Abstract
The tropical montane cloud forest is the most diverse and threatened vegetation type in Mexico. In the last decade, the number of described Ascomycetes species has notably increased, reaching more than 1300 species. This study describes six new species based on their molecular and morphological characteristics. Our results suggest that Mexico has the highest number of described species in the Neotropics. However, many other Mexican lineages still need to be described.
Keywords: Ascomycota, new species, phylogeny, taxonomy, biogeography
1. Introduction
The fungi of the phylum Ascomycota, such as endophytes, mycorrhiza, phytopathogens, and saprobes, have various symbiotic ecological functions, with the latter producing many enzymes that degrade complex polymers such as starch, cellulose, chitin, keratin, and lignin [1]. These functions serve to balance the ecosystem. In tropical regions, species diversity is due to the structural complexity of microclimates and microhabitats. In this sense, the Mexican tropical montane cloud forest (TMCF) has been cataloged as the most diverse per area unit [2,3], which is also reflected by the Ascomycetes group; a significant number of species of this group have been described recently.
Mexico comprises a wide variety of vegetation types. One of the most diverse ecosystems, which is under significant threat, is the TMCF, also known as bosque mesófilo de montaña or bosque nuboso (cloud forest), which is characterized mainly by the presence of clouds at the vegetation level. The TMCF is characterized by high levels of atmospheric humidity, 1500–3000 mm of rainfall, and temperatures of 12–23 °C. The vegetation types in this ecosystem develop in rugged reliefs with a discontinuous distribution pattern, analogous to an archipelago of islands, and in ravines or slopes in the Sierra Madre Occidental to the north of Sinaloa, Nayarit, Jalisco, Colima, and Michoacán; in the Sierra Madre Oriental, from southwestern Tamaulipas to northern Oaxaca, including portions of San Luis Potosí, Hidalgo, Puebla, and Veracruz; and in the Sierra Madre south of Guerrero and Oaxaca. In addition, TMCF is also located in some areas of the Trans-Mexican Volcanic Belt. The flora has geographical links with North America in the tree layer and with South America in the herbaceous and shrub layers; it is closely related to Asian flora. In Mexico, these forests are vital due to their extraordinary biodiversity. Between 2500 and 3000 species of vascular plants inhabit the TMCF of Mexico, representing approximately 10% of its floristic richness, making it the country’s most diverse per area unit [1]. According to Guzmán [4], exhaustive monographic studies of 22 genera of Ascomycota are available.
In 2008, the existence of 1335 species of Ascomycetes was reported in Mexico [5]. These species reportedly belong to 41 orders, 126 families, and 441 genera, including 35% of lichenized ascomycetes and 4.9% marine taxa, without considering the asexual phases. In the same year, Heredia-Abarca et al. [6] registered 1353 anamorph species. Subsequently, Aguirre-Acosta et al. [7] noted that the CONABIO catalog by Cifuentes [8] enlisted 646 species of Ascomycota in Mexico without considering the asexual phases, distributed in 86 families and 275 genera, including lichens. Later, Del Olmo et al. [9] noted that in Mexico, there are 954 Ascomycota species in the TMCF. According to the authors, these fungi are assigned to 10 taxonomic classes: Arthoniomycetes (10 species), Dothideomycetes (125), Eurotiomycetes (35), Geoglossomycetes (2), Lecanoromycetes (167), Leotiomycetes (66), Orbiliomycetes (3), Pezizomycetes (93), Saccharomycetes (1), and Sordariomycetes (333), with 119 incertae sedis.
The main objective of this study was to contribute to the cataloguing of new species of Ascomycetes in the TMCF and update the knowledge on the Ascomycetes richness in this threatened ecosystem type. We aimed to describe, phylogenetically and morphologically, six Ascomycetes species distributed in the Mexican tropical montane cloud forest, an ecosystem in danger of extinction.
2. Material and Methods
2.1. Morphological Studies
Specimens from the “Dr. Gastón Guzmán Huerta” fungal collection at the Herbarium of the Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, Mexico City, Mexico (ENCB), and the “Jose Castillo Tovar” collection at the Instituto Tecnológico de Ciudad Victoria (ITCV) were revised. The color codes follow Kornerup and Wanscher [10] and Bessette et al. [11]. Microscopic observations were made of tissues rehydrated in 5% aqueous KOH and Melzer’s reagent; ascospore dimensions included the ornamentation. The macroscopic features were photographed with a Nikon D7000 camera and the micrographs with a Sony DSCWX350. Additionally, scanning electron microscopy (SEM; Hitachi SU1510, Hitachi, Tokyo, Japan) was used to observe the details of spore walls. The meanings of the taxonomic terms are based on Ulloa and Hanlin [12].
2.2. Amplification and Sequencing
DNA was obtained from herborized exemplars. Genomic DNA was extracted using the CTAB method [13]. The DNA was quantified with a NanoDrop 2000c (Thermo, Waltham, MA, USA). Dilutions were prepared from each sample at 20 ng/µL to amplify 4 regions: internal transcribed spacer rDNA-ITS1 5.8S rDNA-ITS2 (ITS), large nuclear subunit ribosomal DNA (nLSU), the second largest subunit of the RNA polymerase II gene (rpb2), and the region of the small mitochondrial subunit (mtSSU). The reaction mixture for PCR was prepared at a final volume of 15 µL and contained 1× buffer, 0.8 mM dNTPs mix, 20 pmol of each primer, 2 units of GoTaq DNA (Promega, Madison, WI, USA), and 100 ng of template DNA. The PCR products were verified by agarose gel electrophoresis. The gels were run for 1 h at 95 V cm−3 in 1.5% agarose and 1× Tris acetate-EDTA (TAE) buffer. The gels were stained with GelRed (Biotium, Fremont, CA USA), and the bands were visualized in an Infinity 3000 transilluminator (Vilber Lourmat, Eberhardzell, Germany). The amplified products were purified with an ExoSAP purification kit (Affymetrix, Santa Clara, CA, USA), following the manufacturer’s instructions. They were quantified and prepared for sequence reaction using a BigDye Terminator v.3.1 (Applied Biosystems, Foster City, CA, USA). These products were sequenced in both directions with an Applied Biosystems 3730XL DNA analyzer (Applied Biosystems, Foster City, CA, USA) at the Instituto de Biología of the Universidad Nacional Autónoma de México (UNAM). The sequences obtained were compared with the original chromatograms to detect and correct possible reading errors. The sequences of both strands of each gene were analyzed, edited, and assembled using BioEdit v. 7.0.5 [14] to generate a consensus sequence, which was compared with those deposited in GenBank [15] using BLASTN v. 2.2.9 [16].
2.3. Phylogenetic Analysis
Alignment was carried out based on the taxonomic sampling method employed by Pem et al. [17] to explore the phylogenetic relationships of the new species of Holmiella (Table 1). Each gene region was independently aligned using the online version of MAFFT v. 7 [18,19,20]. The alignment was reviewed in PhyDE v.10.0 [21], followed by minor manual adjustments to ensure character homology between taxa. A matrix was formed for ITS with 10 taxa (690 characters) for ITS, 23 taxa (831 characters) for LSU, and 14 taxa (640 characters) for mtSSU. The aligned matrices were concatenated into a single matrix (24 taxa, 2161 characters). Three partitioning schemes were established, one each for the ITS, nLSU, and mtSSU, using the option to minimize the stop codon with Mesquite v3.70 [22].
Table 1.
GenBank accession numbers corresponding to sequences used in phylogenetic analyses of Holmiella hidalgoensis sp. nov. Accessions of new species indicated in bold.
| Species Name | Isolate/Voucher/Strain | GenBank Accessions | ||
|---|---|---|---|---|
| ITS | nrLSU | SSU | ||
| Anisomeridium ubianum (Vain.) R.C. Harris | MPN94 | KY486750 | GU327709 | JN887379 |
| Cryomyces antarcticus Selbmann, de Hoog, Mazzaglia, Friedman & Onofri | CCFEE 536 | ----- | GU250365 | GU250321 |
| Cryomyces minteri Selbmann, de Hoog, Mazzaglia, Friedman & Onofri | CCFEE 5187 | ----- | KC315869 | KC315858 |
| Glyphium elatum (Grev.) H. Zogg | EB 0388 | KM220946 | KM220940 | ----- |
| EB 0342 | KM220945 | KM220938 | KM220935 | |
| EB 0329 | ----- | KM220937 | KM220934 | |
| EB 0365 | ----- | KM220939 | KM220936 | |
| Glyphium grisonense Math. | EB 0376 | ----- | KM220942 | ----- |
| Holmiella hidalgoensis | T. Raymundo 4608 Holotype ENCB | OQ877252 | OQ880481 | OQ878242 |
| Holmiella junipericola Pem, Gafforov, Jeewon & K.D. Hyde | MFLUCC 18-0503 | MH188902 | MH188900 | MH188901 |
| Holmiella juniperi-semiglobosae Pem, Gafforov, Jeewon & K.D. Hyde | MFLUCC 17-1955 | MH188905 | MH188903 | MH188904 |
| Holmiella Sabina (De Not.) Petrini, Samuels & E. Müll. | G.M. 2015-04-29.2 | KY486750 | ----- | ----- |
| Hysteropatella clavispora (Peck.) Hönh. | CBS 247.34 | ----- | AY541493 | AY511483 |
| Hysteropatella elliptica (Fr.) Rehm | G.M. 2013-05-06 01 | ----- | KM220948 | KM220948 |
| CBS 935.97 | ----- | DQ767657 | EF495114 | |
| Hysteropatella prostii (Duby) Rehm | H.B. 9934b | KT876980 | KT876980 | ----- |
| G.M. 2014-05-20 01 | ----- | KM220949 | ----- | |
| Lichenothelia calcarea Henssen | L1324 | ----- | KC015062 | KR045803 |
| Lichenothelia convexa Henssen | L1609 | ----- | KC015071 | KR045805 |
| Patellaria atrata (Hedw.) Fr. | BCC 28877 | KM220950 | GU371829 | ----- |
| BCC 28876 | ----- | KM220950 | ----- | |
| CBS 958.97 | ----- | GU301855 | ----- | |
| Yuccamyces citri Crous | CBS 143161 | MG386043 | MG386096 | |
| Yuccamyces pilosus (R.F. Castañeda) R.F. Castañeda | CBS 579.92 | ----- | MG386097 | ----- |
Alignment was carried out based on the taxonomic sampling method employed by Sun et al. [23] to explore the phylogenetic relationships of the new species of Kirschsteiniothelia (Table 2). Each gene region was independently aligned using the online version of MAFFT v. 7 [18,19,20]. The alignment was reviewed in PhyDE v.10.0 [21], followed by minor manual adjustments to ensure character homology between taxa. A matrix was formed with 23 taxa (695 characters) for ITS and 37 taxa (836 characters) for LSU. The aligned matrices were concatenated into a single matrix (37 taxa, 1534 characters). Two partitioning schemes were established, one each for the ITS and LSU, using the option to minimize the stop codon with Mesquite v3.70 [22].
Table 2.
