Abstract
The sequence of a verocytotoxin 2 (VT2) variant gene that was untypeable by the B subunit PCR and restriction fragment length polymorphism analysis (PCR-RFLP) method described by Tyler et al. (S. D. Tyler, W. M. Johnson, H. Lior, G. Wang, and K. R. Rozee, J. Clin. Microbiol. 29:1339–1343, 1991) was determined and compared with published sequences. It was highly homologous to two recently reported VT2 variant sequences. The PCR-RFLP method described by Tyler et al. was extended to include these new sequences. New VT2 variants were identified in 65 of 359 VT-producing Escherichia coli (VTEC) with newly designed primers (VT2-cm and VT2-f) and were characterized as well by restriction analysis of the amplification products obtained with another VT2-specific primer pair (VT2-e and VT2-f). The VT genes harbored by 64 of these isolates proved to be untypeable by Tyler’s PCR-RFLP method because no amplification was obtained with the primers used with this method (VT2-c and VT2-d). The last isolate harbored the new variant gene in addition to VT2vh-a. None of the isolates harboring these new toxin genes belonged to serogroups O157, O26, O103, O111, and O145. All 65 isolates were negative for the eaeA gene and were significantly less frequently enterohemolytic or positive for the enterohemorrhagic E. coli (EHEC) virulence plasmid than non-O157 VTEC isolates harboring other VT2 genes. They were also less frequently isolated from patients with EHEC-associated symptoms. The extended PCR-RFLP typing method is a useful tool to identify less-virulent VTEC isolates and for VT genotyping in epidemiological studies with non-O157 strains.
Verocytotoxins (VT), also called Shiga-like toxins and Shiga toxins (ST), are cytotoxins produced by some Escherichia coli strains, known as VT-producing E. coli (VTEC), ST-producing E. coli, or enterohemorrhagic E. coli (EHEC) (17). These organisms cause diarrhea, often bloody and sometimes complicated by hemorrhagic colitis and/or hemolytic uremic syndrome (HUS). No consensus about toxin nomenclature has been reached (1, 10), and VT and ST are used synonymously. The most-common serotype of this category of diarrheagenic E. coli is O157:H7, but more than 100 serotypes of E. coli were shown to produce these toxins (13). In addition to the production of cytotoxins, the most pathogenic VTEC strains harbor accessory virulence factors, such as the eaeA gene (needed for the production of attachment-effacement lesions), the production of enterohemolysin, and a high-molecular-weight plasmid called EHEC virulence plasmid (27).
VT were classified in two major classes, VT1 and VT2, based on toxin-neutralization and DNA hybridization tests. Although the VT1 class is very homogeneous, three subtypes of VT2 were identified: VT2, VT2c, and VT2e (32). VT2e, the first-described VT2 variant toxin, is produced mainly by VTEC serotypes associated with edema disease in swine (14) and has rarely been identified in human isolates (26, 35). An important characteristic of this porcine variant toxin is toxicity to Vero cells but not to HeLa cells. Several other VT2 variant toxins, generally not toxic to HeLa cells, were identified in human isolates and were classified as VT2c (18).
Primers can be used to amplify specifically VT1, VT2, or VT2e genes (9, 30). For differentiation of the human VT2 variants, Tyler et al. (38) described a genotyping method based on the restriction fragment length polymorphism (RFLP) analysis of a B-subunit-encoding DNA fragment obtained by PCR. This method classifies human VT2 genes in three groups: VT2, VT2vh-a, and VT2vh-b. The latter two groups correspond to two VT2c genes sequenced by Ito et al. (6), vtx2ha and vtx2hb. We applied Tyler’s typing method successfully to numerous human and animal VTEC isolates (27, 28), but 64 strains positive with primers VT2-a and VT2-b (indicating the presence of a VT2 gene or of one of the human variants) and VT2-v1 and VT2-v2 primers (indicating the presence of a human VT2 variant) could not be further typed because no amplification was obtained with primers VT2-c and VT2-d, which amplify the DNA fragment needed for the RFLP analysis. In this study, the gene present in one human isolate was sequenced and compared to the numerous published sequences of VT genes (2, 6–8, 12, 15, 21–24, 31, 33, 34, 39), and the PCR-RFLP genotyping method of Tyler et al. was expanded by including three new primers. This new PCR-RFLP method was applied to 359 human and animal isolates, including the 64 strains with untypeable VT2 genes. In addition, to evaluate the pathogenicity of the strains harboring the new VT2 genes, the presence of accessory virulence factors was determined for all isolates studied.
