Abstract
The purpose of this study was to evaluate the molecular relatedness of clinical isolates of glycopeptide-resistant Enterococcus faecium isolates collected from hospitals in Michigan. A total of 379 isolates used in this study were all vancomycin-resistant E. faecium isolates collected from 28 hospitals and three extended-care facilities over a 6-year period from 1991 to 1996. For the 379 isolates, there were 73 pulsed-field gel electrophoresis (PFGE) strain types. Within strain types, there were as many as six restriction fragment differences. Most isolates (70%) belonged to six strain types, which were designated M1 (36%), M2 (3%), M3 (18%), M4 (6%), M10 (4%), and M11 (3%). PFGE strain M1 was cultured from 135 patients in 13 hospitals during the period 1993 to 1996. Strain type M2 was cultured from 11 patients in two hospitals during the period 1991 to 1992 and was not observed after 1992. Strain type M3 was cultured from 70 patients in 10 hospitals during the period of 1994 to 1996. Both M4 and M10 were cultured from 23 patients in three hospitals and from 15 patients in two hospitals, respectively, during 1995 to 1996. M11 was cultured from 13 patients in four hospitals during 1996. A total of 23 of 28 hospitals had evidence of clonal dissemination of some isolates. Plasmid content and hybridization analysis done on 103 isolates from one hospital and two affiliated extended-care facilities indicated that the strains contained from one to eight plasmids. Mating experiments indicated transfer of vancomycin resistance from 94 of these isolates into plasmid-free E. faecium GE-1 at transfer frequencies of <10−9 to 10−4. Gentamicin resistance and erythromycin resistance were cotransferred at various frequencies. A probe for the vanA gene hybridized to the plasmids of 23 isolates and to the chromosomes of 72 isolates. A probe for the vanB gene hybridized to the chromosomes of 8 isolates. The results of this study suggest inter- and intrahospital dissemination of vancomycin-resistant E. faecium strains over a 6-year period in southeastern Michigan. The majority of isolates studied belonged to the same few PFGE strains, indicating that clonal dissemination was responsible for most of the spread of resistance that occurred.
Enterococci have emerged in recent years as pathogens in a growing number of serious nosocomial infections including bacteremia and intraabdominal and urinary tract infections (19, 25). Especially worrisome are the increased numbers of enterococcal isolates that are resistant to the aminoglycosides, penicillins, or glycopeptide agents (5–29). The spread of these antibiotic-resistant enterococcal strains has occurred not only within individual hospitals but also between hospitals of various geographic locations across the United States (3–11, 14–19, 22, 24). The proportion of enterococcal isolates that exhibit clinically important multiple antimicrobial resistance has become quite high at some chronic-care facilities, as well as at acute-care hospitals.
A knowledge of the epidemiology of vancomycin-resistant enterococci (VRE) is essential for control of further spread. In this study, contour-clamped homogeneous electric-field (CHEF) electrophoresis (commonly referred to as pulsed-field gel electrophoresis [PFGE]), analysis of plasmid DNA, and hybridization analysis were used to compare DNAs of vancomycin-resistant Enterococcus faecium (VREF) isolates collected from Michigan hospitals over a 6-year period. The purpose of the comparison was to evaluate the molecular relatedness of these isolates to obtain information about geographic dispersion of strains and genes responsible for glycopeptide resistance. This analysis is needed to help determine whether clonal, plasmid, or transposon dissemination or a combination of mechanisms is responsible for some of the resistance that has been observed.
MATERIALS AND METHODS
The 379 isolates used in this study were all VREF isolates collected from 28 hospitals and three extended-care facilities in Michigan over a 6-year period from 1991 through 1996. Isolates from patients who were epidemiologically related and unrelated were evaluated. From one hospital and three extended-care facilities, all VRE isolates from 1991 to 1996 were evaluated. VRE isolates were also collected from 28 of 33 participant institutions as part of a Michigan Department of Community Health (MDCH) surveillance project of VRE epidemiology in Michigan during the period from 1995 to 1996. Rates of resistance to vancomycin were determined by geographic region (Fig. 1). The hospitals selected in the MDCH surveillance study included the two largest hospitals from each of 12 community hospital assessment regions (CHARs) (Fig. 1), and, of the remaining hospitals in Michigan, the 11 largest hospitals. Isolates from 14 hospitals were submitted following suspected outbreaks.
