Abstract
Large amount of electronic clinical data encompasses important information in free text format. To be able to help guide medical decision-making, text needs to be efficiently processed and coded. In this research, we investigate techniques to improve classification of Emergency Department computed tomography (CT) reports. The proposed system uses Natural Language Processing (NLP) to generate structured output from the reports and then machine learning techniques to code for the presence of clinically important injuries for traumatic orbital fracture victims. Topic modeling of the corpora is also utilized as an alternative representation of the patient reports. Our results show that both NLP and topic modeling improves raw text classification results. Within NLP features, filtering the codes using modifiers produces the best performance. Topic modeling shows mixed results. Topic vectors provide good dimensionality reduction and get comparable classification results as with NLP features. However, binary topic classification fails to improve upon raw text classification.
I. Introduction
The first computerized clinical record systems were implemented in mid 1960s [1] and thus far NLP has proven capable in coding electronic medical documents[2]. NLP translates raw text into coded descriptions suitable for computer-aided interpretation and has detected patients with pneumonia, anthrax, tuberculosis, and stroke[3].
Medical Language Extraction and Encoding(MedLEE) is the one of the most widely used NLP software in the medical research community[4], and has successfully interpreted findings from raw text procedure reports such as head CT imaging for stroke and chest radiography for pneumonia [5] [6]. A strength of the structured output of MedLEE is that it provides UMLS codes. UMLS is a repository of many controlled vocabularies in biomedical sciences developed by the US National Library of Medicine[7]. It is a comprehensive thesaurus and ontology of biomedical concepts consisting of 6.4 million unique terms for 1.3 million unique concepts from more than 119 families of biomedical vocabularies. MedLEE matches its findings to Concept Unique Identifiers (CUI)s from UMLS which increases interoperability of the system. CUIs add semantics to the system and fix the problems that could be caused by using different synonyms. This approach coincides with the fact that most biomedical concepts are represented as noun phrases and they do not span across sentences [8]. Other NLP tools, such as Stanford NLP [9] and OpenNLP[10], are not customized for medical terms and they lack modifiers. MetaMap[11] produces mappings to UMLS however it does not create modifiers other than negation. Clinical Text Analysis and Knowledge Extraction System (cTAKES) [12] also provides mappings to UMLS and it can identify the context (probability, history) and negation status; however, MedLEE has a wider range of modifier values.
Weka, or also known as Waikato Environment for Knowledge Analysis, [13] is used for machine learning approaches to classification of raw text and NLP output. Most of the misclassification errors have been shown to be due to the lack of temporal context, lack of location context, and lack of coreference resolution [12]. MedLEE output provides modifiers that help fix this problem. Classification of patient reports using MedLEE has previously been shown to be promising [14].
Topic modeling is an unsupervised technique that can automatically discover topics from a collection of documents. It has been used in variety of fields such as computer vision [15] and biology [16] and in applications such as information retrieval [17] and text segmentation[18]. Stanford Topic Modeling Toolbox [19] is an open source software that provides ways to train and infer topic models for text data. It supports different algorithms such as Latent Dirichlet Allocation (LDA) [20], labeled LDA [21] and Partially Labeled LDA [22].
II. Methodology
The data utilized in this study is from a recently published traumatic orbital fracture project that was a multi-year, multi-center investigation [23]. Using conventional methods, the investigators prospectively collected clinical data and outcomes on over 3,000 patients, including CT imaging reports. The primary approach of our system, as seen in Figure 1, is to take patient reports as input to the MedLEE to tag them with UMLS CUIs and modifiers that show the probability and temporal status. After this tagging process, the output is filtered to exclude findings with low certainties or findings linked with patient’s history or future modifiers. While our original study compared NLP-structured text and raw text, as an alternative approach, reports are also represented as topic vectors. These raw text files, NLP-filtered findings and topic vectors are then combined with their associated outcomes, and passed to the data mining tool WEKA 3.7.5[13] for classification using two well-known classification algorithms (decision tree and Support Vector Machine(SVM)).
