Abstract
The genetic analysis of oocysts recovered from the stools of humans and animals infected with Cryptosporidium parvum has consistently shown the existence of two distinct genotypes. One of the genotypes is found exclusively in some human infections, whereas the other genotype is found in human as well as in animal infections. On the basis of these observations and the results of published epidemiological studies with single polymorphic markers, the existence of two separate transmission cycles has been postulated, one exclusively anthroponotic and the other involving both animals and humans. To test this hypothesis, C. parvum isolates of different geographic and host origins were analyzed by using unlinked genetic polymorphisms. A total of 28 isolates originating from Europe, North and South America, and Australia were examined. Isolates clustered into two groups, one comprising both human and animal isolates and the other comprising isolates only of human origin. The absence of recombinant genotypes is consistent with two reproductively isolated populations within the species C. parvum.
Apicomplexan parasites of the genus Cryptosporidium infect the gastrointestinal or respiratory tracts of a wide range of mammals, birds, reptiles, and fish. Cryptosporidium parvum, a major cause of diarrhea in young livestock, has recently emerged as a widespread enteric pathogen in humans (6, 21). In immunocompetent adults the infection is generally acute and self-limiting, whereas immunocompromised individuals can develop chronic and potentially life-threatening diarrhea. The significant prevalence of C. parvum in patients with opportunistic infections and recent reports of major outbreaks of cryptosporidiosis in the United States and the United Kingdom due to contamination of drinking water supplies (10, 17, 18) indicate that C. parvum is a major public health problem. The absence of effective treatment for cryptosporidiosis highlights the need for preventive measures. To this aim, it is essential to understand the epidemiology of the disease and to identify the transmission routes accounting for human exposure and infections.
Recent studies on the intraspecific genetic variation in C. parvum have shed new light on the population structure of this parasite. Using isoenzyme analysis (1) or different DNA-based techniques (3–5, 11–14, 19), several laboratories, including ours, have found that C. parvum isolates can be divided into two genetically distinct groups, one exclusively associated with human infections and the other associated with both human and animal infections. We refer to these genotypes as H and C, respectively (24). The existence of two genotypes and the apparent lack of recombinants has taxonomical and epidemiological relevance. It implies that C. parvum may not be a uniform species; rather, it may comprise two genetically distinct parasite subpopulations. It also suggests the existence of two independent human transmission cycles.
Most of the C. parvum typing studies carried out to date are based on the analysis of single polymorphisms. A multilocus approach has the potential to better define the structure of the C. parvum population and assess the degree of genetic isolation of the H and C subpopulations. We report herein on the genotyping of 28 C. parvum isolates of various host and geographic origins simultaneously analyzed at up to five polymorphic loci.
MATERIALS AND METHODS
Parasites.
Isolates GCH1, GCH2, GCH3, GCH4, GCH5, H77, H78, P9, P12, P16, P18, P27, P29, A1, S1, Moredun (MD), ICP, UCP, 58EF, 63H, LL, and 740 were described previously (see Table 2). Isolate GCH6 originated from a laboratory worker accidentally infected with C. parvum. Isolates H83, H84, and H87 were isolated from human immunodeficiency virus-negative individuals from Perth, Western Australia, Australia. Isolate UCP has been maintained in calves for more than 8 years at ImmuCell Corp. (Portland, Maine) and was kindly provided by Joe Crabb. Isolate OH originated from a human with an acute infection and was obtained from Lucy Ward, Ohio State University, Wooster. Isolate PC1 was isolated from a captive macaque at the New England Regional Primate Center in Southboro, Mass., and was kindly provided by Keith Mansfield.
TABLE 2.