FIG. 1.
Map showing 12 CHARs in Michigan.
Duplicate isolates from the same patient were excluded. Isolates were from urine (101 isolates), blood (88 isolates), wounds (52 isolates), stool (36 isolates), intraabdominal focus (31 isolates), semiquantitative catheter tip culture (25 isolates), sputum (4 isolates), bone (1 isolate), cerebrospinal fluid (1 isolate), and an unspecified site (40 isolates). Isolates were identified as E. faecium by using biochemical reactions as outlined previously (13). Susceptibility of isolates to vancomycin (Eli Lilly and Co., Indianapolis, Ind.) and teicoplanin (Marion Merrell Dow, Kansas City, Mo.) was determined by microdilution by National Committee for Clinical Laboratory Standard-recommended methods (23).
Genomic DNA was prepared in agarose plugs, digested with the enzyme SmaI (New England BioLabs, Beverley, Mass.), and electrophoresed by using CHEF with a CHEF-DRII apparatus (Bio-Rad Laboratories, Richmond, Calif.) as previously described (12). Total numbers of visible bands were counted for each isolate, and patterns were compared visually. Isolates were considered identical when they had all bands in common. Once isolates were recognized as having identical patterns, a representative isolate of the group was used to compare its pattern with those of other isolates. Isolates were considered closely or possibly related when they differed by changes consistent with one or two genetic events (one to six band differences) as outlined by Tenover et al. (26).
Filter-mating experiments were performed, and transconjugants were screened on brain-heart infusion (BHI) agar plates containing 500 μg of gentamicin per ml and on BHI agar plates containing 25 μg of erythromycin per ml to evaluate whether resistance to these antibiotics cotransferred with vancomycin resistance.
Genomic DNA for restriction enzyme analysis was prepared by previously described methods (2, 21). Plasmid and chromosomal DNAs were separated with a cesium chloride density gradient. Restriction enzyme analysis was performed with EcoRI according to the manufacturer’s recommendations. Samples were run on a 0.7% agarose gel, stained with ethidium bromide, and visualized under UV light. Biotinylated lambda DNA/HindIII fragments were used as size markers (New England BioLabs).
Hybridization experiments for detection of vancomycin resistance determinants for vanA and vanB were done on all isolates under conditions of high stringency. The vanA and vanB gene probes were generated by PCR with the PCR Reagent System (GIBCO-BRL, Gaithersburg, Md.) according to the manufacturer’s recommendations. The oligonucleotide primers chosen for amplification were those described by Clark et al. (9). The vanA gene probe was amplified from E. faecium SF6460, and a 698-bp BamHI fragment was used as the probe. The 433-bp vanB probe was amplified from E. faecium SF6621.
RESULTS
In the hospitals studied, 2% of E. faecalis and 37% of E. faecium isolates were resistant to vancomycin. The highest rates of resistance were in E. faecium isolates from hospitals in CHAR 1 (48% versus 9% of isolates resistant in regions 2 to 12). Analysis of 379 VREF isolates collected over a 6-year period indicated 73 PFGE strain types. Within strain types, there were as many as six restriction fragment differences. The majority of isolates (70%) belonged to six strain types, which were designated M1, M2, M3, M4, M10, and M11 (Fig. 2).
FIG. 2.
PFGE of SmaI-digested genomic DNAs from VREF. Lanes: 1, lambda phage DNA ladder standard; 2, M1, most common strain type (36% of isolates) from CHAR 1; 3, M2 (3% of isolates) from CHAR 1; 4, M3 (18% of isolates) from CHARs 1, 4, and 11; 5, M4 (6% of isolates) from CHAR 1; 6, M10 (4% of isolates) from CHAR 1; 7, M11 (3% of isolates) from CHARs 1 and 4; 8, M20, from CHAR 8; 9, M22, from CHARs 1 and 8; 10, M23, from CHARs 1, 2, and 8; 11, M24 from CHAR 4; 12, M27 from CHAR 4; 13, M66 from CHAR 9; 14, M67 from CHAR 5; 15, M69 from CHAR 11; 4, 7, 9, and 10, strain types found in more than one CHAR; 1 to 6, 9, 10, and 12, strain types involved in interhospital dissemination; 8 and 11, strain types involved in intrahospital dissemination only; 13 to 15, unique strain types not involved in any dissemination.