[Preliminary data were presented as a poster at the 3rd International Symposium and Workshop on Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infections, Baltimore, Md., 22 to 26 June 1997.]
MATERIALS AND METHODS
Bacterial strains.
Three hundred fifty-nine VTEC isolates—97 O157 strains (95 human and 2 animal isolates) and 262 non-O157 strains (185 human and 77 animal isolates)—isolated during several studies performed in Belgium from 1987 to 1996 were analyzed (25, 27–29). Except for four non-O157 isolates from bovine stools, all animal strains were from meat samples. E. coli EH250 is one of the 64 strains whose VT gene was not typeable by Tyler’s PCR-RFLP method. Its O antigen is not typeable, and its flagellar antigen is type H12 (serotype Ount:H12). This strain was isolated in our laboratory in 1995 from the stools of a 9-year-old girl presenting with abdominal cramps but no diarrhea and is deposited under accession number LMG 18459 in the BCCM/LMG bacterium collection, Universiteit Gent, Ghent, Belgium. Polymyxin extracts of this strain prepared and tested as described previously (29) were moderately cytotoxic to Vero cells (50% cytotoxic dose [CD50] in titers per milliliter [20]), 200) but not to HeLa cells (50% cytotoxic dose in titers per milliliter, <100).
Sequencing of the VT gene of one VTEC isolate.
The VT gene of strain EH250 was amplified with two 17-mer oligonucleotide primers fitting all known VT sequences described by Paton et al. (24). The protocol of these authors was used with one modification: the annealing temperature was increased from 46 to 56°C to prevent the appearance of nonspecific DNA fragments. Double-strand sequencing was performed by Eurogentec Bel, Seraing, Belgium, by using an ABI377-based fluorescent sequencing protocol. The resulting sequence was analyzed and compared to other VT sequences with GeneCompar software (Applied Maths, Kortrijk, Belgium). This sequence (VT2d-Ount) and two others that clustered in a distinct group within the VT2 variant sequences (see Results) will be referred to in this study as VT2d genes, also comprising VT2d-OX3a (23), and VT2d-O111 (24).
VT2 variant B subunit gene PCR-RFLP.
The VT genotypes harbored by the VTEC isolates were identified by separate PCRs with specific primers presented in Table 1. Primer VT2-cm, a modification of primer VT2-c, was designed in this study specifically to fit VT2d variant sequences in combination with primer VT2-f. Bacterial suspensions were used without pretreatment in a PCR mixture by denaturation for 5 min at 94°C and subsequently underwent 30 cycles of amplification with denaturation for 25 s at 94°C, annealing for 50 s at 55 or 45°C (for VT2e reactions only), and primer extension for 26 s at 72°C in a GeneAmp PCR system 9600 DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). The amplified DNA was analyzed by electrophoresis through a 2% agarose gel and visualized by ethidium bromide staining. Appropriate positive and negative controls were used in each run. The B subunit genes of strains positive with primers VT2-a and VT2-b—specific for VT2 and its human variants—were subtyped by Tyler’s PCR-RFLP method (38), which identifies VT2, VT2vh-a, and VT2vh-b. In addition, the fragment obtained with primers VT2-e and VT2-f, designed in this study to fit the sequences of VT2 and all human variants, was digested by enzymes HaeIII and PvuII to confirm the identification of VT2d genes and to identify sequences related to VT2d-OX3a and VT2d-Ount. The expected sizes of the restriction fragments obtained by PCR-RFLP are shown in Table 2.
TABLE 1.
