China Postdoctoral Science Foundation 2021M691952, Natural Science Foundation of Shandong Province ZR2021QA024.
Footnotes
SUPPLEMENTARY MATERIALS
The supplementary material for this article is available online.
DATA AVAILABILITY STATEMENT
Restrictions apply to the availability of these data, which were used under license for this study.
References
- 1.Wynn JJ, Distant DA, Pirsch JD, et al. Kidney and pancreas transplantation. American Journal of Transplantation 2004; 4: 72–80. [DOI] [PubMed] [Google Scholar]
- 2.Wang JH, Skeans MA, Israni AK. Current status of kidney transplant outcomes: dying to survive. Advances in chronic kidney disease 2016; 23(5): 281–286. [DOI] [PubMed] [Google Scholar]
- 3.Normand ST, Shahian DM. Statistical and clinical aspects of hospital outcomes profiling. Statistical Science 2007; 22: 206–226. [Google Scholar]
- 4.He K, Kalbfleisch JD, Li Y, Li Y. Evaluating hospital readmission rates in dialysis facilities; adjusting for hospital effects. Lifetime Data Analysis 2013; 19: 490–512. [DOI] [PubMed] [Google Scholar]
- 5.Estes JP, Chen Y, Sentürk D, et al. Profiling dialysis facilities for adverse recurrent events. Statistics in Medicine 2020; 39: 1374–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalbfleisch JD, Wolfe RA. On monitoring outcomes of medical providers. Statistics in Biosciences 2013; 5: 286–302. [Google Scholar]
- 7.Racz M, Sedransk J. Bayesian and frequentist methods for provider profiling using risk-adjusted assessments of medical outcomes. Journal of the American Statistics Association 2010; 105: 48–58. [Google Scholar]
- 8.Ohlssen D, Sharples L, Spiegelhalter D. A hierarchical modelling framework for identifying unusual performance in health care providers. Journal of the Royal Statistical Society A 2007; 170: 865–890. [Google Scholar]
- 9.Salkowski N, Snyder J, Zaun D, Leighton T, Israni A, Kasiske BL. Bayesian methods for assessing transplant program performance. American Journal of Transplantation 2014; 14(6): 1271–1276. [DOI] [PubMed] [Google Scholar]
- 10.Van Pilsum Rasmussen SE, Thomas AG, Garonzik-Wang J, et al. Reported effects of the Scientific Registry of Transplant Recipients 5-tier rating system on US transplant centers: results of a national survey. Transplant International 2018; 31(10): 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wey A, Salkowski N, Kasiske BL, Israni AK, Snyder JJ. A five-tier system for improving the categorization of transplant program performance. Health services research 2018; 53(3): 1979–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Gordon M, Miller J, et al. Capturing Heterogeneity in Repeated Measures Data by Fusion Penalty. Statistics in Medicine 2021; 40: 1901–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox DR. Regression Models and Life Tables. Journal of the Royal Statistical Society B 1972; 34: 187–220. [Google Scholar]
- 14.Tibshirani R, Saunders M, Rosset S, Zhu J, Knight K. Sparsity and smoothness via the fused lasso. Journal of the Royal Statistical Society B 2005; 67: 91–108. [Google Scholar]
- 15.Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. Journal of the American Statistical Association 2001; 96: 1348–1360. [Google Scholar]
- 16.Tibshirani R The Lasso method for variable selection in the Cox model. Statistics in Medicine 1997; 16: 385–395. [DOI] [PubMed] [Google Scholar]
- 17.Ma S, Huang J. A concave pairwise fusion approach to subgroup analysis. Journal of the American Statistical Association 2017; 112: 410–423. [Google Scholar]
- 18.Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Foundations and Trends in Machine Learning 2011; 3: 1–122. [Google Scholar]
- 19.Hastie T, Tibshirani R. Generalized Additive Models. Chapman and Hall, London.. 1990. [DOI] [PubMed] [Google Scholar]
- 20.Gui J, Li H. Penalized Cox Regression Analysis in the High-Dimensional and Low Sample Size Settings, with Applications to Microarray Gene Expression Data. Bioinformatics 2005; 25: 3001–3008. [DOI] [PubMed] [Google Scholar]
- 21..Simon N, Friedman J, Hastie T, Tibshirani R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. Journal of Statistical Software 2011; 39: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Li B, Leng C. Shrinkage tuning parameter selection with a diverging number of parameters. Journal of Royal Statistical Society, Series B 2009; 71: 671–683. [Google Scholar]
- 23.Schwarz C Estimating the dimension of a model. The Annals of Statistics 1978; 6: 461–464. [Google Scholar]
- 24.Rand WM. Objective criteria for the evaluation of clustering methods. Journal of the American Statistics Association 1971; 66: 846–850. [Google Scholar]
- 25.He K, Schaubel DE. Methods for comparing center-specific survival outcomes using direct standardization. Statistics in Medicine 2014; 33(12): 2048–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Hong C, Palmer N, et al. A fast divide-and-conquer sparse Cox regression. Biostatistics 2021; 22: 381–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Lin L. Subgroup analysis for heterogeneous additive partially linear models and its application to car sales data. Computational Statistics and Data Analysis 2019; 138: 239–259. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Restrictions apply to the availability of these data, which were used under license for this study.


