Abstract
This study compared the performances of three human papillomavirus (HPV) detection tests with specimens collected by three alternative procedures. The HPV tests included the Hybrid Capture Tube test (HCT), the microplate-based Hybrid Capture II test (HC II), and the MY09-MY11 L1 consensus primer PCR-based assay. Initial cervical specimens were collected from study subjects with a broom device, and after Papanicolaou smears were made, residual specimens were placed into PreservCyt (PC), a liquid cytology medium. A second specimen was collected from each subject and placed into Digene Specimen Transport Medium (STM). The device for collection of the second specimen alternated with consecutive subjects between a conical cytology brush and a Dacron swab. At the 1.0-pg/ml cutoff, the results of the HC II agreed well with those of the PCR. Specifically, when PCR data were restricted to the types found by the HC II (HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68), there was greater than 90% agreement between the HC II and PCR results with both STM and PC. At a lower cutoff (0.2 pg/ml), HC II-positive results increased further, especially when the test was applied to the PC specimens. However, false-positive HC II results were more often observed at the 0.2-pg/ml cutoff. HC II yielded the highest HPV positivity with specimens placed into PC, followed by specimens collected with a conical brush and placed into STM and, last, by those collected with a Dacron swab and placed into STM. Our results demonstrate the utility of both the STM and PC specimen collection methods and show good agreement between the HC II and PCR.
Certain human papillomaviruses (HPVs) are responsible for the development of cancer and its precursor lesion cervical intraepithelial neoplasia (4, 24, 30). The potential role of HPV testing in cervical cancer screening programs has been under consideration for several years (8–10, 21, 31) and is felt to be promising for women with atypical squamous cells of undetermined significance.
Commercial availability of HPV detection systems in the United States has been limited. The first-generation Hybrid Capture Tube test (HCT; Digene Corporation, Silver Spring, Md.) has been used as an HPV detection test and is the only HPV DNA test approved by the U.S. Food and Drug Administration (5, 23, 29, 33, 35). A second-generation assay with increased analytical sensitivity and a more efficient kit format has recently been developed. This new test, currently under consideration by the U.S. Food and Drug Administration, is named the Hybrid Capture II test (HC II). The hybrid capture technique is based on the formation of RNA-DNA hybrids between HPV DNA that may be present in clinical specimens and complementary unlabeled HPV RNA probes. The RNA-DNA hybrids are captured and immobilized by antihybrid antibodies. Immobilized hybrids are reacted with a monoclonal antibody reagent that is conjugated to alkaline phosphatase, and the complexes are detected via a chemiluminescent substrate reaction. In HCT, a tube luminometer is employed, whereas in HC II, a microplate luminometer reads the light output and displays the assay results as relative light units (RLU). HPV positivity or negativity is based on comparison to a standard positive reference (RLU of a clinical specimen divided by the mean RLU of three positive calibrator references).
The HCT Probe B cocktail detects a limited number of high-risk HPV types, including HPVs 16, 18, 31, 33, 35, 45, 51, 52, and 56, and has been reported to have a diagnostic sensitivity similar to that of the Papanicolaou (Pap) smear (6). HC II detects HPV types at an increased sensitivity compared to that of HCT (23). In addition, probes for HPV types 39, 58, 59, and 68 have been added to the HC II Probe B group. One potential advantage of this test is that it may provide a semiquantitative measure of viral load (8, 9, 11, 16, 23, 31, 32). It has been suggested that viral load may lend prognostic and diagnostic value (13, 20).
To date, investigators in most reported HCT-based studies have used a Dacron swab to collect cervical cells into 1.0 ml of Specimen Transport Medium (STM; Digene Corporation) (31, 34, 35). Recently, a conical cytology brush has been used to collect cervical cells. This modification was made with the expectation that the brush would increase the number of cells obtained and thus provide a specimen with an increased concentration of HPV.
As HPV test formats and specimen collection devices have continued to evolve, new liquid specimen collection media have also become available for routine use in Pap smear screening. Recently, a new technique that employs the collection of cervical cells into PreservCyt (PC) liquid cytology medium, from which ThinPrep (Cytyc, Boxborough, Mass.) thin-layer slides are produced, has been introduced. This method has received approval for clinical use from the U.S. Food and Drug Administration (22, 33). An advantage of using a liquid cytology medium for the collection of cervical specimens is that multiple diagnostic tests can be performed with a single sample. For a cervical diagnosis within the atypical squamous cells of undetermined significance category, HPV testing can be performed on the temporarily stored Pap smear specimen without the cost of a clinical follow-up visit. Accordingly, HPV testing of specimens in PC has recently been approved by the U.S. Food and Drug Administration.
This study compares HPV detection in specimens collected by three different means. These three methods are under consideration as HPV sampling strategies for routine clinical use. HPV detection in these specimens was evaluated by comparing the first-generation HCT, the prototype second-generation HC II, and a widely used PCR assay based on the MY09-MY11-HMB01 primer set. PCR was performed only on the specimens collected in PC. The selected L1 consensus primer PCR-based method detects a broad spectrum of anogenital HPV types that include, but are not limited to, HPVs 6, 11, 16, 18, 26, 31, 33, 34, 35, 39, 45, 51, 52, 53, 54, 55, 56, 57, 58, 59, 61, 62, 64, 66, 67, 68, 70, 72, 73, IS39, MM4 (also called W13B), MM7 (also called P291), MM8 (also called P155), MM9 (also called P238a), CP6108, CP8061, and CP8304 (1–3, 25).
