Abstract
Genomic DNA extracted from 45 vancomycin-resistant Enterococcus faecium (VRE) isolates was cleaved with HindIII and HaeIII and subjected to agarose gel electrophoresis. The ability of this method (restriction endonuclease analysis [REA]) to distinguish strains at the subspecies level was compared with results previously determined by pulsed-field gel electrophoresis (PFGE). Chart reviews were performed to provide a clinical correlation of possible epidemiologic relatedness. A likely clinical association was found for 29 patients as part of two outbreaks. REA found 21 of 21 isolates were the same type in the first outbreak, with PFGE calling 19 strains the same type. In the second outbreak with eight patient isolates, HindIII found six were the same type and two were unique types. HaeIII found three strains were the same type, two strains were a separate type, and three more strains were unique types, while PFGE found three were the same type and five were unique types. No single “ideal” method can be used without clinical epidemiologic investigation, but any of these techniques is helpful in providing focus to infection control practitioners assessing possible outbreaks of nosocomial infection.
Accurate epidemiologic investigation requires an assessment of relatedness between individuals with similar infections in order to determine if person-to-person spread has occurred. In order to accomplish this, one rapid laboratory approach taken has been to determine the presence or absence of genetic identity between microbial strains of the same genus and species affecting persons who may have had a common exposure. For this to be useful, it is desirable to rapidly compare different isolates of an organism in a simple and accurate manner that can demonstrate the presence or absence of important epidemiologic associations (clonality).
Enterococci (especially those carrying vancomycin resistance genes) are now important causes of clinical infections, including endocarditis, urinary tract infection, and superinfection in persons who have received antimicrobial agents (14). Although enterococci are part of normal human gastrointestinal flora and can cause infection from this endogenous source, these organisms can also be spread nosocomially (13, 31). In the past, epidemiologic evaluation of enterococcal infection has been somewhat limited by the lack of a simple and sufficiently discriminatory typing system (2, 11, 13, 16, 31). Recently, however, pulsed-field gel electrophoresis (PFGE) and restriction endonuclease analysis (REA) of genomic DNA were shown to be useful for epidemiologic evaluations of nosocomial enterococcal infections (2, 16). Gordillo et al. compared ribotyping with an rRNA probe derived from Escherichia coli to PFGE for differentiating strains of Enterococcus faecalis and found that PFGE was the superior technique, showing 25 clearly different patterns plus 6 related variants versus 7 ribopattern types (9).
We have used REA of total genomic DNA with success in epidemiologic study of other organisms (6) and have applied this technique to type enterococcal isolates (2). The purposes of this study are (i) to describe our technique, (ii) to report the cataloging of REA types by using two different restriction enzymes from the first 45 vancomycin-resistant Enterococcus faecium isolates at Northwestern Memorial Hospital, and (iii) to compare the results with those previously obtained by PFGE. The comparison of each method’s utility for focusing infection control interventions was assessed in view of the clinical correlation determined by epidemiologic data obtained from comprehensive chart review of the patients involved.
(This report was presented in part at the 35th Annual Meeting, Infectious Diseases Society of America, San Francisco, California, September 1997 [abstract 345].)
MATERIALS AND METHODS
Bacterial isolates.
Forty-five vancomycin-resistant E. faecium isolates from various sites that were obtained from 42 patients hospitalized at Northwestern Memorial Hospital during a 15-month period between July 1992 and October 1993 were recovered from storage at −70°C for this study.
REA typing.
Genomic DNA from the enterococcal isolates was prepared by a modification of the method described by Pitcher and colleagues (21). Colonies from 24-h growth on a blood agar plate were suspended in sufficient 10/1 TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]) to equal that of a no. 2 McFarland standard, centrifuged, resuspended in 0.1 ml of 50-mg/ml lysozyme (Sigma, St. Louis, Mo.) in 10/1 TE buffer, and incubated for 30 min at 37°C. The DNA was harvested by the guanidine thiocyanate-EDTA-Sarkosyl (GES) method. RNase T1 (Gibco BRL, Gaithersburg, Md.) was added to the suspensions. Quantitation of the DNA was made with a Lambda-Bio spectrophotometer and corrected for dilution. Samples were stored at 4°C.
