Abstract
Periodontitis is a common, progressive disease that eventually affects the majority of the population. The local destruction of periodontitis is believed to result from a bacterial infection of the gingival sulcus, and several clinical studies have provided evidence to implicate Porphyromonas gingivalis. If P. gingivalis is a periodontal pathogen, it would be expected to be present in most subjects with disease and rarely detected in subjects with good periodontal health. However, in most previous studies, P. gingivalis has not been detected in the majority of subjects with disease, and age-matched, periodontally healthy controls were not included for comparison. The purpose of the study reported here was to compare the prevalence of P. gingivalis in a group with periodontitis to that of a group that is periodontally healthy. A comprehensive sampling strategy and a sensitive PCR assay were used to maximize the likelihood of detection. The target sequence for P. gingivalis-specific amplification was the transcribed spacer region within the ribosomal operon. P. gingivalis was detected in only 25% (46 of 181) of the healthy subjects but was detected in 79% (103 of 130) of the periodontitis group (P < 0.0001). The odds ratio for being infected with P. gingivalis was 11.2 times greater in the periodontitis group than in the healthy group (95% confidence interval, 6.5 to 19.2). These data implicate P. gingivalis in the pathogenesis of periodontitis and suggest that P. gingivalis may not be a normal inhabitant of a periodontally healthy dentition.
Periodontitis is a common, progressive disease that affects the supporting structures of the teeth, causing loss of attachment to the bone and often resulting in tooth loss (4). Moderate disease eventually affects the majority of persons, and advanced disease is seen in nearly half of individuals over 65 (3). A limited number of bacterial species have been associated with periodontitis, and the strongest evidence has accumulated to implicate Porphyromonas gingivalis. Studies have shown that P. gingivalis occurs with greater frequency and at higher levels at sites which appear to be disease active (15, 17, 18, 21) and that certain periodontal health indicators in individuals are inversely correlated with the presence or levels of P. gingivalis (1, 2, 10, 11, 16, 19). If P. gingivalis is a pathogen, then it would be expected to be detected in most subjects with disease and rarely detected in subjects who are periodontally healthy. However, in most previous studies, P. gingivalis has not been detected in the majority of subjects with disease. Low sensitivities of culturing techniques and sampling of a limited number of oral sites may have caused an underestimate of the organism’s true prevalence in these studies. Information about the presence of P. gingivalis in a healthy population is mostly limited to young subjects who may not yet have evidenced the cumulative destruction used for diagnosis, or from “healthy” sites in mouths with areas of destruction. Little information is available about its prevalence in those individuals who reach middle age without experiencing disease. No good animal model has been identified that would allow the direct testing of Koch’s postulates, but with the advent of molecular techniques for detecting and identifying bacteria, revisions to Koch’s original postulates for establishing microbial pathogenesis that do not require an animal model have been proposed (8). It has been suggested that to establish causality, the nucleic acid sequence belonging to a putative pathogen be detected in most cases of disease and that fewer or no copies should be detected in the absence of disease. Additional supporting evidence would include the disappearance of the pathogen-associated sequence with disease resolution, biological plausibility, and tissue localization (8). Conducting human epidemiologic studies on periodontitis that meet these criteria is challenging, since no direct test for periodontitis exists, and the presence of disease can only be determined by measuring slowly accumulating evidence of past destruction with insensitive and unreliable methods. The purpose of this study was to determine if P. gingivalis meets two major requirements of the revised postulates for sequence-based identification of microbial pathogens. In order to accomplish this, a group of subjects with clear indicators of periodontitis and an age-matched group of subjects who had not exhibited signs of substantial periodontal destruction were identified. A comprehensive sampling strategy and a sensitive and specific DNA sequence-based assay were used to maximize the likelihood of detection of P. gingivalis (14). Detection rates were found to be high in the periodontitis group and low in the healthy group, implicating P. gingivalis in the pathogenesis of periodontitis.
MATERIALS AND METHODS
Study population.
