Abstract
Newcastle disease (ND) is a highly pathogenic viral infection of poultry with significant economic impacts worldwide. Despite the widespread use of vaccines, ND outbreaks continue to occur even within vaccinated poultry farms. Furthermore, novel Newcastle disease virus (NDV) genotypes are emerging in poultry, increasing the need for the development of rapid, accurate, and simple diagnostic methods. We therefore developed two novel sets of visual reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays based on highly conserved regions of the HN and F genes. The limits of detection of the NDV-Common-LAMP assay, for all the NDV strains, were 103.0 EID50/0.1 mL for Kr005 and 102.0 EID50/0.1 mL for Lasota within 35 min. The sensitivity of the NDV-Patho-LAMP assay, used for the strain differentiation of virulent NDV, was 102.0 EID50/0.1 mL for Kr005. No amplification was detected for the non-NDV templates. Next, we probed 95 clinical strains and 7 reference strains with the RT-LAMP assays to assess the feasibility of their use in diagnostics. We observed no cross-reactivity across the 102 strains. Furthermore, there was 100% congruence between the RT-LAMP assays and full-length sequencing of the target genes, indicating the potential for visual RT-LAMP in the identification and differentiation of NDV. These novel RT-LAMP assays are ideally suited for the field or resource-limited environments to facilitate the faster detection and differentiation of NDV, which can reduce or avoid further spread.
Keywords: reverse transcription loop-mediated isothermal amplification, Newcastle disease virus, virulent, differentiation, diagnosis
1. Introduction
Newcastle disease virus (NDV), the etiological agent of Newcastle disease (ND), causes a highly pathogenic viral infection in poultry. NDV is an avian type I paramyxovirus (APMV-1) of the genus Avulavirus, belonging to the family Paramyxoviridae [1,2].
APMV-1 is separated into two distinct clades, class I and class II. Almost all class I viruses are weakly virulent and have been isolated mostly from wild birds (waterfowl and shorebirds) and live bird markets (LBMs) [3]. In contrast, class II viruses are highly virulent and have been found in poultry, pets, and wild birds [4,5].
Class II viruses are further divided into 21 genotypes (genotype I-XXI) and 5 pathotypes (viscerotropic velogenic, neurotropic velogenic, mesogenic, lentogenic, and asymptomatic) based on their clinical signs and pathological lesions [6,7,8]. The virulence of NDV strains is defined using in vivo infections, which measure the intracerebral pathogenicity index (ICPI) in 1-day-old chicks, the mean death time (MDT) in 9-day-old serum pathogen-free (SPF) embryonated chicken eggs, and the intravenous pathogenicity index (IVPI) in 6-week-old SPF chickens [2,9]. These methods are time-consuming, expensive, and laborious and raise ethical considerations for use in routine diagnostics.
The virulence of NDV has been predicted from amino acid sequences at the F protein cleavage site (FCS) using molecular techniques according to the definitions of the World Organization for Animal Health (WOAH) [10]. In most highly virulent velogenic and intermediately virulent mesogenic viruses, the FCS sequence is 112(R/K)-R-Q-(R/K)-R↓F117 (R, arginine; K, lysine; Q, glutamine; F, phenylalanine; arrow, cleavage position; number, residue position in F protein), while the sequence from lentogenic and asymptomatic viruses with low virulence is 112(G/E)-(R/K)-Q-(G/E)-R↓L117 (G, glycine; E, glutamic acid; R, arginine; K, lysine; Q, glutamine; L, leucine; arrow, cleavage position; number, residue position in the F protein).
Most available NDV diagnostic techniques employ variations of reverse-transcription (RT) PCR and real-time quantitative reverse-transcription (qRT) PCR assays, which allow for sensitive detection and pathotype differentiation [11,12,13,14]. Subsequently, PCR products amplified via RT-PCR are analyzed by performing gel electrophoresis, sequencing, a heteroduplex mobility assay, or restriction endonuclease analysis. Additionally, due to the widespread use of NDV vaccine strains and asymptomatic NDV carriage in wild birds, these assays are not suitable for all field samples. Although qRT-PCR allows for fast and high-throughput detection by avoiding a post-PCR processing step, this method strongly relies on trained personnel and expensive laboratory apparatus connected to a stable power supply, limiting its use in many diagnostic laboratories [15,16]. There is therefore a pressing need to develop simple, more rapid, cost-effective, and sensitive detection tools for screening NDV infection.
Loop-mediated isothermal amplification (LAMP) is a nucleotide amplification technique with two or three sets of complementary primers that is conducted within an hour under isothermal conditions [16]. In this study, we successfully developed and evaluated two sets of reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assays, accomplished in one step by adding a reverse transcriptase. The NDV-Common-LAMP assay for NDV and NDV-Patho-LAMP assay for virulent NDV, using highly conserved hemagglutinin-neuraminidase (HN) and fusion (F) NDV sequences, can detect and differentiate NDV for class II.
2. Results
2.1. Optimization of RT-LAMP Assays
Several LAMP primer sets targeting the HN and F genes were screened using varying primer concentrations and incubation times to optimize the RT-LAMP conditions. Increasing the concentrations of the FIP and LF primers targeting the F gene, we generated RT-LAMP products at low RNA template concentrations (Supplementary Figure S1). Furthermore, to successfully amplify and differentiate the genomic RNA of virulent NDV, incubation at 64 °C for more than 35 min was required. After RT-LAMP amplification under isothermal conditions, a color change from pink to yellow indicated a positive reaction. The sequences of the optimized RT-LAMP primer sets are shown in Table 1.
Table 1.
Primer sets optimized for the detection and differentiation of Newcastle disease virus.
