Abstract
Satellite glial cells (SGCs) are among the most abundant non-neuronal cells in dorsal root ganglia (DRGs) and closely envelop sensory neurons that detect painful stimuli. However, little is still known about their homeostatic activities and their contribution to pain. Using single-cell RNA sequencing (scRNA-seq), we were able to obtain a unique transcriptional profile for SGCs. We found enriched expression of the tissue inhibitor metalloproteinase 3 (TIMP3) and other metalloproteinases in SGCs. Small interfering RNA and neutralizing antibody experiments revealed that TIMP3 modulates somatosensory stimuli. TIMP3 expression decreased after paclitaxel treatment, and its rescue by delivery of a recombinant TIMP3 protein reversed and prevented paclitaxel-induced pain. We also established that paclitaxel directly impacts metalloproteinase signaling in cultured SGCs, which may be used to identify potential new treatments for pain. Therefore, our results reveal a metalloproteinase signaling pathway in SGCs for proper processing of somatosensory stimuli and potential discovery of novel pain treatments.
Keywords: Satellite glial cells, neuropathic pain, metalloproteinases, single-cell RNA sequencing
1. Introduction
Sensory neurons in dorsal root ganglia (DRGs) can detect and differentiate diverse sensations, including pain. Pain is a physiological reaction to hazardous stimuli that threaten homeostasis and survival. However, its abnormal processing - due to damage to the nervous system, cancer, diabetes, infection, autoimmune diseases, or chemotherapy - can result in pathological pain conditions (Basbaum et al., 2009; Scholz and Woolf, 2002). These states are often linked to debilitating chronic pain with symptoms like spontaneous pain, hyperalgesia and allodynia. Though sensory neurons and their related pathways have been viewed as main contributors to chronic pain, current clinical treatments targeting neurons have low analgesic efficacy and frequently have adverse effects (Berta et al., 2017; Grosser et al., 2017). It’s now evident that neurons are not the only ones driving various clinical pain states; non-neuronal cells in the nervous system, such as macrophages, T cells, and glial cells, also play important roles (Ji et al., 2013; Ji et al., 2016; Scholz and Woolf, 2007). Among these cells, satellite glial cells (SGCs) that envelop the soma of peripheral sensory neurons, have been overlooked but are gaining attention as novel players in pain and therapeutic targets (Hanani and Spray, 2020).
Satellite glial cells (SGCs) are abundant in peripheral ganglia, such as DRGs, and have multiple roles: providing metabolic and trophic support to sensory neurons, buffering extracellular ions and neurotransmitters, and enabling neuromodulation (Hanani, 2005; Hanani and Spray, 2020). SGCs have characteristics which make them ideal partners with sensory neurons and immune cells: they respond quickly to changes in neuron activity, they express neurotransmitter receptors and GPCRs, and they can respond to the release of mediators such as ATP, TNF, and IL-1β (Berta et al., 2012; Gu et al., 2010; Kim et al., 2016; Suadicani et al., 2010). These communications maintain neuronal homeostasis (Hanani, 2005; Pannese, 2010) and enable optimized responses to external stimuli. However, excessive or inappropriate communication might lead to excessive inflammatory and pain responses after injury (Grace et al., 2014; Ji et al., 2014). Indeed, injury can not only sensitize neurons, but also activate SGCs in sensory ganglia, resulting in exaggerated pain sensitivity and chronic pain (Berta et al., 2017; Ji et al., 2013). SGC mediators have been shown to powerfully modulate neuronal functions in acute and chronic pain, and to reduce the effectiveness of opioid analgesia (Berta et al., 2012). Therefore, a better understanding of SGC biology and functions could lead to new therapeutic options for pain treatment.
Here, we have taken advantage of advances in single cell technologies, which allow us to investigate the transcriptomes of nearly all cell populations in the nervous system (Shapiro et al., 2013), to better understand the biology and functions of SGCs in DRGs. These technologies have already provided valuable insights into the transcriptomes of neuronal and non-neuronal cells in DRGs, but most of them have focused on the heterogeneity of peripheral sensory neurons and neuronal regeneration (Avraham et al., 2020; Renthal et al., 2020; Sharma et al., 2020). What has been largely overlooked in these studies is the heterogeneity of SGCs and the identification of new protein signaling pathways related to pain. Using single-cell RNA sequencing (scRNA-seq), we identified two major subpopulations of SGCs, one of which was enriched in genes associated with metalloproteinase signaling, including the tissue inhibitor metalloproteinase 3 (TIMP3). Changes in TIMP3 expression and function in vivo influenced modulation of somatosensory stimuli and chemotherapy-induced pain, and in vitro modulation of metalloproteinase signaling in SGCs can be used to screen and identify novel drugs for pain. Our findings reveal a previously unknown molecular heterogeneity of SGCs and metalloproteinase signaling for the maintenance of homeostasis in DRG tissues and the discovery of new pain treatments.
2. Materials and Methods
2.1. Animal and human studies
All animal experiments conformed to the guidelines recommended by the National Institute of Health, the International Association for the Study of Pain, and the National Centre for the Replacement, Refinement, and Reduction of Animals in Research ARRIVE guidelines. These experiments were approved by the Institutional Animal Care and Use Committee at the University of Cincinnati (UC). Experiments using mice began at 8–10 weeks of age for both male and female CD1 mice (Cat# 022, Charles River Laboratories, Wilmington, MA). RNAscope experiments used Plp1-tdTomato mice, which were generated by crossing the inducible Plip1-Cre/ERT mice (Cat# 005975) with the Rosa26-LoxP-STOP-LoxP-tdTomato mice (Cat# 007909) from the Jackson Laboratory (Bar Harbor, ME). Recombination was induced by oral delivery of a 200 μl tamoxifen/corn oil solution (20 mg/ml) for three consecutive days one week prior to RNAscope. Mice were housed four per cage at 22 ± 0.5°C under a controlled 14/10 hours light/dark cycle with free access to food and water. Studies involving human DRG tissues were performed on deidentified biospecimens and approved by the UC Institutional Review Boards.
2.2. DRG tissue dissociation
Adult mice were anesthetized with isoflurane and DRGs were pooled from three male mice from the cervical (C1–C8), thoracic (T1–T13), and lumbar (L1–L6) segments. The DRGs were dissociated according to an adapted protocol (Malin et al., 2007). DRGs were bilaterally removed and placed in ice-cold Hank’s balanced salt solution (HBSS, Thermo Fisher Scientific, Waltham, MA) supplemented with 1% pen/strep and 1% HEPES, then finely dissected to remove all nerve roots. The DRGs were incubated in papain in HBSS (60 U/ml, cat# P3125, Millipore Sigma, St. Louis, MO) for 20 min at 37°C, then in collagenase in HBSS (1 mg/ml, cat# C6885, Millipore Sigma) for another 20 min at 37°C. They were triturated with 1 and 200 μl pipettes, passed through a 40-μm filter, and centrifuged through 15% bovine serum albumin (BSA) solution to remove cell doublets and debris. DRG cells were resuspended in PBS-BSA 0.01% and a small portion counterstained with trypan blue to count viable cells with a hemocytometer. Cells were diluted at 1000 cells/μl and considered as passing quality control if >90% were viable and virtually no cell debris was visible.
2.3. Single-cell RNA library preparation, sequencing, and analyses
Single-cell RNA sequencing was conducted by the Gene Expression Core at Cincinnati Children’s Hospital using the 10x Chromium Single Cell Kit (v3, 10x Genomics, Pleasanton, CA). Approximately 15,000 single cells were loaded onto the microfluidic chip. Downstream processes included reverse transcription, cDNA synthesis/amplification, and library preparation according to the manufacturer’s instructions. The library was sequenced on a NovaSeq 6000 System (Illumina, San Diego, CA). An expression matrix was obtained using 10X’s CellRanger pipeline, imported into R, and analyzed with Seurat V3 package (Stuart et al., 2019). Quality control filters included removing cells with a total number of expressed genes outside the range 200–7,000 and more than 10% of reads mapping to mitochondrial genes. A total of 10,172 cells were selected for principal component analysis and visualized by Uniform Manifold Approximation and Projection (UMAP). Clusters were identified at various resolutions using the Louvain modularity optimization as implemented in Seurat V3 package. Optimal clustering resolution was obtained by exploring gene expressions of well-known cellular marker genes (Suppl. Table 1). Differentially expressed genes, defined by having a fold change greater than 0.25 and an adjusted p-value below 0.05, were identified in Seurat V3 package. Subsequently, the data was analyzed for enrichment of GO terms using Enrichr (Kuleshov et al., 2016) and a background gene list including all genes detected in our sequencing (Suppl. Table 1).
2.4. RNAscope (fluorescent in situ hybridization)
Mice were anesthetized with isoflurane and transcardially perfused with 20 ml of PBS followed by 20 ml of 4% paraformaldehyde in PBS at 4 °C. The lumbar L3–L5 DRG were then removed, post-fixed in the same fixative for 2 h, and placed in 30% sucrose solution overnight at 4 °C. Sections of the tissue were cut in a cryostat (12 μm) and mounted onto Superfrost Plus slides (Thermo Fisher Scientific). The slides were dried for 15 min at room temperature and stored at −80 °C until processing. RNA was detected using the RNAscope Multiplex Fluorescent V2 Assay (Advanced Cell Diagnostics, Newark, CA) with the omission of the initial on-slide fixation step. Probes used included Mm-Gja1-C4 (Cat# 486191-C4), Mm-Entpd2-C2 (Cat# 479511), Mm-Timp3-C2 (Cat# 425321-C2), Mm-Mmp14 (Cat# 432001) and Mm-Adam17 (Cat# 418011). After probe incubation and fluorescence signal amplification, slides were incubated in 1:5000 DAPI (Cat# D1306, Thermo Fisher Scientific) in 0.1M PB for 1 min before washing, air-drying, and coverslipping with Prolong Gold Antifade mounting medium (Thermo Fisher Scientific). Images were acquired using a Keyence BZ-X800 microscope and analyzed using NIH Image J open source software (Schindelin et al., 2012).
2.5. Drugs and intrathecal administration
We purchased recombinant human TIMP3 (Cat# 973-TM) and prosaptide Tx14 (Cat# 5151) from R&D Systems (Minneapolis, MN); recombinant mouse TIMP-1 (Cat# 593702) from BioLegend (San Diego, CA); an MMP14 inhibitor (NSC405020, Cat# 444295), MMP2 inhibitor (Cat# 444288), TACE/ADAM17 inhibitor (TAPI-2, Cat# 4444244), and pioglitazone hydrochloride (Cat# E6910) from Millipore Sigma; a Timp3 polyclonal antibody (Cat# ab39184) from Abcam (Cambridge, MA); and a rabbit IgG control (Cat# 12-370) from Millipore Sigma. Additionally, we purchased mouse Timp3-targeting siRNA (Cat# s75166) and non-targeting siRNA (Cat# 4390844) from Thermo Fisher Scientific. These drugs were administered intrathecally (i.t.) via a spinal puncture to deliver reagent into cerebral spinal fluid and DRG tissues, as reported in our previous publications (Lee et al., 2020; Tonello et al., 2019; Tonello et al., 2020). A valid spinal puncture and intrathecal delivery of these drugs was confirmed by a reflexive tail flick after the needle entry into the subarachnoid space.
2.6. Gene knockdown by small interfering RNA (siRNA)
A total of 3 μg of siRNA targeting Timp3 or a non-targeting control siRNA were diluted in a 10 μl solution containing 2.62 μl of in vivo-jetPEI (Cat# 201-10G, Polyplus, New York, NY), and 5% glucose, and injected intrathecally (i.t.) twice (with an interval of 6 h between injections) on day 0. On day 2 following siRNA administration, we assessed the effect of Timp3 knockdown in mice using mechanical and thermal (heat and cold) sensitivity assays, as well as a motor performance test (see section 2.7 Mouse model and behavioral assays). To initially assess the specificity of Timp3 siRNA over other TIMPs in vitro, we transfected SGC cultures (see section 2.9 Satellite glial cell culture) on day 10 with 25 nM of Timp3 or control siRNA for 48h. Transfection was achieved by mixing 10 μM of siRNA with 250 μl OptiMEM media (Cat# 31985062, Thermo Fisher Scientific) containing 7.5 μl TransIT‐X2 (Cat# MIR 6010, Mirus Bio, Madison, WI). To then assess the in vivo specificity and involvement of various downstream pathways associated with mechanical hypersensitivity in Timp3 knockdown mice, we injected an exogenous recombinant TIMP3 (100 ng/site, i.t.), a general endogenous tissue inhibitor of MMPs (TIMP-1, 4 pmol/site, i.t.), MMP2 and MMP14 inhibitors (10 μg/site, i.t.), TACE/ADAM17 inhibitor (TAPI-2, 1 μg/site, i.t.), and a neutralizing antibody for TNF-α (5 μg/site, i.t.).
