Abstract
PURPOSE
We determined whether a large, multianalyte panel of circulating biomarkers can improve detection of early-stage pancreatic ductal adenocarcinoma (PDAC).
MATERIALS AND METHODS
We defined a biologically relevant subspace of blood analytes on the basis of previous identification in premalignant lesions or early-stage PDAC and evaluated each in pilot studies. The 31 analytes that met minimum diagnostic accuracy were measured in serum of 837 subjects (461 healthy, 194 benign pancreatic disease, and 182 early-stage PDAC). We used machine learning to develop classification algorithms using the relationship between subjects on the basis of their changes across the predictors. Model performance was subsequently evaluated in an independent validation data set from 186 additional subjects.
RESULTS
A classification model was trained on 669 subjects (358 healthy, 159 benign, and 152 early-stage PDAC). Model evaluation on a hold-out test set of 168 subjects (103 healthy, 35 benign, and 30 early-stage PDAC) yielded an area under the receiver operating characteristic curve (AUC) of 0.920 for classification of PDAC from non-PDAC (benign and healthy controls) and an AUC of 0.944 for PDAC versus healthy controls. The algorithm was then validated in 146 subsequent cases presenting with pancreatic disease (73 benign pancreatic disease and 73 early- and late-stage PDAC cases) and 40 healthy control subjects. The validation set yielded an AUC of 0.919 for classification of PDAC from non-PDAC and an AUC of 0.925 for PDAC versus healthy controls.
CONCLUSION
Individually weak serum biomarkers can be combined into a strong classification algorithm to develop a blood test to identify patients who may benefit from further testing.
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is one of the most highly lethal diseases resulting in more than 47,000 deaths in the United States annually and nearly 460,000 deaths worldwide.1,2 This number is expected to rise3 with projections that PDAC will be the second leading cause of cancer deaths by 2030.4 Surgical resection remains the mainstay of curative therapy, with 5-year survival rising to 25% for patients who present with resectable disease.5 Advances in pancreatic surgery and the expanded use of neoadjuvant chemotherapy have increased the fraction of patients eligible for surgery and decreased operative morbidity6-8; however, the majority of patients still present with advanced, unresectable disease for which treatments remain ineffective.9 Five-year survival for PDAC remains around 10%, improved from 5% survival rates 20 years ago.10
CONTEXT
Key Objective
To develop a blood-based screening tool to discriminate early-stage pancreatic adenocarcinoma from benign pancreatic disease and healthy subjects.
Knowledge Generated
We demonstrate that a multianalyte panel of serum markers that are weak predictors individually could be combined using machine learning into an effective classification tool. The tool showed high specificity and actionable sensitivity in an independent validation cohort.
Relevance
This approach could be used as the basis of a primary screening test developed for detection of early-stage pancreatic ductal adenocarcinoma to suitably inform the need for secondary screening using imaging, particularly in groups at high risk for developing pancreatic adenocarcinoma.
An accurate, cost-effective screening protocol could substantially increase the number of patients eligible for surgical resection of newly diagnosed PDAC and greatly improve current PDAC-associated oncologic outcomes. Available evidence suggests at least a 5-year period between the development of malignant founder cells and acquisition of metastatic capacity, offering a window of opportunity for identification of early-stage, potentially curable disease.11 Screening for PDAC is a controversial topic, however, with no currently effective screening tool for detecting early tumors or asymptomatic tumors in the general population.12 The US Preventative Services Task Force recently discouraged screening for pancreatic cancer in asymptomatic adults as the risks of harm currently outweigh the benefits.13
A blood-based test may be useful as a primary screening tool for assessing the need for secondary screening by imaging. Limiting screening to groups with high-risk for developing PDAC would increase the pretest probability and reasonably offset the risk of false-positive diagnoses and costs with the survival benefits of early detection. Individual biomarkers, such as carcinoembryonic antigen and cancer antigen (CA) 19-9, lack the accuracy to be used independently for early-stage PDAC screening; however, a blood-based diagnostic panel relying on multiple biomarkers could be developed to improve sensitivity and specificity of a screening assay14,15 and could initially be deployed as a prefilter to inform the need for secondary screening by imaging in high-risk groups.
This study describes the performance of a multianalyte, blood-based primary screening test developed for detection of early-stage PDAC to suitably inform the need for secondary screening using imaging. Potential analytes were assembled from a large body of prior studies that investigated circulating biomarkers in early-stage disease.16-30,31-50,51-71 We used statistical learning to develop classification algorithms using the relationship between classes on the basis of the changes across analytes. A diagnostic algorithm was devised in a large development cohort and subsequently validated in an independent sample set.
MATERIALS AND METHODS
Subjects and Sample Collection
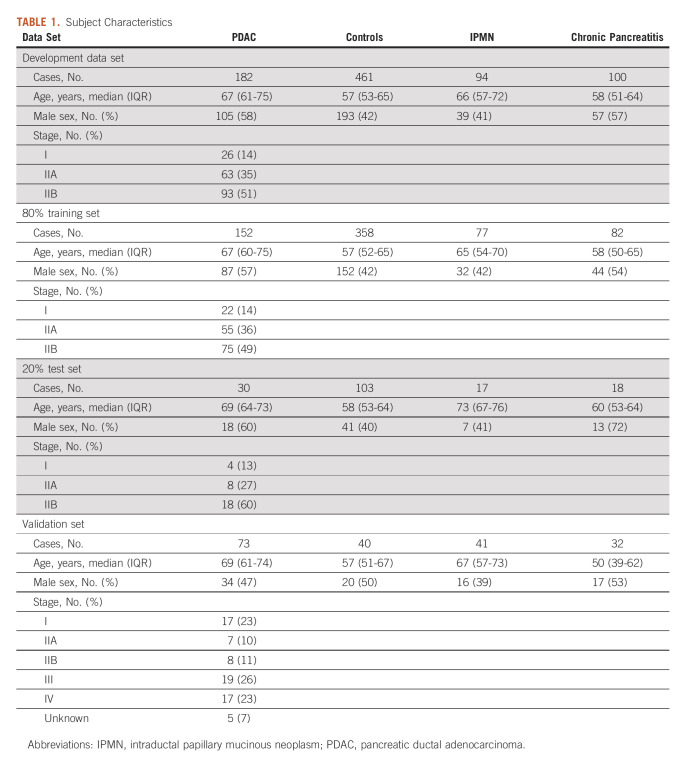
This study retrospectively evaluated a continuous set of prospectively collected samples from cases and controls and used clinical diagnoses as the reference standard. All patients presenting with suspicion of pancreatic disease and accompanying adults to serve as healthy control subjects were approached for enrollment in a research protocol at the Huntsman Cancer Institute between 2005 and 2019. Blood was collected during normal clinic hours from consenting adults and processed for serum, aliquoted, and frozen within 2 hours of collection. Fasting status was not imposed. Serum samples were stored at –80°C for <5 years or –140°C for <10 years before being assayed. Available samples were periodically queried for inclusion in this study, and samples from all patients meeting the inclusion criterion were assayed. Cases were chosen for inclusion if they had cytologically confirmed stage IA, IB, IIA, or IIB PDAC with an available treatment-naïve serum sample. Control samples from patients with benign pancreatic disease included patients with chronic pancreatitis and intraductal papillary mucinous neoplasm (IPMN, n = 94). Samples for IPMN cases were included if the patient had 3 years of subsequent follow-up without intervention for progression. Healthy control samples were included from accompanying adults (independent of the diagnosis of the primary case) and cases obtained from the University of Salerno (n = 39). Additional control cases included excess sera obtained from a regional reference laboratory. Subject characteristics are provided in Table 1. Informed written consent was obtained from each subject enrolled in the research protocol, and all studies were performed with the approval of the Institutional Review Board at the University of Utah in accordance to the principles of Declaration of Helsinki and the US Common Rule.