| Primer Set | Target Gene | Primer | Sequence (5′–3′) | Product | |
|---|---|---|---|---|---|
| Size (bp) | |||||
| NDV-Common-LAMP | HN | Outer | F3 | GGATACCCTCATTCGACATGAG | 188 |
| B3 | TTGCACTCACACTGCAAGA | ||||
| Inner | FIP | TCCGAAGCACACAAGTTATACTG | |||
| CTACATCACAATGTGATA | |||||
| BIP | CATCTCAACAGGGAGGTATCTTCCGATTTTGGGTGTCATC | ||||
| Loop | LF | TGCGAGTGATCTCTGCAACC | |||
| LB | TCTTTTTACTCTGCGTTCCATCAATCT | ||||
| NDV-Patho-LAMP | F | Outer | F3 | GAGGCATACAATAGAACATGAC | 223 |
| B3 | TCTTTAAGCCGGAGGATGTT | ||||
| Inner | FIP | AAGCGTTTTGTCTCCTTCCTCACCC | |||
| CCCTTGGCGATTCCATCG | |||||
| BIP | TGTAGCTCTTGGGGTTGCAACAGC | ||||
| GCAGCATTCTGGTTGGCTTGTAT | |||||
| Loop | LF | CAGACCCTTGTATCCTGCGGAT | |||
| LB | GCACAGATAACAGCAGCCGC | ||||
| NDV-HN-PCR | HN | 1643F | ACCCTAGATCAGATGAGAGCC | 1643 | |
| 1643R | CTACTGTGAGAACTCTGCCTTC | ||||
| 1325F | AATCGGAAGTCTTGCAGTGTG | 1325 | |||
| 1325R | TGTGACTCTGGTAGATGATCTG | ||||
| NDV-F-PCR | F | 1333F | GCTAAGTACTCTGAGCCAAAC | 1333 | |
| 1333R | CAGTATGAGGTGTCGATTCTTCTA | ||||
| 1166F | GGGAACAATCAACTCAGCTCATT | 1166 | |||
| 1166R | GCCATGTGTTCTTTGCTTCTC | ||||
2.2. Specificity of RT-LAMP Assays
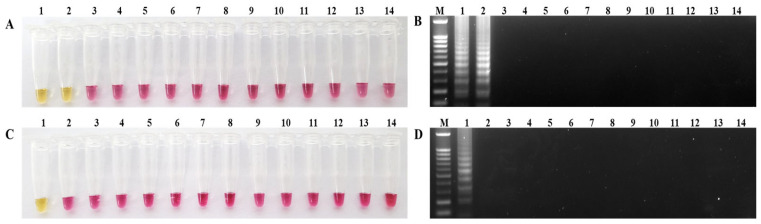
The specificity of the RT-LAMP primers was determined using 50 ng of genomic RNA extracted from APMV-1 (including Kr005 and Lasota), APMV-2, APMV-3, APMV-4, APMV-6, APMV-7, APMV-8, APMV-9, AIV H9N2, AEV, IBDV, IBV, and ARV. Only tubes containing viral RNA from Kr005 and Lasota and the NDV-Common-LAMP primers displayed a color change from pink to yellow, while the mixtures in the other tubes remained pink (Figure 1A). Additionally, the tube containing RNA from Kr005 and the NDV-Patho-LAMP primers displayed a color change from pink to yellow (Figure 1C). No amplification was observed in the other tubes. Positive reactions were also confirmed via electrophoresis on 1.5% TAE agarose gel (Figure 1B,D).
Figure 1.
Specificity of NDV-Common-LAMP and NDV-Patho-LAMP assays. (A) NDV-Common-LAMP and (C) NDV-Patho-LAMP only showed a visual color change from pink to yellow in tubes containing the target genomic RNA. (B) NDV-Common-LAMP and (D) NDV-Patho-LAMP reactions resolved via 1.5% agarose gel electrophoresis. Lane M, 100 bp DNA size marker; lane (tube) 1, APMV-1(Kr005); lane (tube) 2, APMV-1(Lasota); lane (tube) 3, APMV-2; lane (tube) 4, APMV-3; lane (tube) 5, APMV-4; lane (tube) 6, APMV-6; lane (tube) 7, APMV-8; lane (tube) 8, APMV-9; lane (tube) 9, AIV(H9N2); lane (tube) 10, AEV; lane (tube) 11, IBDV; lane (tube) 12, IBV; lane (tube) 13, ARV; lane (tube) 14, negative control.
2.3. Sensitivity of RT-LAMP Assays
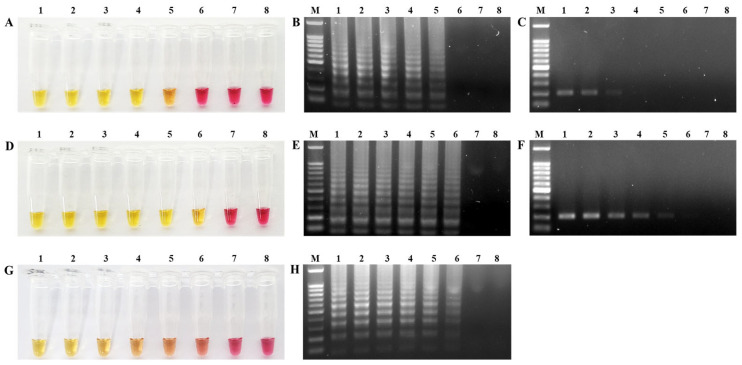
The limit of detection for the NDV-Common-LAMP and RT-PCR assays was determined via the amplification of serially diluted viral RNA from velogenic (Kr005, from 107.0 to 101.0 EID50/0.1 mL) and lentogenic strains (Lasota, from 107.0 to 101.0 EID50/0.1 mL). Positive amplification in the NDV-Common-LAMP assay was confirmed by a color change from pink to yellow and the presence of ladder-like DNA bands on the 1.5% TAE agarose gels. In the NDV-Common-LAMP assay, we detected target genes in the concentration range of 103.0 EID50/0.1 mL for Kr005 and 102.0 EID50/0.1 mL for Lasota under isothermal conditions within 35 min (Figure 2A,B,D,E). In contrast, the sensitivities of the RT-PCR assay using the F3 and B3 primers were 105.0 EID50/0.1 mL for Kr005 and 103.0 EID50/0.1 mL for Lasota (Figure 2C,F). According to the results, the NDV-Common-LAMP assay is 10~100 times more sensitive than RT-PCR for the detection of NDV. The NDV-Patho-LAMP assay amplified the target gene at 102.0 EID50/0.1 mL for Kr005 (Figure 2G,H).
Figure 2.
Detection limits of RT-LAMP and RT-PCR assays. (A,D) Visualization of NDV-Common-LAMP products using Kr005 and Lasota genomic RNA, respectively. (G) Visualization of the NDV-Patho-LAMP assay using Kr005 genomic RNA. LAMP-positive reactions display a color change from pink to yellow. (B,E,H) Agarose gel electrophoresis of LAMP products. (B) NDV-Common-LAMP for Kr005, (E) NDV-Common-LAMP for Lasota, and (H) NDV- Patho-LAMP for Kr005. (C,F) Conventional RT-PCR products resolved via 1.5% agarose gel electrophoresis to compare the sensitivity of RT-PCR with that of the NDV-Common-LAMP assay. Lane M, 100 bp DNA size marker; lane (tube) 1, 107.0 EID50/0.1 mL; lane (tube) 2, 106.0 EID50/0.1 mL; lane (tube) 3, 105.0 EID50/0.1 Ml; lane (tube) 4, 104.0 EID50/0.1 mL; lane (tube) 5, 103.0 EID50/0.1 mL; lane (tube) 6, 102.0 EID50/0.1 mL; lane (tube) 7, 101.0 EID50/0.1 mL; lane (tube) 8, negative control.