2.7. Mouse model of neuropathic pain
To produce an animal model of chemotherapy-induced neuropathic pain (CINP), mice were treated with paclitaxel (cat# T7402, Millipore Sigma), as previously described (Tonello et al., 2019; Tonello et al., 2020). In brief, a 6 mg/mL stock of paclitaxel was diluted with Cremophor EL and 95% dehydrated ethanol (1:1 ratio), and administered intraperitoneally at a dosage of 2 mg/kg in saline, every other day for a total of 4 injections. Control animals received an equivalent volume of a vehicle containing proportional amounts of Cremophor EL and 95% dehydrated ethanol, also diluted in saline. Signs of peripheral neuropathy similar to those found in patients have been validated by numerous investigators in this non-tumor-bearing animal model, including the time-dependent development of mechanical and cold allodynia (Flatters et al., 2017; Li et al., 2015). All experiments in naïve and paclitaxel-treated mice were performed by blinded observers, and no more than three assays were performed each testing day.
2.8. Behavioral assays
Mechanical sensitivity (von Frey assay) - Paw withdrawal thresholds to mechanical stimuli were evaluated using a calibrated von Frey monofilament series (Stoelting Co., Wood Dale, IL). Mice were first acclimated in individual, clear Plexiglas boxes on an elevated wire mesh platform to access the hind paws’ plantar surface. A series of von Frey monofilaments (0.02, 0.07, 0.16, 0.4, 0.6, 1.0, and 1.4 g) was applied perpendicularly to the hindpaw’s plantar surface. The test began with the 0.6 g hair. A positive response was a clear paw withdrawal or shaking. Whenever a positive response occurred, the next lower hair was applied, and whenever a negative response occurred, the next higher hair was applied. The testing consisted of 6 up-and-down stimuli and the pattern of response was converted to a 50% von Frey threshold (Chaplan et al., 1994). The anti-allodynic effect of each drug was quantified with respect to the area under the threshold-time curve (trapezoidal method) over the 24 h post-injection testing period. Data are reported as a percentage of the maximum possible anti-allodynia, calculated as a ratio of actual anti-allodynia to a hypothetical situation where the drug brought withdrawal thresholds to their original baseline at all post-injection time points (Sorge et al., 2015).
Heat sensitivity (Hargreaves assay) - The Plantar Test apparatus (IITC Life Science, Woodland Hills, CA) was used to assess paw withdrawal latency in response to heat stimulation, according to the Hargreaves method (Hargreaves et al., 1988). Animals were first acclimated in plastic observation boxes for 20 minutes. Then, the mid-plantar surface of the mouse hindpaw was exposed to a radiant heat source through a glass floor until paw withdrawal. The intensity of the heat was adjusted to produce a baseline of about 15 seconds in naïve mice, with a maximum cutoff of 20 seconds. Each mouse was tested three times, with at least two minutes between recordings. The average of all recordings was used for statistical analysis.
Cold sensitivity (Dry ice assay) - Mice were acclimated to the Plantar Test apparatus (IITC Life Science) for 20 minutes before cold stimulation with dry ice, according to a previously described protocol (Brenner et al., 2012). A 10 ml syringe, sectioned above the Leur lock and tightly packed with finely crushed dry ice, was pressed firmly on the bottom of the tempered glass directly below the hindpaw. The paw withdrawal latency of the mice was measured with a stopwatch. A standard baseline of 20 seconds was observed in naïve mice, with a maximum cutoff of 30 seconds. All mice were tested three times, with at least 5 minutes between recordings. The average of all recordings was used for statistical analysis.
Motor performance (Rotarod assay) - Motor performance was assessed using a Rotarod Treadmill from IITC Life Science. Mice received two consecutive days of training before testing. The latency to fall was measured with an accelerated rotation speed from 5 to 35 rpm over 3 minutes. All mice were tested three times with at least 10 minutes between recordings and the average of all recordings was used for statistical analysis.
2.9. Satellite glial cell cultures
Freshly dissociated DRG tissues were used to prepare SGC cultures. Mice were anesthetized with isoflurane and lumbar, thoracic and cervical DRG tissues were aseptically removed. DRG tissues were then incubated in papain (60 U/ml, Cat# P3125, Millipore Sigma) for 30 minutes at 37°C followed by collagenase (1 mg/ml, Cat# C6885, Millipore Sigma) for another 30 minutes at 37°C. DRG cells were dissociated using a 1 ml and then a 200 μl pipette tip, filtered through a 40 μm and then a 10 μm cell strainer and cultured in DMEM low glucose media (Cat# 12320032, Thermo Fisher Scientific) with 10% fetal bovine serum, 1% pen/strep and 1% fungizone (Cat# 15290018, Thermo Fisher Scientific). Cultures were maintained at 37°C with 5% carbon dioxide for around 10 days before use. Using this method, we were able to achieve a purity greater than 90% for SGCs. This was assessed by colocalizing glutamine synthetase (GS) with DAPI staining (data not shown).
2.10. Cell culture viability
Cell cultures were assessed for viability 24 hours after exposure to vehicle or paclitaxel using the PrestoBlue Cell Viability Reagent (Cat# A13261, Thermo Fisher Scientific). Briefly, 50 μL was added to the culture media for 30 minutes, and 200 μL of media was collected for absorbance quantification. Absorbance was measured at 600 nm using the EnVision plate reader (PerkinElmer, Waltham, MA).
2.11. Immunofluorescence
DRG tissues and SGC cultures - DRG tissues were dissected and fixed with 4% paraformaldehyde (PFA) for 2 hours. Tissues were then cryoprotected in 30% sucrose and sectioned at 12 μm. Cell cultures were fixed with 4% PFA for 15 minutes and were processed immediately for immunofluorescence. Tissues and cultures were washed in PBS, blocked with 1% BSA, 0.2% Triton X-100 in PBS (BSA solution), and incubated in the BSA solution overnight at 4 °C with the following primary antibodies: TIMP3 Ab (1:500, Cat# MAB973, R&D Systems), FABP7 Ab (1:1000, Cat# PA524949, Thermo Fisher Scientific) or GFAP Ab (1:1000, Cat# MAB360, Millipore Sigma). Tissues and cultures were then incubated with appropriate secondary antibodies conjugated to Alexa Fluor 546 (1:1000, Thermo Fisher Scientific) for 1 hour at room temperature in BSA solution, followed by DAPI (Thermo Fisher Scientific) in 0.1M PB for 1 minute at room temperature. Finally, tissues and cultures were air-dried and coverslipped with Prolong Gold Antifade mounting medium (Thermo Fisher Scientific). Images were acquired using a Keyence BZ-X800 microscope, and images were analyzed using NIH Image J open source software (Schindelin et al., 2012).
Skin tissues – Biopsies from the plantar surface of the hind paws of adult mice were dissected, fixed with 4% PFA for 2 hours, and cryoprotected in 30% sucrose overnight. The tissues were then sliced into 40-μm sections and collected in PBS solution for free-floating staining. Sections were blocked for 1 hour at room temperature in BSA solution and incubated with primary antibodies against the pan-neuronal marker PGP9.5 (1:500; Cat# Z511601-2; Agilent, Santa Clara, CA) overnight at 4°C. Following this, the sections were incubated with the secondary antibody Alexa Fluor 488 (1:1000; Thermo Fisher Scientific) for 1 hour at room temperature, and DAPI was used for counterstaining. A Keyence BZ-X800 microscope was used to acquire images from multiple sections of skin tissues, which were selected at random. Intradermal neuronal fibers (IENFs) - nerve fibers that crossed the dermal/epidermal junction into the epidermis - were analyzed using NIH Image J open source software (Schindelin et al., 2012). The density of the IENFs was determined as the total number of fibers/length of epidermis (IENFs/cm), as previously described (Tonello et al., 2019).
2.12. Western blotting
Western blot was used to validate the tissue-specific knockdown of TIMP3 in both DRG and spinal cord tissues. The tissues were quickly dissected from adult mice, homogenized in RIPA lysis buffer (Millipore Sigma, Cat# 20-188) containing MS-SAFE protease and phosphate inhibitors (Millipore Sigma, Cat# MSSAFE). Protein concentration was measured with Qubit protein assay kit (Thermo Fisher Scientific, Cat# Q33211) and 20 μg of protein was loaded for each lane. Proteins were separated using NuPAGE™ 4–12% Bis-Tris Protein Gel (Thermo Fisher Scientific, Cat# NP0323BOX) and transferred to PVDF membranes. Blocking was done with 2% BSA followed by overnight incubation of TIMP3 antibody (R&D Systems, Cat# MAB973, 1:500) and GAPDH antibody (Cell Signaling, Cat# 3683S, 1:1000) at 4 °C. After that, the membranes were incubated with appropriate HRP-conjugated secondary antibody (Cell Signaling, 1:2000). Blots were detected using chemiluminescence method (SuperSignal™ West Dura Extended Duration Substrate, Thermo Fisher Scientific, Cat# 34075) and scanned with iBrightTM FL1000 Imaging System (Thermo Fisher Scientific). Bands were quantified using NIH software Image J (Schindelin et al., 2012) and relative values of each protein were normalized by GAPDH protein levels.
2.13. Reverse transcription polymerase chain reaction (RT-PCR)
DRG tissues were dissected and stored in TRIzol reagent (Cat# 15596018, Thermo Fisher Scientific). Total RNA was extracted using Direct-zol RNA MiniPrep kit (Cat# R2052, Zymo Research, Irvine, CA). The amount and quality were assessed by SimpliNano UV-Vis Spectrophotometer (General Electric, Boston, MA) and then converted to cDNA using a high-capacity cDNA reverse transcription kit (Cat# 4368814, Thermo Fisher Scientific). Samples were diluted 2:100 for RT-PCR experiments. The following primer pairs were used: mouse Timp3 (forward, 5′-CCGAGGCTTCAGTAAGATGC-3′; reverse, 5′-CCTCTCCACAAAGTTGCACA-3′), mouse Gapdh (forward, 5′-TGAAGGTCGGTGTGAACGAATT-3′; reverse, 5′-GCTTTCTCCATGGTGGTGAAGA-3′), human TIMP3 (forward, 5′-CCTGCTACTACCTGCCTTGC-3′; reverse, 5′-GGTCTGTGGCATTGATGATG-3′) and human GAPDH (forward, 5′-ACCCAGAAGACTGTGGATGG-3′; reverse, 5′-TTCTAGACGGCAGGTCAGGT-3′). The RT-PCR products were visualized on 1.5% agarose gels with SYBR Safe DNA Gel Stain.
2.14. Quantitative Real-Time RT-PCR (qPCR)
Dissected DRG tissues and cell cultures were stored in TRIzol reagent (Cat# 15596018, Thermo Fisher Scientific). Total RNA was extracted, quantified, assessed for quality, and converted into cDNA according to the RT-PCR method. Primers were obtained from PrimerBank (Wang et al., 2012) and are listed in Suppl. Table 3. qPCR was performed on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific) using PowerUp SYBR Green Master Mix (Cat# A25741, Thermo Fisher Scientific). All samples were normalized by Gapdh expression. Relative transcriptional expression ratios per condition were calculated using the Pfaffl method (Pfaffl, 2001).
2.15. Enzyme linked immunosorbent assay (ELISA)
The ELISA kit for mouse TIMP3 was purchased from MyBioSource (Cat# MBS703363) and used according to the manufacturer’s protocol. Cell cultures were harvested 24 hours after incubation with paclitaxel (300nM) alone or in combination with pioglitazone (10μM), and homogenized in RIPA lysis buffer (Cat# 89900, Thermo Fisher Scientific) containing protease and phosphatase inhibitors. Initial protein concentrations were measured using Qubit TM (Thermo Fisher Scientific).
2.16. Statistical analysis
Prism 9.0 (GraphPad, San Diego, CA) was used for statistical analysis. Sample sizes were designed to generate 80% power at two-sided P<0.05. Specific statistical tests used are specified in each figure legend. Generally, an unpaired, two-tailed Student’s t-test was used for analyses between two groups, whereas one-way or two-way ANOVA with a specific multiple-comparisons test was used for multiple groups and/or conditions. Data are presented as mean ± s.e.m., with values derived from independent biological replicates. Technical replicates and data transformations are described in the specific methods (e.g. the description of data transformation for anti-allodynic effect is in the behavioral assays section). Adobe Illustrator 25.0 (Adobe, San Jose, CA) was used for illustrations and figure organization.
3. Results
3.1. Single-cell RNA sequencing can identify the major non-neuronal cell types present in DRG tissues
Research in the past has primarily focused on the diversity and function of DRG neurons (Li et al., 2016; Sharma et al., 2020; Usoskin et al., 2015), yet the diversity and function of non-neuronal cells surrounding these neurons has received less attention. To characterize these non-neuronal cells, we performed single cell RNA sequencing of cervical, thoracic, and lumbar DRGs pooled from three naïve mice, using the Chromium Single Cell Gene Expression Solution (10x Genomics) and unbiased uniform manifold approximation and projection (UMAP) (Becht et al., 2018) analysis (Fig. 1A). A total of 10,172 single DRG cells were considered (see filtering details in Methods and Suppl. Fig. 1) and organized into 12 distinct clusters (c00–c11, Fig. 1B), with a similar number of unique molecular identifiers (UMIs) and an average of around 2000 detected genes for each cluster (Fig. 1C). Each cluster was classified as a cell subtype based on its expression of unique cell markers (Fig. 1D and Suppl. Table 1), which were compared to previous studies that have described canonical markers and functions for individual somatosensory neuron subtypes and characterized non-neuronal cells in the nervous system (Suppl. Fig. 2A and 2B). This cell-type classification is consistent with previous RNA-seq of DRG cells and nuclei (Avraham et al., 2020; Renthal et al., 2020; Zeisel et al., 2018). Mapping of well-known marker genes onto the UMAP plots revealed clusters of both neuronal cells (c00 and c10, expression of Tubb3) and non-neuronal cells (c01–c09 and c11, expression of B2m), including macrophages (c07, expression of Aif1) and glial cells (c01–c03, expression of Sox10) (Fig. 1E).