For restriction endonuclease digestion, genomic DNA (10 to 20 μl) was incubated with restriction endonuclease and digested according to the manufacturer’s instructions (Gibco BRL). All strains were restricted with two enzymes, one used in each of two separate assessments of bacterial relatedness. HindIII was used in one assessment series, and HaeIII was used in the other series. The restricted DNA fragments were separated by agarose gel electrophoresis with 0.6% agarose (Sigma) in TBE buffer (1 M Tris, 0.9 M boric acid, 0.01 M EDTA) at 44 V for 16.5 h. Gels were stained for 2 h in SYBR Green I (Molecular Probes, Eugene, Oreg.) and photographed under UV illumination.
The DNA band patterns for each new isolate digested with a common restriction enzyme were systematically compared according to the method described first by Clabots and colleagues (6). The first isolate in this analysis with a new DNA band pattern was arbitrarily designated a reference REA type. Gels were run so that the molecular weight ladder covered the top 6 cm (60 mm) of the electrophoresis gel from the origin. This was then the portion of the gel used for analysis. Similarities between the new and reference REA types were scored by visual comparison of each 1-mm segment of the top 60 mm of the DNA band patterns run on the same gel. The presence or absence of a DNA band within each segment was assessed. The actual intensity of the band is not part of the similarity scoring system. A similarity index was calculated from the number of identical 1-mm segments expressed as a percentage of the total number of 1-mm segments measured. A pattern with greater than six differences in the 1-mm segments had a similarity index of less than 90% and was designated a new REA type that was used for all future comparisons. For any epidemiologic investigation involving more than 10 isolates of apparently similar types, it is routine to repeat the REA analysis of purified DNA on the same gel to improve pattern matching. Any REA pattern with a similarity index of greater than 90% was included within a type. The types were designated by letters, and a distinct REA pattern within a type (similarity index of >90% but <100%) was designated by a subscript Arabic number indicating a subtype (A0, A1, A2, etc.). For this analysis, all strains within a given type were considered as being possibly related by the typing method.
PFGE typing.
PFGE was performed with the same 45 enterococcal isolates described above at the University of Iowa by the method of Pfaller et al. (20). Restriction digestion of chromosomal DNA was performed with SmaI (New England Biolabs, Inc.). The resultant restriction fragments were resolved in a 1% agarose gel with a CHEF-DRII system (Bio-Rad Laboratories, Richmond, Calif.). The pulse time ramped from 5 to 30 s over 23 h at 13°C and 6 V/cm. PFGE patterns were considered identical if they shared every band, similar (subtype) if they differed from one another by one to three clearly visible bands, and distinct if they differed by over three bands.
Chart reviews.
Detailed review of each of the 42 patients’ charts was completed for the duration of the hospitalization during which they had a culture positive for vancomycin-resistant enterococci (VRE). Data were collected about date of admission and discharge, in-hospital transfers, dates of VRE-positive cultures and body site(s), patient location (nursing unit) within the medical center, any diagnostic testing procedures (location and date), and date(s) seen by various consulting services. Any potentially significant clinical findings such as diarrhea and urinary incontinence were also recorded. Simultaneous location on the same ward, same-day visits by consulting services, same-day common procedures, or presence in the same room within 3 days of another patient with VRE constituted potential relatedness based on clinical assessment. If none of these association criteria were fulfilled, then the patient was not considered epidemiologically related to any other patient. For this report, the grouping into two distinct clusters making up separate potential outbreaks and one group of uniquely unrelated patients was fully based on the epidemiology from the chart review data.
RESULTS
Of these 45 vancomycin-resistant E. faecium isolates, 17 were obtained from rectal swabs as part of ongoing surveillance; 12 were from urine; 6 were from blood; 2 each were obtained from abscesses, catheter tips, and decubitus ulcers; and 1 each was obtained from a surgical wound, a T-tube drainage, hand surveillance, and a rectal biopsy.