Primers used for typing VT genes
| Primer pair | Sequence (5′ to 3′) | Gene specificity | Location within gene | Size of amplified product (bp) | Reference or source |
|---|---|---|---|---|---|
| VT1-a VT1-b | GAA GAG TCC GTG GGA TTA CG AGC GAT GCA GCT ATT AAT AA | VT1 | From nucleotide 860 of A-subunit-coding region to nucleotide 31 of B-subunit-coding region | 130 | Pollard et al. (30) |
| VT2-a VT2-b | TTA ACC ACA CCC ACG GCA GT GCT CTG GAT GCA TCT CTG GT | VT2, VT2vh-a, VT2vh-b, VT2d-Ount, VT2d-OX3a |
From nucleotide 188 to nucleotide 533 of A-subunit-coding region | 346 | Pollard et al. (30) |
| VT2-c VT2-d | AAG AAG ATG TTT ATG GCG GT CAC GAA TCA GGT TAT GCC TC | VT2, VT2vh-a, VT2vh-b | From nucleotide 4 of B subunit region to nucleotide 21 of upstream region | 285 | Tyler et al. (38) |
| VT2v-1 VT2v-2 | CAT TCA GAG TAA AAG TGG CC GGG TGC CTC CCG GTG AGT TC | VT2vh-a, VT2vh-b, VT2d-Ount, VT2d-OX3a | From nucleotide 110 of B-subunit-coding region to nucleotide 225 of upstream region | 385 | Tyler et al. (38) |
| VT2-e VT2-f | AAT ACA TTA TGG GAA AGT AAT A TAA ACT GCA CTT CAG CAA AT | VT2, VT2vh-a, VT2vh-b, VT2d-Ount, VT2d-OX3a |
From nucleotide 883 of A-subunit-coding region to nucleotide 259 of B-subunit-coding region | 348 | This study |
| VT2-cm VT2-f | AAG AAG ATA TTT GTA GCG G TAA ACT GCA CTT CAG CAA AT | VT2d-Ount, VT2d-OX3a | From nucleotide 4 to nucleotide 259 of B subunit region | 256 | This study |
| VTe-a VTe-b | CCT TAA CTA AAA GGA ATA TA CTG GTG GTG TAT GAT TAA TA | VT2e | From nucleotide −25 downstream to nucleotide 205 of A-subunit-coding region | 230 | Johnson et al. (9) |
TABLE 2.
Sizes of the restriction fragments used for RFLP analysis
| Primers used to amplify fragment | Restriction enzyme | Size(s) of fragment(s) in genes coding for B subunit (bp)
|
||||
|---|---|---|---|---|---|---|
| VT2 | VT2c
|
VT2d
|
||||
| VT2vh-a | VT2vh-b | VT2d-Ount | VT2d-OX3a | |||
| VT2-c, VT2-da | HaeIII | 285 | 161, 124 | 161, 124 | ||
| RsaI | 216, 69 | 136, 80 (69)b | 216, 69 | |||
| NciI | 285 | 285 | 159, 126 | |||
| VT2-e, VT2-f | HaeIII | 348 | 216, 132 | 216, 132 | 216, 132 | 167, 132 (49)b |
| PvuII | 323 (25)b | 323 (25)b | 250, 73 (25)b | 200, 120 (28)b | 200, 120 (28)b | |
This primer pair was used according to the method of Tyler et al. (38).
This fragment did not resolve or was too small to be clearly visible under the electrophoresis conditions used.
Accessory virulence factors.
E. coli hemolysins were detected as described before (27) by comparison of hemolysis on CaCl2-washed and unwashed blood agar. Three phenotypes were distinguished as follows: enterohemolysis, alpha-hemolysis, and absence of hemolysis. The eaeA gene was detected by PCR with AE 9 and AE 10 primers (5), and EHEC virulence plasmid sequences were detected with primers MFS1F and MFS1R (4).
Statistical tests.
Statistical analysis was performed by using Yates corrected chi-squared tests or, if an expected value was less than 5, the one-tailed Fisher exact test.
Nucleotide sequence accession number.
The sequence of the VT gene of strain EH250 (VT2d-Ount) has been submitted to the GenBank database under accession no. AF043627.
RESULTS
Sequence of the VT2 gene of strain EH250 and comparison with other VT sequences.
After elimination of the 17 nucleotides at the 5′ and 3′ ends corresponding to the sequences of the primers used to amplify the DNA fragment, a sequence of 1,470 bp (the VT2d-Ount sequence) was obtained. It contained two open reading frames (bp 177 to 1133 and 1149 to 1409) coding for polypeptides of 319 amino acids (putative A subunit) and 87 amino acids (putative B subunit), respectively. Comparison with VT2 sequences available in the GenBank and EMBL databases revealed that VT2d-Ount was more homologous to sequences sltII-OX3a (accession no. X65949) and sltII-O111 (accession no. L11078), called VT2d-OX3a and VT2d-O111, respectively, in this study. By alignment of the A-subunit-coding region of VT2d-Ount with VT2d-OX3a and VT2d-O111, 29- and 30-nucleotide differences, corresponding to 8- and 9-amino-acid changes, respectively, were identified. The B-subunit-coding regions of VT2d-Ount and VT2d-O111 were identical, while only one nucleotide difference was observed with the B-subunit-coding region of VT2d-OX3a, resulting in one amino acid change.