MATERIALS AND METHODS
Study population and collection of specimens.
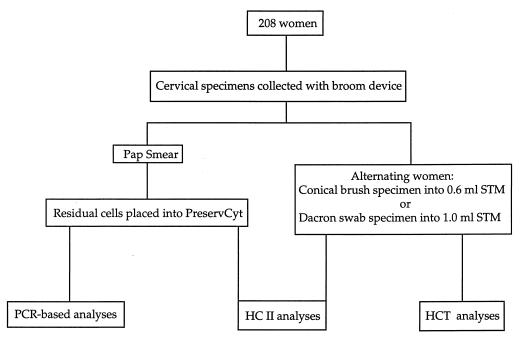
Figure 1 summarizes the cervical specimen collection and HPV testing strategies. Two hundred eight specimens were selected from specimens collected in a prospective population-based natural-history study of 9,175 women conducted in Costa Rica (17). The median age of these women was 37 years. Cervical diagnoses were virtually all within normal limits but included a small number of low-grade abnormalities (n = 10). This cohort consisted of women with past histories or no history of squamous intraepithelial lesions. Initial specimens were collected from all 208 women with a broom device (Cervex Brush; Unimar, Wilton, Conn.) for the Pap smear. Following the preparation of a conventional Pap smear, residual cells on each broom device were placed into ∼20 ml of PC liquid cytology medium (Cytyc). Approximately half of the 208 women had a second cervical specimen collected with a Dacron swab and placed into 1.0 ml of STM. The remaining women had a second cervical specimen collected with a conical brush (Medical Packaging Corporation, Camarillo, Calif.) into 0.6 ml of STM. These two collection methods were alternated between consecutive patients. Specimens collected in PC and STM were stored at ambient temperature and −20°C, respectively, until processing.
FIG. 1.
Flowchart illustrating the cervical specimen collection and HPV testing strategies.
PCR and dot blot-based HPV testing methods.
The PCR and dot blot analyses were performed at the University of New Mexico. Aliquots for PCR testing were taken prior to HC II analyses of these specimens. The laboratory was unaware of the clinical statuses and HCT and HC II results of the study subjects.
A cervical specimen collected in PC was processed by placing a 1.5-ml aliquot into a 1.5-ml Eppendorf tube and centrifuging it for 10 min at 13,000 × g. The supernatant was immediately removed and discarded with a plugged Pasteur pipette. The cell pellet was dried overnight at room temperature. The pellet was resuspended in 150 μl of digestion solution (10 mM Tris, 1 mM EDTA, 200 μg of proteinase K per ml, 0.1% Laureth-12) and digested at 56°C for 1 h. The digestion was followed by a 15-min incubation at 95°C to inactivate the proteinase K. Crude DNA extracts were stored at −20°C until amplification.
Prior to amplification, the crude digests were allowed to reach room temperature and centrifuged briefly. Five microliters of each specimen was amplified per 100 μl of amplification reaction mixture with the MY09-MY11-HMB01 L1 consensus primers and buffer systems (1, 14, 19, 25). AmpliTaq Gold polymerase (Perkin-Elmer, Foster City, Calif.) was used for all reactions. To determine specimen adequacy, the GH20-PC04 human consensus β-globin primer system was coamplified with the HPV consensus primers. Positive and negative PCR controls were included with each amplification. PCRs were conducted in a Perkin-Elmer model 9600 thermocycler with the following amplification profile: an initial 95°C AmpliTaq Gold activation for 9 min; 40 cycles of a 95°C denaturation for 1 min, a 55°C annealing for 1 min, and a 72°C terminal extension for 1 min; and a 72°C final extension for 5 min. Amplimers were analyzed by ethidium bromide-stained agarose gel electrophoresis.
HPV typing analyses were carried out by dot blot hybridization and with biotinylated HPV type-specific oligonucleotide probes as previously described (1). PCR products were denatured and applied to replicate nylon membranes (3.5 μl per well) with dot blot apparatuses (Bio-Rad Laboratories, Hercules, Calif.). A collection of previously characterized HPV PCR products (3.5 μl) were applied as HPV type-specific controls. The membranes were hybridized at 53°C overnight with biotinylated HPV type-specific oligonucleotide probes for HPVs 6 or 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 44, 45, 51, 52, 53, 54, 55, 56, 57, 58, 59, 66, 68, 73 (P238A), MM4 (W13B), MM7 (P291), and MM8 (P155). Probes for HPV types 26 and MM8 and types 40 and 42 were pooled as pairs during hybridization. A β-globin probe was used to assess specimen adequacy. Following hybridization, membranes were washed at 56 to 57°C to remove nonspecifically bound probe. Immobilized probes were detected with streptavidin-horseradish peroxidase (Vector, Burlingame, Calif.) and Enhanced Chemiluminescent Substrate (Amersham, Arlington Heights, Ill.). Blots were exposed to Kodak X-Omat AR 5 film for an initial 10 min, followed by a second 2-h or overnight exposure. The resulting data were analyzed manually by two observers and recorded on a database spreadsheet. Discrepant results were independently arbitrated.
