Abstract
Episodes of extraintestinal salmonellosis treated at a general hospital (1,522 beds) over a 6-year period (1991 to 1996) were characterized by the analysis of phenotypic and genotypic traits of Salmonella organisms and clinical data from medical reports. Extraintestinal salmonellosis accounted for 8% of all salmonellosis episodes. Fifty-two medical reports, dealing with 6 cases of typhoid fever, 32 cases of bacteremia, and 14 focal infections, were reviewed. All cases of typhoid fever except 1, 7 cases of bacteremia, and 5 focal infections were not related to any underlying disease or predisposing factors, while 25 cases of bacteremia and 9 focal infections were associated with some of these risk factors. All typhoid isolates and 65.4% of the nontyphoid isolates were susceptible to antimicrobials. Fifty-one nontyphoid strains were analyzed and assigned to 21 genomic groups, which were defined by serotype, combined ribotype, and combined randomly amplified polymorphic DNA type (each genomic group could include organisms differing in some phenotypic traits). The relationships between genomic groups and clinical presentations were traced. Organisms causing 22 episodes (17 episodes of bacteremia, 2 of pneumonia, 1 of peritonitis, 1 of pyelonephritis, and 1 of cystitis) belonged to a prevalent Salmonella enterica serotype Enteritidis genomic group, which included organisms assigned to four phage types, five biotypes, and four resistance patterns, causing infections in patients with and without risk factors. Seven other genomic groups, 4 Enteritidis groups (associated with both bacteremia and focal infections), 2 Typhimurium groups (one associated with bacteremia and the other with focal infections) and 1 Brandenburg group (associated with bacteremia) included two or more strains, and the remaining 13 genomic groups consisted of only one strain each.
Despite improvements in individual and collective sanitation as well as in the careful monitoring of food processing, both sporadic episodes and outbreaks of salmonellosis continue to occur with high frequency in industrial countries. Factors such as intensive pig and poultry production are contributing to the emergence of new clones of Salmonella pathogenic to humans. Others, such as the increase in international travel, the rapidly growing international food trade between countries with different levels of hygiene in the production and manufacture of foods and with different endemic pathogenic clones, are the cause of their dispersion among different geographic areas. On the other hand, the increase in patients with AIDS, other immunodeficiencies, and chronic disorders, are factors that contribute to putting new groups of people at increased risk of developing severe clinical forms of salmonellosis.
Salmonella is an enteroinvasive bacterium and causes infections that may have one of five different clinical presentations (4, 8). Gastroenteritis is the most common presentation in industrial countries and is considered as an emergent food-borne pathogen-caused disease. It is a self-limited illness of brief duration, usually characterized by diarrhea and fever, in which antibiotic treatment is rarely indicated for immunocompetent patients. Much less frequent but much more severe presentations, usually requiring antibiotic treatment, are bacteremia-septicemia without localized infection and focal infections. Focal infections may affect different sites in the body, causing different disorders, which frequently occur during or after Salmonella bacteremia but may also occur concomitantly with other syndromes. These three clinical presentations may be caused by many Salmonella serotypes considered zoonotic. Although any serotype can cause any of these presentations, certain serotypes have been associated with specific presentations, e.g., Salmonella enterica serotype Enteritidis and serotype Typhimurium are associated with gastroenteritis, while serotype Choleraesuis and serotype Dublin are associated with bacteremia (8, 23). Enteric fever, or typhoid fever, is due to serotype Typhi (which is adapted to human hosts; animals do not serve as a reservoir) but may also be caused by other serotypes (mainly Paratyphi A, B, and C), and antibiotic therapy clearly shortens the duration of the disease. Drinking water disinfection and other sanitation measures have lowered its frequency in industrial countries, but the frequency of this disease remains very high in developing countries (8). A fifth presentation is the chronic carrier state (enteric or urinary), defined as the excretion of salmonellas for months or years after the initial onset of disease, which is most frequent after Typhi infection.
The aim of this study was to carry out an evaluation of extraintestinal salmonellosis cases treated in the Hospital Central de Asturias (HCA), Oviedo, Spain, over a 6-year period (1991 to 1996). It was conducted by analyzing the phenotypic and genotypic traits of Salmonella organisms, clinical and other features of the patients, and the relationships between nontyphoid Salmonella genomic groups and clinical presentations.
MATERIALS AND METHODS
Samples processed and Salmonella characterization.
Blood and other clinical samples from patients with clinically suspected bacteremia and focal infection were processed according to standard techniques (2, 24). The microorganisms from positive cultures were isolated and characterized by standard laboratory methods. The biochemical profile (biotype) was ascertained by the PASCO system (Difco). Serotyping was performed with Bacto-Salmonella antisera for serotypes Enteritidis and Typhimurium (Difco). Assignment to other serotypes, as well as phage typing of Typhimurium and Enteritidis strains was carried out in the Centro Nacional de Microbiología (CNM), Majadahonda, Madrid, Spain, by using established phage-typing systems (1, 25).