Fig. 1. Cellular heterogeneity of the DRG tissues.
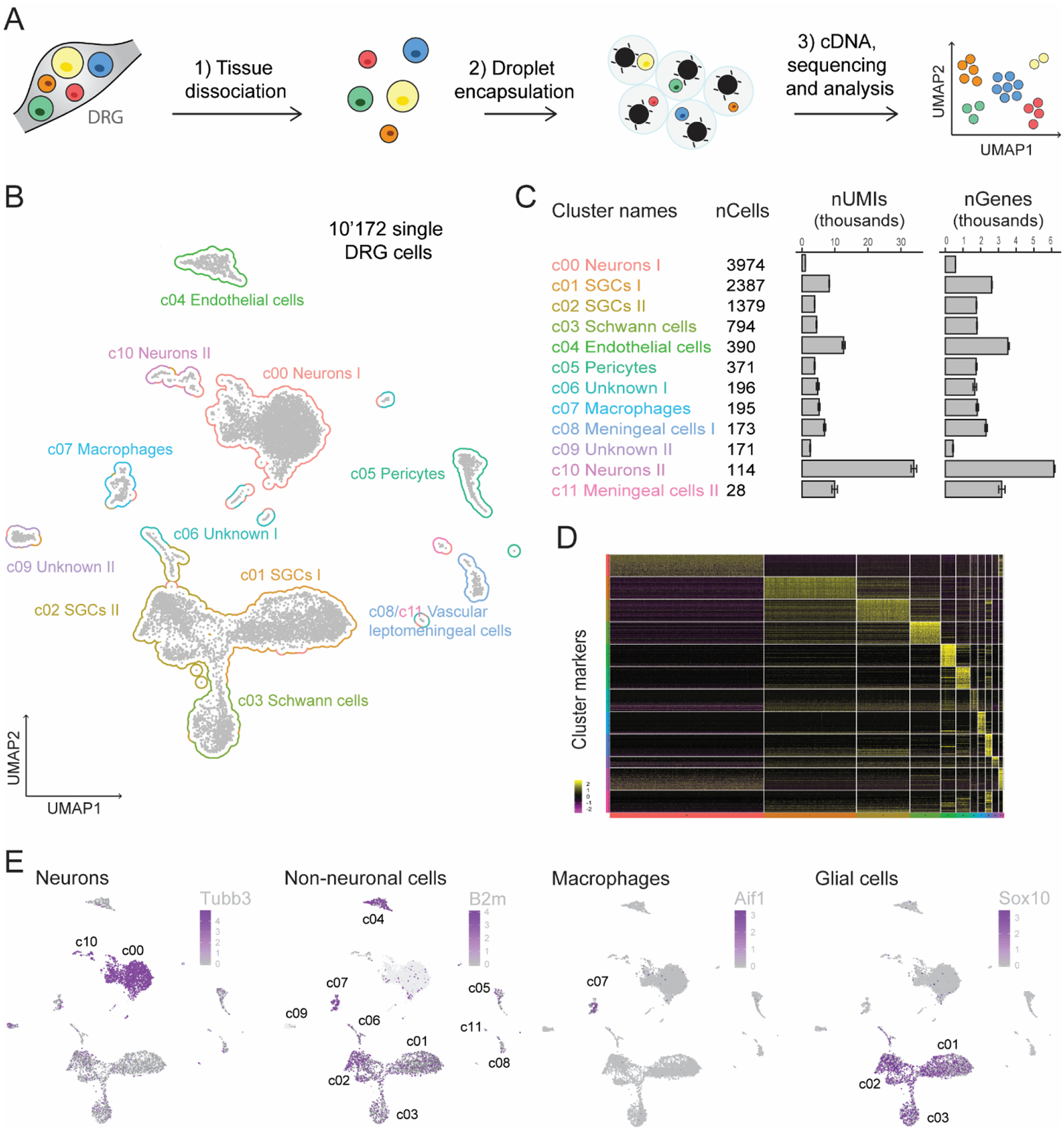
(A) Overview of the experimental strategy is shown. Pooled DRG cell suspensions from three naïve mice were used for scRNA-seq, which was performed using the Chromium droplet encapsulating technology from 10X Genomics. Unsupervised clusters were generated by uniform manifold approximation and projection (UMAP). (B) UMAP plot of 10,172 DRG individual cells is shown. Each point represents an individual cell, and clusters are colored by cell type assignment. (C) Cluster names are followed by the number of cells per cluster, UMIs per cluster, and genes detected per cluster. (D) Heat map of expression of the top 50 most significantly enriched genes for each cluster is shown. (E) UMAP plots for the neuronal marker Tubb3, non-neuronal marker B2m, macrophage marker Aif1, and glial cell marker Sox10 are presented. The scale and color indicate log2 expression of each gene.
3.2. Characterization of molecular and functional heterogeneity of SGCs
The goal of this study was to obtain a comprehensive transcriptome of SGCs, which tightly envelop DRG neuron cell bodies (Fig. 2A) and regulate their environment and function. SGCs represent the largest subtype of glial cells in the DRG, with two distinct clusters (Fig. 1B and 1C). Although SGCs are derived from Schwann cells and share some functions with astrocytes (Hanani and Spray, 2020), both clusters are transcriptionally distinct from Schwann cells and astrocytes (Fig. 2B, Suppl. Fig. 3A and Suppl. Table 2). As previously reported (Avraham et al., 2020), transcripts for Glul/GS - a historically used SGC marker - were found in nearly all DRG cells (data not shown). Closer examination of transcripts in each SGC cluster showed that other historically defined markers, such as Slc1a3, Kcnj10, and Gja1, are enriched in SGC I cluster (Fig. 2C), compared to SGC II cluster, which is characterized by exclusive expression of the enzyme ectonucleoside triphosphate diphosphohydrolase 2 (Entpd2, Fig. 2D). In situ hybridization of Gja1 and Entpd2 confirmed two distinct clusters of SGCs surrounding the same DRG neurons (Fig. 2D). These SGCs, though surrounding the same neurons, presented different biological processes (Fig. 2E). SGC I cluster was enriched in cholesterol and fatty acid process-related genes (e.g., Fabp7, fatty acid binding protein 7, Suppl. Fig. 3B), while SGC II cluster was enriched in genes involved in cellular response to transforming growth factor beta (TGFß) response (e.g., Tgfbr3, TGFß receptor III, Suppl. Fig. 3B) and cell adhesion (e.g., matrix metalloproteinases Mmp2, Mmp14 and disintegrin and metalloproteases Adam17 and Adam23, Suppl. Fig. 3B). These SGC biological processes differed from those associated with Schwann cells and astrocytes (Suppl. Fig. 3C), highlighting unique transcriptional and functional characteristics of the two SGC subtypes.
Fig. 2. Identification of two distinct populations of satellite glial cells (SGCs).
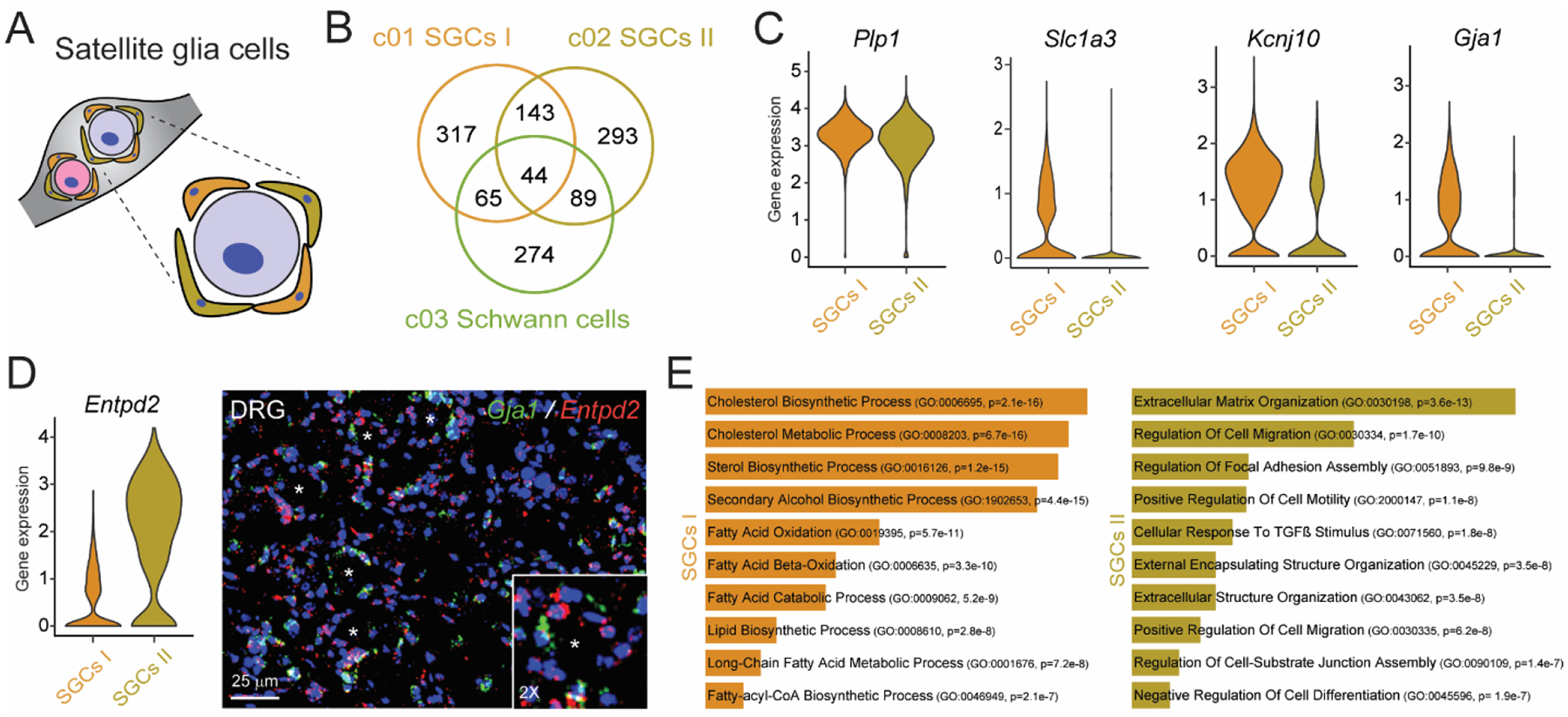
(A) Illustration of SGCs wrapping around sensory neurons in DRGs. (B) Venn diagram of enriched genes of the two populations of SGCs and Schwann cells. (C) Expression levels of Plp1, Slc3a1, Gja1, and Kcnj10 in SGC clusters, as shown by violin plots with a log-normalized y-axis. (D) Violin plots for Entpd2 and representative RNAscope images of a DRG tissue for Gja1 and Entpd2. DAPI (blue) is used as nuclei counterstaining. (E) Gene ontology (GO) analysis of genes expressed in the SGCs I and SGCs II populations.
3.3. Confirmation of TIMP3 expression and metalloprotease signaling in SGCs
The top biological process in the SGCs II cluster is associated with extracellular matrix organization (Fig. 2E). Matrix metalloproteinases (MMPs) and their inhibitors, tissue inhibitors of metalloproteinases (TIMPs), are involved in tissue homeostasis and inflammation by regulating extracellular matrix turnover and proteolytic processes (Khokha et al., 2013). Evidence shows expression of MMP2, MMP9 and TIMP1 in peripheral sensory neurons and SGCs, as well as their importance in pain (Kawasaki et al., 2008; Knight et al., 2019; Tonello et al., 2019). Genes enriched in SGCs I have been explored for their role in pain (Gja1 and Kcnj10) and regeneration (Fabp7), while genes in SGCs II are less known, including Mmp2, Mmp14, and Timp3 (Suppl. Fig. 3B). TIMP3 is unique among TIMPs due to its inhibition of a broad range of substrates (Qi et al., 2003), including disintegrin and metalloproteinases (ADAMs) such as Adam17 (Fig. 3A), also enriched in SGCs II (Suppl. Fig. 3B). Confirming this, fluorescent in situ hybridization showed Mmp14, Timp3, and Adam17 transcripts to be highly expressed in SGCs (Fig. 3B and Suppl. Fig. 4A), and Timp3 co-expressed with Mmp14 and Adam17 in cells surrounding DRG neurons (Fig. 3C). Furthermore, in situ hybridization images from the Allen Mouse Spinal Cord Atlas (https://mousespinal.brain-map.org/), which include DRG tissues, corroborate our data, showing a restricted Timp3 staining pattern in DRG tissues compared to Gja1 and Tubb3 staining patterns detected in both DRG and spinal cord tissues (Suppl. Fig. 4B). Of note, both mouse and human DRG tissues contained TIMP3 mRNA (Fig. 3D), and immunofluorescence showed characteristic SGC circular staining patterns for TIMP3 protein around human DRG neurons (Fig. 3E). Thus, TIMP3 is expressed in SGCs across species, and may control DRG tissue homeostasis by regulating MMP14, ADAM17, and their downstream signaling.
