Abstract
We developed a new cytomegalovirus (CMV) immunoglobulin M (IgM) immunoblot to detect CMV-specific IgM in human sera. The new test contains four viral proteins (vp150, vp82, vp65, and vp28) purified from viral particles and four recombinant proteins (rp150, rp130, rp52, and rp38) purified from Escherichia coli. These antigens were individually loaded onto nitrocellulose strips, and the strips were then used to detect CMV-specific IgM by using a μ-specific conjugate. The new assay was evaluated in parallel with one or two IgM enzyme-linked immunosorbent assays (ELISAs) to test 592 serum samples from different groups of latently or acutely infected individuals. The sensitivity of the new assay with respect to the consensus of two ELISAs was 100%, the specificity was 98.6%, the positive predictive value was 96.9%, and the negative predictive value was 100%. We also evaluated the new test by testing sera from pregnant women and transplant recipients with a known clinical history. Our results suggest that the new test combines high sensitivity with high specificity, characteristics that are mutually exclusive with the other commercially available tests. Furthermore, a statistically significant correlation was observed between the number of IgM-reactive bands and the elevated risk of transmission from CMV-infected pregnant women to their offspring.
Cytomegalovirus (CMV)-specific immunoglobulin M (IgM) is a sensitive and specific indicator of an ongoing or recent CMV infection (2, 6, 23, 26, 33). Serum IgM to CMV can be revealed by a variety of different tests; the most widely used is the enzyme-linked immunosorbent assay (ELISA) (9, 15, 24, 25, 33, 35, 36). Many different ELISAs for CMV IgM are commercially available, and poor agreement has been found among the results obtained by different ELISA kits (16, 20). Western blotting (WB) with viral polypeptides separated from purified viral particles has repeatedly been shown to be an effective method to detect CMV-specific IgM (2, 4, 7, 10, 18), and p150, p82, p65, and p38 are the most immunoreactive antigens.
False-positive WB results have been observed when a serum reacts exclusively with pp150 because two proteins overlap at that molecular weight (ppUL32 and pUL86), and one of them (pUL86) is the herpesvirus group common antigen. To avoid such a false-positive result and therefore to improve WB specificity, a serum sample reacting exclusively with p150 should be confirmed with a p150 recombinant antigen containing significant epitopes of ppUL32.
The sensitivity of the WB assay can also be improved. In fact, at least two CMV nonstructural proteins have been shown to be highly reactive with CMV IgM. These proteins are ppUL44 (pp52), an abundant nuclear phosphoprotein, and pUL57(p130), a DNA-binding protein (5). These proteins are not present in the conventional WB, which contains only viral structural proteins.
Therefore, we have developed an improved version of the WB assay (newWB) (19). Viral structural polypeptides separated by polyacrylamide gel electrophoresis (PAGE) were subjected to WB, along with three recombinant proteins containing significant portions of ppUL32(p150), ppUL44(p52), and ppUL57(p130), as well as two additional control proteins (the carrier CKS protein as a negative control and human μ chain as a positive control).
Despite the good performance obtained with the newWB, we realized that the assay has three limitations. First, it is difficult to correctly interpret the newWB due to the presence of some minor viral reactive bands distinct from p150, p82, p65, p38, and p28. Second, the viral reactive bands on the newWB are not uniform in size due to the various amounts of posttranslational modification of the different viral proteins that occur. For example, the vp150 reactive band is very sharp and thin, whereas the vp65 reactive band is very broad (19). These differences make it difficult to determine the cutoff intensity of the bands accurately. Third, it is difficult to standardize the newWB because the quantitative and qualitative composition of viral proteins present in different preparations of purified viral particles is variable. To overcome these limitations, we developed an improved version of the test (CMV IgM immunoblot) which contains individual purified viral and recombinant antigens uniformly loaded in fixed amounts on nitrocellulose strips. We tested the performance of the new immunoblot with 592 serum samples from different groups of latently or acutely infected individuals.
MATERIALS AND METHODS
Virus and cells.
The Towne strain of CMV was propagated in human embryo fibroblasts by using standard methods. The virus was purified by using a sorbitol cushion followed by a sorbitol gradient as previously described in detail (11, 12).
Viral antigens. (i) Recombinant proteins.