Antimicrobial susceptibility test.
Antimicrobial susceptibility testing was performed according to the guidelines of the National Committee for Clinical Laboratory Standards for the microdilution susceptibility test (17a) by using the commercial antibiotic panel for Spain of the PASCO system (Difco), which includes ampicillin, ampicillin-sulbactam, aztreonam, cefazolin, cefotaxime, cefoxitin, ceftizoxime, ceftriaxone, cefuroxime, imipenem, piperacillin, ticarcillin, chloramphenicol, gentamicin, tobramycin, amikacin, ciprofloxacin, ofloxacin, nalidixic acid, fosfomycin, and trimethoprim-sulfamethoxazole. In addition, streptomycin and sulfadiazine were analyzed by the disk diffusion technique with commercial disks (Difco).
Genetic typing procedures.
Chromosomal DNA isolation and ribotyping were conducted as described in references 10, 11, and 17 by using a DNA fragment carrying the rrnB operon of Escherichia coli as a probe. Hybridization was performed by using the nonradioactive DNA Labelling and Detection Kit of Boehringer GmbH (Mannheim, Germany) according to the manufacturer’s instructions. The different patterns of bands containing all or part of the rRNA genes and flanking sequences (ribotypes) were designated with the first letter (or letters) of the serotype, followed by the first letter(s) of the restriction endonuclease(s) used (H for HincII and PS for the mixture of PstI and SphI) and a number assigned according to the order of detection in our laboratories.
Randomly amplified polymorphic DNA analysis, or RAPD typing, was carried out under the conditions previously described for each of the primers used, OPB-17 (5′AGGGAACGAG) (15) and Universal (5′TCACGATGCA) (27). Minor differences in band intensity, as well as the weak bands, were not considered to define RAPD types, which, in this work, were designated with the first letter of the serotype, followed by the first letter of the primer used (O for OPB-17 and U for Universal) and a number assigned according to the order of detection in our laboratories.
Typhimurium ATCC 14028 and Enteritidis ATCC 13076 were used as control strains in all ribotyping and RAPD typing assays.
Data from medical reports.
Data obtained from the medical reports of patients included age, sex, underlying diseases, portal of entry, predisposing factors, fever, diarrhea, and leukocytosis, or leukopenia, which were categorized according to standard criteria (9, 16). Only two patients were hospitalized simultaneously, but they were in different hospital buildings. During the period 1991 to 1996 there was no suspicion of Salmonella hospital outbreaks, and no investigation into common vehicles of infection or other possible linkages among the patients was recorded.
RESULTS
Microbiological data on extraintestinal salmonellosis.
During 1991 to 1996 in the HCA Microbiology Laboratories, a total of 970 Salmonella isolates were collected, 890 from feces (92%), 59 from blood (6%), and 21 from other clinical samples (2%). These isolates represented 57, 1.3, and 0.025% of bacteria collected from feces, blood, and other clinical samples, respectively. Eighty samples other than feces, from 68 patients, were registered as positive for Salmonella. All but one of the isolates belonged to subspecies 1 and were differentiated into three serogroups (B, C, and D) which included seven serotypes (Typhimurium, Brandenburg, Bredeney, and Derby in serogroup B, Hadar in serogroup C, and Enteritidis and Typhi in serogroup D). No Typhi or Derby organisms were available for further testing, but some data from these organisms were compiled from microbiological reports. Enteritidis and Typhimurium strains were the most frequent; they were isolated from different types of clinical samples and were associated with different clinical presentations (Table 1).
TABLE 1.