GenBank accession numbers corresponding to sequences used in phylogenetic analyses of Kirschsteiniothelia esperanzae sp. nov. Accessions of new species indicated in bold.
| Species Name | Isolate/Voucher/strain | GenBank Accessions | |
|---|---|---|---|
| ITS | nLSU | ||
| Acrospermum adeanum Höhn. | M133 | EU940180 | EU940104 |
| Acrospermum compressum Tode | M151 | EU940161 | EU940084 |
| Acrospermum graminum Lib. | M152 | EU940162 | EU940085 |
| Kirschsteiniothelia aethiops (Sacc.) D. Hawksw. | CBS 109.53 | ----- | AY016361 |
| MFLUCC 16–1104 | MH182583 | MH182589 | |
| S–783 | MH182586 | MH182595 | |
| MFLUCC 15–0424 | KU500571 | KU500578 | |
| Kirschsteiniothelia aquatica Z.L. Luo, K.D. Hyde & H.Y. Su | MFLUCC 17–1685 | MH182587 | MH182594 |
| Kirschsteiniothelia arasbaranica Mehrabi, Hemmati & Asgari | IRAN 2509C | KX621986 | KX621987 |
| IRAN 2508C | KX621983 | KX621984 | |
| Kirschsteiniothelia cangshanensis Z.L. Luo, D.F. Bao, K.D. Hyde & H.Y. Su | MFLUCC 16–1350 | MH182584 | MH182592 |
| Kirschsteiniothelia esperanzae | T. Raymundo 6581 Holotype ENCB | OQ877253 | OQ880482 |
| Kirschsteiniothelia fluminicola Z.L. Luo, K.D. Hyde & H.Y. Su | MFLUCC 16–1263 | MH182582 | MH182588 |
| Kirschsteiniothelia lignicola Boonmee & K.D. Hyde | MFLUCC 10–0036 | HQ441567 | HQ441568 |
| Kirschsteiniothelia nabanheensis Jing W. Liu & Jian Ma | HJAUP C2006 | OQ023274 | OQ023275 |
| HJAUP C2004 | OQ023197 | OQ023273 | |
| Kirschsteiniothelia phoenicis S. N. Zhang & K.D. Hyde | MFLUCC 18–0216 | MG859978 | MG860484 |
| Kirschsteiniothelia rostrata Jing Yang & K.D. Hyde | MFLUCC 15–0619 | KY697280 | KY697276 |
| MFLUCC 16–1124 | ----- | MH182590 | |
| Kirschsteiniothelia submersa Hong Y. Su & K.D. Hyde | MFLUCC 15–0427 | KU500570 | KU500577 |
| S–481 | ----- | MH182591 | |
| S–601 | MH182585 | MH182593 | |
| Kirschsteiniothelia tectonae Doilom, Bhat & K.D. Hyde | MFLUCC 12–0050 | KU144916 | KU764707 |
| Kirschsteiniothelia thailandica Y.R. Su, Yong Wang bis & K.D. Hyde | MFLUCC 20–0116 | MT985633 | MT984443 |
| Kirschsteiniothelia thujina (Peck.) D. Hawksw. | JF 13210 | KM982716 | KM982718 |
| Megalotremis verrucosa (Makhija & Patw.) Aptroot | MPN104 | ----- | GU327718 |
| Phyllobathelium anomalum Lücking | MPN 242 | ----- | GU327722 |
| Phyllobathelium firmum (Stirt.) Věsda | ERP 3175 | ----- | GU327723 |
| Pseudorobillarda eucalypti Tangthir. & K.D. Hyde | MFLUCC 12–0422 | KF827451 | KF827457 |
| Pseudorobillarda phragmitis (Cunnell) M. Morelet | CBS 398.61 | MH858101 | EU754203 |
| Strigula guangxiensis S.H. Jiang, X.L. Wei & J.C. Wei | HMAS-L0138040 | NR146255 | MK206256 |
| Strigula macrocarpa Vain. | HMAS-L0141394 | ----- | MK206240 |
| Strigula nemathora Mont. | MPN 72 | ----- | JN887405 |
| Strigula nitidula Mont. | HMAS-L0139358 | ----- | MN788374 |
| Strigula sinoaustralis S.H. Jiang, X.L. Wei & J.C. Wei | HMAS-L0137204 | ----- | MK206249 |
| Strigula univelbiserialis S.H. Jiang, X.L. Wei & J.C. Wei | HMAS-L0137657 | ----- | MK206243 |
| Tenuitholiascus porinoides S.H. Jiang & J.C. Wei | HMAS-L0139638 | ----- | MK206259 |
| HMAS-L0139639 | ----- | MK206258 | |
| HMAS-L0139640 | ----- | MK206260 | |
Alignment was carried out based on the taxonomic sampling method employed by Healy et al. [24] to explore the phylogenetic relationships of the new species of Microglossum (Table 3). Each gene region was independently aligned using the online version of MAFFT v. 7 [18,19,20]. The alignment was reviewed in PhyDE v.10.0 [21], followed by minor manual adjustments to ensure character homology between taxa. A matrix was formed with 61 taxa (690 characters) for ITS, 23 taxa (831 characters) for LSU, and 22 taxa (670 characters) for the second largest subunit of the RNA polymerase II gene (rpb2). The aligned matrices were concatenated into a single matrix (61 taxa, 2191 characters). Five partitioning schemes were established, one each for the ITS and nLSU and three for the rpb2 gene region, using the option to minimize the stop codon with Mesquite v3.70 [22].
Table 3.
GenBank accession numbers corresponding to sequences used in phylogenetic analyses of Microglossum flavoviride sp. nov. Accessions of new species indicated in bold.
| Species Name | Isolate/Voucher/Strain | GenBank Accessions | ||
|---|---|---|---|---|
| ITS | nLSU | rpb2 | ||
| Microglossum clavatum V. Kučera, Lizoň & Tomšovský | SAV F-11276 | KX382864 | KX382864 | KX382884 |
| SAV F-11272 | KX382841 | ----- | ----- | |
| SAV F-11074 | KX382865 | KX382865 | KX382885 | |
| Microglossum cyanobasis P. Iglesias & Arauzo | AH 43985 | KX371850 | ----- | ----- |
| Microglossum flavoviride | García 18649 Holotype ITCV | OQ877254 | OQ880483 | |
| García 18686 | OQ877255 | OQ880484 | ||
| Microglossum fuscorubens Boud. | ERRO 2012120704 | KX371856 | ----- | ----- |
| ERRO 2012120705 | KX371857 | ----- | ----- | |
| ERRO 2012120706 | KX371858 | ----- | ----- | |
| SAV F-11275 | KX382834 | KX382834 | KX382883 | |
| Microglossum griseoviride V. Kučera, Lizoň & Tomšovský | SAV F-9920 | KX595249 | KC595250 | KX382872 |
| SAV F-10699 | KC595261 | ----- | ----- | |
| SAV F-10696 | KX382857 | ----- | ----- | |
| Microglossum nudipes Boud. | SAV F-11053 | KX382838 | KX382867 | ----- |
| SAV F-11051 | KX382856 | ----- | ----- | |
| SAV F-11274 | KX382836 | KX382836 | KX382888 | |
| SAV F-11285 | KX382859 | KX382869 | KX382887 | |
| SAV F-11271 | KX382837 | ----- | ----- | |
| Microglossum olivaceum (Pers.) Gillet | KM135962 | EU784374 | ----- | ----- |
| KM135599 | EU784373 | ----- | ----- | |
| ERRO 2004110702 | KX371853 | ----- | ----- | |
| Microglossum parvisporum V. Kučera, Lizoň & Tomšovský | SAV F-10998 | KM114901 | KM114901 | KX382879 |
| LE 291852 | KX382839 | ----- | ----- | |
| SAV F-11283 | KM114901 | KM114901 | KX382879 | |
| Microglossum pratense V. Kučera, Tomšovský & Lisoš | SAV F-10024 | KC595259 | KC595260 | KX382880 |
| SAV F-11020 | KJ513006 | KJ513006 | KX382881 | |
| O 64797 | KJ513004 | ----- | ----- | |
| O 294564 | KJ513005 | ----- | ----- | |
| O 170878 | KJ513002 | ----- | ----- | |
| O 270070 | KJ513003 | ----- | ----- | |
| SAV F-11062 | KX382848 | ----- | ----- | |
| LE 294492 | KX382849 | ----- | ----- | |
| LE 294489 | KX382850 | ----- | ----- | |
| SAV F-10568 | KX382851 | ----- | ----- | |
| SAV F-11056 | KX382847 | ----- | ----- | |
| SAV F-11052 | KX382852 | ----- | ----- | |
| Microglossum rufescens (Grelet) Bon | SAV F-9921 | KC595257 | ----- | ----- |
| ERRO 2004110703 | KX371854 | ----- | ----- | |
| ERRO 2011122601 | KX371855 | ----- | ----- | |
| SAV F-11282 | KX382858 | KX382868 | KX382892 | |
| SAV F-11204 | KX382835 | KX382866 | KX382893 | |
| Microglossum rufum (Schwein.) Underw. | Ingo 163 | DQ257360 | ----- | ----- |
| Microglossum tenebrosum V. Kučera, Tomšovský, Lisoš & F. Hampe | SAV F-11273 | KX382842 | ----- | ----- |
| SAV F-11278 | KX382845 | KX382845 | KX382891 | |
| SAV F-11279 | KX382843 | ----- | ----- | |
| SAV F-11070 | KX382846 | KX382846 | KX382890 | |
| SAV F-11072 | KX382844 | KX382844 | KX382889 | |
| Microglossum truncatum V. Kučera, Tomšovský & Lisoš | SAV F-11023 | KJ513009 | KJ513009 | KX382874 |
| SAV F-10720 | KX382840 | ----- | ----- | |
| O 224247 | KJ513010 | ----- | ----- | |
| SAV F-11280 | KX382861 | KX382861 | KX382875 | |
| SAV F-11022 | KJ513011 | ----- | ----- | |
| SAV F-11064 | KX382855 | ----- | ----- | |
| LE 291847 | KX382863 | KX382871 | KX382876 | |
| SAV F-11262 | KX382862 | KX382862 | KX382877 | |
| SAV F-11261 | KX382853 | ----- | ----- | |
| SAV F-11263 | KX382854 | ----- | ----- | |
| Microglossum viride (Schrad. ex J.F. Gmel.) Gillet | SAV F-10249 | KC595253 | KC595254 | KX382873 |
| SAV F-10697 | KC595265 | ----- | ----- | |
| SAV F-10698 | KC595263 | ----- | ----- | |
| KM90199 | EU784375 | ----- | ----- | |
Alignment was carried out based on the taxonomic sampling method employed by [25] to explore the phylogenetic relationships of the new species of Claussenomyces (Table 4). First, the ITS region was aligned using the online version of MAFFT v. 7 [18,19,20]. Next, the alignment was reviewed in PhyDE v.10.0 [21], followed by minor manual adjustments to ensure character homology between taxa. The matrix was composed of 22 taxa (700 characters).
Table 4.
GenBank accession numbers corresponding to sequences used in phylogenetic analyses of Claussenomyces paulinae sp. nov. Accessions of new species indicated in bold.
| Species Name | Isolate/Voucher/Strain | GenBank Accessions |
|---|---|---|
| ITS | ||
| Claussenomyces atrovirens (Pers.) Korf & Abawi | 22FM2A1 | MW709917 |
| Claussenomyces atrovirens (Pers.) Korf & Abawi | LEG25 | MW204926 |
| Claussenomyces atrovirens (Pers.) Korf & Abawi | FC1636 | LC425048 |
| Claussenomyces aff. Atrovirens | GM20144422.1 | MW178207 |
| Claussenomyces aff. Atrovirens | GM20150815.9 | MT949706 |
| Claussenomyces aff. Atrovirens | GM20190817.1 | MT522872 |
| Claussenomyces kirschsteinianus (Kirschst.) G. Marson & Baral | GM20150502.2 | KY689631 |
| Claussenomyces kirschsteinianus (Kirschst.) G. Marson & Baral | GM20141112.2 | KY689629 |
| Claussenomyces kirschsteinianus (Kirschst.) G. Marson & Baral | GM20141108.4 | KY689628 |
| Claussenomyces olivaceus (Fuckel) Sherwood | GM20150423.1 | KY661433 |
| Claussenomyces olivaceus (Fuckel) Sherwood | GM20190729.3 | OP103955 |
| Claussenomyces olivaceus (Fuckel) Sherwood | GM20161231.2 | MW167780 |
| Claussenomyces paulinae | T. Raymundo 7564 Holotype ENCB | OQ877256 |
| Claussenomyces prasinulus (P. Karst.) Korf & Abawi | HB7165a | OM808929 |
| CBS111551 | MN082653 | |
| NBRC 112536 | LC488725 | |
| Collophorina badensis S. Bien & Damm | CBS144833 | NR165902 |
| Collophorina germanica S. Bien & Damm | CBS144831 | NR165903 |
| Collophorina hispanica (Gramaje, Armengol & Damm) Damm & Crous | CBS128569 | MH864962 |
| Collophorina neorubra S. Bien & Damm | CBS144829 | NR165901 |
| Scolecoleotia eriocamporesi H. B. Jiang, Phookamsak & K.D. Hyde | IT3027A | MW981448 |
| Scolecoleotia eriocamporesi H. B. Jiang, Phookamsak & K.D. Hyde | IT3027B | MW981449 |
Alignment was carried out based on the taxonomic sampling method employed by Argnello et al. [26] and Healy et al. [24] to explore the phylogenetic relationships of the new species of Wolfina (Table 5). The ITS region was aligned using the online version of MAFFT v.7 [18,19,20]. The alignment was reviewed in PhyDE v.10.0 [21], followed by minor manual adjustments to ensure character homology between taxa. The matrix was composed of 11 taxa (700 characters).