Summary of isolate and genotypic information for C. parvum samples included in this study
| Isolate | Genotypea | Hostb | Geographic origin | Sourcec | Reference |
|---|---|---|---|---|---|
| H77 | H (5) | Human | Australia | MU | 12 |
| H78 | C (5) | Human | Australia | MU | 12 |
| H83 | H (5) | Human | Australia | MU | |
| H84 | H (5) | Human | Australia | MU | |
| H87 | H (5) | Human | Australia | MU | |
| GCH1 | C (5) | Calf | Massachusetts | TUSVM | 21 |
| GCH2 | H (3) | Human | Connecticut | TUSVM | 4 |
| GCH3 | H (3) | Human | Connecticut | TUSVM | 4 |
| GCH4 | C/H (2) | Human | Massachusetts | TUSVM | 5 |
| GCH5 | C/H (4) | Human | Massachusetts | TUSVM | 5 |
| GCH6 | C (4) | Human | Massachusetts | TUSVM | |
| UCP | C (5) | Calf | Maine | ImmuCell | 4 |
| P9 | C (4) | Human | Wales | PHLS | 19 |
| P12 | H (5) | Human | Wales | PHLS | 19 |
| P16 | H (5) | Human | England | PHLS | 19 |
| P18 | H (5) | Human | England | PHLS | 19 |
| P27 | H (5) | Human | England | PHLS | 19 |
| P29 | H (5) | Human | England | PHLS | 19 |
| PC1 | H (4) | Rhesus monkey | Massachusetts | NERPC | |
| LL | C (5) | Calf | Massachusetts | ImmuCell | 5 |
| 63H | C (4) | Calf | Minnesota | GalaGen | 5 |
| 58EF | C (5) | Calf | Minnesota | GalaGen | 5 |
| 740 | C (5) | Calf | Massachusetts | ImmuCell | 5 |
| MD | C (5) | Sheep | Scotland | MAH | |
| ICP | C (4) | Calf | Idaho | UI | 4 |
| A1 | C (5) | Alpaca | Peru | VFL | 19 |
| S1 | C (5) | Sheep | Spain | VFL | 19 |
| OH | C (3) | Human | Ohio | OSU |
The number of loci assayed is given in parentheses.
Host from which genotyped oocysts were obtained.
PHLS, Public Health Laboratory Service, Clwyd, United Kingdom; MAH, Moredun Animal Health Limited TUSVM, Tufts University School of Veterinary Medicine, North Grafton, Mass.; UT, University of Texas, Houston; VFL, Veterinary Faculty of Leon, Leon, Spain; GalaGen, GalaGen Inc., Arden Hills, Minnesota; Immucell, ImmuCell Corp., Portland, Maine; OSU, Ohio State University, Wooster; UI, University of Idaho, Caldwell; NERPC, New England Regional Primate Center, Southboro, Mass.; MU, Murdoch University, Division of Veterinary and Biomedical Science and State Agricultural Biotechnology Center, Murdoch, Australia.
DNA isolation, PCR, and restriction fragment length polymorphism (RFLP) analysis.
C. parvum genomic DNA (gDNA) was extracted either from purified oocysts or from stool. Briefly, 100 to 200 μl of stool was diluted with equal volume of water and incubated overnight in 0.2% sodium dodecyl sulfate (SDS) and 200 μg of proteinase K per ml. After phenol-chloroform extraction, the gDNA was purified by binding to glass milk (GeneClean; Bio 101). Alternatively, stools were processed as described previously (19). DNA from purified oocysts was released either by three cycles of freezing-thawing or by proteinase K digestion.
Genotypic analysis.
Four of the polymorphic loci used in this study were analyzed by PCR-RFLP assays. The PCR primers used are listed in Table 1. Selected endonucleases recognizing polymorphic cleavage sites within the amplified sequences were used to digest the PCR products and discriminate between alleles on the basis of alternative restriction profiles. PCR-RFLP analyses of the polythreonine [poly(T)] and Cryptosporidium oocyst wall protein (COWP) (16) loci were performed as described previously (5, 19). The gene encoding the thrombospondin-related adhesive protein of Cryptosporidium (TRAP-C1) (20a) was recently found to consist of at least two alleles, TRAP-C1R1 and TRAP-C1R2, differentially associated with animal or human C. parvum isolates (20b). Primers Cp.E and Cp.W were used to amplify a 506-bp fragment from the TRAP-C1-coding sequence. Subsequent digestion of the PCR products with endonuclease RsaI yielded two alternative restriction profiles consisting of two bands for TRAP-C1R1 (455 and 51 bp) and three bands for TRAP-C1R2 (341, 114, and 51 bp). The PCR-RFLP assay for the ribonucleotide reductase (RNR) locus was based on the amplification of a 441-bp fragment of the RNR R1 subunit located at positions 1364 to 1804 (GenBank accession no. AF043243) (24). Digestion was performed with restriction endonuclease Tsp509I in 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 2 mM MgCl2, 0.1% Triton X-100) at 65°C. Extensive sequence polymorphism in the ribosomal internal transcribed spacer region 1 (ITS1) (4) was exploited to design a genotype-specific PCR assay with primers cry8 and cryITS1 (Table 1). Because this polymorphism is based on the presence or absence of PCR products, this assay was always run in parallel with a control PCR amplification.