2.4. Validation of RT-LAMP Assays with NDV Strains
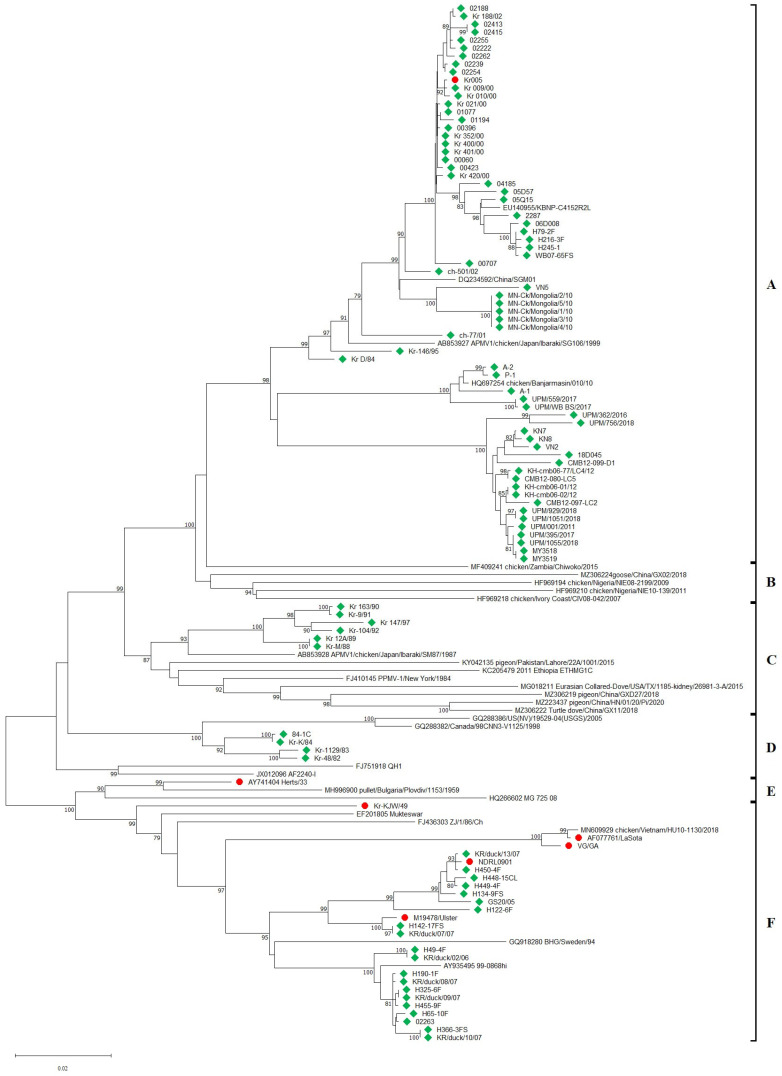
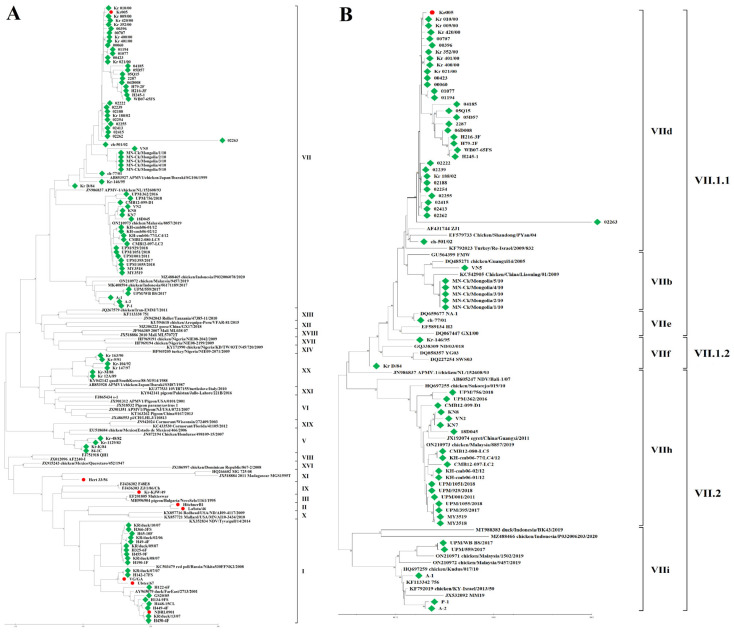
A total of 102 NDV strains were probed using the NDV-Common-LAMP assay. The amplification results were 100% in agreement with those of the NDV-HN-PCR and NDV-F-PCR assays (Table 2). We subsequently analyzed the amplified PCR products via sequencing. Based on the entire F nucleotide sequence of 95 clinical strains, the amino acid sequences (residues 112 to 117) of the FCS were classified into four types: 112RRQKRF117 for 51 strains isolated from Korea, Vietnam, Pakistan, and Malaysia; 112RRRKRF117 for 22 strains isolated from Korea, Mongolia, Vietnam, Cambodia, and Malaysia; 112KRRKRF117 for 2 strains isolated from Malaysia; and 112GKQGRL117 for 20 strains isolated from Korea. Of the 95 field strains, 75 strains were positive in the NDV-Patho-LAMP assay, and these results were 100% congruent with the amino acid sequences of the FCS. No strain with a 112GKQGRL117 FCS was detected using the NDV-Patho-LAMP assay. Phylogenetic analysis of the full-length HN and F genes showed that 95 NDV strains belong to classes A, C, D, E, and F for HN (Figure 3) and are grouped into genotypes V, VII, IX, XI, and XX of class II for F (Figure 4A). Furthermore, genotype VII was divided into the sub-genotypes VII.1.1 (former VIIb, VIId, and VIIe), VII.1.2 (former VIIf), and VII.2 (former VIIh and VIIi) (Figure 4B). The robustness of branching for the sub-genotypes was supported by high bootstrap values.
Table 2.
Comparisons of RT-LAMP assays and sequence analysis of HN and F genes.