Fig. 3. Transcriptional and protein expression of Timp3.
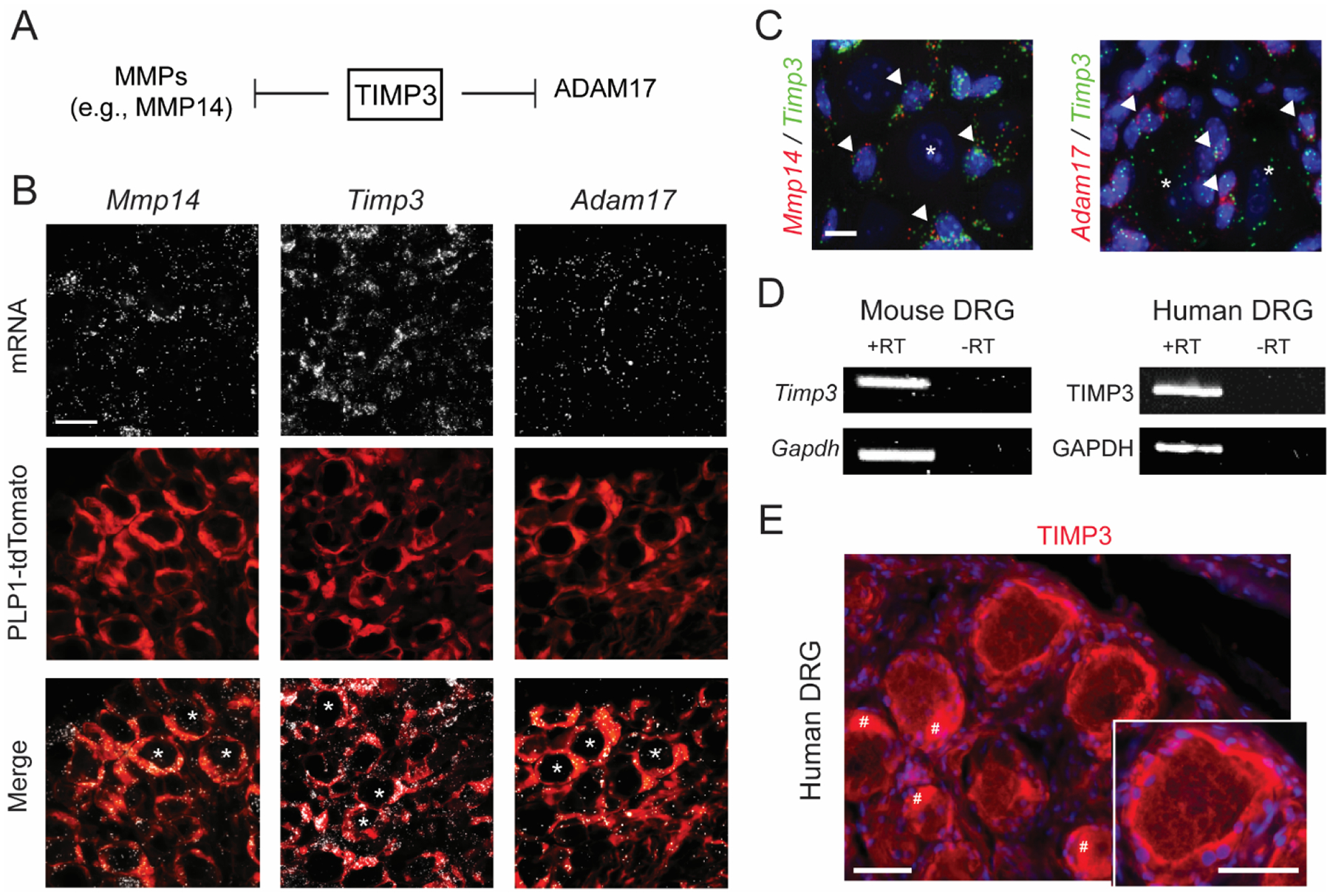
(A) Schematic illustration of TIMP3’s inhibitory activity on matrix metalloproteinases (MMPs) and on disintegrin and metalloprotease 17 (ADAM17), also called TACE (tumor necrosis factor-α-converting enzyme). (B, C) RNAscope localization of Timp3, Mmp14 and Adam17 mRNA expression in DRG of Plp1-Cre/tdTomato mice, wherein Cre recombinase is expressed in satellite glial cells (SGCs) (B), and in naïve CD1 mice (C). Arrowheads indicate mRNA colocalization of Mmp14 and Adam17 with Timp3 in SGCs, * indicates neurons. Scale bars = 25 μm in (B) and 5 μm in (C). (D) PCR in mouse and human DRG tissues. Samples with omitted RT (reverse transcriptase) show no bands, confirming the specificity of the amplification. (E) Immunofluorescence of TIMP3 in human DRG tissue. Scale bars = 50 μm. DAPI was used as counterstain. # indicates the fluorescent signal due to the presence of lipofuscins in human DRG neurons.
3.4. TIMP3 expression modulates response to somatosensory stimuli in naïve mice
TIMPs are secreted proteins known to control tissue homeostasis and function (Khokha et al., 2013), but the role of TIMP3 in DRG tissue remains unknown. To address this, we used a previously described siRNA approach (Tonello et al., 2019) and injected a control siRNA or Timp3 siRNA intrathecally two days prior to behavioral, pharmacological, and biochemical testing (Fig. 4A). We confirmed the tissue-specificity of this approach and showed that the expression levels of TIMP3 protein were significantly decreased in DRG, but not in spinal cord tissues, of mice treated with Timp3 siRNA compared to those treated with control siRNA (Fig. 4B and Suppl. Fig. 5A). Furthermore, we demonstrated through qPCR that Timp3 siRNA specifically and significantly reduced Timp3 mRNA expression levels in SGC cultures compared to other TIMPs (Suppl. Fig. 5B). Since DRGs contain the cell bodies of peripheral sensory neurons transmitting somatosensory information and SGCs can modulate this information (Berta et al., 2017), we tested the innate responses to various somatosensory stimuli in mice treated with Timp3 siRNA and control siRNA. We observed that Timp3 knockdown did not impair motor coordination (Fig. 4C), but caused the development of mechanical allodynia for up to 14 days (Fig. 4D). To a lesser extent, these mice also developed heat hyperalgesia at day 4 and cold allodynia at days 4 and 7 after injection (Fig. 4D).
Fig. 4. Timp3 controls mechanical and thermal sensitivities in naïve mice.
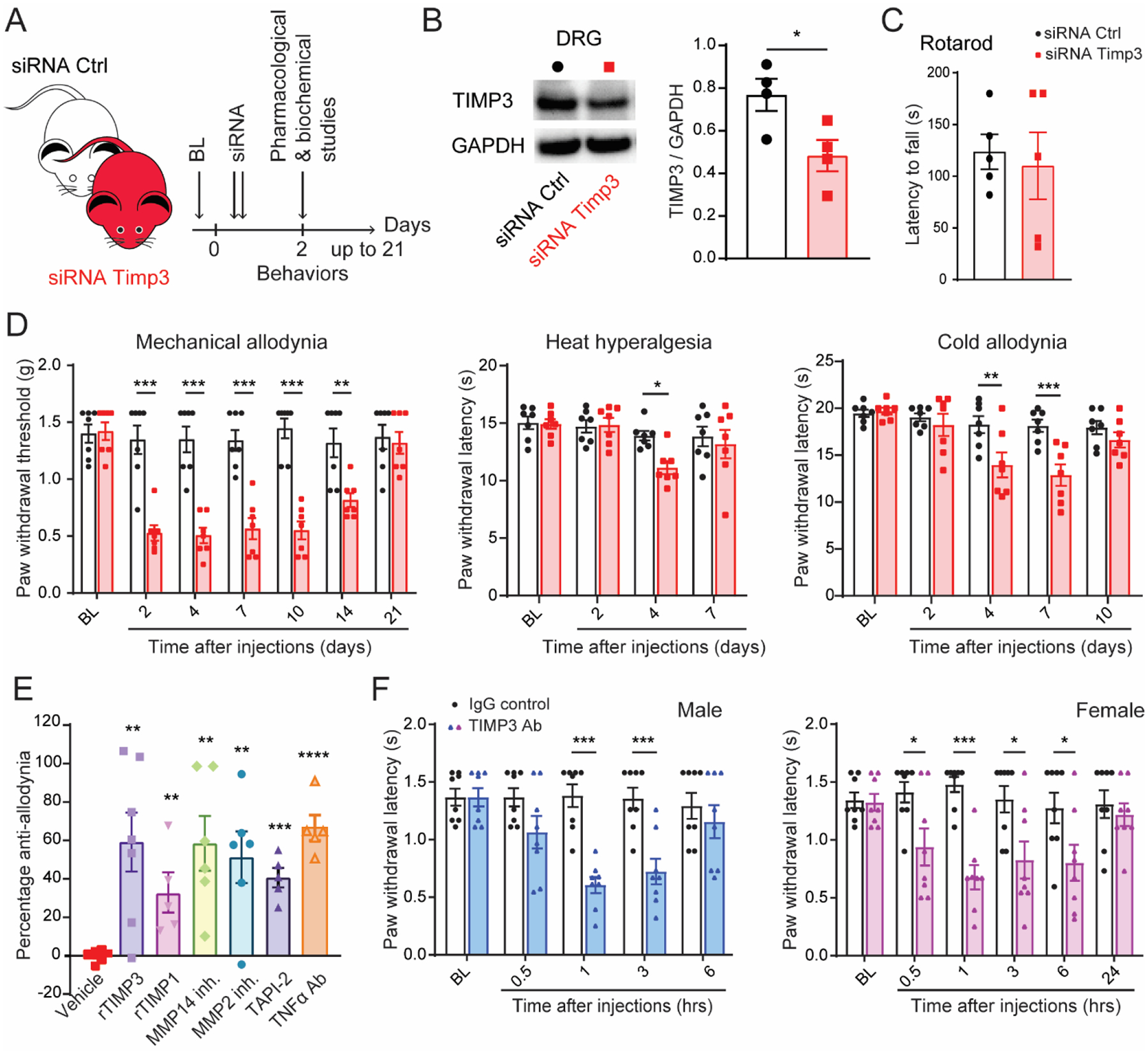
(A) Schematic illustration of the experiment showing the timeline of siRNA injections, pharmacological and biochemical studies. (B) Western blot representative image and quantification show that Timp3 siRNA significantly decreases Timp3 protein levels in DRGs tissues (n=4). (C) Timp3 siRNA injections do not cause locomotor dysfunction in the Rota-rod test (n=5). (D) Mechanical and thermal (von Frey, Hargreaves, and dry ice) allodynia induced by Timp3 siRNA compared to a control (Ctrl) non-targeting siRNA (2 μg of siRNA per delivery in the transfection agent PEI, n=7). (E) Anti-allodynic effect of exogenous recombinant TIMP3 (rTIMP3, 100 ng/site, i.t.), general endogenous tissue inhibitor of MMPs (TIMP-1, 4 pmol/site), MMP2 and MMP14 inhibitors (10 μg/site, i.t.), TACE/ADAM17 inhibitor (TAPI-2, 1 μg/site, i.t.), and a neutralizing antibody for TNF-α (5 μg/site, i.t.) on mechanical allodynia induced by Timp3 siRNA on day 2. (F) Anti-TIMP3 antibody (TIMP3 Ab, 10 μg/site, i.t.) induces mechanical allodynia compared to IgG control in male and female mice. BL = baseline. Data are expressed as mean ± SEM and statistically analyzed by two-tailed t-test (B, C, E) and Two-way ANOVA followed by Sidak’s post hoc test (D, F). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To validate our siRNA approach, we tested whether mechanical allodynia in Timp3 knockdown mice could be rescued by treatment with a TIMP3 recombinant protein (rTIMP3). We found that rTIMP3 (100 ng/site, i.t.) significantly reversed the mechanical allodynia in mice previously injected with Timp3 siRNA (Fig. 4E). It is well-reported that TIMP3 mainly functions as an endogenous inhibitor of MMPs and ADAM17 (Qi et al., 2003). Therefore, we tested the effects of general MMP inhibitors (TIMP1 recombinant protein, rTIMP1, 4 pmol/site) and specific inhibitors of MMP2 (MMP2 inh., 10 μg/site) and MMP14 (MMP14 inh., 10 μg/site), which all significantly reversed the mechanical allodynia in mice previously injected with Timp3 siRNA (Fig. 4E). TIMP3 is known to inhibit tumor necrosis factor (TNF)-a converting enzyme (ADAM17), and TNF-a has a central role in modulating DRG and neuronal homeostasis and somatosensory information (Liu et al., 2002; Mohammed et al., 2004; Qi et al., 2003; Segond von Banchet et al., 2009). In line with this, we found that inhibition of ADAM17 (TAPI-2, 1 μg/site) and TNF-a (neutralizing antibody, 10 μg/site) significantly reversed the mechanical allodynia induced by Timp3 siRNA (Fig. 4E). Importantly, we have demonstrated that injecting mice with a control siRNA does not significantly affect the mechanical threshold of rTIMP3, rTIMP1, and TAPI-2 (Suppl. Fig. 6). This finding rules out the possibility that the reversal of Timp3 siRNA-induced allodynia is due to the indiscriminate analgesic efficacy of these inhibitors.