Figure 1:
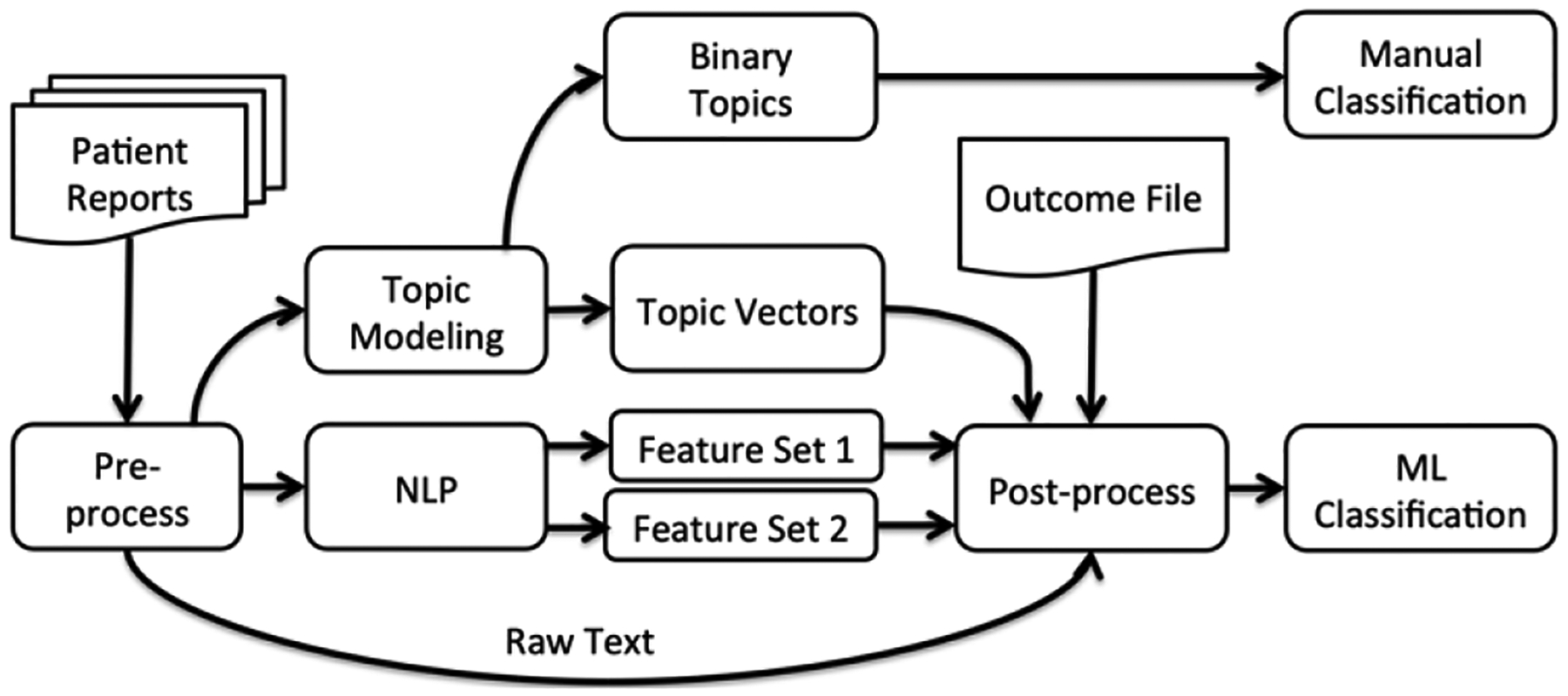
System Overview
In each algorithm, 10-fold stratified cross validation is performed where ten different classifiers are built by partitioning the initial corpus into ten disjoint sets. During each instance, nine sets are used for training and the remaining set for testing. At the end, performance metrics are averaged to get an overall result. By having it stratified, distribution of the class over the test and train sets is kept the same as the distribution of the class over the entire set. This ensures the generality of the classification model.
Topic modeling of the data with two topics is also analyzed as in Figure 1. In this approach, binary topics were assumed to correspond to the binary classes. During post-processing, each report is assigned to the topic with higher probability and no further classification task is needed.
Precision, recall, and F-score are used to evaluate the classification performance, and perplexity is used to compare topic modeling results with a varying number of topics. For binary classification, possible cases are summarized in Table I. Equations 1 and 2 present how these values are obtained based on the predicted class types.
Table I:
Outcomes for Binary Classification
| Predicted class | |||
|---|---|---|---|
| Positive | Negative | ||
| Actual Class | Positive | True Positive (TP) | False Negative (FN) |
| Negative | False Positive (FP) | True Negative (TN) | |
| (1) |
| (2) |
F-score is calculated as an equally weighted harmonic mean of precision and recall (See Equation 3):
| (3) |
Perplexity is calculated as shown in Equation 4:
| (4) |
where q is the learned probability model for an unknown probability distribution p and x1, x2, …, xN is the test set.
A. NLP
A detailed description of how MedLEE works has been previously described [3]. Briefly, for a given a block of raw text, MedLEE preprocessor splits the text into sentences, then does a lexical lookup to identify words, phrases, sentences, and abbreviations. Then, the parser utilizes a grammar to recognize syntactic and semantic patterns and generates intermediate forms, which consist of primary findings and different types of modifiers. Finally, words are composed into phrases and mapped into codes using a table[24]. To adapt MedLEE for other clinical investigations, its lexicon, abbreviations and section names can be changed to reflect the terms and organization seen in the documents to be interpreted. Figure 2 shows sample output.
Figure 2:

MedLEE output
B. Topic Modeling
Stanford Topic Modeling Toolbox (TMT) is used for topic modeling based on Latent Dirichlet Allocation (LDA) [20]. It uses either Gibbs Sampling [25] or collapsed variational Bayes approximation [26] to train topic models. Variational Bayes approximation is faster but requires more memory than Gibbs Sampling. In this research, Variational Bayes is used for training.
C. Text Classification
After considering many different classification algorithms, decision tree and SVM were chosen. Decision tree is preferred due to its explicit rule based output that can be easily evaluated for content validity, whereas SVM is known to perform well in text classification tasks[27]. SVMs are also known to be robust to over-fitting [28] and they are much faster than decision trees.