Table 1 shows PFGE strain types grouped by CHARs. The majority of study isolates (95%) were from southeastern Michigan hospitals (CHAR 1). Most isolates from CHAR 1 (73%) belonged to PFGE strains M1, M2, M3, M4, M10, and M11. Table 2 shows a yearly summary of the number of PFGE strain types and number of isolates separated by type of dissemination. M2 was detected from 1991 to 1992 and was not seen after 1992. M1 was detected from 1993 to 1996; M3 was detected from 1994 to 1996; M4 and M10 were detected from 1995 to 1996; and M11 was detected in 1996. There was a general trend toward an increase in VRE isolates in Michigan during the study period (Table 2). There were 46 PFGE strain types that contained only one isolate. These unique strain types accounted for 63% of total strain types identified and 12% of total isolates. There were 12 PFGE strain types (16%) that contained more than 1 isolate but that were detected in only one hospital. These strains accounted for 8% of the total isolates studied. Table 3 described PFGE strain types that were involved in interhospital dissemination. There were 15 PFGE strain types (21%) that contained 304 isolates (80%). Each of these strain types was found at two or more hospitals. M1 and M3 had the widest ranges of dissemination and were isolated from 13 and 10 hospitals, respectively. For these two strain types, PFGE patterns varied by three bands or fewer for 91 and 71% of isolates, respectively. Isolates in M1 and M3 that varied by four to six bands were from epidemiologically related patients (same hospital) as part of suspected outbreaks. There were five PFGE strain types that were also detected in multiple CHARs (M3, M11, M22, M23, and M27).
TABLE 1.
Results of PFGE strain types by CHARa
| CHAR | PFGE strain type(s) (no. of isolates) | Total no. of isolates |
|---|---|---|
| 1 | M1 (135), -2 (11), -3 (67), -4 (23), -5 (5), -6 (6), -7 (6), -8 (2), -9 (2), -10 (15), -11 (12), -12 (2), -13 (2), -14 (3), -15 (3), -16 (2), -17 (2), -18 (3), -19 (6), -21 (2), -22 (1), -23 (3), -25 (4), -26 (2), -27 to -65 (1 each), -68 (1), -72 (1), -73 (1) | 361 |
| 2 | M23 (2) | 2 |
| 4 | M3 (2), -11 (1), -24 (2), -27 (1), -70 (1) | 7 |
| 5 | M67 (1), -71 (1) | 2 |
| 8 | M20 (2), -22 (1), -23 (1) | 4 |
| 9 | M66 (1) | 1 |
| 11 | M3 (1), -69 (1) | 2 |
See Fig. 1 for region designations.
TABLE 2.
Yearly summary of the number of PFGE strain types and number of isolates involved in no dissemination (unique strains), intrahospital dissemination only, and interhospital dissemination
| Yr | No. of PFGE strain types (no. of isolates)
|
|||
|---|---|---|---|---|
| No dissemination (unique) | Intrahospital dissemination (only) | Interhospital dissemination | Total | |
| 1991 | 1 (1) | 0 | 1 (1) | 2 (2) |
| 1992 | 1 (1) | 1 (2) | 1 (10) | 3 (13) |
| 1993 | 0 | 1 (1) | 1 (10) | 2 (11) |
| 1994 | 1 (1) | 3 (6) | 3 (67) | 7 (73) |
| 1995 | 12 (12) | 4 (11) | 7 (79) | 23 (102) |
| 1996 | 31 (31) | 5 (10) | 13 (137) | 49 (178) |
TABLE 3.