To corroborate the extracellular nature of TIMP3 and our siRNA data, we intrathecally injected mice with a neutralizing antibody (Ab) against TIMP3 (10 μg/site in PBS) or with an IgG control (10 μg/site in PBS). Compared to the control treatment, treatment with TIMP3 Ab induced a significant mechanical allodynia in male and female mice, lasting up to 3 and 6 h, respectively (Fig. 4F). However, this treatment did not induce heat hyperalgesia (Suppl. Fig. 7A) or cold allodynia (Suppl. Fig. 7B), likely due to the transitory inhibition of TIMP3 by the Ab treatment compared to the long-lasting inhibition of the siRNA approach. Together, these data suggest that TIMP3 expression and metalloproteinase signaling (e.g. MMP14 and ADAM17 enriched in SGCs) modulate the responses to mechanical and thermal stimuli.
3.5. TIMP3 expression controls the development of chemotherapy-induced neuropathic pain (CINP).
Metalloproteinases are emerging as major players in neuropathic pain (Ji et al., 2009). We and other groups have reported an increase in expression levels of various MMPs and ADAM17 activity in an animal model of chemotherapy-induced neuropathic pain (Cirrincione et al., 2020; Tonello et al., 2019; Wang et al., 2021; Wu et al., 2015). This condition is often refractory to current treatments. We hypothesized that TIMP3, an endogenous inhibitor for various MMPs and ADAM17, may be regulated and, therefore, offers a novel therapeutic approach for CINP. To test this hypothesis, we used a previously reported CINP mouse model (Li et al., 2015; Tonello et al., 2019). In this model, paclitaxel (PAX, 2 mg/kg) is intraperitoneally injected every other day (days 0, 2, 4, and 6 with a final cumulative dose of 8 mg/kg) to mimic the treatment protocols used in clinic. Neuropathic pain with a similar phenotype to that in patients, including development of mechanical and cold allodynia that peak at 14 days after the first PAX treatment (Fig. 5A), has been validated. We found a significant decrease of TIMP3 expression levels in DRG tissues of mice treated with PAX compared to those treated with a vehicle control (Fig. 5B and C).
Fig. 5. Recombinant TIMP3 protein reverses and prevents mechanical and cold allodynia in a mouse model of chemotherapy-induced neuropathic pain.
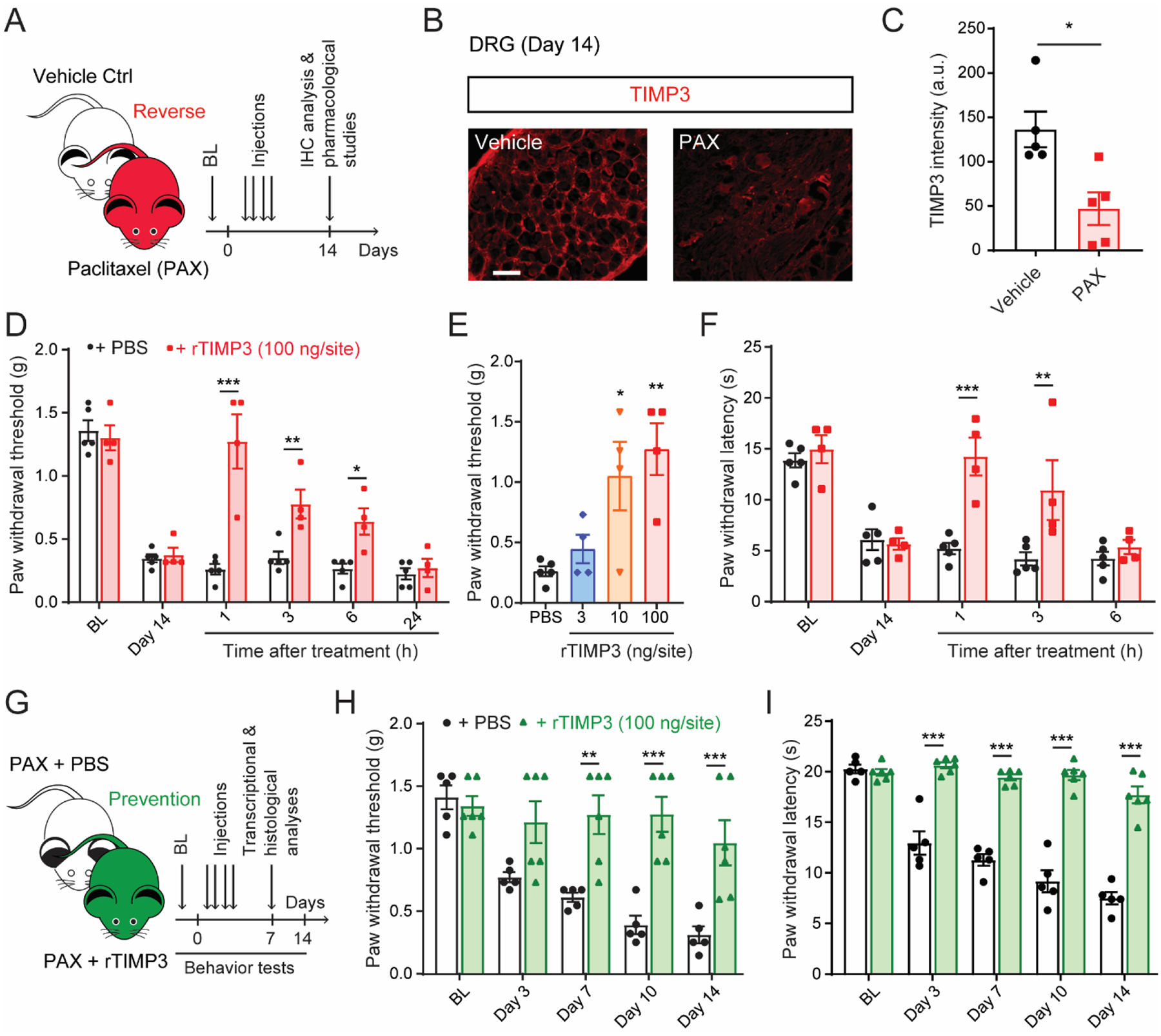
(A) Schematic of the experiment showing the timeline of paclitaxel (PAX) or vehicle injections, pharmacological studies, and immunohistochemistry (IHC) analysis. (B) Representative image and (C) quantification of Timp3 protein in mouse DRG tissue 14 days after first injection of PAX or vehicle control (n = 5). (D, E) Paclitaxel-induced mechanical (von Frey) and (F) cold allodynia (dry ice) are significantly, dose-dependently reversed up to 6 h by single intrathecal administration of recombinant TIMP3 protein (rTIMP3, 3–100 ng/site) delivered at day 14 after chemotherapy injection (n = 4–5). (G) Schematic of experiment showing timeline of PAX injections concomitantly with rTIMP3 or PBS, behavioral tests, transcriptional, and histological analysis. (H, I) Repeated administrations of rTIMP3 (100 ng/site, i.t.) prevent paclitaxel-induced mechanical and cold allodynia (n = 5–6). BL = baseline. Data expressed as mean ± SEM, statistically analyzed by two-tailed t-test (C), two-way ANOVA followed by Sidak’s post hoc test (D, F, H, I), and one-way ANOVA followed by Turkey’s post hoc test (E). *P < 0.05, **P < 0.01, ***P < 0.001.
Since our data demonstrated that treatment with recombinant TIMP3 (rTIMP3) reversed mechanical allodynia in Timp3 knockdown mice (Fig. 4E), we reasoned that similar treatment would reverse paclitaxel-induced allodynia. Indeed, a single intrathecal delivery of rTIMP3 (100 ng/site) significantly reversed both mechanical and cold allodynia (Fig. 5D and F). The analgesic effects of rTIMP3 lasted for few hours and were dose-dependent (3 – 100 ng/site) for the reverse of mechanical allodynia (Fig. 5E). To investigate whether treatments with rTIMP3 at the time of PAX injections can prevent the development of allodynia, repeated intrathecal administrations of rTIMP3 (100 ng/site, delivered every other day for 4 days at the same time as PAX injections) were conducted. Remarkably, this significantly prevented the development of both mechanical and cold allodynia in male mice for up to 14 days (Fig. 5H and I). Similarly, paclitaxel-induced mechanical allodynia was also prevented in female mice (Suppl. Fig. 8A, B).
In line with its role in regulating metalloproteinase signaling, these administrations of rTIMP3 significantly reversed the paclitaxel-induced increases of Mmp14 and Adam17 mRNAs in DRG tissues (Suppl. Fig. 9A). Moreover, pro-inflammatory cytokine Tnf expression levels were significantly decreased, whereas the anti-inflammatory cytokine Tgfb1 expression levels were significantly increased in DRG tissues of mice administered rTIMP3 (Suppl. Fig. 9A). However, there were no changes in the expression levels of the antioxidant gene Sod1 in DRG tissues of mice administered rTIMP3 (Suppl. Fig. 9A). Paclitaxel-treatment is known to reduce intraepidermal nerve fiber (IENF) density in the plantar surface of the paw (Siau et al., 2006), probably due to mitochondrial damages, oxidative stress and high bioenergetically demand of these peripheral nerve (Bennett et al., 2014). Consistent with our previous report (Tonello et al., 2019), paclitaxel treatment reduced the IENF density. However, the administration of rTIMP3 did not reverse the loss of IENF density induced by paclitaxel (Suppl. Fig. 9B). Together, these data indicate that rTIMP3 functions as an analgesic in a preclinical model of CINP by regulating metalloproteinase signaling and decreasing neuroinflammation in DRG tissues.
3.6. Establishment of a potential SGC discovery platform to identify novel treatments for neuropathic pain
Systemic treatments, including paclitaxel, tend to accumulate in DRG tissues due to their rich vascularization with unique endothelial cells that allow drug penetration (Jimenez-Andrade et al., 2008). To test the hypothesis that paclitaxel can directly target SGCs and alter TIMP3 signaling in DRG tissues, we generated SGC cultures and incubated them with a vehicle control solution or PAX at a concentration of 300 nM (Fig. 6A) - similar to concentrations previously reported in rodent DRG tissues (Cavaletti et al., 2000). We found that PAX significantly increased the expression of the glial fibrillary acid protein (GFAP) in SGC cultures (Fig. 6B), indicating that SGCs are directly activated by paclitaxel exposure without impairing their survival (Fig. 6C). We also sought to test whether TIMP3 signaling is altered in these SGC cultures. In line with our observation in DRG tissues from mice treated with PAX (Fig. 5B and C), we found that SGC cultures exposed to PAX showed significant decreases in Timp3 mRNA, as well as significant increases in Mmp2, Mmp14, and Adam17 mRNAs (Fig. 6D). Based on these observations, we hypothesize that drugs that can normalize TIMP3 signaling in SGC cultures exposed to PAX may provide new therapeutic strategies for chemotherapy-induced neuropathic pain.
Fig. 6. Transcriptional analyses of TIMP3 signaling in cultured SGCs after paclitaxel treatment.
