Abstract
Adenoviruses (AdV) cause diseases that range from localized, self-limited illnesses to fatal infections in immunocompromised patients. Culture is assumed to be sensitive but requires viable virus and up to 3 weeks for detection, and it can be inhibited by bacterial contamination. A new PCR method amplifying a region of the hexon gene was developed in order to detect AdV in urine more rapidly and with greater sensitivity than obtainable by culture technology. All 18 serotypes tested were detected. Quantitatively, with optimized urine processing, AdV PCR detected 0.2 PFU/ml (serotype 11) and 10 DNA copies/ml (serotype 2). Serially collected urine samples from human immunodeficiency virus (HIV)-infected patients with concurrent cytomegalovirus retinitis were divided into three groups: AdV culture-positive samples, AdV culture-negative or bacterially contaminated samples from patients with a history of AdV culture-positive urines, and AdV culture-negative samples from patients without a history of AdV culture positivity. Urine samples from healthy adults were also tested by culture and PCR to screen for asymptomatic shedding. Amplification was assessed with and without prior DNA purification. AdV was detected by PCR in 90% of culture-positive urines (100% of unclotted samples, e.g., those culture positive after storage for PCR testing), 71% of culture-negative or bacterially contaminated urines from AdV-infected patients, and 28% from AdV culture-negative patients. Healthy volunteers were culture negative for AdV, and 96% were PCR negative. The new AdV PCR method is rapid and sensitive and can detect viral DNA in samples for which culturing is problematic. The role of AdV replication during HIV infection merits further investigation with sensitive tools such as PCR.
Adenoviruses (AdV) are a significant cause of morbidity and can cause mortality in all age groups. AdV infect and replicate in many cell types and body sites, including the respiratory tract, eye, gastrointestinal tract, urinary tract, and liver. Diseases associated with AdV include epidemic keratoconjunctivitis, pharyngitis, pertussis-like syndrome, pneumonia, acute hemorrhagic cystitis, gastroenteritis, and hepatitis (5).
In immunocompromised patients, AdV-associated case fatality rates have been reported to be as high as 60% for patients with pneumonia and 50% for those with hepatitis, compared with fatality rates of 15% for pneumonia and 10% for hepatitis in immunocompetent patients (6). Fatal disseminated AdV infections have been reported in patients with agammaglobulinemia (13); in bone marrow, renal, and liver transplant recipients (6); and in patients with other immunosuppressive diseases or immunosuppressive therapies (15). On occasion, AdV have been detected in urine prior to dissemination, as occurred in one of our patients, a bone marrow transplant recipient who subsequently died with evidence of disseminated disease (3).
In AIDS patients, organ and tissue distribution of AdV is similar to that in other immunocompromised patients, but the infection rate appears to be higher during AIDS (4, 11, 12). De Jong et al. reported the isolation of AdV from the urine of up to 20% of AIDS patients, even in the absence of evidence of bladder inflammation or bleeding (2).
Current methods for diagnosis of AdV infections have limitations. Culture may be prolonged, electron and immunofluorescence microscopy are relatively insensitive, and assays for antibodies to AdV may yield reactive results due to prior infection (7). False-negative results also occur, especially during late-stage human immunodeficiency virus (HIV) infection. The goals of our study were to develop and optimize a highly sensitive PCR assay for detecting a wide range of AdV types in urine by comparing different methods for DNA preparation and different sets of primers. The method was compared with culture in studies of AdV shedding in urine from AIDS patients and tested for specificity in healthy adults.
MATERIALS AND METHODS
Microbiologic sensitivity and specificity.
