Abstract
A series of 74 Yersinia enterocolitica clinical strains collected in a Spanish region and 10 reference strains, assigned to nine serotypes and five biotypes, were analyzed by ribotyping procedures. Riboprobing, performed separately with HindIII and BglI and using an rrn operon as the probe, generated 13 and 11 ribotypes (discrimination index [DI] = 0.56 and 0.55), respectively. PCR ribotyping, performed with primers complementary to conserved regions of 16S and 23S rRNA genes, generated 13 ribotypes (DI = 0.56). A combination of data from the three procedures allowed for further discrimination into 17 combined ribotypes (DI = 0.83). The dendrogram obtained by cluster analysis of data from riboprobing indicated a high heterogeneity of the ribosomal DNA regions of the strains under study (similarities between 10 and 92%), which were grouped into three clusters at a similarity level of 0.32. The major cluster included 10 branches, and 7 of these formed a subcluster (similarity coefficient, >83%) represented by strains of serotype O:3 and biotype 2, 3, or 4. The second cluster included four branches, represented by strains belonging to seven non-O:3 serotypes, biotypes 1A and 2, and two of these branches included pyrazinamidase-positive as well as pyrazinamidase-negative strains. The remaining three branches, represented by O:3–biotype 4 strains, formed a third cluster weakly related to the others. Data from this study showed that Y. enterocolitica O:3 organisms assigned to a prevalent and endemic lineage and non-O:3 organisms assigned to three other less-frequent lineages are circulating and causing human disease in the Spanish region under study.
Among the enteroinvasive bacteria causing human infections is Yersinia enterocolitica, in which virulence results from a complex interplay between a series of chromosomal and plasmid-borne genes. It is associated mainly with enterocolitis and, less frequently, with a wide variety of clinical and immunologic manifestations (4, 6, 14). Although human carriers are rare, animal carriers are frequent. Humans are infected primarily by the ingestion of fecally contaminated food or water, and over the past two decades an increase in cases of human infection with Y. enterocolitica has been reported in many developed countries (mainly in colder climates) (1, 2, 4, 8, 9, 14). For epidemiological purposes intraspecies differentiation is usually carried out by serotyping and biotyping, which have revealed that only a few clones can be considered primary pathogens (2, 4, 8, 16). They also enable the placement of virulent clones into two extensive groups, the American group (biotype 1B, mainly including serotypes O:8, O:13a, O:13b, O:20, and O:21) and the European group (biotypes 2 to 5, mainly including serotypes O:3, O:5,27, and O:9). However, over the last decade serotype O:3 has emerged as the most frequent in some U.S. countries and states (2, 3). On the other hand, biotype 1A, despite being considered nonpathogenic, has constituted a sizable fraction of strains from patients with enterocolitis in some studies (6, 14), and recently, several other biotypes and serotypes traditionally considered nonpathogenic, which were negative for virulence markers, have been associated with human disease in the Republic of Georgia (15). Both the wide dispersion of the two groups of virulent clones mentioned above and the emergence of clones initially defined as nonvirulent support the use of new epidemiological markers, preferably those that involve direct DNA analysis of the chromosome. A good genetic marker must reveal a high degree of polymorphism, which can be interpreted in terms of genetic variation at specific chromosomal loci. Variation that is selectively neutral and irrelevant to disease processes is best for assessing the genetic relatedness of isolates (7, 17).
The purpose of this study was to explore genetic diversity among contemporary Y. enterocolitica organisms, collected in four hospitals of a Spanish region (the Principality of Asturias), by procedures that analyze the sequence divergence of the rrn loci, which encode the rRNAs. The rRNA genes are organized as polycistronic transcriptional units called rrn operons (5′–promoter–16S–spacer region–23S–5S–3′) and, together with their flanking sequences, form the ribosomal DNA (rDNA) regions. While the rRNA genes are highly conserved, the flanking sequences and the intergenic spacer region (SR) between the 16S and 23S rRNA genes can show a significant degree of variation in length and sequence within a single species, and the differences can be used to discriminate clones and clonal lineages (3, 5, 11–13). The procedures used in this work were (i) analysis of restriction fragment length polymorphisms of rDNA regions generated with two selected restriction endonucleases, HindIII and BglI, and Southern hybridization with an rrn probe and (ii) amplification by PCR and analysis of the SR by using oligonucleotide primers complementary to conserved sequences of the 16S and 23S rRNA genes. The former procedure is usually called ribotyping or riboprobing, and the banding patterns are called ribotypes. The latter procedure is termed PCR ribotyping, and the patterns of amplified fragments are called SR ribotypes. Results from riboprobing and PCR ribotyping were correlated, combined, and used to define combined ribotypes (CRTs), as well as to trace the relationships between ribotypes and serotypes and those between ribotypes and biotypes. In addition, the data were used to ascertain the diversity of types and clonal lineages of Y. enterocolitica causing human disease that have been circulating in the region under study.