All specimens positive for PCR by gel electrophoresis that did not bind to any of the HPV type-specific oligonucleotide probes were subjected to restriction fragment length polymorphism (RFLP) analysis as previously described (3). The resulting RFLP profiles were compared to known HPV type profiles, and the HPV types were designated. The RFLP results were subsequently confirmed by an independent PCR-based detection method (15).
Hybrid capture-based HPV testing methods.
All hybrid capture testing of clinical specimens was performed at Digene Corporation. The laboratory technicians were unaware of the clinical statuses and HPV PCR results of the study subjects.
Cervical specimens collected in STM (0.6 and 1.0 ml) were processed according to the manufacturer’s HCT or HC II package insert. Briefly, one-half volume of NaOH-based denaturation reagent was added to each sample, which was then mixed vigorously and incubated at 65°C for 45 min. Seventy-five microliters of the denatured specimen was removed for analysis by HC II, and 150 μl was removed for HCT. The HC II Probe B cocktail contains RNA probes for high-risk HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68. HPV positivity by HC II was determined with 0.2-, 0.5-, 1.0-, and 10.0-pg/ml detection cutoff limits. HCT used a single fixed 10.0-pg/ml assay cutoff and did not include RNA probes for HPVs 39, 58, 59, and 68.
Specimens collected into PC were analyzed by HC II as follows. A 2-ml aliquot was removed from each specimen and placed into a microcentrifuge tube and centrifuged at 13,000 × g for 15 min. The supernatant was carefully aspirated, and the pellet was resuspended in 50 μl of STM. Twenty-five microliters of denaturation reagent was added, and the specimens were denatured for 45 min at 65°C. From this point, the remainder of the HC II procedure was identical to that applied to the STM specimens.
Data analyses.
Agreement was measured by both absolute agreement and Cohen’s kappa statistic (κ), a measure of the agreement between two methods in excess of that due to chance (12). Approximate 95% confidence intervals (CIs) were computed with an asymptotic variance and critical values from the normal distribution. Proportions were compared with exact P values for the Pearson chi-square test. Statistical significance was achieved when the P value of the test was less than 0.05. All analyses were performed with SAS version 6.12 software (28).
RESULTS
HPV DNA positivity.
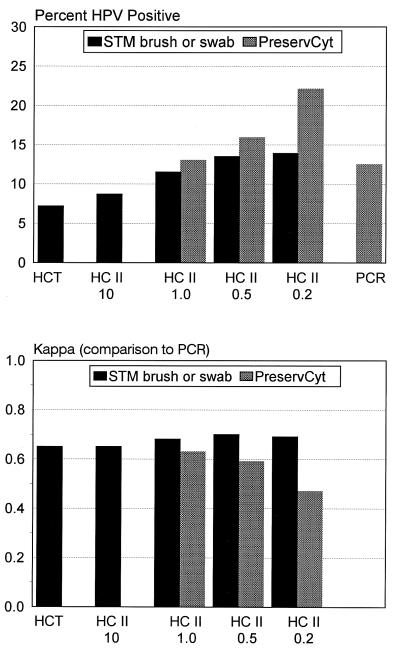
The overall HPV DNA positivity was 24.5% by PCR (any HPV type), and when the test was restricted to the 13 cancer-associated HPV types present in the HC II Probe B cocktail, the HPV DNA positivity was 12.5%. In comparison, the overall HPV DNA positivity was as follows for different media, cutoffs, and tests: 22.1% by HC II of specimens in PC at the 0.2-pg/ml cutoff, 13.9% by HC II of specimens in STM at the 0.2-pg/ml cutoff, 13.0% by HC II of specimens in PC at the 1.0-pg/ml assay cutoff, 11.5% by HC II of specimens in STM at the 1.0-pg/ml cutoff, and 7.2% by HCT testing of specimens in STM.
HCT versus HC II.
Results of the HCT and HC II at the 10.0-pg/ml assay cutoff were equivalent for Dacron swab and conical brush specimens and revealed that, at this assay cutoff, the level of detection of HC II was nearly equivalent to that of the HCT, with a kappa value of 0.90 (95% CIs = 0.79 and 1.00). In addition, HPV detection was only slightly greater by HC II than by HCT (8.7 versus 7.2% positive) and the difference was not statistically significant.
Dacron swab versus conical brush.
HPV DNA detection by HC II was greater for specimens obtained with the conical brush than for specimens obtained with the Dacron swab at 0.2-, 0.5-, and 1.0-pg/ml assay cutoffs (Table 1). However, these differences were not statistically significant. Since specimens were collected alternately with a conical brush and Dacron swab from consecutive patients, differences in HPV positivity may actually reflect chance differences in the prevalence of HPV among the two groups of women, despite randomization. This possible explanation was supported by the results of HC II when it was applied to the PC specimens. A higher HPV positivity was observed with PC specimens from women from whom a second specimen was obtained with a conical brush than with PC specimens from women from whom the second specimen was obtained with a Dacron swab.