To assess microbiologic sensitivity, purified DNA and plaque-quantified virus from two different serotypes were used. Purified AdV serotype 2 DNA was generously provided by Gary Ketner, Johns Hopkins University School of Hygiene and Public Health. AdV serotype 11 was prepared from infected cell lysates obtained from the American Type Culture Collection (ATCC) (Manassas, Va.). The virus was quantified for PFU per milliliter in A549 human lung carcinoma cells, and aliquots of the propagated virus were stored at −70°C. To test the detectability of other AdV, reference serotypes 5, 7, 8, 11, 19, 34, and 35 were purchased from ATCC. Additional AdV isolates including serotypes 1, 2, 3, 4, 7A, 11, 12, 16, 21, 30, 37, 48, and 49, recovered from clinical specimens, were also tested. The types tested were those recovered most frequently from urine samples. We also tested one or more representative isolates from AdV serogroups A, B, C, D, and E. Subsequent to PCR assay, specimens that had been culture positive for AdV prior to storage were recultured to assess the stability of the virus upon storage of the urine sample.
To assess microbiologic specificity, BK virus (BKV), cytomegalovirus (CMV), varicella-zoster virus (VZV), herpes simplex virus type 1 (HSV-1), and HSV-2 (all strains from ATCC); Epstein-Barr virus (EBV) (Sigma, St. Louis, Mo.); and parvovirus B19 (a clinical isolate) were also tested for DNA amplification. Bacteria frequently associated with urinary tract infections were seeded into urine at concentrations of 104–106 CFU/ml. Species tested were Pseudomonas aeruginosa, Staphylococcus aureus, Enterococcus faecalis, and Escherichia coli (all ATCC strains), as well as Proteus vulgaris (a clinical isolate).
In addition, urine samples known to contain BKV, JC virus (JCV), E. coli, E. faecalis, Corynebacterium spp., S. aureus, Proteus mirabilis, P. aeruginosa, and Candida albicans and a sample containing both Citrobacter freundii and Morganella morganii were used to determine the specificity of the AdV PCR primers.
Clinical specimens.
As part of an AIDS-CMV retinitis study (8), 151 HIV-infected patients were enrolled between 1993 and 1997. Urine samples from these patients were cultured for CMV prior to treatment with anti-CMV drugs, at 1 and 3 months after treatment was initiated, and then every 3 months thereafter.
To assess AdV shedding from these AIDS patients, we tested a subset of these urines by PCR. Samples were divided into three groups (see Table 2): group A included 21 AdV culture-positive samples from 14 patients; group B included 2 AdV culture-negative samples and 5 samples that were uninterpretable because of bacterial contamination from patients who had had an AdV culture-positive urine within 2 months of the negative/contaminated sample; and group C included 25 samples, each randomly selected from AIDS patients who had never demonstrated a positive AdV culture. In addition, to assess AdV shedding from healthy adults, urine samples from 23 healthy volunteers (group D) were assayed for AdV by culture and PCR.
TABLE 2.
Detection of AdV in urine by culture and PCR
| Source | Evidence for replicating AdV | Specimen group | No. (%) of specimens
|
|||
|---|---|---|---|---|---|---|
| Total | Culture positive | PCR positive, with DNA purified bya:
|
||||
| Column | Resin | |||||
| AIDS patients | Current | A | 21 | 21 | 19 (90)b | 19b |
| Recentc | B | 7 | 0 | 5 (71) | 5 | |
| Current or recent | A + B | 28 | 21 (75) | 24 (86)b | 24b | |
| None | C | 25 | 0 | 7 (28)d | 2 (8)d | |
| Healthy volunteers | None | D | 23 | 0 | 1 (4) | Not done |
DNA purified from specimens by using QIAamp blood kit column or Chelex resin.
Two samples containing clotted blood, PCR negative by both methods, had deteriorated on storage.
Within 2 months of culture-positive sample.
Total of nine samples were PCR positive.
Sample processing for viral culture.
Clinical urine samples from groups A, B, C, and D were processed within 60 min of collection according to our standard laboratory protocol for viral culture. Briefly, 10 ml of urine was centrifuged at 3,650 × g at 4°C for 30 min. All but 0.5 ml of supernatant and sediment was discarded. One milliliter of viral transport medium (Eagle minimum essential medium with 5% [vol/vol] fetal bovine serum, 50 μg of gentamicin per ml, and 5 μg of amphotericin B per ml) was added to the sediment, and the pH was adjusted to 7.0 with 7.5% (wt/vol) sodium bicarbonate. Culture was performed by inoculating 0.2 ml of the processed sediment into tubes of MRC-5, WI-38, and human foreskin fibroblast cells. The remaining processed sediments were stored in aliquots at −70°C until PCR was performed. Cultures were observed for cytopathologic effect daily for 6 weeks; indirect immunofluorescence with an anti-AdV monoclonal antibody (Bartels Inc., Issaquah, Wash.) was used for confirmation of AdV-specific cytopathic effect.