Table 5.
GenBank accession numbers corresponding to sequences used in phylogenetic analyses of Wolfina molangoensis sp. nov. Accessions of new species indicated in bold.
| Species Name | Isolate/Voucher/Strain | GenBank Accessions |
|---|---|---|
| ITS | ||
| Chorioactis geaster (Peck) Kupfer ex Eckblad | K. Rice s.n. | AY307936 |
| S. Kurogi s.n. | AY307937 | |
| Trichaleurina javanica (Peck) M. Carbone, Agnello & P. Alvarado | KSRF 0019 | MF476196 |
| PL8 | MZ061709 | |
| 20170467 | MK184529 | |
| Trichaleurina sp. | TNS-F-31213 | KF418250 |
| Trichaleurina tenuispora M. Carbone, Yei Z. Wang & Cheng L. Huang | TNM F10376 | NR159000 |
| TNM F20404 | KF418249 | |
| Wolfina aurantiopsis (Ellis) Seaver ex Eckblad | TENN 67128 | KC306744 |
| Wolfina molangoensis | R. Valenzuela 18918 Holotype ENCB | OQ877257 |
| Neournula pouchetii (Berthet & Riousset) Paden | TURA195798 | JX669837 |
Alignment was carried out to resolve the phylogenetic relationships of the new species of Dematophora based on the taxonomic sampling method employed by Wittstein et al. [27] (Table 6). Each gene region was independently aligned using the online version of MAFFT v. 7 [18,19,20]. The alignment was reviewed in PhyDE v.10.0 [21], followed by minor manual adjustments to ensure character homology between taxa. A matrix was formed with 30 taxa (699 characters) for ITS and 18 taxa (836 characters) for LSU. The aligned matrices were concatenated into a single matrix (30 taxa, 1535 characters). Two partitioning schemes were established, one each for the ITS and LSU, using the option to minimize the stop codon with Mesquite v3.70 [22].
Table 6.
GenBank accession numbers corresponding to sequences used in phylogenetic analyses of Dematophora oaxacana sp. nov. Accessions of new species indicated in bold.
| Species Name | Isolate/Voucher/Strain | GenBank Accessions | |
|---|---|---|---|
| ITS | nLSU | ||
| Amphirosellinia fushanensis Y.M. Ju, J.D. Rogers & H.M. Hsieh | HAST 91111209 | GU339496 | ----- |
| Amphirosellinia nigrospora Y.M. Ju, J.D. Rogers & H.M. Hsieh | HAST 91092308 | GU322457 | ----- |
| Coniolarelia limoniispora | MUCL 29409 | MN984615 | MN984624 |
| Dematophora bunodes (Berk. & Broome) C. Lamb., Wittstein & M. Stadler | CBS 123584 | MN984617 | ----- |
| Dematophora bunodes (Berk. & Broome) C. Lamb., Wittstein & M. Stadler | CBS 123585 | MN984618 | ----- |
| Dematophora bunodes (Berk. & Broome) C. Lamb., Wittstein & M. Stadler | CBS 123597 | MN984619 | MN984625 |
| Dematophora buxi (Fabre) C. Lamb., Wittstein & M. Stadler | JDR 99 | GU300070 | ----- |
| Dematophora necatrix R. Hartig | CBS 349.36 | AY909001 | KF719204 |
| Dematophora necatrix R. Hartig | W 97 | DF977487 | DF977487 |
| Dematophora oaxacana | T. Raymundo 6161 Holotype ENCB | OQ877258 | OQ880487 |
| Dematophora oaxacana Sánchez-Flores, R. Valenz. & Raymundo | R. Valenzuela 17218 ENCB | OQ877259 | OQ880488 |
| Dematophora pepo (Pat.) C. Lamb., Wittstein & M. Stadler | CBS 123592 | MN984620 | ----- |
| Entoleuca mammata (Wahlenb.) J.D. Rogers & Y.M. Ju | JDR 100 | GU300072 | ----- |
| Euepixylon sphaeriostomum (Schwein.) Lar.N. Vassiljeva & S.L. Stephenson | JDR 261 | GU292821 | ----- |
| Graphostroma platystomum (Schwein.) Piroz | CBS 270.87 | JX658535 | DQ836906 |
| Hypoxylon fragiforme (Pers.) J. Kickx f. | MUCL 51264 | KC477229 | KM186295 |
| Nemania abortiva J.D. Rogers, Y.M. Ju & Hemmes | BISH 467 | GU292816 | ----- |
| Nemania beaumontii (Berk. & M.A. Curtis) Y.M. Ju & J.D Rogers | HAST 405 | GU292819 | ----- |
| Nemania beaumontii (Berk. & M.A. Curtis) Y.M. Ju & J.D Rogers | FL 0980 | ----- | JQ760608 |
| Nemania bipapillata (Berk. & M.A. Curtis) Pouzar | HAST 90080610 | GU292818 | ----- |
| Podosordaria mexicana Ellis & Holw. | WSP 176 | GU324762 | ----- |
| Podosordaria punctata | CBS 656.78 | KT281904 | KY610496 |
| Rosellinia aquila (Fr.) Ces. & De Not. | MUCL 51703 | KY610392 | KY610460 |
| Rosellinia marcucciana Ces | MUCL 51704 | MN984616 | MN984626 |
| Rosellinia corticium (Schwein.) Sacc. | MUCL 51693 | KY610393 | KY610461 |
| STMA 13324 | MN984621 | MN984627 | |
| STMA 12170-15209 | MN984623 | MN984629 | |
| Rosellinia nectrioides Rehm | CBS 449.89 | MN984622 | MN984628 |
| Xylaria arbuscula Sacc. | CBS 126415 | KY610394 | KY610463 |
| Xylaria hypoxylon (L.) Grev. | CBS 122620 | KY204024 | KY610495 |
| Xylaria bambusicola Y.M. Ju & J.D Rogers | WSP 205 | EF026123 | ----- |
The region was aligned independently using the online version of MAFFT v7 [18,19,20]. The alignments were reviewed in PhyDE [21], followed by minor manual adjustments to maximize character similarity. Phylogenetic inferences were estimated with maximum likelihood in RAxML v. 8.2.10 [28] with a GTR + G model of nucleotide substitution. We ran 1000 rapid bootstrap replicates with the GTRGAMMA model to assess branch support. For Bayesian posterior probability, the best evolutionary model for alignment was sought using PartitionFinder v.2.0 [29,30,31]. Phylogenetic analyses were performed using MrBayes v. 3.2.6 x64 [32]. The information block for matrices included two simultaneous runs, four Monte Carlo chains, temperature set to 0.2 °C, and sampling of 10 million generations (standard deviation ≤0.1) with trees sampled every 1000 generations. The first 25% of samples were discarded as burn-in, and stationarity was checked in Tracer v. 1 [33]. Finally, the trees were visualized and optimized in FigTree v. 1.4.4 [34] and edited in Adobe Illustrator vCS4 (Adobe Systems, Inc., San Jose, CA, USA).
3. Results
3.1. Taxonomy
3.1.1. Dothideomycetes, Patellariales, Patellariaceae
Holmiella hidalgoensis Raymundo, Martínez-González & R. Valenz. sp. nov.
MycoBank: MB842041.
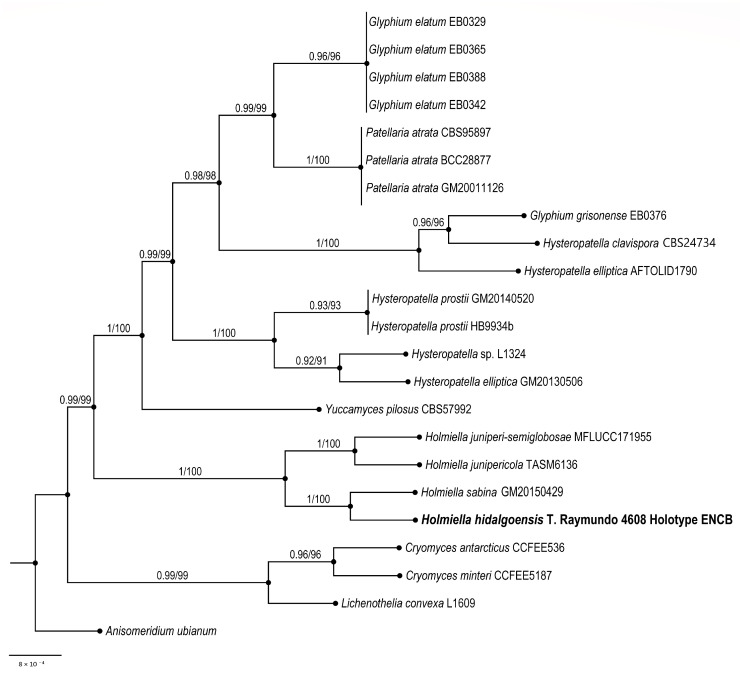
Figures: Figure 1 and Figure 2.
Figure 1.
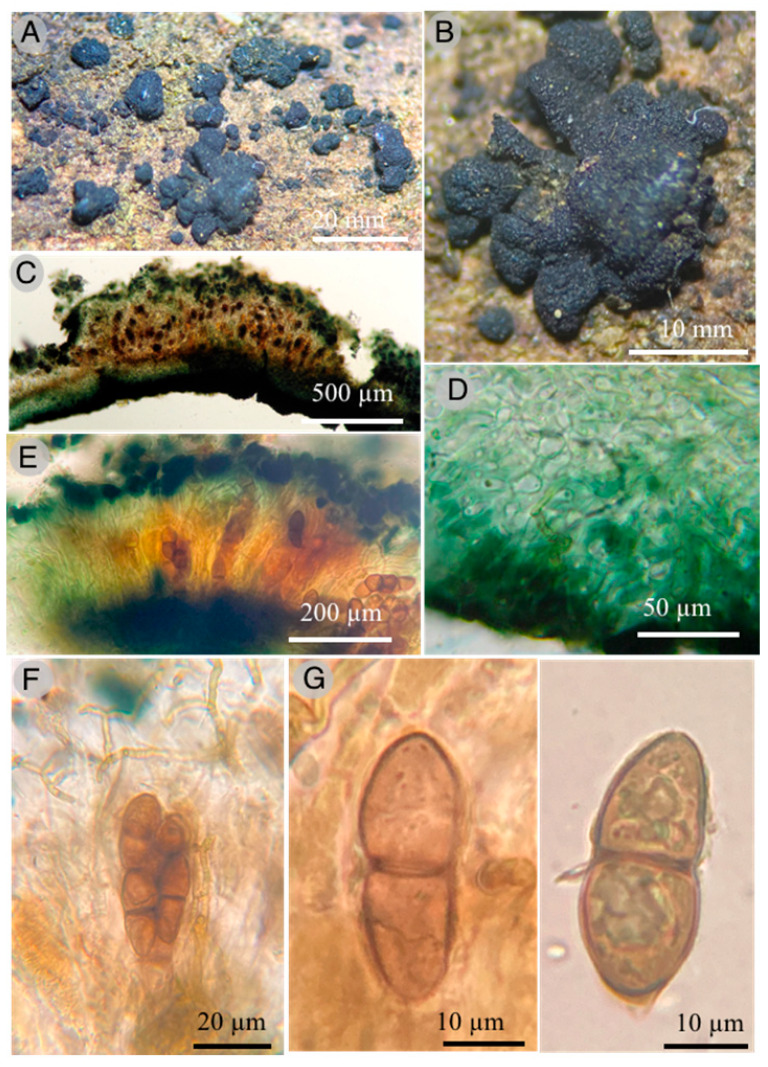
Holmiella hidalgoensis T. Raymundo 4608 Holotipe (A,B) Ascomata; (C) optical microscope images through the ascoma; (D) microscope image of ectal excipulum; (E) optical microscope images of hymenium; (F) optical microscope images of asci with ascospores; (G) optical microscope images of ascospores.
Figure 2.
Bayesian inference phylogram of ITS, LSU, and SSU sequence data. Posterior probability (left of slash) from Bayesian analysis and bootstrap support (right of slash) are given above the nodes. New species Holmiella hidalgoensis is shown in bold.