Microbiological traits of nontyphoid Salmonella causing extraintestinal infections
| GGa | Serotype | Ribotype
|
RAPD type
|
Phenotypic trait
|
Sample of origin | Clinical presentation (no. of episodes) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| HincII | PstI/SphI | OPB | Universal | Biotype | Phage typeb | R patternc | ||||
| I | Enteritidis | E-H1 | E-PS1 | E-O1 | E-U1 | BT1 | PT4 | Susceptible | Blood | Bacteremia (9) |
| Blood/feces | Bacteremia (1) | |||||||||
| BT1 | PT4 | Susceptible | Urine/blood | UTI (2)d | ||||||
| BT1 | PT4 | Susceptible | Peritoneal fluid/feces | Peritonitis (1) | ||||||
| BT1 | PT7 | Susceptible | Blood | Bacteremia (1) | ||||||
| BT1 | PT6 | Ap | Blood | Bacteremia (1) | ||||||
| BT1 | PT4 | C | Pleural fluid/blood | Pneumonia (1) | ||||||
| BT2 | PT4 | Susceptible | Blood | Bacteremia (2) | ||||||
| BT2 | PT6 | Ap | Blood | Bacteremia (1) | ||||||
| BT3 | PT6a | Ap | Pleural fluid | Pneumonia (1) | ||||||
| BT4 | PT6 | Ap | Blood | Bacteremia (1) | ||||||
| BT9 | PT4 | T/S | Blood | Bacteremia (1) | ||||||
| II | Enteritidis | E-H1 | E-PS1 | E-O6 | E-U1 | BT1 | PT6a | Ap | Blood | Bacteremia (1) |
| BT1 | PT6a | Ap | Urine | UTI (1) | ||||||
| BT3 | NC | Susceptible | Needle aspirate | Spondylodiskitis (1) | ||||||
| III | Enteritidis | E-H1 | E-PS2 | E-O1 | E-U1 | BT8 | PT6a | Ap, C | Wound exudate | Wound infection (1) |
| IV | Enteritidis | E-H1 | E-PS37 | E-O1 | E-U1 | BT1 | PT6 | Ap | Tracheal aspirate | Pneumonia (1) |
| V | Enteritidis | E-H1 | E-PS3 | E-O1 | E-U1 | BT2 | PT4 | Susceptible | Peritoneal fluid | Unknown (1)e |
| BT1 | PT1 | C | Blood | Bacteremia (1) | ||||||
| VI | Enteritidis | E-H1 | E-PS38 | E-O1 | E-U1 | BT5 | PT4 | Susceptible | Blood | Bacteremia (1) |
| VII | Enteritidis | E-H1 | E-PS5 | E-O1 | E-U1 | BT1 | PT4 | Susceptible | Blood | Bacteremia (1) |
| VIII | Enteritidis | E-H1 | E-PS10 | E-O1 | E-U1 | BT1 | NT | Susceptible | Blood/abscess | Unknown (1)e |
| IX | Enteritidis | E-H1 | E-PS14 | E-O1 | E-U1 | BT1 | PT4 | Susceptible | Blood | Bacteremia (1) |
| BT1 | PT4 | Susceptible | Urine | UTI (1) | ||||||
| BT10 | PT4 | Susceptible | Blood/feces | Bacteremia (1) | ||||||
| X | Enteritidis | E-H1 | E-PS39 | E-O1 | E-U1 | BT1 | PT1 | Susceptible | Blood | Bacteremia (1) |
| XI | Enteritidis | E-H2 | E-PS1 | E-O1 | E-U1 | BT7 | PT4 | Susceptible | Blood | Bacteremia (1) |
| XII | Enteritidis | E-H1 | E-PS2 | E-O6 | E-U1 | BT1 | PT6a | Ap | Blood | Bacteremia (1) |
| BT6 | PT6a | Ap | Cerebrospinal fluid | Meningitis (1) | ||||||
| XIII | Typhimurium | T-H5 | T-PS2 | T-O1 | T-U1 | BT11 | NT | Susceptible | Blood | Bacteremia (1) |
| BT13 | DT23 | Susceptible | Blood | Bacteremia (1) | ||||||
| XIV | Typhimurium | T-H5 | T-PS1 | T-O1 | T-U1 | BT1 | DT193 | T/S | Urine/feces | UTI (1) |
| XV | Typhimurium | T-H1 | T-PS1 | T-O1 | T-U2 | BT1 | NT | Ap, T/S, C, S | Sputum | Pneumonia (1) |
| BT10 | NT | Ap, C, S | Rectal abscess | Perirectal abscess (1) | ||||||
| XVI | Typhimurium | T-H4 | T-PS1 | T-O1 | T-U1 | BT1 | NT | T/S | Blood | Bacteremia (1) |
| XVII | Typhimurium | T-H5 | T-PS2 | T-O1 | T-U1 | BT12 | NT | Susceptible | Blood | Bacteremia (1) |
| XVIII | Brandenburg | B-H1 | B-PS1 | B-O1 | B-U1 | BT1 | Susceptible | Blood/feces | Bacteremia (1) | |
| BT10 | Susceptible | Blood | Bacteremia (1) | |||||||
| XIX | Hadar | H-H1 | H-PS1 | H-O1 | H-U1 | BT4 | Ap, CFC, S | Blood | Bacteremia (1) | |
| XX | Bredeney | Br-H1 | Br-PS1 | Br-O1 | Br-U1 | BT16 | Susceptible | Blood | Bacteremia (1) | |
| XXI | Subspecies III | S-H1 | S-PS1 | S-O1 | S-U1 | BT15 | Susceptible | Pus | Cholecystitis (1) | |
GG, genomic group defined by biotype, serotypes, and RAPD types.
NC, not conforming; NT, nontypeable.
S, streptomycin; Ap, ampicillin; C, chloramphenicol; T/S, cotrimoxazole; CFC, cefonicid.
UTI, urinary tract infection. One case was pyelonephritis.
Medical report not available.