Subjects for this institutionally approved study were recruited from the clinics of the Ohio State University College of Dentistry. Potential subjects were screened, examined, and selected for participation if they met criteria for periodontal health or disease. These criteria were established to include approximately the most and least healthy one-fourth of the population based on epidemiologic data (13). Probing pocket depths and attachment levels were screened at six sites per tooth by a calibrated examiner with a ball-ended WHO probe with a colored band from 3.5 to 5.5 mm (Hu-Friedy PCP-11.5B). Subjects identified by screening with the WHO probe as meeting inclusion criteria were reexamined with a North Carolina probe (Hu-Friedy PCPUNC15), again measuring six sites per tooth. Subjects were included in the periodontitis group if loss of attachment was greater than or equal to 7 mm at a minimum of three sites and pocket depth was greater than or equal to 6 mm at a minimum of two sites, in order to rule out treated disease. Subjects were included in the healthy group if pocket depths and attachment levels were less than 5.5 mm for all sites. Subjects were excluded from the healthy group if less than 24 teeth remained, to avoid including those with treated disease. The healthy group was age matched to the diseased group.
Bacterial sampling.
Excess saliva was removed with a cotton roll or gauze pad to minimize collection of transient contaminating bacteria from an exogenous source. A sterile, medium endodontic paper point (Caulk-Dentsply) was placed in the mesial sulcus of each tooth for 10 s. All teeth present were sampled to maximize the possibility of detection of P. gingivalis if present at any site. Samples from each individual were pooled in a sterile 2-ml microcentrifuge tube and frozen for later analysis.
Detection of P. gingivalis.
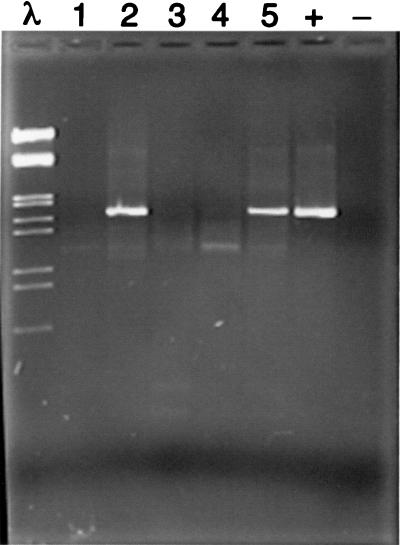
DNA was isolated and samples were analyzed for the presence of P. gingivalis, as previously described (14), with a PCR assay that did not require that the samples be cultured. The target sequence was the ribosomal DNA spacer region between the 16S and 23S ribosomal genes. Briefly, DNA was isolated from the paper points, and a P. gingivalis-specific fragment was generated by nested PCR with first a pair of universal prokaryotic primers followed by amplification with a species-specific primer located in the 16S gene (CGATATACCGTCAAGCTTCCACAG) paired with a universal primer located in the 23S gene. DNA fragments were separated by agarose gel electrophoresis. Samples were scored as positive or negative for P. gingivalis based on a clear band of the expected size (Fig. 1). All second amplifications were repeated and scored a second time. If the results were not in agreement, the amplification was repeated again. The presence of DNA was confirmed in all negative samples by using universal prokaryotic primers.
FIG. 1.
P. gingivalis-specific DNA fragments. The markers in the far left lane are EcoRI and HindIII digestion products of bacteriophage lambda DNA. The far right lane contains a control without template DNA, and next to it is a DNA fragment amplified from P. gingivalis ATCC 33277 which appears at 1.7 kb. Lanes numbered 1 through 5 contain DNA amplified from plaque samples. Of these, lanes 2 and 5 were scored as positive for the presence of P. gingivalis. The image was captured with a model 4910 CCD camera (Cohu, Inc., San Diego, Calif.) and an LG-3 Frame Grabber Board (Scion Corp., Frederick, Md.). Analysis was performed on a Power Macintosh 7100 with the public domain NIH Image program.
Statistical analysis.
The prevalence of P. gingivalis among healthy and diseased subjects and the sex and race distribution were compared by chi-square analysis. The odds ratio and 95% confidence interval were calculated for detection of P. gingivalis in the periodontitis group versus the healthy group. Age comparisons were made by using t tests.