TABLE 1.
Subject Characteristics
Enzyme-Linked Immunosorbent Assay
A total of 837 serum samples were assayed for 31 analytes by enzyme-linked immunosorbent assay. Samples were processed in four batches over the course of 5 years. Commercially available kits (Supplementary Table S1, Data Supplement) were used for analyte quantification according to the manufacturers recommended protocols, including primary antibody optimization, where appropriate. The raw data were log transformed and adjusted for sex, age, and batch (fitting a natural spline with 2 df) using a linear model. The resulting adjusted data were normalized by centering and dividing by the standard deviation of the healthy control data.
Data Analysis
Complete data were available for all predictors, and imputation was not required. Statistical analyses and modeling were performed using R version 3.6.1.72 For diagnostic performance of individual analytes, areas under the receiver operating characteristic (ROC) curve (AUC) were determined. Bootstrap resampling was used to estimate results from repeated analyses. A random sampling with replacement of calibration samples was performed 2000 times, and the range of values was then recorded and compared with the average result. Ensemble models were built using the caretEnsemble package in R73 by stacking using gradient boosting. For modeling, all data with known class in the 837-sample data set were adjusted together before randomly separating into training (80%) and test (20%) sets. Discriminate algorithms were developed from the training data set for dichotomous classification of controls (including healthy, chronic pancreatitis, and benign IPMN patients) and early-stage PDAC cases. Using the training data set, multiple models were sequentially built to optimize AUC, sensitivity, and specificity, and the resulting probabilities averaged for all models. Method selection and tuning parameters for individual modeling methods were evaluated in an intermediate data subset consisting of 74 healthy control, 60 chronic pancreatitis, 76 IPMN, and 122 early-stage PDAC cases. Individual model performance was assessed by 10-fold cross validation. Ensemble classification models involved first optimizing submodels using individual classification methods ('glmnet', 'svmRadial', 'rf', 'nnet', 'knn' packages in R, Supplementary Table S2, Data Supplement) and then optimizing classification by combining the individual methods by stacking (gbm package in R). Again, performance of the ensemble models was assessed by 10-fold cross validation. Ensemble classification models were then used to predict class probabilities to the samples in the test set using a probability of 0.5 as the classification threshold. The final classification model was then applied to the 186-sample validation set after data adjustment.
RESULTS
Analyte Selection and Performance
Potential analytes were identified from prior studies that evaluated serum or plasma biomarkers in premalignant lesions, prediagnostic samples, or early-stage disease (Supplementary Table S3, Data Supplement). Candidate biomarkers were selected for inclusion in the panel if identified in two or more independent experiments from blood products in early-stage PDAC clinical samples. A subset of promising candidates that did not meet this standard were evaluated in pilot studies, and candidate biomarkers were included for further analysis if the analyte was significantly elevated in early-stage PDAC cases relative to healthy control subject (Supplementary Table S4, Data Supplement). A total of 31 analytes were selected for inclusion in the biomarker panel (Table 2). Most of the analytes are secreted and/or function within the extracellular space, and many participate in diverse biological processes associated with cancer (Supplementary Table S5, Data Supplement).
TABLE 2.
Analytes Included in Final Panel
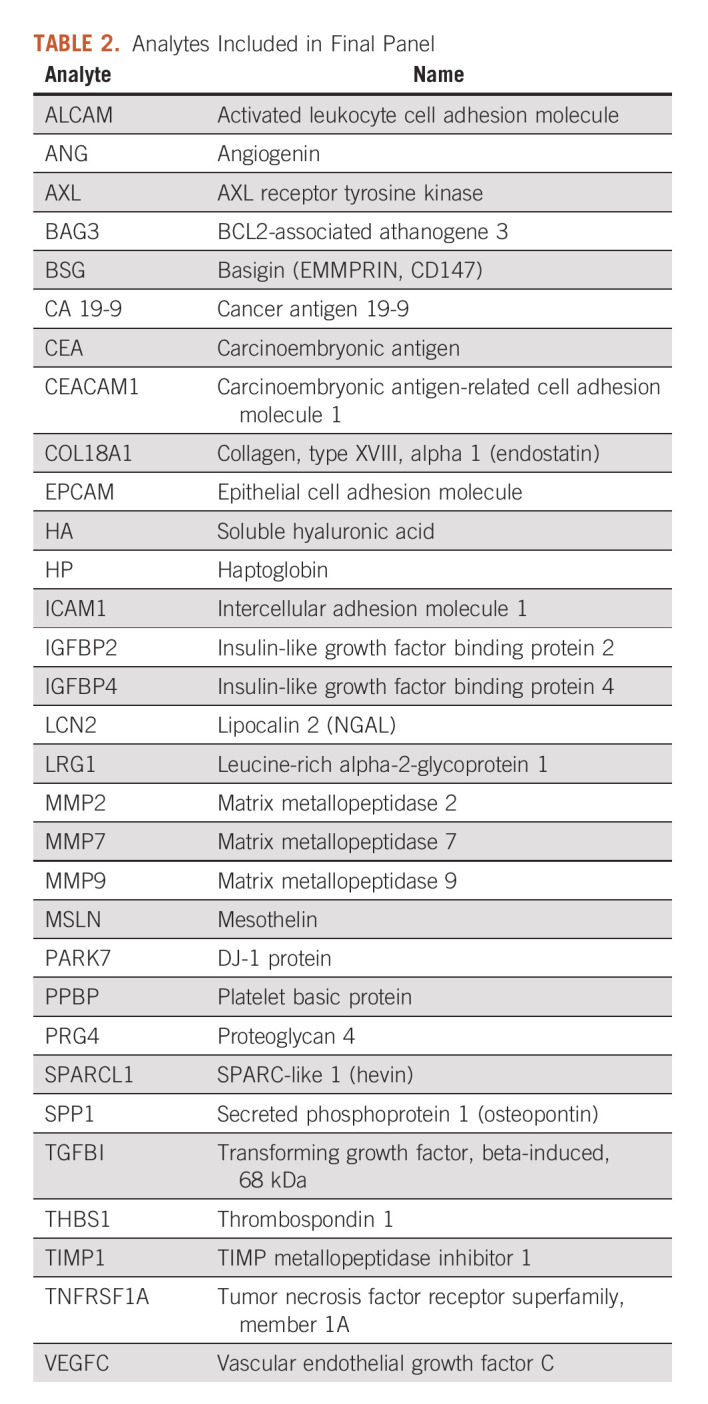
Levels of the selected analytes were measured in a development set of 837 serum samples from treatment-naïve patients with resectable PDAC, subjects with benign pancreatic disease, and subjects who were apparently healthy (Table 1). The diagnostic performance of the individual biomarkers was evaluated by ROC analyses (Supplementary Table S6, Data Supplement). Twenty-nine of the 31 analytes yielded significantly higher area under the ROC curve than predicted by chance for at least one of the comparisons between PDAC and benign IPMN, chronic pancreatitis, or healthy controls demonstrating their potential for diagnostic discrimination of early-stage PDAC. Two analytes (ANG and MMP2) did not yield significant discrimination in this data set despite compelling evidence in the literature. Since the approach allows for interactions between analytes and because elevation of an individual analyte may occur in some PDAC cases, but not reach significance in aggregate data, ANG and MMP2 were included in the data sets for algorithm development, testing, and validation. In general, the individual analytes were weak classifiers for discriminating PDAC from controls.