DNA purification.
Nine approaches to specimen preparation were compared to optimize purification of viral DNA and to remove inhibitors present in urine. These included dilution of the sample in water, Slide-A-Lyser dialysis cassettes (Pierce, Rockford, Ill.), phenol-chloroform extraction, high-speed centrifugation (40,000 × g for 3 or 6 h), chelating ion-exchange resin (Chelex 100; Bio-Rad, Hercules, Calif.), Ultrafree-MC filters (Millipore, Bedford, Mass.), Centricon-100 concentrators (Amicon, Beverly, Mass.), QIAamp blood kit columns (Qiagen, Valencia, Calif.), and QIAamp viral RNA kit columns (Qiagen). Each approach was studied by using urine specimens seeded with serial dilutions of AdV serotype 11 in the presence of components that might interfere with purification or amplification, including hemoglobin, leukocytes, urate crystals, and bacteria.
Two products, Chelex 100 (resin) and QIAamp blood kit (column), were selected for further study because they yielded the highest sensitivity, proved superior for removing inhibitors, and were technically easy to perform. For the resin method, 50 μl of processed sediment was added to 200 μl of 5% (wt/vol) Chelex 100; the mixture was then incubated at 55°C for 15 min, vortexed, boiled for 8 min, and spun for 2 min at 14,000 × g.
For the column method, 200 μl of processed sediment or urine was used. The manufacturer’s instructions were followed except that DNA was eluted with 50 μl of buffer rather than the 200 μl recommended by the manufacturer. We chose 50 μl because it matched the elution volume for the QIAamp RNA kit and proved satisfactory for our assay.
Primers.
Two sets of primers were compared (Table 1): those described by Allard et al. (1), which amplify a 300-bp region of the highly conserved hexon gene (set I), and those described by Yeo et al. (14), which amplify a different, 139-bp region of the hexon gene (set II).
TABLE 1.
Oligonucleotides for PCR amplification and hybridization detection of AdV
| Type (reference) | Designation | Sequence | Amplicon length (bp) |
|---|---|---|---|
| Primer set I (1) | hexAA1885 | 5′-GCCGCAGTGGTCTTACATGCACATC-3′ | 308 |
| hexAA1913 | 5′-CAGCACGCCGCGGATGTCAAAGT-3′ | ||
| Primer set II (14) | Hex 3 | 5′-GACATGACTTTCGAGGTCGATCCCATGGA-3′ | 139 |
| Hex 4 | 5′-CCGGCTGAGAAGGGTGTGCGCAGGTA-3′ | ||
| Probe | Hex 30 | 5′-GACCCCACCCTTCTTTATGTTCTGT-3′ |
Amplification.
For clinical specimens, 10 μl of resin- or column-extracted processed sediment or 2 μl of unextracted processed sediment was added to 40 μl of master mix (final concentrations of 10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 200 μM [each] dNTP, 0.2 μM [each] primer, and 2.5 U of Taq polymerase [Perkin-Elmer, Branchburg, N.J.]), and each reaction mixture was adjusted with water to a final volume of 50 μl. For other specimens (controls and those used for microbiologic specificity studies), 10 μl of the column-extracted sample was added to 40 μl of master mix. Amplification in a thermocycler (Perkin-Elmer 9600) consisted of an initial round at 94°C for 7 min, 55°C for 1 min, and 72°C for 1.5 min, followed by 40 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min.
Detection of amplified products.
The products were separated by electrophoresis in a 4% (wt/vol) NuSieve agarose gel, stained with ethidium bromide, and photographed with UV transillumination and an Eagle Eye II detection system that employs a charge-coupled device (CCD) camera for image intensity enhancement (Strategene, La Jolla, Calif.). Southern blot hybridization was performed on column-extracted samples from groups C and D and on samples used for microbiologic sensitivity and specificity tests.