Summary of PFGE strain types involved in interhospital dissemination
| Strain type | No. of isolates | No. of isolates with the following band differences
|
No. of hospitals | CHARa | Yr(s) cultured | ||
|---|---|---|---|---|---|---|---|
| 0 bands | 1–3 bands | 4–6 bands | |||||
| M2 | 11 | 6 | 3 | 2 | 2 | 1 | 1991–1992 |
| M1 | 135 | 57 | 66 | 12 | 13 | 1 | 1993–1996 |
| M7 | 6 | 0 | 5 | 1 | 5 | 1 | 1994–1995 |
| M3 | 70 | 30 | 20 | 20 | 10 | 1, 4, 11 | 1994–1996 |
| M4 | 23 | 11 | 6 | 6 | 3 | 1 | 1995–1996 |
| M6 | 6 | 2 | 3 | 1 | 3 | 1 | 1995–1996 |
| M19 | 6 | 2 | 0 | 4 | 4 | 1 | 1995–1996 |
| M10 | 15 | 7 | 4 | 4 | 2 | 1 | 1995–1996 |
| M11 | 13 | 4 | 3 | 6 | 4 | 1, 4 | 1996 |
| M12 | 2 | 0 | 0 | 2 | 2 | 1 | 1996 |
| M14 | 3 | 3 | 0 | 0 | 3 | 1 | 1996 |
| M22 | 2 | 0 | 0 | 2 | 2 | 1, 8 | 1996 |
| M23 | 6 | 2 | 4 | 0 | 4 | 1, 2, 8 | 1996 |
| M25 | 4 | 3 | 1 | 0 | 2 | 1 | 1996 |
| M27 | 2 | 0 | 2 | 0 | 2 | 1, 4 | 1996 |
See Fig. 1 for CHAR designations.
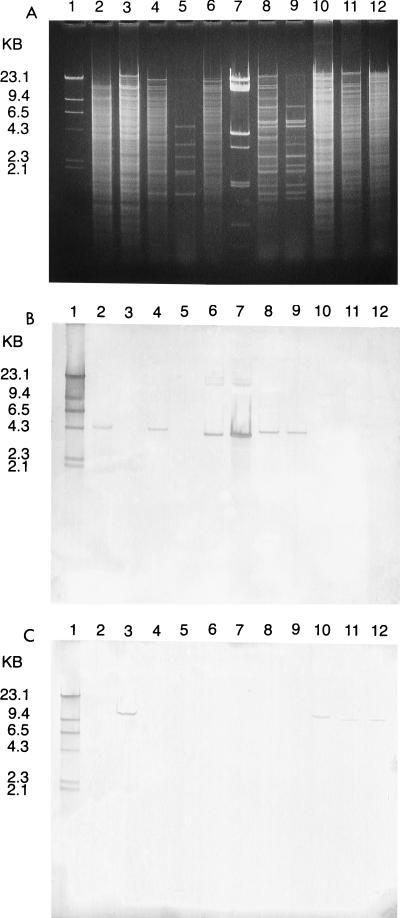
Table 4 and Fig. 3 summarize hybridization studies of 103 VREF isolates collected from a single southeastern Michigan hospital from 1991 to 1996. The vanA gene was found in 95 (92%) of the isolates studied. For 72 of these isolates the vanA gene was located on the chromosome. For the remaining 23 isolates, the vanA gene was located on a plasmid. The largest PFGE group, M1, accounted for 26% of the isolates, for M1 the vanA gene was always located on the chromosome. The second largest PFGE group, M4, accounted for 22% of the isolates and varied according to the location of the vanA gene, with five isolates (24%) having the vanA gene located on the chromosome and 16 isolates (76%) having the vanA gene located on a plasmid (Fig. 4). There was heterogeneity of plasmid content, ranging from one to eight plasmids. The vanB gene was detected in only 8 of the 103 VREF isolates studied. These isolates belonged to six PFGE groups. The vanB gene was located on the chromosome in all isolates. There was a large diversity in plasmid content for the VanB strains as well (one to six plasmids).
TABLE 4.