The following Escherichia coli CMP–2-keto-3-deoxyoctulosonic acid synthetase (CKS) recombinant proteins were used: (i) two ppUL32 regions (amino acids [aa] 595 to 614 and 1006 to 1048) fused together, which can replace the IgM-binding ability of the entire p150 molecule (28); (ii) the carboxy-terminal part of ppUL44 (aa 202 to 434), which contains highly reactive epitopes for IgM and does not contain relevant amino acid sequences cross-reacting with the homologous proteins of other members of the family Herpesviridae (29); (iii) two segments of ppUL57 (aa 540 to 601 and 1144 to 1233) previously shown to be very reactive with serum IgM (22, 37); and (iv) a significant portion of assembly protein ppUL80a (aa 117 to 373) (13). Insoluble CKS-CMV fusion proteins were initially purified after lysis by a combination of detergent washes followed by solubilization in 8 M urea (30). After solubilization, fusion proteins were further purified by Q-Sepharose chromatography (Pharmacia Biotech). Soluble CKS-CMV fusion proteins and soluble CKS protein were purified after cell lysis by preparative sodium dodecyl sulfate-PAGE with a Bio-Rad Prep Cell (Bio-Rad Laboratories, Richmond, Calif.).
(ii) Authentic viral proteins.
The following viral proteins were used: (i) a protein with a molecular mass of 150 kDa (vp150); (ii) a protein with a mass of 82 kDa (vp82); (iii) a protein with a mass of 65 kDa (vp65); and (iv) a protein with a mass of 28 kDa (vp28) (14, 31). Viral particles were purified by using a sorbitol cushion followed by a sorbitol gradient as previously described in detail (11, 12). Purified viral particles were then lysed in PAGE loading buffer by boiling in the presence of β-mercaptoethanol, and structural components were separated and purified by preparative PAGE with a Bio-Rad Prep Cell. Protein purity was checked by PAGE and Coomassie staining, as well as by WB and testing of reactivity with CMV-positive human sera.
CMV serology. (i) Conventional ELISA.
The evaluation of anti-CMV IgG was carried out with two commercial kits: (i) the Enzygnost Anti-CMV/IgG ELISA alpha method (Behring AG, Marburg, Germany) and (ii) Cytomegalovirus IgG EIA WELL (Radim, Rome, Italy). Plates were read on a microELISA automatic reader (Behring AG). Evaluation of anti-CMV IgM was also performed by using two different assays: (i) the Enzygnost Anti-CMV/IgM kit (Behring AG) and (ii) ETI CVTOK-M reverse PLUS (DiaSORIN, Vercelli, Italy). Both kits were used and the results were interpreted as suggested by the manufacturers.
(ii) Serum samples.
In this study, 592 human serum samples we tested from blood donors, healthy adults, pregnant women, and allograft recipients according with our ethical rules. For determination of the immunoblot algorithm, we used 286 IgM-negative and 126 IgM-positive serum samples. The 286 IgM-negative serum samples were obtained from 100 randomly selected blood donors through the courtesy of the Blood Transfusion Center of the St. Orsola General Hospital, Bologna, Italy, and from 186 healthy adults. Two hundred serum samples were CMV IgG positive, and 80 were CMV IgG negative, as determined with two different ELISA kits. Six serum samples gave discordant IgG results and were not included in the study.
The 126 IgM-positive serum samples were from immunocompetent subjects (mainly pregnant women and healthy adults). They were determined to be CMV IgM positive by two different ELISA kits.
Another group of serum samples consisted of 51 from pregnant women (13 were from CMV-uninfected women, 23 were from CMV-infected women who did not transmit the infection, and 15 were from CMV-infected pregnant women who transmitted the infection). All of these sera were obtained between 21 and 24 weeks of gestation.
Another group of samples consisted of 123 serum samples from 35 transplant patients (29 heart and 6 kidney transplant recipients) who underwent human CMV infection during the first 6 months after transplantation. Ten patients had a primary infection, while 25 underwent viral reactivation or reinfection.
We also tested six rheumatoid factor-positive serum samples.
Diagnosis of active CMV infection.
For immunocompromised patients, antigenemia and/or PCR on polymorphonuclear leukocytes (PMNL) was performed. The presence of CMV pp65 (ppUL83) in PMNL (antigenemia) of immunocompromised patients was determined as originally described by van der Biji et al. (34) and modified by Revello et al. (27) by using a CMV pp65-specific pool of two monoclonal antibodies (1C3 and AYM-1 from Argene, Varilhes, France) in indirect immunofluorescence tests. The presence of the CMV genome in PMNL of immunocompromised patients was detected by PCR. Aliquots of 5 × 105 PMNL were used, and the PCR was carried out as previously described (17).
CMV infection in pregnant women was determined by one or more of the following parameters: virus isolation from urine, saliva, or blood; seroconversion for anti-CMV IgM and IgG antibodies; and the presence of low-avidity IgG antibody (21).