TABLE 1.
Polymorphic loci analyzed
| Locus | Primer (sequence) | Typing method | Reference |
|---|---|---|---|
| COWP | cry15 (5′-GTAGATAATGGAAGAGATTGTG-3′) | PCR-RFLP analysis | 19 |
| cry9 (5′-GGACTGAAATACAGGCATTATCTTG-3′) | |||
| poly(T) | cry44 (5′-CTCTTAATCCAATCATTACAAC-3′) | PCR-RFLP analysis | 5 |
| cry39 (5′-GAGTCTAATAATAAACCACTG-3′) | |||
| ITS1 | cry8 (5′-AACATGAACAAGAATTTAATTG-3′) | PCR | 4 |
| cryITS1 (5′-CTT TTCACTTCTTCCTTCCC-3′) | |||
| TRAP-C1 | Cp.E (5′-GGATGGGTATCAGGTAATAAGAA-3′) | PCR-RFLP analysis | This study |
| Cp.W (5′-CAATTCTCTCCCTTTACTTC-3′) | |||
| RNR | SEQ1 (5′-GACCTATTGTTTCAAGTAAC-3′) | PCR-RFLP analysis | 24 |
| UP3 (5′-GTAAAATACCCTTACTCTCAGGCG-3′) |
Target sequences were amplified by 30 to 40 standard PCR cycles with 1 to 5 ng of gDNA/reaction mixture. Endonuclease treatments were carried out in 1× PCR buffer as indicated above. Restriction fragments and PCR products were resolved on 2% agarose or 3.5% MetaPhor agarose (FMC Bioproducts, Rockland, Maine) gels and were visualized by ethidium bromide staining.
Karyotype analysis.
The chromosomal locations of the COWP, RNR, and TRAP-C1 loci were determined by contour-clamped homogeneous electrical field (CHEF) electrophoresis as follows. Agarose blocks containing an amount of C. parvum DNA equivalent to 2 × 107 oocysts of the MD strain or a DNA size marker (Saccharomyces cerevisiae DNA Size Standard; Bio-Rad) were loaded onto a 1% low-gelling-temperature agarose gel made in 0.5× TBE (Tris-borate-EDTA) buffer. Chromosomal DNA was electrophoresed with the CHEF-DR II Pulse Field Electrophoresis System (Bio-Rad) with recirculated 0.5× TBE buffer maintained at 14°C. Electrophoresis was carried out at 120 V with a switch interval of 240 s for 72 h. The gel was then stained with 0.5 μg of ethidium bromide per ml and destained in distilled water, and the chromosomal bands were visualized with a UV transilluminator. The poly(T) locus was mapped by pulsed-field gel electrophoresis (PFGE) with the KSU-1 isolate as described previously (9).
Chromosomal DNA was depurinated by soaking the gel in 0.25 M HCl for 15 min, transferred onto a nylon membrane by capillary absorption, and cross-linked by UV irradiation. The membrane was hybridized sequentially to the 441-bp DNA fragment from the RNR R1 sequence and to the 4,420-bp COWP probe cpMM1 (20). A second membrane was probed with a 1,782-bp DNA fragment spanning most of the coding region of the TRAP-C1 gene (positions 70 to 1851). The poly(T) gene was located by using as a probe the 515-bp PCR fragment defined by primers cry44 and cry37 (5). Probes were labelled by random priming with [32P]dATP. Hybridizations were carried out at 65°C in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s solution–0.1% sodium dodecyl sulfate in the presence of 50 μg of salmon sperm DNA per ml. Washes were performed at 65°C in 0.1× SSC–0.1% sodium dodecyl sulfate. Southern analysis of the poly(T) locus was performed as described previously (8).
RESULTS
Recent studies that used PCR-based methods for the analysis of C. parvum genetic variation at single loci have coherently demonstrated the existence of two main parasite subpopulations characterized by distinct genotypes (3–5, 12–14, 19, 23). The aim of this study was a multilocus analysis of a broad and heterogeneous collection of C. parvum isolates. We chose for our study five genetic markers defined by the PCR primers listed in Table 1. These loci have been shown to consist of at least two alleles differentially associated with animal or human C. parvum isolates. The copy numbers of two markers, RNR and ITS1 (type A), were determined to be 1 and 4, respectively (data not shown; 9).