Diagnostic Development and Performance
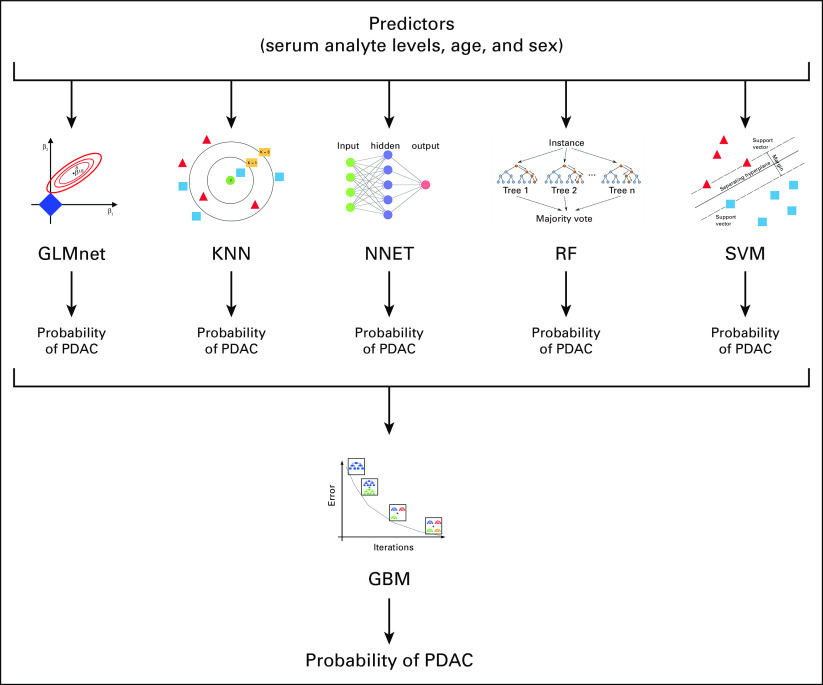
In an attempt to identify a stronger classifier, we used a machine learning approach to combine the information from the individual analytes (Fig 1). The approach proceeded in two steps. First, age, sex, and the 31 analyte levels in samples were provided as predictors to individual machine learning methods. The methods were chosen to represent dissimilar learning techniques,74 and pilot studies indicated that all predictors were used to some extent by one or more of the methods (Supplementary Table S7, Data Supplement). The individual methods were used to assign two-class probabilities (early-stage PDAC or non-PDAC control) for each case by optimizing ROC AUC via bootstrap resampling. Second, the outputs of the individual methods were combined by re-sampling into an ensemble model to assign the final class probabilities. The ensemble model was robust to sequential elimination of the individual methods, but each method contributed to model accuracy when specific combinations were considered (Supplementary Table S8, Data Supplement).
FIG 1.
Machine learning schematic for diagnostic classification. Predictors were fed to individual machine learning methods and used to assign two-class probabilities (early-stage PDAC or non-PDAC control) for each case. Outputs of the individual methods were combined into an ensemble model to assign the final class probabilities. GBM, generalized boosted regression model; GLMnet, lasso and elastic-net regularized generalized linear model; KNN, k-nearest neighbors; NNET, neural network; PDAC, pancreatic ductal adenocarcinoma; RF, random forest; SVM, support vector machines with radial basis function kernel.
The development set of 837 serum samples were randomly split into a training set representing 80% of the cases and a hold-out test set of 20% of cases. The two-step machine learning approach was applied to the training set to develop a diagnostic algorithm. The probabilities assigned by the algorithm to the training set resulted in a sensitivity of 92.8%, correctly identifying 141 of 152 PDAC cases. The algorithm yielded a specificity of 100%, correctly identifying all 358 healthy control cases. For benign pancreatic disease, the algorithm correctly identified all 82 chronic pancreatitis cases but misidentified 1 of 77 IPMN cases for an overall specificity of 99.8% for identification of non-PDAC subjects (Table 3). The high specificity of the algorithm was a desirable result, given the rarity of the disease, but is likely a consequence of the higher ratio of controls to cases rather than a design feature since overall diagnostic accuracy (case versus control) was used to optimize the algorithm.
TABLE 3.
Diagnostic Performance
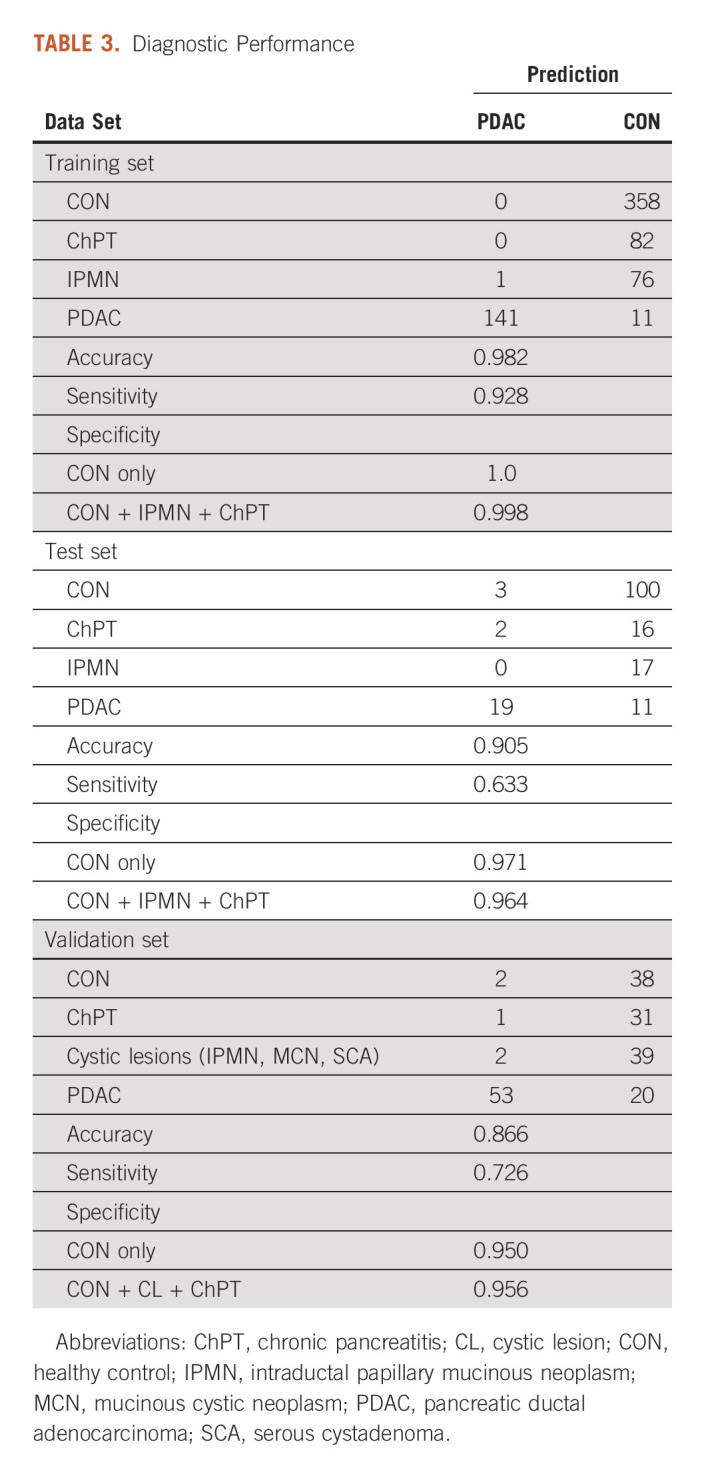
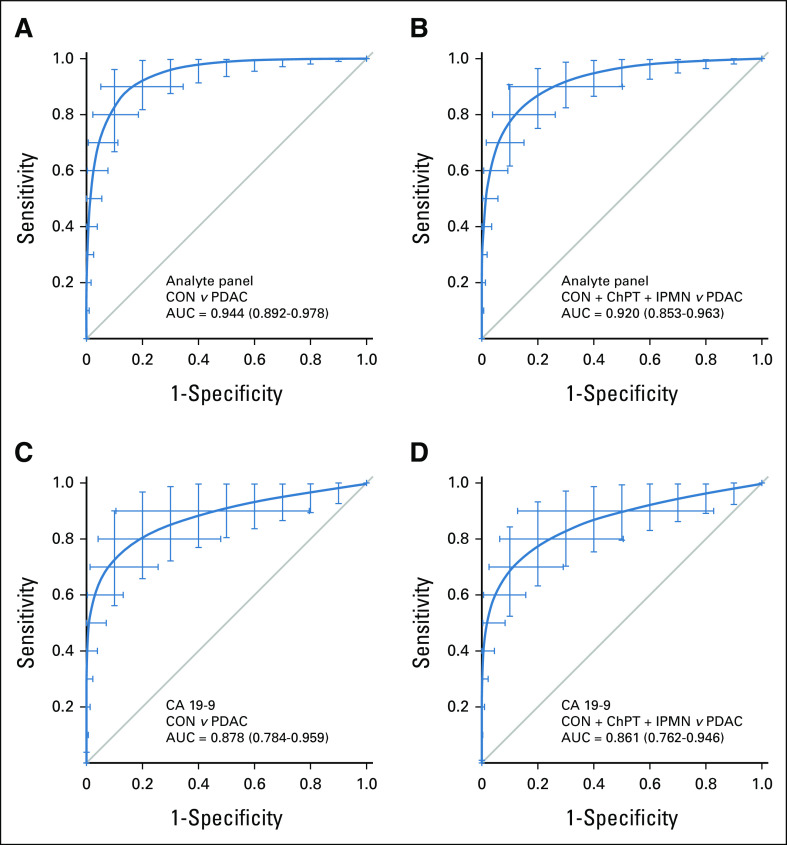
For objective assessment of algorithm accuracy, the resulting algorithm was applied to unused samples from the hold-out test set resulting in a sensitivity of 63.3%, correctly identifying 19 of 30 PDAC cases and a specificity of 97.1% (100 of 101 healthy controls). All 17 IPMN cases were correctly identified, as were 16 of 18 chronic pancreatitis cases, yielding an overall specificity of 96.4% for identification of non-PDAC cases (Table 3). ROC analysis illustrates that the diagnostic ability of the analyte panel for discrimination of PDAC from healthy controls (AUC = 0.944, Fig 2A) was greater than that of CA 19-9 alone (AUC = 0.878, Fig 2C) and for discrimination of PDAC from non-PDAC controls (analyte panel AUC = 0.920, Fig 2B v CA 19-9 AUC = 0.861, Fig 2D).