RESULTS
Approximately 7,000 patient charts were screened, and from among these, approximately 600 individuals were selected for examination. Of these, 311 met inclusion criteria for the study: 130 in the periodontitis group and 181 in the healthy group. Highly statistically significant differences were observed between the prevalence of P. gingivalis in the healthy and periodontitis groups (P < 0.0001). P. gingivalis was detected in only 25% (46 of 181) of the healthy subjects but was found in 79% (103 of 130) of the periodontitis group (Table 1). The odds ratio for being infected with P. gingivalis was 11.2 times greater in the periodontitis group than in the healthy group (95% confidence interval, 6.5 to 19.2).
TABLE 1.
Prevalence of P. gingivalis in healthy and diseased groups
| Subject group | No. of subjects with P. gingivalis detected/no. tested (%)
|
||
|---|---|---|---|
| Diseased | Healthy | Total | |
| All | 103/130 (79) | 46/181 (25) | 149/311 (48) |
| Female | 42/51 (82) | 29/112 (26) | 71/163 (44) |
| Male | 61/79 (77) | 17/69 (25) | 78/148 (53) |
| White | 65/87 (75) | 33/149 (22) | 98/236 (42) |
| African-American | 28/32 (88) | 10/26 (38) | 38/58 (66) |
| Asian-American | 4/5 (80) | 3/5 (60) | 7/10 (70) |
| Other | 6/6 (100) | 0/1 (0) | 6/7 (86) |
The mean age of the periodontitis group was 51.4 years, and the mean age of the healthy group was 49.2 years (Table 2). This difference was not significant by t test (P = 0.18). No relationship was observed for detection of P. gingivalis and age for either group by t test (P = 0.31 for periodontitis group, P = 0.85 for the healthy group). A difference in sex distribution between the two groups was significant by chi-square analysis (P = 0.0002), with more females in the healthy group and fewer in the diseased group (Table 2). However, chi-square analysis showed no significant differences in colonization with P. gingivalis between males and females within groups (P = 0.48 for the periodontitis group and P = 0.85 for the healthy group) (Table 1). A larger number of African-Americans and a smaller number of white subjects were seen in the diseased group than in the healthy group (Table 2). This difference was significant by chi-square analysis (P = 0.02). However, no significant differences in colonization with P. gingivalis were seen within groups for race (P = 0.07 for the healthy group and P = 0.13 for the periodontitis group) (Table 1).
TABLE 2.
Demographics of the healthy and diseased groups
| Periodontal status | Mean age (yr) ± SD | No. (%) of subjects
|
Sample size (no.) | |||||
|---|---|---|---|---|---|---|---|---|
| Female | Male | White | African-American | Asian-American | Other | |||
| Diseased | 51.4 ± 12.8 | 51 (39) | 79 (61) | 87 (67) | 32 (25) | 5 (4) | 6 (5) | 130 |
| Healthy | 49.2 ± 15.2 | 112 (62) | 69 (38) | 149 (82) | 26 (14) | 5 (3) | 1 (1) | 181 |
| Total | 163 | 148 | 236 | 58 | 10 | 7 | 311 | |
DISCUSSION
To investigate the relationship between infection with P. gingivalis and periodontal health, a group of subjects with clear indicators of periodontitis and a healthy, age-matched control group were identified. A comprehensive sampling strategy was employed, and colonization with P. gingivalis was determined with a sensitive and specific PCR-based assay. Samples were taken from 130 adults with periodontitis and 181 controls. By this approach, the presence of P. gingivalis was found to be highly associated with disease, with the odds of detecting it more than 11-fold greater in the periodontitis group than in the healthy group, suggesting that P. gingivalis is a pathogen.
The PCR assay has been shown to be sensitive to as few as 10 bacteria (12), although some efficiency is no doubt lost in sampling and retrieving the bacteria from the paper points. The primer has been shown to be highly specific for P. gingivalis (7, 12), and a search of GenBank failed to find any matches. The spacer region is sufficiently variable that size differences in the fragment generated by PCR are observed among even closely related species, providing a further check for any cross-reactive species. We have analyzed several hundred PCR products by sequencing or heteroduplex analysis and have not found any false positives among the samples that were originally identified as P. gingivalis by PCR and agarose gel electrophoresis (9).