III. Experiments
A. Data Collection
Retrospective chart review of consecutive CT imaging reports for patients suffering traumatic orbital injury was obtained over 26 months from two urban hospitals. Staff radiologists dictated each CT report and the outcome of acute orbital fracture was extracted by a trained data abstractor. A random subset of 511 CT reports were double-coded, and inter-rater analysis revealed excellent agreement with Cohens kappa of 0.97 (95% CI 0.94–0.99). Among the 3,705 reports, 3,242 had negative outcome while 463 had positive.
B. Preprocessing
During preprocessing, all protected health information were removed to meet Institutional Review Board requirements. Medical record numbers from each report were replaced by observation numbers, which are sequence numbers automatically assigned to each report. Frequent words were also removed from the vocabulary to prevent it from getting too large. In addition, these frequent words typically do not add much information; most of them are stop words such as the, a, there, and, them ,are, and with. Other preprocessing tasks such as stemming and lower case conversion were also explored; however, classification performance was not affected. Finally, different weighting options are considered such as term frequency(tf), inverse term frequency(idf) and their combinations. Using the word counts produced slightly better classification results than other options.
C. NLP with MedLEE
We performed a test run of MedLEE to identify areas where modification was needed, and lexical expansion was performed to cover terms specific to the field of orbital fracture findings with their corresponding UMLS codes.