A congenital CMV infection in a newborn was determined by CMV isolation from urine during the first week of life (32).
The new immunoblot.
A sheet of nitrocellulose was inserted into a miniblotter device for blotting of the antigens. Individual suspensions of each of the four viral proteins (vp150, vp82, vp65, and vp28) and the four recombinant proteins (rp150, rp52, rp130, and rp38) in 50 mM Tris-HCl–5 mM EDTA (pH 9), were deposited into the miniblotter wells. Furthermore, two additional control proteins were deposited onto the nitrocellulose. The CKS protein was added as a negative control to monitor for the presence of serum IgM to the bacterial portion of the fusion protein. Human μ chain (IgM) was added as a positive control to monitor the reaction of the conjugate to human IgM. Miniblotters were then gently agitated on a rocking platform overnight at room temperature. The filters were washed briefly in Tris-buffered saline (TBS) and then saturated by incubation with a blocking solution (3% fish gelatin, 1% bovine serum albumin, 5% powdered skim milk, 0.05% Tween 20 in TBS) at room temperature for 1 h. The filters were then cut in 3-mm-wide strips, carrying both viral authentic proteins (at the top) and recombinant polypeptides (at the bottom). The final amounts of viral proteins per strip were 25 ng of vp150, 125 ng of vp65, 50 ng of vp82, 12.5 ng of vp28, 8 ng of rp150, 28.5 ng of rp52, 70 ng of rp130, 15 ng of rp38, 25 ng of CKS, and 25 ng of μ chain.
All of the sequences encoding human CMV products used as recombinant antigens in the assays were submitted to the Blast server of the National Center for Biotechnology Information at the National Institutes of Health for alignment with all known sequences deposited in the GenBank, EMBL, DDBJ, and PDB databases by using BLASTN 2.0.4, the alignment search tool developed by Altschul et al. (1). The results showed no significant homologies between the sequences submitted and the genomic sequences of any of the following organisms deposited in the above databases: herpes simplex virus type 1, herpes simplex virus type 2, human herpes virus 6, human herpes virus 7, Epstein-Barr virus, varicella-zoster virus, rubella virus, hepatitis B virus, hepatitis C virus, and Toxoplasma gondii.
Serum samples were diluted 1:100 in TBS with 10% fetal calf serum and 100-μg/ml CKS. Incubation was performed at room temperature for 1 h. After three washes with phosphate-buffered saline–Tween 20, a goat anti-human μ chain–alkaline phosphatase conjugate (Abbott) diluted 1:1,500 in TBS with 5% fetal calf serum was added and the mixture was incubated at room temperature for 1 h. The filters were subsequently washed three times as before and developed for 15 min with 5-bromo-4-chloro-3-indolyl phosphate toluidinium (salt) (BCIP–nitroblue tetrazolium) (Sigma). After development of the blot, reactive bands were scored.
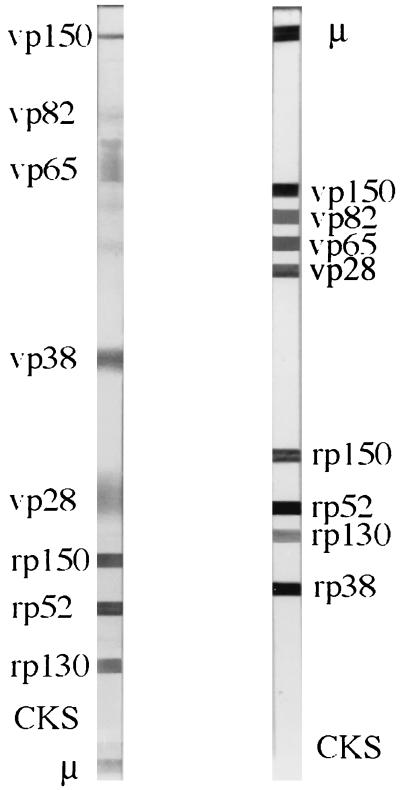
A side-by-side display of the old and new blots is presented in Fig. 1.
FIG. 1.
Side-by-side display of the old (left) and new (right) blots. The same serum sample, at the same dilution, was used for both blots.
Statistics.
Sensitivity, specificity, and positive and negative predictive values were determined as described by Griner et al. (8).
Student’s t test and the Mann-Whitney test were used to estimate the significance of the difference between the numbers of bands observed in serum samples from infected women who transmitted the infection and in those from women who did not transmit CMV to their offspring.
RESULTS
Reactivity with IgM-negative sera.