MATERIALS AND METHODS
Bacterial strains.
This study included 84 strains of Y. enterocolitica (see Table 1). Sixty-six strains were collected from diarrheic human feces from patients attending four Asturian hospitals over the period 1993 through 1995 and were implicated in 66 apparently sporadic episodes of enterocolitis. In fact, they were not associated with outbreaks occurring at different times in different areas and places, and investigation into possible vehicles of infection was not carried out. They represented about 34% of yersiniosis episodes microbiologically diagnosed in these hospitals during this 3-year period. Eight other strains were clinical Asturian strains collected before or during 1992, representing eight different clonal lines previously defined by ribotyping with five enzymes (12). The remaining 10 strains were from other collections, representing pathogenic and nonpathogenic biogroups, and were used as reference strains.
TABLE 1.
Relationships among ribotypes, serotypes, and biotypes of Y. enterocolitica strains
| Ribotype with:
|
CRT | Serotype | Biotype | No. of clinical strains | Yr(s) of isolation | Reference strain designationa | ||
|---|---|---|---|---|---|---|---|---|
| HindIII | BglI | PCR | ||||||
| H1 | B1 | SR1 | I-1 | O:3 | BT4 | 30b | 1984–1995 | CECT 500 |
| O:3 | BT2 | 3 | 1995 | |||||
| O:3 | BT3 | 1 | 1995 | |||||
| SR2 | I-2 | O:3 | BT4 | 5 | 1994–1995 | |||
| O:3 | BT2 | 2 | 1995 | |||||
| O:3 | BT3 | 1 | 1995 | |||||
| SR3 | I-3 | O:3 | BT4 | 2 | 1994–1995 | |||
| O:3 | BT3 | 1 | 1994 | |||||
| SR4 | I-4 | O:3 | BT4 | 1 | 1995 | |||
| SR5 | I-5 | O:3 | BT4 | 1 | 1995 | |||
| SR6 | I-6 | O:3 | BT3 | 1 | 1993 | |||
| H1 | B4 | SR1 | II-1 | O:3 | BT4 | 1b | 1985 | CECT 4055 |
| SR2 | II-2 | O:3 | BT4 | 1 | 1995 | |||
| H1 | B6 | SR1 | III | O:3 | BT4 | 2 | 1995 | |
| O:3 | BT2 | 1 | 1995 | |||||
| H2 | B2 | SR1 | IV | O:3 | BT4 | 1b | 1984 | |
| H3 | B3 | SR7 | V | O:3 | BT4 | 1b | 1984 | |
| H4 | B5 | SR1 | VI | O:3 | BT4 | 1b | 1986 | |
| H5 | B1 | SR1 | VII | O:3 | BT4 | 1b | 1986 | |
| H6 | B1 | SR1 | VIII | O:3 | BT4 | 5b | 1992–1995 | |
| H6 | B4 | SR1 | IX | O:3 | BT4 | 1b | 1992 | |
| H7 | B4 | SR1 | X | O:3 | BT4 | 1 | 1994 | |
| H8 | B1 | SR1 | XI | O:3 | BT4 | 2 | 1995 | |
| H21 | B21 | SR21 | XII-21 | NA | BT1A | 1 | 1993 | |
| O:1,2 | BT2 | 1 | 1993 | |||||
| SR22 | XII-22 | NA | BT1A | 2 | 1993 | |||
| NA | BT2 | 2 | 1993 | |||||
| SR26 | XII-26 | O:5 | BT1A | CLSP:A/95 | ||||
| H21 | B25 | SR24 | XIII-24 | O:9 | BT2 | 1 | 1994 | 2-69 Hellsinkyc |
| SR25 | XIII-25 | O:8 | BT2 | ATCC 27729 | ||||
| SR26 | XIII-26 | O:21 | BT1Ax | Wc188 New Yorkc | ||||
| O:4,32 | BT1Ax | 6249 New Yorkc | ||||||
| H22 | B22 | SR23 | XIV | NA | BT2 | 1 | 1993 | |
| H23 | B23 | SR1 | XV | O:1,2a,3 | BT3 | CECT 559 | ||
| H24 | B24 | SR1 | XVI | O:8 | BT2 | 1 | 1994 | CECT 4054 |
| H25 | B25 | SR25 | XVII | O:13,7 | BT1Ax | 81-Y Barcelonac | ||
CECT, Colección Española de Cultivos Tipo; CLSP, Colección del Laboratorio de Salud Pública del Principado de Asturias.