The nontyphoid isolates were characterized by different phenotypic and genotypic procedures. Isolates from a single patient collected from different samples and showing identical traits were assigned to the same strain. In total, 51 strains were differentiated (Table 1). The phenotypic traits analyzed were the biochemical profile (biotype) and resistance to antimicrobial agents (R pattern) (for all isolates) and the phage type (for Typhimurium and Enteritidis only). The most frequent biotype was BT1 (59%), characterized by fermenting glucose, mannose, saccharose, arabinose, trehalose, sorbitol, melibiose, and rhamnose, producing decarboxylation of lysine and ornithine, using citrate as a carbon source, and growing in the presence of penicillin. The second most frequent was BT2 (8%), differing from BT1 in that it did not ferment saccharose. All Typhi strains had been assigned to BT14, characterized by low biochemical activity (not fermenting arabinose and rhamnose, not producing ornithine decarboxylase, and not using citrate or growing in penicillin).
Thirty-three nontyphoid strains (65%) were susceptible to all antimicrobials tested; the remaining strains were grouped into seven R patterns. The three most frequent R patterns consisted of resistance to only one drug, ampicillin (18%), cotrimoxazole (6%), or chloramphenicol (4%), and only four strains (8%) showed resistance to two or more drugs (for this purpose the trimethoprim-sulfamethoxazole combination was considered a single drug, named cotrimoxazole). The percentage of resistant isolates varied among serotypes; while 57% of Typhimurium isolates were resistant to some antimicrobial, only 33% of Enteritidis isolates had this characteristic. The most frequent resistance was to ampicillin (25.5%), followed by resistance to chloramphenicol (10%) and cotrimoxazole (8%), which are antibiotics used in the control of extraintestinal salmonellosis. All strains were susceptible to quinolones and expanded-spectrum cephalosporins. The six Typhi strains had been reported to be susceptible to all antimicrobials.
Phage typing of Enteritidis revealed the existence of a prevalent phage type, PT4 (61.5%); the second and third most frequent phage types were PT6a (15.4%) and PT6 (10.2%), respectively. All PT6 and PT6a strains were resistant to ampicillin (one was also resistant to chloramphenicol), while all PT4 strains but one were susceptible to all drugs tested. The Typhimurium organisms were assigned to two phage types, DT23 and DT193, but five strains were nontypeable (71.5%). No association between phage type and drug resistance was observed.
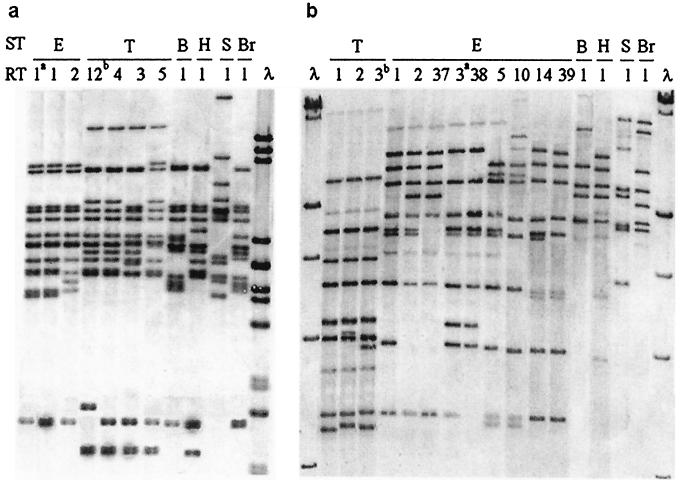
By ribotyping performed with HincII, the nontyphoid isolates were grouped into 9 H ribotypes, and by ribotyping with the PstI-SphI mixture, they were grouped into 15 PS ribotypes (Fig. 1). It is significant that no ribotype included strains of different serotypes and that, while isolates of Enteritidis and Typhimurium were differentiated by both ribotyping procedures, different numbers of types and different distributions of strains into types were found with the two procedures (Table 1). Enteritidis isolates were differentiated into two H ribotypes and nine PS ribotypes (ATCC 13076 showed E-H1 and E-PS3 ribotypes), while Typhimurium strains were differentiated into three H ribotypes and two PS ribotypes (ATCC 14028 showed T-H12 and T-PS3 ribotypes).
FIG. 1.
Ribotypes found in Salmonella strains causing extraintestinal diseases. ST, serotype; E, Enteritidis; T, Typhimurium; B, Brandenburg; H, Hadar; S, Subspecies III; Br, Bredeney. RT, ribotype; the arabic numeral corresponds to the order assigned to the ribotype in our laboratories. a, ATCC 13076; b, ATCC 14028. (a) H ribotypes generated by HincII. λ, lambda DNA cleaved with PstI; sizes of fragments (in kilobases) from top to bottom are 5.08, 4.65, 4.5, 2.84, 2.58, 2.14, 1.98, 1.2, 1.1, 0.8, 0.5, and 0.2. (b) PS ribotypes generated by a mixture of PstI and SphI. λ, lambda DNA cleaved with HindIII; sizes of fragments (in kilobases) from top to bottom are 27.5, 23.1, 9.4, 6.55, 4.36, and 2.32.