TABLE 1.
Percentages of HPV positivity and agreement between HC II results with STM and PC liquid cytology medium specimens
| Collection device(s) | No. of specimens | HC II assay cutoff (pg/ml) | % of specimens that were positive
|
Agreementa
|
|||
|---|---|---|---|---|---|---|---|
| STM HC II | PC HC II | % Total | % Positive | Kappa value (95% CIs) | |||
| Conical brush | 105 | 0.2 | 15.2 | 24.8 | 86.7 | 50.0 | 0.59 (0.40, 0.78) |
| 0.5 | 15.2 | 15.2 | 96.2 | 77.8 | 0.85 (0.71, 0.99) | ||
| 1.0 | 13.3 | 14.3 | 97.1 | 81.3 | 0.88 (0.75, 1.00) | ||
| Dacron swab | 103 | 0.2 | 12.6 | 19.4 | 89.3 | 50.0 | 0.61 (0.40, 0.81) |
| 0.5 | 11.7 | 16.5 | 93.2 | 61.1 | 0.72 (0.53, 0.91) | ||
| 1.0 | 9.7 | 11.7 | 92.2 | 46.7 | 0.59 (0.34, 0.85) | ||
| Conical brush and Dacron swab | 208 | 0.2 | 13.9 | 22.1 | 88.0 | 50.0 | 0.60 (0.46, 0.74) |
| 0.5 | 13.5 | 15.9 | 94.7 | 69.4 | 0.79 (0.67, 0.91) | ||
| 1.0 | 11.5 | 13.0 | 94.7 | 64.5 | 0.75 (0.62, 0.89) | ||
% Total, percentage of the total number of specimens collected in STM and PC that produced identical results; % Positive, percentage of specimens positive in either STM or PC that produced identical results.
Agreement between HC II results obtained from STM and PC specimens was good at the 0.5- and 1.0-pg/ml assay cutoffs, with kappa values of 0.79 and 0.75, respectively (Table 1). At the 0.2-pg/ml HC II assay cutoff, the decrease in agreement (κ = 0.60) was largely attributable to the much greater HPV DNA positivity obtained with the PC specimens (22.1% in PC versus 13.9% in STM). Agreement was better for the group of specimens obtained with a conical brush than for those obtained with a Dacron swab at the 1.0-, 0.5-, and 0.2-pg/ml HC II assay cutoffs. However, the kappa statistics were not significantly different.
HC II versus PCR.
Initial comparisons of HC II and PCR results included PCR detection of only those HPV types included in the HC II Probe B cocktail. When HC II was compared to the PCR method with PC specimens, the kappa values were 0.47 (95% CIs = 0.32 and 0.62), 0.59 (95% CIs = 0.43 and 0.74), and 0.63 (95% CIs = 0.47 and 0.79) for HC II assay cutoff values of 0.2, 0.5, and 1.0 pg/ml, respectively (Fig. 2). Agreement was actually slightly better when results of HC II from STM specimens were compared to the PCR results from PC specimens (kappa values, 0.69, 0.70, and 0.68 for HC II cutoff levels of 0.2, 0.5, and 1.0 pg/ml, respectively). Agreement between HC II and PCR for the group of women sampled with a conical brush was greater than for the group sampled with a Dacron swab, and the kappa values were significantly different at the 0.2-, 0.5-, and 1.0-pg/ml HC II cutoff levels (P = 0.02, 0.04, and 0.03, respectively).
FIG. 2.
Agreement between HPV test results of HCT, HC II, and PCR. The upper panel shows the percentages of specimens positive by HCT, HC II, and PCR. HCT and HC II results for all specimens collected with a conical brush or Dacron swab were combined. HC II and PCR results with PC liquid cytology medium specimens are also shown. In the lower panel, kappa statistics were calculated for HCT and HC II results compared to the PCR results. The results shown in the upper panel were used to derive the values shown in the lower panel. Only data for the HPV types present in the HC II Probe B cocktail (i.e., HPVs 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) are presented.
We performed further analyses of HC II and PCR results in which we considered additional HPV types phylogenetically related to the 13 HPVs included in the HC II Probe B cocktail (Table 2). Absolute agreement and kappa values decreased slightly when related HPV types were included in the comparison. When we used all HPV types detected by PCR (i.e., the 13 HPVs in the Probe B group; HPVs 53, 66, 67, 70, and 73; and the additional HPVs 6, 11, 32, 54, 62, CP6108, CP8061, MM7, and MM8), agreement decreased substantially, especially with STM samples.
TABLE 2.