FIG 2.
Diagnostic performance in the test set. Receiver operating characteristic curves describing the ability of the analyte panel for discrimination of (A) early-stage PDAC cases from (B) healthy controls and all non-PDAC controls including chronic pancreatitis, benign IPMN, and healthy controls (C and D) compared with similar discrimination by CA 19-9 alone in the 20% hold-out test sample set. Bars represent 95% CIs from 2000 bootstrap iterations. AUC, area under the curve; CA, cancer antigen; ChPT, chronic pancreatitis; CON, healthy control; IPMN, intraductal papillary mucinous neoplasm; PDAC, pancreatic ductal adenocarcinoma.
Diagnostic Validation
The experimental design required that clinical information be available for model development, but the validation set was evaluated in the fixed algorithm in a blinded manner. Additional samples, collected over a period of 18 months after the final analyte panel algorithm was constructed using the development set (Table 1), were used to evaluate the diagnostic utility of the algorithm in a real-world setting. The 31 analytes were measured in 186 additional serum samples representing 73 PDAC, 32 chronic pancreatitis, and 41 cystic lesions (27 IPMN, 5 mucinous cystic neoplasm, and 9 serous cystadenoma) cases and 40 samples from healthy control subjects. These data, when supplied to the analyte panel algorithm, correctly identifying 53 of the 73 PDAC cases resulted in a sensitivity of 72.6%, which was better performance than the test set. Classification of controls in the validation set was consistent with results in the test set with a specificity of 95% (38 of 40) for healthy controls and 95.6% (108 of 113) for non-PDAC controls. In a rigorous validation using only those cases that satisfied the same inclusion criterion used for the development set (32 early-stage PDAC, 32 chronic pancreatitis, 40 healthy controls [cystic lesions had less than the required 3 years follow-up]), the algorithm yielded a sensitivity of 81.5% (26 of 32) and specificity of 95% (38 of 40) for healthy controls and 95.8% (69 of 72) for non-PDAC controls.
For the validation set, all cases presenting with pancreatic disease were evaluated. Although the development set only included early-stage resectable PDAC cases, the validation set included >50% late-stage cases (Table 1), reflecting the expected distribution of stages at presentation. For cases with known stage, the analyte panel sensitivity was higher for stage I-II cases (81.3%) than for stage III-IV cases (63.9%), although the comparison was not significant (P = .27 by Fisher's exact test). The development set also had the requirement that IPMN cases have at least 3 years of follow-up with no evidence of progression or high-grade dysplasia, which was not required for the validation set. Thus, cases with cystic lesions in the validation set may have included cases with more advanced disease, which cannot be excluded for the two cystic lesion cases in the validation set, both IPMN, that were classified as PDAC by the algorithm.