A total of 67 (84%) of 80 IgG- and IgM-negative sera were nonreactive to all of the antigens on the new immunoblot. Of the remaining 13 sera, 12 (15%) were reactive only to recombinant proteins and 1 was reactive to a viral protein. The highest reactivity observed in this group of sera was to rp52 (7.5%), followed by rp150 and rp130 (2.5% each). In no case could we detect reactivity with a combination of viral and recombinant proteins.
In addition, 162 (81%) of 200 IgG-positive, IgM-negative sera were nonreactive by the new immunoblot. The remaining 38 sera (19%) gave several different combinations of IgM reactivity. The highest reactivity was observed against vp150 alone (10%), followed by rp52 alone (3.5%). Four sera (2%) reacted with two viral proteins, two sera reacted with two recombinant proteins, and another four sera (2%) reacted with a combination of one viral protein and one recombinant protein.
Reactivity with IgM-positive sera.
The 126 human sera that were determined to be IgM positive by two different ELISA kits were assayed by the new immunoblot. No individual serum reacted exclusively with viral proteins. On the contrary, 11 sera (8.7%) reacted with three or four recombinant proteins only. Many reactivity combinations of viral and recombinant proteins were observed in the remaining 115 (91.3%) of the 126 sera. Of the sera tested, 25% reacted with one viral protein and one or more recombinant proteins. Viral protein vp150 was reactive with all of these sera, with the exception of five sera which were reactive with vp65. In addition, 18 sera (14.3%) reacted with vp150 and rp150 only. Only a few sera (2.4%) showed positive reactivity with all four viral proteins and one or more recombinant proteins. Representative examples of serum reactivity to antigens on the new immunoblot are shown in Fig. 2.
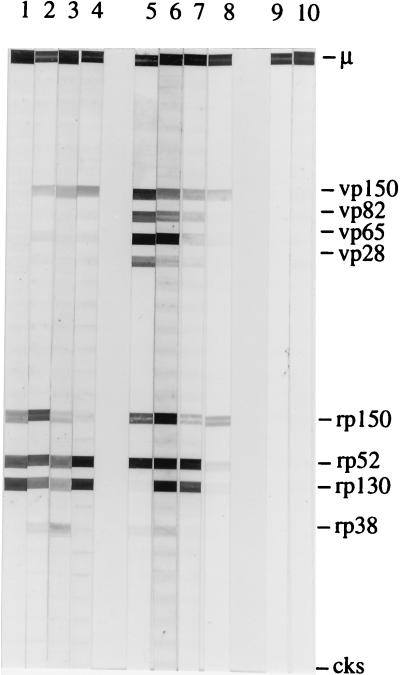
FIG. 2.
Representative examples of serum reactivity with the new immunoblot. Viral and recombinant proteins are identified on the right. CKS is the negative control, and μ is the IgM heavy chain and represents the positive control. Lanes: 1 to 8, IgM-positive sera from pregnant women; 9 and 10, IgM-negative sera from pregnant women. Sera 1 to 4 preferentially reacted with recombinant proteins, while sera 5 to 8 reacted with both viral and recombinant proteins. Sera 5 and 6 were from pregnant women who transmitted the infection.
Algorithm for interpretation of the new immunoblot.
On the basis of the results obtained, we established that the new immunoblot test result was positive when both sections of the strip contained at least one reactive protein band in each section. The test result was also positive when at least three recombinant protein bands were reactive, regardless of the number of reactive protein bands in the viral section of the blot. The new immunoblot test result was negative when there were no viral or recombinant protein reactive bands.
A positive reaction with the human μ-chain band and no reaction with the CKS carrier protein band were necessary to confirm the validity of the assay.
With this algorithm, 4 (1.4%) of 280 serum samples from blood donors were positive for CMV-specific IgM and all 126 IgM-positive sera were positive for CMV-specific IgM with the new immunoblot test. Based on these data, the sensitivity and specificity of the new assay relative to the consensus of two ELISAs were 100 and 98.6%, respectively. The positive and negative predictive values of the assay were 96.8 and 100%, respectively.
By using this algorithm, we tested six sera that were rheumatoid factor positive and had IgG for CMV. No positivity was detected.
Evaluation of the new immunoblot test with sera from pregnant women.