Diagnosis: Ascomata discoidal to ovoid, black, 1–1.5 × 600–800 µm; asci hyaline, pedicellate, bitunicate, 40–45 × 12–16 µm; ascospores ellipsoid to fusoid, golden brown, transverse septae, uniseriate to irregular biseriate, 32–36 × 10–12 µm.
Type: MEXICO: Hidalgo, Zacualtipán de Ángeles municipality, Bosque El Hayal, sobre la desviación a Tlahuelompa, 20°37′34′’ N, 98°37′07′’ W, 2250 m, 2 July 2013, T. Raymundo 4608 (ENCB).
GenBank: ITS OQ877252, nrLSU OQ880481, SSU OQ878242.
Etymology: The epithet indicates that the species grows in Hidalgo.
Ascomata 1–1.5 mm diameter, 600–800 µm thick, solitary to gregarious, erumpent to superficial, rounded to angular, discoidal to powdery, 1 to 1.5 mm diameter, sessile, rough, exposing the asci, black color; basal peridium green to black, 160-200 µm thick; paraphysoids 3–3.5 µm in diameter, filiform, branched, anastomosed, deep green; paraphyses protruding from asci; asci 40–45 × 12–16 µm, bitunicate, cylindrical to clavate, sessile, rounded at the apex, octosporate; ascospores uniseriate to irregularly biseriate in the apical part (28.8–) 32–36 (40) × (9.6–) 10–12 (–12.8) µm broadly ellipsoid to fusiform, bicellular, with one septum and constricted in the middle part, the distal portion slightly larger than the proximal, pale yellowish to dark golden brown when ripe.
Habitat: Gregarious on decaying branches of angiosperms.
Additional specimens examined: MEXICO: Hidalgo, Zacualtipán de Ángeles municipality, Bosque El Hayal, sobre la desviación a Tlahuelompa, 20°37′34″ N, 98°37′07″ W, 2250 m, 2 July 2013, R. Valenzuela 14997 (ENCB, Paratype).
Taxonomical notes: This species is characterized by pulvinated ascomata, black and erumpent, ascospores 32–36 × 10–12 µm, bicellular, with a golden-brown color and a germinative pore. Morphologically and phylogenetically, it is close to Holmiella sabina (De Not.) Petrini, Samuels & E. Müll. However, the former presents ascomata with toothed margins, ascospores of 25–40 × 13–20 µm, bicellular, reddish-brown with two germinative pores. Holmiella juniperi-semiglobosae Pem, Gafforov, Jeewon & K.D. Hyde and H. junipericola Pem, Gafforov, Jeewon & K.D. Hyde are species that are phylogenetically related; however, they grow on Juniperus semiglobosa and J. zerawschanica, respectively, from Uzbekistan [17].
3.1.2. Pleosporales, Kirschteiniotheliaceae
-
2.
Kirschsteiniothelia esperanzae Raymundo, Cobos-Villagrán & R. Valenz. sp. nov.
MycoBank: MB822042.
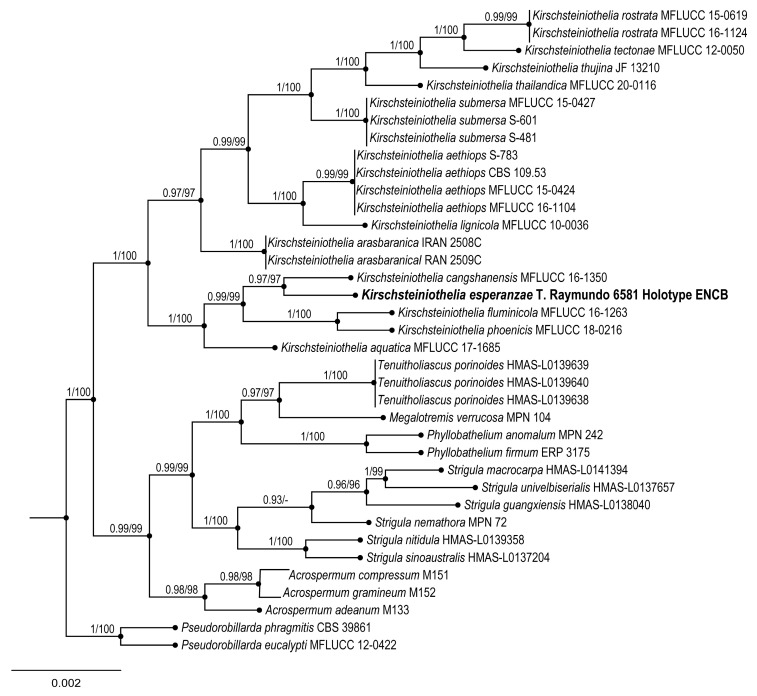
Figures: Figure 3 and Figure 4.
Figure 3.
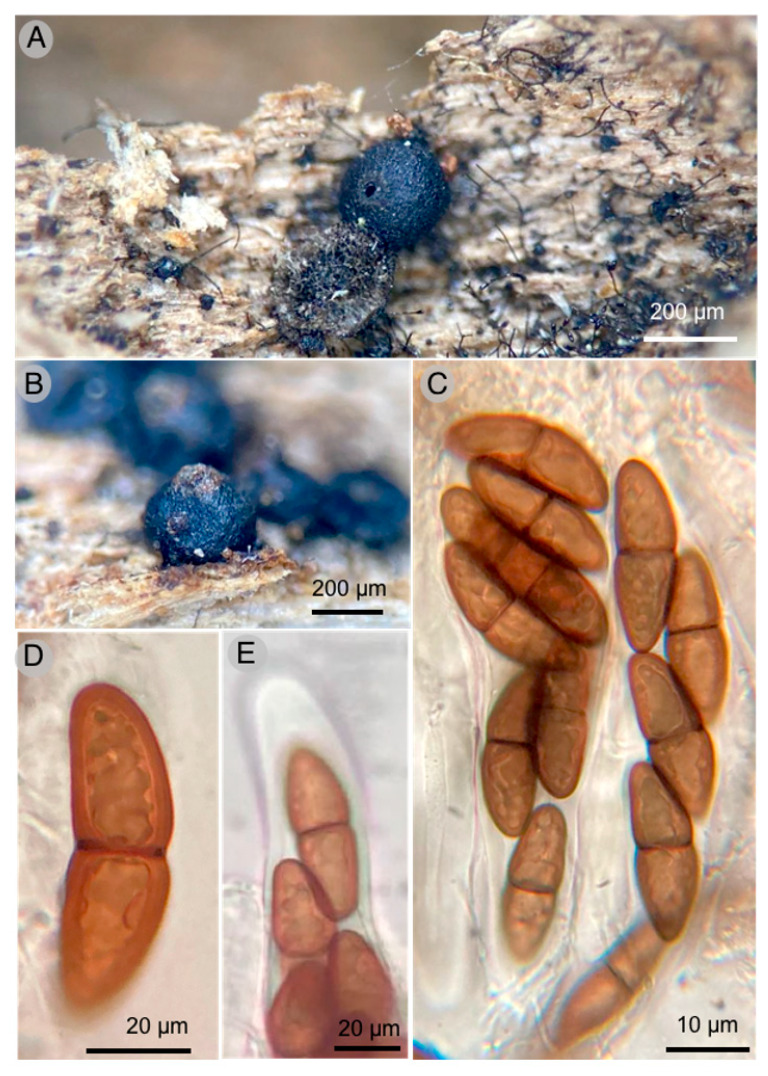
Kirschsteiniothelia esperanzae T. Raymundo 6581 Holotype: (A) pseudothecia showing ostiole; (B) pseudothecia; (C) optical microscope images of asci with ascospores; (D) optical microscope images of ascospore; (E) apical part of asca.
Figure 4.
Bayesian inference phylogram of ITS, LSU, and rpb2 sequence data. Posterior probability (left of slash) and bootstrap support values (right of slash) in Bayesian analysis are given above the nodes. New species Kirschsteiniothelia esperanzae is shown in bold.
Diagnosis: Ascomata 300–700 µm diameter × 600–700 µm high, peridium 55 to 100 μm thick, pseudoperiphyses 5 μm wide, asci (168–)178–203 × 32–35 μm and ascospores 40–50(–53) × 14–17 μm.
Type: MEXICO: Oaxaca, Sierra de Juárez, Ixtlán district, Santiago Comaltepec, La Esperanza, Carretera Oaxaca-Tuxtepec km 51, 17°37′55″ N, 96°22′01″ W, 1600 m, 21 May 2017; T. Raymundo 6581 (ENCB, Holotype).
GenBank: ITS: OQ877253, LSU: OQ880482.
Etymology: The epithet refers to La Esperanza’s locality, where the species was collected.
Pseudothecia 400–700 µm diameter × 600–700 µm high, hemispherical to globose-subglobose, generally aggregated, seldom dispersed, completely superficial when mature, black, with a well-defined brown ostiole; peridium 55–100 µm thick, tapering at the base, 55 µm, laterally to 60 µm and broader at the base of the ostiole, up to 100 µm wide, pseudoparenchymatous, composed of isoradiated cells of (10–) 16–20 × (11–) 12–15 µm, prismatic texture, slightly thick walls, 1–1.5 µm; pseudoparaphysis very thick up to 5 µm wide, hyaline, branched and anastomosed; asci (168–) 178–203 × 32–35 µm, bitunicate, fisitunicate, fusiform to soleiform, with internal apical beak, eight spores; ascospores 40–50 (–53) × 14–17 µm, ellipsoid or soleiform, 1-septate, slightly constricted at the septum, light brown to olive-brown, smooth.
Habitat: Gregarious on decaying wood.
Taxonomical notes: This species is characterized by having larger asci (168–)178–203 × 32–35 µm and ascospores 40–50(–53) × 14–17 µm. This species is morphologically similar to Kirschsteiniothelia thujina (Peck) D. Hawksw. due to the long ascomata (300–600 µm) and ascospores. However, K. thujina has a dark reddish-brown ostiole and an angular texture in the peridium, and the host is Abies balsamea Mill. and Thuja occidentalis L. Kirschsteiniothelia esperanzae has a brown ostiole and peridium cells with prismatic texture, and the host is not identified. This last species was collected in the Oreomunnea mexicana (Standl.) J.-F.Leroy TMCF of Oaxaca. Phylogenetic data confirm that K. esperanzae is a new species, close to K. thujina and K. rostrata Jing Yang & K.D. Hyde. These two species and K. arasbaranica Mehrabi, R. Hemmati & Asgari form a large clade. These three species have the largest ascospores of the group, more than 30 × 15 µm [35,36].
3.1.3. Geoglossomycetes, Geoglossales, Geoglossaceae
-
3.
Microglossum flavoviride Sánchez-Flores, García-Jiménez & Raymundo sp. nov.
MycoBank: MB842043.
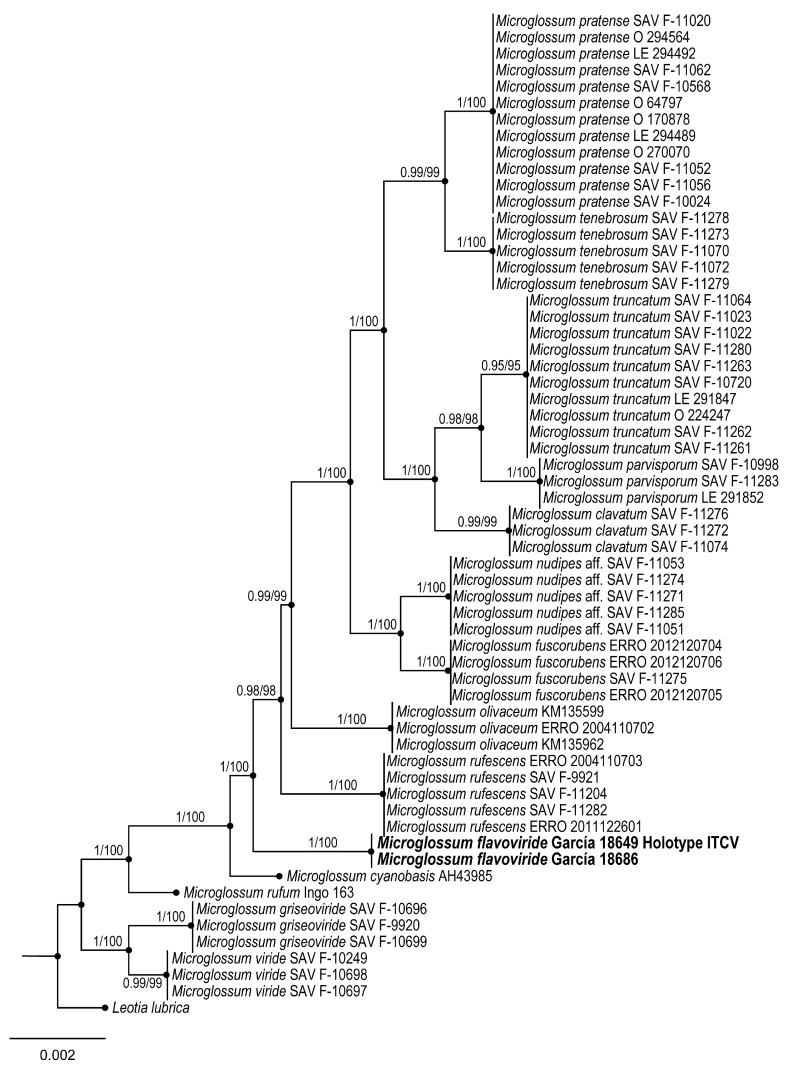
Figures: Figure 5 and Figure 6.