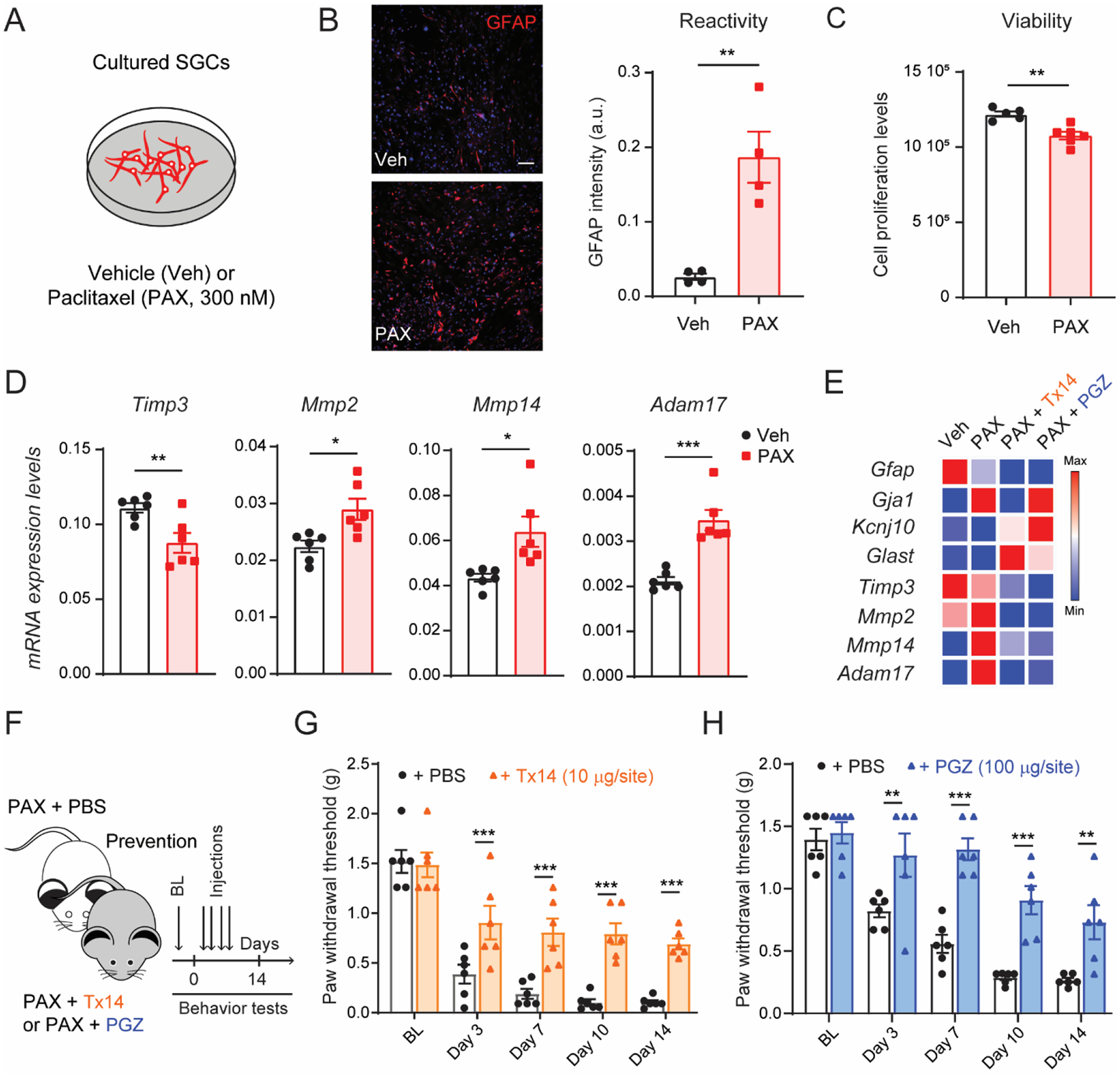
(A) Schematic of the experimental design used in cultured SGCs. (B) Representative image and quantification of immunofluorescence intensity of GFAP protein in SGC culture after 24 h of incubation with paclitaxel (PAX, 300 nM) or vehicle control (Veh; n=4). (C) Quantification of SGC culture viability 24 h after PAX or Veh treatment (n=6). (D) Quantification of mRNA expression levels of Timp3, Mmp2, Mmp14 and Adam17 in SGC culture after PAX or Veh incubation (n=6). (E) Heat map of mRNA expression of SGC and metalloprotease signaling markers in SGC culture after incubation with prosaptide Tx14 (1 μM) or pioglitazone (PGZ, 10 μM) and PAX compared to vehicle (n = 3). (F) Schematic illustrating the timeline of Tx14, PGZ, or PBS concomitantly treated with PAX, and the behavioral assay. Repeated injections of (G) Tx14 (10μg/site, i.t.) or (H) PGZ (100μg/site, i.t.) prevent paclitaxel-induced mechanical allodynia (n=6). BL = baseline. Data are expressed as mean ± SEM and statistically analyzed by two-tailed t-test (B, C, D), and Two-way ANOVA followed by Sidak’s post hoc test (G, H): *P < 0.05, **P < 0.01, ***P < 0.001.
To test this hypothesis, we used the synthetic prosaptide (Tx14) and the anti-diabetic medication pioglitazone (PGZ). Tx14 is reported to have glioprotective properties and to prevent paclitaxel toxicity (Campana et al., 1998; Meyer et al., 2013). PGZ, in addition to acting on peroxisome proliferator-activated receptor γ, has been demonstrated to rescue the expression of TIMP3 in skeletal muscle of diabetic patients (Tripathy et al., 2013). We found that these drugs did not reverse the effects of PAX on canonical SGC genes, but consistently normalized the transcriptional regulation of Mmp14, and Adam17 (Fig. 6E and Suppl. Fig. 10A). Although we surprisingly observed transcriptional downregulation of Timp3 in cultures treated with PAX and PGZ, the TIMP3 protein levels increased in these cultures compared to those treated with only PAX (Suppl. Fig. 10B). This suggests a discrepancy between transcript and protein regulation, but it is also consistent with the protein rescue effect previously reported in the literature (Tripathy et al., 2013). More importantly, administration of Tx14 (10μg/site, i.t.) or PGZ (100μg/site, i.t.) every other day for 4 days, concomitantly with paclitaxel (Fig. 6F), significantly prevented the development of paclitaxel-induced mechanical allodynia in mice (Fig. 6G and H). We also demonstrated that PGZ prevented paclitaxel-induced cold allodynia (Suppl. Fig. 10C). Together, these data suggest a potential cell discovery platform based on the regulation of TIMP3 signaling in SGC cultures exposed to paclitaxel to identify new or repurposed FDA-approved therapeutic drugs.
4. Discussion
Dorsal root ganglia are complex tissues containing neuronal and non-neuronal cells (Ji et al., 2016). However, current analgesic drugs mostly target DRG neurons and are associated with side effects, addiction and limited efficacy (Berta et al., 2017). It is well-known that DRG neurons have unique molecular profiles and are tuned to distinct stimuli, including painful stimuli, yet accumulating evidence suggests non-neuronal cells in DRGs also play a role in neuronal homeostasis and processing of these stimuli. Recently, scRNA-seq revealed diverse non-neuronal cells surrounding sensory neurons (Renthal et al., 2020; Zeisel et al., 2018). A similar strategy uncovered unique molecular profiles in SGCs in DRGs and their roles in development and neuronal regeneration (Avraham et al., 2020; Tasdemir-Yilmaz et al., 2021). SGCs are also known for their activation and function in preclinical models of pain (Hanani and Spray, 2020; Ji et al., 2013). To further characterize the molecular profile of SGCs, we conducted scRNA-seq. We identified two distinct SGC populations and a mechanism by which TIMP3 controls homeostasis and pathological pain through SGC metalloproteinase signaling. This regulation of metalloproteinase signaling in cultured SGCs can offer a novel, reliable platform for non-neuronal and non-addictive pain therapies.
There is a growing body of literature implicating SGCs in neuronal homeostasis and pathological pain (Hanani and Spray, 2020). However, past studies on the role of SGCs in pain have considered them a homogeneous population and focused on a few molecules (Ji et al., 2013). Recently, single-cell sequencing has revealed a more unbiased view of the heterogeneity of glial cells, including SGCs (Avraham et al., 2021; Mapps et al., 2022). In agreement with this, we have identified at least two major SGC subsets in DRGs of naïve mice. We confirmed these distinct subsets through in situ fluorescent hybridization and observed a mosaic distribution around the same DRG neurons, suggesting neuronal-subtype independent distribution of these SGCs. Both subsets (SGCs I and II) may affect neuronal functions and participate in homeostasis and various pathological pain conditions. It is well-documented that SGCs can influence DRG neurons by releasing cytokines and altering expression of proteins such as GLAST1 (encoded by the gene Slc1a3), KIR4.1 (encoded by the gene Kcnj10), and CX43 (encoded by the gene Gja1) (Kim et al., 2016; Retamal et al., 2017; Silva et al., 2017; Takeda et al., 2011). Interestingly, we found an enrichment of these genes in SGCs I, suggesting a major role of this SGC subset in pain. SGCs I also had an enrichment of genes related to fatty acid synthesis and metabolism, which have been linked to neuronal regeneration (Avraham et al., 2020).
Which genes are enriched in SGCs II subset? What are their functions? Functional analysis of SGCs II revealed a potential role in glial cell differentiation, which may support previous reports suggesting SGCs as a population of developmentally arrested Schwann cells that can proliferate after peripheral nerve injury (Donegan et al., 2013; George et al., 2018). However, our data indicated that both SGCs I and SGCs II are transcriptionally distinct from Schwann cells. Whether SGCs proliferate after injury remains a matter of debate (Jager et al., 2020). We found enrichment of genes associated with extracellular matrix organization, including various matrix metalloproteinases and disintegrin and metalloproteinase proteins. These transcripts include Adam10, Adam17, Mmp2, Mmp14, and the tissue inhibitor of metalloproteinase Timp3. TIMP3 is unique for its inhibition of several membrane-bound molecules with sheddase functions, such as MMP14, MMP3, and ADAM17 (which is responsible for the maturation of TNF) (Mohammed et al., 2004; Qi et al., 2003). Due to its plethoric signaling and functions, TIMP3 has become increasingly recognized for its role in homeostasis, neurodegeneration, and inflammation (Mohammed et al., 2004; Qi et al., 2003; Rosenberg, 2009).
We confirmed strong expression of TIMP3 in SGCs surrounding DRG neurons and its colocalization with MMP14 and ADAM17, suggesting a role of TIMP3 in somatosensation by controlling these metalloproteases. To determine whether TIMP3 is involved in somatosensation, we used local knockdown of Timp3 in DRG tissue of naïve mice using siRNA. These mice developed somatosensory hypersensitivities, including robust and long-lasting mechanical allodynia that persisted for up to 14 days. We previously reported that siRNA knockdown lasts about 5–7 days (Berta et al., 2012; Berta et al., 2014; Lee et al., 2018), therefore the mechanical allodynia lasted beyond the knockdown period, suggesting major homeostatic changes in the DRG tissues. This may be due to the involvement of TIMP3 in regulating multiple downstream target proteins, including metalloproteinases and inflammatory mediators. Indeed, pharmacological blockade of various metalloproteases, ADAM17, and extracellular TNF significantly reversed the mechanical allodynia caused by knockdown of TIMP3. TNF is a key mediator of inflammation and pain (Ji et al., 2014). TNF levels are increased in DRGs in animal models of inflammatory and neuropathic pain (Leung and Cahill, 2010). Increased expression and release of TNF are observed in SGCs of mouse models of acute herpetic neuralgia and paclitaxel-induced neuropathic pain (Silva et al., 2017; Wu et al., 2015). Exogenous delivery of TNF in naïve mice elicited mechanical and thermal hypersensitivities, and direct application of TNF to cultured DRG neurons increased their excitability (Jin and Gereau, 2006). Therefore, we suggest TIMP3 in SGCs as a key physiological regulator of neuronal and pain hypersensitivities by controlling metalloproteinase signaling and inflammatory mediators such as TNF.
To assess the role of TIMP3 in pathological pain, we used a paclitaxel-induced mouse model of CINP. This model was used for several reasons. Firstly, paclitaxel increases GFAP expression in SGCs, suggesting their involvement in this type of pain (Warwick and Hanani, 2013). Secondly, it has been reported that paclitaxel increases expression levels of proteins related to metalloprotease signaling, such as MMP2, MMP9, ADAM17 and TNF (Tonello et al., 2019; Wu et al., 2015). Lastly, there is currently no FDA-approved therapy to treat chronic intractable cancer pain (CINP) in cancer patients, which may contribute to the reported misuse of opioids by these patients (Kwon et al., 2013; Reyes-Gibby et al., 2016). Here, we suggest TIMP3 as a novel therapeutic target for CINP, showing that rescuing decreased TIMP3 expression levels in DRGs with an exogenous recombinant TIMP3 protein can reverse and prevent paclitaxel-induced mechanical and cold allodynia in male mice.
Previous studies have revealed sex differences in immune and glial cell signaling that underlies mechanical hypersensitivity (Mogil, 2020). Our results showed that TIMP3 inhibition with a neutralizing antibody induced mechanical allodynia in both male and female naïve mice, implying that SGC signaling is independent of sex. In addition, our lab’s previous data has shown that astrocytic signaling, which is similar to that observed in SGCs and responsible for mechanical allodynia, is also independent of sex. For example, TNF induced similar levels of mechanical hypersensitivity in both naïve males and females (Chen et al., 2018). However, sex-dependent glial signaling has been mostly observed in mice with pathological pain (Sorge et al., 2015; Tansley et al., 2022; Taves et al., 2016), and may not be evident in naïve mice. To address this, we tested and demonstrated that recombinant TIMP3 treatment can prevent paclitaxel-induced mechanical allodynia not only in male, but also in female mice. Overall, SGC signaling seems independent of sex and TIMP3 treatment may prevent neuropathic pain in both male and female mice.
Mechanistically, we have shown that this treatment normalizes TIMP3 signaling by reversing the increased expression of Mmp14 and Adam17 mRNA induced by paclitaxel. Additionally, it potentially reduces neuroinflammation in DRGs by decreasing the transcriptional expression of the pro-inflammatory cytokine Tnf, while increasing the transcriptional expression of the anti-inflammatory mediator Tgfb. Further studies are ongoing to elucidate how TIMP3 regulates neuroinflammation in DRG tissues. In addition to its well-known role in the maturation of pro-inflammatory cytokines (Mohammed et al., 2004), it may also regulate the infiltration of immune cells in DRG through its action on the vascular endothelial growth factor receptor-2 (Qi et al., 2003). Surprisingly, TIMP3 treatment did not rescue the loss of IENF density. Although the loss of IENF density is a common characteristic of chemotherapy-induced neuropathies (Bennett et al., 2014), its contribution to pain remains unclear. Interestingly, pre-clinical and clinical studies have found that paclitaxel-induced pain often precedes deficits in sensory nerve function (Boyette-Davis et al., 2015; Ma et al., 2018), and pain from a high dose of paclitaxel can occur independently from functional alterations of sensory nerves (Flatters and Bennett, 2006). Targeting neuroinflammation in DRGs to treat CIPN is supported by multiple preclinical studies, which have demonstrated that depletion of macrophages, blockade of TNF, and promotion of anti-inflammatory cytokines IL-10 and TGFβ all reduce paclitaxel-induced pain (Krukowski et al., 2016; Tonello et al., 2020; Zhang et al., 2016). Non-steroidal anti-inflammatory drugs are used to treat cancer patients, but their analgesic effects against CIPN require further clinical studies (Desforges et al., 2022).