D. MedLEE Feature Selection
MedLEE output includes problems, findings, and procedures, associated with specific body locations and with modifiers that determine the certainty and the temporal status of each. After MedLEE is run, the output was further processed to include only the relevant information. Several approaches for extracting features from MedLEE output were considered. The first method extracts the problems with body locations with their corresponding UMLS codes that MedLEE had found. In the second approach, only the valid findings are used where status and certainty modifiers are verified.
E. Topic Vectors
As the alternative to structuring the raw text, reports were represented as topic vectors. Different number of topics is analyzed using Stanford TMT. This approach produced greater dimension reduction and made the classification significantly faster.
F. Postprocessing
The raw text of the reports, feature sets from NLP output and topic vectors were compiled into individual files in attribute relation file format (arff), where each line represents one report with its associated outcome. This file can then be loaded into Weka, where it is converted into a word vector representation and classified using the algorithms mentioned in Section II–C.
G. Binary Topic Classification
As an alternative approach to classification via SVM and decision tree, topic modeling with two topics was analyzed assuming each topic may correspond to a class. Data is split into randomly generated training and test sets. Once the model is learned on the training set, topic distributions are inferred on the test set.
IV. Results
Classification results using raw text and two different NLP outputs are compared in Table II. The first NLP output extracts UMLS codes corresponding to findings. The second NLP output filters these codes using the modifiers. Between the two post-processing approaches to creating NLP feature sets, filtered codes produce slightly better results. NLP classification results are similarly excellent using either decision trees or SVM.
Table II:
Classification using Text and NLP features
| Decision Tree | SVM | |||||
|---|---|---|---|---|---|---|
| Text | NLP | Text | NLP | |||
| All | Filtered | All | Filtered | |||
| Precision | 0.947 | 0.955 | 0.97 | 0.959 | 0.968 | 0.971 |
| Recall | 0.948 | 0.956 | 0.97 | 0.960 | 0.969 | 0.971 |
| F-Score | 0.948 | 0.955 | 0.97 | 0.959 | 0.968 | 0.971 |
Results for different number of topics are summarized in Table III and graphically illustrated in Figures 3, 4, and 5. First column shows the dimension reduction achieved over raw text representation. For comparison, there were 1,296 attributes in the bag of words representation of raw text and 1,371 attributes in the NLP feature set. Dimension reduction was calculated as shown in Equation 5:
| (5) |
Table III:
Classification with Topic Vectors
| Number of Topics | Dimension Reduction | Decision Tree | SVM | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F-score | Precision | Recall | F-score | ||
| 2 | 0.999 | 0.766 | 0.875 | 0.817 | 0.766 | 0.875 | 0.817 |
| 3 | 0.998 | 0.901 | 0.908 | 0.903 | 0.905 | 0.912 | 0.907 |
| 5 | 0.997 | 0.93 | 0.93 | 0.93 | 0.935 | 0.936 | 0.935 |