Figure 5.
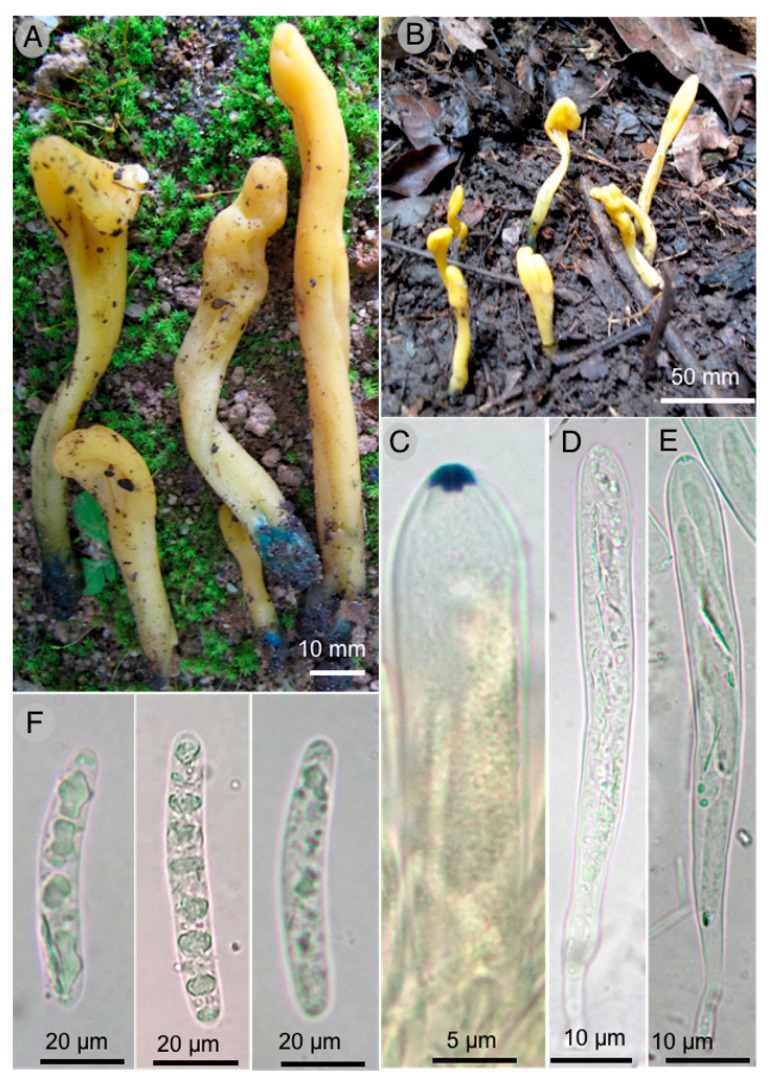
Microglossum flavoviride García 18649 Holotype: (A,B) ascomata; (C) amyloid operculum; (D) immature ascus; (E) mature ascus; (F) ascospores.
Figure 6.
Bayesian inference phylogram of ITS, LSU, and rpb2 sequence data. Posterior probability (left of slash) from Bayesian analysis and bootstrap support (right of slash) are given above the nodes. New species Microglossum flavoviride is shown in bold.
Diagnosis: Ascomata 16–65 × 4–9 mm, gregarious, lanceolate to spatulate, yellowish-green and deep green at the base, asci 111–160 × 11–14 µm, octosporate, hyaline, ascospores (20–) 22–45 × 4–6 (–6.5) µm, bacilliform, cylindrical, with 6–10 septa.
Type: MEXICO: Chiapas, Ocozocoautla municipality, Laguna Bélgica, 16°52′44.12″ N, 93°27′25.64″ W, 1004 m, 16 August 2011, J. García 18649 (ITCV, Holotype).
GenBank: ITS: OQ877254, LSU: OQ880483.
Etymology: It was named flavoviride for the ascoma color.
Ascomata 16–65 mm long, gregarious, lanceolate to spatulate, yellowish-green (30A7) color, deep green (27E8) at the base, cartilaginous consistency, viscous-moist texture; stipe 14–36 mm long, 2 mm wide toward the apex and 4–9 mm wide toward the base, flattened laterally, hollow, turns green when cut, fertile part 10–30 × 4–9 mm; medullar excipulus with intricate texture, formed by hyphae 3–8 µm in diameter, hyaline, indistinguishable subhymen; hymenium 160–185 µm thick; paraphyses 2–4 (–5) µm diameter, filiform, hyaline, septate, bifurcate toward the base, blunt apex, nodulous, irregular to rounded, hook-shaped to straight; asci 111–160 × 11–14 µm, octosporate, hyaline, clavate, amyloid operculum; ascospores (20–) 22–45 × 4–6 (–6.5) µm, bacilliform, cylindrical, slightly allantoic to spindle-shaped, hyaline, multigutulate, 6–10 septa not very visible.
Additional specimens: MEXICO. Chiapas, Ocozocoautla municipality, Laguna Bélgica, 16°52′44.12″ N, 93°27′25.64″ W, 1004 m, 16 August 2011, J. García 18686 (ITCV).
Taxonomical notes: Ascomata 16–65 mm long, yellowish-green and deep green at the base, ascospores (20–) 22–45 × 4–6 (–6.5) µm, bacilliform, cylindrical, slightly allantoic to spindle-shaped, hyaline, 6-10 septa. It can be confused with M. rufum (Schwein.) Underw. due to the color of the ascomata; however, this species presents granulations along the stipe and lacks the green tones at the base, with ascospores of similar size, although slightly smaller (18–) 20–36 (–40) × 4–6 µm, as well as smaller asci 100–135 × 9–12 µm [37]. It can be separated from M. fumosum (Peck) E.J. Durand by the size of the ascospores; the spores of the latter species are broader (16–) 20–40 (–48) × 4–5 µm, and the ascomata are pale yellow, cinnamon brown to reddish ocher [37]. It is distinguished from M. longisporum E.J. Durand by its cinnamon brown ascomata and larger ascospores 40–90 (–100) × 4–6 µm. Macroscopically, it resembles M. cyanobasis P. Iglesias & Arauzo due to the green color at the base of the ascoma; however, ascomata are brown and not yellow as in M. flavoviride, where the ascospores are smaller, 15.4–22.5 × 4.4–6.1 µm, and paraphyses present different forms [38]. Likewise, it has similar shades at the base to M. viride (Schrad. ex J.F. Gmel.) Gillet; however, ascospores of the latter species differ in size and shape, measuring (11–) 18–22 (–25) × (4–) 5–7 µm, and are elliptical to oblong, sometimes curved, and without visible septa [39].
3.1.4. Leotiomycetes, Helotiales, Helotiaceae
-
4.
Claussenomyces paulinae Raymundo
MycoBank: MB842044
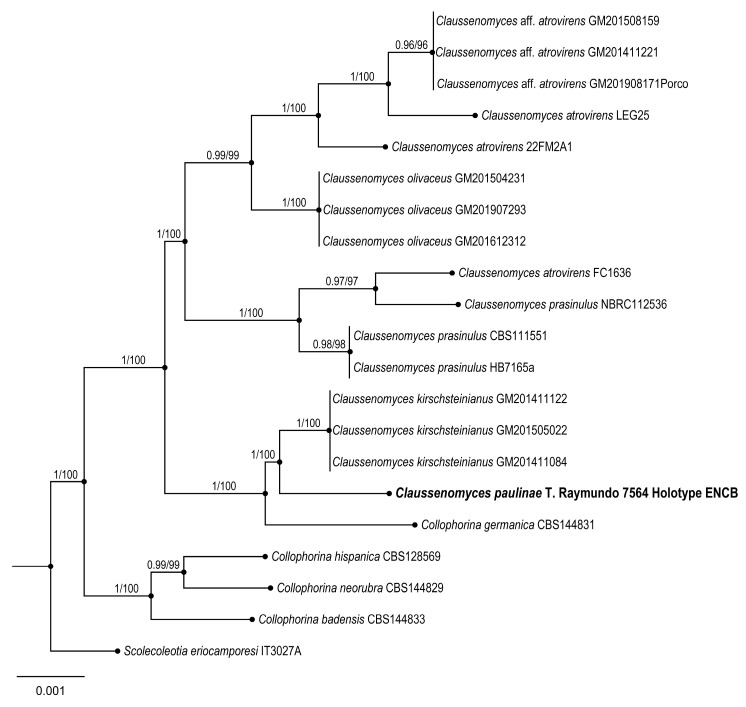
Figures: Figure 7 and Figure 8.
Figure 7.
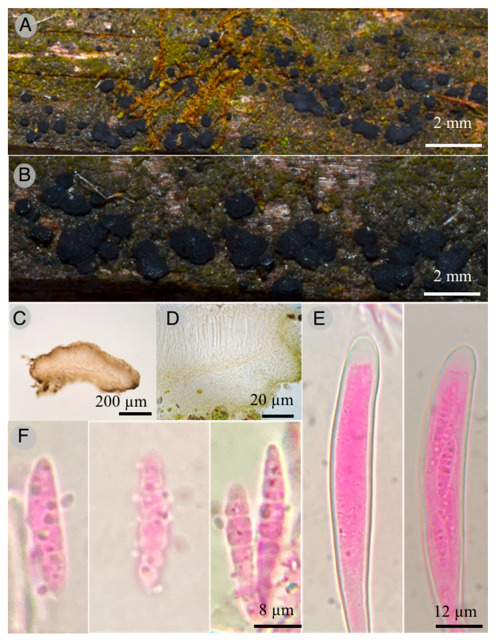
Claussenomyces paulinae T. Raymundo 7564 Holotype: (A,B) apothecia; (C,D) optical microscope images of apothecium; (E) optical microscope images of immature asci and mature asci with ascospores; (F) optical microscope images of ascospores with germination of conidia.
Figure 8.
Bayesian inference phylogram of ITS sequence data. Posterior probability (left of slash) from Bayesian analysis and bootstrap support (right of slash) are given above the nodes. New species Claussenomyces paulinae is shown in bold.
Diagnosis: Apothecia 600–800 μm diameter, discoid to flat, pulvinate, dark, gelatinous consistency; asci 85–120 × 8–10 μm, claviform, septum simple at the base, second basal cell presents crosier; ascospores 18–22 × 3–3.5 μm, fusoid, with three septa and smooth wall, curved, hyaline, some germinating and forming conidia 4 × 2 μm.
Type: MEXICO. Hidalgo: Zacualtipán de Ángeles municipality, El Hayal forest, 20°37′41.6″ N, 98°36′58.4″ W, 2000 m, 30 May 2018, T. Raymundo 7577 (ENCB).
GenBank: ITS: OQ877256.
Etymology: Dedicated to Rosa Paulina Calvillo Medina for her contributions to Mexican mycology.
Diagnosis: Apothecia 600–800 μm in diameter and 600–800 μm in height, flat pulvinate to discoid, bright black color, substipitate, with gelatinous consistency, slightly verrucose texture; ectal excipulum epidermoid to globular, with cells 14–20 μm in diameter, hyaline to pale yellow and green at the margin, thin and smooth walls. Intricate medullar excipulum with swollen hyphae 2 μm in diameter, tapering toward the margin; hymenium hyaline 110 μm thick, filiform paraphyses with capitate apices; asci 85–120 × 8–10 μm, claviform with blunt apices and simple septum at the base; second basal cell presents crosier, biseriate apically when young and uniseriate when mature, obliquely located, octosporic, hyaline; ascospores 18–22 × 3–3.5 μm, fusoid with seven septa and smooth walls, curved, hyaline, some germinating and forming conidia 4 × 2 μm, ovoid, hyaline.