Chemotherapeutic treatments tend to accumulate in DRG tissues due to their rich and fenestrated vascularization (Jimenez-Andrade et al., 2008). Few studies, however, have investigated the direct interaction of paclitaxel with glia cells (Liu et al., 2019; Wu et al., 2015). Our culture studies show that paclitaxel increases the reactivity of SGCs, which is consistent with the transcriptional changes in TIMP3 signaling observed in the DRGs of mice treated with paclitaxel. This culture may be used to screen different drugs foe CINP. As a proof-of-concept, we tested the ability of the glioprotective peptide Tx14 (Campana et al., 1998; Meyer et al., 2013) and antidiabetic medication pioglitazone to reverse the transcriptional changes in TIMP3 signaling in cultured SGCs treated with paclitaxel. We found that transcriptional changes in Mmp14 and Adam17 transcripts were increased in paclitaxel-treated culture, but normalized by both drugs Tx14 and pioglitazone. More importantly, both drugs were able to prevent CINP in mice. We hypothesize that pioglitazone prevents CINP partially through the increase of TIMP3 expression in DRGs, as it can rescue the expression of TIMP3 in skeletal muscle of diabetic patients (Tripathy et al., 2013). However, the mechanisms underlying the regulation of TIMP3 signaling in SGC by pioglitazone and its analgesic effects warrant further investigation. It is also important to consider that pioglitazone can also alleviate neuropathic pain through spinal mechanisms involving peroxisome proliferator-activated receptor γ (Griggs et al., 2016; Griggs et al., 2015). However, a recent study has proposed that activation of PPARγ by pioglitazone may mitigate lupus nephritis symptoms in mice by suppressing miR-21-5p and promoting the increase of TIMP3 (Liu and Zhang, 2021). Interestingly, we observed different regulation of TIMP3 mRNA and protein in SGC cultures treated with pioglitazone. The correspondence between mRNA and protein expression levels is notoriously poor, in part due to different time courses and post-transcriptional regulation (Vogel and Marcotte, 2012). This may also explain the increased level of GFAP protein, but decreased mRNA levels, in SGC cultures treated with paclitaxel. Previously, we showed that voltage-gated sodium channel β2 subunit protein levels were up-regulated in DRG tissues by nerve injury despite no mRNA regulation (Pertin et al., 2005).
This study fully characterized the transcriptional profile of SGCs and identified new therapeutic targets for pain treatment. Despite these strengths, limitations exist. First, the number of genes for most dorsal root ganglion (DRG) neurons was lower than expected in the single-cell RNAseq, preventing us from analyzing their heterogeneity or interactions with other non-neuronal cells. This is likely due to the technical limitation of 10X Genomic microfluidic channels, which are optimized for cells smaller than DRG neurons. Glia and immune cells, which are smaller than DRG neurons, have a number of genes consistent with other single-cell RNAseq studies (Häring et al., 2018; Zeisel et al., 2018). Single-nucleus RNAseq can be used for better transcriptional analysis of various DRG neuronal populations (Renthal et al., 2020). Second, our study focused on metalloproteinase signaling because of its involvement in tissue homeostasis, inflammation and pain (Ji et al., 2009). We propose targeting TIMP3 to alleviate pain since it is expressed in both mouse and human SGCs and is key in regulating various MMPs and pro-inflammatory and algesic mediators, such as TNF. For this purpose, we delivered the recombinant TIMP3 protein to the DRG tissues via intrathecal injection, a commonly used delivery route in pre-clinical studies (Berta et al., 2017). However, we did not confirm whether the TIMP3 protein reached the DRG tissues, and intrathecal injections may not be practical in a clinical setting. Further studies are needed to address these limitations. It is also important to note that the therapeutic potential of targeting metalloproteinase signaling has been hampered by lack of specificity and functional compensation among various MMPs (Khokha et al., 2013; Vandenbroucke and Libert, 2014). Future studies should investigate other potential therapeutic targets such as ion channels and G-protein-coupled receptors. Finally, cultured SGCs may not fully recapitulate their heterogeneity and state in vivo (Avraham et al., 2021; Jager et al., 2022). Nevertheless, we showed that paclitaxel induces similar transcriptional changes in SGC cultures and mouse DRGs, and provided a proof-of-concept for their use to identify new analgesics for CINP.
In summary, pain studies have mainly focused on neurons, yet only few new analgesics have been introduced in the clinic over the past decades. Satellite glial cells are now seen as important players in neuronal homeostasis and disease. Here, we present the transcriptional profile of SGCs and explain a previously unknown mechanism of TIMP3 signaling, which modulates nociception and neuropathic pain. The therapeutic potential of TIMP3 signaling is supported by its expression in human SGCs, role in pain modulation in mice, and development of a potential cellular discovery platform to find new analgesics or repurpose existing FDA-approved drugs.
Supplementary Material
1. BBI highlights.
Single-cell RNA sequencing reveals the diversity of satellite glial cells.
TIMP3 and metalloproteinase signaling are enriched in satellite glial cells.
TIMP3 expression modulates the response to mechanical and thermal stimuli, as well as the development of paclitaxel-induced neuropathic pain.
Metalloproteinase signaling in cultured satellite glial cells incubated with paclitaxel may serve as a platform for therapeutic discovery.
Acknowledgements
This work was funded by the NIH-NINDS grant NS113243 to T.B., as well as supported by the NIH-NINDS grant NS045594 to J.-M.Z. and by the National Research Foundation of Korea grant RS-2023-00253756 to G.C. We also acknowledge the support of institutional grants from the University of Cincinnati (UC) and UC Gardner Neuroscience Institute to T.B. We would like to thank the Single Cell Genomics Core and Bioinformatics Collaborative Services at Cincinnati Children’s Hospital Medical Center for their assistance with sample processing and data analysis of our single-cell RNA sequencing. We also thank Dr. Alexander Chamessian and Dr. Yawar Qadri for fruitful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
Data sharing
Data and materials availability: Single-cell RNA sequencing data is available from the Gene Expression Omnibus under accession number GSE236914. All other data needed to support the conclusions in the paper are available in the paper and/or the Supplementary Materials. Requests regarding protocols and resources should be directed to and will be fulfilled by the lead contact T.B. (temugin.berta@uc.edu).
References
- Avraham O, Deng PY, Jones S, Kuruvilla R, Semenkovich CF, Klyachko VA, Cavalli V, 2020. Satellite glial cells promote regenerative growth in sensory neurons. Nat Commun 11, 4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham O, Feng R, Ewan EE, Rustenhoven J, Zhao G, Cavalli V, 2021. Profiling sensory neuron microenvironment after peripheral and central axon injury reveals key pathways for neural repair. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G, Julius D, 2009. Cellular and molecular mechanisms of pain. Cell 139, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW, 2018. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. [DOI] [PubMed] [Google Scholar]
- Bennett GJ, Doyle T, Salvemini D, 2014. Mitotoxicity in distal symmetrical sensory peripheral neuropathies. Nat Rev Neurol 10, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta T, Liu T, Liu YC, Xu ZZ, Ji RR, 2012. Acute morphine activates satellite glial cells and up-regulates IL-1β in dorsal root ganglia in mice via matrix metalloprotease-9. Mol Pain 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta T, Park CK, Xu ZZ, Xie RG, Liu T, Lü N, Liu YC, Ji RR, 2014. Extracellular caspase-6 drives murine inflammatory pain via microglial TNF-α secretion. J Clin Invest 124, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta T, Qadri Y, Tan PH, Ji RR, 2017. Targeting dorsal root ganglia and primary sensory neurons for the treatment of chronic pain. Expert Opin Ther Targets 21, 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyette-Davis JA, Walters ET, Dougherty PM, 2015. Mechanisms involved in the development of chemotherapy-induced neuropathy. Pain Manag 5, 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner DS, Golden JP, Gereau R.W.t., 2012. A novel behavioral assay for measuring cold sensation in mice. PLoS One 7, e39765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campana WM, Eskeland N, Calcutt NA, Misasi R, Myers RR, O’Brien JS, 1998. Prosaptide prevents paclitaxel neurotoxicity. Neurotoxicology 19, 237–244. [PubMed] [Google Scholar]
- Cavaletti G, Cavalletti E, Oggioni N, Sottani C, Minoia C, D’Incalci M, Zucchetti M, Marmiroli P, Tredici G, 2000. Distribution of paclitaxel within the nervous system of the rat after repeated intravenous administration. Neurotoxicology 21, 389–393. [PubMed] [Google Scholar]
- Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL, 1994. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 53, 55–63. [DOI] [PubMed] [Google Scholar]
- Cirrincione AM, Pellegrini AD, Dominy JR, Benjamin ME, Utkina-Sosunova I, Lotti F, Jergova S, Sagen J, Rieger S, 2020. Paclitaxel-induced peripheral neuropathy is caused by epidermal ROS and mitochondrial damage through conserved MMP-13 activation. Sci Rep 10, 3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desforges AD, Hebert CM, Spence AL, Reid B, Dhaibar HA, Cruz-Topete D, Cornett EM, Kaye AD, Urits I, Viswanath O, 2022. Treatment and diagnosis of chemotherapy-induced peripheral neuropathy: An update. Biomed Pharmacother 147, 112671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donegan M, Kernisant M, Cua C, Jasmin L, Ohara PT, 2013. Satellite glial cell proliferation in the trigeminal ganglia after chronic constriction injury of the infraorbital nerve. Glia 61, 2000–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatters SJL, Bennett GJ, 2006. Studies of peripheral sensory nerves in paclitaxel-induced painful peripheral neuropathy: evidence for mitochondrial dysfunction. Pain 122, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatters SJL, Dougherty PM, Colvin LA, 2017. Clinical and preclinical perspectives on Chemotherapy-Induced Peripheral Neuropathy (CIPN): a narrative review. Br J Anaesth 119, 737–749. [DOI] [PubMed] [Google Scholar]
- George D, Ahrens P, Lambert S, 2018. Satellite glial cells represent a population of developmentally arrested Schwann cells. Glia 66, 1496–1506. [DOI] [PubMed] [Google Scholar]
- Grace PM, Hutchinson MR, Maier SF, Watkins LR, 2014. Pathological pain and the neuroimmune interface. Nat Rev Immunol 14, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs RB, Donahue RR, Adkins BG, Anderson KL, Thibault O, Taylor BK, 2016. Pioglitazone Inhibits the Development of Hyperalgesia and Sensitization of Spinal Nociresponsive Neurons in Type 2 Diabetes. J Pain 17, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs RB, Donahue RR, Morgenweck J, Grace PM, Sutton A, Watkins LR, Taylor BK, 2015. Pioglitazone rapidly reduces neuropathic pain through astrocyte and nongenomic PPARγ mechanisms. Pain 156, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosser T, Woolf CJ, FitzGerald GA, 2017. Time for nonaddictive relief of pain. Science 355, 1026–1027. [DOI] [PubMed] [Google Scholar]
- Gu Y, Chen Y, Zhang X, Li GW, Wang C, Huang LY, 2010. Neuronal soma-satellite glial cell interactions in sensory ganglia and the participation of purinergic receptors. Neuron Glia Biol 6, 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanani M, 2005. Satellite glial cells in sensory ganglia: from form to function. Brain Res Brain Res Rev 48, 457–476. [DOI] [PubMed] [Google Scholar]
- Hanani M, Spray DC, 2020. Emerging importance of satellite glia in nervous system function and dysfunction. Nat Rev Neurosci 21, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves K, Dubner R, Brown F, Flores C, Joris J, 1988. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain 32, 77–88. [DOI] [PubMed] [Google Scholar]
- Häring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lönnerberg P, La Manno G, Sharma N, Borgius L, Kiehn O, Lagerström MC, Linnarsson S, Ernfors P, 2018. Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci 21, 869–880. [DOI] [PubMed] [Google Scholar]
- Jager SE, Pallesen LT, Lin L, Izzi F, Pinheiro AM, Villa-Hernandez S, Cesare P, Vaegter CB, Denk F, 2022. Comparative transcriptional analysis of satellite glial cell injury response. Wellcome Open Res 7, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager SE, Pallesen LT, Richner M, Harley P, Hore Z, McMahon S, Denk F, Vaegter CB, 2020. Changes in the transcriptional fingerprint of satellite glial cells following peripheral nerve injury. Glia 68, 1375–1395. [DOI] [PubMed] [Google Scholar]