Five potential probes were designed and compared for the ability to detect amplified DNA of many AdV serotypes. Probe Hex 30, displayed in Table 1, was selected because of its sensitivity and its ability to detect all serotypes tested. End labeling of probe Hex 30 was performed with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) for 30 min at 37°C. After prehybridization at 42°C for at least 30 min in 3× SSPE (1× SSPE is 0.18 M NaCl, 0.01 M NaOH, 0.01 M sodium phosphate, and 1 mM EDTA [pH 7.4]), 5× Denhardt’s solution (0.1% [wt/vol] Ficoll [type 400], 0.1% [wt/vol] polyvinylpyrrolidine, and 0.1% [wt/vol] bovine serum albumin), 0.5% (wt/vol) sodium dodecyl sulfate, and 0.25 mg of yeast tRNA per ml, 50 ng of labeled probe was added to hybridize at 42°C for 1 h, followed by two washes at room temperature with 6× SSPE and two additional washes at 62°C with 6× SSPE. Signal was detected by exposure to film after autoradiography at −70°C for approximately 16 h.
RESULTS
Amplification.
Both primer sets (I and II) amplified AdV serotypes 1, 2, 3, 4, 5, 7, 7A, 8, 11, 19, 21, 30, 34, 35, and 37. However, we determined that set I would not amplify DNA from isolates of AdV serotype 11, recovered from two different patients, regardless of the modifications employed to achieve amplification. One of those isolates was genotype “c” (restriction fragment length polymorphism-based genotype kindly determined by Adriana Kajon, University of Georgia, Athens), which was recovered from urine, bronchoalveolar lavage fluid, and conjunctivae of a bone marrow transplant recipient who had disseminated AdV infection (3). In contrast, set II amplified AdV DNA from every clinical isolate tested including type 11, which is one of the most common types found in urine and is associated with hemorrhagic cystitis. Serotypes 12, 16, 48, and 49, tested only with set 2, also amplified. Set II was therefore selected for use in this study.
Microbiologic sensitivity and specificity.
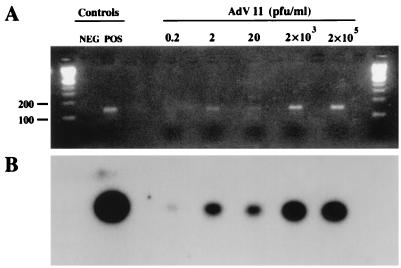
When purified AdV serotype 2 DNA was used as the target, the limits of detection were 102 copies/ml by agarose gel electrophoresis with ethidium bromide staining and 101 copies/ml by Southern blot hybridization. For plaque-quantified AdV serotype 11 seeded into urine, the sensitivity was 2 PFU/ml by gel (Fig. 1A) and 0.2 PFU/ml by hybridization (Fig. 1B). A specific product of 139 bp was not amplified from DNA from BKV, CMV, VZV, HSV-1, HSV-2, parvovirus B19, EBV, P. aeruginosa, S. aureus, E. faecalis, E. coli, or P. vulgaris or from urine samples containing BKV, JCV, E. coli, E. faecalis, Corynebacterium spp., S. aureus, P. mirabilis, P. aeruginosa, C. albicans, or a sample with both C. freundii and M. morganii.
FIG. 1.
Detection by PCR of AdV serotype 11 serially diluted in urine. AdV serotype 11 was propagated and quantified in A549 cells (see Materials and Methods). Serial 100- and 10-fold dilutions of this virus were made in urine from a healthy volunteer (AdV negative by culture and PCR). DNA from each dilution was column purified and then amplified by PCR. Controls for amplification were water (NEG) and 102 copies of AdV serotype 2 purified DNA (POS). (A) CCD-enhanced photography of an ethidium bromide-stained gel. (B) Autoradiogram of DNA in the gel from panel A after Southern blot transfer and hybridization with probe Hex 30 (20-h exposure).