Chromosomal locations of polymorphic markers.
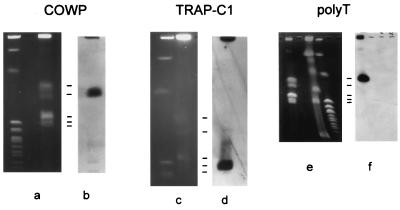
The chromosomal locations of four RFLP markers were determined by Southern analysis of CHEF- or PFGE-separated oocyst DNA (Fig. 1). The chromosomal profiles obtained by these electrophoretic techniques with DNA from the MD and KSU-1 strains (Fig. 1a, c, and e) were analogous to each other and to the profile described by Blunt et al. (2). Accordingly, the five chromosomal bands are numbered from 1 to 5, where band 1 is the largest band (1.54 Mb) and band 5 is the smallest band, which is estimated to be 1.03 Mb. Bands 1 and 3 are thought to comprise multiple comigrating chromosomes (2). Southern blot analysis showed that the markers are located on different chromosomes, namely, COWP and RNR on band 2 (Fig. 1b; data not shown), TRAP-C1 on band 5 (Fig. 1d), and poly(T) on band 1 (Fig. 1f). This analysis demonstrates that our RFLP markers fall into at least three distinct linkage groups.
FIG. 1.
Chromosomal locations of three C. parvum RFLP markers. Oocyst DNA (a, c, and e) was fractionated by CHEF electrophoresis (COWP and TRAP-C1) or PFGE [poly(T)] in parallel with S. cerevisiae DNA standards (a and c, left lanes; e, right lane) and Hansenula wingei chromosomes (e, middle lane). The locations of C. parvum chromosomal bands are indicated with horizontal lines. The molecular sizes of these bands are 1.54, 1.44, 1.24, 1.08, and 1.03 Mb from top to bottom, respectively (2). Panels b, d, and f show the results of Southern analyses with the corresponding probes.
Multilocus genotype analysis.
By using the primers listed in Table 1, 28 C. parvum isolates of disparate geographic and host origins were genotyped. Seventeen isolates originated directly from humans, three isolates (GCH1, LL, and 740) were derived from humans and propagated in calves, and eight isolates were from various animals. Human isolates were both from patients with sporadic cases of infection (H77-87, P12, and GCH2 to GCH6) and from documented outbreaks (P9, P16, P18, P27, and P29). GCH2 and GCH3 were from AIDS patients. Animal isolates included the widely used MD and GCH1 isolates.
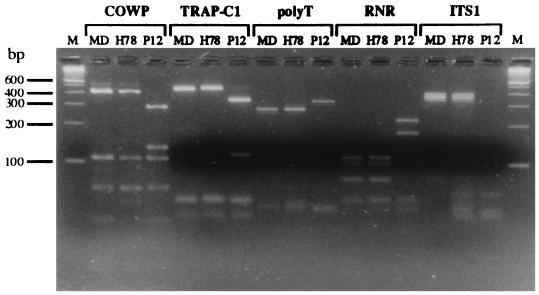
Consistent with previous reports, we observed for each locus two electrophoretic profiles (Fig. 2). The absence of reassorted genotypes is indicative of two reproductively separated subgroups in the species. With the exception of isolate PC1, which originated from a captive macaque, C. parvum of genotype H was exclusively associated with human infections. Genotype C was associated with isolates from animals and a minority of human isolates (LL, 740, H78, GCH6, and P9) (Table 2). The PCR and PCR-RFLP profiles obtained for three representative isolates, two from humans (H78 and P12) and one from an animal (MD), are shown in Fig. 2. For isolates GCH4 and GCH5, RFLP profiles indicative of mixed genotypes at the poly(T) locus were detected (5).
FIG. 2.
PCR and PCR-RFLP fingerprints of three C. parvum isolates obtained with five markers. Isolates MD and H78 are of ovine and human origin, respectively, and are examples of isolates displaying the C genotype, whereas isolate P12 is a human isolate of genotype H. Fractionation of DNA fragments was performed in 3.5% agarose (lane M, 100-bp DNA ladder).