Habitat: Saprotrophic species found on decaying wood of Pinus patula Schiede ex Schltdl. & Cham.
Additional specimens: MEXICO: Hidalgo, Zacualtipán de Ángeles municipality, El Hayal forest, 20°37′41.6″ N, 98°36′58.4″ W, 2000 m, 30 May 2018, R. Valenzuela 18282 (ENCB).
Taxonomical notes: This species has dark gregarious apothecia with jelly consistency, inamyloid asci, and ascospore fragments form secondary spores. Morphologically, it is similar to C. atrovirens (Pers.) Korf & Abawi, which differs by forming dark green apothecia and ascospores with 4–7 septate [40,41]. Another similar species is C. prassinulus (P. Karst.) Korf & Abawi, which has emerald green apothecia with ascospores 13–14 × 3–3.5 μm [42,43]. Phylogenetically, C. paulinae is confirmed as an independent lineage forming an independent branch.
3.1.5. Pezizomycetes, Pezizales, Chorioactidaceae
-
5.
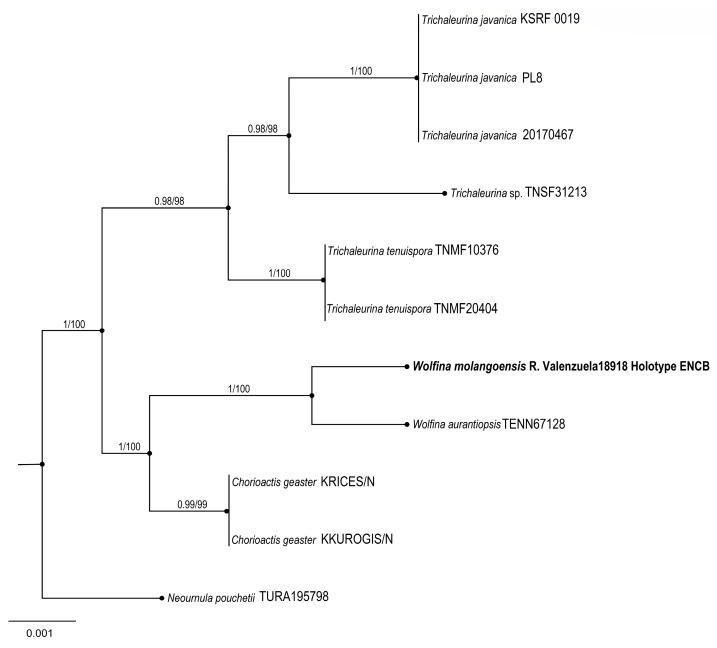
Wolfina molangoensis R. Valenz. & Raymundo
MycoBank: MB842045
Figures: Figure 9 and Figure 10.
Figure 9.
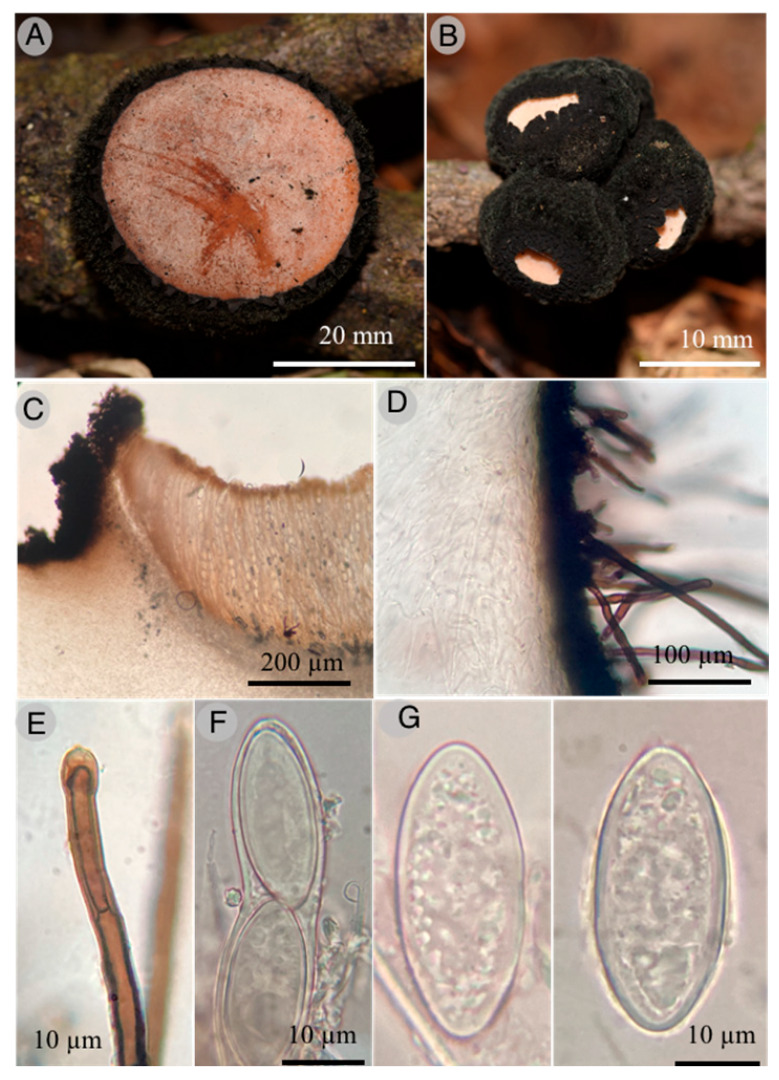
Wolfina molangoensis R. Valenzuela 18918 Holotype: (A) detail of apothecium; (B) apothecia; (C) optical microscope images of hymenium; (D) optical microscope images of ectal excipulum with hairs; (E) optical microscope images of detail of external hair; (F) optical microscope images of asci with ascospores; (G) optical microscope images of ascospores.
Figure 10.
Bayesian inference phylogram of ITS sequence data. Posterior probability (left of slash) from Bayesian analysis and bootstrap support (right of slash) given above the node. New species Wolfina molangoensis is shown in bold. Boldface names indicate samples sequenced for this study.
Diagnosis: Apothecia 20–60 mm diameter, cup-shaped to discoid, external surface black, external hairs velvety; asci 400–450 × 22–24 μm, cylindrical, operculate; ascospores 35–40 × 14–18 μm, elliptical to cylindrical, hyaline, with granular content, smooth, thick-walled.
Type: MEXICO: Hidalgo. Molango municipality, Laguna de Atezca, 20°48′32″ N, 98°44′52″ W, alt. 1281 m, 31 May 2018, R. Valenzuela 18918 (ENCB, Holotype).
GenBank: ITS: OQ877257
Etymology: The name refers to the Molango locality in Hidalgo state.
Apothecia cup-shaped to discoid, sessile, 20–60 mm in diameter; hymenium shallow, pale orange (6A5) to peach (7A4), external surface black, velvety, convoluted, flesh thick, firm, corky when dried; external hairs cylindrical, 4–8 μm diameter, septate, walls up to 1 μm thick, brown, entire, smooth with apex lanceolate; ectal excipulum with pseudoparenchymatous texture, epidermoid cells with thick wall, dark brown; medullar excipulum intricate texture, hyphae hyaline, simple septate, 2–4 μm, wide hyphae; subhymenium of thick texture intricata, septate, 3–4 μm wide hyphae, arranged perpendicular to the asci; paraphyses 3–4 μm diameter, filiform, septate, anastomosing; asci 400–450 × 22–24 μm, cylindrical, operculate, with acute apex, walls up to 2 μm thick, octosporic, hyaline and inamyloid, tapering base and flexuous; ascospores 35–40 × 14–18 μm, elliptical to cylindrical, hyaline with granular content, sharp ends, thick-walled and smooth.
Habitat: Grows on branches of angiosperms.
Additional specimens examined: MEXICO: Hidalgo, Laguna de Atezca, 20°48′32″ N, 98°44′52″ W, 1281 m, 1 June 2018, T. Raymundo 7640 (ENCB, Paratype).
Taxonomical notes: Morphological and phylogenetically, this new species is close to W. aurantiopsis (Ellis) Seaver ex Eckblad; however, W. aurantiopsis forms apothecia 25–45 mm with yellow to ochraceous hymenium and ascospores 25–32 × 10–15 μm, elliptical to cylindrical, hyaline with granular content, rounded ends with thin-walled and striate. Argnello et al. [26] noted that it might be restricted to the eastern USA. We found differences in the size and form of spores between species.
3.1.6. Sordariomycetes, Xylariales, Xylariaceae
-
6.
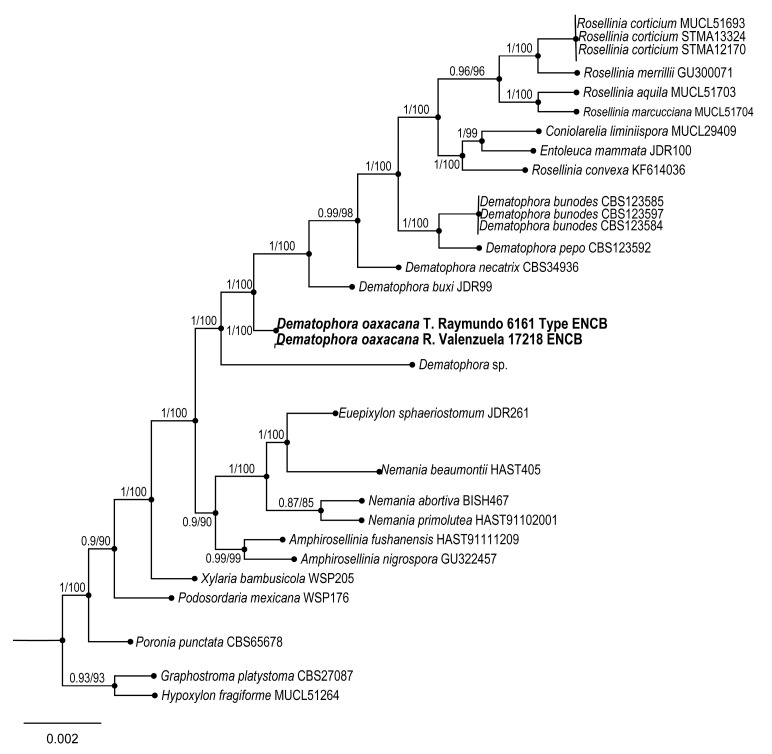
Dematophora oaxacana Sánchez-Flores, R. Valenz. & Raymundo sp. nov.
MycoBank: MB842051.
Figures:Figure 11 and Figure 12.
Figure 11.

Dematophora oaxacana T. Raymundo 6161 Holotype: (A) stromata; (B–D) detail of stromata surface; (E) optical microscope images of ascospores; (F) optical microscope images of ascospores showing germinal line.
Figure 12.
Bayesian inference phylogram of ITS, LSU, and rpb2 sequence data. Posterior probability (left of slash) from Bayesian analysis and bootstrap support (right of slash) given above the nodes. New species Dematophora oaxacana is shown in bold.
Diagnosis: Stromata 500–1100 × 400–700 µm, globose to subglobose, dark, solitary to gregarious, subiculum irregular extension, evanescent, carbonaceous, ascospores 20–29 × 10–13 (–14) µm, ovoid to asymmetrically ellipsoidal, with two cellular appendages, without germ slit.
Type: MEXICO: Oaxaca, Ixtlán de Juárez district, Santiago Comaltepec municipality, km 79 road Tuxtepec-Oaxaca, La Esperanza, Chinantla, 17°37′45″ N, 96°31′33″ W, 1130 m, 22 May 2017, T. Raymundo 6161 (ENCB, Holotype).
GenBank: ITS: OQ877258; nrLSU:OQ889487.
Etymology: The name refers to the state of Oaxaca, where this species was found.