By RAPD typing using the primers OPB-17 and Universal, the series was differentiated into seven RAPD types with each primer, and strains of different serotypes presented different RAPD types with both primers (Fig. 2). Enteritidis strains were differentiated into two RAPD types with OPB-17 but showed a single type with Universal (ATCC 13076 exhibited types E-O2 and E-U1), while Typhimurium strains showed a single type with OPB-17 and two types with Universal (ATCC 14028 showed types T-O1 and T-U1).
FIG. 2.
RAPD types of Salmonella strains causing extraintestinal diseases, obtained by using the primers labelled OPB-17 and Universal. ST, serotype; E, Enteritidis; T, Typhimurium; B, Brandenburg; H, Hadar; S, Subspecies III; Br, Bredeney. AT, amplified type; the arabic numeral corresponds to the order assigned to the RAPD type in our laboratories. a, ATCC 13076; b, ATCC 14028. λ, lambda DNA cleaved with PstI; sizes of fragments (in kilobases) from top to bottom are 5.08, 2.8, 2.1, 1.7, 1.15, 1.1, 0.8, 0.5, and 0.4.
In this work it was considered appropriate to use the data from serotyping and typing methods analyzing DNA traits (ribotyping and RAPD typing) to define genomic groups, and then to incorporate phenotypic traits (biotype, phage type, and R pattern) in order to differentiate clones. According to these criteria, nontyphoid Salmonella isolates were assigned to 21 genomic groups, of which 12 included only Enteritidis strains and 5 included only Typhimurium strains (Table 1). The presence of a prevalent genomic group (genomic group I [GG I], characterized by serotype Enteritidis, ribotypes E-H1 and E-PS1, and RAPD types E-O1 and E-U1) which included nine different clones can be emphasized. The most frequent clone (BT1, PT4, drug susceptible) included organisms to which 13 apparently unrelated episodes (given that the patients affected were hospitalized at different times and in some cases were in hospital wards situated in different buildings and were attended by different personnel) were attributed.
Patient data associated with extraintestinal salmonellosis.
The medical and microbiological reports of only 52 patients with extraintestinal salmonellosis were available for review; it was found that 6 episodes had been diagnosed as typhoid fever, 32 as bacteremia, and 14 as focal infections (5 in the urinary tract, four pleuropulmonary, three abdominal, one in the central nervous system, and one osteoarticular). The most relevant epidemiological and clinical features of the three groups of clinical presentations are summarized in Table 2, and some of these features are discussed here. Typhoid fever and urinary-tract infections were more frequent in females than in males (2:1 and 4:1, respectively), whereas bacteremia and nonurinary focal infections were more frequent in males than in females (2.2:1 and 3:1, respectively). In relation to age groups, all but one of the patients with typhoid fever were under 31 years of age without predisposing conditions. Two of the episodes in the pediatric age group were diagnosed as bacteremia, affecting an 8-month-old boy and an 11-month-old girl, both of whom had no underlying disease but had presented with diarrhea previously. Thirteen bacteremia episodes and one urinary-tract infection affected young adults (20 to 45 years of age), of whom seven had AIDS, one had neoplasia, one had diabetes, one had undergone gastrectomy, and the remaining four had no predisposing conditions but had suffered diarrhea previously. Fifteen cases of bacteremia (46.8%) and 11 focal infections (78.6%) corresponded to the geriatric group (over 61), of whom eight had diabetes.
TABLE 2.
Clinical data from patients with extraintestinal salmonellosis
| Parameter | No. of patients with:
|
||
|---|---|---|---|
| Typhoid fever (n = 6) | Bacteremiaa (n = 32) | Focal infectiona (n = 14) | |
| Sex | |||
| Male | 2 | 22 (9) | 7 (2) |
| Female | 4 | 10 (1) | 7 |
| Age group | |||
| Pediatric (<14 yrs) | 2 | 2 | |
| Adult (15–60 yrs) | 3 | 16 (5) | 2 |
| Geriatric (>61 yrs) | 1 | 14 (5) | 12 (2) |
| Portal of entry | |||
| Unknown | 3 | 14 | 10 |
| Gastrointestinal tract | 3 | 18 | 4 |
| Predisposing factors | |||
| Absent | 5 | 9 | 5 |
| Present | 1 | 23 | 9 |
| Diabetes mellitus | 10 (3) | 5 (2) | |
| Gastrectomy | 4 (2) | ||
| AIDS | 7 (3) | 1 | |
| Corticosteroid therapy | 3 (1) | 1 (1) | |
| Other | 1b | 2c (1) | 2d |
| Clinical data | |||
| Fever (>38°C) | 6 | 27 | 13 |
| Diarrhea | 3 | 17 | 4 |
| Leukocytosis (>10,000/mm3) | 1 | 14 | 8 |
| Leukopenia (<4,500/mm3) | 5 | 6 | 2 |
Number of deaths in parentheses.