Includes a strain collected before 1993 in a hospital of the Principality of Asturias.
Reference strains kindly provided by R. Diaz, Hospital Universitario de Navarra, Pamplona, Spain.
Serotyping and biotyping.
Serotyping was performed on the clinical isolates by the slide agglutination test with commercial antisera for serotypes O:3, O:5, O:8, O:9, and O:1,2 (Denka Seiken Co., Ltd., Tokyo, Japan). Biotyping was performed according to the scheme of Wauters et al. (16).
Chromosomal DNA isolation and ribotyping procedures.
Chromosomal DNA isolation and riboprobing using the restriction endonucleases HindIII and BglI were carried out as described in reference 12. The patterns of bands containing rRNA gene sequences have been designated H ribotypes and B ribotypes, respectively, and the polymorphic restriction sites (PRSs) along the rDNA regions have been inferred by the presence or absence of bands in the totality of ribotypes from each endonuclease. PCR ribotyping was carried out basically as described in reference 11, with primers P1 (5′-TTGTACACACGCCCGTCA-3′) and P2 (5′-GGTACTTAGATGTTTCAGTTC-3′). It is noteworthy that changes in the amplification conditions selected (35 cycles of 1 min at 94–95°C, 1 min at 55–56°C, and 1 to 2 min at 72°C) could produce different amplified DNA band patterns. Minor differences in band intensity were not considered to define SR ribotypes. All preparations and runs were repeated at least three times to evaluate the reproducibility of the method and to prove the stability of the SR ribotypes. Strains showing identical ribotypes with the three procedures were ascribed to the same CRT, and each of these was labelled with a roman numeral (I to XVII) indicating the particular combination between H and B ribotypes found. The roman numeral was followed by an arabic numeral corresponding to the SR ribotype.
Statistical analyses.
The discrimination index (DI), i.e., the probability that two unrelated strains tested from the population would be placed into different typing groups, was calculated by Simpson’s index of diversity (10). For the phylogenetic analysis only riboprobing data were used, and a combined numerical analysis of different banding profiles, revealed with each restriction endonuclease, was performed with a software package as previously described (12, 13).
RESULTS
In a first step, Y. enterocolitica organisms isolated in Asturian hospitals were analyzed by serotyping and biotyping. They were differentiated into serotypes O:1,2, O:3, O:8, and O:9 (or classified as not agglutinating [NA] with the sera used), as well as into biotypes 1A, 2, 3, and 4 (BT1A, BT2, BT3, and BT4) (Table 1). Strains assigned to O:3 belonged to three biotypes: BT4 (56 strains, which produced DNAse but not lipase or indole, reduced nitrate to nitrite, were Voges-Proskauer positive, fermented sucrose and d-trehalose but not d-xylose and salicin, did not hydrolyze esculin, and did not show pyrazinamidase or β-d-glucosidase activity), BT2 (6 strains, which differed from BT4 strains in that they were xylose and indole positive), and BT3 (4 strains that were xylose positive and indole negative, in contrast to BT4 and BT2 strains). Six other strains were NA; of these, three belonged to BT2 and the remaining three to BT1A (differing from BT2 strains in that they were lipase, salicin, esculin, and pyrazinamidase positive). Only one strain each was assigned to the phenotypic groups O:1,2–BT2, O:8–BT2, and O:9-BT2. These data showed that O:3 and NA clinical strains could be differentiated into three (BT4, BT3, and BT2), and two (BT2 and BT1A) biotypes, respectively. Conversely, BT2 included strains of different serotypes (O:3, O:1,2, O:8, and O:9), as well as NA strains. For the assays discussed below, 10 Y. enterocolitica reference strains (Table 1), representing some of these phenotypic groups and five others, were also included in the series.