| Strain | Year of Isolation | Country of Origin | Host | RT-LAMP | Sequencing | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Common | Patho | Common | Patho | GenBank Accession No. | |||||||
| HN | F | F0 Cleavage Site | HN | F | |||||||
| Reference in NCBI |
LaSota/46 | 1946 | - | - | P 1 | N 2 | P | P | GRQGRL | AF077761 | AF077761 |
| HitchnerB1 | 1947 | USA | - | P | N | P | P | GRQGRL | - | JN872151 | |
| Ulster/67 | - | - | - | P | N | P | P | GRQGRL | AY562991 | AY562991 | |
| VG/GA | - | - | - | P | N | P | P | GRQGRL | EU289028 | EU289028 | |
| Hert 33/1956 | - | - | - | P | P | P | P | RRQRRF | AY741404 | AY741404 | |
| Kr-KJW/49 | 1949 | Korea | Chicken | P | P | P | P | RRQKRF | - | AY630409 | |
| Kr005 | 2000 | Korea | Chicken (layer) | P | P | P | P | RRQKRF | - | KY404087 | |
| 02263 | 2002 | Korea | - | P | N | P | P | GKQGRL | OP921680 | OP818810 | |
| GS20/05 | 2005 | Korea | - | P | N | P | P | GKQGRL | OP921630 | OP818760 | |
| KR/duck/02/06 | 2006 | Korea | Duck | P | N | P | P | GKQGRL | OP921712 | OP818842 | |
| KR/duck/07/07 | 2006 | Korea | Duck | P | N | P | P | GKQGRL | OP921639 | OP818769 | |
| KR/duck/08/07 | 2006 | Korea | Duck | P | N | P | P | GKQGRL | OP921640 | OP818770 | |
| KR/duck/09/07 | 2006 | Korea | Duck | P | N | P | P | GKQGRL | OP921641 | OP818771 | |
| KR/duck/10/07 | 2006 | Korea | Duck | P | N | P | P | GKQGRL | OP921642 | OP818772 | |
| KR/duck/13/07 | 2007 | Korea | Duck | P | N | P | P | GKQGRL | OP921691 | OP818821 | |
| H142-17FS | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921624 | OP818754 | |
| H190-1F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921622 | OP818752 | |
| H450-4F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921625 | OP818755 | |
| H455-9F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921626 | OP818756 | |
| H448-15CL | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921627 | OP818757 | |
| H65-10F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921628 | OP818758 | |
| H449-4F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921629 | OP818759 | |
| H49-4F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921634 | OP818764 | |
| H325-6F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921636 | OP818766 | |
| H122-6F | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921637 | OP818767 | |
| H134-9FS | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921638 | OP818768 | |
| H366-3FS | 2007 | Korea | - | P | N | P | P | GKQGRL | OP921632 | OP818762 | |
| Kr-48/82 | 1982 | Korea | Chicken | P | P | P | P | RRQKRF | OP921713 | OP818843 | |
| Kr-1129/83 | 1983 | Korea | - | P | P | P | P | RRQKRF | OP921686 | OP818816 | |
| Kr-M/88 | 1984 | Korea | Quail | P | P | P | P | RRQKRF | OP921718 | OP818848 | |
| Kr-K/84 | 1984 | Korea | Chicken | P | P | P | P | RRQKRF | OP921687 | OP818817 | |
| Kr_D/84 | 1984 | Korea | Peafowl | P | P | P | P | RRQKRF | OP921652 | OP818782 | |
| 84-1C | 1984 | Korea | - | P | P | P | P | RRQKRF | OP921653 | OP818783 | |
| Kr_12A/89 | 1989 | Korea | Chicken | P | P | P | P | RRRKRF | OP921654 | OP818784 | |
| Kr_163/90 | 1990 | Korea | - | P | P | P | P | RRRKRF | OP921655 | OP818785 | |
| Kr-9/91 | 1991 | Korea | - | P | P | P | P | RRRKRF | OP921656 | OP818786 | |
| Kr-104/92 | 1992 | Korea | - | P | P | P | P | RRRKRF | OP921688 | OP818818 | |
| Kr-146/95 | 1995 | Korea | Chicken (broiler) | P | P | P | P | RRQKRF | OP921714 | OP818844 | |
| Kr_147/97 | 1997 | Korea | Chicken (broiler) | P | P | P | P | RRRKRF | OP921657 | OP818787 | |
| Kr_420/00 | 2000 | Korea | Ostrich | P | P | P | P | RRQKRF | OP921658 | OP818788 | |
| Kr_009/00 | 2000 | Korea | Chicken (layer) | P | P | P | P | RRQKRF | OP921659 | OP818789 | |
| Kr_010/00 | 2000 | Korea | Chicken (layer) | P | P | P | P | RRQKRF | OP921660 | OP818790 | |
| Kr_352/00 | 2000 | Korea | Chicken | P | P | P | P | RRQKRF | OP921661 | OP818791 | |
| Kr_400/00 | 2000 | Korea | Chicken (broiler) | P | P | P | P | RRQKRF | OP921662 | OP818792 | |
| Kr_401/00 | 2000 | Korea | Chicken | P | P | P | P | RRQKRF | OP921663 | OP818793 | |
| 00060 | 2000 | Korea | - | P | P | P | P | RRQKRF | OP921664 | OP818794 | |
| 00423 | 2000 | Korea | - | P | P | P | P | RRQKRF | OP921665 | OP818795 | |
| 00707 | 2000 | Korea | - | P | P | P | P | RRQKRF | OP921666 | OP818796 | |
| Kr_021/00 | 2000 | Korea | Chicken (layer) | P | P | P | P | RRQKRF | OP921667 | OP818797 | |
| 01194 | 2001 | Korea | - | P | P | P | P | RRQKRF | OP921669 | OP818799 | |
| 01077 | 2001 | Korea | - | P | P | P | P | RRQKRF | OP921668 | OP818798 | |
| 02188 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921670 | OP818800 | |
| Kr_188/02 | 2002 | Korea | Chicken (broiler) | P | P | P | P | RRQKRF | OP921671 | OP818801 | |
| 00396 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921672 | OP818802 | |
| 02413 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921673 | OP818803 | |
| 02415 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921674 | OP818804 | |
| 02222 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921675 | OP818805 | |
| 02239 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921676 | OP818806 | |
| 02254 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921677 | OP818807 | |
| 02255 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921678 | OP818808 | |
| 02262 | 2002 | Korea | - | P | P | P | P | RRQKRF | OP921679 | OP818809 | |
| 04185 | 2004 | Korea | - | P | P | P | P | RRQKRF | OP921614 | OP818744 | |
| 05D57 | 2005 | Korea | Chicken (broiler) | P | P | P | P | RRQKRF | OP921715 | OP818845 | |
| 05Q15 | 2005 | Korea | - | P | P | P | P | RRQKRF | OP921620 | OP818750 | |
| 06D008 | 2006 | Korea | Chicken (broiler) | P | P | P | P | RRQKRF | OP921618 | OP818748 | |
| 2287 | 2006 | Korea | - | P | P | P | P | RRQKRF | OP921615 | OP818745 | |
| H245-1 | 2007 | Korea | - | P | P | P | P | RRQKRF | OP921619 | OP818749 | |
| H79-2F | 2007 | Korea | - | P | P | P | P | RRQKRF | OP921623 | OP818753 | |
| WB07-65FS | 2007 | Korea | - | P | P | P | P | RRQKRF | OP921631 | OP818761 | |
| H216-3F | 2007 | Korea | - | P | P | P | P | RRQKRF | OP921633 | OP818763 | |