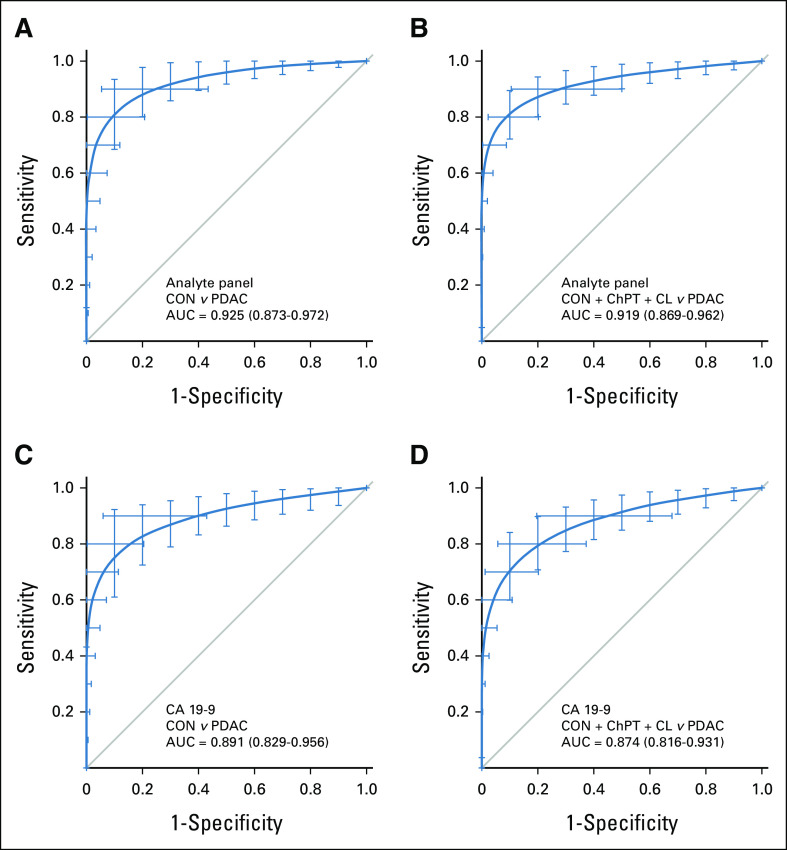
As with the test set, ROC analyses of the validation set showed greater diagnostic ability of the analyte panel for discrimination of PDAC than that of CA 19-9 alone. The analyte panel yielded an AUC of 0.925 for binary discrimination of PDAC from healthy controls (Fig 3A) compared with an AUC of 0.891 for CA 19-9 (Fig 3C). For discrimination of PDAC from all non-PDAC controls, the analyte panel had an AUC of 0.919 (Fig 3B) with CA 19-9 yielding an AUC of 0.874 (Fig 3D). For those PDAC cases that had CA 19-9 levels below the diagnostic threshold of 37 U/mL, the algorithm correctly classified 8 of 25 cases. These results confirm that the algorithm devised by machine learning in the development set was consistent in an independent sample set, show improvement over individual biomarkers, and suggest that the test may be useful for routine screening for early-stage disease in susceptible populations.
FIG 3.
Diagnostic performance in the validation set. Receiver operating characteristic curves describing the ability of the analyte panel for discrimination of (A) early-stage PDAC cases from (B) healthy controls and all non-PDAC controls including chronic pancreatitis, cystic lesions (IPMN, MCN, SCA), and healthy controls (C and D) compared with similar discrimination by CA 19-9 alone in the independent validation sample set. Bars represent 95% CIs from 2000 bootstrap iterations. AUC, area under the curve; CA, cancer antigen; ChPT, chronic pancreatitis; CL, cystic lesion; CON, healthy control; IPMN, intraductal papillary mucinous neoplasm; PDAC, pancreatic ductal adenocarcinoma.
Our approach was to use machine learning to uncover the interactions and structure of the data that would indicate the presence of early-stage disease. Changes in levels in individual analytes because of age or sex could potentially mask this structure. Despite adjusting for these factors in individual analytes, it was still possible that age and sex would contribute as effective predictors in the ensemble model, which seems to be justified by our variable importance subanalysis (Supplementary Table S7, Data Supplement). Diagnostic performance without age/sex preprocessing had slightly poorer overall performance (Supplementary Table S9, Data Supplement), although the results were reasonable and probably actionable.
DISCUSSION
Potential biomarkers evaluated for this study were previously identified as elevated in premalignant or early-stage PDAC and largely validated in an extensive development sample set. The selected biomarkers participate in diverse biological processes, including angiogenesis, apoptosis, immune response, inflammatory response, and extracellular signal transduction. One barrier to diagnostic accuracy is that biomarker elevation may indicate unrelated conditions, but since these processes are also relevant to cancer, a large combined panel should be less ambiguous. A serial screening tool, deployed annually or biannually, would tend to obviate any unrelated acute condition that would be expected to resolve over the course of months unless it was also related to a chronic condition such as cancer. A second barrier to diagnostic accuracy is heterogeneity that may arise from biological, behavioral, and temporal differences between cases and controls. Attempts to find single or small sets of biomarkers would be more prone to the confounding of unrelated conditions and disease heterogeneity.
All predictors that met initial selection criterion were included throughout training, testing, and validation phases with the expectation that different subsets of biomarkers would more effectively discriminate cases and controls by accommodating disease heterogeneity.14,75 The fact that all predictors contributed to diagnostic models supports this expectation (Supplementary Table S7, Data Supplement) and the differential utilization of predictors by the individual methods illustrate that different combinations of biomarkers contributed to discrimination. Derivation of the diagnostic algorithm was designed to be representative, not exhaustive, with the intent of minimizing overfitting and devising an extensible algorithm.
The goal of machine learning is to reveal underlying structure of the data, in this case to assign binary probabilities for designation as either case or control classification. Selection of a single method optimized on one data set may emphasize set-specific or spurious aspects of the data organization that may not faithfully represent the data structure for a different data set. Ensemble methods seek to overcome the limitations of a single machine learning method by using resampling to evaluate performance of multiple machine learning models and then generate a combination of the models that optimizes test performance. This approach has been proven to be as accurate or better than the best individual component.76 A general risk for machine learning approaches is overfitting to a data set that is unrepresentative to the general population; however, we found that our model gave almost identical AUC between test and validation, indicating that this algorithm may be generalizable. That bootstrap accuracy of the test set was 0.920 (AUC), and the accuracy of the independent validation set was nearly identical (AUC = 0.919), suggesting that the tradeoffs between undercomputing and overfitting were reasonable. Consistent with expectation, the algorithm performance was greater than the best individual biomarker, CA 19-9, although CA 19-9 performed very well in our sample sets as CA 19-9 assays achieved the highest range of historical accuracy.60
Ultimately, proof of efficacy will require prospective evaluation in a larger population demonstrating not only improved survival but also decreased mortality. In addition, as this study was largely a single institution investigation, the results will require validation in a multi-institutional cohort to increase external validity. Any screening test for PDAC must be highly specific to avoid false-positive screens leading to increased morbidity, cost, and emotional distress associated with secondary screening tests and treatment.77 The recommendation that screening be limited to high-risk groups78 would increase pretest probability and reduce the number of false-positive diagnoses as PDAC has a far lower prevalence in the general population than other commonly screened malignancies. Screening with imaging is relatively low risk.79,80 However, screening with primary imaging is expensive.81-83 The analytes and algorithms devised in this study may be useful as a primary screening tool for assessing the need for secondary screening by imaging while limiting concerns of unnecessary morbidity caused by false-positive determinations. The analyses routinely yielded the desirable high specificity, although with lower, but actionable sensitivity. In a serial screening program, the test will have the opportunity to increase overall sensitivity by re-evaluation of negative initial tests.
The analyses show that the general approach to PDAC screening is valid, that our samples generate reproducible signals, and that novel analytes contribute to disease classification. This 31-biomarker assay may be helpful in developing a useful tool for identifying early-stage, asymptomatic PDAC, particularly for screening in high-risk patient populations to guide the use of screening imaging. The results also serve to illustrate the proposed analytical methods which allows for interaction between analytes, subsets, and heterogeneous biological response among subjects. Finally, the approach allows for improvement through incorporation of additional biomarkers, including signals from cell-free DNA, circulating tumor cells, and demographic factors.