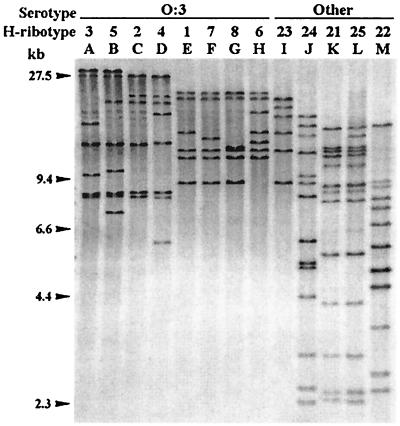
When DNA samples were separately cleaved with HindIII and BglI and were hybridized with the rnnB probe, several ribotypes could be defined (Fig. 1 and 2). With HindIII, 13 different H ribotypes were found in the series, 11 of which were represented by Asturian clinical strains and 8 of which were within serotype O:3. The H ribotypes included 5 to 14 fragments that appeared in the region between 30 and 2.3 kb, and no fragments were common to all 13 H ribotypes. Ribotypes H1 versus H7 and H7 versus H8 differed in the size of absence, respectively, of a single fragment, whereas they showed several fragments that are not present among the remaining H ribotypes. Analysis of the uncommon fragments led to more than 52 PRSs. The H ribotype of the single O:1,2a,3 strain (H23), as well as those of all but two O:3 strains, was composed of seven or fewer fragments, which were larger than 7 kb. Each of the remaining two O:3 strains showed a different ribotype, H4 or H5, that included eight fragments. The H ribotypes established could be visually clustered into two groupings, one comprising H1, H6, H7, H8, and H23 and the other comprising H2, H3, H4, and H5. On the other hand, the H ribotypes from strains assigned to other serotypes included 12 to 14 fragments, some smaller than 6 kb, and formed a third visual grouping (H21, H22, H24, and H25).
FIG. 1.
Ribotypes generated by HindIII in Y. enterocolitica strains of different serotypes and biotypes. Lanes A to D and F to H, ribotypes shown only by O:3–BT4 strains. Lane E, ribotype shown by O:3–BT2, O:3–BT3, and O:3–BT4 strains. Lanes I and J, ribotypes shown by O:1,2a,3–BT3 and O:8–BT2 strains, respectively. Lane K, ribotype shown by O:1,2–BT2, O:4,32–BT1Ax, O:5–BT1A, O:8–BT2, O:9–BT2, O:21–BT1Ax, and NA–BT1A strains. Lanes L and M, ribotypes shown by O:13,7–BT1Ax and NA–BT2 strains, respectively. Data for the clinical and reference strains corresponding to each H ribotype are presented in Table 1.
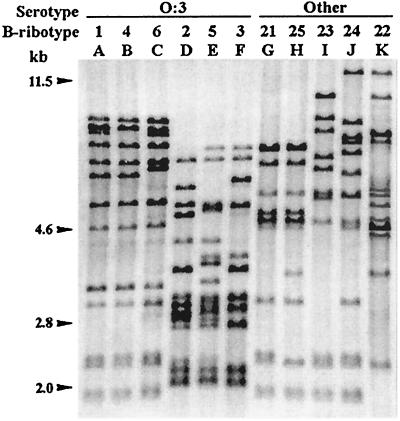
FIG. 2.
Ribotypes generated by BglI in Y. enterocolitica strains of different serotypes and biotypes. Lane A, ribotype shown by O:3–BT2, O:3–BT3, and O:3–BT4 strains. Lanes B to F, ribotypes shown only by O:3–BT4 strains. Lane G, ribotype shown by O:1,2–BT2, O:5–BT1A, NA–BT1A, and NA–BT2 strains. Lane H, ribotype shown by O:4,32–BT1Ax, O:8–BT2, O:9–BT2, O:13,7–BT1Ax, and O:21–BT1Ax strains. Lanes I to K, ribotypes shown by O:1,2a,3–BT3, O:8–BT2, and NA–BT2 strains, respectively. Data for the clinical and reference strains corresponding each B ribotype are presented in Table 1.
With BglI, 11 B ribotypes were found in the series, 10 among the Asturian clinical isolates, and 6 within serotype O:3. The B ribotypes included 10 to 14 fragments along the 14.2- to 1.7-kb region and showed at least 54 PRSs in total. The most frequent ribotype, B1, differed from two other frequent ribotypes, B4 and B6, only in the presence (B4) or absence (B6) of one fragment. All three were found only among O:3 strains. Two other ribotypes, B21 and B25, found among O:1,2, O:5, and NA strains and among O:4,32, O:8, O:9, O:13,7, and O:21 strains, respectively, differed only in two fragments, whereas differences between other B ribotypes affected a higher number of fragments. B ribotypes could be visually clustered into three groupings, two (B1, B4, and B6 and B2, B3, and B5) found only among O:3 organisms and one (B21 to B25) found only among non-O:3 organisms. Despite the relationship between visual grouping and serotype, the B ribotypes falling into the first and the third grouping have more similarities with each other than with members of the second grouping.