Agreement between results of HC II and PCR when HPV types other than those in the HC II Probe B cocktail were included
| Medium | HPV types detected by PCR | HC II cutoff (pg/ml) | % of specimens that were positive
|
Agreementc
|
|||
|---|---|---|---|---|---|---|---|
| HC II | PCRa | % Total | % Positive | Kappa value (95% CIs) | |||
| STM | HC II Probe B types plus related typesb | 0.2 | 13.9 | 91.8 | 59.5 | 0.70 (0.56, 0.83) | |
| 0.5 | 13.5 | 92.3 | 61.0 | 0.71 (0.58, 0.84) | |||
| 1.0 | 11.5 | 92.3 | 59.0 | 0.70 (0.56, 0.84) | |||
| All types | 0.2 | 13.9 | 85.6 | 45.5 | 0.54 (0.40, 0.68) | ||
| 0.5 | 13.5 | 86.1 | 46.3 | 0.56 (0.42, 0.69) | |||
| 1.0 | 11.5 | 86.1 | 44.2 | 0.54 (0.40, 0.68) | |||
| PC | HC II Probe B types plus related typesb | 0.2 | 22.1 | 18.3 | 84.6 | 44.8 | 0.52 (0.38, 0.67) |
| 0.5 | 15.9 | 18.3 | 88.0 | 47.9 | 0.58 (0.43, 0.72) | ||
| 1.0 | 13.0 | 18.3 | 88.9 | 47.7 | 0.58 (0.43, 0.73) | ||
| All types | 0.2 | 22.1 | 24.5 | 81.3 | 42.6 | 0.48 (0.34, 0.62) | |
| 0.5 | 15.9 | 24.5 | 84.6 | 44.8 | 0.53 (0.39, 0.67) | ||
| 1.0 | 13.0 | 24.5 | 84.6 | 41.8 | 0.51 (0.36, 0.65) | ||
The PCR test was done only on PC specimens.
HPV types considered genomically related to HC II Probe B types included HPVs 53, 66, 67, 70, and 73.
% Total, percentage of the total number of specimens tested by HC II and PCR that produced identical results; % Positive, percentage of specimens positive by either HC II or PCR that produced identical results.
A listing of HPV types detected by L1 consensus PCR and those detected by HC II is shown in Table 3. Overall, cervical HPV DNAs were detected by PCR in 24.5% of these women. In this study, HC II detected some infections with HPVs 53, 54, 66, 67, 73, CP6108 (27), and CP8061 (27) as determined by PCR. These HPV types do not have corresponding probes in the HC II Probe B cocktail.
TABLE 3.
HPV types detected by L1 consensus PCR and those detected by HCT and HC II
| PCR HPV type(s)a | No. of specimens | % of total no. of specimens (n = 208) | No. of specimens positive by:
|
|||
|---|---|---|---|---|---|---|
| PC HC II (1.0-pg/ml cutoff) | STM HC II (1.0-pg/ml cutoff) | STM HC II (10.0-pg/ml cutoff) | STM HCT (10.0-pg/ml cutoff) | |||
| 6 or 11 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| 16 | 7 | 3.4 | 6 | 6 | 6 | 6 |
| 16, 54 | 1 | 0.5 | 1 | 1 | 1 | 1 |
| 16, 73 | 1 | 0.5 | 1 | 1 | 1 | 1 |
| 31 | 2 | 1.0 | 1 | 1 | 1 | 1 |
| 31, 53 | 1 | 0.5 | 0 | 1 | 0 | 0 |
| 32 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| 33, 39, MM8 | 1 | 0.5 | 1 | 1 | 1 | 0 |
| 35 | 1 | 0.5 | 1 | 1 | 1 | 1 |
| 51 | 3 | 1.4 | 2 | 1 | 1 | 1 |
| 51, 56 | 1 | 0.5 | 1 | 1 | 1 | 1 |
| 52 | 2 | 1.0 | 1 | 2 | 0 | 0 |
| 53 | 3 | 1.4 | 1 | 1 | 1 | 1 |
| 55 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| 56 | 3 | 1.4 | 2 | 1 | 1 | 1 |
| 56, 66 | 1 | 0.5 | 1 | 1 | 1 | 1 |
| 58 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| 58, MM8 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| 62 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| 66 | 1 | 0.5 | 1 | 1 | 0 | 0 |
| 67 | 1 | 0.5 | 1 | 1 | 1 | 0 |
| 70 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| 73 | 5 | 2.4 | 0 | 2 | 0 | 0 |
| 73, MM8 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| CP6108 | 1 | 0.5 | 1 | 0 | 0 | 0 |
| CP8061 | 2 | 1.0 | 1 | 0 | 0 | 0 |
| MM7 | 5 | 2.4 | 0 | 0 | 0 | 0 |
| MM7, CP8061 | 1 | 0.5 | 0 | 0 | 0 | 0 |
| No HPV DNA detected | 157 | 75.5 | 4 | 1 | 1 | 0 |
Based on dot blot and RFLP analyses of MY09-MY11 PCR amplimers, not sequence data.
Discrepant results between HC II and PCR were ascertained and confirmed by repeat PCR testing of the PC specimens. At the 1.0-pg/ml HC II cutoff, PCR detected high-risk HPVs in eight specimens that were not detected by HC II in PC specimens. In the same analysis, HC II detected four samples positive for high-risk HPV types that were not detected by the PCR test. When we performed an identical analysis of results of PCR and HC II with STM specimens, 11 specimens that were found to contain high-risk HPV types by PCR were detected by HC II. HC II identified one STM specimen with a high-risk HPV type that was not detected by PCR testing.