| ch-77/01 | 2001 | China | Chicken meat | P | P | P | P | RRQKRF | OP921651 | OP818781 | |
| ch-501/02 | 2002 | China | Chicken meat | P | P | P | P | RRQKRF | OP921650 | OP818780 | |
| MN-Ck/Mongolia/1/10 | - | Mongolia | - | P | P | P | P | RRQKRF | OP921699 | OP818829 | |
| MN-Ck/Mongolia/2/10 | - | Mongolia | - | P | P | P | P | RRQKRF | OP921697 | OP818827 | |
| MN-Ck/Mongolia/3/10 | - | Mongolia | - | P | P | P | P | RRQKRF | OP921700 | OP818830 | |
| MN-Ck/Mongolia/4/10 | - | Mongolia | - | P | P | P | P | RRQKRF | OP921701 | OP818831 | |
| MN-Ck/Mongolia/5/10 | - | Mongolia | - | P | P | P | P | RRQKRF | OP921698 | OP818828 | |
| VN5 | 2007 | Vietnam | Chicken | P | P | P | P | RRQKRF | OP921705 | OP818835 | |
| VN2 | 2011 | Vietnam | Chicken | P | P | P | P | RRRKRF | OP921716 | OP818846 | |
| KN7 | 2012 | Vietnam | Chicken | P | P | P | P | RRRKRF | OP921685 | OP818815 | |
| KN8 | 2012 | Vietnam | Chicken | P | P | P | P | RRRKRF | OP921689 | OP818819 | |
| 18D045 | 2018 | Vietnam | - | P | P | P | P | RRRKRF | OP921690 | OP818820 | |
| KH-cmb06-01/12 | 2012 | Cambodia | Chicken | P | P | P | P | RRRKRF | OP921643 | OP818773 | |
| KH-cmb06-02/12 | 2012 | Cambodia | Chicken | P | P | P | P | RRRKRF | OP921644 | OP818774 | |
| KH-cmb06-77/LC4/12 | 2012 | Cambodia | - | P | P | P | P | RRRKRF | OP921645 | OP818775 | |
| CMB12-097-LC2 | 2012 | Cambodia | - | P | P | P | P | RRRKRF | OP921647 | OP818777 | |
| CMB12-080-LC5 | 2012 | Cambodia | - | P | P | P | P | RRRKRF | OP921648 | OP818778 | |
| CMB12-099-D1 | 2012 | Cambodia | - | P | P | P | P | RRRKRF | OP921649 | OP818779 | |
| A-1 | 2016 | Pakistan | Chicken (broiler) | P | P | P | P | RRQKRF | OP921702 | OP818832 | |
| A-2 | 2016 | Pakistan | Chicken (broiler) | P | P | P | P | RRQKRF | OP921703 | OP818833 | |
| P-1 | 2016 | Pakistan | Chicken (broiler) | P | P | P | P | RRQKRF | OP921704 | OP818834 | |
| MY3518 | 2010 | Malaysia | - | P | P | P | P | RRRKRF | OP921616 | OP818746 | |
| MY3519 | 2010 | Malaysia | - | P | P | P | P | RRRKRF | OP921617 | OP818747 | |
| UPM/001/2011 | 2011 | Malaysia | Chicken (broiler) | P | P | P | P | RRRKRF | OP921681 | OP818811 | |
| UPM/362/2016 | 2016 | Malaysia | Chicken (broiler) | P | P | P | P | KRRKRF | OP921682 | OP818812 | |
| UPM/395/2017 | 2017 | Malaysia | Chicken (broiler) | P | P | P | P | RRRKRF | OP921693 | OP818823 | |
| UPM/559/2017 | 2017 | Malaysia | Chicken (broiler) | P | P | P | P | RRQKRF | OP921683 | OP818813 | |
| UPM/WB_BS/2017 | 2017 | Malaysia | Black swan | P | P | P | P | RRQKRF | OP921684 | OP818814 | |
| UPM/756/2018 | 2018 | Malaysia | Chicken (broiler) | P | P | P | P | KRRKRF | OP921692 | OP818822 | |
| UPM/929/2018 | 2018 | Malaysia | Chicken (broiler) | P | P | P | P | RRRKRF | OP921694 | OP818824 | |
| UPM/1051/2018 | 2018 | Malaysia | Chicken (layer) | P | P | P | P | RRRKRF | OP921696 | OP818826 | |
| UPM/1055/2018 | 2018 | Malaysia | Chicken (broiler) | P | P | P | P | RRRKRF | OP921695 | OP818825 | |
1; positive result, 2; negative result.
Figure 3.
Phylogenetic tree based on full-length sequences of the HN gene constructed using the neighbor-joining method with MEGA (version 11.0). The bootstrap values were determined from 1000 replicates of the original data. Values ≥ 70 are indicated on the branches (as percentages). Six genotypes were identified A, B, C, D, E and F. NDV isolates and reference strains are shown by green squares and red circles, respectively.
Figure 4.
Phylogenetic trees based on the complete F gene using the neighbor-joining method with MEGA (version 11.0). Values ≥ 70 are indicated on the branches (as percentages). (A) Phylogenetic analysis of 95 NDV strains (green squares) and seven reference strains (red circles) available in GenBank. (B) Phylogenetic tree showing the sub-classification of genotype VII NDV strains (green squares) and a reference strain (red circle).
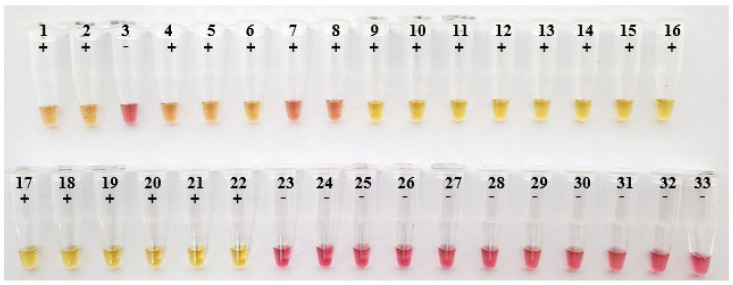
As shown in Figure 5, the practical application of the NDV-Patho-LAMP assay using 32 oropharyngeal (OP) swabs showed seven samples as positive among eight samples in the vaccinated + NDV group. All samples in the non-vaccinated + NDV group appeared as positive in yellow, while all samples in the negative control were yielded as negative in pink.
Figure 5.
Validation of NDV-Patho-LAMP assay using 32 oropharyngeal swabs. The color of LAMP-positive reactions turned yellow, while the color of LAMP-negative reactions remained pink. Tube 1–8, OP swabs collected from a vaccinated + NDV group; tube 9–22, OP swabs collected from a non-vaccinated + NDV group; tube 23–32, OP swabs collected from a negative group; tube 33, negative control.
3. Discussion
ND was first reported in 1926 in Indonesia and has since spread across the globe and is considered the most dangerous disease of poultry [1]. The WOAH classifies NDV as a list ‘A’ disease, and an outbreak of velogenic or mesogenic ND must be immediately reported to the WOAH, which may result in severe trade restrictions and a major economic burden for the global poultry industry [10]. Although intensive vaccination programs and biosecurity practices have been implemented for decades to limit spread of ND, viral outbreaks, especially those caused by sub-genotype VII.2 (VIIh and VIIi) strains, still sporadically occur in the Middle East, America, Europe, and Africa, in addition to Asian countries including China, Mongolia, Vietnam, Malaysia, and Indonesia [1,5,8,17,18,19,20].
In Korea, the last reported case of ND was in 2010 and was caused by virulent strains of the sub-genotype VII.1.1 (VIId), but no further reports of ND outbreaks have emerged since then. The APQA, which was designated as a WOAH reference laboratory for ND in 2010, has regularly performed active and passive surveillance to monitor the presence or absence of NDV in wild birds, LBMs, and poultry farms, which may be important reservoirs for APMV-1. Here, we established two colorimetric RT-LAMP assays to accurately and rapidly detect NDV and vaccine pathotypes for large-scale NDV screening as well as field testing.