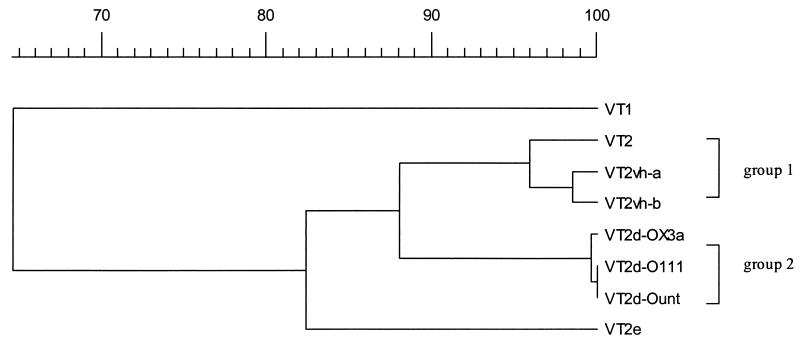
The homology between the sequence of VT2d-Ount and those of the other VT genes most representative of the A- and B-subunit-coding regions is shown in Fig. 1 and 2, respectively. For the A subunit, as expected, VT1 was clearly distant from all VT2 sequences (61.0 to 62.5% homology) and VT2e was 92.0 to 94.4% homologous to the other VT2 sequences. Two groups of highly homologous A subunit sequences were present: group 1 with VT2 and the two VT2c variants (VT2vh-a and VT2vh-b), which were 99.1 to 99.2% homologous to each other; and group 2 with the three VT2d variants, which were 96.9 to 99.9% homologous to each other. The individual sequences of these two groups were 93.4 to 96.0% homologous to the sequences in the other group. For the B subunit sequences, again as expected, VT1 was very distant from the VT2 sequences (64.0 to 65.1% computed homology) and VT2e was only 80.1 to 84.7% homologous to the other VT2 sequences. As for the A subunit, the sequences of the different VT2c and VT2d variants formed two distinct groups, but in contrast to the A subunits, the homology was higher within group 2 (99.6 to 100%) than within group 1 (95.9 to 98.5%). The individual sequences of these two groups were 86.2 to 89.3% homologous to the sequences in the other group. Other published VT2 variant sequences (12, 15, 24, 31) are not shown in the figures but were all related to group 1 for both the A and the B subunits. By Tyler’s PCR-RFLP method they would be assigned to subgroups VT2, VT2vh-a, and VT2vh-b.
FIG. 1.
Dendritic representation of A subunit nucleotide sequence homology. The nucleotide sequence of the A-subunit-coding region of the following genes were compared: VT1 (nucleotide sequence accession no. M16625) (2), VT2 (nucleotide sequence accession no. X07865) (7), VT2vh-a and VT2vh-b (not present in databases) (6), VT2e (nucleotide sequence accession no. M21534) (39), VT2d-OX3a (nucleotide sequence accession no. X65949) (23), VT2d-O11 (nucleotide sequence accession no. L11078) (24), and VT2d-Ount (nucleotide sequence accession no. AF043627) (this study).
FIG. 2.
Dendritic representation of B subunit nucleotide sequence homology. See the legend for Fig. 1 for nucleotide sequence accession number for the sequences containing the B-subunit-protein-coding region.
Detection of VT2d sequences in VTEC isolates.
To include the sequences mentioned above, the PCR-RFLP verocytotoxin B subunit identification method of Tyler et al. (38) was extended as described in Materials and Methods (Tables 1 and 2) by the addition of three primers combined into two pairs. Three hundred forty-four of the 359 VT-producing isolates analyzed in this study could be assigned to a VT genotype or to a combination of VT genotypes, as shown in Table 3. RFLP results for the last 15 isolates did not fit the patterns shown in Table 2 and are referred as VT2vh atypical. New VT2d variants were identified in all 64 strains with previously untypeable genes, in addition to one strain in which VT2d-Ount was present in combination with VT2vh-a. None of the strains previously shown to harbor only a VT1 or VT2e gene was positive for the new VT2 variants.
TABLE 3.