Detection of AdV in urine from AIDS patients.
Among specimens from 151 patients in the CMV retinitis study, 21 samples from 14 patients (9%) were culture positive for AdV. The mean time to isolation of AdV was 24 days, a period longer than expected, probably resulting from cells that were suboptimal for AdV recovery. Samples from groups A, B, and C were tested for AdV DNA by PCR with ethidium bromide staining in agarose gels without Southern blot hybridization (Table 2). The 21 urine samples from group A were tested by PCR without extraction and with resin or column extraction methods. Fifteen (71%) were positive without extraction. In contrast, 19 (90%) were positive by either extraction method; the 2 PCR-negative urines were unique in two regards: they contained clotted blood and, when recultured after storage, they were the only isolates that failed to grow at the time of PCR testing. Among seven specimens from group B, five (71%) were PCR positive with either extraction method. Of the 25 group C samples, 7 (28%) were positive by PCR when column extraction was used; when the resin method was used, 2 different specimens yielded positive results.
Southern blot hybridization was performed to determine if a postamplification step would increase clinical sensitivity and specificity. Hybridization was performed with column-extracted samples from groups C and D. Group A and B specimens had been exhausted before probe Hex 30 was developed. When the results were compared with those from CCD-enhanced photography of ethidium bromide-stained agarose gels, no additional positive specimens were detected.
Detection of AdV in urine from healthy volunteers.
All group D samples were AdV culture negative, and 22 of 23 (96%) were negative by PCR with hybridization. The single PCR-positive sample was from a pregnant women who had sustained severe, AdV culture-positive bilateral conjunctivitis 3 weeks prior to urine collection.
DISCUSSION
Culture has been considered the “gold standard” for laboratory diagnosis of AdV; however, detectable replication typically requires 3 days to 3 weeks, varying with the specimen source and with the concentration of virus in the specimen. Furthermore, culture requires viable virus. AdV can be inactivated by inadequate collection methods, inadequate transport medium, or a prolonged interval between specimen procurement and culture inoculation. Interference with virus isolation can also result from bacterial contamination or toxic effects of the specimen itself. Detecting AdV antigens in clinical samples by enzyme immunoassay has been proven reliable only for enteric types 40 and 41, which may be found in stool in titers high enough to permit detection. Microscopic methods and detection of AdV DNA by direct hybridization have proven to be insensitive (7). Antibody detection has limited value because its use in diagnosis of primary AdV infection is retrospective, because most people have been infected during childhood or as young adults with one or more AdV types, and because a diagnostic rise in titer may fail to develop in immunosuppressed patients. The advantages of detecting viral DNA by PCR include rapidity (1 or 2 days), sensitivity, ability to detect nonviable virus, and potential elimination of toxic effects of the specimen or contaminating microorganisms. Since amplification techniques have been implemented for detecting many different viruses, an increase in sensitivity has been demonstrated, suggesting that culture as the gold standard may require modification.
We developed a sensitive technique for amplifying AdV DNA from urine. PCR showed a decreased time to detection and an increased sensitivity compared to culture. HIV-infected patients were selected for the comparison of AdV detection by PCR versus culture because we and others frequently isolate AdV from these immunocompromised patients (2) and because serially collected surveillance specimens were available for validation studies.
PCR methods of detecting AdV DNA in fecal specimens and conjunctival swabs have been published (1, 9). In these studies, a well-characterized set of primers that amplify part of the hexon gene was used. Because that set did not amplify clinical isolates of the important AdV type 11 from two patients, we were prompted to test a different set of primers (14). Here, we show that our modified PCR method for AdV was highly sensitive, detecting as few as 0.2 PFU/ml in urine, and did not amplify DNA from a broad range of viruses, bacteria, or fungi that might be encountered in urine but did amplify all types of AdV that were likely to be recovered from that source.
When PCR was applied to column- or resin-extracted urine samples known to contain AdV, the sensitivity was high (90%). When samples that were uninterpretable or negative by culture but from patients likely to have replicating AdV were included (group A + B [Table 2]), PCR detected more AdV-positive urines (86%) than did culture (75%).