Hybridization analysis and transfer frequencies of VREF from a single institution in southeastern Michigan
| Strain type(s) | No. of isolates | Vancomycin genotype (location) | Transfer frequencya | Yr(s) cultured |
|---|---|---|---|---|
| M29 | 1 | B (chromosome) | <10−9 | 1991 |
| M30 | 1 | A (chromosome) | 10−4 | 1992 |
| M8 | 2 | B (chromosome) | <10−9 | 1992 |
| M1 | 25 | A (chromosome) | 10−5–10−8 | 1993–1996 |
| M3 | 9 | A (6 on chromosome, 3 on plasmid) | 10−5–10−9 | 1994–1996 |
| M6 | 2 | A (chromosome) | 10−6 | 1995 |
| M4 | 21 | A (5 on chromosome, 16 on plasmid) | 10−5–10−9 | 1995–1996 |
| M10 | 14 | A (chromosome) | 10−4–10−6 | 1995–1996 |
| M7, -22, -33, -40, -41, -42, -43, -49, -51, -54, -58, -61, -62 | 1 (each) | A (chromosome) | 10−5–10−8 | 1995–1996 |
| M23, -34, -60 | 1 (each) | B (chromosome) | <10−9 | 1995–1996 |
| M11 | 4 | A (plasmid) | 10−6–10−7 | 1996 |
| M9 | 2 | B (chromosome) | <10−9 | 1996 |
| M12, -14, -25, -55, -57, -63 | 1 (each) | A (plasmid) | 10−5–<10−9 | 1996 |
Transfer into plasmid-free E. faecium GE-1.
FIG. 3.
(A) Agarose gel electrophoresis of EcoRI-digested genomic and plasmid DNAs from VREF isolates. Lanes: 1, biotin-labeled HindIII-digested lambda phage DNA; 2, chromosomal DNA from a known VanA strain; 3, chromosomal DNA from a known VanB strain; 4 and 5, genomic and plasmid DNA from a group 4 isolate (vanA gene on chromosome); 6 and 7, genomic and plasmid DNA from a group 4 isolate (vanA gene on plasmid); 8 and 9, genomic and plasmid DNA from a group 11 isolate (vanA gene on plasmid); 10 to 12, genomic DNAs from VanB isolates (group 23, group 8, and group 9, respectively). (B) Southern blot of gel shown in panel A probed with biotin-labeled vanA gene. Lanes: 1, biotin-labeled HindIII-digested lambda phage DNA; 2, chromosomal DNA from a known VanA strain positive for the vanA gene; 3, chromosomal DNA from a known VanB strain negative for the vanA gene; 4, genomic DNA from an isolate positive for the vanA gene; 5, plasmid DNA from isolate in lane 4 negative for the vanA gene; 6, genomic DNA from isolate positive for the vanA gene; 7, plasmid DNA from the isolate in lane 6 also positive for the vanA gene; 8, genomic DNA from isolate positive for the vanA gene; 9, plasmid DNA from the isolate in lane 8 positive for the vanA gene; 10 to 12, genomic DNAs from isolates negative for the vanA gene. (C) Southern blot of gel in shown in panel A probed with biotin-labeled vanB gene. Lanes: 1, biotin-labeled HindIII-digested lambda phage DNA; 2, chromosomal DNA from a known VanA strain negative for the vanB gene; 3, chromosomal DNA from a known VanB strain positive for the vanB gene; 4 to 9, genomic and plasmid DNAs from these isolates negative for the vanB gene; 10 to 12, genomic DNAs from these isolates positive for the vanB gene.
FIG. 4.
(A) CHEF electrophoresis of SmaI-digested genomic DNAs from VREF strains. Lanes: 1, lambda phage DNA ladder standard; 2, known VanA strain (vanA gene on the chromosome); 3 and 5, isolates from group 4 with the vanA gene on the chromosome; 4 and 6, isolates from group 4 with the vanA gene on a plasmid; 7 and 8, isolates from group 11 with the vanA gene on a plasmid; 9, isolate from group 3 with the vanA gene on a plasmid; 10, isolate from group 3 with the vanA gene on the chromosome. (B) Southern blot of gel in shown in panel A probed with biotin-labeled vanA gene and biotin-labeled lambda phage DNA. Lanes: 1, lambda phage DNA ladder standard; 2, known VanA isolate positive for the vanA gene; 3, 5, 7, 8, and 10, SmaI-digested genomic DNAs from these isolates positive for the vanA gene (in lanes 7 and 8, the vanA probe hybridizes with a plasmid visible on the CHEF gel); 4, 6, and 9, SmaI-digested genomic DNAs from these isolates negative for the vanA gene (plasmid DNA with the vanA gene is not visible on the CHEF gel).