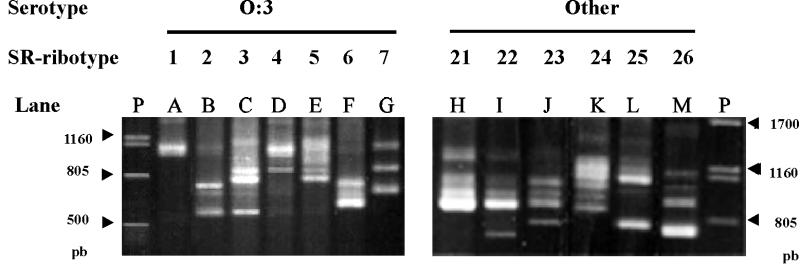
By PCR ribotyping, 13 specific SR ribotypes have been found in the series, 11 among the Asturian clinical strains and 7 among O:3 strains (Fig. 3; Table 1). The SR ribotypes included two to seven well-defined amplified DNA fragments, some stronger than others, but all constant and reproducible under our assay conditions. Some SR ribotype pairs showed one or more common fragments (i.e., SR1 and SR4; SR23 and SR24), but none of the fragments was common to all 13 SR ribotypes. Frequently, weak additional fragments appeared, but they have not been considered here, since we assumed that they had been generated by nonspecific DNA amplification. SR1, defined by only two very closely situated fragments, was the most frequent SR ribotype (59.5% of the series) and appeared among strains assigned to nine H and seven B ribotypes. SR2, SR25, and SR26 each appeared in strains assigned to two different B or H ribotypes, while the remaining SR ribotypes each appeared only in strains assigned to a single, specific H, B, or combined ribotype (Table 1).
FIG. 3.
Ribotypes generated by PCR amplification using primers for conserved regions of 16S and 23S rRNA genes in Y. enterocolitica. Lane A, ribotype shown by O:3–BT2, O:3–BT3, O:3–BT4, O:1,2a,3–BT3, and O:8–BT2 strains. Lane B, ribotype shown by O:3–BT2, O:3–BT3, and O:3–BT4 strains. Lane C, ribotype shown by O:3–BT2 and O:3–BT3 strains. Lanes D, E, and G, ribotype shown by O:3–BT4 strains. Lane F, ribotype shown by one O:3–BT3 strain. Lane H, ribotype shown by O:1,2–BT2 and NA–BT1A strains. Lane I, NA–BT1A and NA–BT2 strains. Lane J, ribotype shown by one NA–BT2 strain. Lane K, ribotype shown by O:9–BT2 strains. Lane L, ribotype shown by O:8–BT2 and O:13,7–BT1Ax strains. Lane M, ribotype shown by O:4,32–BT1Ax and O:21–BT1Ax strains. Lanes P, DNA of phage lambda cleaved with PstI. Data for the clinical and reference strains corresponding to each ribotype are presented in Table 1.
When data from HindIII riboprobing, BglI riboprobing, and PCR ribotyping were compared, a different distribution of strains into types was noted. Thus, strains falling into the most frequent types by a given procedure were differentiated by the two other procedures. Combination of the H and B ribotypes differentiated the series into 17 groups (labelled CRTI to CRTXVII). The addition of SR ribotypes increased the number to 27, because four of the groups were further differentiated (into CRTI-1 to -6; CRTII-1 and -2; CRTXII-21, -22, and -26; and CRTXIII-24 to -26) (Table 1).
The analysis of the relationships between genotypic and phenotypic markers revealed that in the series, each ribotype was associated with a specific serotype, with the following exceptions: ribotypes H21, B21, and B25 were represented by strains assigned to several infrequent serotypes (O:1,2, O:4,32, O:5, O:8, O:9, O:21, and NA for H21; O:1,2, O:5, and NA for B21; O:8, O:9, O:13,7, O:21, and O:4,32 for B25). SR1 was represented by 48 strains assigned to three serotypes: O:3 (45 strains), O:8 (2 strains), and O:1,2a,3 (1 strain). Three other ribotypes, SR21, SR25, and SR26, included strains of different and infrequent serotypes (O:1,2 and NA for SR21; O:8 and O:13,7 for SR25; and O:5, O:21, and O:4,32 for SR26). Four frequent CRTs (CRTI-1 to -3; CRTIII) were represented only by O:3 strains assigned to BT4, BT3, and/or BT2, the only biochemical difference between them being indole production and/or xylose fermentation. Similarly, CRTXII-22 was represented by four NA strains assigned to two different biotypes, BT1A and BT2. In the series, one phenotypic group (O:3–BT4) was prevalent (58 strains; 68.23%); it was subdivided into 8 H ribotypes, 6 B ribotypes, and 7 SR ribotypes, and, by combining data from three procedures, it comprised 17 CRTs or genomic groups. On the other hand, clinical and reference strains expressing biochemical traits not associated with virulence (such as pyrazinamidase activity, esculin hydrolysis, and salicin fermentation) and differing in serotype (O:4,32, O:5, O:13,7, O:21, and NA) and indole production (positive [BT1A] and negative [BT1Ax]) had identical (or not very different) ribotypes. These were also identical to, or not very different from, the ribotypes of strains that did not have these traits, all of which were assigned to BT2 but to different serotypes (O:1,2, O:8, O:9, and NA).