Known and potential pitfalls of diagnostic PCR include inhibitory substances or processing methods that interfere with DNA extraction and subsequent amplification. Initially, when urine samples were tested by PCR without extraction, sensitivity was very low, suggesting that many urine samples contained inhibitory substances. When methods to purify DNA from such sources were tested, sensitivity markedly increased. We designed this PCR assay to take advantage of the existing processing protocol for viral cultures. Unfortunately, two of our processed group A, AdV culture-positive samples were PCR negative and, unlike all of the PCR-positive samples, failed to grow upon reculturing. These specimens were considered to have degraded upon storage. Since these were also the only grossly clotted samples, it was speculated that clotting may have entrapped the virus or that the enzymatic changes associated with clotting may have been associated with degradation of the virus and its DNA. This finding would not be expected to interfere with clinical diagnostic use of PCR technology because deterioration occurred only after storage, the initial cultures having been positive. In diagnostic use, samples would not be stored for extended periods prior to testing. Further, in our hands, grossly hemorrhagic samples, in the absence of clots, have had no apparent effect on our ability to detect viral DNA in clinical samples when the preparative steps described in this study were employed.
Finally, complicated biological fluids such as urine can contain DNA, cells, or microorganisms that lead to false-positive amplification. However, our PCR method proved to be highly specific when tested with viral and bacterial DNAs; clinical samples containing virus, bacteria, yeast, crystals, leukocytes, or erythrocytes; and urine from healthy individuals.
With culture-negative specimens, it is assumed that infectious virions were absent, were at a concentration below the detection threshold, or were inhibited from replicating. In our study, some of these samples (group C) yielded positive PCR results. Because the specificity of this assay appears to be very high and because negative controls were always PCR negative, it is likely that these results truly represent AdV infection or shedding. Further clinical studies using the method described here will help clarify this point. The increased number of positives when column extraction was performed, compared with resin extraction, might be due to concentration of DNA by the column.
A highly sensitive assay might detect asymptomatic AdV shedding in urine from healthy adults. Among 23 specimens that were studied, all were negative for AdV by culture, and 22 (96%) were negative by PCR. Interestingly, the single PCR-positive sample was from a pregnant volunteer who had recently experienced severe adenoviral conjunctivitis. This result may represent continued virus excretion resulting from systemic and subclinical AdV persistence. Additional studies are needed to determine the positive predictive value of our assay for other populations of interest.
In the AIDS study from which our samples were obtained, culture recovery of AdV in urine decreased from 18% in 1993 and 1994 to 5% in 1995 and 1996. This occurred during a period of increasing use of multidrug anti-HIV therapies and was attributed to a general improvement in immunologic function, leading to a diminished incidence of opportunistic infections. However, direct effects of antiviral therapy upon AdV cannot be excluded. The mean concentration of CD4-positive peripheral lymphocytes in AdV culture-positive patients was 21 cells/μl (range, 3 to 81), suggesting that HIV-induced immunosuppression augments the ability of AdV to infect, as suggested by Khoo et al. (11), or to reactivate.
The high frequency of AdV in urine from AIDS patients is not well understood. This PCR assay will provide a highly sensitive and specific tool to better investigate the significance of AdV in the urine of HIV-infected patients. It can be used to determine whether the virus is reactivated and shed without causing pathogenic effects or whether it contributes to the disease manifestations of AIDS.
ACKNOWLEDGMENTS
This work was partially supported by EY grant 10268 from the National Eye Institute, National Institutes of Health.
We thank William Merz for critical review of the manuscript, Julie Knepp for technical advice, Gary Ketner for supplying purified AdV DNA, and Douglas Jabs for providing access to the samples from AIDS patients. J.T. appreciates support from FDA colleagues, especially Steve Gutman and Sharon Hansen.