For VanA isolates, the MICs of vancomycin were 64 to >256 μg/ml and the MICs of teicoplanin were from 16 to >64 μg/ml. Transfer of vancomycin resistance from the VanA isolates into plasmid-free E. faecium GE-1 varied, with frequencies of <10−9 to 10−4. No pattern was found according to PFGE strain type or location of the vanA gene. Gentamicin and erythromycin resistance cotransferred, with frequencies varying from 0 to 100%. For VanB isolates, the MICs of vancomycin and teicoplanin were 32 to 256 and <0.125 to 0.5 μg/ml, respectively. There were no VanB isolates that were teicoplanin resistant. The VanB isolates did not transfer vancomycin resistance into GE-1.
DISCUSSION
In 1986, Le Clerc and colleagues first observed inducible, plasmid-mediated, high-level resistance to both vancomycin and teicoplanin in E. faecium (1). Other reports of vancomycin resistance to E. faecium soon followed (5, 6). Vancomycin resistance to enterococci in the United States was rare prior to 1989. Tenover and coworkers have been monitoring prevalence rates from 97 U.S. hospitals (9, 24). Among these institutions, VREF was reported at approximately 61% during January to March 1994, compared to only 23% in the final quarter of 1993 (5). As part of a long-term surveillance project for antibiotic resistance among pathogens associated with nosocomial infections, the National Nosocomial Surveillance System (NNIS) of the Centers for Disease Control reported a 26-fold increase (0.3 to 7.9%) in VRE from 1989 through 1993. Additionally, among patients in intensive-care units (ICUs) with nosocomial infections, enterococcal isolates resistant to vancomycin increased from 0.4 to 13.6%. Most recent NNIS data indicate rates of VRE of 14.0% in non-ICU patients as well. In general, the types of patients from whom VREF isolates have been recovered are similar to patients with other antibiotic-resistant enterococcal infections. In the United States, cases to date have been nosocomially acquired and involve immunocompromised patients (renal and oncology), elderly patients (>65 years), patients hospitalized in large teaching hospitals for an extended period (>3 weeks), patients with foreign bodies or serious underlying diseases, and patients who had received prolonged antibiotic courses with either vancomycin and/or other antibiotics. In Europe, the epidemiology of VRE is different from that in the United States, with community acquisition of multiple strain types well documented (20, 27, 28).
Molecular epidemiologic techniques have been an essential tool in the study of the epidemiology of nosocomial enterococci. For most epidemiologic evaluations, PFGE has become the method of choice for strain delineation of enterococci. In a recent study from our laboratory, these techniques indicated clonal spread of a vanB isolate between three hospitals in two states (7). Hybridization, restriction mapping, and partial DNA sequence analysis recently demonstrated that a cluster of recent E. faecium isolates containing the vanA gene from the northeastern United States differs from strains initially reported from Europe (17). Isolates from 12 U.S. medical centers analyzed by PFGE indicated both intrahospital and interhospital diversity of strains among multiresistant VanA isolates (24). Another surveillance of isolates from American patients revealed that VanA isolates were primarily from the Northeast but that VanB strains were more geographically dispersed (9). Studies of VREF in Texas conducted by Morena et al. (22) indicated dissemination of the vanB gene by a single strain. However, VanA VREF involved separate strains, as determined by PFGE typing.
Our data showed multiple strain types of VREF from Michigan hospitals over a 6-year period. There were 73 PFGE strain types in 379 isolates. There were 46 PFGE strain types that contained only one isolate, indicating no evidence of dissemination for these isolates. Since over half of the unique strain types (31 of 46) were detected in 1996, dissemination that may have occurred later was not detected. However, a few unique strain types were detected early in the study period (1 in 1991 and 1 in 1992). Both of these strain types as well as many from 1995 (6 of 12) were isolated from the same hospital, in which extensive surveillance was ongoing for the total study period and dissemination would have been detected. These strains support the conclusion that there are some PFGE strain types that appear once or infrequently but that are never involved in spread to other patients or hospitals. While these strain types accounted for the majority of total strain types (63%), they involved only a small percentage of the total isolates studied (12%).
There were 12 PFGE strain types (16%) that contained more than one isolate but were detected in only one hospital, suggesting that these strains were involved in intrahospital transmission only. These strain types also accounted for a small percentage (8%) of the total isolates studied, and for each strain type dissemination was limited to two to five patients.