Matthew A. Firpo
Patents, Royalties, Other Intellectual Property: Patent Pending for Early Detection Biomarker Panel for Pancreatic Cancer (Inst)
Alessandra Rosati
Stock and Other Ownership Interests: Biouniversa, Fibrosys
Liberato Marzullo
Stock and Other Ownership Interests: Biouniversa s.r.l, Fibrosys s.r.l
Margot De Marco
Stock and Other Ownership Interests: Biouniversa, Fibrosys
Antonia Falco
Stock and Other Ownership Interests: Biouniversa
Ignacio Garrido-Laguna
Consulting or Advisory Role: SOTIO, Kanaph Therapeutics, Jazz Pharmaceuticals, OncXerna Therapeutics
Research Funding: Novartis (Inst), Bayer (Inst), Bristol Myers Squibb (Inst), Pfizer (Inst), MedImmune (Inst), Lilly (Inst), Incyte (Inst), GlaxoSmithKline (Inst), Tolero Pharmaceuticals (Inst), BridgeBio Pharma (Inst), Jacobio (Inst), Repare Therapeutics (Inst), Sumitomo Dainippon Pharma Oncology (Inst), Revolution Medicines (Inst)
M. Caterina Turco
Stock and Other Ownership Interests: Biouniversa, Fibrosys
Sean J. Mulvihill
Patents, Royalties, Other Intellectual Property: Intellectual property and patent pending for diagnostic test for pancreatic cancer (Inst), For profit company, Prior Diagnostics, LLC, formed to commercialize diagnostic test for pancreatic cancer
No other potential conflicts of interest were reported.
SUPPORT
Supported in part by research grants from the National Institutes of Health (CA115225, CA151650, CA155586, CA196403, CA200468 to S.J.M. and P30CA042014 to the Huntsman Cancer Institute for support of core facilities), grants from the Huntsman Cancer Institute Gastrointestinal Cancer Research Program and through support from the Huntsman Cancer Foundation (M.A.F. and S.J.M.).
PREPRINT VERSION
Preprint version available on https://www.medrxiv.org/content/10.1101/2022.03.03.22271867v1.
DATA SHARING STATEMENT
Deidentified data generated in this study are available within the article and its supplementary data files.
AUTHOR CONTRIBUTIONS
Conception and design: Matthew A. Firpo, Josh Bleicher, Courtney L. Scaife, Sean J. Mulvihill
Financial support: Sean J. Mulvihill
Administrative support: Sean J. Mulvihill
Provision of study materials or patients: Ignacio Garrido-Laguna
Collection and assembly of data: Matthew A. Firpo, Gayatri D. Khanderao, Alessandra Rosati, Katherine E. Poruk, Sama Kamal, Liberato Marzullo, Margot De Marco, Antonia Falco, Armando Genovese, Jessica M. Adler, Douglas G. Adler, Kajsa E. Affolter, Courtney L. Scaife, M. Caterina Turco, Sean J. Mulvihill
Data analysis and interpretation: Matthew A. Firpo, Kenneth M. Boucher, Josh Bleicher, Vincenzo De Laurenzi, Ignacio Garrido-Laguna, Courtney L. Scaife, M. Caterina Turco
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Matthew A. Firpo
Patents, Royalties, Other Intellectual Property: Patent Pending for Early Detection Biomarker Panel for Pancreatic Cancer (Inst)
Alessandra Rosati
Stock and Other Ownership Interests: Biouniversa, Fibrosys
Liberato Marzullo
Stock and Other Ownership Interests: Biouniversa s.r.l, Fibrosys s.r.l
Margot De Marco
Stock and Other Ownership Interests: Biouniversa, Fibrosys
Antonia Falco
Stock and Other Ownership Interests: Biouniversa
Ignacio Garrido-Laguna
Consulting or Advisory Role: SOTIO, Kanaph Therapeutics, Jazz Pharmaceuticals, OncXerna Therapeutics
Research Funding: Novartis (Inst), Bayer (Inst), Bristol Myers Squibb (Inst), Pfizer (Inst), MedImmune (Inst), Lilly (Inst), Incyte (Inst), GlaxoSmithKline (Inst), Tolero Pharmaceuticals (Inst), BridgeBio Pharma (Inst), Jacobio (Inst), Repare Therapeutics (Inst), Sumitomo Dainippon Pharma Oncology (Inst), Revolution Medicines (Inst)
M. Caterina Turco
Stock and Other Ownership Interests: Biouniversa, Fibrosys
Sean J. Mulvihill
Patents, Royalties, Other Intellectual Property: Intellectual property and patent pending for diagnostic test for pancreatic cancer (Inst), For profit company, Prior Diagnostics, LLC, formed to commercialize diagnostic test for pancreatic cancer
No other potential conflicts of interest were reported.
REFERENCES
- 1.The Global Cancer Observatory Group : Source: Globocan 2018. World Heal Organ 876:2018-2019, 2019 [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2020. CA Cancer J Clin 70:7-30, 2020 [DOI] [PubMed] [Google Scholar]
- 3.Saad AM, Turk T, Al-Husseini MJ, et al. : Trends in pancreatic adenocarcinoma incidence and mortality in the United States in the last four decades; a SEER-based study. BMC Cancer 18:688, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahib L, Smith BD, Aizenberg R, et al. : Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74:2913-2921, 2014 [DOI] [PubMed] [Google Scholar]
- 5.Vincent A, Herman J, Schulick R, et al. : Pancreatic cancer. Lancet 378:607-620, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acher AW, Bleicher J, Cannon A, et al. : Advances in surgery for pancreatic cancer. J Gastrointest Oncol 9:1037-1043, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bockhorn M, Uzunoglu FG, Adham M, et al. : Borderline resectable pancreatic cancer: A consensus statement by the International Study Group of Pancreatic Surgery (ISGPS). Surgery 155:977-988, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Heinrich S, Besselink M, Moehler M, et al. : Opinions and use of neoadjuvant therapy for resectable, borderline resectable, and locally advanced pancreatic cancer: International survey and case-vignette study. BMC Cancer 19:675, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilic M, Ilic I: Epidemiology of pancreatic cancer. World J Gastroenterol 22:9694-9705, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Cancer Institute - Surveillance Epidemiology and End Results Program : Cancer Stat Facts: Pancreatic Cancer. https://seer.cancer.gov/statfacts/html/pancreas.html
- 11.Yachida S, Jones S, Bozic I, et al. : Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467:1114-1117, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan DP, Hong TS, Bardeesy N: Pancreatic adenocarcinoma. N Engl J Med 371:1039-1049, 2014 [DOI] [PubMed] [Google Scholar]
- 13.Owens DK, Davidson KW, Krist AH, et al. : Screening for pancreatic cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. JAMA 322:438-444, 2019 [DOI] [PubMed] [Google Scholar]
- 14.Firpo MA, Boucher KM, Mulvihill SJ: Prospects for developing an accurate diagnostic biomarker panel for low prevalence cancers. Theor Biol Med Model 11:34, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Bamlet WR, Oberg AL, et al. : Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci Transl Med 9:eaah5583, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argani P, Iacobuzio-Donahue C, Ryu B, et al. : Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: Identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE). Clin Cancer Res 7:3862-3868, 2001 [PubMed] [Google Scholar]