For epidemiological purposes the discrimination power of the typing procedures is very important, and this was tested by two different parameters: the number of types generated with each procedure (see discussion above) and the calculation of a DI. The DI values were 0.56, 0.55, and 0.56 for HindIII, BglI, and PCR ribotyping, respectively. The increase in the discrimination power resulting from the combination of data from two or three ribotyping procedures was also calculated, for HindIII and BglI (17 types; DI = 0.67), HindIII and PCR (22 types; DI = 0.76), BglI and PCR (21 types; DI = 0.74), and HindIII, BglI, and PCR (26 types; DI = 0.83). When data from serotyping and biotyping were added, the series was differentiated into 36 groupings or subtypes (DI = 0.86). With the last two options, the 74 Asturian clinical strains were differentiated into 15 CRTs and 30 subtypes, respectively.
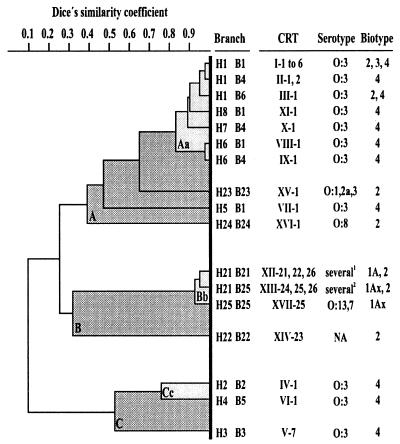
Data from the ribotypes generated with HindIII and BglI were used to trace a similarity dendrogram which showed a high heterogeneity of rDNA regions of the strains under study (similarity between 10 and 98%) and demonstrated that different groupings could be observed at varying levels of similarity (Fig. 4). At a low level (similarity coefficient [S] = 0.32), the strains were distributed into three clusters (A, B, and C), whereas at a high level (S = 0.83), each cluster could be differentiated into a subcluster and one or more branches. Cluster A groups 10 branches comprising strains assigned to 16 CRTs, 4 serotypes, and 3 biotypes. Seven of these branches showed similarities higher than 83%, forming the subcluster Aa, which includes only O:3 strains. Cluster B groups four branches, corresponding to seven CRTs and eight non-O:3 serotypes. It is worthy of note that three of these branches form subcluster Bb, which includes clinical and reference strains assigned to serotypes O:1,2, O:4,32, O:5, O:8, O:9, O:13,7, O:21, and NA, as well as BT1A, BT1Ax, and BT2. Cluster C includes three branches, each corresponding to a single CRT and a single O:3–BT4 clinical strain.
FIG. 4.
Dendrogram obtained from cluster analysis of HindIII and BglI ribotypes of Y. enterocolitica strains by using the Multivariate Statistics Package 2.0a and drawn with KOREL 5.0. A, B, and C and Aa, Bb, and Cc are the clusters and subclusters revealed at similarity coefficients of 0.32 and 0.82, respectively. The CRTs, serotypes, and biotypes falling into each branch are given. Other features and the numbers of strains falling into each branch and cluster are shown in Table 1 and discussed in the text.
DISCUSSION
The usefulness of a trait for typing is related to its stability in a given strain and its diversity among the strains forming one species. Both criteria are fulfilled by the trait analyzed by ribotyping procedures: the rrn operons. In the present work, three ribotyping variants, alone and in combination, were evaluated for Y. enterocolitica typing, and the combination of the three was used to investigate the genetic diversity of a series of Y. enterocolitica clinical and reference strains. In all but three of the O:3 strains tested by HindIII riboprobing in our laboratories (reference 12 and the present work), the H ribotypes included seven or fewer bands, which were larger than 7.5 kb. The three remaining strains showed H ribotypes with eight bands. Our data support the fact that O:3 strains usually have no HindIII cutting points within the rnn operons and that each band usually carries only one rnn operon, although in H ribotypes with fewer than seven bands, some fragments could carry two neighboring rrn operons or, alternatively, could correspond to two different DNA fragments of similar size. On the other hand, all H ribotypes found among strains that did not agglutinate with O:3 serum included 12 or more bands, data supporting the presence of HindIII cutting points within some or all of the rrn operons. Results obtained with BglI could be interpreted in a similar way, since in all the strains tested this enzyme generated ribotypes with 10 or more bands, several of which were smaller than 6.4 kb. This indicates the presence of one or more BglI sites within some or all of the rnn operons. In the strains under study, riboprobing performed with both endonucleases revealed a high level of sequence divergence among the rDNA regions. However, divergences affecting the size of the 16S–23S rRNA SRs were efficiently revealed by PCR ribotyping. The most notable example was that the strains showing H1–B1 ribotypes (grouped as CRTI) were differentiated into six SR ribotypes (CRTI-1 to -6).