Results of VT genotyping
| VT genotype | Total no. | No. and type of isolate in indicated group
|
|||
|---|---|---|---|---|---|
| Serogroup O157
|
Other O serogroups
|
||||
| Human | Animal | Human | Animal | ||
| VT1 | 118 | 0 | 0 | 95 | 23 |
| VT2 | 42 | 16 | 0 | 14 | 12 |
| VT2vh-a | 33 | 27 | 1 | 4 | 1 |
| VT2vh-b | 9 | 0 | 0 | 5 | 4 |
| VT2vh atypicala | 15 | 0 | 0 | 12 | 3 |
| VT2 + VT2vh-a | 32 | 28 | 0 | 3 | 1 |
| VT2 + VT2vh-b | 6 | 0 | 0 | 1 | 5 |
| VT2d-Ount | 23 | 0 | 0 | 15 | 8 |
| VT2d-OX3a | 3 | 0 | 0 | 1 | 2 |
| VT2vh-a + VT2d-Ount | 1 | 0 | 0 | 0 | 1 |
| VT1 + VT2 | 5 | 3 | 0 | 1 | 1 |
| VT1 + VT2vh-a | 21 | 20 | 1 | 0 | 0 |
| VT1 + VT2vh-b | 4 | 0 | 0 | 1 | 3 |
| VT1 + VT2vh-atypicala | 1 | 0 | 0 | 1 | 0 |
| VT1 + VT2d-Ount | 38 | 0 | 0 | 28 | 10 |
| VT1 + VT2 + VT2vh-a | 1 | 1 | 0 | 0 | 0 |
| VT1 + VT2 + VT2vh-b | 1 | 0 | 0 | 1 | 0 |
| VT2e | 6 | 0 | 0 | 4 | 2 |
| Total | 359 | 95 | 2 | 186 | 76 |
Positive with VT2v1 and VT2v-2 primers and negative with VT2-cm and VT2-f primers, but restriction of amplicons obtained with VT2-c and VT2-d primers and with VT2-e and VT2-f primers does not fit predicted RFLP patterns.
Correlation between the presence of VT2d sequences and other virulence factors.
All 65 strains with sequences that were identified as VT2d by the present PCR-RFLP method, alone or in combination with other VT genes, belonged to serogroups other than O157 and were negative for the eaeA gene (Table 3), while 10 (15%) were positive by PCR for the sequences of the EHEC virulence plasmid and 22 (34%) were enterohemolytic. The correlation between the presence of accessory virulence factors and the presence of VT2d sequences in non-O157 VT2-producing isolates is shown in Table 4. An association was found between the presence of a VT2d gene and the absence of the eaeA gene or the EHEC plasmid in human isolates but not in animal isolates. There was also a negative association between the presence of a VT2d gene and EHEC-associated clinical symptoms in 83 of the 90 patients with non-O157 isolates for whom clinical data were available. Twenty-six of 45 VT2-producing isolates (58%) that did not harbor VT2d genes were from patients with either uncomplicated diarrhea (20 patients) or HUS (6 patients), while only 11 of the 38 VT2d-producing isolates (29%) were from patients with uncomplicated diarrhea and none were from patients with HUS (χ2 = 19.7, P = 0.0000093). When only HUS was considered, this difference was still statistically significant (Fisher’s exact test, P = 0.022).
TABLE 4.
Accessory virulence factors in non-O157 isolates positive for any VT2 gene included in this study
| Origin of isolates | Virulence factor | No. of isolates positive for virulence factor/no. of isolates tested (%)
|
χ2 | P | |
|---|---|---|---|---|---|
| With VT2d gene | Without VT2d gene | ||||
| Human | eaeA | 0/44 (0) | 16/46 (35) | 16.31 | 0.000054 |
| EHEC plasmid | 8/44 (18) | 21/46 (46) | 6.56 | 0.010 | |
| Enterohemolysin | 16/44 (36) | 21/46 (46) | 0.46 | NSa | |
| Animal | eaeA | 0/21 (0) | 1/32 (3) | NAb | NS |
| EHEC plasmid | 2/21 (10) | 6/32 (19) | NA | NS | |
| Enterohemolysin | 6/21 (29) | 10/32 (31) | NA | NS | |
NS, not significant.
NA, not applicable; the expected value was less than 5. The Fisher exact test was used to determine the P value.
Strains of neither serogroup O157 (97 isolates) nor other more-pathogenic serogroups such as O26 (20 isolates), O103 (15 isolates), O111 (13 isolates), and O145 (8 isolates) were positive for VT2d sequences.
DISCUSSION
There are no or only a few nucleotide differences between the VT1 genes that were sequenced from different wild-type VTEC strains and the ST gene of Shigella dysenteriae type 1, resulting in toxins that are identical or differ by only a few amino acids (22, 33, 34). However, sequences of toxins of the VT2 class, found in E. coli and in Citrobacter freundii and a few Enterobacter spp. (21, 37), exhibit important natural variation, as shown in Fig. 1 and 2, leading to a number of variant toxins that were classified as VT2, VT2c, and VT2e (1). LeClerc et al. (11) demonstrated high mutation frequencies in O157 EHEC, a factor that could explain the emergence of multiple VT2 genes but that does not explain why VT1 genes are much less variable.