- 17.Bramhall SR, Neoptolemos JP, Stamp GWH, et al. : Imbalance of expression of matrix metalloproteinases (MMPs) and tissue inhibitors of the matrix metalloproteinases (TIMPs) in human pancreatic carcinoma. J Pathol 182:347-355, 1997 [DOI] [PubMed] [Google Scholar]
- 18.Buchholz M, Braun M, Heidenblut A, et al. : Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene 24:6626-6636, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Chen R, Crispin DA, Pan S, et al. : Pilot study of blood biomarker candidates for detection of pancreatic cancer. Pancreas 39:981-988, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen R, Yi EC, Donohoe S, et al. : Pancreatic cancer proteome: The proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology 129:1187-1197, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Coppola D, Szabo M, Boulware D, et al. : Correlation of osteopontin protein expression and pathological stage across a wide variety of tumor histologies. Clin Cancer Res 10:184-190, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Del Villano BC, Brennan S, Brock P, et al. : Radioimmunometric assay for a monoclonal antibody-defined tumor marker, CA 19-9. Clin Chem 29:549-552, 1983 [PubMed] [Google Scholar]
- 23.Ellenrieder V, Alber B, Lacher U, et al. : Role of MT-MMPs and MMP-2 in pancreatic cancer progression. Int J Cancer 85:14-20, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Esposito I, Kayed H, Keleg S, et al. : Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia 9:8-17, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faca VM, Song KS, Wang H, et al. : A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med 5:e123, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falco A, Rosati A, Festa M, et al. : BAG3 is a novel serum biomarker for pancreatic adenocarcinomas. Am J Gastroenterol 108:1178-1180, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Firpo MA, Gay DZ, Granger SR, et al. : Improved diagnosis of pancreatic adenocarcinoma using haptoglobin and serum amyloid A in a panel screen. World J Surg 33:716-722, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gebauer F, Struck L, Tachezy M, et al. : Serum EpCAM expression in pancreatic cancer. Anticancer Res 34:4741-4746, 2014 [PubMed] [Google Scholar]
- 29.Giusti G, Piccinino F, Sagnelli E, et al. : Differential diagnosis between benign and malignant biliary tract obstruction: Discriminative usefulness of some clinical and laboratory data. Acta Hepatogastroenterol (Stuttg) 22:374-379, 1975 [PubMed] [Google Scholar]
- 30.Grutzmann R, Pilarsky C, Ammerpohl O, et al. : Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia 6:611-622, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harsha HC, Kandasamy K, Ranganathan P, et al. : A compendium of potential biomarkers of pancreatic cancer. PLoS Med 6:e1000046, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harvey SR, Hurd TC, Markus G, et al. : Evaluation of urinary plasminogen activator, its receptor, matrix metalloproteinase-9, and von Willebrand factor in pancreatic cancer. Clin Cancer Res 9:4935-4943, 2003 [PubMed] [Google Scholar]
- 33.Hassan R, Laszik ZG, Lerner M, et al. : Mesothelin is overexpressed in pancreaticobiliary adenocarcinomas but not in normal pancreas and chronic pancreatitis. Am J Clin Pathol 124:838-845, 2005 [PubMed] [Google Scholar]
- 34.He XY, Liu BY, Yao WY, et al. : Serum DJ-1 as a diagnostic marker and prognostic factor for pancreatic cancer. J Dig Dis 12:131-137, 2011 [DOI] [PubMed] [Google Scholar]
- 35.Holyoke D, Reynoso G, Chu TM: Carcinoembryonic antigen (CEA) in patients with carcinoma of the digestive tract. Ann Surg 176:559-564, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacobuzio-Donahue CA, Maitra A, Olsen M, et al. : Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol 162:1151-1162, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iacobuzio-Donahue CA, Ryu B, Hruban RH, et al. : Exploring the host desmoplastic response to pancreatic carcinoma: Gene expression of stromal and neoplastic cells at the site of primary invasion. Am J Pathol 160:91-99, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston FM, Tan MC, Tan BR Jr., et al. : Circulating mesothelin protein and cellular antimesothelin immunity in patients with pancreatic cancer. Clin Cancer Res 15:6511-6518, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kakisaka T, Kondo T, Okano T, et al. : Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE): Up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. J Chromatogr B Analyt Technol Biomed Life Sci 852:257-267, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasper HU, Ebert M, Malfertheiner P, et al. : Expression of thrombospondin-1 in pancreatic carcinoma: Correlation with microvessel density. Virchows Arch 438:116-120, 2001 [DOI] [PubMed] [Google Scholar]
- 41.Kendrick ZW, Firpo MA, Repko RC, et al. : Serum IGFBP2 and MSLN as diagnostic and prognostic biomarkers for pancreatic cancer. HPB (Oxford) 16:670-676, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolb A, Kleeff J, Guweidhi A, et al. : Osteopontin influences the invasiveness of pancreatic cancer cells and is increased in neoplastic and inflammatory conditions. Cancer Biol Ther 4:740-746, 2005 [DOI] [PubMed] [Google Scholar]
- 43.Koopmann J, Fedarko NS, Jain A, et al. : Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev 13:487-491, 2004 [PubMed] [Google Scholar]
- 44.Koopmann J, Rosenzweig CNW, Zhang Z, et al. : Serum markers in patients with resectable pancreatic adenocarcinoma: Macrophage inhibitory cytokine 1 versus CA19-9. Clin Cancer Res 12:442-446, 2006 [DOI] [PubMed] [Google Scholar]
- 45.Kuhlmann KF, van Till JO, Boermeester MA, et al. : Evaluation of matrix metalloproteinase 7 in plasma and pancreatic juice as a biomarker for pancreatic cancer. Cancer Epidemiol Biomarkers Prev 16:886-891, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurell H, Bouisson M, Berthelemy P, et al. : Identification of biomarkers of human pancreatic adenocarcinomas by expression profiling and validation with gene expression analysis in endoscopic ultrasound-guided fine needle aspiration samples. World J Gastroenterol 12:3344-3351, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M, Zhai Q, Bharadwaj U, et al. : Cyclophilin A is overexpressed in human pancreatic cancer cells and stimulates cell proliferation through CD147. Cancer 106:2284-2294, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Li YJ, Wei ZM, Meng YX, et al. : Beta-catenin up-regulates the expression of cyclinD1, c-myc and MMP-7 in human pancreatic cancer: Relationships with carcinogenesis and metastasis. World J Gastroenterol 11:2117-2123, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Q, Ozawa F, Friess H, et al. : The anti-apoptotic protein BAG-3 is overexpressed in pancreatic cancer and induced by heat stress in pancreatic cancer cell lines. FEBS Lett 503:151-157, 2001 [DOI] [PubMed] [Google Scholar]