Due to its reproducibility in distributing strains into SR ribotypes, PCR ribotyping has revealed itself as a highly adequate tool for epidemiological purposes, differentiating strains assigned to the most frequent phenotypic groups and also to the most frequent H and B ribotypes. Although in different experiments the amplified profile of a given strain could include one or more bands that were poorly defined or were not always revealed, the profile was still different from those corresponding to other SR ribotypes. In addition, differences in the sizes of certain bands were very slight and did not permit an accurate visual differentiation of common and noncommon bands in the totality of SR ribotypes. Taking both shortcomings into account, we decided to exclude data from PCR ribotyping for tracing the dendrogram of similarity among combined ribotypes. Cluster analysis of data from H and B ribotypes has shown that the Y. enterocolitica strains under study are probably members of different lineages, some of which include strains assigned to different serotypes and/or biotypes. For instance, in the dendrogram some O:3–BT4 strains were separated by a considerable genetic distances, falling into two different clusters. Conversely, clinical and reference strains assigned to different phenotypic groups (non-O:3, non-BT4) yielded identical or not very different ribotypes, forming a subcluster categorized as a single clonal lineage which included both pyrazinamidase-positive and pyrazinamidase-negative strains. The relationships between biotypes and lineages found in the series under study are only partially in line with the relationships among biotypes revealed by multilocus enzyme electrophoresis in other Y. enterocolitica series (7).
The above-mentioned data also led to an increase in our knowledge of the contemporary molecular epidemiology of human yersiniosis in the Principality of Asturias. In this respect, the findings to be emphasized are the following. (i) Ribotyping procedures are useful tools for differentiating individual clinical strains of Y. enterocolitica, as well as for grouping strains with ribotypes that are not very different from each other into clonal lineages and for revealing a high degree of heterogeneity within the rDNA regions of these strains. (ii) Clinical strains collected between 1993 and 1995 were assigned to 18 CRTs, 6 of which could be differentiated by serotyping and/or biotyping. Among these strains, all those assigned to O:3–BT2, O:3–BT3, and O:3–BT4 showed CRTs falling into a single lineage, which can be considered prevalent and endemic because it groups organisms collected over a 12-year period (1984 to 1995) from patients attending four different hospitals sited in an area of about 10,565 km2 with 1,120,000 inhabitants. Some of these CRTs were also shown by O:3–BT4 organisms collected from commercial raw meat products (reference 12 and unpublished data). (iii) Three other CRTs, represented only by O:3–BT4 clinical strains, formed a different lineage, showing a low level of similarity with the CRTs included in the prevalent lineage. These CRTs were infrequently found among strains collected before or during 1992 (reference 12 and unpublished data) and were not represented among strains collected after 1992. (iv) Non-O:3 clinical strains were assigned to five infrequent genomic groups. Three of these strains were BT1A, but their ribotypes were identical to ribotypes of some BT2 clinical strains and were not very different from ribotypes of reference strains representing BT1A, BT1Ax, and BT2. These three BT1A strains were isolated in the same hospital in 1993. One, collected from the feces of a woman, was the only bacterium that could be associated with her acute abdominal pain; the other two were collected from the diarrheic feces of two children, which carried an additional pathogenic species each (Salmonella and Campylobacter species, respectively). It should be pointed out that in previous works (6, 14), clinical evidence showed that some BT1A strains may be adapted to human hosts and were found to be associated with the same clinical symptoms as strains assigned to primary pathogenic biogroups.
All these data show that the polymorphism of the rDNA regions of Y. enterocolitica can be revealed by different ribotyping procedures, and they support the usefulness of CRTs as genetic markers which can be used alone or in addition to conventional phenotypic markers in epidemiological surveys of yersiniosis.