RT-LAMP assays are considered an attractive alternative to PCR-based methods due to their speed, simplicity, specificity, and sensitivity. As RT-LAMP reactions are carried out at a constant temperature of 60–65 °C within an hour, a simple instrument such as a water bath or heat block is sufficient for genomic RNA amplification [21]. Positive results are visually detected as a color change, with no need for agarose gel electrophoresis [22]. Each RT-LAMP assay was optimized under isothermal conditions (64 °C for 35 min) with two sets of primers (six primers each) targeting the HN and F genes. The HN and F genes encode surface glycoproteins of the viral envelope, which are responsible for attachment to cell surface receptors and fusion between the cellular and viral membranes [17,23]. Furthermore, since the amino acid sequence of the FCS has a significant impact on fusogenic activity and proteolytic cleavage, the FCS is a key determinant of viral fusion [1].
Our analysis of the specificities of the NDV-Common-LAMP and NDV-Patho-LAMP assays revealed that the target genes were successfully amplified without cross-reactivity with other avian viral pathogens. The limits of detection for the NDV-Common-LAMP and RT-PCR assays using the F3 and B3 primers were estimated using viral genomic RNA from Kr005 and Lasota. This revealed that the sensitivity of the NDV-Common-LAMP assay was much higher than that of a conventional RT-PCR reaction. Furthermore, the NDV-Common-LAMP assay for Lasota was 100 times more sensitive than conventional RT-PCR [24].
In their previous studies, Pham et al. established a LAMP assay for targeting the F gene in 2005, but this method requires a further 2 h for cDNA synthesis and gene amplification [25]. Li et al. developed a one-step reverse transcription (RT)-LAMP assay, in which reverse transcription and amplification were carried out in a single tube [26]. Kirunda et al. developed a convenient and cheaper alternative of RT-LAMP for NDV detection using tracheal tissues and cloacal and oropharyngeal swab samples [27]. In this study, both the specificity and sensitivity of the NDV-Common-LAMP and NDV-Patho-LAMP assays were superior to those reported previously [26,27]. No color changes were observed for the targeting of Hert 33/1956, Kr-KJW/49, and UPM111 using the previous RT-LAMP primers. We compared the sensitivities of the RT-LAMP assays for detecting NDV using 10-fold serial dilutions of RNA templates extracted from Kr005. The detection limits of the RT-LAMP assays developed by Li et al. and Kirunda et al. [26,27]. were the concentrations of 10−4 and 100 under isothermal conditions, respectively. These observations indicated that, according to the detection limits of the NDV-Patho-LAMP, it is 10~10,000 times more sensitive than the previous RT-LAMP primers. Furthermore, these previously developed RT-LAMP assays could not distinguish virulent NDV, further highlighting the utility of our assays.
To validate the NDV-Common-LAMP and NDV-Patho-LAMP assays, we performed the amplification of 102 NDV strains and compared the results with those of the Sanger sequencing analysis of HN and F. We observed 100% (102/102) positivity in the NDV-Common-LAMP, NDV-HN-PCR, and NDV-F-PCR assays when probing the clinical strains. Among the 95 wild strains, the ratio of virulent NDV positivity to the total samples was 78.9% (75/95) using the NDV-Patho-LAMP assay, which was consistent with the FCS amino acid sequences determined from the NDV-F-PCR amplicons. No false-positive or false-negative results were obtained in the 102 strains using either the NDV-Common-LAMP or NDV-Patho-LAMP assay. Furthermore, we also demonstrated that the NDV-Patho-LAMP assay, using OP swab samples, is suitable for screening field samples.
In conclusion, the newly developed NDV-Common-LAMP and NDV-Patho-LAMP assays targeting the HN and F genes reported in this study are highly sensitive, specific, convenient, and rapid for the detection and differentiation of NDV. Since LAMP reactions can be performed using a battery-driven portable instrument, these RT-LAMP assays have potential utility for on-site diagnosis, even in insufficiently equipped facilities. Our study may assist in the development of prevention and control strategies for NDV in the poultry industry.
4. Materials and Methods
4.1. Primer Design
NDV-Common-LAMP primers for the simultaneous detection of velogenic, mesogenic, lentogenic, and asymptomatic NDV were designed using Clustal W multiple-sequence alignments of the published target genes in GenBank (http://www.ncbi.nlm.nih.gov (accessed on 1 January 2023)). NDV-Patho-primers specific to virulent NDV were designed to target the variable region. Following the analysis of 100 NDV sequences in the NCBI, we selected the HN and F genes as regions for the detection of NDV and differentiation of virulent NDV, respectively. The LAMP primers included the following: a forward outer primer F3, a reverse outer primer B3, a forward inner primer FIP (harboring the F2 region at its 3′-end and the F1c region at its 5′-end), a reverse inner primer BIP (harboring the B2 region at its 3′-end and the B1c region at its 5′-end), a forward loop primer LF, and a reverse loop primer LB. The primers recognized eight conserved regions within their target genes and generated amplification products with sizes of 188 bp for NDV-Common-LAMP and 223 bp for NDV-Patho-LAMP, respectively. To validate the LAMP primers for HN and F, conventional NDV-HN-PCR and NDV-F-PCR primers were designed to amplify full-length sequences of HN and F (Table 1).
4.2. Viral RNA Preparation
Total RNA was extracted from the viral strains APMV-1 (including velogenic Kr005 and lentogenic Lasota), APMV-2 (chicken, Yucaipa, California, 56), APMV-3 (parakeet, Netherlands, 449/75), APMV-4 (duck, Hong Kong, D3/75), APMV-6 (duck, Hong Kong, 18/199/77), APMV-7 (dove, Tennessee, 4/75), APMV-8 (goose, Delaware, 1053/76), APMV-9 (duck, New York, 22/78), avian influenza A (H9N2) virus (AIV H9N2, 16AQ075), avian encephalomyelitis virus (AEV, 14ABQ615), infectious bursal disease virus (IBDV, ATCC VR478), infectious bronchitis virus (IBV, ATCC VR21), and avian reovirus (ARV, 15ABD080) using a QIAamp Viral RNA mini kit (Qiagen, Düsseldorf, Germany), according to the manufacturer’s instructions. Viral RNA was eluted in 50 μL of elution buffer and stored at −70 °C until use.
4.3. Optimization of RT-LAMP Assays
NDV-Common-LAMP and NDV-Patho-LAMP reactions were carried out in a total volume of 25 μL using a WarmStart Colorimetric LAMP 2× Master Mix (NEB, Ipswich, MA, USA). The reaction mixture contained 2 μL of viral RNA, 2× reaction buffer, 2.5 μM of F3 and B3 primers, 22.5 μM of FIP and 20 μM BIP primers, and 12.5 μM of LF and 10 μM LB primers. The reactions were performed under isothermal conditions at 64 °C for 35 min in a heat block.