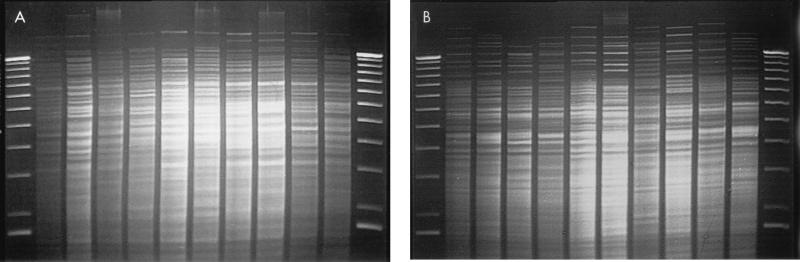
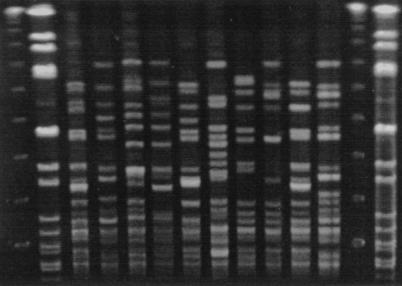
REA with HindIII provided 20 distinct patterns (subtypes) that were categorized into 9 unique types. Isolates cleaved with HindIII yielded between 25 and 35 bands per strain in the 60 mm of the DNA profiles analyzed. REA typing with HaeIII provided 21 subtypes that were categorized into 19 types. Isolates cleaved with HaeIII yielded a similar number of bands per strain in the top 60 mm of the DNA profiles. When these isolates were previously subjected to PFGE, they were found to have 27 distinct subtypes belonging to 21 types. PFGE gave approximately half the number of bands for analysis per strain (typically 12 to 15 bands). Representative isolates are shown that were analyzed by the REA technique with HindIII (Fig. 1A) and HaeIII (Fig. 1B) and by the PFGE technique (Fig. 2).
FIG. 1.
Representative isolates of each type analyzed by the REA technique (HindIII [A] and HaeIII [B]). The lanes, from left to right, represent a 1-kb DNA molecular weight ladder; strains EF18 (HindIII type B2, HaeIII type B2), EF20 (HindIII type C1, HaeIII type D0), EF23 (HindIII type B5, HaeIII type E0), EF27 (HindIII type B6, HaeIII type G0), EF32 (HindIII type B3), EF33 (HindIII type E0, HaeIII type H0), EF36 (HindIII type B4, HaeIII type B4), EF39 (HindIII type D0, HaeIII type J0), EF3 (HindIII type B0, HaeIII type B1), and EF45 (HindIII type C1, HaeIII type D0); and another 1-kb DNA molecular weight ladder standard.
FIG. 2.
Representative isolates of each type analyzed by PFGE. The lanes, from left to right, represent a 48.5-kb lambda DNA molecular weight ladder; a Staphylococcus aureus control digested with SmaI; strains EF18 (type B5), EF20 (type D), EF23 (type E), EF27 (type G), EF32 (type I), EF33 (type J), EF36 (type M), EF39 (type O), EF3 (type B1), and EF45 (type U); another lambda molecular weight ladder; and another S. aureus control.
A likely clinical association was found for 29 patients as part of two distinct outbreaks. REA with HindIII and HaeIII found 21 of 21 isolates were the same type in the first outbreak, with PFGE identifying 19 strains as the same type and 2 isolates as unique types (Table 1). In the second outbreak, represented by eight patient isolates, HindIII found six were the same type and two were unique types. Here, HaeIII found three were the same type, three strains were a separate type, and two more strains were unique types. PFGE found three strains were the same type and five strains were unique types (Table 2). In the seven discrepant isolates from the two outbreaks, HindIII found four types, HaeIII found five types, and PFGE found seven types. Of these seven strains, two appeared clonal by both REA enzymes and clinical association but were not related by PFGE.
TABLE 1.