DISCUSSION
Published studies on C. parvum genotypic heterogeneity rely on the analysis of single loci (3–5, 12, 14, 19, 22). Morgan et al. (13) recently applied two polymorphic markers. From that work, it appears that isolates of C. parvum segregate into two groups, designated H and C.
The use of multiple polymorphisms described in this report confirmed the occurrence of two genotypes. The absence of recombinant isolates indicates reproductive isolation between H and C isolates. This is surprising considering that both genotypes can be found simultaneously in the same host (5) and the fact that C. parvum undergoes an obligatory sexual cycle. With the notable exception of isolate PC1, the host specificities of C. parvum genotypes H and C were confirmed by the present study. PC1, identified as belonging to genotype H, is the first nonhuman H isolate described.
The genomic locations of the RFLP markers on three C. parvum chromosomal bands demonstrate that at least three independent linkage groups were sampled. Because COWP and RNR are both located on chromosomal band 2, and at least two of the five ribosomal genes colocalize with poly(T) and TRAP-C1 (9), the degree of linkage between these loci is unknown.
At first glance, the population structure of C. parvum emerging from this study is reminiscent of that observed in Toxoplasma gondii, in which three clonal lines were identified (7). In apparent contrast to C. parvum, rare mixed genotypes were observed in T. gondii. However, it cannot be excluded that recombinant genotypes will eventually be identified in C. parvum as more isolates are analyzed. Even if such mixed genotypes are identified in future studies, our data suggest that genetic exchange between genotypes is at best a rare event.
Further advances in our understanding of the population structure of C. parvum will come from the application of polymorphisms capable of differentiating among isolates belonging to the same genotypic group. Heterogeneity within each group has been observed. Peng et al. (14) described the presence of three TRAP-C2 alleles defined by five polymorphic nucleotide positions. Similarly, a total of four alleles, each defined by one or several point mutations, were identified within a 145-bp region of the poly(T) locus (23). These observations indicate that genetic fingerprints capable of differentiating among C. parvum isolates or groups of isolates will soon be available and allow us to improve the resolution of C. parvum genotypes. The relevance of this work lies in the potential for identifying clinically relevant markers. From an environmental point of view, high-resolution fingerprints could assist in identifying the source of oocysts found in surface water, facilitating the implementation of measures to reduce the access of oocysts to drinking water.
The potential for each C. parvum genotype to cause different clinical symptoms is unknown. Differences in infectivity for laboratory animals were reported (14, 15) and were also observed among isolates originating from people with AIDS in laboratory animals and in tissue culture (24). Characterization of larger numbers of isolates from humans with chronic and acute infections is in progress. Together with improved fingerprinting methods, this work will lead to a better understanding of cryptosporidiosis.
ACKNOWLEDGMENTS
This work received financial support from USDA (grant 94-371020914), NIH (cooperative agreement U019AI33384 and grant AI26497 from the International Collaborative Infectious Disease Research Program), and the IX AIDS Research Program, Ministero della Sanità, Istituto Superiore di Sanità, Rome, Italy. P.S. is supported by EEC RTD Programme ERBIC 18CT970217 of the European Community.
We sincerely thank all our colleagues who donated samples.