Sequencing of the VT2 gene of strain EH250, one of the 64 strains that could not be typed by Tyler’s method due to lack of amplification with primers VT2c and VT2-d, showed that it was very homologous to two VT2 variant sequences reported by Paton et al. (23, 24). After Tyler’s RFLP-PCR method was extended by the addition of two new primer pairs, all 64 strains with previously untypeable VT genes proved to possess one of the new variant genes, VT2d-Ount or VT2d-OX3a, and one isolate was already positive for VT2vh-a. In addition, the VT2 genes of 15 other strains were atypical: the RFLP patterns of the amplicons obtained with primers VT2-c and VT2-d and primers VT2-e and VT2-f did not fit the predicted patterns. These atypical genes seem to more closely resemble VT2c variants, since amplification was obtained with primers VT2-v1 and VT2-v2 and not with primers VT2-cm and VT2-f. This result confirms that sequence variation in VT2 genes is frequent.
Some data suggest that toxin type could be important in determining the probability of developing HUS. Animal studies have shown that naturally occurring VT2 sequence variation may have a direct impact on the capacity of a given VTEC to cause disease (20). In humans, strains producing VT2 class toxins resulted in HUS more frequently than did VT1 producers (19, 25). However, the VT2d variant toxins we identified in many of our strains are probably less pathogenic for humans. The two VT2d-related variant toxins described by Paton et al. (23, 24) showed a low cytotoxicity to Vero cells, as did the isolate we sequenced. Paton et al. showed also that VT2d-O111 was associated with the lowest oral virulence for streptomycin-treated mice, although VT2d-OX3a showed an oral virulence that was almost as high as that of other VT2 toxins (20). It must also be noted that the new VT2 variant genes were not detected in O157 or the other most-virulent VTEC serotypes, O26, O103, O111, and O145. We compared clinical symptoms in patients with strains harboring the new VT2 variants (all non-O157) to those in patients with strains harboring other non-O157 VT2-positive strains and not with those in patients with non-O157 strains producing only VT1, since the latter were previously shown to harbor accessory virulence factors more frequently (27). VT2d-positive isolates were less frequently associated with diarrhea and HUS. This lower pathogenicity could be due not only to the lower cytotoxicity of these toxins but also to the lack of accessory virulence factors in VT2d-producing strains. The presence of VT2d sequences could indeed be only a marker for less-pathogenic strains. In any case, the absence of known accessory virulence factors should be interpreted cautiously, since it has been shown that alternative virulence factors can be present in some fully pathogenic strains, such as O113:H21, which possesses distinct binding properties (3), or O111:H2, which is enteroaggregative (16).
The extended PCR-RFLP typing method described in this study could be used to identify some less-virulent VTEC isolates. It might be useful as well for subtyping non-O157 strains in epidemiological studies, as has been shown for VT genotyping of O157 strains (36).
ACKNOWLEDGMENTS
This work was partially supported by European Project Community Biomed project contract no. BMH4-CT96-0970 (DG 12 - SSMA) and OZR grant no. 235 (Vrije Universiteit Brussel, Brussels, Belgium).