Description of isolate sources and genomic typing results from the first potential outbreak with 21 VRE strains
| Isolate code | Date isolated (mo/day/yr) | Nursing unit sourcea | Specimen source | Type
|
||
|---|---|---|---|---|---|---|
| REA
|
PFGE | |||||
| HindIII | HaeIII | |||||
| EF3 | 12/2/92 | 15E | Wound | B0 | B1 | B1 |
| EF4 | 12/18/92 | MICU | Blood | B0 | B1 | B1 |
| EF5 | 12/27/92 | 7W | Blood | B0 | B1 | B1 |
| EF6 | 1/6/93 | 14E | Biliary tube | B1 | B1 | B1 |
| EF8 | 2/4/93 | 14W | Rectumb | B2 | B2 | B1 |
| EF10 | 2/5/93 | MICU | Intravenous catheter | B0 | B1 | B1 |
| EF11 | 2/23/93 | SCICU | Rectum | B2 | B3 | B3 |
| EF12 | 3/2/93 | 14W | Rectum | B2 | B3 | B4 |
| EF13 | 3/2/93 | 14W (room 1408) | Toilet seat | B3 | B5 | B5 |
| EF14 | 3/4/93 | SCICU | Hands | B0 | B1 | B1 |
| EF15 | 3/2/93 | 14W (room 1408) | Toilet seat | B2 | B3 | B5 |
| EF16 | 3/2/93 | SCICU | Rectum | B2 | B3 | B1 |
| EF17 | 3/2/93 | SCICU | Rectum | B2 | B3 | B1 |
| EF18 | 2/23/93 | 14W | Rectum | B2 | B2 | B5 |
| EF19 | 2/26/93 | 10E | Chest tube | B2 | B3 | B1 |
| EF25 | 4/20/93 | ER | Urine | B2 | B2 | B1 |
| EF29 | 5/10/93 | MICU | Urine | B2 | B2 | B1 |
| EF30 | 5/14/93 | 10E | Urine | B2 | B2 | B1 |
| EF31 | 5/10/93 | MICU | Intravenous catheter | B2 | B2 | B1 |
| EF32 | 5/22/93 | 7E | Urine | B3 | B3 | I |
| EF36 | 6/2/93 | 8E | Rectal biopsy | B4 | B4 | M |
E and W, East and West Wings, respectively; MICU, Medical Intensive Care Unit; SCICU, Spinal Cord Intensive Care Unit; ER, Emergency Room.
Surveillance isolates.
TABLE 2.
Description of isolate sources and genomic typing results from the second potential outbreak with eight VRE strains
| Isolate code | Date isolated (mo/day/yr) | Nursing unit sourcea | Specimen source | Type
|
||
|---|---|---|---|---|---|---|
| REA
|
PFGE | |||||
| HindIII | HaeIII | |||||
| EF20 | 3/21/93 | 8E | Urine | C1 | D0 | D |
| EF21 | 3/15/93 | 8W | Urine | B2 | B2 | B6 |
| EF22 | 4/7/93 | SICU | Rectumb | B2 | B2 | B7 |
| EF23 | 4/7/93 | SICU | Rectum | B5 | E0 | E |
| EF24 | 4/10/93 | 8E | Urine | D0 | F0 | F |
| EF26 | 4/22/93 | SICU | Blood | B2 | B2 | B1 |
| EF27 | 4/21/93 | 14E | Rectum | B6 | G0 | G |
| EF28 | 4/21/93 | 14W | Rectum | B7 | G1 | H |
For definitions of abbreviations, see Table 1.
Surveillance isolates.
The clinically unrelated patients (Table 3) presented the most diverse genomic groupings. Here, the various methods found from 8 to 14 unique types. Also, there were only two patients who were designated a B type by all three methods, representing a suggestion of clonality in only two (12.5%) of the strains.
TABLE 3.