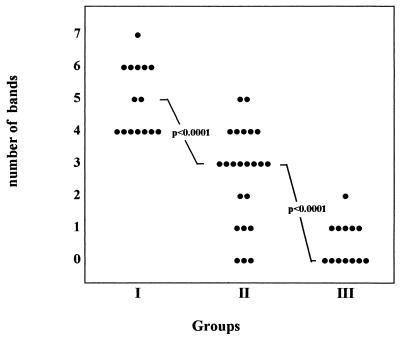
Among 15 CMV-infected pregnant women (group I) who transmitted the infection to their offspring, all were positive by the new immunoblot and 7 were positive by the consensus of two ELISAs. Among 23 CMV-infected pregnant women who did not transmit the infection (group II), 17 were IgM positive by the new immunoblot and 9 were positive by the ELISAs. Among 13 uninfected women (group III), no positivity was observed with either method. Analyzing the strips, we noted a difference between the numbers of IgM-reactive bands in the three groups of women. As shown in Fig. 3, while six or seven bands were reactive with some of the sera from infected pregnant women who transmitted the infection (group I), zero to three bands were reactive with sera from infected pregnant women who did not transmit the infection (group II). Four or five bands were reactive with sera from both groups I and II. Seven sera from uninfected pregnant women (group III) were completely nonreactive, five sera were reactive to one band, and one serum was reactive to two bands. The difference between the numbers of bands observed in groups I and II was statistically significant, with a P of <0.0001 obtained with both the Mann-Whitney test and the Student t test.
FIG. 3.
Distribution of the number of IgM-reactive bands in different groups of pregnant women. Groups: I, infected pregnant women who transmitted the infection; II, infected pregnant women who did not transmit the infection; III, uninfected pregnant women.
Evaluation of the new immunoblot test with sera from transplant recipients.
The new immunoblot and the ELISAs were also compared for the ability to detect CMV-specific IgM in a group of 10 transplant recipients undergoing a primary CMV infection. These patients were also monitored by the antigenemia test. No cases of false positivity were detected before infection (2 to 4 weeks before the first positive antigenemia assay) by either test. At the beginning of the infection (first positive antigenemia assay), 4 of the 10 patients were IgM positive by the new immunoblot and 1 was positive by the consensus of the two ELISAs. Later (second antigenemia-positive assay), 6 to 8 days after the previous test, all 10 patients were IgM positive by both the immunoblot and the ELISAs. A comparison between IgM detection by the new immunoblot and the ELISAs in a follow-up of 25 transplant recipients undergoing a nonprimary CMV infection indicated that no nonspecific reactions were detected by the new immunoblot and by the ELISAs in sera obtained from 20 patients 2 to 4 weeks before infection. One week before infection, two sera were IgM positive by immunoblotting and one of them was also positive by the ELISAs. At the beginning of infection, corresponding to the first positive antigenemia assay, the new immunoblot detected IgM in 10 patients, whereas the ELISAs detected IgM in only 2. Later during infection, the new immunoblot and the ELISAs detected IgM in 24 and 8 patients, respectively.
DISCUSSION
In this report, we describe the development of a new CMV IgM immunoblot test for detection of CMV-specific IgM which contains structural viral proteins purified from viral particles and recombinant structural and nonstructural proteins obtained by molecular biology. The reason we developed the newWB test (19), composed of an upper portion consisting of viral proteins resulting from electrophoretic separation of whole viral particles and a lower portion containing recombinant proteins, was to improve the specificity of the WB assay. Although this assay proved to be much better than the conventional WB, it is difficult to interpret correctly because of the presence of some minor viral bands and the lack of band uniformity. Furthermore, we found difficulties in the standardization of the new test because the quantitative and qualitative composition of viral proteins present in different preparations of purified viral particles is variable. To overcome these limitations, we developed a new version of this assay which differs from the previous one in that the strips contain fixed amounts of purified viral proteins blotted uniformly onto the nitrocellulose. The first part of our study consisted of testing sera that were judged IgM positive or negative by a consensus of two different commercially available ELISA kits, one of which has been proved to be sensitive but nonspecific and the other of which was found to be specific but not sensitive (16). Testing IgM-negative sera from healthy adults and blood donors and IgM-positive sera mainly from pregnant women, we observed that 98.6% of the sera from healthy adults and blood donors either did not show any reactivity to the proteins present in the new immunoblot or showed some reactivity to proteins in one of the two sections of the blot exclusively. On the other hand, 100% of the IgM-positive sera, mainly from pregnant women, reacted with proteins present in both portions of the new immunoblot or with at least three recombinant proteins. For this reason, we decided to assign a positive result exclusively to sera showing reactivity to at least one band in the recombinant section of the blot together with reactivity to at least one band in the viral section of the blot or to at least three recombinant proteins.
With this algorithm, 100% of the IgM-positive sera were shown to be positive by the new immunoblot. The sensitivity of the new assay with respect to the consensus of two ELISAs is 100%, the specificity is 98.6%, the positive predictive value is 96.9%, and the negative predictive value is 100%. Therefore, like the newWB, the new immunoblot also combines high sensitivity with high specificity, characteristics that are mutually exclusive with the other commercially available tests.