Hiatal hernia.
One patient with alcoholism and dementia and another with hiatal hernia.
One colelithiasis and another alcoholism.
Regarding clinical presentation and prognosis, bacteremia and focal infections were more frequent among patients with some underlying disease (78.2 and 64.3%, respectively). However, five of six (83.3%) cases of typhoid fever occurred in patients without underlying disease, and the sixth patient had a nonfatal disease (hiatal hernia). The spectrum of underlying diseases covered a wide range of disorders, but only one was categorized as rapidly fatal: acute leukemia associated with bacteremia and previous gastroenteritis. Twelve patients with bacteremia, two patients with pulmonary infections, and one patient with a urinary infection had ultimately fatal underlying diseases. Of these, seven bacteremia patients and the two patients with pulmonary infections died. The remaining 18 episodes were categorized as having nonfatal prognoses, but three of these patients died. These were three males over the age of 65, of whom two had diabetes and one had undergone gastrectomy. No underlying disease was recorded for seven patients with bacteremia and five with focal infections; none of these patients died.
DISCUSSION
HCA is a general hospital incorporating the former Hospital Covadonga of Oviedo and Hospital General of Asturias, with 1,522 beds and 80% bed occupancy; it serves a population of about 200,000 inhabitants and is the reference center for other hospitals in the Principality of Asturias. In the HCA the overall incidence of bloodstream invasion by nontyphoid salmonellas over the period 1991 to 1996 was 7.63%. The percent Salmonella-positive hemocultures was 1.3%, lower than the percentage reported in the previous decade (24) despite the increase in patients with AIDS and other immunodeficiencies. This is similar to the percentage reported by the Spanish Bacteremia Group (1.58%), which compiled data from six Spanish hospitals (9).
From the present study, three microbiological findings should be discussed. Firstly, in the HCA, over the period 1991 to 1996, the serotypes most frequently recorded as causing human salmonellosis were Enteritidis, Typhimurium, Virchow, and Hadar, with 534, 254, 28, and 8 isolates, respectively; no isolate was identified as Choleraesuis or Dublin. Enteritidis and Typhimurium isolates were also the most frequent causes of extraintestinal salmonellosis treated in the HCA, whereas no episodes attributable to Virchow and only one attributable to Hadar were recorded. These data are in line with those recorded in other Spanish hospitals (3, 18) but differ from those of studies in other countries, in which Choleraesuis (21), Dublin (14), and Virchow (26) were reported as the most frequent causes of bacteremia. A review of Salmonella bacteremia in England and Wales (23) reported that less than 2% of nontyphoid salmonellas isolated from humans were from blood culture and emphasized that while the greatest number of bloodstream isolates were Enteritidis and Typhimurium, the highest incidence of septicemias was attributable to Choleraesuis, Dublin, and Virchow. Secondly, the most frequent genomic group (Enteritidis GG I) causing extraintestinal infections was also the most frequent group associated with intestinal infections. It could be considered endemic in Spain because it has been circulating over the last decade, mainly associated with poultry and hen eggs as the infection vehicles (references 11 and 12 and unpublished data). Thirdly, 35% of the Salmonella isolates were resistant to one (ampicillin, cotrimoxazole, or chloramphenicol) or more (ampicillin and chloramphenicol; ampicillin, chloramphenicol, and cotrimoxazole; or ampicillin and cefonicid) antimicrobial drugs used in salmonellosis control; the rate of resistance to these was similar to the rate reported in other Spanish series (6, 19) but higher than that reported in the United States (13). Not all serotypes had similar frequencies of resistance, and the high percentage of multidrug-resistant Typhimurium isolates is in line with data from other works (5). Another interesting finding is the association of Enteritidis PT6 and PT6a with resistance to ampicillin (78% of the strains tested in our laboratories). These strains carried a 25-MDa plasmid and fell into different genomic groups, while no PT4 strain showed this resistance or carried 25-MDa plasmids.
Regarding the features of the patients with Salmonella bacteremia, the following observations could be noteworthy. (i) Bacteremia was recorded for all age groups but did not show the bimodal distribution, with a higher incidence at the two extremes, which is usually reported (3, 20), although the geriatric group showed the highest frequency (14 patients, most of whom had predisposing factors). The strains collected from geriatric patients belonged to seven genomic groups; one, GG I (Enteritidis), was implicated in eight episodes, and each of the other six groups was implicated in one episode. (ii) Six of the eight patients between 21 and 31 years of age had AIDS. Only salmonellas from four AIDS patients could be characterized, and these were assigned to three genomic groups: two Enteritidis groups (GG I and GG X) and one Typhimurium group (GG XIII). It has been reported that bacteremia is about 100 times more frequent in AIDS patients than in the general population (22). (iii) The most frequent bacterial portal of entry was the gastrointestinal tract (56% of the patients), and presumably most of the remaining patients (44%) could also have been infected by this route. It is also noteworthy that a high percentage of patients (72%) had predisposing factors that could have favored the development of blood infection; other authors report lower percentages, ranging from 27 to 62% (14, 18, 26).