4.4. Specificity and Detection Limits of RT-LAMP Assays
The specificities of the optimized NDV-Common-LAMP and NDV-Patho-LAMP assays were determined using 50 ng of RNA from APMV-1 (including Kr005 and Lasota), APMV -2, APMV-3, APMV-4, APMV -6, APMV -7, APMV -8, APMV -9, AIV H9N2, AEV, IBDV, IBV, and ARV. Each LAMP reaction was performed at 64 °C for 35 min. The limits of detection of the assays were determined by probing 10-fold serial dilutions of viral RNA extracted from 107.0 EID50/0.1 mL KR005 and 107.0 EID50/0.1 mL Lasota. The reaction mixtures were incubated at 64 °C for 35 min. To compare the detection limits of the NDV-Common-LAMP and RT-PCR assays, RT-PCR was performed using F3 and B3 primers (Table 1). PCR amplification was carried out in a 20 μL reaction containing 2 μL of each RNA extract, 2.5 μM of each primer (F3 and B3 of LAMP primers), and 2× reaction buffer (BlackPCR RT-PCR premix; Ventech Science, Daegu, Republic of Korea). The RT-PCR step was 30 min at 45 °C, followed by 5 min at 94 °C. The cycling conditions were 30 cycles of 30 s at 94 °C, 45 s at 55 °C and 72 °C for 30 s, with a final extension at 72 °C for 5 min. The RT-LAMP and RT-PCR products were resolved on 1.5% agarose gel. All dilutions were analyzed in triplicate for the RT-LAMP and PCR assays.
4.5. Validation of RT-LAMP Assays with Wild Strains
In total, 95 wild strains (63 from Korea, 2 from China, 6 from Cambodia, 11 from Malaysia, 5 from Mongolia, 5 from Vietnam, and 3 from Pakistan) and 7 reference strains were collected from the Animal and Plant Quarantine Agency (APQA) in Korea from 1946 to 2018. Of the 95 wild strains, 63 strains were isolated from chicken, duck, quail, peafowl, or ostrich in Korea from 1982 to 2008. The other NDV isolates were obtained from chickens, chicken meat, or black swans in neighboring countries. The reference strains consisted of three velogenic viruses (Hert 33/1956, Kr-KJW/49 and Kr005), two lentogenic viruses (Lasota46 and Hitchner B1), and two asymptomatic viruses (Ulster67 and VG/GA). All wild strains were isolated by inoculating the allantois of 9-day-old specific pathogen-free embryonated chicken eggs [10].
The allantoic fluids with hemagglutination (HA) activity (ranging from 27 to 210) were processed for the extraction of viral RNA using a QIAamp Viral RNA mini kit (Qiagen, Germany). These samples were subsequently subjected to NDV-Common-LAMP, NDV-Patho-LAMP, NDV-HN-PCR, and NDV-F-PCR. To analyze the entire HN and F genes, NDV-HN-PCR and NDV-F-PCR assays were performed in a 20 μL volume containing 2 μL of each RNA extract, 2.5 μM of each primer, and 2× reaction buffer (BlackPCR RT-PCR premix; Ventech Science, Republic of Korea). PCR amplification was performed in a thermal cycler (Eppendorf, Germany) for 1 cycle of 30 min at 45 °C and 5 min at 94 °C, followed by 94 °C for 50 s, 60 °C for 50 s, and 72 °C for 50 s for 37 cycles, before a final extension at 72 °C for 5 min. All amplified fragments were purified using a QIAquick Gel Extraction Kit (Qiagen, Germany), and Sanger sequencing was performed by CosmoGenetech Co. (Seoul, Republic of Korea). The obtained nucleotide sequences were assembled, aligned, and compared using the CLC Main Workbench (Qiagen, Germany). The newly generated datasets were deposited in GenBank (Table 2). Phylogenetic analyses of the complete HN and F genes were performed using the neighbor-joining method [28] in MEGA 11. The statistical significance of the tree was assessed with a bootstrap value of 1000. In addition, OP swabs were collected from 22 artificially infected chickens at 3 days post-challenge (DPC). Of the 22 chickens, 8 chickens were in the vaccinated + NDV group (highly virulent NDV-challenged chickens after vaccination), and 14 chickens were in the non-vaccinated + NDV group (highly virulent NDV challenged chickens), while 10 chickens were negative controls. The genomic RNA was isolated from OP swabs for the NDV-Patho-LAMP assay.
5. Patents
Two patent numbers, 10-2022-0140964 and 10-2022-0140972, for the NDV-Common-LAMP and NDV-Patho-LAMP assays, were approved by the Korean Intellectual Property Office (KIPO) on 28 October 2022.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241813847/s1.
Author Contributions
Conceptualization, H.-S.S.; investigation, H.-S.S. and H.-S.K.; methodology, H.-S.S., H.-S.K. and J.-Y.K.; data curation, H.-S.S.; writing—original draft, H.-S.S.; writing—review and editing, Y.-K.K. and H.-R.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The OP swabs were collected from the experimental animals approved by Animal Ethics Committee of the Animal and Plant Quarantine Agency (Animal Ethics Approval: 2020-556).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received grants from the Animal and Plant Quarantine Agency (grant B-1543084-2021-23) and the Ministry of Agriculture, Food and Rural Affairs of the Republic of Korea.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ganar K., Das M., Sinha S., Kumar S. Newcastle disease virus: Current status and our understanding. Virus Res. 2014;184:71–81. doi: 10.1016/j.virusres.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohaim M.A., Al-Natour M.Q., El Naggar R.F., Abdelsabour M.A., Madbouly Y.M., Ahmed K.A., Munir M. Evolutionary trajectories of avian avulaviruses and vaccines compatibilities in poultry. Vaccines. 2022;10:1862. doi: 10.3390/vaccines10111862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim B.Y., Lee D.H., Kim M.S., Jang J.H., Lee Y.N., Park J.K., Yuk S.S., Lee J.B., Park S.Y., Choi I.S., et al. Exchange of Newcastle disease viruses in Korea: The relatedness of isolates between wild birds, live bird markets, poultry farms and neighboring countries. Infect. Genet. Evol. 2012;12:478–482. doi: 10.1016/j.meegid.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Kim L.M., King D.J., Suarez D.L., Wong C.W., Afonso C.L. Characterization of class I newcastle disease virus isolates from Hong Kong live bird markets and detection using real-time reverse transcription-PCR. J. Clin. Microbiol. 2007;45:1310–1314. doi: 10.1128/JCM.02594-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller P.J., Haddas R., Simanov L., Lublin A., Rehmani S.F., Wajid A., Bibi T., Khan T.A., Yaqub T., Setiyaningsih S. Identification of new sub-genotypes of virulent Newcastle disease virus with potential panzootic features. Infect. Genet. Evol. 2015;29:216–229. doi: 10.1016/j.meegid.2014.10.032. [DOI] [PubMed] [Google Scholar]