Measurable criteria for defining a group with periodontitis and a periodontally healthy group were adopted on the basis of previously reported data about population norms for pocket depth and attachment levels (13). Attachment levels and pocket depths in populations exhibit a continuous frequency distribution rather than a bimodal distribution into “health” and “disease” (13). Since no unequivocal thresholds exist to define periodontal health or disease, criteria were selected to allow the inclusion of no more than the healthiest and most diseased quartiles of the population for study.
Many more than half of the individuals screened for the study were found to fall between the two groups of interest, confirming that the criteria were selective. In particular, it was difficult to identify subjects of a sufficient age who met criteria for periodontal health to match the age of the periodontitis group. Since the subjects were drawn from the patient pool of dental school clinics, it is possible that fewer healthy subjects may have been available in the pool than in the general population. Efforts were made to recruit a larger sample size of healthy subjects, because infection rates were expected to be lower in this group, and similar numbers of infected subjects from each group were needed for future strain analysis.
The healthy group was well matched to the periodontitis group for age, with no significant difference seen. The healthy and periodontitis groups were not matched for sex or race, and there were more males and more African-American subjects in the diseased group than in the healthy group. The racial bias is consistent with previous reports of higher rates of periodontal disease among African-Americans. However, no significant differences in the prevalence of P. gingivalis were seen between sexes or among races in either the healthy or periodontitis groups. Thus, differences in prevalence of P. gingivalis between the healthy and periodontitis groups could not be attributed to age, sex, or race.
Studies have often relied upon dental professionals such as hygienists and dental students as healthy controls. However, we have shown that the prevalence and levels of P. gingivalis are atypically low in this group, probably due to unusually thorough oral hygiene (5). Therefore, the healthy subjects selected for this study were drawn from patient pools to provide a more representative control group.
If P. gingivalis is a pathogen, how can we account for the 25% detection rate among healthy subjects? Several plausible explanations deserve further study. Perhaps the criteria for health were not sufficiently stringent, since pocket depths and attachment loss of up to 5 mm were included. However, stratifying the data by deepest pocket and attachment loss did not show any relationship to prevalence in the healthy group (data not shown). It is also possible that the strains found in the healthy group are not the same as those found in periodontitis. Preliminary analysis to identify and compare strains in the two groups does in fact suggest that different strain groups are found under conditions of health and disease, and further analysis is indicated to address this interesting possibility (9). On the other hand, P. gingivalis was not detected in 21% of the periodontitis group. This could be accounted for by subjects who, although demonstrating evidence of past disease, no longer harbored detectable levels of the pathogen, although it appears to be difficult to eradicate (6, 20). It is also possible that P. gingivalis is not the only organism capable of causing periodontal destruction and that P. gingivalis was not the causative species in these individuals.
In summary, the odds of detecting P. gingivalis were 11.2-fold greater in individuals with periodontitis than in age-matched healthy subjects. This suggests that P. gingivalis is a periodontal pathogen. Investigation to determine if only a subset of strains are associated with disease and longitudinal studies to establish causality are needed.
ACKNOWLEDGMENTS
We gratefully acknowledge Gerard McDonnell for recruiting and sampling subjects and Lourdes Christopher for the examination of subjects.
This work was supported by NIH grant DE10467.