REFERENCES
- 1.Allard A, Girones R, Per J, Wadell G. Polymerase chain reaction for detection of adenoviruses in stool samples. J Clin Microbiol. 1990;28:2659–2667. doi: 10.1128/jcm.28.12.2659-2667.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Jong P J, Valderrama G, Spigland L, Horwitz M S. Adenovirus isolates from urine of patients with acquired immunodeficiency syndrome. Lancet. 1983;i:1293–1296. doi: 10.1016/s0140-6736(83)92411-x. [DOI] [PubMed] [Google Scholar]
- 3.Echavarria, M., S. Ray, P. Charache, R. Ambinder, and J. S. Dumler. PCR detection of adenovirus in a bone marrow transplant recipient: hemorrhagic cystitis as a presenting manifestation of disseminated disease. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 4.Green W R, Graves W L, Frederick W R, Taddesse-Health L. Renal infection due to adenovirus in a patient with human immunodeficiency virus infection. Clin Infect Dis. 1993;18:989–991. doi: 10.1093/clinids/18.6.989. [DOI] [PubMed] [Google Scholar]
- 5.Hierholzer J C. Adenoviruses. In: Lennette E H, Lennette D A, Lennette E T, editors. Diagnostic procedures for viral, rickettsial, and chlamydial infections. 7th ed. Washington, D.C: American Public Health Association; 1995. pp. 169–188. [Google Scholar]
- 6.Hierholzer J C. Adenoviruses in the immunocompromised host. Clin Microbiol Rev. 1992;5:262–274. doi: 10.1128/cmr.5.3.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz M. Adenoviruses. In: Fields B N, editor. Virology. 3rd ed. New York, N.Y: Raven; 1996. pp. 2149–2197. [Google Scholar]
- 8.Jabs D A, Enger C, Dunn J P, Forman M. Cytomegalovirus retinitis and viral resistance: ganciclovir resistance. J Infect Dis. 1998;177:770–773. doi: 10.1086/514249. [DOI] [PubMed] [Google Scholar]
- 9.Jackson R, Morris D J, Cooper R J, Bailey A S, Klapper P E, Cleator G M, Tullo A B. Multiplex polymerase chain reaction for adenovirus and herpes simplex virus in eye swabs. J Virol Methods. 1996;56:41–48. doi: 10.1016/0166-0934(95)01903-0. [DOI] [PubMed] [Google Scholar]
- 10.Janner D, Petru A, Belchisand D, Azimi P. Fatal adenovirus infection in a child with acquired immunodeficiency syndrome. Pediatr Infect Dis J. 1990;9:434–436. doi: 10.1097/00006454-199006000-00013. [DOI] [PubMed] [Google Scholar]
- 11.Khoo S H, Bailey A S, de Jong J C, Mandal B K. Adenovirus infections in human immunodeficiency virus-positive patients: clinical features and molecular epidemiology. J Infect Dis. 1995;172:629–637. doi: 10.1093/infdis/172.3.629. [DOI] [PubMed] [Google Scholar]
- 12.Krilov L R, Rubin L G, Frogel M, Gloster E, Nai D, Kaplan M, Lipson S M. Disseminated adenovirus infection with hepatic necrosis in patients with human immunodeficiency virus infection and other immunodeficiency states. Rev Infect Dis. 1990;12:303–307. doi: 10.1093/clinids/12.2.303. [DOI] [PubMed] [Google Scholar]
- 13.Siegal F, Dikman S H, Arayata R B, Bottone E J. Fatal disseminated adenovirus 11 pneumonia in an agammaglobulinemic patient. Am J Med. 1981;71:1062–1067. doi: 10.1016/0002-9343(81)90343-0. [DOI] [PubMed] [Google Scholar]
- 14.Yeo A C, Cooper R J, Morris D J, Storey C C. Abstracts of the 97th General Meeting the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Development of a multiplex polymerase chain reaction for detection of adenovirus, herpes simplex and Chlamydia trachomatis DNA in eye swabs, abstr. C-416; p. 192. [Google Scholar]
- 15.Zahradnik J M, Spender M, Porter D. Adenovirus infection in the immunocompromised patient. Am J Med. 1980;68:725–732. doi: 10.1016/0002-9343(80)90262-4. [DOI] [PubMed] [Google Scholar]