ACKNOWLEDGMENTS
We thank M. Altwegg for plasmid pKK3535 and R. Díaz (Faculty of Medicine of Navarra) and F. Uruburu of CECT for the Y. enterocolitica reference strains. We thank the staff of the Microbiology Laboratories of the Hospital Central de Asturias, the Hospital San Agustín de Avilés, the Hospital de Jarrio, the Hospital Cabueñes de Gijón, and the Hospital Carmen and Severo Ochoa de Cangas del Narcea for the clinical isolates.
This work has been supported by grants from Oviedo University (Ref. 92/38) and the Fondo de Investigacion Sanitaria (Ref. 95/0030/01).
REFERENCES
- 1.Andersen J K, Sorensen R, Glensbjerg M. Aspects of the epidemiology of Yersinia enterocolitica: a review. Int J Food Microbiol. 1991;13:231–238. doi: 10.1016/0168-1605(91)90007-c. [DOI] [PubMed] [Google Scholar]
- 2.Bisset M L, Powers C, Abbot S L, Janda J M. Epidemiologic investigations of Yersinia enterocolitica and related species: sources, frequency and serogroup distribution. J Clin Microbiol. 1990;28:910–912. doi: 10.1128/jcm.28.5.910-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg H M, Kiehlbauch J A, Wachsmuth I K. Molecular epidemiology of Yersinia enterocolitica O:3 infections: use of chromosomal DNA restriction fragment length polymorphisms of rRNA genes. J Clin Microbiol. 1991;29:2368–2374. doi: 10.1128/jcm.29.11.2368-2374.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 6.Burnens A P, Frey A, Nicolet J. Association between clinical presentation, biogroups and virulence attributes of Yersinia enterocolitica strains in human diarrhoeal disease. Epidemiol Infect. 1996;116:27–34. doi: 10.1017/s0950268800058921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolina M, Peduzzi R. Population genetics of human, animal, and environmental Yersinia strains. Appl Environ Microbiol. 1993;59:442–450. doi: 10.1128/aem.59.2.442-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez-Garcés J L, Wilhelmi I, Cogollos R, Alós J I, Páez M, Balas D, Arribi A, y Delgado-Iribarren A. Factores de patogenicidad, biotipo, serotipo y sensibilidad antimicrobiana de 150 aislamientos clínicos de Yersinia grupo enterocolitica (1992–1994) Enferm Infect Microbiol Clin. 1996;14:596–599. [PubMed] [Google Scholar]
- 9.Gonzalez-Hevia M A, Alvarez Riesgo J A, Mendoza M C. Epidemiological, clinical and microbiological features of Yersinia enterocolitica infections in a community during a four-year period. Eur J Epidemiol. 1990;6:184–190. doi: 10.1007/BF00145792. [DOI] [PubMed] [Google Scholar]
- 10.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostman J R, Edlind T D, LiPuma J J, Stull T L. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J Clin Microbiol. 1992;30:2084–2087. doi: 10.1128/jcm.30.8.2084-2087.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendoza M C, Alzugaray R, Landeras E, González-Hevia M A. Discriminatory power and application of ribotyping of Yersinia enterocolitica O:3 in an epidemiological study. Eur J Clin Microbiol Infect Dis. 1996;15:220–226. doi: 10.1007/BF01591358. [DOI] [PubMed] [Google Scholar]
- 13.Millemann Y, Lesage M-C, Chaslus-Dancla E, Lafont J-P. Value of plasmid profiling, ribotyping, and detection of IS200 for tracing avian isolates of Salmonella typhimurium and S. enteritidis. J Clin Microbiol. 1995;33:173–179. doi: 10.1128/jcm.33.1.173-179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris J G, Jr, Prado V, Ferreccio C, Robins-Browne R M, Bordun A-M, Cayazzo M, Kay B A, Levine M M. Yersinia enterocolitica isolated from two cohorts of young children in Santiago, Chile: incidence of and lack of correlation between illness and proposed virulence factors. J Clin Microbiol. 1991;29:2784–2788. doi: 10.1128/jcm.29.12.2784-2788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulakvelidze A, Dalakishvili K, Barry E, Wauters G, Robins-Browne R, Imnadze P, Morris J G., Jr Analysis of clinical and environmental Yersinia isolates in the Republic of Georgia. J Clin Microbiol. 1996;34:2325–2327. doi: 10.1128/jcm.34.9.2325-2327.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wauters G, Kandolo K, Janssens M. Revised biogrouping scheme of Yersinia enterocolitica. Contrib Microbiol Immunol. 1987;9:14–21. [PubMed] [Google Scholar]
- 17.Whittam S T. Genetic population structure and pathogenicity in enteric bacteria. Symp Soc Gen Microbiol. 1995;52:217–245. [Google Scholar]