- 50.Logsdon CD, Simeone DM, Binkley C, et al. : Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res 63:2649-2657, 2003 [PubMed] [Google Scholar]
- 51.Mahlbacher V, Sewing A, Elsasser HP, et al. : Hyaluronan is a secretory product of human pancreatic adenocarcinoma cells. Eur J Cell Biol 58:28-34, 1992 [PubMed] [Google Scholar]
- 52.Maitra A, Adsay NV, Argani P, et al. : Multicomponent analysis of the pancreatic adenocarcinoma progression model using a pancreatic intraepithelial neoplasia tissue microarray. Mod Pathol 16:902-912, 2003 [DOI] [PubMed] [Google Scholar]
- 53.Melle C, Ernst G, Escher N, et al. : Protein profiling of microdissected pancreas carcinoma and identification of HSP27 as a potential serum marker. Clin Chem 53:629-635, 2007 [DOI] [PubMed] [Google Scholar]
- 54.Missiaglia E, Blaveri E, Terris B, et al. : Analysis of gene expression in cancer cell lines identifies candidate markers for pancreatic tumorigenesis and metastasis. Int J Cancer 112:100-112, 2004 [DOI] [PubMed] [Google Scholar]
- 55.Ohlund D, Ardnor B, Oman M, et al. : Expression pattern and circulating levels of endostatin in patients with pancreas cancer. Int J Cancer 122:2805-2810, 2008 [DOI] [PubMed] [Google Scholar]
- 56.Ordonez NG: Application of mesothelin immunostaining in tumor diagnosis. Am J Surg Pathol 27:1418-1428, 2003 [DOI] [PubMed] [Google Scholar]
- 57.Pan S, Chen R, Crispin DA, et al. : Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling. J Proteome Res 10:2359-2376, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park HD, Kang ES, Kim JW, et al. : Serum CA19-9, cathepsin D, and matrix metalloproteinase-7 as a diagnostic panel for pancreatic ductal adenocarcinoma. Proteomics 12:3590-3597, 2012 [DOI] [PubMed] [Google Scholar]
- 59.Poruk KE, Firpo MA, Scaife CL, et al. : Serum osteopontin and tissue inhibitor of metalloproteinase 1 as diagnostic and prognostic biomarkers for pancreatic adenocarcinoma. Pancreas 42:193-197, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.E Poruk K, Z Gay D, Brown K, et al. : The clinical utility of CA 19-9 in pancreatic adenocarcinoma: Diagnostic and prognostic updates. Curr Mol Med 13:340-351, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qian X, Rothman VL, Nicosia RF, et al. : Expression of thrombospondin-1 in human pancreatic adenocarcinomas: Role in matrix metalloproteinase-9 production. Pathol Oncol Res 7:251-259, 2001 [DOI] [PubMed] [Google Scholar]
- 62.Riethdorf S, Reimers N, Assmann V, et al. : High incidence of EMMPRIN expression in human tumors. Int J Cancer 119:1800-1810, 2006 [DOI] [PubMed] [Google Scholar]
- 63.Schneiderhan W, Diaz F, Fundel M, et al. : Pancreatic stellate cells are an important source of MMP-2 in human pancreatic cancer and accelerate tumor progression in a murine xenograft model and CAM assay. J Cell Sci 120:512-519, 2007 [DOI] [PubMed] [Google Scholar]
- 64.Shimoyama S, Gansauge F, Gansauge S, et al. : Increased angiogenin expression in pancreatic cancer is related to cancer aggressiveness. Cancer Res 56:2703-2706, 1996 [PubMed] [Google Scholar]
- 65.Shimoyama S, Gansauge F, Gansauge S, et al. : Increased angiogenin expression in obstructive chronic pancreatitis surrounding pancreatic cancer but not in pure chronic pancreatitis. Pancreas 18:225-230, 1999 [DOI] [PubMed] [Google Scholar]
- 66.Simeone DM, Ji B, Banerjee M, et al. : CEACAM1, a novel serum biomarker for pancreatic cancer. Pancreas 34:436-443, 2007 [DOI] [PubMed] [Google Scholar]
- 67.Tang RF, Itakura J, Aikawa T, et al. : Overexpression of lymphangiogenic growth factor VEGF-C in human pancreatic cancer. Pancreas 22:285-292, 2001 [DOI] [PubMed] [Google Scholar]
- 68.Xie MJ, Motoo Y, Su SB, et al. : Expression of clusterin in human pancreatic cancer. Pancreas 25:234-238, 2002 [DOI] [PubMed] [Google Scholar]
- 69.Zeh HJ, Winikoff S, Landsittel DP, et al. : Multianalyte profiling of serum cytokines for detection of pancreatic cancer. Cancer Biomarkers 1:259-269, 2005 [DOI] [PubMed] [Google Scholar]
- 70.Zhang W, Erkan M, Abiatari I, et al. : Expression of extracellular matrix metalloproteinase inducer (EMMPRIN/CD147) in pancreatic neoplasm and pancreatic stellate cells. Cancer Biol Ther 6:218-227, 2007 [DOI] [PubMed] [Google Scholar]
- 71.Zhou W, Sokoll LJ, Bruzek DJ, et al. : Identifying markers for pancreatic cancer by gene expression analysis. Cancer Epidemiol Biomarkers Prev 7:109-112, 1998 [PubMed] [Google Scholar]
- 72.R Core Team : R: A Language and Environment for Statistical Computing. Vienna, Austria, R Foundation for Statistical Computing, 2019 [Google Scholar]
- 73.Deane-Mayer ZA, Knowles JE: caretEnsemble: Ensembles of Caret Models. R Package Version 2.0.0, 2016. https://CRAN.R-project.org/package=caretEnsemble [Google Scholar]
- 74.Kuhn M: The Caret Package. https://topepo.github.io/caret/models-clustered-by-tag-similarity.html
- 75.Crawford HC, Wallace MB, Storz P: Early detection and imaging strategies to reveal and target developing pancreatic cancer. Expert Rev Anticancer Ther 20:81-83, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van der Laan MJ, Polley EC, Hubbard AE: Super learner. Stat Appl Genet Mol Biol 6:25, 2007 [DOI] [PubMed] [Google Scholar]
- 77.Poruk KE, Firpo MA, Adler DG, et al. : Screening for pancreatic cancer: Why, how, and who? Ann Surg 257:17-26, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Canto MI, Harinck F, Hruban RH, et al. : International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut 62:339-347, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Canto MI, Goggins M, Hruban RH, et al. : Screening for early pancreatic neoplasia in high-risk individuals: A prospective controlled study. Clin Gastroenterol Hepatol 4:665-681, 2006; quiz 665 [DOI] [PubMed] [Google Scholar]
- 80.Canto MI, Goggins M, Yeo CJ, et al. : Screening for pancreatic neoplasia in high-risk individuals: An EUS-based approach. Clin Gastroenterol Hepatol 2:606-621, 2004 [DOI] [PubMed] [Google Scholar]
- 81.Zubarik R, Gordon SR, Lidofsky SD, et al. : Screening for pancreatic cancer in a high-risk population with serum CA 19-9 and targeted EUS: A feasibility study. Gastrointest Endosc 74:87-95, 2011 [DOI] [PubMed] [Google Scholar]
- 82.Rubenstein JH, Scheiman JM, Anderson MA: A clinical and economic evaluation of endoscopic ultrasound for patients at risk for familial pancreatic adenocarcinoma. Pancreatology 7:514-525, 2007 [DOI] [PubMed] [Google Scholar]
- 83.Rulyak SJ, Kimmey MB, Veenstra DL, et al. : Cost-effectiveness of pancreatic cancer screening in familial pancreatic cancer kindreds. Gastrointest Endosc 57:23-29, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Deidentified data generated in this study are available within the article and its supplementary data files.