REFERENCES
- 1.Alpagot T, Wolff L F, Smith Q T, Tran S D. Risk indicators for periodontal disease in a racially diverse urban population. J Clin Periodontol. 1996;23:982–988. doi: 10.1111/j.1600-051x.1996.tb00524.x. [DOI] [PubMed] [Google Scholar]
- 2.Beck, J. D. 1994. Methods of assessing risk for periodontitis and developing multifactorial models. J. Periodontol. 65(Suppl. 5):468–478. [DOI] [PubMed]
- 3.Brown L J, Brunelle J A, Kingman A. Peridodontal status in the United States, 1988–1991: prevalence, extent, and demographic variation. J Dent Res. 1996;75(Special issue):672–683. doi: 10.1177/002203459607502S07. [DOI] [PubMed] [Google Scholar]
- 4.Brown L J, Oliver R C, Loe H. Periodontal diseases in the U.S. in 1981: prevalence, severity, extent, and role in tooth mortality. J Periodontol. 1989;60:363–370. doi: 10.1902/jop.1989.60.7.363. [DOI] [PubMed] [Google Scholar]
- 5.Christopher L A, Griffen A L, Leys E J. Low incidence of colonization with Porphyromonas gingivalis among dental students. J Dent Res. 1996;75(IADR abstracts):354. [Google Scholar]
- 6.Danser M M, Timmerman M F, van Winkelhoff A J, van der Velden U. The effect of periodontal treatment on periodontal bacteria on the oral mucous membranes. J Periodontol. 1996;67:478–485. doi: 10.1902/jop.1996.67.5.478. [DOI] [PubMed] [Google Scholar]
- 7.Dix K, Watanabe S M, McArdle S, Lee D I, Randolph C, Moncla B, Schwartz D E. Species-specific oligodeoxynucleotide probes for the identification of periodontal bacteria. J Clin Microbiol. 1990;28:319–323. doi: 10.1128/jcm.28.2.319-323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredericks D N, Relman D A. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffen A L, Moeschberger M L, Lyons S R, Leys E J. Identification of virulent strains of Porphyromonas gingivalis. J Dent Res. 1997;76(IADR abstracts):174. [Google Scholar]
- 10.Grossi S G, Zambon J J, Ho A W, Koch G, Dunford R G, Machtei E E, Norderyd O M, Genco R J. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- 11.Haffajee A D, Socransky S S, Smith C, Dibart S. Relation of baseline microbial parameters to future periodontal attachment loss. J Clin Periodontol. 1991;18:744–750. doi: 10.1111/j.1600-051x.1991.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 12.Leys E J, Griffen A L, Strong S J, Fuerst P A. Detection and strain identification of Actinobacillus actinomycetemcomitans by nested PCR. J Clin Microbiol. 1994;32:1288–1294. doi: 10.1128/jcm.32.5.1288-1294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machtei E E, Christersson L A, Grossi S G, Dunford R, Zambon J J, Genco R J. Clinical criteria for the definition of “established periodontitis.”. J Periodontol. 1992;63:206–214. doi: 10.1902/jop.1992.63.3.206. [DOI] [PubMed] [Google Scholar]
- 14.McClellan D L, Griffen A L, Leys E J. Age and prevalence of Porphyromonas gingivalis in children. J Clin Microbiol. 1995;34:2017–2019. doi: 10.1128/jcm.34.8.2017-2019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melvin W L, Assad D A, Miller G A, Gher M E, Simonson L, York A K. Comparison of DNA probe and ELISA microbial analysis methods and their association with adult periodontitis. J Periodontol. 1994;65:576–582. doi: 10.1902/jop.1994.65.6.576. [DOI] [PubMed] [Google Scholar]
- 16.Moore W E, Moore L H, Ranney R R, Smibert R M, Burmeister J A, Schenkein H A. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729–739. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 17.Preus H R, Anerud A, Boysen H, Dunford R G, Zambon J J, Loe H. The natural history of periodontal disease. The correlation of selected microbiological parameters with disease severity in Sri Lankan tea workers. J Clin Periodontol. 1995;22:674–678. doi: 10.1111/j.1600-051x.1995.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 18.Socransky S S, Haffajee A D, Smith C, Dibart S. Relation of counts of microbial species to clinical status at the sampled site. J Clin Periodontol. 1991;18:766–775. doi: 10.1111/j.1600-051x.1991.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 19.Teanpaisan R, Douglas C W, Walsh T F. Characterisation of black-pigmented anaerobes isolated from diseased and healthy periodontal sites. J Periodontal Res. 1995;30:245–251. doi: 10.1111/j.1600-0765.1995.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 20.von Troil-Linden B, Saarela M, Matto J, Alaluusua S, Jousimies-Somer H, Asikainen S. Source of suspected periodontal pathogens re-emerging after periodontal treatment. J Clin Periodontol. 1996;23:601–607. doi: 10.1111/j.1600-051x.1996.tb01831.x. [DOI] [PubMed] [Google Scholar]
- 21.Wolff L F, Aeppli D M, Pihlstrom B, Anderson L, Stoltenberg J, Osborn J, Hardie N, Shelburne C, Fischer G. Natural distribution of 5 bacteria associated with periodontal disease. J Clin Periodontol. 1993;20:699–706. doi: 10.1111/j.1600-051x.1993.tb00694.x. [DOI] [PubMed] [Google Scholar]