DISCUSSION
It has been proposed that HPV DNA testing might be used as an adjunct in the management of women with minor cytological abnormalities (8, 10, 18, 21, 26). This study set out to compare different specimen collection methods for HPV testing. In addition, it was important to compare results of HPV testing of specimens in PC liquid cytology medium by well-established HPV testing methods and to further characterize the newly available HC II HPV method. We compared two hybrid capture methods (HCT and HC II) and L1 consensus PCR for HPV DNA testing, using specimens collected in both STM and PC.
Results by HC II were in excellent agreement with results by HCT at the 10.0-pg/ml cutoff, illustrating the correspondence of the two hybrid capture HPV tests. Also, as expected, when HCT was compared to HC II at the 1.0-, 0.5-, and 0.2-pg/ml assay cutoffs, the agreement progressively decreased. As previously reported (23), a 1.0-pg/ml cutoff can routinely be used in HC II, reflecting an approximately 10-fold increase in sensitivity over that of HCT. It is anticipated that the increased sensitivity may improve the clinical utility of this second-generation HPV hybrid capture test.
The conical brush STM collection method may be an improvement over the Dacron swab STM collection method. This is suggested by both the increase in overall HPV DNA detection and the higher correlation of HC II data from STM conical brush specimens with HC II and PCR data from PC specimens.
Although collection with a conical brush appeared to result in higher HPV DNA detection by HC II than collection with a Dacron swab, further analyses of results stratified by STM sampling device demonstrated a potential problem with subject randomization. When the subjects were separated into two groups based on STM collection device and analyses were limited to those specimens collected in PC, the HC II data showed a higher prevalence of HPV and correlated significantly better with PCR data from the group of women who had an STM sample collected with a conical brush (data not shown). Because the PC specimen was collected prior to the STM specimen, the device used in the second specimen collection should not have affected the initial specimen. Therefore, the apparent difference between these two sampling methods may be due to a chance difference in subjects. However, there was no obvious difference in the ages of and clinics attended by the two groups of women for Pap smear diagnosis. In addition, the samples were collected in a random manner.
Two recent studies reported reasonable correlation between PCR and HCT HPV assay results (7, 34). However, neither of these previous studies used PC. Some of the disagreement between PCR and HC II results may be attributed to the use of separate PC aliquots that were processed and tested at two laboratory sites. In other words, the level of agreement may have been higher if both tests were performed with the same aliquot.
The HC II detection cutoff value represents the relative light unit reading above which samples are considered positive. There is uncertainty as to which HC II cutoff is best in terms of accuracy and clinical utility. The comparison of results of HC II and PCR with PC specimens demonstrated a good correlation at 0.5- and 1.0-pg/ml HC II cutoffs. In contrast, at the 0.2-pg/ml HC II cutoff, the kappa value was only fair (0.47). However, the comparison to PCR results with STM specimens at 0.2 pg/ml resulted in a kappa value of 0.69, indicating good agreement. Our data suggested that, for specimens collected in PC, an HC II cutoff of about 1.0 pg/ml was optimal and that the use of a 0.2-pg/ml HC II cutoff introduced a measurable level of false positivity. This increase in background at the 0.2-pg/ml HC II cutoff was observed only with PC specimens, which implies that there was a higher level of nonspecific detection for samples collected in PC than for those collected in STM.
For specimens collected in STM, use of a 0.2-pg/ml HC II cutoff leads to an apparent increase in the level of HPV positivity compared to the levels produced with the 0.5- and 1.0-pg/ml HC II cutoffs. When using this specimen collection medium, an HC II threshold of less than 1.0 pg/ml might be optimal and suggests that the HPV DNA detection ability of HC II is approaching that of the MY09-MY11-HMB01 PCR method. Sensitivities for PCR and HC II were equivalent when analyses were restricted to the HPV types included in the HC II Probe B cocktail. Additional studies are necessary to evaluate the detection limits of HC II and to compare results with results of sensitive PCR detection methods that distinguish a broad spectrum of HPV types.
Based on the PCR results from matched specimens, cross-reactivity of the HC II Probe B cocktail with phylogenetically related HPV types was observed. When these and additional HPV types were included in HC II and PCR and the results were compared, the kappa statistics and percentages of agreement decreased, partially because HC II detected some of these HPVs. The HPV types not represented by the HC II Probe B group but detected by HC II at a 1.0-pg/ml assay cutoff included HPVs 53, 66, 67, 73, CP6108 (27), and CP8061 (27). In addition, HPVs 53 and 67 were also detected at a 10.0-pg/ml HC II cutoff. It should be noted that limited data are available to assess cervical disease risk for some of these rarer HPV types. Differences in HC II HPV detection endpoints for various cross-reactive HPVs may contribute to the decreases in agreement. These differences in HC II HPV type-specific sensitivities will result in a lower overall comparability with the sensitivity of M409-11-MMB01 PCR with probes that detects these additional HPV types. The clinical relevance of the HC II Probe B cocktail’s cross-reactivities with some low-risk HPVs remains to be fully determined.
ACKNOWLEDGMENTS
This work was funded in part by a contract to C. M. Wheeler from the National Institutes of Health, RFP-NCI-CA-CN-55044-07.
We thank Michele Manos for her continuing helpful advice. We acknowledge the superb Costa Rican study team and the data processing excellence of Julie Buckland and Kay Helgesen at IMS.