Description of isolate sources and genomic typing results with 16 clinically unrelated VRE strains
| Isolate code | Date isolated (mo/day/yr) | Nursing unit sourcea | Specimen source | Type
|
||
|---|---|---|---|---|---|---|
| REA
|
PFGE | |||||
| HindIII | HaeIII | |||||
| EF1 | 7/1/92 | 14E | Blood | A0 | A0 | A |
| EF2 | 9/3/92 | 14W | Blood | B0 | B0 | B1 |
| EF7 | 1/27/93 | Home Health | Urine | B2 | B2 | B2 |
| EF9 | 2/1/93 | 6W | Urine | C0 | C0 | C |
| EF33 | 5/18/93 | 14W | Urine | E0 | H0 | J |
| EF34 | 5/18/93 | 14W | Rectumb | E0 | H1 | K |
| EF35 | 5/18/93 | 14E | Rectum | B2 | B2 | L |
| EF37 | 6/8/93 | 14E | Rectum | E1 | G0 | N |
| EF38 | 6/24/93 | 14E | Blood | F0 | I0 | B1 |
| EF39 | 7/22/93 | 11E | Rectum | D0 | J0 | O |
| EF40 | 7/21/93 | SCICU | Rectum | A1 | K0 | P |
| EF41 | 8/10/93 | 11E | Urine | D1 | L0 | Q |
| EF42 | 7/27/93 | 10W | Urine | F0 | I0 | R |
| EF43 | 8/23/93 | 9W | Skin ulcer | G0 | M0 | S |
| EF44 | 9/27/93 | 12W | Foot ulcer | H0 | N0 | T |
| EF45 | 10/7/93 | 10W | Urine | C1 | D0 | U |
For definitions of abbreviations, see Table 1.
Surveillance isolates.
DISCUSSION
Numerous typing methods have been used by investigators to augment the epidemiologic evaluation of nosocomial infections. Typing methods for enterococci that have been examined include ribotyping (9), biotyping (7, 10, 27), bacteriocin typing (11, 12, 23), phage typing (4, 5, 11–12, 22, 27), and serotyping (25–27). Antimicrobial agent susceptibility testing and determination of plasmid content with or without plasmid digestion patterns have also been used (13, 18, 29–31). None of these methods, however, have proven optimal for typing enterococci. Bacteriophage typing requires access to special reagents and performance of a large number of tests (11). Several investigators have experienced inconsistent plasmid patterns and irreproducible results when using total plasmid content for typing enterococci (9, 16). Recently, PFGE has been shown to be useful for epidemiologic evaluations of nosocomial enterococcal infections (9, 16).
PFGE is used by many different investigators and has shown a great deal of diversity among patterns of epidemiologically unrelated strains (15–17, 19). PFGE has an advantage over traditional agarose gel electrophoresis in that it is possible to separate even very large DNA molecules with as many as 107 nucleotide pairs (1). Ordinary gel electrophoresis fails to separate these molecules because the pores in the gel are too small for the large fragments. The constant electric field can also stretch them into elongated configurations that travel linearly at a rate relatively independent of size. However, frequent alterations in the direction of the electric field force the molecules to reorient in order to move, allowing separation of the large fragments with good resolution. Therefore, restriction endonucleases that have few recognition sites can be used to cleave the DNA, producing fewer fragments that generate more readily visible and easily comparable patterns. The primary disadvantage of PFGE is the relatively lengthy and cumbersome specimen preparation required before running the gel. The equipment required is modest in cost.
Genomic REA analyzes the entire DNA content of a microbe by cleaving the chromosomal DNA and any plasmid DNA into fragments small enough to be separated by electrophoresis on an agarose gel, producing a greater number of bands than PFGE. Although this method is very specific, one disadvantage is that DNA extracted from different isolates needs to be run on the same gel to facilitate pattern comparison because of the large number of bands requiring comparison, and this becomes difficult if an extraordinarily large collection of isolates must be tested. The presence of 30 to 50 bands typically found with REA makes reading of these gels difficult to automate, since no available image analysis system can adequately assess this large number of bands (author’s unpublished observation). The principal advantages of REA are the ease and rapidity of specimen preparation and the minimal amount of equipment required. This technique is also reported to be among the most specific methods of epidemiologic fingerprinting available (1, 28).