The second part of our study consisted of using the new test in combination with a consensus of the two commercially available ELISAs in searching for CMV-specific IgM in sera from subjects whose clinical conditions were known. In a group of CMV-infected pregnant women at 21 to 24 weeks of gestation, the new immunoblot detected 100% of those who transmit the infection to their offspring while the ELISAs missed 53% of them. In a group of CMV-infected pregnant women who did not transmit the infection to their offspring, the new test gave a positive result in 74% of the cases while the ELISAs gave a positive result in 39%. No nonspecific reactivities were detected by the ELISAs or the new immunoblot in non-CMV-infected pregnant women. Interestingly, counting the number of bands recognized by IgM present in sera from CMV-infected women, we observed a similar result for CMV-specific IgM that was already described in the literature by Boppana and Britt for CMV-specific IgG (3). Our data indicate that serum IgM from women who transmit CMV infection reacts with a higher number of bands than does serum IgM from those who do not transmit the infection (P <0.0001), probably reflecting a higher viral load in the former. These results suggest that when the number of reactive bands is four or greater, the risk of transmission of the infection to the fetus is higher. Prenatal diagnosis should be offered in these cases to confirm actual transmission of the virus.
We also followed up a group of 35 transplant recipients undergoing acute CMV infection within the first 6 months after transplantation. Although more transplant recipients should be tested before significant conclusions are reached, it seems that during primary infection, the new immunoblot can detect infection earlier than ELISAs. The ability to detect IgM by the new test was also better than that of the ELISAs, particularly in persons with nonprimary CMV infections, among whom 96% of the cases were detected. On the contrary, the ELISAs were positive in only 32% of the cases. Although serological diagnosis, in general, is less attractive than virological diagnosis because of the immunological disorders occurring in transplant recipients, when performed with a sensitive and specific test, it could represent an additional means of obtaining a diagnosis of ongoing CMV infection.
We believe that this new assay can be considered a reference test for CMV IgM serology, and a field clinical evaluation will begin soon in preparation for commercialization of this new test.
ACKNOWLEDGMENTS
This work was partially supported by the University of Bologna and the Italian Ministry of Education.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basson J, Tardy J C, Aymard M. Pattern of anti-cytomegalovirus IgM antibodies determined by immunoblotting. A study of kidney graft recipients developing a primary or recurrent CMV infection. Arch Virol. 1989;108:259–270. doi: 10.1007/BF01310938. [DOI] [PubMed] [Google Scholar]
- 3.Boppana S B, Britt W J. Antiviral antibody responses and intrauterine transmission after primary maternal cytomegalovirus infection. J Infect Dis. 1995;171:1115–1121. doi: 10.1093/infdis/171.5.1115. [DOI] [PubMed] [Google Scholar]
- 4.Braun W, Weber B, Moell U, Hamann A, Doerr H W. Immunoglobulin A and M patterns to human cytomegalovirus during recurrent infection in patients with AIDS using a modified Western blot. J Virol Methods. 1993;43:65–76. doi: 10.1016/0166-0934(93)90090-e. [DOI] [PubMed] [Google Scholar]
- 5.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny T, Horsnell T, Hutchinson C A, Kouzarides T, Martinetti J A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD 169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 6.Dolan J, Briggs J D, Clements G B. Antibodies to cytomegalovirus in renal allograft recipients: correlation with isolation of virus. J Clin Pathol. 1989;42:1070–1077. doi: 10.1136/jcp.42.10.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gold D, Ashley R, Handsfield H H, Verdon M, Leach L, Mills J, Drew L, Corey L. Immunoblot analysis of the humoral immune response in primary cytomegalovirus infection. J Infect Dis. 1988;157:319–325. doi: 10.1093/infdis/157.2.319. [DOI] [PubMed] [Google Scholar]
- 8.Griner P F, Mayewski R J, Mushlin A I, Greenland P. Selection and interpretation of diagnostic tests and procedures. Principles and applications. Ann Intern Med. 1981;94:557–592. [PubMed] [Google Scholar]