Of the 28 hospitals whose isolates were evaluated in this study, 22 (79%) were found to have PFGE strain types associated with interhospital dissemination. Of the remaining six institutions, one contained a strain involved in intrahospital dissemination only. Five hospitals contained only unique PFGE strain types (not associated with any dissemination); however, three of these submitted only one isolate, and it is thus difficult to draw conclusions about dissemination. Fifteen of the PFGE strain types described in our study were detected in more than one hospital, suggesting interhospital dissemination. While these strain types accounted for only 21% of total strain types, they included 80% of the total isolates studied. These strains appear to pose the most serious threat for spread of vancomycin resistance. Six of the 15 strain types associated with interhospital dissemination accounted for 70% of study isolates (M1 to M4, M10, and M11). Strain type M1 included 135 isolates recovered over a 4-year period from 13 hospitals, all of which were in southeastern Michigan (CHAR 1). M3 included 70 isolates recovered over a 3-year period from 10 hospitals in three CHARs.
To determine the genetic relatedness of VRE isolates that were epidemiologically related, PFGE, hybridization studies, and evaluation of plasmid contents were done on all VRE isolates from a single institution from 1991 to 1996. Hybridization analysis of these isolates (103 isolates from 32 PFGE types) showed that the majority of the isolates (92%) contained the vanA gene. For 70% of these, the vanA gene was located on the chromosome. The VanA isolates accounted for 24 of the 32 PFGE strain types (75%) evaluated. Five of the six most prevalent strain types (M1, -3, -4, -10 and -11) were found to contain the vanA gene, and 11 of the 15 PFGE types involved in interhospital dissemination contained the vanA gene. VanA isolates were detected from 1992 through 1996, with these numbers increasing each year from 1 in 1992 to 35 in 1996. In 1993 and 1994, 100% of the isolates analyzed were found to contain the vanA gene.
Eight of the 103 isolates hybridized were found to contain the vanB gene located on the chromosome. These isolates were contained in six PFGE strain types (Table 4). VanB isolates were detected in 1991 and 1992 and not again until 1995 and 1996. Three of the six PFGE strain types containing the vanB gene were unique strains not associated with any dissemination (M29, M34, and M60). Two PFGE strain types (M8 and M9) were associated only with intrahospital dissemination in this study. M8 was previously shown to be clonally related to isolates from two hospitals in the Chicago, Ill., area (7). One PFGE strain type with the vanB gene (M23) was associated with interhospital dissemination. M23 was isolated from six patients in four hospitals from three CHARs in 1996. This may indicate a reemergence of the vanB gene in Michigan hospitals.
The results of this study provide further evidence for intra- and interhospital spread of some multiply resistant enterococcal isolates. Evaluation of strains from a single institution showed epidemiologically related VanA strains with different PFGE types and plasmid contents. The location of vanA genes on both plasmid and chromosome suggests the possibility of transposon dissemination among these isolates. Evaluation of this possibility requires further study.
ACKNOWLEDGMENTS
This work was supported by the William Beaumont Hospital Research Institute and by Public Health Service grant H50/CCH513220-01 from the Centers for Disease Control and Prevention.
We thank William Hall and Jaime Altamirano for their assistance in this project and Rosalind Smith for assistance in preparation of the manuscript.