One limitation of these genomic digestion techniques is that the degree of relatedness between strains cannot be calculated by the absolute number of bands in common or different. One may not know how to interpret isolates that differ by only a few fragments. Such differences could arise within a single individual from inversions, deletions, or other rearrangements of the chromosome or from the acquisition or loss of a prophage, transposon, insertion sequence, or plasmid. On the other hand, such differences could indicate that isolates are more distantly related (16). In the converse, it also has been illustrated that chromosomal patterns the same as those in tested bacteria can be found in epidemiologically unrelated individuals (8, 24).
In this study, we have analyzed the chromosomal digestion patterns of 45 isolates of VRE cleaved with HindIII and HaeIII and compared these results to those obtained previously by PFGE. On initial assessment, a somewhat surprising diversity appears to exist among the three methods. The two REA studies were discordant in detecting clonality, with HaeIII producing 19 unique clonal types versus 9 produced with HindIII. The same observation was seen when comparing PFGE results. Interestingly, by chart review, the methods were much more concordant in providing an overall epidemiologic interpretation. None of the enzymes produced completely concordant clinical correlation. For example, EF23 was identified as a new type by HaeIII and PFGE, but clinically may have represented nosocomial transmission, because the patients with strains EF22 and EF26 were in rooms adjacent to this patient during the same time period. Conversely, there is no clinical evidence that EF40 and EF1 or EF41 and EF24 should be related, as suggested by HindIII patterns, but not by HaeIII or PFGE. There were also cases in which PFGE categorized two isolates into different types that clinically and by REA (with both enzymes) were the same. For example, EF27 and EF28 were isolated from patients on the same ward on the same day who also had common managing and consulting services and who had even had a Portacath placement within a day of each other. Another such example occurred with strains EF33 and EF34. They were isolated from the same person on the same day, and although from two different sources, they most likely represent the same organism. HindIII found these to be identical, HaeIII classified them as the same type but different subtypes, and PFGE determined them to be different types. Overall, many isolates that were identified as clonal by PFGE and REA had strong clinical data supporting this finding. Apparent discrepancies could be due to errors in visual interpretation of patterns by the investigators and/or poor resolution of some of the bands, or they could be due to actual differences in DNA patterns that are recognized differently by the restriction enzymes used.
Taking a broader view of our two potential outbreak groups and the group of clinically unrelated patients provides an interesting observation. In the 21-patient cluster (Table 1), each method found only one to three types and suggested an epidemiological association in 90 to 100% of cases, indicating a careful infection control investigation would be worthwhile. In potential outbreak 2 (Table 2), the methods identified from three to six types (from a total of eight specimens) and suggested that the largest single clonal group included an association ranging from 38 to 75% of cases. Here, an infection control investigation appears moderately indicated as useful from the typing data. The unrelated patient group was also the most diverse based on all three typing methods. From these 16 specimens, the methods found from 8 to 14 types, with the largest genomic clone (type B) representing only 19% (3 of 16) of the strains by any single method. This result would suggest little likelihood of the ongoing spread of a single, clonal VRE strain between these patients. Therefore, for a clinical application, the three typing approaches were quite concordant in indicating a high, moderate, or low probability of nosocomial spread of clonal VRE from interpretations based on the genomic typing data alone. Supporting our conclusion is the recent report by Bonten and colleagues, who found little genetic variation of VRE within individual patients and that when used as an epidemiologic tool, genetic typing found most strains were either very similar or very different, readily separating related from unrelated isolates (3). They too concluded that typing can be a very powerful tool to evaluate VRE epidemiology.
We believe the data presented show that a genomic typing approach for gathering clonality assessment information can be very useful in focusing the efforts of infection control practitioners when deciding which episodes of nosocomial infection likely represent patient-to-patient spread of a pathogenic microbe. Our results indicate that there is no single “ideal” method that can stand alone without clinical epidemiologic investigation, but all of these techniques are very helpful when reproducibly performed and carefully applied in a timely manner to assess possible outbreaks of nosocomial infection.
ACKNOWLEDGMENTS
This investigation was supported by Northwestern Memorial Hospital and Northwestern University Medical School, Chicago, Ill.