REFERENCES
- 1.Bauer H M, Manos M M. PCR detection of genital human papillomavirus. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C: ASM Press; 1993. pp. 407–413. [Google Scholar]
- 2.Bauer H M, Ting Y, Greer C E, Chambers J C, Tashiro C J, Chimera J, Reingold A, Manos M M. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA. 1991;265:472–476. [PubMed] [Google Scholar]
- 3.Bernard H U, Chan S Y, Manos M M, Ong C K, Villa L L, Delius H, Peyton C L, Bauer H M, Wheeler C M. Identification and assessment of known and novel human papillomaviruses by polymerase chain reaction amplification, restriction fragment length polymorphisms, nucleotide sequence, and phylogenetic algorithms. J Infect Dis. 1994;170:1077–1085. doi: 10.1093/infdis/170.5.1077. [DOI] [PubMed] [Google Scholar]
- 4.Bosch F, Manos M M, Muñoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 5.Brown D R, Bryan J T, Cramer H, Fife K H. Analysis of human papillomavirus types in exophytic condylomata acuminata by hybrid capture and Southern blot techniques. J Clin Microbiol. 1993;31:2667–2673. doi: 10.1128/jcm.31.10.2667-2673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavuslu S, Mant C, Starkey W G, Bible J M, Biswas C, Kell B, Rice P, Best J M, Cason J. Analytic sensitivities of Hybrid-Capture, consensus and type-specific polymerase chain reactions for the detection of human papillomavirus type 16 DNA. J Med Virol. 1996;49:319–324. doi: 10.1002/(SICI)1096-9071(199608)49:4<319::AID-JMV10>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 7.Cope J U, Hildesheim A, Schiffman M H, Manos M M, Lörincz A T, Burk R D, Glass A G, Greer C, Buckland J, Helgesen K, Scott D R, Sherman M E, Kurman R J, Liaw K-L. Comparison of the Hybrid Capture Tube test and PCR for detection of human papillomavirus DNA in cervical specimens. J Clin Microbiol. 1997;35:2262–2265. doi: 10.1128/jcm.35.9.2262-2265.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J T, Lörincz A T, Schiffman M H, Sherman M E, Cullen A, Kurman R J. Human papillomavirus testing by Hybrid Capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 1995;172:946–954. doi: 10.1016/0002-9378(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 9.Cuzick J, Terry G, Ho L, Hollingworth T, Anderson M. Type-specific human papillomavirus DNA in abnormal smears as a predictor of high-grade cervical intraepithelial neoplasia. Br J Cancer. 1994;69:167–171. doi: 10.1038/bjc.1994.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferenczy A. Viral testing for genital human papillomavirus infections: recent progress and clinical potentials. Int J Gynecol Cancer. 1995;5:321–328. doi: 10.1046/j.1525-1438.1995.05050321.x. [DOI] [PubMed] [Google Scholar]
- 11.Ferenczy A, Franco E, Arseneau J, Wright T C, Richart R M. Diagnostic performance of Hybrid Capture human papillomavirus deoxyribonucleic acid assay combined with liquid-based cytologic study. Am J Obstet Gynecol. 1996;175:651–656. doi: 10.1053/ob.1996.v175.a73868. [DOI] [PubMed] [Google Scholar]
- 12.Fleiss J L. Statistical methods for rates and proportions. New York, N.Y: John Wiley and Sons; 1981. pp. 212–236. [Google Scholar]
- 13.Franco E L, Villa L L, Richardson H, Rohan T, Ferenczy A. Epidemiology of cervical human papillomavirus infection. In: Franco E L, Monsonego J, editors. New developments in cervical cancer screening and prevention. World Health Organization/EUROGIN monograph. Oxford, United Kingdom: Blackwell Scientific Publications Ltd.; 1997. pp. 14–22. [Google Scholar]
- 14.Gravitt P E, Manos M M. Polymerase chain reaction-based methods for the detection of human papillomavirus DNA. In: Muñoz N, Bosch F X, Shah K V, Meheus A, editors. The epidemiology of cervical cancer and human papillomavirus. Lyon, France: International Agency for Research against Cancer, Scientific Publications; 1992. pp. 121–133. [PubMed] [Google Scholar]
- 15.Gravitt P E, Peyton C L, Apple R J, Wheeler C M. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall S, Lörincz A, Shah F, Sherman M E, Abbas F, Paull G, Kurman R J, Shah K V. Human papillomavirus DNA detection in cervical specimens by Hybrid Capture: correlation with cytologic and histologic diagnoses of squamous intraepithelial lesions of the cervix. Gynecol Oncol. 1996;62:353–359. doi: 10.1006/gyno.1996.0248. [DOI] [PubMed] [Google Scholar]
- 17.Herrero R, Schiffman M H, Bratti C, Hildesheim A, Balmaceda I, Sherman M E, Greenberg M, Cardenas F, Gomez V, Helgensen K, Morales J, Hutchinson M, Mango L, Alfaro M, Potischman N W, Wacholder S, Swanson C, Brinton L A. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of Costa Rica: the Guanacaste project. Pan Am J Public Health. 1997;1:362–375. doi: 10.1590/s1020-49891997000500005. [DOI] [PubMed] [Google Scholar]