REFERENCES
- 1.Calderwood S B, Acheson D W, Keusch G T, Barrett T J, Griffin P M, Strockbine N A, Swaminathan B, Kaper J B, Levine M M, Kaplan B S, Karch H, O’Brien A D, Obrig T G, Takeda Y, Tarr P I, Wachsmuth I K. Proposed new nomenclature for SLT (VT) family. ASM News. 1996;62:118–119. [Google Scholar]
- 2.Calderwood S B, Auclair F, Donohue Rolfe A, Keusch G T, Mekalanos J J. Nucleotide sequence of the Shiga-like toxin genes of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:4364–4368. doi: 10.1073/pnas.84.13.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dytoc M T, Ismaili A, Philpott D J, Soni R, Brunton J L, Sherman P M. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect Immun. 1994;62:3494–3505. doi: 10.1128/iai.62.8.3494-3505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fratamico P M, Sackitey S K, Wiedmann M, Deng M Y. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33:2188–2191. doi: 10.1128/jcm.33.8.2188-2191.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gannon V P J, Rashed M, King R K, Thomas E J G. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli using polymerase chain reaction. J Clin Microbiol. 1993;31:1268–1274. doi: 10.1128/jcm.31.5.1268-1274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito H, Terai A, Kurazono H, Takeda Y, Nishibuchi M. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb Pathog. 1990;8:47–60. doi: 10.1016/0882-4010(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 7.Jackson M P, Neill M A, O’Brien A D, Holmes R K, Newland J W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol Lett. 1987;44:109–114. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 8.Jackson M P, Newland J W, Holmes R K, O’Brien A D. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb Pathog. 1987;2:147–153. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 9.Johnson W M, Pollard D R, Lior H, Tyler S D, Rozee K R. Differentiation of genes coding for Escherichia coli verotoxin 2 and the verotoxin associated with porcine edema disease (VTe) by the polymerase chain reaction. J Clin Microbiol. 1990;28:2351–2353. doi: 10.1128/jcm.28.10.2351-2353.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karmali M A, Lingwood C A, Petric M, Brunton J, Gyles C. Maintaining the existing phenotype nomenclatures for E. coli cytotoxins. ASM News. 1996;62:167–169. [Google Scholar]
- 11.LeClerc J E, Li B, Payne W L, Cebula T A. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science. 1996;274:1208–1211. doi: 10.1126/science.274.5290.1208. [DOI] [PubMed] [Google Scholar]
- 12.Lin Z, Yamasaki S, Kurazono H, Ohmura M, Karasawa T, Inoue T, Sakamoto S, Suganami T, Takeoka T, Taniguchi Y, et al. Cloning and sequencing of two new Verotoxin 2 variant genes of Escherichia coli isolated from cases of human and bovine diarrhea. Microbiol Immunol. 1993;37:451–459. doi: 10.1111/j.1348-0421.1993.tb03236.x. [DOI] [PubMed] [Google Scholar]
- 13.Lior H. Classification of Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Oxon, United Kingdom: Cab International; 1994. pp. 31–72. [Google Scholar]
- 14.Marques L R M, Peiris J S M, Cryz S J, O’Brien A D. Escherichia coli strains isolated from pigs with edema disease produce a variant of Shiga-like Toxin II. FEMS Microbiol Lett. 1997;44:33–38. [Google Scholar]
- 15.Meyer T, Karch H, Hacker J, Bocklage H, Heesemann J. Cloning and sequencing of a Shiga-like toxin II-related gene from Escherichia coli O157:H7 strain 7279. Int J Med Microbiol Virol Parasitol Infect Dis. 1992;276:176–188. doi: 10.1016/s0934-8840(11)80004-6. [DOI] [PubMed] [Google Scholar]
- 16.Morabito S, Karch H, Mariani-Kurkdjian P, Schmidt H, Minelli F, Bingen E, Caprioli A. Enteroaggregative, Shiga toxin-producing Escherichia coli O111:H2 associated with an outbreak of hemolytic-uremic syndrome. J Clin Microbiol. 1998;36:840–842. doi: 10.1128/jcm.36.3.840-842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oku Y, Yutsudo T, Hirayama T, O’Brien A D, Takeda Y. Purification and some properties of a Vero toxin from a human strain of Escherichia coli that is immunologically related to Shiga-like toxin II (VT2) Microb Pathog. 1989;6:113–122. doi: 10.1016/0882-4010(89)90014-4. [DOI] [PubMed] [Google Scholar]
- 19.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargrett Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 20.Paton A W, Bourne A J, Manning P A, Paton J C. Comparative toxicity and virulence of Escherichia coli clones expressing variant and chimeric Shiga-like toxin type II operons. Infect Immun. 1995;63:2450–2458. doi: 10.1128/iai.63.7.2450-2458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paton A W, Paton J C. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J Clin Microbiol. 1996;34:463–465. doi: 10.1128/jcm.34.2.463-465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton A W, Paton J C, Goldwater P N, Heuzenroeder M W, Manning P A. Sequence of a variant Shiga-like toxin type-I operon of Escherichia coli O111:H−. Gene. 1993;129:87–92. doi: 10.1016/0378-1119(93)90700-d. [DOI] [PubMed] [Google Scholar]