REFERENCES
- 1.Awad-El-Kariem F M, Robinson H A, Dyson D A, Evans D, Wright S, Fox T M, McDonald V. Differentiation between human and animal strains of Cryptosporidium parvum using isoenzyme typing. Parasitology. 1995;110:129–132. doi: 10.1017/s0031182000063885. [DOI] [PubMed] [Google Scholar]
- 2.Blunt D S, Khramtsov N V, Upton S J, Montelone B A. Molecular karyotype analysis of Cryptosporidium parvum: evidence for eight chromosomes and a low-molecular-size molecule. Clin Diagn Lab Immunol. 1997;4:11–13. doi: 10.1128/cdli.4.1.11-13.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnin A, Fourmaux M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygauthier-Toubas D, Gabriel-Pospisil F, Naciri M, Camerlynck P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 4.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carraway M, Tzipori S, Widmer G. New RFLP marker in Cryptosporidium parvum identifies mixed parasite populations and genotypic instability in response to host change. Infect Immun. 1997;65:3958–3960. doi: 10.1128/iai.65.9.3958-3960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guerrant R L. Cryptosporidiosis: an emerging, highly infectious threat. Emerg Infect Dis. 1997;3:51–57. doi: 10.3201/eid0301.970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe D K, Sibley L D. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- 8.Korman S H, Le Blancq S M, Deckelbaum R J, Van der Ploeg L H T. Investigation of human giardiasis by karyotype analysis. J Clin Invest. 1992;89:1725–1733. doi: 10.1172/JCI115774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Blancq S M, Khramtsov N, Zamani F, Upton S J, Wu T W. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 10.McKenzie W R, Hoxie N J, Proctor M E, Gradus M S, Blair K A, Peterson D E, Kazmierzak J J, Addiss D G, Fox K R, Rose J B, Davies J P. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 11.Morgan U M, Constantine C C, O’Donoghue P, Meloni B P, O’Brien P A, Thompson R C A. Molecular characterization of Cryptosporidium isolates from human and other animals using random amplified polymorphic DNA analysis. Am J Trop Med Hyg. 1995;52:559–564. doi: 10.4269/ajtmh.1995.52.559. [DOI] [PubMed] [Google Scholar]
- 12.Morgan U M, Constantine C C, Forbes D A, Thompson R C A. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J Parasitol. 1997;83:825–830. [PubMed] [Google Scholar]
- 13.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertzberg H, Elliot A, Thompson R C A. Molecular characterization of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- 14.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S L, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium parvum isolates: evidence for two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–573. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozio E, Angeles Gomez M A, Mancini Barbieri F, La Rosa G. Cryptosporidium: different behaviour in calves of isolates of human origin. Trans R Soc Trop Med Hyg. 1992;86:636–638. doi: 10.1016/0035-9203(92)90165-9. [DOI] [PubMed] [Google Scholar]
- 16.Ranucci L, Müller H M, La Rosa G, Reckmann I, Gomez Morales M A, Spano F, Pozio E, Crisanti A. Characterization and immunolocalization of a Cryptosporidium protein containing repeated amino acid motifs. Infect Immun. 1993;61:2347–2356. doi: 10.1128/iai.61.6.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith H V, Rose J B. Waterborne cryptosporidiosis. Parasitol Today. 1990;6:8–12. doi: 10.1016/0169-4758(90)90378-h. [DOI] [PubMed] [Google Scholar]
- 18.Solo-Gabriele H, Neumeister S. US outbreaks of cryptosporidiosis. J Am Water Works Assoc. 1996;88:76–86. [Google Scholar]
- 19.Spano F, Putignani L, McLaughlin J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;152:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 20.Spano F, Puri C, Ranucci L, Putignani L, Crisanti A. Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual parasite development. Parasitology. 1997;114:427–437. doi: 10.1017/s0031182096008761. [DOI] [PubMed] [Google Scholar]
- 20a.Spano F, Putignani L, Naitza S, Puri C, Wright S, Crisanti A. Molecular cloning and expression analysis of a Cryptosporidium parvum gene encoding a new member of the thrombospondin family. Mol Biochem Parasitol. 1998;92:147–162. doi: 10.1016/s0166-6851(97)00243-0. [DOI] [PubMed] [Google Scholar]
- 20b.Spano, F., L. Putignani, S. Guida, and A. Crisanti. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp. Parasitol., in press. [DOI] [PubMed]
- 20c.Spano, F., et al. Unpublished data.
- 21.Tzipori S. Cryptosporidiosis in perspective. Adv Parasitol. 1988;27:63–130. doi: 10.1016/S0065-308X(08)60353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasquez J R, Gooze L, Kim K, Gut J, Peterson C, Nelson R G. Potential antifolate resistance determinants and genotypic variation in the bifunctional dihydrofolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol Biochem Parasitol. 1996;79:153–156. doi: 10.1016/0166-6851(96)02647-3. [DOI] [PubMed] [Google Scholar]
- 23.Widmer G. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv Parasitol. 1998;40:224–239. doi: 10.1016/s0065-308x(08)60122-0. [DOI] [PubMed] [Google Scholar]
- 24.Widmer G, Tzipori S, Fichtenbaum C J, Griffiths J K. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J Infect Dis. 1998;178:834–840. doi: 10.1086/515373. [DOI] [PubMed] [Google Scholar]