- Ji RR, Berta T, Nedergaard M, 2013. Glia and pain: is chronic pain a gliopathy? Pain 154 Suppl 1, S10–s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Chamessian A, Zhang YQ, 2016. Pain regulation by non-neuronal cells and inflammation. Science 354, 572–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Xu ZZ, Gao YJ, 2014. Emerging targets in neuroinflammation-driven chronic pain. Nat Rev Drug Discov 13, 533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji RR, Xu ZZ, Wang X, Lo EH, 2009. Matrix metalloprotease regulation of neuropathic pain. Trends Pharmacol Sci 30, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Andrade JM, Herrera MB, Ghilardi JR, Vardanyan M, Melemedjian OK, Mantyh PW, 2008. Vascularization of the dorsal root ganglia and peripheral nerve of the mouse: implications for chemical-induced peripheral sensory neuropathies. Mol Pain 4, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Gereau R.W.t., 2006. Acute p38-mediated modulation of tetrodotoxin-resistant sodium channels in mouse sensory neurons by tumor necrosis factor-alpha. J Neurosci 26, 246–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Xu ZZ, Wang X, Park JY, Zhuang ZY, Tan PH, Gao YJ, Roy K, Corfas G, Lo EH, Ji RR, 2008. Distinct roles of matrix metalloproteases in the early- and late-phase development of neuropathic pain. Nat Med 14, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha R, Murthy A, Weiss A, 2013. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol 13, 649–665. [DOI] [PubMed] [Google Scholar]
- Kim YS, Anderson M, Park K, Zheng Q, Agarwal A, Gong C, Saijilafu, Young L, He S, LaVinka PC, Zhou F, Bergles D, Hanani M, Guan Y, Spray DC, Dong X, 2016. Coupled Activation of Primary Sensory Neurons Contributes to Chronic Pain. Neuron 91, 1085–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight BE, Kozlowski N, Havelin J, King T, Crocker SJ, Young EE, Baumbauer KM, 2019. TIMP-1 Attenuates the Development of Inflammatory Pain Through MMP-Dependent and Receptor-Mediated Cell Signaling Mechanisms. Front Mol Neurosci 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukowski K, Eijkelkamp N, Laumet G, Hack CE, Li Y, Dougherty PM, Heijnen CJ, Kavelaars A, 2016. CD8+ T Cells and Endogenous IL-10 Are Required for Resolution of Chemotherapy-Induced Neuropathic Pain. J Neurosci 36, 11074–11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A, 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JH, Hui D, Chisholm G, Bruera E, 2013. Predictors of long-term opioid treatment among patients who receive chemoradiation for head and neck cancer. Oncologist 18, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Cho PS, Tonello R, Lee HK, Jang JH, Park GY, Hwang SW, Park CK, Jung SJ, Berta T, 2018. Peripheral serotonin receptor 2B and transient receptor potential channel 4 mediate pruritus to serotonergic antidepressants in mice. J Allergy Clin Immunol 142, 1349–1352.e1316. [DOI] [PubMed] [Google Scholar]
- Lee SH, Tonello R, Choi Y, Jung SJ, Berta T, 2020. Sensory Neuron–Expressed TRPC4 Is a Target for the Relief of Psoriasiform Itch and Skin Inflammation in Mice. Journal of Investigative Dermatology 140, 2221–2229.e2226. [DOI] [PubMed] [Google Scholar]
- Leung L, Cahill CM, 2010. TNF-alpha and neuropathic pain--a review. J Neuroinflammation 7, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, Wang SS, Sun MM, Lu YJ, Zhong YQ, Hu XY, Hou R, Zhou BB, Bao L, Xiao HS, Zhang X, 2016. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 26, 83–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Adamek P, Zhang H, Tatsui CE, Rhines LD, Mrozkova P, Li Q, Kosturakis AK, Cassidy RM, Harrison DS, Cata JP, Sapire K, Zhang H, Kennamer-Chapman RM, Jawad AB, Ghetti A, Yan J, Palecek J, Dougherty PM, 2015. The Cancer Chemotherapeutic Paclitaxel Increases Human and Rodent Sensory Neuron Responses to TRPV1 by Activation of TLR4. J Neurosci 35, 13487–13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li H, Brull SJ, Zhang JM, 2002. Increased sensitivity of sensory neurons to tumor necrosis factor alpha in rats with chronic compression of the lumbar ganglia. J Neurophysiol 88, 1393–1399. [DOI] [PubMed] [Google Scholar]
- Liu D, Zhang W, 2021. Pioglitazone Attenuates Lupus Nephritis Symptoms in Mice by Modulating miR-21–5p/TIMP3 Axis: the Key Role of the Activation of Peroxisome Proliferator-Activated Receptor-γ. Inflammation 44, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Liu X, Tonello R, Ling Y, Gao YJ, Berta T, 2019. Paclitaxel-activated astrocytes produce mechanical allodynia in mice by releasing tumor necrosis factor-α and stromal-derived cell factor 1. J Neuroinflammation 16, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Kavelaars A, Dougherty PM, Heijnen CJ, 2018. Beyond symptomatic relief for chemotherapy-induced peripheral neuropathy: Targeting the source. Cancer 124, 2289–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin SA, Davis BM, Molliver DC, 2007. Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc 2, 152–160. [DOI] [PubMed] [Google Scholar]
- Mapps AA, Thomsen MB, Boehm E, Zhao H, Hattar S, Kuruvilla R, 2022. Diversity of satellite glia in sympathetic and sensory ganglia. Cell Rep 38, 110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Giddens MM, Schaefer SA, Hall RA, 2013. GPR37 and GPR37L1 are receptors for the neuroprotective and glioprotective factors prosaptide and prosaposin. Proc Natl Acad Sci U S A 110, 9529–9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil JS, 2020. Qualitative sex differences in pain processing: emerging evidence of a biased literature. Nature Reviews Neuroscience 21, 353–365. [DOI] [PubMed] [Google Scholar]
- Mohammed FF, Smookler DS, Taylor SE, Fingleton B, Kassiri Z, Sanchez OH, English JL, Matrisian LM, Au B, Yeh WC, Khokha R, 2004. Abnormal TNF activity in Timp3−/− mice leads to chronic hepatic inflammation and failure of liver regeneration. Nat Genet 36, 969–977. [DOI] [PubMed] [Google Scholar]
- Pannese E, 2010. The structure of the perineuronal sheath of satellite glial cells (SGCs) in sensory ganglia. Neuron Glia Biol 6, 3–10. [DOI] [PubMed] [Google Scholar]
- Pertin M, Ji RR, Berta T, Powell AJ, Karchewski L, Tate SN, Isom LL, Woolf CJ, Gilliard N, Spahn DR, Decosterd I, 2005. Upregulation of the voltage-gated sodium channel beta2 subunit in neuropathic pain models: characterization of expression in injured and non-injured primary sensory neurons. J Neurosci 25, 10970–10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, Baker A, Anand-Apte B, 2003. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med 9, 407–415. [DOI] [PubMed] [Google Scholar]
- Renthal W, Tochitsky I, Yang L, Cheng YC, Li E, Kawaguchi R, Geschwind DH, Woolf CJ, 2020. Transcriptional Reprogramming of Distinct Peripheral Sensory Neuron Subtypes after Axonal Injury. Neuron 108, 128–144.e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retamal MA, Riquelme MA, Stehberg J, Alcayaga J, 2017. Connexin43 Hemichannels in Satellite Glial Cells, Can They Influence Sensory Neuron Activity? Front Mol Neurosci 10, 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Gibby CC, Anderson KO, Todd KH, 2016. Risk for Opioid Misuse Among Emergency Department Cancer Patients. Acad Emerg Med 23, 151–158. [DOI] [PubMed] [Google Scholar]
- Rosenberg GA, 2009. Matrix metalloproteinases and their multiple roles in neurodegenerative diseases. Lancet Neurol 8, 205–216. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, Woolf CJ, 2002. Can we conquer pain? Nature Neuroscience 5, 1062–1067. [DOI] [PubMed] [Google Scholar]
- Scholz J, Woolf CJ, 2007. The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci 10, 1361–1368. [DOI] [PubMed] [Google Scholar]
- Segond von Banchet G, Boettger MK, Fischer N, Gajda M, Bräuer R, Schaible HG, 2009. Experimental arthritis causes tumor necrosis factor-alpha-dependent infiltration of macrophages into rat dorsal root ganglia which correlates with pain-related behavior. Pain 145, 151–159. [DOI] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S, 2013. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature Reviews Genetics 14, 618–630. [DOI] [PubMed] [Google Scholar]
- Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, Ginty DD, 2020. The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siau C, Xiao W, Bennett G, 2006. Paclitaxel- and vincristine-evoked painful peripheral neuropathies: Loss of epidermal innervation and activation of Langerhans cells. Experimental Neurology 201, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JR, Lopes AH, Talbot J, Cecilio NT, Rossato MF, Silva RL, Souza GR, Silva CR, Lucas G, Fonseca BA, Arruda E, Alves-Filho JC, Cunha FQ, Cunha TM, 2017. Neuroimmune-Glia Interactions in the Sensory Ganglia Account for the Development of Acute Herpetic Neuralgia. J Neurosci 37, 6408–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge RE, Mapplebeck JC, Rosen S, Beggs S, Taves S, Alexander JK, Martin LJ, Austin JS, Sotocinal SG, Chen D, Yang M, Shi XQ, Huang H, Pillon NJ, Bilan PJ, Tu Y, Klip A, Ji RR, Zhang J, Salter MW, Mogil JS, 2015. Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci 18, 1081–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, 2019. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suadicani SO, Cherkas PS, Zuckerman J, Smith DN, Spray DC, Hanani M, 2010. Bidirectional calcium signaling between satellite glial cells and neurons in cultured mouse trigeminal ganglia. Neuron Glia Biol 6, 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Takahashi M, Nasu M, Matsumoto S, 2011. Peripheral inflammation suppresses inward rectifying potassium currents of satellite glial cells in the trigeminal ganglia. Pain 152, 2147–2156. [DOI] [PubMed] [Google Scholar]
- Tansley S, Uttam S, Ureña Guzmán A, Yaqubi M, Pacis A, Parisien M, Deamond H, Wong C, Rabau O, Brown N, Haglund L, Ouellet J, Santaguida C, Ribeiro-Da-Silva A, Tahmasebi S, Prager-Khoutorsky M, Ragoussis J, Zhang J, Salter MW, Diatchenko L, Healy LM, Mogil JS, Khoutorsky A, 2022. Single-cell RNA sequencing reveals time- and sex-specific responses of mouse spinal cord microglia to peripheral nerve injury and links ApoE to chronic pain. Nature Communications 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir-Yilmaz OE, Druckenbrod NR, Olukoya OO, Dong W, Yung AR, Bastille I, Pazyra-Murphy MF, Sitko AA, Hale EB, Vigneau S, Gimelbrant AA, Kharchenko PV, Goodrich LV, Segal RA, 2021. Diversity of developing peripheral glia revealed by single-cell RNA sequencing. Dev Cell 56, 2516–2535.e2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taves S, Berta T, Liu DL, Gan S, Chen G, Kim YH, Van de Ven T, Laufer S, Ji RR, 2016. Spinal inhibition of p38 MAP kinase reduces inflammatory and neuropathic pain in male but not female mice: Sex-dependent microglial signaling in the spinal cord. Brain Behav Immun 55, 70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonello R, Lee SH, Berta T, 2019. Monoclonal Antibody Targeting the Matrix Metalloproteinase 9 Prevents and Reverses Paclitaxel-Induced Peripheral Neuropathy in Mice. J Pain 20, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonello R, Xie W, Lee SH, Wang M, Liu X, Strong JA, Zhang JM, Berta T, 2020. Local Sympathectomy Promotes Anti-inflammatory Responses and Relief of Paclitaxel-induced Mechanical and Cold Allodynia in Mice. Anesthesiology 132, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy D, Daniele G, Fiorentino TV, Perez-Cadena Z, Chavez-Velasquez A, Kamath S, Fanti P, Jenkinson C, Andreozzi F, Federici M, Gastaldelli A, Defronzo RA, Folli F, 2013. Pioglitazone improves glucose metabolism and modulates skeletal muscle TIMP-3-TACE dyad in type 2 diabetes mellitus: a randomised, double-blind, placebo-controlled, mechanistic study. Diabetologia 56, 2153–2163. [DOI] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P, 2015. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18, 145–153. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke RE, Libert C, 2014. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov 13, 904–927. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM, 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Reviews Genetics 13, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shen YJ, Li XJ, Xia J, Sun L, Xu Y, Ma Y, Li D, Xiong YC, 2021. DNMT3b SUMOylation Mediated MMP-2 Upregulation Contribute to Paclitaxel Induced Neuropathic Pain. Neurochem Res 46, 1214–1223. [DOI] [PubMed] [Google Scholar]
- Wang X, Spandidos A, Wang H, Seed B, 2012. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res 40, D1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick RA, Hanani M, 2013. The contribution of satellite glial cells to chemotherapy-induced neuropathic pain. Eur J Pain 17, 571–580. [DOI] [PubMed] [Google Scholar]
- Wu Z, Wang S, Wu I, Mata M, Fink DJ, 2015. Activation of TLR-4 to produce tumour necrosis factor-α in neuropathic pain caused by paclitaxel. Eur J Pain 19, 889–898. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S, 2018. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Y, de Carvalho-Barbosa M, Kavelaars A, Heijnen CJ, Albrecht PJ, Dougherty PM, 2016. Dorsal Root Ganglion Infiltration by Macrophages Contributes to Paclitaxel Chemotherapy-Induced Peripheral Neuropathy. J Pain 17, 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and materials availability: Single-cell RNA sequencing data is available from the Gene Expression Omnibus under accession number GSE236914. All other data needed to support the conclusions in the paper are available in the paper and/or the Supplementary Materials. Requests regarding protocols and resources should be directed to and will be fulfilled by the lead contact T.B. (temugin.berta@uc.edu).