| 10 | 0.995 | 0.941 | 0.943 | 0.942 | 0.954 | 0.955 | 0.954 |
| 15 | 0.992 | 0.953 | 0.954 | 0.953 | 0.957 | 0.958 | 0.957 |
| 20 | 0.989 | 0.952 | 0.951 | 0.952 | 0.961 | 0.962 | 0.962 |
| 25 | 0.987 | 0.948 | 0.948 | 0.948 | 0.959 | 0.96 | 0.96 |
| 30 | 0.984 | 0.953 | 0.954 | 0.954 | 0.964 | 0.965 | 0.965 |
| 50 | 0.973 | 0.943 | 0.942 | 0.943 | 0.962 | 0.963 | 0.962 |
| 100 | 0.946 | 0.941 | 0.941 | 0.941 | 0.968 | 0.968 | 0.968 |
| 150 | 0.919 | 0.945 | 0.946 | 0.94 | 0.968 | 0.969 | 0.968 |
| 200 | 0.892 | 0.942 | 0.943 | 0.943 | 0.967 | 0.968 | 0.967 |
| 250 | 0.865 | 0.939 | 0.938 | 0.938 | 0.965 | 0.966 | 0.966 |
| 300 | 0.838 | 0.94 | 0.94 | 0.94 | 0.962 | 0.962 | 0.962 |
| 400 | 0.784 | 0.938 | 0.938 | 0.938 | 0.961 | 0.961 | 0.961 |
| 500 | 0.73 | 0.935 | 0.935 | 0.935 | 0.961 | 0.962 | 0.962 |
Figure 3:
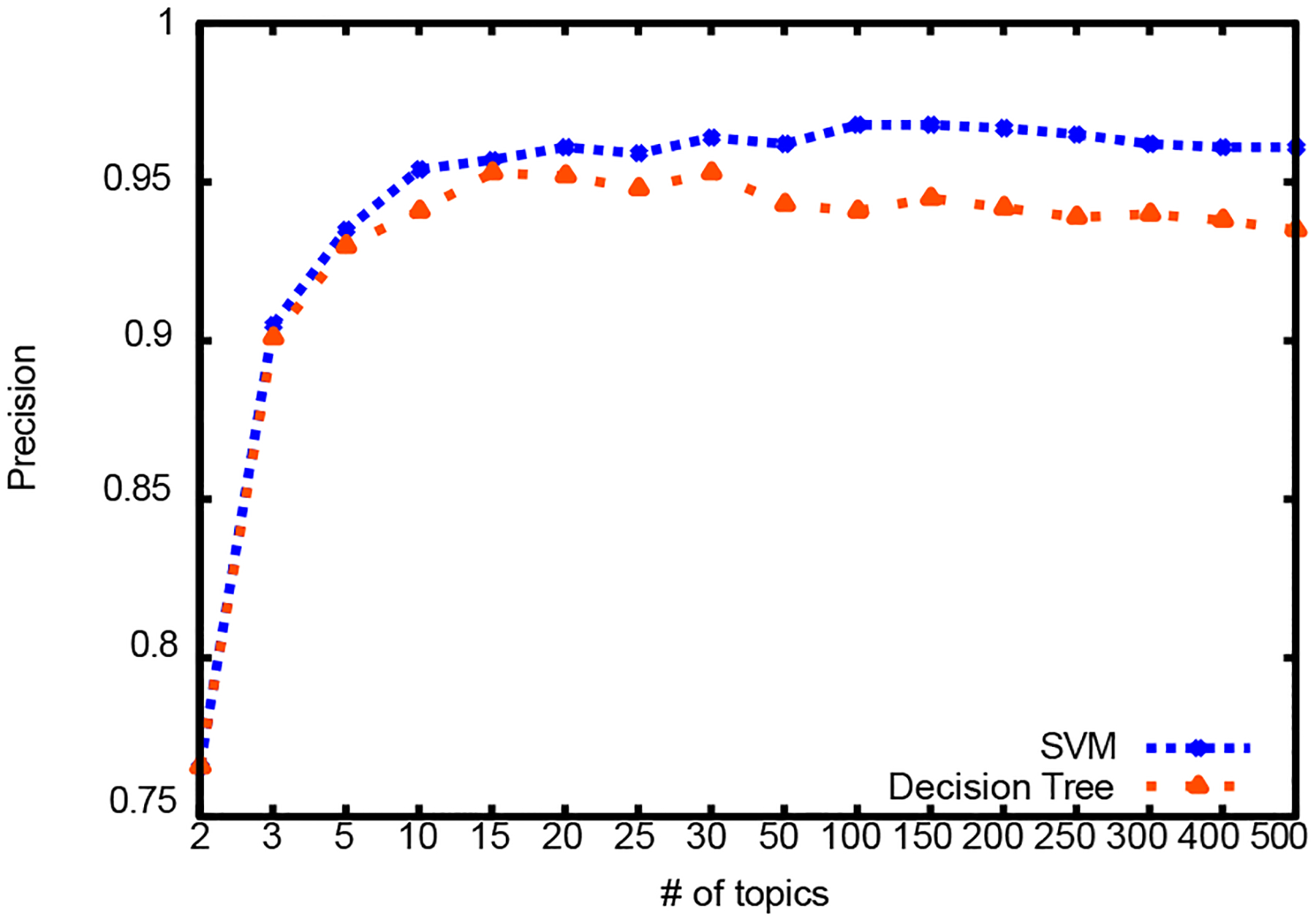
Precision
Figure 4:
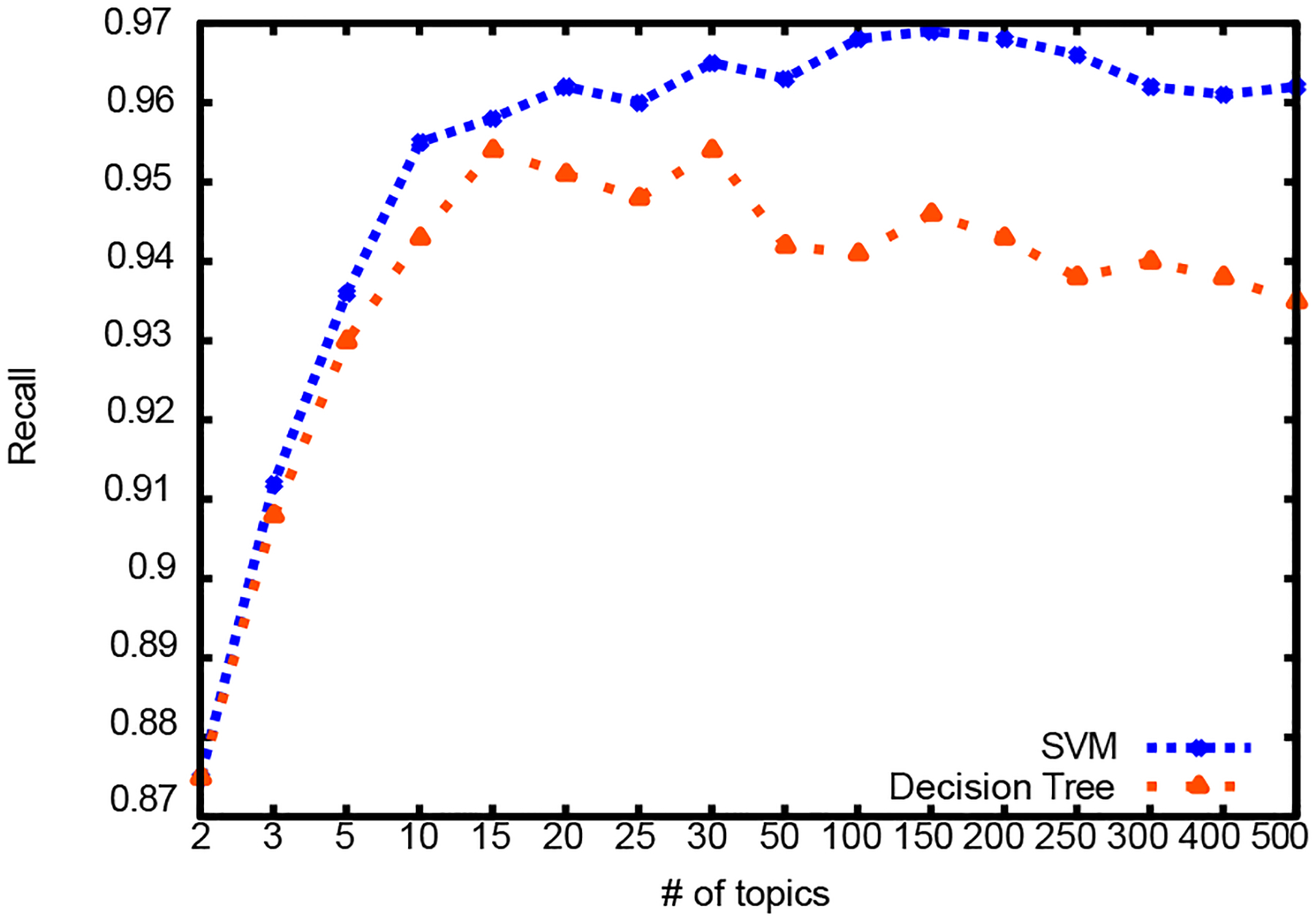
Recall
Figure 5:
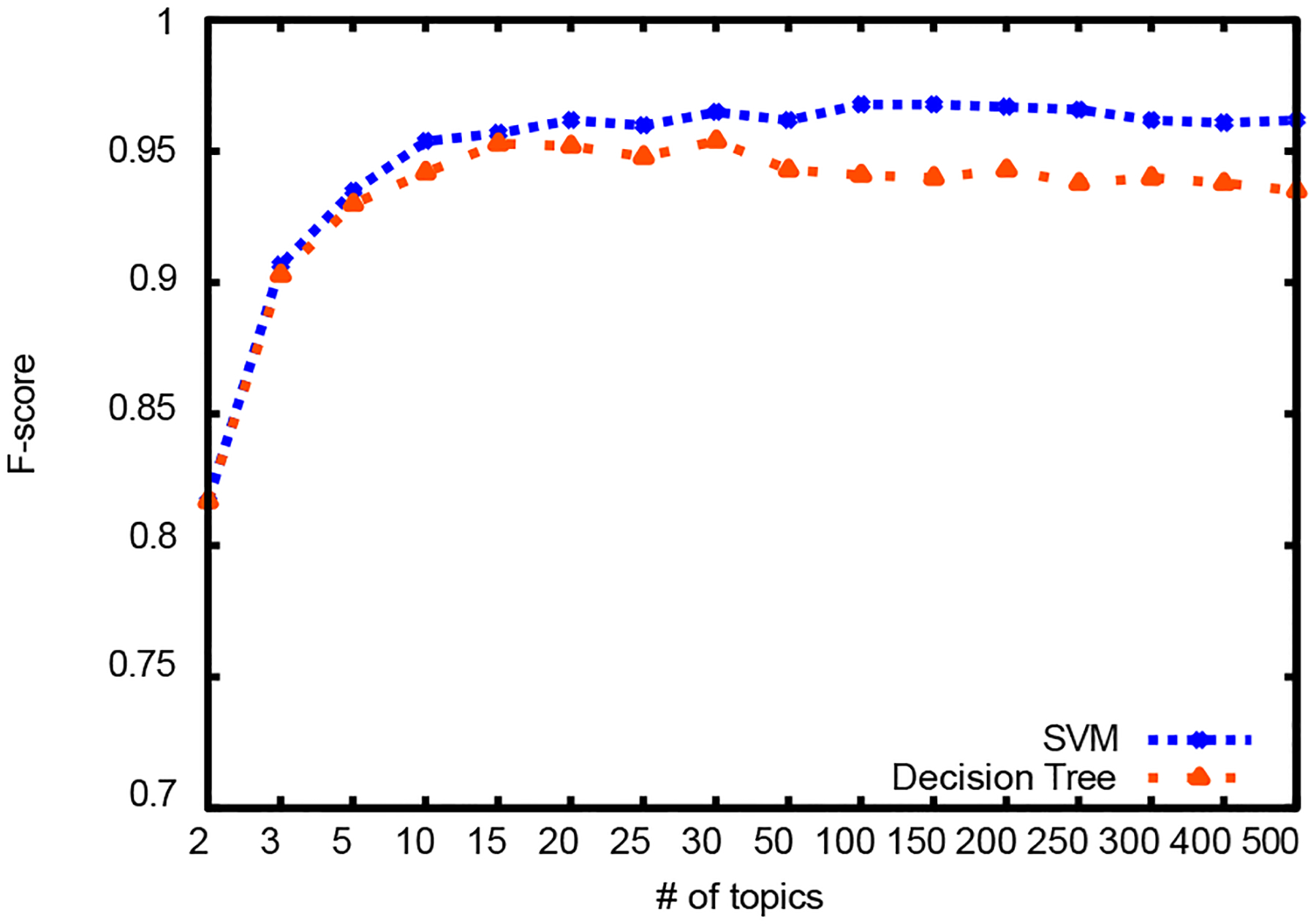
F-score
SVM classification outperforms decision tree classification and it gets comparable results with classification using NLP features. For decision tree classification, topic vectors with 15 and 30 topics produce the best result. For SVM, topic vectors with 100 and 150 topics produce the best result.
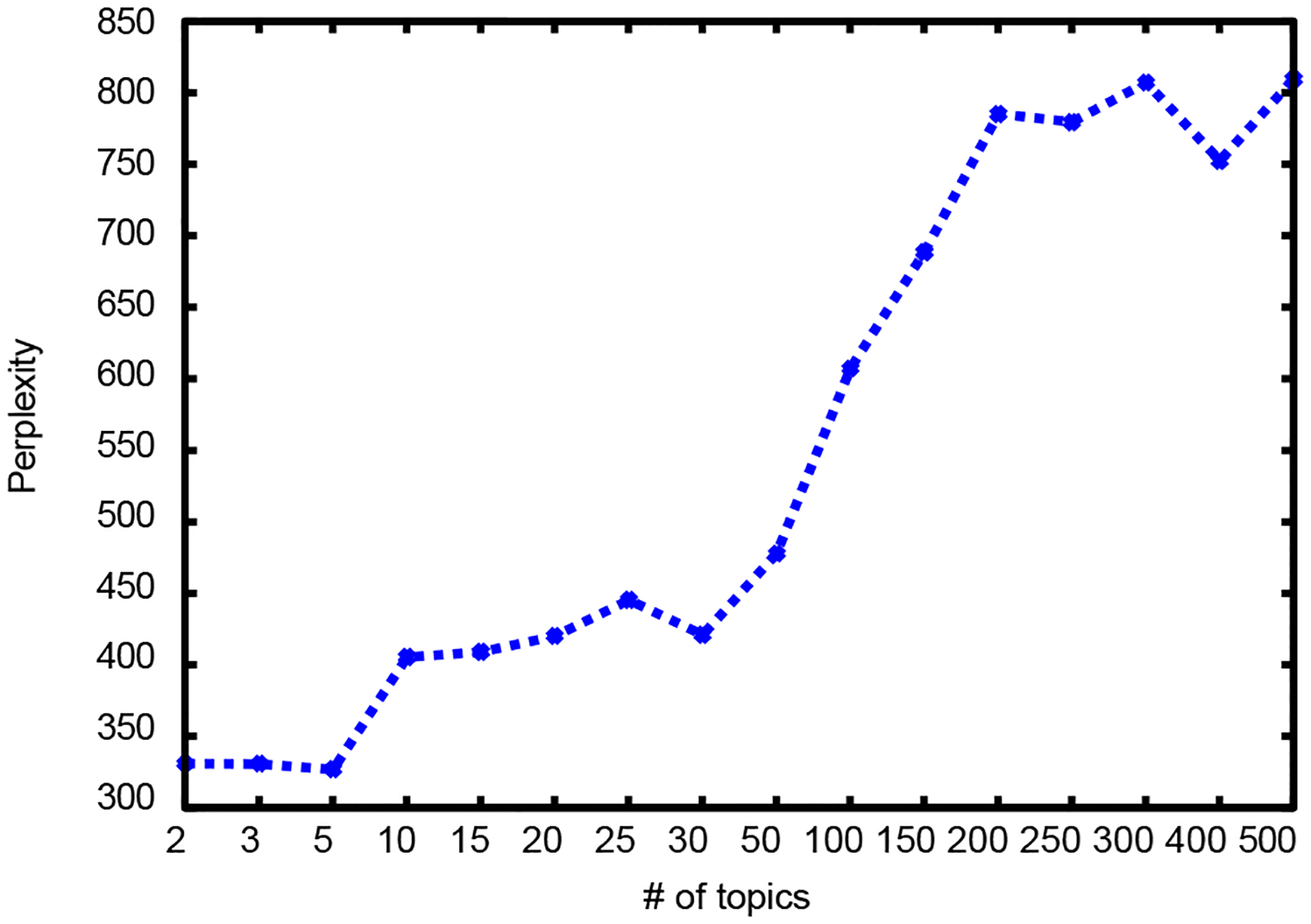
The topic modeling results are summarized in table IV and graphically illustrated in Figure 6. Perplexity is used as a measure to compare within the same corpora and it is expected that the lower the perplexity the better. However, the number of topics that provides the best classification performance does not have the best perplexity.