REFERENCES
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. Molecular biology of the cell. 3rd ed. New York, N.Y: Garland Publishing, Inc.; 1994. [Google Scholar]
- 2.Bodnar U R, Noskin G A, Suriano T, Cooper I, Reisberg B E, Peterson L R. Use of in-house molecular epidemiology and full species identification for controlling spread of vancomycin-resistant Enterococcus faecalis isolates. J Clin Microbiol. 1996;34:2129–2132. doi: 10.1128/jcm.34.9.2129-2132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonten M J M, Hayden M K, Nathan C, Rice T W, Weinstein R A. Stability of vancomycin-resistant enterococcal genotypes isolated from long-term-colonized patients. J Infect Dis. 1998;177:378–382. doi: 10.1086/514196. [DOI] [PubMed] [Google Scholar]
- 4.Brandis H, Plecas P, Andries L. Typing of Streptococcus faecalis and Streptococcus faecium strains by means of bacteriophages. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1985;260:206–215. [PubMed] [Google Scholar]
- 5.Caprioli T, Zaccour F, Kasatiya S S. Phage typing scheme for group D streptococci isolated from human urogenital tract. J Clin Microbiol. 1975;2:311–317. doi: 10.1128/jcm.2.4.311-317.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clabots C R, Johnson S, Bettin K M, Mathie P A, Mulligan M E, Schaberg D R, Peterson L R, Gerding D N. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J Clin Microbiol. 1993;31:1870–1875. doi: 10.1128/jcm.31.7.1870-1875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coudron P E, Mayhall C G, Facklam R R, Spadora A C, Lamb V A, Lybrand M R, Dalton H P. Streptococcus faecium outbreak in a neonatal intensive care unit. J Clin Microbiol. 1984;20:1044–1048. doi: 10.1128/jcm.20.6.1044-1048.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denning D W, Baker C J, Troup N J, Tompkins L S. Restriction endonuclease analysis of human and bovine group B streptococci for epidemiologic study. J Clin Microbiol. 1989;27:1352–1356. doi: 10.1128/jcm.27.6.1352-1356.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordillo M E, Singh K V, Murray B E. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. J Clin Microbiol. 1993;31:1570–1574. doi: 10.1128/jcm.31.6.1570-1574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hussain Z, Kuhn M, Lannigan R, Austin T W. Microbiological investigation of an outbreak of bacteremia due to Streptococcus faecalis in an intensive care unit. J Hosp Infect. 1988;12:263–271. doi: 10.1016/0195-6701(88)90068-0. [DOI] [PubMed] [Google Scholar]
- 11.Kuhnen E, Richter F, Richter K, Andries L. Establishment of a typing system for group D streptococci. Zentralbl Backteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1988;267:322–330. doi: 10.1016/s0176-6724(88)80048-8. [DOI] [PubMed] [Google Scholar]
- 12.Kuhnen E, Rommelsheim K, Andries L. Combined use of phage typing, enterococcinotyping and species differentiation of group D streptococci as an effective epidemiological tool. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1987;266:586–595. doi: 10.1016/s0176-6724(87)80242-0. [DOI] [PubMed] [Google Scholar]
- 13.Luginbuhl L M, Rotbart A, Facklam R R, Roe M H, Elliot J A. Neonatal enterococcal sepsis: case-control study and description of an outbreak. Pediatr Infect Dis J. 1987;6:1022–1030. [PubMed] [Google Scholar]