Diagnosis: Stromata 500–1100 × 400–700 µm, globose to subglobose, dark to dark-brown, solitary, gregarious to cespitose, ostioles finely papillate to punctate; lack of subiculum; ectostroma dark, carbonaceous; endostroma 13–17 µm thick, light orange (5A4); perithecia not collapsed; asci dehiscent in 5% KOH; ascospores 20–29 × 10–13 (–14) µm, ovoid to asymmetrically ellipsoidal, brown, without germ slit, with flat sides ends, two cellular appendages, dehiscent in 5% KOH; external cellular appendage 3–5 µm tall and 5–6 µm wide, subglobose, hyaline; internal cellular appendage 1–2 µm tall and 2–3 µm wide, conical to subglobose, hyaline.
Habitat: Gregarious growing on decaying wood.
Distribution: Only known to be found in the state of Oaxaca.
Additional specimens examined: MEXICO, Oaxaca, Ixtlán de Juárez district, Santiago Comaltepec municipality, El Relámpago, La Esperanza, 17°35′28.1″ N, 96°53′52.2″ W, 1399 m, 29 May 2016, T. Raymundo 6161 (ENCB) and 6164 (ENCB). km 79 road Tuxtepec-Oaxaca, La Esperanza, Chinantla, 17°97′45″ N, 96°31′33″ W, 1130 m, 22 May 2017; 15 May 2015, R. Valenzuela 16111 (ENCB), 16145 (ENCB), T. Raymundo 5710 (ENCB). Loc. cit., 29 May 2016, R. Valenzuela 16667 (ENCB). Loc. cit., 22 May 2017, R. Valenzuela 17218 (ENCB), 17225 (ENCB), 17231 (ENCB), 17243 (ENCB), T. Raymundo 6587 (ENCB). Loc. cit., 23 May 2017, B. Nuñez 4 (ENCB), T. Raymundo 6607 (ENCB). Loc. cit., 30 April 2018, A. Cobos-Villagrán 1134 (ENCB). Paraje San Bernardo, La Esperanza, 17°37′55.4″ N, 96°22′1.5″ W, 25 September 2016, 1600 m, A. Trejo-Arana 17 (ENCB). Villa Alta district, Santiago Camotlán municipality, 5 km of Santiago Camotlán to San Juan Yatzuna, 24 March 2017, T. Raymundo (ENCB). Road Río Blanco, 25 March 2013, Galicia-Ávila 58 (ENCB). Santiago Camotlán, 25 March 2013, Escudero-Leyva 160 (ENCB).
Taxonomical notes: Ascospores measure 20–29.6 × 9.6–12 µm, without germline and double cell appendage. Phylogenetically, this species is close to Dematophora buxi (Fabre) C. Lamb., Wittstein & M. Stadler, differing from the latter in its macro and microscopic characteristics, as a more persistent subicula, with narrower ascospores 19.8–30.1 × 6–8.9 µm, fusoid, with straight germline and rounded apices. It is also similar to D. francisiae (L.E. Petrini) C. Lamb., Wittstein & M. Stadler; however, the latter has a persistent and felted subicula, 29–35 × 8–13 µm, longer ascospores, with a straight germline and rounded apices. Some species of Dematophora were earlier considered under the genus Rosellinia [44].
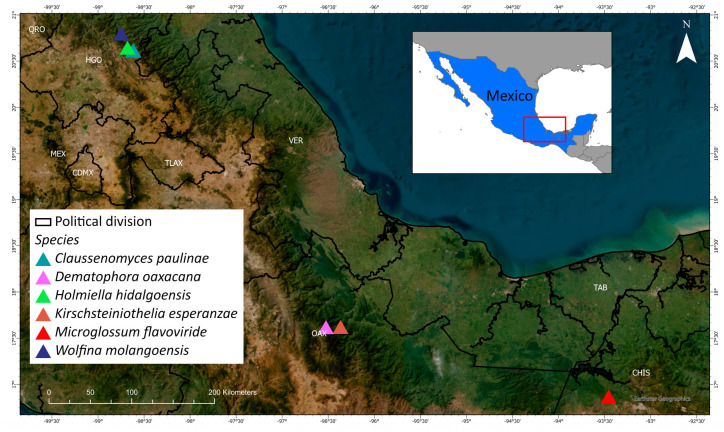
The distribution of the described new species is shown in Figure 13.
Figure 13.
Distribution of new species.
4. Conclusions
The Mexican tropical montane cloud forest (Figure 14) is one of the most diverse ecosystems for fungi. However, databases of other organisms, e.g., plants [45] and birds [46], but not fungi, are available for this ecosystem type. Unfortunately, they have not been extensively studied because of the lack of specialists; so, their representation in herbaria is poor. This study phylogenetically and morphologically describes six new species found in the Mexican TMCF.
Figure 14.

(A) View of Fagus tree in Mexican TMCF from Zacualtipán, Hidalgo. (B) View of Oreomunnea mexicana in La Esperanza, Oaxaca. (C) Tree components of Mexican TMCF in Zacualtipán, Hidalgo.
Characterizing fungal diversity in TMCFs is relevant for forest conservation. These forests provide environmental services such as terrestrial biomass and water degradation and are the source of bioactive secondary metabolites [9].
In 2017, Del Olmo et al. [9] reported 954 Ascomycota species from the Mexican TMCF, and other recent studies added different species to the Mexican TMCF mycobiota. For example, Raymundo et al. [47] described Marthamyces coronadoae, Raymundo et al. [48] described seven species of Hypocreales, Arias et al. [49] registered the asexual phases of 355 species, Medel-Ortiz et al. [50] found seven new records for the TMCF, and Raymundo et al. [41] recorded 10 new species in Mexico. Other studies that recorded new taxa are as follows (in chronological order): Sánchez-Flores et al. [51] described Hymenoscyphus herrerae from Puebla and registered six new species in the country; Raymundo et al. [43] recorded 17 new species from different TMCF localities; and Cobos-Villagrán et al. [52] registered Rhytidhysteron esperanzae and R. mesophila from Oaxaca and Hidalgo, respectively. In Puebla, three studies are relevant: Barbosa-Reséndiz et al. [53] described Daldinia rehmii, Raymundo et al. [54] recorded Unguiculariopsis ravenelii, and Sánchez-Flores et al. [55] described Ionomidotis mesophile. Then, Raymundo et al. [56] described Smardaea isoldae from Hidalgo, and Valenzuela et al. [57] added 10 new records for the TMCF in Oaxaca. In Veracruz, Chacón-Zapata and Gonzalez [58] described Euacanthe renispora, Guzmán-Guillermo et al. [59] described Paruephaedria heimerlii, and Chacón-Zapata and Ramirez-Guillén [60] listed 11 new records of Coronophorales. Finally, de la Fuente et al. [61] described Elaphomyces castilloi from Chiapas. The above information allows us to assume the existence of at least 1389 species inhabiting the Mexican TMCF. As González et al. [5] suggested, the precise number of species is difficult to establish due to nomenclature changes and the imprecision of Ascomycetes species identification.
Among the six new species described in this study, three species are distributed in Hidalgo, Sierra Madre Oriental, a mountainous area characterized by its abrupt topography and high beta diversity. Two were re-collected in Sierra de Juárez (Sierra Norte de Oaxaca), and one in Lagunas de Montebello, Altos de Chiapas, on the southern border with Guatemala. It is worth mentioning that the genera Holmiella and Wolfina are cited for the first time in the country.
Mexico is one of the world’s most diverse areas for fungi; so, it is essential to inventory and describe the fungal species in this type of ecosystem. TMCFs are the most threatened terrestrial ecosystems at the national level and are classified as “habitats in danger of extinction” [62]. In addition, a meta-analysis recently revealed that Mexico is a hotspot for oak species and their ectomycorrhizal mycobionts [63]. Those authors considered that the Mexican oak forests are essential for maintaining biodiversity due to the richness and endemism of fungi, mainly those associated with Fagaceae.
The loss of the TMCF is due to its transformation into grazing land for livestock and agriculture, mainly for avocados and coffee. The fungal abundance is strongly affected by the loss of this ecosystem type. The effects of global warming have not yet been evaluated in the case of these fungi.
Acknowledgments
We appreciate the fine suggestions of the three referees assigned and the authorities of La Esperanza, Tlanchinol, and El Hayal forests for their facilities to study the fungi species.
Author Contributions
T.R., J.G.-J., C.R.M.-G. and R.V. conceived this study. C.R.M.-G. helped with the phylogenetic analyses. T.R., R.V., J.G.-J., A.C.-V., M.S.-F., J.d.l.F., M.M.-P., A.P.-V., J.C.R.-M. and I.L.-V. described the new species. All authors reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare that there are no conflict of interest.
Funding Statement
This research was financed by Instituto Politécnico Nacional (SIP-20230017; SIP-20230642) and CONACYT Project 252934. J. García, J.I. de la Fuente, M. Sánchez-Flores, and I. Luna-Vega thank the Tecnológico Nacional de México-Instituto Tecnológico de Ciudad Victoria and CONACYT project 2015-01-2017 for financial support.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Decon J.W. Fungal Biology. Blackwell Publishing Ltd.; Hoboken, NJ, USA: 2006. p. 371. [Google Scholar]
- 2.Rzedowski J. La Vegetación de México. Limusa; Mexico City, Mexico: 1978. p. 432. [Google Scholar]
- 3.Luna-Vega I., Alcántara-Ayala O., Espinosa D., Morrone J.J. Historical relationships of the Mexican cloud forests: A preliminary vicariance model applying Parsimony Analysis of Endemicity to vascular plant taxa. J. Biogeogr. 1999;26:1299–1305. doi: 10.1046/j.1365-2699.1999.00361.x. [DOI] [Google Scholar]
- 4.Guzmán G. Análisis de los estudios de los macromicetos de México. Rev. Mex. Micol. 2008;28:7–15. [Google Scholar]
- 5.González M., Hanlin R.T. Distribution and occurrence of Ascomycetes in Mexico. North Amer. Fungi. 2008;3:139–145. doi: 10.2509/naf2008.003.0077. [DOI] [Google Scholar]
- 6.Heredia-Abarca G., Arias-Mota R.M., Becerra-Hernández C.I. Análisis de conocimiento de los hongos anamorfos saprobios de México. In: Heredia-Abarca G., editor. Tópicos Sobre Diversidad Ecología y Uso de Los Hongos Microscópicos. Programa Iberomaricano de Ciencia y tecnología para el Desarrollo (CYTED) e Instituto de Ecología, A.C. Xalapa; Xalapa, Mexico: 2008. 386p [Google Scholar]
- 7.Aguirre-Acosta E., Ulloa M., Aguilar S., Cifuentes J., Valenzuela R. Biodiversidad de hongos en México. Rev. Mex. Biodivers. 2014;85:76–81. doi: 10.7550/rmb.33649. [DOI] [Google Scholar]
- 8.Cifuentes J. Capital Natural de México, Vol. 1. Conocimiento actual de la biodiversidad Conabio; Mexico City, Mexico: 2008. Hongos. Catálogo taxonómico de especies de México. [Google Scholar]
- 9.Del Olmo-Ruiz M., García-Sandoval R., Alcántara-Ayala O., Véliz M., Luna-Vega I. Current knowledge of fungi from neotropical montane cloud forests: Distributional patterns and composition. Biodiv. Conserv. 2017;26:1919–1942. doi: 10.1007/s10531-017-1337-5. [DOI] [Google Scholar]
- 10.Kornerup A., Wanscher J.H. Methuen Handbook of Colour. 3rd ed. Eyre Methuen; London, UK: 1978. p. 252. [Google Scholar]
- 11.Bessette A.E., Roody W.C., Bessette A.R. North American Boletes: A Color Guide to the Fleshy PORED Mushrooms. 1st ed. Syracuse University Press; Syracuse, NY, USA: 2000. p. 396. [Google Scholar]
- 12.Ulloa M., Hanlin R.T. Illustrated Dictionary of Mycology. 2nd ed. APS Press; St. Paul, MI, USA: 2012. p. 762. [Google Scholar]
- 13.Martínez-González C.R., Ramírez-Mendoza R., Jiménez-Ramírez J., Gallegos-Vázquez C., Luna-Vega I. Improved method for genomic DNA extraction for Opuntia Mill. (Cactaceae) Plant Methods. 2017;13:82. doi: 10.1186/s13007-017-0234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 15.GenBank National Center of Biotechnology Information. National Library of Medicine. [(accessed on 1 March 2023)];2020 Available online: https://www.ncbi.nlm.nih.gov/genbank.