REFERENCES
- 1.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, et al., editors. Current protocols in molecular biology. 1, unit 2.4. New York, N.Y: John Wiley and Sons, Inc.; 1993. [Google Scholar]
- 3.Bonten M, Slaughter S, Hayden M, Nathan C, Voorhis J V, Rice T, Weinstein R A. Abstracts of the 36th Interscience Conference on Antimicrobial Agents and Chemotherapy, 15 to 17 September 1996, New Orleans, La. Washington, D.C: The American Society for Microbiology; 1996. Patients’ endogenous flora as a source of “nosocomial” vancomycin-resistant enterococci (VRE) [Google Scholar]
- 4.Boyce J M, Opal S M, Chow J W, Zervos M J, Potter-Bynoe G, Sherman C B, Romulo R L C, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 6.Centers for Disease Control. National nosocomial infection surveillance system report. May 1997. Atlanta, Ga: Centers for Disease Control; 1997. [Google Scholar]
- 7.Chow J E, Kuritza A, Shlaes D M, Green M, Sahm D F, Zervos M J. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J Clin Microbiol. 1993;31:1609–1611. doi: 10.1128/jcm.31.6.1609-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coque T M, Arduino R C, Murray B E. High-level resistance to aminoglycosides: comparison of community and nosocomial fecal isolates of enterococci. Clin Infect Dis. 1995;20:1048–1051. doi: 10.1093/clinids/20.4.1048. [DOI] [PubMed] [Google Scholar]
- 9.Clark N C, Cooksey R C, Hill B C, Swenson J M, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dembry L M, Thal L A, Zervos M J. Molecular characterization of a novel glycopeptide resistance phenotype in E. faecium. In Proceedings of the 93rd General Meeting of the American Society for Microbiology, May 1993. Washington, D.C: American Society for Microbiology; 1993. [Google Scholar]
- 11.Dembry L M, Uzokwe K, Zervos M J. Control of endemic glycopeptide-resistant enterococci. Infect Control Hosp Epidemiol. 1996;17:286–292. doi: 10.1086/647297. [DOI] [PubMed] [Google Scholar]
- 12.Donabedian S M, Chow J W, Boyce J M, McCabe R E, Markowitz S M, Coudron P E, Kuritz A, Pierson C L, Zervos M J. Molecular typing of ampicillin-resistant, non-β-lactamase-producing Enterococcus faecium isolates from diverse geographic areas. J Clin Microbiol. 1992;30:2757–2761. doi: 10.1128/jcm.30.11.2757-2761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frieden T R, Munsiff S S, Low D E, et al. Emergence of vancomycin-resistant enterococci in New York City. Lancet. 1993;342:76–79. doi: 10.1016/0140-6736(93)91285-t. [DOI] [PubMed] [Google Scholar]
- 15.Gold H S, Unal S, Cercenado E, Thauvin-Eliopoulos C, Eliopoulos G M, Wennersten C B, Moellering R C., Jr A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob Agents Chemother. 1993;37:1604–1609. doi: 10.1128/aac.37.8.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handwerger S, Raucher B, Altarac D, et al. Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clin Infect Dis. 1993;16:750–755. doi: 10.1093/clind/16.6.750. [DOI] [PubMed] [Google Scholar]
- 17.Handwerger S, Skoble J, Discotto L F, Pucci M. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayden M K, Trenholme G M, Schultz J E, Sahm D F. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J Infect Dis. 1993;167:1224–1227. doi: 10.1093/infdis/167.5.1224. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis, W. R., and W. J. Marton. 1992. Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 29(Suppl. A):19–24. [DOI] [PubMed]
- 20.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerum A M, Aarestrup F M. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 22.Morena F, Grota P, Crisp C, Magnon K, Melcher G P, Jorgensen J H, Patterson J E. Clinical and molecular epidemiology of vancomycin-resistant Enterococcus faecium during its emergence in a city in Southern Texas. Clin Infect Dis. 1995;21:1234–1237. doi: 10.1093/clinids/21.5.1234. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Method for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd ed. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1994. [Google Scholar]
- 24.Sader H S, Tenover M A, Hollis F C, Jones R J, Jones R N. Evaluation and characterization of multiresistant Enterococcus faecium from 12 U.S. medical centers. J Clin Microbiol. 1994;32:2840–2842. doi: 10.1128/jcm.32.11.2840-2842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaberg, D. R. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91(Suppl. 3B):72S–75S. [DOI] [PubMed]
- 26.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van der Auwera P, Pensart N, Korten V, Murray B E, Leclercq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis. 1996;173:1129–1136. doi: 10.1093/infdis/173.5.1129. [DOI] [PubMed] [Google Scholar]
- 28.Witte W, Klare I. Glycopeptide-resistant Enterococcus faecium outside hospitals: a commentary. Microbiol Drug Resist. 1995;3:259–263. doi: 10.1089/mdr.1995.1.259. [DOI] [PubMed] [Google Scholar]
- 29.Zervos M J, Kauffman C A, Therasse P M, Bergman A G, Mikesell T S, Schaberg D R. Nosocomial infection by gentamicin resistant Streptococcus faecalis: an epidemiologic study. Ann Intern Med. 1987;106:687–691. doi: 10.7326/0003-4819-106-5-687. [DOI] [PubMed] [Google Scholar]