- 14.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray B E, Lopardo H A, Rubeglio E A, Frosolono M, Singh K V. Intrahospital spread of a single gentamicin-resistant, β-lactamase-producing strain of Enterococcus faecalis in Argentina. Antimicrob Agents Chemother. 1992;36:230–232. doi: 10.1128/aac.36.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray B E, Singh K V, Markowitz S M, Lopardo H A, Patterson J E, Zervos M J, Rubeglio E, Eliopoulos G M, Rice L B, Goldstein F W, Jenkins S G, Caputo G M, Nasnass R, Moore L S, Wong E S, Weinstock G. Evidence for clonal spread of a single strain of β-lactamase-producing Enterococcus faecalis to six hospitals in five states. J Infect Dis. 1991;163:780–785. doi: 10.1093/infdis/163.4.780. [DOI] [PubMed] [Google Scholar]
- 18.Patterson J E, Masecar B L, Kauffman C A, Schaberg D R, Hierholzer W J, Jr, Zervos M J. Gentamicin resistance plasmids of enterococci from diverse geographic areas are heterogeneous. J Infect Dis. 1988;158:212–216. doi: 10.1093/infdis/158.1.212. [DOI] [PubMed] [Google Scholar]
- 19.Patterson J E, Wanger A, Zscheck K K, Zervos M J, Murray B E. Molecular epidemiology of β-lactamase-producing enterococci. Antimicrob Agents Chemother. 1990;34:302–305. doi: 10.1128/aac.34.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller M A, Hollis R J, Sader H J. Chromosomal restriction fragment analysis by pulsed-field gel electrophoresis. In: Isenberg H D, editor. Clinical microbiology procedures handbook. 2, suppl. 1. Washington, D.C: American Society for Microbiology; 1994. pp. 10.5c.1–10.5.c.12. [Google Scholar]
- 21.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 22.Pleceas P. A phage typing system for Streptococcus faecalis and Streptococcus faecium. In: Parker M T, editor. Pathogenic streptococci—Proceedings of the VIIth International Symposium on Streptococci and Streptococcal Disease. Chertsey, England: Reedbooks; 1979. pp. 264–265. [Google Scholar]
- 23.Pleceas P, Bogdan C, Vereanu A. Enterocine-typing of group D streptococci. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1972;221:173–181. [PubMed] [Google Scholar]
- 24.Prado D, Murray B E, Cleary T G, Pickering L K. Limitations of using the plasmid pattern as an epidemiological tool for clinical isolates of Shigella sonnei. J Infect Dis. 1987;155:314–316. doi: 10.1093/infdis/155.2.314. [DOI] [PubMed] [Google Scholar]
- 25.Sharpe M E. Serological types of Streptococcus faecalis and its varieties and their cell wall type antigen. J Gen Microbiol. 1964;36:151–160. doi: 10.1099/00221287-36-1-151. [DOI] [PubMed] [Google Scholar]
- 26.Sharpe M E, Shattock P M F. The serological typing of group D streptococci associated with outbreaks of neonatal diarrhea. J Gen Microbiol. 1952;6:150–165. doi: 10.1099/00221287-6-1-2-150. [DOI] [PubMed] [Google Scholar]
- 27.Smyth C J, Matthews H, Halpenny M K, Brandis H, Colman G. Biotyping, serotyping and phage typing of Streptococcus faecalis isolated from dental plaque in the human mouth. J Med Microbiol. 1987;23:45–54. doi: 10.1099/00222615-23-1-45. [DOI] [PubMed] [Google Scholar]
- 28.Tompkins L S. The use of molecular methods in infectious diseases. N Engl J Med. 1992;327:1290–1297. doi: 10.1056/NEJM199210293271808. [DOI] [PubMed] [Google Scholar]
- 29.Uttley A H C, George R C, Naidoo J, Woodford N, Johnson A P, Collins C H, Morrison D, Gilfillan A J, Fitch L E, Heptonstall J. High-level vancomycin-resistant enterococci causing hospital infections. Epidemiol Infect. 1989;103:173–181. doi: 10.1017/s0950268800030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodford N, McNamara E, Smyth E, George R C. High-level resistance to gentamicin in Enterococcus faecium. J Antimicrob Chemother. 1992;29:395–403. doi: 10.1093/jac/29.4.395. [DOI] [PubMed] [Google Scholar]
- 31.Zervos M J, Kauffman C A, Therasse P M, Bergman A G, Mikesell T S, Schaberg D R. Nosocomial infection by gentamicin-resistant Streptococcus faecalis: an epidemiologic study. Ann Intern Med. 1987;106:687–691. doi: 10.7326/0003-4819-106-5-687. [DOI] [PubMed] [Google Scholar]