- 9.Kraat Y J, Stals F S, Landini M P, Bruggeman C A. Cytomegalovirus IgM antibody detection: comparison of five assays. New Microbiol. 1993;16:297–307. [PubMed] [Google Scholar]
- 10.Kraat Y J, Stals F S, Christiaans M H L, Lazzarotto T, Landini M P, Bruggeman C A. IgM antibody detection for 38 (ppUL80a) and 150 (ppUL32) kDa proteins by immunoblotting: the earliest parameter for acute cytomegalovirus infection in renal transplant recipients. J Med Virol. 1996;48:289–294. doi: 10.1002/(SICI)1096-9071(199603)48:3<289::AID-JMV13>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Landini M P, Ripalti A. A DNA-nicking activity associated with the nucleocapsid of human cytomegalovirus. Arch Virol. 1982;78:351–356. doi: 10.1007/BF01318089. [DOI] [PubMed] [Google Scholar]
- 12.Landini M P, Rossier E, Schmitz H. Antibodies to human cytomegalovirus structural polypeptides during primary infection. J Virol Methods. 1988;22:309–317. doi: 10.1016/0166-0934(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 13.Landini M P, Guan M X, Jahn G, Lindenmaier W, Mach M, Ripalti A, Necker A, Lazzarotto T, Plachter B. Large-scale screening of human sera with cytomegalovirus recombinant antigens. J Clin Microbiol. 1990;28:1375–1379. doi: 10.1128/jcm.28.6.1375-1379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landini M P. Antibody response to cytomegalovirus proteins. Rev Med Virol. 1992;2:63–72. [Google Scholar]
- 15.Landini M P, Lazzarotto T, Maine G T, Ripalti A, Flanders R. Recombinant mono- and polyantigens to detect cytomegalovirus-specific immunoglobulin M in human sera by enzyme immunoassay. J Clin Microbiol. 1995;33:2535–2542. doi: 10.1128/jcm.33.10.2535-2542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazzarotto T, Dalla Casa B, Campisi B, Landini M P. Enzyme-linked immunoadsorbent assay for the detection of cytomegalovirus-IgM: comparison between eight commercial kits, immunofluorescence and immunoblotting. J Clin Lab Anal. 1992;6:216–218. doi: 10.1002/jcla.1860060409. [DOI] [PubMed] [Google Scholar]
- 17.Lazzarotto T, Furlini G, Re M C, Ramazzotti E, Campisi B, Landini M P. Human cytomegalovirus replication correlates with differentiation in a hematopoietic progenitor cell line and can be modulated by HIV. Arch Virol. 1994;135:13–28. doi: 10.1007/BF01309762. [DOI] [PubMed] [Google Scholar]
- 18.Lazzarotto T, Maine G T, Dal Monte P, Frush H, Shi K, Landini M P. Detection of serum immunoglobulin M to human cytomegalovirus by Western blotting correlates better with virological data than detection by conventional enzyme immunoassay. Clin Diagn Lab Immunol. 1996;3:597–600. doi: 10.1128/cdli.3.5.597-600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazzarotto T, Maine G T, Dal Monte P, Ripalti A, Landini M P. A novel Western blot test containing both viral and recombinant proteins for anticytomegalovirus immunoglobulin M detection. J Clin Microbiol. 1997;35:393–397. doi: 10.1128/jcm.35.2.393-397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazzarotto T, Brojanac S, Maine G T, Landini M P. Search for cytomegalovirus-specific immunoglobulin M: comparison between a new Western blot, conventional Western blot, and nine commercially available assays. Clin Diagn Lab Immunol. 1997;4:483–486. doi: 10.1128/cdli.4.4.483-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazzarotto T, Spezzacatena P, Pradelli P, Abate D A, Varani S, Landini M P. Avidity of immunoglobulin G directed against human cytomegalovirus during primary and secondary infections in immunocompetent and immunocompromised subjects. Clin Diagn Lab Immunol. 1997;4:469–473. doi: 10.1128/cdli.4.4.469-473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maine G T, Lazzarotto T, Chovan L E, Flanders R, Landini M P. The DNA-binding protein pUL57 of human cytomegalovirus: comparison of specific immunoglobulin M (IgM) reactivity with IgM reactivity to other major target antigens. Clin Diagn Lab Immunol. 1996;3:358–360. doi: 10.1128/cdli.3.3.358-360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsano L, Perrillo R P, Flye M W, Hanto D W, Spitzer E D, Thomas J R, Murray P R, Windus D W, Brunt E M, Storch G A. Comparison of culture and serology for the diagnosis of cytomegalovirus infection in kidney and liver transplant recipients. J Infect Dis. 1990;161:454–461. doi: 10.1093/infdis/161.3.454. [DOI] [PubMed] [Google Scholar]