With respect to focal infections, we found that those affecting the urinary tract were the most frequent, including five episodes caused by strains assigned to four genomic groups, three Enteritidis groups (GG I, II, and IX) and one Typhimurium group (GG XIV). The single pyelonephritis episode was caused by one Enteritidis GG I strain in an AIDS patient (a 27-year-old male) with previous and recurrent Salmonella bacteremia. In other reports (7), the most frequent Salmonella focal infections were osteoarticular, while in our series only one patient (a 63-year-old male with diabetes) had a lumbar spondylodiskitis caused by an Enteritidis GG II strain. The four patients with pulmonary infection had illnesses predisposing them to infection; two of these had malignancies. Symptoms and signs were typical of patients presenting with acute bacterial pneumonia, and two patients had Salmonella-positive blood cultures, which supported hematogenous dissemination; these data are in line with the review described in reference 4. Three episodes were caused by Enteritidis (GG I and GG IV), and the fourth was caused by a Typhimurium GG XV strain resistant to ampicillin, cotrimoxazole, chloramphenicol, and streptomycin.
Salmonella meningitis is frequently reported in children, while it is seldom described in adults and AIDS patients. The only episode in our series affected a 65-year-old female without predisposing illness and without a previous episode of gastroenteritis; it was caused by an Enteritidis GG XII strain. Finally, the only three episodes classified as intra-abdominal infections (one case of peritonitis, one of perirectal abscesses, and one of cholecystitis) were caused by three very different strains (Enteritidis GG I, Typhimurium GG XV, and Subspecies IIIb GG XXI, respectively). Infections directly related to the digestive tract had a pathologic significance different from that of the other focal infections by Salmonella and did not indicate severe immunosuppression.
In conclusion, this study revealed that extraintestinal salmonellosis more frequently affected patients with some predisposing factor and/or underlying disease than patients without these features (2:1 ratio). Furthermore, it was caused by genotypically and phenotypically heterogeneous Salmonella organisms, although many of these (43.1%) belong to a prevalent genomic group of Enteritidis. Organisms assigned to this group have also been found frequently in human feces, food samples, and water samples collected over the last decade in different Spanish regions and are considered prevalent and endemic in Spain (11, 12).
ACKNOWLEDGMENTS
We thank M. A. Usera and CNM for the phage typing of the Enteritidis and Typhimurium strains and F. Pérez for the Salmonella isolates.
This work was supported by grants from the Fondo de Investigación Sanitaria (Ref. 95/0030-01 and 98/0296).
REFERENCES
- 1.Anderson E S, Ward L R, de Saxe M J, de Sa J D H. Bacteriophage-typing designations of Salmonella typhimurium. J Hyg. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. [Google Scholar]
- 3.Bassa A, Parras F, Reina J, Villar E, Gil J, Alomar P. Non-Typhi Salmonella bacteraemia. Infection. 1989;17:290–293. doi: 10.1007/BF01650710. [DOI] [PubMed] [Google Scholar]
- 4.Cohen J I, Bartlett J A, Ralph Corey G. Extra-intestinal manifestations of Salmonella infections. Medicine. 1987;66:349–387. doi: 10.1097/00005792-198709000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Cohen M, Tauxe R. Drug-resistant Salmonella in the United States: an epidemiologic perspective. Science. 1986;234:964–969. doi: 10.1126/science.3535069. [DOI] [PubMed] [Google Scholar]
- 6.Galán J C, Varea M, Castillo F J, Clavel A, Gómez-Lus R. Resistencia antibiótica en Salmonella entérica: un problema en aumento. Enferm Infecc Microbiol Clin. 1996;14:528–532. [PubMed] [Google Scholar]
- 7.García-Rodríguez J A, García-Sánchez J E, Muñoz-Bellido J L, García-García M I. Salmonellosis focal en España. Analisis de 14 casos y revisión de la literatura. Enferm Infecc Microbiol Clin. 1990;8:134–142. [PubMed] [Google Scholar]
- 8.Goldberg M B, Rubin R H. The spectrum of Salmonella infection. Infect Dis Clin N Am. 1988;2:571–598. [PubMed] [Google Scholar]