- 16.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 17.Pem D., Gafforv Y., Jeewon R., Hongsanan S., Promputtha I., Doilom M., Hyde K.D. Multigene phylogeny coupled with morphological characterization reveal two new species of Holiella and taxonomic insights within Patellariaceae. Cryptogam. Mycol. 2018;39:193–209. doi: 10.7872/crym/v39.iss2.2018.193. [DOI] [Google Scholar]
- 18.Katoh K., Misawa K., Kuma K.I., Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7, improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller K. SeqState. Appl. Bioinform. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- 22.Maddison W.P., Maddison D.R. Mesquite: A Modular System for Evolutionary Analysis, Version 3.70; 2021. [(accessed on 1 March 2023)]. Available online: http://mesquiteproject.org.
- 23.Sun Y.R., Jayawardena R.S., Hyde K.D., Wang Y. Krischsteiniothelia thailandica sp. nov. (Krischsteiniotheliaceae) from Thailandia. Phytotaxa. 2021;490:172–182. doi: 10.11646/phytotaxa.490.2.3. [DOI] [Google Scholar]
- 24.Healy A.H., Arnold A.E., Bonito G., Huang Y.L., Lemmond B., Pfister D.H., Smith M.E. Endophytism and endolichenism in Pezizomycetes: The exception of the rule? New Phytol. 2021;233:1974–1983. doi: 10.1111/nph.17886. [DOI] [PubMed] [Google Scholar]
- 25.Boonmee S., Wanasinghe D.N., Calabon M.S., Huanraluek N., Chandrasiri S.K.U., Jones G.E.B., Rossi W., Leonardi M., Singh S.K., Rana S., et al. Fungal diversity notes 1387–1511, taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021;111:1–335. doi: 10.1007/s13225-021-00489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argnello C., Carbone M., Braaten C. Wolfina aurantiopsis, a rare species in the family Chorioactidaceae (Pezizales) Ascomycete.Org. 2013;5:39–45. doi: 10.25664/art-0079. [DOI] [Google Scholar]
- 27.Wittstein K., Cordsmeier A., Lambert C., Wendt L., Sir E.B., Weber J., Wurzler N., Petrini L.E., Stadler M. Identification of Rosellinia species as producers of cyclodepsipeptide PF1022 A and resurrection of the genus Dematophora as inferred from polythetic taxonomy. Stud. Mycol. 2020;96:1–16. doi: 10.1016/j.simyco.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML version 8, a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frandsen P.B., Calcott B., Mayer C., Lanfear R. Automatic selection of partitioning schemes for phylogenetic analyses using iterative k-means clustering of site rates. BMC Evol. Biol. 2015;15:13. doi: 10.1186/s12862-015-0283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanfear R., Calcott B., Kainer D., Mayer C., Stamatakis A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014;14:82. doi: 10.1186/1471-2148-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B. Partition Finder 2, new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 32.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 33.Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rambaut A. FigTree, version 1.4.2. 2014. [(accessed on 1 March 2023)]. Available online: http://beast.bio.ed.ac.uk/Tracer.
- 35.Bao D.F., Luo Z.L., Liu J.K., Bhat D.J., Sarunya N., Li W.L., Su H.Y., Hyde K.D. Freshwater fungi in China III: New species and record of Kirshsteiniothelia from northwestern Yunnan province. Mycosphere. 2018;9:755–768. doi: 10.5943/mycosphere/9/4/4. [DOI] [Google Scholar]
- 36.Hyde K.D., Norphanphoun C., Abreu V.P., Bazzicalupo A. Fungal diversity notes 603–708, Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017;87:1–235. doi: 10.1007/s13225-017-0391-3. [DOI] [Google Scholar]
- 37.Mains E.B. North American hyaline-spored species of the Geoglosseae. Mycologia. 1955;47:846–877. doi: 10.1080/00275514.1955.12024501. [DOI] [Google Scholar]
- 38.Iglesias P., Arauzo S. Microglossum cyanobasis un Microglossum nuevo recolectado en Oriñón (Cantabria) Errotari. 2013;10:13–26. [Google Scholar]
- 39.Kučera V., Lizoň P., Tomšovský M., Kučera J., Gaisler J. Re-evaluation of the morphological variability of Microglossum viride and M. griseoviride sp. nov. Mycologia. 2014;106:282–290. doi: 10.3852/106.2.282. [DOI] [PubMed] [Google Scholar]
- 40.Hosoya T. Enumeration of remarkable Japanese Discomycetes (1) Three Helotialean Members New to Japan. Bull. Natn Mus. Tokyo Ser B. 2004;30:155–163. [Google Scholar]
- 41.Raymundo T., Valenzuela R., García-Martínez Y., Bravo-Álvarez M.A., Ramírez-Martínez J.C., Bautista-Hernández S., Palacios-Pacheco M., Luna-Vega I. Ascomycetes (Fungi) from the relic forest of Fagus grandifolia subsp. mexicana in eastern Mexico. Phytotaxa. 2019;418:1–41. doi: 10.11646/phytotaxa.418.1.1. [DOI] [Google Scholar]
- 42.Breitenbach J., Kränzlin F. Fungi of Switzerland. Volume 1. Verlag Mykologia; Luzern, Switzerland: 1984. p. 310. Ascomycetes. [Google Scholar]
- 43.Raymundo T., Valenzuela R., Ramírez-Martínez J.C., Martínez-Pineda M., Cobos Villagrán A., Trejo-Arana A., Sánchez Flores M., Gay-González A.D., Luna-Vega I. New records of Ascomycetes from the tropical montane cloud forests of eastern Mexico. Phytotaxa. 2020;454:161–185. doi: 10.11646/phytotaxa.454.3.1. [DOI] [Google Scholar]
- 44.Petrini L.E. Rosellinia—A World Monograph. Volume 205. Biblioteca Mycologica; Stuttgart, Germany: 2013. p. 410. [Google Scholar]
- 45.Luna-Vega I., Velázquez A., Velázquez E. México. In: Kappelle M., Brown A.D., editors. Bosques Nublados Del Neotrópico. INBio; Santo Domingo de Heredia, Costa Rica: 2001. pp. 183–229. [Google Scholar]
- 46.Sánchez-González L.A., Morrone J.J., Navarro-Singüenza A.G. Distributional patterns of the Neotropical humid montane forest avifaunas. Biol. J. Linn. Soc. 2008;94:175–194. doi: 10.1111/j.1095-8312.2008.00979.x. [DOI] [Google Scholar]
- 47.Raymundo T., Valenzuela R., Esqueda M. Marthamyces coronadoae sp. nov. in a Fagus grandifolia subsp. mexicana forest from Hidalgo State, México. Mycotaxon. 2016;131:521–526. doi: 10.5248/131.521. [DOI] [Google Scholar]
- 48.Raymundo T., Escudero-Leyva E., Soto-Agudelo R., García-Jiménez J., Romero-Bautista L., Valenzuela R. Nuevos registros de Hypocreales (Sordariomycetes, Ascomycota) del bosque mesófilo de monatña de la Sierra Alta Hidalguense en México. Acta Bot. Mex. 2017;120:39–57. doi: 10.21829/abm120.2017.1263. [DOI] [Google Scholar]
- 49.Arias R.M., Heredia-Abarca G., Castañeda-Ruiz R. Checklist of saprobic asexual microfungi from the tropical montane cloud forest of Veracruz, México. Mycotaxon. 2018;132:985. [Google Scholar]
- 50.Medel-Ortiz R., Loera-Hernández F.G., Baeza-Guzmán Y., Palestina-Villa E.N., Belingheri-Lagunes M.E. Ascomicetos asociados a angiospermas en el bosque mesófilo de montaña en el centro de Veracruz, México. Acta Bot. Mex. 2019;126:e1542. doi: 10.21829/abm126.2019.1542. [DOI] [Google Scholar]
- 51.Sánchez-Flores M., Valenzuela R., Hernández-Muñoz M.A., García-Jiménez J., Martínez-Pineda M., Raymundo T. Ascomicetos del bosque mesófilo de montaña de Honey, Puebla de los Ángeles, México. Acta Bot. Mex. 2020;127:e1719. doi: 10.21829/abm127.2020.1719. [DOI] [Google Scholar]
- 52.Cobos-Villagrán A., Valenzuela R., Hernández-Rodríguez C., Calvillo-Medina R.P., Villa-Tanaca L., Mateo-Cid L.E., Pérez-Valdespino A., Martínez-González C.R., Raymundo T. Three new species of Rhytidhysteron (Dothideomycetes, Ascomycota) from Mexico. MycoKeys. 2021;83:123–144. doi: 10.3897/mycokeys.83.68582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbosa-Reséndiz A., Valenzuela R., Sánchez-Flores M., Bautista-Hernández S., Cobos-Villagrán A., Pérez-Valdespino A., Espinoza-Mellado R., Martínez-Pineda M., Raymundo T. El género Daldinia (Sordariomycetes, Ascomycota) en México. Acta Bot. Mex. 2020;127:e1600. doi: 10.21829/abm127.2020.1600. [DOI] [Google Scholar]
- 54.Raymundo T., Martínez-Pineda M., Cobos-Villagrán A., Sánchez-Flores M., Valenzuela R. Primer registro de Unguiculariopsis ravenelii (Leotiomycetes, Ascomycota) en México. Acta Bot. Mex. 2020;127:e1666. doi: 10.21829/abm127.2020.1666. [DOI] [Google Scholar]
- 55.Sánchez-Flores M., Martínez-Pineda M., Raymundo T. Ionomidotis mesophila (Ascomycota, Cordieritidaceae), una especie nueva del bosque de niebla en México. Acta Bot. Mex. 2021;28:e1812. doi: 10.21829/abm128.2021.1812. [DOI] [Google Scholar]
- 56.Raymundo T., Valenzuela R. Smardaea isoldae sp. nov. from a tropical cloud forest in Mexico. Mycotaxon. 2021;136:97–106. doi: 10.5248/136.97. [DOI] [Google Scholar]
- 57.Valenzuela R., Raymundo T., Reyes P., Guzmán-Guillermo J., Acosta S., Ramírez-Martínez J.C., Luna-Vega I. Ascomycetes from the relic forest of Oreomunnea mexicana, Oaxaca, Mexico. Phyotaxa. 2021;528:19–44. doi: 10.11646/phytotaxa.528.1.3. [DOI] [Google Scholar]
- 58.Chacón-Zapata S., González D. Descripción de la segunda especie del género Euacanthe (Scortechiniaceae, Coronophorales), de áreas verdes urbanas y periurbanas de Xalapa, México. Acta Bot. Mex. 2021;128:e1835. doi: 10.21829/abm128.2021.1835. [DOI] [Google Scholar]
- 59.Guzmán-Guillermo J., Raymundo T., Sorcia-Navarrete P., Carvajal-Hernández C.I. Primer registro del hongo briofilo Paruephaedria heimerlii (Dactylosporaceae, Ascomycota) para México. Acta Bot. Mex. 2022;129:e2006. doi: 10.21829/abm129.2022.2006. [DOI] [Google Scholar]
- 60.Chacón-Zapata S., Ramirez-Guillén F. Especies conocidas y nuevos registros de Coronophorales (Ascomycota) en México. Acta Bot. Mex. 2022;129:e2051. doi: 10.21829/abm129.2022.2051. [DOI] [Google Scholar]
- 61.De la Fuente J.I., García-Jiménez J., Raymundo T., Sánchez-Flores M., Valenzuela R., Guevara-Guerrero G., Pérez-Ovando E.C., Ramírez-González C.R. Elaphomyces castilloi (Elaphomycetaceae, Ascomycota) and Entoloma secotioides (Entolomataceae, Basidiomycota), two new sequestrate fungi from tropical montane cloud forest from south Mexico. Mycokeys. 2023;96:127–142. doi: 10.3897/mycokeys.96.98320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Churchill S.P., Balslev H., Forero E., Luteyn J.L., editors. Biodiversity and Conservation of Neotropical Montane Forests; Proceedings of the Neotropical Montane Forest Biodiversity and Conservation Symposium; New York, NY, USA. 21–26 June 1993; No. 26. [Google Scholar]
- 63.García-Guzmán O.M., Garibay-Orijel R., Hernández E., Arellano-Torres E., Oyama K. World-wide meta-analysis of Quercus forests ectomycorrhizal fungal diversity reveals southwestern Mexico as a hotspot. Mycorrhiza. 2017;27:811–822. doi: 10.1007/s00572-017-0793-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.