Table V:
Binary topics
| Topic # | Top words |
|---|---|
| 0 | acute, report, axial, facial, findings, mass, impression, fracture, intact, sinuses |
| 1 | left, right, maxillary, fracture, sinus, orbital, soft, fractures, facial, impression |
Figure 6:
Perplexity
A. Binary Topic Classification
The top ten words for each topic is shown in Table V. Topic #1 is assumed to be corresponding to the positive class and topic #0 is assumed to be corresponding to the negative class. Some words are shared between the topics and some words conflict with their assumed classes. Table VI shows the classification performances using different proportions of training and test sets. Results seem stable between different proportions of training and test sets but do not perform as well as other approaches described in this study.
Table IV:
Perplexity
| # of Topics | Perplexity |
|---|---|
| 2 | 331.28 |
| 3 | 331.08 |
| 5 | 327.29 |
| 10 | 405.86 |
| 15 | 409.40 |
| 20 | 420.44 |
| 25 | 445.69 |
| 30 | 421.42 |
| 50 | 478.33 |
| 100 | 607.61 |
| 150 | 688.97 |
| 200 | 785.48 |
| 250 | 779.81 |
| 300 | 807.37 |
| 400 | 752.77 |
| 500 | 809.88 |
Table VI:
Classification via Topic Modeling
| Test ratio (%) | Precision | Recall | F-Score |
|---|---|---|---|
| 25 | 0.91 | 0.73 | 0.80 |
| 33 | 0.90 | 0.73 | 0.78 |
| 50 | 0.90 | 0.73 | 0.78 |
| Average | 0.90 | 0.73 | 0.80 |
V. Conclusion
In this research, NLP and topic modeling are used to improve raw text classification of CT reports. Using NLP features improves classification results compared to the use of raw text. Within NLP features, filtering the codes using modifiers produces the best performance. Topic modeling shows mixed results. Topic vectors provide good dimensionality reduction and get comparable classification results as with NLP features. However, binary topic classification fails to improve upon raw text classification.
Contributor Information
Efsun Sarioglu, Computer Science Department The George Washington University, Washington, DC, USA.
Hyeong-Ah Choi, Computer Science Department The George Washington University, Washington, DC, USA.
Kabir Yadav, Department of Emergency Medicine The George Washington University, Washington, DC, USA.
References
- [1].Baruch JJ, “Progress In programming for processing English language medical records,” Annals of the New York Academy of Sciences, vol. 126, no. 2, pp. 795–804, 1965. [Online]. Available: 10.1111/j.1749-6632.1965.tb14324.x [DOI] [PubMed] [Google Scholar]
- [2].Stanfill MH, Williams M, Fenton SH, Jenders RA, and Hersh WR, “A systematic literature review of automated clinical coding and classification systems,” Journal of the American Medical Informatics Association, vol. 17, no. 6, pp. 646–651, 2010. [Online]. Available: http://jamia.bmj.com/content/17/6/646.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Friedman C, “A broad-coverage natural language processing system.” Proc AMIA Symp, pp. 270–274, 2000. [PMC free article] [PubMed] [Google Scholar]
- [4].Meystre SM, Savova GK, Kipper-Schuler KC, and Hurdle JF, “Extracting information from textual documents in the electronic health record: a review of recent research.” Yearb Med Inform, pp. 128–144, 2008. [PubMed] [Google Scholar]
- [5].Elkins JS, Friedman C, Boden-Albala B, Sacco RL, and Hripcsak G, “Coding neuroradiology reports for the Northern Manhattan Stroke Study: a comparison of natural language processing and manual review.” Comput Biomed Res, vol. 33, no. 1, pp. 1–10, Feb 2000. [DOI] [PubMed] [Google Scholar]
- [6].Hripcsak G, Kuperman G, Friedman C, and Heitjan D, “A reliability study for evaluating information extraction from radiology reports,” Journal of the American Medical Informatics Association, vol. 6, no. 2, p. 143, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].UMLS, “Unified Medical Language System Home Page,” http://www.nlm.nih.gov/research/umls/, 2012.
- [8].Huang Y, Lowe HJ, Klein D, and Cucina RJ, “Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS specialist lexicon.” J Am Med Inform Assoc, vol. 12, no. 3, pp. 275–285, May-Jun 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stanford NLP, “Stanford Natural Language Processing software,” http://nlp.stanford.edu/software/index.shtml, 2012.