- 9.Grupo de estudio de la bacteriemia. Bacteriemia en seis hospitales españoles. Med Clín. 1986;86:221–232. [PubMed] [Google Scholar]
- 10.Guerra B, Landeras E, Gonzalez-Hevia M A, Mendoza M C. A three-way ribotyping scheme for Salmonella serotype Typhimurium and its usefulness for phylogenetic and epidemiological purposes. J Med Microbiol. 1997;46:307–313. doi: 10.1099/00222615-46-4-307. [DOI] [PubMed] [Google Scholar]
- 11.Landeras E, Mendoza M C. Evaluation of PCR-based typing methods and ribotyping performed with a mixture of PstI and SphI to differentiate strains of Salmonella enterica serotype Enteritidis. J Med Microbiol. 1998;47:427–434. doi: 10.1099/00222615-47-5-427. [DOI] [PubMed] [Google Scholar]
- 12.Landeras E, Gonzalez-Hevia M A, Mendoza M C. Molecular epidemiology of Salmonella serotype Enteritidis. 1998. Relationships between food, environmental and clinical strains. Int. J. Food Microbiol., in press. [DOI] [PubMed] [Google Scholar]
- 13.Lee L A, Puhr N D, Maloney E K, Bean N H, Tauxe R V. Increase in antimicrobial resistant Salmonella infections in the United States, 1989–90. J Infect Dis. 1994;170:128–134. doi: 10.1093/infdis/170.1.128. [DOI] [PubMed] [Google Scholar]
- 14.Lester A, Eriksen N H, Nielsen P B, Friis-Moller A, Bruun B, Scheibel J, Gaarslev K, Kolmos H J. Non-typhoid Salmonella bacteraemia in Greater Copenhagen 1984 to 1988. Eur J Clin Microbiol Infect Dis. 1991;10:486–490. doi: 10.1007/BF01963934. [DOI] [PubMed] [Google Scholar]
- 15.Lin A W, Usera M A, Barrett T J, Goldsby R A. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis. J Clin Microbiol. 1996;34:870–876. doi: 10.1128/jcm.34.4.870-876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCabe W R, Jackson G G. Gram negative bacteremia. I. Etiology and ecology. Arch Intern Med. 1962;110:847–855. [Google Scholar]
- 17.Mendoza M C, Alzugaray R, Landeras E, González-Hevia M A. Discriminatory power and application of ribotyping of Yersinia enterocolitica O:3 in an epidemiological study. Eur J Clin Microbiol Infect Dis. 1996;15:220–226. doi: 10.1007/BF01591358. [DOI] [PubMed] [Google Scholar]
- 17a.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 18.Ramos J M, García-Corbeira P, Aguado J M, Arjona R, Alés J M, Soriano F. Clinical significance of primary vs. secondary bacteremia due to nontyphoid Salmonella in patients without AIDS. Clin Infect Dis. 1994;19:777–780. doi: 10.1093/clinids/19.4.777. [DOI] [PubMed] [Google Scholar]
- 19.Reina J, Gómez J, Serra A, Borrell N. Analysis of the antibiotic resistance detected in 2,043 strains of S. enterica isolates in stool cultures of Spanish patients with acute diarrhoea (1981–1991) J Antimicrob Chemother. 1993;32:765–769. doi: 10.1093/jac/32.5.765. [DOI] [PubMed] [Google Scholar]
- 20.Sai-Cheong L, Pun-Hung Y, Wen-Ben S, Lassarre R. Bacteremia due to non-typhi Salmonella: analysis of 64 cases and review. Clin Infect Dis. 1994;19:693–696. doi: 10.1093/clinids/19.4.693. [DOI] [PubMed] [Google Scholar]
- 21.Saphra I, Winter J W. Clinical manifestations of salmonellosis in man. An evaluation of 7,779 human infections identified at the New York Salmonella Center. N Engl J Med. 1957;256:1128–1134. doi: 10.1056/NEJM195706132562402. [DOI] [PubMed] [Google Scholar]
- 22.Sperber S J, Schleupner C J. Salmonellosis during infection with human immunodeficiency virus. Rev Infect Dis. 1987;9:925–933. doi: 10.1093/clinids/9.5.925. [DOI] [PubMed] [Google Scholar]
- 23.Threlfall E J, Hall M L M, Rowe B. Salmonella bacteraemia in England and Wales, 1981–1990. J Clin Pathol. 1992;45:34–36. doi: 10.1136/jcp.45.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vazquez F, Mendoza M C, Villar M H, Pérez F, Méndez F J. Survey of bacteraemia in a Spanish hospital over a decade (1981–1990) J Hosp Infect. 1994;26:111–121. doi: 10.1016/0195-6701(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 25.Ward L R, de Sa J D H, Rowe B. A phage typing scheme for Salmonella enteritidis. Epidemiol Infect. 1987;99:291–294. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkins E G L, Roberts C. Extraintestinal salmonellosis. Epidemiol Infect. 1988;100:361–368. doi: 10.1017/s095026880006711x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]