- 24.McMahon C A, Dock N L, Lentz E B, Forbes B A, Reinitz E R, Lamberson H V., Jr Detection of cytomegalovirus-specific IgM in renal transplant recipients. J Clin Lab Anal. 1989;3:350–354. doi: 10.1002/jcla.1860030607. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen C M, Hansen K, Andersen H M K, Gerstoft J, Vestergaard B F. An enzyme labelled nuclear antigen immunoassay for detection of cytomegalovirus IgM antibodies in human serum: specific and nonspecific reactions. J Med Virol. 1987;22:67–76. doi: 10.1002/jmv.1890220109. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen S L, Sørensen I, Andersen H K. Kinetics of specific immunoglobulins M, E, A, and G in congenital, primary, and secondary cytomegalovirus infection studied by antibody-capture enzyme-linked immunosorbent assay. J Clin Microbiol. 1988;26:654–661. doi: 10.1128/jcm.26.4.654-661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Revello M G, Zavattoni M, Percivalle E, Grossi P, Gerna G. Correlation between immunofluorescent detection of human cytomegalovirus immediate early antigens in polymorphonuclear leukocytes and viremia. J Infect Dis. 1989;160:159–160. doi: 10.1093/infdis/160.1.159. [DOI] [PubMed] [Google Scholar]
- 28.Ripalti A, Boccuni M C, Campanini F, Bergamini G, Lazzarotto T, Battista M C, Dalla Casa B, Landini M P. Construction of a polyepitope fusion antigen of human cytomegalovirus ppUL32 and detection of specific antibodies by ELISA. Microbiologica. 1994;18:1–12. [PubMed] [Google Scholar]
- 29.Ripalti A, Dal Monte P, Boccuni M C, Campanini F, Bergamini G, Lazzarotto T, Campisi B, Ruan Q, Landini M P. Prokaryotic expression of a large fragment of the most antigenic cytomegalovirus DNA-binding proteins (ppUL44) and its reactivity with human antibodies. J Virol Methods. 1994;46:39–50. doi: 10.1016/0166-0934(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 30.Robinson J M, Pilot-Matias T J, Pratt S D, Patel C B, Bevirt T S, Hunt J C. Analysis of the humoral response to the flagellin protein of Borrelia burgdorferi: cloning of regions capable of differentiating Lyme disease from syphilis. J Clin Microbiol. 1993;31:629–635. doi: 10.1128/jcm.31.3.629-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spaete R R, Gehrz R C, Landini M P. Human cytomegalovirus structural proteins. J Gen Virol. 1994;75:3287–3308. doi: 10.1099/0022-1317-75-12-3287. [DOI] [PubMed] [Google Scholar]
- 32.Stagno S, Pass R F, Reynolds D W, Moore M A, Nahmias A J, Alford C A. Comparative study of diagnostic procedures for congenital cytomegalovirus infection. Pediatrics. 1980;65:251–257. [PubMed] [Google Scholar]
- 33.Stagno S, Tinker M K, Elrod C, Fucillo D, Cloud G, O’Beirne A J. Immunoglobulin M antibodies detected by enzyme-linked immunosorbent assay and radioimmunoassay in the diagnosis of cytomegalovirus infections in pregnant women and newborn infants. J Clin Microbiol. 1985;21:930–935. doi: 10.1128/jcm.21.6.930-935.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Biji W, Torensma R, Son W J, Schirm J, Tegzess A M, The T H. Rapid immunodiagnosis of active cytomegalovirus infection by monoclonal antibody staining of blood leukocytes. J Med Virol. 1988;25:179–188. doi: 10.1002/jmv.1890250208. [DOI] [PubMed] [Google Scholar]
- 35.van Loon N M, Hessen F W A, van der Logt J T M, van der Veen J. Direct enzyme-linked immunosorbent assay that uses peroxidase-labeled antigen for determination of immunoglobulin M antibody to cytomegalovirus. J Clin Microbiol. 1981;13:416–422. doi: 10.1128/jcm.13.3.416-422.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vornhagen R, Plachter B, Hinderer W, The T H, Van Zanten J, Matter L, Schmidt C A, Sonneborg H H, Matter L, Jahn G. Early serodiagnosis of acute human cytomegalovirus infection by enzyme-linked immunosorbent assay using recombinant antigens. J Clin Microbiol. 1994;32:981–986. doi: 10.1128/jcm.32.4.981-986.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vornhagen R, Hinderer W, Sonneborg H H, Bein G, Matter L, The T H, Jahn G, Plachter B. The DNA-binding protein pUL57 of human cytomegalovirus is a major target antigen for the immunoglobulin M antibody response during acute infection. J Clin Microbiol. 1995;33:1927–1930. doi: 10.1128/jcm.33.7.1927-1930.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]