- [10].Apache, “OpenNLP,” http://opennlp.apache.org/, 2012.
- [11].Aronson AR, “Effective mapping of biomedical text to the umls metathesaurus: the metamap program.” Proc AMIA Symp, pp. 17–21, 2001. [PMC free article] [PubMed] [Google Scholar]
- [12].Garla V, III VLR, Dorey-Stein Z, Kidwai F, Scotch M, Womack J, Justice A, and Brandt C, “The Yale cTAKES extensions for document classification: architecture and application.” JAMIA, vol. 18, no. 5, pp. 614–620, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, and Witten IH, “The WEKA data mining software: an update,” SIGKDD Explor. Newsl, vol. 11, no. 1, pp. 10–18, 2009. [Online]. Available: 10.1145/1656274.1656278 [DOI] [Google Scholar]
- [14].Sarioglu E, Yadav K, and Choi H-A, “Classification of Emergency Department CT Imaging Reports using Natural Language Processing and Machine Learning,” in AMIA Proceedings: Summits on Clinical Research Informatics, 2012, p. 150. [Google Scholar]
- [15].Li F-F and Perona P, “A bayesian hierarchical model for learning natural scene categories,” in Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05) - Volume 2 - Volume 02, ser. CVPR ‘05. Washington, DC, USA: IEEE Computer Society, 2005, pp. 524–531. [Online]. Available: 10.1109/CVPR.2005.16 [DOI] [Google Scholar]
- [16].Yeh J-H and Chen C-H, “Protein remote homology detection based on latent topic vector model,” in Networking and Information Technology (ICNIT), 2010 International Conference on, june 2010, pp. 456–460. [Google Scholar]
- [17].Wei X and Croft WB, “Lda-based document models for ad-hoc retrieval,” in Proceedings of the 29th annual international ACM SIGIR conference on Research and development in information retrieval, ser. SIGIR ‘06. New York, NY, USA: ACM, 2006, pp. 178–185. [Online]. Available: http://doi.acm.org/10.1145/1148170.1148204 [Google Scholar]
- [18].Misra H, Yvon F, Jose JM, and Cappe O, “Text segmentation via topic modeling: an analytical study,” in Proceedings of the 18th ACM conference on Information and knowledge management, ser. CIKM ‘09. New York, NY, USA: ACM, 2009, pp. 1553–1556. [Online]. Available: http://doi.acm.org/10.1145/1645953.1646170 [Google Scholar]
- [19].Ramage D and Rosen E, “Stanford Topic Modeling Toolbox,” 2009. [Online]. Available: http://nlp.stanford.edu/software/tmt/tmt-0.3/ [Google Scholar]
- [20].Blei DM, Ng AY, and Jordan MI, “Latent Dirichlet Allocation,” J. Mach. Learn. Res, vol. 3, pp. 993–1022, Mar. 2003. [Online]. Available: http://dl.acm.org/citation.cfm?id=944919.944937 [Google Scholar]
- [21].Ramage D, Hall D, Nallapati R, and Manning CD, “Labeled lda: a supervised topic model for credit attribution in multi-labeled corpora,” in Proceedings of the 2009 Conference on Empirical Methods in Natural Language Processing: Volume 1 - Volume 1, ser. EMNLP ‘09. Stroudsburg, PA, USA: Association for Computational Linguistics, 2009, pp. 248–256. [Online]. Available: http://dl.acm.org/citation.cfm?id=1699510.1699543 [Google Scholar]
- [22].Ramage D, Manning CD, and Dumais S, “Partially labeled topic models for interpretable text mining,” in Proceedings of the 17th ACM SIGKDD international conference on Knowledge discovery and data mining, ser. KDD ‘11. New York, NY, USA: ACM, 2011, pp. 457–465. [Online]. Available: http://doi.acm.org.proxygw.wrlc.org/10.1145/2020408.2020481 [Google Scholar]
- [23].Yadav K, E C, JS H, Z A, V N, and P G. et al., “Derivation of a clinical risk score for traumatic orbital fracture,” 2012, in Press. [DOI] [PubMed]
- [24].Friedman C, Alderson OP, Austin HJ, Cimino JJ, and Johnson S, “A General Natural-Language Text Processor for Clinical Radiology.” Journal of the American Medical Informatics Association, vol. 1, no. 2, pp. 161–174, Mar. 1994. [Online]. Available: 10.1136/jamia.1994.95236146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Griffiths TL and Steyvers M, “Finding scientific topics,” PNAS, vol. 101, no. suppl. 1, pp. 5228–5235, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Asuncion A, Welling M, Smyth P, and Teh Y-W, “On smoothing and inference for topic models,” in UAI, 2009. [Google Scholar]
- [27].Joachims T, “Text categorization with support vector machines: Learning with many relevant features,” 1998.
- [28].Sebastiani F, “Machine learning in automated text categorization,” ACM Comput. Surv, vol. 34, no. 1, pp. 1–47, Mar. 2002. [Online]. Available: http://doi.acm.org/10.1145/505282.505283 [Google Scholar]