- 18.Herrington C S, Evans M F, Charnock F M, Gray W, McGee J O. HPV testing in patients with low grade cervical cytological abnormalities: a follow up study. J Clin Pathol. 1996;49:493–496. doi: 10.1136/jcp.49.6.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hildesheim A, Schiffman M H, Gravitt P E, Glass A G, Greer C E, Zhang T, Scott D R, Rush B B, Lawler P, Sherman M E, Kurman R J, Manos M M. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 20.Ho G Y F, Burk R D, Klein S, Kadish A S, Chang C, Palan P, Basu J, Tachezy R, Lewis R, Romney S. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995;87:1365–1371. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins D, Sherlaw-Johnson C, Gallivan S. Can papillomavirus testing be used to improve cervical cancer screening? Int J Cancer. 1996;65:768–773. doi: 10.1002/(SICI)1097-0215(19960315)65:6<768::AID-IJC10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 22.Lee K R, Ashfaq R, Birdsong G G, Corkill M E, McIntosh K M, Inhorn S L. Comparison of conventional Papanicolaou smears and a fluid-based, thin-layer system for cervical cancer screening. Obstet Gynecol. 1997;90:278–284. doi: 10.1016/S0029-7844(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 23.Lörincz A. Hybrid capture method for detection of human papillomavirus DNA in clinical specimens. Papillomavirus Rep. 1996;7:1–5. doi: 10.1111/j.1447-0756.1996.tb01081.x. [DOI] [PubMed] [Google Scholar]
- 24.Lörincz A. Oncogenic association of specific human papillomavirus types with cervical neoplasia. J Natl Cancer Inst. 1987;79:671–677. [PubMed] [Google Scholar]
- 25.Manos M M, Ting Y, Wright D K, Lewis A J, Broker T R, Wolinsky S M. Use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 26.Mansell M E, Ho L, Terry G, Singer A, Cuzick J. Semi-quantitative human papillomavirus DNA detection in the management of women with minor cytologic abnormalities. Br J Obstet Gynaecol. 1994;101:807–809. doi: 10.1111/j.1471-0528.1994.tb11952.x. [DOI] [PubMed] [Google Scholar]
- 27.Peyton C L, Wheeler C M. Identification of five novel human papillomavirus sequences in the New Mexican triethnic population. J Infect Dis. 1994;170:1089–1092. doi: 10.1093/infdis/170.5.1089. [DOI] [PubMed] [Google Scholar]
- 28.SAS Institute Inc. SAS/STAT software: changes and enhancements through release 6.11. Cary, N.C: SAS Institute Inc.; 1996. pp. 219–230. [Google Scholar]
- 29.Schiffman M H, Kiviat N B, Burk R D, Shah K V, Daniel R W, Lewis R, Kuypers J, Manos M M, Scott D R, Sherman M E, Kurman R J, Stoler M H, Glass A G, Rush B B, Mielzynska I, Lörincz A T. Accuracy and interlaboratory reliability of human papillomavirus DNA testing by hybrid capture. J Clin Microbiol. 1995;33:545–550. doi: 10.1128/jcm.33.3.545-550.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiffman M H, Bauer H M, Hoover R N, Glass A G, Cadell D M, Rush B B, Scott D R, Sherman M E, Kurman R J, Wacholder S, Stanton C K, Manos M M. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993;85:958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]
- 31.Schneider A, Zahm D M, Kirchmayr R K, Schneider V L. Screening for cervical intraepithelial neoplasia grade 2/3: validity of cytologic study, cervicography, and human papillomavirus detection. Am J Obstet Gynecol. 1996;174:1534–1541. doi: 10.1016/s0002-9378(96)70602-6. [DOI] [PubMed] [Google Scholar]
- 32.Shah K V, Solomon L, Daniel R, Cohn S, Vlahov D. Comparison of PCR and Hybrid Capture methods for detection of human papillomavirus in injection drug-using women at high risk of human immunodeficiency virus infection. J Clin Microbiol. 1997;35:517–519. doi: 10.1128/jcm.35.2.517-519.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman M, Schiffman M, Lörincz A, Herrero R, Hutchinson M, Bratti C, Zahniser D, Morales J, Hildesheim A, Helgensen K, Kelly D, Alfaro M, Mena F, Balmaceda I, Mango L, Greenberg M. Cervical specimens collected in liquid buffer are suitable for both cytologic screening and ancillary human papillomavirus testing. Cancer. 1997;81:89–97. [PubMed] [Google Scholar]
- 34.Smits H L, Bollen L J M, Tjong-a-Hung S P, Vonk J, Velden J V D, Ten Kate F J W, Kaan J A, Mol B M, Schegget J T. Intermethod variation in detection of human papillomavirus DNA in cervical smears. J Clin Microbiol. 1995;33:2631–2636. doi: 10.1128/jcm.33.10.2631-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Ferenczy A, Johnson D, Koulos J, Lungu O, Richart R, Wright T. Evaluation of the Hybrid Capture human papillomavirus deoxyribonucleic acid detection test. Am J Obstet Gynecol. 1995;173:1432–1437. doi: 10.1016/0002-9378(95)90629-0. [DOI] [PubMed] [Google Scholar]