- 6.De Leeuw O.S., Koch G., Hartog L., Ravenshorst N., Peeters B.P.H. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin-neuraminidase protein. J. Gen. Virol. 2005;86:1759–1769. doi: 10.1099/vir.0.80822-0. [DOI] [PubMed] [Google Scholar]
- 7.Kgotlele T., Modise B., Nyange J.F., Thanda C., Cattoli G., Dundon W.G. First molecular characterization of avian paramyxovirus-1 (Newcastle disease virus) in Botswana. Virus Genes. 2020;56:646–650. doi: 10.1007/s11262-020-01770-4. [DOI] [PubMed] [Google Scholar]
- 8.Nooruzzaman M., Hossain I., Begum J.A., Moula M., Shamsul Arefin Khaled S.A., Parvin R., Chowdhury E.H., Islam M.R., Diel D.G., Dimitrov K.M. The first report of a virulent Newcastle disease virus of genotype VII.2 causing outbreaks in chickens in Bangladesh. Viruses. 2022;14:2627. doi: 10.3390/v14122627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desingu P.A., Singh S.D., Dhama K., Kumar Vinodh O.R., Singh R., Singh R.K. A rapid method of accurate detection and differentiation of Newcastle disease virus pathotypes by demonstrating multiple bands in degenerate primer based nested RT-PCR. J. Virol. Methods. 2015;212:47–52. doi: 10.1016/j.jviromet.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 10.OIE . Manual of Diagnostic Tests and Vaccines for Terrestrial Animals: Mammals, Birds and Bees. Volume 1. Biological Standards Commission; Paris, France: 2012. Newcastle disease; pp. 555–574. [Google Scholar]
- 11.Seal B.S., King D.J., Bennett J.D. Characterization of Newcastle disease virus isolates by reverse transcription PCR coupled to direct nucleotide sequencing and development of sequence database for pathotype prediction and molecular epidemiological analysis. J. Clin. Microbiol. 1995;33:2624–2630. doi: 10.1128/jcm.33.10.2624-2630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gohm D.S., Thuer B., Hofmann M.A. Detection of Newcastle disease virus in organs and faeces of experimentally infected chickens with RT-PCR. Avian Pathol. 2000;29:143–152. doi: 10.1080/03079450094171. [DOI] [PubMed] [Google Scholar]
- 13.Berinstein A., Sellers H.S., King D.J., Seal B.S. Use of a heteroduplex mobility assay to detect differences in the fusion protein cleavage site coding sequence among Newcastle disease virus isolates. J. Clin. Microbiol. 2001;39:3171–3178. doi: 10.1128/JCM.39.9.3171-3178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creelan J.L., Graham D.A., McCullough S.J. Detection and differentiation of pathogenicity of avian paramyxovirus serotype 1 from field cases with one-step reverse transcriptase-polymerase chain reaction. Avian Pathol. 2002;31:493–499. doi: 10.1080/0307945021000005860. [DOI] [PubMed] [Google Scholar]
- 15.Alves P.A., Oliveira E.G.D., Franco-Luiz A.P.M., Almeida L.T., Gonçalves A.B., Borges I.A., Rocha F.D.S., Rocha R.P., Bezerra M.F., Miranda P., et al. Optimization and clinical validation of colorimetric reverse transcription loop-mediated isothermal amplification, a fast, highly sensitive and specific COVID-19 molecular diagnostic tool that is robust to detect SARS-CoV-2 variants of concern. Front. Microbiol. 2021;18:713713. doi: 10.3389/fmicb.2021.713713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore K.J.M., Cahill J., Aidelberg G., Aronoff R., Bektaş A., Bezdan D., Butler D.J., Chittur S.V., Codyre M., Federici F., et al. Loop-mediated isothermal amplification detection of SARS-CoV-2 and myriad other applications. J. Biomol. Tech. 2021;32:228–275. doi: 10.7171/jbt.21-3203-017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roohani K., Tan S.W., Yeap S.K., Ideris A., Bejo M.H., Omar A.R. Characterisation of genotype VII Newcastle disease virus (NDV) isolated from NDV vaccinated chickens, and the efficacy of LaSota and recombinant genotype VII vaccines against challenge with velogenic NDV. J. Vet. Sci. 2015;16:447–457. doi: 10.4142/jvs.2015.16.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H., Wang J., Ge S., Lv Y., Li Y., Zheng D., Zhao Y., Castellan D., Wang Z. Molecular characterization of new emerging sub-genotype VIIh Newcastle disease viruses in China. Virus Genes. 2019;55:314–321. doi: 10.1007/s11262-019-01651-5. [DOI] [PubMed] [Google Scholar]
- 19.Welch C.N., Shittu I., Abolnik C., Solomon P., Dimitrov K.M., Taylor T.L., Williams Coplin D., Goraichuk I.V., Meseko C.A., Ibu J.O., et al. Genomic comparison of Newcastle disease viruses isolated in Nigeria between 2002 and 2015 reveals circulation of highly diverse genotypes and spillover into wild birds. Arch. Virol. 2019;164:2031–2047. doi: 10.1007/s00705-019-04288-9. [DOI] [PubMed] [Google Scholar]
- 20.Figueroa A., Escobedo E., Solis M., Rivera C., Ikelman A., Gallardo R.A. Outreach efforts to prevent Newcastle disease outbreaks in Southern California. Viruses. 2022;14:1509. doi: 10.3390/v14071509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song H., Bae Y., Park S., Kwon H., Lee H., Joh S. Loop-mediated isothermal amplification assay for detection of four immunosuppressive viruses in chicken. J. Virol. Methods. 2018;256:6–11. doi: 10.1016/j.jviromet.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Glickman R.L., Syddall R.J., Iorio R.M., Sheehan J.P., Bratt M.A. Quantitative basic residue requirements in the cleavage-activation site of the fusion glycoprotein as a determinant of virulence for Newcastle disease virus. J. Virol. 1988;62:354–356. doi: 10.1128/jvi.62.1.354-356.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H., Zhao Y., Zheng D., Lv Y., Zhang W., Xu T., Li J., Wang Z. Multiplex RT-PCR for rapid detection and differentiation of class I and class II Newcastle disease viruses. J. Virol. Methods. 2011;171:149–155. doi: 10.1016/j.jviromet.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 25.Pham H.M., Nakajima C., Ohashi K., Onuma M. Loop-mediated isothermal amplification for rapid detection of Newcastle disease virus. J. Clin. Microbiol. 2005;43:1646–1650. doi: 10.1128/JCM.43.4.1646-1650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q., Xue C., Qin J., Zhou Q., Chen F., Bi Y., Cao Y. An improved reverse transcription loop-mediated isothermal amplification assay for sensitive and specific detection of Newcastle disease virus. Arch. Virol. 2009;154:1433–1440. doi: 10.1007/s00705-009-0464-z. [DOI] [PubMed] [Google Scholar]
- 27.Kirunda H., Thekisoe O., Kasaija P., Kerfua S., Nasinyama G., OpudaAsibo J., Inoue N. Use of reverse transcriptase loop-mediated isothermal amplification assay for field detection of Newcastle disease virus using less invasive samples. Vet. World. 2012;5:206–212. doi: 10.5455/vetworld.2012.206-212. [DOI] [Google Scholar]
- 28.Saitou N., Nei M. The Neighbor-joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.