- 23.Paton A W, Paton J C, Heuzenroeder M W, Goldwater P N, Manning P A. Cloning and nucleotide sequence of a variant Shiga-like toxin II gene from Escherichia coli OX3:H21 isolated from a case of sudden infant death syndrome. Microb Pathog. 1992;13:225–236. doi: 10.1016/0882-4010(92)90023-h. [DOI] [PubMed] [Google Scholar]
- 24.Paton A W, Paton J C, Manning P A. Polymerase chain reaction amplification, cloning and sequencing of variant Escherichia coli Shiga-like toxin type II operons. Microb Pathog. 1993;15:77–82. doi: 10.1006/mpat.1993.1058. [DOI] [PubMed] [Google Scholar]
- 25.Piérard, D., G. Cornu, W. Proesmans, A. Dediste, F. Jacobs, J. Van de Walle, A. Mertens, J. Ramet, and S. Lauwers. Hemolytic uremic syndrome in Belgium: incidence and association with verocytotoxin-producing Escherichia coli infection. Clin. Microbiol. Infect., in press. [DOI] [PubMed]
- 26.Piérard D, Huyghens L, Lauwers S, Lior H. Diarrhoea associated with Escherichia coli producing porcine oedema disease verotoxin. Lancet. 1991;338:762. doi: 10.1016/0140-6736(91)91487-f. [DOI] [PubMed] [Google Scholar]
- 27.Piérard D, Stevens D, Moriau L, Lior H, Lauwers S. Isolation and virulence factors of verocytotoxin-producing Escherichia coli in human stool samples. Clin Microbiol Infect. 1997;3:531–540. doi: 10.1111/j.1469-0691.1997.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 28.Piérard D, Van Damme L, Moriau L, Stevens D, Lauwers S. Virulence factors of verocytotoxin-producing Escherichia coli isolated from raw meats. Appl Environ Microbiol. 1997;63:4585–4587. doi: 10.1128/aem.63.11.4585-4587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piérard D, Van Etterijck R, Breynaert J, Moriau L, Lauwers S. Results of screening for verocytotoxin-producing Escherichia coli in faeces in Belgium. Eur J Clin Microbiol Infect Dis. 1990;9:198–201. doi: 10.1007/BF01963837. [DOI] [PubMed] [Google Scholar]
- 30.Pollard D R, Johnson W M, Lior H, Tyler S D, Rozée K R. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J Clin Microbiol. 1990;28:540–545. doi: 10.1128/jcm.28.3.540-545.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt C K, McKee M L, O’Brien A D. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scotland S M, Smith H R. Vero cytotoxins. In: Sussman M, editor. Escherichia coli: mechanisms of virulence. Cambridge, United Kingdom: Cambridge University Press; 1997. pp. 257–280. [Google Scholar]
- 33.Strockbine N A, Jackson M P, Sung L M, Holmes R K, O’Brien A D. Cloning and sequencing of the genes for Shiga toxin from Shigella dysenteriae type 1. J Bacteriol. 1988;170:1116–1122. doi: 10.1128/jb.170.3.1116-1122.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takao T, Tanabe T, Hong Y M, Shimonishi Y, Kurazono H, Yutsudo T, Sasakawa C, Yoshikawa M, Takeda Y. Identity of molecular structure of Shiga-like toxin I (VT1) from Escherichia coli O157:H7 with that of Shiga toxin. Microb Pathog. 1988;5:57–69. doi: 10.1016/0882-4010(88)90036-8. [DOI] [PubMed] [Google Scholar]
- 35.Thomas A, Cheasty T, Chart H, Rowe B. Isolation of Vero cytotoxin-producing Escherichia coli serotypes O9ab:H- and O101:H-carrying VT2 variant gene sequences from a patient with haemolytic uraemic syndrome. Eur J Clin Microbiol Infect Dis. 1994;13:1074–1076. doi: 10.1007/BF02111832. [DOI] [PubMed] [Google Scholar]
- 36.Thomas A, Smith H R, Rowe B. Use of digoxigenin-labelled oligonucleotide DNA probes for VT2 and VT2 human variant genes to differentiate Vero cytotoxin-producing Escherichia coli strains of serogroup O157. J Clin Microbiol. 1993;31:1700–1703. doi: 10.1128/jcm.31.7.1700-1703.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tschäpe H, Prager R, Streckel W, Fruth A, Tietze E, Bohme G. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: green butter as the infection source. Epidemiol Infect. 1995;114:441–450. doi: 10.1017/s0950268800052158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyler S D, Johnson W M, Lior H, Wang G, Rozee K R. Identification of verotoxin type 2 variant B subunit genes in Escherichia coli by the polymerase chain reaction and restriction fragment length polymorphism analysis. J Clin Microbiol. 1991;29:1339–1343. doi: 10.1128/jcm.29.7.1339-1343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinstein D L, Jackson M P, Samuel J E, Holmes R K, O’Brien A D. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J Bacteriol. 1988;170:4223–4230. doi: 10.1128/jb.170.9.4223-4230.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]